Abstract
There is an urgent need to develop new tumor biomarkers for early cancer detection, but the variability of tumor-derived antigens has been a limitation. Here we demonstrate a novel anti-Tn antibody microarray platform to detect Tn+ glycoproteins, a near universal antigen in carcinoma-derived glycoproteins, for broad detection of cancer. The platform uses a specific recombinant IgG1 to the Tn antigen (CD175) as a capture reagent and a recombinant IgM to the Tn antigen as a detecting reagent. These reagents were validated by immunohistochemistry in recognizing the Tn antigen using hundreds of human tumor specimens. Using this approach, we could detect Tn+ glycoproteins at subnanogram levels using cell lines and culture media, serum, and stool samples from mice engineered to express the Tn antigen in intestinal epithelial cells. The development of a general cancer detection platform using recombinant antibodies for detection of altered tumor glycoproteins expressing a unique antigen could have a significant impact on cancer detection and monitoring.
Keywords: antibody, glycoprotein, microarrays, Tn antigen, tumor-specific carbohydrate antigen
We have developed a novel anti-Tn antigen-based immuno-platform to detect Tn+ glycoproteins expressed by tumor cells as a single reagent to promote early universal cancer detection.
Introduction
The early detection of cancer is paramount to successful therapy, but few biomarker assays are available for detecting specific antigens produced by tumors. Some that have shown promise and are currently used clinically are carbohydrate antigen CA19-9, CA125, prostate-specific antigen (PSA), and carcinoembryonic antigen (CEA; Henry and Hayes 2012; Lee et al. 2020; Giamougiannis et al. 2021; O'Neill and Stoita 2021). However, most of these markers result from distinct changes in individual glycoproteins associated with very specific cancers, but they are also expressed by normal cells and expression can vary widely. Although efforts to combine protein and gene markers have had some success, no single reagent is currently available for universal cancer detection (Cohen et al. 2018). Alterations in posttranslational modifications of proteins, such as those generated by glycosyltransferases, can result in altered glycosylation of multiple glycoprotein targets. In this way, mutation, mis-localization, and other forms of glycosyltransferase dysfunction that accompany neoplastic transformation can result in the expression of an altered, distinct, and specific glycan signature on many different glycoproteins. This results in hundreds if not thousands of glycoproteins that share such a signature, as these would be amplified by each neoplastic cell.
While a variety of tumor-associated carbohydrate antigens have been studied, the most well-known and commonly expressed in human carcinomas is the Tn antigen, GalNAcα1-Ser/Thr (CD175), a glycoprotein-associated carbohydrate antigen consisting of GalNAc linked to Ser and/or Thr residues. Expression of this antigen requires enzymes in the polypeptide αGalNAc-transferase (GALNT) family, of which at least 20 mammalian genes are known (Bennett et al. 2012; de Las Rivas et al. 2019; Beaman et al. 2022; Nielsen et al. 2022). The Tn antigen is a neoantigen, and various studies indicate expression on glycoproteins in most human carcinomas and over half of colorectal cancers (CRC), but not in healthy tissues (Sakai et al. 2010; Kudelka et al. 2015; Stowell et al. 2015; Chia et al. 2016; Kolbl et al. 2016; Sun et al. 2018; Cervoni et al. 2020; Romer et al. 2021). Using specific anti-Tn antibodies to evaluate CRC, one recent study observed 51% of Tn-positivity (n = 140, antibody 5F4), whereas another observed 98% Tn-positivity (n = 43, antibody ReBaGs6; Romer et al. 2021; Dombek et al. 2022). The Tn antigen is a natural biosynthetic precursor that is normally extended by the T-synthase enzyme with addition of galactose to form the Galβ1-3GalNAcα1-Ser/Thr disaccharide (core 1), followed by further elongation by a suite of glycosyltransferases to form the normal repertoire of extended mucin-type O-glycans (Sakai et al. 2010; Ju et al. 2013; Kudelka et al. 2016, 2018). Such normal O-glycans are found in >80% of cell surface and secreted proteins (Steentoft et al. 2013). However, genetic or epigenetic disruption of T-synthase, its specific chaperone Cosmc, mislocalization of polypeptide αGalNAc-transferases, or Zn2+ dysregulation can lead to Tn expression on glycoproteins and a truncated tumor glycocalyx (Xia et al. 2004; Gill et al. 2006; Ju et al. 2008, 2014; Kudelka et al. 2016; Rømer et al. 2023). Because it is a truncated precursor for all O-GalNAc glycans, the Tn antigen is expressed on the majority of cell surface glycoproteins in carcinomas, irrespective of the cellular proteome that defines tissue type. Expression of the Tn antigen in tumor cells has been shown to drive tumorigenesis (Radhakrishnan et al. 2014; Thomas et al. 2019; Hofmann et al. 2021). Thus, the Tn is a highly abundant cell surface marker expressed in many different tumor glycoproteins and important in tumor formation, making it an ideal candidate biomarker for universal cancer detection.
Although all cancers could benefit from early detection, a survival benefit has been demonstrated for CRC, making CRC an ideal test-case for novel cancer detection platforms (Mandel et al. 1993; Thomas et al. 1995; Hardcastle et al. 1996; Mandel et al. 2000; Segnan et al. 2002, 2011; Faivre et al. 2004; Kronborg et al. 2004; Weissfeld et al. 2005; Lindholm et al. 2008; Atkin et al. 2010; Malila et al. 2011; Schoen et al. 2012; Scholefield et al. 2012; Shaukat et al. 2013; Holme et al. 2014; Force et al. 2016). However, current noninvasive fecal assays have poor sensitivities for detecting early lesions (adenoma 1–5 mm: <20%), and invasive approaches, i.e. colonoscopies, have poor adherence (38 versus 69% for fecal occult blood test; Inadomi et al. 2012; Knudsen et al. 2016). This highlights the potential for a stool-based test that detects early disease to have a major impact on patient care. Not only is Tn highly expressed in colorectal adenocarcinomas but also at the earliest signs of cellular atypia, making it an ideal target for early detection (Ju et al. 2013, 2014; Kudelka et al. 2015; Cervoni et al. 2020).
Despite the potential of Tn antigen as a marker of neoplastic disease, reagents capable of specifically detecting this modification on a wide variety of neoplastic lesions have been lacking. The small size of the Tn antigen, potential impact of adjacent protein motifs, and competing structures that may interfere with detection (“Tn-like” structures that terminate in GalNAc, e.g. blood group A) have made it challenging to generate antibodies that specifically recognize Tn antigen irrespective of the glycoprotein and therefore type of neoplastic cell responsible for generating this distinct modification (Sakai et al. 2010; Kudelka et al. 2015; Loureiro et al. 2015). In addition, and most compromising for Tn detection as a tumor biomarker, is the occurrence of the Tn antigen in human IgA1, where it is abundantly expressed in the hinge region in natural IgA1 glycoforms (Mestecky et al. 2008; Lehoux et al. 2014). Although lectins have been used to detect the Tn antigen, such as Helix pomatia agglutinin and Vicia villosa agglutinin (VVA), these lectins bind to other glycans with terminal GalNAc, such as blood group A, limiting their diagnostic application (Borgert et al. 2012). To overcome these obstacles and the potential confounding effect of IgA1 glycoforms, we have exploited 2 novel recombinant monoclonal antibodies we recently developed against the Tn antigen—recombinant human IgG1 Remab6 and recombinant murine IgM ReBaGs6 (Matsumoto et al. 2020). Each has high specificity for Tn in a variety of contexts, yet do not recognize the Tn antigen that occurs in IgA1 glycoforms.
Here we have used these engineered anti-Tn antibodies (Borgert et al. 2012; Matsumoto et al. 2020) to develop an ultrasensitive anti-Tn antibody microarray (ATAM). We demonstrate that these anti-Tn antibodies recognize the most common human tumors but not healthy controls. We validated ATAM using a mouse model engineered to express Tn in the intestinal epithelia and stool samples, and using engineered human cell lines from various tissues, and defined glycoproteins. The results demonstrate subnanogram sensitivity for Tn detection. Thus, ATAM represents a promising approach for early detection of CRC and other carcinomas.
Results
Development of an ATAM
We developed this microarray-based early cancer detection platform ATAM against the Tn antigen (CD175), utilizing recently generated recombinant anti-Tn antibodies (see Material and Methods; Borgert et al. 2012; Matsumoto et al. 2020). Microarrays enable high-throughput, microscale analysis, multi-analyte detection, printing of diverse macromolecules, and easy combination of new tests with existing assays for improved test characteristics. The 2 recombinant anti-Tn antibodies Remab6 human IgG1 and ReBaGs6 murine IgM, each have identical CDRs in their variable domains. Both antibodies recognize Tn clusters rather than single GalNAc residues, which reduces off-target binding and increases tumor specificity (Hirohashi et al. 1985; Borgert et al. 2012). These engineered IgG and IgM isotypes allowed us to optimize printing density/avidity for optimal antibody–carbohydrate interactions (Borgert et al. 2012; Matsumoto et al. 2020). Most importantly, neither of these antibodies bind to human IgA1, where some glycoforms contain the Tn antigen. Since tumor-derived glycoproteins with mucin domains, such as MUC2, contain hundreds of Tn epitopes, a sandwich detection approach can be used with multiple Tn epitopes contained within a single mucin domain (Kudelka et al. 2015, 2016). We empirically evaluated varying concentrations of printed anti-Tn IgG or IgM antibodies (200, 400, and 800 μg/mL) on a nitrocellulose slide. As outlined in Fig. 1a, we found that printing Remab6 at 0.8 mg/mL with detection using biotinylated ReBaGs6 at 5 μg/mL, followed by cyanine-5 streptavidin detection with a microarray scanner, provided optimal sensitivity. In all of our assays, we also printed IgG isotype controls in the same well as Remab6 and included all antibodies in 10 spot replicates in a 16 subwell format (i.e. 16 wells per slide for analysis of 16 individual samples per assay).
Fig. 1.
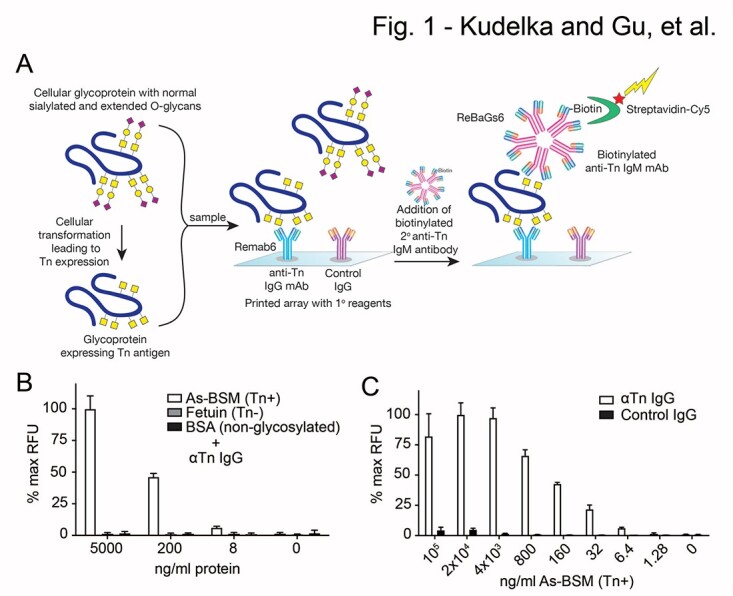
ATAM platform and analysis of defined glycoproteins. a) Tn containing glycoproteins are incubated on the antibody microarray with printed Remab6 and isotype control. Tn antigen captured from the samples is probed with biotinylated ReBaGs6 in a sandwich format and detected with fluorescently labeled streptavidin, which is read in a microarray scanner. Yellow square = GalNAc (Tn). b) ATAM analysis of Tn+ AsBSM, Tn(−) fetuin, and non-glycosylated BSA at various concentrations. c) ATAM analysis of Tn+ AsBSM at various dilutions. Data were normalized to 100 μg/ml printed AsBSM for each well and then to max RFU for each panel. Two independent repeats were performed for b) and c), error bars = ±1 SD.
We assessed this array by incubating defined glycoproteins, including the Tn+ glycoprotein standard asialo-bovine submaxillary mucin (AsBSM), a Tn(−) glycoprotein fetal bovine serum (FBS) fetuin, and a non-glycosylated protein bovine serum albumin (BSA), at 5 μg/mL, 200 ng/mL, 8 ng/mL, and 0 ng/mL. We could detect bound AsBSM, but not the negative controls, at all concentrations (Fig. 1b), demonstrating the utility of the assay to identify Tn+ glycoproteins. To further characterize the sensitivity, we assayed AsBSM at varying concentrations from 100 μg/mL to 1.28 ng/mL in 5-fold dilutions and found that AsBSM was clearly detected as low as 6.4 ng/mL (Fig. 1c). Since we incubated 150 μL of sample per well, we were able to detect Tn antigen at 960 picograms of Tn+ glycoprotein. We conclude that in this format, the sensitivity is in the subnanogram range for Tn+ glycoprotein.
Detection of Tn-containing glycoproteins in feces
We used this platform on complex biological samples by evaluating Tn-containing glycoproteins in feces, a potential source for CRC detection. Feces contain glycosidases that are produced from the microbiome to harvest and utilize host, dietary, and microbial glycans (Gill et al. 2006). These glycosidases could degrade Tn and remove GalNAc from Tn+ glycoproteins, limiting our ability to detect Tn in feces, yet this has not yet been explored. To assess this, we incubated AsBSM or the non-glycosylated protein BSA with WT mouse feces. We evaluated Tn stability by anti-Tn western blot (Fig. S1a) and protein stability with Coomassie Brilliant Blue (CBB) staining after sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. S1b), respectively. Tn staining of AsBSM, as assessed by anti-Tn western blot, decreased after incubation with WT fecal matter (Fig. S1a). Although we could not directly analyze AsBSM protein stability by CBB (CBB poorly stains heavily glycosylated proteins), BSA stability was unperturbed by fecal incubation, suggesting that loss of Tn staining was because of exoglycosidase activity, e.g. α-N-acetylgalactosaminidase (αGalNAcase), rather than general protease activity (Fig. S1b). To determine whether an αGalNAcase was present, we incubated 4MU-αGalNAc with stool (Fig. S1c). 4MU produces a fluorescent signal when released from GalNAc, a property that can be used to assay αGalNAcase activity (Ju et al. 2011). We observed that incubation of 4MU-αGalNAc with feces resulted in increased fluorescence compared with PBS alone, confirming the presence of αGalNAcase activity in feces (Fig. S1c).
To inactivate the fecal αGalNAcase activity, which could potentially interfere with our ability to detect Tn with ATAM, we tested whether mild heat treatment could inactivate the αGalNAcase activity in feces and preserve Tn on fecal glycoproteins for microarray detection. We preincubated WT mouse feces at 95°C for 1 h or 10 min, at 80°C for 10 min, and at 65°C for 1 h or 10 min, and then added the treated material to AsBSM (Fig. S1a), BSA (Fig. S1b), or 4MU-GalNAc (Fig. S1c) to assess αGalNAcase activity and protein stability. All heat treatments prevented loss of Tn from the Tn+ glycoprotein by western blot (Fig. S1a) and substantially reduced αGalNAcase activity by 4MU-GalNAc assay (Fig. S1c). Importantly, these treatments were sufficiently mild to preserve protein stability, as BSA mass was unaltered by CBB (Fig. S1b). In subsequent experiments, we heat-treated feces at 80°C for 10 min, and found that this condition completely eliminated αGalNAcase activity by western blot and fluorescence assay. In summary, we identified optimal conditions to detect Tn+ glycoprotein in feces and avoid degradation by αGalNAcase.
We next validated Tn fecal detection using a physiologic model of Tn expression. Intestinal epithelial cell-specific deletion of Cosmc results in Tn expression throughout the GI-tract (Kudelka et al. 2016; IEC-Cosmc KO mice). Since Cosmc is on the X chromosome, we crossed Cosmcflox/flox with VilCre+ mice to generate VilCre+; Cosmcflox/y KO males, VilCre−; Cosmcflox/y WT males, VilCre+; Cosmcflox/+ mosaic females, and VilCre−; Cosmcflox/+ WT females (Fig. 2a). We previously showed that male IEC-Cosmc KO mice have >90% deletion of Cosmc and expression of Tn in the small and large intestine and that female mosaics have complete deletion of Cosmc and expression of Tn in ~50% of crypts in the small and large bowel because of random X-inactivation (Kudelka et al. 2016).
Fig. 2.
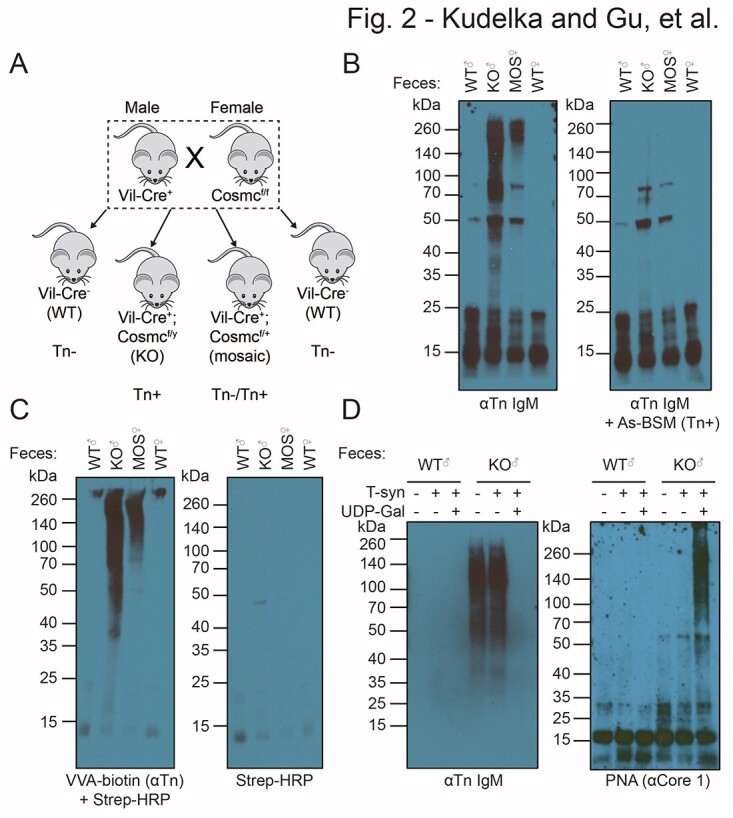
Validation of a genetically engineered mouse with Tn+ feces. a) Breeding strategy to generate intestinal epithelial-specific Cosmc male KO, female mosaic, and WT mice. b) Western blot of male KO, female mosaic, and WT feces stained with ReBaGs6 (left), ReBaGs6 pre-blocked with Tn+ AsBSM (right). c) Biotinylated VVA lectin plus streptavidin-HRP (left), or streptavidin-HRP alone (right). d) Western blot with ReBaGs6 antibody (left) or PNA lectin (right) of WT and KO feces treated with T-synthase enzyme ± UDP-gal. Two independent repeats were performed for b)–d).
Using this physiologic model of Tn expression in the intestine, we assessed whether Tn+ glycoproteins could be detected in feces from these mice. We collected and homogenized feces from IEC-Cosmc KO male mice, mosaic females, and age/gender-matched WT controls and performed western blots with anti-Tn IgM antibody ReBaGs6. IEC-Cosmc KO mice and mosaic fecal lysates had prominent staining > 50 kDa compared with WT feces, with roughly twice the intensity in IEC-Cosmc KO mice as compared with mosaics (Fig. 2b, left). This staining was specific, as it was blocked by preincubating ReBaGs6 with the inhibitor 1 mM Tn+ AsBSM (Fig. 2b, right). Tn antigen was also detected in a similar staining pattern with VVA, which recognizes single Tn/GalNAc residues, in contrast to our anti-Tn mAbs that recognize clusters of at least 2 Tn residues (Fig. 2c).
To further assess the specificity of our anti-Tn mAb CDRs for Tn in IEC-Cosmc KO mice feces, we incubated KO and WT feces with recombinant T-synthase enzyme, which masks Tn by addition of galactose (Fig. 2d). As controls, we either added T-synthase with or without its donor UDP-Gal or did not add T-synthase. Incubating IEC-Cosmc KO mice feces with T-synthase plus UDP-Gal, but not the negative controls, resulted in complete loss of binding of ReBaGs6 as well as increased binding of peanut agglutinin (PNA), which recognizes the T-synthase disaccharide product, core 1 or Galβ1-3GalNAcα1-Ser/Thr (Fig. 2d). These results demonstrate that our anti-Tn mAbs, with identical CDRs, specifically recognize Tn+ glycoproteins in feces of IEC-Cosmc KO mice and mosaic female mice.
We next determined whether we could detect Tn+ glycoproteins from IEC-Cosmc KO mice and mosaic female mice. We collected stool and prepared fecal lysates by vortex and sonication as described in the methods. Feces were stored as lysates at −20°C for up to 1 month, then freshly thawed, heat inactivated, and analyzed by ATAM. To detect Tn from IEC-Cosmc KO mice feces, we pooled 5 KO and separately 5 WT mice, assessed protein concentration by BCA assay, and performed serial dilutions from 1,000 to 0.064 μg/mL (Fig. 3a). IEC-Cosmc KO mice feces were clearly distinguishable from WT feces or printed isotype controls at a concentration ≥ 8 μg/mL (Fig. 3a). Using our earlier observation that this assay can detect AsBSM down to 6.4 ng/mL, the results indicate that Tn+ glycoproteins constitute ~0.1% of IEC-Cosmc KO mice feces. The remaining fecal matter is presumably derived from the host, diet, and microbiome.
Fig. 3.
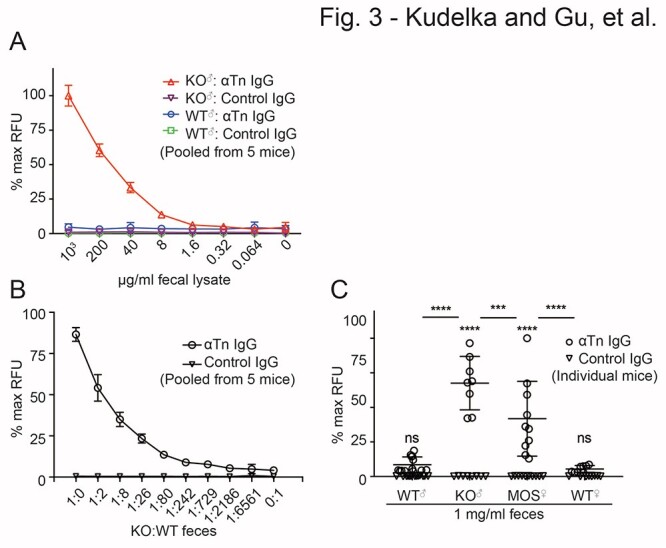
ATAM analysis of Tn+ feces. a) Anti-Tn microarray analysis of WT and KO feces at various dilutions (5 mice pooled/group), b) WT and KO feces mixed at different ratios (5 mice pooled/group prior to mixing), and c) WT male (n = 13), KO (n = 8), mosaic (n = 10), and WT female (n = 8) feces at 1 mg/mL (data from 2 independent experiments pooled and shown on graph). Data were normalized to 100 μg/ml printed AsBSM for each well and then to max RFU for each panel. Two independent repeats from separate groups of mice were performed for a–c). Data in c) were pooled from repeats. Error bars = ±1 SD of spot replicates for a) and b), and of individually analyzed mice for c). For c), 2-way ANOVA and post hoc multiple comparisons with SIDAK correction (**** = P < 0.0001, *** = P = 0.0005, alpha = 0.05); P-value over each genotype compares printed anti-Tn IgG versus isotype control within a genotype; P-value between groups compares anti-Tn expression across genotypes.
Analysis of IEC-Cosmc KO mice feces is limited by the fact that these animals express the Tn antigen on many glycoproteins produced from the intestinal epithelium. To assess the sensitivity of the assay in regard to more patient-relevant samples in which both Tn+ and Tn(−) glycoproteins are mixed, as would be potentially in stool samples from colorectal patients, we pooled feces by mixing IEC-Cosmc KO mice with WT feces at different dilutions ranging from 1:1 to 1:6,561 (Fig. 3b). We could detect Tn+ glycoproteins down to a dilution of 1:2,186 with the WT feces (Fig. 3b). Additionally, female mosaic mice that express Tn in ~50% of crypts provided a unique opportunity to analyze Tn expressed in a fraction of the intestine. We analyzed 1 mg/mL feces of IEC-Cosmc KO, mosaic, and WT mice from 8 to 9 individual mice per group and detected Tn+ glycoproteins in IEC-Cosmc KO mice and mosaic mice as compared with WT and isotype controls (Fig. 3c). Notably, mosaic feces had a ~50% reduction in signal compared with KO mice indicating a dose response (Fig. 3c). These results demonstrate sensitive detection of Tn+ glycoproteins in feces in which there is a high amount of Tn(−) glycoproteins.
Detection of Tn-containing glycoproteins in serum
We next investigated whether ATAM could detect Tn+ glycoproteins in the presence of serum, which could obviously complicate detection of Tn+ glycoproteins, as IgA1 can also carry the Tn antigen. However, we previously showed that the recombinant antibodies ReBaGs6 and Remab6 do not bind Tn antigen on IgA1, because of the unusual nature of the peptide sequence in the hinge region (Matsumoto et al. 2020), thus we predicted that IgA1 would not interfere with our general detection of Tn+ glycoproteins.
For these studies, we utilized human cancer cell lines engineered/selected to express Tn through Cosmc deletion/mutation and their WT counterparts (Ju et al. 2008; Steentoft et al. 2013). We chose cancer cells from diverse tissues, including colorectal, breast, stomach, and kidney, to capture the diversity of Tn expression in carcinomas (Fig. S2). Both the ReBaGs6 (Fig. S2a) and Remab6 (Fig. S2b) antibodies were highly reactive against Tn expressing cells but not their WT counterparts by fluorescence-activated cell sorting (FACS), indicating that both isotypes with identical variable regions recognize Tn in human cells, similar to mouse feces and defined glycoproteins.
As a model to evaluate Tn detection in serum, we analyzed conditioned media from Tn+ and Tn(−) human cancer cells incubated with FBS. We seeded cells to ~20% confluency and collected media at 80–90% confluency. Conditioned media FBS was incubated at varying dilutions on the anti-Tn microarray. Tn+ glycoproteins could be detected in culture media at less diluted conditions with FBS from Tn+ cells but not from their WT Tn(−) counterparts (Fig. 4). Interestingly, different cell lines produced different amounts of Tn+ glycoprotein. Although MKN45 gastric and LS174T colorectal carcinoma cells are GI-derived and highly positive by flow cytometry (Fig. S2), Tn+ glycoproteins could be detected to a 2,000-fold dilution for LS174T but only to a 100-fold dilution for MKN45 (Fig. 4), suggesting that different cell lines and possibly different tissues and/or tumors secrete different quantities of Tn+ glycoprotein into serum. These data demonstrate that ATAM can successfully detect Tn in culture media in the presence of serum.
Fig. 4.
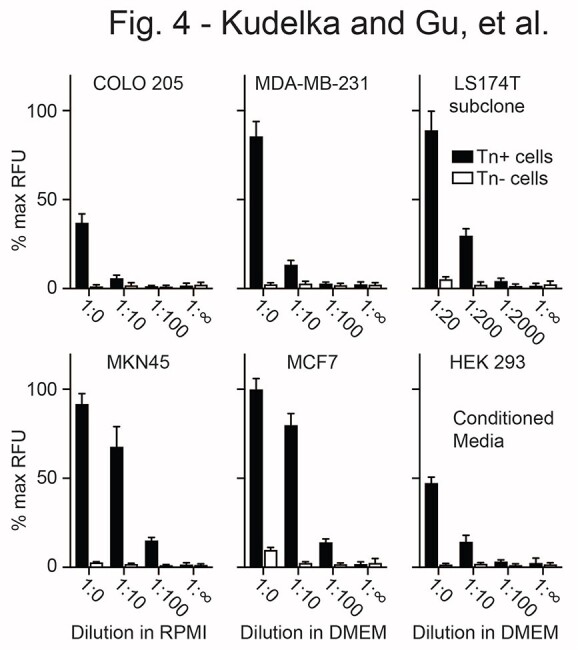
ATAM detection of Tn antigen in conditioned media. WT (Tn(−)) and Cosmc-deficient (Tn+) cell lines were cultured from ~20% to near confluency in media containing 10% FBS. Conditioned media at various dilutions (1:0 = no dilution to 1:2,000) was compared with fresh media (1:∞) by anti-Tn microarray. CRC (COLO-205; LS174T subclone), gastric cancer (MKN45), breast cancer (MCF7, MDA-MB-231), and kidney (HEK293) cell lines were used. Data were normalized to 100 μg/ml printed AsBSM for each well and then to max RFU across all cell lines. Two independent repeats were performed, error bars = ±1 SD.
We then evaluated whether serum factors could inhibit Tn detection by ATAM. Although Tn was previously thought to be expressed exclusively in cancers and not healthy tissues, a significant fraction of IgA1 glycoforms from healthy donors expresses Tn in the IgA1 hinge region (Mestecky et al. 2008; Lehoux et al. 2014). This could potentially confound ATAM detection of Tn+ stool or serum. We previously assessed our anti-Tn antibodies on a glycopeptide array and demonstrated antibody recognition of adjacent Tn glycans in mucin-derived glycopeptides but not in IgA1-derived glycopeptides, presumably because of intervening proline residues in the hinge region (Borgert et al. 2012). To further confirm that full-length IgA1 in serum or feces does not interfere with our assay, we incubated human serum, WT mouse serum, WT mouse feces, or PBS with AsBSM at 200 or 0 ng/mL (Fig. 5a). Neither human serum nor WT mouse serum or feces interfered with AsBSM detection on ATAM, suggesting that soluble IgA1 and other serum/fecal factors do not interfere with Tn detection (Fig. 5a).
Fig. 5.
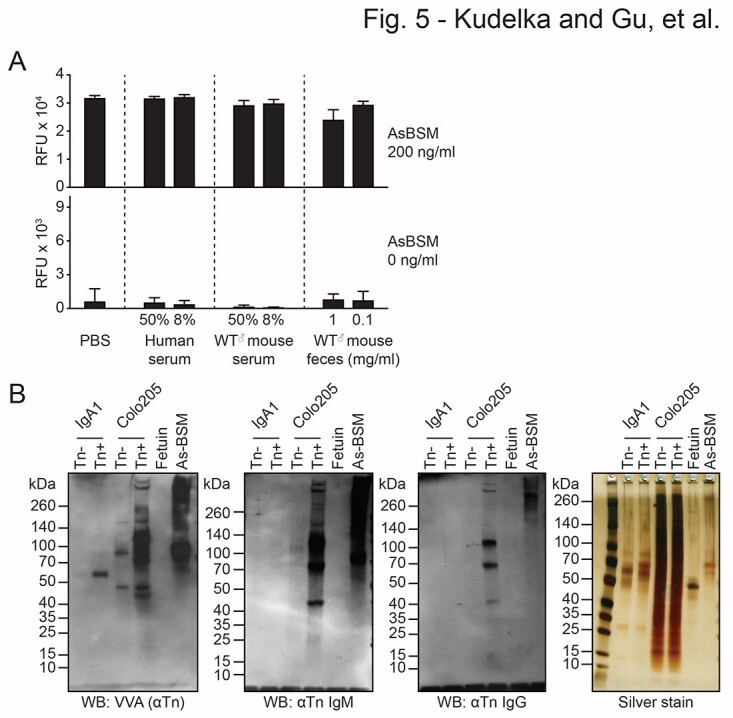
Serum, feces, and IgA1 do not interfere with anti-Tn detection. a) PBS, pooled human serum, pooled WT mouse serum, and pooled WT mouse feces (pooled from 5 mice/humans) at 2 concentrations were incubated with or without Tn+ AsBSM and analyzed by anti-Tn microarray to assess blocking activity. Error bars = ±1 SD. b) Western blot with VVA lectin (left), ReBaGs6 (left middle), and Remab6 (right middle) of Tn(−)/Tn+ IgA1 from parental/Cosmc-KO Dakiki cells, along with cell and protein controls (Tn+/Tn(−) Colo205 lysates, Tn+ AsBSM, and Tn(−) Fetuin) and silver stain loading control (right). Two independent repeats were performed for a) and b).
To further confirm that our anti-Tn antibodies do not react with full-length Tn+ IgA1, we isolated Tn(−) and Tn+ IgA1 from WT and Cosmc-KO Dakiki human B-cell lines and performed western blots with anti-Tn antibodies (Fig. 5b). We used Tn(−) and Tn+ cell lysates from WT and Cosmc-KO Colo205 cells as well as Tn(−) fetuin and Tn+ AsBSM glycoproteins as controls. Silver stain confirmed equal loading. VVA lectin western blot verified that IgA1 from KO cells but not WT Dakiki cells contained Tn antigen, as expected (Fig. 5b). Importantly, neither Remab6 nor ReBaGs6 bound WT or KO IgA1 despite recognizing Tn+ Colo205 cells and Tn+ AsBSM glycoproteins, thus demonstrating that these anti-Tn antibodies do not recognize Tn+ human IgA1. Collectively, these results demonstrate that bovine and human serum, feces, and isolated IgA1 do not interfere with the detection of Tn+ glycoproteins using ATAM.
Detection of Tn-containing glycoproteins in human tumors
Prior studies evaluating Tn expression in human tumors are limited by the use of reagents with poor specificity, for example lectins that cross react with blood group antigens, as well as assessment of limited tumor types. We performed immunohistochemistry with BaGs6 IgM antibody on 236 tumors arising from diverse tissues, including the endometrium, breast, ovary, lung, and colon as well as 30 non-cancer controls from matched tissue types (Table 1 and Fig. S3). The vast majority of these tumors were adenocarcinomas. BaGs6 positively stained 56% of all tumors and 0% of non-cancer controls. When separated by tissue type, >50% of all tumors were positive for Tn with Tn positivity ranging from 51 to 74%. These data demonstrate that our anti-Tn Abs with identical CDRs are able to detect the Tn antigen across a diverse array of the most common human tumors.
Table 1.
Tn antigen expression in neoplastic and normal tissue. Tn antigen was detected with BaGs6 and is specifically expressed in a broad range of neoplastic disease. The percent of all lesions examined for Tn positivity in neoplastic disease or normal is shown with absolute quantities indicated in parentheses as number of positives to total analyzed per tissue. All endometrial, ovarian, and colon lesions were adenocarcinomas. All breast lesions were infiltrating ductal carcinoma of the breast, except one infiltrating lobular carcinoma of the breast. Neoplastic lung lesions were 57% adenocarcinoma and 43% squamous cell carcinoma.
| % neoplasia Tn+ | % normal Tn+ | |
|---|---|---|
| Endometrial | 55 (23/41) | 0 (0/6) |
| Breast | 56 (20/37) | 0 (0/6) |
| Ovarian | 74 (31/43) | 0 (0/6) |
| Lung | 53 (25/48) | 0 (0/6) |
| Colon | 51 (34/67) | 0 (0/6) |
Discussion
Cancer is the second leading cause of mortality worldwide; however, many deaths could be avoided by early detection. To this end, we developed a detection system termed ATAM using tumor-specific anti-carbohydrate antibodies that recognize the Tn antigen, which is expressed in a majority of the most common carcinomas, but not healthy controls. As a proof of concept, we validated our approach for stool-based early CRC detection and serum-based pan-carcinoma detection. Our approach represents the first assay utilizing an individual target antigen for universal cancer detection with specific IgG and IgM antibodies to the Tn antigen (CD175) and highlights the potential of targeting the tumor glycocalyx and secreted glycoproteins for cancer care.
Current cancer biomarkers target individual tumors rather than a variety of tumor types. This is because different tissues express distinct combinations of proteins. Consequently, protein-based biomarkers are typically tissue specific. An alternative approach is to target posttranslational modifications expressed across tissue types. Protein glycosylation is an abundant posttranslational modification shared by all cells; however, prior attempts to identify glycan biomarkers have generally failed to identify universal, tumor-specific structures. However, the Tn antigen (GalNAcα1-Ser/Thr; CD175) is unique in that it is an O-glycan precursor expressed in most cancers on a majority of cell surface and secreted glycoproteins, but not in healthy tissues, except for IgA1. Although Tn is tumor specific, “Tn-like” structures terminating in α-linked GalNAc, such as blood group A, are abundant. We generated a highly sensitive microarray platform that could distinguish Tn in tumors from Tn in the IgA1 hinge region and “Tn-like” structures in cells. Such a technology could transform early cancer detection.
In contrast to specific biomarkers, such as PSA for prostate cancer or fecal occult blood for CRC (Stamey et al. 1987; Hardcastle et al. 1996), few universal biomarkers have been identified. Efforts at universal detection have utilized multiple tumor-type-specific biomarkers or evaluated broad genomic changes (Cohen et al. 2018; Sina et al. 2018). The former only identify tumor types from which the individual biomarkers are derived, but not for less common cancers that are not traditionally included in population-based screening (Ahlquist 2018). Additionally, multi-analyte assays are limited by the properties of the individual biomarkers. For example, CA19-9 and CEA have limited utility individually for screening but have been utilized in combination with other markers. Furthermore, genomic markers require tumor cell shedding, which may not occur at the earliest stages of cancer. The development of ATAM to detect universal alterations in secreted cancer glycoproteins could address these limitations, while being easily combined with other biomarkers for multi-analyte detection.
Some limitations of ATAM are important to note. Although the Tn antigen is highly expressed in most solid tumors, some cancers have infrequent expression (prostate, liver, leukemia, and lymphoma) and thus would not be detected at a frequent rate (Sasaki et al. 1999; Li et al. 2009; Kudelka et al. 2015). We evaluated Tn expression in human adenocarcinomas; however, premalignant lesions would also be important to identify (Cervoni et al. 2020). This is partly addressed in prior studies that have shown Tn expression in precursor lesions (Kudelka et al. 2015). Furthermore, serum and feces contain glycosidases that could inactivate Tn. We demonstrate here, however, that serum does not interfere with ATAM and that fecal glycosidases can be easily inactivated for Tn detection. However, this issue would need to be addressed more fully in the future using sera and fecal samples from multiple donors. Additionally, although intratumor heterogeneity or low but positive Tn expression could lead to lower absolute level of Tn expression for detection, prior studies identified Tn in over 50% of cells in the most common adenocarcinomas with medium or high intensity staining in positive tumors (Romer et al. 2021).
As cancer is a leading cause of death worldwide, and based on the percent of Tn expression across tumors in our study and reported cancer-associated deaths worldwide, we could envision that early detection of cancer by ATAM could significantly impact cancer death rates (Sasaki et al. 1999; Li et al. 2009; Kudelka et al. 2015). In addition to screening, ATAM could monitor disease burden or be applied for personalized Tn-based therapies, for example in preselecting patients to receive CAR-T MUC1-Tn therapy (Posey et al. 2016). In contrast to HER2 expression for Herceptin, the data presented here suggest the frequency and even the specificity of Tn expression is broad and therefore likely to be therapeutically relevant to a wide variety of neoplastic lesions. Thus, we envision ATAM being used for universal cancer screening in a primary care setting, as an assay to guide Tn-targeted therapies through precision medicine, and as a biomarker to follow treatment response.
Materials and methods
Animal and human samples
All mouse and human samples were collected in accordance with the IACUC at Beth Israel Deaconess Medical Center under an approved protocol and the IRB at Emory University under an approved protocol, following the guidelines in all protocols.
Cell lines
Cell lines were a kind gift from Henrik Clausen (WT and Cosmc-KO Colo 205, MDA-MB-231, HEK-293, MCF7, MKN45), from Steven Itzkowitz (LSC, LSB), or derived from Tn+/Tn, LS174T subclones (Sakai et al. 2010). Colo205 and MKN45 were cultured in RPMI. MDA-MB-231, HEK293, MCF7, LSC, LSB, and LS174T were cultured in DMEM. All cells were supplemented with 10% FBS. MCF7 cells were additionally supplemented with 0.01 mg/ml insulin. For Fig. 4, cells were seeded at ~20% confluency and conditioned media was collected several days later (depending on the cell type) at 80–90% confluency.
Antibodies and reagents
ReBaGs6 (anti-Tn mouse IgM) and Remabs6 (anti-Tn human IgG) were reverse engineered from BaGs6 (anti-Tn IgM, a kind gift from Georg Springer) and biotinylated by ProtOn Biotin Antibody and Labeling Kit (Vector Laboratories). Biotinylated VVA and PNA were purchased from Vector Laboratories. Mucin from bovine submaxillary gland (Sigma) was enzymatically desialylated (neuraminidase, Roche) to generate AsBSM.
Antibody microarray printing
Noncontact antibody printing was performed using a sciFLEXARRAYER S11 from Scienion AG. The average spot volume was within 5% variation of 330 pl in phosphate-buffered saline (Corning Cellgro). In all, 10 replicates of 0.8 mg/mL Remabs6 (anti-Tn IgG) and isotype control human IgG were printed in 16-pad Supernova nitrocellulose slides (Grace Bio-Labs). The printed slides were desiccated at 4°C overnight, blocked with Super G Plus Protein Preservative Buffer (Grace Bio-Labs) as directed, and stored at −20°C until use.
Anti-Tn microarray assay
Slides were removed from −20°C storage and dried at room temperature for 10 min. A 16-chamber adaptor (Grace Bio-Labs) was used to partition and seal each subwell. Each subarray was washed 3 times with 200 μl TBST (50 mM Tris-HCl, PH 7.4; 300 mM NaCl and 0.05% Tween 20) for 5 min to remove preservative buffer. 150 μl of sample was added and incubated at 4°C overnight. Sample buffer varied per sample (feces: PBS plus protease inhibitor; protein/glycoprotein/serum: PBS; conditioned media: RPMI or DMEM). After washing 3 times with 200 μl TBST for 10 min each, the slide was blocked with 1% BSA in TBST for 30 min. Block was removed and slides were incubated with 100 μl of 5 μg/mL biotinylated ReBaGs6 (anti-Tn IgM) in 0.5% BSA in TBST 4°C overnight. Slides were then washed 3 times with 200 μl TBST for 10 min each and 100 μl of 5 μg/mL streptavidin-cyanine-5 in 0.5% BSA. TBST was added and incubated for 30 min at room temperature and 2 h at 4°C. The slides were then washed 3 times with 200 μl TBST for 10 min each, washed once with 200 μl MilliQ water. After removing all of the liquid, the adaptor was removed, the slide was dried at room temperature, and the slides were scanned with a GenePix 4300A Scanner (Molecular Devices) microarray scanner at 635 nm excitation. The images were analyzed with the GenePix Pro 7 software.
Collection and preparation of feces
Five mice (age 2–4 months) per genotype were separated into individual clean cages for 3 days. Dried feces were collected and stored at −20°C. All procedures were performed on ice or 4°C. 400 mg feces were preincubated in 4 mL lysis buffer (10 mL PBS plus 1 tablet of protease inhibitor cocktail (Roche)) for 30 min with vortex every 10 min, then sonicated 10 s 3 times each with 20 s of rest, and centrifuged at 4000 rpm for 20 min. The supernatants were collected and removed to 1.5 mL tubes and recentrifuged at 16,000 × G for 10 min. Final supernatants for each mouse were stored in 150 μl aliquots at −20°C. For each set of experiments, a fresh aliquot was thawed, diluted to 2 mg/mL, heat inactivated at 80°C for 10 min, and then centrifuged at 13,000 × G for 2 min.
Western blot
Fecal lysates were separated by SDS-PAGE gels under reducing conditions and transferred to nitrocellulose membranes with high molecular weight protocol (fixed 25 V, 10 min; Bio-Rad Labs). Membranes were blocked in 5% BSA for 1 h at room temperature and probed with primary antibodies (anti-Tn IgG, IgM) or biotinylated lectins (VVA, PNA) followed by incubation with secondary antibodies or streptavidin conjugated with HRP. Proteins were visualized with SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher) and developed.
Immunohistochemistry
Immunohistochemistry for Table 1 was performed with anti-Tn IgM antibody as described (Kudelka et al. 2016; Matsumoto et al. 2020) with some modifications, with staining optimized based on positive (Tn+ LSC) and negative (Tn(−) LSB) controls in formalin-fixed paraffin-embedded cell blocks. Briefly, immunohistochemistry was performed using standard heat-induced epitope retrieval in citrate buffer pH 6. Slides were loaded on the DAKO Autostainer (Dako), exposed to 3% hydrogen peroxide for 5 min, primary antibody for 30 min, labeled with polymer horseradish peroxidase for 30 min (Envision + dual link; Dako), diaminobenzidine as a chromogen for 5 min, and hematoxylin as a counterstain for 5 min. Incubations were performed at room temperature, and between incubations sections were washed with Tris-buffered saline. Human specimens were obtained from banked clinical slides from the Emory University Department of Pathology under appropriate IRB approval. Staining was graded by positive or negative by expert pathologist review, and images were taken at ×40 magnification.
T-synthase enzyme reaction
Fecal lysates were heat inactivated at 80°C for 10 min and dialyzed with a 10 kDa molecular weight cut-off cassette (Thermo Fisher) into 50 mM 2-(N-morpholino) ethanesulfonic acid (MES) buffer, pH 6.8 overnight at 4°C. T-synthase mix (15 μg fecal lysate, 50 mM pH 6.8 MES-NaOH, 20 mM MnCl2, 1 mM UDP-Gal, 1x EDTA-free protease inhibitor table) was added to 12.5 μg/mL T-synthase to a total volume of 50 μl and incubated for 4 h at 37°C. 16.7 μl 4x sample buffer with β-mercaptoethanol was added to the reaction mixture, boiled for 10 min, and analyzed by western blot.
αGalNAcase assay
αGalNAcase activity was assayed by reaction of 10 μl lysate with 40 μl reaction mix (2.5 μl 5 mM GalNAc-α-(4MU), 5 μl 1 M Tris-HCL (pH 7.8), 1 μl 10% Tx-100, and 31.5 μl H2O) at 37°C for 1 h. The reaction was stopped with 100 μl of cold (4°C) 1 M glycine-NaOH (pH 10) and fluorescence was assayed with a fluorescence reader at excitation 355 nm, emission 460 nm. No GalNAc-α-(4MU; replaced with H2O) was used as a background control.
Flow cytometry
Adherent cells were suspended with 0.25% Trypsin and centrifuged at 1,000 rpm for 5 min. 1 × 105 cells/sample tube were incubated with biotinylated ReBaGs6, Remabs6, or isotype control antibodies (5 μg/mL) in 4 individual experiments on ice for 30 min. Cells were centrifuged and washed 3 times with cold PBS, followed by incubation with streptavidin-Cy5 for 30 min. After washing with cold PBS, the cells were analyzed by FACS.
Data analysis
Data were analyzed and plotted with prism software. Statistical tests and data normalization were performed where applicable as described in figure legends. All data points = mean ± 1 SD. Alpha = 0.05. At least 2 independent repeats were performed for all experiments.
Supplementary Material
Acknowledgments
We thank members of the Cummings and Chaikof labs for helpful discussions as well as Melinda Hanes and Rajindra Aryal for providing T-synthase.
Contributor Information
Matthew R Kudelka, Department of Biochemistry, Emory University School of Medicine, Atlanta, GA 30033, United States; Department of Surgery, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA 02215, United States.
Wei Gu, Department of Surgery, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA 02215, United States.
Yasuyuki Matsumoto, Department of Surgery, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA 02215, United States.
Tongzhong Ju, Department of Biochemistry, Emory University School of Medicine, Atlanta, GA 30033, United States.
Richard H Barnes II, Department of Surgery, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA 02215, United States.
Robert J Kardish, Department of Surgery, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA 02215, United States.
Jamie Heimburg-Molinaro, Department of Surgery, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA 02215, United States.
Sylvain Lehoux, Department of Surgery, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA 02215, United States.
Junwei Zeng, Department of Surgery, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA 02215, United States.
Cynthia Cohen, Department of Pathology and Laboratory Medicine, Emory University School of Medicine, Atlanta, GA 30033, United States.
Brian S Robinson, Department of Pathology and Laboratory Medicine, Emory University School of Medicine, Atlanta, GA 30033, United States.
Kinjal S Shah, Department of Pathology and Laboratory Medicine, Emory University School of Medicine, Atlanta, GA 30033, United States.
Elliot L Chaikof, Department of Surgery, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA 02215, United States.
Sean R Stowell, Department of Pathology and Laboratory Medicine, Emory University School of Medicine, Atlanta, GA 30033, United States.
Richard D Cummings, Department of Surgery, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA 02215, United States.
Author contributions
Matthew R. Kudelka (experimental design, conceptualization, performed experiments, generated mice and collected samples, analyzed results, wrote and edited manuscript), Wei Gu (experimental design, conceptualization, performed experiments, generated mice and collected samples, and edited manuscript), Yasuyuki Matsumoto (performed experiments, reviewed manuscript), Tongzhong Ju (experimental design, conceptualization, generated mice and collected samples, supervised experiments, and reviewed manuscript), Richard H. Barnes II (performed experiments, and edited manuscript), Robert J. Kardish (printed microarrays, and reviewed manuscript), Jamie Heimburg-Molinaro (supervised experiments, analyzed results, edited manuscript, reviewed manuscript), Sylvain Lehoux (generated Dakiki KO cells, edited and reviewed manuscript), Junwei Zeng (generated mice and collected samples, and reviewed manuscript), Cynthia Cohen (performed and/or oversaw immunohistochemistry, edited manuscript, reviewed manuscript), Brian S. Robinson (performed and/or oversaw immunohistochemistry, edited manuscript, reviewed manuscript), Kinjal S. Shah (performed and/or oversaw immunohistochemistry, reviewed manuscript), Elliot L. Chaikof (conceptualization, reviewed manuscript), Sean R. Stowell (experimental design, conceptualization, performed experiments, analyzed results, wrote and edited manuscript), Richard D. Cummings (experimental design, conceptualization, supervised experiments, analyzed results, wrote and edited manuscript, provided funding).
Funding
This work was supported by National Institutes of Health [grant numbers U01CA168930 to TJ and RDC, P41GM103694 and R24GM137763 to RDC]; the Burroughs Wellcome Trust Career Award for Medical Scientists; the National Institutes of Health (NIH) Early Independence Grants [DP5OD019892 to SRS]; and Emory Medical Scientist Training Program grant [number T32GM008169 to MRK].
Conflict of interest statement. RHB is currently employed by Penrose TherapeuTx. MRK, YM, ELC, and RDC filed an initial disclosure of intellectual property with the Office of Technology Transfer at Beth Israel Deaconess Medical Center for this work. The other authors declare no conflict of interests.
Data availability
Reagents and original data are available upon request to Richard D. Cummings, or are provided in the paper, or online at: https://ncfg.hms.harvard.edu/ncfg-data/microarray-data.
References
- Ahlquist DA. Universal cancer screening: revolutionary, rational, and realizable. NPJ Precis Oncol. 2018:2(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkin WS, Edwards R, Kralj-Hans I, Wooldrage K, Hart AR, Northover JM, Parkin DM, Wardle J, Duffy SW, Cuzick J, et al. Once-only flexible sigmoidoscopy screening in prevention of colorectal cancer: a multicentre randomised controlled trial. Lancet. 2010:375(9726):1624–1633. [DOI] [PubMed] [Google Scholar]
- Beaman EM, Carter DRF, Brooks SA. GALNTs: master regulators of metastasis-associated epithelial-mesenchymal transition (EMT)? Glycobiology. 2022:32(7):556–579. [DOI] [PubMed] [Google Scholar]
- Bennett EP, Mandel U, Clausen H, Gerken TA, Fritz TA, Tabak LA. Control of mucin-type O-glycosylation: a classification of the polypeptide GalNAc-transferase gene family. Glycobiology. 2012:22(6):736–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgert A, Heimburg-Molinaro J, Song X, Lasanajak Y, Ju T, Liu M, Thompson P, Ragupathi G, Barany G, Smith DF, et al. Deciphering structural elements of mucin glycoprotein recognition. ACS Chem Biol. 2012:7(6):1031–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervoni GE, Cheng JJ, Stackhouse KA, Heimburg-Molinaro J, Cummings RD. O-glycan recognition and function in mice and human cancers. Biochem J. 2020:477(8):1541–1564. [DOI] [PubMed] [Google Scholar]
- Chia J, Goh G, Bard F. Short O-GalNAc glycans: regulation and role in tumor development and clinical perspectives. Biochim Biophys Acta. 2016:1860(8):1623–1639. [DOI] [PubMed] [Google Scholar]
- Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, Douville C, Javed AA, Wong F, Mattox A, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018:359(6378):926–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombek GE, Ore AS, Cheng J, Matsumoto Y, Glickman JN, Fleishman A, Heimburg-Molinaro J, Poylin VY, Fabrizio A, Cataldo T, et al. Immunohistochemical analysis of Tn antigen expression in colorectal adenocarcinoma and precursor lesions. BMC Cancer. 2022:22(1):1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faivre J, Dancourt V, Lejeune C, Tazi MA, Lamour J, Gerard D, Dassonville F, Bonithon-Kopp C. Reduction in colorectal cancer mortality by fecal occult blood screening in a French controlled study. Gastroenterology. 2004:126(7):1674–1680. [DOI] [PubMed] [Google Scholar]
- Force USPST, Bibbins-Domingo K, Grossman DC, Curry SJ, Davidson KW, Epling JW Jr, Garcia FAR, Gillman MW, Harper DM, Kemper AR, et al. Screening for colorectal cancer: US preventive services task Force recommendation statement. JAMA. 2016:315(23):2564–2575. [DOI] [PubMed] [Google Scholar]
- Giamougiannis P, Martin-Hirsch PL, Martin FL. The evolving role of MUC16 (CA125) in the transformation of ovarian cells and the progression of neoplasia. Carcinogenesis. 2021:42(3):327–343. [DOI] [PubMed] [Google Scholar]
- Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006:312(5778):1355–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardcastle JD, Chamberlain JO, Robinson MH, Moss SM, Amar SS, Balfour TW, James PD, Mangham CM. Randomised controlled trial of faecal-occult-blood screening for colorectal cancer. Lancet. 1996:348(9040):1472–1477. [DOI] [PubMed] [Google Scholar]
- Henry NL, Hayes DF. Cancer biomarkers. Mol Oncol. 2012:6(2):140–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirohashi S, Clausen H, Yamada T, Shimosato Y, Hakomori S. Blood group a cross-reacting epitope defined by monoclonal antibodies NCC-LU-35 and -81 expressed in cancer of blood group O or B individuals: its identification as Tn antigen. Proc Natl Acad Sci U S A. 1985:82(20):7039–7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann BT, Picksak AS, Kwiatkowski M, Grupp K, Jucker M, Bachmann K, Mercanoglu B, Izbicki JR, Kahlert C, Bockhorn M, et al. Truncated O-GalNAc glycans impact on fundamental signaling pathways in pancreatic cancer. Glycobiology. 2021:cwab088. 10.1093/glycob/cwab088. [DOI] [PubMed] [Google Scholar]
- Holme O, Loberg M, Kalager M, Bretthauer M, Hernan MA, Aas E, Eide TJ, Skovlund E, Schneede J, Tveit KM, et al. Effect of flexible sigmoidoscopy screening on colorectal cancer incidence and mortality: a randomized clinical trial. JAMA. 2014:312(6):606–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inadomi JM, Vijan S, Janz NK, Fagerlin A, Thomas JP, Lin YV, Munoz R, Lau C, Somsouk M, El-Nachef N, et al. Adherence to colorectal cancer screening: a randomized clinical trial of competing strategies. Arch Intern Med. 2012:172(7):575–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju T, Lanneau GS, Gautam T, Wang Y, Xia B, Stowell SR, Willard MT, Wang W, Xia JY, Zuna RE, et al. Human tumor antigens Tn and sialyl Tn arise from mutations in Cosmc. Cancer Res. 2008:68(6):1636–1646. [DOI] [PubMed] [Google Scholar]
- Ju T, Xia B, Aryal RP, Wang W, Wang Y, Ding X, Mi R, He M, Cummings RD. A novel fluorescent assay for T-synthase activity. Glycobiology. 2011:21(3):352–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju T, Wang Y, Aryal RP, Lehoux SD, Ding X, Kudelka MR, Cutler C, Zeng J, Wang J, Sun X, et al. Tn and sialyl-Tn antigens, aberrant O-glycomics as human disease markers. Proteomics Clin Appl. 2013:7(9–10):618–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju T, Aryal RP, Kudelka MR, Wang Y, Cummings RD. The Cosmc connection to the Tn antigen in cancer. Cancer Biomark. 2014:14(1):63–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen AB, Zauber AG, Rutter CM, Naber SK, Doria-Rose VP, Pabiniak C, Johanson C, Fischer SE, Lansdorp-Vogelaar I, Kuntz KM. Estimation of benefits, burden, and harms of colorectal cancer screening strategies: Modeling study for the US preventive services task Force. JAMA. 2016:315(23):2595–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbl AC, Jeschke U, Friese K, Andergassen U. The role of TF- and Tn-antigens in breast cancer metastasis. Histol Histopathol. 2016:31(6):613–621. [DOI] [PubMed] [Google Scholar]
- Kronborg O, Jorgensen OD, Fenger C, Rasmussen M. Randomized study of biennial screening with a faecal occult blood test: results after nine screening rounds. Scand J Gastroenterol. 2004:39(9):846–851. [DOI] [PubMed] [Google Scholar]
- Kudelka MR, Ju T, Heimburg-Molinaro J, Cummings RD. Simple sugars to complex disease--mucin-type O-glycans in cancer. Adv Cancer Res. 2015:126:53–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudelka MR, Hinrichs BH, Darby T, Moreno CS, Nishio H, Cutler CE, Wang J, Wu H, Zeng J, Wang Y, et al. Cosmc is an X-linked inflammatory bowel disease risk gene that spatially regulates gut microbiota and contributes to sex-specific risk. Proc Natl Acad Sci U S A. 2016:113(51):14787–14792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudelka MR, Nairn AV, Sardar MY, Sun X, Chaikof EL, Ju T, Moremen KW, Cummings RD. Isotopic labeling with cellular O-glycome reporter/amplification (ICORA) for comparative O-glycomics of cultured cells. Glycobiology. 2018:28(4):214–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Las Rivas M, Lira-Navarrete E, Gerken TA, Hurtado-Guerrero R. Polypeptide GalNAc-Ts: from redundancy to specificity. Curr Opin Struct Biol. 2019:56:87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Teng TZJ, Shelat VG. Carbohydrate antigen 19-9 - tumor marker: past, present, and future. World J Gastrointest Surg. 2020:12(12):468–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehoux S, Mi R, Aryal RP, Wang Y, Schjoldager KT, Clausen H, van Die I, Han Y, Chapman AB, Cummings RD, et al. Identification of distinct glycoforms of IgA1 in plasma from patients with immunoglobulin a (IgA) nephropathy and healthy individuals. Mol Cell Proteomics. 2014:13(11):3097–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Anver MR, Butcher DO, Gildersleeve JC. Resolving conflicting data on expression of the Tn antigen and implications for clinical trials with cancer vaccines. Mol Cancer Ther. 2009:8(4):971–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm E, Brevinge H, Haglind E. Survival benefit in a randomized clinical trial of faecal occult blood screening for colorectal cancer. Br J Surg. 2008:95(8):1029–1036. [DOI] [PubMed] [Google Scholar]
- Loureiro LR, Carrascal MA, Barbas A, Ramalho JS, Novo C, Delannoy P, Videira PA. Challenges in antibody development against Tn and Sialyl-Tn antigens. Biomol Ther. 2015:5(3):1783–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malila N, Palva T, Malminiemi O, Paimela H, Anttila A, Hakulinen T, Jarvinen H, Kotisaari ML, Pikkarainen P, Rautalahti M, et al. Coverage and performance of colorectal cancer screening with the faecal occult blood test in Finland. J Med Screen. 2011:18(1):18–23. [DOI] [PubMed] [Google Scholar]
- Mandel JS, Bond JH, Church TR, Snover DC, Bradley GM, Schuman LM, Ederer F. Reducing mortality from colorectal cancer by screening for fecal occult blood. Minnesota colon cancer control study. N Engl J Med. 1993:328(19):1365–1371. [DOI] [PubMed] [Google Scholar]
- Mandel JS, Church TR, Bond JH, Ederer F, Geisser MS, Mongin SJ, Snover DC, Schuman LM. The effect of fecal occult-blood screening on the incidence of colorectal cancer. N Engl J Med. 2000:343(22):1603–1607. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y, Kudelka MR, Hanes MS, Lehoux S, Dutta S, Jones MB, Stackhouse KA, Cervoni GE, Heimburg-Molinaro J, Smith DF, et al. Identification of Tn antigen O-GalNAc-expressing glycoproteins in human carcinomas using novel anti-Tn recombinant antibodies. Glycobiology. 2020:30(12):1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestecky J, Tomana M, Moldoveanu Z, Julian BA, Suzuki H, Matousovic K, Renfrow MB, Novak L, Wyatt RJ, Novak J. Role of aberrant glycosylation of IgA1 molecules in the pathogenesis of IgA nephropathy. Kidney Blood Press Res. 2008:31(1):29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen MI, de Haan N, Kightlinger W, Ye Z, Dabelsteen S, Li M, Jewett MC, Bagdonaite I, Vakhrushev SY, Wandall HH. Global mapping of GalNAc-T isoform-specificities and O-glycosylation site-occupancy in a tissue-forming human cell line. Nat Commun. 2022:13(1):6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill RS, Stoita A. Biomarkers in the diagnosis of pancreatic cancer: are we closer to finding the golden ticket? World J Gastroenterol. 2021:27(26):4045–4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posey AD Jr, Schwab RD, Boesteanu AC, Steentoft C, Mandel U, Engels B, Stone JD, Madsen TD, Schreiber K, Haines KM, et al. Engineered CAR T cells targeting the cancer-associated Tn-glycoform of the membrane mucin MUC1 control adenocarcinoma. Immunity. 2016:44(6):1444–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan P, Dabelsteen S, Madsen FB, Francavilla C, Kopp KL, Steentoft C, Vakhrushev SY, Olsen JV, Hansen L, Bennett EP, et al. Immature truncated O-glycophenotype of cancer directly induces oncogenic features. Proc Natl Acad Sci U S A. 2014:111(39):E4066–E4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romer TB, Aasted MKM, Dabelsteen S, Groen A, Schnabel J, Tan E, Pedersen JW, Haue AD, Wandall HH. Mapping of truncated O-glycans in cancers of epithelial and non-epithelial origin. Br J Cancer. 2021:125(9):1239–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rømer TB, Khoder-Agha F, Aasted MKM, de Haan N, Horn S, Dylander A, Zhang T, Pallesen EMH, Dabelsteen S, Wuhrer M, et al. CRISPR-screen identifies ZIP9 and dysregulated Zn2+ homeostasis as a cause of cancer-associated changes in glycosylation. Glycobiology. 2023:cwad003. 10.1093/glycob/cwad003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai K, Yuasa N, Tsukamoto K, Takasaki-Matsumoto A, Yajima Y, Sato R, Kawakami H, Mizuno M, Takayanagi A, Shimizu N, et al. Isolation and characterization of antibodies against three consecutive Tn-antigen clusters from a phage library displaying human single-chain variable fragments. J Biochem. 2010:147(6):809–817. [DOI] [PubMed] [Google Scholar]
- Sasaki M, Yamato T, Nakanuma Y. Expression of sialyl-Tn, Tn and T antigens in primary liver cancer. Pathol Int. 1999:49(4):325–331. [DOI] [PubMed] [Google Scholar]
- Schoen RE, Pinsky PF, Weissfeld JL, Yokochi LA, Church T, Laiyemo AO, Bresalier R, Andriole GL, Buys SS, Crawford ED, et al. Colorectal-cancer incidence and mortality with screening flexible sigmoidoscopy. N Engl J Med. 2012:366(25):2345–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholefield JH, Moss SM, Mangham CM, Whynes DK, Hardcastle JD. Nottingham trial of faecal occult blood testing for colorectal cancer: a 20-year follow-up. Gut. 2012:61(7):1036–1040. [DOI] [PubMed] [Google Scholar]
- Segnan N, Senore C, Andreoni B, Aste H, Bonelli L, Crosta C, Ferraris R, Gasperoni S, Penna A, Risio M, et al. Baseline findings of the Italian multicenter randomized controlled trial of ``once-only sigmoidoscopy''–SCORE. J Natl Cancer Inst. 2002:94(23):1763–1772. [DOI] [PubMed] [Google Scholar]
- Segnan N, Armaroli P, Bonelli L, Risio M, Sciallero S, Zappa M, Andreoni B, Arrigoni A, Bisanti L, Casella C, et al. Once-only sigmoidoscopy in colorectal cancer screening: follow-up findings of the Italian randomized controlled trial--SCORE. J Natl Cancer Inst. 2011:103(17):1310–1322. [DOI] [PubMed] [Google Scholar]
- Shaukat A, Mongin SJ, Geisser MS, Lederle FA, Bond JH, Mandel JS, Church TR. Long-term mortality after screening for colorectal cancer. N Engl J Med. 2013:369(12):1106–1114. [DOI] [PubMed] [Google Scholar]
- Sina AA, Carrascosa LG, Liang Z, Grewal YS, Wardiana A, Shiddiky MJA, Gardiner RA, Samaratunga H, Gandhi MK, Scott RJ, et al. Epigenetically reprogrammed methylation landscape drives the DNA self-assembly and serves as a universal cancer biomarker. Nat Commun. 2018:9(1):4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamey TA, Yang N, Hay AR, McNeal JE, Freiha FS, Redwine E. Prostate-specific antigen as a serum marker for adenocarcinoma of the prostate. N Engl J Med. 1987:317(15):909–916. [DOI] [PubMed] [Google Scholar]
- Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT, Lavrsen K, Dabelsteen S, Pedersen NB, Marcos-Silva L, et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013:32(10):1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowell SR, Ju T, Cummings RD. Protein glycosylation in cancer. Annu Rev Pathol. 2015:10(1):473–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Ju T, Cummings RD. Differential expression of Cosmc, T-synthase and mucins in Tn-positive colorectal cancers. BMC Cancer. 2018:18(1):827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas W, White CM, Mah J, Geisser MS, Church TR, Mandel JS. Longitudinal compliance with annual screening for fecal occult blood. Minnesota colon cancer control study. Am J Epidemiol. 1995:142(2):176–182. [DOI] [PubMed] [Google Scholar]
- Thomas D, Sagar S, Caffrey T, Grandgenett PM, Radhakrishnan P. Truncated O-glycans promote epithelial-to-mesenchymal transition and stemness properties of pancreatic cancer cells. J Cell Mol Med. 2019:23(10):6885–6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissfeld JL, Schoen RE, Pinsky PF, Bresalier RS, Church T, Yurgalevitch S, Austin JH, Prorok PC, Gohagan JK, PP Team . Flexible sigmoidoscopy in the PLCO cancer screening trial: results from the baseline screening examination of a randomized trial. J Natl Cancer Inst. 2005:97(13):989–997. [DOI] [PubMed] [Google Scholar]
- Xia L, Ju T, Westmuckett A, An G, Ivanciu L, McDaniel JM, Lupu F, Cummings RD, McEver RP. Defective angiogenesis and fatal embryonic hemorrhage in mice lacking core 1-derived O-glycans. J Cell Biol. 2004:164(3):451–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Reagents and original data are available upon request to Richard D. Cummings, or are provided in the paper, or online at: https://ncfg.hms.harvard.edu/ncfg-data/microarray-data.


