Abstract
The gene targeting techniques used to modify chromosomes in mouse embryonic stem cells have had limited success with many other cell types, especially normal primary cells with restricted growth capacity outside the organism. This is due in large part to the technical problems and/or inefficiency of conventional DNA transfer methods, as well as the low rates of homologous recombination obtained in unselected cell populations. We recently described an alternative approach in which adeno-associated virus (AAV) vectors were used to modify homologous chromosomal sequences, and targeting rates close to 1% were observed at the single copy hypoxanthine phosphoribosyl transferase (HPRT) locus in normal human cells (D. W. Russell and R. K. Hirata, Nat. Genet. 18:325–330, 1998). Here we report experiments in which we used a retroviral shuttle vector system to introduce and characterize target loci in human chromosomes, and demonstrate that AAV vectors can correct several types of mutations with high fidelity, independent of chromosomal position. The gene targeting rates varied depending on the type of mutation being corrected, implicating cellular mismatch recognition functions in the reaction. Since AAV vectors can efficiently deliver DNA to many cell types both in vivo and ex vivo, our results suggest that AAV-mediated gene targeting will have wide applicability, including therapeutic gene correction.
Adeno-associated virus (AAV) is a single-stranded, linear DNA virus with a 4.7-kb genome consisting of the viral rep and cap genes flanked by inverted terminal repeat (TR) sequences. AAV vectors containing foreign DNA between the viral TRs can be packaged by rep and cap gene products supplied in trans (10). They can transduce many cell types both by random chromosomal integration (13, 20) and transient gene expression from episomal vector genomes (1). In our previous study, we showed that AAV vectors can also transduce cells by introducing either a 4-bp insertion or a 14-bp deletion mutation at homologous chromosomal loci (12). The gene targeting rates obtained by AAV vectors in normal, unselected human cells (>10−3) were 3 to 5 logs higher than those obtained by conventional targeting methods (2, 17, 19), perhaps due to the efficient nuclear delivery or single-stranded nature of the vector genome (12).
The scientific and therapeutic potential of AAV-mediated gene targeting has not been established yet and will depend in large part on the types of genetic modifications that can be introduced, the accessibility of different chromosomal sites for targeting, and the fidelity of the reaction. A fundamental remaining issue is the structure of targeted loci, including the ends of homology between the target gene and vector genome, since other targeting methods can lead to alterations at these locations (4, 5, 16), and this was not studied in previous AAV experiments. To address these issues, we developed a retroviral shuttle vector system for the introduction and correction of target genes containing a variety of mutations at multiple chromosomal positions. This system allowed us to compare the average correction rates of different mutations located throughout the human genome and to completely determine the structure of the targeted loci at the DNA sequence level.
MATERIALS AND METHODS
Vectors.
The plasmid pLHSNO contains the following sequences in this order (with GenBank accession numbers and nucleotides): pLXSHD retroviral vector backbone (M64753, 1 to 1648); hph gene (K01193, 190 to 1250); simian virus 40 (SV40) early and Tn5 promoters and neo gene (U02434, 3490 to 1929); p15A origin (X06403, 597 to 1409); and pLXSHD backbone (M64753, 3370 to 6374). The various mutations shown in Fig. 1c were introduced into pLHSNO by standard techniques (8, 14) and confirmed by DNA sequencing. Retroviral vectors were produced by calcium phosphate transfection of PG13 packaging cells (9) with pLHSNO and its derivatives, collection of conditioned medium 2 days later, and passage through 0.45-μm-pore-size filters. AAV-SNO648 vector stocks (AAV-2 serotype) were prepared as previously described (12), and their particle numbers were based on the amount of full-length, single-stranded vector DNA detected by alkaline Southern analysis (7).
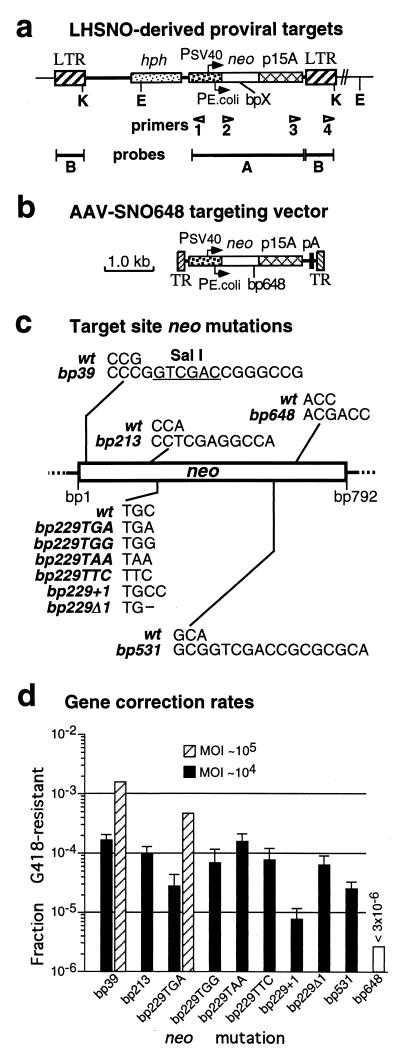
FIG. 1.
Gene correction of retroviral vector target loci. (a) Map of integrated proviral target sites based on LHSNO retroviral vector. The positions of sequencing primers, Southern analysis probes, and KpnI (K) and EcoRI (E) restriction sites are shown. bpX indicates the presence of the neo mutations shown in panel c. (b) Map of AAV-SNO648 targeting vector (12) containing the bp648 neo mutation. The locations of AAV terminal repeats (TR), SV40 (Psv40) and Tn5 (Pe.coli) promoters, transcriptional start sites (arrows), hph and neo genes, p15A plasmid replication origin, polyadenylation site (pA), and retrovirus LTRs (LTR) are shown in panels a and b. (c) Sequences of the wild-type (wt) neo gene present in retroviral vector LHSNO and the different neo mutations present in LHSNO-derived proviral target sites. Mutation names are based on the nucleotide position within the 792-bp neo coding sequence. A SalI restriction site is indicated (underlined bases). (d) Correction rates of the indicated neo mutations in HT-1080 cells containing LHSNO-derived target loci, infected with the AAV-SNO648 vector at an MOI of 104 (solid columns; bars indicate standard errors; n = 3) or 105 (hatched columns; n = 1) vector particles/cell, then plated in the presence or absence of G418. The fraction G418-resistant is the number of G418-resistant colonies/the total number of (unselected) colonies (see Materials and Methods). No G418-resistant colonies were obtained with cells containing the bp648 neo mutation at an MOI of 104 (open column). The reversion rates of all the polyclonal populations were <2 × 10−7, except for those containing the bp229TGG mutation, which had a reversion rate of 2.6 × 10−6 (still below the correction rate).
Cell culture and transduction.
PG13 cells (9), HT-1080 cells (11), and MHF2 normal human fibroblasts (12) were propagated in Dulbecco’s modified Eagle’s medium supplemented with heat-inactivated 10% fetal bovine serum at 37°C. Proviral target sites were introduced into HT-1080 and MHF2 cells (6 × 105 to 8 × 105 cells in 6-cm-diameter dishes) by the addition of LHSNO and LHSNO-derived retroviral vector stocks, followed by selection in hygromycin B (0.2 mg/ml; Calbiochem) for 6 to 9 days. The polyclonal, transduced populations were all derived from >104 independent transduction events, as determined by plating dilutions of the transduced cells in selective medium.
To measure neo gene correction rates (Fig. 1d), HT-1080-derived cells were seeded on day 1 (for a multiplicity of infection [MOI] of 104, 8 × 104 cells/well in 12-well plates [n = 1] or 5 × 104 cells/well in 24-well plates [n = 2]; for an MOI of 105, 5 × 103 cells/well in 96-well plates), infected with AAV-SNO648 vector on day 2 (MOIs assume one population doubling), and then treated with trypsin and plated at different dilutions on day 3. On day 4, G418 was added to the medium (0.7 mg of active compound/ml), except for those dishes used to calculate the total number of unselected CFU. These cells were cultured for 10 to 12 days with media changes every 3 to 4 days until all cells in G418-containing control cultures that did not receive vector were dead. Correction rates were calculated as the number of G418-resistant CFU in the infected culture divided by the total number of CFU. Exposure to vector was not cytotoxic, since plating efficiencies (total CFU/plated cell number) were approximately 70% with or without AAV vector infection. Reversion rates were measured by plating ∼107 uninfected cells from each clone in G418.
Gene correction in MHF2 cells containing LHSNO39 target loci was measured in a similar way (plated at 5 × 103 cells/well in 96-well plates 1 day before infection; MOI, 105), except the cells from each well were transferred to a 6-cm-diameter dish on day 3 and cultured together until day 10 to improve phenotypic expression of the neo gene, dilutions were plated on day 11, and G418 selection (0.7 mg of active compound/ml) began on day 12. The neo reversion rate in these cells was <1.3 × 10−7. Independent, corrected MHF2-LHSNO39 clones targeted by AAV-SNO648 were obtained by plating 104 cells/well in 10 separate 48-well plates on day 1, infecting with AAV-SNO648 (MOI, 5 × 104) on day 2, splitting into multiple 12-well plates (24 wells per original 48 wells) on day 3 to ensure clonality, and selecting with G418 on day 10. After 10 to 12 additional days, the wells containing G418-resistant colonies were identified and expanded for DNA isolation. Assuming 100% plating efficiency, the gene correction rate for this experiment was (2.2 ± 0.7) × 10−4 (G418-resistant colonies per original 48-well culture/total cells per 48-well culture; n = 10). Pools of MHF2-LHSNO39 cells corrected with AAV-SNO648 were obtained the same way except all the cells were plated into one 10-cm-diameter dish on day 11 and expanded to confluence in the presence of G418.
DNA analysis.
Genomic DNA was isolated and analyzed by Southern blots as previously described (14). Integrated retrovirus target loci were rescued by digestion of genomic DNA with EcoRI, circularization with T4 ligase, transfer to Escherichia coli XL1Blue MRF′ (Stratagene), and selection for kanamycin resistance as previously described (13). Three to eight plasmids recovered from each clone were analyzed by restriction digests. Some plasmids (6 of 64) contained an additional EcoRI fragment derived from ligation of unrelated chromosomal DNA. Sequencing was performed with an ABI Prism BigDye terminator cycle sequencing kit (Perkin-Elmer) and an ABI Prism 377 DNA sequencer. Primers used for sequencing were as follows: 9804A (5′-dGGGCGGGACTATGGTTGC-3′) for 5′ homology ends; 9606D (5′-dTTCCTTGCCGCAGCGGTGC-3′) for the neo gene; 9804B (5′-dTGCGCTCCTCCAAGCCAG-3′) for the 3′ homology ends; and 9805A (5′-dGCGCCAGTCTTCCGATAG-3′) for the 3′ long terminal repeat (LTR) chromosomal junction. Unambiguous sequences were obtained from each plasmid from bp −22 to +663 of the neo coding sequence, from 350 bp upstream to 185 bp downstream of the 5′ homology end, from 95 bp upstream to 450 bp downstream of the 3′ homology end, and from −80 bp to at least 300 bp beyond the 3′ LTR chromosomal junction.
RESULTS AND DISCUSSION
The retroviral vector LHSNO (Fig. 1a) contains a hygromycin-B phosphotransferase gene (hph) under the control of the Moloney leukemia virus (MLV) LTR promoter, a neo gene controlled by both the SV40 early and bacterial Tn5 promoters, and a p15A plasmid replication origin. Several mutations, including insertions, deletions, and base substitutions, were introduced into the neo gene of LHSNO to serve as targets for gene correction, each of which disrupted neo gene function (Fig. 1c). Cells transduced by these retroviral vectors can be selected for by growth in hygromycin, and then the mutant neo genes in the integrated vector proviruses can be corrected by gene targeting with AAV vectors and scored by selection in G418. The AAV-SNO648 vector used for gene correction contains 2,376 bp of sequence homology to the target loci, with a 3-nucleotide insertion at position 648 of the neo coding sequence that disrupts gene function (Fig. 1b) (12). Inclusion of the p15A replication origin in the target loci allowed us to recover corrected proviruses as bacterial plasmids for sequence analysis (13).
Polyclonal populations of HT-1080 human fibrosarcoma cells were prepared by transduction with LHSNO-derived retroviral vectors containing different neo mutations and selection in hygromycin. Each population contained >104 independent transduction events, with proviral target sites presumably integrated at different chromosomal locations. These transduced HT-1080 cell populations were infected with the AAV-SNO648 targeting vector, plated at different dilutions, and then cultured in the presence or absence of G418 (Fig. 1d). At an MOI of 104 AAV vector particles/cell, all the neo target site mutations could be corrected except bp648 (which was expected, as this is the same mutation present in the AAV targeting vector). Correction rates of the bp39 and bp229TGA mutations increased about 10-fold at the higher MOI of 105 (to 1.4 × 10−3 in the case of bp39). These correction rates were similar to those observed in HeLa cells containing three integrated neo genes with the bp39 mutation (12), and in both cases, targeting rates increased at higher MOIs. The values shown may underestimate the true targeting rates, since cotransfer of the bp648 mutation in the AAV vector would not produce G418 resistance, and only 85% of the target sites contained a potentially correctable neo gene, based on the fraction of G418-resistant cells present in hygromycin-resistant populations transduced with the parental LHSNO vector (wild-type neo gene). Similar experiments were performed with a polyclonal population of MHF2 normal, male human fibroblasts containing the bp39 mutation, which had a neo gene correction rate of 7.6 × 10−4 at an MOI of 105 AAV-SNO648 particles/cell (see Materials and Methods). This frequency was remarkably close to that obtained with HT-1080 cells (1.4 × 10−3) despite the use of different cell lines with distinct sets of target loci.
The corrected mutations included 1-, 8-, and 14-bp insertions, a 1-bp deletion, three single nucleotide substitutions, and a double nucleotide substitution, which were located at distances of 117 bp (bp531) to 609 bp (bp39) from the bp648 mutation in the AAV targeting vector. Although shorter intermutation distance appeared to decrease correction rates (compare the bp39 and bp531 mutations, both of which were 14-bp insertions), the type of modification being introduced produced more significant effects, with the bp229+1 correction rate being about a log lower than those of other mutations located at the same site. Although more experiments will be required to define the mechanism of gene correction, these findings suggest that mismatch repair enzymes may be involved, since they could recognize specific mutations during heteroduplex formation.
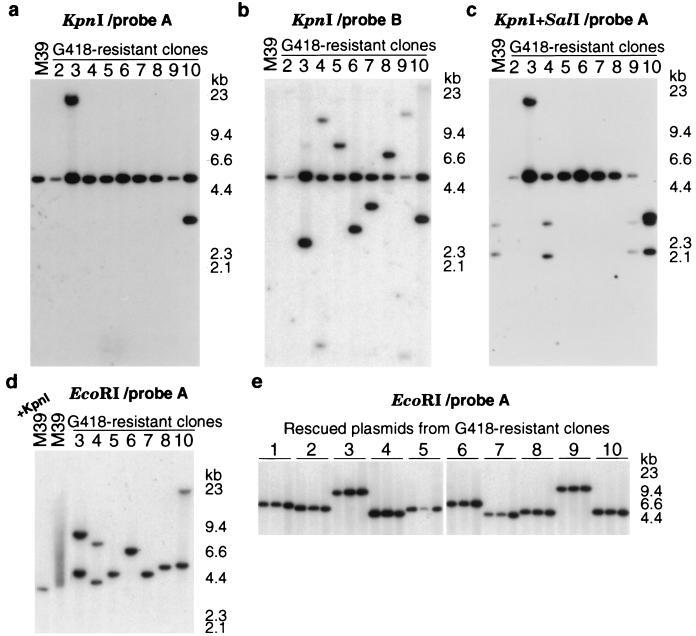
To analyze the structure of targeted loci, 88 independent G418-resistant MHF2 fibroblast clones were generated by neo gene correction of chromosomal LHSNO39 target sites with AAV-SNO648 (MOI, 50,000; neo correction rate of 2.2 × 10−4; see Materials and Methods). Ten of these clones could be expanded to confluence in 6-cm-diameter dishes without undergoing senescence, and these were used to isolate genomic DNA. The proviral target sites in G418-resistant MHF2 clones and control DNA from parental, polyclonal, MHF2 cells containing uncorrected LHSNO39 targets (M39) were analyzed by Southern blots (the limited amount of DNA recovered from clone 1 was inadequate for Southern analysis). Digestion with KpnI was used to cut the proviral LTRs and demonstrate the presence of a neo-hybridizing target locus fragment of the expected 5.1-kb size in each sample, as well as additional fragments in clones 3 and 10 (Fig. 2a). Based on the lack of hybridization to retroviral LTR sequences (Fig. 2b), the extra fragment in clone 3 represents a randomly integrated AAV-SNO648 vector genome, which we also observed previously in some targeted cells (12). The extra fragment in clone 10 hybridized to LTR sequences, and further analysis showed this to be a rearranged retroviral vector provirus (see below). Each clone also contained LTR-hybridizing chromosomal junction fragments of variable sizes, as expected from random integration of the retroviral vector targets (Fig. 2b). Clones 4, 9, and 10 contained two junction fragments, consistent with the presence of two independent target loci. Additional fragments were not observed in the parental M39 cells, due to the polyclonal nature of the cell population. Corrected neo genes were identified by KpnI and SalI double digestion and probing with internal neo sequences (Fig. 2c), since the bp39 mutation contains a SalI site (Fig. 1b). Clones 2 through 9 all contained SalI-resistant, 5.1-kb target loci, demonstrating removal of the bp-39 mutation. Although this fragment was missing in clone 10, further analysis showed that the corrected neo gene was present in the doublet at 2.9 kb (see below). The 2.1- and 2.9-kb fragments in clones 4, 9, and 10 (and parental M39 cells) are the expected sizes of uncorrected, SalI-sensitive second target loci. Taken together, these results show that eight of nine G418-resistant fibroblast clones contained a single corrected target locus with no major rearrangements. In the three clones with two target loci, only one locus underwent gene correction.
FIG. 2.
Southern analysis of targeted loci. (a to d) Genomic DNA from the parental polyclonal population of MHF2 cells transduced with LHSNO39 (M39) and G418-resistant clones 2 to 10 obtained by gene targeting. Genomic DNAs were digested with KpnI (a, b), KpnI and SalI (c), or EcoRI (d) and hybridized with neo probe A (a, c, and d) or LTR probe B (b). Due to the limited amount of genomic DNA obtained, clone 1 was not analyzed in panels a to c, and clones 1, 2, and 9 were not analyzed in panel d. (e) Southern blot of plasmid DNA recovered from the G418-resistant clones 1 to 10. Three plasmids from each clone were linearized with EcoRI and hybridized with probe A.
The corrected neo genes and flanking chromosomal DNA from G418-resistant MHF2 fibroblast clones 1 to 10 were recovered as bacterial plasmids for sequence analysis. Genomic DNAs were digested with EcoRI (which cuts once in the target locus; Fig. 1a), circularized with DNA ligase, and transferred to E. coli as plasmids conferring kanamycin resistance. The plasmids recovered from each fibroblast clone contained a common EcoRI fragment (Fig. 2e; Table 1), and genomic Southern analysis of those clones for which there was enough remaining DNA demonstrated a corresponding EcoRI fragment of the appropriate size (Fig. 2d; Table 1). Clones 3, 4, and 10 also had unrescued fragments based on the presence of either a randomly integrated AAV-SNO648 genome (clone 3) or additional uncorrected target loci (clones 4 and 10). The smear observed in M39 genomic DNA is due to the polyclonal nature of the parental cell population (Fig. 2d).
TABLE 1.
Analysis of corrected LHSNO39 target loci in rescued plasmids
| G418-resistant cellsa | Genomic DNA EcoRI digests on Southern blot (kb)b | Result for rescued plasmids
|
||||
|---|---|---|---|---|---|---|
| EcoRI digest (kb) | Description or sequence of plasmidc
|
|||||
| 5′ End of homology | neo gene | 3′ End of homology | LTR-chromosome junctiond | |||
| Clone 1 | NT | 6.0 | WT | WT | WT | TTTCATAGCTGGGAT |
| Clone 2 | NT | 5.0 | WT | WT | WT | TTTCAAGACGAAAAA |
| Clone 3 | 9.0, 4.6 | 9.0 | WT | WT | WT | TTTCACCCGGAGCCC |
| Clone 4 | 7.3, 4.2 | 4.2 | WT | WT | WT | TTTCAGTACCATCTA |
| Clone 5 | 4.5 | 4.5 | WT | WT | WT | TTTCAGCTTAAGGTA |
| Clone 6 | 6.7 | 6.7 | WT | WT | WT | TTTCATGTAGAACAG |
| Clone 7 | 4.7 | 4.7 | WT | WT | WT | TTTCACTTCCTGATC |
| Clone 8 | 5.0 | 5.0 | WT | WT | WT | TTTCATCTCTTAAAA |
| Clone 9 | NT | 11 | WT | WT | WT | TTTCAGCCTGCATGA |
| Clone 10 | 22, 5.5 | 5.5 | Rearranged | WT | WT | TTTCATGCTGAACTA |
| Pool 1 | Polyclonal | 4.6 | WT | WT | WT | TTTCAAAACCGCCCT |
| Pool 2 | Polyclonal | 13 | WT | WT | WT | TTTCAATGCCACGCA |
All cells derived from MHF2 fibroblasts with LHSNO39 target sites corrected by AAV-SNO648.
NT, not tested due to inadequate amount of genomic DNA.
WT, wild type.
The first 5 nucleotides are from the LTR.
The recovered plasmids from the 10 fibroblast clones described above, as well as two plasmids recovered from pooled cultures of corrected, G418-resistant fibroblasts were used as templates for DNA sequencing (Table 1). The neo genes from all plasmids had the wild-type sequence, demonstrating correction of the bp39 mutation without secondary mutations. The regions at the end of homology between the AAV targeting vector and the retroviral target loci were also sequenced. All 12 plasmids had wild-type sequence identical to the target locus at the 3′ ends of homology. Eleven of 12 plasmids also had wild-type sequence at the 5′ ends of homology. Clone 10 contained a 15- to 18-bp insertion of contiguous, nonhomologous AAV-SNO648 sequence at the 5′ homology end, followed by an inverted LTR-derived sequence from a different proviral integration site (Fig. 3). This suggests that the rearranged homology end in clone 10 involved a recombination event between two retroviral elements, which may have contributed to the incorporation of nonhomologous AAV vector DNA. Taken together, these results show that gene targeting by AAV vectors is a high-fidelity reaction, with 11 of 12 targeted loci having incorporated the intended modification without other mutations or rearrangements. Although these experiments cannot allow us to rule out a selection bias against secondary mutations that disrupt neo function, our previous analysis of HPRT gene targeting in normal human fibroblasts demonstrated that the region surrounding the introduced modification lacks secondary mutations even in the absence of a selection bias (12).
FIG. 3.
Structure of the single rearranged target locus. The rearrangement that occurred at the 5′ end of homology in the corrected locus from clone 10 (see Table 1) is shown. The corrected locus contains 15 to 18 additional bp (there are 3 bp of sequence overlap) of nonhomologous, contiguous AAV vector sequence joined to partial, inverted 3′-LTR sequences from a different retroviral vector provirus. The rearrangement therefore represents a recombination event between the AAV vector genome and two retroviral vector proviruses. The structures of the original, uncorrected proviral target locus, AAV targeting vector, and rearranged, corrected locus are all shown, as is the DNA sequence at the 5′ end of homology. The locations of the hph and neo genes (with bp39 and bp648 mutations), retroviral LTRs, and AAV TRs are indicated.
We also sequenced the human chromosomal DNA flanking the proviral target loci in the recovered plasmids (Table 1). Each flanking sequence was different and consisted of 3′ LTR sequences fused to human DNA in a typical MLV integration pattern (3). For controls, 10 distinct plasmids containing untargeted loci were recovered by using the same methods from the genomic DNA of MHF2 fibroblasts transduced with the LHSNO retroviral vector, which has a functional neo gene. When compared for G+C content, CpG dinucleotide frequency, or the presence of repeated sequences, no significant differences between the flanking human sequences of corrected loci and untargeted control loci were noted (data not shown). Thus, we conclude that AAV-mediated gene targeting occurs at multiple chromosomal positions, without apparent selection for a subset of target sites. Although retroviral integration may preferentially occur in active chromatin (3, 15), most chromosomal regions and expressed genes are accessible (3, 6, 18), so our use of polyclonal cell populations with >104 independent proviral target loci should have minimized position effects and produced average gene correction rates sampled from sites throughout the human genome. Since the target genes were transcribed from the SV40 promoter, we cannot determine if unexpressed loci can be targeted.
In this study, we developed a retrovirus shuttle vector system to characterize chromosomal loci undergoing gene targeting. The advantages of this system are that the structure of both the target locus and targeting vector can be completely controlled, targeting can be studied at many chromosomal positions, the targeted loci can be recovered and sequenced, and targeting rates can be compared in different cell types. Using this approach, we were able to demonstrate that gene targeting by AAV vectors is a high-fidelity reaction involving mismatch recognition that can accurately introduce several types of mutations at homologous target sites present at multiple chromosomal locations. The targeting frequencies were significantly higher than those obtained in human cells by conventional targeting methods and may increase further at higher MOIs (12). The high frequency and fidelity of the process should make possible the precise genetic manipulation of chromosomes in mammalian cells. This will prove useful for introducing subtle mutations into normal cells for research purposes, for creating mutant animal strains, and also for therapeutic gene targeting, where accurate correction of genetic defects will be required.
ACKNOWLEDGMENTS
We thank John Weller for technical assistance.
This work was supported by grants from the March of Dimes Birth Defects Foundation, the Cystic Fibrosis Foundation, and the U.S. National Institutes of Health.
REFERENCES
- 1.Bertran J, Miller J L, Yang Y, Fenimore Justman A, Rueda F, Vanin E F, Nienhuis A W. Recombinant adeno-associated virus-mediated high-efficiency, transient expression of the murine cationic amino acid transporter (ecotropic retroviral receptor) permits stable transduction of human HeLa cells by ecotropic retroviral vectors. J Virol. 1996;70:6759–6766. doi: 10.1128/jvi.70.10.6759-6766.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown J P, Wei W, Sedivy J M. Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science. 1997;277:831–834. doi: 10.1126/science.277.5327.831. [DOI] [PubMed] [Google Scholar]
- 3.Brown P O. Integration of retroviral DNA. Curr Top Microbiol Immunol. 1990;157:19–48. doi: 10.1007/978-3-642-75218-6_2. [DOI] [PubMed] [Google Scholar]
- 4.Doetschman T, Maeda N, Smithies O. Targeted mutation of the Hprt gene in mouse embryonic stem cells. Proc Natl Acad Sci USA. 1988;85:8583–8587. doi: 10.1073/pnas.85.22.8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujita A, Sakagami K, Kanegae Y, Saito I, Kobayashi I. Gene targeting with a replication-defective adenovirus vector. J Virol. 1995;69:6180–6190. doi: 10.1128/jvi.69.10.6180-6190.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hicks G G, Shi E G, Li X M, Li C H, Pawlak M, Ruley H E. Functional genomics in mice by tagged sequence mutagenesis. Nat Genet. 1997;16:338–344. doi: 10.1038/ng0897-338. [DOI] [PubMed] [Google Scholar]
- 7.Inoue N, Russell D W. Packaging cells based on inducible gene amplification for the production of adeno-associated virus vectors. J Virol. 1998;72:7024–7031. doi: 10.1128/jvi.72.9.7024-7031.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 9.Miller A D, Garcia J V, von Suhr N, Lynch C M, Wilson C, Eiden M V. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J Virol. 1991;65:2220–2224. doi: 10.1128/jvi.65.5.2220-2224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muzyczka N. Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr Top Microbiol Immunol. 1992;158:97–129. doi: 10.1007/978-3-642-75608-5_5. [DOI] [PubMed] [Google Scholar]
- 11.Rasheed S, Nelson Rees W A, Toth E M, Arnstein P, Gardner M B. Characterization of a newly derived human sarcoma cell line (HT-1080) Cancer. 1974;33:1027–1033. doi: 10.1002/1097-0142(197404)33:4<1027::aid-cncr2820330419>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 12.Russell D W, Hirata R K. Human gene targeting by viral vectors. Nat Genet. 1998;18:325–330. doi: 10.1038/ng0498-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rutledge E A, Russell D W. Adeno-associated virus vector integration junctions. J Virol. 1997;71:8429–8436. doi: 10.1128/jvi.71.11.8429-8436.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 15.Sandmeyer S B, Hansen L J, Chalker D L. Integration specificity of retrotransposons and retroviruses. Annu Rev Genet. 1990;24:491–518. doi: 10.1146/annurev.ge.24.120190.002423. [DOI] [PubMed] [Google Scholar]
- 16.Thomas K R, Deng C, Capecchi M R. High-fidelity gene targeting in embryonic stem cells by using sequence replacement vectors. Mol Cell Biol. 1992;12:2919–2923. doi: 10.1128/mcb.12.7.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams S R, Ousley F C, Vitez L J, DuBridge R B. Rapid detection of homologous recombinants in nontransformed human cells. Proc Natl Acad Sci USA. 1994;91:11943–11947. doi: 10.1073/pnas.91.25.11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Withers Ward E S, Kitamura Y, Barnes J P, Coffin J M. Distribution of targets for avian retrovirus DNA integration in vivo. Genes Dev. 1994;8:1473–1487. doi: 10.1101/gad.8.12.1473. [DOI] [PubMed] [Google Scholar]
- 19.Yáñez R J, Porter A C. Therapeutic gene targeting. Gene Ther. 1998;5:149–159. doi: 10.1038/sj.gt.3300601. [DOI] [PubMed] [Google Scholar]
- 20.Yang C C, Xiao X, Zhu X, Ansardi D C, Epstein N D, Frey M R, Matera A G, Samulski R J. Cellular recombination pathways and viral terminal repeat hairpin structures are sufficient for adeno-associated virus integration in vivo and in vitro. J Virol. 1997;71:9231–9247. doi: 10.1128/jvi.71.12.9231-9247.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]