Abstract
The coronavirus membrane (M) protein is the key player in virion assembly. One of its functions is to mediate the incorporation of the spikes into the viral envelope. Heterotypic interactions between M and the spike (S) protein can be demonstrated by coimmunoprecipitation and by immunofluorescence colocalization, after coexpression of their genes in eukaryotic cells. Using these assays in a mutagenetic approach, we have mapped the domains in the M protein that are involved in complex formation between M and S. It appeared that the 25-residue luminally exposed amino-terminal domain of the M protein is not important for M-S interaction. A 15-residue deletion, the insertion of a His tag, and replacement of the ectodomain by that of another coronavirus M protein did not affect the ability of the M protein to associate with the S protein. However, complex formation was sensitive to changes in the transmembrane domains of this triple-spanning protein. Deletion of either the first two or the last two transmembrane domains, known not to affect the topology of the protein, led to a considerable decrease in complex formation, but association was not completely abrogated. Various effects of changes in the part of the M protein that is located at the cytoplasmic face of the membrane were observed. Deletions of the extreme carboxy-terminal tail appeared not to interfere with M-S complex formation. However, deletions in the amphipathic domain severely affected M-S interaction. Interestingly, changes in the amino-terminal and extreme carboxy-terminal domains of M, which did not disrupt the interaction with S, are known to be fatal to the ability of the protein to engage in virus particle formation (C. A. M. de Haan, L. Kuo, P. S. Masters, H. Vennema, and P. J. M. Rottier, J. Virol. 72:6838–6850, 1998). Apparently, the structural requirements of the M protein for virus particle assembly differ from the requirements for the formation of M-S complexes.
Enveloped viruses contain a nucleocapsid (NC) surrounded by a lipid bilayer which accommodates the viral membrane proteins. This envelope is formed by budding of the NC through cellular membranes. For most viruses, the viral envelope proteins are incorporated efficiently while host proteins are excluded. The specificity of the virus assembly process is determined by interactions between the viral membrane proteins and with NC or matrix proteins.
Coronaviruses, positive-strand RNA viruses, acquire their envelope by budding of the helical NC into the intermediate compartment between the endoplasmic reticulum (ER) and the Golgi complex (11, 12, 35). The coronavirus envelope contains three or four viral proteins. The membrane (M) glycoprotein is the most abundant envelope protein. It is a triple-spanning membrane protein with a short amino-terminal domain on the outside of the virus (or in the lumen of intracellular organelles) and a long carboxy-terminal domain on the inside (or in the cytoplasm) (reviewed by Rottier [27]). The spike (S) glycoprotein, trimers of which form the virion peplomers, is another major structural protein. It is involved in binding of virions to the host cell and in virus-cell and cell-cell fusion (reviewed by Cavanagh [3]). Some, but not all, coronaviruses contain a third major envelope protein, the hemagglutinin esterase (HE) (reviewed by Brian et al. [2]). Finally, the small envelope (E) protein is a minor, poorly characterized but essential structural component (7, 30, 37).
Lateral interactions between the coronavirus membrane proteins are thought to mediate the formation of the virion envelope. The M protein is obviously the key player in assembly. When expressed alone, it accumulates in the Golgi complex (11, 13) in homomultimeric complexes (15). However, in combination with the E protein, virus-like particles (VLPs) similar to authentic virions in size and shape are assembled, demonstrating that the M and E proteins are the minimal requirements for envelope formation (1, 37). Using the VLP assembly system, we recently showed that mouse hepatitis virus (MHV) particle assembly is critically sensitive to changes in all domains of the M protein. Furthermore, we observed that assembly-competent M protein is able to rescue assembly-incompetent M protein into VLPs, providing evidence for the existence of M-M interactions, which are thought to drive coronavirus envelope assembly (4).
The S protein is dispensable for coronavirus particle assembly. Growth of coronaviruses in the presence of tunicamycin gave rise to the production of spikeless, noninfectious virions (10, 20, 28, 31). Furthermore, temperature-sensitive mutant coronaviruses that fail to incorporate the S protein into particles at the nonpermissive temperature have been described (17, 25). The S protein was also found to be dispensable for VLP formation, although it became incorporated into the particles when present (1, 37). Incorporation of the S protein into the viral envelope is directed by heterotypic interactions with the M protein. These interactions were demonstrated by coimmunoprecipitation, cosedimentation, and immunofluorescence analyses (22, 23). The latter assay made use of the colocalization of the two proteins when coexpressed: under these conditions, the S protein, which is transported to the plasma membrane when on its own, coaccumulates with the M protein in the Golgi complex, the natural residence of M. The M protein was also shown to interact with the other major envelope protein, HE. In cells infected with the bovine coronavirus, which expresses an HE protein, complexes consisting of the M, S, and HE proteins were detected by coimmunoprecipitation (21).
In view of the apparent role of the M protein as the key organizer in envelope assembly and considering the essential functions of the viral spikes, we decided to investigate the interactions between the MHV M and S proteins in more detail. In the present study, we focused on the M protein. Using coimmunoprecipitation and immunofluorescence assays in a mutagenetic analysis, we mapped the M-protein domains involved in M-S interaction.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
The recombinant modified vaccinia virus strain Ankara (MVA) encoding the T7 RNA polymerase (MVA-T7pol) (33) was a kind gift of G. Sutter. OST7-1 cells (obtained from B. Moss) and BHK-21 cells (obtained from the American Type Culture Collection, Manassas, Va.) were maintained as monolayer cultures in Dulbecco’s modified Eagle’s medium containing 10% fetal calf serum, 100 IU of penicillin/ml, and 100 μg of streptomycin/ml (all from Life Technologies). The rabbit polyclonal MHV strain A59 antiserum (K134) (anti-MHV) (28) and the rabbit polyclonal peptide serum raised against the 18 carboxy-terminal amino acids of MHV M (anti-MC) (14) have been described previously. The monoclonal antibody J1.3 against the amino terminus of MHV M (anti-MN) (34) and the monoclonal antibody A3.10 against MHV S (anti-S) (39) were kindly provided by J. Fleming. The rabbit anti-peptide serum 5415 specific for the carboxy terminus of MHV S (anti-SC) was a kind gift of M. Buchmeier. The polyclonal rabbit serum against α-mannosidase II (19) and the antipeptide serum specific for the membrane protein of equine arteritis virus (EAV M) (6) were generously provided by K. Moremen and A. A. F. de Vries, respectively.
Expression vectors and site-directed mutagenesis.
All expression vectors contained the genes under control of bacteriophage T7 transcription regulatory elements. The expression constructs pTUMM and pTUMS contain the MHV A59 M and S genes, respectively, cloned in pTUG31 (37, 38). The construction of M genes coding for the mutant proteins ΔN, ΔC, Δ(a+b), and Δ(b+c) (14) and His, Δ18, and Y211G (4) (Fig. 1) has been described previously. Also, the construct encoding the hybrid protein EAV M+9A has been described previously (5). This hybrid protein has an insertion of 9 amino acids, corresponding to the MHV M amino-terminal sequence, behind the initiating methionine of EAV M. MHV M mutant Δ15 was made by PCR mutagenesis with 5′ internal primer C1 (5′-GTGTATAGATATGAAAGGTACCGTG-3′), corresponding to the region of the M gene that contains the unique KpnI site, and 3′-terminal primer C4 (5′-TTACAGTCGGTAATTTCCGACC-3′), directing the desired mutation. The PCR fragment was cloned into pGEM-T (Promega). The plasmid was digested with KpnI and SpeI, and the resulting fragment was cloned into expression vector pTUMM treated with KpnI and XbaI. This resulted in an M gene coding for a mutant protein that lacks the carboxy-terminal 15 amino acids. Mutant Δ21+2 was made by treating pTUMM with StyI and SmaI, followed by filling in of the StyI site (by using DNA polymerase I, large fragment [Life Technologies]) and religation of the vector with the Ochre Stop HpaI linker (Pharmacia). This resulted in an M gene coding for a mutant protein which lacks the 21 carboxy-terminal amino acids and has an additional Leu residue and Ser residue. In mutant FNM, the amino-terminal domain of MHV M was replaced by that of feline infectious peritonitis virus (FIPV) M. The construct encoding this hybrid protein was generated by splicing overlap extension PCR with 808 (5′-GCAAACTGGAACTTCTCGTTGGGC-3′) and 809 (5′-CAACGAGAAGTTCCAGTTTGCAAGATG-3′), both corresponding to the region coding for a stretch of conserved amino acids in the amino-terminal part of the first transmembrane domain, as inside primers and M13 forward and reverse primers (Promega) as external primers. pALTER-1 (Promega) containing either the MHV M or FIPV M gene was used as the template in the first round of PCR. The PCR products were purified and mixed and then amplified with the external primers. The PCR product obtained in the second round of PCR was digested with BamHI and cloned into expression vector pTUG3 (38) treated with the same enzyme. The construct coding for mutant Sap, which contains a SapI recognition site introduced by silent mutations, was also obtained by splicing overlap extension PCR. This construct was generated by using inside primers 744 (5′-GCATAAGGCTCTTCATCAGGAC-3′) and 745 (5′-CAGTCCTGATGAAGAGCCTTATGC-3′), both corresponding to the region coding for the amino-terminal part of the cytoplasmic domain, introducing the SapI recognition site, and external primers 460 (5′-CCTAGGTTAGTCTTAAGACAC-3′) and 746 (5′-CGTCTAGATTAGGTTCTCAACAATGCGG-3′). Primer 460 corresponds to a region just upstream of the multiple-cloning site in pSFV1 (Life Technologies), while primer 746 corresponds to the 3′ end of the MHV M gene. pSFV1 containing the MHV M gene was used as the template in the first round of PCR. The PCR product obtained in the second round of PCR was cloned into the pNOTA/T7 shuttle vector (5 Prime→3 Prime, Inc.) and subsequently excised from the plasmid by using BamHI and cloned into pLITMUS38 (New England Biolabs). This construct was digested with SapI, treated with mung bean nuclease (Pharmacia), and religated to obtain the construct coding for mutant SapΔ1. In this mutant M gene, the nucleotides coding for Ile at position 110 are deleted, leaving the SapI recognition site intact. The construct was treated with KpnI and XbaI, and the resulting fragment was cloned in expression vector pTUMM digested with the same enzymes. All constructs were verified by sequencing.
FIG. 1.
Overview of mutant M proteins. A schematic representation of the structure of the M protein, with the three transmembrane domains (a, b, and c) indicated, is shown above each set of mutants. Amino acid sequences of the amino-terminal and carboxy-terminal domains and mutations in these domains are shown in panels A and D, respectively. Mutants with deletions in the transmembrane region or in the amphipathic domain are shown in panels B and C, respectively. Gaps represent deletions; the deleted amino acids are indicated. The ability of the different M proteins to interact with the S protein is indicated for each mutant at the right. The coimmunoprecipitation of M and S proteins with anti-S antibodies was taken as a measure of M-S interaction. The semiquantitative scores ++, +, +/−, and − indicate efficient, moderately efficient, inefficient, and undetectable M-S interaction, respectively. The abilities of the different M proteins to support VLP assembly, based on published (4) and unpublished results, are also indicated. The scores + and − indicate whether or not VLPs are synthesized when an M protein is coexpressed with the E protein.
Metabolic labeling and immunoprecipitation.
Subconfluent monolayers of OST7-1 or BHK-21 cells in 10-cm2 tissue culture dishes were inoculated with MVA-T7pol (t = 0 h) and subsequently transfected with plasmid DNA by using Lipofectin (Life Technologies) as described previously (5). At t = 4.5 h, the cells were washed with phosphate-buffered saline and starved for 30 min in cysteine- and methionine-free modified Eagle’s medium containing 10 mM HEPES (pH 7.2) and 5% dialyzed fetal calf serum. The medium was then replaced by 600 μl of similar medium containing 100 μCi of 35S in vitro cell-labeling mixture (Amersham), and the cells were labeled for 1 h. Subsequently, the radioactivity was chased by incubating the cells for 2 h with culture medium containing 2 mM methionine and 2 mM cysteine. Proteins were immunoprecipitated from cell lysates as described previously (23). The immunoprecipitates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE) in 12.5 or 15% polyacrylamide gels. The samples were not boiled before being applied to the gel, except when immunoprecipitates prepared with the anti-M antibodies were analyzed.
Indirect immunofluorescence.
Indirect immunofluorescence experiments were performed with BHK-21 cells grown on 12-mm coverslips. The morphology of these cells makes them more convenient for this assay than OST7-1 cells. At t = 5 h, cycloheximide (0.5 mM) was added to the culture media. Cells were fixed at t = 8 h, permeabilized, and stained for immunofluorescence as described previously (23).
RESULTS
Demonstration of M-S complexes.
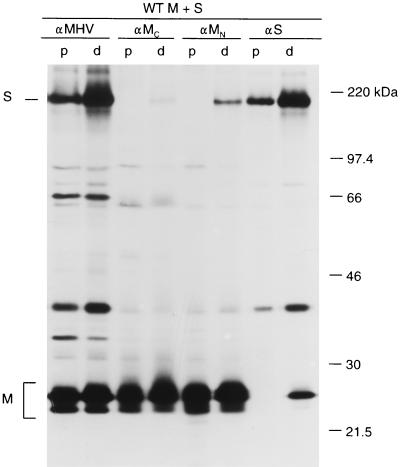
To evaluate the effects of mutations in the M protein on its ability to interact with S, we used the coimmunoprecipitation assay that we developed earlier to demonstrate M-S interaction (23). The principle of the assay is shown for the wild-type (WT) proteins in Fig. 2. In this experiment, the genes coding for M and S were expressed alone or in combination by using the MVA bacteriophage T7 RNA polymerase system in OST7-1 cells. Cells were labeled with 35S-labeled amino acids from 5 to 6 h postinfection (p.i.), and this was followed by a 2 h chase. Cell lysates were prepared and subjected to immunoprecipitation with either the anti-MHV serum, the anti-MC serum, the anti-MN monoclonal antibody, or the anti-S monoclonal antibody. As a control for the specificity of the interactions measured, lysates of cells singly expressing M or S were pooled and subsequently processed similarly for immunoprecipitation (p). The results obtained with the anti-MHV serum showed that M and S were well expressed in single (p) and double (d) expressions. M appears as the well-known set of O-glycosylated forms described previously (13, 35). The first sugar (N-acetylgalactosamine) is added most probably in the Golgi compartment to Thr5 (5, 35). Subsequently, galactose and sialic acid are added in the Golgi complex, sometimes followed by one or two additional, unidentified sugar modifications in the trans-Golgi network (13). The analysis of the lysate from cells coexpressing M and S (d) revealed the formation of M-S complexes. The anti-S-specific antibodies precipitated not only the S protein but also the M protein. Mainly the glycosylated M species were coprecipitated. By using the anti-M-specific antibodies, the S protein was coprecipitated with the M protein. Inspection of the immunoprecipitates from the pooled cell lysates (p) demonstrates the specificity of the coimmunoprecipitation assay. The anti-M and anti-S antibodies precipitated only M or S proteins, respectively; no coimmunoprecipitation was observed. This indicated that the anti-M and anti-S antibodies were indeed specific for either M or S protein and that the observed coimmunoprecipitation was not a nonspecific, postlysis effect.
FIG. 2.
Demonstration of WT M-S complexes. WT M and S genes were expressed in OST7-1 cells, alone or in combination, by using the MVA-T7pol expression system. Cells were labeled for 1 h, and this was followed by a 2-h chase. Cell lysates were prepared and subjected to immunoprecipitation with either the anti-MHV serum (αMHV), the anti-MC serum (αMC), the monoclonal anti-MN antibody (αMN), or the monoclonal anti-S antibody (αS), and the precipitates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. As a control for the double expression (d), lysates of cells singly expressing M or S were pooled and subsequently processed similarly for immunoprecipitation (p). The positions of the S and M proteins are indicated on the left, while the molecular mass marker is indicated on the right.
As a second, independent assay to detect M-S interaction, we used immunofluorescence. This assay is based on the fact that the two proteins localize differently in cells when on their own but colocalize when both are present (23). This is demonstrated in the experiment in Fig. 7. In this experiment, the genes coding for M and S were expressed by using the MVA-T7 system in BHK-21 cells. At 5 h p.i., the cells were treated with cycloheximide for 3 h to block protein synthesis and allow the proteins to reach their destination. The cells were fixed at 8 h p.i. and processed for immunofluorescence with antibodies specific for the S and M proteins. The M protein was found to accumulate in the Golgi complex, as documented previously (11, 13). This localization did not change when the S protein was coexpressed (see Fig. 7B). In contrast, the localization of the S protein was clearly affected by the presence of the M protein. The S protein, when on its own, appeared in an ER-like reticular staining pattern (surface staining was seen when cells were not permeabilized [data not shown]) (see Fig. 7A). When the M protein was coexpressed, the S protein coaccumulated with M in the Golgi complex, although a faint reticular staining pattern was still detectable (see Fig. 7C). The results confirm and extend earlier studies by Opstelten et al. (23) and indicate that coimmunoprecipitation and immunofluorescence assays can be used to demonstrate the existence of M-S complexes.
FIG. 7.
Localization of coexpressed M and S proteins. The gene encoding the S protein was expressed in BHK-21 cells, by using the MVA-T7pol expression system, alone (A) or in combination with the gene encoding WT M (B and C), mutant ΔC (D and E), or mutant Y211G (F and G). At 5 h p.i., cells were treated with cycloheximide for 3 h to block protein synthesis. The cells were fixed at 8 h p.i. and processed for double labeling with the monoclonal anti-S antibody A3.10 (αS; A, C, E, and G) and the peptide serum specific for the carboxy-terminal tail of the M protein (αMC; B, D, and F).
The amino-terminal domain of M is not important for M-S interaction.
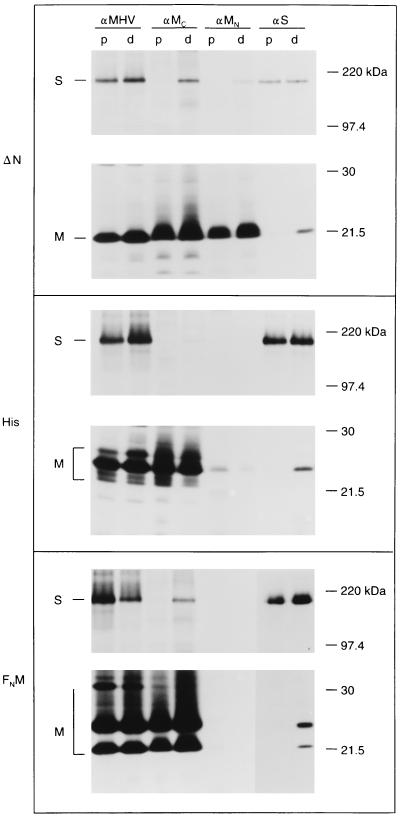
The MHV M protein contains a short amino-terminal domain (25 residues) that is located in the lumen of the intracellular organelles of the secretory pathway. To investigate the role of the amino-terminal domain of the M protein in M-S complex formation, we tested three mutant M proteins for their ability to interact with the S protein. Mutant ΔN lacks almost the entire amino-terminal domain as a result of a deletion of residues A7 through F22 (Fig. 1). Mutant His has an insertion of 6 histidines right behind the initiating methionine. In mutant FNM, the entire amino-terminal domain of MHV M has been replaced by that of FIPV M. A short homologous sequence in the amino-terminal region of the first transmembrane domain (W26 NFS29; MHV M numbering) was selected to fuse the FIPV and MHV sequences. The amino-terminal domain of FIPV M differs significantly from that of MHV M. It is considerably longer, consisting of 53 residues, contains an N-terminal cleavable signal sequence, and has one N-glycosylation site (36).
The mutant M proteins were tested for their ability to form complexes with S by using the coimmunoprecipitation assay described above. In Fig. 3, the relevant parts of the polyacrylamide gels are shown. Immunoprecipitation with the anti-MHV serum showed that the mutant M proteins and the S protein were well expressed in both the single and the double expressions. The anti-M and anti-S antibodies precipitated only mutant M proteins or S protein, respectively, from the pooled cell lysates (p). No coimmunoprecipitation was observed from these lysates. However, M-S complexes were readily detected in the lysates from cells coexpressing mutant M and S proteins (d). Mutant ΔN, which was not glycosylated as a result of the deletion (5), was clearly coprecipitated when the immunoprecipitation was performed with S-specific antibodies and, conversely, S protein was coprecipitated when antibodies to M were used. For mutant His, which became O glycosylated as described previously (5), essentially the same result was obtained. The amount of this mutant protein precipitated by anti-MN was much smaller than that precipitated by anti-MC. This is consistent with earlier observations which showed that the epitope recognized by this antibody is critically dependent on the presence of the serine residues at positions 2 and 3 (4). Thus, the insertion of the histidines between M1 and S2 apparently interferes with the recognition of this epitope. Mutant FNM appeared both in an unglycosylated form (Fig. 3, bottom panel, lower band; about 22 kDa) and as some higher-molecular-mass N-glycosylated species. In addition, due to heterogeneous modifications of the N-linked oligosaccharide, some smearing was also observed in the gel. The presence of the unglycosylated FNM species is indicative of its inefficient transport out of the ER, as was confirmed by immunofluorescence. As expected, this mutant was not recognized by the anti-MN monoclonal antibody. Anti-MC antibodies precipitated both mutant FNM and S protein from the lysate prepared of cells coexpressing these proteins. The anti-S antibodies precipitated, in addition to S protein, both glycosylated and unglycosylated mutant FNM. Since N glycosylation starts in the ER, the latter species presumably represents FNM protein that has not left this compartment. The results with these mutants consistently indicate that the amino-terminal domain of the M protein is not involved in M-S interaction. Deletion, insertion, and complete replacement of this domain did not affect the ability of the protein to associate with S. Also, the absence of O-linked oligosaccharides or the presence of N-linked oligosaccharides on the M protein did not affect M-S interaction.
FIG. 3.
The amino-terminal domain of M is not important for M-S interaction. Expression of M and S genes was performed as described in the legend to Fig. 2. The different M genes tested are indicated on the left. Only the relevant parts of the polyacrylamide gels are shown.
The transmembrane domains of M are necessary for efficient interaction.
The coronavirus M protein transmembrane domains are thought to be important for the formation of homomultimeric M complexes (15). To study whether these transmembrane domains are also important for interaction with the S protein, two mutant M proteins were subjected to a coimmunoprecipitation assay. These mutants have either a deletion of the first and second transmembrane domains [Δ(a+b)] or a deletion of the second and third transmembrane domains [Δ(b+c)], resulting in mutant M proteins with only the third or only the first transmembrane domain, respectively (Fig. 1). These proteins were selected because their membrane topology is the same as that of WT M: amino-terminal domain in the lumen, carboxy terminus in the cytoplasm (14). The results obtained with the anti-MHV serum demonstrated the expression of the mutant M proteins and the S protein (Fig. 4). They also showed that, for unknown reasons, in this particular experiment the expression of the S protein was decreased on coexpression with mutant Δ(a+b). Both M mutants were present mainly in their unglycosylated form even after the 2 h of chase, which is indicative of their inefficient transport out of the ER. Consistently, when the localization of these mutants was assayed by immunofluorescence analysis, they appeared in a reticular (ER-like) staining pattern (data not shown). Analysis of the lysates from cells coexpressing mutant M and S (d) demonstrated that the monoclonal anti-S antibodies precipitated, as well as S protein, very small amounts of the M mutant proteins. Also in those experiments, in which the expression level of the S protein was higher, the amount of coprecipitated mutant Δ(a+b) protein did not increase. The level of coprecipitation of these M mutants was greatly reduced compared to the results obtained with WT M (Fig. 2). Another monoclonal antibody to S (A1.3) (39), recognizing a different epitope, coprecipitated amounts of mutant M similar to those precipitated by anti-S (data not shown). The monoclonal anti-MN antibody clearly precipitated S protein in addition to M. Analysis of the pooled lysates (p) indicated that the observed coimmunoprecipitation was not the result of a nonspecific postlysis effect. The immunofluorescence assay described above was not used here to detect M-S interaction: due to their very inefficient transport, the transmembrane deletion mutants could not be tested for their ability to accumulate S protein in the Golgi complex. The results indicate that although coimmunoprecipitation of M protein transmembrane deletion mutants by anti-S antibodies was affected, the presence of all three transmembrane domains is not an absolute requirement for M-S interaction. Furthermore, M mutants with different transmembrane domains gave similar results, indicating that the “identity” of the transmembrane domain is also not essential for interaction with S.
FIG. 4.
The transmembrane domains of M are necessary for efficient interaction. M and S genes were expressed as described in the legend to Fig. 2.
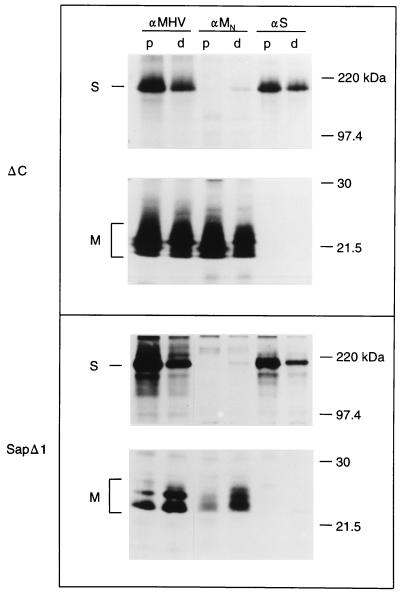
Effects of mutations in the amphipathic domain on M-S interaction.
The carboxy-terminal half of the M protein is located on the cytoplasmic face of the membrane. This domain can be divided into a relatively long amphipathic region and a hydrophilic tail, which is exposed in the cytoplasm. To study the importance of the amphipathic domain in M-S complex formation, two mutants were tested for their ability to interact with S. In mutant ΔC, most of the amphipathic domain is lacking due to a 75-residue deletion, removing residues E121 through D195. Mutant SapΔ1 has a deletion of just 1 amino acid, I110 (Fig. 1). The mutant proteins were expressed singly and in combination with S. As shown in Fig. 5, mutant ΔC was O glycosylated in a pattern similar to that of WT M. Mutant SapΔ1 was O glycosylated less efficiently than mutant ΔC and WT M, consistent with its restricted transport to the Golgi complex as observed by immunofluorescence (data not shown). Analysis of the lysates from cells expressing both mutant ΔC and S proteins demonstrated that although very small amounts of S protein were coprecipitated by specific anti-M antibodies, no ΔC protein was coprecipitated when S-specific antibodies were used. Some coimmunoprecipitation, clearly visible only after prolonged exposure of the gel, was observed with either antibody when mutant SapΔ1 and S were coexpressed. No coprecipitation was observed from the pooled cell lysates, which served as controls. Since mutant ΔC was efficiently transported to the Golgi complex (4), this mutant was also tested in the immunofluorescence assay. As shown in Fig. 7, the reticular ER-like staining pattern of S was not affected by the presence of mutant ΔC (see Fig. 7E). Clearly, the S protein did not colocalize with mutant ΔC to the Golgi complex to an appreciable extent. The results indicated that M proteins with deletions in their amphipathic domain are severely affected in M-S interaction.
FIG. 5.
Effect of mutations in the amphipathic domain on M-S interaction. M and S genes were expressed as described in the legend to Fig. 2.
Mutations in the hydrophilic tail of the M protein.
The extreme carboxy-terminal hydrophilic tail of the M protein plays an essential role in coronavirus particle assembly (4). It was therefore of interest to investigate whether this tail is also in some way involved in M-S interaction. Preliminary experiments showed that short carboxy-terminal truncations, which rendered the protein assembly incompetent (4), did not affect its ability to interact with the S protein (data not shown). Subsequently, several M mutants with larger truncations were tested. Mutants Δ15 and Δ18 have deletions of the 15 and 18 terminal residues, respectively, while mutant Δ21+2 has a deletion of the last 21 amino acids and has two foreign residues (Leu and Ser) introduced due to the construction (Fig. 1). In mutant Y211G, the Tyr residue at position 211 is replaced by a Gly. All these mutant proteins were transported to the Golgi complex when expressed individually (see below); mutants Δ18, Δ21+2, and Y211G could also be detected at the cell surface (data not shown). Each of the mutant M proteins was coexpressed with the S protein, and the interactions were again studied by the coimmunoprecipitation assay (Fig. 6). Immunoprecipitations with the anti-MHV serum again confirmed that all the proteins were well expressed. The M protein deletion mutants were O glycosylated in a pattern similar to that of WT M, with mutant Y211G being glycosylated more efficiently. No coimmunoprecipitation was observed from the pooled lysates (p). Analysis of lysates from cells coexpressing mutant M proteins and S protein (d) demonstrated that mutant Δ15 protein coprecipitated with S and vice versa. Similar to WT M, it was mainly the glycosylated form of the mutant that appeared to be associated with S. Although small amounts of S protein were coprecipitated with mutant Δ18, hardly any coprecipitation of the mutant M protein was detected when S-specific antibodies were used. Surprisingly, when an M mutant with a slightly larger deletion (Δ21+2) was used, coprecipitation of the mutant M protein with S protein was observed again. However, in contrast to WT M and mutant Δ15, mainly the unglycosylated, pre-Golgi form of mutant Δ21+2 was coprecipitated, suggesting that complexes of mutant Δ21+2 and S were compromised in their transport to the Golgi complex. When M-specific antibodies were used, S protein was coprecipitated to a level similar to that observed with mutant Δ15. Finally, mutant Y211G was assayed. The Tyr residue substituted in this mutant is deleted in mutant Δ18 but not in mutant Δ15. It appeared that coprecipitation of mutant Y211G with the S protein, and vice versa, was severely reduced but not absent. In contrast to all other M mutants tested in this study, mutant Y211G was able to assemble into VLPs when coexpressed with the E gene. This allowed us to use the incorporation of S protein into such VLPs as an additional parameter for M-S interaction. The experiment revealed that the S protein was indeed drawn into the VLPs (data not shown), indicating that although the interaction between mutant Y211G and S was apparently decreased, it was not fully abolished.
FIG. 6.
Effect of mutations in the hydrophilic tail on M-S interaction. M and S genes were expressed as described in the legend to Fig. 2.
Because all these carboxy-terminal M mutants were efficiently transported to the Golgi complex, we could use the immunofluorescence assay to independently test for M-S association. Since the deletion mutants are obviously not recognized by the tail-specific peptide serum (anti-MC) used in Fig. 7, these mutants were labeled with the mouse monoclonal antibody to the amino terminus (anti-MN). Fortunately, since the efficiencies of our cotransfections were high (up to 90% of the transfected cells expressed both M and S proteins) and the staining patterns of the ER and Golgi in BHK-21 cells were very typical, we analyzed the localization of M and S in cotransfected cells separately. The availability of rabbit antibodies to the Golgi-resident α-mannosidase II (19) allowed us to mark the Golgi complex. Representative cells were photographed and are shown in Fig. 8. All M carboxy-terminal deletion mutants colocalized with α-mannosidase II, and their localization was not changed by the coexpression of the S protein (Fig. 8B, F, and J). In contrast, the localization of the S protein was clearly affected by the presence of some of the M mutants. Coexpression with mutant Δ15 (as well as with M mutants having shorter truncations) changed the reticular (ER-like) staining pattern of S into a perinuclear (Golgi) pattern. The S protein colocalized with α-mannosidase II just as mutant Δ15 protein did (Fig. 8C and D). However, M mutants with larger deletions were not able to alter the localization of the S protein to the same extent as mutant Δ15. S protein coexpressed with mutant Δ18 maintained its reticular staining pattern (Fig. 8G and H). An intermediate localization pattern was observed when the S protein was coexpressed with mutant Δ21+2. In some cells, the S protein appeared in the typical reticular pattern while some colocalization with α-mannosidase II was detectable (Fig. 8K and L); in other cells, no such costaining was observed, and the protein was present only in its reticular pattern. Finally, when M mutant Y211G and S were coexpressed, the two proteins did not appear to affect each other’s transport. The M mutant was found mainly in the Golgi complex, while for the S protein the characteristic ER-like pattern was observed (Fig. 7F and G), not much different from that of singly expressed S. Taken together, the results of the colocalization assays are consistent with those of the coimmunoprecipitation assays in which the anti-S antibodies were used. They indicate that truncations of up to 15 residues did not severely affect the ability of the M protein to associate with the S protein. Larger truncations were, however, more deleterious for M-S interaction. Complex formation was severely decreased (mutant Δ18) or complexes were inefficiently transported to the Golgi complex (mutant Δ21+2). Complex formation was also strongly impaired by substitution of a single amino acid in the carboxy-terminal domain (Y211G).
FIG. 8.
Localization of S protein coexpressed with M mutants having truncations of the hydrophilic tail. M and S genes were expressed as described in the legend to Fig. 7. Cells were processed for double labeling with either the monoclonal antibody to the amino terminus of M (αMN; B, F, and J) or the monoclonal antibody to S (αS; D, H, and L) and the rabbit serum against the resident Golgi protein α-mannosidase II (A, C, E, G, I, and K).
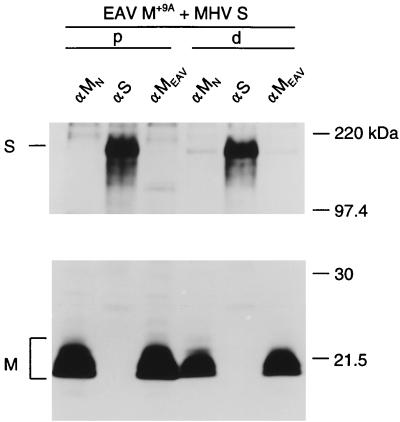
Coimmunoprecipitation assay of S with a control protein.
A complicating factor in the interpretation of our coimmunoprecipitation results was that the observations with the M-specific and the S-specific antibodies were not always mutually confirmatory. In several cases, such as with mutants Δ(a+b), Δ(b+c), ΔC, and Δ18, coimmunoprecipitation of S protein obtained by using M antibodies was much more pronounced than that of M protein obtained by using antibodies to S. To study this discrepancy in more detail, we evaluated the assay by coexpressing the S protein with an unrelated control membrane protein, the chimeric protein EAV M+9A. This protein is simply an N-terminally extended form of the EAV M protein prepared by inserting the 9-residue amino-terminal sequence of the MHV M protein (S2-P10) immediately behind the initiating methionine (5). As a result of this extension, the EAV protein acquired the epitope recognized by the MHV M-specific monoclonal antibody J1.3 (anti-MN) that we used throughout this study. The EAV M protein is a type III membrane protein; it has the same topology as the MHV M protein (6) but is slightly smaller. There are no obvious sequence similarities between the two proteins. The results of the expression experiment are shown in Fig. 9. The EAV M+9A protein was immunoprecipitated by the anti-EAV M serum (anti-EAV) as well as by the monoclonal antibody anti-MN. No coimmunoprecipitation was observed from the pooled lysate (p). Analysis of the lysate from cells coexpressing EAV M+9A and S (d) showed that in addition to EAV M+9A protein, small amounts of S protein were coprecipitated both by the EAV M antiserum and by the monoclonal antibody anti-MN. In contrast, the S antibodies precipitated only S protein. No coimmunoprecipitation of EAV M+9A protein was observed. The results show that the coimmunoprecipitation assay of M-S complexes is highly specific when using the S antibodies but not in its reciprocal format. We cannot exclude an interaction (nonspecific) between the EAV M protein (and some MHV M mutants) and the S protein, which cannot be detected with the S antibodies. However, since the M-S complexes detected with the S-specific antibodies could be confirmed consistently by the independent immunofluorescence assay, we consider these last two assays to be reliable indicators of M-S complex formation.
FIG. 9.
Coimmunoprecipitation of S protein with the control protein EAV M+9A. EAV M+9A and S genes were expressed as described in the legend to Fig. 2. Immunoprecipitations were performed with the monoclonal antibody to the amino terminus of MHV M (αMN), the monoclonal antibody to S (αS), and the antipeptide serum specific for EAV M (αMEAV).
DISCUSSION
The generation of infectious virus in infected cells requires that all essential components be properly collected in the assembled particles. For coronaviruses, particle assembly per se is not dependent on the presence of all these components. Actually, only the two membrane proteins M and E already suffice to create minimal particles, i.e., viral envelopes. Incorporation of the other components—NC and the membrane proteins S and HE—is directed by specific molecular interactions. Inclusion of spikes into virions is driven by heterotypic interactions of the spike protein with the M protein. In this study, we evaluated the structural domains of the M polypeptide involved in association with the S protein. Surprisingly, neither of the membrane-exposed terminal domains of the M molecule were required for interaction with S. In contrast, M-S complex formation was very sensitive to changes in all membrane-associated parts of the molecule. This was most dramatically demonstrated by the effect of a single-residue deletion in the transition domain between the transmembrane triplet and the amphipathic region. Interestingly, the structural requirements of the M protein for the formation of M-S complexes clearly differ from the requirements for viral particle assembly, as illustrated by the opposing sensitivities to changes in the terminal domains.
For the detection of interactions between M and S proteins, we used two independent assays, coimmunoprecipitation and immunofluorescence. The results obtained were fully consistent except for some M mutants, where the immunoprecipitation with the M-specific antibody scored apparently false positive. This interpretation was confirmed when the assay was verified by using an unrelated control protein: while no association was detected between an EAV M protein and coexpressed MHV S protein when S-specific antibodies were used, two different antibodies to the EAV protein coimmunoprecipitated some of the S protein. The reasons for this lack of reciprocity are not clear, but a nonspecific, postlysis effect or cross-reactivity of the antibodies can be excluded since no coimmunoprecipitation was observed from pooled lysates of the separately expressed proteins. It is of note that a similar inconsistency was observed in a study of bovine coronavirus (21). While the existence of M-S complexes in infected cells was convincingly demonstrated in coprecipitation assays with antibodies of either specificity, association could not be unequivocally established when the proteins were coexpressed: coprecipitation was observed only when the M-specific antibodies, not those against S, were used.
The amino-terminal domain of the coronavirus M protein is exposed on the luminal side of intracellular organelles. The function of this domain is not quite clear. It is not actively involved in membrane integration (14), intracellular transport (29), or, probably, interaction with the E protein, since this protein is not appreciably exposed luminally (24). Here we show that it is also not involved in interactions with the S protein. Various mutations, including its complete replacement by the ectodomain of an unrelated coronavirus M protein (mutant FNM), did not affect M-S association. Many of these mutations do, however, interfere with envelope assembly (4). One reason for this might be that particle assembly requires interactions at the level of the ectodomains and that these might be impaired by the mutations. Alternatively, the mutations may have longer-range effects compromising assembly, for instance by interfering with the interactions among M molecules or with E.
Coronavirus M proteins are glycosylated in their luminal domain either by O linkage, as for MHV M, or by N linkage, as for FIPV M (27). The function of M protein glycosylation still remains to be elucidated. Our results indicate that M glycosylation does not play a role in M-S interaction. M proteins that were not glycosylated (mutant ΔN), O glycosylated (WT M), or N glycosylated (FNM) were all efficiently complexed with the S protein. Glycosylation of the M protein is also not required for virus assembly (4). In contrast, N glycosylation of S was found to be essential for the incorporation of spikes into virus particles (10, 20, 28, 31). Obviously, folding of the luminal domain of the S protein is crucially dependent on oligosaccharide addition—the protein aggregates and is arrested when N glycosylation is prevented (unpublished observations)—in contrast to the M protein, which folds and is transported irrespective of its glycosylation state (5).
The mutations in the transmembrane region of M had profound consequences for M-S interactions. Deleting two of the three transmembrane domains reduced complex formation dramatically, although a low level of association could still be detected, regardless of the identity of the remaining transmembrane domain. The presence of all three transmembrane domains is apparently not essential. The transmembrane deletion mutants were retarded in their transport to the Golgi complex, but this is unlikely to account for the decrease in M-S interaction. The MHV M mutant carrying the FIPV M ectodomain (FNM) efficiently associated with the S protein despite its severely impaired intracellular transport. In addition, several studies demonstrated that the S and M proteins engage in interaction in an early compartment, most likely the ER (21, 23). The presence of all three M-protein transmembrane domains is thought to be required for the formation of the large homomultimeric M complexes (15). M mutants lacking one or two transmembrane domains may not be able to assemble into such complexes. This would explain why these proteins fail to assemble into VLPs (4). It might also explain the low level of M-S complex formation, if one assumes that normally the S trimers are accommodated at specific positions within the lattice formed by M molecules.
The amphipathic domain of the coronavirus M protein is located on the cytoplasmic side of the cellular membrane. The structure of this domain is still unresolved. Its resistance to protease may reflect a close association with the membrane surface, but the protein domain might also be folded in an inaccessibly compact conformation and reside outside the membrane. Our present results point to a sensitive role of this domain in M-S interaction. Deletion of the main part of the domain (mutant ΔC) or of just a single amino acid (mutant SapΔ1) had a strong negative effect on M-S complex formation. These mutations were also fatal for envelope assembly (reference 4 and unpublished data). Conceivably, the structural integrity of the amphipathic domain is an important requisite for these molecular interaction processes. It is therefore surprising that deletion of the main part of the domain does not inhibit the transport of the protein to the Golgi apparatus (Fig. 7). Whether this is because this deletion has no effect on homotypic M-M interactions or because these interactions are not required for transport remains to be established.
The extreme carboxy-terminal hydrophilic tail of the M protein, which is located in the cytoplasm, is crucial for envelope assembly. Deletion of only the carboxy-terminal two residues was lethal for the formation of VLPs as well as of MHV virions (4). The effect is probably not due to impairment of lateral interactions between M molecules. Even when the terminal 22 residues were lacking, sucrose gradient analysis showed that the protein was still capable of associating into large oligomeric complexes (15). Such assembly-incompetent truncated forms of M can be consistently rescued into viral particles by assembly-competent molecules (4). Here we showed that for interaction with S, the terminal 15 residues are also not essential. Larger deletions, however, variably affected the association of M with S. This may be due in part to the removal of Y211, since M-S complex formation appeared to be particularly sensitive to changes of this residue. Conversion to a glycine (mutant Y211G) abolished the ability of M to relocate S protein to the Golgi complex. However, M-S interaction seemed not to be completely absent, because some S protein was incorporated into VLPs assembled with this M mutant.
The conclusion that the interactions between M and S occur at the level of the transmembrane and amphipathic domains is also supported by other observations. We have recently demonstrated the incorporation into MHV particles of hybrid S proteins in which the ectodomain had been replaced by that of FIPV S (9). In contrast, the reciprocal hybrid S protein, containing the transmembrane and cytoplasmic domains of FIPV S, was not assembled into MHV particles. These results clearly exclude a role for the ectodomain of the S protein in M-S interaction and implicate such a role for the transmembrane and/or cytoplasmic domain. Furthermore, a peptide serum specific for the cytoplasmic domain of the S protein was unable to precipitate WT M-S complexes. It did, however, precipitate complexes formed by S and the M mutant Δ21+2 to the same extent as the anti-S monoclonal antibodies (unpublished data). This result suggests that the epitope recognized by the peptide serum is “hidden” from the antibody when the protein is complexed with WT M but not with a truncated M protein, supporting the view that interactions occur on the cytoplasmic side of the membrane.
Enveloped viruses have developed different assembly strategies (for a review, see reference 8). A common theme in the assembly of many enveloped viruses is an interaction of the envelope proteins with internal viral components (e.g., matrix proteins) to ensure their efficient coincorporation into the virus. The coronavirus M protein, and especially its amphipathic domain, has several striking features in common with the matrix proteins of minus-strand RNA viruses and retroviruses (16). (i) Matrix proteins are generally quite small amphipathic proteins which bind membranes, although they lack transmembrane sequences. The amphipathic domain of M was also found to associate tightly with membranes by itself, despite the absence of pronounced hydrophobic sequences (18). (ii) Matrix proteins have a tendency to aggregate, which may be indicative of their lattice-forming function during budding. The M protein of MHV was demonstrated to aggregate into large complexes when expressed on its own (15). (iii) The NC-binding properties of the matrix proteins have been convincingly demonstrated. Likewise, the part of the M protein located on the cytoplasmic face of the membrane is the most likely candidate to draw the NC into the envelope. Subviral particles of several coronaviruses, prepared by detergent disruption, still contained M protein associated with the NC (27), while the NC and M proteins of detergent-disrupted virions reassociate at 37°C (32). Furthermore, the M protein was also found on the surface of purified viral cores (26). (iv) Finally, matrix proteins interact with the spike proteins and are responsible for recruiting them into the virus. Similarly, the coronavirus M protein also interacts with the envelope proteins S and HE, thereby mediating their assembly into virus particles (21, 23). Taking these results together, it seems that the coronavirus M protein, and particularly its amphipathic domain, has functions in coronavirus assembly which are similar to the roles played by matrix proteins in the assembly of other enveloped viruses.
ACKNOWLEDGMENTS
We thank our colleagues at the Institute of Virology for helpful discussions.
This research has been financially supported by the Council for Chemical Sciences of the Netherlands Organization for Scientific Research (CW-NWO).
REFERENCES
- 1.Bos E C, Luytjes W, van der Meulen H V, Koerten H K, Spaan W J. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology. 1996;218:52–60. doi: 10.1006/viro.1996.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brian D A, Hogue B G, Kienzle T E. The coronavirus hemagglutinin esterase glycoprotein. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 165–179. [Google Scholar]
- 3.Cavanagh D. The coronavirus surface glycoprotein. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 73–113. [Google Scholar]
- 4.de Haan C A M, Kuo L, Masters P S, Vennema H, Rottier P J M. Coronavirus particle assembly: primary structure requirements of the membrane protein. J Virol. 1998;72:6838–6850. doi: 10.1128/jvi.72.8.6838-6850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Haan C A M, Roestenberg P, de Wit M, de Vries A A F, Nilsson T, Vennema H, Rottier P J M. Structural requirements for O-glycosylation of the mouse hepatitis virus membrane protein. J Biol Chem. 1998;273:29905–29914. doi: 10.1074/jbc.273.45.29905. [DOI] [PubMed] [Google Scholar]
- 6.de Vries A A F, Chirnside E D, Horzinek M C, Rottier P J M. Structural proteins of equine arteritis virus. J Virol. 1992;66:6294–6303. doi: 10.1128/jvi.66.11.6294-6303.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischer F, Stegen C F, Masters P S, Samsonoff W A. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J Virol. 1998;72:7885–7894. doi: 10.1128/jvi.72.10.7885-7894.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garoff H, Hewson R, Opstelten D-J E. Virus maturation by budding. Microbiol Mol Biol Rev. 1998;62:1171–1190. doi: 10.1128/mmbr.62.4.1171-1190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Godeke, G.-J., C. A. M. de Haan, J. W. A. Rossen, H. Vennema, and P. J. M. Rottier. Assembly of spikes into coronavirus particles is mediated by the carboxy-terminal domain of the spike protein. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 10.Holmes K V, Doller E W, Sturman L S. Tunicamycin resistant glycosylation of coronavirus glycoprotein: demonstration of a novel type of viral glycoprotein. Virology. 1981;115:334–344. doi: 10.1016/0042-6822(81)90115-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klumperman J, Krijnse Locker J, Meijer A, Horzinek M C, Geuze H J, Rottier P J M. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J Virol. 1994;68:6523–6534. doi: 10.1128/jvi.68.10.6523-6534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krijnse Locker J, Ericsson M, Rottier P J M, Griffiths G. Characterization of the budding compartment of mouse hepatitis virus: evidence that transport from the RER to the Golgi complex requires only one vesicular transport step. J Cell Biol. 1994;124:55–70. doi: 10.1083/jcb.124.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krijnse Locker J, Griffiths G, Horzinek M C, Rottier P J M. O-glycosylation of the coronavirus M protein. Differential localization of sialyltransferases in N- and O-linked glycosylation. J Biol Chem. 1992;267:14094–14101. doi: 10.1016/S0021-9258(19)49683-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krijnse Locker J, Rose J K, Horzinek M C, Rottier P J M. Membrane assembly of the triple-spanning coronavirus M protein; individual transmembrane domains show preferred orientation. J Biol Chem. 1992;267:21911–21918. doi: 10.1016/S0021-9258(19)36699-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krijnse Locker J M, Opstelten D-J E, Ericsson M, Horzinek C, Rottier P J M. Oligomerization of a trans-Golgi/trans-Golgi network retained protein occurs in the Golgi complex and may be part of its retention. J Biol Chem. 1995;270:8815–8821. doi: 10.1074/jbc.270.15.8815. [DOI] [PubMed] [Google Scholar]
- 16.Lenard J. Negative-strand virus M and retrovirus MA proteins: all in a family? Virology. 1996;216:289–298. doi: 10.1006/viro.1996.0064. [DOI] [PubMed] [Google Scholar]
- 17.Luytjes W, Gerritsma H, Bos E, Spaan W. Characterization of two temperature-sensitive mutants of coronavirus mouse hepatitis virus strain A59 with maturation defects in the spike protein. J Virol. 1997;71:949–955. doi: 10.1128/jvi.71.2.949-955.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayer T, Tamura T, Falk M, Niemann H. Membrane integration and intracellular transport of the coronavirus glycoprotein E1, a class III membrane glycoprotein. J Biol Chem. 1988;263:14956–14963. doi: 10.1016/S0021-9258(18)68131-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moremen K W, Touster O, Robbins P W. Novel purification of the catalytic domain of a Golgi α-mannosidase II: characterization and comparison with the intact enzyme. J Biol Chem. 1991;266:16876–16885. [PubMed] [Google Scholar]
- 20.Mounir S, Talbot P J. Sequence analysis of the membrane protein gene of human coronavirus OC43 and evidence for O-glycosylation. J Gen Virol. 1992;73:2731–2736. doi: 10.1099/0022-1317-73-10-2731. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen V-P, Hogue B G. Protein interactions during coronavirus assembly. J Virol. 1997;71:9278–9284. doi: 10.1128/jvi.71.12.9278-9284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Opstelten D-J E. Envelope glycoprotein interactions in coronavirus assembly. Ph.D. thesis. Utrecht, The Netherlands: Utrecht University; 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Opstelten D-J E, Raamsman M J, Wolfs K, Horzinek M C, Rottier P J M. Envelope glycoprotein interactions in coronavirus assembly. J Cell Biol. 1995;131:339–349. doi: 10.1083/jcb.131.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raamsman, M. J. B., J. Krijnse Locker, A. de Hooghe, A. A. F. de Vries, G. Griffiths, H. Vennema, and P. J. M. Rottier. Characterization of the coronavirus MHV-A59 small membrane protein E. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 25.Ricard C S, Koetzner C A, Sturman L S, Masters P S. A conditional-lethal murine coronavirus mutant that fails to incorporate the spike glycoprotein into assembled virions. Virus Res. 1995;39:261–276. doi: 10.1016/0168-1702(95)00100-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Risco C, Anton I M, Enjuanes L, Carrascosa J L. The transmissible gastroenteritis coronavirus contains a spherical core shell consisting of M and N proteins. J Virol. 1996;70:4773–4777. doi: 10.1128/jvi.70.7.4773-4777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rottier P J M. The coronavirus membrane protein. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 115–139. [Google Scholar]
- 28.Rottier P J M, Horzinek M C, van der Zeijst B A M. Viral protein synthesis in mouse hepatitis virus strain A59-infected cells: effect of tunicamycin. J Virol. 1982;40:350–357. doi: 10.1128/jvi.40.2.350-357.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rottier P J M, Krijnse Locker J, Horzinek M C, Spaan W J M. Expression of MHV-A59 M glycoprotein: effects of deletions on membrane integration and intracellular transport. Adv Exp Med Biol. 1990;276:127–135. doi: 10.1007/978-1-4684-5823-7_19. [DOI] [PubMed] [Google Scholar]
- 30.Siddell S G. The small-membrane protein. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 181–189. [Google Scholar]
- 31.Stern D F, Sefton B M. Coronavirus proteins: structure and function of the oligosaccharides of the avian infectious bronchitis virus glycoproteins. J Virol. 1982;44:804–812. doi: 10.1128/jvi.44.3.804-812.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sturman L S, Holmes K V, Behnke J. Isolation of coronavirus envelope glycoproteins and interaction with the viral nucleocapsid. J Virol. 1980;33:449–462. doi: 10.1128/jvi.33.1.449-462.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sutter G, Ohlman M, Erfle V. Non-replicating vaccinia virus vector efficiently expresses bacteriophage T7 RNA polymerase. FEBS Lett. 1995;371:9–12. doi: 10.1016/0014-5793(95)00843-x. [DOI] [PubMed] [Google Scholar]
- 34.Taguchi F, Fleming J O. Comparison of six different murine coronavirus JHM variants by monoclonal antibodies against the E2 glycoprotein. Virology. 1989;169:233–235. doi: 10.1016/0042-6822(89)90061-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tooze S A, Tooze J, Warren G. Site of addition of N-acetyl-galactosamine to the E1 glycoprotein of mouse hepatitis virus-A59. J Cell Biol. 1988;106:1475–1487. doi: 10.1083/jcb.106.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vennema H, de Groot R J, Harbour D A, Horzinek M C, Spaan W J. Primary structure of the membrane and nucleocapsid protein genes of feline infectious peritonitis virus and immunogenicity of recombinant vaccinia viruses in kittens. Virology. 1991;181:327–335. doi: 10.1016/0042-6822(91)90499-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vennema H, Godeke G-J, Rossen J W A, Voorhout W F, Horzinek M C, Opstelten D-J E, Rottier P J M. Nucleocapsid-independent assembly of coronavirus-like particles by coexpression of viral envelope protein genes. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vennema H, Rijnbrand R, Heijnen L, Horzinek M C, Spaan W J. Enhancement of the vaccinia virus/phage T7 RNA polymerase expression system using encephalomyocarditis virus 5′-untranslated region sequences. Gene. 1991;108:201–209. doi: 10.1016/0378-1119(91)90435-E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weismiller D G, Sturman L S, Buchmeier M J, Fleming J O, Holmes K V. Monoclonal antibodies to the peplomer glycoprotein of coronavirus mouse hepatitis virus identify two subunits and detect a conformational change in the subunit released under mild alkaline conditions. J Virol. 1990;64:3051–3055. doi: 10.1128/jvi.64.6.3051-3055.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]