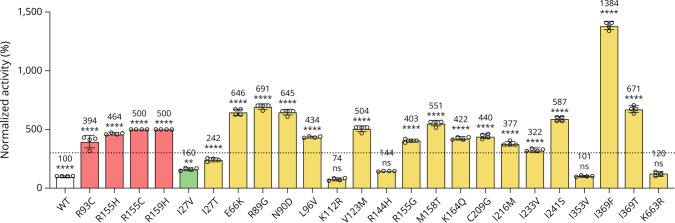
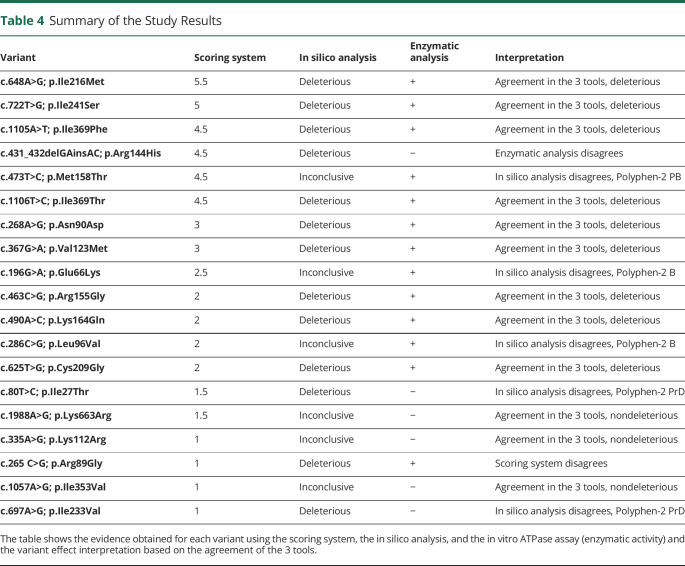
Marianela Schiava
Marianela Schiava, MD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Chiseko Ikenaga
Chiseko Ikenaga, MD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Ana Topf
Ana Topf, PhD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Marta Caballero-Ávila
Marta Caballero-Ávila, MD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Tsui-Fen Chou
Tsui-Fen Chou, PhD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Shan Li
Shan Li, PhD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Feng Wang
Feng Wang, PhD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Jil Daw
Jil Daw, MD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Tanya Stojkovic
Tanya Stojkovic, MD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Rocio Villar-Quiles
Rocio Villar-Quiles, MD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Ichizo Nishino
Ichizo Nishino, MD, PhD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Michio Inoue
Michio Inoue, MD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Yukako Nishimori
Yukako Nishimori, MD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Yoshihiko Saito
Yoshihiko Saito, MD, PhD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Masahisa Katsuno
Masahisa Katsuno, MD, PhD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Seiya Noda
Seiya Noda, MD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Chihiro Ito
Chihiro Ito, MD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Mieko Otsuka
Mieko Otsuka, MD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Sruthi Nahir
Sruthi Nahir, MD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Georgios Manousakis
Georgios Manousakis, MD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
David Walk
David Walk, MD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Colin Quinn
Colin Quinn, MD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Lindsay Alfano
Lindsay Alfano, PhD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Zarife Sahenk
Zarife Sahenk, MD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Giorgio Tasca
Giorgio Tasca, MD, PhD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Mauro Monforte
Mauro Monforte, MD, PhD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Mario Sabatelli
Mario Sabatelli, MD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Giulia Bisogni
Giulia Bisogni, MD, PhD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Anders Oldfors
Anders Oldfors, MD, PhD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Anna Rydeliu
Anna Rydeliu, MD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Endre Pal
Endre Pal, MD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Carmen Paradas
Carmen Paradas, MD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Beatriz Velez
Beatriz Velez, MD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Jan L De Bleecker
Jan L De Bleecker, MD, PhD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Maria Elena Farugia
Maria Elena Farugia, MD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Cheryl Longman
Cheryl Longman, MD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Matthew B Harms
Matthew B Harms, MD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Stuart Ralston
Stuart Ralston, MD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Edmar Zanoteli
Edmar Zanoteli, MD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Andre Macedo Serafim da Silva
Andre Macedo Serafim da Silva, MD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Javier Sotoca
Javier Sotoca, MD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Raul Juntas-Morales
Raul Juntas-Morales, PhD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Jorge Bevilacqua
Jorge Bevilacqua, MD, PhD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Mireya Balart
Mireya Balart, MD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Stuart Talbot
Stuart Talbot, MD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Volker Straub
Volker Straub, MD, PhD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Michela Guglieri
Michela Guglieri, MD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Chiara Marini-Bettolo
Chiara Marini-Bettolo, PhD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,
Jordi Diaz-Manera
Jordi Diaz-Manera, PhD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,*,
Conrad Chris Weihl
Conrad Chris Weihl, MD, PhD
1From the John Walton Muscular Dystrophy Research Centre (M. Schiava, A.T., V.S., M.G., C.M.-B., J.D.-M.), Institute of Genetic Medicine, Centre for Life, Newcastle University and Newcastle Hospitals NHS Foundation Trusts, Newcastle Upon Tyne, United Kingdom; Johns Hopkins University School of Medicine (C. Ikenaga), Baltimore, MD; Unidad de Enfermedades Neuromusculares (M.C.-Á.), Servicio de Neurología, Hospital de la Santa Creu i Sant Pau, Barcelona, Spain; Division of Biology and Biological Engineering (T.-F.C., S.L., F.W.), California Institute of Technology, Pasadena; Department of Neurology (J.D.), Washington University School of Medicine, St. Louis, MO; APHP Centre de référence des maladies neuromusculaires Institut de Myologie Sorbonne Université APHP Hôpital Pitié-Salpêtrière Paris (T.S., R.V.-Q.), France; Department of Neuromuscular Research (I.N., M.I., Y.N., Y.S.), National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP); Departments of Neurology (M.K., S. Noda) and Clinical Research Education (M.K., S. Noda), Nagoya University Graduate School of Medicine; Department of Neurology (M.K., S. Noda), National Hospital Organization Suzuka Hospital; Department of Neurology (C. Ito), Aichi Medical University School of Medicine; Department of Neurology (M.O.), International University of Health and Welfare Hospital, Japan; Department of Neurology Sree Chitra Tirunal Institute for Medical Sciences and Technology (S. Nahir), Thiruvananthapuram, Kerala, India; Department of Neurology (G.M., D.W.), University of Minnesota, Minneapolis; Department of Neurology (C.Q.), University of Pennsylvania, Perelman School of Medicine, Philadelphia; Center for Gene Therapy (L.A., Z.S.), The Abigail Wexner Research Institute at Nationwide Children's Hospital; Department of Pediatrics (L.A., Z.S.), The Ohio State University College of Medicine, Columbus; Unità Operativa Complessa di Neurologia Fondazione Policlinico Universitario A Gemelli IRCCS (G.T., M.M.); Centro clinico NEMO- Fondazione policlinico universitario A. Gemelli IRCCS (M. Sabatelli, G.B.), Rome, Italy; Department of Laboratory Medicine, Institute of Biomedicine, University of Gothenburg (A.O.); Department of Neurology (A.R.), Clinical Sciences Lund, Lund University, Sweden; Departments of Neurology and Neuropathology (E.P.), University of Pécs, Hungary; Neurology Department, Neuromuscular Disorders Unit, Hospital Universitario Virgen del Rocío (C.P., B.V.); Instituto de Biomedicina de Sevilla (C.P.); Centre for Biomedical Network Research on Neurodegenerative Disorders (CIBERNED) Instituto de Salud Carlos III (C.P., B.V.), Madrid, Spain; Neurology Department and Neuromuscular Reference Centre (J.L.D.B.), Gent, Blegium, part of the ERN NMD; Institute of Neurological Sciences (M.E.F.); West Scotland Regional Genetics Service (C.L.), Queen Elizabeth University Hospital, Glasgow, United Kingdom; Columbia University Irving Medical Centre (M.B.H.), New York; Centre for Genomic and Experimental Medicine (S.R.), Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital Edinburgh, United Kingdom; Department of Neurology (E.Z., A.M.S.S.), School of Medicine, Universidade de São Paulo (FMUSP), Brazil; Neurology Service (J.S., R.J.-M.), Neuromuscular Disorders Unit, Hospital Universitari Vall d’Hebron, Barcelona, Spain; Departamento de Neurología y Neurocirugía (J.B.), HCUCH, Departamento de Anatomía y Medicina Legal, Facultad de Medicina, Universidad de Chile; Departamento de Neurología y Neurocirugía Clínica (M.B.), Clínica Dávila, Santiago Chile; Newcastle University (S.T.), Newcastle Upon Tyne, United Kingdom; and Department of Neurology (C.C.W.), Washington University School of Medicine, Saint Louis, MO.
1,*