Abstract
To characterize the sites and nature of binding of influenza A virus matrix protein (M1) to ribonucleoprotein (RNP), M1 of A/WSN/33 was altered by deletion or site-directed mutagenesis, expressed in vitro, and allowed to attach to RNP under a variety of conditions. Approximately 70% of the wild-type (Wt) M1 bound to RNP at pH 7.0, but less than 5% of M1 associated with RNP at pH 5.0. Increasing the concentration of NaCl reduced M1 binding, but even at a high salt concentration (0.6 M NaCl), approximately 20% of the input M1 was capable of binding to RNP. Mutations altering potential M1 RNA-binding regions (basic amino acids 101RKLKR105 and the zinc finger motif at amino acids 148 to 162) had varied effect: mutations of amino acids 101 to 105 reduced RNP binding compared to the Wt M1, but mutations of zinc finger motif did not. Treatment of RNP with RNase reduced M1 binding by approximately half, but even M1 mutants lacking RNA-binding regions had residual binding to RNase-treated RNP provided that the N-terminal 76 amino acids of M1 (containing two hydrophobic domains) were intact. Addition of detergent to the reaction mixture further reduced binding related to the N-terminal 76 amino acids and showed the greatest effect for mutations affecting the RNA-binding regions of basic amino acids. The data suggest that M1 interacts with both the RNA and protein components of RNP in assembly and disassembly of influenza A viruses.
The influenza A virus consists of ribonucleoproteins (RNPs) enclosed in a lipid envelope derived from the host cell membrane. The viral genome is made up of eight separate gene segments of single-stranded, negative-sense RNA coding for at least 10 viral proteins, 3 of which are known to function as polymerases. The viral RNA (vRNA), nucleoprotein (NP), and polymerases are in close association in the RNP (11, 14, 17). Matrix protein (M1) is located between the RNP and the inner surface of the lipid envelope in the intact virion (1, 14, 24). Two major external glycoproteins, hemagglutinin (HA) and neuraminidase (NA), and a small protein serving as a transmembrane channel, M2, are anchored in the viral envelope (13, 26, 32).
In addition to being a structural component of the virion, M1 is integral to many steps in the replication of the influenza virus. During early viral replication, the dissociation of M1 from RNP is required for entry of virus into the host cell cytoplasm (5, 12, 15) and is triggered by transport of H+ ions across the viral membrane by M2 (26). Later in the replication cycle, newly synthesized M1 migrates to the nucleus of the influenza virus-infected cell, where it again is found in close proximity with RNP (5, 19). The interaction of M1 and RNP leads to transport of M1-RNP complexes from the nucleus into the cytoplasm (15). In the maturation of viral particles, the ratio of M1 to NP influences morphologic features and infectivity of the released viruses (22). At the inner cytoplasmic membrane, M1-RNP complexes associate with embedded molecules of HA, NA, and M2 (9, 13, 14). The ability of M1 to interact with lipid membranes as demonstrated by a number of methods may relate to a role for M1 in the budding of newly synthesized virions from the cell surface (4, 10, 23, 30).
The association of M1 and RNP was first reported in the 1970s. When influenza virus HA and NA were removed by proteolytic enzymes, the proteins remaining in the virion were M1, NP, and polymerase (6, 24). The association of M1 with RNP was identified by electrophoretic separation of purified, native RNPs on low-percentage acrylamide gels (21). Depletion of M1 from the RNP rendered the vRNA sensitive to digestion by RNase A, which suggested that RNA is not completely protected by NP alone (20, 29).
Two domains in M1 have been shown to affect the disposition of RNA. One domain residing in a palindromic stretch of basic amino acids (101RKLKR105) has been shown to bind vRNA (8, 27, 29), fulfilling a prediction based on X-ray crystallographic data (25), and to serve also as a nuclear translocation signal for M1 (29). The other domain, containing a zinc finger motif (148C-C---H-H162), has been shown to associate with zinc molecules (7) and to inhibit viral replication (18), but its role in binding RNA is less certain.
Although the critical role of interaction between M1 and RNP in functional and morphologic features of influenza virus replication is apparent, the mechanism of the interaction of M1 and RNP remains unclear. In this report, we describe the association of purified RNPs with 35S-labeled M1 translated from cDNAs coding for either the wild-type (Wt) A/WSN/33 (WSN) M1 or for substitution and deletion mutants of WSN M1. The effects of reconstituting these Wt and mutant M1s with RNP under conditions of altered pH, ionic strength, and added detergent were used to further define the interacting domains of M1 and to explore mechanisms of the M1-RNP interaction.
MATERIALS AND METHODS
Virus and DNA constructs.
Influenza virus WSN was propagated in the allantoic cavities of 9-day-old embryonic eggs at 33°C for 2 days. Virions in the allantoic fluid were concentrated and purified by banding in 15 to 60% sucrose gradients (30).
Plasmids to express the mutant M1 genes with deletions or substitutions were constructed from a Wt WSN M1 gene in a vaccinia virus-based expression vector pTFM21(3) as described previously (3, 29). Briefly, pTFM21 (3) was used to create substitution mutations of WSN M1 corresponding to the RNA-binding amino acids 101RKLKR105 and the zinc finger motif (amino acids 148 to 162). The substitution of 101RKLKR105 by 101SNLNS105 (designated M101-SNLNS-105) was constructed by PCR. The zinc finger motif (148C-151C---159H-162H) in amino acids 148 to 162 was altered to S-C----H-H by PCR using the primer 456-5′GGC CTG GTA TCC GCA ACCT3′-474 and a primer corresponding to the vector sequence. Deletion mutants of the M1 gene were constructed by restriction enzyme digestion and PCR. All of the altered M1 cDNA sequences were confirmed by DNA sequence analysis.
In vitro translation of M protein and purification of RNPs.
In vitro expression of M1 genes was carried out in a coupled transcription-translation system (Promega, Madison, Wis.). Each plasmid (5 μg) was transcribed in vitro in the presence of the T7 phage RNA polymerase and translated in a reticulocyte system in the presence of [35S]methionine. The efficiency of [35S]methionine incorporation into newly synthesized protein was 22%, with a background incorporation of 0.2%. The newly synthesized [35S]methionine-labeled proteins were analyzed by electrophoresis on sodium dodecyl sulfate (SDS)–15% polyacrylamide gels and autoradiography.
Purified RNPs of influenza A virus were obtained by disrupting WSN (0.5 mg/ml) in buffer containing 10 mM Tris-HCl (pH 5.5), 1% Nonidet P-40, 0.6 M NaCl, and 1.25 mM dithiothreitol. After incubation in disruption buffer for 40 min at room temperature, RNPs released from virions in the reaction mixture were pelleted by centrifugation through 25% glycerol with a cushion of 50% glycerol in an SW 55Ti rotor at 120,000 × g for 60 min. The supernatant fluid containing lipids and membrane proteins was discarded, and the pellet containing the RNPs was resuspended in 10 mM Tris-HCl (pH 7.4). Purified RNPs were pooled and stored at −20°C for later use. The protein composition of the RNPs was analyzed by electrophoresis in SDS–12.5% polyacrylamide gels and stained with Coomassie blue.
Protein-RNA interaction assay.
The interaction of M1 with RNA was analyzed as described previously (29). Briefly, WSN RNA was labeled by growing influenza virus in cell cultures containing [32P]orthophosphate (15 μCi/ml; radioactivity concentration, 8 mCi/ml; Amersham). The labeled vRNAs were purified from virus by phenol extraction and stored at −70°C for later use. The M1s used for RNA-binding assays were derived by expression in a coupled transcription-translation system (Promega) from the specific plasmids described above. Bovine serum albumin (Boehringer Mannheim) was used as a negative control. M1s and bovine serum albumin were separated by SDS-polyacrylamide gel electrophoresis (PAGE) before transfer onto nitrocellulose membranes. The transferred proteins were incubated for 90 min at room temperature with 32P-labeled RNA in probing buffer containing 10 mM Tris-HCl (pH 7.5), 50 mM NaCl, 1 mM EDTA, 0.02% Ficoll, and 0.02% polyvinylpyrrolidone. After the completion of the incubation period, the membranes were washed, dried, and exposed to X-ray film. The relative association of RNA with specific M1s was calculated by comparing the density of 32P-labeled RNA bound to mutant M1s with that of labeled RNA bound to WSN Wt M1 protein. The amount of M1 used for the RNA-binding studies was equalized by comparison of the [35S]methionine-labeled mutant M1s with [35S]methionine-labeled Wt M1.
In vitro reconstitution of M1 protein with RNPs.
The analysis of association of M1 and RNP was carried out as follows. Purified RNP (50 μg) was incubated with labeled M1 (10,000 cpm). The negative control containing the same activity of [35S]methionine (10,000 cpm) was derived from the translation product of the same plasmid without M gene insertion. To study the effect of pH on the M1-RNP association, the suspension of M1-RNP complexes was incubated in buffer containing 0.05 M NaCl and 0.05 M Na2HPO4 adjusted to specific pH by addition of 0.1 M citric acid. To study the effect of ionic strength on M1-RNP complex association, the salt concentration was adjusted by adding NaCl to the reaction buffer. To study the effect of detergent on M1-RNP reconstitution, the RNP suspension was mixed in buffer containing 1% Triton X-100 and 0.05 M NaCl and then incubated at 4°C for 2 h. A 20-μl aliquot of the RNP-detergent suspension was centrifuged through a 150-μl cushion of 20% sucrose at 10,000 rpm for 15 min in a 0.5-ml Eppendorf tube, and the resulting pellet was collected. The amount of 35S-labeled M1 associated with RNP was determined by autoradiographic densitometry of proteins separated by SDS-PAGE. The percentage of M1 bound to RNP was calculated from more than three individual experiments by comparing to the total input 35S-labeled M1.
RESULTS
Expression and characterization of substitution and deletion mutants of M1.
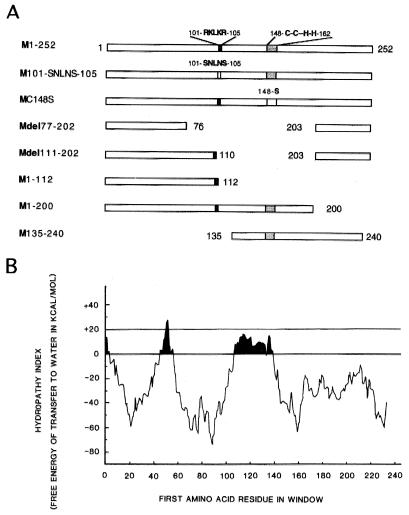
To determine whether M1-RNP complexes involved an M1-RNA interaction, the potential RNA-binding domains of M1 were altered by site-directed mutagenesis. Figure 1A shows schematic diagrams of the M1s translated from constructs containing substitution and deletion mutations; Fig. 1B shows the predicted hydropathy index of WSN Wt M1. Plasmid M101-SNLNS-105 expressed a basic amino acid RNA-binding domain (BAD) altered by replacing amino acids RKLKR by SNLNS; plasmid MC148S contained the DNA sequence coding for an alteration predicted to disrupt the zinc finger motif (16). Both the BAD and the zinc finger motif were deleted from plasmid Mdel77-202. The zinc finger motif was deleted from plasmids Mdel111-202 and M1-112. The coding region for the C-terminal 52 amino acids of plasmid M1-200 was deleted, but it retained both the BAD and the zinc finger motif. The coding regions for the N-terminal 134 and the C-terminal 12 amino acids of plasmid M135-240 were deleted so that the BAD was removed but the zinc finger motif was retained. The hydropathy index of the M1 protein revealed three hydrophobic domains, with two of the domains located in N-terminal 76 amino acids. Except for plasmid M135-240, all of the M1 mutants retained these two hydrophobic domains in the N-terminal 76 amino acids.
FIG. 1.
Schematic models and RNA-binding activities of the M1 proteins translated from the constructs containing substitution and deletion mutants. (A) Expressed amino acid sequences of WSN Wt and mutant M1 constructs; (B) hydropathy index of M1 protein.
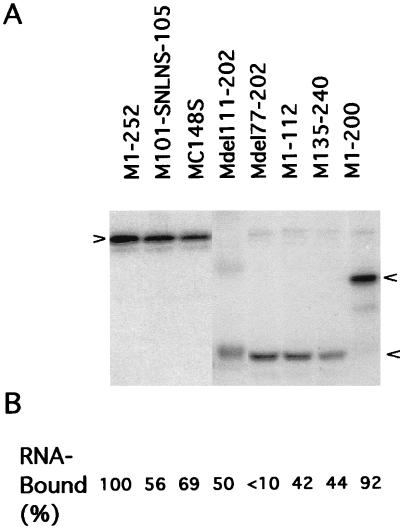
The substituted and truncated M1s were expressed in vitro by using a coupled reticulocyte lysate system (Promega) and were analyzed by autoradiographic densitometry after SDS-PAGE. As shown in Fig. 2, the relative migration of the substituted M1s (from plasmids M101-SNLNS-105 and MC148S) was not considerably affected compared to WSN Wt M1 (M1-252). However, as would be expected, the relative migration of the truncated M1s expressed from the various cDNA constructs was altered in proportion to the length of the translated protein. M1-200 (with 52 amino acids deleted) had the slowest migration among the truncated M1 proteins. Mdel111-202 (with 91 amino acids deleted) migrated slightly behind the truncated M1s with 125 amino acids deleted (Mdel77-202), with 120 amino acids deleted (M1-112), or with 105 amino acids deleted (M135-240). Proteins translated in vitro were further studied by reaction with monoclonal antibodies specific to the M1 protein of WSN virus. Strong reactions of monoclonal antibodies with the substituted and deleted M1 proteins were observed by immunoprecipitation (data not shown) and immunofluorescence (31). Nonspecific bands shown in the translated samples in Fig. 2 represented less than 5% of total translated products and did not react with the monoclonal antibodies to M1 protein. Although the source of these bands is unknown, it is possible that they are nonspecific products from the in vitro translation reaction.
FIG. 2.
(A) SDS-PAGE and autoradiography of in vitro-translated and 35S-labeled M1 proteins from cDNAs coding for WSN Wt and mutants of M1 proteins; (B) RNA-binding activity of WSN Wt and mutants of M1 proteins translated in vitro without addition of [35S]methionine. The percentage of RNA bound to M1 was calculated by comparison to the total input [32P]RNA. The data are means of at least three independent experiments, and the standard deviation of each experimental point was below 10% (data not shown).
The RNA-binding capacity of M1 mutants was studied by a protein-RNA interaction assay previously described (29). The WSN Wt M1 and mutant M1s were translated in vitro, separated by SDS-PAGE, and transferred onto nitrocellulose membranes. RNA-binding activities of WSN Wt (M1-252) and mutant M1 proteins were determined by measuring the binding of 32P-labeled RNA (extracted from WSN virions) to M1 immobilized on nitrocellulose membrane. Figure 2 B shows the relative RNA binding of mutant M1 proteins. Deletion of the C-terminal 52 amino acids (M1-200) had little effect on the RNA-binding capacity of the protein (about an 8% reduction in RNA binding compared to WSN Wt M1). Altering the zinc finger motif (MC148S) resulted in moderate reduction of RNA binding (31%). Substitution of amino acids RKLKR (M101-SNLNS-105) reduced binding activity by 44% compared to WSN Wt M1. Deletions of amino acids that eliminated one of the two RNA-binding domains (Mdel111-202, M1-112, or M135-240) decreased RNA-binding activity by 50 to 58%. However, the M1 expressed from Mdel77-202 missing both the BAD and the zinc finger motif bound RNA only weakly if at all (>90% reduction in RNA binding). These results indicate that in vitro-translated M1s retained RNA-binding capacity provided by one or both of the RNA-binding domains in the construct.
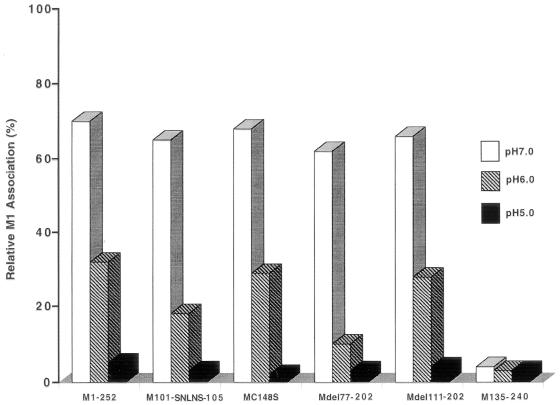
Reassociation of M1 with RNP and effect of low pH.
During viral replication, M1 in incoming viral particles has been shown to dissociate from M1-RNP complexes at the lower pH of the endosomal microenvironment (5). Further studies were conducted to determine whether reassociation of M1 with RNP was possible at low pH. M1s were labeled with [35S]methionine during in vitro translation as described in Materials and Methods. RNPs were purified by disruption of WSN virions with 1% Nonidet P-40 and 0.5 M NaCl at pH 6.5 and then centrifuged through 50% glycerol. RNPs isolated by this method contained two forms of NP (22) and less than 2% residual M1 (Fig. 3). Reconstitution of WSN Wt and mutant M1s with this RNP was carried out at pH 7.0, 6.0, and 5.0 (Fig. 4). M1-RNP complexes were separated from unbound M1 by centrifugation through 20% glycerol and analyzed by SDS-PAGE. The percentage of M1 associated with RNP was calculated as a ratio of the total input M1. The association of M1 with RNP at neutral pH (pH 7.0) was about 70% of total input M1 translated from WSN Wt. Similarly, 60 to 70% of each of the mutant M1s (except M135-240) associated with RNP at pH 7.0. Less than 10% of M1 translated from M135-240 bound to RNP. M135-240 contains the zinc finger motif but is missing the N-terminal portion of M1 including the BAD (101RKLKR105).
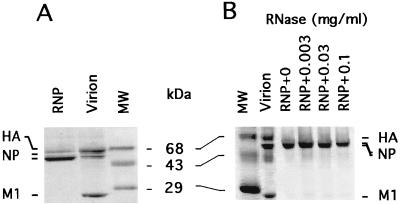
FIG. 3.
SDS-PAGE analysis of purified RNPs and the effect of RNase A treatment on RNP stability. (A) M1-depleted RNPs were purified from virions as described in Materials and Methods. The protein components of RNPs and virions were analyzed by SDS-PAGE (12.5% gel). (B) The RNPs (1 mg/ml) were treated with RNase A at final concentrations of 0.003 to 0.1 mg/ml for 30 min at 37°C. The RNase A-treated RNPs were harvested by centrifugation at 120,000 × g for 60 min through 25% glycerol and a 50% glycerol pad. The resulting pellets were collected and analyzed by SDS-PAGE on a 12.5% gel. Lanes MW, molecular weight markers.
FIG. 4.
Effect of low pH on M1-RNP reconstitution. The reconstitution of M1 and RNP was carried out in buffer containing 0.05 M NaCl at pH 7.0, 6.0, and 5.0 during the reconstitution assay. The RNP-associated M proteins were autoradiographed, and binding reactivity was quantified by densitometry. The percentage of the binding activity from 35S-labeled M1s to RNP was calculated by comparison to the total input 35S-M1 proteins. The data are means of at least three independent experiments, and the standard deviation of each experimental point was below 10% (data not shown).
At pH 6.0, M1 from WSN Wt or from mutations eliminating the zinc finger motif (MC148S and Mdel111-202) was found at 30% relative binding (reduced by approximately 50% compared with results at pH 7.0). Under identical conditions, approximately 15 to 20% relative binding (reduced approximately 80 to 85%) was demonstrated for M1s from mutants lacking the BAD (M101-SNLNS-105 and Mdel77-202). Reconstitution at pH 5.0 essentially abolished M1-RNP association in vitro for all M1s, including Wt M1. As shown in Fig. 4, under no conditions did M1 from M135-240 bind RNP.
These experiments indicate that reassociation of in vitro-translated M1 with RNP is inhibited at lower pH and that M1 can bind to RNP in the absence of both the BAD and the zinc finger motif (Mdel77-202) but not in the absence of the entire N-terminal portion of M1 (M135-240).
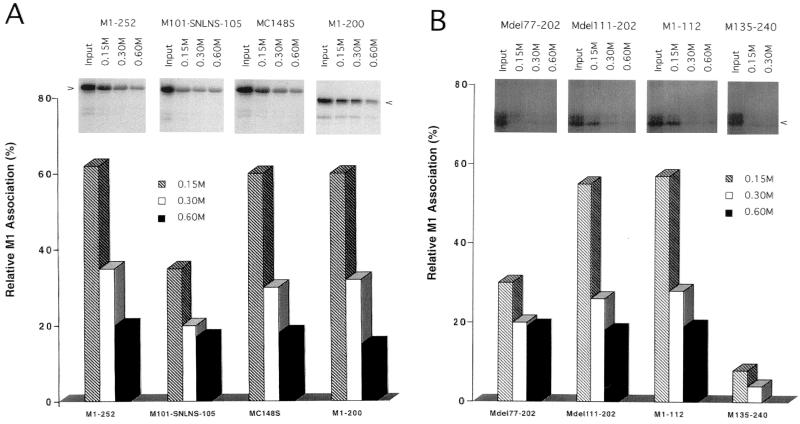
The M1 BAD is affected by ionic strength.
To explore the effects of ionic strength on the interaction of M1 and RNP, reconstitution of 35S-labeled M1 proteins with M1-depleted RNP was carried out in vitro in the presence of various concentrations of NaCl at pH 7.0. As shown in Fig. 5A, approximately 60% of 35S-labeled WSN Wt M1 (M1-252) associated with RNP when reconstitution was carried out in 0.15 M NaCl (approximately physiological salt concentration). Increasing the salt concentration reduced the percentage of M1 bound.
FIG. 5.
Effect of salt on M1-RNP reconstitution of Wt, substitution, and C-terminal truncation M1s (A) and deletion M1 mutants (B). In vitro-translated 35S-labeled M proteins were reconstituted with RNP in the presence of various concentrations of NaCl as specified in Materials and Methods. The percentage of RNP association was quantified by comparison to the total input 35S-M1 proteins. The data show results of a representative binding experiment; the standard deviation of three independent experimental points was below 10% (data not shown).
The RNP-binding activity of M1 with substitution in the BAD (M101-SNLNS-105) was similar to that for WSN Wt M1 at 0.05 M NaCl (Fig. 4) but was reduced by approximately half at 0.15 M NaCl (Fig. 5). Increasing the salt concentration from 0.15 to 0.30 M reduced the association of M1 (M101-SNLNS-105) with RNP from 35 to 20%, but a further increase in salt concentration to 0.60 M NaCl had proportionally little additional effect on RNP binding (17% at 0.60 M NaCl). M1 with substitution in the zinc finger motif (MC148S) or deletion of the C-terminal 52 amino acids (M1-200) had RNP binding similar to that of WSN Wt M1 at all concentrations of NaCl.
Studies of the RNP association of truncated M1s gave results similar to that for substituted M1s. For M1s retaining the BAD but missing the zinc finger motif (Mdel111-202 or M1-112), association with RNP was similar at all salt concentrations to that of WSN Wt M1. However, RNP binding of the M1 expressed from Mdel77-202 (deletion of both the BAD and the zinc finger motif) was reduced to 30% at 0.15 M and to 20% or less at 0.3 to 0.6 M NaCl. Deletion of the N-terminal 134 and the C-terminal 12 amino acids of M1 (M135-240) abolished M1-RNP association. These results suggested that the BAD corresponding to amino acid RKLKR in positions 101 to 105 is involved in RNP association and that the N-terminal region of 76 amino acids of the M protein might also be involved in RNP binding. However, the zinc finger motif corresponding to amino acids 148 to 162 has little effect on RNP association.
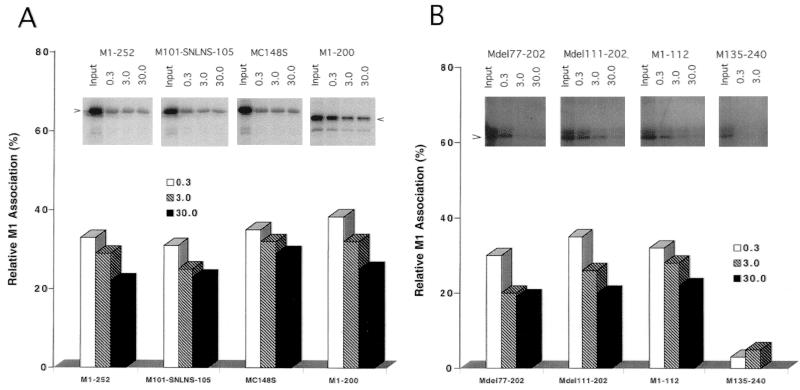
M1-RNP interaction is not related only to RNA binding.
RNA in M1-depleted RNPs of influenza virus can be digested by RNase A, which suggests that NP does not protect all of the RNA of RNPs (20, 29). Therefore, to further understand the role of RNA binding in the M1-RNP association, M1-depleted RNPs of WSN virus were enzymatically treated with RNase A to digest unprotected RNA. Before using RNase A-treated RNP for the reconstitution assay, we examined the effect of incubation with RNase A on both vRNA and RNPs. Although vRNA was digested when the concentration of RNase A was 3 μg/ml (data not shown). RNP cores retained structural integrity after treatment with RNase A at concentrations of up to 100 μg/ml for 30 min at 37°C (Fig. 3B).
To assay association, 35S-labeled WSN Wt and mutant M1s were reconstituted with RNPs that had been preincubated with RNase A. The results of reconstitution of WSN Wt M1 protein (M1-252) and substitution mutants (M101-SNLNS-105 and MC148S) with RNase A-treated RNP are shown in Fig. 6. When RNP was treated with 0.3 μg of RNase A per ml, binding of WSN Wt M1 was only 35% relative to that with untreated RNP (Fig. 6A). At higher concentrations of RNase A, 20 to 25% of WSN Wt M1 protein was bound to RNP. M1s with substitution in the BAD (M101-SNLNS-105) or in the zinc finger motif (MC148S) did not further reduce RNP-binding activity compared to Wt M1 (31 to 35% with RNase A at 0.3 μg/ml and 25 to 32% with RNase A at 3.0 μg/ml), nor did deletion of the C-terminal 52 amino acids (M1-200). M1s with deletion of the C-terminal 140 amino acids (M1-112) or amino acids 77 to 202 (Mdel77-202) also exhibited 30 to 35% association (Fig. 6B). RNP treated with a high concentration of RNase A (30 μg/ml) showed approximately 20% binding in all cases. However, the deletion mutant containing amino acids 135 to 240 (M135-240) exhibited only background RNP binding at all RNase A concentrations. Although RNase A-treated RNP bound less to the Wt or mutant M1s, the persistence of RNP binding after RNase A treatment suggested strongly that M1 binds to RNP not only by way of RNA but also by way of an independent protein-protein interaction.
FIG. 6.
Effect of pretreatment of RNP with RNase A on M1-RNP reconstitution. Relative RNP association of Wt, substitution, and C-terminal truncation M1s (A) and deletion M1 mutants (B). Purified RNPs were treated with RNase A as described in Materials and Methods. The pretreated RNPs were then incubated with 35S-labeled M1 proteins, and the percentage of average RNP association was quantified by comparison to the total input 35S-M1 proteins. The data show results of a representative binding experiment; the standard deviation of three independent experimental points was below 10% (data not shown).
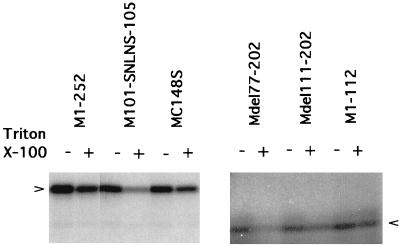
The hydrophobic domains located at the N terminus of M1 protein contribute to RNP-binding activity.
Although the association of M1 with RNPs was reduced by the addition of salt or pretreatment of RNP with RNase A, deletion or substitution in the BAD did not completely abolish RNP-binding activity whereas removal of the N-terminal amino acids including the BAD resulted in complete elimination of RNP binding (M135-240). All of the M1 mutants retaining the N-terminal 76 amino acids exhibited RNP-binding capacity even when the BAD was deleted.
To explore the RNP-binding properties of the N-terminal 76 amino acids, the WSN Wt and mutant M1s were reconstituted with RNP in buffer containing 1% Triton X-100 in 0.05 M NaCl. As shown in Fig. 7, addition of Triton X-100 to the reconstitution mixture resulted in approximately 30% reduction of WSN Wt M1 (M1-252) association to RNP. Substitution in the zinc finger motif (MC148S) did not further reduce the association of M1 with RNP (same as that from WSN Wt; approximately 30% reduction, comparing binding with and without Triton X-100). Other deletions affecting the zinc finger motif (Mdel111-202 or M1-112) did not considerably alter M1-RNP association (10 to 15% reduction). However, substitution in the BAD (M101-SNLNS-105) resulted in an 80% reduction of M1-RNP association in the presence of Triton X-100. The deletion of both the BAD and the zinc finger motif (Mdel77-202) resulted in a similar 85% reduction of RNP association. These data suggested that a detergent-labile element located in N-terminal 76 amino acids of M1 is involved in RNP binding and that it is independent of RNA-binding domains.
FIG. 7.
Effect of detergent on M1-RNP reconstitution. Reconstitution of M1 and RNP was carried out as described in Materials and Methods, with (+) or without (−) addition of 1% Triton X-100.
DISCUSSION
Our data suggest that the RNP-binding activity of M1 is the result of at least two forces, one ionic and the other hydrophobic. Further, the data lead us to postulate that the association of M1 with RNP is through two mechanisms, a protein-RNA interaction and a protein-protein interaction. Since M1 binds nonspecifically to RNA (27), the specificity of the interaction of M1 with RNP may depend on an interaction of M1 and NP. Because the coexpression of M1 and NP genes in transfected cells has not provided evidence of direct binding of M1 with NP alone (2, 33), reassociation of M1 with RNP via a protein-protein interaction suggests that M1 may bind to NP only when NP is already bound to RNA.
According to several separate studies, M1 protein is capable of binding to viral RNA via two sequences, amino acids 90 to 108, containing a nuclear localization domain (RKLKR), and amino acids 135 to 165, containing a zinc finger motif (27–29). Based on this information, we constructed substitution and deletion mutations of the M1 gene of influenza WSN virus for transcription and translation in vitro. Our results confirm that M1 binds to vRNA through the basic amino acid domain (RKLKR) and suggest that binding to RNA may also occur via the zinc finger motif (8, 27, 29). To the contrary, Elster et al. (8), concluded that altering the zinc finger motif in the M1 does not affect its RNA-binding activity. Testing the same substitution construct, Elster et al. used multiple concentrations of M1 and determined a sigmoidal curve for binding. Data on the steep part of the curve were similar to the zinc finger results in our experiment, although Elster et al. indicated that the zinc finger was not involved in RNA binding. Our data are also supported by deletion of the zinc finger region of M1, which independently reduced RNA binding to the same extent as deletion of the BAD. Deletion of both of these RNA-binding domains resulted in the lowest binding activity. Although our study did not show that the zinc finger motif of M1 was involved in RNP-binding activity, the zinc finger motif has been shown to inhibit viral replication in vivo (18).
Influenza virus M1 dissociates from RNP complexes at low pH in vitro (34, 35) and in infected cells (5), but the reassociation of M1 with RNP has not been extensively studied. During the early stages of viral replication, M1 dissociates from the incoming vRNP at the lower pH created by H+ transported from the endosomal environment by M2. Acidic environment may alter the conformation of M1 and results in lower binding affinity for RNP. RNPs subsequently enter the nucleus of the cell, where viral RNAs are transcribed. Late in infection, newly synthesized M1 and RNP bind in the nucleus to form M1-RNP complexes at neutral pH, which results in one-way transport of the complexes from the nucleus (5, 15). It should be noted that the amount of 35S-labeled M1 relative to RNP in our reassociation experiments does not necessarily reflect the ratio of M1 to RNP within virions or cells. However, our studies demonstrate that low pH markedly reduces reassociation of M1 with RNP. In addition, our studies suggest that substitution or deletion of the BAD of M1 increases susceptibility of M1-RNP complexes to the effects of lowered pH (Fig. 4).
Our data regarding the effect of salt concentration on reassociation of M1 with RNP indicate that increasing salt concentrations reduce the binding of M1 with RNP substantially but do not eliminate binding completely. Although the data are not shown, salt concentration has a greater effect on reassociation of input M1 with RNP than on the dissociation of endogenous M1 from M1-RNP complexes. Our data on the effect of salt concentration on the RNP-binding activity of M1 mutants in which the BAD has been altered or deleted provide some insight into the requirement for RNA binding in the process of RNP binding. Substitution of the RNA-binding domain (101RKLKR105) with uncharged amino acids dramatically increases susceptibility to salt disruption and reduces the RNP-binding ability of M1. Similarly deletion of the BAD (Mdel77-202, which also deletes the region containing the zinc finger motif) results in a mutant M1 that binds poorly to RNP. Although these data suggest that RNA binding is involved in RNP binding, the residual RNP binding for M1 with the BAD deleted suggests that RNA binding is not the only means by which M1 binds to RNP. Since the basic amino acid residues (101RKLKR105) also play a role in M1 nuclear localization (31), it is likely that these residues mediate multiple functions of M1. Further studies are needed to assess the contribution of RNA binding to infectivity of the virus.
Since deletion or substitution of the RNA-binding domains of M1 is insufficient to completely abolish binding to RNP, further studies were done to evaluate whether the hydrophobic domains of the N terminus are involved in RNP binding. As noted, the hydropathy index of the amino acid sequence of M1 predicts three major hydrophobic domains, two of which are located within the first 76 amino acids (amino acids 1 to 20 and 45 to 70). The domain located within amino acids 45 to 70 has also been implicated in membrane association (4, 10). The detergent-associated reduction of the RNP-binding activity of the deletion mutant Mdel77-202 (containing the N-terminal 76 amino acids and the C-terminal 50 amino acids but missing potential RNA-binding domains) suggests that a hydrophobic binding domain located in the N-terminal 76 amino acids of M1 is involved in RNP binding independent of RNA binding. The lack of RNP-binding capacity of the mutant M135-202 (missing the N-terminal half of M1) and the persistence of RNP binding after RNase A treatment are also consistent with the hypothesis that an N-terminal hydrophobic domain is involved in RNP binding independent of RNA binding. Although amino acids 45 to 70 have the potential for binding to lipid membranes, we do not know whether the RNP-binding and membrane-binding domains overlap.
The interaction of M1 with RNP is central to assembly and disassembly during the replication cycle of influenza viruses. In addition to acting as a switch to promote the transport of RNP in or out of the nucleus, the RNP binding of M1 has a structural role in final viral assembly. Modification of M1 and corresponding domains on NP may enhance or reduce the affinity of M1s and RNPs, which should be reflected by changes in viral growth kinetics. Further studies of RNP-binding domains may help us complete the understanding of the role of M1 in influenza virus virulence and pathogenesis in a variety of host systems.
ACKNOWLEDGMENTS
We thank Lewis Markoff and C. D. Atreya, Center for Biologic and Evaluation and Research, Food and Drug Administration, for critical reviews of the manuscript.
REFERENCES
- 1.Allen H, McCauley J, Waterfield M, Gething M J. Influenza virus RNA segment 7 has the coding for two polypeptides. Virology. 1980;107:548–551. doi: 10.1016/0042-6822(80)90324-4. [DOI] [PubMed] [Google Scholar]
- 2.Avalos R T, Zhong Y, Nayak D P. Association of influenza virus NP and M1 proteins with cellular cytoskeletal elements in influenza virus-infected cells. J Virol. 1996;71:2947–2958. doi: 10.1128/jvi.71.4.2947-2958.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baylor N W, Li Y, Ye Z, Wagner R R. Transient expression and sequence of the matrix (M1) gene of WSN influenza virus in a vaccinia vector. Virology. 1988;163:618–621. doi: 10.1016/0042-6822(88)90303-0. [DOI] [PubMed] [Google Scholar]
- 4.Bucher D J, Kharitonenkov I G, Zakomirdin J A, Grigoriev V B, Klimenko S M, Davis J F. Incorporation of influenza virus M protein into liposomes. J Virol. 1980;36:586–590. doi: 10.1128/jvi.36.2.586-590.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bui M, Whittaker G, Helenius A. Effect of M1 protein and low pH on nuclear transport of influenza virus ribonucleoproteins. J Virol. 1996;70:8391–8401. doi: 10.1128/jvi.70.12.8391-8401.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Compans R W, Klenk H D. Viral membranes. In: Fraenkel-Conrat H, Wagner R R, editors. Comprehensive virology. Vol. 13. New York, N.Y: Plenum Publishing Corp.; 1979. pp. 293–407. [Google Scholar]
- 7.Elster C, Fourest E, Baudin F, Larsen K, Cusack S, Ruigrok R W. A small percentage of influenza virus M1 protein contains zinc but zinc does not influence in vitro M1-RNA interaction. J Gen Virol. 1994;75:37–42. doi: 10.1099/0022-1317-75-1-37. [DOI] [PubMed] [Google Scholar]
- 8.Elster C, Larsen K, Gagnon J, Ruigrok R W H, Baudim F. Influenza virus M1 protein binds to RNA through its nuclear localization signal. J Gen Virol. 1997;78:1589–1596. doi: 10.1099/0022-1317-78-7-1589. [DOI] [PubMed] [Google Scholar]
- 9.Enami M, Enami K. Influenza virus hemagglutinin and neuraminidase glycoproteins stimulate the membrane association of the matrix protein. J Virol. 1996;70:6653–6657. doi: 10.1128/jvi.70.10.6653-6657.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gregoriades A. Interaction of influenza M protein with viral lipid and phosphatidylcholine vesicles. J Virol. 1980;36:470–479. doi: 10.1128/jvi.36.2.470-479.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heggeness M H, Smith P R, Ulmanen L, Krug R M, Choppin P W. Studies on the helical nucleocapsid of influenza virus. Virology. 1982;118:466–470. doi: 10.1016/0042-6822(82)90367-1. [DOI] [PubMed] [Google Scholar]
- 12.Helenius A. Unpacking of the incoming influenza virus. Cell. 1992;69:577–578. doi: 10.1016/0092-8674(92)90219-3. [DOI] [PubMed] [Google Scholar]
- 13.Lamb R A, Zeebedee S L. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell. 1985;40:627–633. doi: 10.1016/0092-8674(85)90211-9. [DOI] [PubMed] [Google Scholar]
- 14.Lamb R A, Choppin P W. The structure and replication of influenza virus. Annu Rev Biochem. 1983;52:467–506. doi: 10.1146/annurev.bi.52.070183.002343. [DOI] [PubMed] [Google Scholar]
- 15.Martin K, Helenius A. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell. 1991;67:117–130. doi: 10.1016/0092-8674(91)90576-k. [DOI] [PubMed] [Google Scholar]
- 16.Meric C, Goff S P. Characterization of Moloney murine leukemia virus mutants with single-amino-acid substitution in the Cys-His box of the nucleocapsid protein. J Virol. 1989;63:1558–1568. doi: 10.1128/jvi.63.4.1558-1568.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murti K G, Webster R G, Jones I M. Localization of RNP polymerases on influenza viral ribonucleoprotein by immunogold labeling. Virology. 1988;164:562–566. doi: 10.1016/0042-6822(88)90574-0. [DOI] [PubMed] [Google Scholar]
- 18.Nasser E H, Judd A K, Sanchez A, Anastasiou D, Bucher D J. Antiviral activity of influenza virus M1 zinc finger peptides. J Virol. 1996;70:8639–8644. doi: 10.1128/jvi.70.12.8639-8644.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patterson S, Gross J, Oxford J S. The intercellular distribution of influenza virus matrix protein and nucleoprotein in infected cells and their relationship to haemagglutinin in the plasma membrane. J Gen Virol. 1988;69:1859–1872. doi: 10.1099/0022-1317-69-8-1859. [DOI] [PubMed] [Google Scholar]
- 20.Pons M W. Isolation of influenza virus ribonucleoprotein from infected cell. Demonstration of the presence of negative-stranded RNA in viral RNP. Virology. 1971;46:149–160. doi: 10.1016/0042-6822(71)90014-6. [DOI] [PubMed] [Google Scholar]
- 21.Rees P J, Dimmock N J. Kinetics of synthesis of influenza virus ribonucleoprotein structures. J Gen Virol. 1982;59:403–408. doi: 10.1099/0022-1317-59-2-403. [DOI] [PubMed] [Google Scholar]
- 22.Roberts P C, Lamb R A, Compans R W. The M1 and M2 proteins of influenza A virus are important determinants in filamentous particle formation. Virology. 1998;240:127–137. doi: 10.1006/viro.1997.8916. [DOI] [PubMed] [Google Scholar]
- 23.Robertson B H, Bennett C J, Compans R W. Selective dansylation of M protein within intact influenza virus. J Virol. 1982;44:871–876. doi: 10.1128/jvi.44.3.871-876.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schulze I T. The structure of influenza virus. II. A model based on the morphology and composition of subviral particles. Virology. 1972;47:181–196. doi: 10.1016/0042-6822(72)90251-6. [DOI] [PubMed] [Google Scholar]
- 25.Sha B, Luo M. Structure of a bifunctional membrane-RNA binding protein, influenza virus matrix protein M1. Nat Struct Biol. 1997;4:239–244. doi: 10.1038/nsb0397-239. [DOI] [PubMed] [Google Scholar]
- 26.Sugrue R J, Hay A J. Structural of the M2 protein of the influenza A viruses: evidence that it forms a tetrameric channel. Virology. 1991;180:617–624. doi: 10.1016/0042-6822(91)90075-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wakefield L, Brownlee G G. RNA binding properties of influenza virus matrix protein M1. Nucleic Acids Res. 1989;17:8569–8580. doi: 10.1093/nar/17.21.8569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watanabe K, Handa H, Mizumoto K, Nagata K. Mechanisms for inhibition of influenza virus RNA polymerase activity by matrix protein. J Virol. 1996;70:241–247. doi: 10.1128/jvi.70.1.241-247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye Z, Baylor N W, Wagner R R. Transcription-inhibition and RNA-binding domains of influenza virus matrix protein mapped with anti-idiotype antibodies and synthetic peptides. J Virol. 1989;63:3586–3594. doi: 10.1128/jvi.63.9.3586-3594.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye Z, Pal R, Fox J W, Wagner R R. Functional and antigenic domains of the matrix (M1) protein of influenza virus. J Virol. 1987;61:239–246. doi: 10.1128/jvi.61.2.239-246.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ye Z, Robinson D, Wagner R R. Nucleus-targeting domain of the matrix protein (M1) of influenza virus. J Virol. 1995;69:1964–1970. doi: 10.1128/jvi.69.3.1964-1970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zebedee S L, Lamb R A. Influenza A virus M2 protein: monoclonal antibody restriction of virus growth and detection of M2 in virions. J Virol. 1988;62:2762–2772. doi: 10.1128/jvi.62.8.2762-2772.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao H, Ekstrom M, Garoff H. The M1 and NP proteins of influenza A virus form homo- but not heterooligomeric complexes when coexpressed in BHK-21 cell. J Gen Virol. 1998;79:2435–2446. doi: 10.1099/0022-1317-79-10-2435. [DOI] [PubMed] [Google Scholar]
- 34.Zhirnov O P. Solubilization of matrix protein M1/M from virions occurs at different pH for orthomyxo- and paramyxoviruses. Virology. 1990;176:274–279. doi: 10.1016/0042-6822(90)90253-n. [DOI] [PubMed] [Google Scholar]
- 35.Zhirnov O P. Isolation of matrix protein M1 from influenza viruses by acid-dependent extraction with nonionic detergent. Virology. 1992;186:324–330. doi: 10.1016/0042-6822(92)90090-c. [DOI] [PubMed] [Google Scholar]