Abstract
We recently reported that a cationic peptide, T22 ([Tyr5,12, Lys7]-polyphemusin II), specifically inhibits human immunodeficiency virus type 1 (HIV-1) infection mediated by CXCR4 (T. Murakami et al., J. Exp. Med. 186:1389–1393, 1997). Here we demonstrate that T22 effectively inhibits replication of T-tropic HIV-1, including primary isolates, but not of non-T-tropic strains. By using a panel of chimeric viruses between T- and M-tropic HIV-1 strains, viral determinants for T22 susceptibility were mapped to the V3 loop region of gp120. T22 bound to CXCR4 and interfered with stromal-cell-derived factor-1α–CXCR4 interactions in a competitive manner. Blocking of anti-CXCR4 monoclonal antibodies by T22 suggested that the peptide interacts with the N terminus and two of the extracellular loops of CXCR4. Furthermore, the inhibition of cell-cell fusion in cells expressing CXCR4/CXCR2 chimeric receptors suggested that determinants for sensitivity of CXCR4 to T22 include the three extracellular loops of the coreceptor.
Viral receptors facilitate the invasion of target cells by viral particles. CD4 is a primary receptor for attachment of human immunodeficiency virus type 1 (HIV-1), and a member of the chemokine receptor family is a coreceptor for target cell entry (21, 43). HIV-1 isolates can be grouped into three general phenotypic categories based on their cell tropism: T-cell line-tropic (T-tropic) HIV-1 strains can infect peripheral blood T lymphocytes and leukemic T-cell lines and can arise late in the evolution of AIDS. Macrophage-tropic (M-tropic) strains of HIV-1 (2, 18, 25) can infect peripheral blood T lymphocytes and macrophages but not T-cell lines. Dual-tropic HIV-1 isolates can infect peripheral blood T lymphocytes, T-cell lines, and macrophages. In terms of coreceptor usage, T-tropic HIV-1 strains primarily use a CXC chemokine receptor, CXCR4 (29), whereas M-tropic HIV-1 strains use mainly CCR5 (2, 14, 18, 25). Dualtropic HIV-1 strains can use CXCR4 and CCR5, as well as CCR3 and CCR2b (14, 24). Recently, several other seven transmembrane-spanning proteins that are structurally similar to chemokine receptors, most of which are orphan receptors, have been found to be able to mediate HIV-1 entry into CD4+ T cells (13, 19, 24, 27, 30, 38, 51, 54). In addition, CD4-independent, chemokine receptor-dependent infection with some HIV-1 or HIV-2 strains has been reported (26, 28, 31), suggesting a major role of the chemokine receptor in HIV entry. Therefore, the chemokine receptors constitute an important target for the development of anti-HIV drugs. It has also been documented that specific ligands for these receptors are able to inhibit HIV infection. Pre-B-cell growth-stimulating factor/stromal-cell-derived factor (SDF-1) (47, 66), the ligand for CXCR4, inhibits entry of T-tropic, but not M-tropic, HIV-1 strains (7, 49). In contrast, natural ligands for CCR5, regulated upon activation, normal T cell expressed and secreted (RANTES), macrophage inflammatory protein-1α (MIP-1α), and MIP-1β, inhibit entry of M-tropic HIV-1 but not T-tropic viruses (15, 67, 71). These findings suggest a novel strategy for the design of anti-HIV drugs. Recently, RANTES derivatives have been shown to be specific inhibitors of infection with M-tropic HIV-1 as a CCR5 antagonist (6, 58). We recently found that T22 ([Tyr5,12, Lys7]-polyphemusin II), a synthetic derivative of basic peptides isolated from blood cells of horseshoe crabs, is a specific antagonist for both T-tropic HIV-1 infection and signal transduction through ligand binding (45). Two other small molecular CXCR4 antagonists have also been reported recently (23, 56). It is important to understand the mechanism by which CXCR4 inhibitors act and to develop improved chemokine receptor inhibitors. In the present study, we show that T22 directly binds to extracellular loops of CXCR4 and inhibits its functions in binding and signaling of SDF-1α, as well as coreceptor activity for T-tropic HIV-1 strains.
MATERIALS AND METHODS
Reagents.
T22, tachyplesin I, polyphemusin II, and 4Ala-T-I were produced by using Fmoc (9-fluorenylmethoxycarbonyl) solid-phase or solution-phase peptide synthesis as described elsewhere (40, 59, 64). The synthetic peptides were purified by high-performance liquid chromatography (HPLC) and gel filtration and were identified by mass spectroscopy and amino acid analyses after acid hydrolysis and LAP digestion. SDF-1α was synthesized by stepwise disulfide-forming reactions as described previously (62). 125I-labeled human recombinant SDF-1α was obtained from Dupont-NEN (Boston, Mass.). The labeling was performed by using a lactoperoxidase procedure and the [125I]SDF-1α is purified by reversed-phase HPLC.
Construction of chimeric HIV-1 clones.
Two molecular clones of HIV-1, NL4-3 and JR-CSF, were described previously (1, 35). Most HIV-1 chimeras were constructed by using fragments from NL4-3 and JR-CSF. Chimeric viruses CNC-DX, CNC-MX, NCN-SN, NCN-SM, and NCN-MN were made as described previously (12). Viruses CNC-AX, CNC-AD, NCN-AX, and NCN-DX were made by similar methods. NL-CSFV3, in which the V3 region of NL4-3 was replaced by that of JR-CSF, was constructed as follows. A pair of synthetic oligonucleotides containing StuI, MluI, XbaI, and NheI sites were inserted into pNL4-3-10-17 to create MluI and XbaI sites. The StuI-to-MluI (region 6822 to 7102) fragment from NL4-3 (obtained by PCR), the MluI-to-XbaI (position 7199) fragment from JR-CSF (synthetic oligonucleotides), and the XbaI-to-NheI (position 7250) fragment from NL4-3 (synthetic oligonucleotides) were sequentially substituted into the modified pNL4-3-10-17 to form a full-length chimeric plasmid DNA. To confirm that the above manipulations do not affect viral infectivity and cell tropism, the MluI-to-XbaI fragment of NL-CSFV3 was replaced by that of NL4-3. The reconstituted NL4-3 was indistinguishable from the original NL4-3 in infectivity and cell tropism. Viral stocks were prepared after electroporation of COS cells and titrated in phytohemagglutinin (PHA) (Seikagaku Corp., Tokyo, Japan)-stimulated human peripheral blood mononuclear cells (PBMC) as previously described (34).
Construction of chimeric receptors.
Chimeric receptors composed of CXCR4 and CXCR2 were constructed by the PCR-ligation-PCR approach as described previously (39).
Cells and culture conditions.
HeLa S3, MT-2, MT-4, and MOLT-4 cells were maintained in RPMI 1640 supplemented with 10% fetal bovine serum (FBS). HeLa cells were maintained in Dulbecco minimal essential medium (DMEM) containing 10% FBS. QT6 cells stably expressing CD4 containing a cytoplasmic tail deletion were provided by A. Koito (University of Tsukuba, Tsukuba, Japan). These cells were maintained in DMEM containing 10% FBS and 300 μg of G418 per ml. PBMC were isolated from human immunodeficiency virus (HIV) seronegative healthy donors by using Ficoll-Hypaque (Pharmacia Biotech, Uppsala, Sweden) and grown in RPMI 1640 containing 20% FBS and 100 U of recombinant interleukin-2 (IL-2) (Shionogi Co., Osaka, Japan) per ml after stimulation with 1 μg of PHA per ml for 2 to 3 days. Culture media were supplemented with 100 U of penicillin and 100 μg of streptomycin per ml.
Monocyte-derived macrophage (MDM) culture.
Monocytes were enriched from PBMC by adherence to plastic (100-mm-diameter dishes) coated with human AB serum (Flow Laboratory, McLean, Va.) in RPMI 1640 medium containing 20% FBS at 37°C for 24 h. One day later, nonadherent cells were removed, and the adherent cells were cultured for 2 days in RPMI 1640 containing 15% FBS and 5% giant cell tumor supernatant (IGEN, Rockville, Md.). Adherent macrophages were treated with 0.5 mM EDTA in phosphate-buffered saline (PBS) for 10 min to transfer cells to 24-well plates at a density of 2.5 × 105 cells/well and then used for infection experiments.
Viruses.
The origins of the primary clinical isolates were as follows. TMD-1 and TMD-2 were simultaneously isolated from PBMC or plasma, respectively, of a patient with AIDS (73). YU-1 and YU-2 were isolated from a patient with AIDS-related complex and an HIV-1-positive healthy carrier, respectively (75). YU-9 and YU-10 were isolated from two other patients with AIDS (74). All of the primary clinical isolates were obtained by a coculture procedure by using HIV-1 seronegative PBMC. SF13 (10, 11) and SF162 (57) were provided by T. Shioda (University of Tokyo, Tokyo, Japan). Ba-L was obtained from the AIDS Research and Reference Reagent Program. All viruses were propagated in PHA-stimulated PBMC cultures from a single donor. At 7 or 10 days after infection, culture supernatants containing the viruses were recovered, filtered through a membrane (0.22 μm [pore size]), and the amount of HIV-1gag p24 was measured by enzyme-linked immunosorbent assay (ELISA) (Cellular Products, Buffalo, N.Y.). Aliquots of the viral stocks were stored at −80°C. The titer of each virus stock was determined by endpoint titration of fivefold limiting dilution in triplicate on PHA-stimulated PBMC from a single donor. The 50% tissue culture infectious dose (TCID50) was calculated by the method of Reed and Muench (52).
HIV-1 infection.
PBMC, MDM, MT-2, and MT-4 cells were exposed to virus suspensions at a multiplicity of infection (MOI) of 0.001. After 2 h (PBMC and MDM cells) or 1 h (MT-2 cells and MT-4 cells) of adsorption at 37°C, the cells were washed three times with serum-free RPMI 1640 medium, and fresh medium was then added. Amounts of HIV-1gag p24 in the culture supernatants were measured by ELISA 7 days (PBMC and MT-4 cells) or 14 days (MDM cells and MT-2 cells) after infection.
Cell fusion assay with fluorescent dye.
The use of recombinant vaccinia virus (RVV) to transiently express CD4 and HIV-1 gp120/gp41 was as described previously (33). A spinner culture of HeLa S3 cells was infected with gp120/gp41 encoding RVV (vR10EN) and monolayer HeLa cells were infected with human CD4 encoding RVV (vRT4) at an MOI of 10. After 1 h of adsorption of these viruses, the cells were washed with PBS without calcium and magnesium [PBS(−)] and cultured for 15 to 20 h at 37°C. Lipid mixing and content mixing were measured by the fusion assay as described previously (20). Briefly, gp120/gp41-expressing HeLa S3 cells were washed with PBS(−) and resuspended in PBS(−) at a density of 2 × 105/ml. The cells were labeled with the lipophilic fluorescent probe, octadecyl rhodamine B chloride (R18; 1 mg/ml; Molecular Probes, Inc., Eugene, Oreg.). After being washed with PBS(−) containing 1% bovine serum albumin and 0.02% EDTA, the cells were resuspended in PBS(−) containing 0.5 mM CaCl2 [PBS(+)] and overlaid on CD4-expressing HeLa cells. Transfer of the fluorescent dye from HeLa S3 cells into adherent HeLa cells was observed through membrane fusion (lipid mixing) after 1 h of cocultivation. The appearance of multinuclear giant cells was observed through migration of nuclei (content mixing) after 5 h of cocultivation. Cells were fixed with PBS(−) containing 3% paraformaldehyde for 15 min at room temperature and were then observed under a 16× objective lens of a fluorescence microscope.
Cell-cell fusion assay.
A previously described β-galactosidase-based gene reporter assay that used α-complementation of the enzyme was modified to quantitate cell-cell fusion events. Briefly, the α-subunit of β-galactosidase and the HIV-1 IIIB envelope protein were introduced into effector HeLa S3 cells by RVV. Target QT6 cells stably transfected with a form of CD4 lacking the cytoplasmic tail were infected with RVV encoding the ω-subunit of β-galactosidase and transfected with chimeric receptors by using Fugene 6 (Boehringer Mannheim). After overnight incubation, effector cells were added to the target cells to initiate fusion in the presence or absence of anti-HIV peptides. At 16 to 20 h after this mixing, the cells were lysed and assayed for β-galactosidase activity.
Inhibition of MAb binding to MOLT-4 cells by T22 and its lead peptides.
One million MOLT-4 cells were treated with various concentrations of T22 and 3 μM 4Ala-T-I for 1 h at 37°C. The cells were washed three times with PBS(−) and stained for 30 min with 5 μg of the anti-CXCR4 monoclonal antibody (MAb), 12G5 (28), or the anti-CXCR4 rat MAb R6-48 (65) per ml. The cells were then stained for 30 min with fluorescein isothiocyanate-conjugated anti-mouse immunoglobulin G (IgG) or anti-rat IgG at 4°C. The mean fluorescence of stained cells was measured by use of a FACScalibur apparatus (Becton Dickinson).
Measurement of CXCR4 downregulation.
The downregulation of CXCR4 on MOLT-4 cells was determined as described previously (3). Briefly, one million MOLT-4 cells were treated with various concentrations of SDF-1α or with 200 nM of T22 or 4Ala-T-I for 30 min at 37°C. The cells were washed with acidic glycine buffer to remove the bound peptides and stained with 12G5, and CXCR4 expression was measured by fluorescence-activated cell sorter analysis.
SDF-1α radioligand binding assay.
MOLT-4 cells (106) were incubated at 4°C for 1 h in the presence of 0.4 nM 125I-labeled SDF-1α and increasing concentrations of unlabeled SDF-1α (0.1 to 1,000 nM) in 100 μl of RPMI medium containing 1% bovine serum albumin (binding buffer). The cells were washed three times with the binding buffer. The amount of bound 125I-labeled SDF-1α was determined by using a gamma counter.
Chemotaxis assay.
The chemotaxis assay was carried out as described previously (4). Briefly, 106 PHA and interleukin-2 (IL-2)-activated PBMC were washed with PBS(−) and suspended in RPMI 1640 containing 0.25% human albumin (Sigma) in the presence or absence of various concentrations of SDF-1α and T22 and 3 μM 4Ala-T-I. They were then added to the top chamber of a 5-μm-pore-size polycarbonate Transwell culture insert (Costar, Cambridge, Mass.) and pretreated for 30 min at 37°C in a CO2 incubator. The bottom of the chamber contained 100 nM SDF-1α. The cells were cultured at 37°C for 3 h, and the number of cells that transmigrated into the lower chamber was then counted microscopically.
RESULTS
Restriction of T-tropic HIV-1 infection by T22.
We previously reported that T22 blocks infection by laboratory-adapted HIV-1 strains such as HTLV IIIIB and NL4-3 (48). For the purpose of evaluating anti-HIV therapeutic potential, we felt it was important to analyze clinical isolates. Thus, we examined the effect of T22 on the infectivity of primary clinical HIV-1 isolates and other molecularly cloned strains. These included three T-tropic (NL4-3, TMD-1, and YU-10), five M-tropic (JR-CSF, SF162, Ba-L, TMD-2, and YU-9), and one dual-tropic (SF13), and two uncharacterized HIV-1 strains (YU-1 and YU-2). YU-1, YU-2, YU-9, YU-10, TMD-1, and TMD-2 are primary clinical isolates. T22 blocked infection of PBMC with primary T-tropic HIV-1 strains such as TMD-1 and YU-10 with similar efficiency as NL4-3 (Table 1), whereas T22 failed to inhibit infection by non-T-tropic HIV-1 strains, even at concentrations as high as 3 μM. T22 did not interfere with the infectivity of a dual-tropic strain, SF13, in PBMC, although this peptide efficiently inhibited the infection of SF13 in MT-4 cells (data not shown). Thus, T22 is effective against all T-tropic HIV-1 strains tested, including primary clinical isolates.
TABLE 1.
Host range, cytopathic properties, and sensitivity to T22 of various HIV-1 isolates
| Virus strain | HIVgag p24 (ng/ml)
|
Cytopathicity in MT-2a | IC50 (μM) of T22 in PBMCb | |||
|---|---|---|---|---|---|---|
| PBMC | MT-4 | MT-2 | Macrophage | |||
| NL4-3 | 69 | 2,391 | 914 | <0.1 | ++ | 0.04 |
| TMD-1 | 135 | 64 | 24 | <0.1 | + | 0.03 |
| YU-10 | 24 | 756 | 20 | <0.1 | + | 0.08 |
| JR-CSF | 18 | <0.1 | <0.1 | 1.2 | − | >3 |
| SF162 | 20 | <0.1 | <0.1 | 66 | − | >3 |
| Ba-L | 88 | <0.1 | <0.1 | 58 | − | >3 |
| TMD-2 | 87 | <0.1 | <0.1 | 13 | − | >3 |
| YU-9 | 172 | <0.1 | <0.1 | 11 | − | >3 |
| YU-1 | 18 | <0.1 | <0.1 | <0.1 | − | >3 |
| YU-2 | 16 | <0.1 | <0.1 | <0.1 | − | >3 |
| SF13 | 29 | 1,173 | 1,727 | 2 | + | >3 |
HIV-1-induced cytopathicity in MT-2 cells was assessed by measuring the formation of syncytia 14 days after infection. +, ∼20% cells; ++, >80% cells showing syncytia.
Anti-HIV activity of T22 was determined by measuring HIV-1gag p24 antigen in culture supernatant of PBMC at 7 days postinfection. Results are representative of at least two independent infection experiments.
The V3 loop region of gp120 is a determinant of T22 sensitivity.
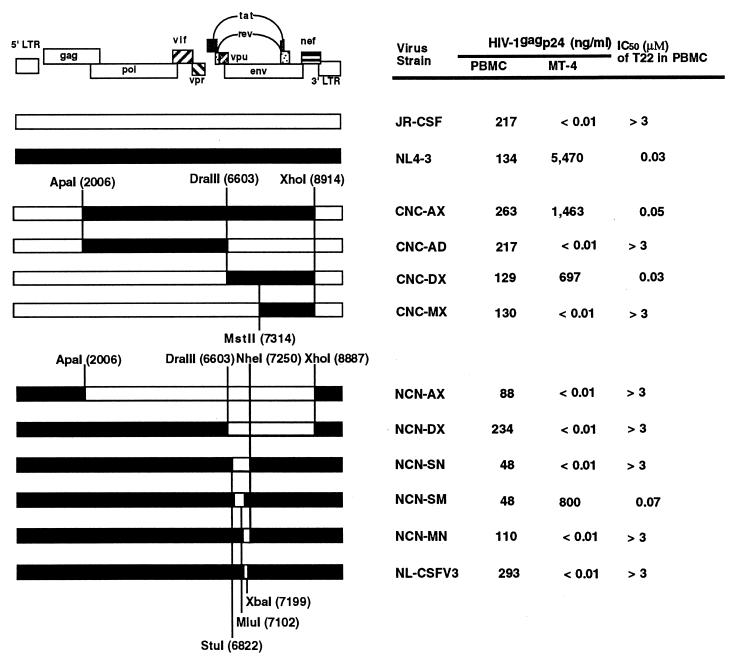
To localize viral determinants responsible for sensitivity to T22 treatment, we used a variety of infectious chimeric viral DNA clones composed of complementary segments of NL4-3 (T-tropic) and JR-CSF (M-tropic). Determination of the 50% inhibitory concentration (IC50) of T22 against chimeric viruses indicated that the sensitivity of the chimeras to T22 is determined by the presence of the DraIII-MstII (positions 6603 to 7314) fragment of NL4-3 (Fig. 1). T22 efficiently inhibited infection with CNC-DX in which most of the env regions of JR-CSF were replaced by those of NL4-3. In contrast, the CNC-MX chimeras, in which the latter half of env (not including the V3 loop region) of JR-CSF was replaced with that of NL4-3, was not sensitive to T22. Thus, the env region from V1 to V3 seems to be responsible for viral sensitivity to T22 treatment. Figure 1 also showed that loss of the V3 loop region from T-tropic virus caused resistance to T22 treatment. T22 did not inhibit non-T-tropic NL-CSFV3 in which only the region encoding the 33-amino-acid NL4-3 V3 loop was substituted with that of JR-CSF. A clear correlation was observed between T-cell-line tropism and the sensitivity to T22 treatment. Thus, these data indicate that viral sensitivity to T22 treatment is conferred by the V3 loop region.
FIG. 1.
Comparison of HIV-1 replication in PHA-stimulated PBMC and MT-4 cells by using infectious HIV-1 molecular clones NL4-3, JR-CSF, and chimeric constructs of these clones. Mean values are shown for HIV-1gag p24 in the culture supernatant at 7 days postinfection. Anti-HIV activity of T22 was also determined for each clone in PBMC, and the IC50 is expressed. The results are representative of three independent experiments.
To confirm that resistance of M-tropic HIV-1 strains to T22 treatment is achieved at the entry phase of viral replication, we analyzed viral DNA synthesis by using a semiquantitative PCR assay at 24 h after infection. DNA synthesis by T-tropic NL4-3 and CNC-DX was blocked by T22 treatment in a dose-dependent manner. In contrast, the level of viral DNA synthesis after infection by non-T-tropic strains such as JR-CSF and NL-CSFV3 was not affected by T22 (data not shown).
T22 inhibits lipid mixing of membrane in HIV-1 Env induced cell-cell fusion.
As reported previously, we found that T22 exerts its anti-HIV activity as a CXCR4 antagonist. Therefore, we attempted to determine the stage at which T22 acts. By using a fluorescent lipid marker, we were able to distinguish two stages, lipid and content mixing, in a coculture of HIV-1 gp120/gp41 (Env)-expressing and CD4-expressing cells. One hour after cocultivation, redistribution of the fluorescent lipid marker R18 was observed, but multinuclear giant cells were not formed yet, indicating that lipid mixing took place within 1 h after cocultivation. A 30 nM concentration of T22 significantly suppressed lipid mixing. The IC50 of T22 on lipid mixing was estimated to be 12 nM by counting more than 200 cells. T22 also blocked content mixing and syncytium formation between both cells subsequent to lipid mixing (data not shown). These results show that T22 inhibits HIV-1 induced cell-cell fusion at the stage of lipid mixing.
T22 inhibits SDF-1α function through binding to CXCR4.
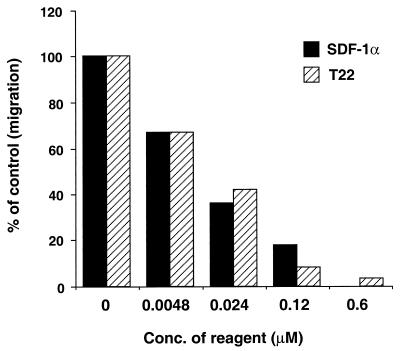
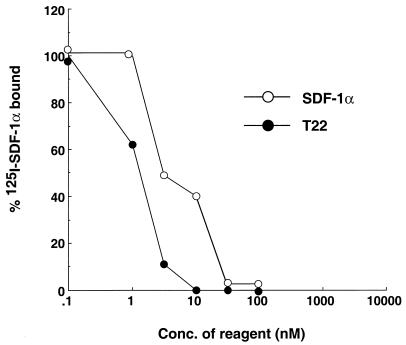
We previously showed that T22 specifically inhibits Ca2+ mobilization induced by the ligand of CXCR4, SDF-1α. To extend this finding, the effect of T22 on chemotaxis of PHA, and IL-2-stimulated PBMC induced by SDF-1α was examined. T22 effectively inhibited SDF-1α-induced chemotaxis in a dose-dependent manner with an IC50 of 30 nM. SDF-1α, as a positive control, showed desensitization of chemotaxis with an efficiency similar to that of T22 (Fig. 2). 4Ala-T-I (NH2-KWAFRVAYRGIAYRRAR-CONH2), a negative control peptide, did not affect chemotaxis at a 3 μM concentration (data not shown). This inhibitory mechanism for the effect of T22 appeared to be through its binding to CXCR4. [125I]SDF-1α binding to MOLT-4 cells was inhibited by T22 in a dose-dependent manner (Fig. 3). The IC50 of unlabeled SDF-1α was 10 nM. T22 apparently inhibited the binding with a higher efficacy than did SDF-1α. 4Ala-T-I did not affect ligand binding to CXCR4 even at 3 μM (data not shown). These results suggest that T22 interferes with HIV-1–CXCR4 interactions by binding to CXCR4. Binding of T22 to CXCR4 did not downregulate the receptor at a T22 concentration of 200 nM, whereas SDF-1α downregulated CXCR4 in a dose-dependent fashion as described previously (data not shown). These results suggest that the anti-HIV mechanism of T22 was mediated via receptor competition with HIV-1.
FIG. 2.
Effect of T22 on the chemotactic response of PHA-stimulated PBMC induced by SDF-1α. A total of 106 PHA- and IL-2-activated PBMC were washed with PBS(−) and suspended in RPMI 1640 containing 0.25% human albumin in the presence or absence of various concentrations of SDF-1α and T22 and 3 μM 4Ala-T-I. They were then added to the top chamber of a 5-μm (pore-size) polycarbonate Transwell culture insert and incubated for 30 min at 37°C. The bottom chamber contained 100 nM SDF-1α. The cells were cultured at 37°C for 3 h. The number of cells that transmigrated into the lower chamber was counted microscopically. The results are representative of three independent experiments.
FIG. 3.
Inhibition of 125I-labeled SDF-1α binding to CXCR4 by T22. MOLT-4 cells (106) were incubated at 4°C for 1 h in the presence of 0.4 nM 125I-labeled SDF-1α and increasing concentrations of T22 or unlabeled SDF-1α in 100 μl of RPMI medium containing 1% BSA (binding buffer). The cells were washed three times with the binding buffer. The amount of bound 125I-labeled SDF-1α was determined by using a gamma counter. Specific binding (70,000 cpm on average in the absence of inhibitors) was determined by subtracting from each datum point the amount of nonspecific binding (10,000 cpm on average) observed in the presence of 1 μM unlabeled SDF-1α. The results are representative of three independent experiments.
T22 binding region of CXCR4.
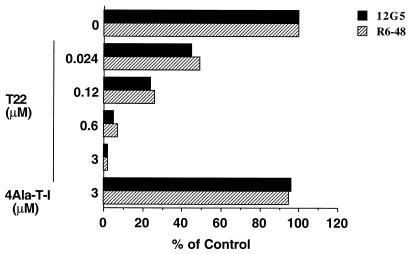
Next, we attempted to identify the T22 binding region of CXCR4. Cells were pretreated with T22 and the binding activity of CXCR4-specific MAbs to CXCR4 was measured. Two MAbs, R6-48 and 12G5, were used. 12G5 binds the first extracellular loop (ECL1) and ECL2 of CXCR4 (28); the binding epitope of R6-48 was determined to be at the N terminus of CXCR4 by analyzing a panel of chimeric receptors and by using a peptide inhibition assay (data not shown). Pretreatment with T22 efficiently blocked binding of both antibodies to the target cells (Fig. 4). A negative control peptide, 4Ala-T-I, did not inhibit the binding of either antibody, even at a 3 μM concentration. This inhibition appeared to be specific for CXCR4, since T22 treatment did not affect the binding of Leu-3a (an MAb that recognizes the gp120 binding site on CD4) and OKT4 (another CD4-specific MAb) (data not shown). These results suggest that T22 directly binds extracellular regions of CXCR4 which are at least overlapping with epitopes recognized by R6-48 (N terminus), and 12G5 (ECL1 and ECL2).
FIG. 4.
Effect of T22 on reactivity of anti-CXCR4 MAbs with MOLT-4 cells. MOLT-4 cells (106) were treated with various concentrations of T22 or else 3 μM 4Ala-T-I for 1 h at 37°C. The cells were washed and stained with anti-CXCR4 MAbs as described in Materials and Methods. Antibody binding is indicated as a percentage of the mean fluorescence of stained cells observed in the absence of the peptides. Results are representative of at least two independent experiments.
Three extracellular loops are involved in the sensitivity of HIV-1 to T22.
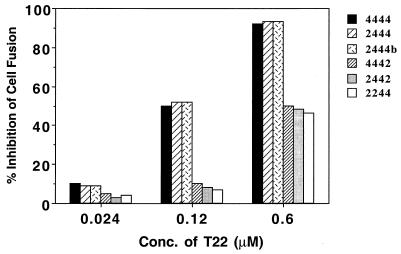
To further define determinants of CXCR4 that are involved in its interaction with T22, we used a cell-cell fusion assay between HIV-1 IIIB Env-expressing HeLa cells and QT6 cells expressing CD4 and CXCR4 or chimeric receptors composed of complementary regions of CXCR4 and CXCR2. First, we confirmed the fusion efficiency of each chimeric receptor. We observed a similar fusion efficiency compared to the previous report with other T-tropic HIV-1 Env (39), indicating that all extracellular loops contributed to CXCR4 coreceptor function in HIV-1 IIIB Env-mediated cell-cell fusion (data not shown). We then investigated the inhibitory activity of T22 on IIIB Env-mediated cell-cell fusion. We used chimeric receptors, 2444, 2444b, 4442, 2442, and 2244, which showed more than 20% CXCR4 fusion activity. A 0.6 μM concentration of T22 completely inhibited the fusion mediated by 4444, 2444, and 2444b, whereas the inhibition of the fusion mediated by 4442, 2442, and 2244 was only partial (Fig. 5). These results indicated that ECL1, ECL2, and ECL3 of CXCR4 contributed to the sensitivity of CXCR4 coreceptor activity to T22.
FIG. 5.
Anti-HIV activity of T22 on fusion between cells expressing HIV-1 IIIB Env and cells expressing CD4 and either CXCR4 or CXCR2/CXCR4 chimeras. Cell-cell fusion was measured as follows. The α-subunit of β-galactosidase and HIV-1 IIIB envelope protein were introduced into effector HeLa S3 cells by RVV. Target QT6 cells stably transfected with a form of CD4 lacking the cytoplasmic tail were infected with RVV encoding the ω-subunit of β-galactosidase and transfected with chimeric receptors. After overnight incubation, both target and effector cells were preincubated with various concentrations of T22 for 30 min before the cells were mixed. Then, 16 to 20 h after being mixed, the cells were lysed and assayed for β-galactosidase activity. The results shown are representative of two independent experiments. The nomenclature of the CXCR4 chimeras is as follows: 2442, for example has the N terminus and the third extracellular regions of CXCR2 and the first and second extracellular regions of CXCR4.
DISCUSSION
In this study, we demonstrated that T22 inhibits replication of T-tropic strains of HIV-1, including primary isolates, but not of non-T-tropic strains in PHA-stimulated PBMC (Table 1). Viral determinants for T22 susceptibility were mapped to the V3 loop region of gp120 by using a panel of chimeric viruses between T-tropic NL4-3 and M-tropic JR-CSF (Fig. 1). We also showed that T22 interfered with SDF-1α–CXCR4 interactions (Fig. 2 and 3) without downregulating CXCR4 surface expression. Inhibition of anti-CXCR4 MAb (R6-48 and 12G5) binding by T22 suggested that the peptide interacts with the N terminus and two ECLs of CXCR4 (Fig. 4). These findings that T22 blocks SDF-1α and MAb binding to CXCR4 closely parallel previous observations made with AMD3100 and ALX40-4C (22, 23, 37). Moreover, T22-induced inhibition of cell-cell fusion in cells expressing CXCR4/CXCR2 chimeric receptors suggested that determinants for sensitivity of CXCR4 to the peptide include all three ECLs of CXCR4 (Fig. 5).
The bicyclam AMD3100, a small CXCR4 antagonist, has recently been demonstrated to interact with aspartic acids of CXCR4 ECL2 (37). It is interesting to compare the mechanism of T22 action with other CXCR4 inhibitors and helpful to develop ideal CXCR4 inhibitors. The data mentioned above are very reminiscent of those obtained with AMD3100. Do these compounds with different structures have the same anti-HIV mechanism? The analysis of target sites for AMD3100 focused on ECL2, since this extracellular loop seems to be important for the HIV-1 coreceptor activity. However, we attempted to identify the regions on which T22 acts for two reasons: (i) even though ECL2 is crucial for coreceptor activity for T-tropic HIV-1, we cannot rule out the possibility that T22 also acts on other extracellular regions, and (ii) it has recently been found that charged residues in ECL2 of CXCR4 do not contribute to coreceptor activity with dual-tropic and T-tropic HIV-1 Env, a result different from that reported in the AMD3100 study (70).
Initially, we investigated the inhibitory activity of T22 on the binding of two anti-CXCR4 MAbs, which have different epitopes. T22 inhibited the binding of both MAbs, one of which recognizes the N terminus and the other of which binds ECL1 and 2 with the same efficacy (Fig. 4). There are two possible explanations for the above results: (i) T22 binds to the N terminus and ECL1 and/or ECL2 and (ii) T22 binding to CXCR4 leads to a global conformational change of CXCR4 which affects the R6-48 and 12G5 binding epitopes. As a result, neither antibody binds to CXCR4 after T22 treatment. Considering our results with chimeric coreceptors, the first explanation is likely, but we as yet cannot rule out the second possibility.
To further examine determinants of CXCR4 binding to T22, we used cell-cell fusion between HIV-1 IIIB Env-expressing cells and cells expressing CD4 and CXCR4 or CXCR4/CXCR2 chimeric receptors. Our study with chimeric receptors suggested that T22 acts at least on ECL1 and ECL3 of CXCR4 (Fig. 5). Although T22 was capable of inhibiting the coreceptor activity of chimeras that lacked ECL1 or ECL3, it was not possible to implicate ECL2 in the loss-of-function experiments because all of the active chimeras contained this domain of CXCR4. The exact site(s) of T22 binding in CXCR4 remains unclear; a more precise mapping of the T22 binding site(s) will require the use of 125I-labeled T22 and a number of additional chimeric and mutant forms of CXCR4. Very recently Arakaki et al. reported that T134, a T22 derivative, effectively inhibits replication of AMD3100-resistant HIV-1 isolates, suggesting that the action sites of T22 and AMD3100 are not the same (5).
The viral determinant for T22 inhibition was mapped to the V3 loop region of gp120, and HIV-1 replication was blocked at an early step of infection (Fig. 1). Considering previous reports that the V3 loop region is essential for the determination of HIV-1 coreceptor usage, the above results are consistent with our recent observation that T22 is an antagonist for CXCR4 functions as both an HIV-1 coreceptor and a chemokine receptor. This finding also supports the notion that T22 is a CXCR4 inhibitor because the role of the V3 loop in the interaction of gp120 with coreceptors has been strongly suggested by a number of studies (16, 67, 71). However, the recently reported crystal structure of an HIV-1 gp120 core protein in complex with CD4 and a neutralizing antibody suggested that a conserved region adjacent to the V3 loop is important for chemokine receptor binding (36, 53, 72). Furthermore, a previous study in which escape mutants of AMD3100 had sequence changes in gp120 both within and outside the V3 loop also demonstrates the importance of gp120 regions outside the V3 loop (17). It was previously suggested that a β-sheet-type II β-turn-β-sheet (antiparallel β-sheet) structure and the position and number of basic amino acids in T22 are crucial for its anti-HIV activity (61, 63). The conformation of the V3 loop in solution, which is reported as a turn-β-strand-β-turn-β-strand helix (8, 9, 68, 69), and the existence of basic amino acids at certain positions are critical for virus infectivity (32, 50). Given the structural similarity between T22 and the V3 loop of T-tropic HIV-1 described above, we hypothesize that T22 binds to CXCR4 by mimicking the V3 loop of T-tropic HIV-1. The recent report that a cyclic V3 peptide effectively inhibits infection by T-tropic HIV-1 by binding to CXCR4 supports this hypothesis (55).
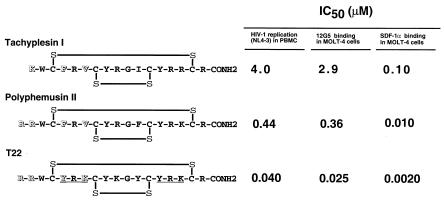
We have recently reported pharmacophore identification of T22 (60). The number of Arg residues near the N terminus of T22 is closely related to its antiviral activity. Tyr-Arg-Lys in the N-terminal portion is also closely associated with anti-HIV activity, probably by maintaining the β-structure of the peptide. To illustrate this point, inhibition of T-tropic HIV-1 replication, anti-CXCR4 MAb and SDF-1α binding among T22 and its lead peptides, tachyplesin I and polyphemusin II, are summarized in Fig. 6. The anti-HIV activities (IC50) of tachyplesin I, polyphemusin II, and T22 in PBMC are 4.0, 0.44, and 0.040 μM, respectively. The anti-HIV activity of these peptides is well correlated with the inhibitory activity of these peptides on 12G5 and SDF-1α binding. Furthermore, polyphemusin II showed a similar inhibition pattern with lower activity in cell-cell fusion experiments with CXCR4 chimeras, suggesting that polyphemusin II also acts on ECL1 and ECL3 of CXCR4 with lower affinity (data not shown). Thus, we demonstrated that the pharmacophore of T22 in anti-HIV activity is attributed to its affinity to certain regions of CXCR4, which is recognized by its natural ligand SDF-1α and the MAb 12G5.
FIG. 6.
Amino acid structure, inhibitory activity against HIV-1 replication, 12G5 binding, and SDF-1α binding of tachyplesin I, polyphemusin II, and T22. HIV-1 replication, 12G5 binding, and SDF-1α binding were measured as described in Table 1, Fig. 4, and Fig. 3, respectively. The amino acid differences which are critical for anti-HIV-1 activity are highlighted (open letters). The Tyr-Arg-Lys repeat in T22 is underlined.
The site of T22 inhibition was shown to be the early stage of virus-cell fusion. By monitoring the lipid mixing stage separately from the later stages by using a lipophilic fluorescent probe, we could show that T22 completely inhibits lipid mixing, resulting in the blocking of content mixing and of syncytium formation. It has been reported that amphipathic peptides, including a lead compound of T22, exert their antibacterial activity by permeabilizing the plasma membrane and perturbing the metabolism of treated cells (41, 42). This was not the cause of inhibition of HIV-1 infectivity, since T22 inhibits HIV-1 replication at the concentration much lower than toxic one. Moreover, virions pseudotyped with the vesicular stomatitis virus G protein or the amphotropic murine leukemia virus envelope protein were not influenced by T22 treatment (data not shown). Thus, we conclude that the inhibition of virus-cell or cell-cell fusion by T22 is HIV Env specific. It is interesting to note that a gp41 peptide, which can act on the stage of virus-cell fusion, also inhibits lipid mixing more efficiently than content mixing (44).
We demonstrated that T22 inhibits the infection of PBMC by all T-tropic HIV-1 strains examined, including clinical primary isolates, and impairs infection of a T-cell line by a dual-tropic HIV-1 strain, SF13 (Table 1). These results imply that T22 might reduce virus spread in vivo by blocking CXCR4 coreceptor activity. Several other chemokine receptors, such as Bonzo/STRL33, Bob/GPR15, GPR1, US28, and APJ, most of which are orphan receptors, have been found to be able to mediate entry of HIV-1, HIV-2, and simian immunodeficiency virus (13, 19, 24, 27, 30, 38, 51, 54). It is interesting and important to know whether T22 also acts on those receptors, although we know that T22 does not block MCP-1-induced Ca2+ mobilization in CCR2b-transfected cells, RANTES-induced Ca2+ mobilization in CCR5-transfected cells, and CCR5-mediated entry of HIV-1 JR-CSF (46).
In summary, we showed here that a cationic peptide, T22, specifically inhibits replication of T-tropic HIV-1 by acting on extracellular regions of CXCR4. Further studies, discussed above, will help to develop more potent anti-HIV drugs which target the HIV coreceptors.
ACKNOWLEDGMENTS
We thank E. O. Freed for critical review of the manuscript.
This work was supported by grants from the Ministry of Public Health and Welfare and the Ministry of Biotechnology and Science in Japan. Y.K. and N.Y. were sponsored by the Japan Health Sciences Foundation. N.Y. was also supported by Priority Areas from the Ministry of Education, Sports and Culture, and CREST (Core Research for Evolutional Sciences and Technology) of Japan Science and Technology Corporation. N.Y. was also supported by the Program for Promotion of Fundamental Studies in Health Sciences of the Organization for Drug ADR Relief, R&D Promotion, and Products Review of Japan. S.C.P. was supported by National Institutes of Health grant A141396 and the Agnes Brown Duggan Endowment.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 3.Amara A, Le Gall S, Schwartz O, Salamero J, Montes M, Loetscher P, Baggiolini M, Virelizier J-L, Arenzana-Seisdedos F. HIV coreceptor downregulation as a antiviral principle: SDF-1α-dependent internalization of the chemokine receptor CXCR4 contributes to inhibition of HIV replication. J Exp Med. 1997;186:139–146. doi: 10.1084/jem.186.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arai M, Ohashi T, Tsukahara T, Murakami T, Hori T, Uchiyama T, Yamamoto N, Kannagi M, Fujii M. Human T-cell leukemia virus type 1 Tax protein induces the expression of lymphocyte chemoattractant SDF-1/PBSF. Virology. 1998;241:298–303. doi: 10.1006/viro.1997.8968. [DOI] [PubMed] [Google Scholar]
- 5.Arakaki R, Tamamura H, Premanathan M, Kanbara K, Ramanan S, Mochizuki K, Baba M, Fujii N, Nakashima H. T134, a small-molecule CXCR4 inhibitor, has no cross-drug resistance with AMD3100, a CXCR4 antagonist with a different structure. J Virol. 1999;73:1719–1723. doi: 10.1128/jvi.73.2.1719-1723.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arenzana-Seisdedos F, Virelizer J-L, Rousset D, Clark-Lewis I, Loetscher P, Moser B, Baggiolini M. HIV blocked by chemokine antagonist. Nature. 1996;383:400. doi: 10.1038/383400a0. [DOI] [PubMed] [Google Scholar]
- 7.Bleul C C, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer T A. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–832. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 8.Catasti P, Bradbury E M, Gupta G. Structure and polymorphism of HIV-1 third variable loops. J Biol Chem. 1996;271:8236–8242. doi: 10.1074/jbc.271.14.8236. [DOI] [PubMed] [Google Scholar]
- 9.Catasti P, Fontenot J D, Bradbury E M, Gupta G. Local and global structural properties of the HIV-MN V3 loop. J Biol Chem. 1995;270:2224–2232. doi: 10.1074/jbc.270.5.2224. [DOI] [PubMed] [Google Scholar]
- 10.Cheng M C, Seto D, Tateno M, Levy J A. Biologic features of HIV-1 that correlate with virulence in the host. Science. 1988;240:80–82. doi: 10.1126/science.2832945. [DOI] [PubMed] [Google Scholar]
- 11.Cheng M C, Shioda T, Levy J A. Host range, replicative, and cytopathic properties of human immunodeficiency virus type 1 are determined by very few amino acid changes in tat and gp120. J Virol. 1991;65:6931–6941. doi: 10.1128/jvi.65.12.6931-6941.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chesebro B, Nishio J, Perryman S, Cann A, O’Brien W, Chen I S, Wehrly K. Identification of human immunodeficiency virus envelope gene sequences influencing viral entry into CD4-positive HeLa cells, T-leukemia cells, and macrophages. J Virol. 1991;65:5782–5789. doi: 10.1128/jvi.65.11.5782-5789.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choe H, Farzan M, Konkel M, Martin K, Sun Y, Marcon L, Cayabyab M, Berman M, Dorf M E, Gerard N, Gerard C, Sodroski J. The orphan seven-transmembrane receptor apj supports the entry of primary T-cell-line-tropic and dualtropic human immunodeficiency virus type 1. J Virol. 1998;72:6113–6118. doi: 10.1128/jvi.72.7.6113-6118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 15.Cocchi F, DeVico A L, Garzino D A, Arya S K, Gallo R C, Lusso P. Identification of RANTES, MIP-1α and MIP-1β as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 16.Cocchi F, DeVico A L, Garzino D A, Cara A, Gallo R C, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 17.de Vreese K, Kofler-Mongold V, Leutgeb C, Weber V, Vermeire K, Schacht S, Anne J, De Clercq E, Datema R, Werner G. The molecular target of bicyclams, potent inhibitors of human immunodeficiency virus replication. J Virol. 1996;70:689–696. doi: 10.1128/jvi.70.2.689-696.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di M P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 19.Deng H K, Unutmaz D, KewalRamani V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 20.Dimitrov D S, Golding H, Blumenthal R. Initial stages of HIV-1 envelope glycoprotein-mediated cell fusion monitored by a new assay based on redistribution of fluorescent dyes. AIDS Res Hum Retroviruses. 1991;7:799–805. doi: 10.1089/aid.1991.7.799. [DOI] [PubMed] [Google Scholar]
- 21.Doms R W, Peiper S C. Unwelcomed guests with master keys: how HIV uses chemokine receptors for cellular entry. Virology. 1997;235:179–190. doi: 10.1006/viro.1997.8703. [DOI] [PubMed] [Google Scholar]
- 22.Donzella G A, Schols D, Lin S W, Este J A, Nagashima K A, Maddon P J, Allaway G P, Sakmar T P, Henson G, De Clercq E, Moore J P. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat Med. 1998;4:72–77. doi: 10.1038/nm0198-072. [DOI] [PubMed] [Google Scholar]
- 23.Doranz B J, Grovit-Ferbas K, Sharron M P, Mao S H, Goetz M B, Daar E S, Doms R W, O’Brien W A. A small-molecule inhibitor directed against the chemokine receptor CXCR4 prevents its use as an HIV-1 coreceptor. J Exp Med. 1997;186:1395–1400. doi: 10.1084/jem.186.8.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 25.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 26.Dumonceaux J, Nisole S, Chanel C, Quivet L, Amara A, Baleux F, Briand P, Hazan U. Spontaneous mutations in the env gene of the human immunodeficiency virus type 1 NDK isolate are associated with a CD4-independent entry phenotype. J Virol. 1998;72:512–519. doi: 10.1128/jvi.72.1.512-519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edinger A L, Hoffman T L, Sharron M, Lee B, Yi Y, Choe W, Kolson D L, Mitrovic B, Zhou Y, Faulds D, Collman R G, Hesselgesser J, Horuk R, Doms R W. An orphan seven-transmembrane domain receptor expressed widely in the brain functions as a coreceptor for human immunodeficiency virus type 1 and simian immunodeficiency virus. J Virol. 1998;72:7934–7940. doi: 10.1128/jvi.72.10.7934-7940.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau H B, Choe S, Vance P J, Wells T N, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 29.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 30.Hoffman T L, Stephens E B, Narayan O, Doms R W. HIV type I envelope determinants for use of the CCR2b, CCR3, STRL33, and APJ coreceptors. Proc Natl Acad Sci USA. 1998;95:11360–11365. doi: 10.1073/pnas.95.19.11360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoxie J A, LaBranche C C, Endres M J, Turner J D, Berson J F, Doms R W, Matthews T J. CD4-independent utilization of the CXCR4 chemokine receptor by HIV-1 and HIV-2. J Reprod Immunol. 1998;41:197–211. doi: 10.1016/s0165-0378(98)00059-x. [DOI] [PubMed] [Google Scholar]
- 32.Ivanoff L A, Dubay J W, Morris J F, Roberts S J, Gutshall L, Sternberg E J, Hunter E, Matthews T J, Petteway S J. V3 loop region of the HIV-1 gp120 envelope protein is essential for virus infectivity. Virology. 1992;187:423–432. doi: 10.1016/0042-6822(92)90444-t. [DOI] [PubMed] [Google Scholar]
- 33.Jin N Y, Funahashi S, Shida H. Constructions of vaccinia virus A-type inclusion body protein, tandemly repeated mutant 7.5 kDa protein, and hemagglutinin gene promoters support high levels of expression. Arch Virol. 1994;138:315–330. doi: 10.1007/BF01379134. [DOI] [PubMed] [Google Scholar]
- 34.Kawano Y, Tanaka Y, Misawa N, Tanaka R, Kira J I, Kimura T, Fukushi M, Sano K, Goto T, Nakai M, Kobayashi T, Yamamoto N, Koyanagi Y. Mutational analysis of human immunodeficiency virus type 1 (HIV-1) accessory genes: requirement of a site in the nef gene for HIV-1 replication in activated CD4+ T cells in vitro and in vivo. J Virol. 1997;71:8456–8466. doi: 10.1128/jvi.71.11.8456-8466.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koyanagi Y, Miles S, Mitsuyasu R T, Merrill J E, Vinters H V, Chen I S. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science. 1987;236:819–822. doi: 10.1126/science.3646751. [DOI] [PubMed] [Google Scholar]
- 36.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Labrosse B, Brelot A, Heveker N, Sol N, Schols D, De Clercq E, Alizon M. Determinants for sensitivity of human immunodeficiency virus coreceptor CXCR4 to the bicyclam AMD3100. J Virol. 1998;72:6381–6388. doi: 10.1128/jvi.72.8.6381-6388.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao F, Alkhatib G, Peden K W, Sharma G, Berger E A, Farber J M. STRL33, A novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J Exp Med. 1997;185:2015–2023. doi: 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu Z, Berson J F, Chen Y, Turner J D, Zhang T, Sharron M, Jenks M H, Wang Z, Kim J, Rucker J, Hoxie J A, Peiper S C, Doms R W. Evolution of HIV-1 coreceptor usage through interactions with distinct CCR5 and CXCR4 domains. Proc Natl Acad Sci USA. 1997;94:6426–6431. doi: 10.1073/pnas.94.12.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masuda M, Nakashima H, Ueda T, Naba H, Ikoma R, Otaka A, Terakawa Y, Tamamura H, Ibuka T, Murakami T, et al. A novel anti-HIV synthetic peptide, T-22 ([Tyr5,12, Lys7]-polyphemusin II) Biochem Biophys Res Commun. 1992;189:845–850. doi: 10.1016/0006-291x(92)92280-b. [DOI] [PubMed] [Google Scholar]
- 41.Matsuzaki K, Fukui M, Fujii N, Miyajima K. Interactions of an antimicrobial peptide, tachyplesin I, with lipid membranes. Biochim Biophys Acta. 1991;1070:259–264. doi: 10.1016/0005-2736(91)90173-6. [DOI] [PubMed] [Google Scholar]
- 42.Matsuzaki K, Yoneyama S, Fujii N, Miyajima K, Yamada K, Kirino Y, Anzai K. Membrane permeabilization mechanisms of a cyclic antimicrobial peptide, tachyplesin I, and its linear analog. Biochemistry. 1997;36:9799–9806. doi: 10.1021/bi970588v. [DOI] [PubMed] [Google Scholar]
- 43.Moore J P, Trkola A, Dragic T. Co-receptors for HIV-1 entry. Curr Opin Immunol. 1997;9:551–562. doi: 10.1016/s0952-7915(97)80110-0. [DOI] [PubMed] [Google Scholar]
- 44.Munoz-Barroso I, Durell S, Sakaguchi K, Appella E, Blumenthal R. Dilation of the human immunodeficiency virus-1 envelope glycoprotein fusion pore revealed by the inhibitory action of a synthetic peptide from gp41. J Cell Biol. 1998;140:315–323. doi: 10.1083/jcb.140.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murakami T, Nakajima T, Koyanagi Y, Tachibana K, Fujii N, Tamamura H, Yoshida N, Waki M, Matsumoto A, Yoshie O, Kishimoto T, Yamamoto N, Nagasawa T. A small molecule CXCR4 inhibitor that blocks T cell line-tropic HIV-1 infection. J Exp Med. 1997;186:1389–1393. doi: 10.1084/jem.186.8.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murakami, T., and N. Yamamoto. Unpublished results.
- 47.Nagasawa T, Kikutani H, Kishimoto T. Molecular cloning and structure of a pre-B-cell growth-stimulating factor. Proc Natl Acad Sci USA. 1994;91:2305–2309. doi: 10.1073/pnas.91.6.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakashima H, Masuda M, Murakami T, Koyanagi Y, Matsumoto A, Fujii N, Yamamoto N. Anti-human immunodeficiency virus activity of a novel synthetic peptide, T22 ([Tyr-5,12, Lys-7]polyphemusin II): a possible inhibitor of virus-cell fusion. Antimicrob Agents Chemother. 1992;36:1249–1255. doi: 10.1128/aac.36.6.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J-L, Arenzana-Seisdedos F, Schwartz O, Heard J-L, Clark-Lewis I, Legler D F, Loetsher M, Baggiolini M, Moser B. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 50.Page K A, Stearns S M, Littman D R. Analysis of mutations in the V3 domain of gp160 that affect fusion and infectivity. J Virol. 1992;66:524–533. doi: 10.1128/jvi.66.1.524-533.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pleskoff O, Treboute C, Brelot A, Heveker N, Seman M, Alizon M. Identification of a chemokine receptor encoded by human cytomegalovirus as a cofactor for HIV-1 entry. Science. 1997;276:1874–1878. doi: 10.1126/science.276.5320.1874. [DOI] [PubMed] [Google Scholar]
- 52.Reed L J, Muench H. A simple method of estimated fifty percent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 53.Rizzuto C D, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 54.Rucker J, Edinger A L, Sharron M, Samson M, Lee B, Berson J F, Yi Y, Margulies B, Collman R G, Doranz B J, Parmentier M, Doms R W. Utilization of chemokine receptors, orphan receptors, and herpesvirus-encoded receptors by diverse human and simian immunodeficiency viruses. J Virol. 1997;71:8999–9007. doi: 10.1128/jvi.71.12.8999-9007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sakaida H, Hori T, Yonezawa A, Sato A, Isaka Y, Yoshie O, Hattori T, Uchiyama T. T-Tropic human immunodeficiency virus type 1 (HIV-1)-derived V3 loop peptides directly bind to CXCR-4 and inhibit T-tropic HIV-1 infection. J Virol. 1998;72:9763–9770. doi: 10.1128/jvi.72.12.9763-9770.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schols D, Struyf S, Van Damme J, Est J A, Henson G, De Clercq E. Inhibition of T-tropic HIV strains by selective antagonization of the chemokine receptor CXCR4. J Exp Med. 1997;186:1383–1388. doi: 10.1084/jem.186.8.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shioda T, Levy J A, Cheng M C. Macrophage and T cell-line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature. 1991;349:167–169. doi: 10.1038/349167a0. [DOI] [PubMed] [Google Scholar]
- 58.Simmons G, Clapham P R, Picard L, Offord R E, Rosenkilde M M, Schwartz T W, Buser R, Wells T, Proudfoot A E. Potent inhibition of HIV-1 infectivity in macrophages and lymphocytes by a novel CCR5 antagonist. Science. 1997;276:276–279. doi: 10.1126/science.276.5310.276. [DOI] [PubMed] [Google Scholar]
- 59.Tamamura H, Ikoma R, Niwa M, Funakoshi S, Murakami T, Fujii N. Antimicrobial activity and conformation of tachyplesin I and its analogs. Chem Pharmacol Bull. 1993;41:978–980. doi: 10.1248/cpb.41.978. [DOI] [PubMed] [Google Scholar]
- 60.Tamamura H, Imai M, Ishihara T, Masuda M, Funakoshi H, Oyake H, Murakami T, Arakaki R, Nakashima H, Otaka A, Ibuka T, Waki M, Matsumoto A, Yamamoto N, Fujii N. Pharmacophore identification of a chemokine receptor (CXCR4) antagonist, T22 ([Tyr(5,12),Lys7]-polyphemusin II), which specifically blocks T cell-line-tropic HIV-1 infection. Bioorg Med Chem. 1998;6:1033–1041. doi: 10.1016/s0968-0896(98)00061-3. [DOI] [PubMed] [Google Scholar]
- 61.Tamamura H, Kuroda M, Masuda M, Otaka A, Funakoshi S, Nakashima H, Yamamoto N, Waki M, Matsumoto A, Lancelin J M, et al. A comparative study of the solution structures of tachyplesin I and a novel anti-HIV synthetic peptide, T22 ([Tyr5,12, Lys7]-polyphemusin II), determined by nuclear magnetic resonance. Biochim Biophys Acta. 1993;1163:209–216. doi: 10.1016/0167-4838(93)90183-r. [DOI] [PubMed] [Google Scholar]
- 62.Tamamura H, Matsumoto F, Sakano K, Otaka A, Ibuka T, Fujii N. Unambiguous synthesis of stromal cell-derived factor-1 by regioselective disulfide bond formation using a DMSO-aqueous HCl system. Chem Commun. 1998;1:151–152. [Google Scholar]
- 63.Tamamura H, Murakami T, Masuda M, Otaka A, Takada W, Ibuka T, Nakashima H, Waki M, Matsumoto A, Yamamoto N, et al. Structure-activity relationships of an anti-HIV peptide, T22. Biochem Biophys Res Commun. 1994;205:1729–1735. doi: 10.1006/bbrc.1994.2868. [DOI] [PubMed] [Google Scholar]
- 64.Tamamura H, Otaka A, Takada W, Terakawa Y, Yoshizawa H, Masuda M, Ibuka T, Murakami T, Nakashima H, Waki M, Matsumoto A, Yamamoto N, Fujii N. Solution-phase synthesis of an anti-human immunodeficiency virus peptide, T22 ([Tyr5,12,Lys7]-polyphemusin II), and the modification of Trp by the p-methoxybenzyl group of Cys during trimethylsilyl trifluoromethanesulfonate deprotection. Chem Pharm Bull. 1995;43:12–18. doi: 10.1248/cpb.43.12. [DOI] [PubMed] [Google Scholar]
- 65.Tanaka, Y. Unpublished results.
- 66.Tashiro K, Tada H, Heilker R, Shirozu M, Nakano T, Honjo T. Signal sequence trap: a cloning strategy for secreted proteins and type I membrane proteins. Science. 1993;261:600–603. doi: 10.1126/science.8342023. [DOI] [PubMed] [Google Scholar]
- 67.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng M C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 68.Vranken W F, Budesinsky M, Fant F, Boulez K, Borremans F A. The complete consensus V3 loop peptide of the envelope protein gp120 of HIV-1 shows pronounced helical character in solution. FEBS Lett. 1995;374:117–121. doi: 10.1016/0014-5793(95)01086-t. [DOI] [PubMed] [Google Scholar]
- 69.Vranken W F, Budesinsky M, Martins J C, Fant F, Boulez K, Gras M H, Borremans F A. Conformational features of a synthetic cyclic peptide corresponding to the complete V3 loop of the RF HIV-1 strain in water and water/trifluoroethanol solutions. Eur J Biochem. 1996;236:100–108. doi: 10.1111/j.1432-1033.1996.00100.x. [DOI] [PubMed] [Google Scholar]
- 70.Wang Z X, Berson J F, Zhang T Y, Cen Y H, Sun Y, Sharron M, Lu Z H, Peiper S C. CXCR4 sequences involved in coreceptor determination of human immunodeficiency virus type-1 tropism. Unmasking of activity with M-tropic Env glycoproteins. J Biol Chem. 1998;273:15007–15015. doi: 10.1074/jbc.273.24.15007. [DOI] [PubMed] [Google Scholar]
- 71.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 72.Wyatt R, Kwong P D, Desjardins E, Sweet R W, Robinson J, Hendrickson W A, Sodroski J G. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 73.Yamashita A, Yamamoto N, Matsuda J, Koyanagi Y. Cell type-specific heterogeneity of the HIV-1 V3 loop in infected individuals: selection of virus in macrophages and plasma. Virology. 1994;204:170–179. doi: 10.1006/viro.1994.1521. [DOI] [PubMed] [Google Scholar]
- 74.Yoshiyama, H. Unpublished results.
- 75.Yoshiyama H, Harada S, Kajii T, Yamamoto N. Narrow host range of AIDS-related retroviruses (YU-1, 2, 3, 4) isolated from Japanese hemophiliacs: inability to infect H9, Molt-4, and MT-4 cells. Jpn J Cancer Res. 1986;77:514–516. [PubMed] [Google Scholar]