Abstract
Most eukaryotic initiation factors (eIFs) are required for internal translation initiation at the internal ribosome entry site (IRES) of picornaviruses. eIF4B is incorporated into ribosomal 48S initiation complexes with the IRES RNA of foot-and-mouth disease virus (FMDV). In contrast to the weak interaction of eIF4B with capped cellular mRNAs and its release upon entry of the ribosomal 60S subunit, eIF4B remains tightly associated with the FMDV IRES during formation of complete 80S ribosomes. Binding of eIF4B to the IRES is energy dependent, and binding of the small ribosomal subunit to the IRES requires the previous energy-dependent association of initiation factors with the IRES. The interaction of eIF4B with the IRES in 48S and 80S complexes is independent of the location of the initiator AUG and thus independent of the mechanism by which the small ribosomal subunit is placed at the actual start codon, either by direct internal ribosomal entry or by scanning. eIF4B does not greatly rearrange its binding to the IRES upon entry of the ribosomal subunits, and the interaction of eIF4B with the IRES is independent of the polypyrimidine tract-binding protein, which enhances FMDV translation.
The internal ribosome entry sites (IRES) of picornaviruses mediate the internal initiation of translation on an RNA (12, 29). During replication of picornaviruses and the distantly related hepatitis C virus and pestiviruses, the strategy of internal initiation allows these viruses to shut down the cap-dependent cellular translation, whereas synthesis of the picornavirus polyprotein is initiated cap independently from the IRES element located far downstream from the 5′ end of the viral RNA (6, 11).
The IRES elements fold into highly organized conserved secondary and probably tertiary structures that guide the ribosome to an internal site of the RNA at the IRES 3′ end (11, 16). The prototype IRES elements of picornaviruses are classified into three groups, those of the enteroviruses and rhinoviruses (including poliovirus), the cardioviruses and aphthoviruses (including foot-and-mouth disease virus [FMDV]), and hepatitis A virus. They contain a conserved core in their 3′ region (16) and two cis elements at the IRES 3′ end, an oligopyrimidine tract followed by an AUG triplet. In cardioviruses and aphthoviruses, translation is usually initiated at this AUG. In FMDV, a second start site 84 nucleotides downstream is utilized after scanning (5). In contrast, the conserved AUG in poliovirus is silent for translation and the actual polyprotein start site further downstream is reached by scanning. Pilipenko et al. (33) proposed a starting-window concept, implying that the small ribosomal subunit encounters the RNA directly 3′ to the IRES element and is either guided directly to the authentic AUG codon in the starting window to initiate translation or starts scanning to find it.
To support internal translation initiation, picornaviruses use essentially the translational apparatus of the host cell but with some modifications. On the one hand, picornaviruses recruit unconventional cellular proteins in addition to standard initiation factors, like the 57-kDa polypyrimidine tract-binding protein (PTB). PTB binds to several picornavirus IRES elements and actively enhances the translation of FMDV (17, 24, 25) and the related encephalomyocarditis virus (EMCV) (7, 12, 14). On the other hand, almost the complete set of eukaryotic initiation factors (eIFs) appears to be essential in picornavirus internal initiation (3, 28, 30, 31), except the cap-binding protein eIF4E.
During the translation initiation on normal cellular mRNAs, a 48S complex that includes the mRNA and the ribosomal 43S preinitiation complex is formed. The RNA helicase eIF4A and its stimulating cofactor eIF4B bind weakly to the mRNA 5′ untranslated region and are assumed to melt secondary structures (35) to allow processive scanning of the ribosomal preinitiation complex from the RNA 5′ end to the AUG start codon. When the preinitiation complex arrests on the initiator AUG, all initiation factors dissociate and the large ribosomal subunit joins the 48S initiation complex (27).
The initiation factor eIF4B has three domains and appears to be a multipurpose adapter involved in several interactions during translation initiation. It can bind to 18S rRNA (20), self-dimerize and mediate its contact with the small ribosomal subunit by interacting with the ribosome-bound eIF3 (21), and stimulate the eIF4A helicase activity (19). Moreover, eIF4B has an RNA-annealing activity that is able to catalyze the hybridization of two complementary single-stranded RNAs (1). eIF4B is involved in internal initiation from the EMCV IRES (4, 9), and previous work in our group has demonstrated that eIF4B directly contacts the FMDV IRES 3′ region including stem-loop 4 (22). Several subdomains in the FMDV IRES 3′ region are essential both for binding of eIF4B and for translation, while the AUG start codon is not required for binding (36), indicating that eIF4B is functionally involved in internal initiation by directly contacting the IRES core upstream of the coding sequence. However, until now it has not been clear in which way eIF4B exerts its function on the IRES. We do not know the stages at which eIF4B is actually involved in the formation of initiation complexes, and we do not know whether unconventional factors like PTB support the function of eIF4B.
In this study, we investigated whether eIF4B is incorporated in ribosomal initiation complexes with the FMDV IRES and the stages of formation of initiation complexes at which eIF4B is involved. The interaction of eIF4B with the IRES RNA was studied in two functionally different situations of initiation that also occur naturally in FMDV. The actual initiator AUG was either located directly in the starting window at the IRES 3′ border or reached by scanning after internal entry of the ribosome. Moreover, we analyzed whether the interaction of eIF4B in the initiation complexes is dependent on the unconventional factor PTB.
MATERIALS AND METHODS
Plasmids.
pSP912 (25) contains the FMDV O1K IRES and coding sequences from positions 363 to 912 (thereby including 108 nucleotides of coding sequence). The dicistronic expression vector pD12 was derived from pD128 (25). It contains the chloramphenicol acetyltransferase (CAT) gene and the FMDV IRES (positions 185 to 805) with the 11th ATG of FMDV fused to the luciferase gene. pD13 resembles pD12 but has FMDV N-terminal coding sequences extending to the 12th authentic ATG (position 889), fused to the luciferase gene. Thus, pD13 contains both authentic initiator ATGs of FMDV. pD14 was derived from pD13 but has the 11th ATG (position 805) mutated to TGG (introduced by a fragment derived from pFMDV-L′ [8]) and thus provides only the 12th ATG of FMDV. pM12, pM13, and pM14 are monocistronic expression vectors corresponding to pD12, pD13, and pD14, respectively, with the CAT gene removed.
Preparation of RNAs and translation.
pSP912 was linearized with SacI 3′ to the FMDV sequence, and pM12, pM13, and pM14 were linearized with BsiWI 152 bp downstream of the luciferase ATG. Labeled RNAs were synthesized as described before (25) with SP6 RNA polymerase in the presence of 2.5 μM [α-32P]CTP (400 Ci/mmol; Amersham) plus 15 μM unlabeled CTP. RNAs were separated from unincorporated nucleotides by gel filtration on Sephadex G-50 columns (Pharmacia). Dicistronic plasmids were linearized with BamHI downstream of the luciferase gene, and mRNA was synthesized in the presence of 500 μM unlabeled nucleotides. A 1-μg portion of dicistronic mRNA was used in a 20-μl reaction mixture containing 10 μl of rabbit reticulocyte lysate (RRL; Promega), 0.7 μl of [35S]methionine, and KCl added to the total K+ concentration in addition to the endogenous potassium-acetate as indicated; the reaction mixture was incubated for 45 min at 30°C, and 2 μl was analyzed by gel electrophoresis and autoradiography.
UV cross-linking assays.
UV cross-linking assays were performed with 3.3 μl of RRL and 0.2 pmol of IRES RNA labeled with either [α-32P]ATP, [α-32P]CTP, [α-32P]GTP, or [α-32P]UTP as indicated in a volume of 10 μl, incubated 10 min at 30°C, and irradiated with UV light for 5 min. Excess RNA was digested with RNase A at 0.1 mg/ml at 37°C for 60 min, proteins were separated on sodium dodecyl sulfate (SDS)–10% polyacrylamide gels and analyzed by autoradiography.
Initiation complex formation, sedimentation analyses, and UV cross-linking.
Initiation complex formation and separation were performed essentially as described before (25). Either standard RRL or PTB-depleted RRL (25) was used. Binding-reaction mixtures contained 50 μl of the respective RRL, 15 mM Tris-Cl [pH 7.5], 0.5 mM MgCl2, 1 mM dithiothreitol (DTT), 100 mM added KCl, and protease inhibitors as described previously (25) in a volume of 150 μl. The total added K+ concentration was 138 mM. If indicated, GMP-PNP (Sigma) was used 4 mM and anisomycin was used at 0.17 mM. RRL treated with anisomycin was preincubated for 5 min at 30°C. Then 2 to 5 pmol of IRES RNA was added, and the samples were incubated for 10 min at 30°C (unless otherwise indicated) for initiation complex formation and then irradiated for 5 min at 0°C with a 6-W UV lamp at 500 μW/cm2 (254 nm).
Each sample was loaded onto a 10 to 35% sucrose gradient containing 50 mM Tris-Cl (pH 8.4), 6 mM MgCl2, 60 mM NaCl, 10 mM DTT, and protease inhibitors as in reference (25), centrifuged for 4.5 h at 200,000 × g at 4°C, and fractionated into 24 fractions of 500 μl. Four-microliter aliquots thereof were subjected to scintillation counting. The fractions were treated with RNase A at 0.1 mg/ml at 37°C for 60 min. Proteins were precipitated in the presence of 10% trichloroacetic acid (TCA) and 5 μg gelatin for 30 min. After centrifugation, pellets were washed twice with ethanol, air dried, and redissolved in protein sample buffer containing 3 M urea. The samples were separated on SDS–12.5% polyacrylamide gels and analyzed by autoradiography.
Initiation complex formation with RRL devoid of ribosomes.
For some experiments, RRL devoid of ribosomes was prepared. A 1-ml volume of RRL was adjusted to 250 mM potassium acetate, and ribosomes were pelleted for 2 h at 200,000 × g and 4°C. The ribosomes were resuspended in 80 μl of 15 mM Tris-Cl (pH 7.5)–0.5 mM MgCl2–1 mM DTT. A 50-μl volume of RRL devoid of ribosomes was used as before in a binding reaction, adjusted to 138 mM potassium acetate, for 10 min at the temperature indicated. After UV cross-linking at 0°C for 5 min, a 4-μl volume of the resuspended ribosomes was added and incubated for another 10 min at either 0 or 30°C as indicated. The reactions were separated on sucrose gradients and analyzed as above.
Immunoprecipitations.
Protein A-coated polyacrylate beads (Fluka) were washed in phosphate-buffered saline (PBS; 8 mM Na2HPO4, 2 mM NaH2PO4, 140 mM NaCl) plus 0.1 mg of tRNA per ml and 0.1 mg of bovine serum albumin (BSA) per ml (PBS-tRNA-BSA). Then 25 μl of either anti-eIF4B antiserum (kindly provided by N. Sonenberg) or preimmune serum was added to the beads in PBS-tRNA-BSA and incubated for 1 h. Excess antiserum was removed by washing with PBS-tRNA-BSA. The gradient fractions from the 80S, 48S, and 20S peaks were pooled after RNase treatment as indicated. To these pools, an equal volume of neutralization buffer (45 mM HEPES [pH 6.5], 200 mM NaCl, 1% Nonidet P-40, 20 mM β-mercaptoethanol) was added. Immunoprecipitation with the antibody-coated beads was performed for 2 h at 4°C. The beads were collected by centrifugation, and the supernatants were saved. The beads were washed in PBS-tRNA-BSA–0.5% Nonidet P-40–10 mM β-mercaptoethanol once for 1 min and twice for 1 h at 4°C. The beads were resuspended in 500 μl of PBS with 1 mM CaCl2, 0.1 mg of RNase A per ml, and 300 U of S7 nuclease per ml and incubated at 37°C for 30 min. After the beads were washed another three times in PBS, they were resuspended in protein sample buffer containing 3 M urea. Proteins form the supernatants saved after immunoprecipitation were concentrated by TCA precipitation, washed with ethanol, collected by centrifugation, and air dried. Pellets were redissolved in protein sample buffer containing 3 M urea. All samples were analyzed by separation on SDS–12.5% polyacrylamide gels and autoradiography.
RNA-protein fingerprint assay.
Binding reactions, UV cross-linking reactions with [α-32P]CTP-labeled FMDV IRES and RRL, and sucrose gradient centrifugation were performed as above. The fractions were treated with RNase, and proteins were concentrated by TCA precipitation. The fractions corresponding to either the 80S, 48S, or 20S peaks were pooled, and proteins were separated by SDS-polyacrylamide gel electrophoresis. Gel slices containing the labeled eIF4B protein were excised after autoradiography. The crumbled gel pieces were incubated for 24 h either with 5 mg of BNPS-skatole (Sigma) in 500 μl of 75% acetic acid at 37°C (18) or with 10 mg of cyanogen bromide in 700 μl of 70% formic acid at room temperature in the dark (26). BNPS-skatole was removed by repeated extraction with ethyl acetate. After the samples were lyophilized, the chemical treatment was repeated to achieve complete cleavage as far as possible. BNPS-skatole was again removed with ethyl acetate, the samples were lyophilized several times, peptides were separated on peptide gels (37), and radiolabeled peptides were analyzed by autoradiography.
RESULTS
eIF4B is incorporated into both 48S and 80S ribosomal initiation complexes with FMDV IRES RNA.
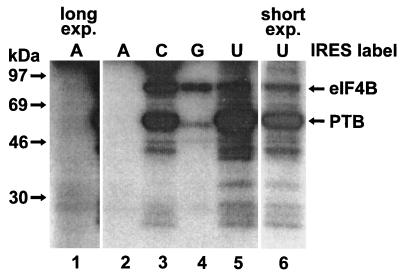
To investigate at which stages eIF4B is involved in the formation of ribosomal initiation complexes with the FMDV IRES, we used the UV cross-linking assay, since in a Western blot all of the eIF4B molecules in the RRL would be detected, not only those which are directly bound to the IRES RNA. First, we tested all four different nucleotides for labeling of IRES RNA (Fig. 1). With CTP, both eIF4B (apparent molecular mass of 80 kDa) and PTB (57 kDa) were labeled well in the UV cross-linking assay (lane 3), whereas with UTP, mainly PTB was labeled (lanes 5 and 6). With GTP, labeling of eIF4B was weaker (lane 4), and with ATP, almost no protein was labeled (lanes 1 and 2). Therefore, [α-32P]-CTP-labeled IRES RNA was used for the detection of both eIF4B and PTB in all the following UV cross-linking assays.
FIG. 1.
Detection of eIF4B in RRL by using FMDV IRES RNA labeled with different nucleotides. UV cross-linking assays were performed with RRL and IRES RNA in the presence of either [α-32P]ATP (A), [α-32P]CTP (C), [α-32P]GTP (G), or [α-32P]UTP (U) as indicated. The molecular masses of marker proteins and the positions of eIF4B and PTB are indicated. Lane 1 is a threefold-longer exposure of lane 2, and in lane 6 50% of a sample corresponding to that in lane 5 was subjected to a slightly shorter exposure.
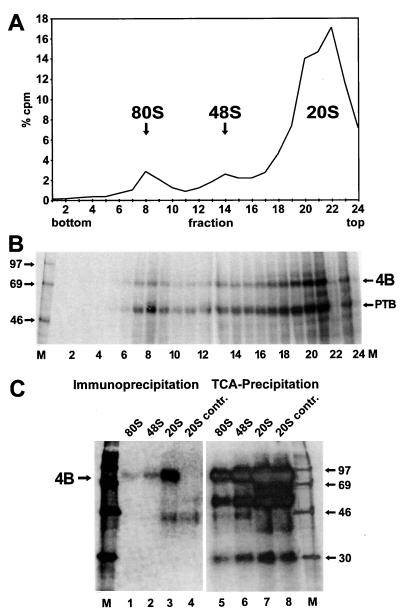
For analyzing the presence of eIF4B in initiation complexes with the FMDV IRES, the RNA was incubated with RRL for 10 min at 30°C, allowing the binding of proteins and ribosomal subunits to the IRES (25). The reaction mixture was transferred to 0°C and irradiated with UV light. By that, we captured the proteins bound to the IRES after 10 min and then detected them after completion of the experiment. Initiation complexes were then separated on a sucrose gradient (Fig. 2A). Both 80S and 48S initiation complexes (fractions 7 to 9 and fractions 13 to 15, respectively) were formed with the IRES RNA. RNA not associated with ribosomal subunits remained in the upper part of the gradient (fractions 18 and above).
FIG. 2.
Detection of eIF4B in ribosomal initiation complexes with FMDV IRES RNA. (A) Radioactivity profile of the gradient. [α-32P]CTP-labeled IRES-912 RNA was incubated with RRL for 10 min at 30°C. Samples were irradiated with UV, and initiation complexes were separated on a 10 to 35% sucrose gradient. Fractions were collected from the bottom (fraction 1), and aliquots were used for scintillation counting. The amount of radioactivity in each fraction relative to the total amount of radioactivity in the entire gradient (% cpm) is plotted against the fraction number. The 80S, 48S, and 20S peaks are indicated. (B) UV-cross-linked proteins from the gradient shown in panel A. Fractions were treated with RNase A, and proteins were precipitated with TCA, resolved on SDS-polyacrylamide gels, and visualized by autoradiography. 4B, eIF4B. (C) Identification of eIF4B in ribosomal initiation complexes by immunoprecipitation. The binding reaction with RRL and IRES RNA, UV cross-linking, and gradient run were performed as in panel A. The fractions containing either the 80S, 48S, or 20S peak were pooled and treated with RNase A. The 20S pool was divided into two portions. eIF4B was immunoprecipitated from the 80S pool, from the 48S pool, and from 50% of the 20S pool with eIF4B antiserum (left). From the other 50% of the 20S pool, the control with preimmune serum (20S contr.) was obtained. Proteins from the supernatants were concentrated by TCA precipitation (right). Proteins were resolved on an SDS-polyacrylamide gel and visualized by autoradiography. Molecular masses of marker proteins (M) are given in kilodaltons.
Proteins bound to the IRES RNA were analyzed by UV cross-linking, RNase treatment, and SDS-polyacrylamide gel electrophoresis (Fig. 2B). Essentially two proteins, associated both with the ribosomal complexes and with the material remaining at the top of the gradient, were visible. The 57-kDa protein was PTB (25), and the protein migrating with an apparent molecular mass of 80 kDa was eIF4B as identified by immunoprecipitation (see below). Remarkably, eIF4B was associated with the IRES RNA not only in 48S complexes (fractions 12 to 14) but also in 80S complexes (fractions 7 to 9). This was not expected, since during initiation of translation from normal cellular mRNAs the initiation factors are supposed to dissociate from the 48S complex upon joining of the large ribosomal subunit (27). Almost no material representing IRES 912 RNA loaded with two ribosomes sedimenting faster than the 80S complexes was detected, most probably due to the very short extension of only 23 nucleotides of the RNA downstream of the second AUG. In addition, the efficiency of formation of RNA 912 loaded with two ribosomes can be expected to be very low, since it amounts to the square of the efficiency of formation of RNA loaded with one ribosome.
The identity of the eIF4B protein interacting with the IRES RNA in the initiation complexes was verified by immunoprecipitation (Fig. 2C). By using the UV cross-linking assay, only eIF4B molecules in the RRL that were actually bound to the IRES were detected. The experiment was performed as before, but the fractions containing 80S complexes, 48S complexes, or material from the top of the gradient (termed 20S) were pooled. eIF4B was immunoprecipitated with an anti-eIF4B antiserum (Fig. 2C, left), and the supernatants after immunoprecipitation were collected by TCA precipitation and analyzed as a control (right panel). From the 20S material, eIF4B was immunoprecipitated as a strongly labeled band (lane 3). Also from the ribosomal initiation complexes, eIF4B was clearly identified, both from 80S complexes (lane 1) and from 48S complexes (lane 2). The ratio of the intensities of the eIF4B bands immunoprecipitated from the initiation complexes and the 20S material is similar to the relationship of the respective peaks in the gradient profile. Although the efficiency of the immunoprecipitation was low in terms of yield due to the dilution of the sample, the immunoprecipitation was specific, since eIF4B was not precipitated at all from the 20S material with preimmune serum (20S contr., lane 4), and a 30-kDa protein (lanes 5 to 8) was not precipitated at all (lanes 1 to 4). Accordingly, the association of some weak bands like the 45 kDa protein (lanes 3 and 4) with the beads must be regarded non unspecific. This band did not completely disappear after the beads were washed extensively after immunoprecipitation.
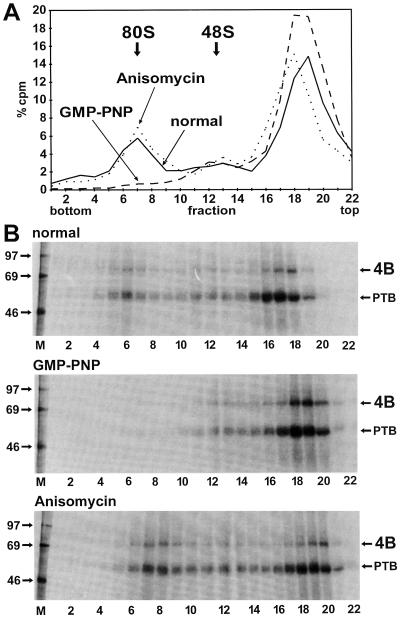
The identity of the ribosomal initiation complexes was confirmed by using stage-specific translation inhibitors. GMP-PNP competes with GTP incorporated into the ternary complex; it inhibits the release of eIF2 from the small ribosomal subunit and thus prevents association of the 60S subunit. By using GMP-PNP, the formation of 80S complexes was completely abolished while the 48S complexes were slightly enriched (Fig. 3A). Neither eIF4B nor PTB appeared in the gradient fractions corresponding to the 80S peak (Fig. 3B, middle panel). Only minute amounts of material, perhaps representing RNA loaded with two small ribosomal subunits, sedimented beyond the 48S position. However, these complexes are expected to appear only in small amounts, since only 5 to 10% of the free RNA was incorporated into 48S complexes. For RNA loaded with two 40S subunits, this efficiency must be squared, giving 0.25 to 1% of the total RNA. Moreover, RNA 912 extends only a little way downstream of the 12th AUG, leading to inefficient binding of ribosomes to this AUG. The second inhibitor, anisomycin, inhibits the ribosomal peptidyltransferase and freezes the ribosomes on the RNA immediately after association of the 60S subunit. By using this antibiotic, the relative amount of RNA associated with 80S complexes was enriched (Fig. 3A) and more eIF4B and PTB were associated with 80S complexes (Fig. 3B, bottom panel) compared to the standard reaction (top panel). Thus, eIF4B and PTB are indeed associated with ribosomal complexes.
FIG. 3.
Enrichment of eIF4B in initiation complexes with IRES RNA by stage-specific translation initiation inhibitors. (A) Gradient profiles. The binding reactions with RRL and labeled IRES-912 RNA were performed as in Fig. 1 (solid line), in the presence of 4 mM GMP-PNP (dashed line), or in the presence of 0.17 mM anisomycin (dotted line). (B) UV-cross-linked proteins from RNase-treated fractions of the gradients shown in panel A were collected by TCA precipitation, resolved on SDS-polyacrylamide gels, and visualized by autoradiography. Molecular masses of marker proteins (M) are given in kilodaltons. 4B, eIF4B.
Kinetics of the entry of eIF4B into ribosomal initiation complexes.
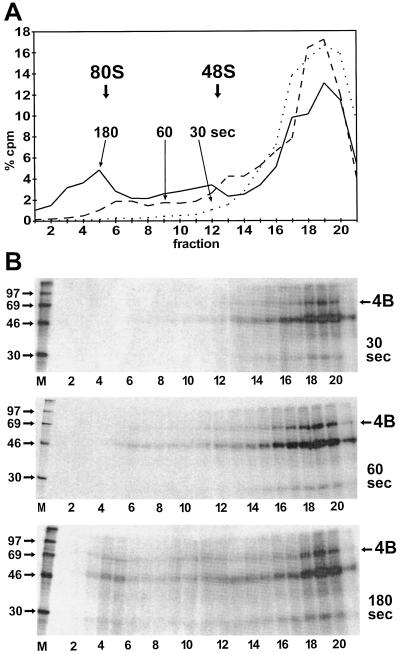
To investigate the time dependency of the incorporation of eIF4B together with the IRES RNA into the ribosomal initiation complexes, RRL was incubated with IRES RNA for a limited time at 30°C to permit binding of the initiation factors and the ribosomal subunits to the RNA. All other steps such as UV cross-linking and gradient loading were performed at 0°C to prevent further binding of ribosomes to IRES RNA. The results are shown in Fig. 4. After 30 s of incubation at 30°C, no 80S initiation complexes could be detected and no separate 48S peak was resolved. Nevertheless, small amounts of 48S complexes could be present in the forward-trailing part of the large 20S peak (Fig. 4A). After 60 s, two separate peaks of 48S and 80S complexes were detected, and only after 180 s had normal amounts of initiation complexes been formed, similar to the reaction incubated for 10 min (compare with Fig. 2 and 3), indicating that the formation of initiation complexes was completely finished. The time required for formation of maximal amounts of initiation complexes can be concluded to be between 1 and 3 min, in accordance with the range of 2 to 5 min found previously (25).
FIG. 4.
Kinetics of the entry of FMDV IRES RNA and eIF4B into ribosomal initiation complexes. (A) Gradient profiles. The binding reactions were performed with RRL as in Fig. 2, but the incubation time at 30°C for binding of the ribosomal subunits to the IRES RNA was limited to either 30 s (dotted line), 60 s (dashed line), or 180 s (solid line). All other steps, including UV cross-linking and gradient loading, were performed at 0°C. Each binding reaction was analyzed on a separate gradient. (B) UV-cross-linked proteins from the RNase-treated gradient fractions from panel A were collected by TCA precipitation, resolved on SDS-polyacrylamide gels, and visualized by autoradiography. Molecular masses of marker proteins (M) are given in kilodaltons. 4B, eIF4B.
The gradient fractions were next analyzed for the presence of eIF4B. After 30 s, eIF4B was detectable in the upper part of the gradient but only minute amounts of eIF4B were detected in the fractions in which 48S ribosomal complexes would appear (Fig. 4B, top panel). After 60 s, the total amount of cross-linked eIF4B was increased, and trace amounts of eIF4B appeared in the ribosomal complexes (middle panel). Only after 180 s were considerable amounts of eIF4B detected in the complexes (bottom panel). The relationship of the amounts of eIF4B in the 80S complexes to the 48S complex correlates with the relationship of the corresponding peaks in the radioactivity profile of the gradient (Fig. 4A). Thus, eIF4B is incorporated into the initiation complexes strictly in parallel with the IRES RNA that initiates complex formation. Moreover, the total amount of eIF4B detected in the gradients increases with longer incubation at 30°C. This suggests that eIF4B first contacts the IRES temperature dependently and then is incorporated into the ribosomal complexes.
Order of binding of eIF4B and ribosomes to the IRES RNA.
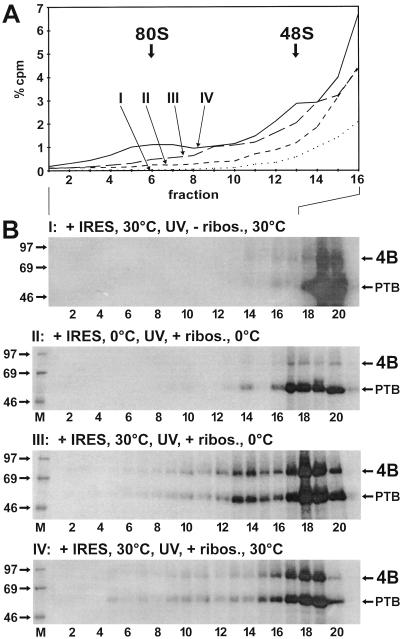
To determine the order of binding steps in which eIF4B and the ribosomes bind to the FMDV IRES and to determine the temperature sensitivity of these interactions, the ribosomes were separated from the soluble fraction of the reticulocyte lysate containing the initiation factors. First the RRL was adjusted to 250 mM potassium acetate to dissociate initiation factors from the ribosomes, and then the ribosomes were pelleted. This ensured that sufficient amounts of initiation factors, particularly eIF4B, were present in the soluble fraction. The binding of initiation factors to the FMDV IRES RNA in the absence of ribosomes was performed at either 30 or 0°C to determine the temperature sensitivity of this step. After that, UV cross-linking was performed at 0°C to covalently cross-link the bound proteins to the RNA. In this way, the ability of the proteins to bind to the IRES RNA in the absence of ribosomes was detected. After the UV cross-linking step, the ribosomes were added back and the reaction mixture was incubated again at either 30 or 0°C for the association of the ribosomes. The reaction products were then analyzed on sucrose gradients as above.
When this RRL cleared of ribosomes was used in a binding reaction with FMDV IRES at 30°C, and no ribosomes were added after UV cross-linking for the second incubation at 30°C, no initiation complexes were formed (Fig. 5A, reaction I), and both eIF4B and PTB were found UV cross-linked to the IRES only in the top fractions of the gradient (Fig. 5B, reaction I). This indicates that (i) the cytoplasm cleared of ribosomes was indeed devoid of ribosomes and (ii) eIF4B and PTB bound to the FMDV IRES in the complete absence of ribosomes.
FIG. 5.
Order and temperature dependency of binding events. Binding reactions and gradient runs were performed as in Fig. 4, but binding of initiation factors, UV cross-linking, and binding of ribosomes were performed separately and at different temperatures. (A) Gradient profiles of fractions 1 to 16. (B) UV-cross-linked proteins from the gradients shown in panel A. RRL devoid of ribosomes was incubated with RNA 912 (+ IRES) for 10 min at 30 or 0°C as indicated. The reaction mixtures were then irradiated with UV light at 0°C to covalently cross-link the RNA to the initiation factors. Only after that were ribosomes added in panels II to IV (+ ribos.), but not in panel I (− ribos.), and the reaction mixtures were again incubated for 10 min at 30 or 0°C as indicated. After that, the reaction products were loaded onto sucrose gradients and analyzed as in Fig. 4. 4B, eIF4B.
In reaction II, the temperature sensitivity of the binding of eIF4B to the FMDV IRES was analyzed. IRES RNA was incubated with the RRL devoid of ribosomes at 0°C. After UV cross-linking, the ribosomes were added back and the reaction mixture was incubated again for 10 min at 0°C. In this reaction, only small amounts of 48S complexes were formed and PTB appeared in the top fractions but eIF4B bound only marginally (Fig. 5A and B, II). This indicates that the binding of eIF4B to the IRES is largely temperature dependent but that binding of PTB is temperature independent, as found previously (25). In addition, very small amounts of 48S complexes had formed also at the low temperature (fraction 14), indicating that 48S complex formation can occur at the low temperature only at a largely reduced rate.
In reaction III, IRES RNA was incubated with the RRL devoid of ribosomes at 30°C for 10 min, and after UV cross-linking, ribosomes were added back and the reaction mixture was incubated for another 10 min at 0°C (Fig. 5B, reaction III). In this reaction, large amounts of eIF4B were cross-linked to the IRES, confirming that the binding of eIF4B to the IRES occurs energy dependently in the absence of ribosomes. Moreover, a large amount of eIF4B was present in the 48S peak, indicating that the binding of the ribosomal 40S subunit to the IRES preloaded with initiation factors is not energy dependent. However, only minute amounts of 80S complexes were formed, indicating that this step involves energy-dependent reactions.
In reaction IV, the incubation was performed in the presence of the ribosomes at 30°C. In this case, not only 48S but also 80S complexes were formed, as in a normal binding reaction. This indicates that the joining of the large ribosomal 60S subunit requires energy-dependent reactions, like the hydrolysis of the eIF2-bound GTP. In addition, this experiment confirms that all components required for initiation complex formation were present in the extracts used for the above reconstitution experiments.
The interaction of eIF4B is independent of the location of the initiator AUG.
The first authentic initiator AUG of the FMDV IRES (the 11th AUG in strain O1K) is located at a conserved distance downstream of the pyrimidine tract (17). This AUG is assumed to be located within the starting window (33) at the 3′ end of the IRES, implying that the small ribosomal subunit is placed directly adjacent to this AUG. However, a second AUG 84 nucleotides downstream of the 11th AUG (the 12th in FMDV O1K) is used after scanning of the small ribosomal subunit (5). Thus, it could be assumed that the interaction of eIF4B with the FMDV IRES upon utilization of this 12th AUG is different from its interaction upon utilization of the 11th AUG, since the ribosomal subunit reaches the two AUGs by different mechanisms. During initiation at the 12th AUG, the action of eIF4B may resemble that in cap-dependent initiation, where a eIF4A-eIF4B complex acts as an unwindase, removing RNA secondary structures, and eIF4B dissociates from the RNA upon entry of the 60S subunit (27).
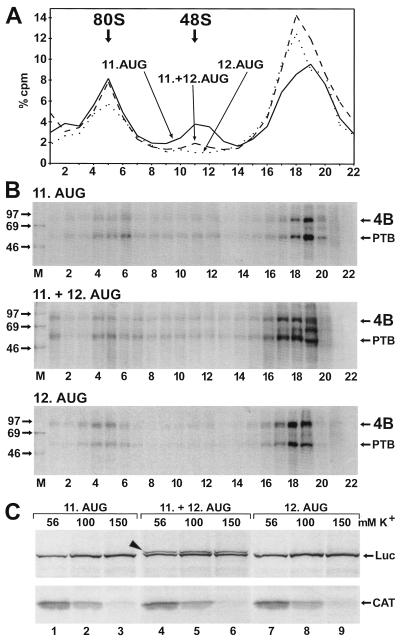
To detect such a possible change in the interaction of eIF4B with the FMDV IRES between these different start situations, we used three different RNAs. pM12 RNA contains the FMDV IRES including only the 11th AUG. pM13 IRES RNA includes both the 11th and the 12th AUGs. In contrast, pM14 IRES RNA contains only the 12th AUG, and the 11th AUG was mutated. All three RNAs contain a sufficiently long tail of some 150 nucleotides downstream of the respective AUGs to ensure efficient ribosome binding. With pM12 IRES RNA containing the 11th AUG, the formation of ribosomal complexes (Fig. 6A) and the incorporation of eIF4B into these complexes (Fig. 6B, top panel) was as before with pSP912 RNA. Also with pM13 RNA (middle panel) and even with pM14 RNA containing only the 12th AUG (bottom panel), eIF4B was detected in the ribosomal 80S complexes. Thus, eIF4B was incorporated into the 80S complexes irrespective of whether the initiator AUG was reached directly by guiding the small ribosomal subunit to the starting window (pM12 IRES) or whether it was reached by scanning (pM14 IRES). This result demonstrates that the interaction of eIF4B with this picornavirus IRES is independent of the location of the actual initiator AUG. In contrast, smaller amounts of 48S complexes were detected with pM13 IRES RNA containing both the 11th and 12th AUGs and almost no 48S complexes were detected with pM14 IRES RNA containing only the 12th AUG. Correspondingly, smaller amounts of eIF4B were detected in the fractions corresponding to the 48S peak with pM14 RNA (Fig. 6B, bottom panel) compared to pM12 or pM13 RNA (top and middle panels). These reduced amounts of 48S complexes obtained with pM14 IRES RNA may be due to a faster conversion of 48S complexes into 80S complexes.
FIG. 6.
Effect of initiator AUG position. (A) Gradient profiles. The binding reactions with RRL and labeled IRES RNAs were performed as in Fig. 2 but with pM12 IRES RNA containing only the 11th AUG of FMDV (solid line), pM13 IRES with both the 11th and the 12th AUG (dashed line), or pM14 IRES with only the 12th AUG but the 11th AUG mutated to UGG (dotted line). (B) UV cross-linked proteins from the gradients shown in panel A. Molecular masses of marker proteins (M) are given in kilodaltons. 4B, eIF4B. (C) Translation from the two AUGs of the FMDV IRES. Dicistronic mRNAs transcribed from pD12 (11. AUG), pD13 (11. +12. AUG), and pD14 (12. AUG) were translated in RRL in the presence of [35S]methionine at the total K+ concentrations as indicated. The translated CAT and luciferase (Luc) proteins are indicated on the right, and the additional luciferase protein with a N-terminal extension derived from pD13 is indicated by an arrowhead.
With pM13 RNA containing both AUGs, a considerable amount of complexes sedimenting faster than 80S was detected (Fig. 6B, middle panel, lane 1), perhaps representing RNAs that are loaded with two 80S ribosomes. Dicistronic mRNAs were used to analyze the translation products driven by corresponding constructs containing either the 11th AUG, both AUGs, or only the 12th AUG. With pD13 containing both AUGs, a second luciferase protein with an N-terminal extension was produced (Fig. 6C, top panels, lanes 4 to 6) in addition to the normal luciferase protein produced by pD12 (lanes 1 to 3) or pD14 containing only the 12th AUG (lanes 7 to 9). Thus, both authentic AUGs contained in this IRES variant are used for translation, in accordance with the finding that two 80S ribosomes are probably bound to it simultaneously. The translation of luciferase proteins was optimal at salt concentrations between 100 and 150 mM total K+, confirming that our binding reactions with 138 mM K+ were performed in the optimal K+ concentration range, while the efficiency of the 5′-dependent translation CAT protein decreased with increasing salt concentrations (Fig. 6C, bottom panels).
eIF4B does not rearrange its contacts to the IRES RNA upon ribosome entry.
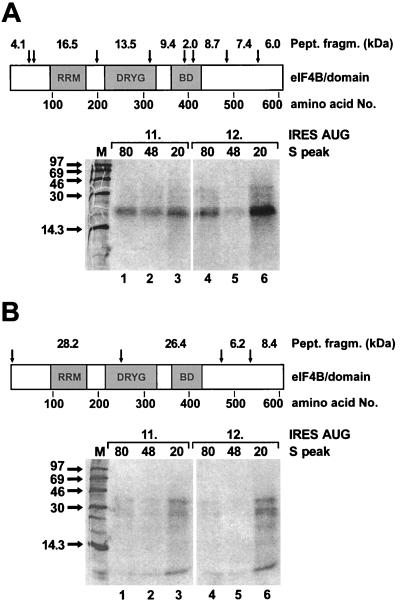
To test for a possible change of the contact sites within the eIF4B protein that bind the IRES during formation of 48S and transition to 80S initiation complexes, we performed an RNA-protein fingerprint assay in which the protein was fragmented after being UV cross-linked to the RNA. RRL including the native intact eIF4B protein was used in a binding reaction with the IRES RNA as above. After UV irradiation, initiation complexes were separated on sucrose gradients, the gradient fractions were treated with RNase, and proteins were TCA precipitated. The gel-purified labeled eIF4B protein from the 80S, 48S, and 20S peaks was subjected to chemical fragmentation by either BNPS-skatole (cleaving after tryptophane) or CNBr (cleaving after methionine), and labeled peptides were analyzed on peptide gels. In this assay, both the eIF4B protein and the IRES RNA are native and complete during the binding reaction and UV cross-linking. Only after that is excess RNA digested and the protein fragmented. Thus, the radiolabeled peptides obtained represent the contact sites between native eIF4B protein and complete IRES RNA.
When the contact of eIF4B to the IRES was analyzed by using BNPS-skatole, a peptide of about 20 kDa was radioactively labeled (Fig. 7A). Since the amounts of the labeled peptide were too small for subsequent identification, we can only speculate about its nature. Most probably it represents the 16.5-kDa peptide migrating more slowly due to the attached RNA oligonucleotide(s), suggesting that the N-terminal RNA recognition motif domain of eIF4B is involved in IRES binding. Only small amounts of other cleavage products with higher molecular masses, presumably representing partially cleaved products, are visible. These did not disappear even after two cycles of chemical treatment, suggesting that the corresponding cleavage sites are not accessible to complete chemical cleavage.
FIG. 7.
RNA-protein fingerprint assay for detection of eIF4B peptides contacting the FMDV IRES in ribosomal initiation complexes. The binding reaction and UV cross-linking with RRL and either labeled pM12 RNA (11. AUG) or pM14 RNA (12. AUG) was performed as in Fig. 2, and initiation complexes were separated on sucrose gradients. The fractions corresponding to either the 80S, 48S, or 20S peaks were pooled and treated with RNase, and the proteins were TCA precipitated. The eIF4B protein labeled by UV cross-linked RNA fragments was purified on an SDS-polyacrylamide gel, excised, and treated with either BNPS-skatole, which cleaves after tryptophan residues (A), or CNBr, which cleaves after methionine residues (B). The corresponding cleavage sites in the eIF4B protein (arrows), the calculated sizes of the peptide fragments after cleavage, and the domains of eIF4B are shown. RRM, RNA recognition motif domain; DRYG, DRYG domain (21); BD, basic domain. Molecular masses of marker proteins (M) are given in kilodaltons.
The pattern of labeled peptide fragments did not change upon entry of the small ribosomal subunit forming the 48S complexes (lane 2) or even after entry of the large subunit in the 80S complexes (lane 1) compared with the 20S peak not containing ribosomal subunits (lane 3). Moreover, the pattern of labeled peptides was the same irrespective of whether the small ribosomal subunit was placed directly adjacent to the 11th AUG in the starting window on pM12 IRES RNA (lanes 1 to 3) or the 12th AUG was reached by scanning on pM14 IRES RNA (lanes 4 to 6). Corresponding to the smaller amounts of 48S complexes formed with pM14 RNA (compare with Fig. 6B, bottom panel), less eIF4B cleavage products were obtained from the ribosomal complexes with pM14 RNA.
In a similar experiment, the eIF4B protein contacting the FMDV IRES in ribosomal complexes was cleaved with CNBr (Fig. 7B). The bands obtained from the ribosomal complexes are very weak. Nevertheless, they allow the conclusion that the pattern of labeled peptide fragments did not change upon transition of eIF4B from the 20S peak to the 48S complexes and to the 80S complexes or with the RNA used. Also in this experiment, no identification of the labeled peptides was possible due to their very small amounts. These results indicate that the eIF4B protein does not grossly rearrange its binding to the FMDV IRES during the transition to 48S and 80S initiation complexes.
The presence of eIF4B in 48S and 80S complexes is independent of PTB.
As a supporting noncanonical factor, the cellular RNA-binding protein PTB had been found to stimulate FMDV translation (24). Moreover, PTB is incorporated into 48S and 80S initiation complexes with the FMDV IRES (25). Thus, we asked whether the interaction of eIF4B with the IRES in the ribosomal initiation complexes would be supported by PTB.
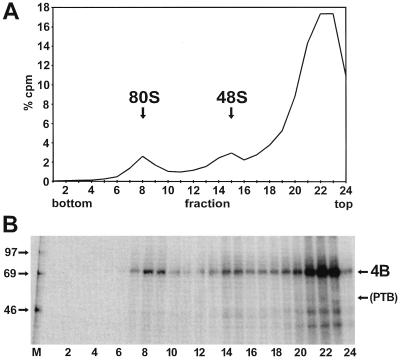
To answer this question, we removed the endogenous PTB from RRL, resulting in a lysate that is still translation competent for 5′-dependent and IRES-dependent translation and still contains eIF4B. In the PTB-depleted RRL, the FMDV IRES activity decreased by a factor of 2 to 3 due to the missing enhancing activity of PTB (24, 25). This PTB-depleted lysate was then used to investigate the association of eIF4B with initiation complexes formed with the FMDV IRES (Fig. 8). Both 48S and 80S complexes were formed (Fig. 8A), as with untreated RRL (compare with Fig. 2), indicating that PTB is not essential for complex formation. When the proteins associated with the ribosomal complexes were analyzed in the UV cross-linking assay (Fig. 8B), only eIF4B, not PTB, was detected. The amounts of eIF4B cross-linked to the IRES in the fractions containing the initiation complexes were as large as with untreated RRL (compare with Fig. 2B), and the presence of eIF4B could be clearly assigned to the positions of the 48S and 80S complexes as before. Thus, the interaction of eIF4B with the FMDV IRES and its probably important role in translation initiation is not supported by the action of PTB.
FIG. 8.
eIF4B in initiation complexes with PTB-depleted RRL. (A) Radioactivity profile of the gradient. The binding reaction with labeled IRES-912 RNA was performed as in Fig. 2, but PTB-depleted RRL was used instead of standard RRL. (B) UV-cross-linked proteins from the gradient shown in panel A. Molecular masses of marker proteins (M) are given in kilodaltons. 4B, eIF4B.
DISCUSSION
In this study, we show that eIF4B interacts directly with the FMDV IRES in ribosomal initiation complexes, pointing to an important role of eIF4B during the assembly of initiation complexes with the IRES. Several subdomains in the IRES 3′ region are indispensable for both the direct contact of eIF4B with the FMDV IRES and FMDV translation (36), and a sequence element within one of these subdomains is highly conserved in the cardiovirus-aphthovirus group (11). This indicates that eIF4B plays an important role in guiding the ribosome to the IRES, consistent with early reports that eIF4B is required for translation from the EMCV IRES (4, 9) and also with the finding that 48S complex formation with the EMCV IRES was enhanced threefold in the presence of eIF4B (30).
We used the UV cross-linking assay to detect only eIF4B molecules that are directly bound to the IRES and functionally incorporated into ribosomal initiation complexes. In contrast, in a Western blot all of the eIF4B molecules in the extract would be detected, including those that may be associated with ribosomes but do not necessarily have direct contact with the IRES RNA. eIF4B-IRES complexes are present in the soluble material at the top of the gradient, and a fraction of these are incorporated into ribosomal complexes. The distribution of these eIF4B molecules directly bound to the IRES differs from the distribution found for the totality of the eIF4B molecules. Less than 10% of the total cellular eIF4B were purified from the soluble postribosomal supernatant, while most of the eIF4B was associated with ribosomes (39), consistent with the distribution of eIF4B in gradients detected by an anti-eIF4B antibody in a Western blot (26a). Thus, only a subfraction of the eIF4B molecules are actively involved in forming functional initiation complexes and hence are in direct contact with the initiation complex forming viral IRES RNA.
eIF4B appears to be involved in the initial phase of FMDV IRES-dependent initiation. It binds temperature dependently to the IRES independently of ribosomes, since eIF4B was UV cross-linked to the IRES in the complete absence of ribosomes, and in kinetic experiments eIF4B first appears in the gradient fractions not containing ribosomal subunits. Thus, eIF4B first binds to the IRES, and only then does the ribosome enter this RNA-protein complex. This binding of eIF4B to the IRES is dependent on ATP (22), while eIF2 hydrolyzes GTP. Only after the energy-dependent binding of eIF4B (and probably other initiation factors) to the IRES is the small ribosomal subunit able to enter the IRES region. This entry of the small ribosomal subunit is energy independent. In contrast, the joining of the 60S subunit is again energy dependent, most probably due to the hydrolysis of the eIF2-bound GTP residue prior to entry of the 60S subunit. The formation of initiation complexes with the FMDV IRES appears to be a rather slow process. The time required for the formation of 48S and 80S complexes found here and also previously (25) was between 2 and 3 min. This is considerably longer than that reported for cellular mRNAs, which was less than 1 min (2, 23). With α- and β-globin mRNA, 48S complexes were formed rapidly, while the subsequent joining of the 60S subunit required more time (2). This suggests that during IRES-dependent initiation, binding of initiation factors and entry of the small ribosomal subunit to the complex IRES structure involve RNA-protein and protein-protein interactions that require more time than binding of the cap-binding complex to the cap of cellular mRNAs.
The interaction of eIF4B with the FMDV IRES in the complete 80S ribosomes contrasts with the widely accepted idea that in cap-dependent translation all initiation factors dissociate from the 48S complex upon joining of the large ribosomal subunit (27) and thus are not associated with complete 80S ribosomes. For example, another initiation factor, eIF4G, was found associated with 43S and 48S initiation complexes with cellular mRNA (13). This indicates that eIF4G is associated with the small ribosomal subunit independently of mRNA. However, eIF4G was not found in complete 80S ribosomes, suggesting that it dissociates from the RNA upon joining of the 60S subunit. The situation with the FMDV IRES is clearly different, since eIF4B definitely remains bound to the IRES in the 80S complexes as well. The incorporation of eIF4B into these complexes together with the IRES must be regarded as functional, since its binding to the IRES is temperature and time dependent and since the eIF4B molecules that are UV cross-linked to the IRES RNA are in direct physical contact with the RNA in the initiation complexes.
Comparison of IRES elements has revealed the presence of a common core structure in their 3′ region (16). By interacting with this core structure, eIF4B may be involved in directing the small ribosomal subunit to the starting window (33) at the 3′ end of the IRES. Nevertheless, the interaction of eIF4B with the FMDV IRES does not change, irrespective of whether an AUG is used as the actual initiator start codon in the starting window or whether the actual start codon is reached after scanning of the small subunit. No principal differences were observed in the binding of eIF4B to the IRES between these two different situations of initiation. Moreover, by comparing the patterns of contact sites in the eIF4B protein that bind to the IRES RNA either without engagement of ribosomes in the 48S complexes or in the 80S complexes, we found that eIF4B does not essentially change its contacts with the IRES RNA. Thus, the action of eIF4B does not involve substantial rearrangements of the contacts between eIF4B and IRES after the entry of ribosomal subunits. In turn, after binding of eIF4B, subsequent steps of protein binding and ribosomal entry may occur on a core of eIF4B (and possibly other initiation factors) bound to the 3′ region of the IRES. However, this does not, of course, exclude the possibility that any additional eIF4B molecules are involved in the unwinding of secondary structures between the IRES and the second initiator AUG during scanning.
Concerning the binding of other proteins to the IRES, eIF4B could be suspected to mediate the stimulatory action of the noncanonical factor PTB, which enhances FMDV translation efficiency. Although the mere binding of PTB to the IRES is temperature independent, its incorporation into 48S and 80S complexes with the IRES occurs in a similar way to that of eIF4B (25). Here we demonstrate that the initiation complexes were formed with the FMDV IRES in the absence of PTB, while eIF4B still bound well to the IRES. This indicates that the interaction of eIF4B with the IRES in the initiation complexes is independent of PTB. The stimulating activity of PTB and the binding of eIF4B are two different processes in FMDV translation initiation which are obviously not mutually dependent but which both add to the total activity.
The above data point to a key role of eIF4B in picornavirus internal initiation. The interaction of eIF4B with the IRES may introduce a second multivalent adapter in addition to eIF4G (10), thereby providing additional links between the IRES and ribosome. One link provided by eIF4B may be the bridge IRES-eIF4B-eIF3-40S, facilitated by protein-protein interactions between IRES-bound eIF4B and the 170-kDa subunit of the ribosome-bound eIF3 (21). A second link may be an IRES-eIF4B-18S rRNA interaction. eIF4B has two separate RNA-binding domains (20) and can therefore connect two RNA molecules. This may be even enhanced by eIF4B dimer formation (21). Another link between IRES and the ribosomal 40S subunit may be provided by an interaction between eIF4B and eIF4G. eIF4G was found not only in 48S complexes with a cellular mRNA but also in 43S complexes (13) and thus is associated with the small ribosomal subunit independently of the presence of mRNA. Since binding of the central domain of eIF4G to the EMCV IRES is enhanced in the presence of eIF4B (32), the stimulation of formation of 48S complexes with the EMCV IRES RNA by eIF4B may be caused by an interaction between IRES-bound eIF4B and ribosome-bound eIF4G.
Moreover, some observations point to a possible enzymatic function of eIF4B in positioning the starting window of the IRES to the appropriate site on the ribosome. One observation is that eIF4B binding to the FMDV IRES is ATP dependent (22), suggesting that the ATP-dependent RNA helicase eIF4A is involved as an interacting cofactor. Indeed, a direct binding of both eIF4A and eIF4B to the IRES has been demonstrated for EMCV (15). The second observation is that eIF4B has RNA-annealing activity (1), which could help to adjust the start codon on the ribosome by a local hybridization between the RNA which is translated and the 18S rRNA. Like prokaryotic translation initiation signals, picornavirus IRES elements have conserved cis elements within their pyrimidine tracts which are located in a conserved distance upstream of a conserved AUG codon at the IRES 3′ border (34, 38). By alternating annealing and melting events, an eIF4A-eIF4B complex may help to hybridize these sequences in the IRES pyrimidine tract to complementary sequences in the 18S rRNA and thus adjust the IRES initiator AUG to the correct position on the small ribosomal subunit.
ACKNOWLEDGMENTS
We thank N. Sonenberg for kindly providing the eIF4B antiserum and E. Beck for helpful discussions.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (SFB 535).
REFERENCES
- 1.Altmann M, Wittmer B, Méthot N, Sonenberg N, Trachsel H. The Saccharomyces cerevisiae translation initiation factor Tif3 and its mammalian homologue, eIF-4B, have RNA annealing activity. EMBO J. 1995;14:3820–3827. doi: 10.1002/j.1460-2075.1995.tb00051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthony D D, Merrick W C. Analysis of 40 S and 80 S complexes with mRNA as measured by sucrose density gradients and primer extension inhibition. J Biol Chem. 1992;267:1554–1562. [PubMed] [Google Scholar]
- 3.Anthony D D, Merrick W C. Eukaryotic initiation factor (eIF)-4F. Implications for a role in internal initiation of translation. J Biol Chem. 1991;266:10218–10226. [PubMed] [Google Scholar]
- 4.Baglioni C, Simili M, Shafritz D A. Initiation activity of EMC virus RNA, binding to initiation factor eIF-4B and shut-off of host cell protein synthesis. Nature. 1978;275:240–243. doi: 10.1038/275240a0. [DOI] [PubMed] [Google Scholar]
- 5.Belsham G J. Dual initiation sites of protein synthesis on foot-and-mouth disease virus RNA are selected following internal entry and scanning of ribosomes in vivo. EMBO J. 1992;11:1105–1110. doi: 10.1002/j.1460-2075.1992.tb05150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belsham G J, Sonenberg N. RNA-protein interactions in regulation of picornavirus RNA translation. Microbiol Rev. 1996;60:499–511. doi: 10.1128/mr.60.3.499-511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borovjagin A, Pestova T, Shatsky I. Pyrimidine tract binding protein strongly stimulates in vitro encephalomyocarditis virus RNA translation at the level of preinitiation complex formation. FEBS Lett. 1994;351:299–302. doi: 10.1016/0014-5793(94)00848-5. [DOI] [PubMed] [Google Scholar]
- 8.Cao X, Bergmann I E, Füllkrug R, Beck E. Functional analysis of the two alternative translation initiation sites of foot-and-mouth disease virus. J Virol. 1995;69:560–563. doi: 10.1128/jvi.69.1.560-563.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Golini F, Thach S S, Birge C H, Safer B, Merrick W C, Thach R E. Competition between cellular and viral mRNAs in vitro is regulated by a messenger discriminatory initiation factor. Proc Natl Acad Sci USA. 1976;73:3040–3044. doi: 10.1073/pnas.73.9.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hentze M W. eIF4G: a multipurpose ribosome adapter? Science. 1997;275:500–501. doi: 10.1126/science.275.5299.500. [DOI] [PubMed] [Google Scholar]
- 11.Jackson R J, Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 12.Jang S K, Wimmer E. Cap-independent translation of encephalomyocarditis virus RNA: structural elements of the internal ribosomal entry site and involvement of a cellular 57-kD RNA-binding protein. Genes Dev. 1990;4:1560–1572. doi: 10.1101/gad.4.9.1560. [DOI] [PubMed] [Google Scholar]
- 13.Joshi B, Yan R, Rhoads R E. In vitro synthesis of human protein synthesis initiation factor 4 gamma and its localization on 43 and 48 S initiation complexes. J Biol Chem. 1994;269:2048–2055. [PubMed] [Google Scholar]
- 14.Kaminski A, Hunt S L, Patton J G, Jackson R J. Direct evidence that polypyrimidine tract binding protein (PTB) is essential for internal initiation of translation of encephalomyocarditis virus RNA. RNA. 1995;1:924–938. [PMC free article] [PubMed] [Google Scholar]
- 15.Kolupaeva V G, Pestova T V, Hellen C U, Shatsky I N. Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J Biol Chem. 1998;273:18599–18604. doi: 10.1074/jbc.273.29.18599. [DOI] [PubMed] [Google Scholar]
- 16.Le S Y, Siddiqui A, Maizel J V., Jr A common structural core in the internal ribosome entry sites of picornavirus, hepatitis C virus, and pestivirus. Virus Genes. 1996;12:135–147. doi: 10.1007/BF00572952. [DOI] [PubMed] [Google Scholar]
- 17.Luz N, Beck E. A cellular 57 kDa protein binds to two regions of the internal translation initiation site of foot-and-mouth disease virus. FEBS Lett. 1990;269:311–314. doi: 10.1016/0014-5793(90)81182-n. [DOI] [PubMed] [Google Scholar]
- 18.Martenson R E, Deibler G E, Kramer A J. Reaction of peptide 89-169 of bovine myelin basic protein with 2-(2-nitrophenylsulfenyl)-3-methyl-3′-bromoindolenine. Biochemistry. 1977;16:216–221. doi: 10.1021/bi00621a009. [DOI] [PubMed] [Google Scholar]
- 19.Méthot N, Pause A, Hershey J W, Sonenberg N. The translation initiation factor eIF-4B contains an RNA-binding region that is distinct and independent from its ribonucleoprotein consensus sequence. Mol Cell Biol. 1994;14:2307–2316. doi: 10.1128/mcb.14.4.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Méthot N, Pickett G, Keene J D, Sonenberg N. In vitro RNA selection identifies RNA ligands that specifically bind to eukaryotic translation initiation factor 4B: the role of the RNA recognition motif. RNA. 1996;2:38–50. [PMC free article] [PubMed] [Google Scholar]
- 21.Méthot N, Song M S, Sonenberg N. A region rich in aspartic acid, arginine, tyrosine, and glycine (DRYG) mediates eukaryotic initiation factor 4B (eIF4B) self-association and interaction with eIF3. Mol Cell Biol. 1996;16:5328–5334. doi: 10.1128/mcb.16.10.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer K, Petersen A, Niepmann M, Beck E. Interaction of eukaryotic initiation factor eIF-4B with a picornavirus internal translation initiation site. J Virol. 1995;69:2819–2824. doi: 10.1128/jvi.69.5.2819-2824.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson E M, Winkler M M. Regulation of mRNA entry into polysomes. Parameters affecting polysome size and the fraction of mRNA in polysomes. J Biol Chem. 1987;262:11501–11506. [PubMed] [Google Scholar]
- 24.Niepmann M. Porcine polypyrimidine tract-binding protein stimulates translation initiation at the internal ribosome entry site of foot-and-mouth-disease virus. FEBS Lett. 1996;388:39–42. doi: 10.1016/0014-5793(96)00509-1. [DOI] [PubMed] [Google Scholar]
- 25.Niepmann M, Petersen A, Meyer K, Beck E. Functional involvement of polypyrimidine tract-binding protein in translation initiation complexes with the internal ribosome entry site of foot-and-mouth disease virus. J Virol. 1997;71:8330–8339. doi: 10.1128/jvi.71.11.8330-8339.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nikodem V, Fresco J R. Protein fingerprinting by SDS-gel electrophoresis after partial fragmentation with CNBr. Anal Biochem. 1979;97:382–386. doi: 10.1016/0003-2697(79)90089-7. [DOI] [PubMed] [Google Scholar]
- 26a.Ochs, K. Unpublished data.
- 27.Pain V M. Initiation of protein synthesis in eukaryotic cells. Eur J Biochem. 1996;236:747–771. doi: 10.1111/j.1432-1033.1996.00747.x. [DOI] [PubMed] [Google Scholar]
- 28.Pause A, Méthot N, Svitkin Y, Merrick W C, Sonenberg N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 1994;13:1205–1215. doi: 10.1002/j.1460-2075.1994.tb06370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 30.Pestova T V, Hellen C U, Shatsky I N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pestova T V, Shatsky I N, Fletcher S P, Jackson R J, Hellen C U. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pestova T V, Shatsky I N, Hellen C U. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pilipenko E V, Gmyl A P, Maslova S V, Belov G A, Sinyakov A N, Huang M, Brown T D, Agol V I. Starting window, a distinct element in the cap-independent internal initiation of translation on picornaviral RNA. J Mol Biol. 1994;241:398–414. doi: 10.1006/jmbi.1994.1516. [DOI] [PubMed] [Google Scholar]
- 34.Pilipenko E V, Gmyl A P, Maslova S V, Svitkin Y V, Sinyakov A N, Agol V I. Prokaryotic-like cis elements in the cap-independent internal initiation of translation on picornavirus RNA. Cell. 1992;68:119–131. doi: 10.1016/0092-8674(92)90211-t. [DOI] [PubMed] [Google Scholar]
- 35.Rozen F, Edery I, Meerovitch K, Dever T E, Merrick W C, Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol Cell Biol. 1990;10:1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rust R C, Ochs K, Meyer K, Beck E, Niepmann M. Interaction of eukaryotic initiation factor eIF4B with the internal ribosome entry site of foot-and-mouth disease virus is independent of the polypyrimidine tract-binding protein. J Virol. 1999;73:6111–6113. doi: 10.1128/jvi.73.7.6111-6113.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 38.Scheper G C, Voorma H O, Thomas A A. Basepairing with 18S ribosomal RNA in internal initiation of translation. FEBS Lett. 1994;352:271–275. doi: 10.1016/0014-5793(94)00975-9. [DOI] [PubMed] [Google Scholar]
- 39.Thomas A, Goumans H, Amesz H, Benne R, Voorma H O. A comparison of the initiation factors of eukaryotic protein synthesis from ribosomes and from the postribosomal supernatant. Eur J Biochem. 1979;98:329–337. doi: 10.1111/j.1432-1033.1979.tb13192.x. [DOI] [PubMed] [Google Scholar]