Summary
Background
Brugada syndrome (BrS) is a cardiac channelopathy that can result in sudden cardiac death (SCD). SCN5A is the most frequent gene linked to BrS, but the genotype–phenotype correlations are not completely matched. Clinical phenotypes of a particular SCN5A variant may range from asymptomatic to SCD. Here, we used comparison of induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs) derived from a SCN5A mutation-positive (D356Y) BrS family with severely affected proband, asymptomatic mutation carriers (AMCs) and healthy controls to investigate this variation.
Methods
26 iPSC lines were generated from skin fibroblasts using nonintegrated Sendai virus. The generated iPSCs were differentiated into cardiomyocytes using a monolayer-based differentiation protocol.
Findings
D356Y iPSC-CMs exhibited increased beat interval variability, slower depolarization, cardiac arrhythmias, defects of Na+ channel function and irregular Ca2+ signaling, when compared to controls. Importantly, the phenotype severity observed in AMC iPSC-CMs was milder than that of proband iPSC-CMs, an observation exacerbated by flecainide. Interestingly, the iPSC-CMs of the proband exhibited markedly decreased Ca2+ currents in comparison with control and AMC iPSC-CMs. CRISPR/Cas9-mediated genome editing to correct D356Y in proband iPSC-CMs effectively rescued the arrhythmic phenotype and restored Na+ and Ca2+ currents. Moreover, drug screening using established BrS iPSC-CM models demonstrated that quinidine and sotalol possessed antiarrhythmic effects in an individual-dependent manner. Clinically, venous and oral administration of calcium partially reduced the malignant arrhythmic events of the proband in mid-term follow-up.
Interpretation
Patient-specific and genome-edited iPSC-CMs can recapitulate the varying phenotypic severity of BrS. Our findings suggest that preservation of the Ca2+ currents might be a compensatory mechanism to resist arrhythmogenesis in BrS AMCs.
Funding
National Key R&D Program of China (2017YFA0103700), National Natural Science Foundation of China (81922006, 81870175), Natural Science Foundation of Zhejiang Province (LD21H020001, LR15H020001), National Natural Science Foundation of China (81970269), Key Research and Development Program of Zhejiang Province (2019C03022) and Natural Science Foundation of Zhejiang Province (LY16H020002).
Keywords: iPSC-CMs, Brugada syndrome, SCN5A, Asymptomatic mutation carriers, Calcium current
Research in context.
Evidence before this study
Brugada syndrome (BrS) is an inherited cardiac arrhythmic disorder characterized by ST-segment elevation in the right precordial electrocardiogram leads and tightly associated with increased risk of sudden cardiac death (SCD). SCN5A is the most frequent gene linked to BrS, but the genotype–phenotype correlations are not completely matched. Clinical phenotypes of a particular SCN5A variant may range from asymptomatic to SCD.
Added value of this study
Patient-specific and genome-edited induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs) can recapitulate the varying phenotypic severity of BrS. Preservation of the Ca2+ currents might be a compensatory mechanism to resist arrhythmogenesis in BrS asymptomatic mutation carriers (AMCs).
Implications of all the available evidence
The present study demonstrates that iPSC-CM model can serve as an adjuvant tool for diagnosing the pathogenicity of a particular BrS variant, for functional stratification of BrS AMCs, and for screening of therapeutic drugs against specific mutations for personalized medicine. These findings could help elucidate the mechanisms underlying the varying phenotypic severity of BrS and aid in the development of novel therapeutic strategies.
Introduction
Brugada syndrome (BrS), initially described in 1992, is an inherited cardiac channelopathy characterized by a distinctive electrocardiogram (ECG) pattern of coved ST segment elevation in the right precordial leads (V1-V3) and increased risk of sudden cardiac death (SCD) due to ventricular fibrillation (VF).1,2 SCD may be the first indication that the patient is affected by BrS. The prevalence of BrS is believed to range from 1 in 5000 to 1 in 200, and it is 8–10 times more prevalent in men than women.1 BrS is a major cause of SCD in healthy young individuals with a notable impact on patients with structurally normal hearts. It is responsible for 4–12% of all SCD cases and approximately 20% of individuals with structurally normal hearts.3,4 The only proven effective therapeutic strategy for the prevention of SCD in BrS patients is limited to implantable cardioverter-defibrillator (ICD), which is invasive and life-altering therapy, especially for those of younger ages.1,5,6
At least 19 genes have been identified as BrS-associated or modifier genes, which mainly encode sodium (Na+), potassium (K+) or calcium (Ca2+) channels as well as proteins associated with these channels.7,8 SCN5A, the first gene linked to BrS, was identified in 1998 and noted to encode the alpha subunit of the cardiac sodium channel.9 It has been reported that BrS patients with SCN5A mutations exhibit increased conduction abnormalities on ECG and possess higher risk for cardiac events than those patients lacking such mutations.10 However, BrS is often genetically undetermined and the genotype–phenotype correlations are not completely matched, even for SCN5A mutation–positive BrS families.11
BrS is an autosomal dominant disease characterized by incomplete penetrance. To date, more than 300 pathogenic variants have been reported, most of which show affected SCN5A.1 However, only 30–35% of clinically diagnosed cases are genetically diagnosed and the yield of SCN5A-related variants, the most common BrS-associated gene, is less than 30%.1,9 The clinical phenotype of a particular variant in the SCN5A gene may show differing levels of penetrance among individuals even within the same family, thus complicating BrS diagnosis and risk stratification, especially for asymptomatic mutation carriers (AMCs).
The invention of induced pluripotent stem cells (iPSCs) has enabled many studies that have shed new light on areas of translational and regenerative medicine in the cardiovascular field.12, 13, 14 As iPSCs can now be robustly differentiated into cardiomyocytes, this provides the opportunity to generate patient- and disease-specific iPSC-derived cardiomyocytes (iPSC-CMs).15,16 Sharing a high homology with specific human's primary cardiomyocytes and expressing the major cardiac ion channels naturally found in the human heart, iPSC-CM models provide a greater opportunity for comprehensive assessment of ion channel function, action potential features, and characteristics of arrhythmias, than do alternative models such as animal cardiomyocyte models or heterologous transfections.17
In this study, we recruited a ten-member BrS family cohort carrying the variant D356Y in the SCN5A gene, which was not previously reported. This family included the severely affected proband, AMCs (the proband's mother and younger brother), and healthy controls (the proband's father and six other members). We first combined the iPSC-CM model with CRISPR/Cas9-mediated genome editing technology to determine the pathogenicity of the identified variant. Secondly, we investigated the variation in the phenotypic severity of this BrS variant and examined its underlying molecular mechanisms using comprehensive characterization of control, AMC and severely affected BrS iPSC-CMs. Finally, we screened the therapeutic effects of clinically antiarrhythmic drugs using the established BrS iPSC-CM models.
Methods
Patient recruitment
The proband, who was clinically diagnosed as BrS, was initially recruited for this study. Standard 12-lead ECGs were performed using a TC30 digital ECG recorder. Two higher intercostal space (ICS) ECGs were obtained by placing V1-V3 leads at the third and second ICS. Transthoracic ultrasound echocardiograms were obtained in all the participants. The proband underwent programmed electrophysiology study (PES) to assess ventricular tachycardia (VT)/VF inducibility. The stimulation protocol used was as previously reported.18 A minimum coupling interval of premature ventricular beats was set to 200 ms for S2 and S3 and to refractoriness for S4. The inducibility protocol was performed from the apex of the right ventricle and from the right ventricular outflow tract unless VT or VF had been induced at the first location.19 Mapping and catheter ablation were performed when the proband had recurrent episodes of VT/VF. Follow-up visits were scheduled for every 6 months. Skin biopsies were obtained from ten members in the family, who all signed informed consents. Considering BrS is a rare disease, we tried to enroll every possible family member irrespective of gender. We recorded the gender of the participants when collecting the ECGs and other clinical data. For those members who were shown in full pedigree but not enrolled in our study, the gender was reported by the proband. This study was approved by the Ethics Committees of the First Affiliated Hospital, Zhejiang University School of Medicine (2014-326).
Whole exome sequencing
Genomic DNA was isolated from peripheral blood with CWE9600 Automated Nucleic Acid Extraction System using CWE2100 Blood DNA Kit V2 (CWBiotech, CW2553). 750 ng genomic DNA was fragmented into 200–300 bp length by Scientz08-III Ultrasonic Homogenizer (SCIENTZ). The DNA fragments were then processed by end-repairing, A-tailing and adaptor ligation using KAPA Library Preparation Kit (Illumina, KR0453, v3.13), followed by an 8-cycle pre-capture PCR amplification. The amplified DNA sample was then captured in the Agilent SureSelect XT2 Target Enrichment System (Agilent Technologies, Inc.). Captured DNA fragments were purified by Dynabeads MyOne Streptavidin T1 (Invitrogen) and amplified by 13 cycle post-capture PCR. The final products were purified by Agencourt AMPure XP (Beckman Coulter, Inc.) and quantitated with Life Invitrogen Qubit 3.0 by Qubit dsDNA HS Assay Kit (Invitrogen). Eventually, quantified DNA was sequenced with 150-bp paired-end reads on Illumina Novaseq 6000 platform (Illumina, Inc.) according to the standard manual. The raw sequencing data produced on Novaseq platform were filtered and aligned against the human reference genome (hg19) using the BWA Aligner (http://bio-bwa.sourceforge.net/) after evaluated according to Illumina Sequence Control Software (SCS). The single-nucleotide polymorphisms (SNPs) were called by using GATK software (Genome Analysis ToolKit) (www.broadinstitute.org/gatk). Variants were annotated using ANNOVAR (annovar.openbioinformatics.org/en/latest/). The pathogenic level of SNP loci were evaluated according to ACMG guideline.20
Skin biopsies and maintenance of fibroblasts
Freshly isolated skin biopsies were rinsed with Dulbecco's Phosphate-Buffered Saline (DPBS) (Gibco, C14190500BT) and transferred into a 1.5-mL tube. Tissue was minced in collagenase I (1 mg/mL in Dulbecco's modified Eagle medium (DMEM), Gibco, C11995500BT) and allowed to digest for 6 h at 37 °C. Dissociated dermal fibroblasts were plated and maintained with DMEM containing 10% FBS (Gibco, 10091148), 100 U/mL Penicillin and 100 μg/mL Streptomycin (Gibco, 15140122) at 37 °C, 95% air and 5% CO2 in a humidified incubator. All cells were used for reprogramming within 5 passages.
Generation of iPSC lines
Somatic reprogramming was used to generate iPSC lines from skin fibroblasts using CytoTune-iPS 2.0 Sendai Reprogramming Kit following the manufacturer's instructions (Invitrogen, A16517).
Culture and maintenance of iPSCs
The iPSCs were cultured in feeder-free mTeSR (STEMCELL Technologies, 85850) media on matrigel-coated (Corning, 354277) plates at 37 °C with 5% (vol/vol) CO2. The media were daily changed and cells were passaged every 3–4 days using Accutase (STEMCELL Technologies, 07920). All cultures were routinely tested for mycoplasma.
Genetic screening and analysis at the iPSC level
The iPSCs were cultured on Matrigel-coated 6-well plates with mTeSR media, and harvested at 80–90% confluence for subsequent analysis. Genomic DNA was extracted using a commercial DNA isolation kit (TIANGEN, DP304-03). Polymerase chain reaction (PCR) was carried out on EasyCycler 96 (Analytik Jena). SNP loci within SCN5A gene (c.1066G > T/p.D356Y) was amplified and analyzed by direct sequencing, and then confirmed by sub-cloning. The forward and primer reverse sequences for SCN5A are 5′-AGCACGAACAAAGTCACGGA-3' (forward) and 5′-CCAGGGGTTCAAATAGCCAT-3' (reverse), respectively.
Karyotyping
Chromosome preparation and karyotype analysis were performed according to standard protocols by the Department of Prenatal Diagnosis (Screening) Center of Hangzhou Women's Hospital (Hangzhou Maternity and Child Health Care Hospital). G-band staining was applied for the preparation of the chromosome specimens. At least 30 metaphases were counted and 6 metaphases were karyotyped for each iPSC line.
Alkaline phosphatase staining
Alkaline phosphatase (ALP) staining was performed using the VECTOR Blue Alkaline Phosphatase Substrate Kit (Vector Laboratories, SK-5300) following the manufacturer's instructions.
Cardiac differentiation
The iPSC-CMs were generated using a 2D monolayer differentiation protocol as previously described.21,22 Briefly, ∼105 undifferentiated cells were dissociated and re-plated into matrigel-coated 6-well plates. Cells were cultured and expanded to 85% cell confluence, and then treated for 2 days with 6 μM CHIR99021 (Axon Medchem, 1386) in RPMI 1640 (Gibco, C11875500BT) with B27 supplement minus insulin (Gibco, A1895601, USA) (RPMI + B27-Insulin) to activate Wnt signaling pathway. On day 2, cells were placed in RPMI + B27-Insulin with CHIR99021 removal. On days 3–4, cells were treated with 5 μM IWR-1 (Sigma–Aldrich, 681669) to inhibit Wnt signaling pathway. On days 5–6, cells were removed from IWR-1 treatment and placed in RPMI + B27-Insulin. From day 7 onwards, cells were placed and cultured in RPMI 1640 and B27 supplement with insulin (Gibco, 17504044) (RPMI + B27+Insulin) until beating was observed. Cells were glucose-starved for 3 days with RPMI + B27+Insulin for the purification.23 Cardiomyocytes of day 30–40 after cardiac differentiation were utilized for downstream functional assays.
Immunofluorescent staining
Cells were fixed with 4% Paraformaldehyde (PFA) (Maokang Biotechnology, MM1504) for 15 min, permeabilized with 0.1% Triton X (Sangon Biotech, A110694) for 5 min, and blocked with 3% BSA (Sigma–Aldrich, A1933) for 1 h. Cells were subsequently stained with appropriate primary antibodies and AlexaFluor conjugated secondary antibodies including AlexaFluor® 647 (Abcam, ab150079, 1:500, RRID: AB_2722623), AlexaFluor® 594 (Abcam, ab150080, 1:500, RRID: ab150080; Abcam, ab150108, 1:500, RRID: AB_2732073) and AlexaFluor® 488 (Abcam, ab150113, 1:500, RRID: AB_2576208; Invitrogen, A11008, 1:500, RRID: AB_143165). Nuclei were stained with DAPI (Roche Diagnostics). For the staining of pluripotency markers, the primary antibodies were a goat anti-human OCT4 (Cell Signaling Technology, 2750S, 1:200, RRID: AB_823583), a rabbit anti-human NANOG (Santa Cruz Biotechnology, sc-33759, 1:200, RRID: AB_2150401), a mouse anti-human SSEA-4 (Abcam, ab16287, 1:200, RRID: AB_778073) and a rabbit anti-human SOX2 (Abcam, ab171380, 1:200, RRID: AB_2732072). For the staining of cardiac-specific markers, the primary antibodies were a mouse anti-human TNNT2 (Abcam, ab8295, 1:500, RRID: AB_306445) and a rabbit anti-human α-actinin (Cell Signaling Technology, 6487P, 1:100, RRID: AB_11179206). Pictures were taken with 60 × objective on confocal microscope (Nikon, A1) using NIS-Elements AR software (Nikon).
Patch clamp recordings from iPSC-CMs
The monolayer iPSC-CMs were mechanically and enzymatically dissociated to obtain single cells, which were seeded on matrigel-coated glass coverslips (Warner Instruments). Cells with spontaneous beatings were selected and action potentials were recorded using an EPC-10 patch clamp amplifier (HEKA). Continuous extracellular solution perfusion was achieved using a rapid solution exchanger (Bio-logic Science Instruments). Data were acquired using PatchMaster software (HEKA) and digitized at 1 kHz. Data analyses were performed using Igor Pro (Wavemetrics) and Prism (Graphpad). A TC-344B heating system (Warner Instruments) was used to maintain the temperature at 35.5–37 °C. Tyrodes solution was used as the external solution containing 140 mM NaCl, 5.4 mM KCl, 1 mM MgCl2, 10 mM glucose, 1.8 mM CaCl2 and 10 mM HEPES (pH 7.4 with NaOH). The internal solution contained 120 mM KCl, 1 mM MgCl2, 10 mM HEPES, 3 mM Mg-ATP, and 10 mM EGTA (pH 7.2 with KOH). Key action potential parameters were quantified, including maximal diastolic potential (MDP), overshoot, action potential amplitude (APA), action potential duration at 90% repolarization (APD90), maximal upstroke velocity (Vmax) and beating rate. Ventricular-like iPSC-CMs were distinguished based on the action potential morphology and action potential parameters, which exhibit a clear plateau phase, larger APA and Vmax values, more negative MDP values, APD30–40/APD70–80> 1.5 and APD90/APD50≤ 1.3.
Different ionic currents were recorded from single iPSC-CMs using the whole-cell patch clamp technique with conventional voltage clamp protocols. Na+ current was recorded at 35–37 °C. The bath solution contained: 50 mM NaCl, 110 mM CsCl, 1.8 mM CaCl2, 1 mM MgCl2, 10 mM glucose, 10 mM HEPES and 0.001 mM Nifedipine (pH 7.4 with CsOH). Pipette solutions contained: 10 mM NaCl, 135 mM CsCl, 2 mM CaCl2, 5 mM MgATP, 5 mM EGTA, and 10 mM HEPES (pH 7.2 with CsOH). Currents was elicited by a 40-ms long depolarizing test pulse from −80 to 60 mV in 5-mV increments at holding potential of −80 mV. To measure steady-state inactivation, a pre-pulse applying the inactivation voltage from −110 to 0 mV in steps of 10-mV for 400 ms followed by 40-ms long pulses of 0 mV to elicit current. Ca2+ current was recorded at room temperature, and the bath solution contained: 160 mM TEA-Cl, 5 mM CaCl2, 1 mM MgCl2, 10 mM glucose, 10 mM HEPES, 0.01 mM TTX, 2 mM 4-AP (pH 7.4 with CsOH). Pipette solutions contained: 145 mM CsCl, 5 mM NaCl, 1 mM CaCl2, 5 mM MgATP, 5 mM EGTA, and 10 mM HEPES (pH 7.2 with CsOH). With a holding potential of −80 mV, a 3-s pre-pulse was applied to −50 mV to voltage inactivate Na+ and T-type Ca2+ channels then applying a 100-ms long test pulse from −60 to 60 mV in 10-mV increments to elicit current. To measure steady-state inactivation, a pre-pulse to between −50 and −10 mV in step of 5-mV for 3 s followed by 400-ms long pulses to 0 mV to activate Ca2+ current. All currents were normalized to cell capacitance to obtain current density. Steady-state activation/inactivation curves were fitted by using a Boltzmann equation: I/Imax = A/{1.0 + exp [(V1/2 - V)/k]}, in which V1/2 is half-maximum activation potential and k is slope factor.
Multi-electrode array (MEA) recordings
For cell preparation, a 20 μl droplet of coating solution (Matrigel) was applied on the area of the electrodes of 1-well or 6-well MEA probes (60MEA200/30iR-Ti-gr or 60-6well MEA200/30iR-Ti-tcr, Multi Channel Systems), which were incubated at 37 °C in 5% CO2 for at least 1 h. The iPSC-CMs were then dissociated from the 6-well plates using TrypLE, then reseeded onto recording wells at a density of 1–1.5 × 105 cells in 20 μl bead of the cell suspension per well. After incubation with 5% CO2 at 37 °C for 2 h to promote adhesion, each well was filled with culture media to a final volume (1-well was 1 mL and 6-well was 500 μl). Culture medium was changed after 48 h, afterwards be sure to exchange medium every two or three days throughout 5–7 days of culturing period. Field potentials were recorded from spontaneously beating iPSC-CMs using the MEA2100 data acquisition system (Multi Channel Systems) with sampling at 10 kHz. All experiments were performed at 37 °C and began after a 20-min equilibration period. The data were analyzed with Cardio 2D+ software (Multi Channel Systems). Coefficient of variation (CV) was used to quantify rhythm irregularity for beat intervals.
Ca2+ imaging using Fluo-4 AM
The iPSC-CMs grown on coverslips were loaded with RPMI 1640 medium without Phenol Red (Gibco, 11835030) supplemented with 5 μM Fluo-4 AM (Invitrogen, F14201) for 15 min in the dark at room temperature. After washing with pre-warmed DPBS and RPMI 1640 two times, the cells were then immersed in imaging buffer for 10 min before imaging experiments. For imaging, cells were placed in a chamber equipped with a temperature-controller under constant perfusion of 37 °C imaging buffer. Calcium signaling was made by recording the fluorescence of cells using an Ultra High Speed Wavelength Switcher (Lambda DG-4, Sutter Instruments) with a CCD camera (Zyla, Andor) mounted on an inverted microscope (Eclipse Ti, Nikon). Data were acquired using NIS-Elements software (Nikon) and were quantified as the extracellular background signal subtracted fluorescence intensity (F) and then normalized to the baseline fluorescence (F0). Transient amplitude was presented as a relative scale (ΔF/F0).
Ca2+ imaging using Fura-2 AM
The iPSC-CMs grown on coverslips were loaded with RPMI 1640 medium without phenol red (Gibco, 11835030) supplemented with 5 μM Fura-2 AM (Invitrogen, F1221) for 30 min in the dark at room temperature. After washing with pre-warmed DPBS and RPMI 1640 for two times, before experiment the cells were immersed in imaging buffer for 30 min. Ca2+ signaling was made by recording the fluorescence of cells using an Ultra High Speed Wavelength Switcher (Lambda DG-4, Sutter Instruments) with a CCD camera (Zyla, Andor) mounted on an inverted microscope (Eclipse Ti, Nikon). Data were acquired using NIS-Elements software (Nikon). Fluorescent signals were obtained upon excitation at 340 nm (F340) and 380 nm (F380). Amplitude of Ca2+ transient is defined as the ratio of F340/F380.
Assessment of sarcoplasmic reticulum Ca2+ load and NCX activity
The iPSC-CMs were incubated with 5 μM Ca2+ sensitive dye Fluo-4 AM (ThermoFisher Scientific) for 15 min at room temperature in phenol red-free RPMI 1640. Following incubation, the indicator-containing medium was replaced with phenol red-free RPMI 1640 for an additional 30 min at room temperature to allow de-esterification of the indicator. The iPSC-CMs were next transferred to a chamber with a pair of parallel electrodes mounted an epifluorescence microscope and field-stimulated at 0.5 Hz in normal Tyrode's (NT) solution (140 mM NaCl, 5.4 mM KCl, 1.0 mM MgCl2, 5 mM HEPES, 1.8 mM CaCl2, 10 mM glucose, pH 7.4) until a steady-state Ca2+ transient was observed. Steady-state sarcoplasmic reticulum (SR) Ca2+ content was estimated by rapid application of 10 mM caffeine after pacing. NCX activity (1/τ) was calculated during caffeine application as the inverse of the rate of decrease of cytosolic Ca2+ (τ).
RNA sequencing
Total RNA was extracted from the tissue using TRIzol® Reagent according the manufacturer's instructions (Invitrogen) and genomic DNA was removed using DNase I (TaKara). RNA sequencing (RNA-Seq) transcriptome libraries were prepared following TruSeqTM RNA sample preparation Kit from Illumina (San Diego, CA), using 1 μg of total RNA. Shortly, messenger RNA was isolated with polyA selection by oligo (dT) beads and fragmented using fragmentation buffer. cDNA synthesis, end repair, A-base addition and ligation of the Illumina-indexed adaptors were performed according to Illumina's protocol. Libraries were then size selected for cDNA target fragments of 200–300 bp on 2% Low Range Ultra Agarose followed by PCR amplified using Phusion DNA polymerase (NEB) for 15 PCR cycles. After quantified by TBS380, Paired-end libraries were sequenced with the Illumina HiSeq PE 2 × 151bp read length. The raw paired end reads were trimmed and quality controlled by Trimmomatic with default parameters (http://www.usadellab.org/cms/uploads/supplementary/Trimmomatic). Clean reads were then separately aligned to human reference genome with orientation mode using tophat software (http://tophat.cbcb.umd.edu/). Identification of DEGs (differential expression genes) between different samples was performed by using DEseq2 software (https://bioconductor.org/packages/release/bioc/html/DESeq2.html). A hierarchical clustering was conducted based on gene expression using pheatmap package (https://www.rdocumentation.org/packages/pheatmap/versions/1.0.12/topics/pheatmap). Gene Ontology (GO) functional enrichment was conducted upon the DEGs using clusterProfiler package (bioconductor.org/packages/release/bioc/html/clusterProfiler.html). The raw sequencing data of the transcriptome have been deposited to NCBI SRA database with the accession number PRJNA985079.
Western blot
The iPSC-CMs in a well of 6-well plate were detached with Trypsin–EDTA (Gibco, 25200056), and then pelleted at 300 g for 3–5 min at 4 °C. After washing with DPBS, the pellets were re-suspended in 50–100 μl lysis buffer. Lysates were placed on ice for 30 min and the supernatants were collected after centrifuging at 12,000 rpm for 5 min. Protein concentration was measured using a BCA kit (Pierce, 23227). Western blot was performed with the following antibodies: SERCA2a (Santa Cruz Biotechnology, sc-53010, 1:200, RRID: AB_630230), RYR2 (abcam, ab2868, 1:500, RRID: AB_2183051), sodium/calcium exchanger 1 (NCX1) (ProteinTech, 55075-1-AP, 1:500, RRID: AB_2881262), Cav 1.2 (ProteinTech, 21774-1-AP, 1:2000, RRID: 21774-1-AP), total phospholamban (PLN) (Cell Signaling Technology, 14562S, 1:1000, RRID: AB_2798511), phosphorylated PLN (Cell Signaling Technology, 8496S, 1:1000, RRID: AB_10949102), Nav1.5 (Alomone Labs, ASC-005, 1:500, RRID: AB_2040001), and GAPDH (Abmart, M20006, 1:5000, RRID: AB_2737054). Intensity values for each band were determined as the integrated density (sum of pixel values) within a fixed area using Quantity One software (Biorad).
Surface expression of Nav1.5
HEK293 cells transiently transfected with WT or mutant Nav1.5 were washed with DPBS at pH 7.4. Membrane proteins were biotinylated by incubating cells with 2.5 mg/mL of EZ-LinkTM Sulfo-NHS-SS-Biotin (Thermo Scientific, 21331) in DPBS for 30 min at 4 °C. Cells were washed 3 times with 100 mM glycine in DPBS, then washed with DPBS containing 20 mM glycine, and scrapped in Triton X-100 lysis buffer (1% Triton X-100, 50 mM Tris/HCl pH 7.4, 150 mM NaCl, 1 mM EDTA and Complete Protease Inhibitor Cocktail (Roche, 04693116001)). Lysates were obtained after 1 h rotating at 4 °C. Insoluble materials were removed by centrifugation. After protein BCA quantification, 10% of supernatants were mixed with SDS-PAGE loading buffer and heated for 5 min at 70 °C (total protein). The others were incubated with High Capacity Neutravidin Agarose Resin (Thermo Scientific, 29202) overnight at 4 °C. The beads were precipitated and washed with Triton X-100 lysis buffer for 5 times. Precipitated beads were re-suspended in SDS-PAGE loading buffer and heated for 5 min at 70 °C (membrane protein). Total and surface expression was quantified by Western blot with a rabbit anti-human Nav1.5 antibody (Alomone Labs, ASC-005, 1:500, RRID: AB_2040001) and a rabbit antibody against Na+/K+-ATPase (Abcam, ab7671, 1:500, RRID: AB_306023), which was used as a membrane protein loading control.
Gene correction of SCN5A D356Y mutation by CRISPR/Cas9
The genomic sequence of SCN5A D356Y was selected for guide RNA (gRNA) design and its design was according to the CRISPR online design tool: http://crispr.mit.edu/. The sequences of a pair of oligos for gRNA synthesis are listed as followed: Forward, 5′-CACCGGCAGGCGAGAACCCCGACCA-3’; Reverse, 5′-AAACTGGTCGGGGTTCTCGCCTGCC-3’. The oligos were annealed and ligated into a linearized lentiCRISPRv2 vector for generating a CRISPR plasmid as previously described. Correction of D356Y mutation in BrS proband iPSCs via homologous recombination with a single-stranded oligodeoxynucleotide (ssODN) was carried out as previously described with slight modifications. The sequence of ssODN used to repair mutant allele are listed as followed: 5′-CTGTGTGTGGCTGCAGGACATGTCCGGAGGGCTACCGGTGCCTAAAGGCCGGCGAAAATCCAGATCACGGCTACACTAGTTTCGATTCCTTTGCCTGGGCCTTTCTTGCACTCTTCCGCCTGATG-3’. In brief, BrS proband iPSCs were dissociated with Accutase and seeded on matrigel with mTeSR medium containing ROCK inhibitor Y-27632 (Selleck, S1049) overnight. For Lipofectamine transfection, Opti-Mem (Gibco, 31985062) containing ssODNs, CRISPR plasmids and Lipofectamine 3000 was added into the culture medium of the iPSCs. One day after transfection, medium was removed and replaced with mTeSR containing puromycin. After additional 2 days, medium was changed for mTeSR without puromycin. One week later, puromycin-resistant clones were picked and then verified by genomic PCR and DNA sequencing. Primers used for genomic PCR are listed as followed: Forward, 5′-AGCACGAACAAAGTCACGGA-3’; Reverse, 5′-CCAGGGGTTCAAATAGCCAT-3’. The positive iPSC clones were expanded and used for further experiments.
Compounds and solutions
All the chemicals used in the electrophysiological experiments were purchased from Sigma–Aldrich. Nifedipine was purchased from Sigma–Aldrich, and stock solutions were prepared in 1 mM in DMSO. 4-aminopyridine (4-AP) was purchased from Sigma–Aldrich, and stock solutions were prepared in 1 M in distilled water. Tetrodotoxin (TTX) was purchased from Absin, and stock solutions were prepared in 10 mM in distilled water. The solutions of caffeine (Sigma–Aldrich, C0750) were freshly prepared when using. Quinidine and verapamil were purchased from Sigma–Aldrich, and stock solutions were both prepared in 10 mM in DMSO. Sotalol and cilostazol were purchased from Selleck, and stock solutions were both prepared in 10 mM in DMSO. Bepridil was purchased from Tocris, and stock solutions were prepared in 1 mM in DMSO.
Statistical analysis
Statistical significance was determined by unpaired two-tailed Student's t-tests to compare two groups and by One-way ANOVA to compare multiple groups. Data were shown as mean ± sem and analyzed using GraphPad Prism 6 (GraphPad Software). More Methods are provided in the Supplementary Material.
Role of funders
The funders had no role in study design, data collection, data analysis, interpretation, writing of report, or in the decision to submit the paper.
Results
Clinical characteristics of the BrS family
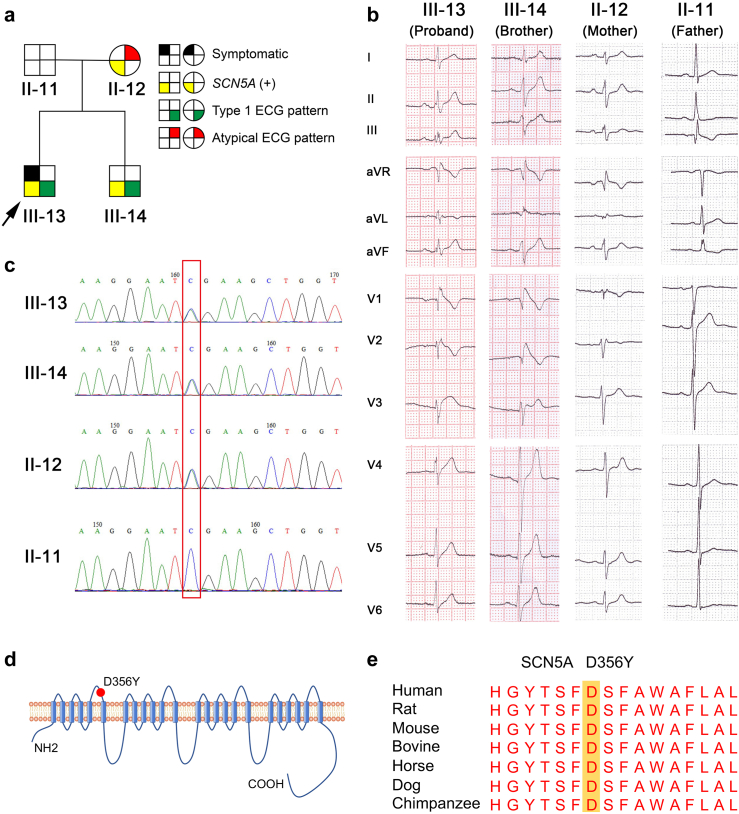
The proband presented to local hospital with recurrent syncope and seizure at 20 years old (Fig. 1a and Supplementary Table S1). He was initially misdiagnosed as an epileptic and no treatment was prescribed. The next year, the proband had a syncope episode and VF was recorded in the ambulance. He was then diagnosed with BrS according to the history of unexplained syncope and typical type 1 Brugada wave pattern on the second ICS ECG (Fig. 1a and Supplementary Figure S1 and S2), and a single-chamber ICD was implanted in 2008. The ICD became depleted three times due to recurrent shocks and the ICD generators were replaced in 2012, 2016 and 2020. PES and catheter ablation procedures were also performed in 2013, 2015 and 2020. The younger brother of the proband showed typical type 1 Brugada pattern on the second and third ICS ECG, but remained completely asymptomatic over the 6-year follow-up (Fig. 1a and b and Supplementary Table S1). The parents of the proband were otherwise healthy except the mother showing a minimally abnormal ECG pattern in the second ICS ECG (Fig. 1a and b and Supplementary Table S1). Other family members were all healthy and showed normal ECGs (Supplementary Figure S1–S3). Upon genetic screening, the proband was found to have a missense variant (c.1066G > T/p.D356Y) in the SCN5A gene (Fig. 1c). The variant was found to be located in the pore loop region of domain I and was noted as highly conserved among multiple species when cross referenced with GERP software (http://mendel.stanford.edu/sidowlab/downloads/gerp/index.html) (Fig. 1d and e). Results of analysis using in silico prediction algorithms, SIFT (http://sift.jcvi.org) and PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2), indicated the variation as ‘damaging’. The variant was also highlighted as not previously reported and classified as “likely pathogenic” or “variant of undetermined significance” according to the American College of Medical Genetics and Genomics (ACMG) guidelines.20 The mother and brother of the proband were confirmed to carry the same mutation by Sanger sequencing, but the mutation was confirmed as absent in the father and other family members (Fig. 1C and Supplementary Figure S4).
Fig. 1.
Clinical characteristics of a BrS family. a. The major pedigree of the BrS family recruited in this study. The arrow indicates the proband. Females are denoted by circles, and males are denoted by squares. Black section indicates arrhythmias such as ventricular tachycardia or fibrillation related to BrS. Yellow section indicates the presence of D356Y variant in SCN5A gene. Green section indicates the presence of type 1 BrS ECG pattern. Red section indicates the presence of minimal changes on precordial leads. b. The representative ECGs from the proband (III-13), proband's brother (III-14), mother (II-12) and father (II-11). c. Sanger verification of missense variant D356Y in SCN5A gene identified by whole exome sequencing in the pedigree. d. Schematic representation of the SCN5A-encoded sodium channel (Nav1.5). D356Y variant locates at domain I pore loop indicated by the red solid circle. e. Amino acid sequence conservation of SCN5A gene from various species in the region surrounding amino acid 356 (highlighted).
Generation and characterization of iPSC-CM lines in the BrS family
Skin biopsies were obtained from the severely affected proband (III-13) and the proband's unaffected father (II-11), asymptomatic mother (II-12), brother (III-14), and six other healthy control members (II-16, II-17, II-22, III-19, III-23 and III-25). Skin fibroblasts were cultured and expanded (Supplementary Figure S5a), and iPSCs were generated from primary fibroblasts using non-integrated Sendai-viral transduction of reprogramming factors (Supplementary Table S2). The generated iPSCs lines exhibited typical human embryonic stem cell morphology (Supplementary Figure S5b), showed alkaline phosphatase (ALP) activity (Supplementary Figure S5c), stained positive for pluripotent markers (Supplementary Figure S6), and showed normal karyotypes (Supplementary Figure S7). Moreover, genetic screening confirmed the presence of SCN5A mutation D356Y in iPSCs of the proband and his brother and mother, but absence in iPSCs of his father and other healthy controls (Supplementary Figure S8). Different iPSCs were cultured and passaged until passage 20 and were subsequently differentiated into cardiomyocytes using a small molecule-based differentiation protocol, which can give rise to a purification of >90% through FACS analysis (Supplementary Figure S9). Spontaneously contracting foci began to appear after 10–14 days of cardiac differentiation and the generated iPSC-CMs demonstrated positive staining of the cardiac specific markers TNNT2 and α-actinin (Supplementary Figure S10). We observed no significant morphological changes between different iPSC-CM lines. Patch clamp recordings revealed that majority of the generated iPSC-CMs were ventricular-like myocytes (Supplementary Figure S11). We further performed RNA-Seq using samples of different iPSC-CMs. A hierarchical clustering showed that the global expression profiles were overall separated between healthy control (father, III-23, III-25) and BrS (proband, brother, mother) iPSC-CMs (Supplementary Figure S12a). There were 210 genes reached the criteria of DEGs (Supplementary Figure S12b). Through functional enrichment of these DEGs, several GO terms were identified to relate with ion channel activity (Supplementary Figure S12c).
Arrhythmic phenotype in D356Y iPSC-CMs
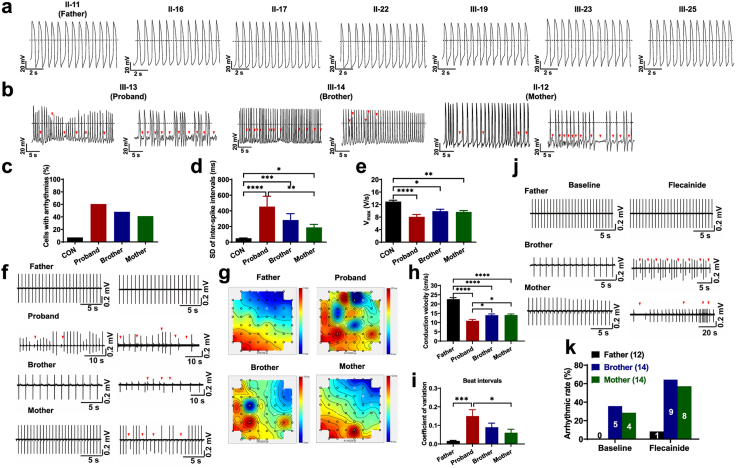
Clinically, cardiac electrical dysfunction is a hallmark of patients with BrS who experience severe arrhythmias and exhibit abnormal ECGs.1 To investigate if such arrhythmic predisposition could be observable in vitro, we conducted single-cell patch clamp recordings to characterize the action potential profile of iPSC-CMs from different individuals in the same BrS family affected by SCN5A D356Y. Ventricular-like myocytes were studied and key action potential parameters were quantified (Supplementary Figure S13 and Supplementary Table S3). The data collected from iPSC-CMs derived from the seven different controls (father and six other control members) were averaged as “CON”. We observed that the iPSC-CMs from seven healthy control subjects exhibited normal action potential profiles (Fig. 2a). However, a large subfraction of proband iPSC-CMs was seen to exhibit arrhythmic waveforms, when compared to CON iPSC-CMs (CON: 7.2%; proband: 60.5%) (Fig. 2b and c). Surprisingly, the arrhythmic phenotype was also noted in the iPSC-CMs from the asymptomatic brother and mother, but at a lower level than in proband iPSC-CMs (brother: 47.9%; mother: 41.2%) (Fig. 2b and c). Notably, we observed markedly increased inter-spike interval variability and reduced maximal upstroke velocity (Vmax) in proband, brother and mother iPSC-CMs, when compared to CON iPSC-CMs (Fig. 2d and e). Moreover, we observed significantly depolarized maximal diastolic potential (MDP) and decreased action potential amplitude (APA) in the iPSC-CMs of the proband and his brother, but not in those of the mother (Supplementary Figure S13a and b). The action potential duration at 90% repolarization (APD90) and beating rate were comparable between CON and BrS iPSC-CMs (Supplementary Figure S13c and d).
Fig. 2.
Arrhythmic phenotype in D356Y iPSC-CMs. a–b. Representative action potential tracings recorded from control (proband’ father (father) and other six healthy family members) and BrS (proband, proband's brother (brother), proband's mother (mother)) iPSC-CMs. Red arrows indicate arrhythmias. Dash lines indicate 0 mV. c–e. Bar graphs to compare arrhythmic incidence, SD of inter-spike intervals (ISIs), and Vmax between CON, proband, brother and mother iPSC-CMs by One-way ANOVA (Tukey method). Reduced Vmax indicates slower depolarization. n = 20–138. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 and ∗∗∗∗P < 0.0001. f. Representative field potential (FP) tracings recorded from father, proband, brother and mother iPSC-CMs. Red arrows indicate arrhythmogenic activities. g. Representative conduction heatmaps of father, proband, brother and mother monolayer iPSC-CMs. Blue color represents the start of the heartbeat and different colors indicate the propagation delay times over the MEA. Black lines indicate the isochrones. h–i. Bar graphs to compare conduction velocity and normalized SD of inter-beat intervals between different groups in f by One-way ANOVA (Tukey method). n = 9–15. ∗P < 0.05, ∗∗∗P < 0.001 and ∗∗∗∗P < 0.0001. j. Representative FP tracings recorded from father, brother and mother iPSC-CMs before and after acute treatment of 1 μM flecainide. Red arrows indicate flecainide-induced arrhythmogenic activities. k. Bar graph to compare the percentage of cells with arrhythmias between different groups in j.
We also recorded extracellular field potentials from monolayer iPSC-CMs by MEA and identified the arrhythmic phenotype of BrS iPSC-CMs at a multicellular level. The presence of arrhythmogenic activities was observed in 10 of 15 independent MEA studies of the iPSC-CMs from the proband (66.7%) but 0 of 12 in those of the father (Fig. 2f). Consistently, a milder arrhythmic phenotype was noted in studies of the iPSC-CMs of the brother and mother, demonstrating incidence of 35.7% (5 of 14 independent MEA studies) and 28.6% (4 of 14 independent MEA studies), respectively (Fig. 2f). Conduction velocity was significantly lower in the iPSC-CMs of the proband, brother and mother in comparison to that of the father (Fig. 2g and h). Moreover, the coefficient of variation (CV) of beat intervals was significantly higher in the iPSC-CMs of the proband as compared to those of the father, indicating rhythmic irregularity (Fig. 2i).
It has been previously been shown that testing using the Na+ channel blocker flecainide is a valuable tool in diagnosing SCN5A-related BrS.24 We therefore sought to evaluate whether flecainide stimulation may help uncover the nature and cause of the arrhythmic phenotype in AMC iPSC-CMs in vitro. For this, when rhythmic and nonarrhythmic recordings of brother and mother iPSC-CMs were observed at baseline levels, acute stimulation of these cells with flecainide (1 μM) was then applied. The presence of arrhythmias was observed in the majority studied iPSC-CMs of the brother and mother upon flecainide stimulation (brother: 9 of 14 independent MEA studies; mother: 8 of 14 independent MEA studies) (Fig. 2j and k). Conversely, the iPSC-CMs of the father were more tolerant to flecainide stimulation where flecainide-induced arrhythmogenic activities were only observed in 1 of 12 of the independent MEA studies (Fig. 2j and k). Taken together, these results demonstrate that D356Y iPSC-CMs exhibit an arrhythmic burden and abnormal electrophysiological profile, but that the phenotype shows less severity in AMC iPSC-CMs than in those of the severely affected proband.
Defect of Na+ channel function in D356Y iPSC-CMs
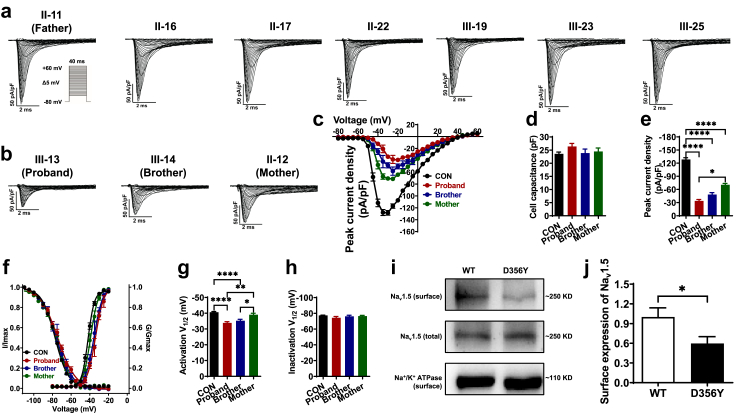
To assess if SCN5A D356Y may result in Na + channel dysfunction, we performed voltage clamp recordings to isolate Na+ currents from iPSC-CMs at the single-cell level (Fig. 3a–c and Supplementary Table S4). No significant differences were observed in cell capacitance (20–30 pF), indicating that differences in sodium currents were not due to differences in cell size (Fig. 3d). Importantly, the iPSC-CMs from the proband and his brother showed dramatically lower Na+ current density as compared to CON iPSC-CMs. However, the lower Na+ current density was less evident in the iPSC-CMs of the mother (CON: 128.5 ± 3.9 pA/pF; proband: 33.9 ± 3.2 pA/pF; brother: 48.5 ± 4.4 pA/pF; mother: 70.6 ± 3.2 pA/pF) (Fig. 3e). Accordingly, proband and brother iPSC-CMs exhibited a significant positive shift in steady-state activation (SSA) by 5–6 mV, while the shift of SSA curve in the iPSC-CMs of the mother were milder (CON: −40.6 ± 0.5 mV; proband: −33.9 ± 0.8 mV; brother: −35.2 ± 0.9 mV; mother: −39.1 ± 0.9 mV) (Fig. 3f and g). The steady-state inactivation curves were comparable between CON and BrS iPSC-CMs (Fig. 3f and h). To assess if the reduction of Na+ current density could be due to altered expression of the Na+ channel protein (Nav1.5), we employed a cell surface protein biotinylation assay in HEK293 cells expressing WT or D356Y channels (Fig. 3i). We observed that the surface expression of Nav1.5 was significantly lower in cells expressing the D356Y channel, compared to that of their WT counterparts (Fig. 3j). These results point to a defect of Na+ channel function in D356Y iPSC-CMs, but that the phenotypic severity of this is milder in the iPSC-CMs of the asymptomatic mother.
Fig. 3.
Defect of Na+channel function in D356Y iPSC-CMs. a–b. Representative Na+ current tracings isolated from control, BrS or AMC iPSC-CMs. c. Comparison of Na+ current–voltage relationship curve (IV curve) between CON, proband, brother and mother iPSC-CMs. d. Bar graph to compare the cell capacitance between CON, proband, brother and mother iPSC-CMs by One-way ANOVA (Tukey method). e. Bar graph to compare the peak Na+ current density at −30 mV between CON, proband, brother and mother iPSC-CMs by One-way ANOVA (Tukey method). n = 14–102. ∗P < 0.05 and ∗∗∗∗P < 0.0001. f. Comparison of steady-state activation and inactivation of Na+ current between CON, proband, brother and mother iPSC-CMs. g–h. Bar graphs to compare the V1/2 of steady-state activation or inactivation between CON, proband, brother and mother iPSC-CMs by One-way ANOVA (Tukey method). n = 12–102. ∗P < 0.05, ∗∗P < 0.01 and ∗∗∗∗P < 0.0001. i. Western blot analysis of total and membrane expression of WT and D356Y Nav1.5 transiently transfected in HEK293 cells. Na+/K+-ATPase is used for the loading control. j. Bar graph to compare the normalized surface expression of Nav1.5 between WT and D356Y by unpaired two-tailed Student's t-test. n = 6. ∗P < 0.05.
Irregular Ca2+ signaling in D356Y iPSC-CMs
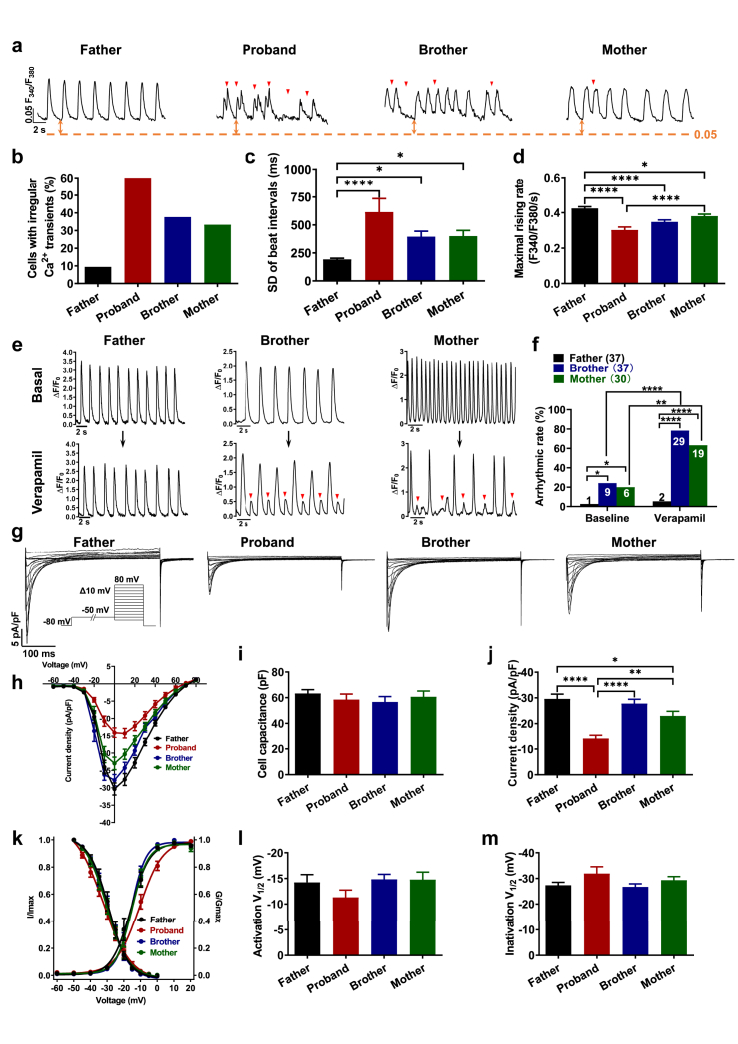
Ca2+ plays a key role in normal cardiac function, in particular, excitation-contraction coupling and normal electric rhythms, the abnormal alteration of which can lead to various types of cardiac arrhythmias including BrS.25, 26, 27 We therefore performed Ca2+ imaging using fura-2 AM dye to ratiometrically record Ca2+ transients in iPSC-CMs (Supplementary Table S5). The iPSC-CMs of the father exhibited a normal Ca2+ transient profile (Fig. 4a and b and Supplementary Figure S14a). By contrast, irregular Ca2+ transients were identified in the iPSC-CMs of the proband and his brother and mother, mirroring the arrhythmic phenotype seen in action potential studies (Fig. 4a and b and Supplementary Figure S14a). We also observed significantly increased beat interval variation and reduced maximal rising rate in the iPSC-CMs of the proband, brother and mother, whereas the diastolic intracellular Ca2+ [Ca2+]i and decay 90 remained unchanged (Fig. 4c and d and Supplementary Figure S14b and c). Ca2+ amplitude and peak Ca2+ were both significantly decreased in proband iPSC-CMs but not in the iPSC-CMs of the brother and mother (Supplementary Figure S14d and e). We further assessed the SR Ca2+ load by addition of a high-concentration caffeine (10 mM) in iPSC-CMs (Supplementary Figure S15a). Proband iPSC-CMs showed a significantly larger SR Ca2+ load than father iPSC-CMs (Supplementary Figure S15b). Notably, the NCX activity, indicated by the time constant of Ca2+ removal, was greatly decreased in proband iPSC-CMs as compared to father, brother and mother iPSC-CMs (Supplementary Figure S15c). Moreover, the expression levels of key Ca2+-handling proteins (SERCA2a, RYR2, sodium/calcium exchanger 1 (NCX1), Cav1.2 and phospholamban (PLN)) remained unchanged between father, mother, proband and brother iPSC-CMs (Supplementary Figure S16). Collectively, these results indicate irregular Ca2+ signaling in D356Y iPSC-CMs.
Fig. 4.
Irregular Ca2+signaling in D356Y iPSC-CMs. a. Representative Ca2+ transient tracings recorded from father, proband, brother and mother iPSC-CMs. Arrhythmia-like abnormal Ca2+ transient events are indicated by red arrows. Red dash lines indicate 0.05 F340/F380. b–d. Bar graphs to compare the percentage of cells exhibiting abnormal Ca2+ transient events, SD of beat intervals, and maximal rising rate between father, proband, brother and mother iPSC-CMs by One-way ANOVA (Tukey method). n = 52–84. ∗P < 0.05 and ∗∗∗∗P < 0.0001. e. Representative Ca2+ transient tracings before and after 100 nM verapamil stress in father, brother or mother iPSC-CMs. Arrhythmia-like abnormal Ca2+ transient events are indicated by red arrows. f. Bar graph to compare the percentage of cells with arrhythmias between different groups in e by Fisher's test. ∗P < 0.05, ∗∗P < 0.01 and ∗∗∗∗P < 0.0001. g. Representative Ca2+ current tracings isolated from father, proband, brother and mother iPSC-CMs. h. Comparison of Ca2+ IV curve between CON, proband, brother and mother iPSC-CMs. i–j. Bar graphs to compare the cell capacitance and the peak Ca2+ current density at 0 mV between different groups in h by One-way ANOVA (Tukey method). n = 11–15. ∗P < 0.05, ∗∗P < 0.01 and ∗∗∗∗P < 0.0001. k. Comparison of steady-state activation and inactivation of Ca2+ current between CON, proband, brother and mother iPSC-CMs. l–m. Bar graphs to compare the V1/2 of steady-state activation and inactivation between different groups in k by One-way ANOVA (Tukey method). n = 10–13.
Reduced Ca2+ current in the iPSC-CMs of the proband
It has been reported that the antiarrhythmic drug verapamil, that serves as an L-type Ca2+ channel blocker (CCB), can be utilized in clinical setting to clarify BrS patterns.1 We hypothesized that such an in vivo electrophysiological study could be recapitulated in vitro. Accordingly, baseline recordings from brother and mother iPSC-CMs showing regular Ca2+ transient patterns were selected, and acute administration of 100 nM verapamil was then applied to induce arrhythmia-like waveforms, thus exacerbating the aberrant cellular phenotypes (Fig. 4e and f and Supplementary Figure S17). Moreover, we isolated Ca2+ currents from different iPSC-CMs using a single-cell voltage clamp (Supplementary Table S6). The iPSC-CMs of the proband showed significantly reduced Ca2+ current density in comparison to those of the father, while the SSA and steady-state inactivation (SSI) remained unchanged (Fig. 4g–m). Importantly, the Ca2+ current density showed a lesser reduction in brother and mother iPSC-CMs as compared to proband iPSC-CMs (Fig. 4g–j). Altogether, these results suggest that the Ca2+ currents are impaired in proband iPSC-CMs but are preserved in AMC iPSC-CMs.
Rescuing BrS phenotypes of proband iPSC-CMs by genetic correction of SCN5A D356Y
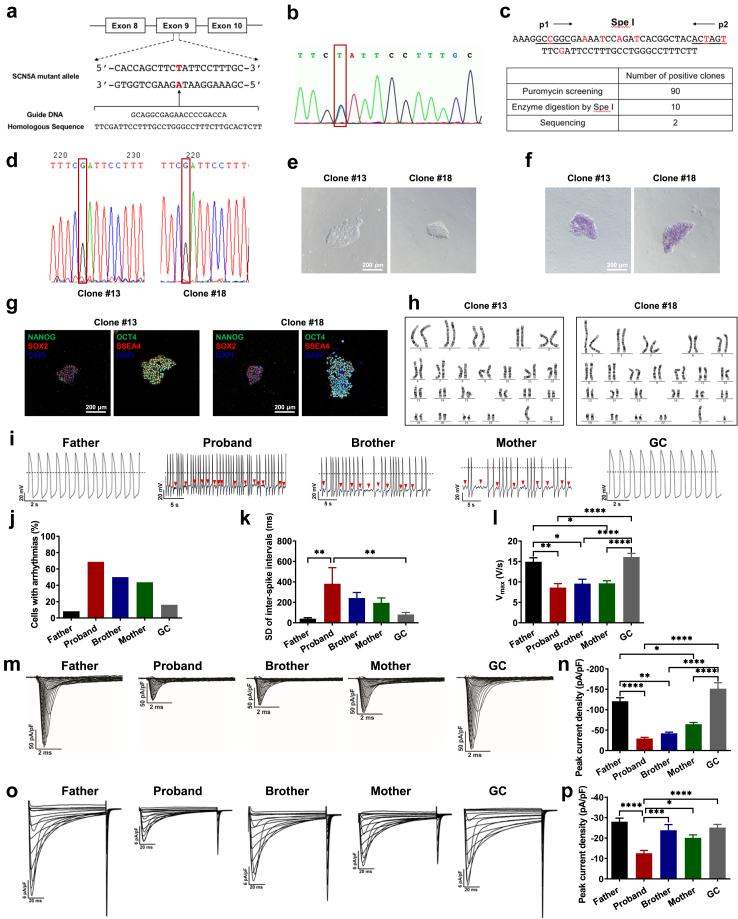
To determine the extent to which SCN5A D356Y accounts for the arrhythmic phenotype in this BrS variant, the mutation in the iPSCs of the proband was corrected using CRISPR/Cas9-mediated genome editing technology (Fig. 5a). Two positive clones were obtained and site-specific gene correction was confirmed by genomic sequencing (Fig. 5b–d). The generated gene-corrected (GC) iPSCs displayed typical human embryonic stem cell morphology, stained positive for ALP and pluripotent markers, and retained a normal karyotype (Fig. 5e–h). Upon differentiation, action potential recordings demonstrated that the arrhythmic phenotype was rescued in GC iPSC-CMs (Fig. 5i and j). GC iPSC-CMs also showed improved beat interval variation, Vmax, MDP and APA (Fig. 5k and l and Supplementary Figure S18). The Nav1.5 expression and the sodium channel function were restored in GC iPSC-CMs, resembling those from the iPSC-CMs of the father (Fig. 5m and n and Supplementary Figure S19 and S20). Consistent with the observation in father iPSC-CMs, flecainide-induced arrhythmogenic activities in GC iPSC-CMs were only observed in 1 of 9 of the independent MEA studies (Supplementary Figure S21). Gene correction of SCN5A D356Y rescued the reduced Ca2+ current phenotype, normalized the SR Ca2+ load, and restored the NCX activity identified in proband iPSC-CMs (Fig. 5o and p, Supplementary Figure S15 and S22). These results indicate that D356Y is a pathogenic mutation of BrS, and that editing of D356Y is sufficient to rescue BrS phenotypes.
Fig. 5.
Rescuing BrS phenotypes of proband iPSC-CMs by genetic correction of SCN5A D356Y. a. Strategy of correcting SCN5A D356Y mutation. The mutation site is marked in red. b. Confirmation of the existence of SCN5A D356Y mutation in proband iPSCs. c. Spe I restriction digestion of PCR products before and after gene correction, and summary of number of positive clones after puromycin screening (90 positive clones), enzyme digestion by Spe I (10 positive clones) and DNA sequencing (2 positive clones), respectively. d. DNA sequencing demonstrating the correction of 1066G > T (D356Y) mutation, and two gene-corrected clones were obtained (clone #13 and clone #18). e. Typical morphology of GC iPSCs. Scale bar, 200 μm. f. Alkaline Phosphatase (ALP) staining of GC iPSCs. Scale bar, 200 μm. g. Pluripotent staining of GC iPSCs using NANOG (green), SOX2 (red), OCT4 (green) and SSEA4 (red). DAPI indicates nuclear staining (blue). Scale bar, 200 μm. h. Representative graphs of karyotype of GC iPSCs. i. Representative action potential tracings recorded from father, proband, brother, mother and GC iPSC-CMs. Red arrows indicate arrhythmias. Dash lines indicate 0 mV. j–l. Bar graphs to compare arrhythmic incidence, SD of ISIs, and Vmax between different groups in i by One-way ANOVA (Tukey method). ∗P < 0.05, ∗∗P < 0.01 and ∗∗∗∗P < 0.0001. m. Representative Na + current tracings isolated from father, proband, brother, mother and GC iPSC-CMs. n. Bar graph to compare the peak Na+ current density at −30 mV between different groups in m by One-way ANOVA (Tukey method). ∗P < 0.05, ∗∗P < 0.01 and ∗∗∗∗P < 0.0001. o. Representative Ca2+ current tracings isolated from father, proband, brother, mother and GC iPSC-CMs. p. Bar graph to compare the peak Ca2+ current density at 0 mV between different groups in o by One-way ANOVA (Tukey method). n = 8–15. ∗P < 0.05, ∗∗∗P < 0.001 and ∗∗∗∗P < 0.0001.
Personalized drug screening
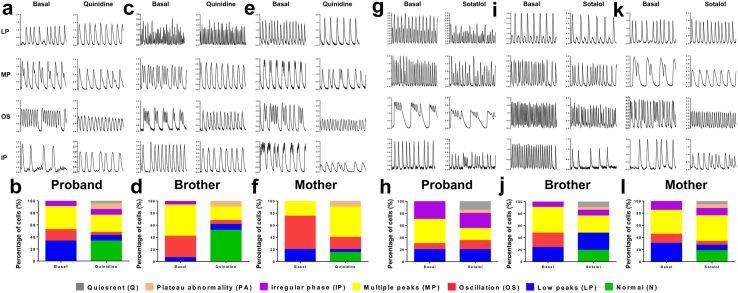
Having established the BrS human iPSC-CM models, we next used these “disease in a dish” models to evaluate drugs that may possess therapeutic benefits for different BrS individuals in the same family. 4 drugs (quinidine, sotalol, cilostazol and bepridil), previously reported as clinical medications for pharmacological therapy of BrS, were selected for testing using Fluo-4 AM-based Ca2+ imaging assays with suppressed arrhythmogenicity as the readout.28,29 A diverse pattern of irregular Ca2+ transients was seen in BrS iPSC-CMs at baseline, which was classified into 4 subtypes including low peaks (LP), multiple peaks (MP), oscillation (OS) and irregular phase (IP). Plateau abnormality (PA) and quiescent (Q) patterns were also observed upon drug induction. Importantly, 1 μM quinidine application significantly eliminated the arrhythmias, resulting in an increased fraction of cells exhibiting a normal pattern (N) of Ca2+ transients (proband: 7 out of 21 cells; brother: 16 out of 31 cells; mother: 3 out 20 cells) (Fig. 6a–f). The rescue effect of quinidine was reflected by targeting LP and OS in the iPSC-CMs of the proband or mother, whereas OS and MP abnormalities were greatly alleviated in those of the brother (Fig. 6a–f). Moreover, no anti-arrhythmic effect was noted by sotalol treatment (3 μM) in proband iPSC-CMs (Fig. 6g and h). Interestingly, sotalol application significantly abolished the arrhythmias by normalizing OS abnormalities in the iPSC-CMs of the brother, or LP/OS abnormalities in those of the mother (brother: 4 out of 21 cells; mother: 6 out of 33 cells) (Fig. 6i–l). On the other hand, we observed no antiarrhythmic effects resulting from the application of the other 2 drugs, cilostazol (1 μM) and bepridil (100 nM) (Supplementary Figure S23 and S24). Altogether, these results suggest that quinidine and sotalol are therapeutically effective for BrS with a SCN5A D356Y genotype in an individual-dependent manner.
Fig. 6.
Personalized drug screening. a, c and e. Representative Ca2+ transient tracings recorded from proband, brother and mother iPSC-CMs by Ca2+ imaging using Fluo-4 AM before and after 1 μM quinidine application. b, d and f. Bar graphs to compare percentage of cells exhibiting normal (N) or abnormal Ca2+ transient pattern including low peaks (LP), multiple peaks (MP), oscillation (OS) and irregular phase (IP). Plateau abnormality (PA) and quiescent (Q) before and after quinidine application in proband, brother and mother iPSC-CMs. n = 20–31. g, i and k. Representative Ca2+ transient tracings recorded from proband, brother and mother iPSC-CMs by Ca2+ imaging using Fluo-4 AM before and after 3 μM sotalol application. h, j and l. Bar graphs to compare percentage of cells exhibiting normal (N) or abnormal Ca2+ transient pattern including low peaks (LP), multiple peaks (MP), oscillation (OS) and irregular phase (IP). Plateau abnormality (PA) and quiescent (Q) before and after sotalol application in proband, brother and mother iPSC-CMs. n = 20–33.
Clinical follow-up
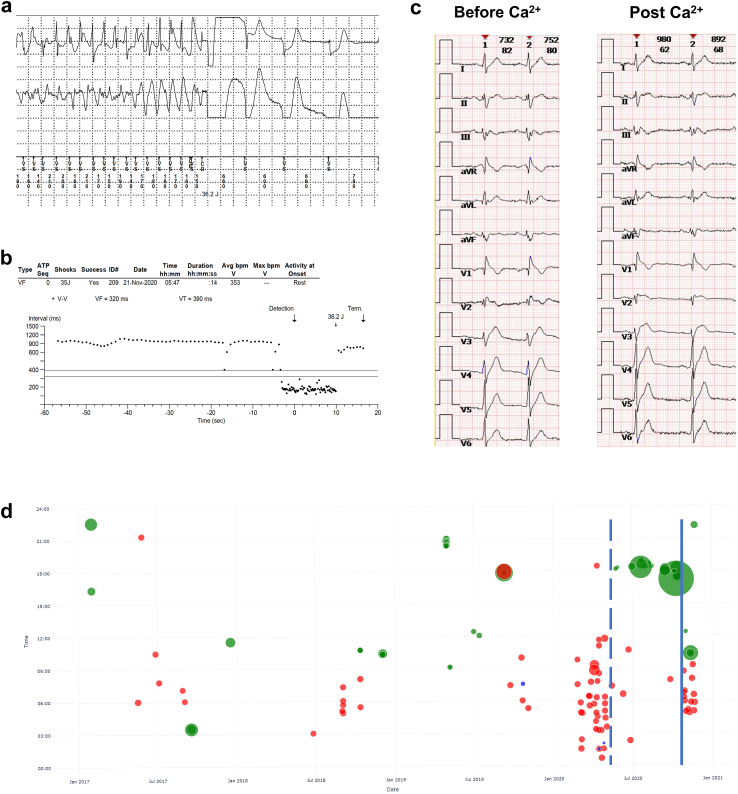
The proband was regularly followed up for 6 years. Multiple ventricular episodes including VT and VF were recorded even after three catheter ablation procedures (Fig. 7a and b). Considering limited available therapies and recurrent ventricular arrhythmias, intravenous and oral application of calcium was administrated and this seemed to partially reduce the ventricular arrhythmias. The intravenous loading dose and oral dose were calcium gluconate 2000 mg once and calcium carbonate 600 mg/Vitamin D3 125 IU twice a day, respectively, according to the upper limitation of dietary recommendation for calcium supplementation.30 The surface ECGs before and immediately post the loading dose of calcium are shown in Fig. 7c. The corrected QT (QTc) intervals on V5 lead were 383 ms and 373 ms, respectively. The T waves on precordial leads showed more intensive trend after calcium application. Most events occurred during midnight to noon, often in the summer season. In February 2020, the proband moved from his hometown (Location: 29°30′ northern latitude, 114°25′ east longitude) to the city of Guangzhou (Location: 23°6′ northern latitude, 113°15′ east longitude) for a 4-month temporary job. On oral calcium supplement, lethal ventricular episodes were significantly reduced. However, calcium therapy was discontinued by the proband himself and ventricular episodes increased again after calcium discontinuation (Fig. 7d, Supplementary Figure S25 and S26).
Fig. 7.
Malignant arrhythmic events and timeline of therapies. a. Representative ventricular fibrillation episode terminated by direct current cardioversion. b. R–R interval plot of ventricular fibrillation. c. The timeline of malignant ventricular events and the therapies. The green circles, blue circles and red dots indicate ventricular arrhythmias with frequency <200 bpm, 200–300 bpm, and events terminated by ICD shocks, respectively. The area of the dots indicates the duration of ventricular arrhythmias. Dashed and solid blue lines indicate the start and withdrawal of oral calcium supplement. d. The ECG tracings before and immediately after the application of intravenous calcium. The QTc intervals on V5 lead were 383 ms and 373 ms, respectively. The T wave on precordial leads showed more intensive trend after calcium application.
Discussion
In this study, by utilizing iPSC-CM models combined with CRISPR/Cas9-mediated genome editing technology, we determined that the identified variant (SCN5A D356Y) that was not previously reported is a pathogenic loss-of-function mutation of BrS. We demonstrated that D356Y iPSC-CMs recapitulated numerous aspects of the BrS phenotype including increased beat interval variability, slower depolarization, cardiac arrhythmias, defect of Na+ channel function and irregular Ca2+ signaling. Importantly, the phenotypic severity observed in AMC iPSC-CMs was milder than that of proband iPSC-CMs, an observation that was exacerbated by the introduction of flecainide stress. Interestingly, we identified that proband iPSC-CMs exhibited markedly decreased Ca2+ current in comparison with both control and AMC iPSC-CMs. Gene correction of D356Y in proband iPSC-CMs effectively rescued the arrhythmic phenotype and restored the Na+ and Ca2+ currents. Moreover, drug screening using established iPSC-CM models demonstrated that quinidine and sotalol possessed antiarrhythmic effects in an individual-dependent manner. Clinically, venous and oral administration of calcium partially reduced malignant arrhythmic events of the proband in mid-term follow-up. Our study demonstrates a platform for the conduction of comprehensive investigations of the cellular abnormalities of a BrS family cohort with the SCN5A mutation, enhancing the potentials for accurate diagnosis and risk stratification for the disease. Based on our functional evidence, the proband's younger brother and mother, who are BrS AMCs, are likely to be predisposed to malignant arrhythmias and therefore require close follow-up.
BrS is a primary electrical disorder characterized by typical ECG signs and malignant ventricular arrhythmias. It has been recognized worldwide as a major cause of SCD in otherwise healthy patients.31 Currently, as SCD is often the first indication that the patient is affected by BrS, options for earlier detection are a critical requirement. Also, most BrS patients remain asymptomatic for extended periods, yet the risk stratification strategy for this population remains unresolved. Some existing studies have been attempted which generated iPSC models of BrS.32, 33, 34, 35, 36, 37, 38, 39 Most of the reported BrS iPSC models were generated from patients carrying the SCN5A variants, and the findings are similar among these reports.32, 33, 34, 35,39, 40 Our previous study demonstrated that iPSC-CMs from BrS patients carrying the SCN5A variants exhibited increased triggered activity, reduced sodium current density, decreased Vmax, and aberrant Ca2+ handling in comparison with healthy control iPSC-CMs, and genomic correction of the variant in BrS iPSC-CMs could reverse the abnormal cellular phenotype.32 Besides the SCN5A gene, iPSC-CMs with variants of other sodium channel genes (SCN1B and SCN10A) also showed a sodium channel deficiency phenotype.37,38 There are also published data from the iPSC-CM models with rare BrS-related genes. Belbachir et al. generated a BrS iPSC model with the RAAD variant, which exhibited reduced sodium current density, decreased Vmax, prolonged APD, and increased trigger activities.36 Cerrone et al. reported an iPSC model from a BrS patient carrying the PKP2 variant, and the BrS iPSC-CMs showed a decreased sodium current density compared to healthy control iPSC-CMs.41 However, functional assessments have been only based on 1–2 sporadic patient(s) and unrelated healthy control(s) in the aforementioned studies. Our study facilitated the recruitment of a large BrS family cohort carrying SCN5A D356Y consisting of one severely affected BrS patient, two AMCs and seven controls, which enabled the comparison of functional characterization between different individuals of the same family. It is notable that we demonstrate that the varying phenotypic severities of different BrS patient individuals (symptomatic or asymptomatic) bearing the same mutation can be evaluated by the iPSC model, thus bypassing the deficiencies of cellular transfection models.
One noteworthy finding that has not been previously reported is the variation in L-type Ca2+ channel (LTCC) currents among different individuals with BrS from the same family. Alongside reduced Na+ current, the iPSC-CMs from the proband displayed significantly decreased Ca2+ current density. However, the iPSC-CMs from AMCs largely maintained their LTCC functionality.
The reduction in Ca2+ current density holds great physiological significance. Impaired Na + channel function alone impedes the upstroke slope of action potentials, and when coupled with a simultaneous decrease in Ca2+ currents, it further limits or eliminates the plateau phase of action potentials. This dual abnormality disrupts the repolarization process of cardiomyocytes, elevating the risk of inadequate early repolarization and ventricular fibrillation. It is well known that mutations in Ca2+ channel genes are associated with BrS, leading to defect of Ca2+ channel function.42,43 Furthermore, it is well established that Ca2+ entry through LTCC serves as the trigger for the fine-tuned SR Ca2+ release during each contractile cycle in cardiomyocytes, a process known as calcium-induced calcium release (CICR).44,45 With the trigger being smaller, SR Ca2+ release is reduced, leading to an increased proportion of Ca2+ trapped within the SR. On the other hand, the SR may need to maintain its Ca2+ load to compensate for the reduced trigger and to meet systemic metabolic demands. Indeed, our findings by the rapid caffeine perfusion protocol revealed a significant augmentation in SR Ca2+ load in proband over control iPSC-CMs. This compensatory SR Ca2+ “overload”, however, increasing the likelihood of spontaneous Ca2+ release from the SR and predisposing individuals to arrhythmogenic events.46,47 SR Ca2+ overload can give rise to the generation of large spontaneous Ca2+ release events, referred to as Ca2+ waves, from the SR into the cytosol. These Ca2+ waves can activate inward sodium–calcium exchange (NCX) currents, resulting in delayed afterdepolarizations (DADs). DADs can trigger premature action potentials and potentially initiate arrhythmias. Thus, reduced LTCC activity may exacerbate arrhythmias in the proband through various mechanisms. Moreover, Verapamil is utilized as a provocative drug during diagnostic testing for BrS because of its capability to reveal the distinctive ECG pattern linked with the syndrome. Our findings may provide further support for the rationale behind this approach.
Additionally, the rapid caffeine perfusion experiments revealed a significant decrease in the activity of the NCX (Supplementary Figure S14). Considering that the primary physiological function of NCX is to remove extracellular Ca2+ entry out of the cell, this reduction could be an adaptive response to the decreased LTCC activity. While the diminished NCX activity alone may alleviate DADs induced by Ca2+ waves, it may not be sufficient to fully counterbalance the detrimental consequences of SR Ca2+ overload.
At present, we do not have a definitive answer regarding the underlying mechanism behind the reduction in Ca2+ current. However, our Western blot data, although not statistically significant, indicated a reduction of over 20% in LTCC protein expression, which could have functional significance. Additionally, our biotin labeling experiment revealed that the D356Y mutation in Na+ channels may hinder their proper integration into the plasma membrane. The delivery and insertion of ion channels from cargo vesicles into the sarcolemma are crucial for their proper functioning. Considering that both Na+ channels and Ca2+ channels are typically localized in close proximity and have an excessive channel repertoire stored in vesicles beneath the plasma membrane, it is plausible to hypothesize that this hindrance may also contribute to a decreased density of Ca2+ current due to the similar regulatory mechanisms of Ca2+ channels.48,49
Currently we are still conducting in-depth analysis of the whole exome sequencing data and transcriptomic data to explore other potential mechanisms. However, our main focus at this stage is to accurately describe the observed phenomena and provide plausible explanations based on our current understanding.
The pharmacological treatment of BrS has been recommended in the case of electrical storms as an adjunct to ICD and as preventative therapy for currently asymptomatic patients who may be at risk for the development of life-threatening arrhythmic events.18 Using the unique “clinical trial in a dish” model, we demonstrated that quinidine and sotalol exhibited antiarrhythmic effects for the SCN5A D356Y genotype in an individual-dependent manner, whereas the other 2 tested drugs, cilostazol and bepridil, had no such effect. This highlighted the ability of iPSC models to screen tailored therapeutic drugs of different BrS individuals (even in the same family) for personalized medicine.
Radiofrequency ablation for preventing VF recurrence following arrhythmic storm was recently reported.50,51 The proband in this study underwent three bouts of endo-epicardial mapping and ablation to reduce VF episodes.52 However, in this case VF episodes still occurred irregularly. This could be partially explained by the trend of the firing of spontaneous arrhythmias as shown in our iPSC-CM model. We could speculate that the arrhythmogenic triggers underlying VF in BrS patients exist in general ventricular cardiomyocytes, rather than in localized regions such as right ventricular outflow tract. These triggers could not be completed eliminated and therefore such procedures showed limited effectiveness in these patients. Considering the lack of accessibility of quinidine in China and the arrhythmic effects of CCB confirmed by the in vitro study, calcium supplementation was prescribed for the proband to suppress the frequent ICD shocks after the third ablation procedure. Interestingly, the loading dose of calcium shortened QTc interval and partially reduced ICD shocks over a several-month span. During this period, the onset time and ventricular rate of the residual arrhythmias also changed where the onset time of events mainly occurred in the afternoon, the ventricular rate decreased, and most events could terminate spontaneously. These findings implied that an imbalance in calcium had participated in the arrhythmogenesis of BrS. Although the use of CCBs have been reported to be safe over long-term follow-up periods in a few clinical trials,53,54 these agents have not yet been routinely recommended for the treatment of BrS. Our study provides increasing evidence for the effectiveness of calcium supplement in BrS, but more investigations are still warranted.
Another interesting result was the possible relationship between the climate and the arrhythmic events. The proband was shocked multiple times and most of these events occusrred in summer, probably related to higher environmental temperature. When the proband moved to a lower latitude city, the higher temperature might induce clinical arrhythmic events. Regarding temperature, whilst the relationship between BrS and fever has been well-established by previous studies,55, 56, 57 any relationship between environmental temperature and arrhythmic events in BrS has yet to be presented. In our study, the events showed distinct pattern related to seasons and/or latitude. More follow-up data is required to confirm this finding.
One limitation to our study is that the in vitro phenotypes and clinical characteristics among BrS patients were variable and the mechanism of these heterogeneity remain to be unveiled. Furthermore, engineered heart tissue (EHT) and transgenic mice models are arranged for the future investigations.
In conclusion, we demonstrate that the iPSC model can serve as an adjuvant tool for diagnosing the pathogenicity of a particular variant, for functional stratification of BrS AMCs, and for the screening therapeutic drugs against specific mutations for personalized medicine. Our results also suggest that preservation of the Ca2+ currents might be a compensatory mechanism to resist arrhythmogenesis in BrS AMCs. The concepts described in this study may hold the potential to be broadened and extended to incorporate investigations of other complex and fatal genetic cardiac diseases beyond that of BrS. This may enable the application of precision medicine into early diagnosis, management and prevention for such conditions.
Contributors
P.L. and C.J. designed and supervised the study. Y.S., J.S., X.W., J.W., F.G., H.Q., H.F., D.C., H.W., M.L., W.W., Y.F., G.F. and T.G. performed the experiments and analyzed data. P.L. and W.W. wrote the manuscript. All authors read and approved the final version of the manuscript. Y.S., J.S., X.W., P.L. and C.J. directly accessed and verified the underlying data reported in the manuscript. All authors read and approved the final version of the manuscript.
Data sharing statement
The data described in this article are available from the corresponding authors upon reasonable request.
Declaration of interests
The authors declare no conflict of interests.
Acknowledgements
We would like to thank the core facility of Zhejiang University Institute of Translational Medicine for assistance with flow cytometry and confocal microscopy experiments. This work was supported by National Key R&D Program of China (2017YFA0103700) (P.L.), National Natural Science Foundation of China (81922006, 81870175) (P.L.), Natural Science Foundation of Zhejiang Province (LD21H020001, LR15H020001) (P.L.), National Natural Science Foundation of China (81970269) (C.J.), Key Research and Development Program of Zhejiang Province (2019C03022) (C.J.) and Natural Science Foundation of Zhejiang Province (LY16H020002) (Y.S.).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104741.
Contributor Information
Ping Liang, Email: pingliang@zju.edu.cn.
Chenyang Jiang, Email: cyjiang@zju.edu.cn.
Appendix A. Supplementary data
References
- 1.Coppola G., Corrado E., Curnis A., et al. Update on brugada syndrome 2019. Curr Probl Cardiol. 2021;46(3) doi: 10.1016/j.cpcardiol.2019.100454. [DOI] [PubMed] [Google Scholar]
- 2.Brugada P., Brugada J. Right bundle branch block, persistent ST segment elevation and sudden cardiac death: a distinct clinical and electrocardiographic syndrome. J Am Coll Cardiol. 1992;20(6):1391–1396. doi: 10.1016/0735-1097(92)90253-j. [DOI] [PubMed] [Google Scholar]
- 3.Wu Y., Ai M., Bardeesi A.S.A., et al. Brugada syndrome: a fatal disease with complex genetic etiologies–still a long way to go. Forensic Sci Res. 2017;2(3):115–125. doi: 10.1080/20961790.2017.1333203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antzelevitch C., Brugada P., Brugada J., Brugada R., Towbin J.A., Nademanee K. Brugada syndrome: 1992–2002. J Am Coll Cardiol. 2003;41(10):1665–1671. doi: 10.1016/s0735-1097(03)00310-3. [DOI] [PubMed] [Google Scholar]
- 5.Sieira J., Dendramis G., Brugada P. Pathogenesis and management of Brugada syndrome. Nat Rev Cardiol. 2016;13(12):744–756. doi: 10.1038/nrcardio.2016.143. [DOI] [PubMed] [Google Scholar]
- 6.Curcio A., Santarpia G., Indolfi C. The brugada syndrome―from gene to therapy. Circ J. 2017;81(3):290–297. doi: 10.1253/circj.CJ-16-0971. [DOI] [PubMed] [Google Scholar]
- 7.Juang J.-M.J., Horie M. Genetics of brugada syndrome. J Arrhythmia. 2016;32(5):418–425. doi: 10.1016/j.joa.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antzelevitch C., Patocskai B. Brugada syndrome: clinical, genetic, molecular, cellular, and ionic aspects. Curr Probl Cardiol. 2016;41(1):7–57. doi: 10.1016/j.cpcardiol.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Q., Kirsch G.E., Zhang D., et al. Genetic basis and molecular mechanism for idiopathic ventricular fibrillation. Nature. 1998;392(6673):293–296. doi: 10.1038/32675. [DOI] [PubMed] [Google Scholar]
- 10.Yamagata K., Horie M., Aiba T., et al. Genotype-phenotype correlation of SCN5A mutation for the clinical and electrocardiographic characteristics of probands with Brugada syndrome: a Japanese multicenter registry. Circulation. 2017;135(23):2255–2270. doi: 10.1161/CIRCULATIONAHA.117.027983. [DOI] [PubMed] [Google Scholar]
- 11.Jenewein T., Beckmann B., Rose S., et al. Genotype-phenotype dilemma in a case of sudden cardiac death with the E1053K mutation and a deletion in the SCN5A gene. Forensic Sci Int. 2017;275:187–194. doi: 10.1016/j.forsciint.2017.02.038. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi K., Tanabe K., Ohnuki M., et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 13.Yu J., Vodyanik M.A., Smuga-Otto K., et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 14.Shi Y., Inoue H., Wu J.C., Yamanaka S. Induced pluripotent stem cell technology: a decade of progress. Nat Rev Drug Discov. 2017;16(2):115–130. doi: 10.1038/nrd.2016.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burridge P.W., Keller G., Gold J.D., Wu J.C. Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell. 2012;10(1):16–28. doi: 10.1016/j.stem.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshida Y., Yamanaka S. Induced pluripotent stem cells 10 Years later: for cardiac applications. Circ Res. 2017;120(12):1958–1968. doi: 10.1161/CIRCRESAHA.117.311080. [DOI] [PubMed] [Google Scholar]
- 17.Sallam K., Li Y., Sager P.T., Houser S.R., Wu J.C. Finding the rhythm of sudden cardiac death: new opportunities using induced pluripotent stem cell–derived cardiomyocytes. Circ Res. 2015;116(12):1989–2004. doi: 10.1161/CIRCRESAHA.116.304494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Priori S.G., Gasparini M., Napolitano C., et al. Risk stratification in brugada syndrome. J Am Coll Cardiol. 2012;59(1):37–45. doi: 10.1016/j.jacc.2011.08.064. [DOI] [PubMed] [Google Scholar]
- 19.Antzelevitch C., Brugada P., Borggrefe M., et al. Brugada syndrome: report of the second consensus conference: endorsed by the heart rhythm society and the European heart rhythm association. Circulation. 2005;111(5):659–670. doi: 10.1161/01.CIR.0000152479.54298.51. [DOI] [PubMed] [Google Scholar]
- 20.Richards S., Aziz N., Bale S., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of medical genetics and genomics and the association for molecular pathology. Genet Med. 2015;17(5):405–423. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo F., Sun Y., Wang X., et al. Patient-specific and gene-corrected induced pluripotent stem cell-derived cardiomyocytes elucidate single-cell phenotype of short QT syndrome. Circ Res. 2019;124(1):66–78. doi: 10.1161/CIRCRESAHA.118.313518. [DOI] [PubMed] [Google Scholar]
- 22.Tang L., Wang H., Dai B., et al. Human induced pluripotent stem cell-derived cardiomyocytes reveal abnormal TGFbeta signaling in type 2 diabetes mellitus. J Am Coll Cardiol. 2020;142:53–64. doi: 10.1016/j.yjmcc.2020.03.016. [DOI] [PubMed] [Google Scholar]
- 23.Tohyama S., Hattori F., Sano M., et al. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell. 2013;12(1):127–137. doi: 10.1016/j.stem.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 24.Meregalli P.G., Ruijter J.M., Hofman N., Bezzina C.R., Wilde A.A.M., Tan H.L. Diagnostic value of flecainide testing in unmasking SCN5A-related brugada syndrome. J Cardiovasc Electrophysiol. 2006;17(8):857–864. doi: 10.1111/j.1540-8167.2006.00531.x. [DOI] [PubMed] [Google Scholar]
- 25.Kistamás K., Veress R., Horváth B., Bányász T., Nánási P.P., Eisner D.A. Calcium handling defects and cardiac arrhythmia syndromes. Front Pharmacol. 2020;11:72. doi: 10.3389/fphar.2020.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monasky M.M., Pappone C., Piccoli M., Ghiroldi A., Micaglio E., Anastasia L. Calcium in Brugada syndrome: questions for future research. Front Pharmacol. 2018;9:1088. doi: 10.3389/fphys.2018.01088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bers D.M. Calcium cycling and signaling in cardiac myocytes. Annu Rev Physiol. 2008;70(1):23–49. doi: 10.1146/annurev.physiol.70.113006.100455. [DOI] [PubMed] [Google Scholar]
- 28.Glatter K.A., Wang Q., Keating M., Chen S., Chiamvimonvat N., Scheinman M.M. Effectiveness of sotalol treatment in symptomatic Brugada syndrome. Am J Cardiol. 2004;93(10):1320–1322. doi: 10.1016/j.amjcard.2004.02.023. [DOI] [PubMed] [Google Scholar]
- 29.Belhassen B., Michowitz Y., Brodie O.T. Pharmacological therapy in brugada syndrome. Arrhythm Electrophysiol Rev. 2018;7(2):135. doi: 10.15420/aer.2018.21.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grossman D.C., Curry S.J., Owens D.K., et al. Vitamin D, calcium, or combined supplementation for the primary prevention of fractures in community-dwelling adults. JAMA. 2018;319(15):1592. doi: 10.1001/jama.2018.3185. [DOI] [PubMed] [Google Scholar]
- 31.Snir A.D., Raju H. Current controversies and challenges in brugada syndrome. Eur Cardiol Rev. 2019;14(3):169. doi: 10.15420/ecr.2019.12.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang P., Sallam K., Wu H., et al. Patient-specific and genome-edited induced pluripotent stem cell-derived cardiomyocytes elucidate single-cell phenotype of brugada syndrome. J Am Coll Cardiol. 2016;68(19):2086–2096. doi: 10.1016/j.jacc.2016.07.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayano M., Makiyama T., Kamakura T., et al. Development of a patient-derived induced pluripotent stem cell model for the investigation of SCN5A-D1275N-related cardiac sodium channelopathy. Circ J. 2017;81(12):1783–1791. doi: 10.1253/circj.CJ-17-0064. [DOI] [PubMed] [Google Scholar]
- 34.Ma D., Liu Z., Loh L.J., et al. Identification of an I Na-dependent and I to-mediated proarrhythmic mechanism in cardiomyocytes derived from pluripotent stem cells of a Brugada syndrome patient. Sci Rep. 2018;8(1):1–11. doi: 10.1038/s41598-018-29574-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selga E., Sendfeld F., Martinez-Moreno R., et al. Sodium channel current loss of function in induced pluripotent stem cell-derived cardiomyocytes from a Brugada syndrome patient. J Mol Cell Cardiol. 2018;114:10–19. doi: 10.1016/j.yjmcc.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belbachir N., Portero V., Al Sayed Z.R., et al. RRAD mutation causes electrical and cytoskeletal defects in cardiomyocytes derived from a familial case of Brugada syndrome. Eur Heart J. 2019;40(37):3081–3094. doi: 10.1093/eurheartj/ehz308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.El-Battrawy I., Albers S., Cyganek L., et al. A cellular model of Brugada syndrome with SCN10A variants using human-induced pluripotent stem cell-derived cardiomyocytes. EP Europace. 2019;21(9):1410–1421. doi: 10.1093/europace/euz122. [DOI] [PubMed] [Google Scholar]
- 38.El-Battrawy I., Müller J., Zhao Z., et al. Studying Brugada syndrome with an SCN1B variants in human-induced pluripotent stem cell-derived cardiomyocytes. Front Cell Deve Biol. 2019;7:261. doi: 10.3389/fcell.2019.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li W., Stauske M., Luo X., et al. Disease phenotypes and mechanisms of iPSC-derived cardiomyocytes from Brugada syndrome patients with a loss-of-function SCN5A mutation. Front Cell Deve Biol. 2020;8:1181. doi: 10.3389/fcell.2020.592893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kosmidis G., Veerman C.C., Casini S., et al. Readthrough-promoting drugs gentamicin and PTC124 fail to rescue Nav1.5 function of human-induced pluripotent stem cell-derived cardiomyocytes carrying nonsense mutations in the sodium channel gene SCN5A. Circ Arrhythm Electrophysiol. 2016;9(11) doi: 10.1161/CIRCEP.116.004227. [DOI] [PubMed] [Google Scholar]
- 41.Cerrone M., Lin X., Zhang M., et al. Missense mutations in plakophilin-2 cause sodium current deficit and associate with a Brugada syndrome phenotype. Circulation. 2014;129(10):1092–1103. doi: 10.1161/CIRCULATIONAHA.113.003077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Antzelevitch C., Pollevick G.D., Cordeiro J.M., et al. Loss-of-function mutations in the cardiac calcium channel underlie a new clinical entity characterized by ST-segment elevation, short QT intervals, and sudden cardiac death. Circulation. 2007;115(4):442–449. doi: 10.1161/CIRCULATIONAHA.106.668392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burashnikov E., Pfeiffer R., Barajas-Martinez H., et al. Mutations in the cardiac L-type calcium channel associated with inherited J-wave syndromes and sudden cardiac death. Heart Rhythm. 2010;7(12):1872–1882. doi: 10.1016/j.hrthm.2010.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fabiato A. Calcium-induced release of calcium from the cardiac sarcoplasmic reticulum. Am J Physiol. 1983;245(1):C1–C14. doi: 10.1152/ajpcell.1983.245.1.C1. [DOI] [PubMed] [Google Scholar]
- 45.Bers D.M. Cardiac excitation-contraction coupling. Nature. 2002;415(6868):198–205. doi: 10.1038/415198a. [DOI] [PubMed] [Google Scholar]
- 46.Chen W., Wang R., Chen B., et al. The ryanodine receptor store-sensing gate controls Ca2+ waves and Ca2+-triggered arrhythmias. Nat Med. 2014;20(2):184–192. doi: 10.1038/nm.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou Q., Xiao J., Jiang D., et al. Carvedilol and its new analogs suppress arrhythmogenic store overload-induced Ca2+ release. Nat Med. 2011;17(8):1003–1009. doi: 10.1038/nm.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murfitt L., Whiteley G., Iqbal M.M., Kitmitto A. Targeting caveolin-3 for the treatment of diabetic cardiomyopathy. Pharmacol Ther. 2015;151:50–71. doi: 10.1016/j.pharmthera.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 49.Balse E., Steele D.F., Abriel H., Coulombe A., Fedida D., Hatem S.N. Dynamic of ion channel expression at the plasma membrane of cardiomyocytes. Physiol Rev. 2012;92(3):1317–1358. doi: 10.1152/physrev.00041.2011. [DOI] [PubMed] [Google Scholar]
- 50.Nademanee K., Veerakul G., Chandanamattha P., et al. Prevention of ventricular fibrillation episodes in Brugada syndrome by catheter ablation over the anterior right ventricular outflow tract epicardium. Circulation. 2011;123(12):1270–1279. doi: 10.1161/CIRCULATIONAHA.110.972612. [DOI] [PubMed] [Google Scholar]
- 51.Pappone C., Brugada J., Vicedomini G., et al. Electrical substrate elimination in 135 consecutive patients with brugada syndrome. Circ Arrhythm Electrophysiol. 2017;10(5) doi: 10.1161/CIRCEP.117.005053. [DOI] [PubMed] [Google Scholar]
- 52.Zhang P., Tung R., Zhang Z., et al. Characterization of the epicardial substrate for catheter ablation of Brugada syndrome. Heart Rhythm. 2016;13(11):2151–2158. doi: 10.1016/j.hrthm.2016.07.025. [DOI] [PubMed] [Google Scholar]
- 53.Kamakura T., Wada M., Ishibashi K., et al. Feasibility evaluation of long-term use of beta-blockers and calcium antagonists in patients with Brugada syndrome. Europace. 2018;20(FI1):f72–f76. doi: 10.1093/europace/eux198. [DOI] [PubMed] [Google Scholar]
- 54.Yoshikawa T., Izumi C., Kaitani K., Nakagawa Y. A case of Brugada syndrome coexisting with vasospastic angina: caution should be taken when using calcium channel blockers. J Cardiol Cases. 2011;4(3):e143–e147. doi: 10.1016/j.jccase.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mizusawa Y., Morita H., Adler A., et al. Prognostic significance of fever-induced Brugada syndrome. Heart Rhythm. 2016;13(7):1515–1520. doi: 10.1016/j.hrthm.2016.03.044. [DOI] [PubMed] [Google Scholar]
- 56.Michowitz Y., Milman A., Sarquella-Brugada G., et al. Fever-related arrhythmic events in the multicenter survey on arrhythmic events in brugada syndrome. Heart Rhythm. 2018;15(9):1394–1401. doi: 10.1016/j.hrthm.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 57.Chen X., Zhao H., Sun L., Zhu W., Zhang F. Electrocardiogram characteristics and arrhythmic events during fever in patients with fever-induced brugada syndrome. Cardiology. 2020;145(3):130–135. doi: 10.1159/000505642. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.