Abstract
Background
Immunogenic cell death (ICD) triggers adaptive immune responses that aid in anticancer therapy. However, the significance of ICD-associated genes (ICDAGs) in clinical applications and their potential impact on the tumor microenvironment (TME) remains unclear.
Methods
The TCGA cohort was divided into different ICD clusters using the method of Consensus clustering. We assessed the clinical results and TME features of various ICD clusters. GSVA quantified the activation of hallmark gene sets. To establish an ICD molecular subtypes-related prognostic model (ICDRPM), we performed LASSO Cox regression analysis on the differentially expressed genes (DEGs) among ICD subtypes. We evaluated the assessment of risk groups by analyzing the proportion of immune cells, the TME, differences in genomic mutation, the efficacy of immunotherapy, and drug sensitivity. To enhance the clinical effectiveness of the ICDRPM, a nomograph was developed.
Results
Two distinct molecular subtypes were identified, and changes in ICDRGs were associated with clinical outcomes and TME characteristics of patients. A total of 1162 differentially expressed genes (DEGs) were obtained from two ICD clusters, and an ICDRPS was then developed to predict overall survival (OS). During both internal and external validation, patients classified as high-risk exhibited significantly poorer overall survival compared to those classified as low-risk. Additionally, the ICDRPS (ICD_score) was identified as an independent prognostic indicator for patients with BC, demonstrating excellent predictive performance. Afterward, we constructed a dependable nomogram to improve the practicality of the ICD_score. Furthermore, low-risk individuals showed stronger immunocyte infiltration, higher immune checkpoint expression, and higher IPS-PD-1 combined IPS-CTLA4 scores, indicating a greater response to immune checkpoint inhibitors (ICIs). Moreover, individuals categorized as having low or high risk exhibited contrasting sensitivity to anticancer medications.
Conclusions
The model constructed for genes related to ICD provided meaningful clinical implications for immunotherapy, and facilitated individualized treatment for BC patients.
Keywords: Bladder cancer, Immunogenic cell death, Tumor microenvironment, Immunotherapy, Molecular subtype
1. Introduction
Bladder cancer (BC), a common urological malignancy, ranks 10th among all diagnosed cancers [1]. According to the status of muscular invasion, it could be stratified as non-muscle-invasive bladder cancer (NMIBC) and muscle-invasive bladder cancer (MIBC) [2]. Despite neoadjuvant and adjuvant chemotherapy, MIBC has an unfavorable outlook and frequently experiences recurrence following the initial resection [3]. Recently, the treatment of advanced BC has been transformed by cancer immunotherapy, which involves the use of immune checkpoint inhibitors (ICIs) [4,5]. Nevertheless, ICIs only elicit a response in a small fraction of patients [[4], [5], [6]]. Increasing evidence has suggested that the therapeutic response of ICIs may be related to the tumor immune microenvironment (TIME) [7]. Therefore, it is vital to determine immune-specific biomarkers for targeting immunotherapy in BC.
The body's innate defense mechanism against cancer cells is known as anti-tumor immunity. Multiple mechanisms collaborate to recognize and eradicate tumors. The following are some of the primary mechanisms involved in the immune response against tumors: (1) Immune cells: The immune system consists of specialized cells that can identify and eliminate cancerous cells. The types of cells present are T cells, B cells, NK cells, and dendritic cells [[8], [9], [10]]. (2) Tumor-associated antigens (TAAs): TAAs are molecules produced by cancer cells that are recognized by the immune system as foreign. Immune cells can then attack cancer cells that display these antigens [[11], [12], [13]]. (3) Tumor-infiltrating lymphocytes (TILs): TILs are immune cells that have migrated into the tumor microenvironment and can directly attack cancer cells [[14], [15], [16], [17]]. (4) Cytokines: Cytokines are molecules that immune cells produce for signaling purposes. By facilitating the attraction of immune cells to the tumor site, they can enhance the immune response [18]. (5) Checkpoint inhibitors: Checkpoint inhibitors, also known as immune checkpoint inhibitors, are medications that inhibit specific molecules on immune cells to prevent cancer cells from deactivating them. This has the potential to boost the body's immune system in fighting against cancer [19]. By collaborating, these mechanisms function in unison to detect and eradicate malignant cells. Nevertheless, on occasion, cancerous cells can elude the immune system's monitoring mechanisms, resulting in the development and advancement of tumors. Scientists are exploring methods to boost the body's ability to fight against tumors to enhance the effectiveness of cancer treatment.
Cancer development and progression heavily rely on the tumor microenvironment. Additionally, it impacts the reaction of cancer cells toward immunotherapy [20]. These key factors that contribute to tumor resistance to immunotherapy include tumor heterogeneity, immunosuppressive cells, inflammatory cytokines, tumor-associated fibroblasts, and genetic alteration [[21], [22], [23], [24]]. Developing successful cancer treatments relies heavily on comprehending the mechanisms behind tumor resistance to immunotherapy. Scientists are currently exploring methods to overcome these obstacles and improve the immune system's ability to fight against cancerous cells. This involves the creation of novel treatments that focus on the tumor microenvironment, along with the discovery of biomarkers that can anticipate the reaction to immunotherapy.
Immunogenic cell death (ICD) is a kind of antitumor immunity that alters the immunological microenvironments of tumors by emitting warning signals, which is beneficial for immunotherapy [25,26]. Numerous studies have explored that ICD was majorly triggered and mediated by damage-associated molecular patterns (DAMPs). Chronic exposure to DAMPs may activate autoimmunity and advance immune-mediated elimination in the TME [27]. ICD enhances antitumor immune responses by transforming dying cancer cells into therapeutic immunizations [28]. Higher ICD propensity tumors may evoke a more robust antitumor immune response, helping to impede tumor proliferation. Nevertheless, studies on the clinical applications of ICD are limited. Therefore, further studies on BC patients should be explored in the clinical setting to validate the potential of ICD in tumor immunity. In a nutshell, the intervention of ICD may provide a novel research direction for the prognosis and therapy of patients with BC. Hence, we developed an innovative predictive framework utilizing the ICD molecular classification to forecast overall survival and provide improved treatment recommendations for individuals diagnosed with BC.
2. Materials and methods
2.1. Public data collecting and processing
In this study, we retrieve the raw read count of RNA sequencing from 412 individuals diagnosed with Bladder Urothelial Carcinoma (BLCA) and 19 healthy bladder tissues. Additionally, we collect mutation data and clinical information of 407 BLCA patients from The Cancer Genome Atlas (TCGA) repository. Simultaneously, data on the expression profiles of microarrays (GSE13507 for MIBC (n = 62) and NMIBC (n = 103) and GSE32894 for MIBC (n = 51)), along with clinical details of bladder cancer samples, were acquired from the Gene Expression Omnibus (GEO) database. To decrease the likelihood of non-cancer-related fatalities, we eliminated 6 patients diagnosed with BC who had a survival period of less than 30 days. Supplementary Table 1 displays the clinicopathological features of patients with bladder cancer in the TCGA and GEO cohorts.
2.2. Consensus clustering analysis of ICDRGs
34 ICDRGs in total (Supplementary Table 2) were extracted from earlier publications [29]. The “ConsensusClusteringPlus” package was utilized for performing consensus clustering analysis. Patients with BC in the TCGA dataset were categorized into various clusters. The best categorization was determined by analyzing the cumulative distribution function (CDF) curve and the correlations among clusters. After that, the “ggplot2” and “Rtsne” R packages were utilized to perform principal component analysis (PCA) and t-distributed stochastic neighbor embedding (t-SNE). The prognostic predictive performance of the ICD cluster was assessed using Kaplan-Meier curves. The heatmap was created using the R software package called “pheatmap”, and the distinctions were detected utilizing the R package “limma”.
2.3. Relationship of molecular patterns with TME in BC
To measure the amount of immune cells infiltrating in various ICD clusters, we conducted single-sample gene set enrichment analysis (ssGSEA) using the R package “GSEABase” [30]. Subsequently, the analysis of gene set variation (GSVA) enrichment was utilized to uncover the potential biological functions among different subtypes of ICD.
2.4. Identification of DEGs and functional enrichment analysis
Further investigation was conducted using genes that exhibited differential expression (DEGs) (with a false discovery rate (FDR) below 0.05 and a log|FC| exceeding 1) among various subtypes. For the analysis of DEGs, we utilize the R package called “clusterProfiler” to perform Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis.
2.5. Development of the ICD-related prognostic model (ICDRPM)
Using univariate Cox regression analysis, we acquired the prognostic DEGs linked to ICD clusters from the aforementioned outcomes. Later, we conducted Lasso Cox regression analysis with the assistance of the R package called “glmnet”. The TCGA training cohort was used to develop the ICDRPM after selecting Candidate DEGs through multivariate Cox analysis. The ICDRPM was used to calculate the ICD_score for every BC patient. Subsequently, the patients in both the training cohort and validation cohort were categorized into low- and high-risk groups according to the median ICD_score. The ICD_score equation is as follows: ICD_score = .
The coefficient and expression of each prognostic DEG are represented by coef and exp, respectively. To ascertain the distribution of patients in various risk groups, the distribution of patients in different risk groups was determined using the R package called “Rtsne” through PCA and tSNE analyses. The disparities in overall survival between these two groups were examined using K-Meier curves. The performance of ICDRPM was evaluated by calculating the receiver operating characteristic (ROC) curve with the assistance of the “survcomp” and “survivalROC” packages.
2.6. Clinical significance analysis of the prognostic ICD_score
We verified the independent prognostic performance of ICD_score by integrating patients’ clinical information and ICD_score for uniCox and multivariate Cox analysis, after excluding patients with incomplete data. The operating system of subgroups was established through K-M analysis utilizing the “survminer” software package. Moreover, the association between ICD_score and clinical characteristics was investigated utilizing the “limma” software package, encompassing factors such as sex, age, grade, and TNM stage.
2.7. Establishment of a predictive nomogram
The nomogram was plotted using the “rms” package in conjunction with the patients' ICD_core and other clinicopathological characteristics [31]. To examine the prognostic predictive precision of the nomogram, especially in forecasting the patients’ 1-, 3-, and 5-year overall survival (OS), the ROC curve was generated. To confirm the accuracy of the column line graphs, calibration curves were employed.
2.8. Assessment of the immune status between the two risk groups
The CIBERPORT algorithm was employed for calculating the abundance of immune infiltrating cells. To quantify the tumor microenvironment, we utilized the ESTIMATE algorithm [32] to compute the stromal score, immune score, and ESTIMATE score (which is the sum of stromal and immune scores) for each sample. Furthermore, a comparison was made between the risk groups regarding the levels of expression of immune checkpoints that are commonly observed. As an immune checkpoint inhibitors (ICIs) response indicator, the Immunophenotypescore (IPS) is positively correlated with tumor immunogenicity; a high IPS indicates an effective response to ICIs therapy. To compare the responses to immune checkpoint inhibitors (ICIs) among various risk groups, the BLCA cohort's IPS was obtained from The Cancer Immune Atlas (TCIA) website (https://www.tcia.at/home).
2.9. Genomic mutation and drug sensitivity analysis
To illustrate the distribution of gene variations between two risk groups, we utilized the R package called “maftools” to generate a waterfall diagram. In light of the significance of tumor mutation burden (TMB) in guiding immunotherapy [33], we additionally assessed the TMB score in two different risk categories. Additionally, a stratified survival analysis was conducted to investigate the association between TMB and the ICD_score. To assess the ability of ICD_score in predicting clinical drug therapy sensitivity in BC patients, we calculated the IC50 values for each patient for 198 drugs using the “oncoPredict” package [34].
2.10. Statistical analysis
Version 4.2.0 of R was utilized for all statistical analyses. Different group comparisons were conducted using the Wilcoxon and Kruskal-Wallis tests. The Kaplan-Meier analysis utilized log-rank tests. The symbols * represent a significance level of P ≤ 0.05, ** represent a significance level of P ≤ 0.01, *** represent a significance level of P ≤ 0.001, and **** represent a significance level of P ≤ 0.0001. Significance was observed among them, with a statistically significant difference at P < 0.05.
3. Results
3.1. Generation of ICD subtypes in BC
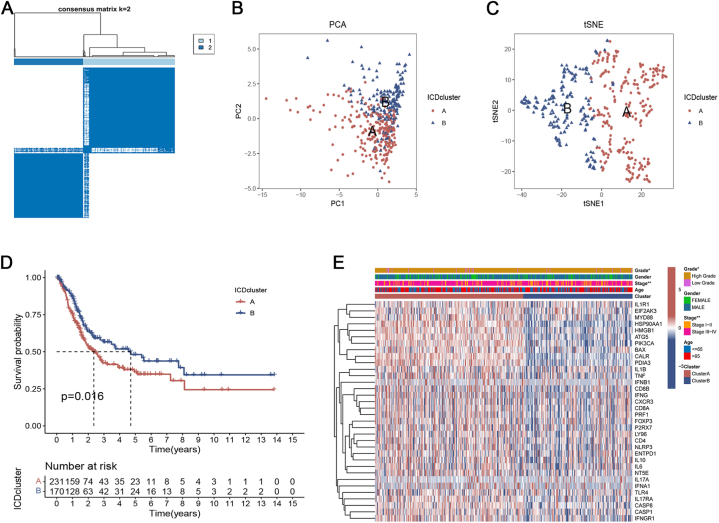
To further determine the correlation between ICD and BC, consensus clustering was performed to stratify BC samples into different subtypes based on the expression levels of 34 ICDRGs. We found that the intragroup correlations were the highest and the intergroup correlations were the lowest when k = 2 by increasing the clustering variable (k) from 2 to 9. Therefore, 401 patients in the TCGA cohort were stratified into two subtypes, namely ICD clusters A (n = 231) and B (n = 170) (Fig. 1A). PCA and t-SNE analyses analysis showed the reliability of the grouping (Fig. 1B and C). According to the survival analysis, Fig. 1D shows that the OS of subtype B patients was remarkably higher than that of subtype A patients. Furthermore, ICD cluster A exhibited elevated levels of gene expression related to ICD and was linked to a more advanced AJCC stage and lower grade (Fig. 1E).
Fig. 1.
Generation of ICD subtypes in bladder cancer patients. (A) Consensus matrix heatmaps at optimal k = 2 and their related regions. (B, C) PCA and t-SNE analyses reflected huge differences in the transcriptome between the two ICD subtypes. (D) The overall survival curve of bladder cancer patients in two ICD subtypes. (E) The heatmap visualized the correlation between the clinical features of patients and two ICD subtypes.
3.2. Characteristics of TME in different subtypes
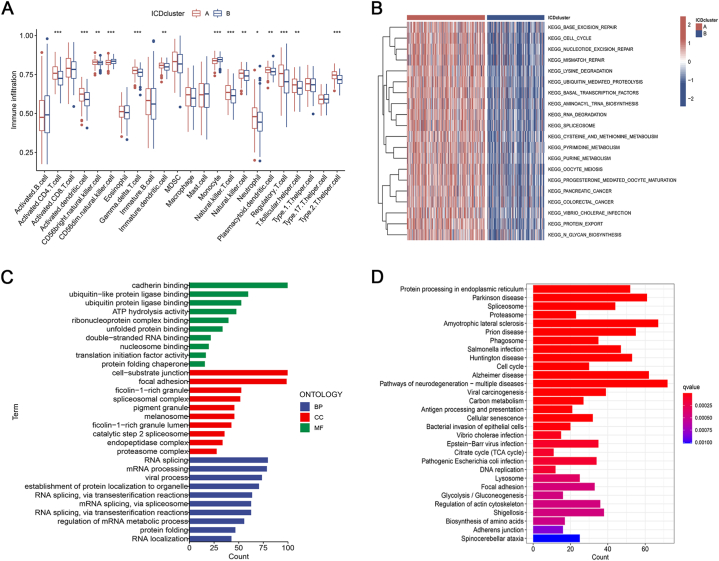
We observed that the percentage of most infiltrated immune cells varied across ICD clusters based on the ssGSEA algorithm (Fig. 2A). The levels of activated CD4 T cells, CD56bright natural killer cells, gamma delta T cells, activated dendritic cells, natural killer T cells, natural killer cells, neutrophils, plasmacytoid dendritic cells, regulatory T cells, T follicular helper cells, and Type 2 T helper cells of subtype A were significantly higher compared to subtype B. On the other hand, CD56dim natural killer cells and mast cells of subtype A were noticeably lower than subtype B. Furthermore, we conducted GSVA analysis to compare the disparities in ICD-related enrichment pathways between the two subgroups. The analysis revealed that cell cycle, pancreatic cancer, colorectal cancer, and metabolism were enriched in ICD cluster A (Fig. 2B). The findings suggested that various categories of ICD had distinct impacts on the physiological functioning of individuals diagnosed with BC.
Fig. 2.
Characteristics of TME and biological function analysis. (A) Correlation between the two subtypes and 23 human immune cell subgroups. (B) GSVA analyses of the two ICD subtypes. (C) GO enrichment analysis of DEGs between two distinct subtypes. (D) KEGG enrichment analysis of DEGs between two distinct subtypes.
3.3. The potential biological function of DEGs
To examine the possible biological characteristics of ICD subtypes, we acquired 1162 DEGs associated with ICD between cluster A and cluster B through the utilization of the “limma” package. Following this, we conducted GO and KEGG analyses. According to the GO terms, the differentially expressed genes (DEGs) were predominantly enriched in RNA splicing, focal adhesion, and binding to cadherin (Fig. 2C). KEGG analysis showed significant enrichment in cell cycle, focal adhesion, antigen processing and presentation, cellular senescence, glycolysis/gluconeogenesis, etc (Fig. 2D).
3.4. Establishment and validation of the ICDRPM
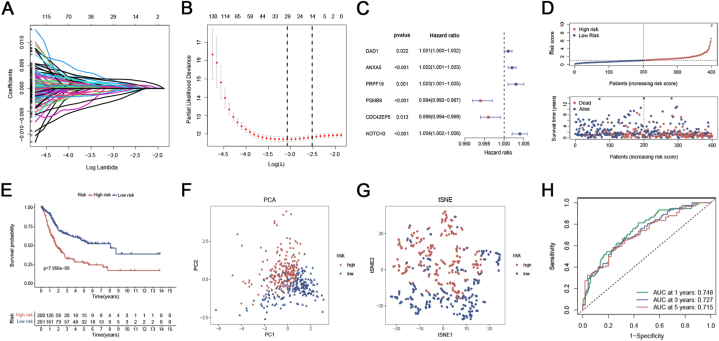
Using uniCox analysis, we assessed the predictive significance of DEGs linked to ICD subtypes in the training set and identified a total of 219 genes associated with prognosis at p < 0.05 (Supplementary Table 3). Subsequently, LASSO and multiCox analyses for these prognosis-related DEGs were performed to develop an optimal prognostic model (Fig. 3A–C). Finally, 6 genes (DAD1, ANXA5, PRPF19, PSMB9, CDC42EP5, and NOTCH3) were obtained in accordance with the Akaike Information Criteria (AIC) values. The ICD_score was constructed using the findings from multiCox analysis.
Fig. 3.
Establishment of the ICDRPM in the TCGA training cohort. (A) The changing track of the Lasso regressed independent variable, the abscissa represented the logarithm of the independent variable lambda, and the ordinate represented the coefficient of the independent variable. (B) The confidence interval under each Lambda of Lasso. (C) Multivariable Cox regression analysis of OS. Data are presented as hazard ratio (HR) ± 95% confidence interval [CI]. (D) The distribution of ICD_score and survival status of bladder cancer patients with increasing ICD risk scores. (E) Kaplan-Meier curve of OS according to the ICDRPM in the TCGA cohort. (F, G) PCA and t-SNE analyses of high and low-risk groups. (H) ROC curve for predicting OS at 1, 3, and 5 years.
ICD_score = (0.001160 * expression of DAD1) + (0.002261 * expression of ANXA5) + (0.003111 * expression of PRPF19) + (−0.005631 * expression of PSMB9) + (−0.003542 * expression of CDC42EP5) + (0.004057 * expression of NOTCH3). All individuals were categorized into low- or high-risk groups based on the median ICD_score, and OS in the training cohort was compared. As the ICD_score increased, the patients' OS decreased and the mortality rate showed a gradual increase (Fig. 3D). In the training cohort, patients at high risk exhibited poor overall survival outcomes (Fig. 3E). The patients in the two risk groups demonstrated a strong ability to be distinguished according to the results of PCA and t-SNE analyses (Fig. 3F and G). In the training cohort, the model accurately predicted patients’ OS at 1, 3, and 5 years with AUC values of 0.748, 0.727, and 0.715, respectively, as shown in Fig. 3H.
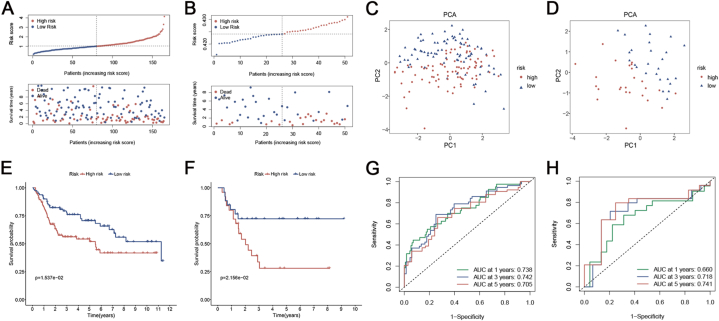
The ICD_score in the GSE13507 BCE cohort (including NMIBC and MIBC) and GSE32894 MIBC cohort was calculated using the identical coefficient and formula to ensure the validity of the signature. The TCGA training cohort stratified all BC patients into low- and high-risk groups using identical cut-off values. Likewise, as the ICD_score rose, the patients’ overall survival declined, and the mortality rate steadily rose in both GSE13507 and GSE32894 groups (Fig. 4A and B). From PCA, the ICD_score exhibited good discriminating power with two distinct clusters (Fig. 4C and D). In both GSE13507 and GSE32894 cohorts, the Kaplan-Meier analysis demonstrated that patients with a low risk had superior overall survival compared to patients with a high risk (Fig. 4E and F). Additional examination of ROC curves verified the strong predictive ability of the signature in both the GSE13507 and GSE32894 groups (Fig. 4G and H). When combined, the ICDRPM demonstrated robust efficacy in predicting survival in the TCGA training dataset as well as two GEO validation datasets.
Fig. 4.
Validation of the ICDRPM in the GSE13507 cohort. (A) The distribution of ICD_score and survival status of bladder cancer patients with increasing ICD risk scores. (B) PCA analysis of high and low-risk groups. (C) Kaplan-Meier curve of OS according to the ICDRPM in the validation cohort. (D) ROC curve for predicting OS at 1, 3, and 5 years.
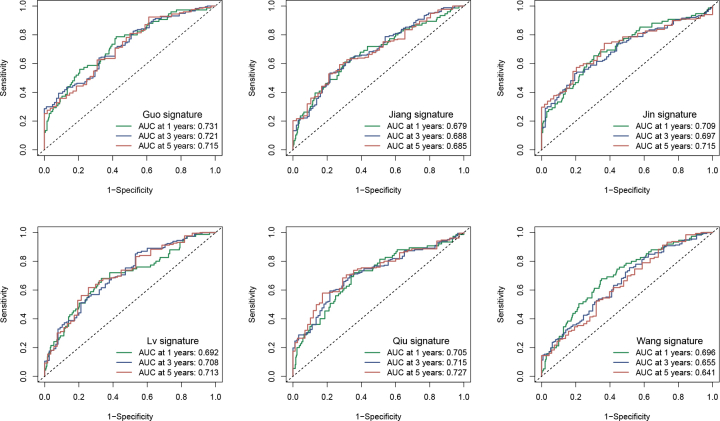
Afterward, we chose six immune signatures that had been previously published for bladder cancer to compare with our signature (Fig. S1). According to the results, the area under the curve (AUC) of our signature was considerably better than that of other clinical features (Fig. S1). Additionally, our signature surpassed the performance of the other six immune signatures previously published.
3.5. Clinical correlation analysis of the prognostic ICD_score
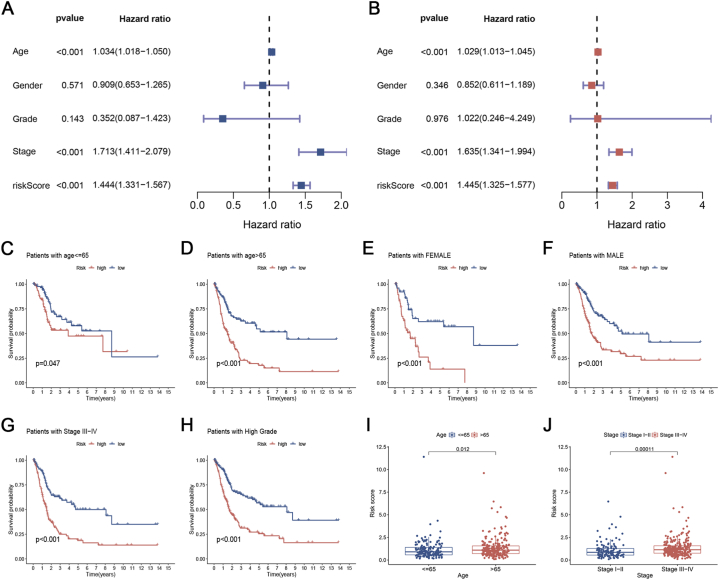
To assess the autonomous predictive significance of ICD_score (Fig. 5A and B), we conducted both uniCox and multiCox examinations. According to the forest plot, the ICD_score was identified as a separate prognostic determinant. To calculate the ICD_score within the subgroups, we additionally performed survival analysis on the subgroups. Individuals were categorized based on different clinical traits. According to the data presented in Fig. 5C–H, individuals classified as low-risk had longer lifespans across different age groups (≤65 or > 65 years), gender groups (female or male), stage III-IV, and high-grade groups when compared to those classified as high-risk. Furthermore, to investigate the connection between ICD_score and clinical characteristics, we examined the association between clinical data and observed that older individuals exhibited an elevated ICD_score (Fig. 5I). Furthermore, a greater risk score was observed in the advanced TNM stage (Fig. 5J).
Fig. 5.
Clinical correlation analysis of the prognostic ICD_score. (A, B) Univariate and multivariate Cox regression analyses of ICD_score and other clinical variables. Data are presented as hazard ratio (HR) ± 95% confidence interval [CI]. (C–H) Kaplan-Meier curve of OS according to the ICDRPM in the TCGA subgroup with age younger than 65 (C), age older than 65 (D), female (E), male (F), stage III-IV (G), and high-grade (H). (I, J) Association of ICD_score with patients' age (I) and TNM stage (J).
3.6. Construction and evaluation of nomograph
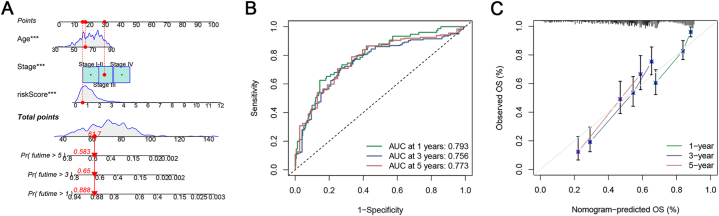
A line graph was drawn depicting the factors involved in the construction of the nomogram, which included ICD_score, stage, and age. The line graphs could predict the 1-, 3- and 5-year OS of patients with BC based on the total score (Fig. 6A). The ROC curve reflects the high accuracy of the line graph in predicting OS (Fig. 6B). The calibration curves showed strong fitting between predictions and observations (Fig. 6C). The findings indicate that the utilization of ICD_score in conjunction with additional clinical factors can greatly enhance the capacity to forecast patient prognosis.
Fig. 6.
Construction and evaluation of nomograph. (A) Development of a nomogram by combining ICD_score with age, and TNM stage to predict the survival probability. (B) ROC curves of the nomogram. (C) Calibration plots of the nomogram.
3.7. Assessment of TIME and immunotherapeutic response in distinct groups
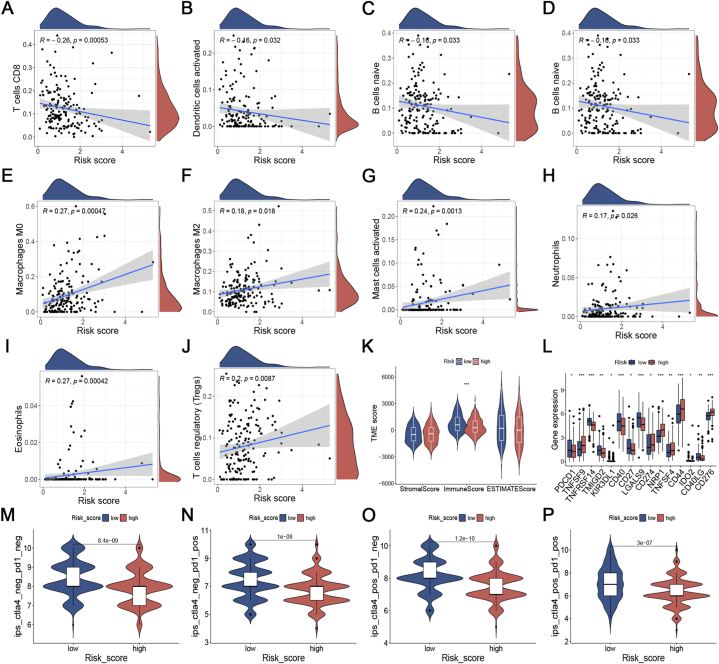
Distinct ratios of immune cells were observed in the two risk groups (Fig. 7A–J). The ICD_score exhibited a negative correlation with CD8+ T cells, activated dendritic cells, B cells naïve, and plasma cells. Conversely, M0 and M2 Macrophages, Neutrophils, Eosinophils, T cells regulatory, and activated mast cells showed a positive correlation with the ICD_score. Later on, it was discovered that the immune score of the low-risk category showed a clear tendency to rise in comparison to the high-risk category (Fig. 7K). These results demonstrated that ICDRPM could accurately predict TIME in the different risk populations.
Fig. 7.
Assessment of tumor immune microenvironment and immunotherapeutic response in distinct groups. (A–J) Correlation of ICD_score with 28 immune cells. (K) Comparison of immune score in high-risk and low-risk patients. (L) Differential expression of immune checkpoint molecules in the high- and low-risk groups. (M − P) Differential levels of IPS with CTLA-4 (pos/neg)/PD-1 (pos/neg) in different score groups. pos: positive, neg: negative. IPS: immunophenoscore.
ICI is regarded as a significant advancement in the field of cancer therapy. Consequently, we conducted a comparison of 35 prevalent ICPs in two distinct risk categories, including PD-1, PD-L1, and TNFSF families. The expression levels of these ICPs exhibited notable variations between the two risk subgroups (Fig. 7L). In the assessment of IPS for patients in the risk groups, we discovered that those classified as low risk exhibited elevated IPS scores when treated with anti-PD-1 therapy, anti-CTLA4 therapy, or a combination of both (Fig. 7M − P).
3.8. Mutation and drug susceptibility analysis
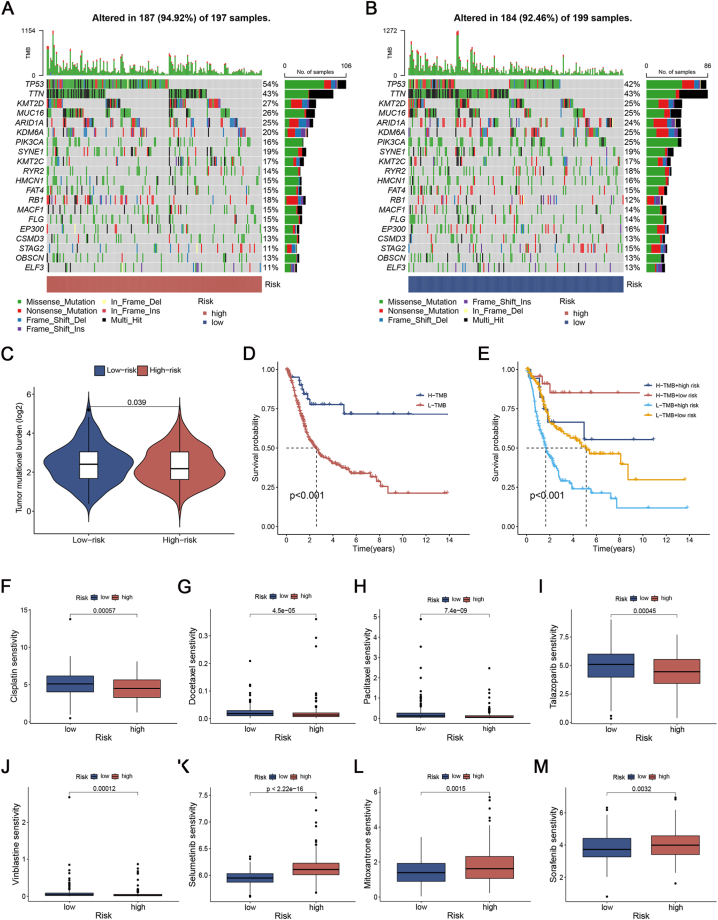
Fig. 8A and B displayed the genomic mutation landscape of the two risk categories in TCGA-BLCA. According to the data presented in Fig. 8A and B, the group with the highest score exhibited elevated mutation rates. From the low-risk to the high-risk groups, the TP53 gene mutation rates showed a more notable increase (42% vs. 54%; Fig. 8A and B) compared to other genes. After conducting additional analysis, we observed that the TMB of the high-risk group was comparatively lower than that of the low-risk group (Fig. 8C). Afterward, we categorized the patients into two TMB groups based on the TMB score. According to Fig. 8D, the group with a high TMB exhibited a greater rate of survival compared to the group with a low TMB. Fig. 8E illustrated that patients with a low ICD_score had a significantly better OS compared to those with a high ICD_score, regardless of their TMB level.
Fig. 8.
Mutation and drug susceptibility analysis. (A, B) SNV waterfall of top 20 (mutation frequency) genes in high-risk group (A) and low-risk group (B). (C) Comparison of TMB between the high- and low-risk groups. (D, E) Survival analysis of TMB (D) and TMB combined with the ICD_score (E). TMB: tumor mutation burden. (F–M) IC50 box diagram of the 8 drugs with the significant difference in drug sensitivity in the high- and low-risk groups respectively.
To investigate the potential clinical application of the two risk groups, the GDSC database was used to predict the IC50 value of 198 drugs, indicating their sensitivity to medication. Of which 84 drugs exhibited significant differences between the two risk groups. Patients with high ICD_score may have positive responses to Cisplatin, Docetaxel, Paclitaxel, Talazoparib, and Vinblastine, while patients with low ICD_score may have positive responses to Selumetinib, Mitoxantrone, and Sorafenib, etc (Fig. 8F-M).
4. Discussion
ICD, which was identified in recent decades, sheds light on the pertinent conversation between dying malignant cells and adaptive immunocytes in cancer treatment [35]. ICD modifies the tumor immunological microenvironment by emitting danger signals or DAMPs, which may benefit immunotherapy [35]. Consequently, ICD-related biomarkers may aid in identifying BC patients who may benefit from antitumor therapy. At present, most studies focus on one or two ICDRGs or one type of immune cell. However, the characteristics of TMEs and the overall impact of the combined action of numerous ICDRGs have not been thoroughly explained. Hence, the objective of this research was to systematically investigate ICDRGs and develop a precise and user-friendly clinical scoring framework using ICD molecular subtypes. This framework will assist in accurately forecasting the prognosis of individuals with BC and providing personalized treatment recommendations.
In the current research, we assessed the manifestation of 34 ICDRGs in BC and healthy tissues, resulting in the categorization of patients into two distinct molecular subcategories. The two subtypes exhibited notable variations in clinical outcomes and immune infiltrating cell. Furthermore, there is a strong correlation between the differentially expressed genes (DEGs) of the two ICD subtypes and the pathways associated with immunity and tumors. Next, we built a robust ICDRPM and a quantitative nomogram that significantly improves its performance. The high and low ICD_score groups exhibited notable variations in clinical characteristics, prognosis, tumor microenvironment (TME), immune checkpoint expression, mutation profile, response to immunotherapy, and sensitivity to drugs. The ICDRPM allows for more precise forecasts and categorizations of patients’ prognoses, which hold immense importance in clinical practice and also showcase individual treatment results.
BC is a type of cancer that triggers an immune response due to its high tumor mutation burden (TMB) and the presence of neoantigens [36]. Additionally, it interacts with immune cells in the tumor microenvironment (TME). The prognosis of BC patients is significantly influenced by numerous immune cell types in the TME [37,38]. Therefore, we examined the correlation between ICD_score and the immune landscape and found that the high-risk category displayed a characteristic tumor microenvironment with immunosuppression. In particular, there was a notable decrease in the prevalence of CD8+ T cells and activated dendritic cells among the high-risk group, whereas there was an increase in the prevalence of Tregs, M0, and M2 macrophages. Moreover, the immune score of the high-risk cohort was significantly inferior to that of the low-risk cohort. Favorable overall survival was observed in correlation with an augmentation of infiltrating CD8+ T cells [7]. B cells can inhibit cancer progression by killing cancer cells directly, secreting immunoglobulins, or promoting T cell response [39]. In contrast, Tregs play a crucial role in preserving immune tolerance by not only suppressing effector cells in the tumor but also serving as vital participants in the peritumor, stroma, vasculature, and lymphatics to restrict anti-tumor immune responses [40]. Additionally, M0 macrophages can transform into M2 macrophages, thereby facilitating immune evasion [41]. Furthermore, mast cells play a crucial role in controlling both inflammation and immunosuppression [42]. In addition to inhibiting the immune response against tumors through the release of anti-inflammatory cytokines, they also can promote tumor growth by controlling the formation of new blood vessels [43,44]. The low-risk subgroup showed an increased presence of immune cells infiltrating the tumors, leading to their classification as “hot tumors” and indicating significant potential for fighting cancer. The presence of an immunosuppressive TIME in the high-risk subgroup of patients could potentially account for the significantly poorer survival rate observed in high-risk BC patients.
Clinical outcomes and response to immunotherapy have been demonstrated to be associated with TMB. Multiple research studies have indicated that elevated TMB demonstrates efficacy in forecasting rates of objective remission and progression-free survival; however, its capacity to predict overall survival is restricted [45]. Through our research, we discovered a strong correlation between TMB and the ICD_score. By incorporating both TMB and the score into a comprehensive predictive model, we were able to enhance survival prognosis, thereby offering valuable insights for immunotherapy guidance. The success of immune-checkpoint blockade has greatly contributed to the effectiveness of immunotherapy as a significant clinical approach in cancer treatment. The safety and effectiveness of some ICIs have been confirmed by clinical studies [46,47]. During this investigation, we noticed a notable disparity in the manifestation of PD-L1 and PD-1 among the various rating categories. The IPS demonstrated improved treatment outcomes in the low score category irrespective of the PD-1 and CTLA-4 status, offering a valuable reference for immunotherapy.
Taken together, we systematically explored ICDRGs and established a novel clinical scoring system that would facilitate clinical application, especially in combination with clinical features. These findings may contribute to further understanding of the role of ICDRGs in BC progression as well as drug treatment responses, which highlights the potential of this model in prognosis prediction and targeted therapy of BC. However, there are several shortcomings in this study. First, the data utilized in this research are obtained from publicly accessible sources, and it is necessary to validate them further by incorporating data from a prospective and multicenter study. Second, the interaction between the genes involved in constructing the model and tumor immunity needs to be tested by experimental investigations in vitro and in vivo. Third, several key clinical variables, such as surgery and neoadjuvant chemoradiotherapy are not available, which may affect prognosis and the prognostic state of immune response.
5. Conclusions
We developed and verified a robust and potent prognostic signature associated with ICD to anticipate the survival results and immunotherapy response in BC patients. This signature also aids in optimizing individualized therapy, rendering it highly valuable for clinical application and translation.
Author contribution statement
Lei Gu: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Gang Hu: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Juan Hu: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Fei Wen: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
Not applicable.
Data availability statement
All transcriptome and expression profile data were obtained from The Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov/) and Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/). These data are publicly available.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors are grateful for the help of all colleagues and the public database.
Footnotes
Supplementary data to this article can be found online at 10.1016/j.heliyon.2023.e18848.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
figs1.
Comparison of our signature with other immune signatures. The ROC curves for the Guo, Jiang, Jin, Lv, Qiu, and Wang risk models at 1, 3, and 5 years of OS.
References
- 1.Witjes J.A., Bruins H.M., Cathomas R., Compérat E.M., Cowan N.C., Gakis G., et al. European association of urology guidelines on muscle-invasive and metastatic bladder cancer: summary of the 2020 guidelines. Eur. Urol. 2021 Jan;79(1):82–104. doi: 10.1016/j.eururo.2020.03.055. [DOI] [PubMed] [Google Scholar]
- 2.Grayson M. Bladder cancer. Nature. 2017 Nov 8;551(7679):S33. doi: 10.1038/551S33a. [DOI] [PubMed] [Google Scholar]
- 3.Shelley M.D., Jones G., Cleves A., Wilt T.J., Mason M.D., Kynaston H.G. Intravesical gemcitabine therapy for non-muscle invasive bladder cancer (NMIBC): a systematic review. BJU Int. 2012 Feb;109(4):496–505. doi: 10.1111/j.1464-410X.2011.10880.x. [DOI] [PubMed] [Google Scholar]
- 4.Rosenberg J.E., Hoffman-Censits J., Powles T., van der Heijden M.S., Balar A.V., Necchi A., et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016 May 7;387(10031):1909–1920. doi: 10.1016/S0140-6736(16)00561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma P., Retz M., Siefker-Radtke A., Baron A., Necchi A., Bedke J., et al. Nivolumab in metastatic urothelial carcinoma after platinum therapy (CheckMate 275): a multicentre, single-arm, phase 2 trial. Lancet Oncol. 2017 Mar;18(3):312–322. doi: 10.1016/S1470-2045(17)30065-7. [DOI] [PubMed] [Google Scholar]
- 6.Patel V.G., Oh W.K., Galsky M.D. Treatment of muscle-invasive and advanced bladder cancer in 2020. CA A Cancer J. Clin. 2020 Sep;70(5):404–423. doi: 10.3322/caac.21631. [DOI] [PubMed] [Google Scholar]
- 7.Fridman W.H., Zitvogel L., Sautès-Fridman C., Kroemer G. The immune contexture in cancer prognosis and treatment. Nat. Rev. Clin. Oncol. 2017 Dec;14(12):717–734. doi: 10.1038/nrclinonc.2017.101. [DOI] [PubMed] [Google Scholar]
- 8.Xiao M., Xie L., Cao G., Lei S., Wang P., Wei Z., et al. CD4(+) T-cell epitope-based heterologous prime-boost vaccination potentiates anti-tumor immunity and PD-1/PD-L1 immunotherapy. J. Immunother. Canc. 2022 May;10(5) doi: 10.1136/jitc-2021-004022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park J., Hsueh P.C., Li Z., Ho P.C. Microenvironment-driven metabolic adaptations guiding CD8(+) T cell anti-tumor immunity. Immunity. 2023 Jan 10;56(1):32–42. doi: 10.1016/j.immuni.2022.12.008. [DOI] [PubMed] [Google Scholar]
- 10.Marciscano A.E., Anandasabapathy N. The role of dendritic cells in cancer and anti-tumor immunity. Semin. Immunol. 2021 Feb;52 doi: 10.1016/j.smim.2021.101481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi I.K., Wang Z., Ke Q., Hong M., Paul D.W., Jr., Fernandes S.M., et al. Mechanism of EBV inducing anti-tumour immunity and its therapeutic use. Nature. 2021 Feb;590(7844):157–162. doi: 10.1038/s41586-020-03075-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu G., Zhang Q., Zhang J., Liu F. Targeting tumor-associated antigen: a promising CAR-T therapeutic strategy for glioblastoma treatment. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.661606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laureano R.S., Sprooten J., Vanmeerbeerk I., Borras D.M., Govaerts J., Naulaerts S., et al. Trial watch: dendritic cell (DC)-based immunotherapy for cancer. OncoImmunology. 2022;11(1) doi: 10.1080/2162402X.2022.2096363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie Y., Xie F., Zhang L., Zhou X., Huang J., Wang F., et al. Targeted anti-tumor immunotherapy using tumor infiltrating cells. Adv. Sci. 2021 Nov;8(22) doi: 10.1002/advs.202101672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang S.S., Liu W., Ly D., Xu H., Qu L., Zhang L. Tumor-infiltrating B cells: their role and application in anti-tumor immunity in lung cancer. Cell. Mol. Immunol. 2019 Jan;16(1):6–18. doi: 10.1038/s41423-018-0027-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tay C., Tanaka A., Sakaguchi S. Tumor-infiltrating regulatory T cells as targets of cancer immunotherapy. Cancer Cell. 2023 Mar 13;41(3):450–465. doi: 10.1016/j.ccell.2023.02.014. [DOI] [PubMed] [Google Scholar]
- 17.Duong E., Fessenden T.B., Lutz E., Dinter T., Yim L., Blatt S., et al. Type I interferon activates MHC class I-dressed CD11b(+) conventional dendritic cells to promote protective anti-tumor CD8(+) T cell immunity. Immunity. 2022 Feb 8;55(2):308. doi: 10.1016/j.immuni.2021.10.020. 23.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ozga A.J., Chow M.T., Luster A.D. Chemokines and the immune response to cancer. Immunity. 2021 May 11;54(5):859–874. doi: 10.1016/j.immuni.2021.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geuijen C., Tacken P., Wang L.C., Klooster R., van Loo P.F., Zhou J., et al. A human CD137×PD-L1 bispecific antibody promotes anti-tumor immunity via context-dependent T cell costimulation and checkpoint blockade. Nat. Commun. 2021 Jul 21;12(1):4445. doi: 10.1038/s41467-021-24767-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bejarano L., Jordāo M.J.C., Joyce J.A. Therapeutic targeting of the tumor microenvironment. Cancer Discov. 2021 Apr;11(4):933–959. doi: 10.1158/2159-8290.CD-20-1808. [DOI] [PubMed] [Google Scholar]
- 21.Vesely M.D., Zhang T., Chen L. Resistance mechanisms to anti-PD cancer immunotherapy. Annu. Rev. Immunol. 2022 Apr 26;40:45–74. doi: 10.1146/annurev-immunol-070621-030155. [DOI] [PubMed] [Google Scholar]
- 22.Vitale I., Shema E., Loi S., Galluzzi L. Intratumoral heterogeneity in cancer progression and response to immunotherapy. Nat. Med. 2021 Feb;27(2):212–224. doi: 10.1038/s41591-021-01233-9. [DOI] [PubMed] [Google Scholar]
- 23.Kumari N., Choi S.H. Tumor-associated macrophages in cancer: recent advancements in cancer nanoimmunotherapies. J. Exp. Clin. Cancer Res. 2022 Feb 19;41(1):68. doi: 10.1186/s13046-022-02272-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenkins L., Jungwirth U., Avgustinova A., Iravani M., Mills A., Haider S., et al. Cancer-associated fibroblasts suppress CD8+ T-cell infiltration and confer resistance to immune-checkpoint blockade. Cancer Res. 2022 Aug 16;82(16):2904–2917. doi: 10.1158/0008-5472.CAN-21-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garg A.D., Agostinis P. ER stress, autophagy and immunogenic cell death in photodynamic therapy-induced anti-cancer immune responses. Photochem. Photobiol. Sci. : Off. J. Eur. Photochem. Ass. Eur. Soci. Photobiol. 2014 Mar;13(3):474–487. doi: 10.1039/c3pp50333j. [DOI] [PubMed] [Google Scholar]
- 26.Kepp O., Senovilla L., Vitale I., Vacchelli E., Adjemian S., Agostinis P., et al. Consensus guidelines for the detection of immunogenic cell death. OncoImmunology. 2014 Oct;3(9) doi: 10.4161/21624011.2014.955691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou J., Wang G., Chen Y., Wang H., Hua Y., Cai Z. Immunogenic cell death in cancer therapy: present and emerging inducers. J. Cell Mol. Med. 2019 Aug;23(8):4854–4865. doi: 10.1111/jcmm.14356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duan X., Chan C., Lin W. Nanoparticle-mediated immunogenic cell death enables and potentiates cancer immunotherapy. Angew Chem. Int. Ed. Engl. 2019 Jan 14;58(3):670–680. doi: 10.1002/anie.201804882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garg A.D., De Ruysscher D., Agostinis P. Immunological metagene signatures derived from immunogenic cancer cell death associate with improved survival of patients with lung, breast or ovarian malignancies: a large-scale meta-analysis. OncoImmunology. 2016 Feb;5(2) doi: 10.1080/2162402X.2015.1069938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foroutan M., Bhuva D.D., Lyu R., Horan K., Cursons J., Davis M.J. Single sample scoring of molecular phenotypes. BMC Bioinf. 2018 Nov 6;19(1):404. doi: 10.1186/s12859-018-2435-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harrell F.E., Jr., Lee K.L., Mark D.B. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat. Med. 1996 Feb 28;15(4):361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 32.Yoshihara K., Shahmoradgoli M., Martínez E., Vegesna R., Kim H., Torres-Garcia W., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mariathasan S., Turley S.J., Nickles D., Castiglioni A., Yuen K., Wang Y., et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018 Feb 22;554(7693):544–548. doi: 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maeser D., Gruener R.F., Huang R.S. oncoPredict: an R package for predicting in vivo or cancer patient drug response and biomarkers from cell line screening data. Briefings Bioinf. 2021 Nov 5;(6):22. doi: 10.1093/bib/bbab260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krysko D.V., Garg A.D., Kaczmarek A., Krysko O., Agostinis P., Vandenabeele P. Immunogenic cell death and DAMPs in cancer therapy. Nat. Rev. Cancer. 2012 Dec;12(12):860–875. doi: 10.1038/nrc3380. [DOI] [PubMed] [Google Scholar]
- 36.Kandoth C., McLellan M.D., Vandin F., Ye K., Niu B., Lu C., et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013 Oct 17;502(7471):333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Z., Zhou Q., Wang Z., Zhang H., Zeng H., Huang Q., et al. Intratumoral TIGIT(+) CD8(+) T-cell infiltration determines poor prognosis and immune evasion in patients with muscle-invasive bladder cancer. J. Immunother. Canc. 2020 Aug;8(2) doi: 10.1136/jitc-2020-000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zirakzadeh A.A., Sherif A., Rosenblatt R., Ahlén Bergman E., Winerdal M., Yang D., et al. Tumour-associated B cells in urothelial urinary bladder cancer. Scand. J. Immunol. 2020 Feb;91(2) doi: 10.1111/sji.12830. [DOI] [PubMed] [Google Scholar]
- 39.Tokunaga R., Naseem M., Lo J.H., Battaglin F., Soni S., Puccini A., et al. B cell and B cell-related pathways for novel cancer treatments. Cancer Treat Rev. 2019 Feb;73:10–19. doi: 10.1016/j.ctrv.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scott E.N., Gocher A.M., Workman C.J., Vignali D.A.A. Regulatory T cells: barriers of immune infiltration into the tumor microenvironment. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.702726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roulleaux Dugage M., Nassif E.F., Italiano A., Bahleda R. Improving immunotherapy efficacy in soft-tissue sarcomas: a biomarker driven and histotype tailored review. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.775761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang B., Lei Z., Zhang G.M., Li D., Song C., Li B., et al. SCF-mediated mast cell infiltration and activation exacerbate the inflammation and immunosuppression in tumor microenvironment. Blood. 2008 Aug 15;112(4):1269–1279. doi: 10.1182/blood-2008-03-147033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varricchi G., Galdiero M.R., Loffredo S., Marone G., Iannone R., Marone G., et al. Are mast cells MASTers in cancer? Front. Immunol. 2017;8:424. doi: 10.3389/fimmu.2017.00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiong Y., Liu L., Xia Y., Qi Y., Chen Y., Chen L., et al. Tumor infiltrating mast cells determine oncogenic HIF-2α-conferred immune evasion in clear cell renal cell carcinoma. Cancer Immunol. Immunother. 2019 May;68(5):731–741. doi: 10.1007/s00262-019-02314-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li R., Han D., Shi J., Han Y., Tan P., Zhang R., et al. Choosing tumor mutational burden wisely for immunotherapy: a hard road to explore. Biochim. Biophys. Acta Rev. Canc. 2020 Dec;1874(2) doi: 10.1016/j.bbcan.2020.188420. [DOI] [PubMed] [Google Scholar]
- 46.Riley R.S., June C.H., Langer R., Mitchell M.J. Delivery technologies for cancer immunotherapy. Nat. Rev. Drug Discov. 2019 Mar;18(3):175–196. doi: 10.1038/s41573-018-0006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y., Zhang Z. The history and advances in cancer immunotherapy: understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell. Mol. Immunol. 2020 Aug;17(8):807–821. doi: 10.1038/s41423-020-0488-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All transcriptome and expression profile data were obtained from The Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov/) and Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/). These data are publicly available.