Abstract
The regions of the simian virus 40 (SV40) core origin that are required for stable assembly of virally encoded T antigen (T-ag) and the T-ag origin binding domain (T-ag-obd131–260) have been determined. Binding of the purified T-ag-obd131–260 is mediated by interactions with the central region of the core origin, site II. In contrast, T-ag binding and hexamer assembly requires a larger region of the core origin that includes both site II and an additional fragment of DNA that may be positioned on either side of site II. These studies indicate that in the context of T-ag, the origin binding domain can engage the pentanucleotides in site II only if a second region of T-ag interacts with one of the flanking sequences. The requirements for T-ag double-hexamer assembly are complex; the nucleotide cofactor present in the reaction modulates the sequence requirements for oligomerization. Nevertheless, these experiments provide additional evidence that only a subset of the SV40 core origin is required for assembly of T-ag double hexamers.
Little is known about eukaryotic origins of replication or the protein-DNA interactions that take place at these sites. One reason for this situation is that sequences that constitute higher eukaryotic origins of replication have not been extensively characterized (10, 25). An additional reason is that although many of the proteins that interact with eukaryotic origins of replication have been identified (reviewed in references 6, 36, and 82), little is known about the structures of these molecules. Therefore, well-defined viral model systems are being used to provide insights into the protein-DNA interactions that occur at eukaryotic origins. The goal of these studies is to define, in molecular terms, the steps that take place during initiation of DNA replication and the regulation of these events.
To address these issues, we and others have been analyzing the protein-DNA interactions that occur at the simian virus 40 (SV40) origin of replication. What is known about T antigen (T-ag), the SV40 origin, and their interactions has been thoroughly reviewed (2, 7, 29); therefore, only a brief overview of this topic, which emphasizes recent developments in this field, is provided. T-ag is a 708-amino-acid phosphoprotein that catalyzes many of the steps necessary for the initiation of SV40 DNA replication. For instance, it binds to the core origin, catalyzes origin-specific DNA unwinding, and recruits cellular proteins required for the initiation process (for reviews, see references 2, 7, and 29). The functional complexity of T-ag is mirrored in its organization. It is composed of several structural domains including a J domain (11, 41, 72), a DNA binding domain (for reviews see references 7 and 29), and a region containing the ATPase activity (12, 31, 88). The best-defined and most extensively studied domain of T-ag is the origin binding domain (T-ag-obd). This domain is necessary and sufficient for sequence-specific binding to the SV40 core origin at levels comparable to that of the full-length T-ag (reviewed in reference 7).
The 64-bp SV40 core origin is composed of three separate regions (19, 21, 58). The central region, termed site II, contains four GAGGC pentanucleotides, arranged as inverted pairs; each pentanucleotide serves as a binding site for T-ag (24, 38, 48, 77, 78). Site II is flanked by a 17-bp adenine-thymine (AT)-rich domain and a second region termed the early palindrome (EP). Upon T-ag binding to the core origin, the EP is the site of initial melting (4, 58). Regarding the AT-rich region, it includes a sequence of eight adenines, which induce a bend in the DNA structure (20, 34, 44). It has also been reported that this region activates the DNA helicase function of T-ag bound to the origin (58) and subsequent unwinding (45).
In the presence of ATP, T-ag forms a bilobed structure on the SV40 core origin (17, 18, 50). Each lobe consists of a hexamer of T-ag encircling the DNA segment to which it is bound (14, 50, 59, 65, 81). Experiments indicate that hexamer formation is initiated by the binding of a T-ag monomer to a single pentanucleotide (14, 38), followed by the assembly, via protein-protein interactions, of five additional T-ag monomers (14, 35, 50). DNase I-based footprinting techniques revealed that the double-hexamer complex formed in the presence of ATP covers the entire core origin complex (3, 22). Recent studies indicate that protein-protein interactions between hexamers are mediated through the T-ag-obd and an N-terminal phosphorylated region (83). Moreover, it has been reported that preformed hexamers do not bind to DNA (14, 35, 67). It is interesting that T-ag molecules encoded by a number of papovaviruses are highly homologous (70). Furthermore, the architectural features of the SV40 core origin are very similar to those in the replication origins of other polyomaviruses (e.g., BK, JC, and SA12 papovaviruses [19, 45]). Thus, studies of T-ag assembly events on the SV40 core origin are thought to represent similar protein-DNA interactions on a number of viral origins.
In recent studies we have attempted to define more fully the protein-DNA interactions required for T-ag assembly on the SV40 origin. Advances made in understanding this process include the determination of the solution structure of the purified T-ag-obd131–260 (48). The structure of this domain revealed that the A1 and B2 regions (the T-ag subdomains that mediate binding to the pentanucleotides [69, 89]) are situated in a pair of loops that are located together in space (48). In related studies, the interactions of the T-ag-obd131–260 with the core origin and with core origin molecules containing transition mutations in various pentanucleotides, were examined (39). It was observed that only two of the four GAGGC binding sites within site II are required for stable T-ag-obd131–260 binding (39). Subsequent studies demonstrated that the formation of T-ag double hexamers on the SV40 core origin also requires only two of the four pentanucleotides located within site II (38). The biological relevance of T-ag double hexamers formed on “active pairs” of pentanucleotides was demonstrated by their ability to catalyze a set of previously described structural alterations in the AT and EP regions (38). In view of these experiments, it was proposed that the second pair of pentanucleotides is required at some point between T-ag assembly and DNA-unwinding events (38).
To further characterize initiation of SV40 replication, we have examined what region(s), in addition to site II, is necessary for assembly of either T-ag-obd131–260 or T-ag. These studies reflect, in part, our interest in establishing which subdomains of T-ag are interacting with particular subregions of the core origin. An additional motivation for these studies is our long-term goal of establishing how eukaryotic helicases, such as T-ag (32, 73), are assembled and subsequently activated. Results of these studies are presented herein.
MATERIALS AND METHODS
Commercial supplies of proteins, chemical reagents, and oligonucleotides.
Both T4 polynucleotide kinase and HaeIII were purchased from Gibco-BRL. Thrombin was obtained from Haematologic Technologies Inc. ATP, ADP, adenylyl imidodiphosphate (AMP-PNP), and hexokinase were obtained from Boehringer Mannheim Biochemicals. Oligonucleotides were synthesized on an Applied Biosystems 394 DNA synthesizer at the protein chemistry facility at Tufts University, purified by electrophoresis through 10% polyacrylamide–8% urea gels, and isolated as previously described (63). Dideoxy sequencing reactions (64) were performed with a kit supplied by Amersham Life Sciences, Inc.
Purification of T-ag, T-ag-obd131–260, and T-ag-obd112–260.
SV40 T-ag was generated by using a baculovirus expression vector containing the T-ag-encoding SV40 A gene (57) and isolated by immunoaffinity techniques with the PAb 419 monoclonal antibody (27, 68, 85). To isolate T-ag-obd131–260 and T-ag-obd112–260, Escherichia coli BL21 cells were transformed with the expression vectors pGEX-T-ag-obd131–260 and pGEX-T-ag-obd112–260 and the desired proteins were isolated by published procedures (39). Purified T-ag, T-ag-obd131–260, and T-ag-obd112–260 were dialyzed against T-ag storage buffer (20 mM Tris-HCl [pH 8.0], 50 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol [DTT], 0.1 mM phenylmethylsulfonyl fluoride, 0.2 μg of leupeptin per ml, 0.2 μg of antipain per ml, 10% glycerol) and frozen at −80°C until use.
Oligonucleotide purification and band shift assays.
Double-stranded oligonucleotides were formed by incubating complementary fragments of DNA in hybridization buffer as described by Kadonaga and Tjian (40) and 32P labeled by standard procedures (63). 32P-labeled oligonucleotides were electrophoresed in neutral 10% polyacrylamide gels, and the desired DNA fragments were eluted in oligonucleotide extraction buffer (63). Following ethanol precipitation, an 80% ethanol wash, and drying, the labeled oligonucleotides were resuspended, as needed, in deionized H2O to 25 fmol/μl.
Band shift reactions were conducted under SV40 replication conditions (85) as previously described (17, 49, 55). The reaction mixtures (20 μl) contained 7 mM MgCl2, 0.5 mM DTT, 4 mM ATP (or, where indicated, AMP-PNP, ADP, or no nucleotide), 40 mM creatine phosphate (pH 7.6), 0.48 μg of creatine phosphate kinase (omitted from reaction mixtures containing ADP), 5 μg of bovine serum albumin, 0.8 μg of HaeIII-digested pBR322 DNA (∼2.5 pmol; used as a nonspecific competitor), and 25 fmol of the indicated double-stranded oligonucleotide (labeled as described above). To ensure that trace amounts of ATP were removed from experiments conducted in the presence of ADP, these reactions were conducted in the presence of 1 U of hexokinase and 10 mM glucose (75). Finally, various amounts of protein (purified T-ag or T-ag-obd) were added to the reaction mixtures. After a 20-min incubation at 37°C, glutaraldehyde (0.1% final concentration) was added, and the reaction mixtures were further incubated for 5 min. The reactions were stopped by the addition of 5 μl of 6× loading dye II (15% Ficoll, 0.25% bromophenol blue, 0.25% xylene cyanol) (63) to the reaction mixtures. Samples (∼5,000 cpm/lane) were then applied to a 4 to 12% gradient polyacrylamide gel(s) (acrylamide/bisacrylamide ratio, 19:1) and electrophoresed in 0.5× Tris-borate-EDTA (TBE) for ∼2 h (∼500 V, 20 mA, and 10 W). The gel(s) was dried, subjected to autoradiography, and placed in a PhosphorImager cassette. The band shift reactions were quantitated with a Molecular Dynamics PhosphorImager.
KMNO4 footprinting.
Structural alterations in the core origin regions flanking site II upon protein binding were monitored by use of the KMNO4 footprinting technique (4). Reactions in 30-μl mixtures) were conducted under replication conditions (85), as previously described (38), with the SV40 core origin containing plasmid pSV01ΔEP as the DNA substrate (8). As in previous studies (38), oligonucleotide 1 (5′ TGAGCGGATACATATTTG 3′), 5′-end labeled with [γ-32P]ATP and T4 polynucleotide kinase (63), was used in the primer extension reactions. Upon completion of the reactions, the samples were ethanol precipitated, washed with 80% ethanol, and electrophoresed for ∼3 h at 1,500 V and 40 mA on a 7% polyacrylamide gel containing 8 M urea. A dideoxy sequencing ladder (64), formed with oligonucleotide 1 as the primer, was used to establish the locations of the modified residues.
Nitrocellulose filter binding of SV40 T-ag–DNA complexes.
The nitrocellulose filter assay for T-ag binding was based on previously published methods (3, 47, 51). Reaction mixtures (20 μl) contained 7 mM MgCl2, 0.5 mM DTT, 40 mM creatine phosphate (di-Tris salt [pH 7.6]), 0.48 μg of creatine phosphate kinase (omitted from reaction mixtures containing ADP), 0.2 mg of bovine serum albumin per ml, 0.8 μg of HaeIII-digested pBR322 DNA, 25 fmol of a given oligonucleotide (∼0.6 × 106 cpm/pmol), the indicated nucleotide cofactor and T-ag. Reactions conducted in the presence of ADP also included 1 U of hexokinase and 10 mM glucose. After incubation for 20 min at 37°C, the mixtures were filtered under suction through alkali-treated nitrocellulose filters (Millipore type HAWP; pore size, 0.45 μm; stored in 100 mM Tris · HCl [pH 7.5]). The filters were then washed with 5 ml of 100 mM Tris · HCl (pH 7.5), dried, and counted in a Beckman LS 3801 scintillation counter.
RESULTS
Determination of the minimal region of the SV40 core origin that enables stable T-ag-obd binding.
The 64-bp SV40 core origin (see Fig. 3, diagram 1) is necessary and sufficient for origin-specific unwinding and DNA replication (15, 21, 23, 26, 44, 56, 74). There is, however, some uncertainty about the sequence requirements for T-ag assembly events. For instance, it has been reported that all three SV40 core origin domains are necessary for the assembly of T-ag into double hexamers (1, 59, 81). In contrast, other studies indicate that the entire core origin is not required for T-ag assembly events (38, 58, 61). Therefore, a series of site II-based oligonucleotides were synthesized and used to investigate the minimal sequence requirements for T-ag assembly on the core origin.
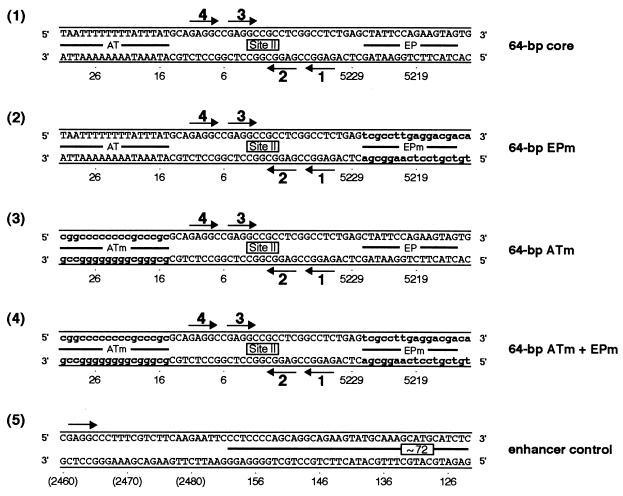
FIG. 3.
Sequences of the 64-bp core oligonucleotide and mutant forms of this molecule. The locations of the AT-rich regions, site II, and the EP regions are depicted. As in Fig. 1, the arrows depict the four GAGGC pentanucleotides within site II that serve as binding sites for T-ag. Lowercase boldface letters represent transition mutations (m) introduced into the indicated regions. The names of the individual oligonucleotides are given to the right of their sequences.
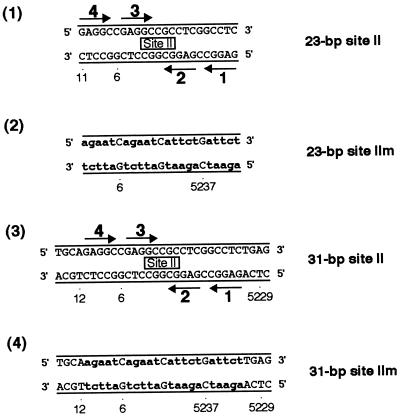
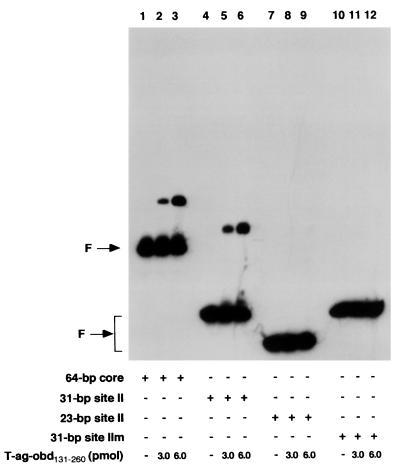
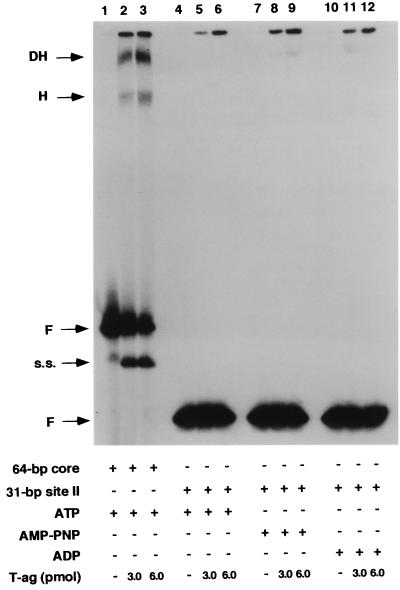
In an initial series of experiments, gel mobility shift assays (see Materials and Methods) were used with the site II-based oligonucleotides (Fig. 1) and the T-ag-obd131–260 (Fig. 2). As a positive control, the interaction of the T-ag-obd131–260 with the 64-bp core oligonucleotide (see Fig. 3, diagram 1) was examined. Inspection of Fig. 2 (lanes 2 and 3) confirms previous reports (39) that T-ag-obd131–260 binds to the 64-bp core oligonucleotide and forms one very distinct band shift species. Comparison of lanes 5 and 6 with lanes 2 and 3 demonstrates that the 31-bp site II oligonucleotide (Fig. 1, diagram 3) supports T-ag-obd131–260 binding at levels equivalent to that of the 64-bp core origin. In contrast, inspection of lanes 8 and 9 reveals that a fragment of DNA containing just site II, the 23-bp site II oligonucleotide (Fig. 1, diagram 1), did not support T-ag-obd131–260 binding. As a control for non-sequence-specific interactions, band shift reactions were conducted with the 31-bp site IIm (mutant) oligonucleotide (Fig. 1, diagram 4); this DNA fragment did not support detectable levels of binding (Fig. 2, lanes 11 and 12). Additional band shift reactions were conducted with the 23-bp site IIm oligonucleotide (Fig. 1, diagram 2). As expected, binding of T-ag-obd131–260 to this molecule was not detected (data not shown). The reactions in Fig. 2, lanes 1, 4, 7, and 10, were conducted with the indicated oligonucleotides in the absence of protein. When these reactions were repeated in the presence of ADP or AMP-PNP or in the absence of exogenous nucleotides, identical results were obtained (data not shown), as expected given that T-ag-obd131–260 does not contain the nucleotide binding domain (5, 13). These experiments demonstrate that T-ag-obd131–260 is sufficient for nucleotide cofactor-independent binding to site II, provided that this region is flanked on either side by a small number of additional base pairs (∼4 bp per side).
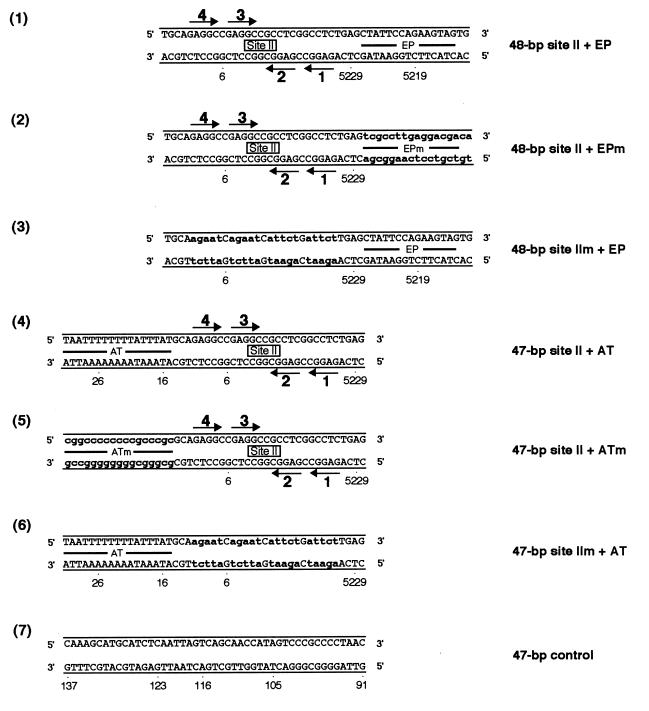
FIG. 1.
Sequences of the site II-based oligonucleotides. Diagrams 1 and 3 depict oligonucleotides containing site II, while diagrams 2 and 4 represent control oligonucleotides. The arrows depict the four GAGGC pentanucleotides within site II that are recognition sequences for T-ag, numbered as previously described (43). The names of the oligonucleotides are given to the right of each figure. Lowercase boldface letters in the control oligonucleotides represent the transition mutations (m) introduced into individual GAGGC pentanucleotides. SV40 sequences are numbered as described elsewhere (79).
FIG. 2.
Representative gel mobility shift assay used to establish the minimal sequence requirement for T-ag-obd131–260 binding to site II. Lanes: 2 and 3, products of band shift assays conducted with the 64-bp core oligonucleotide and 3 or 6 pmol, respectively, of T-ag-obd131–260; 5 and 6, products of band shift assays conducted with the 31-bp site II oligonucleotide and 3 or 6 pmol, respectively, of T-ag-obd131–260; 8 and 9, products of band shift assays conducted with the 23-bp site II oligonucleotide and 3 or 6 pmol, respectively, of T-ag-obd131–260. As a control for nonspecific binding, band shift assays were conducted with the 31-bp site IIm control oligonucleotide and either 3 or 6 pmol of T-ag-obd131–260 (lanes 11 and 12). Lanes 1, 4, 7, and 10 contain the products of band shift assays conducted in the absence of protein with the indicated oligonucleotides. The input or free duplex DNA (F) is indicated by the arrow. The protein-to-oligonucleotide ratios with 3 and 6 pmol of T-ag-obd131–260 and 25 fmol of oligonucleotide are 120:1 and 240:1, respectively.
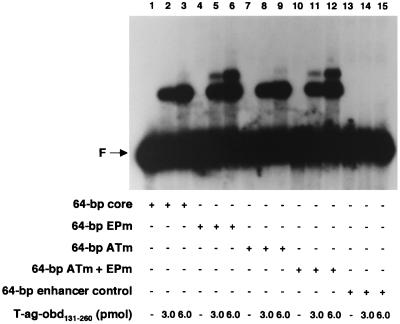
To test whether the flanking sequences modulate T-ag-obd131–260 binding to site II, we examined whether mutations in the AT-rich and EP regions influenced T-ag-obd131–260 binding to the core origin. To conduct these studies, gel mobility shift assays were performed under replication conditions with T-ag-obd131–260 and with oligonucleotides containing either the complete core origin or core origin derivatives with transition mutations in either the AT-rich region, the EP region, or both (Fig. 3, diagrams 1 to 4). Inspection of Fig. 4 demonstrates that relative to the 64-bp core oligonucleotide (lanes 2 and 3), the 64-bp EPm (lanes 5 and 6), the 64-bp ATm (lanes 8 and 9), and the 64-bp ATm + EPm (lanes 11 and 12) oligonucleotides supported nearly identical levels of T-ag-obd131–260 assembly. To assay for nonspecific binding, the reactions in lanes 14 and 15 were conducted with the 64-bp enhancer control oligonucleotide (Fig. 3, diagram 5). These latter experiments confirm that in absence of site II, T-ag-obd131–260 makes limited contacts with DNA (39). The reactions in lanes 1, 4, 7, 10, and 13 were conducted with the indicated oligonucleotides in the absence of protein. Identical results were obtained when these reactions were repeated in the presence of AMP-PNP or ADP or in the absence of exogenous nucleotide cofactors (data not shown). We concluded that T-ag-obd131–260 binds to site II via a mechanism that is largely independent of the sequence composition of the flanking regions. However, a second band was generated in reactions involving oligonucleotides with a mutant EP (lanes 5, 6, 11 and 12). The molecular basis for the formation of this additional reaction product is not understood.
FIG. 4.
Gel mobility shift assays used to establish whether mutant forms of the flanking sequences influence T-ag-obd131–260 binding to site II. As a positive control, gel mobility shift assays were conducted with the 64-bp core oligonucleotide and either 3 or 6 pmol of T-ag-obd131–260 (lanes 2 and 3). In related reactions, T-ag-obd131–260 (3 or 6 pmol) was incubated with the 64-bp EPm (lanes 5 and 6), the 64-bp ATm (lanes 8 and 9), the 64-bp ATm + EPm (lanes 11 and 12), or the 64-bp enhancer control (lanes 14 and 15). The reactions in lanes 1, 4, 7, 10, and 13 were conducted in the absence of protein. The input or free duplex DNA (F) is indicated by the arrow. The protein-to-oligonucleotide ratios used in these reactions are given in the legend to Fig. 2. Quantitation with a PhosphorImager revealed that with 6 pmol of T-ag-obd131–260, the percentage of input DNA shifted into the major band shift product formed with the 64-bp core, 64-bp EPm, 64-bp ATm, and 64-bp ATm + EPm oligonucleotides was 22.4, 25.7, 24.0, and 23.1%, respectively.
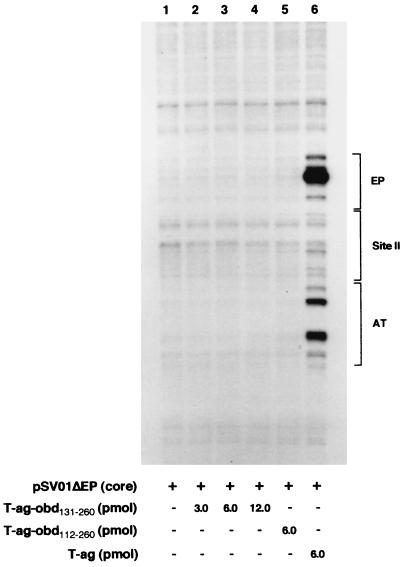
On circular DNA templates, T-ag binding to the SV40 core origin is associated with structural changes in the AT-rich tract and melting of approximately 8 bp within the EP (4, 38, 58). To determine whether binding of T-ag-obd131–260 to the core origin resulted in similar structural alterations, KMnO4 oxidation assays (4) were performed (Fig. 5). As expected, when pSV01ΔEP(core) (8) was treated with KMnO4 in the absence of protein, distortions in the AT-rich and EP regions were not detected (Fig. 5, lane 1). However, the previously described structural alterations were detected in the presence of T-ag (lane 6). In the presence of 3, 6, or 12 pmol of the T-ag-obd131–260, oxidation within the flanking regions was not detected (lanes 2 to 4, respectively). Furthermore, structural alterations were not detected in pSV01ΔEP(core) when a slightly larger version of T-ag-obd131–260, i.e., T-ag-obd112–260 (39), was used in the KMnO4 assays (lane 5). We concluded that binding of T-ag-obd131–260 or T-ag-obd112–260 to site II does not result in detectable structural alterations in the AT-rich and EP regions. Collectively, the studies presented in Fig. 2, 4, and 5 indicate that purified forms of T-ag-obd bind to site II but do not make additional contacts with the flanking sequences. This conclusion is consistent with studies indicating that T-ag-obd131–260 has a single GAGGC-specific binding site for DNA (reviewed in reference 7). It is also consistent with previous phenanthroline-copper footprinting studies that demonstrated that a subregion of site II, but not the flanking sequences, was protected upon T-ag-obd131–260 binding to the core origin (38, 39).
FIG. 5.
Determination of whether purified forms of T-ag-obd catalyze structural changes in the core origin. As controls, the SV40 origin-containing plasmid pSV01ΔEP was incubated under replication conditions in the absence (lane 1) or presence (lane 6) of T-ag. The reactions in lanes 2, 3, and 4 were conducted in the presence of the indicated amounts of T-ag-obd131–260, while the reaction in lane 5 was conducted in the presence of 6 pmol of T-ag-obd112–260 (39). After treatment with KMnO4, the sites of oxidation were probed by primer extension reactions with 32P-labeled oligonucleotide 1 (see Materials and Methods). The primer extension products were analyzed by electrophoresis on a 7% polyacrylamide gel containing 8 M urea. The locations of the EP, site II, and AT-rich sequence elements are indicated on the right.
Determination whether T-ag has the same sequence requirements for assembly as T-ag-obd131–260.
Given that T-ag-obd is normally a subdomain of T-ag, one might predict that site II-based fragments of DNA would also support T-ag assembly events. Initial band shift experiments with the 23-bp site II oligonucleotide (Fig. 1, diagram 1) revealed that T-ag did not assemble on this oligonucleotide (data not presented), a result consistent with a previous study conducted in the absence of ATP (54). Related experiments (Fig. 6) demonstrated that in contrast to T-ag-obd131–260, the 31-bp site II oligonucleotide (Fig. 1, diagram 3) did not support T-ag assembly in the presence of ATP (lanes 5 and 6), AMP-PNP (lanes 8 and 9), or ADP (lanes 11 and 12). The reactions in lanes 2 and 3 were conducted with the 64-bp core oligonucleotide and served as a positive control. Previous characterization of the two products formed in these assays revealed that they consisted of T-ag hexamers and double hexamers (14, 59, 81). The reactions in lanes 1, 4, 7, and 10 were conducted in the absence of T-ag.
FIG. 6.
Gel mobility shift assay used to establish whether T-ag can assemble on the 31-bp site II oligonucleotide. As a positive control, 3 or 6 pmol of T-ag was incubated with the 64-bp core oligonucleotide (lanes 2 and 3) in the presence of ATP. T-ag (3 or 6 pmol) was also incubated with 25 fmol of the 31-bp site II oligonucleotide in the presence of different analogs of ATP; the reactions in lanes 5 and 6 were conducted in the presence of ATP, those in lanes 8 and 9 were conducted in the presence of AMP-PNP, and those in lanes 11 and 12 were conducted in the presence of ADP. The reactions in lanes 1, 4, 7, and 10 were conducted in the absence of protein. The input or free duplex DNA (F) is indicated by the arrow. Single-stranded DNA (s.s.), the product of the T-ag helicase activity, is present in elevated amounts in lanes 2 and 3. DH, double hexamer; H, hexamer. As in previous examples, the protein-to-oligonucleotide ratios used in these reactions are given in the legend to Fig. 2.
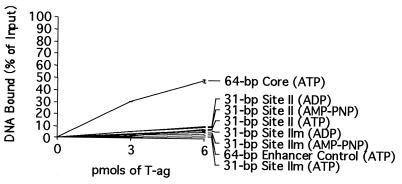
As an additional assay of the ability of T-ag to interact with site II-based oligonucleotides, nitrocellulose filter binding assays (see Materials and Methods) were conducted; the results of these studies performed under glutaraldehyde-free conditions are presented in Fig. 7. It is apparent from these experiments that relative to the ability of T-ag to bind to the SV40 core origin (top line), T-ag has little or no ability to interact with the 31-bp site II oligonucleotide (Fig. 1, diagram 3), or the 31-bp site IIm oligonucleotide (diagram 4), regardless of the analogue of ATP present in the reaction mixture. It is concluded that although a fragment of DNA consisting primarily of site II is able to support T-ag-obd131–260 binding, this DNA fragment does not support T-ag assembly.
FIG. 7.
Filter binding assays used to measure the ability of T-ag to interact with the 31-bp site II oligonucleotide under replication conditions. The interaction of T-ag (0, 3, or 6 pmol) with 25 fmol of the 31-bp site II oligonucleotide was measured by nitrocellulose filter binding assays in the presence of the indicated nucleotide cofactors. The percentage of input oligonucleotide bound to a given filter was determined by scintillation counting. As a positive control, the interaction of T-ag (0, 3, or 6 pmol) with the 64-bp core oligonucleotide was measured in the presence of ATP. Additional controls were conducted with the 64-bp enhancer control and the 31-bp site IIm and the indicated nucleotide cofactors.
Determination of the core origin sequences, in addition to site II, that are required for T-ag assembly events.
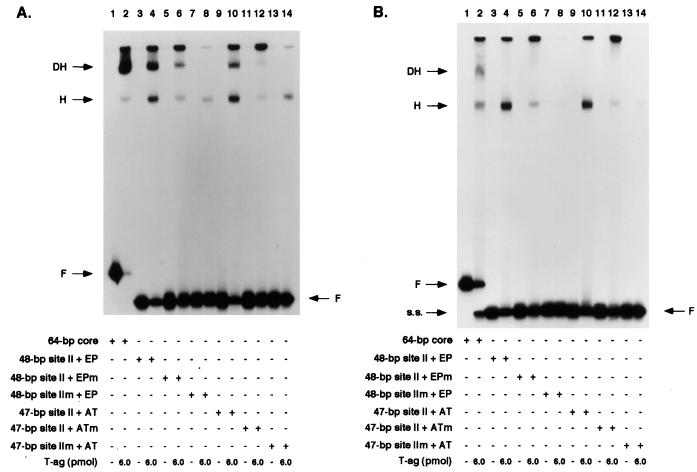
To establish what additional sequences are required for site II-dependent T-ag assembly events, band shift experiments were conducted with a set of oligonucleotides termed the asymmetric extensions of site II (Fig. 8). An initial set of experiments was conducted with AMP-PNP (Fig. 9A); this nonhydrolyzable analog of ATP prevents the loss of T-ag–DNA complexes caused by the helicase activity of T-ag (16, 32, 71, 73, 84) and enables an accurate measurement of hexamer and double-hexamer formation. As a positive control, band shift reactions were conducted with the 64-bp core oligonucleotide (Fig. 3, diagram 1); the products of this reaction (Fig. 9A, lane 2) included T-ag hexamers and double hexamers. (For PhosphorImager-based quantitation of the experiments in Fig. 9, see Table 1.) The results of reactions conducted with the 48-bp site II + EP oligonucleotide (Fig. 8, diagram 1) are displayed in lane 4. The products formed in this reaction included species that comigrated with T-ag hexamers and double hexamers; verification that these are hexamers and double hexamers was provided by native gel electrophoresis techniques (references 14, 46, 59, and 60 and data not shown). Whether the wild-type sequence of the EP region is critical for T-ag assembly events was tested by using the 48-bp site II + EPm oligonucleotide (Fig. 8, diagram 2); this DNA fragment supported hexamer and double-hexamer formation but at reduced levels relative to the wild type (Fig. 9A, lane 6) (see also Table 1 and Fig. 10). The critical role of site II in the assembly process was confirmed by the results obtained with the 48-bp site IIm + EP oligonucleotide (Fig. 8, diagram 3); T-ag assembly on this molecule was limited to hexamer formation (Fig. 9A, lane 8). Hexamer formation on this molecule probably reflects both sequence-independent assembly events (see below) and interactions of T-ag with the EP (58). Additional experiments were performed to determine the extent to which the AT-rich region enables assembly on site II. Figure 9A, lane 10, reveals that the 47-bp site II + AT oligonucleotide (Fig. 8, diagram 4) supports the formation of T-ag hexamers and double hexamers. Additional reactions conducted with the 47-bp site II + ATm oligonucleotide (Fig. 8, diagram 5) indicate that, relative to the wild-type flanking sequence, this molecule supports reduced amounts of hexamer and double-hexamer formation (Fig. 9A, lane 12) (see also Table 1 and Fig. 10). Figure 9A, lane 14, containing the products of a reaction formed with the 47-bp site IIm + AT oligonucleotide (Fig. 8, diagram 6), revealed that this molecule is capable of supporting hexamer formation. In Fig. 9A, the odd-numbered lanes contained the products formed when the band shift experiments were conducted in the absence of T-ag. Finally, as a control for non-sequence-specific interactions, band shift reactions were conducted with the 47-bp control oligonucleotide (Fig. 8, diagram 7); this molecule supported low levels of hexamer formation (see Table 1). Non-sequence-specific binding events may explain, at least in part, assembly on the 48-bp site IIm + EP and 47-bp site IIm + AT oligonucleotides. Collectively, these experiments demonstrate that in the presence of AMP-PNP, T-ag binds to site II provided that this core origin subdomain is flanked on at least one side by additional sequences. Preferential binding to molecules containing wild-type sequences indicates that sequence-specific interactions with the flanking sequences promote T-ag assembly. However, it is also possible that wild-type flanking sequences adopt a conformation that promotes T-ag binding. Moreover, significant contact with the sugar-phosphate backbone is suggested by the assembly events on oligonucleotides containing mutations in the flanking sequences.
FIG. 8.
The set of oligonucleotides collectively termed the asymmetric extensions of site II; the names of the individual oligonucleotides are given to the right of their sequences. The locations of the AT-rich region, site II, and the EP region are depicted. As in previous examples, the arrows depict the four GAGGC pentanucleotides within site II that serve as recognition sites for T-ag. Diagram 1 presents the sequence of the oligonucleotide containing site II and the EP region, while diagram 4 presents the sequence of the oligonucleotide-containing site II and the AT region. Control oligonucleotides, containing transition mutations in the EP and AT regions, are depicted in diagrams 2 and 5, respectively. Additional control oligonucleotides, containing the wild-type EP and AT-rich regions and transition mutations in the pentanucleotides are depicted in diagrams 3 and 6, respectively. As in previous examples, lowercase boldface letters represent transition mutations (m) introduced into the indicated regions. Finally, the sequence of the 47-bp control oligonucleotide, used to measure non-sequence-specific binding, is presented in diagram 7.
FIG. 9.
Representative gel mobility shift assays used to assess the ability of T-ag to interact with oligonucleotides containing site II and either of the flanking sequences. (A) The experiments were performed in the presence of AMP-PNP with 6 pmol of T-ag and 25 fmol of the indicated oligonucleotide. As a positive control, the reaction in lane 2 was conducted with T-ag and the 64-bp core oligonucleotide. Reaction products formed with T-ag and the 48-bp site II + EP, the 48-bp site II + EPm, and the 48-bp site IIm + EP oligonucleotides are shown in lanes 4, 6, and 8, respectively. Reactions conducted with the 47-bp site II + AT, the 47-bp site II + ATm, and the 47-bp site IIm + AT oligonucleotides are shown in lanes 10, 12, and 14, respectively. The products of band shift reactions conducted in the absence of T-ag and the indicated oligonucleotides are shown in the odd-numbered lanes. (B) The experiments are identical to those in panel A, except that they were conducted in the presence of ATP. In both panels the arrows indicate the positions of T-ag hexamers (H), T-ag double hexamers (DH), and free DNA (F). Single-stranded DNA (s.s.), formed owing to the helicase activity of T-ag, is present in lane 2. The protein-to-oligonucleotide ratio with 6 pmol of T-ag and 25 fmol of oligonucleotide is 240:1.
TABLE 1.
Quantitation of T-ag hexamer and double-hexamer formation on the set of oligonucleotides termed the asymmetric extensions of site IIa
| Oligonucleotide and hexamer type H and DH | % Formation of hexamer and double hexamer in presence ofb:
|
|||
|---|---|---|---|---|
| AMP-PNP | ADP | ATP | No nucleotide | |
| 64-bp core | ||||
| DH | 62.78 | 66.83 | 11.36 | 7.74 |
| 2.39 | ||||
| 2.59 | ||||
| H | 4.17 | 8.08 | 6.94 | 3.75 |
| Total | 66.95 | 74.91 | 18.30 | 16.46 |
| 48-bp site II + EP | ||||
| DH | 23.49 | 18.01 | 0.21 | 2.72 |
| 0.50 | ||||
| 0.26 | ||||
| H | 13.13 | 28.69 | 15.68 | 0.14 |
| Total | 36.62 | 46.70 | 15.90 | 3.62 |
| 47-bp site II + AT | ||||
| DH | 14.49 | 14.87 | 0.11 | 0.01 |
| 2.57 | ||||
| H | 19.58 | 40.32 | 13.77 | 2.89 |
| Total | 34.06 | 55.19 | 13.87 | 5.47 |
| 48-bp site II + EPm | ||||
| DH | 7.65 | 6.68 | 0.18 | |
| 0.25 | ||||
| 0.10 | ||||
| H | 4.46 | 7.37 | 4.26 | 0.09 |
| Total | 12.11 | 14.05 | 4.26 | 0.61 |
| 47-bp site II + ATm | ||||
| DH | 3.85 | 2.30 | ||
| 0.13 | ||||
| 0.07 | ||||
| H | 2.78 | 4.89 | 2.61 | 0.34 |
| Total | 6.63 | 7.19 | 2.61 | 0.54 |
| 47-bp site IIm + AT | ||||
| DH | 0.49 | |||
| H | 6.20 | 5.85 | 0.99 | 0.27 |
| Total | 6.20 | 6.34 | 0.99 | 0.27 |
| 48-bp site IIm + EP | ||||
| DH | 0.19 | |||
| H | 3.52 | 3.01 | 1.52 | 0.04 |
| Total | 3.52 | 3.19 | 1.52 | 0.04 |
| 47-bp control | ||||
| DH | 0.14 | 0.05 | ||
| H | 0.75 | 2.17 | 0.49 | 0.38 |
| Total | 0.89 | 2.17 | 0.49 | 0.43 |
The experiments in Fig. 9A (performed in the presence of AMP-PNP), Fig. 9B (performed in the presence of ATP), similar experiments conducted in the presence of ADP or in the absence of exogenous nucleotide (data not shown), and control experiments conducted with the 47-bp control oligonucleotide (Fig. 8, diagram 7, and data not shown) were quantitated with a Molecular Dynamics PhosphorImager.
For a given lane, the percentage of DNA in the hexamer (H) and double-hexamer (DH) species were determined. In those reactions conducted in the absence of nucleotide, intermediate species were detected running between the hexamer and double-hexamer bands. Numbers in italics indicate the percentage of DNA in these intermediate species.
FIG. 10.
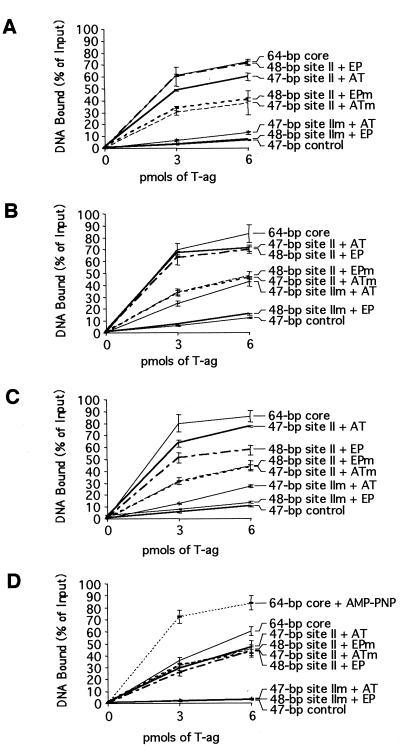
Filter binding assays used to measure the ability of T-ag to interact with the asymmetric extensions of site II set of oligonucleotides. The amount of oligonucleotide bound to T-ag was established by nitrocellulose filter binding assays and scintillation counting. The interactions of T-ag (0, 3, and 6 pmol) with these oligonucleotides were measured in the presence of ATP (A), AMP-PNP (B), or ADP (C) and in the absence of nucleotide (D). The names of the individual oligonucleotides are shown to the right of the figure.
The band shift experiments in Fig. 9A, conducted with AMP-PNP as the nucleotide cofactor, were repeated in the presence of ATP instead of AMP-PNP (Fig. 9B). As a positive control, a reaction was conducted with T-ag and the 64-bp core oligonucleotide. As expected, the products included T-ag hexamers and double hexamers (Fig. 9B, lane 2). The reaction products that formed when T-ag was incubated with the set of oligonucleotides termed the asymmetric extensions of site II (Fig. 8), are presented in Fig. 9B, lanes 2, 4, 6, 8, 10, and 12. In every case, T-ag assembled into hexamers but not double hexamers. Molecules containing wild-type flanking sequences (e.g., the 48-bp site II + EP and the 47-bp site II + AT oligonucleotides) supported higher levels of hexamers than did molecules containing mutant sequences (e.g., the 48-bp site IIm + EP and 47-bp site IIm + AT oligonucleotides) (quantitated in Table 1). As in Fig. 9A, the products of band shift reactions conducted in the absence of T-ag are presented in the odd-numbered lanes. As a control for non-sequence-specific DNA interactions, band shift reactions were conducted with the 47-bp control oligonucleotide (Fig. 8, diagram 7, and data not shown); this molecule supported very low levels of hexamer formation (see Table 1). These experiments confirm that T-ag binds to molecules containing asymmetric extensions of site II. Furthermore, as with the studies conducted in the presence of AMP-PNP, these experiments indicate that T-ag binding is promoted by sequence-specific, or conformation-dependent, interactions with the flanking sequences. However, since double hexamers are detected on full-length core origin-containing oligonucleotides in the presence of ATP but not on the asymmetric extensions of site II, ATP binding or hydrolysis appears to increase the sequence requirements for double-hexamer formation or stability.
To further characterize T-ag assembly, the experiments in Fig. 9A and B were repeated in the presence of ADP and in the absence of any exogenous nucleotide (autoradiograms not shown; however, see Table 1 for a quantitation of the results of these experiments). In general, the products formed in the presence of ADP were similar to those formed in the presence of AMP-PNP. Moreover, in the absence of exogenously added nucleotide, T-ag assembly was clearly defective, a result consistent with previous studies (3, 17, 22, 50, 59).
The band shift experiments conducted in the presence of AMP-PNP (Fig. 9A), ATP (Fig. 9B), and ADP and in the absence of nucleotide were quantitated with a PhosphorImager. The results of these analyses are presented in Table 1, which shows that regardless of the nucleotide cofactor present in the reaction mixture, the relative binding ability of the individual oligonucleotides is nearly the same. Indeed, Table 1 is arranged such that the best substrate for T-ag binding in the presence of AMP-PNP, the 48-bp site II + EP oligonucleotide, is at the top of the table while the weakest substrate, the 47-bp control oligonucleotide, is at the bottom. Further inspection of Table 1 reveals that similar amounts of hexamer and double hexamer formed in the presence of AMP-PNP and ADP and confirms that in the presence of ATP, double hexamers were not detected on the asymmetric extensions of site II, a result reflected in the relatively low yields of T-ag assembled on this set of oligonucleotides. The data in Table 1 also confirms that in the absence of exogenous nucleotides, binding to this set of oligonucleotides was greatly reduced. Finally, since T-ag bound the 48-bp site IIm + EP and 47-bp site IIm + AT oligonucleotides at levels similar to those for the 47-bp control oligonucleotide, assembly events on these molecules are largely sequence nonspecific (a conclusion also supported by data presented in Fig. 10).
Band shift reactions conducted with T-ag require cross-linking with glutaraldehyde (see Materials and Methods). Therefore, to confirm the conclusions drawn from the band shift experiments in Fig. 9 and Table 1, nitrocellulose filter binding assays were conducted with T-ag and the asymmetric extensions of site II (Fig. 8), in the presence of different nucleotide cofactors. The results of these cross-linking-independent studies are presented in Fig. 10. Figures 10A to C, showing results of experiments conducted in the presence of ATP, AMP-PNP, and ADP, respectively, reveals that the 48-bp site II + EP and the 47-bp site II + AT oligonucleotides bound T-ag at levels nearly identical to those for the 64-bp core oligonucleotide. Consistent with the experiments presented in Fig. 9, the 48-bp site II + EPm and the 47-bp site II + ATm oligonucleotides bound T-ag at reduced levels relative to molecules containing wild-type flanking sequences. These experiments provide additional evidence that sequence-specific, or conformation-dependent, interactions with the flanking sequences are necessary for optimal assembly of T-ag. As expected from the results in Fig. 9, binding to oligonucleotides containing mutant forms of site II was greatly reduced. Moreover, to quantitate non-sequence-specific binding of T-ag to DNA, reactions were conducted with the “47-bp control” oligonucleotide; virtually background levels of binding were detected, regardless of the nucleotide cofactor present in the reaction mixture. Of considerable interest is that in the absence of exogenously added nucleotide cofactors (Fig. 10D), the level of binding to molecules containing the wild-type flanking sequences (i.e., the 48-bp site II + EP and the 47-bp site II + AT oligonucleotides) was reduced to levels supported by molecules containing mutant flanking sequences (i.e., the 48-bp site II + EPm and the 47-bp site II + ATm oligonucleotides). These experiments indicate that in the absence of nucleotide cofactors, T-ag cannot distinguish between wild-type and mutant flanking sequences. Thus, a nucleotide cofactor bound to T-ag may be necessary for sequence-specific interactions with the flanking sequences.
DISCUSSION
T-ag-obd131–260 is sufficient for site-specific binding to site II, provided that this region is flanked by a small number of base pairs. The presence or absence of different ATP analogues had no effect on the binding of T-ag-obd131–260 to site II. Furthermore, the AT-rich or EP regions, or mutant forms of these sequences, did not modulate the binding of T-ag-obd131–260 to site II. Moreover, when bound to site II, T-ag-obd131–260 did not induce structural changes in either the EP or AT-rich regions. Based on these observations, it is concluded that the interactions of T-ag-obd131–260 with the SV40 core origin are limited to site II. Regarding the locations within site II bound by T-ag-obd131–260, previous studies conducted with core origin length oligonucleotides (64 bp) demonstrated that monomers of the origin binding domain preferentially assembled on pentanucleotides 1 and 3 but that binding to pentanucleotides 2 and 4 occurred if pentanucleotides 1 and 3 were mutated (39).
Similar analyses of the core origin and nucleotide requirements for T-ag assembly events were previously conducted. It was concluded that T-ag has relatively few sequence-specific interactions with the core origin regions flanking site II (1, 4, 24, 37, 58, 66), although contacts with the EP were noted (e.g., with residues 5225 and 5228 to 5230) (4, 58, 66). It was also reported that T-ag interacts with linear DNA fragments (33, 52) or circular plasmids (58) containing just site II. Consistent with these observations, it was proposed that the primary function of the AT-rich and EP regions is not to promote binding but to undergo structural changes required for the initiation of DNA replication (1).
In view of our T-ag-obd131–260 studies and the results summarized in the previous paragraph, it was anticipated that T-ag might also bind to an oligonucleotide containing just site II. Nevertheless, the 31-bp site II oligonucleotide did not support T-ag assembly, regardless of the ATP analog used in the reaction. Thus, in the context of the whole T-ag molecule, T-ag-obd is not able to bind a subfragment of the core origin that was bound by purified T-ag-obd131–260. To establish which sequences, in addition to site II, are required for T-ag assembly events, we analyzed binding to larger subfragments of the core origin. These experiments revealed that an asymmetric extension of site II is essential for hexamer formation and nucleotide cofactor-dependent double-hexamer assembly (see below). The observation that mutant forms of the flanking sequences support T-ag assembly, albeit at lower levels compared to wild-type sequences, indicates that both sequence-specific and non-sequencespecific interactions take place during T-ag assembly, a result consistent with conclusions drawn in previous studies (59, 66). Additional evidence that non-sequence-specific interactions are important for T-ag assembly on duplex DNA was provided by experiments conducted with a 17-bp oligonucleotide containing site I. Site I is thought to be involved mainly in autoregulation of early transcription and contains two pentanucleotides arranged as direct repeats (29). T-ag did not interact with the 17-bp site I-containing oligonucleotide. However, when site I was flanked by 15 bp of a randomly chosen sequence, it became a substrate for T-ag binding (39). These and related studies (62) indicate that T-ag cannot stably bind to the GAGGC sequences in either site I or site II unless it makes contacts with sequences flanking the pentanucleotides. Additional sequence-specific interactions are likely to promote complex stability or subsequent steps necessary for origin-specific DNA unwinding (9, 16, 28, 86).
Given that either of the flanking sequences enables T-ag binding to site II, it was of interest to determine whether the flanking sequences need to be covalently linked to site II or whether they can complement binding to site II in trans. In an additional series of band shift experiments, we established that an oligonucleotide containing the EP did not allow binding to an oligonucleotide containing site II (data not shown). This observation is consistent with entropy-based arguments that intramolecular reactions are favored over intermolecular reactions (30): if two origin subfragments were able to support T-ag binding, instead of one, there would be an energetically unfavorable increase in order.
Concerning the requirements for double-hexamer formation, it is apparent that subfragments of the core origin support double-hexamer assembly provided that the reactions are conducted in the presence of AMP-PNP or ADP. As with hexamer assembly, double-hexamer formation is promoted by wild-type flanking sequences (Table 1). However, it is also apparent from Fig. 9 and Table 1 that the core origin subfragments support hexamer but not doublehexamer formation when ATP is used as the nucleotide cofactor. In the presence of ATP, double hexamers can be detected only on oligonucleotides containing the full-length core origin. Why hexamers, but not double hexamers, are detected on the core origin subfragments, in the presence of ATP, is not understood. One possibility is that ATP binding, or perhaps hydrolysis, alters hexamer-hexamer interactions such that additional T-ag–origin binding events are necessary for complex formation or stability. Regardless of the explanation, the observation that nucleotide cofactors govern the sequence requirements for T-ag assembly on the SV40 origin is probably relevant to minor differences in the conclusions drawn in certain previous studies (1, 58, 59, 61).
Regarding the domains of T-ag that may be engaging the flanking regions, since purified T-ag-obd131–260 does not interact with the EP and AT-rich regions, it is likely that non-T-ag-obd residues are required for these binding events. Consistent with this hypothesis, it was reported that T-ag residues 121 to 135 are required for interactions with the AT-rich region while a second, poorly defined region is necessary for interactions with the EP (87). Moreover, it was previously concluded that melting of the EP domain is an activity of T-ag that is distinct from its pentanucleotide binding and DNA helicase activities (58). While it is not known which non-T-ag-obd region of T-ag is interacting with the EP, it is known that the T-ag domain responsible for binding to single-stranded DNA is also separate from T-ag-obd (39, 88). Furthermore, it was reported that the C11A T-ag mutation, Pro522 → Ser, is markedly reduced in its ability to bind single-stranded DNA and partial duplex helicase substrates (53). In light of these studies, it will be interesting to determine if the non-T-ag-obd regions that interact with the flanking sequences and single-stranded DNA overlap and if they are in close proximity to the nucleotide binding site. This latter possibility is of interest since in several prokaryotic helicases, the single-stranded-DNA- and nucleotide-binding sites are near each other (see, e.g., references 42, 76, and 80). Finally, it is noted that at relatively high concentrations, T-ag can melt the EP in the absence of other origin sequences (58). In view of the evidence that T-ag has an EP-specific DNA binding site, this interaction may account for site II-independent binding to the EP and subsequent melting events.
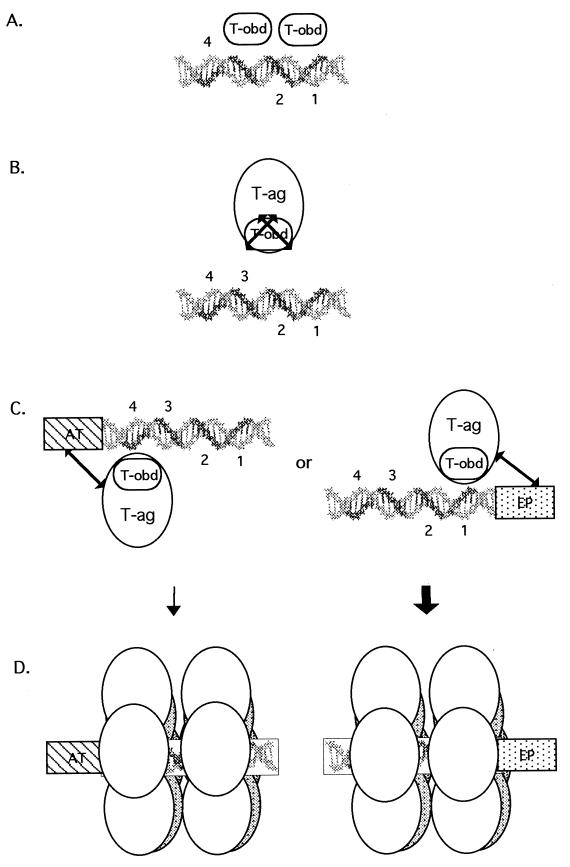
A model of T-ag assembly events on the core origin is presented in Fig. 11. T-ag-obd131–260 is necessary and sufficient for binding to the pentanucleotides in site II (step A). In light of results presented in a previous study (39), purified T-ag-obd131–260 is depicted bound to site II as a dimer to pentanucleotides 1 and 3. Upon binding to site II, T-ag-obd131–260 does not make additional contacts with the AT-rich or EP region. In contrast, in the context of T-ag, T-ag-obd is not able to interact with site II-based fragments of DNA (step B). Why the T-ag-obd within T-ag cannot stably bind site II is not clear; therefore, arrows are used to symbolize that this region is obstructed. However, when site II is extended on either side by flanking sequences, the block to T-ag binding, and therefore to the T-ag-obd/site II interaction, is removed (step C). The extended arrows symbolize the poorly defined interactions between the non-T-ag-obd region(s) and the flanking sequences that are essential for binding. Owing to the importance of the flanking sequences in the assembly process, T-ag is depicted binding to pentanucleotides proximal to the flanking sequences (pentanucleotide 1 or 4); however, this has not yet been confirmed. Once a T-ag hexamer forms on a given pentanucleotide, cooperative interactions (see, e.g., references 55 and 83) are likely to promote the formation of a second hexamer on “active pairs” of pentanucleotides (i.e., 1 and 3 or 2 and 4) (38) (step D). It was previously suggested that double-hexamer formation on active pairs of pentanucleotides may block further assembly events on the unoccupied pentanucleotides (38, 39). Regarding the role(s) of the nucleotide cofactor in the assembly process, as noted above, the interaction between T-ag-obd131–260 and site II is nucleotide independent. Therefore, a nucleotide cofactor bound to T-ag is necessary at some other step in the assembly process. One possible step is suggested by the data in Fig. 10; the ability of T-ag to make sequence-specific contacts with the flanking sequences was nucleotide dependent. Therefore, sequence-specific interactions between the flanking sequences and the non-T-ag-obd DNA binding site(s) may be nucleotide dependent. In addition, assembly studies conducted in the presence of ATP indicate that nucleotide cofactors may modulate the protein-protein interactions that take place during double-hexamer formation.
FIG. 11.
Model depicting the requirements for binding of T-ag-obd131–260 and T-ag to site II and the asymmetric extensions of site II. (A) A slightly elongated version of site II is necessary and sufficient for binding of T-ag-obd131–260 (abbreviated T-obd in this model). The GAGGC pentanucleotides are shown in bold and numbered as previously described (43). Based on results obtained in a previous study (39), T-ag-obd131–260 is depicted bound to pentanucleotides 1 and 3 as a dimer (B). In contrast, T-ag and therefore the T-ag-obd present in this molecule cannot bind to oligonucleotides containing just site II. The arrows covering T-ag-obd are used to symbolize that in the context of T-ag, this domain cannot bind to site II. (C) In the presence of either of the flanking sequences, the obstacle(s) to the interactions of T-ag with site II are removed. It is proposed that the flanking sequences enable additional T-ag/origin contacts, depicted by the extended arrows, and that these interactions result in conformational changes that expose T-ag-obd. (D) Upon binding of a T-ag monomer to a pentanucleotide, protein-protein interactions give rise to hexamers; in turn, hexamers promote the cooperative assembly of double hexamers on active pairs of pentanucleotides. The bold arrow is used to show that assembly events involving the EP region are preferred over those involving the AT-rich region (see the text). Finally, while subfragments of the core origin support double-hexamer formation in the presence of AMP-PNP or ADP, only hexamers are detected in the presence of ATP. Therefore, ATP or ATP hydrolysis expands the repertoire of T-ag/core interactions necessary for double-hexamer formation or stability.
Given that either flanking sequence can be used to initiate T-ag assembly on the core origin subfragments, an interesting question is whether one is preferentially used in the context of the core origin. It was previously reported that hexamers preferentially form on the early half of the core origin and stimulate the assembly of hexamers on the late half of the origin (59). Furthermore, 1,10-phenanthroline-copper footprinting studies demonstrated that under replication conditions, T-ag double hexamers preferentially bound to pentanucleotides 1 and 3 (38). Preferential assembly of double hexamers on the early side of the core origin, symbolized by the thick arrow in Fig. 11, is also consistent with the observation that in the presence of AMP-PNP, missing purine or pyrimidine bases had deleterious effects on T-ag assembly only when they were missing from the EP element (66). It is also consistent with studies indicating that the EP and site II cooperate to enhance the binding of T-ag (58). Collectively, these studies indicate that T-ag assembly is initiated via interactions with the EP region and that double hexamers initially occupy pentanucleotides 1 and 3 (38).
Once double hexamers form, they are positioned in a manner that facilitates subsequent interactions with regions of the core origin not used in the assembly process (i.e., the unoccupied pair of pentanucleotides [38] and the second flanking region). Interactions with these sequences may depend upon the hydrolysis of ATP or binding of an essential cofactor for initiation [e.g., HSSB (RPA)]. Low but detectable protection of pentanucleotides 2 and 4 in previous footprinting studies, conducted in the presence of ATP, may reflect nucleotide-dependent remodeling (38), a possibility currently under investigation.
In summary, our studies demonstrate that the interactions of T-ag with the SV40 core origin are complex. Binding depends upon associations between the T-ag-obd and site II and on interactions between non-T-ag-obd residues and the flanking regions. Clearly, much remains to be learned about the protein-DNA interactions that take place at the SV40 origin, the structural consequences of these interactions, and the way these processes are regulated. It will be interesting to establish the extent to which similar protein-DNA interactions take place at other eukaryotic origins.
ACKNOWLEDGMENTS
H.Y.K. and B.A.B. contributed equally to this work.
We thank W. W. Bachovchin, D. G. Sanford and B. S. Schaffhausen for comments on this manuscript and for helpful discussions.
This study was funded by grants from NIH (9R01GM55397 and 5R01GM53618).
REFERENCES
- 1.Borowiec J A. Inhibition of structural changes in the simian virus 40 core origin of replication by mutation of essential origin sequences. J Virol. 1992;66:5248–5255. doi: 10.1128/jvi.66.9.5248-5255.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borowiec J A, Dean F B, Bullock P A, Hurwitz J. Binding and unwinding—how T antigen engages the SV40 origin of DNA replication. Cell. 1990;60:181–184. doi: 10.1016/0092-8674(90)90730-3. [DOI] [PubMed] [Google Scholar]
- 3.Borowiec J A, Hurwitz J. ATP stimulates the binding of the simian virus 40 (SV40) large tumor antigen to the SV40 origin of replication. Proc Natl Acad Sci USA. 1988;85:64–68. doi: 10.1073/pnas.85.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borowiec J A, Hurwitz J. Localized melting and structural changes in the SV40 origin of replication induced by T-antigen. EMBO J. 1988;7:3149–3158. doi: 10.1002/j.1460-2075.1988.tb03182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradley M K, Smith T F, Lathrop R H, Livingston D M, Webster T A. Consensus topography in the ATP binding site of the simian virus 40 and polyomavirus large tumor antigens. Proc Natl Acad Sci USA. 1987;84:4026–4030. doi: 10.1073/pnas.84.12.4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brush G S, Kelly T J. Mechanisms for replicating DNA. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 1–43. [Google Scholar]
- 7.Bullock P A. The initiation of simian virus 40 DNA replication in vitro. Crit Rev Biochem Mol Biol. 1997;32:503–568. doi: 10.3109/10409239709082001. [DOI] [PubMed] [Google Scholar]
- 8.Bullock P A, Joo W S, Sreekumar K R, Mellow C. Initiation of SV40 DNA replication in vitro: analysis of the role played by sequences flanking the core origin on initial synthesis events. Virology. 1997;227:460–473. doi: 10.1006/viro.1996.8347. [DOI] [PubMed] [Google Scholar]
- 9.Bullock P A, Seo Y S, Hurwitz J. Initiation of simian virus 40 DNA replication in vitro: pulse-chase experiments identify the first labeled species as topologically unwound. Proc Natl Acad Sci USA. 1989;86:3944–3948. doi: 10.1073/pnas.86.11.3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burhans W C, Huberman J A. DNA replication origins in animal cells: a question of context? Science. 1994;263:639–640. doi: 10.1126/science.8303270. [DOI] [PubMed] [Google Scholar]
- 11.Campbell K S, Mullane K P, Aksoy I A, Stubdal H, Pipas J M, Silver P A, Roberts T M, Schaffhausen B S, DeCaprio J A. DnaJ/hsp40 chaperone domain of SV40 large T antigen promotes efficient viral DNA replication. Genes Dev. 1997;11:1098–1110. doi: 10.1101/gad.11.9.1098. [DOI] [PubMed] [Google Scholar]
- 12.Clark R, Peden K, Pipas J M, Nathans D, Tjian R. Biochemical activities of T-antigen proteins encoded by simian virus 40 A gene deletion mutants. Mol Cell Biol. 1983;3:220–228. doi: 10.1128/mcb.3.2.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clertant P, Gaudray P, May E, Cuzin F. The nucleotide binding site detected by affinity labeling in the large T proteins of polyoma and SV40 viruses is distinct from their ATPase catalytic site. J Biol Chem. 1984;259:15196–15203. [PubMed] [Google Scholar]
- 14.Dean F B, Borowiec J A, Eki T, Hurwitz J. The simian virus 40 T antigen double hexamer assembles around the DNA at the replication origin. J Biol Chem. 1992;267:14129–14137. [PubMed] [Google Scholar]
- 15.Dean F B, Borowiec J A, Ishimi Y, Deb S, Tegtmeyer P, Hurwitz J. Simian virus 40 large tumor antigen requires three core replication origin domains for DNA unwinding and replication in vitro. Proc Natl Acad Sci USA. 1987;84:8267–8271. doi: 10.1073/pnas.84.23.8267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dean F B, Bullock P, Murakami Y, Wobbe C R, Weissbach L, Hurwitz J. Simian virus 40 (SV40) DNA replication: SV40 large T antigen unwinds DNA containing the SV40 origin of replication. Proc Natl Acad Sci USA. 1987;84:16–20. doi: 10.1073/pnas.84.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dean F B, Dodson M, Echols H, Hurwitz J. ATP-dependent formation of a specialized nucleoprotein structure by simian virus 40 (SV40) large tumor antigen at the SV40 replication origin. Proc Natl Acad Sci USA. 1987;84:8981–8985. doi: 10.1073/pnas.84.24.8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dean F B, Lee S H, Kwong A D, Bullock P, Borowiec J A, Kenny M K, Seo Y S, Eki T, Matsumoto T, Hodgins G, Hurwitz J. SV40 DNA replication in vitro. UCLA Symp Mol Cell Biol New Ser. 1989;127:315–326. [Google Scholar]
- 19.Deb S, DeLucia A L, Baur C-P, Koff A, Tegtmeyer P. Domain structure of the simian virus 40 core origin of replication. Mol Cell Biol. 1986;6:1663–1670. doi: 10.1128/mcb.6.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deb S, DeLucia A L, Koff A, Tsui S, Tegtmeyer P. The adenine-thymine domain of the simian virus 40 core origin directs DNA bending and coordinately regulates DNA replication. Mol Cell Biol. 1986;6:4578–4584. doi: 10.1128/mcb.6.12.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deb S, Tsui S, Koff A, DeLucia A L, Parsons R, Tegtmeyer P. The T-antigen-binding domain of the simian virus 40 core-origin of replication. J Virol. 1987;61:2143–2149. doi: 10.1128/jvi.61.7.2143-2149.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deb S P, Tegtmeyer P. ATP enhances the binding of simian virus 40 large T antigen to the origin of replication. J Virol. 1987;61:3649–3654. doi: 10.1128/jvi.61.12.3649-3654.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeLucia A L, Deb S, Partin K, Tegtmeyer P. Functional interactions of the simian virus 40 core origin of replication with flanking regulatory sequences. J Virol. 1986;57:138–144. doi: 10.1128/jvi.57.1.138-144.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeLucia A L, Lewton B A, Tjian R, Tegtmeyer P. Topography of simian virus 40 A protein-DNA complexes: arrangement of pentanucleotide interaction sites at the origin of replication. J Virol. 1983;46:143–150. doi: 10.1128/jvi.46.1.143-150.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 26.DiMaio D, Nathans D. Cold-sensitive regulatory mutants of simian virus 40. J Mol Biol. 1980;140:129–146. doi: 10.1016/0022-2836(80)90359-9. [DOI] [PubMed] [Google Scholar]
- 27.Dixon R A F, Nathans D. Purification of simian virus 40 large T antigen by immunoaffinity chromatography. J Virol. 1985;53:1001–1004. doi: 10.1128/jvi.53.3.1001-1004.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dodson M, Dean F B, Bullock P, Echols H, Hurwitz J. Unwinding of duplex DNA from the SV40 origin of replication by T antigen. Science. 1987;238:964–967. doi: 10.1126/science.2823389. [DOI] [PubMed] [Google Scholar]
- 29.Fanning E, Knippers R. Structure and function of simian virus 40 large tumor antigen. Annu Rev Biochem. 1992;61:55–85. doi: 10.1146/annurev.bi.61.070192.000415. [DOI] [PubMed] [Google Scholar]
- 30.Fersht A. Enzyme structure and mechanism. 2nd ed. New York, N.Y: W. H. Freeman & Co.; 1984. [Google Scholar]
- 31.Giacherio D, Hager L P. A poly(dT)-stimulated ATPase activity associated with simian virus 40 large T antigen. J Biol Chem. 1979;254:8117–8120. [PubMed] [Google Scholar]
- 32.Goetz G S, Dean F B, Hurwitz J, Matson S W. The unwinding of duplex regions in DNA by the simian virus 40 large tumor antigen-associated DNA helicase activity. J Biol Chem. 1988;263:383–392. [PubMed] [Google Scholar]
- 33.Gottlieb P, Nasoff M S, Fisher E F, Walsh A M, Caruthers M H. Binding studies of SV40 T-antigen to SV40 binding site II. Nucleic Acids Res. 1985;13:6621–6634. doi: 10.1093/nar/13.18.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hertz G Z, Young M R, Mertz J E. The A+T-rich sequence of the simian virus 40 origin is essential for replication and is involved in bending of the viral DNA. J Virol. 1987;61:2322–2325. doi: 10.1128/jvi.61.7.2322-2325.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang S G, Weisshart K, Gilbert I, Fanning E. Stoichiometry and mechanisms of assembly of SV40 T antigen complexes with the viral origin of DNA replication and DNA polymerase α-primase. Biochemistry. 1998;37:15345–15352. doi: 10.1021/bi9810959. [DOI] [PubMed] [Google Scholar]
- 36.Hurwitz J, Dean F B, Kwong A D, Lee S-H. The in vitro replication of DNA containing the SV40 origin. J Biol Chem. 1990;265:18043–18046. [PubMed] [Google Scholar]
- 37.Jones K A, Myers R M, Tjian R. Mutational analysis of simian virus 40 large T antigen DNA binding sites. EMBO J. 1984;3:3247–3255. doi: 10.1002/j.1460-2075.1984.tb02286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joo W S, Kim H Y, Purviance J D, Sreekumar K R, Bullock P A. Assembly of T-antigen double hexamers on the simian virus 40 core origin requires only a subset of the available binding sites. Mol Cell Biol. 1998;18:2677–2687. doi: 10.1128/mcb.18.5.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joo W S, Luo X, Denis D, Kim H Y, Rainey G J, Jones C, Sreekumar K R, Bullock P A. Purification of the SV40 T-antigen DNA binding domain and characterization of its interactions with the SV40 origin. J Virol. 1997;71:3972–3985. doi: 10.1128/jvi.71.5.3972-3985.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kadonaga J T, Tjian R. Affinity purification of sequence-specific DNA binding proteins. Proc Natl Acad Sci USA. 1986;83:5889–5893. doi: 10.1073/pnas.83.16.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kelly W L, Landry S J. Chaperone power in a virus? Trends Biol Sci. 1994;19:277–278. doi: 10.1016/0968-0004(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 42.Korolev S, Hsieh J, Gauss G H, Lohman T M, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 43.Lewton B A, DeLucia A L, Tegtmeyer P. Binding of simian virus 40 A protein to DNA with deletions at the origin of replication. J Virol. 1984;49:9–13. doi: 10.1128/jvi.49.1.9-13.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J J, Peden K W C, Dixon R A F, Kelly T. Functional organization of the simian virus 40 origin of DNA replication. Mol Cell Biol. 1986;6:1117–1128. doi: 10.1128/mcb.6.4.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li L, Li B L, Hock M, Wang E, Folk W R. Sequences flanking the pentanucleotide T-antigen binding sites in the polyomavirus core origin help determine selectivity of DNA replication. J Virol. 1995;69:7570–7578. doi: 10.1128/jvi.69.12.7570-7578.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lorimer H E, Reynisdottir I, Ness S, Prives C. Unusual properties of a replication-defective mutant SV40 large T-antigen. Virology. 1993;192:402–414. doi: 10.1006/viro.1993.1055. [DOI] [PubMed] [Google Scholar]
- 47.Lorimer H E, Wang E H, Prives C. The DNA-binding properties of polyomavirus large T antigen are altered by ATP and other nucleotides. J Virol. 1991;65:687–699. doi: 10.1128/jvi.65.2.687-699.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo X, Sanford D G, Bullock P A, Bachovchin W W. Structure of the origin specific DNA binding domain from simian virus 40 T-antigen. Nat Struct Biol. 1996;3:1034–1039. doi: 10.1038/nsb1296-1034. [DOI] [PubMed] [Google Scholar]
- 49.Lusky M, Hurwitz J, Seo Y-S. Cooperative assembly of the bovine papilloma virus E1 and E2 proteins on the replication origin requires an intact E2 binding site. J Biol Chem. 1993;268:15795–15803. [PubMed] [Google Scholar]
- 50.Mastrangelo I A, Hough P V C, Wall J S, Dodson M, Dean F B, Hurwitz J. ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature (London) 1989;338:658–662. doi: 10.1038/338658a0. [DOI] [PubMed] [Google Scholar]
- 51.McEntee K, Weinstock G M, Lehman I R. recA protein-catalyzed strand assimilation: stimulation by Escherichia coli single-stranded DNA-binding protein. Proc Natl Acad Sci USA. 1980;77:857–861. doi: 10.1073/pnas.77.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McVey D, Woelker B, Tegtmeyer P. Mechanisms of simian virus 40 T-antigen activation by phosphorylation of threonine 124. J Virol. 1996;70:3887–3893. doi: 10.1128/jvi.70.6.3887-3893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mohr I J, Fairman M P, Stillman B, Gluzman Y. Large T-antigen mutants define multiple steps in the initiation of simian virus 40 DNA replication. J Virol. 1989;63:4181–4188. doi: 10.1128/jvi.63.10.4181-4188.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Müller D, Ugi I, Ballas K, Reiser P, Henning R, Montenarh M. The AT-rich sequence of the SV40 control region influences the binding of SV40 T antigen to binding sites II and III. Virology. 1987;161:81–90. doi: 10.1016/0042-6822(87)90173-5. [DOI] [PubMed] [Google Scholar]
- 55.Murakami Y, Hurwitz J. DNA polymerase α stimulates the ATP-dependent binding of simian virus tumor T antigen to the SV40 origin of replication. J Biol Chem. 1993;268:11018–11027. [PubMed] [Google Scholar]
- 56.Myers R M, Tjian R. Construction and analysis of simian virus 40 origins defective in tumor antigen binding and DNA replication. Proc Natl Acad Sci USA. 1980;77:6491–6495. doi: 10.1073/pnas.77.11.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O’Reilly D R, Miller L K. Expression and complex formation of simian virus 40 large T antigen and mouse p53 in insect cells. J Virol. 1988;62:3109–3119. doi: 10.1128/jvi.62.9.3109-3119.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parsons R, Anderson M E, Tegtmeyer P. Three domains in the simian virus 40 core origin orchestrate the binding, melting, and DNA helicase activities of T antigen. J Virol. 1990;64:509–518. doi: 10.1128/jvi.64.2.509-518.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parsons R E, Stenger J E, Ray S, Welker R, Anderson M E, Tegtmeyer P. Cooperative assembly of simian virus 40 T-antigen hexamers on functional halves of the replication origin. J Virol. 1991;65:2798–2806. doi: 10.1128/jvi.65.6.2798-2806.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reynisdottir I, Lorimer H E, Friedman P N, Wang E H, Prives C. Phosphorylation and active ATP hydrolysis are not required for SV40 T antigen hexamer formation. J Biol Chem. 1993;268:24647–24654. [PubMed] [Google Scholar]
- 61.Roberts J. Simian virus 40 (SV40) large tumor antigen causes stepwise changes in SV40 origin structure during initiation of DNA replication. Proc Natl Acad Sci USA. 1989;86:3939–3943. doi: 10.1073/pnas.86.11.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ryder K, Vakalopoulou E, Mertz R, Mastrangelo I, Hough P, Tegtmeyer P, Fanning E. Seventeen base pairs of region I encode a novel tripartite binding signal for SV40 T antigen. Cell. 1985;42:539–548. doi: 10.1016/0092-8674(85)90111-4. [DOI] [PubMed] [Google Scholar]
- 63.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 64.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.San Martin M C, Gruss C, Carazo J M. Six molecules of SV40 large T antigen assemble in a propeller-shaped particle around a channel. J Mol Biol. 1997;268:15–20. doi: 10.1006/jmbi.1997.0952. [DOI] [PubMed] [Google Scholar]
- 66.SenGupta D J, Borowiec J A. Strand and face: the topography of interactions between the SV40 origin of replication and T-antigen during the initiation of replication. EMBO J. 1994;13:982–992. doi: 10.1002/j.1460-2075.1994.tb06343.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.SenGupta D J, Borowiec J A. Strand-specific recognition of a synthetic DNA replication fork by the SV40 large tumor antigen. Science. 1992;256:1656–1661. doi: 10.1126/science.256.5064.1656. [DOI] [PubMed] [Google Scholar]
- 68.Simanis V, Lane D P. An immunoaffinity purification procedure for SV40 large T antigen. Virology. 1985;144:88–100. doi: 10.1016/0042-6822(85)90308-3. [DOI] [PubMed] [Google Scholar]
- 69.Simmons D T, Loeber G, Tegtmeyer P. Four major sequence elements of simian virus 40 large T antigen coordinate its specific and nonspecific DNA binding. J Virol. 1990;64:1973–1983. doi: 10.1128/jvi.64.5.1973-1983.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Simmons D T, Wun-Kim K, Young W. Identification of simian virus 40 T-antigen residues important for specific and nonspecific binding to DNA and for helicase activity. J Virol. 1990;64:4858–4865. doi: 10.1128/jvi.64.10.4858-4865.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Smelkova N V, Borowiec J A. Dimerization of simian virus 40 T-antigen hexamers activates T-antigen DNA helicase activity. J Virol. 1997;71:8766–8773. doi: 10.1128/jvi.71.11.8766-8773.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Srinivasan A, McClellan A J, Vartikar J, Marks I, Cantalupo P, Li Y, Whyte P, Rundell K, Brodsky J L, Pipas J M. The amino-terminal transforming region of simian virus 40 large T and small t antigens functions as a J domain. Mol Cell Biol. 1997;17:4761–4773. doi: 10.1128/mcb.17.8.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stahl H, Droge P, Knippers R. DNA helicase activity of SV40 large tumor antigen. EMBO J. 1986;5:1939–1944. doi: 10.1002/j.1460-2075.1986.tb04447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stillman B, Gerard R D, Guggenheimer R A, Gluzman Y. T antigen and template requirement for SV40 DNA replication in vitro. EMBO J. 1985;4:2933–2939. doi: 10.1002/j.1460-2075.1985.tb04026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Studwell P S, O’Donnell M. Processive replication is contingent on the exonuclease subunit of DNA polymerase III holoenzyme. J Biol Chem. 1990;265:1171–1178. [PubMed] [Google Scholar]
- 76.Subramanya H S, Bird L E, Brannigan J A, Wigley D B. Crystal structure of a DExx box DNA helicase. Nature. 1996;384:379–383. doi: 10.1038/384379a0. [DOI] [PubMed] [Google Scholar]
- 77.Tegtmeyer P, Lewton B A, DeLucia A L, Wilson V G, Ryder K. Topography of simian virus 40 A protein-DNA complexes: arrangement of protein bound to the origin of replication. J Virol. 1983;46:151–161. doi: 10.1128/jvi.46.1.151-161.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tjian R. The binding site on SV40 DNA for a T-antigen related protein. Cell. 1978;13:165–179. doi: 10.1016/0092-8674(78)90147-2. [DOI] [PubMed] [Google Scholar]
- 79.Tooze J. DNA tumor viruses. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1981. [Google Scholar]
- 80.Velankar S S, Soultanas P, Dillingham M S, Subramanya H S, Wigley D B. Crystal structures of complexes of PcrA DNA Helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 81.Virshup D M, Russo A A R, Kelly T J. Mechanism of activation of simian virus 40 DNA replication by protein phosphatase 2A. Mol Cell Biol. 1992;12:4883–4895. doi: 10.1128/mcb.12.11.4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu Rev Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 83.Weisshart K, Taneja P, Jenne A, Herbig U, Simmons D T, Fanning E. Two regions of simian virus 40 T antigen determine cooperativity of double-hexamer assembly on the viral origin of DNA replication and promote hexamer interactions during bidirectional origin DNA unwinding. J Virol. 1999;73:2201–2211. doi: 10.1128/jvi.73.3.2201-2211.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wessel R, Schweizer J, Stahl H. Simian virus 40 T-antigen DNA helicase is a hexamer which forms a binary complex during bidirectional unwinding from the viral origin of DNA replication. J Virol. 1992;66:804–815. doi: 10.1128/jvi.66.2.804-815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wobbe C R, Dean F, Weissbach L, Hurwitz J. In vitro replication of duplex circular DNA containing the simian virus 40 DNA origin site. Proc Natl Acad Sci USA. 1985;82:5710–5714. doi: 10.1073/pnas.82.17.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wold M S, Li J J, Kelly T J. Initiation of simian virus 40 DNA replication in vitro: large-tumor-antigen- and origin-dependent unwinding of the template. Proc Natl Acad Sci USA. 1987;84:3643–3647. doi: 10.1073/pnas.84.11.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wold M S, Weinberg D H, Virshup D M, Li J J, Kelly T J. Identification of cellular proteins required for simian virus 40 DNA replication. J Biol Chem. 1989;264:2801–2809. [PubMed] [Google Scholar]
- 88.Wu C, Edgil D, Simmons D T. The origin DNA-binding and single-stranded DNA-binding domains of simian virus large T antigen are distinct. J Virol. 1998;72:10256–10259. doi: 10.1128/jvi.72.12.10256-10259.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wun-Kim K, Upson R, Young W, Melendy T, Stillman B, Simmons D T. The DNA-binding domain of simian virus 40 tumor antigen has multiple functions. J Virol. 1993;67:7608–7611. doi: 10.1128/jvi.67.12.7608-7611.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]