Abstract
Traditional Chinese medicine (TCM) not only maintains the health of Asian people but also provides a great resource of active natural products for modern drug development. Herein, we developed a Database of Constituents Absorbed into the Blood and Metabolites of TCM (DCABM-TCM), the first database systematically collecting blood constituents of TCM prescriptions and herbs, including prototypes and metabolites experimentally detected in the blood, together with the corresponding detailed detection conditions through manual literature mining. The DCABM-TCM has collected 1816 blood constituents with chemical structures of 192 prescriptions and 194 herbs and integrated their related annotations, including physicochemical, absorption, distribution, metabolism, excretion, and toxicity properties, and associated targets, pathways, and diseases. Furthermore, the DCABM-TCM supported two blood constituent-based analysis functions, the network pharmacology analysis for TCM molecular mechanism elucidation, and the target/pathway/disease-based screening of candidate blood constituents, herbs, or prescriptions for TCM-based drug discovery. The DCABM-TCM is freely accessible at http://bionet.ncpsb.org.cn/dcabm-tcm/. The DCABM-TCM will contribute to the elucidation of effective constituents and molecular mechanism of TCMs and the discovery of TCM-derived drug-like compounds that are both bioactive and bioavailable.
Introduction
Traditional Chinese medicine (TCM) not only plays an important role in maintaining the health of people in Asia1 but also provides a great resource of bioactive natural products for modern drug development. Many successful western drugs are derived from TCMs, including artemisinin extracted from Artemisia annua L. for malaria (for which the discoverer won the 2015 Nobel Prize in Physiology or Medicine) and ephedrine from Ephedra sinicaStapf for asthma treatment. Because TCM often functions through the synergistic action of multiple components and targets (take Realgar–Indigo naturalis formula as an example. By comparing with the effects of mono- or biagents, Wang et al. have proved that the combination of its active ingredients tetraarsenic tetrasulfide, indirubin, and tanshinone IIA separately from its compositive herbs realgar, Indigo naturalis, and Salvia miltiorrhiza produces synergy during the treatment of human acute promyelocytic leukemia by hitting multiple targets2), TCM research can also inspire the treatment of complex diseases, such as cancers and diabetes, caused by abnormal interactions among multiple molecules and pathways (for example, the abnormal interaction between PD-1 and PD-L1 is responsible for cancer immune escape, promoting tumor development, and PD1/PD-L1 blockade therapy has been used to treat multiple solid tumors3).4 However, owing to TCM’s diverse constituents and their complex interactions with the human body, it is still challenging to clarify its effective constituents and molecular mechanisms. This greatly hinders TCM’s more widespread application and TCM-based modern drug development.
Most constituents in TCM produce pharmacological effects only after they are absorbed into the bloodstream.5 These “constituents detected in the blood” (also denoted as “blood constituents” in this paper) include the original TCM constituents directly absorbed into the blood (i.e., “constituents absorbed into the blood” or “prototypes”) and the “metabolites” of the original constituents. “Prototypes” are the original TCM constituents detected in the blood. Although they may have passed through the gastrointestinal tract or liver, they remain unchanged after they reach the bloodstream. “Metabolites” are transformed from original constituents by chemical reactions in vivo (in the gastrointestinal tract or liver). Compared with the ordinary TCM constituents (also called “original constituents” in this paper) generally detected in vitro, these constituents that reach the bloodstream, including prototypes and metabolites, are more likely to be the active ones responsible for the pharmacological effects. The identification of blood constituents greatly avoids the false positives and negatives in the identification of TCM effective constituents, where the false positives refer to those active in vitro but cannot be absorbed or become inactive after metabolism, and the false negatives refer to those inactive in vitro but become active after metabolism.
The systematic collection of these constituents of TCMs detected in the blood is important for (1) the elucidation of the effective material basis and molecular mechanism of TCMs; (2) the selection of quality markers for herbs;6 (3) the discovery of TCM-derived drug-like compounds that are both bioactive and absorbable; and (4) the study of the properties of natural compounds that can be absorbed, such as structural and physicochemical features. Compared with ordinary constituents of TCMs generally detected in vitro, identifying blood constituents is a great step forward for all the issues mentioned above.
Recently, blood constituents for increasing TCM prescriptions and herbs were identified using liquid chromatography coupled with tandem mass spectrometry (LC–MS/MS). However, these data are scattered in various publications. Moreover, the blood constituents of a prescription/herb were often detected by multiple studies. In these studies, the detection conditions could have been different, leading to the detection of different blood constituents. It is necessary to collect and compile these data systematically.
However, none of the existing public databases have recorded blood constituent information of TCM prescriptions and herbs. Currently, there have been many great TCM databases/platforms. For example, ETCM,7 SymMap,8 TCMID,9 TCM-ID,10 TCM Database@Taiwan,11 TCM-Mesh,12 YaTCM,13 and TCMSP14 all have provided comprehensive TCM prescription/herb-ingredient-target-pathway/disease associations, as well as other related information, where SymMap has also uniquely provided associations between TCM symptom, herbs, and symptoms used in modern medicine and TCMSP has also given absorption, distribution, metabolism, excretion (ADME) properties of TCM ingredients. In addition, multiple integrated resources for herbs used in other countries have emerged, such as ETM-DB for Ethiopian traditional medicines,15 TM-MC for Northeast Asian (including China, Korea, and Japan) traditional medicines,16 IMPPAT for Indian medicinal plants,17 and PharmDB-K for Korean traditional medicines.18 CMAUP has been developed for a broader range of useful plants.19 However, all these databases/platforms have only collected ordinary constituents generally detected in vitro and have not covered constituents detected in the blood. HIM, the herbal ingredients in vivo metabolism database, has recorded in vivo metabolic information of herb-derived single compounds.20 Unlike HIM, herein, we focused on the prescription/herb as a whole. Compared with HIM, the collection of blood constituents of prescriptions or herbs has unique advantages in the elucidation of the effective constituents and the multicomponent synergistic molecular mechanism of a prescription/herb, herbal quality control, the study of the properties of absorbable natural products, and TCM symptom-inspired discovery of lead compounds derived from TCM (i.e., phenotype-based drug discovery).8
Therefore, we developed a Database of Constituents Absorbed into the Blood and Metabolites of Traditional Chinese Medicine (DCABM-TCM) (http://bionet.ncpsb.org.cn/dcabm-tcm/), the first database systematically collecting blood constituents of TCM prescriptions and herbs together with the corresponding detailed detection conditions by manual literature mining. In addition, the DCABM-TCM was also designed to support two TCM blood constituent-based data analysis functions: TCM molecular mechanism elucidation and TCM-based drug discovery.
Materials and Methods
Core Data Collection and Curation
The core data of the DCABM-TCM are constituents (including prototype components and metabolites) of TCM herbs and prescriptions detected in the blood using various experimental techniques, such as LC–MS/MS. In addition, considering that the experimental conditions have an important impact on the detected blood constituents of a prescription or herb, we also recorded the corresponding detailed detection conditions, including the extraction method, experimental animal and animal model, administration method and dose, blood collection time and location, and the corresponding source publication.
All these data were manually mined from papers from PubMed (https://pubmed.ncbi.nlm.nih.gov/) and China National Knowledge Infrastructure (CNKI, https://www.cnki.net/). CNKI hosts articles published in Chinese. As shown in Figure 1, first, potentially related papers were identified by searching PubMed and CNKI abstracts (published till December 2019) with two keyword lists using a self-developed bioentity recognizer (which has been successfully used to establish the AAgAtlas database21). For English papers from PubMed, the first keyword list included “Traditional Chinese Medicine”, “TCM”, “Chinese herbal medicine”, “CHM”, “herb”, “formula”, “prescription”, and their various variations. The other list included 61 phrases that potentially suggested the “constituent detected in the blood”, such as “constituent absorbed into the blood”, “absorbed component”, “detected in plasma”, “serum fingerprint”, and “serum pharmacochemistry”. For Chinese papers from CNKI, corresponding keywords in Chinese were used. Potentially related papers were those that simultaneously contained these two classes of keywords in their abstracts. Subsequently, we conducted a preliminary screening by manually browsing all the titles and abstracts. Then, we manually checked the full texts and supplementary materials of the qualified papers to extract the core data of the DCABM-TCM mentioned in the previous paragraph. Chinese information was simultaneously translated into English. The manually extracted data fields mainly included source publication, herb or prescription name, compositive herb list of the prescription, chemical names and molecular formulas of blood constituents, and the corresponding detailed detection conditions described in the previous paragraph. In addition, we recorded which blood constituents were prototypes and which were metabolites when available. Every piece of manually curated information was checked by at least three different experts to ensure data quality.
Figure 1.
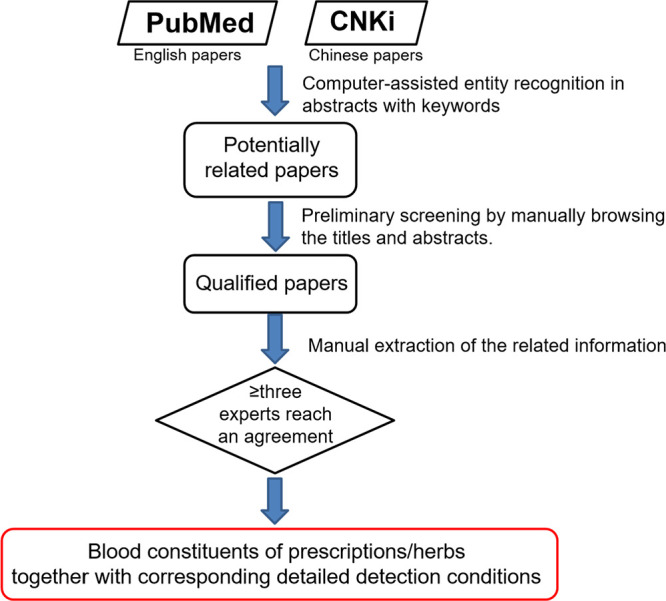
Data curation workflow of the DCABM-TCM core data.
In the DCABM-TCM, blood constituents were uniformly mapped to PubChem compound identifiers (CIDs)22 with the help of PubChemPy23 or by manual based on compound names (after necessary cleaning and standardization) together with molecular formulas when available. Then, we manually scanned related source publications for those that still do not have corresponding PubChem CIDs and tried to extract their structural information. Two-dimensional chemical structure graphs of blood constituents were collected and converted into the InChI identifiers using ChemDraw.24 These blood constituents were further mapped to PubChem CIDs based on their InChI identifiers. In summary, in the DCABM-TCM, blood constituents were divided into three types: (a) those that have mapped PubChem CIDs (and thus also have chemical structures from PubChem), (b) those without PubChem CIDs but with chemical structures (directly from source publications), and (c) those that have neither mapped PubChem CIDs nor chemical structures. For this type, there is a special case that the original literature only identified the chemical formula of the blood constituent (by analyzing the MS data), not the name. For such blood constituents, we recorded their name as “unknown”.
Annotation Data Collection
We also collected various annotation data around the core data to help the TCM molecular mechanism elucidation and TCM-based drug discovery. Indication information of an herb was obtained from the TCMID database.9 For a blood constituent with a PubChem CID, its basic information, including structure, chemical abstracts service (CAS) number, cross-reference, and chemical and physical properties, was obtained directly from the PubChem database22 with the help of PubChemPy.23 For a blood constituent without a PubChem CID but with a chemical structure (directly from source publication), its chemical and physical properties were computed using the PaDEL–Descriptor software25 and XLOGP program26 based on its chemical structure. Herein, only six classical physicochemical properties were considered, including molecular weight, the logarithm of the partition coefficient between n-octanol and water (logP),27 topological polar surface area (TPSA), number of hydrogen bond donors in the structure, number of hydrogen bond acceptors, and number of rotatable bonds, which are important for computationally evaluating the “drug-likeness”.28 Like the DrugBank database,29 the 22 absorption, distribution, metabolism, excretion, and toxicity (ADMET) properties of the blood constituents, including human intestinal absorption, blood–brain barrier penetration, Caco-2 permeability, P-glycoprotein substrate and inhibitor, CYP450 substrate and inhibitor (CYP1A2, 2C9, 2D6, 2C19, and 3A4), hERG inhibitors, AMES mutagenicity, carcinogens, and rat acute toxicity, were predicted using admetSAR.28 The hierarchical classification of blood constituents was given by ClassyFire,30 which provides a rule-based structural classification for compounds.
Furthermore, blood constituent–target gene associations were obtained from BATMAN-TCM,31 including known and predicted ones. A target gene’s basic annotation information, such as synonyms, full name, and cross-references, was obtained from the NCBI Entrez Gene database (downloaded on 05/14/2020).32 Target gene–Gene Ontology (GO) term association data were also obtained from the Entrez Gene database (downloaded on 05/14/2020); biological pathway data were obtained from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (downloaded on 02/26/2020).33 Target gene–disease associations were obtained from the Online Mendelian Inheritance in Man (OMIM) database34 and the comparative toxicogenomics database (CTD).35 For OMIM, we directly extracted gene–OMIM disease associations from Swiss-Prot (downloaded on 05/15/2020).36 For CTD, we only used its gene–disease associations with direct evidence (version: Jul 6, 2017).
Implementation of the Two Analysis Functions
To help users better utilize the core data and explore its value, the DCABM-TCM supports two blood constituent-based analysis functions.
The network pharmacology analysis function of a prescription/herb/blood constituent was implemented with the aid of the BATMAN-TCM.31 The BATMAN-TCM is the first online bioinformatics analysis tool for the molecular mechanism of TCM previously developed by us. The analyses supported by the BATMAN-TCM mainly include (1) target prediction of blood constituents based on a similarity-based target prediction method (which ranks potential drug–target interactions based on their similarity to the known drug–target interactions);31 (2) functional analyses of targets, including biological pathway, GO functional term, and disease enrichment analyses; and (3) the visualization of a blood constituent–target–pathway/disease association network. Concerning the BATMAN-TCM released in 2016, in the DCABM-TCM, the background data supporting the related analyses, mainly including human gene–GO term/disease associations and pathway data, were updated to the later version as described above.
In the function of “target/pathway/disease-based screening of candidate blood constituents, herbs, and prescriptions”, the candidate blood constituents targeting a target gene were determined with the help of the BATMAN-TCM. Furthermore, we thought that the significantly enriched prescriptions/herbs among these candidate blood constituents were potential candidate prescriptions/herbs targeting this target gene. Similarly, for a pathway/disease, we thought that significantly enriched blood constituents among the member genes of the pathway/the known related genes of the disease were potential candidate blood constituents targeting this pathway/disease. Moreover, significantly enriched herbs/prescriptions among these candidate blood constituents were considered as potential candidate herbs/prescriptions targeting this pathway/disease. The enrichment analyses were implemented based on the hypergeometric cumulative distribution test, and the multiple testing correction was based on the Benjamini-Hochberg correction method.37
Database Implementation
The foundation of the DCABM-TCM was a MongoDB database. On top of this database, the analysis application was implemented in Perl and Node.js, and the web presentation application was written in JavaScript and CSS. The structural similarity search was implemented based on Open Babel,38 where the FP2 fingerprint and the Tanimoto coefficient were used to compute the structural similarity of the two compounds. FP2 is a path-based fingerprint generated by analyzing all possible linear fragments of the molecule structure with a length ranging from one to seven atoms.38 The DCABM-TCM web server is compatible with Chrome, Firefox, and Opera browsers for Windows and Chrome, Firefox, Opera, and Safari for the Mac operating system.
Results
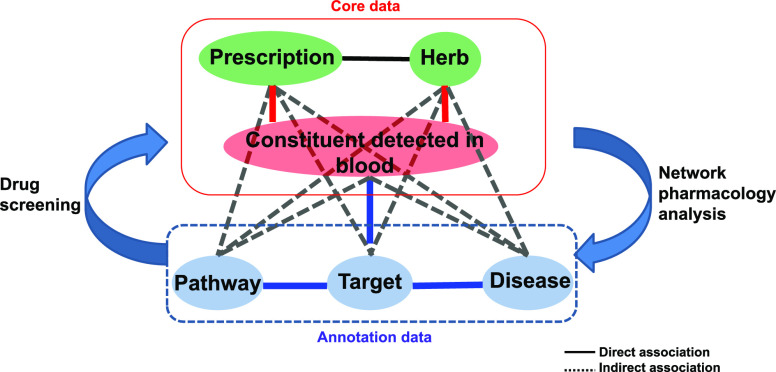
Overview of the DCABM-TCM
The DCABM-TCM can be freely accessed at http://bionet.ncpsb.org.cn/dcabm-tcm/. Its core data are constituents absorbed into the blood and metabolites of TCM prescriptions and herbs, together with the corresponding detailed detection conditions. Around the core data, annotation data include physicochemical properties, ADMET properties of the blood constituents and the associated targets, GO functional terms, pathways, and diseases. Therefore, the DCABM-TCM has six types of entities: prescriptions, herbs, blood constituents, targets, pathways, and diseases (Figure 2). For every entity, the DCABM-TCM provides a detailed annotation page presenting the related core and annotation information. The DCABM-TCM supports data browse and search for each of the six types of entities. The DCABM-TCM also supports two blood constituent-based data analysis functions: (1) the network pharmacology analysis for a prescription/herb/blood constituent to reveal its potential molecular mechanism based on its blood constituent(s) and (2) the screening of candidate blood constituents and further herbs/prescriptions (to which the blood constituents belong) potentially targeting a target/pathway/disease to help TCM-derived drug discovery. The two analysis functions and results are presented on the detailed annotation page of the corresponding entity type. In addition, data download and submission are also supported by the DCABM-TCM.
Figure 2.
Overview of the DCABM-TCM database.
Data Statistics and Analyses
Currently, the DCABM-TCM has recorded 1816 blood constituents with chemical structures (among which 1740 can be mapped to PubChem CIDs) of 192 prescriptions and 194 herbs, including 970 prototypes (among which 961 can be mapped to PubChem CIDs) and 385 metabolites (323 with PubChem CIDs) (Table 1) (Additional files 1–3, which can also be downloaded on the “Download” page of the DCABM-TCM). In addition, the DCABM-TCM has also recorded 1893 blood constituents without chemical structures (most of them are metabolites), which is counted based on the non-redundant “name (chemical formula)”. These blood constituents without structures could be structurally redundant with each other and with the ones with structures. In our data, only about a fourth of the source publications have stated which are prototypes and which are metabolites among the detected blood constituents of a prescription or herb. In addition, 3970 target genes, 333 KEGG pathways, and 4006 CTD diseases were associated with the constituents detected in the blood (Table 1).
Table 1. Statistics of the DCABM-TCM Data.
| data type | number |
|---|---|
| prescriptions | 192 |
| herbs | 194 |
| constituents with PubChem CIDs detected in the blood (including prototypes and metabolites) | 1740 |
| constituents with chemical structures detected in the blood (including prototypes and metabolites) | 1816 |
| prototypes with PubChem CIDs | 961 |
| prototypes with chemical structures | 970 |
| metabolites with PubChem CIDs | 323 |
| metabolites with chemical structures | 385 |
| target genes of blood constituents (including the known and predicted ones with scores ≥10) | 3970 |
| blood constituent–target gene associations | 55,967 |
| KEGG pathways involving the target genes | 333 |
| target gene–KEGG pathway associations | 17,538 |
| CTD diseases associated with the target genes | 4006 |
| target gene–CTD disease associations | 73,616 |
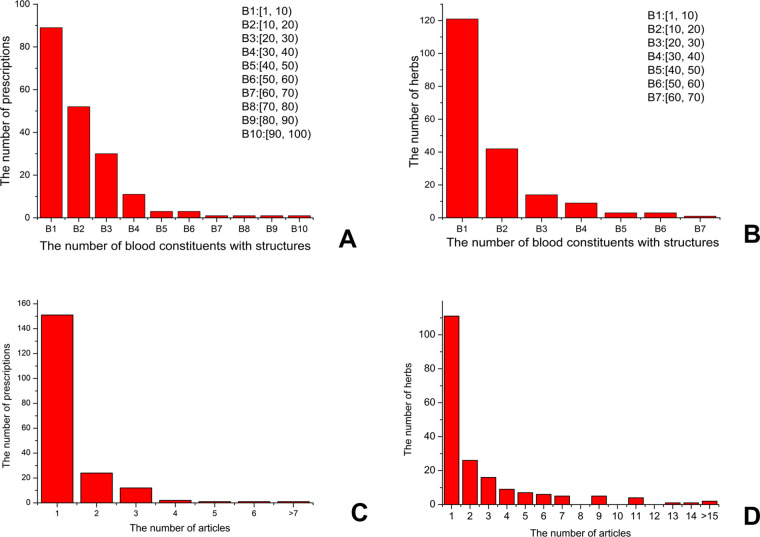
The median (average) number of blood constituents with structures was 11 (15.1) for a prescription and 6.5 (10.6) for an herb (Figure 3A,B). The number of the detected blood constituents of a prescription was often lower than the sum of the detected blood constituents of its compositive herbs. Most blood constituents detected in a prescription were also the blood constituents of its compositive herbs, while sometimes, a few new blood constituents appeared, which might be produced by the interactions between its compositive herbs, such as those produced in the decocting process of the prescription. These data of constituents detected in the blood were mined from 448 papers. In our data, for most prescriptions and herbs, their blood constituents were studied from only one paper, while some were studied from multiple papers. On average, the blood constituents of a prescription and herb were studied from 1.4 papers and 2.6 papers, respectively (Figure 3C,D).
Figure 3.
Distributions of the DCABM-TCM data. Distributions of the number of blood constituents with chemical structures of prescriptions (A) and herbs (B). Distributions of the number of papers that studied the blood constituents of a prescription (C) and herb (D).
The recorded detection conditions mainly included the extraction method, experimental animal and animal model, administration method and dose, and the blood collection time and location. The extraction methods included water, ethanol, and methanol extraction. Experimental animals included rats, mice, and rabbits. For the animal model, most of the studies used normal models, and the minority used disease models, such as rheumatoid arthritis, acute heart failure, and cerebral ischemia–reperfusion injury models. For the administration method, the vast majority of studies adopted the intragastrical administration (i.e., ig), and some studies adopted other methods, including intravenous injection (i.e., iv), intraperitoneal injection (i.e., ip), intravenous drip, intraduodenal administration, and intestinal circulatory perfusion. The administration dose was recorded typically in three forms: (1) “XX g/kg”, representing a single administration; (2) “XX g/kg, XX times”, representing multiple dosing in a short period; and (3) “XX g/kg, XX times/day, XX days”, representing a consecutive administration for XX days. The blood collection time was generally 0.5–3 h after the last administration, divided into single and multiple time points. Finally, blood collection locations mainly included the postorbital venous plexus, the abdominal aorta, the hepatic portal vein, the eyeball, and the fosse orbital vein.
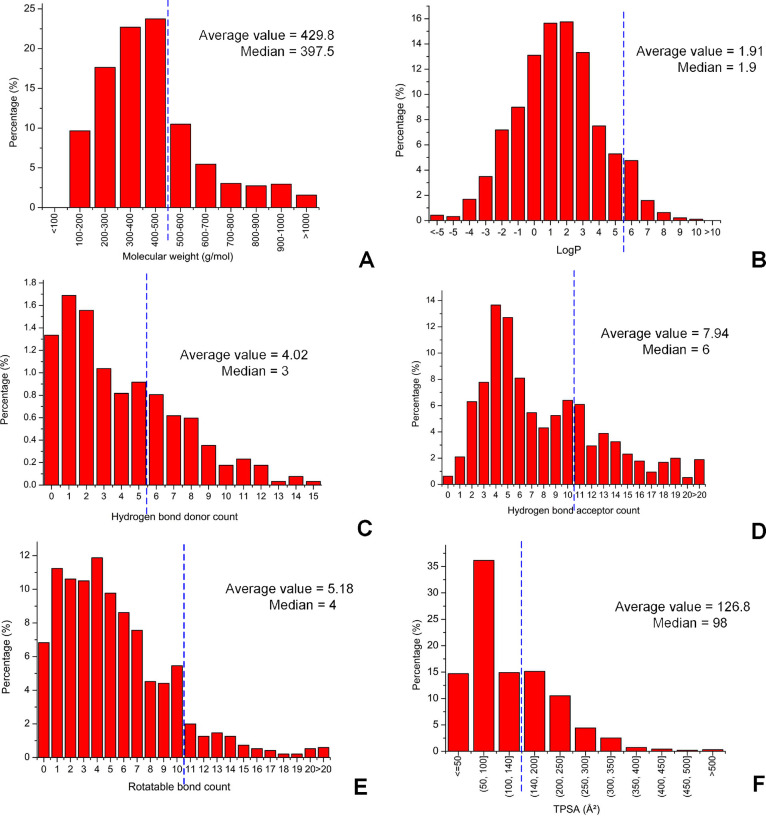
Finally, we described the physicochemical property distributions of 953 absorbed prototype constituents with structures of the prescriptions and herbs via oral administration (i.e., intragastrical administration) (Figure 4). We observed that most absorbed constituents via oral administration satisfied the traditional rule for the drug-like molecule screening (molecular weight ≤500, logP ≤ 5, hydrogen bond donor count ≤5, hydrogen bond acceptor count ≤10, rotatable bond count ≤10, TPSA ≤ 14039), but many still did not satisfy the rule, suggesting the imperfection of the traditional rule in estimating the absorption and permeation of molecules.
Figure 4.
Distributions of physicochemical properties of orally absorbed prototype constituents with structures. (A) Molecular weight. (B) logP (the logarithm of the partition coefficient between n-octanol and water). (C) Hydrogen bond donor count. (D) Hydrogen bond acceptor count. (E) Rotatable bond count. (F) TPSA (topological polar surface area). The average values and medians of the properties are given on the graphs. The property cutoff of the traditional rule of the drug-like molecule screening is marked on each of the graphs by a dotted line.
Usage of the DCABM-TCM
The DCABM-TCM supports data browsing, searching, downloading, and submission. In addition, it also supports two data analysis functions, as described in the next section.
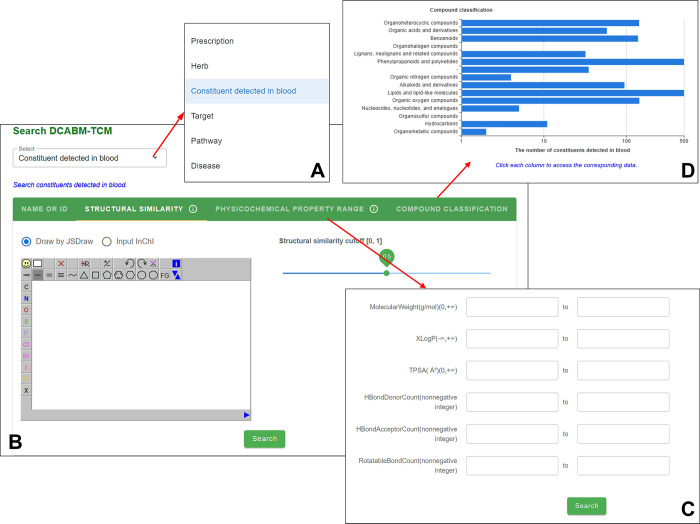
For each of the six types of annotation pages, including prescriptions, herbs, constituents detected in the blood, targets, pathways, and diseases, the DCABM-TCM supports browsing and searching. Among all the six types of entities that are searchable (Figure 5A), for the prescription or herb, users can search by English name or Pinyin name; for the target, by Entrez Gene ID, Gene symbol, Gene full name or by protein sequence similarity; for the disease, by disease name or CTD disease ID; and for the pathway, by KEGG pathway name or ID. Finally, for the constituent detected in the blood, the DCABM-TCM supports search by (1) compound name or PubChem CID, (2) structural similarity, (3) physicochemical property range, or (4) compound classification. When searching by structural similarity, users input a compound with the InChI identifier or draw the structure of a compound with the help of JSDraw40 and set the structural similarity cutoff (Figure 5B); then, structurally similar blood constituents in the order of decreasing similarity scores (≥cutoff) are returned. When searching by the property range, users can specify the range of the physicochemical properties of the returned blood constituents, including the molecular weight, logP, TPSA, hydrogen bond donor count, hydrogen bond acceptor count, and rotatable bond count (Figure 5C). All blood constituents with structures in the DCABM-TCM are divided into 14 superclasses, given by a histogram graph on the “Compound classification” Search page of the blood constituent. Selecting a column returns blood constituents belonging to that superclass (Figure 5D).
Figure 5.
Search modes supported by the DCABM-TCM. (A) Users can search the database by prescriptions, herbs, blood constituents, targets, pathways, or diseases. For blood constituents, in addition to routine search by the name and PubChem CID, the DCABM-TCM also supports search by structural similarity (B), physicochemical property range (C), and compound classification (D).
Blood constituents of all prescriptions/herbs can be downloaded on the Download page. In addition, RESTful access to the DCABM-TCM core data is also supported, through which users can access a prescription/herb blood constituent and the corresponding detailed detection conditions by its Pinyin name and can also access the prescription and herb list to which a blood constituent belongs by its PubChem CID or name.
On the submission page, users can submit the constituents of a prescription or herb detected in the blood derived from their study or other publications to the DCABM-TCM by simply filling in the information in some required and optional fields. Periodically, after manual verification, we will integrate them into the DCABM-TCM.
Annotation Page
For every entity of the six types, including prescriptions, herbs, blood constituents, targets, pathways, and diseases, the DCABM-TCM provides a detailed annotation page presenting the related core and annotation information and the corresponding interactive analysis functions and analysis results.
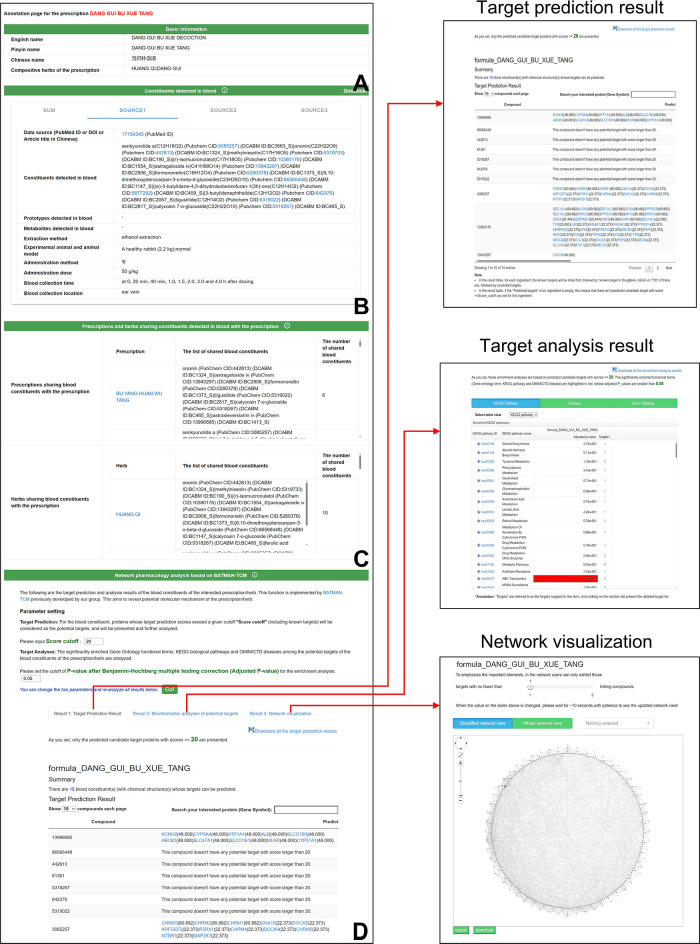
For a prescription/herb, we give its basic information on its detailed annotation page, including the Chinese/English/Pinyin/Latin name and compositive herbs for a prescription (Figure 6A). Subsequently, the constituents detected in the blood are provided, which can be downloaded using the “Download” button on the upper-right corner (Figure 6B). We provide the constituents detected in the blood (as well as the prototypes and metabolites if provided by the corresponding source publication) and their corresponding detailed detection conditions and source publications of the prescription/herb. Considering the potential difference of the blood constituents detected by different studies due to the differences in the detection conditions, we provide the blood constituent-related information from different source publications, respectively, which can be browsed by the label switch of “Source1”, “Source2”, and so on. The content of the label “Sum” is the sum of blood constituents from different sources. In the next column, other prescriptions and herbs in the DCABM-TCM that share blood constituents with the interested prescription/herb together with the shared blood constituents are listed (Figure 6C). In the DCABM-TCM, each record of the blood constituent information of an herb/prescription is supported by direct literature evidence from a certain wet experiment under a certain experimental condition. We do not directly infer the blood constituents of an herb as those of the prescription to which the herb belongs. The reason is that an herb’s blood constituents may not be detected in the blood when the prescription (the herb belongs to) is administrated due to administration dosage, different experimental conditions, or other reasons. We also do not directly infer the blood constituents of a prescription as those of its compositive herb. This is because we do not know which herb these blood constituents belong to, and some blood constituents of a prescription may not be blood constituents of any of its compositive herbs as they might be produced by the interactions between its compositive herbs, such as those produced in the decocting process of the prescription. We estimate whether a prescription/herb and another prescription/herb share blood constituents based on their respective blood constituent records (directly supported by a publication; only blood constituents with chemical structures are considered). Although an herb is a compositive herb of a prescription, we do not directly consider that this herb and this prescription share blood constituents. Finally, at the end of the annotation page, the network pharmacology analysis function and analysis results are presented (Figure 6D). This analysis is implemented based on the BATMAN-TCM, a bioinformatics analysis tool for the molecular mechanism of TCM previously developed by us.31 In this function, known and predicted targets of the blood constituents of the prescription/herb, the enriched GO functional terms, KEGG biological pathways, and CTD/OMIM diseases among the targets are analyzed. Moreover, the blood constituent–target–pathway/disease association network is visualized. The target prediction score cutoff and the P-value of the enrichment analysis after multiple testing corrections can be interactively changed, and then, the results are reanalyzed. All analysis results and the network graph/file can be downloaded. This function aims to reveal the potential molecular mechanism of the prescription/herb based on its blood constituents. We believe that compared with ordinary constituents (detected in vitro) of a prescription/herb, the results of the network pharmacology analysis based on its blood constituents are more reliable for revealing its potential molecular mechanism. See Supplementary information for more detailed network pharmacology analysis function tutorials.
Figure 6.
Prescription annotation page. (A) Basic information. (B) Constituents detected in the blood. This column gives the blood constituents of the prescription (including prototypes and metabolites) together with the detailed detection conditions. (C) Other prescriptions and herbs in the DCABM-TCM that share blood constituents with the prescription. (D) Network pharmacology analysis. Users can specify the target prediction score cutoff and P-value of enrichment analyses after multiple testing corrections. The analysis results include target prediction of blood constituents of the prescription, GO/pathway/disease enrichment analysis, and the blood constituent–target–pathway/disease association network visualization.
All blood constituents can be browsed and searched, but their annotation pages are divided into two types. One is for those with chemical structures (with/without PubChem CIDs), and the other is for the remaining ones without chemical structures. For the former, its annotation page gives basic information (including name, molecular formula, PubChem CID, CAS number, structure, and cross-references), physicochemical properties, compound structural classification, ADMET properties, the lists of prescriptions and herbs the blood constituent belongs to, and the network pharmacology analysis results for the blood constituent. For the latter, the annotation page is relatively simple, on which data fields only include compound name, chemical formula, the herbs and prescriptions to which the blood constituent belongs, and type (prototype/metabolite).
For a target/pathway/disease, the content of its annotation page is only divided into two sections (Figure 7). One gives the basic information of the entry, including name, ID, cross-references, GO function terms a target entry belongs to, pathways a target entry participates in, diseases a target entry is associated with, member genes a pathway entry contains, and disease entry-related genes (Figure 7A). The other gives the results for the analysis function of the candidate prescriptions, herbs, and blood constituents targeting the target/pathway/disease (Figure 7B). The candidate blood constituents of a target are given based on known and predicted drug–target associations provided by the BATMAN-TCM. The candidate blood constituents potentially targeting a pathway/disease are the significantly enriched blood constituents among the pathway member genes/disease-related genes. Furthermore, the candidate prescriptions/herbs potentially targeting a target/pathway/disease are significantly enriched ones among its candidate blood constituents (see Materials and Methods). Users can specify the target prediction score cutoff and P-value of the enrichment analyses after multiple testing corrections. All the above mentioned analysis results can be downloaded. This function aims to help the target/pathway/disease-based screening of candidate blood constituents, herbs, and prescriptions. See Supplementary information for more detailed tutorials of this function.
Figure 7.
Target annotation page. (A) Basic information. (B) Candidate blood constituents (i), prescriptions (ii), and herbs (iii) targeting the target gene. In this analysis, users can specify the target prediction score cutoff and the P-value of enrichment analyses after multiple testing corrections.
In addition, for user-friendliness, on each of the annotation pages, we provide various hyperlinks to external databases, such as DrugBank,29 ChEBI,41 KEGG,33 DGIdb,42 OMIM,34 and CTD;35 to internal pages; and to the Help documents in proper positions. Especially, recently developed/updated several databases are closely related to the contents of the DCABM-TCM. For example, DrugMAP provides interacting molecule information for >30,000 drugs/drug candidates;43 DRESIS provides drug resistance information;44 VARIDT mines various variability data of drug transporters;45 TTD provides comprehensive drug targets as well as various related information;46 NPCDR provides comprehensive information of natural product-based drug combinations;47 and INTEDE gives interactome of drug-metabolizing enzymes.48 In DCABM-TCM, we also provide hyperlinks to these databases, comprehensively describing the pharmaceutical/biological characteristics of the blood constituents/targets in DCABM-TCM.
Discussion
The vast majority of TCM constituents can produce pharmacological effects only after they reach the bloodstream. Compared with the ordinary constituents detected in vitro, which can be obtained from many existing TCM databases, the constituents of a TCM detected in the blood, which no previous database has recorded, are more likely to be the effective ones responsible for its pharmacological effects. Herein, we manually collected the most comprehensive experimentally detected blood constituents of TCM prescriptions and herbs so far, together with the corresponding detailed detection conditions, and we integrated these blood constituents’ associated annotations, such as physicochemical properties; ADMET properties; targets; and further associated GO terms, pathways, and diseases to establish the DCABM-TCM. In addition to data browsing, search, download, and submission, the DCABM-TCM was also designed to support two typical applications of these blood constituents: (1) potential molecular mechanism elucidation of a prescription or herb by the network pharmacology analysis function and (2) TCM-based drug discovery based on the function of the prioritization of candidate drugs (including blood constituents and further herbs and prescriptions the blood constituents belong to) potentially targeting a target/pathway/disease.
In addition to the blood constituents with chemical structures, DCABM-TCM has also recorded many blood constituents without chemical structures, and even some of them only have names/chemical formulas. This is mainly because the corresponding source publication has not provided the chemical structures/chemical formulas/names. For such blood constituents, the main principle of DCABM-TCM literature mining is to “record and present them as they are given in the source publication”, because we think that “it’s more informative than giving nothing”, and at least for future accurate identification (i.e., knowing the chemical structure) of blood constituents, the temporarily incomplete information about the blood constituent can provide reference for later researchers to verify their findings to some extent. As more related studies emerge, we will update the database, and the missing structures will be gradually refined.
In summary, the DCABM-TCM is designed to be a specialized database for collecting a completely new type of TCM data, aiming to provide a new dimension of TCM information. Its unique core data, together with related annotations and analysis functions around the core data, can benefit researchers interested in TCM modernization and a wider range of drug researchers. Compared with previous ordinary TCM constituents detected in vitro, the blood constituents are a great step forward not only for elucidating the TCM molecular mechanism and quality control of herbs but also for TCM-inspired modern drug discovery, which are more likely to be effective/bioactive and bioavailable.
Acknowledgments
We would like to thank Yun Tang and Chaofeng Lou from East China University of Science and Technology for their help in using admetSAR and Jian Wang, Dan Wang, and Jiangyong Gu for the fruitful discussion.
Glossary
Abbreviations
- ADMET
absorption, distribution, metabolism, excretion and toxicity
- CAS
Chemical Abstracts Service
- CID
compound identifier
- CNKI
China National Knowledge Infrastructure
- CTD
comparative toxicogenomics database
- DCABM-TCM
Database of Constituents Absorbed into the Blood and Metabolites of Traditional Chinese Medicine
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LC–MS/MS
liquid chromatography coupled with tandem mass spectrometry
- logP
the logarithm of the partition coefficient between n-octanol and water
- OMIM
Online Mendelian Inheritance in Man
- TCM
traditional Chinese medicine
- TPSA
topological polar surface area
Data Availability Statement
The database can be accessed at http://bionet.ncpsb.org.cn/dcabm-tcm/, and the web server is compatible with Chrome, Firefox, and Opera browsers for Windows and Safari, Chrome, Firefox, and Opera browsers for the Mac operating system.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jcim.3c00365.
Author Contributions
◆ X.L., J.L., B.F. and R.C. contributed equally to this work.
Author Contributions
Z.L. designed the database and wrote the manuscript. D.L. and F.X. provided guidance and revised the manuscript. B.F., J.L., R.L., L.X., L.Y., X.C., J.Z., Y.L., and B.Y. performed the data curation. X.L., R.C., J.J., H.C., H.L., S.G., F.G., J.G., and Y.Q. implemented the database.
This work was supported by the National Key Research and Development Program of China (2017YFC1700105, 2020YFE0202200, and 2021YFA1301603), National Natural Science Foundation of China (31601064 and 32088101), Beijing Nova Program (Z171100001117117), and State Key Laboratory of Proteomics of China (SKLPO202010).
The authors declare no competing financial interest.
Supplementary Material
References
- Cheung F. TCM: made in China. Nature 2011, 480, S82–S83. 10.1038/480S82a. [DOI] [PubMed] [Google Scholar]
- Wang L.; Zhou G. B.; Liu P.; Song J. H.; Liang Y.; Yan X. J.; Xu F.; Wang B. S.; Mao J. H.; Shen Z. X.; Chen S. J.; Chen Z. Dissection of mechanisms of Chinese medicinal formula Realgar-Indigo naturalis as an effective treatment for promyelocytic leukemia. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 4826–4831. 10.1073/pnas.0712365105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y.; Liu D.; Li L. PD-1/PD-L1 pathway: current researches in cancer. Am. J. Cancer Res. 2020, 10, 727–742. [PMC free article] [PubMed] [Google Scholar]
- Kim H. U.; Ryu J. Y.; Lee J. O.; Lee S. Y. A systems approach to traditional oriental medicine. Nat. Biotechnol. 2015, 33, 264–268. 10.1038/nbt.3167. [DOI] [PubMed] [Google Scholar]
- Wang X.Preface. In: Wang X.; Zhang A.; Sun H., Eds.; Serum Pharmacochemistry of Traditional Chinese Medicine; Academic Press: London, 2017. [Google Scholar]
- Li Y.; Xie Y.; He Y.; Hou W.; Liao M.; Liu C. X. Quality markers of traditional Chinese medicine: concept, progress, and perspective. Engineering 2019, 5, 888–894. 10.1016/j.eng.2019.01.015. [DOI] [Google Scholar]
- Xu H. Y.; Zhang Y. Q.; Liu Z. M.; Chen T.; Lv C. Y.; Tang S. H.; Zhang X. B.; Zhang W.; Li Z. Y.; Zhou R. R.; Yang H. J.; Wang X. J.; Huang L. Q. ETCM: an encyclopaedia of traditional Chinese medicine. Nucleic Acids Res. 2019, 47, D976–D982. 10.1093/nar/gky987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.; Zhang F.; Yang K.; Fang S.; Bu D.; Li H.; Sun L.; Hu H.; Gao K.; Wang W.; Zhou X.; Zhao Y.; Chen J. SymMap: an integrative database of traditional Chinese medicine enhanced by symptom mapping. Nucleic Acids Res. 2019, 47, D1110–D1117. 10.1093/nar/gky1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L.; Xie D.; Yu Y.; Liu H.; Shi Y.; Shi T.; Wen C. TCMID 2.0: a comprehensive resource for TCM. Nucleic Acids Res. 2018, 46, D1117–D1120. 10.1093/nar/gkx1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.; Zhou H.; Liu Y. B.; Wang J. F.; Li H.; Ung C. Y.; Han L. Y.; Cao Z. W.; Chen Y. Z. Database of traditional Chinese medicine and its application to studies of mechanism and to prescription validation. Br. J. Pharmacol. 2006, 149, 1092–1103. 10.1038/sj.bjp.0706945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. Y. TCM Database@Taiwan: the world’s largest traditional Chinese medicine database for drug screening in silico. PLoS One 2011, 6, e15939 10.1371/journal.pone.0015939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R. Z.; Yu S. J.; Bai H.; Ning K. TCM-Mesh: the database and analytical system for network pharmacology analysis for TCM preparations. Sci. Rep. 2017, 7, 2821. 10.1038/s41598-017-03039-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B.; Ma C.; Zhao X.; Hu Z.; Du T.; Xu X.; Wang Z.; Lin J. YaTCM: yet another traditional Chinese medicine database for drug discovery. Comput. Struct. Biotechnol. J. 2018, 16, 600–610. 10.1016/j.csbj.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ru J.; Li P.; Wang J.; Zhou W.; Li B.; Huang C.; Li P.; Guo Z.; Tao W.; Yang Y.; Xu X.; Li Y.; Wang Y.; Yang L. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J. Cheminf. 2014, 6, 13. 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultum L. E.; Woyessa A. M.; Lee D. ETM-DB: integrated Ethiopian traditional herbal medicine and phytochemicals database. BMC Complementary Altern. Med. 2019, 19, 212. 10.1186/s12906-019-2634-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. K.; Nam S.; Jang H.; Kim A.; Lee J. J. TM-MC: a database of medicinal materials and chemical compounds in NorthEast Asian traditional medicine. BMC Complementary Altern. Med. 2015, 15, 218. 10.1186/s12906-015-0758-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivek-Ananth R. P.; Mohanraj K.; Sahoo A. K.; Samal A. IMPPAT2.0:An Enhanced and Expanded Phytochemical Atlas of Indian Medicinal Plants. ACS Omega 2023, 8, 8827–8845. 10.1021/acsomega.3c00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H.; Park K. M.; Han D. J.; Bang N. Y.; Kim D. H.; Na H.; Lim S.; Kim T. B.; Kim D. G.; Kim H. J.; Chung Y.; Sung S. H.; Surh Y. J.; Kim S.; Han B. W. PharmDB-K: integrated bio-pharmacological network database for traditional Korean medicine. PLoS One 2015, 10, e0142624 10.1371/journal.pone.0142624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X.; Zhang P.; Wang Y.; Qin C.; Chen S.; He W.; Tao L.; Tan Y.; Gao D.; Wang B.; Chen Z.; Chen W.; Jiang Y. Y.; Chen Y. Z. CMAUP: a database of collective molecular activities of useful plants. Nucleic Acids Res. 2019, 47, D1118–D1127. 10.1093/nar/gky965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H.; Tang K.; Liu Q.; Sun Y.; Huang Q.; Zhu R.; Gao J.; Zhang D.; Huang C.; Cao Z. HIM-herbal ingredients in-vivo metabolism database. Aust. J. Chem. 2013, 5, 28. 10.1186/1758-2946-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D.; Yang L.; Zhang P.; LaBaer J.; Hermjakob H.; Li D.; Yu X. AAgAtlas 1.0: a human autoantigen database. Nucleic Acids Res. 2017, 45, D769–D776. 10.1093/nar/gkw946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.; Chen J.; Cheng T.; Gindulyte A.; He J.; He S.; Li Q.; Shoemaker B. A.; Thiessen P. A.; Yu B.; Zaslavsky L.; Zhang J.; Bolton E. E. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019, 47, D1102–D1109. 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matt S.PubChemPy documentation. https://pubchempy.readthedocs.io/en/latest/#. Accessed March 3, 2022, 10.1073/pnas.2203904119. [DOI]
- ChemDraw, a product of PerkinElmer. https://www.cambridgesoft.com/software/overview.aspx. Accessed March 3, 2022.
- Yap C. W. PaDEL-descriptor: an open source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 2011, 32, 1466–1474. 10.1002/jcc.21707. [DOI] [PubMed] [Google Scholar]
- Wang R.; Fu Y.; Lai L. A new atom-additive method for calculating partition coefficients. J. Chem. Inf. Comput. Sci. 1997, 37, 615–621. 10.1021/ci960169p. [DOI] [Google Scholar]
- Cheng T.; Zhao Y.; Li X.; Lin F.; Xu Y.; Zhang X.; Li Y.; Wang R.; Lai L. Computation of octanol-water partition coefficients by guiding an additive model with knowledge. J. Chem. Inf. Model. 2007, 47, 2140–2148. 10.1021/ci700257y. [DOI] [PubMed] [Google Scholar]
- Cheng F.; Li W.; Zhou Y.; Shen J.; Wu Z.; Liu G.; Lee P. W.; Tang Y. admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. J. Chem. Inf. Model. 2012, 52, 3099–3105. 10.1021/ci300367a. [DOI] [PubMed] [Google Scholar]
- Wishart D. S.; Feunang Y. D.; Guo A. C.; Lo E. J.; Marcu A.; Grant J. R.; Sajed T.; Johnson D.; Li C.; Sayeeda Z.; Assempour N.; Iynkkaran I.; Liu Y.; Maciejewski A.; Gale N.; Wilson A.; Chin L.; Cummings R.; Le D.; Pon A.; Knox C.; Wilson M. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djoumbou; Feunang Y.; Eisner R.; Knox C.; Chepelev L.; Hastings J.; Owen G.; Fahy E.; Steinbeck C.; Subramanian S.; Bolton E.; Greiner R.; Wishart D. S. Classyfire: automated chemical classification with a comprehensive, computable taxonomy. J. Cheminf. 2016, 8, 61. 10.1186/s13321-016-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.; Guo F.; Wang Y.; Li C.; Zhang X.; Li H.; Diao L.; Gu J.; Wang W.; Li D.; He F. BATMAN-TCM: a bioinformatics analysis tool for molecular mechANism of traditional Chinese medicine. Sci. Rep. 2016, 6, 21146. 10.1038/srep21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglott D.; Ostell J.; Pruitt K. D.; Tatusova T. Entrez gene: gene-centered information at NCBI. Nucleic Acids Res. 2011, 39, D52–D57. 10.1093/nar/gkq1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M.; Furumichi M.; Tanabe M.; Sato Y.; Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberger J. S.; Bocchini C. A.; Schiettecatte F.; Scott A. F.; Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015, 43, D789–D798. 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattingly C. J.; Rosenstein M. C.; Colby G. T.; Forrest Jr J. N.; Boyer J. L. The Comparative Toxicogenomics Database (CTD): a resource for comparative toxicological studies. J. Exp. Zool., A 2006, 305, 689–692. 10.1002/jez.a.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet E.; Lieberherr D.; Tognolli M.; Schneider M.; Bairoch A. UniProtKB/Swiss-Prot. Methods Mol. Biol. 2007, 406, 89–112. [DOI] [PubMed] [Google Scholar]
- Benjamini Y.; Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995, 57, 289–300. 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- O’Boyle N. M.; Banck M.; James C. A.; Morley C.; Vandermeersch T.; Hutchison G. R. Open Babel: an open chemical toolbox. J. Cheminf. 2011, 3, 33. 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitty A.; Zhong M.; Viarengo L.; Beglov D.; Hall D. R.; Vajda S. Quantifying the chameleonic properties of macrocycles and other high-molecular-weight drugs. Drug Discovery Today 2016, 21, 712–717. 10.1016/j.drudis.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JSDraw – A Javascript Framework for Cheminformatics and Bioinformatics. http://elncloud.com/jsdrawapp/jsdraw/. Accessed April 25, 2020, 10.12688/f1000research.27158.2.
- Degtyarenko K.; de Matos P.; Ennis M.; Hastings J.; Zbinden M.; McNaught A.; Alcántara R.; Darsow M.; Guedj M.; Ashburner M. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008, 36, D344–D350. 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotto K. C.; Wagner A. H.; Feng Y. Y.; Kiwala S.; Coffman A. C.; Spies G.; Wollam A.; Spies N. C.; Griffith O. L.; Griffith M. DGIdb 3.0: a redesign and expansion of the drug-gene interaction database. Nucleic Acids Res. 2018, 46, D1068–D1073. 10.1093/nar/gkx1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F.; Yin J.; Lu M.; Mou M.; Li Z.; Zeng Z.; Tan Y.; Wang S.; Chu X.; Dai H.; Hou T.; Zeng S.; Chen Y.; Zhu F. DrugMAP: molecular atlas and pharma-information of all drugs. Nucleic Acids Res. 2023, 51, D1288–D1299. 10.1093/nar/gkac813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X.; Zhang Y.; Li H.; Zhou Y.; Shi S.; Chen Z.; He X.; Zhang H.; Li F.; Yin J.; Mou M.; Wang Y.; Qiu Y.; Zhu F. DRESIS: the first comprehensive landscape of drug resistance information. Nucleic Acids Res. 2023, 51, D1263–D1275. 10.1093/nar/gkac812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu T.; Li F.; Zhang Y.; Yin J.; Qiu W.; Li X.; Liu X.; Xin W.; Wang C.; Yu L.; Gao J.; Zheng Q.; Zeng S.; Zhu F. VARIDT 2.0: structural variability of drug transporter. Nucleic Acids Res. 2022, 50, D1417–D1431. 10.1093/nar/gkab1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Zhang S.; Li F.; Zhou Y.; Zhang Y.; Wang Z.; Zhang R.; Zhu J.; Ren Y.; Tan Y.; Qin C.; Li Y.; Li X.; Chen Y.; Zhu F. Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 2020, 48, D1031–D1041. 10.1093/nar/gkz981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X.; Zhang Y.; Zhou Y.; Lian X.; Yan L.; Pan T.; Jin T.; Xie H.; Liang Z.; Qiu W.; Wang J.; Li Z.; Zhu F.; Sui X. NPCDR: natural product-based drug combination and its disease-specific molecular regulation. Nucleic Acids Res. 2022, 50, D1324–D1333. 10.1093/nar/gkab913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J.; Li F.; Zhou Y.; Mou M.; Lu Y.; Chen K.; Xue J.; Luo Y.; Fu J.; He X.; Gao J.; Zeng S.; Yu L.; Zhu F. INTEDE: interactome of drug-metabolizing enzymes. Nucleic Acids Res. 2021, 49, D1233–D1243. 10.1093/nar/gkaa755. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The database can be accessed at http://bionet.ncpsb.org.cn/dcabm-tcm/, and the web server is compatible with Chrome, Firefox, and Opera browsers for Windows and Safari, Chrome, Firefox, and Opera browsers for the Mac operating system.