Abstract
The retrovirus SL3 induces T-cell lymphomas in mice. The transcriptional enhancer in the long terminal repeat (LTR) of SL3 contains two 72-bp repeats. Each repeat contains a binding site for the transcription factor CBF (also called AML1). The CBF binding sites are called core elements. SAA is a mutant that is identical to SL3 except for the presence of a single-base-pair substitution in each of the two core elements. This mutation significantly attenuates viral lymphomagenicity. Most lymphomas that occur in SAA-infected mice contain proviruses with reversions or second-site suppressor mutations within the core element. We examined the selective pressures that might account for the predominance of the reversions and suppressor mutations in tumor proviruses by analyzing when proviruses with altered core sequences became abundant during the course of lymphomagenesis. Altered core sequences were easily detected in thymus DNAs by 4 to 6 weeks after SAA infection of mice, well before lymphomas were grossly evident. This result is consistent with the hypothesis that viruses with the core sequence alterations emerged because they replicated more effectively in mice than SAA. The number of 72-bp tandem, repeats in the viral LTR was found to vary, presumably as a consequence of reverse transcriptase slippage during polymerization. Proviruses with two repeats predominated in the thymuses of SAA- and SL3-infected mice before lymphomas developed, although LTRs with one or three repeats were also present. This suggested that two was the optimal number of 72-bp repeats for viral replication. However, in lymphomas, proviruses with three or four repeats usually predominated. This suggested that a late step in the process of lymphomagenesis led to the abundance of proviruses with additional repeats. We hypothesize that proviruses with additional 72-bp repeats endowed the cells containing them with a selective growth advantage.
Murine leukemia viruses (MuLVs) are retroviruses that can induce hematopoietic tumors in mice. Binding sites for the transcription factor CBF are crucial for lymphomagenicity of MuLVs (11, 19, 23). These binding sites, called core elements, are present in the long terminal repeats (LTRs) of MuLVs and other mammalian C-type retroviruses (9). Frequently, the cores lie within a region of 50 to 100 bp that is tandemly duplicated within the LTR.
Mutations within the core element strongly affect the pathogenicity of MuLVs. Mutation of both cores of Moloney MuLV increased the latency period before disease onset and altered the cell type specificity of the disease from thymic lymphoma to erythroid leukemia (23). SL3, a potent MuLV that induces T-cell lymphomas in mice, contains two 72-bp repeats in the LTR. Each repeat contains two different CBF binding sites. The core I site of SL3, referred to here as the core of this virus, was found to be significant for T-cell lymphomagenicity of the virus (10, 19). Mutation of core II by itself had little effect on lymphomagenicity, although mutation of both cores simultaneously had a greater effect on lymphomagenicity than mutation of core I alone (10). The core exhibited about a fivefold-higher binding affinity for CBF than a second CBF binding site in each 72-bp repeat known as core II (25). Mutations in the SL3 core element reduced transcription in T cells about fourfold in most T-cell lines (3, 17, 19, 24, 30). Mutations of core II by itself reduced transcription twofold or less in T cells (30). However, mutations of both the core and the core II elements reduced transcriptional activity more than the mutation of the core alone (30). Thus, the effects of the core mutations on viral lymphomagenicity paralleled the effects on transcription (10, 19).
The sequence of the SL3 core (TGTGGTTAA) differs from that of the related nonleukemogenic virus called Akv (TGTGGTCAA) by 1 bp (the difference is underlined). Akv is an endogenous ecotropic MuLV from AKR mice that is relatively weakly pathogenic (15). Although the Akv core bound CBF in electrophoretic mobility shift assays about twofold less efficiently than the SL3 core (29), a mutant of SL3 containing the Akv sequence in both enhancer cores had significantly reduced lymphomagenicity and transcriptional activity in T cells (17, 19). The mutant virus, referred to as SAA, was identical to SL3 except for the T-to-C change in the enhancer cores of the LTR in both 72-bp repeats. SAA exhibited an increased latency period prior to the appearance of lymphomas relative to SL3, and/or the incidence of the disease was decreased, depending on the mouse strain utilized (17, 19). Thus, the precise sequence of the SL3 core element was crucial for maximum pathogenicity of the virus.
Analysis of proviruses in SAA-induced tumors demonstrated that reversions were present in proviruses in approximately 70% of the tumors. In addition, about 20% of the mice had tumor proviruses that retained the T-to-C core mutation but had acquired a second mutation in the core (19). Viral constructs with cores called So (TGCGGTCAA) or T* (TGTGGTCTA) that contained the second-site mutations were engineered. The mutations in the So and T* cores were found to be second-site suppressor mutations, because viruses containing them were significantly more pathogenic than the SAA virus (17). The suppressor mutations also restored transcriptional activity of the viral LTR to levels comparable to that of SL3 in T cells. In addition, the mutations in the So and T* cores also restored CBF binding activity to SL3 levels (17). Thus, viruses with either reversions or suppressor mutations within the core were selected during the course of lymphomagenesis by SAA, presumably as a function of increased transcriptional activity in T cells.
In this study, experiments were designed to examine the selective pressures that account for the predominance of proviruses with reversions or second-site suppressors in lymphomas of SAA-infected mice. One possibility was that the reversions and suppressor mutations affect viral replication. Even a modest advantage in viral replication can be strongly selected if there is a sufficient number of rounds of viral replication (2, 5). Another possibility was that the ability of an inserted provirus to activate an adjacent proto-oncogene might be increased by the core changes, thereby leading to the preferential outgrowth of clones from cells containing proviruses with altered cores. We reasoned that if viral replication was a key selective pressure, then proviruses with altered core sequences would appear early on, prior to tumor outgrowth. Gross lymphomas do not appear until a few months after viral inoculation into newborn mice. If tumor cell proliferation was a selective pressure, then core mutations would not be selected until late in the disease process, concurrent with tumor outgrowth. Tissue samples were collected at various time points after infection of neonatal AKR mice with SAA and analyzed to see when reversions and suppressor mutations came to predominate.
MATERIALS AND METHODS
Tumorigenicity assays.
Newborn AKR/J mice (<1.5 days old) were injected intraperitoneally with 0.1 ml of SAA virus (103 XC PFU). Infectious SAA viral stock was prepared previously by Morrison et al. (19). The structure of SAA is summarized in Fig. 1. The SAA virus contained the complete SL3 genome except that the Akv core sequence was present in both enhancer repeats. SAA-infected mice were sacrificed at 2, 4, 6, 8, and 12 weeks after inoculation and necropsied. As a control, additional SAA-infected mice were not sacrificed until lymphomas developed. Gross pathological examination of lymphomatous mice always revealed an enlarged thymus, spleen, peripheral lymph nodes, mesenteric lymph nodes, and/or liver. Enlarged organs were stored frozen at −80°C until DNA was prepared from them. Southern blotting with a T-cell receptor β probe was used to test whether the tumors were of T-cell origin (1, 12).
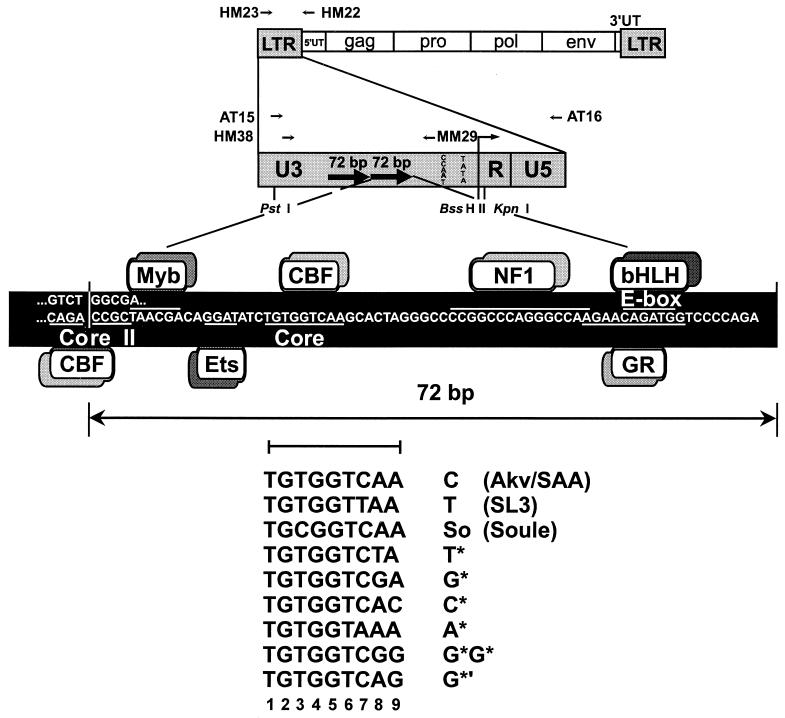
FIG. 1.
Structures of the SAA virus and its LTR. The top portion of the diagram shows the viral genome and the positions of primers used for PCR amplification and for sequencing. The bottom of the diagram represents one 72-bp repeat in the viral LTR enhancer, with transcription factor binding sites indicated. Sequences of core elements in proviruses from SAA-infected mice are depicted. The numbers used to designate each position within the core are shown below the core sequences. UT, untranslated region.
Analysis of viral enhancer sequences in infected cells by direct sequencing.
LTR DNAs were amplified from tissue DNA samples by using PCR primers HM38 (5′ AAGGCTTAGCCAGCTAACTGCAGTAACGCC 3′) at positions −466 to −436 and HM22 (5′ GATGCCGGCACACACACACACACTCTCCC 3′) at positions +272 to +244 relative to the transcriptional initiation site (Fig. 1). PCR was with a 30-cycle program of 1 min at 94°C, 1 min at 64°C, and 2 min at 72°C. PCR products were electrophoretically resolved on a 5% nondenaturing polyacrylamide gel (19). The individual bands differed by multiples of 72 bp. Each band was excised, and the DNA was isolated by using Qiaex II (Qiagen). The bands were reamplified by PCR under the same conditions with primer pair AT 15 (5′ TCGACGCGTCTGCAGTAACGCCATTTTGC 3′) and AT 16 (5′ AACCCCCGAGCAGGCCCGATCGATCA 3′) (positions −458 to −429 and +143 to +168, respectively, relative to the transcription initiation start site) (27). The resulting PCR products were isolated by using QIAquick (Qiagen) and sequenced directly by using primer MM29 (5′ TCATCTGGGGAACCTTGAGAC 3′) at positions −136 to −115 relative to the transcription initiation start site (Fig. 1).
Analysis of viral enhancer sequences in infected cells by plasmid subcloning.
Proviral LTR sequences in tissue DNA samples were PCR amplified with HM38 and HM22 as described above. The LTR sequences were subcloned into plasmid pCR 2.1 from the Invitrogen TA cloning kit. Random clones were selected for DNA sequencing with primer MM29 as described above.
RESULTS
Infection of mice with SAA.
Neonatal AKR/J mice were infected with SAA virus. SAA virus was identical to SL3 except for the 1-bp T-to-C change within both enhancer cores of both LTRs (Fig. 1). AKR/J mice were selected to observe the time course of appearance of mutations because 100% of SAA-inoculated mice of this strain developed tumors over a relatively synchronous period of 12 to 18 weeks postinoculation. Previous studies revealed that 80% of SAA-inoculated AKR/J mice contained reversions or second-site suppressor mutations (19). Groups of SAA-infected mice were sacrificed at 2, 4, 6, 8, and 12 weeks postinfection, and DNA was prepared from thymuses of individual animals. By examining the LTR enhancers of proviruses in DNA rather than enhancers in tissue RNA, we could be confident of not detecting alterations in viral sequences until viruses with the alterations had been selected by some mechanism to become a predominant fraction of all of the proviruses in that tissue. Additional mice were sacrificed when obvious lymphomas appeared, rather than at a particular time point, to serve as positive controls.
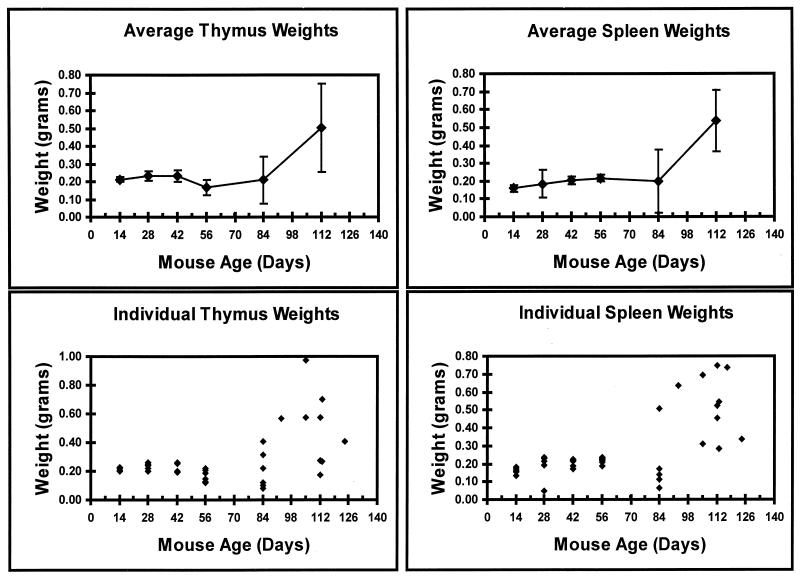
SAA specifically induces T-cell lymphomas (19). Tumors were characterized by gross enlargement of the thymus, spleen, and/or additional organs. Organ weights are a useful, simple marker for the presence of frank lymphomas. Thus, the thymus and spleen of each SAA-infected animal were collected and weighed (Fig. 2). Thymic weights increased normally with mouse development (4), reaching the maximum size at 6 weeks of age (Fig. 2). By 8 weeks, normal thymic involution (4) was observed, indicated by the decrease in thymic weight (Fig. 2). At 12 weeks postinfection, some animals displayed enlarged thymuses (Fig. 2). Mice kept to the end point of frank lymphoma usually contained a grossly enlarged thymus (Fig. 2). Spleen weights were also observed to increase with time (Fig. 2) through 12 weeks of age as seen in normal mouse development (13). A grossly enlarged spleen was initially detected around 12 weeks. As frank lymphomas appeared after 12 weeks, greatly enlarged spleens were evident in all animals (Fig. 2).
FIG. 2.
Weights of thymuses and spleens in SAA-infected mice. The top panels represent the mean weights over time. The bottom panels show the weights of the organs in individual animals at the specified times.
Variations in the number of 72-bp enhancer repeats.
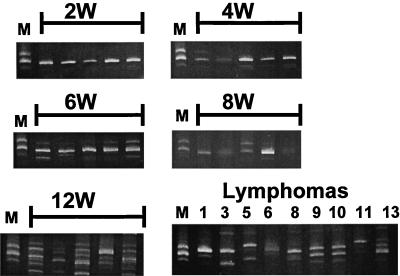
Proviruses with one, two, and three 72-bp enhancer repeats were commonly seen in lymphomas induced by SL3 or SAA virus (17, 19–21). Combinations of proviruses with different repeat numbers could frequently be found in tumors (19). The changes in repeat number were confirmed by Southern blotting and found not to be PCR artifacts (19). It was previously observed that virus generated by transfection of a molecular clone of a provirus with two enhancer repeats quickly became a mixture of isoforms upon replication in NIH 3T3 cells. These contained one, two, or three repeats (17, 19, 20). The isoform with two repeats was the predominant form during passage in NIH 3T3 fibroblasts (19). Even when a clone with three repeats was used for transfection, proviruses with two repeats quickly became predominant (17). This was interpreted to mean that genomes with two 72-bp repeats had a selective advantage in cultured fibroblasts, but due to reverse transcriptase jumps, viral genomes with altered number of repeats frequently form. To examine the enhancer sequences of the viral genomes in SAA-infected mice, DNA was extracted from thymic tissues from each animal at individual time points. LTR sequences were PCR amplified with a primer specific to the 5′ untranslated region of SAA and a primer specific for U3 portion of the LTR (Fig. 1). The specificity of the 5′ untranslated region primer ensured that variants of the SAA virus, but not endogenous retroviruses found in AKR/J mice, would be amplified. Due to the presence of proviruses with multiple 72-bp repeats, the PCR products were electrophoresed on high-resolution polyacrylamide gels to determine what isoforms were present at each time point (Fig. 3). At 2 weeks, only bands with two enhancer repeats were easily observed, although proviruses with one repeat could be detected (Fig. 3). The 4- and 6-week samples retained the two-repeat structure as the predominant isoform; however, bands corresponding to enhancers with one and three repeats were also evident (Fig. 3). Four of the five 8-week samples contained two 72-bp repeats as the predominant isoform. One sample gave an approximately equal mix of bands containing two or three 72-bp repeats (Fig. 3). At the 12-week time point and in the later lymphoma samples, multiple variants were always present, and the principal isoforms differed from sample to sample (Fig. 3). Bands ranging from one to five repeats were detected. Occasionally, bands corresponding to non-unit-length repeats were detected, as seen previously (Fig. 3) (6, 19). These were particularly pronounced at the 12-week time point. We conclude that proviruses with multiple numbers of enhancer repeats were present throughout the lives of the SAA-infected mice. However, proviruses with two repeats predominated until late in the disease process.
FIG. 3.
PCR-amplified LTR fragments from proviral DNA present in thymic tissue of SAA-inoculated mice. Amplified DNAs from mice at 2, 4, 6, 8, and 12 weeks and lymphomas after electrophoresis on a 5% polyacrylamide gel are shown. In the lymphomatous samples, the numbers correspond to numbers of the mice in Table 3. The marker (lanes M) is DNA amplified from a tumor induced by SL3 virus with the So core where proviruses were present that were known from sequencing studies to have one, two, and three 72-bp repeats.
Appearance of proviruses with core element mutations.
To determine when during the disease process proviruses with altered core sequences became predominant, the PCR-amplified viral enhancers were resolved on polyacrylamide gels and directly sequenced. Direct sequencing of PCR products was the same approach that was previously used to detect proviruses with altered enhancers in most SAA-induced lymphomas (19) and allowed comparison of the results between the two studies. Only the abundant bands were analyzed. When two abundant bands were present, as in the tumor samples, both were analyzed. The core sequences were determined for each 72-bp repeat, and a summary of the results is shown in Table 1. The five mice at the 2-week time point all retained the original sequence found in the SAA virus, i.e., two Akv cores (Table 1). In both the 4- and 6-week mouse groups, one of five mice had a reversion to the SL3 sequence present in at least one proviral enhancer core (Table 1). In the 8-week group, four of the five mice contained reversions in at least one repeat, while the core sequences from one animal contained Akv at both positions (Table 1). In the 12-week group, none of the animals retained the original SAA virus sequence. Three of the samples contained reversions, and two animals had additional mutations. One of these, referred to here as A*, TGTGGTAAA (Fig. 1), had a change (underlined) that matched the nucleotide at the corresponding position of Moloney MuLV (22). A novel core sequence, TGTGGTCAC, was also observed. This sequence is referred to as C* (Fig. 1) and was previously observed in MuLVs (9). Of the 11 lymphomas from the SAA-inoculated mice examined in this study, none of the samples yielded a sequence that retained both Akv core sequences as in the parental SAA virus (Table 1). Ten of the lymphoma samples contained at least one reverted core. Two of the mice had proviruses with the So core that contains a second suppressor mutation (Table 1). One mouse had provirus with the T* core that also contains a suppressor mutation (Table 1). Another sequence, TGTGGTCGA, referred to as G*, was also detected. This sequence was previously observed in a tumor provirus in another mouse (19). It was also previously found in the core of an endogenous xenotropic MuLV of NFS mice (14).
TABLE 1.
Core elements detected by direct sequencing of viral enhancer fragments that were PCR amplified from prelymphomatous tissues and lymphomas
| Wk | No. of micea | No. of mice with the following core sequenceb:
|
||||
|---|---|---|---|---|---|---|
| Cc | T | So | T* | Otherd | ||
| 2 | 5 | 5 | 0 | 0 | 0 | 0 |
| 4 | 5 | 4 | 1 | 0 | 0 | 0 |
| 6 | 5 | 4 | 1 | 0 | 0 | 0 |
| 8 | 5 | 1 | 4 | 0 | 0 | 0 |
| 12 | 5 | 0 | 3 | 0 | 0 | 2 |
| Le | 11 | 0 | 10 | 2 | 1 | 1 |
DNA was isolated from thymuses of the indicated number of SAA-infected mice at each point.
To be scored as positive, at least one repeat unit in a virus from an individual animal had the indicated core sequence.
C, all of the enhancer cores had the sequence of the Akv core.
Other core sequences detected were as follows: G*, TGTGGTCGA; C*, TGTGGTCAC; or A*, TGTGGTAAA.
L, Lymphomas. Lymphomas in several mice produced amplified viral products that contained a mixture of core elements. Thus, the total number of core elements detected was greater than the number of mice analyzed.
Sequencing of enhancer regions from individual proviruses in prelymphomatous tissues.
The direct sequencing of PCR products showed that by the time tumors occurred, proviruses with altered cores were detectable in all of the individual mice in this study. Revertants could be detected as a substantial fraction of the proviruses in the thymus as early in the lymphomatous process as 4 weeks postinfection. However, only 20% of the mice evidenced an altered core before the 8-week time point (Table 1). A more sensitive approach was used to detect proviruses with altered cores at early time points. PCR products were cloned into a plasmid vector, and individual clones were sequenced (Table 2). Sequencing of subcloned LTR PCR products also allowed us to tell what the core sequences were in each 72-bp repeat of individual proviruses. The six clones of proviruses from two mice analyzed for the 2-week time point all retained the original sequence present in the SAA virus, i.e., two Akv cores (Table 2). At 4 weeks postinfection, one animal contained a provirus with a reverted core and the other animal had the Akv sequence in each core. Two weeks later, at 6 weeks, reversions and second-site suppressor mutations were detected in proviruses in two of three mice examined (Table 2). One mouse (no. 4) contained proviruses with Akv, SL3, So, and/or G* cores (Table 2). The So and G* cores were found in a single provirus with two repeats (Table 2). Four of five mice at 8 weeks had a provirus with a reversion or a second-site mutation. Two novel core sequences were detected, TGTGGTCAG (G*′) and TGTGGTCGG (G*G*) at the 8-week time point. All of the 8-week mice had at least one clone that retained the original SAA viral sequence (Table 2). We conclude that proviruses with altered cores were abundant in mice by 4 to 6 weeks after the mice were infected. Moreover, most clones from mice at the 2-, 4-, 6-, and 8-week time points came from proviruses with two 72-bp repeats, although clones with one and three units were detected (Table 2).
TABLE 2.
Core sequences detected in individual clones of PCR-amplified enhancer fragments from prelymphomatous thymus tissues of SAA-infected mice
| Wk | Mouse no.a | No. of enhancers tested | Enhancer sequence(s)b
|
||
|---|---|---|---|---|---|
| 1 repeat | 2 repeats | 3 repeats | |||
| 2 | 1 | 3 | C,C (3) | ||
| 2 | 3 | C,C (3) | |||
| 4 | 3 | 4 | C,C (2); C,T (1) | C,C,C (1) | |
| 4 | 3 | C (1) | C,C (1) | C,C,C (1) | |
| 6 | 1 | 3 | T (1) | C,C (1); C,T (1) | |
| 4 | 4 | T,C (1); C,C (1); So, G* (1) | C,C,C (1) | ||
| 5 | 3 | C (2) | C,C (1) | ||
| 8 | 1 | 2 | C,C (1); G*G*,G*′ (1) | ||
| 3 | 10 | T (1) | C,C (4) | T,T,G* (1); T,T,C (4) | |
| 4 | 7 | T (2) | T,T (3); C,C (2) | ||
| 5 | 3 | T,T (2); C,C (1) | |||
Mouse numbers correspond to samples in the PCR assay shown left to right at each time point in Fig. 3.
The sequences of the enhancer repeat regions are represented such that the letter represents the core element detected in a single enhancer repeat unit. Core elements are abbreviated as in Fig. 1. When multiple repeats were present, the first letter represents the core in the repeat distal to the viral LTR promoter and the last letter represents the core in the promoter-proximal repeat. The numbers in parentheses indicate the number of individual clones with a particular structure.
Sequencing of enhancers of individual proviruses in tumors.
We also examined more thoroughly the variety of cores present in lymphomas by sequencing individual clones of PCR products (Table 3). Almost every provirus analyzed had one or more core sequences that were altered relative to the Akv core of the inoculated SAA virus. Mice 4 and 11 were the only ones that had proviruses with two Akv cores as in the parental SAA virus. Thus, the cells that gave rise to tumors likely acquired most of their proviruses after viruses with altered cores became abundant in the mice.
TABLE 3.
Core sequences detected in individual clones of PCR-amplified fragments from thymic lymphomas in SAA-infected mice
| Mouse no.a | No. of enhancers tested | Enhancer sequence(s)b
|
|||
|---|---|---|---|---|---|
| 1 repeat | 2 repeats | 3 repeats | ≥4 repeats | ||
| 1 | 4 | T,T (2); T,C (1) | T,T,C (1) | ||
| 3 | 3 | T (1) | T,T,C (2) | ||
| 4 | 3 | T,T (2); C,C (1) | |||
| 5 | 6 | So,So,So (3); T,T,C (2) | T,T,T,C (1) | ||
| 6 | 3 | T,T,C (1) | T,T,T,C (1); T,T,T,T,C (1) | ||
| 9 | 3 | T,T (1); T,C (2) | |||
| 10 | 3 | T,T,T,T (1); T,T,T,C (1); T,C,T,C (1) | |||
| 11 | 3 | T (1) | T,T (1) | T,T,T (1) | |
| 12 | 3 | T*,T*,T* (1); T*,T*,C (1) | C,T*,T,T (1) | ||
| 13 | 4 | C,C (1) | C*,C*,C (2) | C,C,C,C (1) | |
Mouse numbers correspond to samples in the PCR assay shown left to right at each time point in Fig. 3.
The sequences of the enhancer repeat regions are represented such that the letter represents the core element detected in a single enhancer repeat unit. Core elements are abbreviated as in Fig. 1. When multiple repeats were present, the first letter represents the core in the repeat distal to the viral LTR promoter and the last letter represents the core in the promoter-proximal repeat. The numbers in parentheses indicate the number of individual clones with a particular structure.
Most proviruses in tumors had three or four 72-bp repeats (Table 3). Proviruses with four or more repeats were found in mice 5, 6, 10, 12, and 13 (Table 3). Lymphomas in mice 1, 3, 4, 6, 9, and 10 contained proviruses with SL3 or Akv cores. SL3 cores predominated (Table 3). Second-site suppressor mutations (17) were found in mouse 5 (So) and mouse 12 (T*). Another novel altered core sequence, C* (TGTGGTCAC), was detected in mouse 13 (Table 3). This core was also detected in a 12-week animal by direct sequencing (Table 1). Thus, most proviruses in tumors had either reversions or one of the two confirmed second-site suppressor mutations, So or T*. All 11 lymphomas that were examined had at least one detectable provirus with an altered core (Table 3).
The sequencing of subcloned PCR products allowed an analysis of cores present in individual proviruses that was not possible in previous studies (17, 19), where the PCR products were directly sequenced. One interesting observation from these studies (Tables 2 and 3) is that two different core elements could be detected in different 72-bp repeats of a single provirus. Mouse 6 at 6 weeks, mouse 3 at 8 weeks, and mouse 12 in the lymphoma samples all had individual proviruses with two different core alterations. The last mouse is particularly interesting because it had four 72-bp repeats, including two with reversions to the SL3 sequence and a third with the confirmed T* suppressor mutation (17). These results show that core mutations can accumulate successively in an individual virus. Either two successive point mutations occurred in the core elements of individual viruses or recombination between viruses with two independent core mutations occurred.
Time course analysis of proviruses in SL3-infected mice.
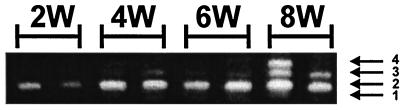
The structures of LTR enhancers of proviruses in mice infected with SL3 virus were also analyzed (Fig. 4) to determine if the process paralleled the results for SAA-infected mice. Lymphomas appear in SL3-infected NIH/Swiss mice beginning at about 9 weeks of age (8, 10, 17, 19–21). Newborn NIH/Swiss mice were inoculated with SL3 virus, and the animals were sacrificed at 2, 4, 6, and 8 weeks. DNA was extracted from the animals, LTR sequences were PCR amplified, and the products were resolved by electrophoresis (Fig. 4). As seen in the SAA-inoculated mice (Fig. 3), the proviruses detected at 2, 4, and 6 weeks predominantly contained two enhancer repeat structures (Fig. 4). At 8 weeks postinfection, just before grossly obvious lymphomas started to appear, proviruses with three 72-bp repeats were starting to become relatively abundant (Fig. 4). As previously shown, proviruses in lymphomas in SL3-infected mice contained either two or three repeats as the predominant form (19). Proviral DNAs from mice at 8 weeks and from SL3-induced lymphomas were cloned into plasmids and sequenced. Three clones from lymphomas in three different mice at each time point were sequenced (16). As previously observed (19), no mutations were detected in the cores (16).
FIG. 4.
PCR-amplified LTR fragments from proviral DNA present in thymic tissue of SL3-infected mice. Proviral DNAs from mice at 2, 4, 6, and 8 weeks resolved on a 5% polyacrylamide gel are shown. Arrows indicate the positions of the fragments with one, two, three, or four 72-bp repeats.
DISCUSSION
Infection of mice with SAA virus resulted in the appearance of viruses with altered core elements. The most common alteration observed was the reversion of the Akv core from TGTGGTCAA to TGTGGTTAA, the original core of SL3. Enhancer cores with the confirmed suppressor mutations So and T* (17) were also detected. The T* core sequence was not as prevalent in this study as in the earlier study (1 of 11 mice [9%] [Table 1] compared to 6 of 39 mice [16%] [19]). The combined data from the two studies produced the results that the So core was detected in 5 of 50 lymphomatous mice (10%) and T* was detected in 7 of 50 (14%) (Table 1). Core sequences called G* (TGTGGTCGA), C* (TGTGGTCAC), A* (TGTGGTAAA), G*G* (TGTGGTCGG), and G*′ (TGTGGTCAG) with other alterations (underlined) were also detected. Thus, mutations are strongly selected during the lymphomagenic process in mice following infection with the SAA virus. In a series of studies with a 3-bp mutation of the SL3 core, a second mechanism of suppression was also observed (6–8, 10). No alterations of the core were observed, presumably due to more effective inactivation of CBF binding by the 3-bp mutation than by the 1-bp mutation in SAA virus. Instead, deletions of the NF1 binding site in the 72-bp repeats were repeatedly detected (6). These deletions partially restored transcriptional activity of the viral LTR in T cells and increased viral lymphomagenicity (7). We also detected a few proviruses with NF1 site deletions in our present study (16), and these were likely suppressor mutations.
We previously showed that the mutations in the So and T* cores increased binding of CBF and increased transcriptional activity of the viral LTR (17). We hypothesize that the other core alterations had the same effect. However, since A*, G*G*, and G*′ were detected only once each in 50 mice, we do not know yet if these mutations are repeatedly selected. We interpret the repeated occurrence of the same mutation as evidence supporting the hypothesis that the core changes represent suppressor mutations within the core. The G* mutation was seen in two prelymphomatous mice (Table 2), one lymphomatous mouse in this study (Table 1), and one lymphomatous mouse in a previous study (19). The C* mutation was observed in a 12-week mouse and in a lymphomatous mouse. The independent appearance of both the G* and C* core mutations in multiple mice makes them likely candidates for being second-site suppressor mutations. Second-site mutations of the core were detected less frequently than reversions and were first detected later in the lymphomagenic process than reversions. Most likely, this indicates that second-site mutations do not restore the same level of viral replicative activity as the reversion does. Eight different core mutations were detected in all of our studies. Positions 3, 7, 8, and 9 (Fig. 1) within the core were the only sites that varied as a result of these mutations. Perhaps the reason that core positions 1, 2, 4, 5, and 6 (Fig. 1) were not detected is that mutations at those positions might not increase CBF binding or may even inhibit binding. Mutagenesis and DNA binding analyses indicated that substitutions at those positions did inhibit CBF binding (18, 25, 26). In addition to affecting CBF binding, the nucleotides within the core element may affect the ability of other transcription factors to interact with CBF and the viral LTR enhancer.
The reversions and known suppressor mutations increase CBF binding and transcriptional activity of the viral LTR relative to SAA/Akv. Presumably, the other second-site mutations in the core increase CBF binding and T-cell transcription, as was demonstrated for So and T* cores (17). We hypothesize that these effects allow the virus to replicate more effectively in the target tissues of mice. Our time course study supports this argument. Alterations of the core sequences were not detected at 2 weeks following SAA infection of mice. However, by 4 to 6 weeks postinoculation, changes in the core were easily detected. This may correlate with the types of cells that are the main targets of SL3 infection at different times after infection. One study found that in the first few weeks after infection, SL3 was found predominantly in dendritic cells and macrophages in the thymuses of infected mice (28). Infection of T cells was detected starting about 4 weeks after the inoculation of the virus into mice (28). Thus, the appearance of proviruses with altered cores starting at about 4 weeks of age might reflect the fact that this is the period when substantial infection of T cells begins. By 8 weeks, core mutations were detected in every mouse examined. Time points between 2 and 8 weeks represent the period prior to the appearance of obvious frank lymphomas in the thymus and spleen (Fig. 2). Since proviruses with altered cores were detected relatively early in the disease process, it appears that the primary selective pressure that results in their abundance is that the altered viruses outgrow the parental SAA virus.
Proviruses with variable number of 72-bp enhancer repeats could be detected throughout the course of disease, from 2 weeks postinfection to end-stage lymphomas. However, during the prelymphomatous phase, 8 weeks or less for SAA and 6 weeks or less for SL3, genomes with two 72-bp repeats predominated (Fig. 3 and 4). This was interpreted to mean that the presence of two 72-bp repeats is more efficient for viral replication. Perhaps the presence of three or more 72-bp repeats increases transcription but is disadvantageous for viral replication, possibly by interfering with viral RNA packaging. Although viral genomes with two repeats may replicate more effectively, slippage by reverse transcriptase may frequently regenerate genomes with altered numbers of repeats. Thus, these isoforms are continuously detected.
It was not until grossly obvious lymphomas appeared that proviruses with more than two repeats generally predominated (Fig. 3 and Table 3 versus Table 2). This suggests that the additional repeats offer a selective advantage late in the lymphomagenic process. Reversions and known suppressor mutations were the predominant core alterations in proviruses with three and four 72-bp repeats in lymphomas (Table 3). These mutations probably occurred before the repeat number expanded and resulted in a viral LTR enhancer with higher transcriptional activity. We hypothesize that the additional repeats are present in the subset of the proviruses that are integrated adjacent to cellular proto-oncogenes in the tumor cell genome. Indeed, in our earlier study (19) we found that five of six proviruses adjacent to c-myc or pim-1 had three 72-bp repeats. We also hypothesized that the extra repeat may result in a higher level of transcription of the oncogene than for a provirus with fewer repeats. This in turn may provide a growth advantage to clones of cells that have acquired a provirus with the additional repeats.
In summary, we hypothesize that both viral replication and clonal proliferation of tumor cells provide selective pressures that affect the structures of the LTR enhancer. Viruses with alterations in the core sequences are likely to have a selective replicative advantage over the parental SAA virus that allows them to become abundant relatively early in the lymphomagenic process. When a provirus with three or more repeats forms adjacent to a cellular proto-oncogene, the cell containing it may clonally outgrow a cell with a provirus containing fewer repeats integrated near a proto-oncogene. This results in the predominance of proviruses with more than two 72-bp repeats even though two repeats may be optimal for viral replication.
ACKNOWLEDGMENTS
We thank Su Mei, Eleanore Kim, Angel Nieves, Joseph Pantginis, and Lillie Lopez for help with these studies.
This work was supported by NIH grants CA44822 and CA57337 to J.L. and by American Cancer Society grant RPG-94-012-VM to L.S.L. M.J.M. was supported by NIH training grant GM07491. NIH Cancer Center grant CA13330 to the Albert Einstein College of Medicine supported core facilities for oligonucleotide synthesis and DNA sequencing.
REFERENCES
- 1.Athas G, Choi B, Prabhu S, Lobelle-Rich P, Levy L S. Genetic determinants of feline leukemia virus-induced multicentric lymphomas. Virology. 1995;214:431–438. doi: 10.1006/viro.1995.0053. [DOI] [PubMed] [Google Scholar]
- 2.Batschelet E, Domingo E, Weissmann C. The proportion of revertant and mutant phage in a growing population, as a function of mutation and growth rate. Gene. 1976;1:27–32. doi: 10.1016/0378-1119(76)90004-4. [DOI] [PubMed] [Google Scholar]
- 3.Boral A L, Okenquist S A, Lenz J. Identification of the SL3-3 virus enhancer core as a T-lymphoma cell-specific element. J Virol. 1989;63:76–84. doi: 10.1128/jvi.63.1.76-84.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clarke A G, MacLennan K A. The many facets of thymic involution. Immunol Today. 1986;7:204–205. doi: 10.1016/0167-5699(86)90105-2. [DOI] [PubMed] [Google Scholar]
- 5.Coffin J M. Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. J Gen Virol. 1979;42:1–26. doi: 10.1099/0022-1317-42-1-1. [DOI] [PubMed] [Google Scholar]
- 6.Ethelberg S, Hallberg B, Lovmand J, Luz A, Grundström T, Pedersen F S. Second-site proviral enhancer alterations in lymphomas induced by enhancer mutants of SL3-3 murine leukemia virus: negative effect of nuclear factor 1 binding site. J Virol. 1997;71:1196–1206. doi: 10.1128/jvi.71.2.1196-1206.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ethelberg S, Lovmand J, Schmidt J, Luz A, Pedersen F S. Increased lymphomagenicity and restored disease specificity of AML 1 site (core) mutant SL3-3 murine leukemia virus by a second-site enhancer varient envolved in vivo. J Virol. 1997;71:7273–7280. doi: 10.1128/jvi.71.10.7273-7280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ethelberg S, Sorensen A B, Schmidt J, Luz A, Pedersen F S. An SL3-3 murine leukemia virus enhancer variant more pathogenic than the wild type obtained by assisted molecular evolution in vivo. J Virol. 1997;71:9796–9799. doi: 10.1128/jvi.71.12.9796-9799.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Golemis E A, Speck N A, Hopkins N. Alignment of U3 sequences of mammalian type C viruses: identification of highly conserved motifs and implications for enhancer design. J Virol. 1990;64:534–542. doi: 10.1128/jvi.64.2.534-542.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hallberg B, Schmidt J, Luz A, Pedersen F S, Grundström T. SL3-3 enhancer factor 1 transcriptional activators are required for tumor formation by SL3-3 murine leukemia virus. J Virol. 1991;65:4177–4181. doi: 10.1128/jvi.65.8.4177-4181.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hallberg B, Thornell A, Holm M, Grundström T. SEF1 binding is important for T cell specific enhancers of genes for T cell receptor-CD3 subunits. Nucleic Acids Res. 1992;20:6495–6499. doi: 10.1093/nar/20.24.6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hedrick S M, Cohen D I, Nielsen E A, Davis M M. Isolation of cDNA clones encoding T cell-specific membrane-associated proteins. Nature. 1984;308:149–153. doi: 10.1038/308149a0. [DOI] [PubMed] [Google Scholar]
- 13.Hummel K P, Richardson F L, Fekete E. Anatomy. In: Green E L, editor. Biology of the laboratory mouse. New York, N.Y: McGraw-Hill; 1966. pp. 247–307. [Google Scholar]
- 14.Khan A S, Martin A M. Endogenous murine leukemia proviral long terminal repeats contain a unique 190-base-pair insert. Proc Natl Acad Sci USA. 1983;80:2699–2703. doi: 10.1073/pnas.80.9.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lovmand J, Sorensen A B, Schmidt J, Ostrowski M C, Luz A, Pedersen F S. B-cell lymphoma induction by Akv murine leukemia viruses harboring one or both copies of the tandem repeat in the U3 enhancer. J Virol. 1998;72:5745–5756. doi: 10.1128/jvi.72.7.5745-5756.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martiney, M. J., and J. Lenz. 1999. Unpublished results.
- 17.Martiney M J, Levy L, Lenz J. Suppressor mutations within the core binding factor (CBF/AML1) binding site of a T-cell lymphomagenic retrovirus. J Virol. 1999;73:2143–2152. doi: 10.1128/jvi.73.3.2143-2152.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melnikova I N, Crute B E, Wang S, Speck N A. Sequence specificity of the core-binding factor. J Virol. 1993;67:2408–2411. doi: 10.1128/jvi.67.4.2408-2411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morrison H L, Soni B, Lenz J. Long terminal repeat enhancer core sequences in proviruses adjacent to c-myc in T-cell lymphomas induced by a murine retrovirus. J Virol. 1995;69:446–455. doi: 10.1128/jvi.69.1.446-455.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nieves A, Levy L S, Lenz J. Importance of a c-Myb binding site for lymphomagenesis by the retrovirus SL3-3. J Virol. 1997;71:1213–1219. doi: 10.1128/jvi.71.2.1213-1219.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pantginis J, Beaty R M, Levy L S, Lenz J. The feline leukemia virus long terminal repeat contains a potent determinant of T-cell lymphomagenicity. J Virol. 1997;71:9786–9791. doi: 10.1128/jvi.71.12.9786-9791.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shinnick T M, Lerner R A, Sutcliffe J G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981;293:543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- 23.Speck N A, Renjifo B, Golemis E, Fredrickson T, Hartley J, Hopkins N. Mutation of the core or adjacent LVb elements of the Moloney murine leukemia virus enhancer alters disease specificity. Genes Dev. 1990;4:233–242. doi: 10.1101/gad.4.2.233. [DOI] [PubMed] [Google Scholar]
- 24.Thornell A, Hallberg B, Grundström T. Differential protein binding in lymphocytes to a sequence in the enhancer of the mouse retrovirus SL3-3. Mol Cell Biol. 1988;8:1625–1637. doi: 10.1128/mcb.8.4.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thornell A, Hallberg B, Grundstöm T. Binding of SL3-3 enhancer factor 1 transcriptional activators to viral and chromosomal enhancer sequences. J Virol. 1991;65:42–50. doi: 10.1128/jvi.65.1.42-50.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thornell A, Holm M, Grundström T. Purification of SEF1 proteins binding to transcriptional enhancer elements active in T lymphocytes. J Biol Chem. 1993;268:21946–21954. [PubMed] [Google Scholar]
- 27.Trubetskoy A M, Okenquist S A, Lenz J. R region sequences in the long terminal repeat of a murine retrovirus specifically increase expression of unspliced RNAs. J Virol. 1999;73:3477–3483. doi: 10.1128/jvi.73.4.3477-3483.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uittenbogaart C, Law W, Leenen P, Bristol G, van Ewijk W, Hays E. Thymic dendritic cells are primary targets for the oncogenic virus SL3-3. J Virol. 1998;72:10118–10125. doi: 10.1128/jvi.72.12.10118-10125.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zaiman A L, Lewis A F, Crute B E, Speck N A, Lenz J. Transcriptional activity of core binding factor α (AML 1) and β subunit on murine leukemia virus enhancer cores. J Virol. 1995;69:2898–2906. doi: 10.1128/jvi.69.5.2898-2906.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaiman A L, Nieves A, Lenz J. CBF, Myb, and Ets binding sites are important for activity of the core I element of the murine retrovirus SL3-3 in T lymphocytes. J Virol. 1998;72:3129–3137. doi: 10.1128/jvi.72.4.3129-3137.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]