Abstract
Small RNAs (sRNAs) are a class of short, non-coding regulatory RNAs that have emerged as critical components of defense regulatory networks across plant kingdoms. Many sRNA-based technologies, such as host-induced gene silencing (HIGS), spray-induced gene silencing (SIGS), virus-induced gene silencing (VIGS), artificial microRNA (amiRNA) and synthetic trans-acting siRNA (syn-tasiRNA)-mediated RNA interference (RNAi), have been developed as disease control strategies in both monocot and dicot plants, particularly in crops. This review aims to highlight our current understanding of the roles of sRNAs including miRNAs, heterochromatic siRNAs (hc-siRNAs), phased, secondary siRNAs (phasiRNAs) and natural antisense siRNAs (nat-siRNAs) in disease resistance, and sRNAs-mediated trade-offs between defense and growth in crops. In particular, we focus on the diverse functions of sRNAs in defense responses to bacterial and fungal pathogens, oomycete and virus in crops. Further, we highlight the application of sRNA-based technologies in protecting crops from pathogens. Further research perspectives are proposed to develop new sRNAs-based efficient strategies to breed non-genetically modified (GMO), disease-tolerant crops for sustainable agriculture.
Keywords: Small RNAs, HIGS, Crop diseases, RNAi, RNAi-based technology
Introduction
In nature, plants constantly face diverse biotic stresses, including bacteria, fungi, oomycetes, nematodes and viruses. Pathogen infection causes approximately 30% of global crop losses annually worldwide. Therefore, disease control is vital for assuring food security worldwide. During long coevolution with pathogens, plants have armed with various defense tools to prevent pathogens, which constitute a two-tiered immune machinery to detect and prevent pathogen invasion. The first layer is pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) governed by cell surface pattern recognition receptors (PRRs). In order to circumvent PTI, pathogens evolve effector proteins to suppress host PTI response, which is known as effector triggered susceptibility (ETS). As a counter-defense to prevent further infection, plants have then evolved highly polymorphic nucleotide-binding site (NBS) and leucine-rich repeat (LRR) domain-containing (NLR) resistance (R) proteins that directly or indirectly recognize pathogen effectors and form homo- or hetero-NLR complexes or resistosomes to trigger effector-triggered immunity (ETI) (Alves et al. 2014; Cui et al. 2015; Dangl et al. 2013; Ma et al. 2020; Spoel and Dong 2012; Wang et al. 2019a, b; Zhou and Zhang 2020). The second layer of immunity is more robust than PTI, which usually confers high resistance against pathogens. Therefore, NLR immune receptors are the major breeding targets for disease resistance in crops (Li et al. 2020a; Deng et al. 2020). PTI and ETI are interconnected and play synergistic roles to induce a set of downstream defense responses such as the generation of reactive oxygen species (ROS) and global transcriptional reprogramming for defense (Ngou et al. 2021; Yuan et al. 2021).
Small RNAs (sRNAs) are a class of 18–30 nt, non-coding RNAs, which play vital roles in regulating gene expression and maintaining the genome stability. Based on the different biogenesis pathway and the divergent modes of action, plant sRNAs can be cataloged into two major classes, microRNAs (miRNAs) and small interfering RNAs (siRNAs). MiRNAs and siRNAs are generated from miRNA genes (MIRs) and double-stranded RNAs (dsRNAs) by the cleavage activity of Dicer-like (DCL) proteins, respectively (Borges and Martienssen 2015; Chen 2009; Ghildiyal and Zamore 2009). Plant primary miRNAs (pri-miRNAs) are transcribed from MIRs by RNA polymerase II (Pol II) and trimmed into precursor miRNAs (pre-miRNAs) by DCL1, and pre-miRNAs are further processed by DCL1 to generate mature miRNAs (Kurihara and Watanabe 2004). Mature miRNAs, after stabilized by HEN1, a methyltransferase that catalyzes 2′-O-methylation, are loaded onto ARGONAUTE (AGO) proteins and other components to form the RNA-induced silencing complex (RISC) to silence targets by mRNAs cleavage or translational inhibition at post-transcriptional level, or directing DNA methylation that represses transcription at transcriptional level (Padmanabhan et al. 2009; Song et al. 2019; Voinnet 2009; Wu et al. 2010). siRNAs in plants are generated by DCL2-4 from diverse endogenous and exogenous dsRNAs precursors. An expanding world of plant siRNAs has been discovered, such as heterochromatic siRNAs (hc-siRNAs), phased, secondary siRNAs (phasiRNAs) and natural antisense siRNAs (nat-siRNAs) (Baulcombe 2004; Bologna and Voinnet 2014; Borges and Martienssen 2015; Castel and Martienssen 2013; Fei et al. 2013; Ghildiyal and Zamore 2009). Hc-siRNAs are derived from single-stranded RNA (ssRNA) transcribed from transposable elements and other repeat regions by Pol IV. The ssRNA transcripts are converted into dsRNAs by RNA-dependent RNA polymerase 2 (RDR2), and hc-siRNAs are subsequently produced by DCL3-mediated processing (Li et al. 2006; Pontes et al. 2006). The hc-siRNAs can direct DNA methylation in an AGO4-dependent pathway, which is known as RNA-directed DNA methylation (RdDM) (Chan et al. 2004). phasiRNA biogenesis requires the activity of miRNA-containing RISC, which cleaves the transcripts derived from phasiRNA-producing loci (PHAS loci). After cleavage, the transcripts are converted to dsRNAs, which produce 21- or 24-nt phasiRNAs by the activity of DCLs (Fei et al. 2013). Trans-acting siRNAs (tasiRNAs) are the first reported phasiRNAs in plants. A noncoding transcript originated from tasiRNA locus (namely TAS genes) that is converted into dsRNAs by RDR6 and then dsRNAs are cleaved into 21-nt siRNAs by DCL4 (Gasciolli et al. 2005; Peragine et al. 2004). phasiRNAs can be loaded onto AGOs and exert their repressive roles to their targets in trans (Fei et al. 2013). Nat-siRNAs are produced by the activity of DCL1/2, RDR6 and HEN1 from dsRNA precursors, which originate from bidirectional transcription of two partially overlapping genes (cis-natsiRNAs) or highly complementary transcripts derived from different loci in the genome (trans-natsiRNAs) (Katiyar-Agarwal et al. 2006). These siRNAs, like miRNAs, also play essential regulatory roles in plant development and abiotic/biotic stress by modulating target gene expression either transcriptionally through RdDM or post-transcriptionally by cleaving target mRNAs and/or repressing gene translation (Bologna and Voinnet 2014; Borges and Martienssen 2015; Castel and Martienssen 2013; Ghildiyal and Zamore 2009). Therefore, sRNAs actively regulate gene and protein expression, thereby imposing important impacts on various physiological processes.
In the following section, we will summarize recent findings and current progresses on sRNA involvement and roles in crop disease resistance, with aspects of sRNAs in crop-fungi, crop-oomycetes, crop-bacterial, and crop-virus interactions. We then propose research perspectives in sRNAs-based technologies in crop disease control.
Overview of sRNAs involved in PTI and ETI
Accumulative evidences have uncovered important roles of miRNAs in regulating immune responses in plants (Fei et al. 2016; Huang et al. 2019; Katiyar-Agarwal and Jin 2010; Padmanabhan et al. 2009; Ruiz-Ferrer and Voinnet 2009; Wang & Galili 2019; Wang et al. 2019a, b; Weiberg et al. 2014) (Fig. 1a). The first miRNA reported to modulate plant immunity is Arabidopsis miR393. miR393 is induced by the PAMP peptide flg22 and guides the cleavage of the mRNAs coding the F-box auxin receptors TIR1, AFB2, and AFB3 to attenuate the auxin signaling pathway, which is required for PTI response (Navarro et al. 2006). In addition to miR393, miR167 and miR160, induced by Pseudomonas syringae pv. tomato DC3000 hrcC that triggers PTI but not ETI in Arabidopsis, also target auxin-response factors (ARFs) to regulate auxin signaling (Zhang et al. 2011). Upon flg22 treatment, miR160a, miR398b and miR773 regulated callose deposition (Li et al. 2010).
Fig. 1.
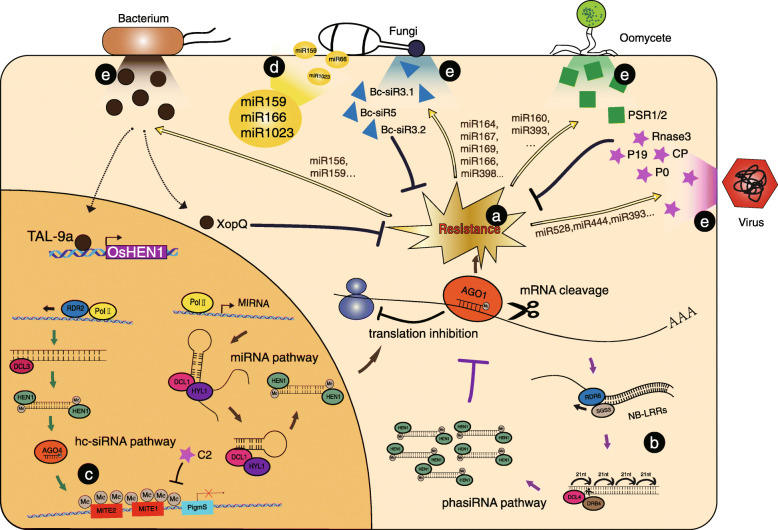
Small RNAs regulate plant defense against-pathogen. In response to pathogen attacks, plants accurately fine-tune the expression of endogenous gene through sRNAs-trigged mRNA cleavage, translational repression as well as DNA methylation. a sRNAs regulate plant resistance to various pathogens through mRNA cleavage or translational repression. miR164 (Wang et al. 2018), miR167 (Zhao et al. 2020) and miR169 (Li et al. 2017) which directly target transcription factors play negative roles in plant response to M. oryzae while miR166 (Salvador-Guirao et al. 2018) and miR398 (Li et al. 2019a) are positive regulators. miR160 (Natarajan et al. 2018) and miR393 (Wong et al. 2014) regulate the interactions between crop and oomycetes. Some other small RNAs such as miR156 (Liu et al. 2019) and miR159 (Zhao et al. 2015b) modulate the disease resistance against bacterial pathogens. Upon virus infection, some miRNAs target essential components of RNAi to regulate plant immunity, like miR168 (Du et al. 2011). b Hc-siRNA-mediated DNA methylation at the MITE region of PigmS promoter represses PigmS expression (Deng et al. 2017). c Plant small RNAs not only function in host cells, but also move into invasive enemies. For example, miR159, miR166 (Zhang et al. 2016c) and miR1023 (Jiao and Peng 2018) trigger the silencing of fungal virulence genes. d PhasiRNA pathway fine-tunes the expression level of R gene in the absence of pathogen (González et al. 2015). e During the long-term arm-race between crops and pathogens, pathogens can secrete specific proteins or small RNAs into plant cells to enhance plant susceptibility to ensure their own virulence. For instance, B. cinerea delivers Bc-siRNAs to plant cell to hijack host RNAi pathway (Weiberg et al. 2013). TAL-effectors such as Tal9a from genus Xanthomonas can bind to host specific promoter motifs and activate host genes expression (Moscou and Bogdanove 2009). A non-TAL effector XopQ can up-regulates host sRNAmiR1876 through an unknown mechanism (Jiang et al. 2020). Some proteins such as PSR1 and PSR2 (Xiong et al. 2014) from P. sojae and P19 (Silhavy et al. 2002), P0 (Li et al. 2019b) , CP (Karran and Sanfaçon 2014), RNase 3 (Cuellar et al. 2009; Kreuze et al. 2005), C2 (Yang et al. 2013) from different kinds of viruses can disrupt host immune response by suppressing RNA silencing pathways in plants
More importantly, miRNAs fine-tune the transcription of NLR immune receptors to efficiently balance defense and growth in plants (Fei et al. 2016; Huang et al. 2019; Wang et al. 2019a, b), since at the absence of pathogens, high expression of NLR genes may result in the fitness cost in plants (Purrington 2000; Richard et al. 2018; Tian et al. 2019). miRNAs such as miR482/2118 family (Fei et al. 2016), Bra-miR1885 (He et al. 2008), id47, id97, id113 (Carra et al. 2009), Pta-miR946 (Lu et al. 2007), and Md-miRLn11 (Ma et al. 2014) mediate cleavage of NLR transcripts in Arabidopsis, Brassica rapa, Vitis vinifera, Pinus taeda, Citrus trifoliata and woody plants, respectively.
Besides miRNAs, siRNAs also function as key regulators of plant immunity (Fig. 1a). For example, nat-siRNAATGB2, which originate from the overlapping region of transcripts of a small GTP-binding protein ATGB2 and a pentatricopeptide repeat-like (PPRL) gene, is induced upon infection by P. syringae carrying effector avrRpt2, and silences PPRL that negatively regulates RPS2-mediated disease resistance (Katiyar-Agarwal et al. 2006). miR482/miR2118-mediated phasiRNAs can also fine-tune the expression of the cognate NLR genes in divergent plant species such as Arabidopsis (Borrelli et al. 2018), tomato (Solanum lycopersicum) (Canto-Pastor et al. 2019), Medicago (Medicago truncatula) (Zhai et al. 2011) and soybean (Glycine max) (Zhao et al. 2015a). In the absence of pathogens, the miR482/2118-phasiRNA cascade favors growth by suppressing NLR expression (Fig. 1b). Upon pathogen infection, the phasiRNAs-mediated suppression of NLRs is released to ensure ETI (González et al. 2015). In dcl4 and ago1 mutant plants, the NLR gene SNC1 is constitutively activated and multiple R genes in RPP5 locus are upregulated (Yi and Richards 2007).
sRNAs modulate resistance against fungal pathogens in crops
Fungal pathogens are a major group of plant invaders that cause many notorious plant diseases. Among them, Magnaporthe oryzae (M. oryzae) causes rice (Oryza sativa) blast and is the most destructive fungal pathogen worldwide (Dean et al. 2012; Zhang et al. 2016b). Several miRNAs have been reported to negatively affect blast resistance. These miRNAs usually mediate transcriptional reprogramming to regulate immune responses through directly targeting transcription factors (Fig. 1a). For example, Osa-miR164a targets OsNAC60 and negatively regulates its activity, which suppresses rice blast resistance. Moreover, the Osa-miR164a/NAC60 module plays a conserved regulatory role in plant resistance to rice sheath blight, tomato late blight, and soybean root and stem rot diseases (Wang et al. 2018). Osa-miR169 suppresses transcription factor NF-YAs and enhances rice susceptibility to M. oryzae (Li et al. 2017). Similarly, Osa-miR167d also negatively regulates rice immunity against M. oryzae (Zhao et al. 2020). When infected by M. oryzae, rice plants accumulate higher levels of Osa-miR319, which guides cleavage of OsTCP21 mRNA. The Osa-miR319-mediated suppression of OsTCP21 results in stronger disease symptoms by reducing cellular ROS and jasmonic acid (JA) levels (Zhang et al. 2018). miR396 is a highly conserved miRNA family targeting Growth Regulating Factor (OsGRF) genes. The miR396-OsGRF module plays a vital role in balancing growth and immunity against the blast fungus. Overexpressing of a miR396-resistant version of OsGRF or blocking miR396 expression not only enhances rice resistance to M. oryzae but also improves yield traits (Chandran et al. 2018). A target mimic of miR156fhl-3p (MIM156-3p) indirectly increases the expression of squamosa promoter-binding-like transcription factor OsSPL14 to enhance rice blast disease resistance by reducing the abundance of miR156-5p, which fine-tunes the tradeoff between blast disease resistance and yield (Zhang et al. 2020). Most recently, Osa-miR439 is reported to negatively regulate rice blast resistance through inhibiting the induction of defense-related genes and accumulation of H2O2 (Lu et al. 2021).Osa-miR168 can target OsAGO1 and suppression of Osa-miR168 by a target mimic (MIM168) improves yield, flowering time and immunity to M. oryzae, which suggest the potential application of miRNAs in coordinating plant immunity with growth and development (Wang et al. 2021). Interestingly, some miRNAs play positive roles in rice blast resistance. Overexpression of miR160a in transgenic rice plants enhances resistance to the blast fungus. Overexpressing MIR166k-h modulates EIN2 expression and therefore enhances rice resistance to M. oryzae and Fusarium fujikuroi (Salvador-Guirao et al. 2018). Osa-miR398b suppresses the expression of several targets, Cu/Zn-Superoxidase Dismutase (CSD1/2), Superoxide Dismutase X (SODX), and Copper Chaperone for Superoxide Dismutase (CCSD). csd1/2 and sodx mutants display increased H2O2 accumulation and enhanced blast resistance, while ccsd mutants show enhanced blast susceptibility with lower levels of H2O2 (Li et al. 2019a). A novel rice miRNA, Osa-miR7695, positively regulates resistance to M. oryzae by targeting Natural resistance-associated macrophage protein 6 (OsNramp6), a Fe transporter. Interestingly, Osa-miR7695 appears to be subjected to subspecies-specific selection. Overexpression of Osa-miR162a induces defense genes expression and the accumulation of H2O2, and increases blast resistance in transgenic rice (Li et al. 2020b).
Importantly, the RdDM pathway plays an important role in regulating immune responses against the blast fungus (Deng et al. 2017) (Fig. 1c). In rice, Pigm locus fine-tunes blast resistance and the trade-off between defense and yield. Among NLRs of the Pigm cluster, PigmR confers broad-spectrum resistance, whereas PigmS, whose expression is modulated by RdDM pathway, competitively suppresses PigmR homodimerization to suppress resistance. RNAi-mediated silencing of the RdDM pathway genes, OsRDR2, OsDCL3a, and OsAGO4a, in Pigm background reduces hc-siRNA accumulation and methylation levels of CHH sites at the MITE (Miniature Inverted-Repeat Transposable Elements) region of PigmS promoter and subsequently facilitates PigmS expression level (Deng et al. 2017). Blocking the expression of OsAGO4a in Pigm background increases blast susceptibility, consistent with the attenuation of PigmR-mediated resistance by PigmS (Deng et al. 2017). This DNA methylation-mediated regulation of the PigmR-PigmS NLR pair might provide a molecular niche that alleviates selection pressure for mutations against PigmR recognition, ensuring durable resistance to M. oryzae. Mostly recently, a novel MITE-derived microRNA, Osa-miR812w, is reported to positively regulate rice blast resistance through directing DNA methylation at target genes in cis and trans (Campo et al. 2021). In addition to M. oryzae, some sRNAs regulate the defense against other fungal pathogens in rice. The necrotrophic fungal pathogen Rhizoctonia solani (R. solani) represses the expression of siR109944 in rice. One candidate target of siR109944 is F-Box domain and LRR-containing protein 55 (FBL55), which is a transport inhibitor response 1 (TIR1)-like protein. Transgenic plants disrupting siR109944 biogenesis or overexpressing FBL55 display enhanced resistance to R. solani, probably attributing to the altered auxin homeostasis by FBL55 (Qiao et al. 2020). In wheat (Triticum aestivum L.), miR408 positively regulates plant resistance to wheat stripe rust fungus by guiding the cleavage of TaCLP1 mRNA (Feng et al. 2013). In barley (Hordeum vulgare L.), a novel miR9863 family triggers the biogenesis of 21-nt phasiRNAs and miR9863-phasiRNA cascade forms a feed-forward regulatory machinery to suppress the immune signaling mediated by group I Mildew resistance locus a (Mla) alleles in response to barley powdery mildew fungus. Overexpression of miR9863 members specifically attenuates disease resistance and cell death triggered by MLA1 but not MLA10 (Liu et al. 2014). Interestingly, Mla as well as Rom1 (restoration of Mla resistance 1) negatively regulate accumulation levels of hvu-miR398 which represses HvSOD1 accumulation and influences ETI in response to the powdery mildew fungus (Xu et al. 2014). In cotton (Gossypium hirsutum L.), the ghr-miR477-silencing lines display decreased resistance to Verticillium dahlia, while knockdown of its target CaM-binding protein GhCBP60A increases plant resistance by up-regulating isochorismate synthase GhICS1 expression to increase salicylic acid (SA) level (Hu et al. 2020).
Surprisingly, recent studies have revealed that sRNAs can move within plant cells through plasmodesmata and phloem, and transport between plant cells and pathogens by vesicles (Cai et al. 2019; Cai et al. 2018a; Chitwood and Timmermans 2010; Devers et al. 2020; Dunoyer et al. 2010). The bidirectional cross-kingdom movements of sRNAs between plants and pathogens have two different effects on plant immunity. On one hand, some host sRNAs can be transported into invading pathogens and suppress their virulence (Fig. 1d). Upon the infection of Verticillium dahliae, the production of cotton miR166 and miR159 is increased, and both miRNAs are transported to the fungus to specifically silence virulence genes, Ca2+-dependent cysteine protease (Clp-1) and isotrichodermin C-15 hydroxylase (HiC-15), respectively (Zhang et al. 2016c). Similarly, wheat miR1023 can silence the alpha/beta hydrolase gene of F. graminearum to suppress its invasion (Jiao and Peng 2018). These miRNAs may be secreted by exosome-like extracellular vesicles at the infection sites and taken up by the fungal cells to induce silencing of fungal genes associated with pathogenicity, similar to the role of sRNAs (TAS1c-siR483, TAS2-siR453, and IGN-siR1) secreted by Arabidopsis cells in defense against Botrytis cinerea (B. cinerea) (Cai et al. 2018a, b). Above all, the export of specific host miRNAs and siRNAs to inhibit the expression of virulence genes in pathogens may be a conserved and efficient host defense strategy against fungal pathogens.
On the other hand, some sRNAs can serve as effectors to suppress host immunity by hijacking host RNA interference pathways (Fig. 1e). After B. cinerea infection, mitogen activated protein kinase 1/2 (MPK1/2), peroxiredoxin (PRXIIF) and cell wall-associated kinase (WAK) mRNAs, are targeted by Bc-siR3.2, Bc-siR3.1 and Bc-siR5, respectively. B. cinerea dcl1 dcl2 double mutant that cannot produce Bc-sRNAs displays reduced pathogenicity (Weiberg et al. 2013), whereas dcl1 or dcl2 single mutant still produces sRNA effectors to maintain virulence on plants, supporting that sRNA effectors are essential for B. cinerea pathogenicity (Wang et al. 2016b; Weiberg et al. 2013). B. cinerea can also deliver small RNA effectors Bc-siR37 into host cells to suppress host immunity by targeting AtWRKY7, AtPMR6, and AtFEI2 (Wang et al. 2017). Fol-milR1 from Fusarium oxysporum f.sp. lycopersici (Fol), which is exported into tomato cells after infection and sequentially loaded onto tomato ARGONAUTE 4a (SlyAGO4a), targets the CBL-interacting protein kinase SlyFRG4. slyfrg4 mutant plants exhibit enhanced disease susceptibility to Fol, while slyago4a knock-down plants display enhanced resistance to Fol (Ji et al. 2021).
sRNAs regulate the interaction between crops and oomycetes
Oomycetes, one of the two most important groups of eukaryotic plant pathogens, are classified in the kingdom Protoctista and are evolutionally related to heterokont, biflagellate, golden-brown algae (Thines 2018). Several recent reports have revealed the important regulatory roles of sRNAs in plant resistance to two important oomycetes, Phytophthora infestans (P. infestans) and Phytophthora sojae (P. sojae) (Fig. 1a).
In potato (Solanum chacoense and Solanum tuberosum cv. Désirée) plants infected by P. infestans, miR160 is induced in both local and systemic leaves. miR160 knockdown plants fail to elicit systemic acquired resistance (SAR). miR160 targets and mediates the cleavage of StARF10 mRNA. StARF10 protein can bind to the promoter of StGH3.6, a key hub in SA-auxin cross-talk, suggesting the important roles of miR160-StARF10-StGH3.6 module in the antagonistic cross-talk between SA-mediated pathogen defense processes and auxin-mediated growth (Natarajan et al. 2018). In soybean infected by P. sojae, several miRNAs, such as miR1510, miR393, miR1507 and miR2109, regulate plant defense responses. Upon the infection, miR1510 expression is repressed, along with the increased accumulation level of its target Glyma.16G135500, which encodes a classic type of plant disease resistance-associated gene. The overexpression of gma-miR1510a/b in hairy roots enhances susceptibility, suggesting that miR1510 may serve as a negative regulator in plant defense against P. sojae. However, miR393 confers enhanced resistance to P. sojae by up-regulating the expression of 2-hydroxyisoflavanone dehydratase GmHID1 and isoflavone synthase GmIFS1 in isoflavonoid biosynthetic pathway (Wong et al. 2014). miR1507 and miR2109 trigger the production of phasiRNAs to regulate the expression of defense-associated R genes during P. sojae infection (Wong et al. 2014). These RNA silencing pathways can be suppressed by two members of conserved RxLR family effectors, PSR1 and PSR2, which are secreted by P. sojae (Qiao et al. 2013; Qiao et al. 2015; Xiong et al. 2014) (Fig. 1e). Above all, our knowledge on the roles of sRNAs in regulating the crosstalk between host and oomycetes remains largely elusive. Further systemic studies are needed to uncover the potential regulatory roles of small RNAs in defending crops against important oomycetes pathogens.
sRNAs alter crop resistance against bacterial pathogens
Diverse classes of plant endogenous sRNAs, including miRNAs and hc-siRNAs, have been reported to modulate crop immune responses to bacterial pathogens (Fig. 1a). The bacterial blight caused by Xanthomonas oryzae pv. Oryzae (Xoo), the most devasting vascular diseases in rice, causes severe crop loss each year. Downregulation of Osa-miR156 or overexpression of its targets, Ideal Plant Architecture1 (IPA1) and transcription factor OsSPL7, lead to enhanced resistance against Xoo at the expense of rice yield (Liu et al. 2019). To ensure rice resistance to Xoo without yield penalty, specific promoter is selected and applied in plant genetic engineering. In Xanthomonas, TAL (transcription activator-like) effectors bind to specific motifs in host gene promoters and activate the expression of host genes (Moscou and Bogdanove 2009). Among these effectors, Tal9a can bind to the promoter of OsHEN1 and activate its expression, which increases the accumulation levels of many small RNAs and enhances plant susceptibility (Liu et al. 2019) (Fig. 1e). Overexpression of IPA1 driven by OsHEN1 promoter can fine-tune fitness penalty and disease resistance against Xoo (Liu et al. 2019). Besides, the expression of Osa-miR159 is repressed by Xoo invasion, which activates the expression of Osa-miR159 targets GAMYB and OsLRR-RLK2 to modulate plant responses to Xoo (Zhao et al. 2015b). Interestingly, in maize (Zea mays) treated by Bacillus velezensis FZB42, four miRNAs, Zma-miR169a-5p, Zma-miR169c-5p, Zma-miR169i-5p and Zma-miR395b-5p, are repressed, which might fine-tune the activity of NF-Y transcription factors to activate the induced systemic resistance (ISR) to enhance plant defense response against pathogen infections (Xie et al. 2019).
In addition to the posttranscriptional activity of miRNAs, some miRNAs and hc-siRNAs can regulate plant resistance to bacterial pathogens through RdDM pathways. During Xoo infection, OsNBS8R expression is induced by PAMPs, e.g., flagellin and chitin, which activates downstream signaling in PTI to increase rice resistance. However, the non-TAL effector XopQ from Xoo up-regulates Osa-miR1876 and suppresses the transcription of OsNBS8R through DNA methylation, which partly underlies the ETS in rice-Xoo arm-race (Jiang et al. 2020). TE-siR815, a hc-siRNA generated from WRKY45-1 locus, reduces rice resistance to Xoo through repressing the expression of ST1 that encodes an LRR-type protein by RdDM. Suppression of ST1 abolishes WRKY45-mediated resistance, thereby leading to disease susceptibility (Zhang et al. 2016a).
Diverse sRNAs participate in crop-viruses interaction
In the battle between plants and viruses, virus-derived siRNAs (vsiRNA) as well as plant endogenous virus-activated miRNAs or siRNAs are loaded onto antiviral AGOs to inhibit the invasion of DNA or RNA viruses. These siRNAs may repress viral RNA through mRNA cleavage or translational inhibition, silence viral DNA through RdDM pathway, or affect host resistance (Carbonell and Carrington 2015; Prasad et al. 2019).
Some miRNAs in rice, such as Osa-miR528, Osa-miR444 and Osa-miR393, have been reported to positively or negatively regulate virus pathogenicity or host resistance (Fig. 1a). Osa-miR528 can be activated by the transcription factor OsSPL9 and negatively regulate the expression of its target L-Ascorbate Oxidase (AO) mRNA in rice. Loss-of-function of spl9 causes decreased Osa-miR528 accumulation and a substantial increase of AO, resulting in enhanced plant resistance to Rice stripe virus (RSV) (Wu et al. 2017). Osa-miR444 has a pivotal function in the cross talk of nitrate signaling and the antiviral response. Without RSV infection, the expression of OsRDR1, an important component in RNA silencing pathway, is repressed by OsMADS23, OsMADS27a and OsMADS57. Upon virus infection, Osa-miR444 is induced to diminish the expression of OsMADS members and subsequently activates the OsRDR1-dependent RNA silencing pathway, which confers plant resistance against virus infection by silencing both viral RNAs and host genes (Wang et al. 2016a). In addition to the antibacterial role (Navarro et al. 2006), Osa-miR393 also regulates plant antiviral responses by repressing auxin signaling. Transgenic rice plants overexpressing Osa-miR393 display increased susceptibility to Rice black streaked dwarf virus (RBSDV) due to the repression of auxin signaling (Zhang et al. 2019). In Brassica, miR1885 fine-tunes plant growth and immunity through different mechanisms. During vegetative stage, miR1885 is maintained at low levels to ensure normal development and basal immunity. After infected by Turnip mosaic virus (TuMV), silencing of BraCP24 mediated by miR1885 is enhanced to initiate precocious flowering, whereas the repression of R gene BraTNL1 by miR1885-phasiRNAs cascade is antagonized by TuMV-induced BraTNL1 expression (Cui et al. 2020). In tobacco, miR-6019 and miR-6020 cleave N gene to generate phased siRNAs, which compromise resistance to Tobacco mosaic virus (TMV) by repressing the N gene expression under normal circumstances. TMV infection diminishes the accumulation levels of miRNA-6019/6020 and phasiRNAs, thus releasing the repression of N gene and limiting the virus spread (Deng et al. 2018). These reports have revealed the complicated roles of miRNAs and phasiRNAs in plant resistance against virus.
vsiRNAs or plant siRNAs-mediated RNAi pathways are major antiviral defense machinery in plant model systems. Among the essential components of RNAi, RDR6 acts as a positive regulator in the resistance against viruses. Upon Rice Dwarf Phytoreovirus (RDV) infection, the expression of RDR6 is downregulated. Besides, the accumulation of RDV vsiRNAs is reduced in the osrdr6 knockdown transgenic plants, which results in increased susceptibility in rice (Hong et al. 2015). Besides RDR6, multiple AGOs function in crop antiviral defense, such as AGO1/18 in rice, AGO1/2/4 in Nicotiana benthamiana (Carbonell and Carrington 2015). In rice, AGO1 and AGO18 synergistically modulate antiviral defense. AGO18 induced by virus competes with AGO1 for binding Osa-miR168 to release miR168-mediated suppression of AGO1 upon viral infection. Overexpression of miR168-resistant AGO1 rescues the deficiency of ago18 in viral resistance (Du et al. 2011).
Like oomycetes, fungal and bacterial pathogens, viruses have evolved multiple virulence or effector proteins to disrupt host immune responses or RNA silencing pathways (Fig. 1e). Polerovirus P0 interacts with E3 ligase S-phase kinase regulated protein 1 (SKP1) to enhance the degradation of multiple AGOs by 26S proteasome system and autophagy pathways before RISC assembly (Li et al. 2019b). Tomato ringspot virus (ToRSV) suppressor coat protein (CP) binds to AGO1, suppresses its translational inhibitory activity and further enhances AGO1 degradation through autophagy (Karran and Sanfaçon 2014). RNase 3, a VSR encoded by Sweet potato chlorotic stunt crinivirus (SPCSV), cuts 21-24-nt vsiRNAs into 14 bp inactive products, thus effectively precluding the formation of antiviral RISC (Cuellar et al. 2009; Kreuze et al. 2005). C2 protein, encoded by DNA virus Beet severe curly top virus, is an effector that counteracts antiviral defense by interfering with gene silencing and metabolic defense responses. C2 mediates a decrease in DNA methylation levels of promoter regions from where reduced siRNAs derived, thereby upregulating the expression of the viral coding genes (Yang et al. 2013). The virus 19 kDa protein (P19) of tombusviruses inhibits post-transcriptional gene silencing by specifically binding to double-stranded siRNAs in tobacco (Silhavy et al. 2002). VsiRNA1 can suppress the expression of wheat thioredoxin-like (TaAAED1) gene which negatively regulates the production of ROS (Liu et al. 2021). Transgenic amiRNA1 plants in wheat confers a broad-spectrum disease resistance to Chinese wheat mosaic virus, Barley stripe mosaic virus, and Puccinia striiformis f. sp. tritici (Liu et al. 2021).
sRNA-based technologies in engineering disease resistance
Based on the above knowledge on the roles of small RNAs in plant immunity, many sRNA-based technologies have been developed to protect plants against pathogens, such as artificial microRNA (amiRNA) and synthetic trans-acting siRNA (syn-tasiRNA)-mediated RNAi, host-induced gene silencing (HIGS), spray-induced gene silencing (SIGS) and virus-induced gene silencing (VIGS) (Fig. 2a-d). These strategies are applied to breed crops with stable disease resistance or fine-tune the trade-off between plant immunity and yield.
Fig. 2.
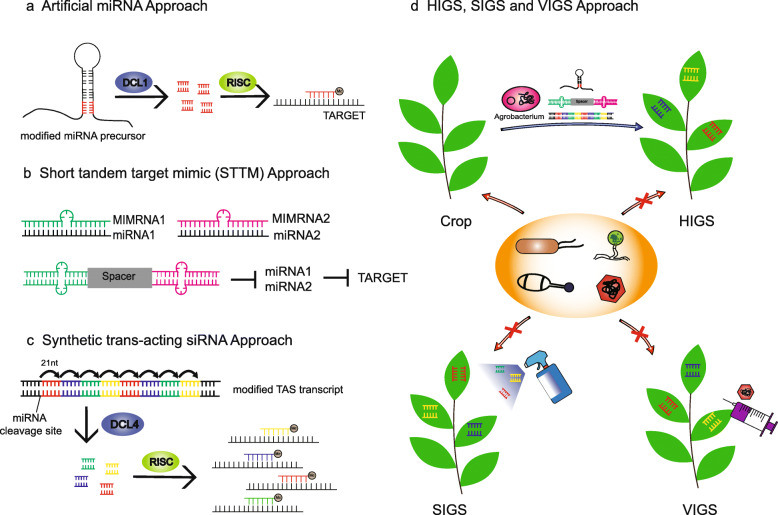
sRNA-based technologies in protecting crop plants. Many sRNA-based technologies have been developed to protect plants from pathogens. a Artificial microRNA (amiRNA) approach. For example, the transgenic barley lines that carry a polycistronic amiRNA precursor construct (VirusBuster171) expressing three amiRNAs simultaneously under the control of a constitutive promoter, display enhanced resistance to Wheat dwarf virus (WDV) (Kis et al. 2016). b Short tandem target mimic (STTM) approach in modulating the activity of miRNAs. STTM is composed of two miRNA binding sites which have mismatches at the mRNA cleavage sites. The two mimic sequences are usually separated by a spacer linker. In soybean, inhibition of miR1507 as well as miR482 by STTM compromises their suppression of NBS-LRR genes (Bao et al. 2018). c Synthetic trans-acting siRNA approach. MiRNAs target TAS loci and produce phased, secondary siRNAs with the help of DCL4. Thus, TAS genes are engineered to express multiple synthetic ta-siRNAs (syn-tasiRNAs) that target multiple viruses at diverse genomic position. d Host-induced gene silencing (HIGS), spray-induced gene silencing (SIGS) and virus-induced gene silencing (VIGS). Based on that plant sRNAs can be transferred to organisms colonizing or feeding on the plant, scientists engineer transgenic plants which produce sRNAs targeting pathogen sequences to avoid infection. Meanwhile, VIGS, which use the virus expression vector as the medium, and SIGS, which directly use pathogen-gene-targeting dsRNAs or sRNAs, are two other strategies. These new approaches not only control plant disease but also have the advantages of simplicity, high specificity, flexibility and stability
amiRNA-induced silencing approaches are widely used to protect plants against viruses (Fig. 2a). A. thaliana transgenic plants which express modified miR159 precursor-based amiRNAs targeting two viral suppressors P69 and Hc-Pro simultaneously, exhibit resistance against both Turnip yellow mosaic virus (TYMV) and Turnip mosaic virus (TuMV) (Niu et al. 2006). Similarly, using Arabidopsis pre-miRNA159a as the backbone, overexpressing amiRNAs targeting the replicase gene of Watermelon silver mottle virus (WSMoV) in N. benthamiana enhance plant defense even 21 days post viral inoculation (Kung et al. 2012). These findings prove the possibility of employing the amiRNA approach to breed crops with broad-spectrum resistance to tospoviruses as well as other viruses (Mitter et al. 2016). The transgenic barley lines that carry a polycistronic amiRNA precursor construct (VirusBuster171) expressing three amiRNAs simultaneously under the control of a constitutive promoter, display enhanced resistance to Wheat dwarf virus (WDV) (Kis et al. 2016). Besides amiRNA technology, short tandem target mimic (STTM) technology is also used to repress the function of miRNAs (Fig. 2b). Inhibition of miR1507a, miR1507c, miR482a, miR168a and miR1515a by STTM compromise Soybean mosaic virus (SMV) infection efficiency in soybean (Bao et al. 2018). Tomato STTM482/2118b lines display enhanced resistance to infection with the oomycete and bacterial pathogens (Canto-Pastor et al. 2019). TAS genes are also engineered to express multiple synthetic ta-siRNAs (syn-tasiRNAs) that target multiple viruses at diverse genomic positions (Fig. 2c). For example, The Arabidopsis TAS3a gene is used as backbone to generate syn-tasiRNAs targeting the genome of TuMV and Cucumber mosaic virus (CMV). Transgenic Arabidopsis plants expressing these syn-tasiRNAs show elevated resistance to both viruses (Chen et al. 2016). In tomato, expression of a syn-tasiRNA construct that can produce four different syn-tasiRNAs against Tomato spotted wilt virus (TSWV) enhance plant antiviral resistance (Carbonell et al. 2019).
Besides RNAi technologies based on amiRNAs, STTM and syn-tasiRNAs, many new RNA-based approaches, such as HIGS (Nowara et al. 2010), VIGS (Cooper and Campbell 2017; Ranjan et al. 2018), and SIGS (Koch et al. 2016), have been developed to control plant disease by targeting host susceptibility factors, pathogen-derived RNAs or pathogen effectors (Fig. 2d). HIGS technology has been widely used to defend plants against fungal pathogens. Expressing sRNAs that target Bc-DCL1 and Bc-DCL2 in tomato silence Bc-DCL genes and attenuate fungal pathogenicity and growth, suggesting the bidirectional cross-kingdom RNAi and sRNA trafficking between plants and fungi (Wang et al. 2016b). Expressing CYP3 dsRNA in barley leaves produces sRNAs to silence fungal cytochrome P450, which inhibit the growth and alter fungal morphology of F. graminearum at the incubation site (Koch et al. 2013). In barley and wheat, the accumulation of dsRNAs or antisense RNA targeting fungal transcripts affects the development of the powdery mildew fungus Blumeria graminis (Nowara et al. 2010). Surprisingly, dsRNAs have efficacy to target pathogens even directly sprayed on plants, which is known as SIGS (Sang and Kim 2020). Foliar application of dsRNA is effective in reducing disease symptoms in Brassica napus infected by Sclerotinia sclerotiorum or B. cinerea (McLoughlin et al. 2018). Similar to HIGS, direct spraying detached barley leaves with CYP3 dsRNAs prior to F. graminearum infection also effectively inhibits disease development (Koch et al. 2016). SIGS of F. graminearum AGO and DCL genes protects barley leaves from F. graminearum infection (Werner et al. 2020). Moreover, SIGS has been adopted to compromise Verticillium wilt in tomato (Song and Thomma 2018) and late blight disease in potato (Kalyandurg et al. 2021). It is needed to note that the efficiency of SIGS for disease control largely depends on the efficiency of RNA uptake by pathogens and the secondary amplification of siRNA machinery (Qiao et al. 2021; Song and Thomma 2018).
Another approach to enhance plant resistance to pathogens is VIGS which takes advantage of plant sRNAs-mediated antiviral defense mechanism. VIGS is nowadays widely used to engineer knockdown plants due to its easy to operate and high efficiency. VIGS mainly involves three steps: design vectors derived from viruses carrying viral target genes to be silenced, infect the plant hosts by Agrobacterium tumefaciens and silence the target genes to activate defense against pathogen infection (Becker and Lange 2010). The viral vectors used in VIGS are mainly derived from positive-strand RNA viruses such as Potato virus X (PVX), TMV, and Tobacco rattle virus (TRV) (Purkayastha and Dasgupta 2009). The Barley stripe mosaic virus (BSMV), a tripartite RNA virus, which can infect many important crops like barley, wheat, rice and maize, is also modified for VIGS (Purkayastha and Dasgupta 2009). Many studies have revealed that VIGS is a powerful tool for identifying functional genes that confer disease resistance in crops. For example, knocking-down TaBON1 or TaBON3 by VIGS enhances wheat disease resistance to powdery mildew by upregulating defense responses in wheat (Zou et al. 2018). The reduced expression of TaDIR1-2 through the BSMV-mediated VIGS system contributes to the wheat defense response against Puccinia striiformis f. sp. tritici (Ahmed et al. 2017; Scofield et al. 2005). The role of agmatinecoumaroyl transferase TaACT in Fusarium head blight (FHB) resistance is validated by VIGS in wheat (Kage et al. 2017). Through VIGS strategy, the regulatory role of the TaMED25-TaEIL1-TaERF1 module in bread wheat defense against powdery mildew is identified (Liu et al. 2016). In tomato, reverse genetic studies using VIGS technology reveal that SlMAPKKKε, a positive regulator of cell death, is required for disease resistance against Pst and Xanthomonas campestris pv. vesicatoria (Xcv) (Melech-Bonfil and Sessa 2010).
Concluding remarks and perspectives
Due to the ever-lasting arms race between plants and pathogens, there are no permanent disease control strategies which are absolute effective or completely without risk (including off-target effects, selection between ideal and specific target sites). As reviewed above, small RNAs mediate multilayer regulation in crop immune responses against pathogens, including post-transcriptional gene silencing by guiding mRNA cleavage/degradation or translational repression, transcriptional gene silencing by direct DNA methylation or chromatin modification (Fig. 1 and Table 1). A series of evidence indicate that these pathogen-responsive small RNAs may fine-tune or reprogram gene expression by silencing negative regulators or inducing positive regulators of immune responses.
Table 1.
small RNAs involved in crop disease resistance
| Pathogen | Crop | Small RNA | Influence (in plant resistance) | Target | Reference· | |
|---|---|---|---|---|---|---|
| Fungi | Magnaporthe oryzae | rice | miR156 | negative | SPL14 | (Zhang et al. 2020) |
| Magnaporthe oryzae | rice | miR160 | positive | ARF16 | ||
| Magnaporthe oryzae | rice | miR162 | positive | DCL1a | (Li et al. 2020b) | |
| Magnaporthe oryzae | rice | miR164a | negative | OsNAC60 | (Wang et al. 2018) | |
| Magnaporthe oryzae | rice | miR166 | positive | EIN2 | (Salvador-Guirao et al. 2018) | |
| Magnaporthe oryzae | rice | miR167d | negative | ARF12 | (Zhao et al. 2020) | |
| Magnaporthe oryzae | rice | miR168 | negative | AGO1 | (Wang et al. 2021) | |
| Magnaporthe oryzae | rice | miR169 | negative | NF-YAs | (Li et al. 2017) | |
| Magnaporthe oryzae | rice | miR319 | negative | TCP21 | (Zhang et al. 2018) | |
| Magnaporthe oryzae | rice | miR396 | negative | OsGRFs | (Chandran et al. 2018) | |
| Magnaporthe oryzae | rice | miR398 | positive | CSD1, CSD2, SODX | (Li et al. 2019a) | |
| Magnaporthe oryzae | rice | miR439 | negative | / | (Lu et al. 2021) | |
| Magnaporthe oryzae | rice | miR812 | positive | ACO3, CIPK10, LRR | (Campo et al. 2021) | |
| Magnaporthe oryzae | rice | miR7695 | positive | OsNramp6 | (Campo et al. 2013) | |
| Magnaporthe oryzae | rice | MITE1/2-siRNAs | positive | PigmS | (Deng et al. 2017) | |
| Fusarium fujikuroi | rice | miR166 | positive | EIN2 | (Salvador-Guirao et al. 2018) | |
| Rhizoctonia solani | rice | siR109944 | negative | FBL55 | (Qiao et al. 2020) | |
| Blumeria graminis f. sp.hordei | Barley | miR398 | negative | HvSOD1 | (Xu et al. 2014) | |
| Blumeria graminis f. sp.hordei | Barley | miR9863 | negative | MLA1 | (Liu et al. 2014) | |
| Verticillium dahlia | cotton | miR159 | positive | HiC-15 | (Zhang et al. 2016c) | |
| Verticillium dahlia | cotton | miR166 | positive | Clp-1 | (Zhang et al. 2016c) | |
| Verticillium dahlia | cotton | miR477 | positive | CBP60A | (Hu et al. 2020) | |
| Puccinia striiformis f. sp. tritici | wheat | miR408 | negative | CLP1 | (Feng et al. 2013) | |
| Fusarium graminearum | wheat | miR1023 | positive | FGSG_03101 | (Jiao and Peng 2018) | |
| Oomycete | Phytophthora infestans | potato | miR160 | unknown | ARF10 | (Natarajan et al. 2018) |
| Phytophthora sojae | soybean | miR393 | positive | / | (Wong et al. 2014) | |
| Phytophthora sojae | soybean | miR1507 | unknown | Glyma04g29220, Glyma06g39720, Glyma06g39720 | (Wong et al. 2014) | |
| Phytophthora sojae | soybean | miR219 | unknown | Glyma06g39720, Glyma01g06750 | (Wong et al. 2014) | |
| Phytophthora sojae | soybean | miR1510 | negative | Glyma.16G135500 | (Cui et al. 2017) | |
| Bacterial | Xanthomonas oryzae pv. Oryzae | rice | miR156 | negative | IPA1, OsSPL7 | (Liu et al. 2019) |
| Xanthomonas oryzae pv. Oryzae | rice | miR159 | unknown | GAMYB1, OsLRR-RLK2 | (Zhao et al. 2015b) | |
| Xanthomonas oryzae pv. Oryzae | rice | TE-siR815 | negative | ST1 | (Zhang et al. 2016a) | |
| Xanthomonas oryzae pv. Oryzae | rice | miR1876 | negative | NBS8R | (Jiang et al. 2020) | |
| Bacillus velezensis FZB42 | maize | miR169 | unknown | NF-Y | (Xie et al. 2019) | |
| miR395 | unknown | |||||
| Virus | Tobacco rattle virus (TRV)/Cucumber mosaic virus (CMV) | tomato | miR482 | negative | LRR1, LRR2 | (Shivaprasad et al. 2012) |
| Rice black streaked dwarf virus (RBSDV) | rice | miR393 | negative | TIR1 | (Zhang et al. 2019) | |
| Rice stripe virus (RSV) | rice | miR168 | negative | AGO1 | (Du et al. 2011) | |
| Rice stripe virus (RSV) | rice | miR444 | positive | RDR1, MADS | (Wang et al. 2016a) | |
| Rice stripe virus (RSV) | rice | miR528 | negative | AO |
(Wu et al. 2017) (Yao et al. 2019) |
|
| Tobacco mosaic virus (TMV) | tobacoo | miR6019, miR6020 | negative | N | (Li et al. 2012) | |
| Turnip mosaic virus (TuMV) | Brassica napus | miR1885 | negative | BraTIR1, BraTNL1 | (Cui et al. 2020) | |
| Soybean mosaic virus (SMV) | soybean | miR168 | negative | AGO1 | (Bao et al. 2018) | |
| Soybean mosaic virus (SMV) | soybean | miR1515 | / | DCL2 | ||
| Soybean mosaic virus (SMV) | soybean | mIR1507, miR482 | negative | NBS-LRR | ||
Despite the advantages of simplicity, high specificity, flexibility and stability, there are some limitations of RNAi-based approaches. For example, a major hurdle in the practical application of SIGS is the rapid degradation of naked RNAs. To overcome this problem, nanomaterials such as chitosan-complexed single-walled carbon nanotubes (Demirer et al. 2019; Kwak et al. 2019) and layered double hydroxide (LDH) clay nanosheets (Qiao et al. 2021), may be used to deliver and stabilize sRNAs. These new approaches not only facilitate biomolecule transport into plant cells with high efficiency and without toxicity or tissue damage but also protect RNA cargo from nuclease degradation. Other candidate protective delivery systems such as artificial extracellular vesicle (EVs) or liposomes mimic plant EV may also be used (Regente et al. 2017). However, our knowledge of plant EV is rather limited. Whether artificial EVs can function in vivo still needs to be investigated. Despite efficient dsRNA uptake in many fungal plant pathogens have been achieved, the efficiency of dsRNAs uptake in other fungi still need to be resolved (Qiao et al. 2021; Rosa et al. 2018). Moreover, strategies for control bacteria-induced diseases are rather limited.
To breed non-genetically modified (GMO), disease-tolerant crops, SIGS will be a promising technology to enhance crop resistance to disease. sRNAs-based strategies may be synergistically applied with other molecular approaches. Combined with the use of newly developed multi-transgene stacking toolkit containing marker/marker-excision cassette (Zhu et al. 2017), HIGS may be optimized to silence multiple pathogens through various RNA constructs targeting different genes in pathogens. Moreover, multiple viral vectors-based transient reprogramming has been used to trigger alterations of agronomic traits, such as flowering time, plant height or drought tolerance (Torti et al. 2021). Similar viral vectors-based RNAi may be developed to manage plant pathogen pandemics. To date, there are almost no RNAi-based field applications for managing plant diseases caused by bacteria and fungi, although there are already many field trials and even approval for RNAi-based targeting of plant pathogens (Rosa et al. 2018). More economic, high efficiency, low toxicity agricultural development will be the perpetual goals. RNAi-based technology will play a profound role in protecting plant from pathogens in the future.
Acknowledgments
Due to space limitations, we apologize to our colleagues whose important work are not cited in this review.
Adherence to national and international regulations
Not applicable.
Abbreviations
- Small RNAs (sRNAs)
A class of 18-30 nt, non-coding RNAs, including microRNAs and small interfering RNAs in plants, which modulate gene expression and maintain the genome stability
- Pathogen-associated molecular patterns (PAMPs)
Small molecules from microbes that can be recognized by host pattern recognition receptors
- Pattern recognition receptors (PRRs)
A class of receptors responsible for recognizing the PAMPs from microbes
- Pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI)
The immune responses that are initiated by the recognition of PAMPs by PRRs
- Effector-triggered immunity (ETI)
A cascade of defense responses that are initiated by the direct or indirect recognition of pathogen effectors by host resistance (R) proteins
- RNA-induced silencing complex (RISC)
A complex containing ARGONAUTE (AGO) proteins, single-stranded small RNAs and other proteins, which can silence target gene expression by mRNAs cleavage, translational inhibition, or DNA methylation-mediated transcriptional inhibition
- RNA-directed DNA methylation (RdDM)
The de novo cytosine methylation in plants that is guided by small interfering RNAs (siRNAs)
- Systemic acquired resistance (SAR)
The systemic immune responses triggered by an initial localized infection, which confer long-lasting and broad-spectrum protection in plants
- Artificial microRNAs (amiRNAs)
A class of artificial small RNAs generated from synthetic miRNA precursors using endogenous pre-miRNA as the backbone
- Synthetic trans-acting siRNAs (syn-tasiRNAs)
Artificial siRNAs produced from the engineered TAS locus
- Short tandem target mimic (STTM)
A potent technology for blocking the activity of small RNAs, which triggers the degradation or sequestration of target small RNAs.
- Host-induced gene silencing (HIGS)
An RNAi-based technology in which sRNAs are produced from dsRNAs expressed in hosts to induce silencing of pathogen genes, which protect plants from pathogens
- Spray-induced gene silencing (SIGS)
A newly developed technology used to increase plant resistance against pathogens by direct spraying dsRNAs on plants
- Virus-induced gene silencing (VIGS)
An efficient method that is applied to block host gene expression through modified virus-based vectors carrying sequences derived from corresponding host genes
Authors’ contributions
J.L. and Z.H. planned and designed this work. The original draft was prepared by J.T. and X.G. and edited by J.L. and Z.H. The author(s) read and approved the final manuscript.
Funding
This work was supported by funding from the National Natural Science Foundation of China (91940301 to Z. H., 32070564 and 31600207 to J. L.) and Yunnan Fundamental Research Projects (202101AW070002 to J. L.).
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
Author Z.H. is a member of the Editorial Board and was not involved in the journal's review of, or decisions related to, this manuscript.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jun Tang and Xueting Gu contributed equally to this work.
Contributor Information
Junzhong Liu, Email: liujunzhong@ynu.edu.cn.
Zuhua He, Email: zhhe@cemps.ac.cn.
References
- Ahmed SM, Liu P, Xue Q, Ji C, Qi T, Guo J, Guo J, Kang Z. TaDIR1-2, a wheat ortholog of lipid transfer protein AtDIR1 contributes to negative regulation of wheat resistance against Puccinia striiformis f. sp. tritici. Front Plant Sci. 2017;8:521. doi: 10.3389/fpls.2017.00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves MS, Dadalto SP, Goncalves AB, de Souza GB, Barros VA, Fietto LG. Transcription factor functional protein-protein interactions in plant defense responses. Proteomes. 2014;2:85–106. doi: 10.3390/proteomes2010085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao D, Ganbaatar O, Cui X, Yu R, Bao W, Falk BW, Wuriyanghan H. Down-regulation of genes coding for core RNAi components and disease resistance proteins via corresponding microRNAs might be correlated with successful soybean mosaic virus infection in soybean. Mol Plant Pathol. 2018;19:948–960. doi: 10.1111/mpp.12581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- Becker A, Lange M. VIGS – genomics goes functional. Trends Plant Sci. 2010;15:1–4. doi: 10.1016/j.tplants.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Bologna NG, Voinnet O. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu Rev Plant Biol. 2014;65:473–503. doi: 10.1146/annurev-arplant-050213-035728. [DOI] [PubMed] [Google Scholar]
- Borges F, Martienssen RA. The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol. 2015;16:727–741. doi: 10.1038/nrm4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrelli GM, Mazzucotelli E, Marone D, Crosatti C, Michelotti V, Valè G, Mastrangelo AM. Regulation and evolution of NLR genes: a close interconnection for plant immunity. Int J Mol Sci. 2018;19:1662. doi: 10.3390/ijms19061662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, He B, Jin H. A safe ride in extracellular vesicles - small RNA trafficking between plant hosts and pathogens. Curr Opin Plant Biol. 2019;52:140–148. doi: 10.1016/j.pbi.2019.09.001. [DOI] [PubMed] [Google Scholar]
- Cai Q, He B, Kogel KH, Jin H. Cross-kingdom RNA trafficking and environmental RNAi-nature's blueprint for modern crop protection strategies. Curr Opin Microbiol. 2018;46:58–64. doi: 10.1016/j.mib.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Qiao L, Wang M, He B, Lin FM, Palmquist J, Huang SD, Jin H. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science. 2018;360:1126–1129. doi: 10.1126/science.aar4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo S, Peris-Peris C, Siré C, Moreno AB, Donaire L, Zytnicki M, Notredame C, Llave C, San Segundo B. Identification of a novel microRNA (miRNA) from rice that targets an alternatively spliced transcript of the Nramp6 (natural resistance-associated macrophage protein 6) gene involved in pathogen resistance. New Phytol. 2013;199:212–227. doi: 10.1111/nph.12292. [DOI] [PubMed] [Google Scholar]
- Campo S, Sánchez-Sanuy F, Camargo-Ramírez R, Gómez-Ariza J, Baldrich P, Campos-Soriano L, Soto-Suárez M, Segundo BS (2021) A novel transposable element-derived microRNA participates in plant immunity to rice blast disease. Plant Biotechnol J. 10.1111/pbi.13592 [DOI] [PMC free article] [PubMed]
- Canto-Pastor A, Bamc S, Valli AA, Summers W, Schornack S, Baulcombe DC. Enhanced resistance to bacterial and oomycete pathogens by short tandem target mimic RNAs in tomato. Proc Natl Acad Sci U S A. 2019;116:2755–2760. doi: 10.1073/pnas.1814380116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell A, Carrington JC. Antiviral roles of plant ARGONAUTES. Curr Opin Plant Biol. 2015;27:111–117. doi: 10.1016/j.pbi.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell A, Lisón P, Daròs JA. Multi-targeting of viral RNAs with synthetic trans-acting small interfering RNAs enhances plant antiviral resistance. Plant J. 2019;100:720–737. doi: 10.1111/tpj.14466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carra A, Mica E, Gambino G, Pindo M, Moser C, Pè ME, Schubert A. Cloning and characterization of small non-coding RNAs from grape. Plant J. 2009;59:750–763. doi: 10.1111/j.1365-313X.2009.03906.x. [DOI] [PubMed] [Google Scholar]
- Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW, Zilberman D, Xie Z, Johansen LK, Carrington JC, Jacobsen SE. RNA silencing genes control de novo DNA methylation. Science. 2004;303:1336. doi: 10.1126/science.1095989. [DOI] [PubMed] [Google Scholar]
- Chandran V, Wang H, Gao F, Cao XL, Chen YP, Li GB, Zhu Y, Yang XM, Zhang LL, Zhao ZX, Zhao JH, Wang YG, Li S, Fan J, Li Y, Zhao JQ, Li SQ, Wang WM. miR396-OsGRFs module balances growth and rice blast disease-resistance. Front Plant Sci. 2018;9:1999. doi: 10.3389/fpls.2018.01999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Cheng X, Cai J, Zhan L, Wu X, Liu Q, Wu X. Multiple virus resistance using artificial trans-acting siRNAs. J Virol Methods. 2016;228:16–20. doi: 10.1016/j.jviromet.2015.11.004. [DOI] [PubMed] [Google Scholar]
- Chen X. Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol. 2009;25:21–44. doi: 10.1146/annurev.cellbio.042308.113417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood DH, Timmermans MC. Small RNAs are on the move. Nature. 2010;467:415–419. doi: 10.1038/nature09351. [DOI] [PubMed] [Google Scholar]
- Cooper B, Campbell KB. Protection against common bean rust conferred by a gene-silencing method. Phytopathology. 2017;107:920–927. doi: 10.1094/PHYTO-03-17-0095-R. [DOI] [PubMed] [Google Scholar]
- Cuellar WJ, Kreuze JF, Rajamäki ML, Cruzado KR, Untiveros M, Valkonen JP. Elimination of antiviral defense by viral RNase III. Proc Natl Acad Sci U S A. 2009;106:10354–10358. doi: 10.1073/pnas.0806042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Tsuda K, Parker JE. Effector-triggered immunity: from pathogen perception to robust defense. Annu Rev Plant Biol. 2015;66:487–511. doi: 10.1146/annurev-arplant-050213-040012. [DOI] [PubMed] [Google Scholar]
- Cui C, Wang J-J, Zhao J-H, Fang Y-Y, He X-F, Guo H-S, Duan C-G. A Brassica miRNA regulates plant growth and immunity through distinct modes of action. Mol Plant. 2020;13:231–245. doi: 10.1016/j.molp.2019.11.010. [DOI] [PubMed] [Google Scholar]
- Cui X, Yan Q, Gan S, Xue D, Dou D, Guo N, Xing H. Overexpression of gma-miR1510a/b suppresses the expression of a NB-LRR domain gene and reduces resistance to Phytophthora sojae. Gene. 2017;621:32–39. doi: 10.1016/j.gene.2017.04.015. [DOI] [PubMed] [Google Scholar]
- Dangl JL, Horvath DM, Staskawicz BJ. Pivoting the plant immune system from dissection to deployment. Science. 2013;341:746–751. doi: 10.1126/science.1236011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean R, Van Kan JA, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, Rudd JJ, Dickman M, Kahmann R, Ellis J, Foster GD. The top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol. 2012;13:414–430. doi: 10.1111/j.1364-3703.2011.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirer GS, Zhang H, Matos JL, Goh NS, Cunningham FJ, Sung Y, Chang R, Aditham AJ, Chio L, Cho MJ, Staskawicz B, Landry MP. High aspect ratio nanomaterials enable delivery of functional genetic material without DNA integration in mature plants. Nat Nanotechnol. 2019;14:456–464. doi: 10.1038/s41565-019-0382-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Wang J, Tung J, Liu D, Zhou Y, He S, Du Y, Baker B, Li F. A role for small RNA in regulating innate immunity during plant growth. PLoS Pathog. 2018;14:e1006756. doi: 10.1371/journal.ppat.1006756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Zhai K, Xie Z, Yang D, Zhu X, Liu J, Wang X, Qin P, Yang Y, Zhang G, Li Q, Zhang J, Wu S, Milazzo J, Mao B, Wang E, Xie H, Tharreau D, He Z. Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science. 2017;355:962–965. doi: 10.1126/science.aai8898. [DOI] [PubMed] [Google Scholar]
- Deng YW, Ning YS, Yang DL, Zhai KR, Wang GL, He ZH (2020) Molecular basis of disease resistance and perspectives on breeding strategies for resistance improvement in crops. Mol Plant 13:1402–1419. 10.1016/j.molp.2020.09.018 [DOI] [PubMed]
- Devers EA, Brosnan CA, Sarazin A, Albertini D, Amsler AC, Brioudes F, Jullien PE, Lim P, Schott G, Voinnet O. Movement and differential consumption of short interfering RNA duplexes underlie mobile RNA interference. Nat Plants. 2020;6:789–799. doi: 10.1038/s41477-020-0687-2. [DOI] [PubMed] [Google Scholar]
- Du P, Wu J, Zhang J, Zhao S, Zheng H, Gao G, Wei L, Li Y. Viral infection induces expression of novel phased microRNAs from conserved cellular microRNA precursors. PLoS Pathog. 2011;7:e1002176. doi: 10.1371/journal.ppat.1002176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P, Schott G, Himber C, Meyer D, Takeda A, Carrington JC, Voinnet O. Small RNA duplexes function as mobile silencing signals between plant cells. Science. 2010;328:912–916. doi: 10.1126/science.1185880. [DOI] [PubMed] [Google Scholar]
- Fei Q, Xia R, Meyers BC. Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell. 2013;25:2400–2415. doi: 10.1105/tpc.113.114652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Q, Zhang Y, Xia R, Meyers BC. Small RNAs add zing to the zig-Zag-zig model of plant defenses. Mol Plant Microbe Interact. 2016;29:165–169. doi: 10.1094/MPMI-09-15-0212-FI. [DOI] [PubMed] [Google Scholar]
- Feng H, Zhang Q, Wang Q, Wang X, Liu J, Li M, Huang L, Kang Z. Target of tae-miR408, a chemocyanin-like protein gene (TaCLP1), plays positive roles in wheat response to high-salinity, heavy cupric stress and stripe rust. Plant Mol Biol. 2013;83:433–443. doi: 10.1007/s11103-013-0101-9. [DOI] [PubMed] [Google Scholar]
- Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol. 2005;15:1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González VM, Müller S, Baulcombe D, Puigdomènech P. Evolution of NBS-LRR gene copies among dicot plants and its regulation by members of the miR482/2118 superfamily of miRNAs. Mol Plant. 2015;8:329–331. doi: 10.1016/j.molp.2014.11.013. [DOI] [PubMed] [Google Scholar]
- He XF, Fang YY, Feng L, Guo HS. Characterization of conserved and novel microRNAs and their targets, including a TuMV-induced TIR-NBS-LRR class R gene-derived novel miRNA in Brassica. FEBS Lett. 2008;582:2445–2452. doi: 10.1016/j.febslet.2008.06.011. [DOI] [PubMed] [Google Scholar]
- Hong W, Qian D, Sun R, Jiang L, Wang Y, Wei C, Zhang Z, Li Y. OsRDR6 plays role in host defense against double-stranded RNA virus, Rice dwarf Phytoreovirus. Sci Rep. 2015;5:11324. doi: 10.1038/srep11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Hao M, Wang L, Liu J, Zhang Z, Tang Y, Peng Q, Yang Z, Wu J. The cotton miR477-CBP60A module participates in plant defense against Verticillium dahlia. Mol Plant Microbe Interact. 2020;33:624–636. doi: 10.1094/MPMI-10-19-0302-R. [DOI] [PubMed] [Google Scholar]
- Huang CY, Wang H, Hu P, Hamby R, Jin H. Small RNAs - big players in plant-microbe interactions. Cell Host Microbe. 2019;26:173–182. doi: 10.1016/j.chom.2019.07.021. [DOI] [PubMed] [Google Scholar]
- Ji HM, Mao HY, Li SJ, Feng T, Zhang ZY, Cheng L, Luo SJ, Borkovich KA, Ouyang SQ (2021) Fol-milR1, a pathogenicity factor of Fusarium oxysporum, confers tomato wilt disease resistance by impairing host immune responses. New Phytol. 10.1111/nph.17436 [DOI] [PMC free article] [PubMed]
- Jiang G, Liu D, Yin D, Zhou Z, Shi Y, Li C, Zhu L, Zhai W. A rice NBS-ARC gene conferring quantitative resistance to bacterial blight is regulated by a pathogen effector-inducible miRNA. Mol Plant. 2020;13:1752–1767. doi: 10.1016/j.molp.2020.09.015. [DOI] [PubMed] [Google Scholar]
- Jiao J, Peng D. Wheat microRNA1023 suppresses invasion of Fusarium graminearum via targeting and silencing FGSG_03101. J Plant Interact. 2018;13:514–521. doi: 10.1080/17429145.2018.1528512. [DOI] [Google Scholar]
- Kage U, Karre S, Kushalappa AC, McCartney C. Identification and characterization of a fusarium head blight resistance gene TaACT in wheat QTL-2DL. Plant Biotechnol J. 2017;15:447–457. doi: 10.1111/pbi.12641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyandurg PB, Sundararajan P, Dubey M, Ghadamgahi F, Zahid MA, Whisson S, Vetukuri RR (2021) Spray-induced gene silencing as a potential tool to control potato late blight disease. Phytopathology. 10.1094/PHYTO-02-21-0054-SC [DOI] [PubMed]
- Karran RA, Sanfaçon H. Tomato ringspot virus coat protein binds to ARGONAUTE 1 and suppresses the translation repression of a reporter gene. Mol Plant Microbe Interact. 2014;27:933–943. doi: 10.1094/MPMI-04-14-0099-R. [DOI] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Jin H. Role of small RNAs in host-microbe interactions. Annu Rev Phytopathol. 2010;48:225–246. doi: 10.1146/annurev-phyto-073009-114457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas A, Jr, Zhu JK, Staskawicz BJ, Jin H. A pathogen-inducible endogenous siRNA in plant immunity. Proc Natl Acad Sci U S A. 2006;103:18002–18007. doi: 10.1073/pnas.0608258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kis A, Tholt G, Ivanics M, Várallyay É, Jenes B, Havelda Z. Polycistronic artificial miRNA-mediated resistance to wheat dwarf virus in barley is highly efficient at low temperature. Mol Plant Pathol. 2016;17:427–437. doi: 10.1111/mpp.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch A, Biedenkopf D, Furch A, Weber L, Rossbach O, Abdellatef E, Linicus L, Johannsmeier J, Jelonek L, Goesmann A, Cardoza V, McMillan J, Mentzel T, Kogel K-H. An RNAi-based control of Fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS Pathog. 2016;12:e1005901. doi: 10.1371/journal.ppat.1005901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch A, Kumar N, Weber L, Keller H, Imani J, Kogel K-H. Host-induced gene silencing of cytochrome P450 lanosterol C14 alpha-demethylase-encoding genes confers strong resistance to Fusarium species. Proc Natl Acad Sci U S A. 2013;110:19324–19329. doi: 10.1073/pnas.1306373110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuze JF, Savenkov EI, Cuellar W, Li X, Valkonen JP. Viral class 1 RNase III involved in suppression of RNA silencing. J Virol. 2005;79:7227–7238. doi: 10.1128/JVI.79.11.7227-7238.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung YJ, Lin SS, Huang YL, Chen TC, Harish SS, Chua NH, Yeh SD. Multiple artificial microRNAs targeting conserved motifs of the replicase gene confer robust transgenic resistance to negative-sense single-stranded RNA plant virus. Mol Plant Pathol. 2012;13:303–317. doi: 10.1111/j.1364-3703.2011.00747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci U S A. 2004;101:12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak SY, Lew TTS, Sweeney CJ, Koman VB, Wong MH, Bohmert-Tatarev K, Snell KD, Seo JS, Chua NH, Strano MS. Chloroplast-selective gene delivery and expression in planta using chitosan-complexed single-walled carbon nanotube carriers. Nat Nanotechnol. 2019;14:447–455. doi: 10.1038/s41565-019-0375-4. [DOI] [PubMed] [Google Scholar]
- Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SW, Lagrange T, Pikaard CS, Jacobsen SE. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell. 2006;126:93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- Li F, Pignatta D, Bendix C, Brunkard JO, Cohn MM, Tung J, Sun H, Kumar P, Baker B. MicroRNA regulation of plant innate immune receptors. Proc Natl Acad Sci U S A. 2012;109:1790–1795. doi: 10.1073/pnas.1118282109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XP, Ma XC, Wang H, Zhu Y, Liu XX, Li TT, Zheng YP, Zhao JQ, Zhang JW, Huang YY, Pu M, Feng H, Fan J, Li Y, Wang WM. Osa-miR162a fine-tunes rice resistance to Magnaporthe oryzae and yield. Rice (N Y) 2020;13:38. doi: 10.1186/s12284-020-00396-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Cao XL, Zhu Y, Yang XM, Zhang KN, Xiao ZY, Wang H, Zhao JH, Zhang LL, Li GB, Zheng YP, Fan J, Wang J, Chen XQ, Wu XJ, Zhao JQ, Dong OX, Chen XW, Chern M, Wang WM. Osa-miR398b boosts H2O2 production and rice blast disease-resistance via multiple superoxide dismutases. New Phytol. 2019;222:1507–1522. doi: 10.1111/nph.15678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sun Q, Zhao T, Xiang H, Zhang X, Wu Z, Zhou C, Zhang X, Wang Y, Zhang Y, Wang X, Li D, Yu J, Dinesh-Kumar SP, Han C. Interaction between Brassica yellows virus silencing suppressor P0 and plant SKP1 facilitates stability of P0 in vivo against degradation by proteasome and autophagy pathways. New Phytol. 2019;222:1458–1473. doi: 10.1111/nph.15702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang Q, Zhang J, Wu L, Qi Y, Zhou JM. Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol. 2010;152:2222–2231. doi: 10.1104/pp.109.151803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhao SL, Li JL, Hu XH, Wang H, Cao XL, Xu YJ, Zhao ZX, Xiao ZY, Yang N, Fan J, Huang F, Wang WM. Osa-miR169 negatively regulates rice immunity against the blast fungus Magnaporthe oryzae. Front Plant Sci. 2017;8:2. doi: 10.3389/fpls.2017.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Deng YW, Ning YS, He ZH, Wang GL (2020a) Exploiting broad-spectrum disease resistance in crops: from molecular dissection to breeding. Annu Rev Plant Biol 71:575–603. 10.1146/annurev-arplant-010720-022215 [DOI] [PubMed]
- Liu J, Cheng X, Liu D, Xu W, Wise R, Shen QH. The miR9863 family regulates distinct Mla alleles in barley to attenuate NLR receptor-triggered disease resistance and cell-death signaling. PLoS Genet. 2014;10:e1004755. doi: 10.1371/journal.pgen.1004755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Shi Z, Zhang X, Wang M, Zhang L, Zheng K, Liu J, Hu X, Di C, Qian Q, He Z, Yang DL. Inducible overexpression of ideal plant Architecture1 improves both yield and disease resistance in rice. Nat Plants. 2019;5:398–400. doi: 10.1038/s41477-019-0383-2. [DOI] [PubMed] [Google Scholar]
- Liu P, Zhang X, Zhang F, Xu M, Ye Z, Wang K, Liu S, Han X, Cheng Y, Zhong K, Zhang T, Li L, Ma Y, Chen M, Chen J, Yang J (2021) A virus-derived siRNA activates plant immunity by interfering with ROS scavenging. Mol Plant. 10.1016/j.molp.2021.03.022 [DOI] [PubMed]
- Liu J, Zhang T, Jia J, Sun J. The wheat mediator subunit TaMED25 interacts with the transcription factor TaEIL1 to negatively regulate disease resistance against powdery mildew. Plant Physiol. 2016;170:1799. doi: 10.1104/pp.15.01784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Yang X, Chen J, Li T, Li Y. Osa-miR439 negatively regulates rice immunity against Magnaporthe oryzae. Ric Sci. 2021;28:156–165. doi: 10.1016/j.rsci.2021.01.005. [DOI] [Google Scholar]
- Lu S, Sun YH, Amerson H, Chiang VL. MicroRNAs in loblolly pine (Pinus taeda L.) and their association with fusiform rust gall development. Plant J. 2007;51:1077–1098. doi: 10.1111/j.1365-313X.2007.03208.x. [DOI] [PubMed] [Google Scholar]
- Ma C, Lu Y, Bai S, Zhang W, Duan X, Meng D, Wang Z, Wang A, Zhou Z, Li T. Cloning and characterization of miRNAs and their targets, including a novel miRNA-targeted NBS-LRR protein class gene in apple (Golden delicious) Mol Plant. 2014;7:218–230. doi: 10.1093/mp/sst101. [DOI] [PubMed] [Google Scholar]
- Ma S, Lapin D, Liu L, Sun Y, Song W, Zhang X, Logemann E, Yu D, Wang J, Jirschitzka J, Han Z, Schulze-Lefert P, Parker JE, Chai J. Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme. Science. 2020;370:eabe3069. doi: 10.1126/science.abe3069. [DOI] [PubMed] [Google Scholar]
- McLoughlin AG, Wytinck N, Walker PL, Girard IJ, Rashid KY, de Kievit T, Fernando WGD, Whyard S, Belmonte MF. Identification and application of exogenous dsRNA confers plant protection against Sclerotinia sclerotiorum and Botrytis cinerea. Sci Rep. 2018;8:7320. doi: 10.1038/s41598-018-25434-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melech-Bonfil S, Sessa G. Tomato MAPKKKε is a positive regulator of cell-death signaling networks associated with plant immunity. Plant J. 2010;64:379–391. doi: 10.1111/j.1365-313x.2010.04333.x. [DOI] [PubMed] [Google Scholar]
- Mitter N, Zhai Y, Bai AX, Chua K, Eid S, Constantin M, Mitchell R, Pappu HR. Evaluation and identification of candidate genes for artificial microRNA-mediated resistance to tomato spotted wilt virus. Virus Res. 2016;211:151–158. doi: 10.1016/j.virusres.2015.10.003. [DOI] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- Natarajan B, Kalsi HS, Godbole P, Malankar N, Thiagarayaselvam A, Siddappa S, Thulasiram HV, Chakrabarti SK, Banerjee AK. MiRNA160 is associated with local defense and systemic acquired resistance against Phytophthora infestans infection in potato. J Exp Bot. 2018;69:2023–2036. doi: 10.1093/jxb/ery025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- Ngou BPM, Ahn HK, Ding P, Jones JDG. Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature. 2021;592:110–115. doi: 10.1038/s41586-021-03315-7. [DOI] [PubMed] [Google Scholar]
- Niu QW, Lin SS, Reyes JL, Chen KC, Wu HW, Yeh SD, Chua NH. Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat Biotechnol. 2006;24:1420–1428. doi: 10.1038/nbt1255. [DOI] [PubMed] [Google Scholar]
- Nowara D, Gay A, Lacomme C, Shaw J, Ridout C, Douchkov D, Hensel G, Kumlehn J, Schweizer P. HIGS: host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell. 2010;22:3130–3141. doi: 10.1105/tpc.110.077040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan C, Zhang X, Jin H. Host small RNAs are big contributors to plant innate immunity. Curr Opin Plant Biol. 2009;12:465–472. doi: 10.1016/j.pbi.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O, Li CF, Costa Nunes P, Haag J, Ream T, Vitins A, Jacobsen SE, Pikaard CS. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126:79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- Prasad A, Sharma N, Muthamilarasan M, Rana S, Prasad M. Recent advances in small RNA mediated plant-virus interactions. Crit Rev Biotechnol. 2019;39:587–601. doi: 10.1080/07388551.2019.1597830. [DOI] [PubMed] [Google Scholar]
- Purkayastha A, Dasgupta I. Virus-induced gene silencing: a versatile tool for discovery of gene functions in plants. Plant Physiol Biochem. 2009;47:967–976. doi: 10.1016/j.plaphy.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Purrington CB. Costs of resistance. Curr Opin Plant Biol. 2000;3:305–308. doi: 10.1016/S1369-5266(00)00085-6. [DOI] [PubMed] [Google Scholar]
- Qiao L, Lan C, Capriotti L, Ah-Fong A, Nino Sanchez J, Hamby R, Heller J, Zhao H, Louise Glass N, Judelson HS, Mezzetti B, Niu D, Jin H (2021) Spray-induced gene silencing for disease control is dependent on the efficiency of pathogen RNA uptake. Plant Biotechnol J. 10.1094/PHYTO-02-21-0054-SC [DOI] [PMC free article] [PubMed]
- Qiao L, Zheng L, Sheng C, Zhao H, Jin H, Niu D. Rice siR109944 suppresses plant immunity to sheath blight and impacts multiple agronomic traits by affecting auxin homeostasis. Plant J. 2020;102:948–964. doi: 10.1111/tpj.14677. [DOI] [PubMed] [Google Scholar]
- Qiao Y, Liu L, Xiong Q, Flores C, Wong J, Shi J, Wang X, Liu X, Xiang Q, Jiang S, Zhang F, Wang Y, Judelson HS, Chen X, Ma W. Oomycete pathogens encode RNA silencing suppressors. Nat Genet. 2013;45:330–333. doi: 10.1038/ng.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y, Shi J, Zhai Y, Hou Y, Ma W. Phytophthora effector targets a novel component of small RNA pathway in plants to promote infection. Proc Natl Acad Sci U S A. 2015;112:5850–5855. doi: 10.1073/pnas.1421475112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan A, Jayaraman D, Grau C, Hill JH, Whitham SA, Ané JM, Smith DL, Kabbage M. The pathogenic development of Sclerotinia sclerotiorum in soybean requires specific host NADPH oxidases. Mol Plant Pathol. 2018;19:700–714. doi: 10.1111/mpp.12555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regente M, Pinedo M, Clemente HS, Balliau T, Jamet E, de la Canal L. Plant extracellular vesicles are incorporated by a fungal pathogen and inhibit its growth. J Exp Bot. 2017;68:5485–5495. doi: 10.1093/jxb/erx355. [DOI] [PubMed] [Google Scholar]
- Richard MMS, Gratias A, Meyers BC, Geffroy V. Molecular mechanisms that limit the costs of NLR-mediated resistance in plants. Mol Plant Pathol. 2018;19:2516–2523. doi: 10.1111/mpp.12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa C, Kuo YW, Wuriyanghan H, Falk BW. RNA interference mechanisms and applications in plant pathology. Annu Rev Phytopathol. 2018;56:581–610. doi: 10.1146/annurev-phyto-080417-050044. [DOI] [PubMed] [Google Scholar]
- Ruiz-Ferrer V, Voinnet O. Roles of plant small RNAs in biotic stress responses. Annu Rev Plant Biol. 2009;60:485–510. doi: 10.1146/annurev.arplant.043008.092111. [DOI] [PubMed] [Google Scholar]
- Salvador-Guirao R, Hsing YI, San Segundo B. The polycistronic miR166k-166h positively regulates rice immunity via post-transcriptional control of EIN2. Front Plant Sci. 2018;9:337. doi: 10.3389/fpls.2018.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang H, Kim J-I. Advanced strategies to control plant pathogenic fungi by host-induced gene silencing (HIGS) and spray-induced gene silencing (SIGS) Plant Biotechnol Rep. 2020;14:1–8. doi: 10.1007/s11816-019-00588-3. [DOI] [Google Scholar]
- Scofield SR, Huang L, Brandt AS, Gill BS. Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway. Plant Physiol. 2005;138:2165–2173. doi: 10.1104/pp.105.061861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad PV, Chen HM, Patel K, Bond DM, Santos BA, Baulcombe DC. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell. 2012;24:859–874. doi: 10.1105/tpc.111.095380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy D, Molnár A, Lucioli A, Szittya G, Hornyik C, Tavazza M, Burgyán J. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 2002;21:3070–3080. doi: 10.1093/emboj/cdf312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Li Y, Cao X, Qi Y. MicroRNAs and their regulatory roles in plant-environment interactions. Annu Rev Plant Biol. 2019;70:489–525. doi: 10.1146/annurev-arplant-050718-100334. [DOI] [PubMed] [Google Scholar]
- Song Y, Thomma B. Host-induced gene silencing compromises Verticillium wilt in tomato and Arabidopsis. Mol Plant Pathol. 2018;19:77–89. doi: 10.1111/mpp.12500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Dong X. How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol. 2012;12:89–100. doi: 10.1038/nri3141. [DOI] [PubMed] [Google Scholar]
- Thines M. Oomycetes. Curr Biol. 2018;28:R812–R813. doi: 10.1016/j.cub.2018.05.062. [DOI] [PubMed] [Google Scholar]
- Tian W, Hou C, Ren Z, Wang C, Zhao F, Dahlbeck D, Hu S, Zhang L, Niu Q, Li L, Staskawicz BJ, Luan S. A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature. 2019;572:131–135. doi: 10.1038/s41586-019-1413-y. [DOI] [PubMed] [Google Scholar]
- Torti S, Schlesier R, Thümmler A, Bartels D, Römer P, Koch B, Werner S, Panwar V, Kanyuka K, Wirén NV, Jones JDG, Hause G, Giritch A, Gleba Y. Transient reprogramming of crop plants for agronomic performance. Nat Plants. 2021;7:159–171. doi: 10.1038/s41477-021-00851-y. [DOI] [PubMed] [Google Scholar]
- Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- Wang H, Jiao X, Kong X, Hamera S, Wu Y, Chen X, Fang R, Yan Y. A signaling cascade from miR444 to RDR1 in rice antiviral RNA silencing pathway. Plant Physiol. 2016;170:2365–2377. doi: 10.1104/pp.15.01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Li Y, Chern M, Zhu Y, Zhang LL, Lu JH, Li XP, Dang WQ, Ma XC, Yang ZR, Yao SZ, Zhao ZX, Fan J, Huang YY, Zhang JW, Pu M, Wang J, He M, Li WT, Chen XW, Wu XJ, Li SG, Li P, Li Y, Ronald PC, Wang WM. Suppression of rice miR168 improves yield, flowering time and immunity. Nat Plants. 2021;7:129–136. doi: 10.1038/s41477-021-00852-x. [DOI] [PubMed] [Google Scholar]
- Wang J, Hu M, Wang J, Qi J, Han Z, Wang G, Qi Y, Wang HW, Zhou JM, Chai J. Reconstitution and structure of a plant NLR resistosome conferring immunity. Science. 2019;364:eaav5870. doi: 10.1126/science.aav5870. [DOI] [PubMed] [Google Scholar]
- Wang J, Wang J, Hu M, Wu S, Qi J, Wang G, Han Z, Qi Y, Gao N, Wang HW, Zhou JM, Chai J. Ligand-triggered allosteric ADP release primes a plant NLR complex. Science. 2019;364:eaav5868. doi: 10.1126/science.aav5868. [DOI] [PubMed] [Google Scholar]
- Wang M, Weiberg A, Dellota E, Jr, Yamane D, Jin H. Botrytis small RNA Bc-siR37 suppresses plant defense genes by cross-kingdom RNAi. RNA Biol. 2017;14:421–428. doi: 10.1080/15476286.2017.1291112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Weiberg A, Lin FM, Thomma BP, Huang HD, Jin H. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat Plants. 2016;2:16151. doi: 10.1038/nplants.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Galili G. Tuning the orchestra: miRNAs in plant immunity. Trends Plant Sci. 2019;24:189–191. doi: 10.1016/j.tplants.2019.01.009. [DOI] [PubMed] [Google Scholar]
- Wang Z, Xia Y, Lin S, Wang Y, Guo B, Song X, Ding S, Zheng L, Feng R, Chen S, Bao Y, Sheng C, Zhang X, Wu J, Niu D, Jin H, Zhao H (2018) Osa-miR164a targets OsNAC60 and negatively regulates rice immunity against the blast fungus Magnaporthe oryzae. Plant J:584–597. 10.1111/tpj.13972 [DOI] [PubMed]
- Weiberg A, Wang M, Bellinger M, Jin H. Small RNAs: a new paradigm in plant-microbe interactions. Annu Rev Phytopathol. 2014;52:495–516. doi: 10.1146/annurev-phyto-102313-045933. [DOI] [PubMed] [Google Scholar]
- Weiberg A, Wang M, Lin FM, Zhao H, Zhang Z, Kaloshian I, Huang HD, Jin H. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science. 2013;342:118–123. doi: 10.1126/science.1239705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner BT, Gaffar FY, Schuemann J, Biedenkopf D, Koch AM. RNA-spray-mediated silencing of Fusarium graminearum AGO and DCL genes improve barley disease resistance. Front Plant Sci. 2020;11:476. doi: 10.3389/fpls.2020.00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J, Gao L, Yang Y, Zhai J, Arikit S, Yu Y, Duan S, Chan V, Xiong Q, Yan J, Li S, Liu R, Wang Y, Tang G, Meyers BC, Chen X, Ma W. Roles of small RNAs in soybean defense against Phytophthora sojae infection. Plant J. 2014;79:928–940. doi: 10.1111/tpj.12590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Yang R, Yang Z, Yao S, Zhao S, Wang Y, Li P, Song X, Jin L, Zhou T, Lan Y, Xie L, Zhou X, Chu C. ROS accumulation and antiviral defence control by microRNA528 in rice. Nat Plants. 2017;3:16203. doi: 10.1038/nplants.2016.203. [DOI] [PubMed] [Google Scholar]
- Wu L, Zhou H, Zhang Q, Zhang J, Ni F, Liu C, Qi Y. DNA methylation mediated by a microRNA pathway. Mol Cell. 2010;38:465–475. doi: 10.1016/j.molcel.2010.03.008. [DOI] [PubMed] [Google Scholar]
- Xie S, Yu H, Li E, Wang Y, Liu J, Jiang H. Identification of miRNAs involved in Bacillus velezensis FZB42-activated induced systemic resistance in maize. Int J Mol Sci. 2019;20:5057. doi: 10.3390/ijms20205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Q, Ye W, Choi D, Wong J, Qiao Y, Tao K, Wang Y, Ma W. Phytophthora suppressor of RNA silencing 2 is a conserved RxLR effector that promotes infection in soybean and Arabidopsis thaliana. Mol Plant Microbe Interact. 2014;27:1379–1389. doi: 10.1094/MPMI-06-14-0190-R. [DOI] [PubMed] [Google Scholar]
- Xu W, Meng Y, Wise RP. Mla- and Rom1-mediated control of microRNA398 and chloroplast copper/zinc superoxide dismutase regulates cell death in response to the barley powdery mildew fungus. New Phytol. 2014;201:1396–1412. doi: 10.1111/nph.12598. [DOI] [PubMed] [Google Scholar]
- Yang LP, Fang YY, An CP, Dong L, Zhang ZH, Chen H, Xie Q, Guo HS. C2-mediated decrease in DNA methylation, accumulation of siRNAs, and increase in expression for genes involved in defense pathways in plants infected with beet severe curly top virus. Plant J. 2013;73:910–917. doi: 10.1111/tpj.12081. [DOI] [PubMed] [Google Scholar]
- Yao S, Yang Z, Yang R, Huang Y, Guo G, Kong X, Lan Y, Zhou T, Wang H, Wang W, Cao X, Wu J, Li Y. Transcriptional regulation of miR528 by OsSPL9 orchestrates antiviral response in rice. Mol Plant. 2019;12:1114–1122. doi: 10.1016/j.molp.2019.04.010. [DOI] [PubMed] [Google Scholar]
- Yi H, Richards EJ. A cluster of disease resistance genes in Arabidopsis is coordinately regulated by transcriptional activation and RNA silencing. Plant Cell. 2007;19:2929–2939. doi: 10.1105/tpc.107.051821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Jiang Z, Bi G, Nomura K, Liu M, Wang Y, Cai B, Zhou JM, He SY, Xin XF. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature. 2021;592:105–109. doi: 10.1038/s41586-021-03316-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Jeong DH, De Paoli E, Park S, Rosen BD, Li Y, Gonzalez AJ, Yan Z, Kitto SL, Grusak MA, Jackson SA, Stacey G, Cook DR, Green PJ, Sherrier DJ, Meyers BC. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011;25:2540–2553. doi: 10.1101/gad.177527.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Tan X, Li L, He Y, Hong G, Li J, Lin L, Cheng Y, Yan F, Chen J, Sun Z. Suppression of auxin signalling promotes rice susceptibility to Rice black streaked dwarf virus infection. Mol Plant Pathol. 2019;20:1093–1104. doi: 10.1111/mpp.12814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Tao Z, Hong H, Chen Z, Wu C, Li X, Xiao J, Wang S (2016a) Transposon-derived small RNA is responsible for modified function of WRKY45 locus. Nat Plants 2:16016. 10.1038/nplants.2016.16 [DOI] [PubMed]
- Zhang LL, Li Y, Zheng YP, Wang H, Yang X, Chen JF, Zhou SX, Wang LF, Li XP, Ma XC, Zhao JQ, Pu M, Feng H, Fan J, Zhang JW, Huang YY, Wang WM. Expressing a target mimic of miR156fhl-3p enhances rice blast disease resistance without yield penalty by improving SPL14 expression. Front Genet. 2020;11:327. doi: 10.3389/fgene.2020.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Luo J, Rossman AY, Aoki T, Chuma I, Crous PW, Dean R, de Vries RP, Donofrio N, Hyde KD, Lebrun MH, Talbot NJ, Tharreau D, Tosa Y, Valent B, Wang Z, Xu JR (2016b) Generic names in Magnaporthales. IMA Fungus 7:155–159. 10.5598/imafungus.2016.07.01.09 [DOI] [PMC free article] [PubMed]
- Zhang T, Zhao YL, Zhao JH, Wang S, Jin Y, Chen ZQ, Fang YY, Hua CL, Ding SW, Guo HS (2016c) Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat Plants 2:16153. 10.1038/nplants.2016.153 [DOI] [PubMed]
- Zhang W, Gao S, Zhou X, Chellappan P, Chen Z, Zhou X, Zhang X, Fromuth N, Coutino G, Coffey M, Jin H. Bacteria-responsive microRNAs regulate plant innate immunity by modulating plant hormone networks. Plant Mol Biol. 2011;75:93–105. doi: 10.1007/s11103-010-9710-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Bao Y, Shan D, Wang Z, Song X, Wang Z, Wang J, He L, Wu L, Zhang Z, Niu D, Jin H, Zhao H. Magnaporthe oryzae induces the expression of a microRNA to suppress the immune response in rice. Plant Physiol. 2018;177:352–368. doi: 10.1104/pp.17.01665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Cai C, Zhai J, Lin F, Li L, Shreve J, Thimmapuram J, Hughes TJ, Meyers BC, Ma J. Coordination of microRNAs, phasiRNAs, and NB-LRR genes in response to a plant pathogen: insights from analyses of a set of soybean Rps gene near-isogenic lines. Plant Genome. 2015;8:eplantgenome2014.2009.0044. doi: 10.3835/plantgenome2014.09.0044. [DOI] [PubMed] [Google Scholar]
- Zhao YT, Wang M, Wang ZM, Fang RX, Wang XJ, Jia YT. Dynamic and coordinated expression changes of rice small RNAs in response to Xanthomonas oryzae pv. Oryzae. J Genet Genomics. 2015;42:625–637. doi: 10.1016/j.jgg.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Zhao ZX, Feng Q, Cao XL, Zhu Y, Wang H, Chandran V, Fan J, Zhao JQ, Pu M, Li Y, Wang WM. Osa-miR167d facilitates infection of Magnaporthe oryzae in rice. J Integr Plant Biol. 2020;62:702–715. doi: 10.1111/jipb.12816. [DOI] [PubMed] [Google Scholar]
- Zhou JM, Zhang Y. Plant immunity: danger perception and signaling. Cell. 2020;181:978–989. doi: 10.1016/j.cell.2020.04.028. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Yu S, Zeng D, Liu H, Wang H, Yang Z, Xie X, Shen R, Tan J, Li H, Zhao X, Zhang Q, Chen Y, Guo J, Chen L, Liu YG. Development of “purple endosperm rice” by engineering anthocyanin biosynthesis in the endosperm with a high-efficiency transgene stacking system. Mol Plant. 2017;10:918–929. doi: 10.1016/j.molp.2017.05.008. [DOI] [PubMed] [Google Scholar]
- Zou B, Ding Y, Liu H, Hua J. Silencing of copine genes confers common wheat enhanced resistance to powdery mildew. Mol Plant Pathol. 2018;19:1343–1352. doi: 10.1111/mpp.12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.