Abstract
We previously identified two open reading frames (ORFs) of murine cytomegalovirus (MCMV), M83 and M84, which are putative homologs of the human cytomegalovirus (HCMV) UL83 tegument phosphoprotein pp65 (L. D. Cranmer, C. L. Clark, C. S. Morello, H. E. Farrell, W. D. Rawlinson, and D. H. Spector, J. Virol. 70:7929–7939, 1996). In this report, we show that unlike the M83 gene product, the M84 protein is expressed at early times in the infection and cannot be detected in the virion. To elucidate the functional differences between the two pp65 homologs in acute and latent MCMV infections, we constructed two MCMV K181 mutants in which either the M83 or M84 ORF was deleted. The resultant viruses, designated ΔM83 and ΔM84, respectively, were found to replicate in NIH 3T3 cells with kinetics identical to those of the parent strain. Western blot analysis demonstrated that except for the absence of M83 or M84 protein expression in the respective mutants, no global perturbations of protein expression were detected. When ΔM83 and ΔM84 were inoculated intraperitoneally (i.p.) into BALB/c mice, both viruses showed similar attenuated growth in the spleen, liver, and kidney. However, only ΔM83 was severely growth restricted in the salivary glands, a phenotype that was abolished upon restoration of the M83 ORF. ΔM83’s growth was similarly restricted in the salivary glands of the resistant C3H/HeN or highly sensitive 129/J strain, as well as in the lungs of all three strains following intranasal inoculation. Using a nested-PCR assay, we found that both ΔM83 and ΔM84 established latency in BALB/c mice, with slightly decreased levels of ΔM83 and ΔM84 genomic DNAs, relative to K181, observed in the salivary glands and lungs. Immunization of BALB/c mice with 105 PFU of K181, ΔM83, or ΔM84 i.p. provided similar levels of protection against lethal challenge. Although immunization with 200 PFU of ΔM83 also provided complete protection, this dose allowed both the immunizing and challenge viruses to establish latency in the spleen. Our results show that the two MCMV pp65 homologs differ in their expression kinetics, virion association, and influence on viral tropism and/or dissemination.
Studies of immunity to human cytomegalovirus (HCMV) are essential for the development of vaccines and adoptive transfer therapies for the populations most at risk for HCMV infection and disease. Although the vigorous antibody responses generated by the virus during infection may limit the spread of recurrent infection, the cell-mediated responses to the virus appear to play the dominant role in control of the acute infection and suppression of reactivation (46, 47, 52). Investigations into the viral gene products targeted by the host immune responses have demonstrated that the HCMV 65-kDa tegument phosphoprotein pp65 (UL83) is the target of strong antibody (30), cytotoxic T-lymphocyte (CTL) (9, 39, 63), and lymphoproliferative (5, 61) responses. Moreover, by limiting-dilution analysis of CTLs from seropositive individuals, it was found that the frequency of pp65-specific CTL precursors was between 1 in 12,000 and 1 in 28,000, thereby representing a significant fraction of the total HCMV-specific CTL precursor frequency, which ranges from 1 in 7,500 to 1 in 19,000 (9). Results of another such study measuring the lysis of fibroblasts infected with wild-type HCMV or a pp65 deletion mutant suggested that between 70 and 90% of HCMV-specific CTL precursors were pp65 specific (63). Although these results implicate pp65 as a major target of CTLs, its role in protective immunity following acute infection remains to be proven.
The function of pp65 in HCMV replication is not known. The pp65 phosphoprotein is an especially abundant component of dense bodies, but it is also found in the tegument of infectious virions and noninfectious enveloped particles (2). A unique bipartite nuclear localization signal causes virion-associated pp65 to be rapidly translocated to the nucleus upon infection (17, 55), suggesting that this protein may play a role in very early events in gene regulation. However, no transcriptional activity has been demonstrated for this protein. A kinase activity has been found associated with pp65 in several studies (10, 56), and although Polo-like kinase 1 was recently found to bind to pp65 in infected cells (18), pp65 itself has not been definitively shown to be a kinase.
An HCMV deletion mutant lacking the UL83 open reading frame (ORF) encoding pp65 has been successfully generated by Schmolke and coworkers (56). This UL83 mutant, designated RVAd65, shows wild-type growth in cultured fibroblasts, demonstrating that pp65 is not required for viral replication in cell culture. Maturation of mutant virions in infected human foreskin fibroblasts appeared to be delayed compared to the parent virus when examined at 6 days postinfection (p.i.), though both viruses found in the supernatant were equally infectious. When the virion-associated kinase activities of the parent and mutant viruses were examined in vitro, the lower-molecular-weight virion proteins of RVAd65 were found to be underphosphorylated compared to those of the wild-type virus.
Experiments examining HCMV antigen presentation supported the existence of a pp65-associated kinase activity and, more notably, suggested a novel role for pp65 in immune evasion (20). When primary fibroblasts were infected with HCMV AD169 or RVAd65 and subjected to in vitro lysis by immediate-early 1 (IE1)-specific CTL clones, only the cells infected with RVAd65 were specifically lysed. The data indicated that increased phosphorylation of IE1 threonine residues occurred in the presence of pp65 and that this was responsible for the lack of presentation of IE1. These results describe an immune evasion mechanism distinct from several others described for HCMV (for a review, see reference 26), and it will be important to determine whether this mechanism of CTL evasion occurs in vivo.
Because of the limits of examining HCMV pathogenesis in humans, the murine cytomegalovirus (MCMV) model of infection has provided an experimental model for pathogenesis and immunity studies (28). Due to the immunodominance of pp65 in HCMV infection, our laboratory has previously identified and described the homologs of pp65 in MCMV (13). We found that both M83, the positional homolog of UL83 in the MCMV genome, and the adjacent M84 ORF exhibit homology to UL83, with the deduced amino acid sequence of M84 showing slightly stronger UL83 homology. However, M84 also exhibits significant amino acid homology to its positional homolog, UL84, a nonstructural protein possibly involved in negative regulation of the IE2 transactivator (19) and implicated in promoting viral DNA replication (54). We also demonstrated that like pp65, the M83 protein is a late, virion-associated protein that is the target of humoral responses during the infection.
In this study, we began to assess the roles of M83 and M84 during MCMV infection in order to provide some insights into the role of HCMV pp65. We constructed two MCMV mutants in which one of these ORFs was deleted and replaced with a selectable marker cassette. The viruses in which M83 and M84 were deleted, designated ΔM83 and ΔM84, respectively, were found to grow in cultured fibroblasts with kinetics equivalent to those of the wild-type virus, demonstrating that each of these genes is dispensable for growth in culture. However, when either virus was inoculated into three inbred mouse strains by either the intraperitoneal (i.p.) or intranasal (i.n.) route, its replication was found to be attenuated. ΔM83, in particular, was severely restricted for growth in the salivary glands and lungs. This phenotype was reversed upon restoration of M83 expression. Both mutants were found to establish latency in the spleen, salivary glands, and lungs of BALB/c mice, where latency is defined as the presence of viral DNA detectable by PCR in the absence of detectable infectious virus. We also examined the immunity generated against the mutant viruses by using them to immunize BALB/c mice prior to lethal challenge with the virulent MCMV strain K181. We found that responses to both mutant viruses provided protection, equal to that of the parent strain, against subsequent lethal challenge. An i.p. dose of 200 PFU of ΔM83 was fully protective against replication of the challenge virus in the spleen and salivary glands, but this dose was still sufficient to allow the immunizing ΔM83 virus as well as the challenge virus to establish latency.
MATERIALS AND METHODS
Cell culture and virus preparation.
NIH 3T3 cells (ATCC CRL 1658) were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (vol/vol) heat-inactivated calf serum (CS) and (per milliliter) 0.29 mg of l-glutamine, 200 U of penicillin, 0.2 mg of streptomycin, 0.05 mg of gentamicin, and 1.5 μg of amphotericin B (DMEM + 10% CS). Mouse embryonic fibroblasts (MEFs) were grown in the above-described medium with 10% heat-inactivated fetal bovine serum being substituted for the CS. The preparation of salivary gland-derived MCMV strain K181 and tissue culture-derived MCMV has been previously described (13, 15). Virus was stored at −80°C until use. For analysis of virion proteins by Western blotting, MCMV was purified by density gradient centrifugation of tissue culture supernatants as previously described (12).
Mice, in vivo infections, organ harvests, and MCMV plaque assay.
Female BALB/c (H-2d) mice were obtained from Harlan Sprague Dawley, Inc., The Jackson Laboratory, or Simonsen Laboratories, Inc., at 5 to 6 weeks of age. Female C3H/HeN (H-2k) mice were obtained from Simonsen Laboratories at 5 to 6 weeks of age. Female 129/J (H-2b) mice were obtained from The Jackson Laboratory at 6 to 9 weeks of age. Mice were housed in microisolator-covered cages in a vivarium (University of California, San Diego) and given food and water ad libitum.
For i.p. infections, either salivary gland- or tissue culture-derived MCMV K181 was diluted in phosphate-buffered saline (PBS) such that 0.5 ml of virus was injected. For i.n. inoculations, mice were lightly anesthetized with Metofane (methoxyflurane) before 50 μl of tissue culture-derived virus diluted in DMEM+10% CS was instilled into the nares. On various days after infection, mice were sacrificed and organs were removed, homogenized in a Dounce homogenizer, and stored for MCMV titer determination as previously described (21).
Plasmid constructions.
Restriction endonucleases, T4 DNA ligase, T4 DNA polymerase, calf intestinal alkaline phosphatase (CIP), and the Klenow fragment of DNA polymerase were purchased from BRL Life Technologies, Inc. (Bethesda, Md.). Phosphorylated oligonucleotide linkers and Escherichia coli SCS110 competent cells were purchased from Stratagene (La Jolla, Calif.). DNA fragments were gel purified by using either GeneClean (Bio101) or Ultrafree-MC 0.45-μm centrifugal filter devices (Millipore). The subcloning methods employed below were carried out as described by Sambrook et al. (53). Unless otherwise specified, plasmids were propagated in Escherichia coli DH5α and purified by anion-exchange chromatography, using Qiagen Maxiprep or Miniprep kits. Construction of the lacZ/gpt plasmid pON855 was previously described (62).
The enhanced green fluorescent protein–puromycin-N-acetyltransferase resistance (EGFP-puro) fusion construct was constructed by combining the pPUR vector (Clontech, Palo Alto, Calif.) with the pEGFP-C1 C-terminal protein fusion vector (Clontech) in a fashion similar to that recommended by M. Prichard and G. Pari (45a). Specifically, both vectors were first prepared from E. coli SCS110 so that Dam-sensitive sites could be cleaved. The unmethylated pPUR DNA was digested with BsiWI and XbaI, and the 660-bp fragment containing the pPUR ORF from the BsiWI site 62 bp downstream of the AUG codon to the XbaI site downstream of the stop codon was gel purified. To provide the sequences containing the HCMV immediate-early (IE) promoter-enhancer driving the EGFP ORF, pEGFP-C1 was cleaved with BglII and XbaI and the 4.7-kbp vector fragment was gel purified. The BglII end of this fragment is located in the multiple cloning site at the 3′ end of the EGFP ORF and allows the addition of C-terminal fusions. To regenerate the 5′ 62 bp of the puro ORF and allow ligation of this ORF to the 3′ end of EGFP, a double-stranded, synthetic adapter was synthesized (Integrated DNA Technologies, Inc., Coralville, Iowa). This adapter was made by annealing two 5′-phosphorylated deoxyribonucleotides (sense, 5′-GAT CTA TGA CCG AGT ACA AGC CCA CGG TGC GCC TCG CCA CCC GCG ACG ACG TCC CCC GGG CC-3′; antisense, 5′-GTA CGG CCC GGG GGA CGT CGT CGC GGG TGG CGA GGC GCA CCG TGG GCT TGT ACT CGG TCA TA-3′) to provide a BglII site at the 5′ end, the first 62 bp of the puro ORF, and a BsiWI site at the 3′ end which connects to the remainder of the puro ORF. In a triple ligation, the XbaI-BglII EGFP vector was ligated to the BglII-to-BsiWI puro ORF 5′ adapter and the BsiWI-XbaI puro ORF 3′ fragment. This ligation product was used to transform SCS110 cells, and resulting transformants were screened by restriction digestion for the presence of the adapter and the puro 3′ fragment. Positive clones were sequenced by the dideoxy method (Sequenase) across both ligation joints, and the plasmid pEGFP.C1/puro was found to contain the puro ORF in frame with the EGFP ORF. This plasmid was cleaved with AseI and MluI and blunted with the Klenow fragment, and BamHI linkers were ligated to the blunt ends. The 2.3-kbp fragment containing the HCMV IE promoter, the EGFP-puro ORF, and the simian virus 40 polyadenylation site (derived from pEGFP-C1) was gel purified and then ligated to BamHI-digested pGEM-3zf(+) vector to yield pGEM-EGFP/puro.
To construct ΔM83, a deletion-substitution plasmid was generated by flanking the lacZ/gpt cassette with 0.8 kbp of MCMV genome sequence from the M84 region and 1.15 kbp of M82-containing sequence. The vector M83(Stu-Xho)-pBS contains the entire M83 ORF on a 4.35-kbp XhoI (nucleotide 120962)-to-StuI (nucleotide 116614) fragment subcloned into the XhoI and EcoRV sites of pBluescript II KS(+) (Stratagene, La Jolla, Calif.), with restriction site numbers in parentheses indicating the nucleotide number from the published complete DNA sequence of MCMV Smith strain (GenBank accession no. U68299) (48). This vector was digested with PstI to release the M83 ORF-containing sequence from 14 bp upstream of the 3′ end of M84 to 260 bp upstream from the 3′ end of the M83 ORF and then recircularized to yield pM83-5′. The single NotI and SstI sites in the pBluescript II multiple cloning site of pM83-5′ were cleaved, and a 1.15-kbp NotI (117590)-to-SstI (116438) genomic fragment, derived from the plasmid H3C(RV)-Gem (13) (which contains the M82 ORF on a 3-kbp EcoRV fragment), was directionally ligated in to generate pM83-flank. pM83-flank was digested with NotI to linearize the vector in between flanking regions. The lacZ/gpt cassette was released from pON855 by BamHI digestion, the ends were filled in with the Klenow fragment, and phosphorylated NotI linkers were ligated to the filled-in ends. After linker addition and NotI digestion, the 4.8-kbp lacZ/gpt NotI fragment was agarose gel purified and ligated to the NotI-digested pM83-flank vector. Resulting transformants were analyzed by restriction enzyme analysis, and the final vector, pΔM83KO8, was selected because the transcription of the gpt gene proceeded in the same direction as that of M84 and M82. Prior to electroporation, pΔM83KO8 was linearized with StuI and VspI to release vector sequences and facilitate recombination.
To generate ΔM83-2, an M83 deletion mutant with a reconstructed 3′ end of the M84 ORF, an oligonucleotide adapter was constructed by annealing the 28-mer M84-3′ sense (5′-GCA GAA CAT CTG ATA GAA TAA AGC TTG C-3′) and the 36-mer M84-3′ antisense (5′-GGC CGC AAG CTT TAT TCT ATC AGA TGT TCT GCT GCA-3′) oligonucleotides. This adapter contains the sequence encoding the carboxy-terminal 5 amino acids of M84 followed by two in-frame termination codons and a polyadenylation signal. This adapter was then ligated to NotI- and PstI-digested pM83-flank, and dideoxy sequencing confirmed that the 3′ coding sequence for M84 was regenerated in the resulting clone, pΔM83+link. The NotI-ended lacZ/gpt cassette described above was then ligated to NotI-digested pΔM83+link, and a resulting clone, pΔM83-2, was chosen because gpt, M84, and M82 transcription proceeded in the same direction. Prior to virus generation, pΔM83-2 was digested as described above for pΔM83KO8.
To generate the recombination construct for ΔM84, the HindIII C fragment clone of MCMV (40) was used to isolate M84 flanking sequence on the M85 side of the ORF. The HindIII C clone was digested with NarI and EcoRI, blunt ended with the Klenow fragment, and ligated to phosphorylated PstI linkers. After overnight ligation, the NarI fragments of HindIII C were digested with PstI and SphI and a 1.07-kbp SphI (122951)-to-PstI (NarI at site 121876) fragment was gel purified. The plasmid pM84-5′ was generated by ligating this fragment to SphI- and PstI-digested pGEM-1 (Promega). To isolate M84 flanking sequence from the M83 region of the genome, M83(Stu-Xho)-pBS was cut with SmaI and a 4.75-kbp SmaI fragment was gel purified and blunt-end ligated with EcoRI linkers. The linked plasmid was digested with EcoRI and PstI, and a 0.99-kbp PstI (120098)-to-EcoRI (SmaI at site 119105) fragment was gel purified and ligated to PstI- and EcoRI-cut pGEM-1 to yield pM84-3′. The inserts from pM84-5′ and pM84-3′ were released by digestion with the appropriate enzymes, gel purified, and triple ligated with SphI- and EcoRI-digested pGEM-4Z (Promega) to yield pM84-5′,3′. The lacZ/gpt cassette, which had PstI linkers ligated onto Klenow fragment-filled BamHI ends, was ligated into PstI-digested pM84-5′,3′ to yield pΔM84KO2. The pΔM84KO2 clone was selected by virtue of gpt transcription proceeding in the same direction as that of M83 and M85. To restore the TATA box of M83 into this vector, the vector pGEM-iM84/2 (13) was digested with ApaI and blunt ended with T4 polymerase, and PstI linkers were ligated to the blunted ends. After digestion with PstI, a 473-bp PstI fragment from ApaI (120571) to PstI (120098) was gel purified. The pΔM84KO2 vector was partially digested with PstI (1), and singly cut plasmid (9.62 kbp) was gel purified and CIP treated. The 473-bp TATA-containing PstI fragment was ligated into the CIP-treated, singly PstI-cut pΔM84KO2, and the ligation mixture was transformed into E. coli. Recombinant plasmids were screened by EcoRI and NarI digestion and agarose gel electrophoresis, and a clone which contained the TATA fragment in the correct PstI site and in the correct orientation was selected. This plasmid, pΔM84KOT-5, was digested with BamHI and HindIII to remove vector sequences prior to electroporation.
To construct rΔM83—the rescued ΔM83 virus—a plasmid containing a rescue cassette was constructed such that the EGFP-puro cassette and M83 ORF could be inserted into the ΔM83 genome to yield a puromycin-selectable intermediate virus with the lacZ/gpt and EGFP-puro cassettes flanked by M84 sequences. To construct the rescue cassette vector, pON855 was digested with SstI and BamHI and the 3.3-kbp SstI-BamHI gpt-containing fragment gene was gel purified. The EGFP-puro cassette was released from pGEM-EGFP/puro by BamHI digestion, and the 2.3-kbp insert was gel purified. The EGFP-puro BamHI fragment and the SstI-BamHI fragment of gpt were triple ligated to SstI- and BamHI-digested pSP72 (Promega). Because of problems encountered with subcloning gpt sequences in E. coli strains such as DH5α and XL-1 Blue, the WB-1 (gpt-negative) strain (57) was used for subcloning and propagating gpt-containing plasmids. WB-1 cells were electroporated with the triple-ligation mixture, using a BTX ECM-600 electroporator in accordance with the manufacturer’s recommendations, and transformants were grown on M9 minimal medium agar plates (1) supplemented with 100 μg of ampicillin and 100 μg of xanthine per ml. A clone, pSP-gpt/EGFP, containing both gpt and EGFP-puro genes in the same orientation was identified by restriction digestion. pSP-gpt/EGFP was amplified in M9 minimal medium supplemented as described above, and Maxiprep plasmid DNA was digested with HindIII and then CIP treated. To isolate MCMV genome sequence containing M83 and flanking ORFs, the MCMV sequences of pM83X5.5 were used. The plasmid pM83X5.5 was constructed by digesting the MCMV HindIII C plasmid with XhoI, HindIII, and DraI, gel purifying the 5.5-kbp XhoI (115429-to-120962) fragment, and ligating it to XhoI-digested, CIP-treated pGEM-7Zf(+) (Promega). pM83X5.5 was digested with XhoI and blunt ended with the Klenow fragment, and HindIII linkers were ligated to the ends. After HindIII digestion, the 5.5-kbp HindIII-linked M83 sequence was gel purified and ligated to HindIII-digested pSP-gpt/EGFP. The final rescue clone, pM83Res13.6, was selected after restriction digestion analysis showed that gpt, EGFP-puro, and the MCMV M84 and M82 ORFs were transcribed in the same direction. Before electroporation, pM83Res13.6 was digested with SstI to release vector sequences.
For the generation of a specific antiserum to M32 protein, the 5′ end of the ORF was reconstructed to allow in-frame fusion with the glutathione S-transferase (GST) ORF in pGEX-KG. The 5′ 222 bp of the M32 ORF was amplified by PCR, in the process placing a BamHI site just 5′ of the initiating methionine codon. The template for amplification was the plasmid pBS-4.2 H3B(rev), which contains the entire M32 ORF on a 4.3-kbp SstI fragment (nucleotides 38900 to 43192) in pBluescript II KS(+), and the primers were 5′-M32 (5′-GCG CGG ATC CAT GTC CGC TCG AGG GCG CGC-3′) and 3′-M32 (5′-GAG CTT CTC GTG GTA CCT GAG CCA GAG GAC-3′) (Integrated DNA Technologies, Inc.). Standard PCR mixtures, supplemented with 2 mM MgCl2, were prepared with and without template DNA, using materials supplied with the GeneAmp PCR reagent kit (Perkin-Elmer) and following the manufacturer’s recommendations. Thirty cycles of PCR were carried out (1 min at 94°C, 1 min at 55°C, and 2 min at 72°C), followed by a 10-min extension at 72°C. An approximately 250-bp product synthesized only in the presence of template DNA was cut with BamHI and KpnI, isolated, and ligated to BamHI- and KpnI-cut pGEM-4Z (Promega), yielding pGEM-5′M32. Both strands of the PCR product were sequenced. A single transition mutation (G to A) was found at a position 222 nucleotides from the first nucleotide of the initiating methionine codon, resulting in a change in codon 74 of the ORF from valine to isoleucine. This change was judged to be irrelevant, and pGEM-5′M32 was cleaved with BamHI and EcoRI and the ∼250-bp fragment was isolated and ligated to BamHI- and EcoRI-cut pGEX-KG, yielding pGEX-5′M32. A 1.8-kbp KpnI-HindIII fragment of pBS-4.2 H3B(rev) was isolated and ligated to KpnI- and HindIII-cleaved pGEX-5′M32, yielding pGEX-M32.
For production of the M84 protein as a GST fusion, the 2-kbp BamHI-EcoRI fragment of pGEM-M84 (13) was subcloned into BamHI- and EcoRI-cut pGEX-KG to yield pGEX-M84.
Electroporation and mutant MCMV selection.
All mutant MCMVs were generated in NIH 3T3 cells by homologous recombination between linearized plasmids containing selectable markers, and the K181 genome was introduced by infection as described by Vieira and colleagues (62). To generate the ΔM83, M83-2, and ΔM84 viruses, 30 μg of linearized plasmid was electroporated into 4 × 106 NIH 3T3 cells by using a Gene Pulser II apparatus (Bio-Rad). DNA and cells were added in 0.4 ml to a 4-mm-gap cuvette and pulsed with 0.22 kV and 975 μF, using the measure capacitance function. After being pulsed, cells were seeded in 10 ml of DMEM + 10% CS in a 10-cm-diameter tissue culture dish and incubated overnight. The following day, the electroporated cells were infected with MCMV K181 at a multiplicity of infection (MOI) of 3 for 6 h, the inoculum was removed, and 10 ml of fresh medium was added. Three days p.i., the supernatant from the infected cells was harvested and clarified by low-speed centrifugation, and 50 to 500 μl of the clarified supernatant was used to infect 75% confluent NIH 3T3 cells in a T-75 flask. At 3 to 4 h p.i., virus was selected as described elsewhere (62), except that 150 μg of xanthine per ml was used. Recombinant viruses were propagated two to three times under gpt selection conditions and then subjected to three rounds of limiting dilution and plaque purification until virus homogeneity was achieved.
To generate rΔM83, SstI-digested pM83Res13.6 was electroporated into NIH 3T3 cells, using a BTX ECM-600 electroporator (Genetronics, Inc.) in accordance with the manufacturer’s protocol for C3H fibroblasts (protocol no. PR038). In brief, two identical electroporations were carried out with 3.2 × 106 NIH 3T3 cells in 0.8 ml of DMEM (without CS). Cells and 30 μg of DNA were added to 4-mm-gap cuvettes, the suspensions were mixed, and each was pulsed at 300 V, 72 Ω, and 2,500 μF. After both cuvettes were electroporated, the cells were combined with 4 ml of DMEM+10% CS, seeded into a 60-mm-diameter dish, and incubated at 37°C and 10% CO2 overnight. The following day, the electroporated cells were infected with ΔM83 at an MOI of 3.5 for 4 h, the inoculum was removed, and 5 ml of fresh medium was added. At 3 days p.i., the supernatant was removed and clarified, and 50 or 100 μl of the clarified supernatant was used to infect 75% confluent NIH 3T3 cells in a 60-mm-diameter dish for 4 h. The following day, the medium was replaced with 4 ml of DMEM+10% CS containing 5 μg of puromycin (Clontech) per ml. At 3 days p.i., 0.5 ml of cleared supernatant from the first selection was used to infect another 60-mm-diameter dish of NIH 3T3 cells for the second selection. After three rounds of puromycin selection, plaques resulting from infection of cells without puromycin were observed under fluorescence and EGFP-positive plaques were picked and used to infect fresh monolayers. Resulting plaques were stained by overlaying with DMEM+2% CS supplemented with 0.5% agarose and 0.3 mg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) per ml. After X-Gal staining for 5 h, clear plaques were picked and subjected to two to three rounds of limiting dilution and plaque purification until only EGFP-negative, lacZ-negative virus remained.
Production and purification of antisera.
A rabbit antiserum against GST-M84 purified from E. coli inclusion bodies was generated in a naive seronegative male New Zealand White rabbit by previously described methods (13). The resultant GST-M84 antiserum was subjected to caprylic acid precipitation and ammonium sulfate precipitation as described by Harlow and Lane (23). To further purify M84-specific antibodies from the serum, portions of serum were affinity purified with nitrocellulose membrane-immobilized M84 produced by COS-7 cells transiently expressing pcDNA3-M84.
To generate immune rabbit serum against GST-M32, a naive female New Zealand White rabbit that was seronegative for GST-M32 was immunized with a GST-M32 fusion protein derived from bacterial inclusion bodies as for GST-M84. The resulting GST-M32 antiserum was prepared and adsorbed to an E. coli DH5α-derived acetone powder as previously described (13).
To produce a pp89-specific antiserum, BALB/c mice were intradermally immunized three times in 2 weeks with 30 μg of a plasmid DNA vaccine vector (pcDNA3-pp89) expressing the full-length pp89 cDNA from the HCMV IE promoter-enhancer (21). Ten weeks following the first immunization, mice were bled and serum fractions were prepared and stored at −20°C until use.
Southern and Western blot analyses.
Genomic DNA from NIH 3T3 cells infected with wild-type or plaque-purified recombinant viruses was prepared by using a commercial kit (Qiagen Blood Kit) in accordance with the manufacturer’s recommendations. Restriction digestion and Southern blot analysis were performed on these DNAs by standard procedures (53). MCMV genomic probes were isolated from restriction enzyme-digested HindIII C plasmid (40), 32P labeled by random priming (Prime-It II; Stratagene), purified by Sephadex G-50 chromatography, and hybridized to UV-cross-linked DNA blots in Rapid-Hyb buffer (Amersham) according to the manufacturer’s recommendations.
For Western blot analysis of viral protein expression, NIH 3T3 cells were infected at an MOI of 3 to 3.5 with the various mutants, and at various times p.i., cells were collected and solubilized at 42°C for 10 min in Laemmli sample buffer. Solubilization at this temperature was critical for prevention of aggregation of the M83 and M84 proteins. Proteins were separated by electrophoresis on sodium dodecyl sulfate (SDS)–7.5% acrylamide gels, electroblotted to nitrocellulose, and subjected to Western analysis as previously described (13). M83 (13) and M32 proteins were detected with their respective rabbit polyclonal antisera (diluted 1:2,000). M84 was detected with the rabbit anti-GST-M84 antiserum (affinity purified as described above), and pp89 was detected with a pcDNA3-pp89-immunized mouse serum (diluted 1:1,000). Bound antibodies were detected by horseradish peroxidase-linked anti-rabbit- or anti-mouse immunoglobulin G whole antibodies (Amersham) and enhanced chemiluminescence (SuperSignal Substrate; Pierce Chemical Co.).
To examine the temporal expression of the M84 protein, MCMV-infected NIH 3T3 cells were also subjected to treatment with cycloheximide (CHX) or phosphonoacetic acid (PAA) as described previously (12). Briefly, cells were infected in the presence of CHX and at 8 h p.i. were washed three times with PBS, fed with medium without CHX, and incubated until being harvested at 12 h p.i. PAA-treated cells were infected and incubated in the presence of drug until being harvested at 48 h p.i.
Growth kinetics of MCMV in cell culture.
For single-cycle growth analysis, triplicate cultures of NIH 3T3 cells in 24-well dishes were infected at an MOI of 3 with tissue culture-derived MCMV isolates. After adsorption, cells were washed twice with PBS and then fed with DMEM + 10% CS. On each day p.i., supernatants were harvested, cleared of cellular debris as described above, and stored at −80°C in 1% (vol/vol) dimethyl sulfoxide. For multicycle growth analysis, triplicate cultures were infected at an MOI of 0.05 and supernatants were harvested and stored as described above. Viral titers for entire growth experiments were determined together in plaque assays on NIH 3T3 cells as described above.
PCR detection of latent MCMV DNA.
BALB/c mice were infected i.p. with tissue culture-derived wild-type or mutant MCMV and then housed for 5 to 10 months to allow the resolution of the acute infection. To confirm the absence of persistent infectious virus, portions of the spleens of these mice were homogenized, sonicated, and diluted prior to incubation with MEFs or NIH 3T3 cells as previously described (44). Cells were observed for cytopathic effects (CPE) for 10 to 14 days, and the establishment of latency was defined as the absence of infectious virus concomitant with the presence of MCMV DNA sequences as determined by PCR. To detect latent MCMV genomes, DNA was extracted from the tissues of uninfected or latently infected mice by using a commercial kit (Qiagen Tissue Kit) according to the manufacturer’s instructions for preparation of RNA-free DNA. To prevent cross-contamination of viral DNA, dissections were performed with autoclaved instruments and in a BioGuard hood not used for the handling of MCMV or acutely infected mice. In addition, the groups of mice were sacrificed on consecutive days. Spleen DNA was extracted from only stromal fractions prepared by sedimentation as previously described (41), whereas lung and salivary gland DNAs were prepared from minced whole tissues. DNAs were quantified by measurement of the optical density at 260 nm, and agarose gel electrophoresis was used to confirm DNA concentrations and integrity.
MCMV DNA was detected by nested PCR of the IE1 region as described by Koffron et al. (32). In the first reaction, serial dilutions containing either 1, 0.1, or 0.01 μg of tissue DNA were amplified in 50-μl reaction volumes containing 1× PCR Buffer II (Perkin-Elmer), 3 mM MgCl2, 40 pmol each of the SY1 and SY2 primers (32), and 200 μM each deoxynucleoside triphosphate. The manual hot-start technique was employed by withholding the 0.25 U of AmpliTaq (Perkin-Elmer) from the reaction until the thermocycler (PTC100; MJ Research) reached 85°C. Following an initial denaturation step of 96°C for 3 min, reaction mixtures underwent 20 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s followed by a final extension of 72°C for 8 min. This reaction amplifies a 549-bp product that spans introns 2 and 3 of IE1. One microliter of this reaction mixture was reamplified for 30 cycles in a second 50-μl reaction volume containing standard buffer conditions, primers CH16 and CH17 (27), and 2.5 mM MgCl2, using the hot-start method and the same cycling parameters as were employed for the first reaction. This second reaction amplifies a 310-bp region of the first reaction product. Five microliters of the reaction 2 product was analyzed by agarose gel electrophoresis on 2% agarose (three parts NuSieve agarose [FMC BioProducts] and one part ultrapure agarose [GIBCO Life Sciences])-Tris-borate-EDTA (TBE) gels and ethidium bromide staining. Images were recorded by UV illumination and by using a Gel-Doc imaging system (Bio-Rad Laboratories).
To serve as a control for sensitivity of the nested PCR, the MCMV HindIII L plasmid (40) was digested with HindIII, phenol-chloroform extracted, and ethanol precipitated, and the MCMV genomic insert was quantified by agarose gel electrophoresis. Serial dilutions of the plasmid DNA were made first in elution buffer AE (Qiagen) and then in AE containing uninfected-mouse genomic DNA such that 5 μl, containing 25 to 0.2 copies of the MCMV HindIII L insert and 1 μg of mouse DNA (from the same type of tissue as that under analysis), was added to each reaction. Optimization trials showed that the sensitivity of the assay was always about the same regardless of the tissue or the mass of genomic DNA (0.01, 0.1, or 1 μg) (data not shown) added. Negative controls contained the appropriate tissue DNA from uninfected mice.
To specifically detect the ΔM83 genome, primers homologous to the gpt-selectable marker were designed for nested-PCR amplification. The first reaction amplified a 599-bp fragment of gpt, using the primers gpt-1S (5′-ACAGGCTGGGACACTTCACA-3′) and gpt-1A (5′-CAGGGTTTCGCTCAGGTTTG-3′), and the second reaction used primers gpt-2S (5′-TGGCGCGTGAACTGGGTATT-3′) and gpt-2A (5′-TCCCACGGCTGTTCAATCCA-3′) to amplify a 283-bp region of the first reaction product. Amplification reactions and cycling conditions were as described for IE1 (see above), except that the MgCl2 concentration in both reactions was 1.5 mM and the annealing temperature for the first reaction was 58°C. The sensitivity of the nested reaction was monitored by amplification of a linearized gpt-containing plasmid in genomic DNA from uninfected mice as described above.
To confirm that the genomic DNAs were undegraded and free of PCR inhibitors, β-actin sequences were amplified from the same DNA dilutions that were negative for MCMV DNA. A single 50-μl reaction mixture containing primers BA-1 and BA-2 (27), 2.5 mM MgCl2, and the composition described above was amplified by the hot-start procedure followed by 35 to 40 cycles of the conditions described above for IE1.
RESULTS
M84 protein is expressed at early times p.i. and is not detectable in the virion.
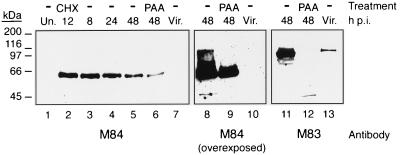
We previously showed by Northern blot analysis that the M83 gene product was likely encoded by a 5-kb mRNA that was detectable at 24 to 48 h p.i. and that M83 protein was expressed with late gene kinetics and phosphorylated in vivo (13). We also showed that the M84 gene product was likely encoded by a 6.9-kb transcript that was directed by either of the TATA boxes located just 5′ of the M84 ORF (13). This transcript was detectable at 8 h p.i., suggesting that it belonged to the early class of genes. To determine the kinetics of M84 protein expression, we used Western blot analysis and a rabbit polyclonal antiserum to GST-M84 to examine whole-cell lysates prepared from NIH 3T3 cells infected with MCMV at a high MOI and harvested at 8, 24, and 48 h p.i. We found that solubilization of cell lysates at 100°C in Laemmli buffer resulted in aggregation of more than 99% of the M84 protein into an insoluble mass (data not shown). The M83 protein was also found to aggregate somewhat upon boiling, resulting in M83-immunoreactive species migrating to positions corresponding to 125, 105, and 70 kDa and often to greater than 200 kDa (13). However, when lysates were solubilized at 42°C, single immunoreactive 105- and 65-kDa species were detected with the M83- and M84-specific antisera, respectively (Fig. 1).
FIG. 1.
Kinetics of M84 protein expression and absence of virion association. NIH 3T3 cells were infected with K181 at an MOI of 3, and at 8, 24, and 48 h p.i., cells were harvested and whole-cell lysates were prepared as described in Materials and Methods. Cells were also treated for the first 8 h of infection with CHX before the 12-h-p.i. harvest or were infected and incubated in the presence of PAA until the 48-h-p.i. harvest. Western blots of lysates and purified virions (Vir.) were probed with an affinity-purified rabbit antiserum to GST-M84 or a GST-M83-specific antiserum. Each panel depicts the same blot, and lanes are numbered at the bottom. Lanes 8 to 10 depict lanes 5 to 7 after prolonged exposure, and lanes 11 to 13 show lanes 5 to 7 after the blot was stripped and reprobed with the M83 antiserum. Un., uninfected. Positions of molecular mass markers are shown on the left.
As seen in Fig. 1, a high level of M84 protein was detected at 8 h p.i., with a subsequent steady decline over the 48-h infection period observed. Infection for 8 h in the presence of CHX and a subsequent 4-h release from the drug resulted in an M84 protein expression level (lane 2) at least as high as the level found at 8 h p.i. without the drug (lane 3). When cells were infected in the presence of PAA, which limits expression to IE and early proteins, M84 protein was detectable at a level approximately 50% of that seen at 48 h p.i. in the absence of the drug (lane 6).
To determine whether M84 was associated with virions, we purified virions from the tissue culture supernatant of MCMV-infected NIH 3T3 cells by density gradient centrifugation and analyzed the associated proteins by Western blotting with the anti-GST-M84 antiserum as a probe. Figure 1 shows that no M84-specific protein was detected in the virions (lane 7), even when the blot was overexposed (lane 10) or analyzed with more-sensitive detection reagents, such as SuperSignal Blaze (Pierce Chemical Co.) (data not shown). As a control, we documented that M83 protein was readily detected on the same blot with an M83-specific rabbit antiserum (lane 13). The results presented here, coupled with those described previously (13), show that although both M83 and M84 exhibit significant amino acid homology to the HCMV pp65 matrix protein (UL83), their properties are quite different. The M83 protein is expressed at late times in the infection cycle and is incorporated into the virion, while M84 is a nonstructural protein expressed at early times p.i.
Construction of MCMV deletion mutants ΔM83 and ΔM84 and rescued virus rΔM83.
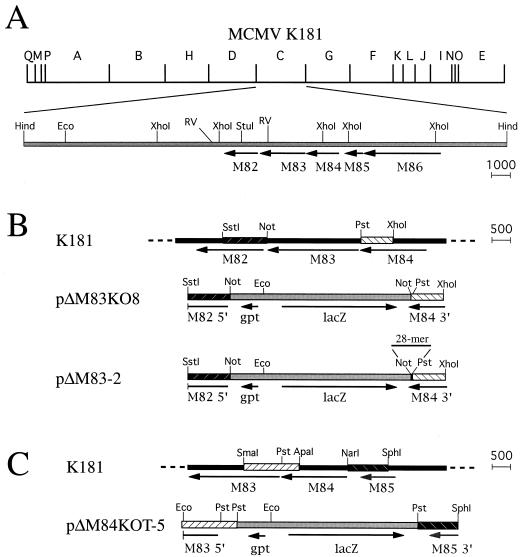
To begin characterizing the functions of M83 and M84 in vivo, single-deletion MCMV mutants were constructed. Insertional mutagenesis of the MCMV HindIII C region (Fig. 2A) was performed by replacing the M83 or M84 ORF with the lacZ/gpt cassette (Fig. 2B and C, respectively). As described in detail in Materials and Methods, recombinant plasmids were constructed such that the marker cassette was flanked by 0.9 to 1.4 kbp of genomic sequence surrounding the region of the genome to be deleted. Following restriction digestion, these plasmids were electroporated into NIH 3T3 fibroblasts, which were subsequently infected with wild-type MCMV K181. Viruses that underwent homologous recombination between genome and plasmid sequences were isolated by mycophenolic acid selection and plaque purification.
FIG. 2.
Construction of the ΔM83, ΔM83-2, and ΔM84 deletion mutants of MCMV. (A) HindIII restriction map of MCMV K181 and location of ORFs M82 to M86 in the HindIII C region. Arrows indicate the transcription direction and sizes of the ORFs. All ORF lengths are to scale, with the scale (in base pairs) indicated on the right. Also shown are the positions of restriction sites important for plasmid construction. Abbreviations: Hind, HindIII; Eco, EcoRI; RV, EcoRV; Pst, PstI; Not, NotI. (B) Construction of ΔM83 and ΔM83-2 by homologous recombination between either the pΔM83KO8 or pΔM83-2 insert, respectively, and the K181 genome. Identical shading and cross-hatching indicate homologous regions, while dashed lines indicate continuation of genome sequences. Indicated by a horizontal bar above pΔM83-2 is the 28 bp provided as a NotI-to-PstI oligonucleotide adapter as described in Materials and Methods. (C) Construction of ΔM84 by recombination of homologous sequences of K181 and the insert from pΔM83KOT-5. The restriction sites at the borders of some homologous regions have been changed in the plasmid to facilitate subcloning.
To restore the M83 ORF in the ΔM83 virus, we initially used a protocol in which NIH 3T3 cells were electroporated with a plasmid containing the M83 ORF flanked by 1 to 2 kbp of upstream and downstream sequences and then infected with ΔM83. Resultant viruses were plaque purified, and the plaques were stained with X-Gal to identify virus which had lost the lacZ/gpt cassette by incorporating the M83 ORF, thereby generating the wild-type genome. However, after several repetitions of electroporation and infection, and following the staining of several thousand plaques, it became apparent that identifying a low-frequency clear plaque in a background of blue ΔM83 plaques was not practical, particularly since an unacceptably high percentage of ΔM83 plaques did not stain blue at the same time as the others.
To solve this problem, we devised a protocol which involved constructing a viral intermediate that contained the wild-type M83 gene plus another selectable marker. Using a method described by M. Prichard and G. Pari (45a), we constructed a cassette expressing the EGFP and puromycin resistance genes as a single fusion protein for efficient generation of CMV mutants (see Materials and Methods). We then assembled a rescue plasmid, pM83Res13.6 (Fig. 3B), such that homologous recombination would introduce the EGFP-puro cassette and the M82, M83, and M84 ORF sequences into the ΔM83 genome (Fig. 3C). The resultant lacZ+ EGFP+ intermediate M83 rescue virus was expected to have a relatively high rate of homologous recombination of M84 direct-repeat sequences (one endogenous and one introduced on the rescue cassette) (Fig. 3C), and intramolecular recombination of these sequences in the intermediate virus would delete the intervening DNA sequences. As a consequence, this virus would revert back to a LacZ− EGFP− phenotype and the wild-type HindIII C region would be restored (Fig. 3D). The intermediate LacZ+ EGFP+ virus was first generated by electroporation of NIH 3T3 cells with linearized pM83Res13.6, infection with ΔM83, and puromycin selection. EGFP+ plaques were picked and used to infect fresh cultures in the absence of drugs. Viral plaques from these cultures were stained with X-Gal, and LacZ− EGFP− plaques were isolated and further plaque purified to homogeneity.
FIG. 3.
Construction of the M83 revertant rΔM83, using the EGFP-puro cassette. (A) DNA sequences of the ΔM83 genome were recombined in NIH 3T3 cells with homologous sequences (indicated by identical shading and cross-hatching) in the rescue plasmid cassette pM83Res13.6 (B). Selected restriction sites used for subcloning or for reference are indicated with abbreviations as in Fig. 2. Bam, BamHI. (C) Recombination between both homologous regions yields an rΔM83 intermediate virus in which the EGFP-puro cassette and full-length M83 ORF have been inserted into the ΔM83 genome as indicated. Intramolecular recombination between M84 direct-repeat sequences in the rΔM83 intermediate would be expected to delete intervening marker gene sequences (bracketed in panel C), resulting in an EGFP− LacZ− virus with a wild-type (wt) HindIII C region (D).
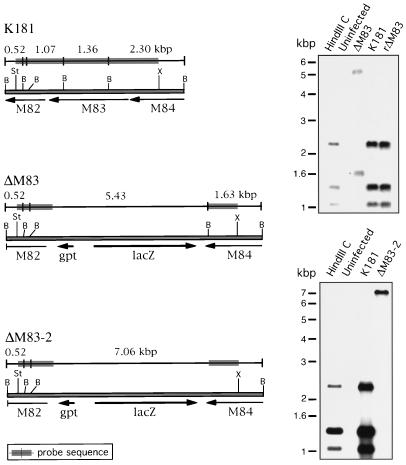
Genomic DNAs from uninfected cells and from cells infected with the mutant or revertant viruses were purified for Southern blot analysis. For analysis of the ΔM83 and rΔM83 genomes, genomic or HindIII C plasmid DNAs were digested with BamHI, electrophoresed, and blotted to a nylon membrane. A 4.4-kbp StuI-to-XhoI genomic probe was isolated from the HindIII C plasmid, 32P labeled by random priming, and used to probe the blot. For analysis of the ΔM84 genome and further confirmation of the rΔM83 genome structure, genomic and HindIII C plasmid DNAs were digested with SstI and XhoI and fragments were detected with a 7.6-kbp EcoRV-to-NotI genomic probe. Southern blot analysis of the HindIII C regions of the ΔM83, ΔM84, and rΔM83 viruses showed restriction patterns indicative of the insertion or removal of the lacZ/gpt cassette (Fig. 4 and 5). The common 0.52-kbp band expected in the plasmid or viral lanes in Fig. 4 was detected upon prolonged exposure of the blot (data not shown). No wild-type-specific restriction fragments were detected in the ΔM83 and ΔM84 lanes after overexposure of the blots, indicating that the virus stocks were free from contaminating wild-type virus. Similarly, no bands specific for the rΔM83 intermediate or ΔM83 viruses were detectable in the rΔM83 lanes upon overexposure of the blots, indicating that the stock of revertants was also homogeneous. In addition, Southern analysis of the resulting rΔM83 virus showed the wild-type structure when two different restriction digestion schemes and genomic probes were used (Fig. 4 and 5).
FIG. 4.
Genomic analysis of ΔM83, ΔM83-2, and rΔM83 viruses by restriction digestion and Southern blotting. (Left panels) The restriction maps of the portions of the HindIII C regions under analysis are shown for the K181, ΔM83, and ΔM83-2 viruses, with arrows indicating the positions, lengths, and directions of transcription of ORFs. B, BamHI; St, StuI; X, XhoI. Above each restriction map are shown the lengths (in kilobase pairs) and positions of restriction fragments detected by the genomic probe (indicated by the shaded regions). (Right panels) Genomic DNA was purified from uninfected cells or cells infected with the virus indicated. Genomic and HindIII C plasmid DNAs were digested with BamHI, electrophoresed, and blotted. The 4.4-kbp StuI-to-XhoI genomic probe was isolated from the HindIII C plasmid, 32P labeled by random priming, and used to probe the blot. The expected 0.52-kbp band in both panels was visible upon prolonged exposure of the blots.
FIG. 5.
Genomic analysis of the ΔM84 and rΔM83 viruses by restriction digestion and Southern blotting. Details of genomic maps and DNA preparation are as described in the legend to Fig. 4. Ss, SstI; RV, EcoRV; N, NotI; X, XhoI. Genomic and HindIII C plasmid DNAs were digested with SstI and XhoI prior to electrophoresis, blotting, and detection with the 7.6-kbp EcoRV-to-NotI genomic probe.
Replication of ΔM83 and ΔM84 in cell culture.
The roles of M83 and M84 proteins in MCMV replication were examined by single- and multicycle replication experiments using ΔM83 and ΔM84. Single-cycle replication experiments measured the accumulation of extracellular tissue culture virus in NIH 3T3 fibroblasts infected with K181, ΔM83, or ΔM84, or rΔM83. We found that the ΔM84 (Fig. 6A), ΔM83 (Fig. 6B), and rΔM83 (Fig. 6B and C) viruses replicated with wild-type kinetics and that their peak titers were comparable. When the combined levels of cell-associated and extracellular virus were measured in a separate experiment, the growth kinetics and final titers of all three viruses were also indistinguishable (data not shown). NIH 3T3 cells were also infected at a low MOI in order to examine the replication efficiency and cell-to-cell spread of the mutant viruses. Extracellular virus titers measured after infection with either ΔM83 or ΔM84 at an MOI of 0.05 were identical to those of the parent strain (Fig. 6D). This finding was consistent with the similar CPE, plaque sizes, plaque morphologies, and final viral titers obtained during several simultaneous preparations of K181, ΔM83, and ΔM84 stocks (data not shown). Taken together, these data indicate that the M83 and M84 ORFs are dispensable for replication in cultured fibroblasts.
FIG. 6.
Growth of mutant and revertant MCMVs in NIH 3T3 cells. All titers presented are the means of the log10 PFU/ml of the extracellular tissue culture medium of triplicate cultures, with error bars indicating the standard deviations (SD). (A) Single-cycle growth of K181 and ΔM84 after infection at an MOI of 3. (B) Single-cycle growth of K181, ΔM83, and rΔM83 in NIH 3T3 cells as described for panel A. (C) Single-cycle growth of K181, ΔM83, ΔM83-2, and rΔM83 as described for panel A. (D) Multicycle growth of K181, ΔM83, and ΔM84 in NIH 3T3 cells infected at an MOI of 0.05.
Protein expression from mutant and revertant MCMVs.
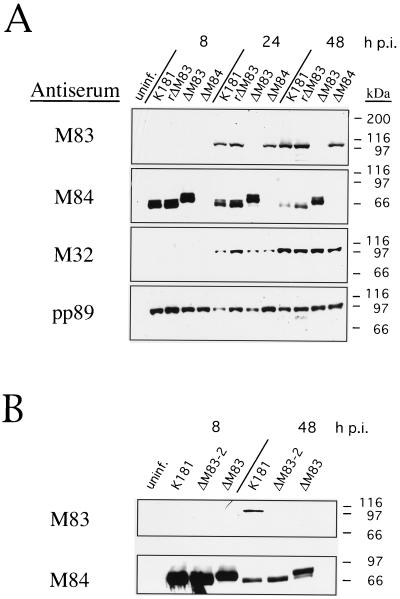
The kinetics of protein expression from the various MCMV isolates were analyzed to determine whether expression of selected proteins representing the three temporal classes of gene products was grossly affected in any of the mutants as well as to confirm that the appropriate MCMV gene products were not expressed in their respective mutants. For these experiments, NIH 3T3 cells were infected with either K181, ΔM83, ΔM84, or rΔM83 at an MOI of 3.5, and at 8, 24, and 48 h p.i., cells were harvested and whole-cell extracts were prepared as described above.
Western blot analysis using the rabbit M83 antiserum demonstrated the absence of detectable M83 protein in ΔM83-infected cells. M83 in cells infected with K181, ΔM84, or rΔM83 was detectable by 24 h p.i., and its levels were similar in all of these infected cells (Fig. 7A), although the rΔM83 infection proceeded slightly faster than did the others as judged by CPE formation and earlier detection of the various proteins examined. Thus, M83 expression kinetics and levels in the M84 deletion mutant and in the M83 revertant virus were not significantly affected by the genetic manipulations. We previously showed that M83 is a late gene, with the corresponding protein being undetectable until 48 h p.i. in infected NIH 3T3 cells by Western blotting (13). Improvements in detection sensitivity and variations in actual MOI may account for the earlier detection of M83, at 24 h p.i., in these studies.
FIG. 7.
Western blot analysis of mutant and revertant MCMVs. Whole-cell lysates were prepared from uninfected or MCMV-infected NIH 3T3 cells harvested at 8, 24, or 48 h p.i. Lysates were electrophoresed on 7.5% polyacrylamide–SDS gels, and separated proteins were electroblotted onto nitrocellulose. M83, M84, and M32 proteins were detected with rabbit polyclonal antisera as described in Materials and Methods. pp89 protein was detected with an antiserum from BALB/c mice immunized with intradermal injections of pcDNA3-pp89. Bound antibodies were detected with horseradish peroxidase-coupled anti-mouse or anti-rabbit antibodies as in Fig. 1. (A) Expression of M83, M84, M32, and pp89 proteins in NIH 3T3 cells after infection with K181, rΔM83, ΔM83, or ΔM84. (B) An independent experiment showing M83 and M84 expression 8 or 48 h after infection with K181, ΔM83, or ΔM83-2. uninf., uninfected. The positions of molecular mass markers are shown on the right.
Protein species reactive to the M84 antiserum were found in all of the viruses except ΔM84. Similar to the data shown in Fig. 1, high levels of M84 protein were detected at 8 h p.i. in the wild-type, ΔM83, and rΔM83 viruses, and M84 protein levels tapered off through the 48-h-p.i. time point. In the case of the wild-type and rΔM83 viruses, the protein appeared to be a doublet, 64 and 66 kDa in size. However, we noted that the M84 protein in the ΔM83 virus appeared to be slightly larger. This aberration was explored by sequencing the M84 gene-lacZ junction in the plasmid used to construct the ΔM83 virus, pΔM83KO8. It was found that the M84 ORF extended beyond the PstI site junction, 75 bp into the vector sequence used for construction of the recombination plasmid. The stop codon and the coding sequence for the C-terminal 3 amino acids of M84 were therefore replaced with 35 amino acids of the ORF until the first termination codon was reached. This allowed fusion of an extra 3 kDa of vector-encoded protein onto the C terminus of the M84 protein, consistent with the migration pattern observed by Western blotting. In this experiment and others, we also noted that the level of the M84 fusion protein in the ΔM83-infected cells was at least twofold higher at 48 h p.i. than the levels in cells infected with K181 or ΔM83.
Because ΔM83 was found to express a mutant form of M84, we were concerned that we would not be able to determine unambiguously whether any phenotype found in this virus was due to the absence of the M83 gene product, the presence of the M84 fusion protein, or both. We therefore constructed a M83 deletion mutant, ΔM83-2, which contained the wild-type 3′ coding sequence of the M84 ORF. The recombinant vector pΔM83-2 (Fig. 2B) was constructed by inserting an oligonucleotide adapter into the NotI and PstI sites at the M84 gene and lacZ borders in order to provide the coding sequence for the carboxy terminus of M84 as well as termination codons. This vector was used for in vivo homologous recombination with MCMV K181 as described for ΔM83 and ΔM84, and Southern blot analysis showed the expected restriction pattern for the replacement of M83 with the lacZ/gpt cassette (Fig. 4). In addition, the single-cycle replication kinetics of ΔM83-2 in NIH 3T3 cells were found to be identical to those of K181, rΔM83, and the first M83 mutant ΔM83 (Fig. 6C). Finally, Western blot analysis was performed to determine whether the M84 gene product in ΔM83-2 was wild type with respect to both its relative mobility on SDS-polyacrylamide gels and its expression kinetics. As with ΔM83, M83 protein was not detectable at any time p.i. in cells infected with ΔM83-2 (Fig. 7B). Unlike ΔM83, however, the expression level and relative mobility of M84 protein in ΔM83-2-infected cells were indistinguishable from those found in K181-infected cells (Fig. 7B). Thus, ΔM83-2 appeared to express wild-type M84 protein in the absence of M83 gene product.
Western blot analyses of the products of an IE gene and a late gene, IE1-pp89 and the M32 gene product, respectively, showed wild-type kinetics and levels of expression of these gene products in all of the viruses (Fig. 7A). Taken together, the results showed that neither IE and late gene expression nor the production of infectious virus was significantly affected by the absence of the M83 or M84 gene product.
ΔM83 and ΔM84 are both attenuated for replication in BALB/c mice, with ΔM83 showing a specific defect for growth in salivary glands and lungs.
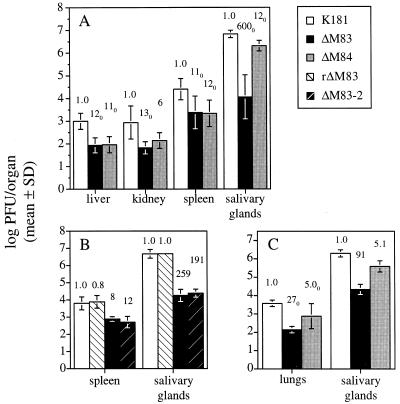
The roles of M83 and M84 in viral replication in the host were examined by infection of inbred strains of mice with the mutant viruses by two different inoculation routes. We first examined the replication of the mutant viruses in the susceptible BALB/c strain following i.p. infection. Five mice per group were infected with 5 × 105 PFU of tissue culture-derived K181, ΔM83, or ΔM84, and on day 4 p.i. the livers, spleens, and kidneys were harvested. At the time of sacrifice, both the K181- and, to a lesser degree, the ΔM84-infected mice exhibited ruffled fur but no hunching or lethargy, while the ΔM83-infected mice displayed no morbidity. As seen in Fig. 8A, titers of ΔM83 and ΔM84 in the spleen were reduced more than 11- and 12-fold, respectively, relative to that of the parent strain. Because virus was sometimes undetectable in one or more organs of a particular group (as denoted by the subscript 0 in Fig. 8 and 10), the titers of those organs were arbitrarily set to the limit of assay sensitivity, generally 102 PFU/organ, for mean calculations. Thus, fold reductions of titers in those groups are likely underestimates. In the liver, similar 11- to 12-fold reductions relative to wild-type titers were seen for both mutant viruses (Fig. 8A). The titer of ΔM84 in the kidneys was 6-fold lower than that of K181, while ΔM83 titers were at least 13-fold below the wild-type level. Taken together, compared to the wild type, tissue culture-passaged MCMV, both ΔM83 and ΔM84 appear to be growth attenuated in the abdominal target organs.
FIG. 8.
Growth of mutant and revertant MCMVs in BALB/c mice following i.p. or i.n. inoculation. (A) Five BALB/c mice per group were inoculated i.p. with 5 × 105 PFU of tissue culture-derived virus, and resultant viral titers in the livers, kidneys, and spleens 4 days postinoculation, or in the salivary glands 10 days postinoculation, are shown. Titers represent the means of the log10 PFU/organ values, with the standard deviations (SD) being shown by error bars. Values above each bar indicate the fold reductions of the nonlogarithmic mean titers (i.e., PFU/organ) of that group relative to the corresponding K181-infected controls. The subscript 0 indicates that in at least one organ in that group, virus was undetectable, and that the titer of that organ was set to the limit of detection for calculation purposes. (B) Day 4 spleen and day 10 salivary gland titers of four mice per group inoculated i.p. with K181, rΔM83, ΔM83, or ΔM83-2 as described for panel A. (C) MCMV titers in the lungs and salivary glands of four mice per group 14 days following inoculation i.n. with 1.5 × 105 PFU of virus.
FIG. 10.
Growth of mutant MCMV in 129/J mice following i.p. or i.n. inoculation. (A) MCMV titers of four 129/J mice per group following i.p. inoculation with 5 × 105 PFU of tissue culture-derived virus. Organ harvest days, titer value calculations, and fold reduction values above each bar are as described in the legend to Fig. 8. (B) MCMV titers in the lungs and salivary glands of four mice per group 14 days following i.n. inoculation with 2.5 × 105 PFU of virus. SD, standard deviation.
Replication of ΔM83 in the salivary glands, however, was severely attenuated, and these titers were 600-fold lower than those of K181 on day 10 p.i. (Fig. 8A). This decrease in virus titer in the salivary glands was not due to the expression of the M84 fusion protein, since the ΔM83-2 virus, which is wild type with respect to M84 protein expression, as determined by Western blotting, replicates in the spleen and salivary glands to levels similar to those of ΔM83 (Fig. 8B). By comparison, the level of replication of ΔM84 in salivary glands was approximately 12-fold lower than that of the parent strain. Independent experiments in which the inoculation dose of ΔM83 was 105 PFU or lower showed that the level of attenuation of replication in the salivary glands varied between 150- and 600-fold (Fig. 8B and data not shown). Following injection of 5 × 105 PFU of ΔM83, the titers were still reduced by approximately 600-fold, but by increasing the i.p. inoculation dose 5-fold to 2.5 × 106 PFU for each of the viruses, titers of ΔM83 in the salivary glands were reduced only 25-fold compared to the K181 titer (data not shown). As expected, we found that the rescue of M83 expression in the rΔM83 virus restored wild-type replication levels in the spleen and salivary glands following i.p. infection (Fig. 8B).
Because the route of administration of MCMV has been shown to be important in terms of the nature and severity of the infection (28), we assessed the replicative abilities of the mutant viruses following infection by a nonparenteral route. Four BALB/c mice per group were given i.n. inoculations of 1.5 × 105 PFU of tissue culture-derived virus, and on day 14 p.i., lungs and salivary glands were harvested and assayed for infectious MCMV. As shown in Fig. 8C, the replication of ΔM83 in salivary glands was attenuated approximately 90-fold relative to that of K181, while ΔM84 titers were on average 5-fold lower than those of the wild type. Similarly, virus titers in the lungs of the ΔM83-infected mice were at least 27-fold lower than those of the K181-infected mice, and ΔM84 replication in the lung was reduced approximately 5-fold compared to that of the wild type. Thus, following inoculation of BALB/c mice by either a parenteral or mucosal route, replication of ΔM83 in lung and salivary gland tissues was significantly reduced below wild-type levels.
ΔM83 and ΔM84 show patterns of restricted replication in C3H/HeN and 129/J mice similar to those seen in BALB/c mice.
The ability of MCMV to replicate in vivo has been found to depend on the strain of mouse examined (22). We therefore tested the growth of the MCMV mutants in an inbred mouse strain highly resistant to MCMV. Four C3H/HeN female mice per group were infected with ΔM83 or ΔM84 by inoculation of either 1.5 × 106 PFU (i.p.) or 1.5 × 105 PFU (i.n.) of tissue culture-derived viruses. Because C3H/HeN mice are able to effectively limit MCMV replication in the abdominal organs, and because only tissue culture-derived virus was used for these experiments, salivary glands were the only tissue examined for viral replication in this strain.
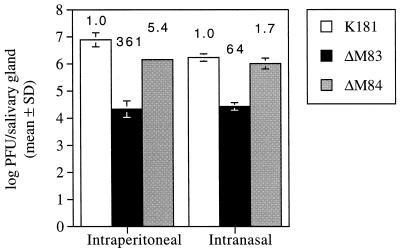
As expected, we observed no morbidity in the C3H/HeN mice after infection. K181 titers in the salivary glands following i.p. inoculation reached an average of 106.9 PFU/organ, while titers resulting from ΔM83 and ΔM84 infection were approximately 360- and 5-fold lower, respectively (Fig. 9). Moreover, replication of ΔM83 in the salivary glands following i.n. inoculation resulted in a titer that was approximately 64-fold lower than that resulting from K181 infection. The average ΔM84 titer, by comparison, was only 1.7-fold below the wild-type level. Therefore, the relative levels of attenuation of the mutant viruses in the salivary glands of C3H/HeN and BALB/c strains of mice appear similar.
FIG. 9.
Growth of mutant MCMV in the salivary glands of C3H/HeN mice. Shown are MCMV titers 10 days following i.p. inoculation with 1.5 × 106 PFU of tissue culture-derived virus and 14 days following i.n. inoculation with 1.5 × 105 PFU of virus. Means, standard deviations (SD), and fold-reduction values above each bar are as described in the legend to Fig. 8.
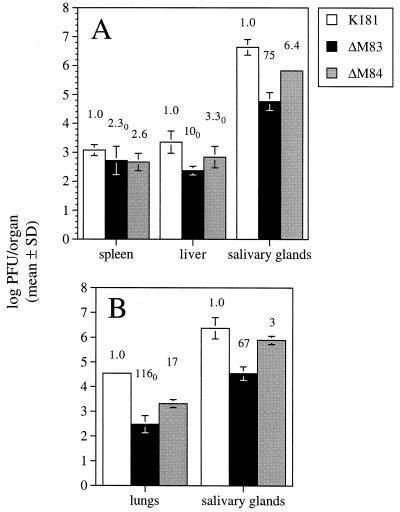
Lastly, we examined the replication of the mutants in the MCMV-sensitive inbred mouse strain 129/J. We have found the 50% lethal dose LD50 of salivary gland-derived MCMV in the 129/J female mice to be roughly 10-fold lower than that in the moderately sensitive BALB/c strain (data not shown). Four 129/J female mice per group were inoculated i.p. with 5 × 105 PFU of the mutant viruses. On day 2 p.i., the mice infected with K181 or ΔM84 exhibited ruffled fur, but no signs of morbidity were observed by day 3. On day 4, mice were sacrificed and organs were removed for MCMV titer determination. Spleens of mice infected with K181 were noticeably smaller and more necrotic than those from the ΔM83- or ΔM84-infected mice, a phenomenon seen after infection of various inbred strains of mice with virulent MCMV (42). MCMV titers in the 129/J spleens were 103.1 PFU/spleen for the K181-infected mice and only two- to threefold lower in the ΔM83 and ΔM84 groups (Fig. 10A). In contrast, a greater than 10-fold reduction in ΔM83 titer compared to that of K181 was seen in the liver. A greater than threefold reduction in titer below that of the wild type was seen for ΔM84 in this organ. More importantly, 10 days after i.p. infection, titers of ΔM83 in the salivary glands were 75-fold lower than those observed in K181-infected mice (Fig. 10A). As with the other strains of mice tested, ΔM84 replicated to an approximately sixfold lower level than the parent virus.
When 129/J mice were inoculated i.n. with 2.5 × 105 PFU of the three viruses, ΔM83 replication was reduced 67-fold in the salivary glands and at least 116-fold in the lung relative to that of the wild-type virus (Fig. 10B). In contrast, the titers of ΔM84 in salivary gland and lung tissues were reduced only 17- and 3-fold, respectively, below those of the wild-type controls. When all of the data on virus replication in organs are taken together, both deletion-mutant viruses appear to be attenuated 2- to 10-fold in replication in abdominal organs compared to the parent virus. More importantly, ΔM83 is markedly attenuated 25- to 600-fold in the salivary glands following i.p. or i.n. infection, and following i.n. inoculation, 27- to 116-fold attenuation is observed in the lungs. ΔM84, in contrast, consistently replicates in the lungs and salivary glands to levels only 5- to 10-fold lower than those of the parent strain.
Levels of MCMV DNA in BALB/c mice latently infected with ΔM83 and ΔM84.
Having found that M83 plays a role in dissemination to or replication in the lung and salivary glands during the acute infection of mice, we sought to determine whether M83 or M84 is involved in the establishment of latency. For these experiments, groups of BALB/c mice were inoculated i.p. with 105 PFU of tissue culture-derived wild-type or mutant MCMV. These mice were then housed for 10 months to allow sufficient time for the resolution of the acute and persistent infections and the establishment of latency, as defined by the presence of detectable viral DNA in the absence of infectious virus. Since spleen and lungs appear to be preferred reservoirs for MCMV during latency (4, 11, 34, 41), and since replication of ΔM83 in salivary glands was impaired, we focused on those three organs for analysis of latent virus. In addition, because previous studies have shown that the spleen cells that harbor the latent MCMV genome are located in the stroma (41, 45), spleens were fractionated and genomic DNA was harvested only from the stromal cells.
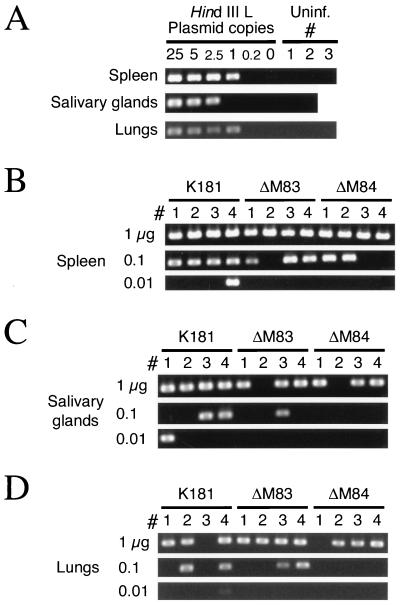
MCMV DNA in these tissues was detected semiquantitatively by nested-PCR amplification and endpoint dilution of target DNAs. Primers spanning introns 2 and 3 of IE1 were chosen in order to distinguish amplification of genomic IE1 sequences from that of IE1 cDNA sequences found on plasmid vectors commonly used in the laboratory. As a control for the sensitivity of each set of nested reactions, linearized HindIII L plasmid containing the IE1 target was diluted in elution buffer containing genomic DNA from uninfected mice. The results of representative control reactions containing 0 to 25 copies of plasmid DNA are shown in Fig. 11A. DNAs from tissues of uninfected mice were routinely negative for MCMV genomic DNA. The sensitivity of the assay was consistent among the various sources of background DNA, since a single copy of HindIII L DNA was detected in approximately two-thirds of the control reactions. This frequency correlates well with the expected Poisson frequency of 63% for a single copy. Signal from the 2.5-copy dilution was found in 90 to 95% of the control reactions, while signal from the 0.2-copy dilution was rarely detected.
FIG. 11.
PCR amplification of MCMV DNA in latently infected BALB/c mice. (A) Control nested PCRs in which IE1-specific primer pairs were used to amplify serial dilutions of linearized HindIII L plasmid. Dilutions were made such that 25 to 0.2 copies of plasmid and 1 μg of uninfected mouse genomic DNA purified from the organs shown were amplified. The negative controls contained 1 μg of the organ DNA from uninfected mice, and PCR results for two to three individual uninfected mice are shown. All positive reactions produced only a single 310-bp product (shown) upon ethidium bromide staining, and representative results for the plasmid dilutions are shown (see Results). (B) Nested-PCR amplification of IE1 in the splenic stromal DNA isolated from four mice (no. 1 to 4) per group infected with 105 PFU of tissue culture-derived K181, ΔM83, or ΔM84. Serial 10-fold dilutions of organ DNA were amplified independently. (C) Nested PCR of serial dilutions of salivary gland DNAs from mice 1 to 4 in each group as described for panel B. (D) Nested PCR of serial dilutions of lung DNAs from mice 1 to 4 as described for panel B.
In Fig. 11B, we show that the IE1 target sequences from all four mice latently infected with either K181, ΔM83, or ΔM84 were consistently amplified when 1 μg of splenic stromal DNA was subjected to the nested PCR. Therefore, the ability of the ΔM83 and ΔM84 mutants to establish latency was apparently intact. To assess whether there were vast differences in the latent-DNA load established by the various MCMVs, we subjected serial 10-fold dilutions of organ DNAs to PCR amplification. As seen in Fig. 11B, using stromal DNA, the number of positive PCRs decreased similarly with all three viruses upon dilution of the DNA. In the salivary glands (Fig. 11C) and lungs (Fig. 11D), the number of positive signals from the mice infected with ΔM84 appeared to decrease more readily upon DNA dilution than did that for the ΔM83-infected mice or the K181-infected controls. When these experiments were repeated, some negative organ DNA dilutions alternatively scored positive, but the overall number of positive mice remained relatively constant for each virus type (data not shown). Lung and salivary gland DNA samples in which MCMV DNA was never detected were subjected to a control amplification reaction of a mouse gene, β-actin, to ensure that the DNA was amplifiable. β-Actin sequences were detected in all of the reactions that were negative for MCMV DNA (data not shown). Taken together, these data show that both ΔM83 and ΔM84 establish latency in the spleen, salivary glands, and lungs, with the amount of latent ΔM84 DNA being slightly lower in the salivary glands and lungs than that of the parent strain.
Immunization with either ΔM83 or ΔM84 virus confers wild-type protection against subsequent lethal challenge.
Several studies examining the HCMV proteins targeted by CTLs have demonstrated the prevalence of CTLs specific for the UL83 gene product pp65 and have thus implicated this protein as being an important protective antigen (9, 39, 63). Because both the M83 and M84 proteins of MCMV possess homology to the UL83 gene product, we sought to determine if these proteins were necessary for the generation of a protective response. We immunized four BALB/c mice per group by i.p. inoculation of 105 PFU of tissue culture-derived K181, ΔM83, or ΔM84. Control mice were mock immunized with PBS. Four weeks after immunization, all of the mice were challenged i.p. with 5 × 105 PFU (two LD50s) of salivary gland-derived K181. Spleens and salivary glands were harvested on days 6 and 10 postchallenge, respectively, for MCMV titer determination. We found that the spleens of the mock-immunized mice all contained high levels of virus, with a mean titer (n = 4) of 106.2 PFU/spleen (Table 1). In contrast, only one of four mice immunized with either K181 or ΔM83 had viral titers above the limits of detection, and the spleens of the ΔM84 mice had no detectable virus. While all four of the mock-immunized mice succumbed to the infection prior to day 10 postchallenge, only one K181-immunized mouse had detectable virus in the salivary glands, and no ΔM83- or ΔM84-immunized mice had detectable virus. These results indicate that the deletion of either the M83 or M84 ORF does not compromise the protective immunity generated, since similar levels of virus were found in mice immunized with mutant or wild-type virus after a high dose of challenge virus. To assess the genotype of the virus recovered from the spleen of the ΔM83-immunized mouse, we stained the plaques that appeared in the assay. After overnight staining with X-Gal, none of the plaques stained blue (data not shown), indicating that the challenge virus, not the immunizing virus, was actively replicating at the time of sacrifice.
TABLE 1.
MCMV replication in BALB/c mice immunized with mutant virus and challenged i.p. with two LD50s of MCMV K181a
| Immunizing virus | No. of MCMV-positive organs and MCMV titer in:
|
|||
|---|---|---|---|---|
| Spleen
|
Salivary glands
|
|||
| No. of MCMV positive organs/total | MCMV titerb | No. of MCMV positive organs/total | MCMV titer | |
| None (PBS) | 4/4 | 6.2c | —d | — |
| K181 | 1/4 | 2.0 | 1/4 | 3.9 |
| ΔM83 | 1/4 | 2.5 | 0/4 | <2e |
| ΔM84 | 0/4 | <2 | 0/4 | <2 |
Naive BALB/c female mice were injected i.p. with PBS or 105 PFU of tissue culture-derived MCMV and were challenged i.p. with 5 × 105 PFU of salivary gland-derived K181 at 4 weeks postimmunization. Spleens and salivary glands were harvested at days 6 and 10, respectively, postchallenge.
Log PFU of MCMV-positive organ(s).
Value is the mean.
—, all mice died prior to harvest day.
<2, no virus detected.
Immunization with lower doses of ΔM83 confers protection against lethal challenge but does not prevent the establishment of latency of the immunizing or challenge virus.
Prior infection with either mutant virus was able to provide wild-type immunity when mice were challenged with a lethal dose of virus. We further tested the protective ability of the salivary gland-attenuated ΔM83 virus by titrating the immunizing dose to determine the minimum dose necessary for protection from acute or latent infection. BALB/c mice were immunized i.p. with either 30 or 200 PFU of tissue culture-derived ΔM83. Four weeks after immunization, mice were either challenged i.p. with a lethal dose of virulent K181, as described above, or left unchallenged. Spleens and salivary glands were harvested for plaque assay on the same days as indicated above. Table 2 shows that ΔM83 provided a similar level of protection in the spleen across the range of immunizing doses tested. Virus was undetectable in the group immunized with 30 PFU, and 103.7 PFU was found in only one of the mice immunized with 200 PFU. Virus was more frequently detected in the salivary glands at 10 days postchallenge, with two of the mice immunized with 30 PFU having titers of more than 102 PFU/organ. As indicated above, upon X-Gal staining of resultant plaques, no plaques stained blue, showing that only challenge virus was detected (data not shown). Taken together, the titration data suggest that immunizations with 30, 200, or 105 PFU of ΔM83 provide similar protection against lethal challenge.
TABLE 2.
MCMV replication in BALB/c mice immunized with low doses of ΔM83 and challenged i.p. with two LD50s of MCMV K181a
| Immunizing ΔM83 dose (PFU) | No. of MCMV-positive organs and MCMV titer in:
|
|||
|---|---|---|---|---|
| Spleen
|
Salivary glands
|
|||
| No. of MCMV-positive organs/total | MCMV titerb | No. of MCMV-positive organs/total | MCMV titer(s) | |
| 30 | 0/4 | <2c | 2/4 | 3.3, 3.8 |
| 200 | 1/4 | 3.7 | 0/4 | <2 |
Naive BALB/c female mice were injected i.p. with tissue culture-derived ΔM83 and were challenged i.p. with 5 × 105 PFU of salivary gland-derived K181 at 4 weeks postimmunization. Spleens and salivary glands were harvested at days 6 and 10, respectively, postchallenge.
Log PFU of MCMV-positive organ(s).
<2, no virus detected.
Since the protective responses generated with smaller immunizing doses of ΔM83 were able to almost completely suppress the replication of a 5 × 105 PFU challenge of highly virulent virus, we proceeded to assess whether the immunizing virus was able to establish latency. We also determined whether the protective responses against ΔM83 were able to prevent the challenge virus from establishing latency. Groups of the mice immunized with 200 PFU of ΔM83 (either challenged or unchallenged) from the titration experiment described above were housed to establish latency. Five months after the mice were challenged with K181 or mock challenged, their spleens were removed and genomic DNA was prepared from the stromal fractions as described above. To confirm that no persistent virus remained, approximately one-quarter to one-third of each spleen from two unchallenged mice and two challenged mice was homogenized, sonicated, and used to infect cultures of MEFs or NIH 3T3 cells as previously described (44). No CPE developed in any of the cultures, indicating the absence of persistent virus.
We then subjected splenic stromal DNA to nested PCR for the detection of IE1 DNA sequences as described above. The first set of reactions, with 1 μg of DNA from mice infected with only 200 PFU of ΔM83, gave spurious positive results for any given spleen. Since the sensitivity controls routinely showed the detection of a single plasmid copy with 1 μg of background organ DNA, we suspected that the amounts of latent viral genome in these mice were near the limit of detection. We therefore subjected the spleens of all mice to multiple independent nested PCRs with 1 μg of input DNA in order to provide an estimate of the low level of latent DNA.
Our results showed that while IE1 sequences were never detected in uninfected mice, three of four mice immunized with 200 PFU ΔM83 had detectable MCMV IE1 DNA in at least one reaction (Table 3). The levels of viral DNA in the mice that established latency were relatively low, since only one or two of six PCR amplifications yielded the IE1 amplification product. In contrast, five of six mice that were immunized with ΔM83 and given a lethal challenge dose of K181 had detectable viral DNA in all reactions. One of the dually infected mice, mouse no. 3, had no detectable IE1 sequences in a total of 13 PCRs. This stromal DNA was resubjected to column purification to remove any copurifying PCR inhibitors. However, no MCMV was detectable in mouse no. 3 DNA after repurification, and two to three copies of the control plasmid were routinely amplified when added to 1 μg of mouse no. 3 genomic DNA (data not shown). In addition, β-actin sequences were amplified similarly in 10-fold dilutions of mouse 3 and mouse 1 of the same group (data not shown), indicating that the DNA was amplifiable and free of inhibitors.
TABLE 3.
Detection of latent MCMV in BALB/c mice immunized with 200 PFU of ΔM83 and challenged with a lethal dose of MCMV K181a
| Gene amplifiedb | No. of positive PCRs/no. of reactions for individual mice infected with the following virusesc:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Uninf.
|
ΔM83 only
|
ΔM83 + K181
|
||||||||||
| Mouse 1 | Mouse 2 | Mouse 1 | Mouse 2 | Mouse 3 | Mouse 4 | Mouse 1 | Mouse 2 | Mouse 3 | Mouse 4 | Mouse 5 | Mouse 6 | |
| IE1 | 0/6 | 0/6 | 2/6 | 1/6 | 0/6 | 2/6 | 6/6 | 6/6 | 0/13 | 6/6 | 4/4 | 4/4 |
| gpt | 0/4 | 0/4 | 1/4 | 1/4 | 1/4 | 2/4 | 0/4 | 1/4 | 0/4 | 0/4 | 2/4 | 0/4 |
The same BALB/c mice described in Table 2 that were i.p. immunized with 200 PFU of tissue culture-derived ΔM83 were either challenged with K181 (see Table 2) or left unchallenged. Five months after the challenge day, mice were sacrificed, spleens were harvested, splenic stromal fractions were prepared, and genomic DNA was isolated for nested PCR.
The same genomic DNA preparations were used for nested-PCR detection of both MCMV IE1 sequences or gpt sequences from ΔM83.
Virus inoculation groups: Uninf., uninfected; ΔM83 only, ΔM83 immunization only; ΔM83 + K181, ΔM83 immunization and K181 challenge.
Because the ΔM83 virus used for immunization and K181 used for the challenge both contain the IE1 target sequence, the above results reflect the total amount of MCMV DNA without differentiating between the two viruses inoculated. Although the mice that were injected with both viruses received a 2,500-fold higher dose of virulent K181 than of tissue culture-attenuated ΔM83, we could not exclude the possibility that the high load of latent genome in these mice contained a significant fraction of ΔM83 genome. We therefore designed nested-PCR primer sets that would specifically detect ΔM83 genome by targeting the gpt selectable marker gene. As with the IE1 nested PCR, the sensitivity of the gpt PCR was sufficient to detect a single copy of linearized, gpt-containing plasmid in a background of 1 μg of uninfected stromal DNA (data not shown).
When the same stromal DNAs that were analyzed by IE1 amplification were subjected to amplification of gpt, we detected gpt sequences in the spleens of all four of the mice infected with ΔM83 alone (Table 3). Similar to the IE1-specific PCRs, the target sequence was detected in only one or two of four gpt amplification reactions for each of four mice, indicating that the ΔM83 genome level was near the limit of sensitivity. No gpt sequences were amplified in any of the reactions using stromal DNA from uninfected mice, indicating that amplification was specific for the gpt sequences in the ΔM83 virus. In contrast to the detection of IE1 DNA, PCR amplification of gpt sequences in the K181-challenged mice showed only low levels of these sequences. Only two of six mice had detectable gpt after four independent reactions, and only one or two of the four reactions were scored positive, suggesting again that the amount of gpt DNA was near the limit of detection. Mouse no. 3 of the group of dually immunized mice again had no detectable MCMV DNA.
Taken together, the PCR results for the singly and dually infected mice suggest that i.p. infection with 200 PFU of ΔM83 leads to the establishment of a very low latent-genome level in the spleen. Mice that subsequently received a high dose of virulent K181 had substantially higher loads of latent MCMV genome, but the ΔM83 genome appeared to constitute only a minority of that DNA since the gpt gene was barely detectable. Of note, one of the six mice receiving both viruses (ΔM83 + K181 mouse no. 3) did not contain detectable MCMV DNA, as determined when both IE1 and gpt sequences were amplified from stroma, suggesting that both viruses failed to establish latency in this mouse. In addition, when IE1 sequences were amplified from the salivary gland DNA of four ΔM83 + K181 mice (mice 1 to 4), IE1 product was detected in mice 1, 2, and 4 after 1 μg of DNA was amplified and in mice 2 and 4 when 0.1 μg of DNA was amplified (data not shown). Thus, MCMV DNA was not detectable in mouse 3 after PCR amplification of DNA isolated from either the spleen or salivary glands.
DISCUSSION
Because the results of several immunological studies strongly implicate the HCMV tegument phosphoprotein pp65 as being both a target of protective CTLs and a selective immune evasion protein, parallel studies in a relevant animal model are imperative for investigation of these roles in vivo. We have constructed two MCMV mutants, each lacking one of the ORFs that exhibit amino acid homology to pp65, in order to further characterize the relationships between the UL83, UL84, M83, and M84 proteins in vivo. While the M83 protein was previously shown to be similar to UL83/pp65 by virtue of its late expression, in vivo phosphorylation, and virion association (13), we demonstrated here that the properties of M83 and M84 are quite different in vivo despite the amino acid homology exhibited by these proteins. When the M84 protein was analyzed by Western blotting, we found that M84 was expressed at early times p.i. and was not detectable in the virion. Consistent with the UL84 protein being detectable in HCMV-infected cells at 6 h p.i. (25), we detected high levels of the MCMV homolog, M84, at 8 h p.i. Upon construction of MCMV mutants with deletions of the pp65 homolog genes, we found that like UL83 of HCMV (56), M83 is dispensable for viral replication in cultured fibroblasts, with replication levels and protein expression kinetics indistinguishable from their respective parent viruses.
We demonstrated that like the M83 ORF, M84, the HCMV UL84 positional homolog in MCMV, is also dispensable for replication in cultured fibroblasts. Although we found no differences in replication levels or protein expression levels in culture, ΔM84 grew to 5- to 10-fold-reduced levels in the target organs of three mouse strains. The UL84 protein has been shown to associate with the HCMV IE2-86 transactivator during lytic infection (58) and has since been shown by cotransfection of permissive cells to be a trans-dominant inhibitor of IE2-86 transcriptional activation (19). Results from immunoprecipitation analyses of cotransfected COS cells suggest that binding of pUL84 to IE2-86 is required for IE2-86 inhibition (19). Furthermore, transient or stable expression of pUL84 at IE times in infection was shown to inhibit the expression of early HCMV genes and viral replication in U373 cells (19). While overexpression of the M84 protein at IE times in the infection of permissive mouse cells may inhibit MCMV replication similarly, its absence from the infected cells does not appear to dysregulate the infection process in vitro. The UL84 gene product has also been implicated as having an essential function in the formation of replication compartments and oriLyt-dependent DNA replication in cotransfection assays (54). However, the wild-type replication of ΔM84 in culture suggests either that these cotransfection assays are not indicative of the true conditions in the infected cell or that the functions of UL84 and M84 differ. The construction and characterization of a UL84 deletion mutant of HCMV may help to clarify this issue.
In these studies, we found that the absence of M83 expression results in low viral titers in both salivary gland and lung tissues. Both H-2 and non-H-2-linked genes have been shown to determine the ability of mice to control MCMV (22), and it has been shown that susceptible and resistant strains utilize different effector subsets for clearance of the acute infection (36). However, we found similar levels of restricted growth of ΔM83 in inbred mouse strains with very high, moderate, and low levels of susceptibility to MCMV infection. We also found that growth of ΔM83 in 21-day-old BALB/c weanlings, which do not yet possess fully active MCMV defenses (8, 24), showed a pattern of attenuation similar to that seen in adult BALB/c mice (data not shown). Taken together, it appears that the reduced level of replication of ΔM83 in the salivary gland is not mediated by host immune responses.
Although we cannot exclude the possibility that the expression of the lacZ or gpt transgene contributed at least partially to the attenuation of the ΔM83 and ΔM84 viruses in vivo in our experiments, we consider it very unlikely. There is only one report which suggests that the defective replication of an MCMV deletion mutant in salivary glands might be due to the expression of lacZ (60). In contrast, Farrell et al. recently showed that an MCMV deletion mutant that was constructed with the lacZ/gpt cassette used in our study replicated in the salivary gland to the same high level as the wild-type virus (16). These differential results may be due to the fact that different promoters were used for lacZ expression; the former used the HCMV IE1/IE2 promoter-enhancer (60), while we and Farrell et al. used the weaker human/rat β-actin promoter (62). In another study, Crnković-Mertens and coworkers also examined the effect of lacZ expression on MCMV replication in vivo. In their experiments, they constructed an MCMV deletion mutant that expressed lacZ from the Rous sarcoma virus promoter and then used this mutant to construct a lacZ-deficient virus in which the marker gene was excised (14). Upon infection of newborn BALB/c mice, lacZ+ virus and lacZ deletion mutants were found to replicate to similar levels. Finally, since in our studies both ΔM83 and ΔM84 viruses contained the same marker cassette, the marked difference in the replication of these mutants in the salivary glands and lungs makes it unlikely that lacZ or gpt accounts for the replication defect of ΔM83 in these organs. Our data also indicate that the fusion of 35 amino acids of vector-encoded sequence to the M84 protein in ΔM83 is not responsible for the observed defect in replication in the salivary glands. Upon restoration of the M84 ORF sequence and stop codon in the M83 deletion mutant ΔM83-2, we found that replication in the spleen and salivary glands was attenuated similarly to that of ΔM83. Taken together, our results suggest that it is primarily the deletion of M83, and not the addition of marker genes or a M84 fusion protein, that accounts for the marked attenuation of ΔM83 in salivary glands and lungs.
A key question from these experiments is how the lack of M83 leads to a change in tropism or dissemination. It is possible that the M83 defect inhibits spread of the virus to the salivary glands and lungs, entry into permissive cells, or full replication inside infected cells. Delivery of ΔM83 directly into the lungs by i.n. inoculation consistently resulted in the same low levels of viral replication in this organ and in salivary glands as were found following systemic infection. These results suggested that the spread of ΔM83 to these secondary organs of infection was not significantly affected, although measurement of virally infected leukocytes in mice infected with parent or mutant viruses may help address this point. The relative attenuation of ΔM83 in the salivary gland was found to be dose dependent, and we found that systemic inoculation of a very high viral dose (2.5 × 106 PFU i.p.) was able to at least partially overcome the defect in virus replication in the salivary glands (data not shown). Thus, it appears either that a defect in ΔM83 entry may be compensated by maximal viral seeding or that the efficiency of ΔM83 replication in the salivary gland cells is multiplicity dependent.
Several other mutants of MCMV have been characterized that fail to replicate or replicate poorly in the salivary glands. The Vancouver strain of MCMV, which was isolated after multiple passages of the Smith strain in tissue culture, could not be recovered from the salivary glands of CD1 mice following i.p. inoculation of 106 PFU (7). This tissue culture-adapted strain was found to contain an insertion in the EcoRI K region and a deletion of the XbaI I/L junction in the HindIII E region. Another mutant, RM868, was constructed by insertion of the lacZ/gpt cassette into ORF m133 in the HindIII J region (35). While the RM868 virus was found to replicate to wild-type levels in the spleen, adrenals, kidneys, and liver of BALB/c mice, its level of replication in the salivary gland was reduced by 4 orders of magnitude following i.p. inoculation of 106 PFU of tissue culture-derived virus. Because of the selective defect in replication in the salivary gland, the m133 ORF was designated sgg1 (for salivary gland growth 1) (35, 38). As with ΔM83, the replication of another sgg1 mutant, RQ401, was found to be similarly attenuated following either i.n. or i.p. inoculation (38). However, both sgg1 mutants appear to be more defective than ΔM83, since the dose-dependent defect in virus replication in the salivary gland never reduced ΔM83 titers to more than 3 orders of magnitude below the parental-strain level. Because neither the Vancouver strain nor the sgg1 mutation map to the HindIII C region, M83 appears to be an additional MCMV gene that is specifically involved with maximal replication in the salivary gland, and perhaps the lung. This is not surprising due to the importance of salivary gland tropism for viral amplification, virulent-virus production (31), and, most likely, horizontal transmission. The observation that titers of 104 PFU per salivary gland still result following deletion of M83 suggests that the M83 gene product plays a smaller role in replication in salivary glands than do the other sgg ORF products described. Perhaps the sgg1 β and M83 γ genes cooperate during infection of salivary gland cells in order to help produce the 108-PFU titers observed in that organ.
Both MCMV deletion mutants were found to be competent in establishing latency in the organs that usually harbor latent MCMV genomes. The levels of latent ΔM83 and ΔM84 genomes in the salivary glands and lungs of BALB/c mice may have been lower than those of the parent strain, although our detection methods were only semiquantitative. While the levels of replication of both mutants were lower in these organs during the acute infection, the relative amounts of infectious ΔM83 and ΔM84 viruses did not correlate with their respective latent-genome levels. For example, although ΔM83 replication was severely restricted in the salivary glands, the amount of latent viral genome in this organ was nearly as large as that of K181. In addition, ΔM84 infection resulted in the lowest level of latent viral DNA in the salivary gland and lung, even though ΔM84 consistently replicated to higher levels than ΔM83 in these organs. These findings are consistent with previous studies that showed that while the overall viral load during acute infection generally affects the resultant latent-genome levels (49), local virus production is not linked to the level of latent DNA in a particular organ (3). In addition, MCMV mutants that are replication defective in vivo but establish latency have been described (6), indicating that replication and latency establishment may not be directly linked. While the levels of latent ΔM83 and ΔM84 DNA were similar to those of the parent strain, their relative abilities to reactivate into productive infections are not known. It was previously found that one MCMV mutant that initially replicated to 106-fold lower levels than the parent strain was able to reactivate with wild-type kinetics in cocultures of splenic fragments (60).
CTLs specific for pp65 are abundant in HCMV-seropositive individuals across diverse HLA genetic backgrounds (63), suggesting a key role for this protein in the protective response. In these studies, we found that i.p. immunization of BALB/c mice with either mutant provided the same level of protection against subsequent lethal challenge as did immunization with the parent strain. This suggests that the responses to M83 or M84 during infection of this strain are not essential for the development of protection in this strain. While the CTL responses to the IE1 pp89 protein have been shown previously to be immunodominant in BALB/c mice (33), there is evidence that there are CTL responses to other viral targets which may contribute to complete immunity (29, 50, 51). We have found that after genetic immunization of mice with plasmids expressing either the M83 or M84 protein, only the immune responses to M84 conferred protection against replication in the spleen, and only in the BALB/c H-2d strain (43). Although CTLs specific to M83 may be generated during natural infection, our DNA immunization assay indicates that the BALB/c mouse successfully targets M84 instead of M83 for the generation of protective immunity.
Having found that prior immunization with the attenuated ΔM83 virus could protect against a subsequent lethal challenge, we sought to determine if there was a minimum dose of immunizing virus that could provide complete protection without establishing latency. Although a 200-PFU i.p. dose of ΔM83 prevented replication of the challenge virus in the spleen and salivary gland, both the immunizing and challenge viruses established latency. The amount of latent ΔM83 genome in the spleen was near the limit of sensitivity for the assay used, and it is not known if the level of latent genome would have been further diminished or become zero following immunization with 30 PFU of virus. Other attenuated MCMV mutants have been described that are able to induce protective immunity while not significantly replicating or establishing latency. One such mutant, recently described, could not be reactivated from splenic explants after it was administered subcutaneously but was able to significantly delay the onset and reduce the overall frequency of reactivation of challenge virus (37). However, the risk of reactivation of the challenge virus was not eliminated by immunization with this virus. Prevention of MCMV latency by augmentation of antiviral immune effectors has been a difficult prospect, since it has been found that adoptive transfer of up to 107 CD8+ lymphocytes from MCMV-immune mice cannot prevent the establishment of latency of a challenge virus, although the levels of latent DNA genome are reduced (59). A further understanding of the immune mechanisms responsible for clearance of the acute infection and minimizing the levels of resulting latent virus is needed to define the conditions necessary for preventing the establishment of latency.
These results further demonstrate the similarities between the homologous gene products of HCMV and MCMV, and they raise new questions about the functions of the UL83 and UL84 gene products during infection of the host. Because studies of HCMV pathogenesis have practical limitations, the continued understanding of the functions of HCMV gene products will continue to rely on parallel studies in the mouse model. We hope that together these studies will provide the basis for designing a safe and effective vaccine against HCMV infection and disease.
ACKNOWLEDGMENTS
We thank Chuck Clark for constructing the EGFP-puro cassette, Billy Krauss for excellent technical assistance, and members of this laboratory for critical reading of the manuscript.
This work was supported by research grant no. 6-FY97-0409 and 6-FY98-0650 from the March of Dimes Birth Defects Foundation and by NIH grant AI20954. L. D. Cranmer was supported in part by a grant from the Life and Health Insurance Medical Research Fund and by NIH-NIGMS predoctoral training grant GM07198.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1993. [Google Scholar]
- 2.Baldick C J, Jr, Shenk T. Proteins associated with purified human cytomegalovirus particles. J Virol. 1996;70:6097–6105. doi: 10.1128/jvi.70.9.6097-6105.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balthesen M, Dreher L, Lucin P, Reddehase M J. The establishment of cytomegalovirus latency in organs is not linked to local virus production during primary infection. J Gen Virol. 1994;75:2329–2336. doi: 10.1099/0022-1317-75-9-2329. [DOI] [PubMed] [Google Scholar]
- 4.Balthesen M, Messerle M, Reddehase M J. Lungs are a major organ site of cytomegalovirus latency and recurrence. J Virol. 1993;67:5360–5366. doi: 10.1128/jvi.67.9.5360-5366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beninga J, Kropff B, Mach M. Comparative analysis of fourteen individual human cytomegalovirus proteins for helper T cell response. J Gen Virol. 1995;76:153–160. doi: 10.1099/0022-1317-76-1-153. [DOI] [PubMed] [Google Scholar]
- 6.Bevan I S, Sammons C C, Sweet C. Investigation of murine cytomegalovirus latency and reactivation in mice using viral mutants and the polymerase chain reaction. J Med Virol. 1996;48:308–320. doi: 10.1002/(SICI)1096-9071(199604)48:4<308::AID-JMV3>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 7.Boname J M, Chantler J K. Characterization of a strain of murine cytomegalovirus which fails to grow in the salivary glands of mice. J Gen Virol. 1992;73:2021–2029. doi: 10.1099/0022-1317-73-8-2021. [DOI] [PubMed] [Google Scholar]
- 8.Booss J, Wheelock E F. Correlation of survival from murine cytomegalovirus infection with spleen cell responsiveness to concanavalin A. Proc Soc Exp Biol Med. 1975;149:443–445. doi: 10.3181/00379727-149-38824. [DOI] [PubMed] [Google Scholar]
- 9.Boppana S B, Britt W J. Recognition of human cytomegalovirus (HCMV) gene products by HCMV-specific cytotoxic T cells. Virology. 1996;222:293–296. doi: 10.1006/viro.1996.0424. [DOI] [PubMed] [Google Scholar]
- 10.Britt W J, Auger D. Human cytomegalovirus virion-associated protein with kinase activity. J Virol. 1986;59:185–188. doi: 10.1128/jvi.59.1.185-188.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins T, Pomeroy C, Jordan M C. Detection of latent cytomegalovirus DNA in diverse organs of mice. J Infect Dis. 1993;168:725–729. doi: 10.1093/infdis/168.3.725. [DOI] [PubMed] [Google Scholar]
- 12.Cranmer L D, Clark C, Spector D H. Cloning, characterization, and expression of the murine cytomegalovirus homologue of the human cytomegalovirus 28-kDa matrix phosphoprotein (UL99) Virology. 1994;205:417–429. doi: 10.1006/viro.1994.1662. [DOI] [PubMed] [Google Scholar]
- 13.Cranmer L D, Clark C L, Morello C S, Farrell H E, Rawlinson W D, Spector D H. Identification, analysis, and evolutionary relationships of the putative murine cytomegalovirus homologs of the human cytomegalovirus UL82 (pp71) and UL83 (pp65) matrix phosphoproteins. J Virol. 1996;70:7929–7939. doi: 10.1128/jvi.70.11.7929-7939.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crnković-Mertens I, Messerle M, Milotić I, Szepan U, Kučić N, Krmpotić A, Jonjić S, Koszinowski U H. Virus attenuation after deletion of the cytomegalovirus Fc receptor gene is not due to antibody control. J Virol. 1998;72:1377–1382. doi: 10.1128/jvi.72.2.1377-1382.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elliott R, Clark C, Jaquish D, Spector D H. Transcription analysis and sequence of the putative murine cytomegalovirus DNA polymerase gene. Virology. 1991;185:169–186. doi: 10.1016/0042-6822(91)90765-4. [DOI] [PubMed] [Google Scholar]
- 16.Farrell H E, Vally H, Lynch D M, Fleming P, Shellam G R, Scalzo A A, Davis-Poynter N J. Inhibition of natural killer cells by a cytomegalovirus MHC class I homologue in vivo. Nature. 1997;386:510–514. doi: 10.1038/386510a0. [DOI] [PubMed] [Google Scholar]
- 17.Gallina A, Percivalle E, Simoncini L, Revello M G, Gerna G, Milanesi G. Human cytomegalovirus pp65 lower matrix phosphoprotein harbours two transplantable nuclear localization signals. J Gen Virol. 1996;77:1151–1157. doi: 10.1099/0022-1317-77-6-1151. [DOI] [PubMed] [Google Scholar]
- 18.Gallina A, Simoncini L, Garbelli S, Percivalle E, Pedrali-Noy G, Lee K S, Erikson R L, Plachter B, Gerna G, Milanesi G. Polo-like kinase 1 as a target for human cytomegalovirus pp65 lower matrix protein. J Virol. 1999;73:1468–1478. doi: 10.1128/jvi.73.2.1468-1478.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gebert S, Schmolke S, Sorg G, Flöss S, Plachter B, Stamminger T. The UL84 protein of human cytomegalovirus acts as a transdominant inhibitor of immediate-early-mediated transactivation that is able to prevent viral replication. J Virol. 1997;71:7048–7060. doi: 10.1128/jvi.71.9.7048-7060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilbert M J, Riddell S R, Plachter B, Greenberg P D. Cytomegalovirus selectively blocks antigen processing and presentation of its immediate-early gene product. Nature. 1996;383:720–722. doi: 10.1038/383720a0. [DOI] [PubMed] [Google Scholar]
- 21.González Armas J C, Morello C S, Cranmer L D, Spector D H. DNA immunization confers protection against murine cytomegalovirus infection. J Virol. 1996;70:7921–7928. doi: 10.1128/jvi.70.11.7921-7928.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grundy J E, Mackenzie J S, Stanley N F. Influence of H-2 and non-H-2 genes on resistance to murine cytomegalovirus infection. Infect Immun. 1981;32:277–286. doi: 10.1128/iai.32.1.277-286.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 24.Hayashi K, Eizuru Y, Sato S, Minamishima Y. The role of NK cell activity in age-dependent resistance of mice to murine cytomegalovirus infection. Microbiol Immunol. 1985;29:939–950. doi: 10.1111/j.1348-0421.1985.tb02958.x. [DOI] [PubMed] [Google Scholar]
- 25.He Y S, Xu L, Huang E-S. Characterization of human cytomegalovirus UL84 early gene and identification of its putative protein product. J Virol. 1992;66:1098–1108. doi: 10.1128/jvi.66.2.1098-1108.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hengel H, Koszinowski U H. Interference with antigen processing by viruses. Curr Opin Immunol. 1997;9:470–476. doi: 10.1016/s0952-7915(97)80097-0. [DOI] [PubMed] [Google Scholar]
- 27.Henry S C, Hamilton J D. Detection of murine cytomegalovirus immediate early 1 transcripts in the spleens of latently infected mice. J Infect Dis. 1993;167:950–954. doi: 10.1093/infdis/167.4.950. [DOI] [PubMed] [Google Scholar]
- 28.Ho M. Cytomegalovirus biology and infection. 2nd ed. New York, N.Y: Plenum Medical Book Co.; 1992. [Google Scholar]
- 29.Holtappels R, Podlech J, Geginat G, Steffens H-P, Thomas D, Reddehase M J. Control of murine cytomegalovirus in the lungs: relative but not absolute immunodominance of the immediate-early 1 nonapeptide during the antiviral cytolytic T-lymphocyte response in pulmonary infiltrates. J Virol. 1998;72:7201–7212. doi: 10.1128/jvi.72.9.7201-7212.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jahn G, Scholl B C, Traupe B, Fleckenstein B. The two major structural phosphoproteins (pp65 and pp150) of human cytomegalovirus and their antigenic properties. J Gen Virol. 1987;68:1327–1337. doi: 10.1099/0022-1317-68-5-1327. [DOI] [PubMed] [Google Scholar]
- 31.Jordan M C, Takagi J L. Virulence characteristics of murine cytomegalovirus in cell and organ cultures. Infect Immun. 1983;41:841–843. doi: 10.1128/iai.41.2.841-843.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koffron A J, Hummel M, Patterson B K, Yan S, Kaufman D B, Fryer J P, Stuart F P, Abecassis M I. Cellular localization of latent murine cytomegalovirus. J Virol. 1998;72:95–103. doi: 10.1128/jvi.72.1.95-103.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koszinowski U H, Reddehase M J, Del Val M. Principles of cytomegalovirus antigen presentation in vitro and in vivo. Semin Immunol. 1992;4:71–79. [PubMed] [Google Scholar]
- 34.Kurz S, Steffens H-P, Mayer A, Harris J R, Reddehase M J. Latency versus persistence or intermittent recurrences: evidence for a latent state of murine cytomegalovirus in the lungs. J Virol. 1997;71:2980–2987. doi: 10.1128/jvi.71.4.2980-2987.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lagenaur L A, Manning W C, Vieira J, Martens C L, Mocarski E S. Structure and function of the murine cytomegalovirus sgg1 gene: a determinant of viral growth in salivary gland acinar cells. J Virol. 1994;68:7717–7727. doi: 10.1128/jvi.68.12.7717-7727.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lathbury L J, Allan J E, Shellam G R, Scalzo A A. Effect of host genotype in determining the relative roles of natural killer cells and T cells in mediating protection against murine cytomegalovirus infection. J Gen Virol. 1996;77:2605–2613. doi: 10.1099/0022-1317-77-10-2605. [DOI] [PubMed] [Google Scholar]
- 37.MacDonald M R, Li X-Y, Stenberg R M, Campbell A E, Virgin H W., IV Mucosal and parenteral vaccination against acute and latent murine cytomegalovirus (MCMV) infection by using an attenuated MCMV mutant. J Virol. 1998;72:442–451. doi: 10.1128/jvi.72.1.442-451.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manning W C, Stoddart C A, Lagenaur L A, Abenes G B, Mocarski E S. Cytomegalovirus determinant of replication in salivary glands. J Virol. 1992;66:3794–3802. doi: 10.1128/jvi.66.6.3794-3802.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McLaughlin-Taylor E, Pande H, Forman S J, Tanamachi B, Li C R, Zaia J A, Greenberg P D, Riddell S R. Identification of the major late human cytomegalovirus matrix protein pp65 as a target antigen for CD8+ virus-specific cytotoxic T lymphocytes. J Med Virol. 1994;43:103–110. doi: 10.1002/jmv.1890430119. [DOI] [PubMed] [Google Scholar]
- 40.Mercer J A, Marks J R, Spector D H. Molecular cloning and restriction endonuclease mapping of the murine cytomegalovirus genome (Smith strain) Virology. 1983;129:94–106. doi: 10.1016/0042-6822(83)90398-7. [DOI] [PubMed] [Google Scholar]
- 41.Mercer J A, Wiley C A, Spector D H. Pathogenesis of murine cytomegalovirus infection: identification of infected cells in the spleen during acute and latent infections. J Virol. 1988;62:987–997. doi: 10.1128/jvi.62.3.987-997.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mims C A, Gould J. Splenic necrosis in mice infected with cytomegalovirus. J Infect Dis. 1978;137:587–591. doi: 10.1093/infdis/137.5.587. [DOI] [PubMed] [Google Scholar]
- 43.Morello, C. S., L. D. Cranmer, and D. H. Spector. Unpublished data.
- 44.Pollock J L, Virgin H W., IV Latency, without persistence, of murine cytomegalovirus in the spleen and kidney. J Virol. 1995;69:1762–1768. doi: 10.1128/jvi.69.3.1762-1768.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pomeroy C, Hilleren P J, Jordan M C. Latent murine cytomegalovirus DNA in splenic stromal cells of mice. J Virol. 1991;65:3330–3334. doi: 10.1128/jvi.65.6.3330-3334.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45a.Prichard, M., and G. Pari. Personal communication.
- 46.Quinnan G V, Jr, Kirmani N, Esber E, Saral R, Manischewitz J F, Rogers J L, Rook A H, Santos G W, Burns W H. HLA-restricted cytotoxic T lymphocyte and nonthymic cytotoxic lymphocyte responses to cytomegalovirus infection of bone marrow transplant recipients. J Immunol. 1981;126:2036–2041. [PubMed] [Google Scholar]
- 47.Quinnan G V, Jr, Kirmani N, Rook A H, Manischewitz J F, Jackson L, Moreschi G, Santos G W, Saral R, Burns W H. Cytotoxic T cells in cytomegalovirus infection: HLA-restricted T-lymphocyte and non-T-lymphocyte cytotoxic responses correlate with recovery from cytomegalovirus infection in bone-marrow-transplant recipients. N Engl J Med. 1982;307:7–13. doi: 10.1056/NEJM198207013070102. [DOI] [PubMed] [Google Scholar]
- 48.Rawlinson W D, Farrell H E, Barrell B G. Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol. 1996;70:8833–8849. doi: 10.1128/jvi.70.12.8833-8849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reddehase M J, Balthesen M, Rapp M, Jonjic S, Pavic I, Koszinowski U H. The conditions of primary infection define the load of latent viral genome in organs and the risk of recurrent cytomegalovirus disease. J Exp Med. 1994;179:185–193. doi: 10.1084/jem.179.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reddehase M J, Bühring H-J, Koszinowski U H. Cloned long-term cytolytic T-lymphocyte line with specificity for an immediate-early membrane antigen of murine cytomegalovirus. J Virol. 1986;57:408–412. doi: 10.1128/jvi.57.1.408-412.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reddehase M J, Keil G M, Koszinowski U H. The cytolytic T lymphocyte response to the murine cytomegalovirus. II. Detection of virus replication stage-specific antigens by separate populations of in vivo active cytolytic T lymphocyte precursors. Eur J Immunol. 1984;14:56–61. doi: 10.1002/eji.1830140111. [DOI] [PubMed] [Google Scholar]
- 52.Reusser P, Riddell S R, Meyers J D, Greenberg P D. Cytotoxic T-lymphocyte response to cytomegalovirus after human allogeneic bone marrow transplantation: pattern of recovery and correlation with cytomegalovirus infection and disease. Blood. 1991;78:1373–1380. [PubMed] [Google Scholar]
- 53.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 54.Sarisky R T, Hayward G S. Evidence that the UL84 gene product of human cytomegalovirus is essential for promoting oriLyt-dependent DNA replication and formation of replication compartments in cotransfection assays. J Virol. 1996;70:7398–7413. doi: 10.1128/jvi.70.11.7398-7413.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmolke S, Drescher P, Jahn G, Plachter B. Nuclear targeting of the tegument protein pp65 (UL83) of human cytomegalovirus: an unusual bipartite nuclear localization signal functions with other portions of the protein to mediate its efficient nuclear transport. J Virol. 1995;69:1071–1078. doi: 10.1128/jvi.69.2.1071-1078.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmolke S, Kern H F, Drescher P, Jahn G, Plachter B. The dominant phosphoprotein pp65 (UL83) of human cytomegalovirus is dispensable for growth in cell culture. J Virol. 1995;69:5959–5968. doi: 10.1128/jvi.69.10.5959-5968.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spaete R R, Mocarski E S. Insertion and deletion mutagenesis of the human cytomegalovirus genome. Proc Natl Acad Sci USA. 1987;84:7213–7217. doi: 10.1073/pnas.84.20.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spector D J, Tevethia M J. Protein-protein interactions between human cytomegalovirus IE2-580aa and pUL84 in lytically infected cells. J Virol. 1994;68:7549–7553. doi: 10.1128/jvi.68.11.7549-7553.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Steffens H-P, Kurz S, Holtappels R, Reddehase M J. Preemptive CD8 T-cell immunotherapy of acute cytomegalovirus infection prevents lethal disease, limits the burden of latent viral genomes, and reduces the risk of virus recurrence. J Virol. 1998;72:1797–1804. doi: 10.1128/jvi.72.3.1797-1804.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stoddart C A, Cardin R D, Boname J M, Manning W C, Abenes G B, Mocarski E S. Peripheral blood mononuclear phagocytes mediate dissemination of murine cytomegalovirus. J Virol. 1994;68:6243–6253. doi: 10.1128/jvi.68.10.6243-6253.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Zanten J, Harmsen M C, van der Meer P, van der Bij W, van Son W J, van der Giessen M, Prop J, de Leij L, The T H. Proliferative T cell responses to four human cytomegalovirus-specific proteins in healthy subjects and solid organ transplant recipients. J Infect Dis. 1995;172:879–882. doi: 10.1093/infdis/172.3.879. [DOI] [PubMed] [Google Scholar]
- 62.Vieira J, Farrell H E, Rawlinson W D, Mocarski E S. Genes in the HindIII J fragment of the murine cytomegalovirus genome are dispensable for growth in cultured cells: insertion mutagenesis with a lacZ/gpt cassette. J Virol. 1994;68:4837–4846. doi: 10.1128/jvi.68.8.4837-4846.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wills M R, Carmichael A J, Mynard K, Jin X, Weekes M P, Plachter B, Sissons J G P. The human cytotoxic T-lymphocyte (CTL) response to cytomegalovirus is dominated by structural protein pp65: frequency, specificity, and T-cell receptor usage of pp65-specific CTL. J Virol. 1996;70:7569–7579. doi: 10.1128/jvi.70.11.7569-7579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]