Abstract
All of the previously reported recombinant RNA-dependent RNA polymerases (RdRp), the NS5B enzymes, of hepatitis C virus (HCV) could function only in a primer-dependent and template-nonspecific manner, which is different from the expected properties of the functional viral enzymes in the cells. We have now expressed a recombinant NS5B that is able to synthesize a full-length HCV genome in a template-dependent and primer-independent manner. The kinetics of RNA synthesis showed that this RdRp can initiate RNA synthesis de novo and yield a full-length RNA product of genomic size (9.5 kb), indicating that it did not use the copy-back RNA as a primer. This RdRp was also able to accept heterologous viral RNA templates, including poly(A)- and non-poly(A)-tailed RNA, in a primer-independent manner, but the products in these cases were heterogeneous. The RdRp used some homopolymeric RNA templates only in the presence of a primer. By using the 3′-end 98 nucleotides (nt) of HCV RNA, which is conserved in all genotypes of HCV, as a template, a distinct RNA product was generated. Truncation of 21 nt from the 5′ end or 45 nt from the 3′ end of the 98-nt RNA abolished almost completely its ability to serve as a template. Inclusion of the 3′-end variable sequence region and the U-rich tract upstream of the X region in the template significantly enhanced RNA synthesis. The 3′ end of minus-strand RNA of HCV genome also served as a template, and it required a minimum of 239 nt from the 3′ end. These data defined the cis-acting sequences for HCV RNA synthesis at the 3′ end of HCV RNA in both the plus and minus senses. This is the first recombinant HCV RdRp capable of copying the full-length HCV RNA in the primer-independent manner expected of the functional HCV RNA polymerase.
Hepatitis C virus (HCV) is a positive-sense single-stranded RNA virus causing acute and chronic hepatitis in humans, frequently leading to liver cirrhosis and hepatocellular carcinoma (14, 31). The HCV genomic RNA is 9.5 kb in length and consists of a long open reading frame (ORF), which is flanked by highly conserved untranslated regions (UTRs) at both the 5′ and 3′ ends (6, 7, 20, 38, 41, 44, 46). The ORF encodes a polyprotein of approximately 3,000 amino acids, which is processed into at least 10 polypeptides by cellular and viral proteases (25). The 5′-UTR of 341 nucleotides (nt) contains an internal ribosomal entry site (IRES), which consists of four stem-loop structures followed by a translational initiation codon for the polypeptide (6, 7, 41, 44). The 3′-UTR of 200 to 300 nt contains (i) a short variable sequence of approximately 40 nt; (ii) a poly(U) region of variable length; (iii) a polypyrimidine poly(U/C) tract; and (iv) a highly conserved 98-nt region, termed the X region (20, 38, 46). The 98-nt forms three stable stem-loop structures and binds a cellular protein, polypyrimidine-tract-binding protein (PTB) (16, 40). The PTB-binding sites were mapped to stem-loop structures 2 and 3 of the X region (16). The X region has been shown to enhance HCV translation, possibly through interaction with the 5′ end (17). Conceivably, the 3′-end sequence may also be important for HCV RNA replication, because it likely includes the cis-acting signals for RNA replication.
The HCV NS5B is at the C terminus of the HCV polyprotein and is the last viral protein to be translated from the HCV genome. It contains motifs shared by RNA-dependent RNA polymerases (RdRps), such as the GDD motif (21). Indeed, RdRp activity has been demonstrated with the recombinant NS5B expressed in insect cells (4, 23) and the C-terminus-truncated NS5B expressed in Escherichia coli (10, 47). All of these reported recombinant HCV RdRps utilize a wide range of RNAs as a template without preference, although they do prefer certain homopolyribonucleotides to others. All of them also require a separate RNA or a folded-back 3′ end of the template as a primer. In only one report was the HCV RdRp shown to be able to use the full-length HCV RNA as a template, and it generated an 18-kb RNA product, probably by a copy-back priming mechanism (23). The purified NS5B from the insect cells was associated with a terminal transferase (TNTase) activity, which might add extra nucleotides to the end of the template and, in turn, result in snap-back initiation of RNA synthesis (4, 23). HCV RdRps without a TNTase activity have also been reported (47); however, their template specificity and the nature of their product have not been examined. A primer-dependent RNA synthesis using a snap-back RNA template is not expected to yield an authentic RNA product, because the 3′ end of the RNA template will inevitably be excluded from the product, or extra nucleotides will be included in the product. Similar snap-back RNA synthesis was observed with the poliovirus RdRp, 3Dpol, purified from infected HeLa cells (24, 49), and recombinant RdRps of rabbit hemorrhagic disease virus and bovine viral diarrhea virus expressed in E. coli (43, 50). RdRps from some animal viruses were able to synthesize template-size products, but only in the presence of an artificial primer (30, 32, 43). In contrast, RdRps from most of the plant RNA viruses can recognize the structured 3′ end of plant viral RNA as a promoter and initiate RNA synthesis de novo. For example, brome mosaic virus RdRp was able to initiate the synthesis of minus-strand RNA by recognition of the tRNA-like structure at the 3′ end of the genome (9), and many other plant viral RdRps showed relatively stringent template specificity in vitro in a primer-independent manner (29, 35, 36).
The replication of the plus-strand RNA viral genome consists of two steps: synthesis of complementary minus-strand RNA with the plus-strand genomic RNA as a template and the subsequent synthesis of the plus-strand RNA genome with the minus-strand RNA as a template. Thus, the cis-acting sequences required for the replication of the viral genome are generally thought to be located at the 3′ end of both plus- and minus-strand RNAs and act as promoters for the initiation of RNA synthesis by RdRp. Promoter analysis using defective interfering (DI) RNAs and in vitro transcription systems for a number of viruses revealed that the 3′ end sequence and structure on both plus and minus strands of viral RNA genome are required for RNA replication (3, 12, 22, 27, 33, 34, 36, 37).
In this study, we have expressed an enzymatically active HCV NS5B in E. coli, which can copy HCV RNA into a full-length RNA product of 9.5 kb in the absence of an added primer. It does not have intrinsic terminal transferase activity. It uses the 3′ end sequence of both plus and minus strands of HCV RNA as templates. Truncation of these end sequences resulted in the loss of template function, thus defining the cis-acting sequences essential for the synthesis of plus- and minus-strand HCV RNA in vitro. This recombinant HCV RdRp has the properties expected of an authentic HCV polymerase and thus will provide an excellent target for screening antiviral agents.
MATERIALS AND METHODS
Cloning, expression, and purification of recombinant HCV NS5B from E. coli.
The cDNA for the NS5B ORF from amino acids 2420 to 3011 of an HCV genotype 1b isolate (48) was cloned into pCR2.1 (Invitrogen) and subcloned into an expression vector, pTrcHisB (Invitrogen), under the control of the E. coli trc promoter to obtain pThNS5B. The resulting expression vector encodes NS5B with a (His)6 and several extra amino acids at the N terminus. The NS5B mutant containing a substitution of the first aspartate of the GDD motif with histidine (NS5BD318H) was generated by site-directed mutagenesis with a sequential PCR method (8).
NS5B was expressed in E. coli BL21 (F− ompT hsdSB [rB−mB−] gal dcm; Novagen) by the following procedures. E. coli BL21 transformed with pThNS5B was grown in Luria-Bertani medium containing 100 μg of ampicillin per ml to an optical density at 600 nm (OD600) of 0.8 at 37°C, and protein expression was induced at 25°C for 6 h by addition of 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cell pellets obtained from 1-liter cultures were washed once with phosphate-buffered saline (PBS) and resuspended in 20 ml of binding buffer (50 mM Na-phosphate [pH 8.0], 300 mM NaCl, 10 mM imidazole, 10 mM β-mercaptoethanol, 10% glycerol, 1% Nonidet P-40) supplemented with 1 mM phenylmethylsulfonyl fluoride (Sigma) and 10 mM leupeptin (Boehringer Mannheim). After undergoing freezing and thawing once, cells were sonicated on ice, and the cleared lysate was obtained by centrifugation at 35,000 × g for 15 min. The (His)6-tagged NS5B was bound to Ni-nitrilotriacetic acid (NTA)-Sepharose resin (Qiagen) preequilibrated with the binding buffer and washed with the binding buffer containing 50 mM imidazole. The bound NS5B was eluted with the binding buffer containing imidazole in a step-gradient manner (ca. 100 mM to 500 mM). The NS5B protein peaks, eluted with 250 to 350 mM imidazole, were combined and dialyzed against buffer A (50 mM Tris-HCl [pH 8.0], 1 mM dithiothreitol [DTT], 50 mM NaCl, 5 mM MgCl2, 10% glycerol), followed by freezing at −80°C in a small aliquot. Protein concentrations were determined by using a Bio-Rad protein assay kit with a bovine serum albumin standard. The enzyme could be kept for 2 to 3 months without a noticeable drop in the enzymatic activity.
Micrococcal nuclease treatment of the NS5B-active fraction.
To remove any RNA or DNA fragments in the NS5B preparation that might act as primers, the NS5B-active fractions were treated with 1 U of micrococcal nuclease per μl (Boehringer Mannheim) in the presence of 2 mM calcium acetate at 30°C for 30 min, and EGTA was then added to a final concentration of 5 mM to stop the reaction. The nuclease-treated NS5B fraction was used to analyze the kinetics of HCV full-length RNA synthesis.
Western blot analysis.
The anti-NS5B antibody generated in rabbits (15) and human HCV genotype 1b-infected patient serum were used to detect NS5B. Fractions from the Ni-NTA resin were resolved on a sodium dodecyl sulfate (SDS)–10% polyacrylamide gel, transferred to a nitrocellulose membrane, blocked with 5% nonfat dry milk in PBS, and reacted first with rabbit anti-NS5B polyclonal antibody and then with a goat anti-rabbit antibody conjugated to horseradish peroxidase (American Qualex). Membrane-bound antibodies were detected with the ECL enhanced chemiluminescence kit (Amersham).
Preparation of in vitro transcripts for RdRp templates.
For preparation of the full-length HCV genomic RNA, an HCV 1b cDNA clone, pCV-J4L6S (48), was digested with XbaI, and the linearized plasmid was used for the synthesis of in vitro transcripts by using T7 RNA polymerase (Promega) as described previously (16). The in vitro runoff transcripts are expected to have an extra 4 nt derived from the vector sequence. pDE25 plasmid linearized with XbaI was used to obtain the mouse hepatitis virus (MHV) DI RNA transcripts (26); the in vitro runoff transcripts are expected to have a poly(A) tail followed by 9 nt derived from the vector sequence at the 3′ end of the transcripts. For a non-poly(A)-tailed RNA template, SmaI-digested pT7TCVms containing the full-length cDNA of turnip crinkle virus (TCV) was used to generate in vitro transcripts (28).
For generation of 98-nt RNA templates representing the X region of the HCV 3′ end and its deletion mutants, primer sets previously described (16) were used for PCR amplification with pCR-HCV-X(+) and its derivatives (16) as templates and Vent DNA polymerase (New England Biolabs). For preparation of minus-strand RNA templates complementary to the HCV 5′-UTR region, reverse oligonucleotides T7(−)341 (5′-TAATACGACTCACTATAGGGTGCACGGTCTACGAG-3′), T7(−)239 (5′-TAATACGACTCACTATAGGGGCACGCCCAAATCTC-3′), T7(−)122 (5′-TAATACGACTCACTATAGGGTCCTGGAGGCTGCAC-3′), T7(−)84 (5′-TAATACGACTCACTATAGGGCTAGACGCTTTCTGC-3′), and T7(−)44 (5′-TAATACGACTCACTATAGGGAGTGATCTATGGTGG-3′) were used with a forward oligonucleotide, HCV1a5′end (5′-GCCAGCCCCCTGATGGGG-3′), to amplify the DNA templates by using Vent DNA polymerase. The T7 RNA polymerase promoter sequence is underlined, and the sequence complementary to the HCV 5′-UTR is shown in boldface and italic. The PCR-amplified products were gel purified, and transcripts were synthesized with T7 RNA polymerase.
After in vitro transcription with T7 RNA polymerase, DNA templates were digested by treating the transcription mixture with RQ1 DNase (Promega) for 15 min at 37°C. The runoff transcripts were then phenol-chloroform extracted twice and chloroform extracted once to remove T7 RNA polymerase, followed by precipitation with 2.5 volumes of 5 M ammonium acetate and isopropanol (1:5). For preparation of the 98-nt template used for TNTase activity assay, free ribonucleotides present in the transcripts were removed by using a Sephadex G-25 column, and the flowthrough fractions were precipitated with ammonium acetate and isopropanol. The concentration of transcripts was determined by measuring the OD260.
RNA-dependent RNA polymerase and TNTase activity assays.
In vitro RdRp activity was determined in a total volume of 25 μl containing 50 mM Tris-HCl (pH 8.0); 50 mM NaCl; 5 mM MgCl2; 100 mM potassium glutamate; 1 mM DTT; 10% glycerol; 20 μg of actinomycin D per ml (Sigma); 20 U of RNase inhibitor (Promega); 0.5 mM each ATP, CTP, and GTP; 5 μM UTP; 10 μCi of [α-32P]UTP (3,000 Ci/mmol; NEN Research Products); 5 μg of HCV, MHV DI DE25, or TCV RNA; and about 200 ng of the purified NS5B. For small templates, 200 ng of RNA was routinely used. The reaction mixture was incubated at 25°C for 2 h unless otherwise specified. After RdRp reactions, 35 μl of double-distilled H2O containing 20 μg of glycogen (Boehringer Mannheim) and 60 μl of acidic phenol emulsion (phenol-chloroform [Ambion]–10% SDS–0.5 M EDTA [1:1:0.2:0.04]) were added to the reaction mixture to terminate the reactions. The RNAs were then precipitated with 2.5 volumes of 5 M ammonium acetate-isopropanol (1:5), followed by washing with 70% ethanol. The products were resuspended in a denaturing loading buffer containing 95% formamide with 10 mM EDTA and 0.025% (each) xylene cyanol and bromophenol blue. After heat denaturation and quick chilling on ice, the products were resolved on a 5 or 20% sequencing gel (19:1 acrylamide-bisacrylamide) containing 8 M urea in 1× Tris-borate-EDTA buffer or on a 1% agarose gel. The denaturing sequencing gel was prerun at 25 mA for 3 h before samples were loaded. The gels were stained with ethidium bromide, photographed to locate the template positions, and then dried after fixing. The dried gels were exposed to X-ray film for autoradiography. For RdRp reactions with homopolymeric RNAs, 1 μg of homopolymeric RNA [poly(A), poly(C), poly(G), and poly(U); Pharmacia] was added to the reaction mixture as described above, except that 10 μM UTP, GTP, CTP, or ATP was included in the reaction with 10 μCi of [32P]UTP, [32P]GTP, [32P]CTP, or [32P]ATP (3,000 Ci/mmol; NEN Research Products) for poly(A), poly(C), poly(G), and poly(U) template, respectively. The RdRp reactions were conducted in the absence or presence of primer [10 pmol of oligonucleotide (U)20, (G)20, (C)20, and (A)20 for poly(A), poly(C), poly(G), and poly(U), respectively]. After the reactions, the products were precipitated with 5% trichloroacetic acid (TCA) in the presence of 10 μg of calf thymus DNA and applied to a GF/C glass filter (Whatman). The unincorporated nucleotides were washed twice with 5 ml of ice-cold 5% TCA and twice with 5 ml of ice-cold 1% TCA. Finally, the filters were washed with 5 ml of 95% ethanol to remove any remaining TCA and dried. The amount of radioactivity present in each filter was then determined with a liquid scintillation counter (LC600 IC; Beckman).
For the TNTase assay, 10 μCi of [32P]UTP, [32P]GTP, [32P]CTP, or [32P]ATP mixed with 10 μM cold UTP, GTP, CTP, or ATP, respectively, was used as a single ribonucleotide triphosphate (rNTP) in the RdRp reaction mixtures with the 98-nt RNA as a template. Alternatively, oligonucleotide (A)20 or (U)20 (20 pmol each) was used as a template to assay TNTase, with 10 μCi of [32P]UTP, [32P]GTP, [32P]CTP, or [32P]ATP, without cold nucleotide, as the single rNTP.
Nuclease S1 treatment of RdRp products.
The RdRp products were resuspended in nuclease S1 digestion buffer (50 mM NaOAc [pH 4.6], 1 mM ZnSO4, 5% glycerol) containing a low (50 mM) or high (500 mM) concentration of NaCl and digested with 200 U of nuclease S1 per ml (Promega) at 37°C for 30 min. The nuclease S1-treated samples were extracted with phenol-chloroform and chloroform sequentially and precipitated with ethanol. The pellets were resuspended in the denaturing loading buffer and heat denatured at 96°C for 5 min, followed by quick chilling on ice before loading of samples onto the gels.
RESULTS
Expression and purification of recombinant HCV NS5B.
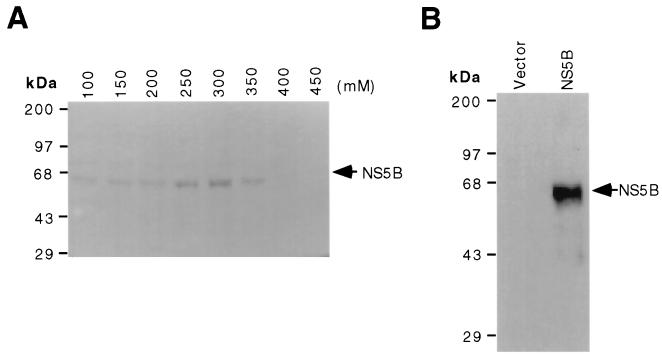
To investigate the biochemical functions of HCV RdRp, the NS5B-encoding cDNA from HCV genotype 1b was PCR amplified and cloned under the control of the E. coli trc promoter. This system avoids the potential contamination by T7 RNA polymerase and allows convenient single-step purification with the Ni-NTA affinity column. Even though it has previously been reported that the overexpressed NS5B usually formed inclusion bodies in E. coli (10, 47), we were able to obtain soluble, enzymatically active full-length HCV NS5B as a fusion protein containing a (His)6 at the N terminus when protein expression was induced at 25°C. The soluble form of full-length NS5B was purified by Ni-NTA affinity chromatography. An NS5B protein of about 65 kDa was obtained with 250 to 350 mM imidazole (Fig. 1A). The authenticity of the recombinant HCV NS5B was confirmed by Western blot analysis with anti-NS5B antibody generated in rabbits (15), detecting a 65-kDa recombinant NS5B protein, which was not present in the corresponding fraction obtained from the E. coli transformed with the expression vector alone (Fig. 1B). This protein was also detected with HCV patient sera (data not shown). The fractions (250 to 450 mM eluates) containing NS5B were pooled, dialyzed, and used for RdRp activity assays.
FIG. 1.
Expression and purification of (His)6-tagged HCV NS5B from E. coli. (A) Imidazole elution profile of the NS5B expressed in E. coli. Fractions eluted from Ni-NTA resin by different concentrations of imidazole were run in an SDS–10% polyacrylamide gel and stained with Coomassie brilliant blue G-250. (B) Western blot analysis of NS5B. Fractions eluted with 200 mM imidazole were used for immunoblotting. Vector and NS5B indicate the fractions obtained from the soluble extracts of E. coli transformed with the expression vector alone and the NS5B-expression vector, pThNS5B, respectively. Arrowheads indicate the positions of NS5B. The sizes of protein markers (Bethesda Research Laboratories) are indicated in kilodaltons.
Template-dependent RNA synthesis by the wild-type and mutant NS5B.
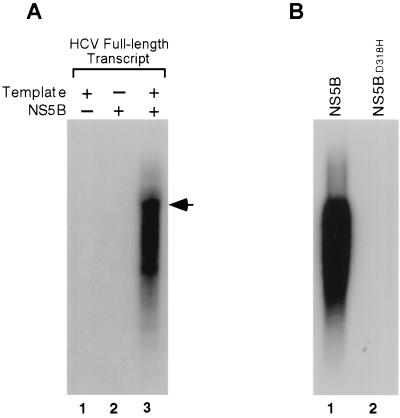
To test the template dependency of NS5B, RdRp activity assays were performed in the absence and presence of an exogenous template. We used the full-length HCV RNA in vitro runoff transcripts as a template. The purified NS5B was able to synthesize an RNA product with a size equivalent to that of the exogenous HCV full-length RNA template without the requirement of an exogenous primer (Fig. 2A, lane 3). In the absence of added RNA, no RNA products were obtained, indicating that the NS5B preparation was not contaminated with any RNA that can serve as a template (Fig. 2A, lane 2). Finally, in the absence of NS5B, no products were produced, indicating that no residual T7 RNA polymerase activity, which can label RNA templates by extension of the 3′ end or transcribe RNA template itself under certain conditions (2, 39), was present in the RNA template used for the RdRp reactions (Fig. 2A, lane 1). Furthermore, the NS5BD318H mutant, which has a histidine at the position of the first aspartate of the GDD motif (21), was completely inactive as an RdRp (Fig. 2B, lane 2). These results indicate that NS5B is the enzyme responsible for the synthesis of HCV full-length RNA in vitro and that the RNA synthesis carried out by NS5B is template dependent.
FIG. 2.
Template dependency of RNA synthesis by the wild-type and mutant NS5B. (A) In vitro RdRp assays were conducted by using full-length HCV RNA as a template under the conditions indicated above the gel. + and − indicate the presence and absence, respectively, of the template and NS5B in the RdRp reactions. An arrowhead indicates the position of the template on 1% agarose gel. (B) Enzyme activity of wild-type (NS5B) and mutant (NS5BD318H) NS5B by using the full-length HCV RNA as a template.
Kinetics of synthesis of full-length HCV RNA by NS5B.
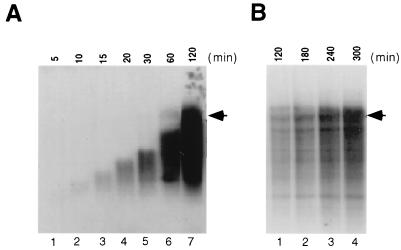
With the full-length HCV genome used as a template for the RdRp activity assay, the time course of RNA synthesis was analyzed to understand the mechanism of RdRp reactions. The results showed that the size of products at the 10-min time point was approximately 1 kb and steadily increased until between 1 and 2 h, when the product reached the template size (Fig. 3A). To eliminate the possibility that the NS5B preparation contained small fragments of RNA or DNA that could serve as primers, the NS5B-active fractions were treated with micrococcal nuclease. The nuclease-treated NS5B still showed similar kinetics of HCV RNA synthesis (data not shown). These results strongly suggest that the RdRp reaction initiating de novo to synthesize an RNA product, rather than initiated from a snapped-back 3′ end of the template; the latter mechanism would have generated RNA products starting from 9.5 kb and ending at 19 kb, as seen previously (23). From the kinetics of the increase in size of the RNA products, the transcription elongation rate is estimated to be approximately 150 nt/min.
FIG. 3.
Kinetics of RNA synthesis by HCV NS5B with full-length HCV RNA as a template. (A) In vitro RdRp reactions were terminated at the time points indicated above the gel in minutes and analyzed on a 1% agarose gel after the products had been heat denatured. (B) Longer RdRp reactions up to 5 h. Exposure time was controlled to visualize the discrete bands for late time points (2, 3, 4, and 5 h). Arrowheads indicate the positions of the RNA template.
Further incubation resulted in accumulation of all of these products, particularly the RNA product of template size. In addition, an RNA product longer than the template size also appeared at late time points (Fig. 3B, lanes 1 to 4). The nature of this product is not yet known, partly because the size of this RNA could not be determined accurately. Both the longer-than-template and template-size products accumulated up to 5 h. Since the longer-than-template product appeared only after a 1-h incubation, it may represent an extension of the initial RdRp product, probably extended on a snapped-back RNA product. Shorter exposure of the gel to X-ray film (Fig. 3B) also revealed several distinct products smaller than the template size. The profile of these products remained unchanged from 2 to 5 h, while they increased in amount. They may represent strong transcription-pausing sites on the template, which are commonly observed in RNA polymerase reactions.
Nuclease S1 digestion of RdRp products.
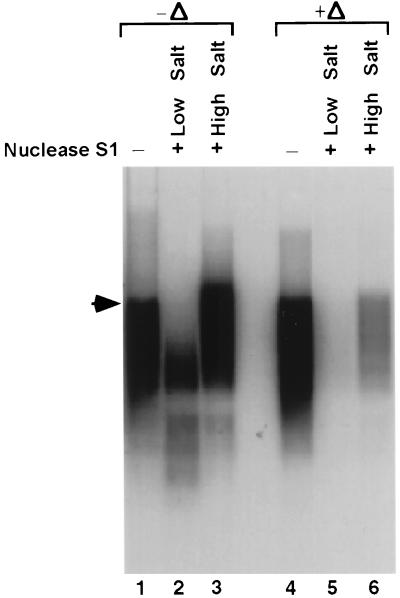
To characterize the RNA products, we first analyzed whether the products synthesized from the HCV full-length RNA template are in the single- or double-stranded form. The RNA products were treated with nuclease S1 under either low (50 mM NaCl) or high (500 mM NaCl) salt conditions. The results showed that at a high salt concentration, the products were almost completely resistant to nuclease S1 digestion (Fig. 4, lane 3), suggesting that the products are in double-stranded form. However, at a low salt concentration, the products were partially digested (lane 2). After the products were heat denatured at 96°C for 5 min and quick-chilled on ice prior to nuclease S1 treatment under a low salt concentration, the products were completely susceptible to nuclease S1 digestion (lane 5). In the presence of a high concentration of salt, the amounts of the products were also significantly reduced, although there were still some nuclease S1-resistant RNA products (lane 6). These results indicate that most of products were in double-stranded RNA form, probably forming a stable RNA duplex with the template or with the product itself, especially under the high salt conditions. The RNA template was completely digested by nuclease S1 under all of the conditions tested above (as determined by ethidium bromide staining); this is not surprising, since the RNA template used was in vast excess relative to the RNA products.
FIG. 4.
Nuclease S1 treatment of the products synthesized by NS5B by using the full-length HCV RNA template. RdRp-synthesized products were digested by nuclease S1 in 50 mM NaCl (low salt) or 500 mM NaCl (high salt). The Δ represents heat denaturation of the products at 96°C for 5 min and quick chilling on ice prior to the nuclease S1 treatment. Products were resolved on 1% agarose gel after heat denaturation of the samples at 96°C for 5 min. The arrowhead indicates the position of the RNA template.
In vitro RdRp reactions using heterologous viral RNA templates.
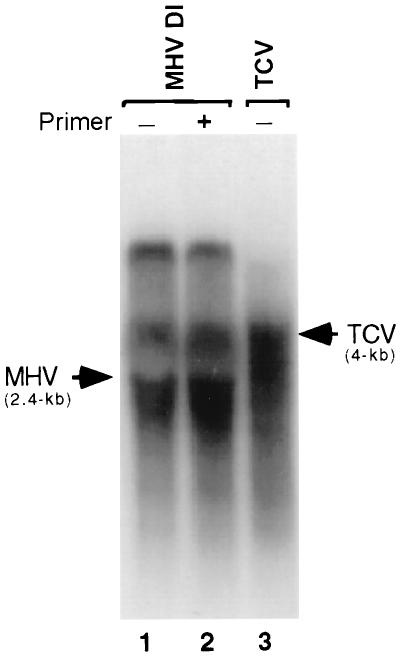
To examine the template specificity and primer dependency of NS5B, we tested several heterologous viral RNAs to examine their ability to serve as templates for NS5B. MHV-associated DI RNA was used as a poly(A)-tailed viral RNA. NS5B was able to transcribe this template without the requirement of an exogenous primer (Fig. 5, lane 1). However, the product was considerably more heterogeneous than when HCV RNA was used as the template (Fig. 2). In addition to the product corresponding to the template size, several heterogeneous products longer than the template size were synthesized (Fig. 5, lane 1). Interestingly, addition of oligo(U)20 did not enhance RNA synthesis, nor did it change the pattern of products synthesized (lane 2), again suggesting that RNA synthesis does not utilize an exogenous primer. When a non-poly(A)-tailed RNA, TCV genomic RNA, which contains a structured 3′ end, was used as a template, a distinct product of the genome size was obtained, suggesting a primer-independent RNA synthesis (lane 3). A minor longer-than-template-size product and several distinct smaller products were also obtained (lane 3). The properties of these products were analogous to those obtained when HCV RNA was used as a template. These data suggest that NS5B of HCV can also recognize the heterologous templates in vitro in a primer-independent manner, with preference for the RNAs with a structured 3′ end.
FIG. 5.
RdRp reactions with heterologous RNAs as templates. MHV DI RNA (2.4 kb) and TCV RNA (4 kb) were used for the RdRp reactions. The primer used for the transcription of MHV DI RNA was oligo(U)20. Products were resolved on a 1% agarose gel after heat denaturation of the samples at 96°C for 5 min. Arrowheads indicate the template positions of the MHV DI and TCV RNA templates.
In vitro RdRp reactions using homopolymeric RNA templates.
Since we observed primer-independent RNA synthesis by using both HCV and heterologous viral RNA templates, we tested primer dependency and template specificity of NS5B by using homopolymeric RNAs. RdRp assays were performed with homopolymeric RNA templates in the absence and presence of a primer, (rN)20, complementary to the templates. The incorporation of 32P-ribonucleotides into TCA-precipitable products was determined (Table 1). As previously shown in the other report (23), very little RNA synthesis was detected in the absence of the primer. However, in the presence of complementary oligonucleotides, significant 32P incorporation was observed when poly(A) or poly(C) RNA was used as the template. In contrast, poly(G) and poly(U) templates were very weak templates even in the presence of a primer. These data show that even though NS5B was able to replicate HCV and other viral RNA templates in a primer-independent manner (Fig. 5), the homopolymer templates require primers, and NS5B has a certain template preference among these linear homopolymeric RNAs. Thus, HCV NS5B appears to be able to discriminate different RNA templates based on sequence and structure.
TABLE 1.
NS5B polymerase activity on homopolymeric templatesa
| Template | Primer | Incorporation (103 cpm) |
|---|---|---|
| Poly(A) | − | 35 |
| (U)20 | 1,807 | |
| Poly(C) | − | 46 |
| (G)20 | 1,402 | |
| Poly(G) | − | 15 |
| (C)20 | 137 | |
| Poly(U) | − | 15 |
| (A)20 | 29 |
RdRp activity of NS5B was measured by using 1 μg of homopolymeric RNAs in the absence (−) and presence of 10 pmol of primer complementary to the templates. The labeled products were collected on a GF/C glass filter after precipitation with 5% TCA, and radioactivities were measured.
The cis-acting sequences at the 3′-UTR for RNA synthesis initiated from the plus-strand HCV genome.
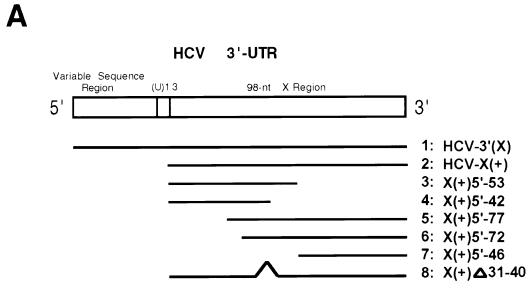
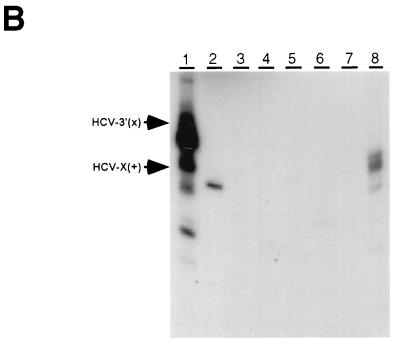
Since the 98-nt X region at the 3′ end of the HCV genome is very conserved in sequence and has a stable secondary structure (16, 20, 38), it has been proposed that the X region may be involved in viral RNA synthesis (16, 20, 38). Therefore, we examined whether the 98-nt X region and/or its upstream sequence serves as the cis-acting sequence for HCV RNA synthesis. The results showed that when the 98-nt RNA [HCV-X(+)] was used, a distinct RNA product which migrated slightly faster than the template was obtained, indicating that this RNA provides HCV RdRp a very homogeneous starting site. Interestingly, all of the deletion mutants tested, except the mutant [X(+)Δ31–40], lost the template activity almost completely, suggesting that the integrity of sequence and/or structure of the X region is important for NS5B to initiate RNA synthesis. The mutant [X(+)Δ31–40], which has a 10-nt deletion on the loop region of stem-loop 2 (16), yielded several RNA products longer than the template size (Fig. 6B, lane 8). This result suggests that the sequences on the loop region of stem-loop 2 may contribute to the specificity of initiation. These data combined indicate that NS5B may recognize specific sequences at the 3′ end of HCV RNA to start RNA synthesis. When an artificial full-length 3′-UTR of HCV, including the X region, (U)13 tract, and upstream variable sequence region [HCV-3′(+)], was used (16), the amounts of RNA products were significantly enhanced. The major product was slightly smaller than the template size, but several other heterogeneous RNA products were also detected (Fig. 6B, lane 1). This result suggests that the variable sequence region and/or (U)13 tract might act as an enhancer element for transcription initiation. We have performed preliminary characterization of the products made by the X region and the full-length 3′-UTR. Both products were very resistant to digestion with nuclease S1, and their electrophoretic mobility did not change after heat denaturation (data not shown), suggesting that they may be in a very stable RNA duplex structure.
FIG. 6.
cis-acting sequences for RNA synthesis on the 3′ end of plus-strand HCV RNA by NS5B. (A) The structure of RNA templates used for RdRp assays. (B) The products were run on an 8 M urea–5% polyacrylamide gel. The positions of the 98-nt template [HCV-X(+)] and the full-length 3′-UTR HCV [HCV-3′(X)] are indicated with arrowheads. The templates used for the reactions are indicated above the gel.
The cis-acting sequences necessary for RNA synthesis from the minus-strand HCV genome.
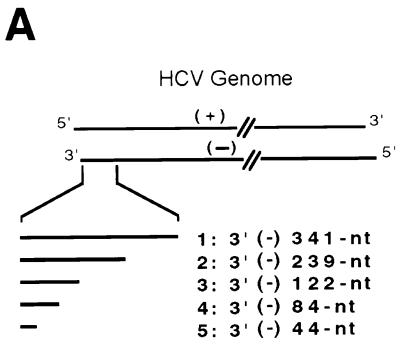
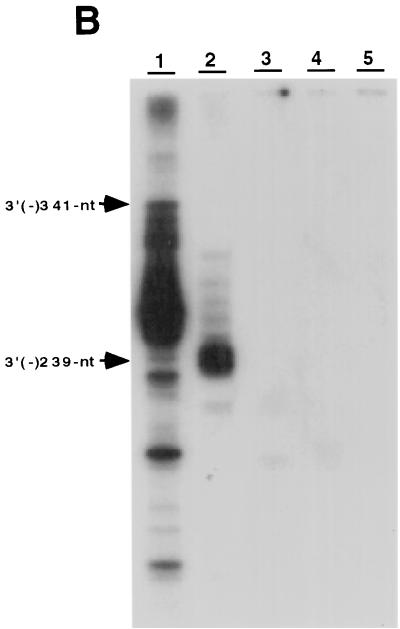
The results presented above showed that NS5B recognizes the 3′ end of HCV RNA for minus-strand RNA synthesis. We then determined whether the 3′ end of the HCV minus-strand genome contains the cis-acting sequence for plus-strand RNA synthesis. The RNA templates representing different lengths of the 3′ end of the minus-strand HCV genome (Fig. 7A) were used for the RdRp reactions. The results showed that both 341- and 239-nt templates served as efficient templates and yielded a major product equivalent to the template size (Fig. 7B, lanes 1 and 2). In contrast, the 122-nt and smaller RNAs did not have any template activity (Fig. 7B, lanes 3 to 5). This result suggests that the promoter sequence for NS5B is located approximately between nt 122 and 239 from the 3′ end of the minus-strand HCV genome.
FIG. 7.
cis-acting sequences for RNA synthesis on the 3′ end of plus-strand HCV RNA. (A) The structure of RNA templates used. (B) Products were analyzed on an 8 M urea–5% polyacrylamide gel. The positions of the 341-nt [3′(−) 341-nt] and 239-nt [3′(−) 239-nt] templates are indicated with arrowheads. The templates used for the reactions are indicated above the gel.
No TNTase activity associated with the recombinant HCV NS5B.
It has been shown that TNTase activity is associated with the purified NS5B expressed in insect cells using recombinant baculoviruses (4, 23). The origin of the TNTase is not known. Since TNTase may elongate the template from the 3′-OH residue and generate an artificial primer acting in cis, the presence of TNTase associated with RdRp may obscure the real properties of RdRp. Therefore, we tested if the E. coli-expressed NS5B has such TNTase activity after Ni-NTA chromatography purification. Using the X region as a template, TNTase activity was tested with four different [32P]rNTPs to see if there was any end-labeling activity at the 3′ end of the template. As shown in Fig. 8, no 32P-labeled products were detected with any of the [32P]rNTPs, indicating that our NS5B preparation did not have TNTase activities. Furthermore, the NS5B mutant (NS5BD318H) did not exhibit TNTase activity under the same conditions (data not shown). We also used oligonucleotides (A)20 and (U)20 in the presence of a single one of four different [32P]rNTPs at high specific activity (undiluted with cold rNTPs) to test the TNTase activity of NS5B on these homopolymeric RNAs. No TNTase activity was detected (data not shown).
FIG. 8.
TNTase activity assay of the recombinant NS5B. RdRp reactions were conducted with a single rNTP (10 μM each ribonucleotide with 10 μCi of [32P]rNTP) by using the 98-nt RNA as a template. Products were analyzed on an 8 M urea–20% polyacrylamide gel. The template (98-nt) and free ribonucleotide (Free rNTP) positions are indicated on the right side of the autoradiograph.
DISCUSSION
In this study, we have expressed a recombinant RdRp of HCV, namely, a full-length NS5B with an (His)6 at the N terminus, as a soluble protein in E. coli. By expression of NS5B at low temperature and extraction with a nonionic detergent, Nonidet P-40, in combination with a high concentration of salt, we were able to obtain a soluble, full-length, and enzymatically active HCV NS5B. Our recombinant HCV RdRp has several prominent properties which make it more closely resemble the functional HCV RdRp in vivo than any of the previously reported HCV RdRps (4, 10, 23, 47). (i) It can synthesize a full-length HCV cRNA with the HCV genomic RNA used as a template. (ii) It does not require an exogenous primer or a snapped-back RNA as a primer. (iii) It recognizes the specific sequences at the 3′ end of both plus- and minus-strand HCV RNAs for the initiation of RNA synthesis. (iv) It does not contain a TNTase, thus allowing unequivocal demonstration of RdRp activities. (v) Finally, it contains the full-length sequence of NS5B, in contrast to previous studies (10, 47), which reported that the full-length NS5B is insoluble in E. coli. Thus, our recombinant NS5B most closely reflects the predicted properties of a functional HCV RdRp.
Several pieces of evidence suggest that our recombinant HCV NS5B was able to carry out the synthesis of full-length HCV RNA in a primer-independent manner. First, the kinetics of RNA synthesis by this RdRp using the full-length HCV RNA as a template indicated that the NS5B-mediated RNA synthesis is not initiated from the snapped-back template, since the initial RNA products were very small and they gradually increased to the full-length template size (Fig. 3A, lanes 1 to 7). If RNA synthesis were started by a snap-back mechanism, the predominant transcription product would be longer than the template size, even at the early stage of transcription, as previously observed with the NS5B expressed by recombinant baculoviruses (23), since the product will be extended from the 3′ end of the template. The predominant RNA product in our reaction corresponded to the full-length HCV RNA in size. Thus, our recombinant NS5B is the first HCV RdRp capable of synthesizing full-length HCV RNA, although we cannot yet ascertain whether the ends of RNA products faithfully mirror those of the RNA template. Our kinetics studies also showed that longer incubation (even after NS5B copied the full-length template completely) generated products that are longer than the template, which may represent the products extended from the newly synthesized full-length products by a snap-back priming mechanism, since this product was not observed until the full-length product was detected (Fig. 3). This possibility raised the interesting question of whether NS5B can accomplish cyclic replication in vitro, namely, synthesis of minus-strand cRNA by using the plus-strand genome, followed by synthesis of plus-strand RNA, in turn, from cRNA. Previously, cucumber mosaic virus and flock house virus RdRps have been shown to be able to accomplish complete replication in vitro (13, 45). For flock house virus RdRp, cyclic transcription was observed when glycerophospholipids (GPLs) were added to RdRp, suggesting a role of GPLs in the induction or stimulation of single-stranded RNA synthesis. For most viral RdRps, the RNA products are usually in double-stranded form. However, the single-stranded form of replicative intermediates has been observed in turnip yellow mosaic virus-infected cells by using electron microscopy (11). For Qβ RdRp, the product synthesized by the polymerase was also in the single-stranded form; the double-stranded form of Qβ RNA could not be used as a template (5). Based on nuclease S1 digestion results (Fig. 4), the products synthesized by HCV NS5B in vitro are mainly in the double-stranded form, which may represent annealing of the products to the template. Alternatively, as suggested by the products that are longer than the full-length RNA, a second round of synthesis may have been carried out by HCV NS5B to generate double-stranded RNA products. Nevertheless, NS5B may require other viral proteins or cellular proteins to accomplish a faithful cyclic replication.
The recombinant HCV NS5B was also found to use non-HCV viral RNAs as templates, such as MHV DI and TCV RNA, in the absence of an exogenous primer (Fig. 5). For the poly(A)-tailed template, MHV DI RNA, NS5B synthesized multiple RNA species, including those of template size and longer than template size, in a primer-independent manner (Fig. 5, lanes 1 and 2). Addition of oligo(U)20 primer did not abolish the synthesis of longer products, and the profile of RNA products was not affected either (Fig. 5, lanes 1 and 2), implying that RNA synthesis initiated from the sequence within the template, rather than from poly(A). Since no primer [oligo (U)20]-dependent transcription was observed with MHV DI RNA despite the presence of a poly(A) tail, in contrast to the primer-dependent transcription when poly(A) homopolymer was used as a template, the primer dependency of the poly(A)-tailed template may be superseded by the flanking sequences. In any case, the RNA products made from this polyadenylated RNA (MHV DI) were more heterogeneous than those produced with HCV RNA as the template. In this regard, it is interesting to note that the 3′-end sequence of MHV DI RNA shows over 50% sequence identity with the 3′ end of the HCV X region (data not shown). In contrast, TCV RNA, which does not have a poly(A) tail but, instead, has a stem-loop structure, yielded a relatively homogeneous full-length product of template size, almost similar to the products made from HCV RNA. Thus, it appears that HCV NS5B may recognize a stable structure at the 3′ end of RNA. The need for a structured element in RNA templates for HCV NS5B is underscored by the finding that homopolymeric RNAs cannot serve as a template unless there is a primer. Furthermore, NS5B exhibited different template preferences for different homopolymeric RNAs, poly(A)>poly(C)>>poly(G)>poly(U), confirming the previous reports (23). Thus, NS5B appears to be able to discriminate between different RNA templates based on RNA sequence as well as structure.
Despite the finding that HCV RdRp was able to use many different RNAs as templates in vitro, there appeared to be a specific sequence requirement for cis-acting signals for RNA synthesis when HCV RNA was used. We showed that the X region is a minimal cis-acting sequence required for HCV minus-strand RNA synthesis. Similarly, the cis-acting sequence required for HCV positive-strand RNA synthesis is located at 239 to 122 nt from the 3′ end of the minus-strand HCV genome. Surprisingly, sequence alignment between both cis-acting elements showed 53% sequence identity (data not shown). The 3′ end of minus-strand RNA also showed stable stem-loop structures (data not shown). Additional studies are required to precisely determine the sequence and structure required for RNA replication at the two ends. Nevertheless, this is the first recombinant HCV RdRp that has been shown to exhibit a requirement for a specific sequence on the HCV RNA template. Several other viral RdRps have also been shown to recognize the RNA template specifically. For example, alfalfa mosaic virus RdRp requires 133 and 163 nt from the 3′ end for initiation of minus-strand synthesis in vitro (42). TCV RdRp requires an internal cis-acting sequence proximal to the 3′ end for plus-strand RNA synthesis (36). Nevertheless, these RdRps also can use other RNAs as templates, similar to HCV RdRp. Interestingly, the variable sequence and (UC)-rich sequence upstream of the X region have a significant enhancer effect on RNA synthesis carried out by the HCV RdRp. Since this sequence is variable among HCVs of different genotypes (20, 38, 46), it is conceivable that this sequence from different genotypes may determine the efficiency of viral RNA synthesis and thus the biology of different HCV genotypes. However, the products synthesized by using the full-length 3′-UTR template were much more heterogeneous than those synthesized with the X region alone; the majority of RNA products migrated faster than the template, and smeared bands around the template were detected. These might be due to the slippage of polymerase along the poly(U) tract or premature termination of transcription after copying the 98 nt. Slippage of polymerase on homopolymer templates has been described for E. coli and yeast RNA polymerases (18, 19).
The existence of cis-acting sequences on the 3′ end of both plus and minus strands of HCV RNA indicates that NS5B exhibits a sequence and/or structure preference for the HCV genome. However, we cannot rule out the possibility that other viral and/or cellular proteins are involved in the determination of specificity and regulation of transcription. In this regard, it is interesting to note that PTB interacts with the X region at the 3′ end of the plus-strand HCV genome (16). However, so far, the possible role of PTB in the regulation of HCV RNA synthesis has not been demonstrated, although PTB has been shown to be involved in translational regulation by interacting with either the 5′ IRES or 3′ X region of the HCV genome (1, 17). It will be interesting to see if there are any cellular proteins interacting with the 3′ end of the minus-strand HCV genome.
ACKNOWLEDGMENTS
We thank J. Bukh at NIH and A. E. Simon at University of Massachusetts, Amherst, for the infectious clones of HCV and TCV, respectively. We are also grateful to S. B. Hwang for the anti-NS5B polyclonal antibody.
This work was supported by research grant AI40038 from the National Institutes of Health. J.-W.O. is a Research Associate and M.M.C.L. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Ali N, Siddiqui A. Interaction of polypyrimidine tract-binding protein with the 5′ noncoding region of the hepatitis C virus RNA genome and its functional requirement in internal initiation of translation. J Virol. 1995;69:6367–6375. doi: 10.1128/jvi.69.10.6367-6375.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnaud-Barbe N, Cheynet-Sauvion V, Oriol G, Mandrand B, Mallet F. Transcription of RNA templates by T7 RNA polymerase. Nucleic Acids Res. 1998;26:3350–3354. doi: 10.1093/nar/26.15.3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ball L A. Replication of the genomic RNA of a positive-strand RNA animal virus from negative-sense transcripts. Proc Natl Acad Sci USA. 1994;91:12443–12447. doi: 10.1073/pnas.91.26.12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behrens S-E, Tomei L, De Francesco R. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 1996;15:12–22. [PMC free article] [PubMed] [Google Scholar]
- 5.Blumenthal T, Carmichael G G. RNA replication: function and structure of Qβ-replicase. Annu Rev Biochem. 1979;48:525–548. doi: 10.1146/annurev.bi.48.070179.002521. [DOI] [PubMed] [Google Scholar]
- 6.Brown E A, Zhang H, Ping L H, Lemon S M. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 1992;20:5041–5045. doi: 10.1093/nar/20.19.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen P-J, Lin M-H, Tai K-F, Liu P-C, Lin C-J, Chen D-S. The Taiwanese hepatitis C virus genome: sequence determination and mapping of the 5′ termini of viral genomic and antigenomic RNA. Virology. 1992;188:102–113. doi: 10.1016/0042-6822(92)90739-c. [DOI] [PubMed] [Google Scholar]
- 8.Cormack B. Introduction of a point mutation by sequential PCR steps. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley & Sons; 1994. pp. 8.5.7–8.5.9. [Google Scholar]
- 9.Dreher T W, Hall T C. Mutational analysis of the sequences and structural requirements in brome mosaic virus RNA for minus strand promoter activity. J Mol Biol. 1988;201:31–44. doi: 10.1016/0022-2836(88)90436-6. [DOI] [PubMed] [Google Scholar]
- 10.Ferrai E, Wright-Minogue J, Fang J W S, Baroudy B M, Lau J Y N, Hong Z. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J Virol. 1999;73:1649–1654. doi: 10.1128/jvi.73.2.1649-1654.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garaier M, Mamoun R, Bove J M. TYMV RNA replication in vivo: replicative intermediate is mainly single stranded. Virology. 1980;104:357–374. doi: 10.1016/0042-6822(80)90340-2. [DOI] [PubMed] [Google Scholar]
- 12.Gargouri-Bouzid R, David C, Haenni A-L. The 3′ promoter region involved in RNA synthesis directed by the turnip yellow mosaic virus genome in vitro. FEBS Lett. 1991;294:56–58. doi: 10.1016/0014-5793(91)81342-6. [DOI] [PubMed] [Google Scholar]
- 13.Hayes R I, Buck K W. Complete replication of a eukaryotic virus RNA in vitro by a purified RNA-dependent RNA polymerase. Cell. 1990;63:363–368. doi: 10.1016/0092-8674(90)90169-f. [DOI] [PubMed] [Google Scholar]
- 14.Houghton M. Hepatitis C viruses. In: Fields B N, Knipe D M, Howley P M, et al., editors. Virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1035–1058. [Google Scholar]
- 15.Hwang S B, Park K-J, Kim Y-S, Sung Y C, Lai M M C. Hepatitis C virus NS5B protein is a membrane-associated phosphoprotein with a predominantly perinuclear localization. Virology. 1997;227:439–446. doi: 10.1006/viro.1996.8357. [DOI] [PubMed] [Google Scholar]
- 16.Ito T, Lai M M C. Determination of the secondary structure of and cellular protein binding to the 3′-untranslated region of the hepatitis C virus RNA genome. J Virol. 1997;71:8698–8706. doi: 10.1128/jvi.71.11.8698-8706.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito T, Tahara S M, Lai M M C. The 3′-untranslated region of hepatitis C virus RNA enhances translation from an internal ribosomal entry site. J Virol. 1998;72:8789–8796. doi: 10.1128/jvi.72.11.8789-8796.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacques J P, Kolakofsky D. Pseudo-template transcription by Escherichia coli RNA polymerase at a mutant promoter. Genes Dev. 1991;5:707–713. doi: 10.1101/gad.4.10.1801. [DOI] [PubMed] [Google Scholar]
- 19.Jeong S W, Lang W H, Reeder R H. The yeast transcription terminator for RNA polymerase I is designed to prevent polymerase slippage. J Biol Chem. 1996;271:16104–16110. doi: 10.1074/jbc.271.27.16104. [DOI] [PubMed] [Google Scholar]
- 20.Kolykhalov A A, Feinstone S M, Rice C M. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J Virol. 1996;70:3363–3371. doi: 10.1128/jvi.70.6.3363-3371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koonin E V. The phylogeny of RNA-dependent RNA polymerase of positive-strand RNA viruses. J Gen Virol. 1991;72:2197–2206. doi: 10.1099/0022-1317-72-9-2197. [DOI] [PubMed] [Google Scholar]
- 22.Lin Y-J, Liao C-L, Lai M M C. Identification of the cis-acting signal for minus-strand RNA synthesis of a murine coronavirus: implications for the role of minus-strand RNA in RNA replication and transcription. J Virol. 1994;68:8131–8140. doi: 10.1128/jvi.68.12.8131-8140.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lohmann V, Körner F, Herian U, Bartenschlager R. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J Virol. 1997;71:8416–8428. doi: 10.1128/jvi.71.11.8416-8428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lubinski J M, Ransone L J, Dasgupta A. Primer-dependent synthesis of covalently linked dimeric RNA molecules by poliovirus replicase. J Virol. 1987;61:2997–3003. doi: 10.1128/jvi.61.10.2997-3003.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Major M E, Feinstone S M. The molecular virology of hepatitis C. Hepatology. 1997;25:1527–1538. doi: 10.1002/hep.510250637. [DOI] [PubMed] [Google Scholar]
- 26.Makino S, Lai M M C. High-frequency leader switching during coronavirus defective interfering RNA replication. J Virol. 1989;63:5285–5292. doi: 10.1128/jvi.63.12.5285-5292.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller W A, Bujarski J J, Dreher T W, Hall T C. Minus-strand initiation by brome mosaic virus replicase within the 3′ tRNA-like structure of native and modified RNA templates. J Mol Biol. 1986;187:537–546. doi: 10.1016/0022-2836(86)90332-3. [DOI] [PubMed] [Google Scholar]
- 28.Oh J-W, Kong Q, Song C, Carpenter C D, Simon A E. Open reading frames of turnip crinkle virus involved in satellite symptom expression and incompatibility with Arabidopsis thaliana ecotype Dijon. Mol Plant-Microbe Interact. 1995;8:979–987. doi: 10.1094/mpmi-8-0979. [DOI] [PubMed] [Google Scholar]
- 29.Osman T A M, Buck K W. Complete replication in vitro of tobacco mosaic virus RNA by a template-dependent, membrane-bound RNA polymerase. J Virol. 1996;70:6227–6234. doi: 10.1128/jvi.70.9.6227-6234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plotch S J, Palant O, Gluzman Y. Purification and properties of poliovirus RNA polymerase expressed in Escherichia coli. J Virol. 1989;63:216–225. doi: 10.1128/jvi.63.1.216-225.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saito I, Miyamura T, Ohbayashi A, Harada H, Katayama T, Kikuchi S, Watanabe Y, Koi S, Onji M, Ohta Y, Choo Q-L, Houghton M, Kuo G. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci USA. 1990;87:6917–6921. doi: 10.1073/pnas.87.17.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sankar S, Porter A G. Expression, purification, and properties of recombinant encephalomyocarditis virus RNA-dependent RNA polymerase. J Virol. 1991;65:2993–3000. doi: 10.1128/jvi.65.6.2993-3000.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarnow P, Bernstein H D, Baltimore D. A poliovirus temperature-sensitive mutant located in a noncoding region of the genome. Proc Natl Acad Sci USA. 1986;83:571–575. doi: 10.1073/pnas.83.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarnow P. Role of 3′-end sequences in infectivity of poliovirus transcripts made in vitro. J Virol. 1989;63:467–470. doi: 10.1128/jvi.63.1.467-470.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh R N, Dreher T W. Turnip yellow mosaic virus RNA-dependent RNA polymerase: initiation of minus strand synthesis in vitro. Virology. 1997;233:430–439. doi: 10.1006/viro.1997.8621. [DOI] [PubMed] [Google Scholar]
- 36.Song C, Simon A E. RNA-dependent RNA polymerase from plants infected with turnip crinkle virus can transcribe (+)- and (−)-strands of virus-associated RNAs. Proc Natl Acad Sci USA. 1994;91:8792–8796. doi: 10.1073/pnas.91.19.8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanaka T, Kato N, Cho M-J, Sugiyama K, Shimotohno K. Structure of the 3′ terminus of the hepatitis C virus genome. J Virol. 1996;70:3307–3312. doi: 10.1128/jvi.70.5.3307-3312.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Triana-Alonso F J, Dabrowski M, Wadzack J, Nierhaus K H. Self-coded 3′-extension of run-off transcripts produces aberrant products during in vitro transcription with T7 RNA polymerase. J Biol Chem. 1995;270:6298–6307. doi: 10.1074/jbc.270.11.6298. [DOI] [PubMed] [Google Scholar]
- 40.Tsuchihara K, Tanaka T, Hijikata M, Kuge S, Toyoda H, Nomoto A, Yamamoto N, Shimotohno K. Specific interaction of polypyrimidine tract-binding protein with the extreme 3′-terminal structure of the hepatitis C virus genome, the 3′X. J Virol. 1997;71:6720–6726. doi: 10.1128/jvi.71.9.6720-6726.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomota A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Kury A C, Langereis K, Houwing C J, Jaspars E M J, Bol J F. cis-acting elements involved in replication of alfalfa mosaic virus RNAs in vitro. Virology. 1990;176:346–354. doi: 10.1016/0042-6822(90)90004-b. [DOI] [PubMed] [Google Scholar]
- 43.Vazquez A L, Alonso J M M, Casais R, Boga J A, Parra F. Expression of enzymatically active rabbit hemorrhagic disease virus RNA-dependent RNA polymerase in Escherichia coli. J Virol. 1998;72:2999–3004. doi: 10.1128/jvi.72.4.2999-3004.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang C, Sarnow P, Siddiqui A. A conserved helical element is essential for internal initiation of translation of hepatitis C virus RNA. J Virol. 1994;68:7301–7307. doi: 10.1128/jvi.68.11.7301-7307.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu S-H, Kaesberg P. Synthesis of template-sense, single-strand flockhouse virus RNA in a cell-free replication system. Virology. 1991;183:392–396. doi: 10.1016/0042-6822(91)90153-3. [DOI] [PubMed] [Google Scholar]
- 46.Yamada N, Tanihara K, Takada A, Yorihuzi T, Tsutsumi M, Shimomura H, Tsuji T, Date T. Genetic organization and diversity of the 3′ noncoding region of the hepatitis C virus genome. Virology. 1996;223:255–261. doi: 10.1006/viro.1996.0476. [DOI] [PubMed] [Google Scholar]
- 47.Yamashita T, Kaneko S, Shirota Y, Qin W, Nomura T, Kobayashi K, Murakami S. RNA-dependent RNA polymerase activity of the soluble recombinant hepatitis C virus NS5B protein truncated at the C-terminal region. J Biol Chem. 1998;273:15479–15486. doi: 10.1074/jbc.273.25.15479. [DOI] [PubMed] [Google Scholar]
- 48.Yanagi M, Claire M S, Shapiro M, Emerson S U, Purcell R H, Bukh J. Transcripts of a chimeric cDNA of hepatitis C virus genotype 1b are infectious in vivo. Virology. 1998;244:161–172. doi: 10.1006/viro.1998.9092. [DOI] [PubMed] [Google Scholar]
- 49.Young D C, Tuschall D M, Flanegan J B. Poliovirus RNA-dependent RNA polymerase and host cell protein synthesize product RNA twice the size of poliovirion RNA in vitro. J Virol. 1985;54:256–264. doi: 10.1128/jvi.54.2.256-264.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhong W, Gutshall L L, Del Vecchio A M. Identification and characterization of an RNA-dependent RNA polymerase within the nonstructural protein 5B region of bovine viral diarrhea virus. J Virol. 1998;72:9365–9369. doi: 10.1128/jvi.72.11.9365-9369.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]