Abstract
dUTPase is a ubiquitous and essential enzyme responsible for regulating cellular levels of dUTP. The dut gene exists as single, tandemly duplicated, and tandemly triplicated copies. Crystallized single-copy dUTPases have been shown to assemble as homotrimers. dUTPase is encoded as an auxiliary gene in a number of virus genomes. The origin of viral dut genes has remained unresolved since their initial discovery. A comprehensive analysis of dUTPase amino acid sequence relationships was performed to explore the evolutionary dynamics of dut in viruses and their hosts. Our data set, comprised of 24 host and 51 viral sequences, includes representative sequences from available eukaryotes, archaea, eubacteria cells, and viruses, including herpesviruses. These amino acid sequences were aligned by using a hidden Markov model approach developed to align divergent data. Known secondary structures from single-copy crystals were mapped onto the aligned duplicate and triplicate sequences. We show how duplicated dUTPases might fold into a monomer, and we hypothesize that triplicated dUTPases also assemble as monomers. Phylogenetic analysis revealed at least five viral dUTPase sequence lineages in well-supported monophyletic clusters with eukaryotic, eubacterial, and archaeal hosts. We have identified all five as strong examples of horizontal transfer as well as additional potential transfer of dut genes among eubacteria, between eubacteria and viruses, and between retroviruses. The evidence for horizontal transfers is particularly interesting since eukaryotic dut genes have introns, while DNA virus dut genes do not. This implies that an intermediary retroid agent facilitated the horizontal transfer process between host mRNA and DNA viruses.
dUTPase is the enzyme that catalyzes the conversion of dUTP to dUMP and PPi. This activity controls dUTP concentration and provides dUMP as a precursor for the TTP biosynthesis pathway. dUTPase is critical to cell survival because excess dUTP is incorporated into DNA, leading to excision repair, DNA breakage, and death. The presence of dUTPase has been demonstrated as necessary for the survival of organisms such as Escherichia coli (14) and Saccharomyces cerevisiae (21). The gene for dUTPase (dut) is ubiquitous in eukaryotes, eubacteria, and archaea. dut is also found in a number of retroviruses and DNA viruses, where viral dUTPases may help control local dUTP levels during replication and also enhance viral replication in nondividing host tissues (56, 68). dUTPases are characterized by a series of five conserved amino acid motifs. This common set of conserved sequence motifs was initially identified by sequence comparison in herpesviruses (3, 11, 45), retroviruses (40), and poxviruses (49, 60), although the function was not known. Comparison of these motifs with a later-characterized E. coli dUTPase sequence revealed their identity as dUTPase motifs (44). The importance of these conserved motifs for dUTPase function has been demonstrated by mutagenesis (59, 70–74). Furthermore, X-ray crystallography of dUTPases from E. coli (6, 32), feline immunodeficiency virus (FIV) (55), equine infectious anemia virus (EIAV) (9), and Homo sapiens (50) confirmed that the five conserved motifs comprise the active site of the enzyme.
The evolutionary origin of the viral dut gene was subject to speculation even before its function was identified (40). Many questions remain unresolved, such as whether the dut genes of mouse mammary tumor virus (MMTV) and nonprimate lentiviruses are of separate cellular origin or the result of horizontal transfer between these two retroviral lineages (41, 44). The origin and history of dut in DNA viruses are similarly obscure. In order to address these issues, we have performed a comprehensive analysis of dUTPase sequences. This is the first study to date that has attempted to include alpha- and gammaherpesvirus dUTPases and contains representative dUTPase sequences from all of the available eubacterial, eukaryotic, archaeal, and viral sequences.
Host (24) and viral (51) dUTPase amino acid sequences (Table 1) were collected and aligned, and the phylogenetic relationships were inferred. To aid in the alignment, we considered the five conserved active site motifs discussed above as an ordered series of motifs (OSM). An OSM is comprised of amino acid motifs conserved in sequence and order (38). Known crystal structures were coordinated with the three observed motif arrangements (single, tandemly duplicated, and tandemly triplicated). Subunit folding and assembly in dUTPases without known structures is hypothesized based on the alignment presented here. At least five strong cases for horizontal transfer between eukaryotic and archaeal hosts and viral pathogens are identified, as well as possible cases for transfer among eubacteria, between eubacteria and viruses, and between retroviruses. The intronless structure of the dut locus in some DNA viruses implies acquisition of a host message derived cDNA via a reverse transcriptase-encoding agent.
TABLE 1.
Number and distribution of sequences useda
| Type and no. of sequences (taxon)b | Code(s) | Motif arrangement | Genome type | Locationc (reference) |
|---|---|---|---|---|
| Eukaryotes | ||||
| 3 (mammals) | MMUS,d HSAP, RNOR | A | DNA | ND |
| 1 (invertebrate [3]) | CELEA/M/Ce | C | DNA | ND |
| 1 (plant) | LESC | A | DNA | ND |
| 2 (unicellular) | SCER, CALB | A | DNA | Variable |
| Eukaryotic viruses | ||||
| 11 (nonprimate lentiviruses) | 1FIV, OVLV, FIVZ, MLIK, FISD, CAEV, FIPT, VISN, FIPL, EIAV, PLPP | A | RNA | gag, dut, protease (40) (Fig. 2) |
| 12 (MMTV-related retroviruses) | MMTV, HER1, MMTC, HER2, SARV, SHIA, SIV2, CHIA, MPMV, MIAP, JSRV, SMRV | A | RNA | RH, dut, IN (40) (Fig. 2) |
| 6 (alphaherpesvirus [12]) | HH1A/C, BH1A/C, EH1A/C, EH4A/C, SH1A/C, HH3A/C | B | DNA | RDPRI, dut, primase (24) (Fig. 2) |
| 6 (gammaherpesvirus [12]) | HH4A/C, HH8A/C, AH1A/C, EH2A/C, MHUA/C, SH2A/C | B | DNA | RDPRI, primase, dutd (Fig. 2) |
| 2 (unclassified [fish] herpesviruses) | IHER | A | DNA | ORF48, dut, ORF50 (10) |
| SHER | A | DNA | ORF48, dut, ORF57/58 (10) | |
| 6 (poxviruses) | VACW, CPOX, VACL, SPOX, VARI, ORFV | A | DNA | Variable (37) (Fig. 2) |
| 1 (baculovirus) | ONPH | A | DNA | Conotoxin, dut, RnRLSf |
| 1 (African swine fever virus) | ASFV | A | DNA | ORF J18L, dut, ORF K2R |
| 1 (adenovirus) | AVAD | A | DNA | ORF15, dut (75) |
| 1 (phycodnavirus) | PBCV | A | DNA | a548L, dut, a553L (34) |
| Eubacteria | ||||
| 7 (proteobacteria) | A | DNA | Variableg (Fig. 2) | |
| Alpha | BJAP | |||
| Beta | NGON | |||
| Gamma | ECOL, HINF, PAER, CBUR | |||
| Epsilon | HPYL | |||
| 6 (firmicutes) | A | DNA | Variable | |
| Clostridium group | CDIF, CACE, BSUB, MLEP | |||
| Actinomycetes | MTUB, SCOE | |||
| 1 (Chlamydiales) | CTRA | A | DNA | accD, dut, ptsN |
| 1 (Spirochaetales) | TPAL | A | DNA | HIMP,hdut, pnp |
| Eubacterial viruses (bacteriophage), | ||||
| 3 (siphoviruses) | BPRT, BPT5, BPSP | A | DNA | Variable (28, 69) |
| Archaea, 2 (archaea) | DAMB, MJAN | A | DNA | Variable |
| Archaeal virus, 1 (bacillovirus) | SIRV | A | DNA | ND |
Sequence codes are defined in the Appendix. For specific motif arrangements (A, B, and C), see Fig. 1.
The number of duplicated and triplicated copies of dut in a taxon are indicated in brackets.
Genome locations are designated as “variable” in a group of taxa if the genes adjacent to dut are different. ND, not determined.
Sequence determined by alignment of six M. musculus cDNA contigs available in GenBank and by conceptual translation.
C. elegans cDNA and nuclear sequences available in GenBank are consistent with a tandem triplication.
RnRLS, Ribonucleotide reductase large subunit.
P. aeruginosa upstream and downstream sequences were conceptually translated, and homologs were identified by using tBLASTn.
HIMP, hypothetical integral membrane protein.
MATERIALS AND METHODS
Hardware and software.
All analyses were conducted on SUN Ultras (1/140 and 1/170) or SPARCstations (4, 5, or 10/514MP) running SunOS release 5.5 or 5.6. Programs were used as follows: for sequence collection, tBlastn (2); for alignment, ClustalW version 1.6 (25) and SAM 2.0 (27, 29); and for phylogenetic analysis, Phylip 3.57c (15) and Ancestor (17).
Sequence collection.
All of the dUTPase protein sequences in this study were acquired from the GenBank sequence database or from other public online genome databases (63–66). The National Center for Biotechnology’s tBLASTn program was implemented with the PAM 250 matrix (12) as a search method to identify amino acid sequences with similarity to known dUTPases. Duplicated and triplicated dUTPases were analyzed as two and three separate sequences, respectively.
Pairwise alignment and clustering.
An in-house program was used for preliminary alignment of the collected sequences and to calculate approximate pairwise distances by using the Needleman-Wunsch algorithm (51) with a Dayhoff matrix (12). Another in-house program was used to cluster sequences hierarchically according to pairwise distance values. The hierarchical clustering identified five sequence subclasses, within which all sequences are at least 60% identical. We have developed a hidden Markov model (HMM) strategy for subclass alignment that incorporates the groupings of similar sequences (42).
Application of HMMs to sequence alignment.
An HMM is a stochastic production model representing the sequences used in a training set. In its simplest form a model is initialized a priori for: (i) the transition into a match, deletion, or insertion state and (ii) the occurrence of a given amino acid in a match or insert state. Each node in the model, comprised of these probabilities, represents a column in the alignment. By using the initial model and all training sequences, all possible paths for each sequence through the model are evaluated to obtain new estimates of the parameters that will increase the likelihood of the model. This process is repeated until the model converges. Surgery is performed to add or delete nodes in the model to improve this likelihood. A multiple alignment is generated by computing the negative logarithm of the probability of the single most likely path through the model for a particular sequence, given all the possible paths generated by the training sequences. Sequences are aligned to the model in this way, rather than to one another. The SAM modeling software conveniently provides an automated method for accomplishing these steps.
Construction of initial subclass HMMs.
In the SAM package the designation of special node types within the model is allowed. The special nodes are immune to model surgery. Two types of special nodes are used in the studies presented here to constrain the OSM within a model. Type A nodes are invariant and cannot undergo further training. Type K nodes undergo transition training but not match or insert training. The core amino acid residues of a motif are assigned type A nodes, while the amino and carboxyl residues of the motif are designated type K. This designation allows for the transition training into and out of the type A nodes representing the OSM.
In the subclass models the OSM is constrained by the designation of type A and K nodes at the same positions in each model. Preliminary alignments for each subclass were performed by using the PAM series matrices option in ClustalW to identify conserved regions and positions corresponding to the OSM. By using “modelfromalign” (SAM), nodes corresponding to the observed amino acid frequencies and delete and insert transition probabilities for the OSM were created based on the ClustalW alignment. Five initial subclass HMMs containing the OSM nodes were constructed with “buildmodel” (SAM). The number of generic nodes inserted between motifs corresponded to the longest stretch of amino acids in each motif-intervening region (MIR). A MIR is the region between each motif of the OSM. The generic nodes for each subclass model were initialized to reflect the overall proportion of amino acids present in each subclass data set. The generic nodes were then trained by use of subclass weighting (see below). The end result was a set of subclass models with amino acid probabilities at nodes representing the OSM and MIRs (39).
Weighting.
Weighting allows each HMM to be trained by using information from all of the sequences in the data set to different degrees. Two levels of weighting were used: within subclasses and between subclasses. The distribution of sequences was normalized within each subclass based on the initial pairwise distance values by assigning more weight to distant sequences than to overrepresented sequences. In this way the distribution of sequences in a subclass was normalized. These within-class weights comprised 75% of the influence of sequences on their own subclass model. Between-class weights (from the other four subclasses) comprised the remaining 25% influence. Including weights from the other subclasses in this way improves MIR alignment between subclasses (42).
Training of subclass HMMs.
Given the subclass sequences, initial HMMs, and sequence weights as described above, five subclass HMMs were each trained using buildmodel (SAM). This step simulates training the subclass models in parallel because each subclass is influenced by all of the sequences according to the weights assigned above. All models were run at the default parameter settings except: Nmodels = 5, del_jump_conf = 50, match_jump_conf = 50, ins_jump_conf = 50, and insconf = 100,000 (43). The same random seed was used within each subclass for reproducibility in every run. To prevent surgery while training, Nsurgery was set to 0. Five final HMMs were produced, one from each subclass, in which the OSM and MIR nodes contained frequencies corresponding to each column of the alignment.
Generation of multiple alignment.
Sequences from each subclass were aligned to their final automated HMM, generating a subclass alignment. All five subclass alignments were then merged by an in-house program (a2mmerge) and displayed as a single comprehensive multiple alignment. Even though the alignment generated by the strategy described above was a vast improvement over the initial ClustalW alignment, refinements were still required. Manual adjustments were subsequently introduced either when obvious regions of identity or similarity were not detected by the strategy or when alternative gapping would either produce more consistent local region relationships or minimize the mutational events required to align one set of sequences to another.
Phylogenetic analysis.
The divergent amino terminus was trimmed from all sequences to minimize the effect of noise in the form of insertions and deletions. This region is considered relatively unimportant to the basic function of the protein and can experience differential splicing. For example, the human dut gene is processed differently depending on its cellular destination; the amino terminus of the mitochondrial copy is 69 residues longer than the nuclear copy (31). A pairwise distance matrix (Table 2) was constructed from the trimmed alignment with a formula accounting for sequence composition by using Dayhoff matrix distance values (16). The additive distance, unrooted Fitch-Margoliash algorithm (18), FITCH (Phylip), was used to analyze the distance matrix. The input order was jumbled and reanalyzed 100 times to ensure that the search tree was not influenced by taxon order, as per Phylip distance method instructions. The resulting best search tree was compared to one slightly shorter in which the retrovirus dUTPases are monophyletic. Bootstrap replicates (100 total) of the alignment were constructed by using SEQBOOT (Phylip), and individual distance matrices were generated for each replicate. All 100 distance matrices were run as a multiple data set in FITCH (Phylip), jumbling each 10 times. The bootstrap outputs were compiled with CONSENSE (Phylip) and mapped onto the modified search tree. For comparison, the alignment was also analyzed with the Ancestor program. The Ancestor implementation backtranslates proteins and performs the Fitch-Margoliash algorithm on the hypothesized nucleotides. The shortest Phylip distance tree was input as the starting tree. The final Ancestor tree was compared with the Phylip tree for distance. In addition, the Ancestor search tree was analyzed with Phylip to ascertain its length according to the Phylip implementation of the same algorithm. As well as analyzing the data set as a whole, OSM (78 residues) and MIR (185 residues) regions were partitioned. Separate analyses were performed as described above to ascertain how each region type contributes to the overall phylogeny estimate.
TABLE 2.
Average pairwise distance values between dUTPases from various groups of organisms
| Taxon | Avg pairwise distancea (SD) of:
|
||||||
|---|---|---|---|---|---|---|---|
| HSAPb [1] | ORFV [1] | Poxvirusc [5] | AVAD [1] | PBCV [1] | Eubacteriad [15] | Archaeae [2] | |
| HSAP [1] | 0.00 (0.00) | ||||||
| Rodentsf [2] | 4.53 (0.68) | ||||||
| Mammalsg [3] | 34.50 (0.68) | 37.63 (5.68) | 34.48 (1.62) | 58.99 (1.55) | 101.95 (11.52) | 194.22 (33.27) | |
| Eukaryotesh [6] | 46.65 (6.91) | 59.14 (7.93) | 67.74 (5.30) | 54.86 (6.21) | 56.47 (9.04) | 97.98 (10.62) | 196.66 (24.60) |
Distance values were calculated as described in Materials and Methods and used to produce the phylogeny reported in Fig. 4. The number of organisms is given in brackets. dUTPase abbreviations are as defined in the text. Standard deviations are in parentheses.
Human dUTPase (HSAP) is provided as a reference point for comparison with dUTPases from rodents and nonmammalian eukaryotes.
Poxvirus dUTPases excluding that of ORF virus (VACW, VARI, CPOX, VACL, and SPOX).
All 15 eubacterial sequences as listed in Table 1.
Archaeal dUTPases (MJAN and DAMB).
Rodent dUTPases (MMUS and RNOR).
Mammalian dUTPases (HSAP, MMUS, and RNOR).
Eukaryote dUTPases excluding those of mammals (CELEA, CELEM, CELEC, LESC, CALB, and SCER).
RESULTS
Variations on motif order.
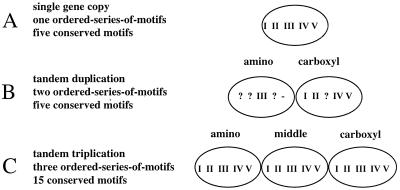
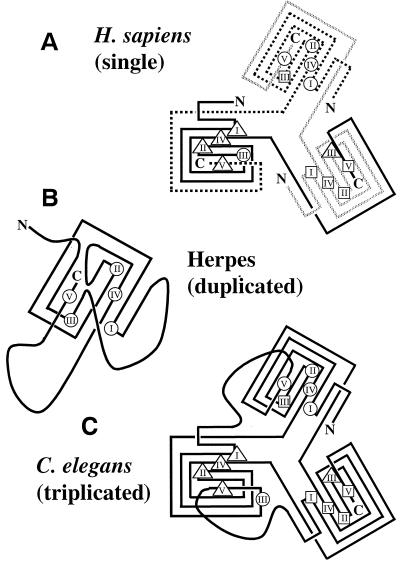
The majority of dUTPases considered in this study (62 of 75 total) display a common arrangement of five accepted motifs, referred to as I, II, III, IV, and V. Fish herpesviruses (10) and humans, for example, have a single-unit length dUTPase with the common arrangement of motifs (Fig. 1A). In alpha- and gammaherpesviruses, however, the dUTPase is roughly twice as long as the common arrangement (45). In the amino portion of the dUTPase of these viruses, a clearly recognizable motif III is present, but only residual motifs I, II, and IV appear. The carboxy portion of the herpesvirus dUTPase has recognizable motifs I, II, IV, and V, while a residual motif III is located between II and IV (Fig. 1B). Caenorhabditis elegans has a triplication of the single arrangement such that there are 15 complete motifs (26) (Fig. 1C). When the cDNA and nuclear sequences were compared, introns were detected within the coding regions. These introns are the same size and in the same place in each copy (data not shown).
FIG. 1.
Three types of motif arrangements. (A) Of the 75 dUTPases in this study, 62 consist of a single set of five conserved motifs (I to V). In addition to this common arrangement, two other variations are known. (B) All of the available alpha- and gammaherpesvirus sequences (six each) encode a dUTPase that has a single set of motifs but is twice as long as the common single arrangement. Motif III is conserved only in the amino portion of the protein, while motifs I, II, IV, and V are conserved in the carboxy portion. Possible remnants of motifs which imply a duplication of a single copy in the ancestor of these lineages and a subsequent loss of duplicate motifs are indicated by a question mark. The conservation of these remnants is variable within and between taxa (data not shown). Of the 12 available sequences, 10 have no amino acid residues in the position where the remnant of motif V would be; this is indicated by a dash (–). Sequences with this arrangement were analyzed in two sections: amino and carboxy. (C) The C. elegans sequence contains a triplicated dut colinearly transcribed and translated. Each copy of the triplicated protein has a complete set of the five motifs found in the common single copy arrangement of the gene. Each copy (amino, middle, and carboxy) was analyzed separately.
Genomic location of dut and adjacent genes.
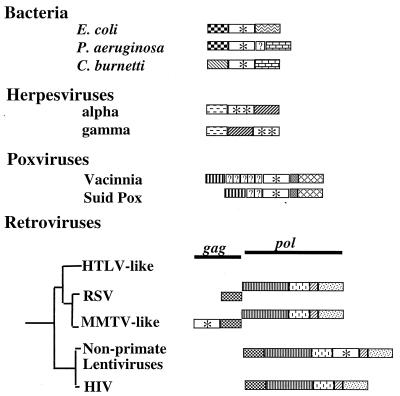
The location of the dut gene is variable among even closely related lineages of eubacteria and RNA and DNA viruses (Fig. 2). The proteobacteria E. coli, Pseudomonas aeruginosa, and Coxiella burnetii are closely related, but each encodes a different pair of genes flanking dut. In alphaherpesviruses dut is present between the ribonucleotide reductase-related protein and the primase genes, while in gammaherpesviruses dut is distal to the primase gene. The number and sequences of genes present between dut and the transcription initiation factor is variable between vaccinia virus and suid poxvirus. dut is found in the gag region in one lineage of retroviruses and in the pol region of another lineage (Fig. 2). These patterns demonstrate that the genomic location of dut is labile in viruses as well as in eubacteria.
FIG. 2.
The location of the dut gene is variable in the genomes of eubacteria, DNA viruses, and retroviruses. Closely related proteobacteria, E. coli, P. aeruginosa, and C. burnetii, all encode a dUTPase as do their more distant relatives. A eubacterial gene map comparison indicates that the type and number of genes surrounding dut varies. The DNA regions bounding the dut gene of P. aeruginosa were determined by conceptual translation. An additional potential ORF of unknown homology is indicated by a question mark. Alpha- and gammaherpesviruses both encode dUTPase, but the dut gene is not in the same position relative to the primase in these two lineages (24). Poxviruses have substituted genes upstream of dut (37). Between dut and the ribonucleotide reductase there is a single homologous but unidentified reading frame in both vaccinia virus and suid poxvirus. Between the transcription initiation factor gene and dut, however, neither of the two ORFs in suid poxvirus are homologous to any of the five ORFs in vaccinia virus. Two lineages of retroviruses (MMTV-like and nonprimate lentiviruses) transcribe dut as part of the gag and pol polycistrons, respectively. dUTPase is not encoded by close relatives of MMTV and nonprimate lentiviruses, neither human T-lymphotropic virus-like retroviruses, Rous sarcoma virus, nor HIV. Gene region and gene abbreviations are as follows. Single-copy dut genes are designated by an asterisk and duplicated dut genes by two asterisks. Symbols are as follows. Eubacteria:  , the DNA synthesis flavoprotein;
, the DNA synthesis flavoprotein;  , unknown function;
, unknown function;  , the phosphomannomutase;
, the phosphomannomutase;  , the outer membrane protein PIB. Herpesvirus genes:
, the outer membrane protein PIB. Herpesvirus genes:  , the ribonucleotide reductase-related protein;
, the ribonucleotide reductase-related protein;  , the primase. Poxviruses:
, the primase. Poxviruses:  , the transcription initiation factor;
, the transcription initiation factor;  , the ribonucleotide reductase. Retroviruses:
, the ribonucleotide reductase. Retroviruses:  , the protease;
, the protease;  , the reverse transcriptase;
, the reverse transcriptase;  , the ribonuclease H;
, the ribonuclease H;  , the integrase. Unidentified homologous reading frames are indicated (
, the integrase. Unidentified homologous reading frames are indicated ( ). Unidentified nonhomologous reading frames are indicated with a question mark.
). Unidentified nonhomologous reading frames are indicated with a question mark.
Alignment.
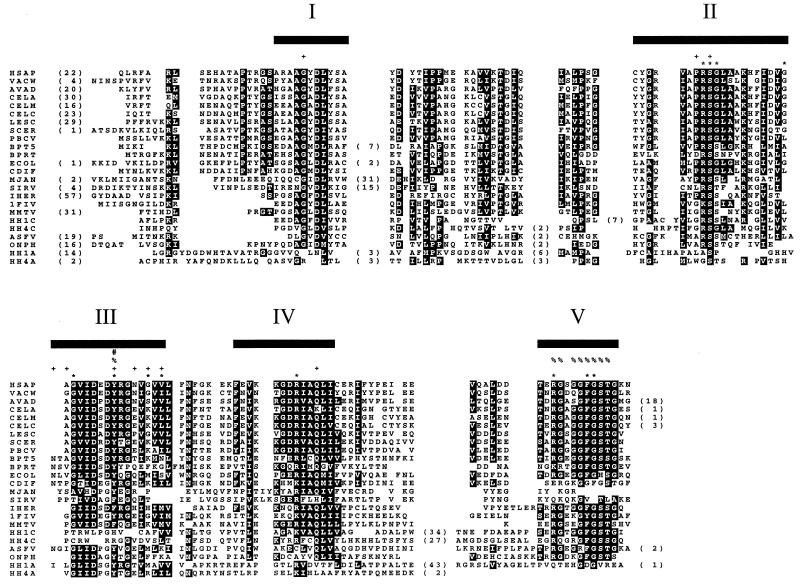
All 89 dUTPase amino acid sequence copies were aligned. Duplicated and triplicated dUTPases were analyzed as two and three individual copies, respectively. The resulting alignment (Fig. 3) corroborates the conservation of putative functional residues, as indicated by two of the crystal structures that included substrate analogs (32, 50). Columns of highly conserved residues correspond to the locations of substrate-binding residues. For example, a substrate-binding serine (S) is conserved in motif II (Fig. 3). Functional analysis of E. coli (ECOL), EIAV, and FIV (1FIV) dUTPases has independently confirmed the importance of the tyrosine (Y) in motif III for substrate binding (59, 72–74). Bacteriophage (BPT5 and BPRT), eubacteria (ECOL and CDIF), and African swine fever virus (ASFV) share a conserved asparagine (N) residue at the beginning of motif III that is not present in any of the other sequences. This residue is implicated in substrate binding in E. coli (32) and may indicate a structural variation in known dUTPases. Only a few exceptions are apparent in the conservation of the motifs. Motif III is not conserved in the carboxyl portion of alpha- and gammaherpesvirus dUTPases. Residues corresponding to motif V are absent in the amino portion of all alpha- and gammaherpesviruses with the exception of HH1A and HH3A, in which they are very divergent (Fig. 3). Archaeal and archaeal virus dUTPases (MJAN and SIRV) are also very divergent in motif V. In addition, there is a large insertion (21 to 38 residues) between motifs IV and V in carboxy copies of alpha- and gammaherpesvirus dUTPases.
FIG. 3.
Representative alignment of sequences analyzed in this study. OSM regions are indicated with bars and Roman numerals. MIRs include all residues between motifs I, II, III, IV, and V and those carboxyl to motif V (Arabic numbers in parentheses indicate residues removed for display). dUTPase residues involved in substrate binding as determined by X-ray crystallography are indicated above the alignment: E. coli (ECOL) by a plus symbol (+) (32) and H. sapiens (HSAP) by an asterisk (∗) (50). Residues implicated in dUTPase function by mutagenesis are also indicated above the alignment: FIV (1FIV) by a pound symbol (#) (74), E. coli also by a pound symbol (#) (73), and EIAV by a percent symbol (%) (59, 72). The carboxy copy of alpha- and gammaherpesvirus dUTPases (HH1C and HH4C) have no recognizable motif III. The amino copies of these dUTPases (HH1A and HH4A) are very divergent in motifs I, II, and IV. Motif V is truncated or divergent in HH1A, HH4A, and archaeal/archaeal virus dUTPases (MJAN/SIRV). There is a large insertion (21 to 38 residues) in the carboxy alpha- and gammaherpesvirus dUTPases (HH1C and HH4C) between motifs IV and V. Amino acid residues in a column are highlighted when more than half are identical or highly similar: (FY), (ILMV), (ST), (AG), (DE), (NQ), and (KR). The number of residues trimmed from the amino terminus of each sequence are indicated in parentheses at the beginning of the alignment. For a list of all sequence codes, see the Appendix.
Pairwise distances.
A pairwise distance matrix based on Dayhoff matrix distance values was constructed from the alignment and used for phylogenetic inference. Average pairwise distances (and standard deviations) between selected groups of organisms are shown in Table 2. For reference, average distances are shown for the human dUTPase sequence (HSAP) compared with dUTPases of rodents and with dUTPases of nonmammalian eukaryotes. The lowest possible distance is that of a sequence to itself (i.e., 0). HSAP has an average distance of 4.53 to dUTPases of rodents and an average distance of 46.65 to dUTPases of nonmammalian eukaryotes. At the other extreme, the average pairwise distance between eukaryotic and eubacterial dUTPases is close to 100 and between eukaryotic and archaeal dUTPases is close to 200 (Table 2).
Several interesting patterns are apparent in this matrix. dUTPase sequences from certain viruses with eukaryotic hosts show strikingly low pairwise distances to eukaryotic dUTPases. For example, ORF virus dUTPase (ORFV) has an average pairwise distance of 34.5 to dUTPases of mammals but, when compared with dUTPases from nonmammalian eukaryotes, the average pairwise distance increases to 59.14. The low distance of ORFV to mammalian dUTPases is particularly interesting given that the distance of ORFV to other poxvirus dUTPases is on average 56.74 (standard deviation [SD] of 1.65). dUTPases from poxviruses (excluding ORFV) and avian adenovirus (AVAD) also have low pairwise distances to mammalian dUTPases. dUTPase from a chlorella virus isolated from a paramecium (PBCV), however, does not show less difference from mammalian dUTPases than from nonmammalian eukaryotic dUTPases (Table 2).
Phylogeny reconstruction.
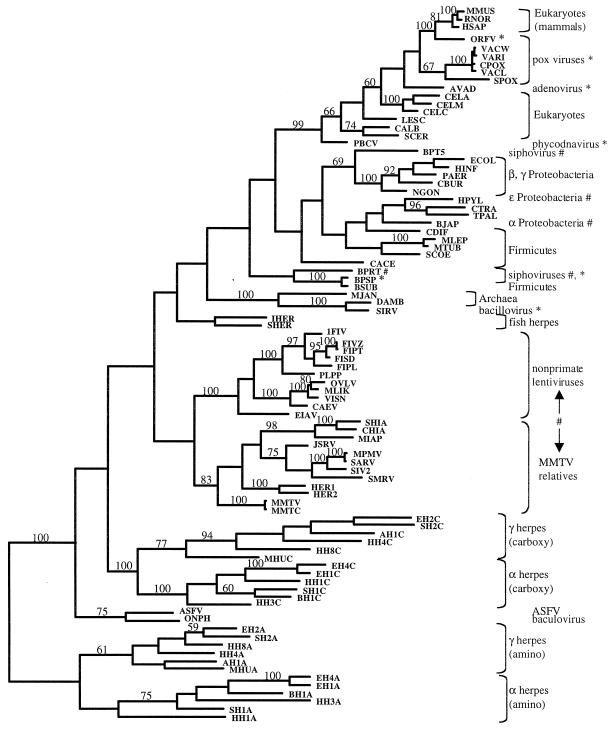
The complete pairwise distance matrix was used to reconstruct the phylogenetic relationships of the available dUTPases (Fig. 4). Distance analysis was employed because of the large numbers and sizes of insertions and deletions present in the data set (Fig. 3). Both FITCH (Phylip) and Ancestor implementations of the Fitch-Margoliash algorithm were used. Results from both programs were very similar with the exception of a few closely related taxa within clades. For convenience, only bootstraps greater than 50% are shown in Fig. 4. From the presence or lack of high bootstrap values we can see which parts of the phylogeny are well or poorly resolved. The topology indicates some unusual features of dUTPase similarities and relationships.
FIG. 4.
Shortest unrooted phylogenetic tree consistent with additive distances (Fitch-Margoliash algorithm). Poxvirus and avian adenovirus dUTPases are most similar to those of vertebrates. Chlorella virus (PBCV) dUTPase clusters with those of the eukaryotes. The bacteriophage r1t (BPRT) dUTPase clusters with those of the firmicutes. dUTPase from bacteriophage SPβ (BPSP) clusters with the sequence from host, B. subtilis (BSUB). Bacteriophage T5 (BPT5) also has a dUTPase similar to that of its proteobacterial host, E. coli (ECOL). Archaeal virus SIRV encodes a dUTPase similar to that of D. ambivalens (DAMB). Two proteobacterial dUTPases H. pylori (HPYL) and B. japonicum (BJAP) cluster with those of the groups Firmicutes, Chlamydiales, and Spirochaetales. MMTV relatives and nonprimate lentiviruses encode similar dUTPases. Strong cases for horizontal transfer supported by high bootstraps are indicated in this tree by an asterisk (∗). Other potential but less-well-supported cases are indicated with a pound symbol (#). Branch lengths are roughly proportional to distances, with the exception of very short branches, which are exaggerated for visibility. Groups of taxa corresponding to those listed in Table 1 are indicated with brackets and labels. Bootstrap values of 50% and higher are shown above supported branches. All taxon identifiers are listed in the Appendix.
The amino portion of alpha- and gammaherpesvirus dUTPases contains the fewest conserved functional and structural residues relative to the rest of the samples (Fig. 1, 3, and 4). For this reason, the amino herpesvirus dUTPase sequences are separated from the other dUTPases in the data set with 100% bootstrap support. The carboxyl portion of the duplicated herpesvirus dUTPases, grouped together with a 100% bootstrap, is different from other dUTPases due to the divergence in the region of motif III and the MIR between motifs I and II. Divergence is found in the MIRs throughout the data set, but in the case of herpesvirus dUTPases, there is additional divergence in part of the normally conserved OSM in each copy (amino and carboxyl). In contrast, the amino, middle, and carboxy copies of the C. elegans dUTPase have diverged very little since their duplication, and all three copies cluster together with a 100% bootstrap. For both the amino and carboxy copies of herpesvirus dUTPases, the alpha and gamma lineages form distinct monophyletic clades (Fig. 4). Trout herpesvirus (IHER) and salmon herpesvirus (SHER [not shown in Fig. 3]) dUTPases are similar to each other, but their single-copy arrangements make it difficult to determine their relationship to the duplicated dUTPases of alpha- and gammaherpesviruses. This is due to the fact that motif III and its flanking MIR are in a different place in the duplicated protein. The different dUTPase arrangement is consistent with the fact that fish herpesviruses are known to be a highly divergent lineage among herpesviruses as a group (10).
Monophyly of retroviral dUTPases was examined with the available data. The FITCH (Phylip) implementation found a tree which did not support monophyly of retroviral dUTPases (sum of squares [SS] = 226.22, %SD = 16.10; not shown). When this topology was rearranged such that the retroviral dUTPases were monophyletic, the result was a shorter, although not statistically different, topology (SS = 201.33, %SD = 16.03; Fig. 4). Ancestor was supplied with the alignment (Fig. 3) and the shortest FITCH (Phylip) topology (Fig. 4) as the initial tree. The shortest tree found by Ancestor was longer by FITCH (Phylip) standards (SS = 289.89, %SD = 19.24), but also supported monophyly of retroviral dUTPases (not shown).
The five motifs comprising the OSM are highly conserved relative to the MIRs (Fig. 3). Parallelism or convergence in a catalytic region (OSM) alone could generate a misleading topology. In order to ascertain the resolution and degree of the phylogenetic signal from conserved motifs and divergent nonmotif regions, OSM and MIR sequences were analyzed separately. Both data sets indicate the presence of the monophyletic groups of taxa described above (data not shown). The ability of divergent MIRs alone to recover the same monophyletic groups of taxa demonstrates that the recovered topology is not due simply to parallelism or convergence in the OSM. In addition, there are clear examples of functional convergence in unrelated dUTPases isolated from T4 phage and Leishmania sp. which show no detectable sequence convergence or similarity to the five classic motifs described here (4, 76). These two lines of evidence support the likelihood that evolution and horizontal transfer are responsible for the topology generated, rather than convergence and parallelism.
Well-supported cases for horizontal transfer between viruses and their hosts.
The usual diagnostic pattern for horizontal transfer of a gene is the discordant phylogeny it produces. Among the dUTPase amino acid sequences analyzed here, several examples of such a discordant phylogeny are apparent. In particular, there are several groups of sequences which are unexpectedly and consistently monophyletic (Fig. 4) no matter what distance method or data partition was used (not shown). The observed patterns indicate probable cases of horizontal transfer of the dut gene between viruses and hosts (eukaryotic, eubacterial, and archaeal). The data supporting specific instances of horizontal transfer are addressed below.
Between eukaryotes and viruses.
Four eukaryotic virus lineages encode dUTPase sequences that are monophyletic with those of eukaryotes with a bootstrap of 99% (Fig. 4). Several poxviruses encode dUTPase sequences (ORFV, VACW, VARI, CPOX, VACL, and SPOX). All of these viruses infect mammals, and their dUTPases cluster with the three mammalian sequences (MMUS, RNOR, and HSAP). Particularly striking is the fact that ORF virus dUTPase (ORFV) clusters with mammalian dUTPases with a 100% bootstrap rather than with other poxviruses. This is because the average pairwise distance between ORFV and mammalian dUTPases (34.50 [Table 2]) is lower than between ORFV and other poxvirus dUTPases (56.74, SD = 1.65).
Avian adenovirus is the only adenovirus to encode a dUTPase, and its sequence (AVAD) groups with those of animals with a 60% bootstrap. There is no avian dUTPase sequence available for comparison. The dUTPase isolated from a chlorella-like alga inside Paramecium bursaria (PBCV) also groups with those of eukaryotes. No paramecium or chlorella sequence is available for comparison. All of these viral dUTPases group with those of eukaryotes with a high bootstrap value (99%), and in some cases with the subset that specifically includes their hosts.
Between eubacteria and viruses.
dUTPase in bacteriophage SPβ (BPSP) clusters with the host Bacillus subtilis sequence (BSUB) with a 100% bootstrap. This virus has been found in the host genome as an integrated prophage (22). The bnrdE and bnrdF genes in this virus also resemble host analogs nrdE and nrdF (22, 33).
Between archaea and viruses.
The only sequence in this study of a dUTPase from an archaeal virus, SIRV, is monophyletic with that of archaeon Desulfurolobus ambivalens with a bootstrap value of 100%. This result is consistent with the phylogeny reported by Prangisvili et al. (54). The small archaeal cluster including the Methanococcus jannaschii dUTPase sequence is also very well defined with a bootstrap of 100%.
DISCUSSION
This study, containing the largest number of dUTPase amino acid sequences to date, allows us to examine the evolution of the dut gene in detail. In addition to the organisms listed in Table 1, dut has also been detected in rhesus monkeys, dogs, cows, rabbits, and chickens (46). dUTPase is a beneficial component of the viral replication machinery but is nonessential in host dividing tissues for herpesviruses (35, 56, 57), poxviruses (19), and nonprimate lentiviruses (36, 52, 53). All currently known functional dUTPases have the motifs described here (Fig. 1 and 3), with the exception of unrelated dCTPase-dUTPase bifunctional enzymes in phage (76) and an unrelated dUTP-specific protein in Leishmania sp. (4). Given the widespread distribution of obligatory dut genes in host organisms, the beneficial nature of the gene for viruses in some host tissues, and the similarity of many viral dUTPases to host dUTPases (Fig. 4), it is likely that several viruses have acquired dut genes from their hosts.
Our phylogeny (Fig. 4) clearly reveals this pattern. The poxviruses included in this study (Table 1) encode a dUTPase, whereas molluscum contagiosum virus (MCV) does not. Based on other proteins, recent studies observed similar relationships among the poxviruses studied here and that the MCV sequences are basal to the group (58). All six poxvirus dUTPases studied here cluster with those of eukaryotes by using either the phylogenetic method and OSM, MIR, or the combined data set. Five poxvirus dUTPases (VACW, VARI, CPOX, VACL, and SPOX) group together with a bootstrap value of 67% (100% excluding SPOX). This implies a common eukaryotic origin for these five poxvirus dUTPases. Thus, it appears that a host dut gene was acquired subsequent to the divergence of MCV but prior to that of swinepox, vaccinia, variola, and cowpox viruses. In addition, ORF virus dUTPase (ORFV) clusters with mammalian dUTPases (100%) and is more similar to mammalian dUTPases than to dUTPases from other poxviruses (Table 2). This pattern is due mostly to similarity in the OSM rather than the MIR (data not shown), which may imply extreme divergence from dUTPases of other poxviruses combined with convergence to the host OSM. Alternatively, a separate transfer may have resulted in the observed ORF virus dut gene. The dUTPase sequence from a baculovirus infecting gypsy moths and its similarity to dUTPases of poxviruses has been recently published (30). Our preliminary analysis of this sequence shows that it has close similarity with eukaryotic dUTPases in general, indicating that it is another possible example of horizontal transfer (data not shown).
It has been suggested that avian adenovirus dut (AVAD) is related to an analogous unidentified reading frame in human adenoviruses (75). The avian adenovirus sequence groups with animal dUTPases with a bootstrap of 60%, with multicellular eukaryotic dUTPases at 66%, and with eukaryotic dUTPases in general at 99%. This result strongly implies a eukaryotic dut transfer as the origin of this gene. If this gene is in fact related to the unidentified reading frame in human adenoviruses, it must have diverged considerably in that lineage after acquisition.
P. bursaria chlorella virus dUTPase (PBCV) also clearly clusters with eukaryotic dUTPases with a 99% bootstrap, supporting monophyly. All other single-copy dUTPases from eukaryotic DNA viruses (IHER, SHER, ASFV, and ONPH) are scattered about the topology, very distant from the eukaryotic dUTPases (Fig. 4). If they also originally acquired dut genes from eukaryotes, sufficient divergence has occurred to obscure this fact. Bacteriophage SPβ encodes a dUTPase (BPSP) that clusters with host B. subtilis dUTPase (BSUB) with a bootstrap of 100%. Archaeal virus dUTPase (SIRV) clusters with archaeal dUTPases (MJAN and DAMB) with a high bootstrap (100%). Although this phylogeny is consistent with that reported by Prangisvili et al. (54), no assertions of horizontal transfer have been made for this viral gene until now.
Additional potential cases for horizontal transfer.
One siphovirus (bacteriophage) dUTPase sequence clusters with those of eubacteria no matter which of the two phylogenetic methods or data sets (OSM, MIR, or combined) is used. Bacteriophage T5 (BPT5) was isolated from E. coli, a member of the gamma subclass of the class proteobacterium (28), and its sequence clusters basal to those of gamma and beta proteobacteria (ECOL, HINF, PAER, CBUR, and NGON) with a bootstrap value of 69% (Fig. 4). Bacteriophage r1t was isolated from firmicute host Lactococcus lactis (69), and its dUTPase sequence (BPRT) clusters with that of firmicute B. subtilis (BSUB). The BPRT sequence is 97% identical to a recently deposited L. lactis dUTPase sequence. Preliminary analysis indicates a bootstrap of more than 96% for monophyly of the L. lactis and BPRT dUTPase sequences (data not shown). The dUTPase of bacteriophage phi PVL isolated from S. aureus, was also recently deposited in GenBank. Preliminary analysis of this sequence shows similarity to dUTPases of eubacteria (data not shown).
The chlamydial (CTRA) and spirochete (TPAL) dUTPase sequences cluster together with a bootstrap of 96%. Chlamydiales and Spirochaetales comprise eubacterial groups that are entirely separate from firmicutes and proteobacteria. Although not supported with a 50% bootstrap, epsilon and alpha proteobacterial dUTPases (HPYL and BJAP) cluster with those of Chlamydiales and Spirochaetales, within dUTPases of firmicutes. This is inconsistent with the organismal phylogeny, as epsilon and alpha proteobacteria are more closely related to other (beta and gamma) proteobacteria (13). It is possible that dUTPase sequences are insufficient to resolve these relationships, given the few representative sequences from these taxa currently available. Alternatively, members of these groups may have exchanged dut genes. Several other genes of Helicobacter pylori (67) and Chlamydia trachomatis (61) are suspected to have been acquired horizontally from other eubacteria, as well as eukaryotes. More dUTPase sequences from members of underrepresented taxa will be necessary to resolve this issue.
Two retrovirus lineages contain a complete dut gene (MMTV-like and nonprimate lentiviruses). The gene is located in a different genomic region in each lineage (Fig. 2) (41), yet the protein sequence is sufficiently similar between these lineages to imply a possible common origin relative to other dUTPase protein sequences available. The shortest distance tree supports monophyly of the dUTPase sequences in retroviruses, although this topology is not significantly shorter than the search tree in which the nonprimate lentivirus dUTPases are polyphyletic with respect to those of MMTV relatives. There are close relatives to each lineage that do not encode a dUTPase (Fig. 2). While our best tree is consistent with horizontal transfer between retroviral lineages, the lack of statistical significance between the two trees prevents a definitive claim for horizontal transfer in this case.
Alpha- and gammaherpesviruses encode a recognizable dUTPase, while the related betaherpesviruses encode an analogous reading frame containing no recognizable motifs. dUTPase activity has not yet been assayed in betaherpesviruses. If the gene is not present in betaherpesviruses, then perhaps alpha- and gammaherpesviruses are more closely related to each other and inherited a duplicated dut gene from a common ancestor. This evolutionary scenario would be consistent with a reported herpesvirus polymerase phylogeny (24). Channel catfish virus and salmonid herpesvirus also encode a dUTPase, but it is a single copy and quite different from those reported in vertebrates. Fish herpesviruses are known to be the most distant members of the herpesviruses (10). These viruses may have acquired the gene independently or early in the evolution of herpesviruses, before the duplication observed in alpha- and gammaherpesvirus lineages. Either scenario is consistent with reported herpesvirus phylogenies (24).
Genome location.
The fact that the location of dut is variable in the closely related genomes of viruses and eubacteria (Fig. 2) demonstrates that it has moved. Despite its variable position, dut was observed adjacent to other genes involved in nucleotide metabolism such as ribonucleotide reductase, transcription initiation factors, primase, and DNA synthesis flavoprotein (Fig. 2). In eubacteria the proximity of dut to other genes needed for similar functions might be beneficial. There is evidence that genes coding for related biochemical functions in eubacteria frequently occur adjacent to one another (62). In retroviruses the location of dut affects its level of expression. The gag portion of the gag/pol polycistron is translated approximately 20 times more than the entire polycistron (7). Thus, in MMTV relatives one would expect a 20-fold-greater level of dut expression than in nonprimate lentiviruses.
Mechanisms of horizontal transfer.
Retroviruses may acquire sequences relatively easily from their hosts or from each other because of viral recombination or incorporation of host mRNA into the retroviral genome (23). A mature dut RNA message could theoretically be copackaged in a retrovirus and then incorporated into its genome. This might explain why none of the retroviral dut sequences have introns even though their vertebrate counterparts do; a pattern also observed in c- and v-oncogenes, for example c-myc (7). dUTPase is encoded in a different region in two different retrovirus lineages (MMTV relatives and nonprimate lentiviruses), despite its absence in close relatives of these lineages. It is most reasonable to assume a horizontal transfer between the two lineages or independent acquisition of the gene, whereas a loss in all other relatives is highly unlikely. If convergence or parallelism were operating because both dUTPases function in a retroviral background, one might expect the generally conserved OSM to reflect this. It is the MIRs that support monophyly of the MMTV and nonprimate lentivirus dUTPases rather than the OSM (data not shown). In addition, a spuma-related retrovirus encodes a dUTPase in a third unique location (8). We have another study in progress to determine the relationship of this third type of retroviral dUTPase to the others. Due to the rapid rate of evolution in RNA genomes and consequent high sequence divergence, the source of these genes (host or retroviral) may be undeterminable. It has recently been proposed that the outer domain of gp120 in primate lentivirus human immunodeficiency virus (HIV) also originated as a host dUTPase sequence (1). While it is possible that gp120 evolved from a dUTPase-like sequence, the extreme lack of conserved dUTPase residues between and within the dUTPase OSM in gp120 makes the identity of the original protein impossible to confirm.
Lack of introns in DNA virus dUTPases implies a retroviral intermediate.
Some hosts have introns in their dut genes, although viral dut genes do not. For example, the human dut gene spans about 14 kb and contains approximately five introns (30a). Mouse (30a) and rhesus monkey (46) dut genes are also reported to have introns. In our study, dut in C. elegans was found to have two introns in each of its three copies (data not shown). One of the introns in C. elegans dut corresponds exactly to the position of an intron in H. sapiens dut in motif IV. To explain the presence of intronless dut genes in DNA viruses, we hypothesize that these viruses acquired a cDNA of an RNA message analogous to the mechanism that resulted in v-oncogenes. The agent responsible for mediating this event must encode a reverse transcriptase. Transfer may have occurred in different cellular compartments, since poxviruses replicate in the cytoplasm, whereas adenoviruses replicate in the nucleus. In contrast, the process by which a eukaryotic nucleus might acquire a retroviral or DNA virus dut gene would not necessarily require an intermediary retroid agent. Introns would be inserted subsequent to the transfer. Since the viral, not the eukaryotic, topology is discordant, and given the presence of dut introns in eukaryotes, it is most likely that the viruses acquired dut from their eukaryotic hosts rather than vice versa. Convergence or parallelism of viral copies with host genes is unlikely because MIRs alone support the monophyly of viral dUTPases with their hosts (data not shown) nearly as well as the entire sequence (Fig. 4). If convergence or parallelism were operating, one would expect the OSM to support monophyly and the MIRs to contribute noise or a different topology. In addition, functional but unrelated dUTPases in T4 phage and Leishmania spp. are a clear example of functional convergence in the absence of sequence homology (4, 76).
Implications for folding and subunit assembly.
While the location of structural and functional residues has been determined by X-ray crystallography in single-copy dUTPases (9, 32, 50, 55), structures have not been reported for any dUTPases with other motif arrangements. On the basis of the conserved structural and functional residues in the alignment (Fig. 3), it is possible to hypothesize the type of folding and assembly that might occur in duplicated and triplicated dUTPases (Fig. 5). Alpha- and gammaherpesviruses have a duplication such that the first copy conserves motif III and the second copy conserves I, II, IV, and V (Fig. 1 and 3). The available evidence indicates that herpesvirus dUTPases function as monomers (5). Some residues involved in secondary structure (50) are conserved as well, implying that much of the structure surrounding motifs I, II, IV, and V is similar (data not shown). The structure of the amino portion of the alpha- and gammaherpesvirus dUTPase remains ambiguous, although motif III is likely present in an active site as is found in homotrimers comprised of single copies. The large 21- to 38-residue insertion between motifs IV and V in carboxy copies of herpesvirus dUTPases may aid this by allowing the flexible tail (containing motif V) as found in E. coli (70) to double back to complete the active site in the monomer (Fig. 5).
FIG. 5.
Assembly of dUTPase subunits. The four known dUTPase crystals (FIV, EIAV, E. coli, and H. sapiens) indicate a homotrimer folding to form three separate active sites in which each of the five conserved motifs is present. (A) H. sapiens (single). It was observed in the H. sapiens dUTPase that each subunit contributes at least one motif to each active site (50). In this diagram, the characteristic beta-barrel has been flattened to show the components of the trimer fold. Due to this flattening, the position of motif III is misleading. In the three-dimensional structure, motif III on the solid-gray chain associates with motifs I, II, and IV of the solid-black chain and motif V of the dashed chain. To help clarify these relationships, each active site is indicated by a shape (triangle, square, and circle) surrounding the motifs of which it is comprised (based on the published structure and assembly of H. sapiens dUTPase, [50]). (B) Herpesvirus (duplicated). Based on the H. sapiens structure and our alignment (Fig. 3), a possible structure for an alpha- and a gammaherpesvirus monomer is proposed. Motif III is present in the amino portion of the repeat instead of between motifs II and IV as in H. sapiens (Fig. 1). (C) C. elegans (triplicated). The dUTPase of C. elegans is the only dUTPase example to date of a tandem triplication comprising of three complete single copies (Fig. 1). This protein could potentially fold into a single monomeric dUTPase of the approximate size of the H. sapiens homotrimer. Straight lines indicate conserved structural regions inferred from the alignment. Curved lines indicate that the structure between motif V of one copy and motif I of the next is unknown. Each subunit is indicated by solid-black, dashed, or solid-gray lines. The amino (N) and carboxy (C) termini of each subunit are labeled. Motifs are labeled I to V in their approximate positions in the dUTPase alignment (Fig. 3).
Duplicated genes typically undergo one of several well-documented fates. Generally, one gene will retain its function while the other diverges into a pseudogene or evolves a new function. Alternatively, both copies may retain similar functions, as in the globin family. Fused tandem copies functioning in concert, each with a complete OSM, have been observed in aspartic acid proteases. The fate of alpha- and gammaherpesvirus dut genes is unusual in that it is the only example to our knowledge of fused tandem copies each potentially contributing a subset of motifs to the catalytic site.
The dUTPase of C. elegans has a novel arrangement in that it is tandemly triplicated such that a single peptide could theoretically fold into a shape similar to that of a single-copy homotrimer (Fig. 5). The way in which the peptide may fold between motif V of one copy and motif I of the next is ambiguous. There are two introns of the same size and in the same position within each copy of the C. elegans dut gene, one of which interrupts the coding regions. The DNA sequences of the three copies are fairly similar (60% identical in a three-way comparison of all three coding regions), although the introns are more divergent (data not shown). This is consistent with the 100% bootstrap for the cluster containing the amino acid sequences of each copy (Fig. 4). The conservation of gene organization and sequence in these copies may indicate that the triplication is fairly recent.
The patterns of dut horizontal transfer observed between archaea, eubacteria, and eukaryotes and their viruses have important implications for clinical and genomic research. In terms of clinical implications, other DNA and RNA viruses may acquire the dut gene and thereby expand their pathology to include nondividing tissues. The dUTPase protein has been proposed as a target for antiviral drugs and cancer chemotherapy (47, 48). It is thought that primate lentiviruses such as HIV, which do not encode dUTPase, may activate and use dut genes in human endogenous retroviruses (HERVs) (48). Uncontrolled pools of dUTP are considered toxic to rapidly dividing tissue because they lead to incorporation of dUTP in DNA, excessive DNA repair, and cell death. Already a common target for chemotherapy, thymidylate synthase is an enzyme downstream of dUTPase in the biochemical pathway for conversion of dUTP to TTP. Targeting the dUTPase protein may likewise have potential for controlling the rapid growth of tissues (47). Understanding the nature and relationships of native and HERV dUTPases in humans will be important for identifying specific areas of the protein to target, and such studies are in progress.
In this study we have established a compelling case for at least five horizontal acquisitions of viral dut genes from eukaryotic, eubacterial, and archaeal hosts. These include dUTPases from: poxviruses, avian adenovirus, Paramecium bursaria chlorella virus, bacteriophage SPβ, and archaeal virus SIRV. A sixth potential case is that ORF virus acquired its dUTPase separately from those of other poxviruses. We have identified potential transfer events among several eubacteria, two between eubacteria and their viruses, and one among retroviruses. We have demonstrated that dut is present in variable genomic locations in eubacteria and RNA and DNA viruses. Finally, potential structures are hypothesized by mapping primary structures of various uncrystallized motif arrangements onto known tertiary structures of known dUTPase crystals. dut provides an example of a gene undergoing ubiquitous horizontal transfer relative to viral core genes, with different arrangements of the same conserved motifs functioning in different genetic background. This study illustrates that both sequence similarity and genomic location need to be considered when reconstructing the evolutionary history of individual genes and the genomes in which they are found. Future comparative genomic studies will reveal how many more genes move as often and how many other dUTPase motif arrangements may exist.
ACKNOWLEDGMENTS
We acknowledge the following sources for dUTPase sequences unavailable from GenBank: Fraser et al. (22) for the use of their T. pallidum sequence; the Gonococcal Genome Sequencing Project and B. A. Roe et al. for the use of their N. gonorrhoeae sequence (64); the Chlamydia Genome Project of R. S. Stephens et al. for the use of their C. trachomatis sequence (63); the Genome Therapeutics/The Microbial Genome Project for the use of their C. acetobutylicum sequence (65); and the Pseudomonas Genome Project, a collaborative effort of the University of Washington Genome Center, The PathoGenesis Corporation, and the U.S. Cystic Fibrosis Foundation for the use of their P. aeruginosa sequence (66). Finally, we thank J. Kowalski and J. Liu for assistance in HMM construction and technical assistance, I. K. Jordan and S. Corro for advice and comments on the manuscript, and R. Ladner and J. Alfano for stimulating discussion.
This work was supported by NIH grant AI28309 and a Research Career Development Award to M.A.M.
Appendix
Eukaryotic sequence codes are: MMUS, Mus musculus; RNOR, Rattus norvegicus; HSAP, Homo sapiens; CELA/M/C, Caenorhabditis elegans amino, middle, and carboxy copies; LESC, Lycospersion esculentum; SCER, Saccharomyces cerevisiae; and CALB, Candida albicans. Nonprimate lentivirus sequence codes are: 1FIV, FIV strain Oma; FIVZ, strain Z1; FISD, an isolate from San Diego; FIPT, strain Petaluma; FIPL, an unknown strain; PLPP, puma lentivirus 14; OVLV, ovine lentivirus; MLIK, Maedi-visna-like virus; CAEV, caprine arthritis encephalitis virus; VISN, visna virus; and EIAV, equine infectious anemia virus. MMTV-related retrovirus sequence codes are: MMTV, mouse mammary tumor virus, strain unknown; MMTC, MMTV strain C3H; SARV, simian AIDS retrovirus; SIV2, simian SRV-2 retrovirus; MPMV, Mason-Pfizer monkey virus; JSRV, Jaagsiekte sheep retrovirus; SMRV, squirrel monkey retrovirus; HER1, human endogenous retrovirus K10; HER2, an unnamed HERV-K; SHIA, syrian hamster intracisternal particle; CHIA, Chinese hamster intracisternal particle; and MIAP, mouse intracisternal particle. A/C indicates the amino/carboxy copies of each duplicated alpha- and gammaherpesvirus sequence. Alphaherpesvirus sequence codes are: HH1A/C, human herpesvirus 1; HH3A/C, human herpesvirus 3; BH1A/C, bovine herpesvirus 1; EH1A/C, equine herpesvirus 1; EH4A/C, equine herpesvirus 4; and SH1A/C, suid herpesvirus 1. Gammaherpesvirus sequence codes are: HH4A/C, human herpesvirus 4; HH8A/C, human herpesvirus 8; AH1A/C, alcelaphine herpesvirus 1; EH2A/C equine herpesvirus 2; MHUA/C, murine herpesvirus; and SH2A/C, saimiriine herpesvirus 2. The fish herpesvirus sequence codes are: IHER, ictalurid (channel catfish) herpesvirus, and SHER, salmonid (rainbow trout) herpesvirus. Poxvirus sequence codes are: VACW, vaccinia virus, strain WR; VACL, vaccinia virus, strain Liverpool; VARI, variola virus; CPOX, cowpox virus; SPOX, swinepox virus; and ORFV, ORF virus. Other eukaryotic DNA virus sequence codes are: ONPH, Orygia pseudotsugata nuclear polyhedrosis virus; ASFV, African swine fever virus; AVAD, avian adenovirus; and PBCV, Paramecium bursaria chlorella virus 1. Eubacterial sequence codes are: ECOL, Escherichia coli; HINF, Haemophilus influenzae; PAER, Pseudomonas aeruginosa; CBUR, Coxiella burnetii; HPYL, Helicobacter pylori; NGON, Neisseria gonorrhoeae; BJAP, Bradyrhizobium japonicum; MLEP, Mycobacterium leprae; MTUB, Mycobacterium tuberculosis; CDIF, Clostridium difficile; CACE, Clostridium acetobutylicum; SCOE, Streptomyces coelicolor; CTRA, Chlamydia trachomatis; TPAL, Treponema pallidum; and BSUB, Bacillus subtilis. Bacteriophage (siphovirus) sequence codes are: BPRT, bacteriophage r1t; BPT5, bacteriophage T5; and BPSP, bacteriophage SPβ. Archaeal sequence codes are: DAMB, Desulfurolobus ambivalens, and MJAN, Methanococcus jannaschii. The archaeal virus (bacillovirus) sequence code is SIRV, Sulfobulus islandicus rod-shaped virus 1.
REFERENCES
- 1.Abergel C, Robertson D L, Claverie J-M. “Hidden” dUTPase sequence in human immunodeficiency virus type 1 gp120. J Virol. 1999;73:751–753. doi: 10.1128/jvi.73.1.751-753.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Baer R, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatfull G, Hudson G S, Satchwell S C, Seguin C. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 4.Camacho A, Arrebola R, Peña-Diaz J, Ruiz-Pérez L M, González-Pacanowska D. Description of a novel eukaryotic deoxyuridine 5′-triphosphate nucleotidohydrolase in Leishmania major. Biochem J. 1997;325:441–447. doi: 10.1042/bj3250441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caradonna S J, Adamkiewicz D M. Purification and properties of the deoxyuridine triphosphate nucleotidohydrolase enzyme derived from HeLa S3 cells. J Biol Chem. 1984;259:5459–5464. [PubMed] [Google Scholar]
- 6.Cedergren-Zeppezauer E S, Larsson G, Nyman P O, Dauter Z, Wilson K S. Crystal structure of a dUTPase. Nature. 1992;355:740–743. doi: 10.1038/355740a0. [DOI] [PubMed] [Google Scholar]
- 7.Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. 1st ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 8.Cordonnier A, Casella J-F, Heidmann T. Isolation of novel human endogenous retrovirus-like elements with foamy virus-related pol sequence. J Virol. 1995;69:5890–5897. doi: 10.1128/jvi.69.9.5890-5897.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dauter Z, Persson R, Rosengren A M, Nyman P O, Wilson K S, Cedergren-Zeppezauer E S. Crystal structure of dUTPase from equine infectious anaemia virus: active site metal binding in a substrate analogue complex. J Mol Biol. 1999;285:655–673. doi: 10.1006/jmbi.1998.2332. [DOI] [PubMed] [Google Scholar]
- 10.Davison A J. The genome of salmonid herpesvirus 1. J Virol. 1998;72:1974–1982. doi: 10.1128/jvi.72.3.1974-1982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davison A J, Scott J E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986;67:1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 12.Dayhoff M O, Schwartz R M, Orcutt B C. A model of evolutionary change in proteins. In: Dayhoff M O, editor. Atlas of protein sequence and structure, vol. 5, suppl. 3. Washington, D.C: National Biomedical Research Foundation; 1978. pp. 345–352. [Google Scholar]
- 13.Eisen J A. The RecA protein as a model molecule for molecular systematic studies of bacteria: comparison of trees of RecAs and 16S rRNAs from the same species. J Mol Evol. 1995;41:1105–1123. doi: 10.1007/BF00173192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El-Hajj H H, Zhang H, Weiss B. Lethality of a dut (deoxyuridine triphosphatase) mutation in Escherichia coli. J Bacteriol. 1988;170:1069–1075. doi: 10.1128/jb.170.3.1069-1075.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 16.Feng D F, Doolittle R F. Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J Mol Evol. 1987;25:351–360. doi: 10.1007/BF02603120. [DOI] [PubMed] [Google Scholar]
- 17.Fitch W M, Farris J S. Evolutionary trees with minimum nucleotide replacements from amino acid sequences. J Mol Evol. 1974;3:263–278. doi: 10.1007/BF01796042. [DOI] [PubMed] [Google Scholar]
- 18.Fitch W M, Margoliash E. Construction of phylogenetic trees. Science. 1967;155:279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- 19.Fleming S B, Lyttle D J, Sullivan J T, Mercer A A, Robinson A J. Genomic analysis of a transposition-deletion variant of ORF virus reveals a 3.3 kbp region of non-essential DNA. J Gen Virol. 1995;76:2969–2978. doi: 10.1099/0022-1317-76-12-2969. [DOI] [PubMed] [Google Scholar]
- 20.Fraser C M, Norris S J, Weinstock G M, White O, Sutton G G, Dodson R, Gwinn M, Hickey E K, Clayton R, Ketchum K A, Sodergren E, Hardham J M, McLeod M P, Salzberg S, Peterson J, Khalak H, Richardson D, Howell J K, Chidambaram M, Utterback T, McDonald L, Artiach P, Bowman C, Cotton M D, Fujii C, Garland S, Hatch B, Horst K, Roberts L, Watthey L, Janice W, Smith H O, Venter C J. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 21.Gadsen M, McIntosh E, Game J, Wilson P, Haynes R. dUTP pyrophosphatase is an essential enzyme in Saccharomyces cerevisiae. EMBO J. 1993;12:4425–4431. doi: 10.1002/j.1460-2075.1993.tb06127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghim S-Y, Choi S-K, Shin B-S, Park S-H. An 8 kb nucleotide sequence at the 3′ flanking region of the sspC gene (184°) on the Bacillus subtilis 168 chromosome containing an intein and an intron. DNA Res. 1998;5:121–126. doi: 10.1093/dnares/5.2.121. [DOI] [PubMed] [Google Scholar]
- 23.Hajjar A M, Linial M L. A model system for nonhomologous recombination between retroviral and cellular RNA. J Virol. 1993;67:3845–3853. doi: 10.1128/jvi.67.7.3845-3853.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hannenhalli S, Chappey C, Koonin E V, Pevzner P A. Genome sequence comparison and scenarios for gene rearrangements: a test case. Genomics. 1995;30:299–311. doi: 10.1006/geno.1995.9873. [DOI] [PubMed] [Google Scholar]
- 25.Higgins D, Bleasby A J, Fuchs R. Clustal V: improved software for multiple sequence alignment. CABIOS. 1991;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 26.Hill F, Loakes D, Brown D M. Polymerase recognition of synthetic oligodeoxyribonucleotides incorporating degenerate pyrimidine and purine bases. Proc Natl Acad Sci USA. 1998;95:4258–4263. doi: 10.1073/pnas.95.8.4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hughey R, Krogh A. Hidden Markov models for sequence analysis: extension and analysis of the basic method. Comput Appl Biosci. 1996;12:95–107. doi: 10.1093/bioinformatics/12.2.95. [DOI] [PubMed] [Google Scholar]
- 28.Kaliman A V. Identification of the bacteriophage T5 dUTPase by protein sequence comparisons. DNA Seq. 1996;6:347–350. doi: 10.3109/10425179609047573. [DOI] [PubMed] [Google Scholar]
- 29.Krogh A, Brown I, Mian S, Sjolander K, Haussler D. Hidden Markov models in computational biology: applications to protein modeling. J Mol Biol. 1994;235:1501–1531. doi: 10.1006/jmbi.1994.1104. [DOI] [PubMed] [Google Scholar]
- 30.Kuzio J, Pearson M N, Harwood S H, Funk J C, Evans J T, Slavicek J M, Rohrmann G F. Sequence and analysis of the genome of a baculovirus pathogenic for Lymantria dispar. Virology. 1999;253:17–34. doi: 10.1006/viro.1998.9469. [DOI] [PubMed] [Google Scholar]
- 30a.Ladner, R. Personal communication.
- 31.Ladner R D, McNulty D E, Carr S A, Roberts G D, Caradonna S J. Characterization of distinct nuclear and mitochondrial forms of human deoxyuridine triphosphate nucleotidohydrolase. J Biol Chem. 1996;271:7745–7751. doi: 10.1074/jbc.271.13.7745. [DOI] [PubMed] [Google Scholar]
- 32.Larsson G, Svensson L A, Nyman P O. Crystal structure of the Escherichia coli dUTPase in complex with a substrate analogue (dUTP) Nat Struct Biol. 1996;3:532–538. doi: 10.1038/nsb0696-532. [DOI] [PubMed] [Google Scholar]
- 33.Lazarevic V, Soldo B, Düsterhöft A, Hilbert H, Mauël C, Karamata D. Introns and intein coding sequence in the ribonucleotide reductase genes of Bacillus subtilis temperate bacteriophage SPbeta. Proc Natl Acad Sci USA. 1998;95:1692–1697. doi: 10.1073/pnas.95.4.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Lu Z, Sun L, Ropp S, Kutish G F, Rock D L, van Etten J L. Analysis of 74 kb of DNA located at the right end of the 330-kb chlorella virus PBCV-1 genome. Virology. 1997;237:360–377. doi: 10.1006/viro.1997.8805. [DOI] [PubMed] [Google Scholar]
- 35.Liang X, Tang M, Manns B, Babiuk L A, Zamb T J. Identification and deletion mutagenesis of the bovine herpesvirus 1 dUTPase gene and a gene homologous to herpes simplex virus UL49.5′. Virology. 1993;195:42–50. doi: 10.1006/viro.1993.1344. [DOI] [PubMed] [Google Scholar]
- 36.Lichtenstein D L, Rushlow K E, Cook R F, Raabe M L, Swardson C J, Kociba G J, Issel C J, Montelaro R C. Replication in vitro and in vivo of an equine infectious anemia virus mutant deficient in dUTPase activity. J Virol. 1995;69:2881–2888. doi: 10.1128/jvi.69.5.2881-2888.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Massung R F, Jayarama V, Moyer R W. DNA sequence analysis of conserved and unique regions of swinepox virus: identification of genetic elements supporting phenotypic observations including a novel G protein-coupled receptor homologue. Virology. 1993;197:511–528. doi: 10.1006/viro.1993.1625. [DOI] [PubMed] [Google Scholar]
- 38.McClure M A. Evolution of retroposons by acquisition or deletion of retrovirus-like genes. Mol Biol Evol. 1991;8:835–856. doi: 10.1093/oxfordjournals.molbev.a040686. [DOI] [PubMed] [Google Scholar]
- 39.McClure M A, Hudak J, Kowalski J M. Low identity, low similarity protein sequences: independent modeling of the ordered-series-of-motifs and motif-intervening-regions. In: Miyano S, Takagi T, editors. Genome informatics 1998. 1998. pp. 183–192. Universal Academy Press, Inc., Tokyo, Japan. [PubMed] [Google Scholar]
- 40.McClure M A, Johnson M S, Doolittle R F. Relocation of a protease-like gene segment between two retroviruses. Proc Natl Acad Sci USA. 1987;84:2693–2697. doi: 10.1073/pnas.84.9.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McClure M A, Johnson M S, Feng D-F, Doolittle R F. Sequence comparisons of retroviral proteins: relative rates of change and general phylogeny. Proc Natl Acad Sci USA. 1988;85:2469–2473. doi: 10.1073/pnas.85.8.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McClure M A, Kowalski J. The effects of ordered-series-of-motifs anchoring and subclass modeling on the generation of HMMs representing highly divergent protein sequences. In: Altman R B, Dunker A K, Hunter L, Klein T E, Lauderdale K, editors. Proceedings of the Pacific Symposium on Biocomputing, 1999. River Edge, N.J: World Scientific; 1999. pp. 162–170. [DOI] [PubMed] [Google Scholar]
- 43.McClure M A, Raman R. Parameterization studies of hidden Markov models representing highly divergent protein sequences. In: Hunter L, Shriver B D, editors. Proceedings of the 28th Hawaii International Conference on System Sciences. V. Washington, D.C: IEEE Computer Society Press; 1995. pp. 184–193. [Google Scholar]
- 44.McGeoch D J. Protein sequences, comparisons show that the ‘pseudoproteases’ encoded by poxviruses and certain retroviruses belong to the deoxyuridine triphosphatase family. Nucleic Acids Res. 1990;18:4105–4110. doi: 10.1093/nar/18.14.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 46.McIntosh E M, Ager D D, Gadsden M H, Haynes R H. Human dUTP pyrophosphatase: cDNA sequence and potential biological importance of the enzyme. Proc Natl Acad Sci USA. 1992;89:8020–8024. doi: 10.1073/pnas.89.17.8020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McIntosh E M, Haynes R H. dUTP pyrophosphatase as a potential target for chemotherapeutic drug development. Acta Biochim Pol. 1997;44:159–171. [PubMed] [Google Scholar]
- 48.McIntosh E M, Haynes R H. HIV and human endogenous retroviruses: an hypothesis with therapeutic implications. Acta Biochim Pol. 1996;43:583–592. [PubMed] [Google Scholar]
- 49.Mercer A A, Fraser K M, Stockwell P A, Robinson A J. A homologue of retroviral pseudoproteases in the paraoxvirus, ORF virus. Virology. 1989;172:665–668. doi: 10.1016/0042-6822(89)90212-2. [DOI] [PubMed] [Google Scholar]
- 50.Mol C D, Harris J M, McIntosh E M, Tainer J A. Human dUTP pyrophosphatase: uracil recognition by a beta hairpin and active sites formed by three separate subunits. Structure. 1996;4:1077–1092. doi: 10.1016/s0969-2126(96)00114-1. [DOI] [PubMed] [Google Scholar]
- 51.Needleman S B, Wunsch C D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 52.Petursson G, Turelli P, Sigridur M, Georgsson G, Andresson O S, Torsteinsdottir S, Vigne R, Andresdottir V, Gunnarsson E, Agnarsdottir G, Querat G. Visna virus dUTPase is dispensable for neuropathogenicity. J Virol. 1998;72:1657–1661. doi: 10.1128/jvi.72.2.1657-1661.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Phillips T R, Prospero-Garcia O, Wheeler D W, Wagaman P C, Lerner D L, Fox H S, Whalen L R, Bloom F E, Elder J H, Henriksen S J. Neurologic dysfunctions caused by a molecular clone of feline immunodeficiency virus, FIV-PPR. J Neurovirol. 1996;2:388–396. doi: 10.3109/13550289609146904. [DOI] [PubMed] [Google Scholar]
- 54.Prangisvili D, Klenk H-P, Jakobs G, Schmiechen A, Hanselmann C, Holz I, Zillig W. Biochemical and phylogenetic characterization of the dUTPase from the archaeal virus SIRV. J Biol Chem. 1998;273:6024–6029. doi: 10.1074/jbc.273.11.6024. [DOI] [PubMed] [Google Scholar]
- 55.Prasad S G, Stura E A, McRee D E, Laco G S, Hasselkus-Light C, Elder J H, Stout C D. Crystal structure of dUTP pyrophosphatase from feline immunodeficiency virus. Protein Sci. 1996;5:2429–2437. doi: 10.1002/pro.5560051205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pyles R B, Sawtell N M, Thompson R L. Herpes simplex virus type 1 dUTPase mutants are attenuated for neurovirulence, neuroinvasiveness, and reactivation from latency. J Virol. 1992;66:6706–6713. doi: 10.1128/jvi.66.11.6706-6713.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ross J, Williams M, Cohen J I. Disruption of the varicella-zoster virus dUTPase and the adjacent ORF9A gene results in impaired growth and reduced syncytia formation in vitro. Virology. 1997;234:186–195. doi: 10.1006/viro.1997.8652. [DOI] [PubMed] [Google Scholar]
- 58.Senkevich T G, Koonin E V, Bugert J J, Darai G, Moss B. The genome of Molluscum contagiosum virus: analysis and comparison with other poxviruses. Virology. 1997;233:19–42. doi: 10.1006/viro.1997.8607. [DOI] [PubMed] [Google Scholar]
- 59.Shao H, Robek M D, Threadgill D S, Mankowski L S, Cameron C E, Fuller F J, Payne S L. Characterization and mutational studies of equine infectious anemia virus dUTPase. Biochem Biophys Acta. 1997;1339:181–191. doi: 10.1016/s0167-4838(96)00229-4. [DOI] [PubMed] [Google Scholar]
- 60.Slabaugh M B, Roseman N A. Retroviral protease-like gene in the vaccinia virus genome. Proc Natl Acad Sci USA. 1989;86:4152–4155. doi: 10.1073/pnas.86.11.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stephens R S, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov R L, Zhao Q, Koonin E V, Davis R W. Genome sequence of a obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 62.Tamames J, Casari G, Ouzounis C, Valencie A. Conserved clusters of functionally related genes in two bacterial genomes. J Mol Evol. 1997;44:66–73. doi: 10.1007/pl00006122. [DOI] [PubMed] [Google Scholar]
- 63.The Chlamydia Genome Project. [Online.] University of California, Berkeley. http://chlamydia-www.berkeley.edu:4231. [20 January 1999, last date accessed.]
- 64.The Gonococcal Genome Sequencing Project. [Online.] http://dna1.chem.ou.edu/gono.html. [20 January 1999, last date accessed.]
- 65.The Microbial Genome Project. [Online.] Genome therapeutics. http:// www.genomecorp.com/genesequences/clostridium/clospage.html. [20 January 1999, last date accessed.]
- 66.The Pseudomonas Genome Project. [Online.] University of Washington Genome Center/PathoGenesis Corp./U.S. Cystic Fibrosis Foundation. http://www.pseudomonas.com. [20 January 1999, last date accessed.]
- 67.Tomb J-F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzgerald L M, Lee N, Adams M D, Hickey E K, Berg D E, Gocayne J D, Utterback T R, Peterson J D, Kelley J M, Cotton M D, Weidman J M, Fuji C, Bowman C, Watthey L, Wallin E, Hayes W S, Borodovsky M, Karp P D, Smith H O, Fraser C M, Venter C J. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 68.Turelli P, Guiguen F, Mornex J-F, Vigne R, Querat G. dUTPase-minus Caprine arthritis-encephalitis virus is attenuated for pathogenesis and accumulates G-to-A substitutions. J Virol. 1997;71:4522–4530. doi: 10.1128/jvi.71.6.4522-4530.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Sinderen D, Karsens H, Kok J, Terpstra P, Ruiters M H J, Venema G, Nauta A. Sequence analysis and molecular characterization of the temperate lactococcal bacteriophage r1t. Mol Microbiol. 1996;19:1343–1355. doi: 10.1111/j.1365-2958.1996.tb02478.x. [DOI] [PubMed] [Google Scholar]
- 70.Vertessy B G. Flexible glycine rich motif of Escherichia coli deoxyuridine triphosphate nucleotidohydrolase is important for functional but not for structural integrity of the enzyme. Proteins. 1997;28:568–579. [PubMed] [Google Scholar]
- 71.Vertessy B G, Larsson G, Persson T, Bergman A-C, Persson R, Nyman P O. The complete triphosphate moiety of non-hydrolyzable substrate analogues is required for a conformational shift of the flexible C-terminus in Escherichia coli dUTP pyrophosphatase. FEBS Lett. 1998;421:83–88. doi: 10.1016/s0014-5793(97)01545-7. [DOI] [PubMed] [Google Scholar]
- 72.Vertessy B G, Persson R, Rosengren A-M, Zeppezauer M, Olof N P. Specific derivatization of the active site tyrosine in dUTPase perturbs ligand binding to the active site. Biochem Biophys Res Commun. 1996;219:294–300. doi: 10.1006/bbrc.1996.0226. [DOI] [PubMed] [Google Scholar]
- 73.Vertessy B G, Zalud P, Nyman P O, Zeppezauer M. Identification of tyrosine as a functional residue in the active site of Escherichia coli dUTPase. Biochem Biophys Acta. 1994;1205:146–150. doi: 10.1016/0167-4838(94)90103-1. [DOI] [PubMed] [Google Scholar]
- 74.Wagaman P C, Hasselkus-Light C S, Henson M, Lerner D L, Phillips T R, Elder J H. Molecular cloning and characterization of deoxyuridine triphosphatase from feline immunodeficiency virus (FIV) Virology. 1993;196:451–457. doi: 10.1006/viro.1993.1501. [DOI] [PubMed] [Google Scholar]
- 75.Weiss R S, Lee S S, Prasad B V V, Javier R T. Human adenovirus early region 4 open reading frame 1 genes encode growth-transforming proteins that may be distantly related to dUTP pyrophosphatase enzymes. J Virol. 1997;71:1857–1870. doi: 10.1128/jvi.71.3.1857-1870.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wheeler L J, Ray N F, Ungermann C, Hendricks S P, Bernard M A, Hanson E S, Mathews C K. T4 phage gene 32 protein as a candidate organizing factor for the deoxyribonucleoside triphosphate synthetase complex. J Biol Chem. 1996;271:11156–11162. doi: 10.1074/jbc.271.19.11156. [DOI] [PubMed] [Google Scholar]