Abstract
Epstein-Barr virus (EBV) is implicated in different central nervous system syndromes. The major cellular receptor for EBV, complement receptor type 2 (CR2) (CD21), is expressed by different astrocyte cell lines and human fetal astrocytes, suggesting their susceptibility to EBV infection. We demonstrated the infection of two astrocyte cell lines, T98 and CB193, at low levels. As infection was mediated by CR2, we used two stable CR2 transfectant astrocyte cell lines (T98CR2 and CB193CR2) to achieve a more efficient infection. We have monitored EBV gene expression for 2 months and observed the transient infection of T98 and T98CR2 cells and persistent infection of CB193 and CB193CR2 cells. The detection of BZLF1, BALF2, and BcLF1 mRNA expression suggests that the lytic cycle is initiated at early time points postinfection. At later time points the pattern of mRNA expressed (EBER1, EBNA1, EBNA2, and LMP1) differs from latency type III in the absence of LMP2A transcription and in the expression of BALF2 and BcLF1 but not BZLF1. A reactivation of the lytic cycle was achieved in CB193CR2 cells by the addition of phorbol esters. These studies identify astrocyte cell lines as targets for EBV infection and suggest that this infection might play a role in the pathology of EBV in the brain.
Epstein-Barr virus (EBV) is one of the eight known human herpesviruses (45). About 90% of the world’s adult population is estimated to be infected with EBV. Primary infection with EBV normally occurs through salivary exchange in the oropharynx, and this is believed to result in virus replication in stratified squamous epithelial cells and in the subsequent infection of trafficking B lymphocytes. Following primary infection, either symptomatic or silent, the virus persists in the healthy host throughout the entire life span by mechanisms that are not fully understood. There is strong evidence implicating EBV in the pathogenesis of human tumors of B-cell origin (endemic Burkitt lymphoma) or of other origin, such as nasopharyngeal carcinoma (an epithelial malignancy), some cases of Hodgkin’s disease, and some subsets of T-cell lymphoma (44).
A variety of neurological syndromes can occur with primary EBV infection, clinically manifested as infectious mononucleosis (25, 50). The central nervous system (CNS) syndromes include diffuse or focal encephalitis (12, 23), aseptic meningitis, Guillain-Barré syndrome, Bell’s palsy, acute cerebellar ataxia, transverse myelitis, and peripheral neuropathy (48). Although such cases are rare, encephalitis represents a serious and potentially fatal complication of infectious mononucleosis. Furthermore the role of EBV in demyelinating disease or subacute or chronic meningoencephalitis such as Rasmussen’s encephalitis has been suspected but has not been investigated in a systematic manner (5, 56, 57). EBV DNA was detected by PCR in brain biopsy samples obtained from patients in whom EBV was suspected to play a role in neurological syndromes like encephalitis, CNS lymphoma (22, 26), fatal disseminated lymphoproliferation, progressive multifocal leukoencephalopathy, multiple sclerosis, or encephalopathy. In situ assays will be required to demonstrate which and how many cells of the brain may harbor the virus (40). It seems that EBV-carrying B lymphocytes in the brain are the most likely source of the positive PCR results, but those studies could not rule out the possibility of a direct infection of brain resident cells. EBV DNA was also found in almost all cases of AIDS-related primary lymphoma of the CNS (20) early in the course of the disease, even preceding the detection of lymphoma by radiological examination. Cinque and coworkers and D’Arminio Monforte and coworkers were able to directly detect EBV DNA in cerebrospinal fluid (CSF) by PCR (11, 13). This might originate from a leakage of neoplastic cells containing EBV into CSF, from virions liberated through cell necrosis, or from an active lytic EBV infection (11). The presence of EBV in CSF suggests that the virus can get in contact with neural cells. Thus, to study the interaction between EBV and astrocytes is interesting for several reasons.
First, astrocytes constitute the most numerous cell type in the brain. Over the last decades, it has become increasingly evident that these cells perform a variety of important cerebral functions (36). Hence, any disruption of astrocyte function would have devastating consequences on brain function. Second, it was reported that astrocytes may be infected by HIV in vivo (3, 38). The life cycle of the virus and the mode of infection are unique for this cell type, with a low production of viral particles and a high level of the transcription of the regulatory genes nef and tat (29, 54). Even the interaction of envelope proteins of human immunodeficiency virus type 1 (HIV-1) with astrocytes seems to disturb their cellular functions (38). HIV-1-restricted infection could interfere with the normal astrocyte functions and lead to a dysregulation of astrocytic interactions with other neural cell types. Despite the nonpermissive, restricted nature of HIV-1 infection, it was possible to show that astrocytes can produce HIV-1 and can contribute to its spreading under the influence of appropriate signals delivered by permissive cells (like activated mononuclear phagocytes and lymphocytes) (14). Factors which modulate the viral load of HIV-1 could influence disease progression either positively or negatively. The coinfection of cells (or individuals) with other viruses, particularly herpesviruses, has the potential to effect such modulations (24). Until now, there has been no report of a possible interaction between EBV and astrocytes or HIV-infected astrocytes found in the brains of AIDS patients suffering from EBV DNA-positive primary cerebral lymphoma. Third, the most important data which convinced us to study the interaction of astrocytes and EBV were contributed by Gasque and coworkers (21). Astrocyte cell lines and human fetal astrocytes were described as the only brain cells expressing complement receptor type 2 (CR2) (21). CR2 is the major known cellular receptor for EBV (1, 39). This is a 145-kDa glycoprotein, first described as the receptor for the C3d component of complement expressed on human B lymphocytes (19). The binding sites for EBV and C3dg are contained within the two amino-terminal short consensus repeats (SCR) of 60 amino acids (SCR1 and SCR2) (8). A truncated CR2 comprising these two SCRs, the transmembrane, and intracytoplasmic region is sufficient to mediate EBV binding and infection (8, 9). Astrocyte-CR2 functionality was evaluated on astrocyte cell lines and human fetal astrocytes by the specific binding of C3d and gp340, recombinant protein of EBV surface protein gp350, which could be blocked specifically with polyclonal anti-CR2 antibodies (21). These data suggest that at least astrocyte cell lines and human fetal astrocytes can interact with EBV. The possibility of astrocyte infection by EBV remains to be demonstrated and constitutes the purpose of our study.
We have studied the expression of different genes during the 2 months following the infection of two different astrocyte cell lines. We have decided to develop an astrocyte culture model based on a model already described by Li and coworkers to study the EBV infection of epithelial cells (30). We have constructed two stably CR2-transfected astrocyte cell lines. The validity of this model is shown here, in that we obtained the same EBV transcript pattern for the CR2 transfectant and wild-type cell lines.
MATERIALS AND METHODS
Cell culture and viruses.
The human cell line T98-G was obtained from the American Type Culture Collection (ATCC) (Manassas, Va.; CRL-1690) and presented characteristics of glioblastoma multiforme. A second cell line used in this study, CB193, was a glioma grade III, kindly provided by B. Delpech (Centre Henri Becquerel, Rouen, France). T98CR2 and CB193CR2 cell lines were obtained by the stable transfection of those two astrocyte cell lines with CR2 cDNA (34). Cell lines were cultivated in Ham’s F-12 medium (J. Bio, Paris, France) supplemented with l-glutamine (2 mM), penicillin (100 U/ml), streptomycin (100 μg/ml), and 10% fetal calf serum (FCS) (Biologic Industries, Kibbutz Beth Haemek, Israel). All B-cell lines, Raji (ATCC; CCL-86), Ramos (ATCC; CRL-1596), and B95-8 (ATCC; CRL 1612), established from marmoset B cells by immortalization with EBV in throat washings from an individual suffering from infectious mononucleosis, were propagated in complete RPMI 1640 medium (Bio-Whittaker, Verviers, Belgium) supplemented as described above. The cell lines were routinely screened by the Hoechst 33258 DNA staining method to ensure that they were free of mycoplasma.
MAbs, flow cytometry analysis, and sorting.
Monoclonal antibodies (MAbs) used for the screening and the analysis of epitopes expressed by transfected cells were HB5 (immunoglobulin G2a [IgG2a]) (ATCC; HB135) (52), purified from ascites fluid. The isotype control was VD3 (IgG2a) (42).
For flow cytometry analysis, 106 cells were washed with fluorescence-activated cell sorting (FACS) buffer (phosphate-buffered saline [PBS]–0.5% bovine serum albumin–0.1% NaN3) and incubated with MAb at a saturating concentration (2 μg/ml in FACS buffer) for 30 min on ice and, after washing with cold FACS buffer, with fluorescein isothiocyanate (FITC)-conjugated anti-mouse F(ab′)2 fragments (Dakopatts, Glostrup, Denmark) diluted 1:50 in FACS buffer. After being washed, the cells were analyzed with a FACSCalibur apparatus (Becton Dickinson, Mountain View, Calif.).
Virus production.
Infection was done with virus produced by the B95-8 cell line. After removal of B95-8 cells by centrifugation at 3,000 × g for 15 min, supernatants of cell cultures were concentrated 500-fold by ultracentrifugation at 25,000 × g for 1.5 h at 18°C. The virus pellet was resuspended in Ham’s F-12–2% FCS and stored in aliquots at −70°C. For infection, concentrated virus was diluted 1:10 in Ham’s F-12–2% FCS. To inactivate EBV, viral preparations were boiled for 45 min at 95°C. For cocultivation experiments, B95-8 cells were irradiated from a 137Cs γ-source with 30 Gy.
EBV particle quantification and infection procedures.
DNA from 0.5 ml of viral preparation was prepared by using the Qiagen Blood kit (Qiagen, Hilden, Germany) according to the instructions of the manufacturer. Finally the DNA was resuspended in 200 μl of water, and 5 μl was used for each PCR assay. The number of EBV particles was quantified with a competitive DNA PCR established by Jäger et al. (28). As an internal standard, pGEM 3Z-30, a construct containing a sequence from the BRLF2 reading frame with a 30-bp deletion (kind gift of M. Jäger, University of Regensburg, Regensburg, Germany) was added to the amplification mixture in 10-fold dilutions from 6 × 106 to 6 × 102 molecules.
For the infection of astrocyte cell lines in monolayer cultures, approximately 106 cells were washed two times with PBS and incubated with 1 ml of viral preparation or with 106 irradiated B95-8 cells at 37°C for 2 to 3 h. Cells were then washed several times with PBS and cultivated with 15 ml of complete Ham’s F-12 medium.
To evaluate the role of CR2 in astrocyte infection, cells were preincubated with or without MAb FE8, which blocks infection of B lymphocytes with EBV (kindly given by W. Prodinger [43]), at 20 μg/ml for 60 min at 4°C. After three washes with PBS, cells were incubated with EBV as described before. At different time points after infection a part of the cells was harvested and assayed for the presence of EBV DNA and RNA.
DNA extraction and EBV DNA detection.
Cells were harvested 48 h after the addition of EBV by trypsinization and pelleted. Cellular DNA extraction and EBV DNA detection by PCR were performed as described by Martin et al. (33). This resulted eventually in the amplification of a 527-bp fragment (positions 14256 to 14783 of the EBV genome), a segment which is within the BamHI W repeat. PCR conditions were as follows: an initial denaturation at 94°C for 3 min, 30 cycles (94°C for 1 min, 55°C for 1 min, and 72°C for 1 min), and 72°C for 10 min. For GAPDH (glyceraldehyde-3-phosphate dehydrogenase), 5′-GTG AAG GTC GGA GTC AAC G-3′ and 5′-GGT GAA GAC GCC AGT GGA CTC-3′ were used. Conditions for PCRs were as follows: an initial denaturation at 94°C for 3 min, 30 cycles (94°C for 1 min, 52°C for 1 min, and 72°C for 1 min), and 72°C for 10 min. Fifteen microliters of each reaction mixture was analyzed by electrophoresis through a 2% agarose gel, stained with ethidium bromide, and blotted onto nylon membranes (Bio-Rad, Richmond, Calif.) for Southern blotting. Filters were hybridized with 32P-labeled probes under stringent conditions. As a hybridization probe the PCR product from the positive control B95-8 cell line was labeled by a standard procedure. The hybridization signal was quantified with a phosphorimager (Fuji model BS 100).
Detection of EBV RNA.
Cells were harvested 3, 6, 13, 22, 34, 43, and 62 days after the addition of EBV by trypsinization and pelleted. Total RNA was prepared from different cell lines by using the Qiagen RNeasy kit (Qiagen) according to the instructions of the manufacturer. RNA integrity was confirmed by electrophoresis. RNA preparations were treated with DNase I (Boehringer; 3 U/μg of RNA abolishes all DNA traces), and control samples were prepared by treatment of an aliquot of the RNA preparation with DNase I and RNase A (40 μg/ml for 5 μg of RNA; Boehringer).
Prior to PCR steps, the reverse transcription (RT) was carried out at 37°C for 60 min in a 25-μl final volume with 2 μg of total RNA, 60 U of RNasin (Promega), 1 mM concentrations of deoxynucleoside triphosphates (dNTPs), 300 pmol of oligo(dT) (MWG Biotech, Ebersberg, Germany), and 400 U of Moloney murine leukemia virus-RT (Boehringer) in the reaction buffer (10 mM Tris-HCl, 15 mM KCl, 0.6 mM MgCl2 and 5 mM dithiothreitol).
The transcription of EBV genes was assayed by RT and PCR analysis with a set of EBV-specific oligonucleotide primers (Table 1). PCR (except for EBER1 amplification) was carried out with 4 μl of reverse-transcribed RNA mixture (with 5 μl of the first PCR product for the nested PCR), in a final volume of 50 μl with 20 pmol of primers for the first PCR (50 pmol for the nested PCR) in a 1× buffer (Promega), 200 μM concentrations of dNTPs, 1.5 mM MgCl2, and 1 U of Taq DNA polymerase (Promega). For the EBER1 amplification, 0.5 μg of mRNA were reverse transcribed directly by using 50 pmol of each primer with 60 U of RNasin (Promega), 1 mM concentrations of dNTPs, and 400 U of Moloney murine leukemia virus-RT (Boehringer) in the reaction buffer (10 mM Tris-HCl, 15 mM KCl, 0.6 mM MgCl2). The RT was carried out at 37°C for 60 min and 5 min at 95°C to inactivate enzymes. Then 1 U of Taq DNA polymerase (Promega) was added to the mixture.
TABLE 1.
Sequences of primers used for RT-PCR and nested PCR and their positions in the genome of the EBV B95-8 strain
| Gene target | Oligonucleotide sequence (5′ to 3′) | Coordinates (nt) |
|---|---|---|
| EBER1 5′a | AAAACATGCGGACCACCAGC | 6776–6795 |
| EBER1 3′ | AGGACCTACGCTGCCCTAGA | 6648–6629 |
| EBNA1 5′RTb | CTTAGGAAGCGTTTCTTGAGCTT | 67482–67504 |
| EBNA1 3′RT | GGGTCTCCGGACACCATCTC | 108168–108149 |
| EBNA1 5′nested | GAGCGTTTGGGAGAGCTGAT | 67514–67533 |
| EBNA1 3′nested | CATTTCCAGGTCCTGTACCT | 107987–107967 |
| LMP2A 5′RTb | CACCGCTTATGAGGACCCA | 166729–166749 |
| LMP2A 3′RT | CTGCGGCCAGCAATGAAAC | 434–415 |
| LMP2A 5′nested | ATGACTCATCTCAACACATA | 166874–166893 |
| LMP2A 3′nested | CATGTTAGGCAAATTGCAAAA | 380–361 |
| LMP1 5′RTc | ACACACTGCCCTGAGGATGG | 169471–169490 |
| LMP1 3′RT | ATACCTAAGA/CAAGTAAGCA | 168956–168965 (or 169042–169051) |
| LMP1 5′nested | CTTCAGAAGAGACCTTCTCT | 169243–169262 |
| LMP1 3′nested | ACAATGCCTGTCCGTGCAAA | 169100–169081 |
| EBNA2 5′RTa | AGAGGAGGTGGTAAGCGGTCC | 14802–14822 |
| EBNA2 3′RT | TGACGGGTTTCCAAGACTATCC | 48584–48563 |
| EBNA2 5′nested | AGCGGTTCACCTTCAGGGCC | 14815–14832 (or 47761–47762) |
| EBNA2 3′nested | TGTAGGCATGATGGCGGCAG | 48512–48493 |
| BALF2 5′RT | GTCAAGATGTTCAAGGACGTGG | 163072–163094 |
| BALF2 3′RT | CTCATAGCACATACAGATGGGC | 162878–162856 |
| BALF2 5′nested | GGTCAAGAGCTGCTACCACG | 162978–162998 |
| BALF2 3′nested | GGAGATGTCCTGCAGGATGG | 162901–162881 |
| BZLF1 5′RTd | ATTGCACCTTGCCGGCCACCTTTG | 103180–103194 |
| BZLF1 3′RT | CGGCATTTTCTGGAAGCCACCCGA | 102486–102462 |
| BZLF1 5′nested | GACCAAGCTACCAGAG TCTAT | 103057–103077 |
| BZLF1 3′nested | CAGAATCGCATTCCTCCAGCCA | 102530–102512 (or 102657–102655) |
| BcLF1 5′RT | TATGCCCAATCCCAAGTACACG | 136210–136231 |
| BcLF1 3′RT | TGGACGGGTGGAGGAAGTCTTC | 135888–135867 |
| BcLF1 5′nested | ACACAGCAGCTACCGGTGGA | 136137–136176 |
| BcLF1 3′nested | TGTTGCAGGGAGTAGGTCTC | 136014–135995 |
The PCR conditions used for all amplifications (except for EBER1 amplification) were as follows: 94°C for 3 min, 35 cycles (94°C for 30 s, 55°C for 45 s, and 72°C for 1 min), and 72°C for 10 min. The conditions used for EBER1 were as follows: 94°C for 3 min, 40 cycles (94°C for 30 s, 50°C for 1 min, and 72°C for 1 min), and 72°C for 10 min.
For each amplification B95-8 cDNA was used as a positive control, and the PCR mixture without DNA or with cDNA of uninfected astrocytes was used as a negative control. Ten microliters from the PCR mixture was separated by electrophoresis on a 2% agarose gel.
LMP1 detection by immunofluorescence assay.
For latent membrane protein 1 (LMP1) antigen detection, cells were directly fixed after trypsinization with 4% paraformaldehyde (in PBS) during 20 min at 37°C on a slide. After air drying, primary MAb anti-LMP1 CS1-4 (46) (Dakopatts; diluted 1:100 in PBS–1% FCS–0.2% saponin) was applied to the sections, and the slides were incubated at 37°C for 60 min. The slides were then subjected to two 5-min washes in PBS–1% FCS, followed by an additional 60-min incubation with FITC-conjugated goat anti-mouse IgG (Dakopatts) diluted 1:50 in PBS–1% FCS–0.2% saponin. Washed slides were mounted with glycerol and examined by using an Olympus fluorescence microscope.
EBNA2 and BZLF1 protein detection by FACS analysis.
MAb PE2 was used for EBNA2 antigen detection (59) (Dakopatts), and MAb BZ1 was used for BZLF1 detection (60) (Dakopatts). To increase the specificity of labeling, MAbs were biotin labeled by a standard procedure. IgG1 was used as an isotype control. For flow cytometry analysis, 106 cells were fixed with cold acetone. To diminish the background level, rabbit IgG (serum diluted 1:10 in PBS–1% FCS–0.2% saponin) was incubated with cells on ice for 30 min. Cells were incubated with biotin-MAb (diluted 1:100 in PBS–1% FCS–0.2% saponin) for 30 min on ice and with phycoerythrin-conjugated streptavidin (diluted 1:1,000 in PBS–1% FCS–0.2% saponin) for 30 min on ice. After washing, the cells were analyzed with a FACSCalibur apparatus.
Lytic cycle induction.
For EBV lytic cycle induction, CB193CR2 cells at day 60 postinfection (p.i.) were exposed to 5 × 10−8 M 12-tetradecanoyl-phorbol-13-acetate (TPA). After three further days, the cells were harvested, and mRNA was extracted for BZLF1-mRNA detection by RT-PCR as described above.
RESULTS
Detection of CR2 in astrocyte cell lines.
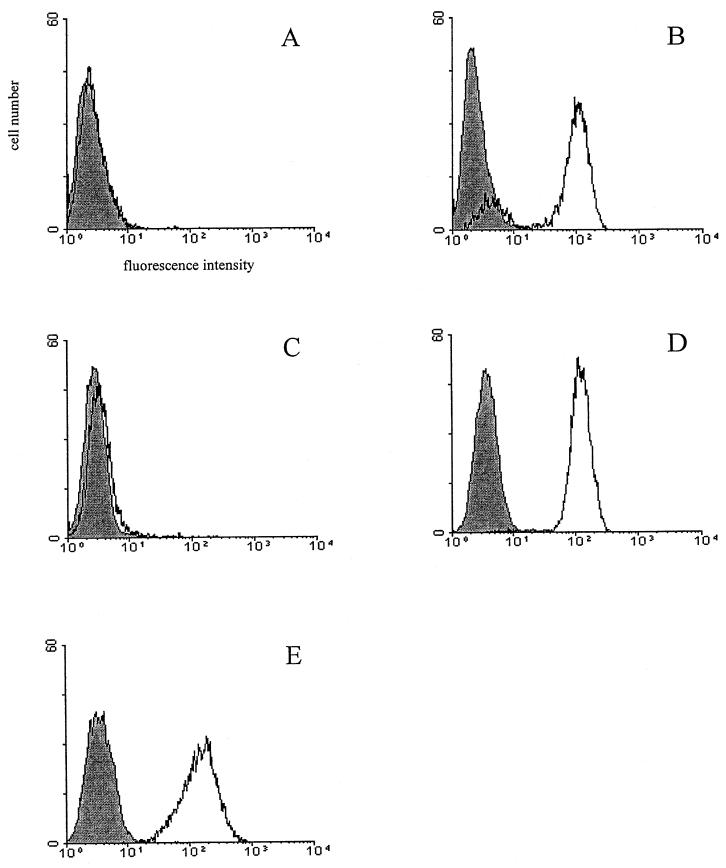
CR2 mRNA could be detected after RT-PCR analysis in the two parental astrocyte cell lines, T98 and CB193 (data not shown), confirming a previous report by Gasque and coworkers (21). However, CR2 antigen was not observed by flow cytometry analysis (Fig. 1 and Table 2) with the anti-CR2 MAb HB5. The lack of detection of surface CR2 protein with this MAb had also been reported by Gasque et al. (21). Nevertheless, they showed CR2 expression at low levels when a monospecific polyclonal anti-CR2 antibody and other MAbs like OKB7 were used.
FIG. 1.
Analysis of CR2 expression by using MAb HB5 on wild-type and CR2-transfected astrocyte lines: T98WT (A), T98CR2 (B), CB193WT (C), CB193CR2 (D), and Raji (positive control) (E). The cells were stained with anti-CR2 MAb HB5 (open area) or with an isotype control (shaded area) and FITC-labeled goat F(ab′)2 anti-mouse IgG, as described in Materials and Methods.
TABLE 2.
Flow cytometric analysis results for CR2 expression
| Cell line | MFI |
|---|---|
| T98WT | 7.84 |
| T98CR2 | 228.54 |
| CB193WT | 7.01 |
| CB193CR2 | 163.63 |
| Raji | 172.55 |
Transfection of astrocytic cell lines with CR2 expression vector and expression of transfected CR2.
The low level of CR2 on the two parental astrocyte cell lines (T98 and CB193) is probably the first barrier to studies of the influence of EBV interaction with astrocytes. To overcome this, these cell lines were transfected with the CR2 expression vector pSFFV-CR2. The transfection procedure is presented elsewhere (34). Figure 1 presents the CR2 expression after sorting out. The cell population of CB193CR2 was homogeneous, while T98CR2 still exhibited much broader CR2 expression, even after several sorting cycles. As shown in Table 2, however, the level of CR2 expression in CB193CR2 and T98CR2 was comparable to that in Raji cells. Transfected cells are morphologically identical to the untransfected parent cell line and display essentially the same growth features (data not shown).
In addition, HB5 or FE8 MAbs immunoprecipitated the 145-kDa CR2 protein from 125I-surface-labeled T98CR2, CB193CR2, and Raji cells and only on the CB193 untransfected cell line after a long exposure (data not shown). The cell membrane localization of CR2 on the transfected cell lines was confirmed by immunohistochemistry (data not shown).
Functionality of the CR2-transfected cell lines: detection of EBV DNA and BZLF1 mRNA in CR2-expressing and wild-type cell lines after EBV infection.
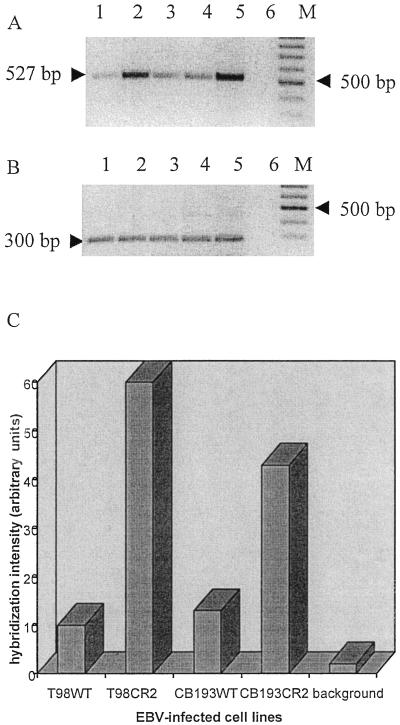
We further determined whether the CR2 protein expressed on wild-type and transfectant cell lines was able to support infection of the cells with EBV. Cells were exposed to intact virus, cultivated for 48 h in a complete medium, and then analyzed for BZLF1 expression as well as for the presence of EBV DNA. For a more sensitive determination of cellular infection, a PCR assay was used to detect a 527-bp fragment of the BamHI W segment of the EBV genome. We succeeded in testing all four cell lines, wild types and transfectants: after Southern blotting, hybridization with 32P-labeled probe, and semiquantification of EBV DNA with a phosphorimager the higher quantity of EBV DNA in the CR2 transfectant cell lines was revealed (Fig. 2).
FIG. 2.
Detection of EBV DNA on astrocyte lines 48 h after infection. DNA from each cell line was prepared and used as a template for PCR amplification of a 527-bp fragment of the BamHI W fragment of the EBV genome. (A) PCR amplification products were resolved on a 2% agarose gel and detected by staining with ethidium bromide (inverted picture). A negative control that lacked template DNA (lane 6) and a positive control that used B95-8 DNA as a template (lane 5) were included. Lane 1, T98WT; lane 2, T98CR2; lane 3, CB193WT; lane 4, CB193CR2; M, molecular weight marker (100-bp ladder). (B) The same DNA preparation was amplified for the GAPDH gene by using specific primers. (C) Semiquantification of the EBV DNA PCR signal. After Southern blotting, the membrane was hybridized with a specific 32P-labeled probe. The hybridization signal was analyzed with a phosphorimager and calculated as arbitrary units. The background lane shows the hybridization signal from the negative control. The signal obtained for the positive control was too strong, and the quantification was impossible.
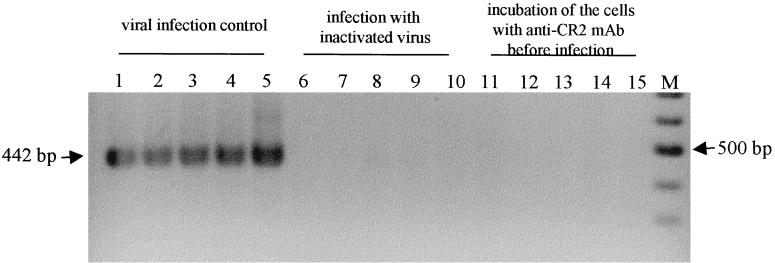
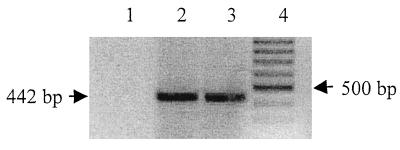
To confirm that the EBV DNA signal was due to intracellular EBV genome and not to virions attached to the surface of the target cells, we also examined at the same time point the mRNA expression of the early transcribed gene BZLF1. After total RNA extraction and DNase I treatment, an RT-PCR followed by a nested PCR and a control treatment with RNase A were performed as described in Materials and Methods. As indicated by the presence of the 442-bp product derived from spliced BZLF1 mRNA, gene expression was observed in all four cell lines tested at this time point (Fig. 3). By this approach we could not observe differences in the quantities in the four infected cell lines. Cells incubated with boiled EBV remained uninfected (Fig. 3, lanes 6 to 10). We then tested the capacity of these cell lines to support infection with EBV in the presence of blocking anti-CR2 MAb FE8, which was shown to block EBV infection of resting B cells by binding to SCR1 and SCR2 of CR2 (43). Cells were preincubated in PBS–1% FCS with FE8 (final concentration, 20 μg/ml) before EBV exposure. As shown in Fig. 3 (lanes 11 to 14), BZLF1 mRNA could not be detected in any of the four cell lines.
FIG. 3.
Expression of spliced BZLF1 mRNA 2 days after infection. Cells were infected with EBV prepared from B95-8 supernatant and harvested 2 days p.i. RNAs were extracted and submitted to PCR amplification with specific primers for BZLF1 mRNAs (their sequences are shown in Table 1). PCR amplification products were detected by staining with ethidium bromide (inverted picture). Different virus preparations were used: EBV concentrated from B95-8 supernatant and diluted in Ham’s F-12 medium supplemented with 2% FCS (lane 1, T98WT; lane 2, T98CR2; lane 3, CB193WT; lane 4, CB193CR2; the B-cell line Ramos was used as a positive control for the infection procedure in lane 5), virus inactivated by boiling for 45 min (lane 6, T98WT; lane 7, T98CR2; lane 8, CB193WT; lane 9, CB193CR2; lane 10, Ramos), or cells preincubated 1 h at 4°C with anti-CR2 MAb FE8 (20 μg/ml) before infection with active viral preparation (lane 11, T98WT; lane 12, T98CR2; lane 13, CB193WT; lane 14, CB193CR2; lane 15, Ramos). M, molecular weight marker (100-bp ladder).
Analysis of viral gene transcription in astrocyte cell lines.
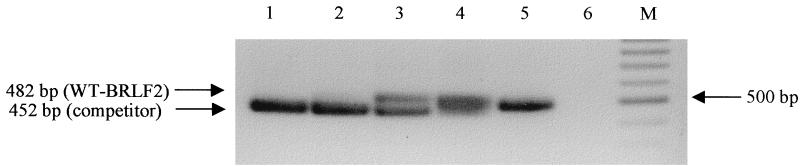
To establish the transcription profile, we infected 106 astrocytes with EBV. The multiplicity of infection was estimated by using the PCR quantification system developed by Jäger et al. (28) and is shown in Fig. 4. Briefly, this assay was done by a quantitative DNA PCR for transcripts of the BLRF2 open reading frame encoding viral protein p23 with a competitor construct that contained the corresponding target sequence of BLRF2 with a deletion of 30 bp. Using this protocol we were able to quantitatively determine that around 106 virions were added to 106 astrocytes. Since during DNA extraction a part of the viral DNA was certainly lost, this probably represents an underestimation of EBV particles present, given also that the calculated number of EBV genomes probably does not correspond with the number of infectious virions. Taken together, our data demonstrated that a low multiplicity of infection (around 1) could be used efficiently for the infection of all cell lines in this study.
FIG. 4.
Competitive DNA PCR for quantification of viral particles in EBV preparations used for infection. DNA was extracted from the EBV preparation and used for BLRF2 PCR. For quantification a competitor DNA with a 30-bp deletion was added in 10-fold dilutions ranging from 15 × 105 to 15 × 101 molecules per reaction (lanes 1 to 5). When competitor and viral DNAs were present in similar concentrations (here approximately 15 × 103 [lane 3]) both PCR products were visible in similar amounts. Lane 6 shows the PCR negative control. M, molecular weight marker (100-bp ladder). WT, wild type.
To study the kinetics of EBV gene expression, the cells were cultivated up to 62 days p.i. under the same conditions, and total cellular RNAs were extracted at several time points. Figure 5 and Table 3 present the time course of EBV gene expression in the four astrocyte cell lines. We could detect some EBV transcripts in T98 until day 22 p.i. for the wild type and day 43 for the CR2 transfectant cell line. So, it seems likely that the infection was transient in this cell line. On the contrary CB193WT (the wild type) and CB193CR2 cell lines still expressed EBV mRNA after 2 months p.i. and were still infected 3 months p.i. (data not shown).
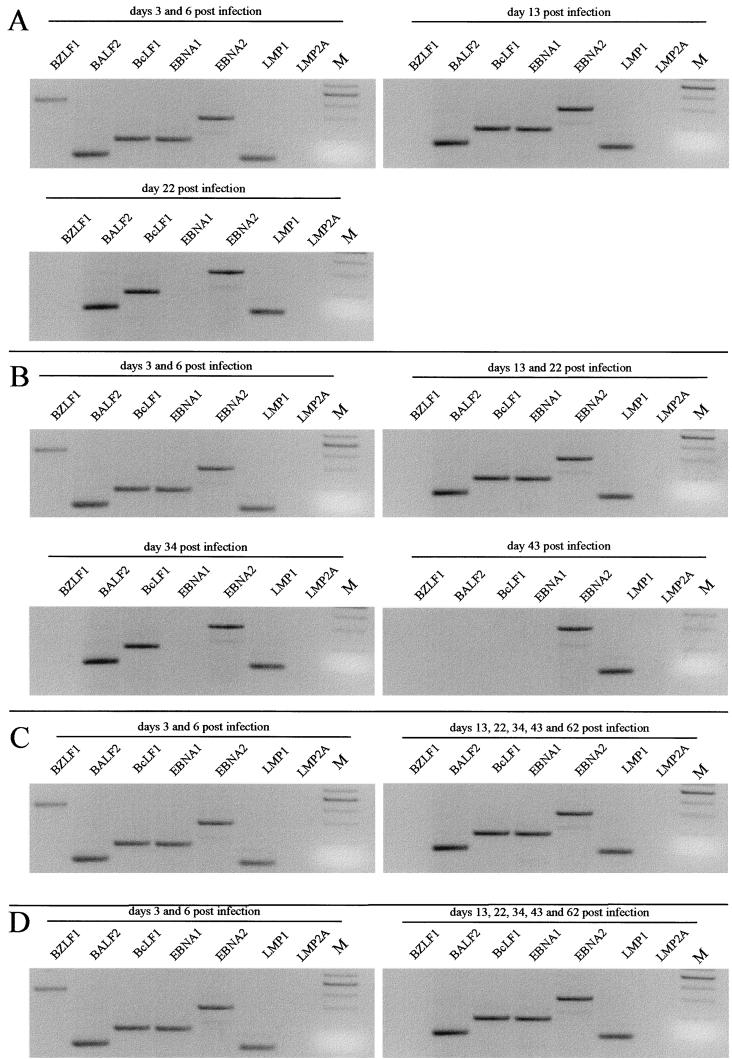
FIG. 5.
PCR analysis of BALF2 and BcLF1 mRNAs and spliced BZLF1, EBNA1, EBNA2, LMP1, and LMP2A mRNAs in T98WT (A), T98CR2 (B), CB193WT (C), and CB193CR2 (D) cell lines at different time points after infection. After nested PCR with specific primers the PCR products were detected by staining with ethidium bromide (inverted picture). The sizes expected are 442 bp for BZLF1, 117 bp for BALF2, 181 bp for BcLF1, 172 bp for EBNA1, 300 bp for EBNA2, and 280 bp for LMP2A (see Materials and Methods for details). M, 100-bp ladder with the prominent band of 500 bp.
TABLE 3.
Summary of EBV transcripts detected by RT-PCR in astrocyte lines at different time points
| Cell line | Gene | Expression at day p.i.a
|
||||||
|---|---|---|---|---|---|---|---|---|
| 3 | 6 | 13 | 22 | 34 | 43 | 62 | ||
| T98WT | BZLF1 | + | + | − | − | − | − | ND |
| BALF2 | + | + | + | + | − | − | ND | |
| BcLF1 | + | + | + | + | − | − | ND | |
| EBER1 | + | + | + | + | − | − | ND | |
| EBNA1 | + | + | + | − | − | − | ND | |
| EBNA2 | + | + | + | + | − | − | ND | |
| LMP1 | + | + | + | + | − | − | ND | |
| LMP2A | − | − | − | − | ND | ND | ND | |
| T98CR2 | BZLF1 | + | + | − | − | − | − | − |
| BALF2 | + | + | + | + | + | − | − | |
| BcLF1 | + | + | + | + | + | − | − | |
| EBER1 | + | + | + | + | + | + | − | |
| EBNA1 | + | + | + | + | − | − | − | |
| EBNA2 | + | + | + | + | + | + | − | |
| LMP1 | + | + | + | + | + | + | − | |
| LMP2A | − | − | − | − | ND | ND | ND | |
| CB193WT | BZLF1 | + | + | − | − | − | − | − |
| BALF2 | + | + | + | + | + | + | + | |
| BcLF1 | + | + | + | + | + | + | + | |
| EBER1 | + | + | + | + | + | + | + | |
| EBNA1 | + | + | + | + | + | + | + | |
| EBNA2 | + | + | + | + | + | + | + | |
| LMP1 | + | + | + | + | + | + | + | |
| LMP2A | − | − | − | − | ND | ND | ND | |
| CB193CR2 | BZLF1 | + | + | − | − | − | − | − |
| BALF2 | + | + | + | + | + | + | + | |
| BcLF1 | + | + | + | + | + | + | + | |
| EBER1 | + | + | + | + | + | + | + | |
| EBNA1 | + | + | + | + | + | + | + | |
| EBNA2 | + | + | + | + | + | + | + | |
| LMP1 | + | + | + | + | + | + | + | |
| LMP2A | − | − | − | − | ND | ND | ND | |
+, expression; −, no expression; ND, not determined.
Since we could not find any data about astrocyte infection, we have tested several genes implicated in different steps of the viral cycle in B cells. The specificity of the various oligonucleotide-primer combinations for individual EBV transcripts has been already documented and confirmed in a series of RT-PCR assays carried out with RNA from cell line B95-8. EBNA1, EBNA2, LMP1, LMP2A, and BZLF1 oligonucleotides specifically amplify cDNA obtained from spliced mRNA. With BALF2 and BcLF1 oligonucleotides it is not possible to distinguish between cDNA and genomic DNA. Therefore, the pretreatment of RNA preparations with DNase I was obligatory. For all the PCR amplifications presented here RNA preparations were pretreated with DNase I. The amplification specificity was assessed by DNase I and RNase A pretreatment.
As shown in Fig. 5 and Table 3, we observed the transient expression of BZLF1 mRNA in all four cell lines at days 3 and 6 p.i., but the signal was lost at the later time points tested. The finding that BZLF1 mRNA is expressed suggested that a productive phase of the viral life cycle could occur within the infected astrocyte lines. To test this hypothesis, we examined mRNA expression of the early gene BALF2 and the late gene BcLF1. Interestingly, we observed BALF2 mRNA not only at the same time points as BZLF1 mRNA but even later when BZLF1 mRNA were not detected any longer. BALF2 mRNA expression was not detected at day 43 for T98CR2, although the cells were still infected as other EBV transcripts were present at this time. We then examined the mRNA expression of the major capsid protein gene BcLF1 as a late-expressed gene. The same expression profile as for BALF2 mRNA was obtained.
To investigate mRNA expression of some genes expressed in latently infected cells, we targeted the small noncoding EBER1 RNA for amplification, since this is an abundant EBV latent transcript common to all known forms of latency. The results obtained by hybridization with a 32P-labeled probe are reported in Table 3. EBER1 RNAs were expressed until day 22 p.i. for T98WT, until day 43 for T98CR2, and at all the time points tested for CB193WT and CR2-transfected cell lines.
EBNA1 mRNA is also a common product of all forms of latency. Two forms of EBNA1 mRNA with different splice structures have been described: BamHI Q/U/K splice structure, expressed in particular in Burkitt’s lymphoma cell lines and nasopharyngeal carcinoma cells, and BamHI Y3/U/K splice structure, characteristic of lymphoblastoid cell lines (LCLs) (7). The primers were chosen in the exons U and K to examine EBNA1 mRNA expression in astrocyte lines. As illustrated in Fig. 5 and Table 3, EBNA1 mRNA expression was observed in CB193WT and CB193CR2 at all the time points. The expression pattern was different for T98WT and T98CR2. Indeed, EBNA1 mRNA expression was detected at the early time points but was lost at day 22 for T98WT and at day 34 for T98CR2, although the cells were still infected as evaluated by EBNA2 and LMP1 mRNA expression (Table 3).
EBNA2 mRNA expression is restricted at the latency type III. Its functions are important in infected B cells. EBNA2 mRNA expression was monitored with primers situated in different exons (one primer covers an exon boundary). The 300-bp product was easily detected by nested PCR until day 22 for T98WT and until day 43 for T98CR2. The same expression profile for EBNA1 was observed for the CB193 cell line (Fig. 5 and Table 3).
For LMP1 mRNA expression, the signal observed was weak after the first amplification (data not shown), and so we decided to improve the sensitivity of detection by using a second round of amplification with a pair of nested primers located within the first and second exons, leading to a product of 104 bp for RNA amplification in contrast to 182 bp for EBV genomic DNA. After the second round of amplification, the expected 104-bp product could be easily detected on an agarose gel (Fig. 5). LMP1 mRNA expression was observed at all time points for CB193WT and CB193CR2 and lost at day 34 for T98WT and day 62 for T98CR2 (Table 3). As LMP1 mRNAs are very abundant in LCLs (more than EBNA2 mRNA), immunofluorescence experiments were done to observe LMP1 protein. Indirect immunostaining was performed by using the CS1-4 MAb, and strongly fluorescent patches could be observed in the persistently infected cell line CB193 (Fig. 6) in some cells. EBNA2 protein was detected by FACS analysis with MAb PE2. Because the staining level was very low for B95-8 cell line, we decided to amplify specific staining by biotin labeling the MAb and then revealed it with phycoerythrin-conjugated streptavidin. Nonspecific staining was abolished by preincubating cells with rabbit IgG. Cell staining was evaluated by the measurement of mean fluorescence intensity (MFI) (Table 4). The persistently infected cell line CB193CR2 shows an MFI three times higher than that of the uninfected CB193CR2 cell line.
FIG. 6.
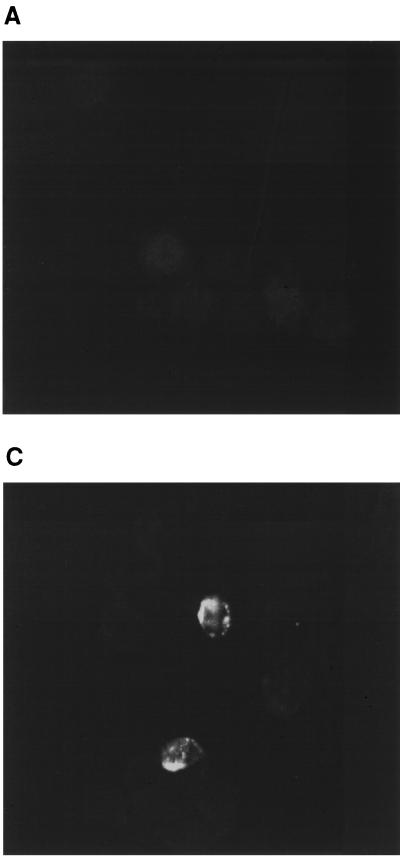
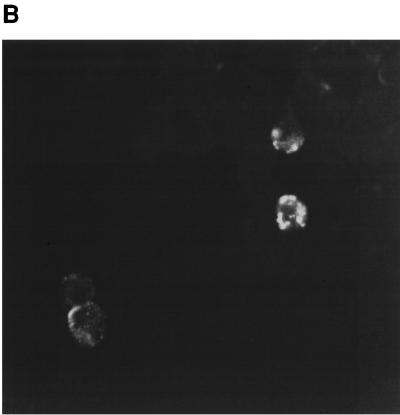
Detection of LMP1 by immunofluorescence. EBV-infected CB193CR2 cells were fixed in 4% paraformaldehyde for 20 min and incubated for 60 min at room temperature with MAb CS1-4, followed by goat anti-mouse IgG. (A) LMP1 staining of uninfected cells. (B) Staining of cells after infection with EBV prepared from B95-8 cell line. (C) Staining of cells 20 days after infection with γ-irradiated B95-8 cells (magnification, ×63).
TABLE 4.
Flow cytometric analysis results for EBNA2 and BZLF1 expression in cell lines at day 60 p.i.a
| Cell line | MFI for:
|
|
|---|---|---|
| EBNA2 | BZLF1 | |
| B95-8 | 21.53 | 19.79 |
| CB193CR2 | 3.12 | 2.89 |
| CB193CR2-EBV | 10.99 | 12.73 |
Cell lines were stimulated for 3 days with TPA before BZLF1 detection (see Materials and Methods).
Unlike the situation in LCLs, no amplification of the 280-bp product indicating LMP2A mRNA was detected for all the cell lines after nested PCR at each time point. We tried to enhance the sensitivity of detection with a Southern blot hybridization of PCR fragments, but no positive result was obtained.
Taken together these results show that the infection of the T98 cell line is abortive and that EBV seems to be lost subsequently to the loss of EBNA1 after approximately 1 month, while CB193 is persistently infected.
Furthermore, it was possible to reinduce BZLF1 mRNA expression in the CB193 cell line 70 days p.i. by the addition of TPA. Indeed, EBV lytic replication in LCLs can be induced by different means. Treatment with phorbol esters efficiently induces lytic replication (17, 18, 51). As shown in Fig. 7, EBV-infected CB193 cells responded well to TPA induction and spontaneously expressed BZLF1 mRNA at 3 and 6 days poststimulation. BZLF1 protein could be detected at day 3 poststimulation (Table 4). The staining level is very low, but the system biotin-conjugated MAb BZ1 allows specific labeling.
FIG. 7.
Expression of BZLF1 mRNA 3 and 6 days after stimulation of CB193CR2 cell line (infected with EBV 70 days before) with TPA (5 × 10−8 M final concentration). RNA was extracted and submitted to PCR amplification with specific primers for BZLF1 mRNAs (the sequences are shown in Table 1). PCR amplification products were detected by staining with ethidium bromide (inverted picture). RT-PCR results from EBV-infected CB193CR2 cells without stimulation (lane 1), after 3 days of stimulation (lane 2), or after 6 days of stimulation (lane 3) are shown. M, molecular weight marker (100-bp ladder).
Another mode of viral infection of CB193 cell lines.
The possibility that EBV infects astrocytes and persists in these cells convinced us to try to infect the cells directly by coculture with B95-8 cell line to mimic the in vivo situation. We exposed the four cell lines to γ-irradiated B95-8 cells to address the possibility of infection of astrocytes by direct contact to EBV harboring peripheral/circulating B cells or by the virus liberated by B-cells. The same number of γ-irradiated B95-8 cells were put for 2 h on astrocytes which were then extensively washed, trypsinized, and centrifuged to remove B cells and cell fragments. In parallel, γ-irradiated B95-8 cells were cultivated in a normal medium to observe the viability of this population. After 15 days of culture it was not possible to observe living irradiated B95-8 cells. EBV infection of astrocytes was assessed by the detection of LMP1 protein by immunofluorescence assay 20 days p.i. As shown in Fig. 6, it was possible to observe EBV infection in CB193 cells. For T98 cell lines, the LMP1 staining was too weak to assess the EBV infection of these cells. The clear size difference between astrocytes and B cells abolished the possibility that the staining obtained was due to B cells that were still alive.
DISCUSSION
EBV DNA was detected by PCR in brain biopsy samples and in the CSF obtained from patients in whom EBV was suspected to play a role in neurological syndromes. So, it could make contact with cells expressing its receptor. CR2 is the major cellular receptor known for EBV (1, 39), although recently another one was described for gastric carcinoma cells (58). Gasque et al. have examined the distribution of CR2 in different types of brain resident cells and found a low-level expression of CR2 in different astrocyte lines and human fetal astrocytes (21). Even if CR2 expression remains at a low level, the physiologic role for astrocyte CR2 has not been defined yet. In accordance with the B-cell system, CR2 expressed on astrocytes could be involved in the EBV infection of this cell type. Here, we demonstrated that (i) two different astrocyte cell lines, T98 and CB193, could be infected by EBV, (ii) virus entry into the cells was mediated by CR2, (iii) EBV infects CR2-transfected T98 and CB193 cells at a higher level, and (iv) the T98 cell line is transiently infected and CB193 is persistently infected by EBV.
The program of EBV gene expression varies, depending on the lineage of host cells and the time after infection, and we wanted to characterize it in astrocytes. In epithelial cells, the lytic cycle is frequently induced, leading to the production of progeny virus. The infection of B lymphocytes with EBV establishes a predominantly latent infection.
Data on the lytic replication of EBV in B cells have been obtained predominantly after reactivation with chemical treatment or by superinfection of the cells (45). Lytic DNA replication proceeds from the origin oriLyt and results in a 100- to 1,000-fold amplification of the EBV genome via concatemeric intermediates. The activation of the replicative cycle of EBV starts with the expression of the immediate-early transactivators Zta, Rta, and Mta encoded by the BZLF1, BRLF1, and BMLF1 genes, respectively. Transactivators cooperate in the activation of further early genes and finally in the expression of late viral proteins (32). EBV encodes a large number of proteins involved in viral DNA synthesis. Fixman et al. demonstrated that six proteins are essential for viral replication, including BALF2, which encodes a single-stranded DNA binding protein (16). The transcription of BcLF1, which encodes the major capsid protein of EBV, permitted evaluation of the possible production of virus. The transcription pattern obtained at the early time points (days 3 and 6 p.i.), and not later, suggests that the lytic cycle in astrocytes starts with replication of the viral genome, expression of late proteins (structural proteins of the virus), and probably also with virus production. The key role of the BZLF1 product, Zta, is maintenance of the balance between latent and lytic replication (41, 45). The BZLF1 promoter region was reported to contain several binding motifs for positive and negative regulatory factors (18, 35, 49). Thus, we suggest that transcriptional control of Zta in astrocytes occurs and furthermore, similarly to the reactivation of the BZLF1 gene in B cells, that transcriptional activation may be achieved by using phorbol esters (17, 18, 51). The coexpression of proteins observed in the latency of EBV in B cells at these time points was referred to in the study of Alfieri et al., who have shown expression of EBNA2, EBNA1, and LMP1 in the early times after infection of B lymphocytes (2).
The latent transcription pattern detected in astrocytes was similar to latency type III in LCLs (32) with the simultaneous expression of EBNA1, EBNA2, LMP1, and EBER1, except for the lack of LMP2A mRNA in our cell lines. Of note, the expression of all the latent genes observed in LCLs was not tested in this study, and this issue is currently under investigation. It is known that in LCLs different EBNAs are encoded by individual mRNAs generated by differential splicing of the same long primary transcripts expressed from either the BamHI C promoter (Cp) or the BamHI W promoter (Wp) (32), while EBNA1 expressed in Burkitt lymphoma cells results from a promoter localized in the BamHI F/Q region of the viral genome (61). The coexpression of EBNA1 and EBNA2 suggests that these transcripts are driven from Cp or Wp promoter in astrocytes.
In latently infected B cells, multiple copies of the viral genome are maintained predominantly as episomes that are replicated in synchrony with cellular DNA. Latent replication proceeds from ori P. EBNA1 is the only virus-encoded protein required for replication of the episomal EBV genome, whereas all other proteins are provided by the host cell (44). We observed the loss of EBNA1 mRNA expression in T98 cell lines and subsequently the loss of the virus. The major function of EBNA1 protein is the maintenance of the EBV genome in an episomal state in latently infected cells (32), and therefore we propose that ori P is active in these cells and that the loss of EBNA1 expression leads to the loss of all traces of virus in the cells. So, we can qualify the infection of T98 cells as a transient event. For CR2-transfected T98CR2, the disappearance of infection occurred later, suggesting a greater number of infected cells in the first phase of infection. In contrast, we could observe EBV mRNA expression 9 weeks p.i. in CB193 astrocyte lines, indicating that the viral genome is stable in these cells. The viral genome is transcribed, and so the infection is active and persistent. T98 and CB193 cell lines exhibit different levels of differentiation, and of these two cell lines CB193 shares more features with native astrocytes, such as cell surface markers like CR1, and morphological characteristics which render this cell line closer to the in vivo situation.
The EBNA2 protein expression level is low in the EBV-infected CB193CR2 cell line. We used for this experiment the supernatant from ascites fluid and not purified MAb; furthermore, the study of Brink et al. revealed that PE2 MAb is not very sensitive (6). These two reasons could explain why the labeling level for the B95-8 cell line, as well as for CB193CR2 cells persistently infected by EBV, is so low. EBNA2 protein expression by EBV-infected astrocyte lines could dysregulate cellular metabolism as in B cells (32). Furthermore, studies have shown that EBV, like other herpesviruses, can activate the HIV-1 long terminal repeat and in particular EBNA2 transactivates the HIV-1 long terminal repeat and enhances HIV-1 replication in T cells (62). On the contrary, HIV-1 expression seems to be downregulated in partially purified B cells coinfected with EBV in vitro (27). This difference in HIV-1 expression by the EBV-infected cells could depend on the permissiveness of the cells for EBV replication: EBNA2 may downregulate HIV-1 expression in B cells permissive for EBV replication and upregulate HIV-1 expression in T cells which are nonpermissive for EBV replication (62). Until now, there has been no report about a possible interaction between EBV and HIV-infected astrocytes found in the brains of AIDS patients suffering from EBV DNA-positive primary cerebral lymphoma.
Interestingly the high-level expression of LMP1 protein in infected astrocyte lines was observed by immunofluorescence assay. Recent reports, focused on the essential role of this protein (10, 31), have shown that LMP1 plays a central role in the transformation process of human B lymphocytes in vitro. It can mimic members of the family of tumor necrosis factor receptors and thereby transmit growth signals from the cell membrane to the nucleus through cytoplasmic tumor necrosis factor receptor-associated factors (TRAFs). LMP1-bound proteins activate NF-κB and thereby cause cell proliferation. Liebowitz et al. showed the interaction between the EBV-transforming protein LMP1 and the TRAF-1 and TRAF-3 signal transduction molecules in some lymphomas, in addition to the activation of NF-κB in these tumors (31). The high-level expression of LMP1 by infected astrocytes could induce cellular changes that may be of interest for the interaction of EBV with normal astrocytes in vivo.
An uncommon feature of EBV gene expression in astrocytes is the constitutive expression of early genes (virus replicative) and late genes (BALF2 and BcLF1, respectively) in the absence of the transactivator Zta. Since Zta is essential for the lytic cycle in B cells and this gene is not present in any of our cell lines, we cannot explain this observation by the coexistence of one population latently infected with a transcription pattern close to the LCL pattern and another infected lytically (15, 41, 47, 55). Recently, Nakamura et al. (37) demonstrated that EBV-infected MT2 clones express proteins associated with the latent viral cycle (including EBNA1 and LMP1) and that constitutive transcription from early replicative and late genes occurs. In their model the BZLF1 protein was observed in most of the EBV-infected MT2 clones and could activate transcription of those genes. In contrast the same experiment carried out on the human B-cell lines BJAB and Louckes did not result in the expression of early genes. We assume that persistently infected astrocytes, like MT2 clones, express at the same time some latent, early, and late genes. In this case the transcriptional control of early genes would be independent of Zta, whereas another transcription transactivator may be involved in this process. In accordance with the results from the study of Bogedain et al. we propose that in astrocytes, as in LCLs, where the influence of Zta expression is less efficient than in cells derived from Burkitt lymphomas, Rta could play an important transactivator role (4).
In conclusion the investigations presented provide evidence that two astrocyte lines tested express functional CR2, the major EBV receptor. The virus binds specifically to these cells, infects them, and induces the expression of several EBV genes. The presence of some EBV proteins (LMP1, EBNA2, and BZLF1) in some infected cells demonstrates that the virus is expressed and suggests that astrocyte metabolism is modified after infection. The lytic cycle seems to take place in the early stages p.i., and later a switch to a latent pattern occurs with an uncommon transcription profile. The infection of T98 is transient, but the CB193 cell line is persistently infected. To facilitate further studies, we used two CR2 transfectant astrocyte lines. Although the presence of the transfected plasmid in CB193CR2 and T98CR2 could affect the transcription of the EBV genome, we have demonstrated that the transcriptional pattern in the transfectant cell lines was similar to that in parental cell lines. Thus, our studies validate the use of our transfectant model to obtain a more efficient infection of astrocytes with EBV. Moreover, we were able to infect these cell lines with cell-free virus and also directly via γ-irradiated B95-8 cells. Thus, this study contributes to a better understanding of the interaction between EBV and astrocytes and thereby of the pathogenesis of EBV infection in vivo.
ACKNOWLEDGMENTS
We thank Laco Kacani for critically discussing the manuscript and Marie T. Schouft and Heidi Recheis for excellent technical assistance. We are grateful to Fritz Schwarzmann and Hans Wolf, University of Regensburg, for helpful advice during the progress of this work.
This work was supported in part by grants from the Austrian Science Fund F202 and by the Biomed 2 Programme of the European Commission BMH4-CT96-1005, travel grant Amadee (Action Integrees). Anne Menet is supported by a grant from the French Ministry of Science (contrat 9636).
REFERENCES
- 1.Ahearn J M, Fearon D T. Structure and function of the complement receptors, CR1 (CD35) and CR2 (CD21) Adv Immunol. 1989;46:183–219. doi: 10.1016/s0065-2776(08)60654-9. [DOI] [PubMed] [Google Scholar]
- 2.Alfieri C, Birkenbach M, Kieff E. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology. 1991;181:595–608. doi: 10.1016/0042-6822(91)90893-g. [DOI] [PubMed] [Google Scholar]
- 3.Bagasra O, Lavi E, Bobroski L, Khalili K, Pestaner J P, Tawadros R, Pomerantz R J. Cellular reservoirs of HIV-1 in the central nervous system of infected individuals: identification by the combination of in situ polymerase chain reaction and immunohistochemistry. AIDS. 1996;10:573–585. doi: 10.1097/00002030-199606000-00002. [DOI] [PubMed] [Google Scholar]
- 4.Bogedain C, Alliger P, Schwarzmann F, Marschall M, Wolf H, Jilg W. Different activation of Epstein-Barr virus immediate-early and early genes in Burkitt lymphoma cells and lymphoblastoid cell lines. J Virol. 1994;68:1200–1203. doi: 10.1128/jvi.68.2.1200-1203.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bray P F, Culp K W, McFarlin D E, Panitch H S, Torkelson R D, Schlight J P. Demyelinating disease after neurologically complicated primary Epstein-Barr virus infection. Neurology. 1992;42:278–282. doi: 10.1212/wnl.42.2.278. [DOI] [PubMed] [Google Scholar]
- 6.Brink A A, Dukers D F, Van der Brule A J, Oudejans J J, Middeldorp J M, Miejer C J, Jiwa M. Presence of Epstein-Barr virus latency type III at the single cell level in post-transplantation lymphoproliferative disorders and AIDS related lymphomas. J Clin Pathol. 1997;50:911–918. doi: 10.1136/jcp.50.11.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brooks L, Yao Q Y, Rickinson A B, Young L S. Epstein-Barr virus latent gene transcription in nasopharyngeal carcinoma cells: coexpression of EBNA1, LMP1, and LMP2 transcripts. J Virol. 1992;66:2689–2697. doi: 10.1128/jvi.66.5.2689-2697.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carel J C, Frazier B, Ley T J, Holers V M. Analysis of epitope expression and the functional repertoire of recombinant complement receptor 2 (CR2/CD21) in mouse and human cells. J Immunol. 1989;143:923–930. [PubMed] [Google Scholar]
- 9.Carel J C, Myones B L, Frazier B, Holers V M. Structural requirements for C3d,g/Epstein-Barr virus receptor (CR2/CD21) ligand binding, internalization, and viral infection. J Biol Chem. 1990;265:12293–12299. [PubMed] [Google Scholar]
- 10.Cheung S T, Leung S F, Lo K W, Chiu K W, Tam J S, Fok T F, Johnson P J, Lee J C, Huang D P. Specific latent membrane protein 1 gene sequences in type 1 and type 2 Epstein-Barr virus from nasopharyngeal carcinoma in Hong Kong. Int J Cancer. 1998;76:399–406. doi: 10.1002/(sici)1097-0215(19980504)76:3<399::aid-ijc18>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 11.Cinque P, Brytting M, Vago L, Castagna A, Parravicini C, Zanchetta N, D’Arminio Monforte A, Wahren B, Lazzarin A, Linde A. Epstein-Barr virus DNA in cerebrospinal fluid from patients with AIDS-related primary lymphoma of the central nervous system. Lancet. 1993;342:398–401. doi: 10.1016/0140-6736(93)92814-a. [DOI] [PubMed] [Google Scholar]
- 12.Cleary T G, Henle W, Pickering L K. Acute cerebellar ataxia associated with Epstein-Barr virus infection. JAMA. 1980;243:148–149. [PubMed] [Google Scholar]
- 13.D’Arminio Monforte A, Cinque P, Vago L, Rocca A, Castagna A, Gervasoni C, Terreni M R, Novati R, Gori A, Lazzarin A, et al. A comparison of brain biopsy and CSF-PCR in the diagnosis of CNS lesions in AIDS patients. J Neurol. 1997;244:35–39. doi: 10.1007/pl00007727. [DOI] [PubMed] [Google Scholar]
- 14.Di Rienzo A M, Aloisi F, Santarcangelo C, Palladino C, Olivetta E, Genovese D, Verani P, Levi G. Virological and molecular parameters of HIV-1 infection of human embryonic astrocytes. Arch Virol. 1998;143:1599–1615. doi: 10.1007/s007050050401. [DOI] [PubMed] [Google Scholar]
- 15.Farrell P J, Rowe D T, Rooney C M, Kouzarides T. Epstein-Barr virus BZLF1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J. 1989;8:127–132. doi: 10.1002/j.1460-2075.1989.tb03356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fixman E D, Hayward G S, Hayward S D. trans-acting requirements for replication of Epstein-Barr virus ori-Lyt. J Virol. 1992;66:5030–5039. doi: 10.1128/jvi.66.8.5030-5039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flemington E, Speck S H. Autoregulation of Epstein-Barr virus putative lytic switch gene BZLF1. J Virol. 1990;64:1227–1232. doi: 10.1128/jvi.64.3.1227-1232.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flemington E, Speck S H. Identification of phorbol ester response elements in the promoter of Epstein-Barr virus putative lytic switch gene BZLF1. J Virol. 1990;64:1217–1226. doi: 10.1128/jvi.64.3.1217-1226.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujisaku A, Harley J B, Frank M B, Gruner A, Frazier B, Holers V M. Genomic organization and polymorphisms of the human C3d/Epstein-Barr virus receptor. J Biol Chem. 1989;264:2118–2125. [PubMed] [Google Scholar]
- 20.Gaidano G, Carbone A, Dalla Favera R. Pathogenesis of AIDS-related lymphomas: molecular and histogenetic heterogeneity. Am J Pathol. 1998;152:623–630. [PMC free article] [PubMed] [Google Scholar]
- 21.Gasque P, Chan P, Mauger C, Schouft M T, Singhrao S, Dierich M P, Morgan B P, Fontaine M. Identification and characterization of complement C3 receptors on human astrocytes. J Immunol. 1996;156:2247–2255. [PubMed] [Google Scholar]
- 22.Geddes J F, Bhattacharjee M B, Savage K, Scaravilli F, McLaughlin J E. Primary cerebral lymphoma: a study of 47 cases probed for Epstein-Barr virus genome. J Clin Pathol. 1992;45:587–590. doi: 10.1136/jcp.45.7.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilbert J W, Culebras A. Cerebellitis in infectious mononucleosis. JAMA. 1972;220:727–735. [PubMed] [Google Scholar]
- 24.Griffiths P D. Studies of viral co-factors for human immunodeficiency virus in vitro and in vivo. J Gen Virol. 1998;79:213–220. doi: 10.1099/0022-1317-79-2-213. [DOI] [PubMed] [Google Scholar]
- 25.Grose C, Henle W, Henle G, Feorino P M. Primary Epstein-Barr virus infections in acute neurologic diseases. N Engl J Med. 1975;292:392–395. doi: 10.1056/NEJM197502202920804. [DOI] [PubMed] [Google Scholar]
- 26.Hamilton Dutoit S J, Raphael M, Audouin J, Diebold J, Lisse I, Pedersen E, Oksenhendler E, Marelle L, Pallesen G. In situ demonstration of Epstein-Barr virus small RNAs (EBER 1) in acquired immunodeficiency syndrome-related lymphomas: correlation with tumor morphology and primary site. Blood. 1993;82:619–624. [PubMed] [Google Scholar]
- 27.Henderson E E, Yang J Y, Zhang R D, Bealer M. Altered HIV expression and EBV-induced transformation in coinfected PBLs and PBL subpopulations. Virology. 1991;182:186–198. doi: 10.1016/0042-6822(91)90662-u. [DOI] [PubMed] [Google Scholar]
- 28.Jäger M, Prang N, Mitterer M, Larcher C, Huemer H P, Reischl U, Wolf H, Schwarzmann F. Pathogenesis of chronic Epstein-Barr virus infection: detection of a virus strain with a high rate of lytic replication. Br J Haematol. 1996;95:626–636. doi: 10.1046/j.1365-2141.1996.d01-1969.x. [DOI] [PubMed] [Google Scholar]
- 29.Kleinschmidt A, Neumann M, Moller C, Erfle V, Brack Werner R. Restricted expression of HIV1 in human astrocytes: molecular basis for viral persistence in the CNS. Res Virol. 1994;145:147–153. doi: 10.1016/s0923-2516(07)80016-1. [DOI] [PubMed] [Google Scholar]
- 30.Li Q X, Young L S, Niedobitek G, Dawson C W, Birkenbach M, Wang F, Rickinson A B. Epstein-Barr virus infection and replication in a human epithelial cell system. Nature. 1992;356:347–350. doi: 10.1038/356347a0. [DOI] [PubMed] [Google Scholar]
- 31.Liebowitz D. Epstein-Barr virus and a cellular signaling pathway in lymphomas from immunosuppressed patients. N Engl J Med. 1998;338:1413–1421. doi: 10.1056/NEJM199805143382003. . (Comment, 338:1461–1463.) [DOI] [PubMed] [Google Scholar]
- 32.Longnecker R. Molecular biology of Epstein-Barr virus. In: McCance D J, editor. Human tumor viruses. Washington, D.C: American Society for Microbiology; 1998. pp. 135–174. [Google Scholar]
- 33.Martin D R, Marlowe R L, Ahearn J M. Determination of the role for CD21 during Epstein-Barr virus infection of B-lymphoblastoid cells. J Virol. 1994;68:4716–4726. doi: 10.1128/jvi.68.8.4716-4726.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Menet, A., P. Chan, M. Schwendinger, W. M. Prodinger, M. P. Dierich, and M. Fontaine. 1999. Unpublished data. [DOI] [PMC free article] [PubMed]
- 35.Montalvo E A, Shi Y, Shenk T E, Levine A J. Negative regulation of the BZLF1 promoter of Epstein-Barr virus. J Virol. 1991;65:3647–3655. doi: 10.1128/jvi.65.7.3647-3655.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murphy S. Astrocytes, pharmacology and function. San Diego, Calif: Academic Press; 1993. [Google Scholar]
- 37.Nakamura H, Iwakiri D, Ono Y, Fujiwara S. Epstein-Barr-virus-infected human T-cell line with a unique pattern of viral-gene expression. Int J Cancer. 1998;76:587–594. doi: 10.1002/(sici)1097-0215(19980518)76:4<587::aid-ijc23>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 38.Nath A, Power C, Geiger J D. Interactions of the human immunodeficiency with astrocytes. Perspect Drug Discov. 1996;5:30–42. [Google Scholar]
- 39.Nemerow G R, Wolfert R, McNaughton M E, Cooper N R. Identification and characterization of the Epstein-Barr virus receptor on human B lymphocytes and its relationship to the C3d complement receptor (CR2) J Virol. 1985;55:347–351. doi: 10.1128/jvi.55.2.347-351.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pedneault L, Katz B Z, Miller G. Detection of Epstein-Barr virus in the brain by the polymerase chain reaction. Ann Neurol. 1992;32:184–192. doi: 10.1002/ana.410320210. [DOI] [PubMed] [Google Scholar]
- 41.Prang N, Wolf H, Schwarzmann F. Epstein-Barr virus lytic replication is controlled by posttranscriptional negative regulation of BZLF1. J Virol. 1995;69:2644–2648. doi: 10.1128/jvi.69.4.2644-2648.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prodinger W M, Larcher C, Schwendinger M, Dierich M P. Ligation of the functional domain of complement receptor type 2 (CR2, CD21) is relevant for complex formation in T cell lines. J Immunol. 1996;156:2580–2584. [PubMed] [Google Scholar]
- 43.Prodinger W M, Schwendinger M, Schoch J, Köchle M, Larcher C, Dierich M P. Characterization of the C3dg binding to a recess formed between short consensus repeats 1 and 2 of complement receptor type 2 (CR2, CD21) J Immunol. 1998;161:4604–4610. [PubMed] [Google Scholar]
- 44.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Raven Press; 1996. pp. 2397–2446. [Google Scholar]
- 45.Roizman B. Herpesviridae. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Raven Press; 1996. pp. 2221–2230. [Google Scholar]
- 46.Rowe M, Evans H S, Young L S, Hennessy K, Kieff E, Rickinson A B. Monoclonal antibodies to the latent membrane protein of Epstein-Barr virus reveal heterogeneity of the protein and inducible expression in virus-transformed cells. J Gen Virol. 1987;68:1575–1586. doi: 10.1099/0022-1317-68-6-1575. [DOI] [PubMed] [Google Scholar]
- 47.Sarisky R T, Gao Z, Lieberman P M, Fixman E D, Hayward G S, Hayward S D. A replication function associated with the activation domain of the Epstein-Barr virus Zta transactivator. J Virol. 1996;70:8340–8347. doi: 10.1128/jvi.70.12.8340-8347.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schooley R T, Dolin R. Epstein-Barr virus (infectious mononucleosis) In: Mandell G L, Douglas R G J, Bennett J E, editors. Principles and practice of infectious diseases. New York, N.Y: Churchill-Livingstone; 1990. pp. 1172–1185. [Google Scholar]
- 49.Schwarzmann F, Prang N, Reichelt B, Rinkes B, Haist S, Marschall M, Wolf H. Negatively cis-acting elements in the distal part of the promoter of Epstein-Barr virus trans-activator gene BZLF1. J Gen Virol. 1994;75:1999–2006. doi: 10.1099/0022-1317-75-8-1999. [DOI] [PubMed] [Google Scholar]
- 50.Silverstein A, Steinberg G, Nathanson M. Nervous system involvement in infectious mononucleosis. Arch Neurol. 1972;26:353–358. doi: 10.1001/archneur.1972.00490100083009. [DOI] [PubMed] [Google Scholar]
- 51.Speck S H, Chatila T, Flemington E. Reactivation of Epstein-Barr virus: regulation and function of the BZLF1 gene. Trends Microbiol. 1997;5:399–405. doi: 10.1016/S0966-842X(97)01129-3. [DOI] [PubMed] [Google Scholar]
- 52.Tedder T F, Clement L T, Cooper M D. Expression of C3d receptors during human B cell differentiation: immunofluorescence analysis with the HB-5 monoclonal antibody. J Immunol. 1984;133:678–683. [PubMed] [Google Scholar]
- 53.Tierney R J, Steven N, Young L S, Rickinson A B. Epstein-Barr virus latency in blood mononuclear cells: analysis of viral gene transcription during primary infection and in the carrier state. J Virol. 1994;68:7374–7385. doi: 10.1128/jvi.68.11.7374-7385.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tornatore C, Meyers K, Atwood W, Conant K, Major E. Temporal patterns of human immunodeficiency virus type 1 transcripts in human fetal astrocytes. J Virol. 1994;68:93–102. doi: 10.1128/jvi.68.1.93-102.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Urier G, Buisson M, Chambard P, Sergeant A. The Epstein-Barr virus early protein EB1 activates transcription from different responsive elements including AP-1 binding sites. EMBO J. 1989;8:1447–1453. doi: 10.1002/j.1460-2075.1989.tb03527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walter G F, Renella R R. Epstein-Barr virus in brain and Rasmussen’s encephalitis. Lancet. 1989;i:279–280. doi: 10.1016/s0140-6736(89)91292-0. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 57.Warner H B, Carp R I. Multiple sclerosis etiology—an Epstein-Barr virus hypothesis. Med Hypotheses. 1988;25:93–97. doi: 10.1016/0306-9877(88)90024-2. [DOI] [PubMed] [Google Scholar]
- 58.Yoshiyama H, Imai S, Shimizu N, Takada K. Epstein-Barr virus infection of human gastric carcinoma cells: implication of the existence of a new virus receptor different from CD21. J Virol. 1997;71:5688–5691. doi: 10.1128/jvi.71.7.5688-5691.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Young L S, Alfieri C, Hennessy K, Evans H S, O’Hara C, Anderson K C. Expression of Epstein-Barr virus transformation-associated genes in tissues of patients with EBV lymphoproliferative disease. N Engl J Med. 1989;321:1080–1085. doi: 10.1056/NEJM198910193211604. [DOI] [PubMed] [Google Scholar]
- 60.Young L S, Lau R, Rowe M, Niedobitek G, Packham G, Shanahan F. Differentiation-associated expression of the Epstein-Barr virus BZLF1 transactivator protein in oral hairy leukoplakia. J Virol. 1991;65:2868–2874. doi: 10.1128/jvi.65.6.2868-2874.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Young L S, Rowe M. Epstein-Barr virus, lymphomas and Hodgkin’s disease. Semin Cancer Biol. 1992;3:273–284. [PubMed] [Google Scholar]
- 62.Zhang R D, Guan M, Park Y, Tawadros R, Yang J Y, Gold B, Wu B, Henderson E E. Synergy between human immunodeficiency virus type 1 and Epstein-Barr virus in T lymphoblastoid cell lines. AIDS Res Hum Retroviruses. 1997;13:161–171. doi: 10.1089/aid.1997.13.161. [DOI] [PubMed] [Google Scholar]