Abstract
The mouse hepatitis virus (MHV) spike glycoprotein, S, has been implicated as a major determinant of viral pathogenesis. In the absence of a full-length molecular clone, however, it has been difficult to address the role of individual viral genes in pathogenesis. By using targeted RNA recombination to introduce the S gene of MHV4, a highly neurovirulent strain, into the genome of MHV-A59, a mildly neurovirulent strain, we have been able to directly address the role of the S gene in neurovirulence. In cell culture, the recombinants containing the MHV4 S gene, S4R22 and S4R21, exhibited a small-plaque phenotype and replicated to low levels, similar to wild-type MHV4. Intracranial inoculation of C57BL/6 mice with S4R22 and S4R21 revealed a marked alteration in pathogenesis. Relative to wild-type control recombinant viruses (wtR13 and wtR9), containing the MHV-A59 S gene, the MHV4 S gene recombinants exhibited a dramatic increase in virulence and an increase in both viral antigen staining and inflammation in the central nervous system. There was not, however, an increase in the level of viral replication in the brain. These studies demonstrate that the MHV4 S gene alone is sufficient to confer a highly neurovirulent phenotype to a recombinant virus deriving the remainder of its genome from a mildly neurovirulent virus, MHV-A59. This definitively confirms previous findings, suggesting that the spike is a major determinant of pathogenesis.
The Coronaviridae family of viruses is studied widely for its ability to generate diseases in a number of different animal hosts. Mouse hepatitis virus (MHV), a member of this family, provides a useful model system for studying both acute and chronic virus-induced neurologic disease. Intracranial or intranasal inoculation of susceptible mice with neurotropic strains of MHV can result in a range of outcomes from acute encephalomyelitis to chronic demyelinating disease (1, 24). The outcome of infection is determined by a number of factors, including the viral strain, dose, route of inoculation, host strain, age, and immune status (9). The role of the viral strain in determining the severity of the acute disease is well established, but the viral determinants which explain these differences are not known. Two strains which differ markedly in their neurovirulence are MHV4 and MHV-A59. Infection of weanling mice with low doses of MHV type 4 (MHV4, a strain JHM isolate), a highly neurovirulent strain, produces a severe and often fatal encephalitis (10), while much higher doses of MHV-A59, a mildly neurovirulent strain, are required to produce a mild encephalitis (28, 29).
The lack of an infectious molecular clone of MHV has limited the study of viral determinants of pathogenesis to the comparison of different strains and mutant viruses. From these comparisons, however, it has become evident that the MHV spike (S) glycoprotein plays a key role in pathogenesis. The importance of the S protein in pathogenesis is consistent with its biologic function in both viral entry and viral spread (8, 46). When expressed on the virion envelope, S binds to the cellular receptor and induces the fusion of viral and cell membranes during viral entry. Subsequent to infection, S protein expressed on the plasma membrane of infected cells induces cell-cell fusion. S also plays a role in the immune response to viral infection, as a target for neutralizing antibodies (8) and as an inducer of a cell-mediated immunity (4, 7).
The S glycoprotein, arranged in a homo-oligomeric structure on the virion surface, forms the characteristic spikes protruding from the virion envelope. For most strains, the S protein is cleaved posttranslationally into an amino-terminal (S1) and a carboxy-terminal (S2) subunit (16, 44). S1 is believed to make up the globular head of the spike and has been demonstrated to exhibit a receptor-binding activity (12, 26). The S2 subunit, containing a transmembrane domain and two heptad repeat regions, is believed to form the stalk portion of the spike and to mediate membrane fusion (12, 34).
From the study of numerous variant viruses, selected for resistance to neutralizing monoclonal antibodies, an association has been made between various mutations or deletions in S and neuroattenuation (10, 15, 19, 37, 45). Interestingly, many of these viruses display mutations or deletions in a region of S1 termed the hypervariable region (HVR), or in and around the heptad repeat regions of S2. In general, it is not known how mutations in the HVR or in S2 could affect neurovirulence, but it has been suggested that they may affect fusogenicity, cytotoxicity, viral spread, or receptor binding (13, 17, 40).
Until recently, the technology to introduce specific mutations into the MHV genome was not available, and thus pathogenesis studies had to rely on associations drawn from the comparison of nonisogenic variant viruses. With the development of targeted recombination, the 3′ end of the MHV genome containing the structural genes has been made available for genetic analysis (14, 31). Using this technique, we have generated isogenic recombinant viruses that contain either the S gene of the highly neurovirulent MHV4 or the S gene of the mildly neurovirulent MHV-A59. With these pairs of recombinant viruses we have been able to directly address the role of the S gene in neurovirulence. Recombinant viruses, containing the MHV4 S gene, displayed a dramatic increase in virulence, while control recombinant viruses, containing the wild-type MHV-A59 S gene, displayed a pathogenesis similar to that of wild-type MHV-A59. This increase in virulence was associated with an increase in viral antigen expression and inflammation, but not in the extent of viral replication in the central nervous system (CNS).
MATERIALS AND METHODS
Virus and cells.
MHV-A59 was obtained from Lawrence Sturman (Albany, N.Y.). MHV4 was obtained from Michael J. Buchmeier (La Jolla, Calif.). Alb4 (obtained from Paul Masters, Albany, N.Y.) contains an 87-nucleotide deletion (resulting in a 29-amino-acid in-frame deletion) in a nonessential spacer region in the N (nucleocapsid) gene; it produces small plaques at the nonpermissive temperature (39°C) and is thermolabile (25). Murine L2 cells and 17Cl-1 cells were maintained on plastic tissue culture flasks in Dulbecco minimal essential medium (DMEM) with 10% fetal bovine serum (FBS). Spinner cultures of L2 cells were maintained in Joklik minimal essential medium with 10% FBS at densities of between 2 × 105 and 2 × 106 cells per ml.
Plasmids and PCR mutagenesis.
The construction of pMH54 (obtained from Paul Masters) involved the extension of the pFV1 vector (14) to include the 3′ 1.1-kb of the hemagglutinin esterase gene of MHV-A59 (26a) (see Fig. 1). The inclusion of the 3′ portion of the hemagglutinin esterase gene in the pMH54 plasmid allowed us to select for viruses which had recombined 5′ to the S gene, and thus contained the entire recombinant spike gene. The sequence of the S gene in pMH54 is the same as that of wild-type MHV-A59 (23) except for the introduction of AvrII and SbfI restriction sites. The AvrII site was created by introducing two silent mutations in codons 12 and 13 of S. The SbfI site was generated in the intergenic region between gene S and gene 4 by making three nucleotide substitutions at 12, 15, and 17 bp past the S gene termination codon. The AvrII/SbfI sites allowed us to easily introduce different S genes into the background of pMH54 and to identify recombinant viruses containing full-length recombinant spike genes. For RNA transcription, the plasmid was linearized just 3′ to the poly(A) tail by digestion with PacI.
FIG. 1.
Schematic diagram of targeted recombination and the generated recombinant viruses. The synthetic RNA transcribed from the vector pMH54-S4, encoding the MHV4 S gene, or pMH54, encoding the MHV-A59 S gene, and the corresponding 3′ end of the Alb4 genome, derived from MHV-A59, are shown. Viral genes and the sites of the Alb4 deletion are indicated. The curved line between the genome and the synthetic RNA pMH54-S4/pMH54 indicates the region in which a crossover must have occurred. S4R22 and 21 represent the genomes of the MHV4 S gene recombinants, and wtR13 and 9 represent the genomes of the wild-type recombinants. The deletion in the MHV-A59 S gene is indicated. Open bars indicate the MHV-A59 sequence, and closed bars indicate the MHV4 sequence.
The pGEM4Z plasmid containing the MHV4 S gene, pSwt (18), was obtained from Michael J. Buchmeier. To replace the MHV-A59 S gene in pMH54 with the MHV4 S gene, we introduced the AvrII and SbfI restriction sites into the MHV4 S gene. This was done by PCR mutagenesis (22) with Vent polymerase and the primers listed in Table 1. A restriction fragment containing the AvrII site (and a HindIII cloning site) was generated by amplification with primers JJ2 and A59-4. The 1,185-bp PCR fragment was gel purified, digested with restriction enzymes HindIII and PflmI, and then cloned into the corresponding sites in pSwt. This clone was designated pG-MHV4-S1, and the entire 728-bp fragment was sequenced. A restriction fragment containing the SbfI restriction site (and a BamHI cloning site) was also generated by amplification with primers wzl-15 and JJ3. The 875-bp PCR fragment was gel purified, digested with the restriction enzymes BsmI and BamHI, and then cloned into the corresponding sites in pG-MHV4-S1. This clone was designated pG-MHV4-S2, and the entire 670-bp fragment was sequenced. The pG-MHV4-S2 plasmid was digested with restriction enzymes AvrII and SbfI, and then cloned into the corresponding sites in pMH54. This clone was designated pMH54-S4, and sequencing confirmed that no secondary site mutations had been introduced.
TABLE 1.
Primers used for mutagenesis of pSwt and for recombinant genome sequencing
| Primer | Sequence (5′ to 3′)a | MHV-A59 genome location (nucleotide no.)b |
|---|---|---|
| FIJ-79 | GCGAATTATAGTGGTGGCACAC | 944–966 (HE ORF) |
| JJ2 | cgcgaagcttgaatccTAGGGTATA TTGGTGATTTTAGATGTATCC | 36–73 (S gene) |
| RS221 | GTTTATCCGCATAGTACGCAT | 661–641 (S gene) |
| A59-4 | TCAACTTGCCTACTTCGGGG | 1205–1186 (S gene) |
| WZL-15 | AGCAAAAGCCCAGATAGA | 3114–3131 (S gene) |
| JJ3 | gcggatccaagtCCtGcAGgGGCTG TGATAGTCAATCCTCATGAGA | 3994–3961 (S gene) |
Restriction sites introduced into primers are shown in lowercase letters.
Reverse numbering indicates a negative-strand primer. HE ORF, hemagglutin esterase open reading frame.
Targeted RNA recombination and selection of recombinants.
Targeted RNA recombination was carried out between Alb4 and synthetic capped RNAs as described previously (31, 36) except that the template for RNA transcription was pMH54 or pMH54-S4. The synthetic RNAs contained the entire 3′ end of the MHV genome beginning with the 3′ portion of the hemagglutinin esterase gene extending through S, genes 4 and 5, M, and N. For recombination, L2 cells in spinner culture were harvested and infected at a multiplicity of infection of 1 PFU of Alb4 per cell at 33°C. At 2 h postinfection, the cells were washed in phosphate-buffered saline (PBS) without calcium and magnesium and then transfected by electroporation with the synthetic RNA by using two consecutive pulses from a Bio-Rad Gene Pulser apparatus set at 0.3 kV and 960 μF. Infected and transfected cells were plated onto a monolayer of 17Cl-1 cells, and released virus was harvested at approximately 24 h postinfection. The selection of wild-type recombinants, in which the wild-type MHV-A59 S gene has been reintroduced, relied on selection against the thermolabile, temperature-sensitive, small-plaque phenotype of Alb4 (14, 31). Briefly, this selection strategy employed heat treatment for 24 h at 40°C in 50 mM Tris-maleate (pH 6.5)–100 mM EDTA–10% FBS, plating on L2 monolayers at 39°C, and identification of viruses exhibiting a large-plaque morphology (compared to the parental Alb4). This strategy was modified to select MHV4 S gene recombinants after initial attempts to select recombinant viruses by using heat treatment were unsuccessful. Neutralizing monoclonal antibodies, A2.1 and A2.3, specific for the MHV-A59 S protein (obtained from John O. Fleming, Madison, Wis.) were used to select against viruses that did not express the full-length MHV4 S protein. After recombination, putative recombinant viruses were incubated for 1 h at 37°C in the presence of a 1:10 dilution of A2.1 and A2.3 and then plaqued at 39°C on L2 cells. Candidate recombinants were further analyzed. As described previously, we analyzed for repair of the Alb4 deletion by detecting size differences in PCR fragments generated from the N gene (31). The recombinant viruses were then screened for the presence of the 5′ portion of the recombinant MHV4 or MHV-A59 S gene. The screen depends on the presence of the AvrII site mentioned above. A 988-bp fragment surrounding the AvrII site was amplified from the genomes of the recombinant viruses, MHV-A59, and Alb4 by using the primers FIJ-79 and RS-221. Digestion of this fragment with AvrII followed by electrophoresis demonstrated that 19 of 24 MHV4 S gene recombinant viruses contained the 5′ end of the S gene from pMH54-S4. The selected recombinants were plaque purified once again, and viral stocks were grown on 17Cl-1 cells for further analysis (14).
S gene sequencing.
For sequencing of the S gene, reverse transcriptase PCR amplification was carried out by cytoplasmic RNA extracted from virus-infected L2 cells as templates. The oligonucleotides listed in Table 1 and described previously (31) were used for amplification of the designated regions. Double-stranded PCR products were analyzed by automated sequencing by the Taq dye terminator procedure according to the manufacturer’s protocol (Taq DyeDeoxy Terminator Cycle Sequencing Kit; Applied Biosystems). The primers used for amplification were also used for sequencing, and each fragment was sequenced in both directions.
Viral growth curves.
L2 cell monolayers were prepared in 12-well plates in DMEM with 10% FBS. Confluent monolayers were infected with each virus (2 PFU/cell) in duplicate wells and incubated for 1 h at 37°C. After adsorption, the cells were washed with Tris-buffered saline three times and then fed with 1.5 ml of DMEM–10% FBS. At the times indicated, the cells were lysed by three cycles of freeze-thawing, and the supernatants were removed and titered by plaque assay on L2 cells as previously described (21).
Inoculation of mice.
All animal experiments used 4-week-old MHV-free C57BL/6 mice (The Jackson Laboratory, Bar Harbor, Maine). Mice were anesthetized with methoxyflurane (Metofane; Pittman-Moore, Mundelein, Ill.). For intracranial inoculation, the amount of virus designated in each experiment was diluted in PBS containing 0.75% bovine serum albumin, and a total volume of 20 μl was injected into the left cerebral hemisphere. Mock-infected controls were inoculated similarly but with an uninfected cell lysate at a comparable dilution.
Virulence assays.
Fifty-percent lethal dose (LD50) assays were carried out as described previously (23). Mice were inoculated intracranially with four- or fivefold serial dilutions of wild-type or recombinant viruses. In two independent experiments, a total of 5 to 15 animals per dilution per virus were analyzed. Mice were examined for signs of disease or death on a daily basis for up to 21 days postinfection. LD50 values were calculated by the Reed-Muench method (41, 43).
Virus replication in mice.
For the measurement of virus replication in the brain and liver, at selected times postinfection, mice were sacrificed and perfused with 10 ml of PBS, and the brains and livers were removed. The left half of each brain was placed directly into 2 ml of isotonic saline with 0.167% gelatin (gel saline), and the central lobe of the liver was placed in 2 ml of gel saline (39). The right half of the brain and a small portion of the liver was used for histology and viral antigen staining as described below. All organs were weighed and stored frozen at −80°C until virus titers were determined. Organs were homogenized, and virus titers were determined by plaque assay on L2 cell monolayers (23).
To determine the virus titers after a viral dose of 1 to 2 LD50s (Table 2), mice were inoculated with either 5,000 PFU of MHV-A59, wtR13, and wtR9 or 10 PFU of MHV4, S4R22, and S4R21, and the virus titers were determined on days 1, 3, 4, 5, and 7 postinfection (n = 4). For the kinetic analysis (see Fig. 3), mice were inoculated with 10 PFU of S4R22 or wtR13, and virus titers were determined on days 1, 4, 5, and 7 postinfection (n = 4).
TABLE 2.
Virulence of recombinant viruses and viral titers in the CNS after intracranial inoculation
| Virus | Virulence, log10(LD50)a | Viral titer, log10(PFU/g)b |
|---|---|---|
| MHV-A59 | 3.67 | 6.45 ± 0.43 |
| wtR13 | 3.83 | 6.33 ± 0.11 |
| wtR9 | 3.04 | 6.80 ± 0.13 |
| MHV4 | 0.44 | 6.70 ± 0.07 |
| S4R22 | 1.02 | 4.88 ± 0.49 |
| S4R21 | 0.94 | 4.81 ± 0.30 |
LD50 assays were carried out as described previously (23).
Viral replication in the brain was measured at 1, 3, 4, 5, and 7 days postinfection, and the peak viral titers are shown at 5 days postinfection for all viruses except wtR13, which was at 3 days postinfection. Animals were infected with 1 to 2 LD50s (5,000 PFU) of MHV-A59, wtR13, and wtR9 and with 1 to 2 LD50s (10 PFU) of MHV4, S4R22, and S4R21.
FIG. 3.
Viral replication in the brains and livers of animals after intracranial inoculation. C57BL/6 weanling mice were infected with 10 PFU of MHV4 (●), S4R22 (▴), S4R21 (▵), and wtR13 (■). Animals were sacrificed at the indicated times, and virus titers in the brains (A) or livers (B) were determined by plaque assay. The limit of detection was 500 PFU/g of tissue. The data shown represent the means (and standard deviations) of the titers from eight (S4R22) or four (all other viruses) animals.
Histology and immunohistochemistry.
For the analysis of inflammation and viral antigen expression, mice were sacrificed at selected times postinfection and perfused with 10 ml of PBS, and the brains, spinal cords, and livers were removed. The right half of the brain, spinal cord, and a small portion of the liver were fixed in formalin overnight. (The rest of the brain and part of the liver were used for the virus titration as described above.) Formalin-fixed tissue was embedded in paraffin, sectioned, and left unstained for immunohistochemistry or stained with hematoxylin and eosin (H&E) for histological analysis.
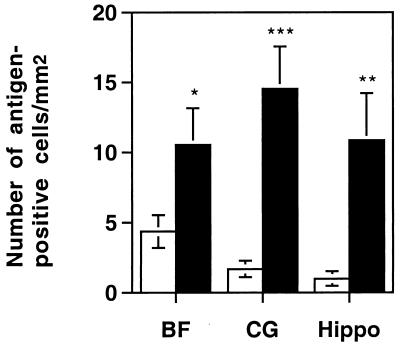
Immunohistochemical analysis was performed by the avidin-biotin-immunoperoxidase technique (Vector Laboratories, Burlingame, Calif.) by using diaminobenzadine tetrahydrochloride as a substrate, and a 1:500 dilution of rabbit anti-MHV-A59 serum made against detergent-disrupted MHV-A59. Control slides were incubated in parallel with preimmune rabbit serum, and sections from mock-infected animals were incubated with rabbit anti-MHV-A59 serum. All slides were read in a blinded manner. For each of four mice per virus, three sagittal brain sections, separated by 50 μm, were examined, and the number of viral-antigen-positive cells per square millimeter of tissue was determined. For each section, similar regions of the brain were studied, and a total of 4 mm2 of tissue in the basal forebrain and 2 mm2 of tissue in the hippocampus and cingulate gyrus were examined. The number of positively stained cells per square millimeter (see Fig. 6) represents the average number of antigen-positive cells per square millimeter from all of the sections examined.
FIG. 6.
Histopathology of the brains of C57BL/6 mice infected with recombinant viruses. Mice were infected intracranially as for Fig. 3 and sacrificed at 7 days postinfection. Brains were removed, fixed, sectioned, and stained with H&E as described in Materials and Methods. (A) S4R22-infected brain shows multiple large foci of inflammation in the midbrain and pons. (B) wtR13-infected brain shows a few small foci of inflammation in the midbrain and pons. (C) Higher magnification of the S4R22-infected brain shown in panel A demonstrates severe perivascular cuffing. (D) Higher magnification of the wtR13-infected brain shown in panel B shows moderate perivascular cuffing. Magnification: A and B, ×190; C and D, ×380.
H&E-stained histological sections were blinded and independently read by two investigators. The scoring of encephalitis was based on a previously described scale (42). One to two sagittal sections from each of four to five mice inoculated with 10 PFU of S4R22 or wtR13 were examined on day 7 postinfection. The intensity of inflammation was scored as follows: 0, normal tissue; 1, mild; 2, moderate; and 3, severe. The extent of spread, or the number of regions involved in inflammation, was scored as follows: 0, normal tissue; 1, mild (less than five regions); 2, moderate (between six and seven regions); and 3, severe (more than eight brain regions involved). The specific areas of the brain that were examined included the olfactory bulb, basal forebrain, hippocampus, diencephalon, cingulate gyrus and periventricular areas, neocortex, midbrain, pons, and cerebellum. The final score was the sum of the intensity and extent scores (maximum score of 6). Scores were averaged between the two investigators and presented as the means ± standard deviation. Statistical analysis was done by using the two-sided t test.
RESULTS
Generation of viruses with recombinant spike genes.
In order to study determinants of pathogenesis within the S gene, we used targeted recombination to replace the S gene of MHV-A59, a mildly neurovirulent strain of MHV, with that of a highly neurovirulent strain, MHV4 (Fig. 1). Targeted recombination was carried out (as described in Materials and Methods) between synthetic RNAs transcribed from pMH54-S4 and the recipient virus Alb4, a thermolabile, temperature-sensitive mutant of MHV-A59. The thermolability and temperature-sensitive plaque morphology of Alb4 is conferred by a deletion in the N gene which may be repaired by recombination with a wild-type N gene; thus, recombinant viruses were initially selected by resistance to heat treatment (14, 31). Attempts to select recombinant viruses containing the MHV4 S gene by using heat treatment, however, were unsuccessful since the MHV4 S gene conferred some degree of thermolability to the recombinant viruses (data not shown). Thus, we used a modified version of the selection strategy described previously (14, 31). Briefly, neutralizing monoclonal antibodies specific for the MHV-A59 S protein, A2.1 and A2.3 (11, 20), were used to select against viruses that expressed the MHV-A59 S protein derived from Alb4 (as described in Materials and Methods). The remaining plaques were therefore enriched for MHV4 S gene recombinants. Because Alb4 is thermolabile and does not replicate efficiently in animals, it was important to select recombinant viruses with the wild-type MHV-A59 S and N genes, in the Alb4 background, to be used as controls instead of Alb4. Wild-type recombinants were generated with synthetic RNA transcribed from pMH54, and recombinant viruses were selected by using temperature sensitivity, thermolability, and plaque size as described previously (14, 31).
The potential recombinants were plaque purified and screened by using PCR amplification and gel electrophoresis for the presence of the 5′ portion of the recombinant S gene and repair of the N gene deletion (14, 31). This screen was facilitated by the introduction of an AvrII restriction site in the signal sequence of the recombinant S gene (as described in Materials and Methods). For both the MHV4 S gene-containing and MHV-A59 S gene-containing recombinants, we characterized two independently derived recombinants isolated from separate recombination events. The MHV4 S gene recombinants were named S4R22 and S4R21, and the wild-type recombinants were named wtR13 and wtR9.
To verify that no secondary site mutations had been introduced into the S gene, we sequenced the 3′ portion of the hemagglutinin esterase open reading frame through to the 3′ end of the S gene. The sequencing of the S genes of the four recombinant viruses revealed that only one silent mutation (C to T) had been introduced at nucleotide 2159 of S4R21. The sequence of the S gene from the MHV4 S gene recombinants and the cloning plasmid used to generate them, pSwt, was identical to the previously published sequence of the MHV4 S gene (37) except for a previously noted silent mutation at nucleotide 3990 (18) and a three-nucleotide mutation (CTG in published sequence to GCT in cloned DNA), which resulted in an amino acid change at position 255 (leucine to alanine). Recombinant viruses that contained the two restriction sites, AvrII and SbfI, and were repaired in the N gene were likely the result of single recombination events 5′ to the S gene. Three of the four recombinant viruses contained all three markers, but analysis of wtR9 revealed that it contained the AvrII restriction site and the repaired N gene but not the 3′ SbfI restriction site. Thus, wtR9 underwent at least three recombination events, one of which was within the S gene. Since the sequence of the S gene of Alb4 is identical to that of MHV-A59, we were not able to determine the exact location of the recombination event within S.
In vitro growth characteristics of the recombinant viruses.
In cell culture, MHV4 and MHV-A59 show very different replication properties. While MHV-A59 replicates to a high titer in a number of cell lines, it has been shown that MHV4 replicates to a lower titer and displays higher levels of fusion and cytotoxicity (5, 19, 40). To determine whether the recombinant viruses, differing in S, exhibited altered replication in L2 cells, we compared their replication to that of MHV4 and MHV-A59. Single-step growth curves were performed, and the results (Fig. 2) indicate that the pattern of replication segregated with the S gene. The recombinant viruses containing the MHV4 S gene (S4R22 and S4R21) and wild-type MHV4 demonstrated a decrease in the extent of viral replication compared to viruses containing the MHV-A59 S gene, wtR13, wtR9, and MHV-A59. In addition, the S gene also determined the plaque morphology. At 48 h after the infection of L2 monolayers, MHV-A59 and the wild-type recombinants exhibited a large-plaque phenotype (>2 mm), while MHV4 and the MHV4 S gene recombinants exhibited a small-plaque phenotype (<1 mm) (data not shown). Thus, in cell culture the S gene of MHV4 conferred an alteration in viral replication and plaque morphology.
FIG. 2.
Time course of recombinant virus production in L2 cell cultures. L2 cells were infected in duplicate with MHV-A59 (○), wtR13 (■), wtR9 (□), MHV4 (●), S4R22 (▴), and S4R21 (▵) at a multiplicity of infection of 2 PFU/cell. At the indicated times, cells and cell culture supernatant were freeze-thawed three times and clarified, and virus titers were then determined by plaque assay. Each point represents the mean titer of duplicate samples.
Virulence of recombinant viruses.
Using isogenic recombinant viruses that differ exclusively in the S gene, we sought to determine whether the S gene of the highly neurovirulent MHV4 virus was sufficient to alter the virulence of the recombinant viruses. Thus, mice were inoculated intracranially with serial dilutions of S4R22 and S4R21, containing the MHV4 spike, wtR13 and wtR9, containing the MHV-A59 spike, wild-type MHV4, and wild-type MHV-A59. Infected mice were observed for lethality and an LD50 was calculated for each virus. The results are shown in Table 2. The recombinant viruses containing the MHV4 S gene (S4R22 and S4R21) displayed a dramatically lower LD50 value or a more virulent phenotype than the wild-type recombinants (wtR13 and wtR9). As expected, wtR13 and wtR9 displayed an LD50 similar to that of MHV-A59. The LD50 value for wild-type MHV4 was similar to that of the MHV4 S gene recombinants. Thus, the MHV4 S gene was able to confer a significantly more virulent phenotype.
Clinical signs also segregated with the spike gene. Mice inoculated with approximately 1 to 2 LD50s, 5,000 PFU, of the MHV-A59 spike-containing viruses (wtR13, wtR9, or MHV-A59) exhibited delayed weight gain and growth early in infection (data not shown). Mice inoculated with approximately 1 to 2 LD50s, 10 PFU, of the MHV4 spike-containing viruses (S4R22, S4R21, or MHV4) appeared normal up until days 4 to 6 postinfection, when all mice began to exhibit clinical signs such as hunched posture, ruffled fur, and abnormal gait (data not shown).
Recombinant virus replication in the brains and livers of infected mice.
The recombinants containing the MHV4 S gene (S4R22 and S4R21) were dramatically more virulent than the recombinants containing the MHV-A59 S gene (wtR13 and wtR9). To determine whether the location or extent of viral replication was altered for the recombinant viruses, we infected animals intracranially with virus and determined the titers of the virus in the brains and livers at various times postinfection as described in Materials and Methods.
Initially, we compared the viral replication in the brain of S4R22, S4R21, MHV4, wtR13, wtR9, and MHV-A59 by using approximately 1 to 2 times the LD50 for each virus. Infection at this dose allowed us to compare the replication patterns of each virus at a dose which resulted in roughly equal mortality. Mice were inoculated intracranially with 10 PFU of the MHV4 spike-containing viruses (S4R22, S4R21, and MHV4) and 5,000 PFU of the MHV-A59 spike-containing viruses (wtR9, wtR13, and MHV-A59). The peak viral titers are shown in Table 2. As expected, the kinetics and final extent of replication in the brain of wtR13, wtR9, and MHV-A59 were similar. Infection with only 10 PFU of MHV4 also resulted in a similar final extent of viral replication. Despite having a similar virulence, the MHV4 S gene recombinants, S4R22 and S4R21, exhibited significantly lower levels of viral replication than wild-type MHV4.
To compare the abilities of the MHV4 spike- and MHV-A59 spike-containing viruses to replicate in the brain and liver, we infected animals intracranially with 10 PFU of either S4R21, S4R22, wtR13, or wild-type MHV4. At this dose, more than half of the mice infected with a MHV4 S gene-containing virus would be expected to die, whereas none of the mice infected with wtR13 would be expected to die. The viral titers in the brains and livers were determined at various times postinfection (Fig. 3). In the brain, the MHV4 spike- and MHV-A59 spike-containing recombinant viruses exhibited nearly identical replication both in terms of the extent of replication and the kinetics. The wild-type MHV4 virus, which shares only the spike gene with the MHV4 S gene recombinants, replicated to a higher titer than did the recombinant viruses. At this dose, viral replication in the liver was at or below the limit of detection (500 PFU/g), and histologic examination of the livers revealed little to no hepatitis (data not shown) for both the MHV4 S gene and wild-type recombinants. Thus, despite the striking difference in virulence after inoculation with 10 PFU of virus, the MHV4 S gene recombinants and the wild-type recombinants showed similar levels of viral replication in the brain and minimal replication in the liver.
Distribution of viral antigen in the brains of infected animals.
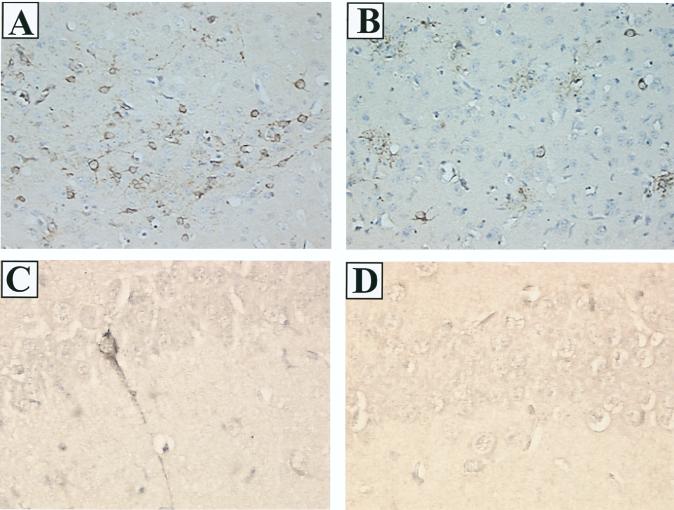
To examine the possibility that the MHV4 S gene recombinants had an altered pattern of viral spread within the brain, we used immunohistochemistry to determine the distribution of viral antigen. Brain sections from animals infected with 10 PFU of S4R22, S4R21, wtR13, or wild-type MHV4 at day 3 or 4, day 5, and day 7 postinfection were stained with polyclonal antisera against MHV proteins, blinded, and examined. At the peak of viral antigen expression, on day 5 postinfection, brain sections from a total of four animals per virus were examined and representative sections are shown in Fig. 4. The MHV4 S gene-containing viruses, S4R22, S4R21, and wild-type MHV4 exhibited similar patterns of antigen staining which differed in two ways from those seen after infection with the MHV-A59 spike-containing virus, wtR13. In some areas of the brain, such as the basal forebrain (Fig. 4A and 4B), the number of antigen-positive cells per region was greater for the MHV4 S gene recombinants than for the wild-type recombinant, wtR13. In other regions of the brain, such as the hippocampus (Fig. 4C and D), the MHV4 S gene recombinants exhibited positive antigen staining, whereas similar brain regions from wtR13-infected animals were negative for viral antigen staining. To quantify the differences in the intensity and extent of positive antigen staining that were observed between the MHV4 S gene- and the MHV-A59 S gene-containing viruses, the number of antigen-positive cells per square millimeter of brain tissue was determined in the basal forebrain, cingulate gyrus, and hippocampus (Fig. 5). These three regions were selected since they reflected the range of differences which were seen between the two recombinant viruses. In the cingulate gyrus and the hippocampus, the number of antigen-positive cells was significantly greater for the animals infected with the MHV4 S gene recombinant, S4R22, than for the animals infected with the wild-type recombinant, wtR13 (two-tailed t test; P < 0.001 and P < 0.01, respectively). In the basal forebrain there was less disparity between the two recombinant viruses, yet the difference was still statistically significant (two-tailed t test, P < 0.05). As determined by morphology, the MHV4 S gene- and MHV-A59 S gene-containing viruses, including both wild-type and recombinant viruses, exhibited positive antigen staining of neuronal and nonneuronal cell types with no obvious differences in cell type.
FIG. 4.
Immunohistochemistry of the brains of C57BL/6 mice infected with recombinant viruses. Mice were infected as described for Fig. 3 and then sacrificed at 5 days postinfection. Brains were removed, fixed, and sectioned, and MHV proteins were detected by immunolabeling with rabbit anti-MHV-A59 serum. (A) S4R22-infected brain shows severe viral antigen staining in the basal forebrain. (B) wtR13-infected brain shows moderate viral antigen staining in the basal forebrain. In both panels A and B antigen staining is evident in the cytoplasm of neurons. High-power magnification of the hippocampus demonstrates viral antigen staining of a neuron in S4R22-infected brain (C) and the absence of antigen staining in wtR13-infected brain (D). Magnification: A and B, ×190; C and D, ×380.
FIG. 5.
Viral-antigen-positive cells in the hippocampus, cingulate gyrus, and basal forebrain of mice infected with recombinant viruses. Mice were infected as for Fig. 3 and sacrificed at 5 days postinfection. Brains were removed, fixed, and sectioned, and MHV proteins were detected by immunolabeling with rabbit anti-MHV-A59 serum. All sections were blinded, and the numbers of viral-antigen-positive cells per square millimeter of tissue in the basal forebrain (BF), cingulate gyrus (CG), and hippocampus (Hippo) were determined by averaging the number of positive cells per square millimeter in three sagittal sections from each of four animals per virus. Error bars indicate the standard error of the mean. The S4R22 virus-infected mice (closed bars) exhibited significantly more antigen-positive cells than did the wtR13 virus-infected mice (open bars). ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001 (two-sided t test).
Distribution and extent of histopathology in the brains of infected animals.
The results of immunohistochemical examination suggested that infection with the MHV4 spike-containing recombinants resulted in a more intense and extensive infection of the CNS compared to infection with the MHV-A59 spike-containing recombinants. To determine whether the inflammatory response to infection with the MHV4 spike-containing recombinants was altered, we examined the severity of encephalitis induced by the recombinant viruses. Brain and spinal cord sections from infected mice were obtained on days 3 or 4, day 5, and day 7 postinfection, and stained with H&E to assess the severity of inflammation.
At the peak of inflammation 7 days postinfection with 10 PFU of virus, the MHV4 spike-containing viruses (S4R22, S4R21, and MHV4) caused a more intense and extensive inflammatory response than did the MHV-A59 spike-containing virus, wtR13. Representative sections from a total of four (MHV4 and S4R21), five (wtR13), or eight (S4R22) infected animals (Fig. 6) illustrate that the MHV4 S gene-containing viruses induced a dramatic degree of perivascular cuffing and inflammation (Fig. 6A and C), while infection with wtR13 resulted in less severe inflammation (Fig. 6B and D). The overall distribution of inflammation was as described in previous reports for MHV4, another JHM strain (MHV-JHM), and MHV-A59 (2, 13, 28, 30). The regions most consistently involved included the olfactory system, the piriform and entorhinal cortices, basal forebrain structures such as the septal nuclei and piriform cortices, the amygdala complex, regions of the diencephalon and basal ganglia, the periventricular areas and cingulate gyrus, regions of the hippocampus, the neocortex, and the brain stem.
In order to provide a more quantitative interpretation of these data, histologic sections from mice infected with 10 PFU of S4R22, containing the MHV4 spike, or wtR13, containing the MHV-A59 spike, were blinded and scored by two independent reviewers (as described in Materials and Methods). The histologic score, representing both the intensity of inflammation and the extent, or the number of brain regions exhibiting inflammation, was significantly greater for S4R22 (4.8 ± 0.6) compared to wtR13 (3.5 ± 0.9) (two-tailed t test; P < 0.001).
To determine whether infection with a high dose of the MHV-A59 spike-containing viruses could reproduce the intense inflammation seen with 10 PFU of the MHV4 spike-containing viruses, mice were infected with a dose of 1 to 2 LD50s, 5,000 PFU, of the MHV-A59 spike-containing viruses (wtR13, wtR9, and MHV-A59). Once again, the intensity and extent of inflammation was decreased relative to that induced by the MHV4 spike-containing viruses, yet the location of inflammation was similar (data not shown). The peak of inflammation was slightly earlier at this dose (day 5 or 7 postinfection). Thus, the intensity of inflammation seen with a dose of approximately 1 to 2 LD50s is greater for the MHV4 S gene recombinants than for the MHV-A59 S gene recombinants. Overall, these data demonstrate that the MHV4 spike-containing viruses exhibited an increase in the intensity and the extent of inflammation relative to the MHV-A59 spike-containing viruses.
DISCUSSION
Previous studies have suggested a correlation between mutations in S and alterations in virulence; however, these studies have been limited by the lack of technology to isolate mutations in the same genetic background. The use of targeted recombination has allowed us to begin to definitively map specific phenotypic properties to the S protein. In our recent study (31), a specific mutation in the S gene, introduced by targeted recombination, was found to alter the pathogenesis of the virus. We have used the targeted recombination technique here to directly address the role of the MHV4 S gene in pathogenesis and virulence. By comparing recombinant viruses, which differ exclusively in the S gene, we have been able to characterize the effect of S on neurovirulence.
For each desired recombinant virus, we studied at least two independently isolated recombinants to ensure that spurious mutations introduced during viral replication did not interfere with our interpretations. In cell culture and in vivo, the two MHV4 S gene recombinant viruses, S4R22 and S4R21, exhibited identical phenotypic properties. The wild-type recombinants also demonstrated phenotypic properties similar to each other and to MHV-A59. Due to the similarity in pathogenic properties within each group of recombinant viruses and the striking differences between MHV-A59 and MHV4 spike-containing viruses, we attributed differences in pathogenesis between the MHV4 S gene recombinants and the wild-type recombinants to alterations in the S gene.
One of the wild-type recombinants, wtR9, showed a slightly lower LD50 than did MHV-A59. This is most likely the result of a mutation(s) outside of S, acquired during recombination or subsequent viral passage. To verify that wtR9 was an exception and that wtR13 was a representative wild-type recombinant, we determined the LD50 of an additional wild-type recombinant, wtR2, and found it to be similar to both wtR13 and MHV-A59, with a log10(LD50) of 3.4. In addition, a previously characterized wild-type recombinant, wtR10, displayed virulence identical to that of MHV-A59 (31).
After intracranial inoculation, the pathogenesis of the recombinant viruses containing the MHV4 S gene was clearly different from that of the recombinants containing the MHV-A59 S gene. As measured by LD50, the MHV4 S gene conferred a dramatic increase in virulence, and this virulence was associated with increased viral antigen expression and inflammation in the CNS and viral replication in the brain but not in the liver. Comparison of the viral replication in the brain of the MHV4 spike- and MHV-A59 spike-containing viruses, however, revealed that they replicated with similar kinetics and to a similar final extent (Fig. 3). Thus, the neurovirulence of the MHV4 S gene-containing recombinants relative to the MHV-A59 S gene-containing recombinants could not be attributed to a higher load of infectious virus in the brain. In support of this observation that neurovirulence is not correlated with replication levels, peak viral titers for wild-type MHV4 and the MHV4 S gene recombinants were nearly 100-fold different despite a similar virulence. This variation in viral replication between the recombinant viruses and wild-type MHV4 must be attributable to genetic differences outside of the S gene.
There are several possible mechanisms by which the MHV4 spike may confer an increase in neurovirulence. To begin it is helpful to compare the primary structure of the S glycoprotein of MHV4 (37) and MHV-A59 (35). Within S2 the amino acid sequence identity is quite high, 96%, but in S1 there is more variation, with less than 89% identity. In addition, a single amino acid difference in the cleavage site of MHV-A59 relative to MHV4 has been shown to decrease the efficiency of cleavage of the S protein (6). The most striking difference between the two S genes, however, is a large deletion of 52 amino acids in the HVR of MHV-A59 (37).
One possible explanation for the difference in neurovirulence between the MHV4 S gene recombinants and the MHV-A59 S gene recombinants is that the MHV4 S protein is more cytotoxic. An increase in viral cytotoxicity, particularly of nonrenewable cells such as neurons, could dramatically affect host survival. From the study of neuroattenuated mutant viruses an association has been made between deletions in the HVR and diminished cytopathic effects in tissue culture (19). Furthermore, it has been suggested that the fusion potential of the spike is related to its ability to induce cytotoxicity and that this cytotoxicity may be due in part to intracellular fusion (40). Thus, the more fusogenic MHV4 spike would induce more cytotoxicity than the less fusogenic MHV-A59 spike. An additional mechanism of cell death, virus-induced apoptosis, has been associated with neurovirulence in Sindbis virus (32). Although the role of apoptosis in MHV neurovirulence is not known, MHV-induced apoptosis has been reported in macrophages (3).
A second possible mechanism to explain the neurovirulence conferred by the MHV4 S gene is related to the rate or extent of the spread of the virus and stems from the study of a neuroattenuated mutant of MHV4, V5A13.1, which was selected for resistance to neutralizing monoclonal antibodies (10). Characterization of this variant virus in vivo revealed that, relative to MHV4, it had a decreased rate of spread and decreased viral replication in the brain (13). Although the mechanism of decreased spread is not known, it is speculated that decreased fusogenicity may play a role (19). The MHV4 S gene recombinants exhibited a greater amount of viral antigen expression than did the MHV-A59 S gene recombinants. However, we did not detect a difference in the extent of viral replication in the brain. One possible explanation is that the MHV4 S gene-containing recombinants spread more quickly from cell to cell yet produced less infectious virus per cell than did the MHV-A59 spike-containing viruses.
The outcome of viral infection is determined by the complex interaction between the virus and the host immune system. Thus, a study of MHV pathogenesis must take into consideration the immune response to the virus. The S protein is a target of both neutralizing and cell-mediated immunity, and studies with the JHM strain of MHV have identified two CD8+ T-cell epitopes in the S glycoprotein (4, 7). Interestingly, the more immunodominant epitope, encompassing amino acids 510 to 518, within the HVR of S1, is deleted in MHV-A59. Thus, it is possible that the immune response to this epitope contributes to the intense inflammation and overall neurovirulence of the MHV4 S gene. Alternatively, the MHV4 S protein may stimulate an altered immune response that is ultimately more neurotoxic. As has been shown with a number of viruses, including MHV and Sindbis virus, the stimulation of specific immune modulators can mediate disease severity (27, 33, 38). Thus, either global immune stimulation or stimulation of specific immune mediators may play a role in determining the neurovirulence of various S proteins.
We have shown, by using targeted homologous recombination, that the MHV4 S gene contains determinants of neurovirulence. This finding demonstrates unambiguously what has been suggested by earlier studies associating mutations in S with alterations in neurovirulence. Future studies will be directed at precisely defining which functional regions of the MHV4 S gene are necessary and sufficient to confer this neurovirulent phenotype. In addition, we will investigate the immune response to the recombinant viruses. With this knowledge we hope to gain a better understanding of the mechanism of MHV-induced neurologic disease.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants NS-30606 and NS-21954 (S.R.W.) and a National Multiple Sclerosis Society grant RG-2615A1/2 (E.L.). J.J.P. was supported in part by training grant GM-07229.
We thank Paul Masters for pMH54, Alb4, and advice throughout the project; Michael J. Buchmeier for pSwt and MHV4; and John O. Fleming for monoclonal antibodies A2.1 and A2.3. We thank Jean Tsai for kindly providing wtR13, Su-hun Seo for excellent technical assistance, and Luis M. Schang, Jean Tsai, and Henry Teng for critical reading of the manuscript.
REFERENCES
- 1.Bailey O T, Pappenheimer A M, Cheever F S, Daniels J B. A murine virus (JHM) causing disseminated encephalomyelitis with extensive destruction of myelin. II. Pathology. J Exp Med. 1949;90:195–212. doi: 10.1084/jem.90.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnett E M, Cassell M D, Perlman S. Two neurotropic viruses, herpes simplex virus type 1 and mouse hepatitis virus, spread along different neural pathways from the main olfactory bulb. Neuroscience. 1993;57:1007–1025. doi: 10.1016/0306-4522(93)90045-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belyavskyi M, Belyavskaya E, Levy G A, Leibowitz J L. Coronavirus MHV-3-induced apoptosis in macrophages. Virology. 1998;250:41–49. doi: 10.1006/viro.1998.9356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergmann C C, Yao Q, Lin M, Stohlman S A. The JHM strain of mouse hepatitis virus induces a spike protein-specific Db-restricted cytotoxic T cell response. J Gen Virol. 1996;77:315–325. doi: 10.1099/0022-1317-77-2-315. [DOI] [PubMed] [Google Scholar]
- 5.Bond C W, Leibowitz J L, Robb J A. Pathogenic murine coronaviruses: II. Characterization of virus-specific proteins of murine coronaviruses JHMV and A59V. Virology. 1979;94:371–384. doi: 10.1016/0042-6822(79)90468-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bos E C W, Heijnen L, Luytjes W, Spaan W J M. Mutational analysis of the murine coronavirus spike protein: effect on cell-to-cell fusion. Virology. 1995;214:453–463. doi: 10.1006/viro.1995.0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castro R F, Perlman S. CD8+ T-cell epitopes within the surface glycoprotein of a neurotropic coronavirus and correlation with pathogenicity. J Virol. 1995;69:8127–8131. doi: 10.1128/jvi.69.12.8127-8131.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins A R, Knobler R L, Powell H, Buchmeier M J. Monoclonal antibodies to murine hepatitis virus-4 (strain JHM) define the viral glycoprotein responsible for attachment and cell-cell fusion. Virology. 1982;119:358–371. doi: 10.1016/0042-6822(82)90095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Compton S R, Barthold S W, Smith A L. The cellular and molecular pathogenesis of coronaviruses. Lab Anim Sci. 1993;43:15–28. . [Erratum 43:203.] [PubMed] [Google Scholar]
- 10.Dalziel R G, Lampert P W, Talbot P J, Buchmeier M J. Site-specific alteration of murine hepatitis virus type 4 peplomer glycoprotein E2 results in reduced neurovirulence. J Virol. 1986;59:463–471. doi: 10.1128/jvi.59.2.463-471.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daniel C, Anderson R, Buchmeier M J, Fleming J O, Spaan W J M, Wege H, Talbot P J. Identification of an immunodominant linear neutralization domain on the S2 portion of the murine coronavirus spike glycoprotein and evidence that it forms part of complex tridimensional structure. J Virol. 1993;67:1185–1194. doi: 10.1128/jvi.67.3.1185-1194.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeGroot R J, Luytjes W, Horzinek M C, van der Zeijst B A M, Spaan W J M, Lenstra J A. Evidence for a coiled-coil structure in the spike proteins of coronaviruses. J Mol Biol. 1987;196:963–966. doi: 10.1016/0022-2836(87)90422-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fazakerley J K, Parker S E, Bloom F, Buchmeier M J. The V5A13.1 envelope glycoprotein deletion mutant of mouse hepatitis virus type-4 is neuroattenuated by its reduced rate of spread in the central nervous system. Virology. 1992;187:178–188. doi: 10.1016/0042-6822(92)90306-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer F, Stegen C F, Koetzner C A, Masters P S. Analysis of a recombinant mouse hepatitis virus expressing a foreign gene reveals a novel aspect of coronavirus transcription. J Virol. 1997;71:5148–5160. doi: 10.1128/jvi.71.7.5148-5160.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fleming J O, Trousdale M D, El-Zaatari F A K, Stohlman S A, Weiner L P. Pathogenicity of antigenic variants of murine coronavirus JHM seleced with monoclonal antibodies. J Virol. 1986;58:869–875. doi: 10.1128/jvi.58.3.869-875.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frana M F, Behnke J N, Sturman L S, Holmes K V. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: host-dependent differences in proteolytic cleavage and cell fusion. J Virol. 1985;56:912–920. doi: 10.1128/jvi.56.3.912-920.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallagher T M. A role for naturally occurring variation of the murine coronavirus spike protein in stabilizing association with the cellular receptor. J Virol. 1997;71:3129–3137. doi: 10.1128/jvi.71.4.3129-3137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallagher T M, Escarmis C, Buchmeier M J. Alteration of pH dependence of coronavirus-induced cell fusion: effect of mutations in the spike glycoprotein. J Virol. 1991;65:1916–1928. doi: 10.1128/jvi.65.4.1916-1928.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallagher T M, Parker S E, Buchmeier M J. Neutralization-resistant variants of a neurotropic coronavirus are generated by deletions within the amino-terminal half of the spike glycoprotein. J Virol. 1990;64:731–741. doi: 10.1128/jvi.64.2.731-741.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilmore W, Fleming J O, Stohlman S A, Weiner L P. Characterization of the structural proteins of the murine coronavirus strain A59 using monoclonal antibodies. Proc Soc Exp Biol Med. 1987;185:177–186. doi: 10.3181/00379727-185-42532. [DOI] [PubMed] [Google Scholar]
- 21.Gombold J L, Hingley S T, Weiss S R. Fusion-defective mutants of mouse hepatitis virus A59 contain a mutation in the spike protein cleavage signal. J Virol. 1993;67:4504–4512. doi: 10.1128/jvi.67.8.4504-4512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higuchi R. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. pp. 177–183. [Google Scholar]
- 23.Hingley S T, Gombold J L, Lavi E, Weiss S R. MHV-A59 fusion mutants are attenuated and display altered hepatotropism. Virology. 1994;200:1–10. doi: 10.1006/viro.1994.1156. [DOI] [PubMed] [Google Scholar]
- 24.Houtman J J, Fleming J O. Pathogenesis of mouse hepatitis virus-induced demyelination. J Neurovirol. 1996;2:361–376. doi: 10.3109/13550289609146902. [DOI] [PubMed] [Google Scholar]
- 25.Koetzner C A, Parker M M, Ricard C S, Sturman L S, Masters P S. Repair and mutagenesis of the genome of a deletion mutant of the murine coronavirus mouse hepatitis virus by targeted RNA recombination. J Virol. 1992;66:1841–1848. doi: 10.1128/jvi.66.4.1841-1848.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kubo H, Yamada Y K, Taguchi F. Localization of neutralizing epitopes and the receptor-binding site within the amino-terminal 330 amino acids of the murine coronavirus spike protein. J Virol. 1994;68:5403–5410. doi: 10.1128/jvi.68.9.5403-5410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a.Kuo, L., G.-J. Godeke, M. J. B. Raamsman, P. S. Masters, and P. J. M. Rottier. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host species barrier. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 27.Lane T E, Fox H S, Buchmeier M J. Inhibition of nitric oxide synthase-2 reduces the severity of mouse hepatitis virus-induced demyelination: implications for NOS2/NO regulation of chemokine expression and inflammation. J Neurovirol. 1999;5:48–54. doi: 10.3109/13550289909029745. [DOI] [PubMed] [Google Scholar]
- 28.Lavi E, Fishman P S, Highkin M K, Weiss S R. Limbic encephalitis after inhalation of a murine coronavirus. Lab Invest. 1988;58:31–36. [PubMed] [Google Scholar]
- 29.Lavi E, Gilden D H, Highkin M K, Weiss S R. The organ tropism of mouse hepatitis virus strain A59 is dependent on dose and route of inoculation. Lab Anim Sci. 1986;36:130–135. [PubMed] [Google Scholar]
- 30.Lavi E, Murray E M, Makino S, Stohlman S A, Lai M M C, Weiss S R. Determinants of coronavirus MHV pathogenesis are localized to 3′ portions of the genome as determined by ribonucleic acid-ribonucleic acid recombination. Lab Invest. 1990;62:570–578. [PubMed] [Google Scholar]
- 31.Leparc-Goffart I, Hingley S T, Chua M M, Phillips J, Lavi E, Weiss S R. Targeted recombination within the spike gene of murine coronavirus mouse hepatitis virus-A59: Q159 is a determinant of hepatotropism. J Virol. 1998;72:9628–9636. doi: 10.1128/jvi.72.12.9628-9636.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis J, Wesselingh S L, Griffin D E, Hardwick J M. Alphavirus-induced apoptosis in mouse brains correlates with neurovirulence. J Virol. 1996;70:1828–1835. doi: 10.1128/jvi.70.3.1828-1835.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang X H, Goldman J E, Jiang H H, Levine B. Resistance of interleukin-1B-deficient mice to fatal Sindbis encephalitis. J Virol. 1999;73:2563–2567. doi: 10.1128/jvi.73.3.2563-2567.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo Z, Weiss S R. Roles in cell-to-cell fusion of two conserved hydrophobic regions in the murine coronavirus spike protein. Virology. 1998;244:483–494. doi: 10.1006/viro.1998.9121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luytjes W, Sturman L S, Bredenbeck P J, Charite J, van der Zeijst B A M, Horzinek M C, Spaan W J M. Primary structure of the glycoprotein E2 of coronavirus MHV-A59 and identification of the trypsin cleavage site. Virology. 1987;161:479–487. doi: 10.1016/0042-6822(87)90142-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masters P S, Koetzner C A, Kerr C A, Heo Y. Optimization of targeted RNA recombination and mapping of a novel nucleocapsid gene mutation in the coronavirus mouse hepatitis virus. J Virol. 1994;68:328–337. doi: 10.1128/jvi.68.1.328-337.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parker S E, Gallagher T M, Buchmeier M J. Sequence analysis reveals extensive polymorphism and evidence of deletions within the E2 glycoprotein gene of several strains of murine hepatitis virus. Virology. 1989;173:664–673. doi: 10.1016/0042-6822(89)90579-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pope M, Marsden P A, Cole E, Sloan S, Fung L S, Ning Q, Ding J W, Leibowitz J L, Phillips M J, Levy G A. Resistance to murine hepatitis virus strain 3 is dependent on production of nitric oxide. J Virol. 1998;72:7084–7090. doi: 10.1128/jvi.72.9.7084-7090.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramig R F. Isolation and genetic characterization of temperature sensitive mutants of simian rotavirus SA11. Virology. 1982;120:93–135. doi: 10.1016/0042-6822(82)90009-5. [DOI] [PubMed] [Google Scholar]
- 40.Rao P V, Gallagher T M. Intracellular complexes of viral spike and cellular receptor accumulate during cytopathic murine coronavirus infections. J Virol. 1998;72:3278–3288. doi: 10.1128/jvi.72.4.3278-3288.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reed L J, Muench H. A simple method of estimating fifty per cent points. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 42.Sasseville V G, Smith M M, Mackay C R, Pauley D R, Mansfield K G, Ringler D J, Lackner A A. Chemokine expression in simian immunodeficiency virus-induced AIDS encephalitis. Am J Pathol. 1996;149:1459–1467. [PMC free article] [PubMed] [Google Scholar]
- 43.Smith A L, Barthold S W. Methods in viral pathogenesis. In: Nathanson N, editor. Viral pathogenesis. Philadelphia, Pa: Lippincott-Raven; 1997. pp. 483–506. [Google Scholar]
- 44.Sturman L S, Ricard C S, Holmes K V. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: activation of cell fusing activity of virions by trypsin and separation of two different 90K cleavage fragments. J Virol. 1985;56:904–911. doi: 10.1128/jvi.56.3.904-911.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wege H, Winter J, Meyermann R. The peplomer protein E2 of coronavirus JHM as a determinant of neurovirulence: definition of critical epitopes by variant analysis. J Gen Virol. 1988;69:87–98. doi: 10.1099/0022-1317-69-1-87. [DOI] [PubMed] [Google Scholar]
- 46.Williams R K, Jiang G S, Holmes K V. Receptor for mouse hepatitis virus is a member of the carcinoembryonic antigen family of glycoproteins. Proc Natl Acad Sci USA. 1991;88:5533–5536. doi: 10.1073/pnas.88.13.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]