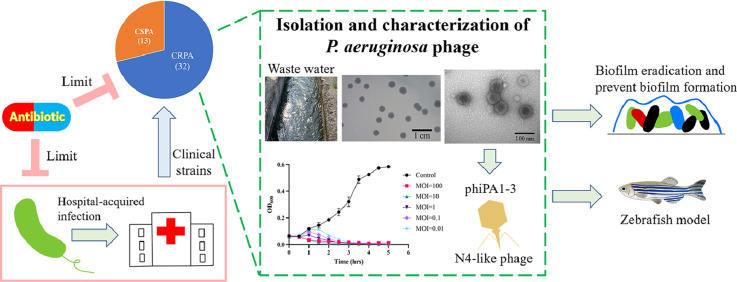
Highlights
-
•
Bacteriophage vB_PaP_phiPA1-3 was isolated and characterized in terms of its phenotypical, biological, and genomic characteristics against carbapenem-resistant Pseudomonas aeruginosa (CRPA).
-
•
The host range spectrum revealed that phiPA1-3 is a moderate host range phage that infects 20% of clinically isolated strains of P. aeruginosa, including CRPA.
-
•
Biofilm eradication and phage rescue of animals from bacterial infections were assessed to demonstrate the potential efficacy of phiPA1-3.
Keywords: Carbapenem-resistant P. aeruginosa, Anti-biofilm, Phage therapy, N4-like phages, Schitoviridae
Abstract
Multiple drug-resistant (MDR) Pseudomonas aeruginosa commonly causes severe hospital-acquired infections. The gradual emergence of carbapenem-resistant P. aeruginosa has recently gained attention. A wide array of P. aeruginosa-mediated pathogenic mechanisms, including its biofilm-forming ability, limits the use of effective antimicrobial treatments against it. In the present study, we isolated and characterized the phenotypic, biological, and genomic characteristics of a bacteriophage, vB_PaP_phiPA1-3 (phiPA1-3). Biofilm eradication and phage rescue from bacterial infections were assessed to demonstrate the efficacy of the application potential. Host range spectrum analysis revealed that phiPA1-3 is a moderate host range phage that infects 20% of the clinically isolated strains of P. aeruginosa tested, including carbapenem-resistant P. aeruginosa (CRPA). The phage exhibited stability at pH 7.0 and 9.0, with significantly reduced viability below pH 5.0 and beyond pH 9.0. phiPA1-3 is a lytic phage with a burst size of 619 plaque-forming units/infected cell at 37 °C and can effectively lyse bacteria in a multiplicity of infection-dependent manner. The genome size of phiPA1-3 was found to be 73,402 bp, with a G+C content of 54.7%, containing 93 open reading frames, of which 62 were annotated as hypothetical proteins and the remaining 31 had known functions. The phage possesses several proteins similar to those found in N4-like phages, including three types of RNA polymerases. This study concluded that phiPA1-3 belongs to the N4-like Schitoviridae family, can potentially eradicate P. aeruginosa biofilms, and thus, serve as a valuable tool for controlling CRPA infections.
Graphical abstract
1. Introduction
Pseudomonas aeruginosa is a bacterium commonly found in soil, water, and plants. However, it is also a notorious opportunistic pathogen that can lead to severe infections in humans, especially in individuals with severe wounds, resulting in sepsis in immunocompromised patients, chronic lung infections in patients with cystic fibrosis, chronic obstructive pulmonary disease, bladder catheter-associated chronic infections in the urinary tract, and serious ventilator-associated pneumonia. It is responsible for significant mortality rates and imposes billions of dollars in healthcare expenses (Blanchette et al., 2017; Morales et al., 2012). This bacterium is equipped with several adhesion factors, such as flagella, pili, and biofilms; and can survive in water, on different surfaces, and on medical devices, causing various infections. The development of biofilms using P. aeruginosa as a model has been reviewed previously (Sauer et al., 2022), which has highlighted the need to investigate how biofilms are associated with clinical problems (Lichtenberg et al., 2022; Tolker-Nielsen, 2014; Yin et al., 2022). In most cases, treatment of P. aeruginosa is very challenging owing to its multiple mechanisms to evade treatment by current antibiotics; carbapenem-resistant P. aeruginosa (CRPA) has especially been categorized as a ‘critical’ pathogen by the World Health Organization (Tacconelli et al., 2018). In this era in which antibiotic resistance is considered a major worldwide concern, controlling these infections is often difficult. Owing to the current shortage of research on novel antibiotics that can combat multidrug-resistant (MDR) P. aeruginosa, phages have emerged as a potential solution to address this issue by serving as a new antibacterial therapeutic approach. Phages with specific targeting mechanisms against the Pseudomonas genus were discovered in the mid-twentieth century, marking a paradigm shift in the development of therapeutic approaches (Chegini et al., 2020). The use of phages to inhibit P. aeruginosa has garnered significant attention because of their high impact on nosocomial infections and antibiotic resistance. Phage cocktails are typically more effective in inhibiting bacterial infections, thus making them a potential solution for combating P. aeruginosa (Jault et al., 2019; Li et al., 2021; Vaitekenas et al., 2021). Hence, Pseudomonas phage isolation and characterization are needed not only for in vitro bactericidal studies, but also in vivo animal and human experiments. In comparison to the literature from the rest of the world, there are only a limited number of studies available on Pseudomonas phages isolated in Taiwan. Therefore, the isolation of additional phages with activity against P. aeruginosa is crucial to counter antibiotic resistance and advance phage applications. In this study, we describe the phenotypic, biological, and genotypic properties of a newly isolated phage phiPA1-3 that we isolated from wastewater around a hospital in eastern Taiwan, and demonstrated its lytic efficiency against CRPA, as well as, in vivo therapeutic efficacy. These results indicated that phiPA1-3 can be used as an antibiotic substitute for the treatment of clinical CRPA infections or for environmental decontamination.
2. Materials and methods
2.1. Bacterial strains and growth conditions
P. aeruginosa strain PA001 was isolated from the patient's sputum and identified using 16S rRNA gene sequence analysis. For phage host range testing, additional clinical isolates, including 44 P. aeruginosa, 3 Klebsiella pneumoniae, 3 Enterobacter cloacae, and 2 Escherichia coli, were also tested. All bacterial strains were routinely grown in Luria-Bertani broth (LB) or agar (1.5%) (LA) medium, at 37 °C, under aerobic conditions. To establish a bacterial lawn, we used a double-layer agar technique with 0.75% agar (Kropinski et al., 2009).
2.2. Antibiotic-susceptibility profile of P. aeruginosa
Nine antibiotics were used to assess the antibiotic-susceptibility profile of P. aeruginosa clinical isolates using the Kirby–Baur disc diffusion method, according to the guidelines of the Clinical and Laboratory Standards Institute. MDR strains were defined as those that were resistant to three or more classes of antibiotics. The antibiotics used in this study included 10 μg Imipenem, 10 μg Meropenem, 10 μg Ertapenem, 10 μg Streptomycin, 10 μg Gentamycin, 30 μg Kanamycin, 5 μg Ciprofloxacin, 10 μg Colistin, and 15 μg Erythromycin. P. aeruginosa BCRC11633 was used as a quality control strain.
2.3. Phage isolation, propagation, purification, and titer determination
Phages were isolated from a 9 mL wastewater sample taken from a sewer at Tzu Chi Hospital, Hualien, Taiwan, using a modified traditional method. The sample was filtered using a 0.45 μm filter and supplemented with 1 mL of mid-log phase culture of PA001, followed by incubation at 37 °C for 24 h. The presence of phages in the supernatant was confirmed by means of spot-testing on a PA001 lawn. A single phage plaque was selected with a toothpick using the double-agar layer method and purified in three consecutive steps. The purified phage was named vB_PaP_phiPA1-3 (shortened form: phiPA1-3) and propagated in 50 mL of early log-phase cells until complete lysis (Adriaenssens and Brister, 2017; Van Twest and Kropinski, 2009). The culture was then centrifuged and the supernatant obtained was passed through a 0.45 μm membrane filter. Phage particles in the supernatant were pelleted by means of centrifugation (3 h at 19,000 rpm in a Beckman Avanti JXN-26, Beckman Coulter Inc., Brea, CA, USA), resuspended in 1 mL of SM buffer [50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 8 mM MgSO4, and 0.01% gelatin), and purified by banding on the block gradient of CsCl representing 1.15, 1.45, 1.50, and 1.70 g/cm3 (1.5 mL for each block) in ultracentrifugation (30,000 rpm for 5 h, at 4 °C, with the SW41Ti rotor in a Beckman XPN-100). The collected phage band was dialyzed against the SM buffer and then stored at 4 °C until further use. The phage titers were obtained by serially diluting the phage suspensions. Subsequently, 100 µL of the optimal dilution was taken, mixed with overnight host bacterial culture on the molten top agar and poured onto the bottom agar layer consisting of LB medium to obtain a countable number of individual plaques. The number of plaque-forming units (PFU) was calculated for 1 mL of concentrated suspension. The resulting high-titer suspension (1012 PFU/mL) was used for further experiments.
2.4. Host range and efficiency of plating (EOP) determination
To determine the host range of the phage phiPA1-3, a concentrated phage suspension was spotted on the exponentially growing P. aeruginosa, K. pneumoniae, E. cloacae, and E. coli clinical isolates. Briefly, 50 µL of tested overnight bacterial cultures grown in LB medium at 37 °C were mixed with 5 mL of soft agar and poured onto LA. Following that, 3 μL of phage suspension (108 PFU/mL) was spotted on the soft agar lawns and incubated at 37 °C for 16–18 h. Spot clear zones were used to check the sensitivity of the bacteria towards the phage. This assay was repeated thrice. For the colonies testing positive, plating efficiency was determined as previously described (Mirzaei and Nilsson, 2015), with some modifications. Briefly, the selected isolates were grown overnight at 37 °C and 50 μL of each culture was used in double-layer agar with 100 µL of diluted phage lysate. The phage lysate was 106-fold diluted from the phage stock. The plates were incubated overnight (18 h) at 37 °C and the number of PFU was determined. The EOP was calculated (average PFU on target bacteria/average PFU on host bacteria) along with the standard deviation (± SD) for three measurements.
2.5. Phage biology experiments
The phage adsorption assay was performed as previously described (Lin et al., 2010), with minor modifications. Briefly, exponentially growing PA001 cells were mixed with a phage suspension with multiplicity of infection (MOI)=0.0005 and incubated at 37 °C. Samples of 1 mL were collected from time=0 min to 10 min, at intervals of 1 and 2 min. After centrifugation, the supernatants were titrated at different time intervals to further determine the unabsorbed phages using the plaque assay method.
The one-step phage growth curve and burst size (average number of phages released by each bacterium after lysis) were determined as previously described (Lin et al., 2010), with modifications. Briefly, 1 mL of host bacterial cells [mid-log phase=1.25 × 108 colony-forming units (CFU)/mL] was harvested. The bacterial cells were infected with the phage at an MOI of 0.0001. The phages were allowed to adsorb on ice for 15 min. The mixture was then centrifuged at 12,000 rpm, 4 °C, for 5 min, to remove the unadsorbed phage particles and the pellet was resuspended in 50 mL of LB medium, and incubated at 37 °C, with shaking. Aliquots of 1 mL were sampled from zero time to 70 min, at 10 min intervals. Samples were centrifuged (12,000 rpm, 2 min) and the PFU of the supernatant was determined using double-layer agar. The latency period was defined as the time between infection and the shortest incubation time that allowed for phage production. Burst size was calculated as the ratio of the number of phage particles released at the plateau level to the initial number of infected bacterial cells. All experiments were performed thrice.
The bacteriolytic ability of the phiPA1-3 phage was demonstrated in exponential growth phase cultures of the P. aeruginosa PA001 isolate, as described previousl (Horváth et al., 2020), with minor modifications. Briefly, a 96-well microtiter plate (Thermo Fisher Scientific Inc., Waltham, MA, USA) was partitioned into parallel sections, each containing 3 identical columns. Each row contained 200 μL bacterial culture (1.5 × 108 CFU/mL). Bacteria not inoculated with phages served as positive controls. To all other rows, 20 μL phage suspensions were added with different phage dilutions: 3 × 109 (MOI=100), 3 × 108 (MOI=10), 3 × 107 (MOI=1), 3 × 106 (MOI=0.1), 3 × 105 (MOI=0.01). The microtiter plate was put at 37 °C for 6 h at 30 min intervals. Experiments were performed in triplicates and reproduced in three independent trials.
For the thermal-stability assay, phage suspensions (108 PFU/mL) were incubated at 4 °C, 25 °C, 30 °C, 37 °C, 50 °C, and 65 °C, and aliquots were taken after 1 h of incubation. For the pH stability assay, a phage suspension (1 × 108 PFU/mL) was inoculated in a series of tubes containing fresh LB broth at pH 3.0, 5.0, 7.0, 9.0, and 11.0, and incubated at 37 °C. Aliquots were collected after 1 h of incubation. Phage titers were determined using P. aeruginosa PA001 as host cells, by means of the double-layer agar method. All procedures were repeated in triplicate, and the results were averaged.
2.6. Electron microscopy
The morphology of the phages was examined using negative-stain transmission electron microscopy (TEM). Briefly, 10 μL of purified phage suspension (1012 PFU/mL) was placed on 400 mesh carbon-coated grids with 2% uranyl acetate, followed by rinsing with distilled water several times, and then examined in a Hitachi H-7500 transmission electron microscope operated at 80 kV (Hitachi Company, Tokyo, Japan). At least 20 phage images were used to assess particle dimensions.
2.7. Pulsed-field gel electrophoresis
Pulsed-field gel electrophoresis was performed using a CHEFDRIII System (Bio-Rad Laboratories, Hercules, CA, USA) at 6 V/cm and 14 °C, with pulse ramps from 5 to 20 s, for 18 h (for the undigested phage DNA), in 0.5 × Tris-borate-EDTA buffer. A Midrange PFG Marker (New England BioLabs, Beverly, MA, USA) was used as a molecular size standard.
2.8. Phage structural proteins analysis
Purified phage particles were mixed with sample buffer (62.5 mM Tris-HCl pH 6.8, containing 5% 2-mercaptoethanol, 2% sodium dodecyl sulfate, 10% glycerol, and 0.01% Bromophenol Blue) and heated in a boiling water bath for 15 min, followed by separation of the structural proteins on a gradient SDS-polyacrylamide gel (4%–20%). Protein bands were detected by means of Coomassie Blue staining. After staining, the bands were excised from the gel and digested with trypsin. Identification was performed using liquid chromatography-tandem mass spectrometry. Raw data were searched using MASCOT (https://www.matrixscience.com/search_form_select.html), against a local database of all possible peptide spectra deduced from the phiPA1-3 genome sequence.
2.9. Anti-biofilm activity measurement
To monitor the timing of biofilm-formation by P. aeruginosa PA001, the bacteria were grown in 6-well plates on cover slides in LB medium, at a concentration of 2 × 107 CFU in 2 mL medium. Biofilms were observed at different time-points (every 2 h) for 24 h. To visualize the biofilm, the medium was removed and the biofilm was washed with phosphate-buffered saline (PBS) and stained with 0.1% Crystal Violet for 30 min, at room temperature (RT). After staining, the biofilms were washed with sterile water and observed under a light microscope. The experiments were performed using three independent colonies.
In the co-culture assay, the bacteria were mixed with different MOIs (0.01, 0.1, 1, 10, and 100) of phiPA1-3 and grown in 6-well plates on cover slides in LB medium, at a concentration of 2 × 107 CFU in 2 mL medium, for 6 h. After 6 h, the medium was removed and the wells were washed with PBS. The biofilms were then stained with 0.1% Crystal Violet for 30 min, at RT, washed with sterile water, and observed under a light microscope, as described above.
For quantification of the inhibition of biofilm-formation upon co-culturing of phiPA1-3 with P. aeruginosa PA001, 160 μL of mid-log phase cells (1.25 × 108 CFU/mL) in LB medium were mixed with 40 μL of different MOIs of phiPA1-3 (0.01, 0.1, 1, 10, and 100) and grown in 96-well plates, at 37 °C, for 6 h. To determine the amount of biofilm, the supernatants were removed; the wells were washed twice with PBS, and then fixed with methanol for 15 min, at RT. After removing methanol, 0.1% Crystal Violet was added, and the mixture was left at RT for 30 min. Thereafter, the Crystal Violet was removed and the wells were washed twice with PBS. Finally, the Crystal Violet was dissolved in 95% ethanol, and the OD570 was measured.
To remove preformed biofilms, P. aeruginosa PA001 was cultured in LB medium in 6-well plates on cover slides, at 2 × 107 CFU in 2 mL for 6 h. Different concentrations of phiPA1-3 (2 × 109, 2 × 108, and 2 × 107 PFU) or SM buffer were used as controls in 6-well plates, for 18 h. The slides were then removed from the wells and washed twice with PBS. Thereafter, the samples were fixed on ice in 2.5% glutaraldehyde/0.1 M sodium cacodylate buffer for 60 min, and then washed twice with 5% sucrose/0.1 M sodium cacodylate buffer on ice for 15 min. After washing, the post-fixed sample was treated with 1% osmium/0.1 M sodium cacodylate buffer for 60 min, at RT, and then washed twice with 5% sucrose/0.1 M sodium cacodylate buffer for 15 min, at RT. In the next step, the samples were dehydrated in a graded ethanol series (50%, 70%, and 95%) and chemically dried twice using 50% ethanol/50% hexamethyldisilazane (Sigma-Aldrich), for 15 min. Finally, the samples were chemically dried twice using 100% hexamethyldisilazane, for 15 min. The specimens were mounted on a metal stub and gold-coated before examining the surface morphology of the biofilms under high vacuum using a scanning microscope (HITACHI S-4700).
To quantify the eradicated biofilms, P. aeruginosa PA001 was cultured in LB medium in 6-well plates on cover slides, at a concentration of 2 × 107 CFU in 2 mL medium, for 6 h. Subsequently, different concentrations of phiPA1-3 phage (2 × 109, 2 × 108, and 2 × 107 PFU) and the same volume of SM buffer as a control were added to each well and incubated for another 6 h before quantification. To determine the amount of biofilm formed, the supernatants were removed, and the wells were washed twice with PBS and then fixed with methanol for 15 min, at RT. After removing methanol, 0.1% Crystal Violet was added, and the mixture was left at RT for 30 min. The Crystal Violet was then removed and the wells were washed twice with PBS. Finally, Crystal Violet was dissolved in 95% ethanol, and the OD570 was measured.
2.10. Genome sequencing and analysis
The genomic sequence of phiPA1-3 was determined by BioTools Company, Taiwan, Taipei. Total genomic DNA was extracted using the Phage DNA Isolation Kit (Norgen Biotek Corporation, Thorold, Ontario, Canada), and the DNA concentration was determined using a Qubit® 4.0 fluorometer (Thermo Scientific, USA). Briefly, 1 μg of phage genomic DNA was purified and sent for library construction (SQKLSK109) and sequencing (both by Oxford Nanopore Technologies, UK). After sequencing, the raw data were decoded using Guppy, checked using NanoPack (v1.1.0) (De Coster et al., 2018; Wick et al., 2019), and assembled using Canu (v2.1.1) (Koren et al., 2017), to obtain primary contigs. The primary contigs were processed using Racon (v1.4.22) (Vaser et al., 2017), and model correction was performed using Medaka (v1.2.3) (Oxford Nanopore Technologies). Finally, the contigs were automatically corrected using homologous sequences extracted from closely related genomes using Homopolish (v0.2) (Huang et al., 2021). To evaluate the quality of assemblies and completeness of the genome, fully processed contigs were analyzed using QUAST (v5.0.2) (Gurevich et al., 2013), BUSCO (v5.0.0) (Manni et al., 2021) Prokka (v1.14.5) (Seemann, 2014), with default settings to predict open reading frames (ORFs) and locate tRNA and rRNA regions. InteroProScan (https://www.ebi.ac.uk/interpro/search/sequence/) and HHpred (https://toolkit.tuebingen.mpg.de/tools/hhpred) were used to improve the predictions. We also referred to the ORFs using Rapid Annotation with Subsystem Technology (https://rast.nmpdr.org/). A genome map was generated using SnapGene Viewer (www.snapgene.com). Functional predictions were made using BLASTP. Conserved domains were predicted using the NCBI Conserved Domains (https://www.ncbi.nlm.nih.gov/Strecture/cdd/wrpsb.cgi). Potential tRNA and tmRNA genes were predicted using Aragorn (Laslett and Canback, 2004) and tRNAScanSE (Chan and Lowe, 2019). ResFinder (Bortolaia et al., 2020) and VirulenceFinder (Kleinheinz et al., 2014) were used to predict the presence of antibiotic resistance genes and virulence factors. Phylogenetic analysis was performed using MEGA11 (Tamura et al., 2021) software, with default settings. Comparative genomic analysis was performed using Easyfig 2.2.3 (Sullivan et al., 2011).
2.11. Zebrafish infection and phage rescue experiments
For in vivo phage prevention and rescue experiments, zebrafish (Danio rerio) wild type AB lines were used. The fish were maintained at the Tzu Chi University Fish Core Facility, according to standard protocols. Mixed male and female populations of zebrafish were kept in 9 L tanks at 28 °C and maintained in a 14 h light/10 h dark cycle. Infection experiments were performed as previously described (Shahed-Al-Mahmud et al., 2021), with some modifications.
Upon centrifuging (15,000 × g, 30 min) the phage suspensions, the resulting suspension had a titer of 1011 PFU/mL. The logarithmic P. aeruginosa PA001 culture was centrifuged and the pellets were resuspended in 0.85% NaCl. The OD600 was set to 10, to obtain a final suspension of 1010 CFU/mL.
Adult zebrafish (>4 months-old) were grouped as follows (n=20 per group). One group of fish was anesthetized with 0.2% tricaine and injected with 20 μL (1.4∼2.4 × 107 CFU) of P. aeruginosa PA001 suspended in 0.85% NaCl through the cloaca, with an insulin needle. The other group was infected with the same dose of P. aeruginosa PA001 strain and injected with a dose of 2 × 108 PFU phiPA1-3 into the cloaca, half an hour later. In addition, to assess the potential acute toxicity of phiPA1-3, zebrafish were injected with 2 × 108 PFU of phiPA1-3 or 0.85% NaCl alone in the absence of bacterial infection. After treatment, the zebrafish were transferred to separate tanks and monitored at least five times daily, for 3 d. The number of surviving fish on day 3 was plotted, and Fisher's Exact Test was used to evaluate the therapeutic efficacy of phiPA1-3. Using SPSS software, Kaplan–Meier survival curves (Jung et al., 2018) were plotted to show the cumulative probability of survival over the 3-day period using the log-rank test or generalized Wilcoxon test for statistics. Laboratory protocols were performed by well-trained scientists and approved by the Institutional Animal Care and Use Committee of Tzu Chi University, Hualien, Taiwan (IACUC approval no. 111091).
2.12. Statistical analysis
Data were analyzed according to the experimental design. To achieve approximate normality, the counts were log-transformed after adding a value of 1 to avoid taking logs of zero. The effects were considered significant at p=0.05, and pair-wise differences were assessed using t-test. All analyses were performed with the statistical software package Statistica v.10 (www.statsoft.com).
2.13. Nucleotide sequence accession number
The annotated sequence has been submitted to GenBank, with the accession number OQ378339.
3. Results
3.1. Host antibiotic-susceptibility profile
The forty-five clinical P. aeruginosa isolates (Table 1) were subjected to antibiotic profile analysis, which confirmed that thirty-two (71%) of those were CRPA according to the Clinical and Laboratory Standards Institute guidelines. To clarify the genetic variability of these clinical isolates, the random amplified polymorphic DNA (RAPD)-PCR-based technique was performed with primers (ERIC-1F, ATGTAAGCTCCTGGGGATTCAC; ERIC-2R, AAGTAAGTGACTGGGGTGAGCG). All the clinical strains of P. aeruginosa collected in this study had different RAPD patterns, indicating high genetic diversity amongst them. In addition, phylogenetic analysis of the RAPD profiles did not match the phage-susceptible effect (Fig. S1). The broad-spectrum of drug resistance and genetic diversity highlights CRPA as a serious problem in the clinical environment in Taiwan.
Table 1.
Host range of phiPA1-3 against P. aeruginosa and other species.
| Bacterial Strains | Antibiotic Resistance Profilea | EOP (% ± SD)b |
|---|---|---|
| P. aeruginosa | ||
| PA001 | CRPA | 100±2.16 |
| LCL12 | CRPA | – |
| LCL13 | CRPA | – |
| LCL14 | CRPA | – |
| PS-1 | CRPA | – |
| PS-2 | CRPA | – |
| PS-3 | CRPA | – |
| PS-4 | CRPA | – |
| PS-5 | CRPA | – |
| PS-6 | CRPA | – |
| PS-7 | CRPA | – |
| PA10 | CRPA | – |
| PA13 | CRPA | – |
| PA18 | CRPA | – |
| PA20 | CRPA | 0.67±6.84 |
| PA22 | CRPA | – |
| PA25 | CRPA | – |
| PA75 | CRPA | 0.001±0.47 |
| PA76 | CRPA | – |
| PA77 | CRPA | – |
| PA78 | CRPA | – |
| PA79 | CRPA | – |
| PA80 | CSPA | – |
| PA81 | CRPA | – |
| PA82 | CSPA | – |
| PA83 | CSPA | – |
| PA84 | CRPA | – |
| PA85 | CRPA | – |
| PA86 | CRPA | – |
| PA87 | CSPA | – |
| PA88 | CRPA | – |
| PA89 | CSPA | – |
| PA90 | CSPA | – |
| PA91 | CRPA | – |
| PA92 | CSPA | – |
| PA002 | CSPA | 35.41±1.88 |
| PA005 | CRPA | – |
| PA006 | CSPA | – |
| PA009 | CSPA | 16.56±5.35 |
| PA010 | CRPA | – |
| PA011 | CRPA | – |
| PA022 | CSPA | 0.01±2.16 |
| PA023 | CSPA | 0.008±3.29 |
| PA024 | CRPA | 0.0008±5.43 |
| PA025 | CSPA | 178.12±9.20 |
| K. pneumoniae | ||
| 98,035 | ND | – |
| 95,509 | ND | – |
| 96,111 | ND | – |
| E. coli | ||
| 78,030 | MDR | – |
| 70,751 | MDR | – |
| E. cloacae | ||
| 57,546 | CRE | – |
| 48,742 | CRE | – |
| 55,269 | CRE | – |
aCRPA, carbapenem-resistant P. aeruginosa; CSPA, carbapenem-sensitive P. aeruginosa; ND, not detected; MDR, multidrug-resistant; CRE, carbapenem-resistant E. cloacae.
bEOP: efficiency of plating, -: could not be infected with phiPA1-3.
3.2. Plaque and phage morphology of phiPA1-3
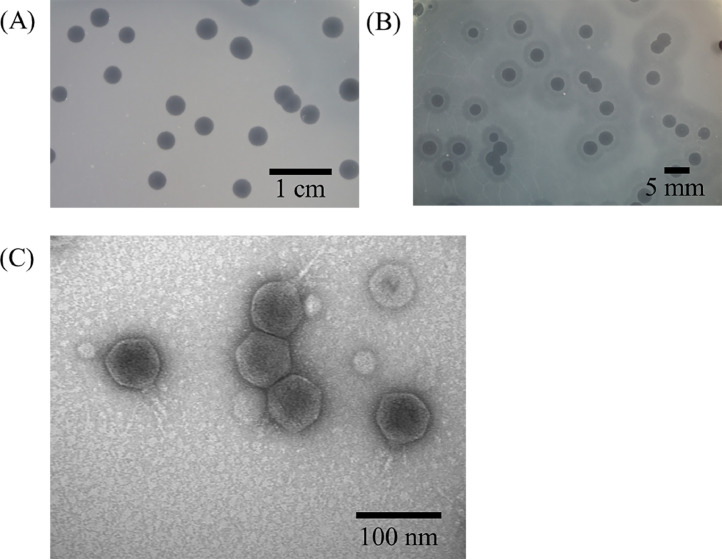
The clinically isolated strain, PA001 (CRPA), was used as an indicator host to isolate phages from wastewater samples collected near Tzu Chi Hospital in Hualien, Taiwan. One phage, phiPA1-3, was isolated by means of spot-testing against P. aeruginosa PA001 using an enrichment method. On double-layer agar plates, phiPA1-3 formed large and clear plaques with an average diameter of 3.5±0.5 mm (n=20), and an expanding halo around the plaques during storage at RT, while the lysis zones remained constant (Fig. 1A and B).
Fig. 1.
The plaque and phage morphology of phiPA1-3. When the phage was incubated with P. aeruginosa strain PA001, clear plaques with an expanding halo were observed (A). Pure phage plaques were visible on the first day, and the halo around the plaques expanded over time at room temperature (B). Examination of the phage morphology using transmission electron microscopy revealed a head diameter of 65.7 ± 2.6 nm, tail length of 47.3 ± 2.7 nm, and small appendages at the end of the tail (C).
To characterize the phage, phiPA1-3 was repeatedly purified by means of the plaque assay, and a stock of 1012 PFU/mL was prepared and subjected to ultracentrifugation in a CsCl gradient. To determine the morphology of the phages, a CsCl-purified phage sample was negatively stained and observed under a transmission electron microscope. The electron micrograph showed that the phage had a head diameter of 65.7±2.6 nm and tail length of 47.3±2.7 nm, in addition to small appendages at the tail (Fig. 1C). Based on the short, non-contractile tail observed in the TEM images, phiPA1-3 was classified as a member of the Caudoviricetes according to the latest version of International Committee on Taxonomy of Viruses (Turner et al., 2023)
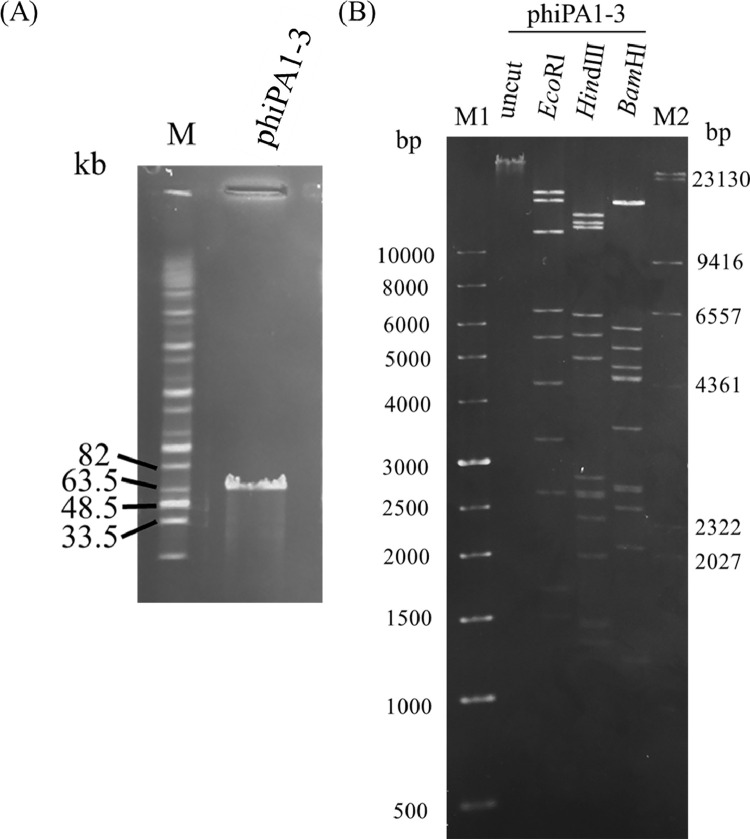
3.3. Restriction analysis of phage DNA
Pulsed-field gel electrophoresis performed to estimate the size of the phage genome revealed a size of approximately 70 kb (Fig. 2A). When EcoRI, HindIII, and BamHI were used to digest the DNA, multiple bands were observed on a resolved gel (Fig. 2B), which proved that phiPA1-3 is a double-stranded DNA phage that is digestible by restriction enzymes.
Fig. 2.
Restriction analysis of phage genomic DNA. (A) Uncleaved phage DNA. Lane M: Midrange PFG marker; (B) Restriction enzyme-digested fragments of the phage phiPA1-3. Lane M1: 1 Kb DNA ladder; M2: λ/HindIII marker; uncut: uncleaved phiPA1-3 DNA; the lanes labeled EcoRI, HindIII, and BamHI represent restriction fragments that have been cleaved by the respective enzymes.
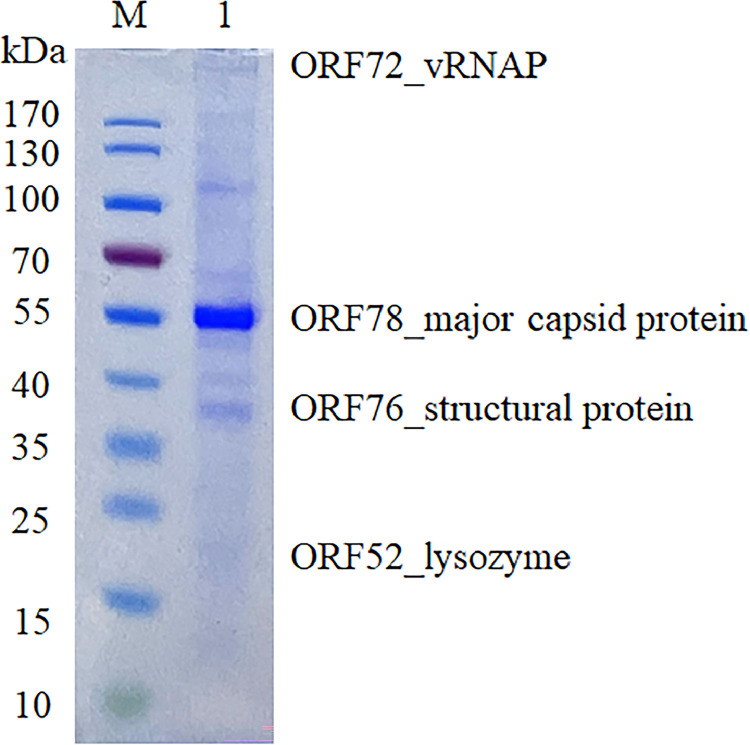
3.4. Structural proteins profile of phage phiPA1-3
To confirm the structural proteins of phiPA1-3, 1.7 × 1010 PFU of CsCl-purified particles were subjected to SDS-PAGE separation, which presented at least eight distinct protein bands with molecular weights ranging from 35 to 170 kDa upon visualization by staining the gels with Coomassie Brilliant Blue (Fig. 3). The most abundant protein band in the gel was approximately 55 kDa, presumed to be the major capsid protein of phiPA1-3. A band with a molecular weight greater than 170 kDa was observed near the well. Liquid chromatography-tandem mass spectroscopy identified all visible bands, and only four bands matched the annotated proteins in the phiPA1-3 genome (shown next to the gel in Fig. 3), one of which was the major coat protein, as expected. In addition to the identified proteins, some bands were heavily contaminated with capsid proteins and were difficult to identify.
Fig. 3.
SDS-PAGE analysis of the structural proteins of phipA1-3. Proteins were visualized on a 4%–20% (w/v) gradient gel and identified by means of LC-MS/MS analysis. The identified proteins have been indicated.
3.5. Host spectrum determination and EOP analysis
Among the 45 clinically isolated P. aeruginosa strains, nine, including host PA001, were sensitive to phiPA1-3, indicating a moderate host range (20%) for this phage. Notably, phiPA1-3 could infect carbapenem-resistant as well as -sensitive P. aeruginosa strains. Four of the nine phage-sensitive strains were CRPA. Some phage-sensitive strains had higher EOP than the host strain PA001 (Table S1). This indicates highly efficient lytic activity against susceptible strains. Interestingly, when phiPA1-3 was infected with different susceptible host strains, the plaques displayed different sizes and morphologies. Some particles were large, small, and clear, whereas others were small and turbid (Fig. S2). To confirm that these plaques were indeed caused by phiPA1-3 infection, we used primers specific to phiPA1-3 for PCR. All plaques had detectable phiPA1-3 amplicons after gel electrophoresis, indicating that this phage formed different plaque morphologies due to infection with different susceptible strains. No infectivity with other species of gram-negative bacteria was detected, suggesting that phiPA1-3 is specific to certain P. aeruginosa.
3.6. Analysis of the biological properties and stability of phiPA1-3
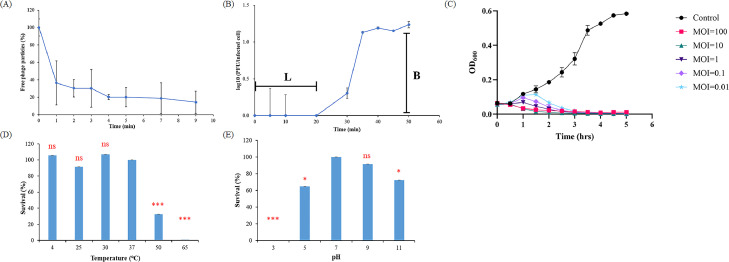
Adsorption analysis (Fig. 4A) showed that more than 60% of the phage absorbed to the host within 1 min, while more than 90% adsorbed within 10 min, at 37 °C, indicating that phiPA1-3 has a high adsorption efficiency to its host cell. One-step growth showed that phiPA1-3 has a latent phase of approximately 20 min and a burst size of 619 PFU/infected cell at 37 °C (Fig. 4B). The bacteriolytic activity of the phage was monitored at different MOIs using PA001 as the host (Fig. 4C). The absorbance of the phage-containing culture gradually increased during the first hour and then decreased significantly from 1.5 h to approximately 5 h, in an MOI-dependent manner, indicating that the host cells were killed. In addition, lower MOIs took longer to achieve bactericidal effects than higher MOIs. Inoculation with an MOI of 0.01 was sufficient to prevent host growth. These findings suggested that the phage effectively prevents bacterial growth and may help control CRPA infections.
Fig. 4.
Biological properties and stability of phage phiPA1-3. (A) Adsorption assays of phiPA1-3 with host PA001. (B) One-step growth curve of phiPA1-3 in the culture broth of PA001. (C) Lysis kinetics of phiPA1-3 on host PA001, at MOIs of 0.01, 0.1, 1, 10, and 100. Phages were incubated for 1 h under different temperatures (D) and pH levels (E), after which the stability of phiPA1-3 was determined in terms of plaque-formation on host lawns. Changes in PFUs were calculated relative to the control. The results are represented as mean ± standard deviation of triplicate experiments. MOI, multiplicity of infection.
In the phage thermal-stability experiment, the phage suspension was incubated at six different temperatures (4 °C, 25 °C, 30 °C, 37 °C, 50 °C, and 65 °C) for 1 h, and the phage titer of the 4 °C group was used as the control. These results demonstrated that phiPA1-3 is stable at temperatures below 37 °C. However, the titer of the phages was significantly reduced after 1 h of exposure at 50 °C, while the phage almost completely lost infectivity activity after 1 h of exposure at 60 °C (Fig. 4D).
For the pH stability experiment, phage phiPA1-3 was incubated in buffers with different pH values ranging from 3 to 11, for 1 h. phiPA1-3 was found to be the most stable at pH levels of 7.0 and 9.0, and no significant loss in titer was observed. However, phage viability was significantly reduced below pH 5.0 and beyond pH 9.0. In particular, the phage almost completely lost its infectivity at pH 3.0 (Fig. 4E). Stability analysis showed that storage of phiPA1-3 below 37 °C and in buffers with pH between 7 and 9 can maintain its optimal infectious activity.
3.7. Anti-biofilm activity of phiPA1-3
Biofilms comprise complex bacterial communities. P. aeruginosa naturally forms strong biofilms that are extremely difficult to eliminate. To assess the anti-biofilm activity of phiPA1-3, we first observed the kinetics of biofilm-formation by P. aeruginosa PA001 on coverslips in 6-well plates. Formation of mature biofilms was observed after 6 h, which covered the entire slide. Moreover. the biofilms became denser and more enriched over time (Fig. S3). Therefore, we selected mature biofilms after 6 h of culture as the target for co-culturing phages with bacteria, to prevent biofilm-formation and eradication assays for preformed biofilms.
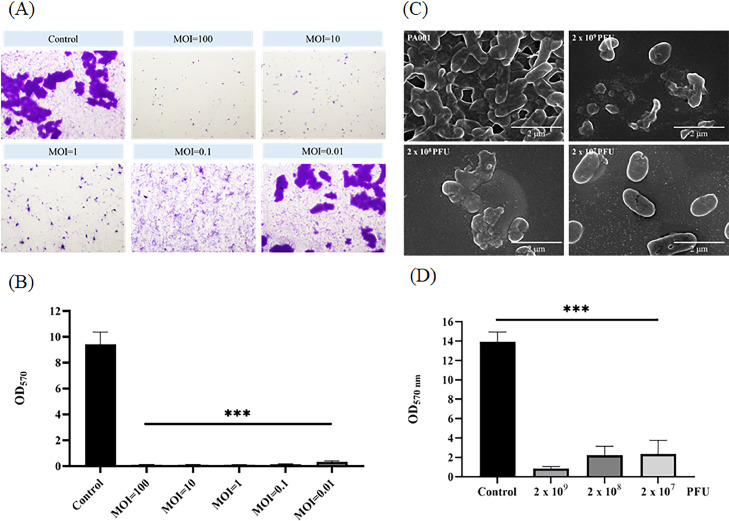
For the co-culture assay, P. aeruginosa PA001 was cultivated in 6-well plates and mixed with different MOIs of phiPA1-3 at the same time as the host, for 6 h. The MOI levels were 0.01, 0.1, 1, 10, and 100, and the control group was not treated with the phages. At a high dose of phages (MOI=100), there was almost no visible biofilm on the surface of the slide; at an MOI lower than 0.01, the results were comparable to those of the control group; and at an MOI of 0.1, only a thin biofilm layer was formed. This suggests that when a higher MOI (>0.1) was applied, there was significant prevention of biofilm-formation by phiPA1-3 on the glass surface (Fig. 5A).
Fig. 5.
Anti-biofilm activity measurement. (A) Co-culturing of phage and bacteria for biofilm-formation inhibition assay. P. aeruginosa PA001 was cultivated on 6-well plates and mixed with different multiplicities of infection (0.01, 0.1, 1, 10, and 100) of phiPA1-3 for 6 h. The control was no phage added. Each experiment was performed in triplicate. (B) Quantification of the results for the assay in (A). (C) Scanning electron micrography of the biofilm-eradication test of phiPA1-3. PA001 was cultured on a 6-well plate for 6 h to obtain a mature biofilm, following which varying amounts of phiPA1-3 (2 × 109, 2 × 108, and 2 × 107 PFU) were added, and the samples were cultured for an additional 18 h. (D) Quantification of biofilm-eradication test; all means were calculated from three independent measurements. “***” indicates p<0.001.
To quantify the prevention efficiency of phiPA1-3, we used a 96-well culture plate and co-cultured the PA001 host with different MOI levels of phiPA1-3. All MOIs effectively inhibited biofilm-formation (Fig. 5B).
To perform a biofilm eradication assay, we grew PA001 on a 6-well plate for 6 h, to obtain a mature biofilm. Following that, varying amounts of phiPA1-3 (2 × 109, 2 × 108, and 2 × 107 PFU) were added, and the samples were cultured for an additional 18 h. After 18 h, the samples were prepared for scanning electron microscopy (SEM) observation, which showed that high doses of phage treatment significantly inhibited biofilm formation (Fig. 5C).
To quantify the remaining biofilm after phiPA1-3 treatment, we grew a 6 h biofilm and followed the method described above. Upon quantification, it was found that all MOI levels exhibited a significant difference compared to the control group without the addition of phages (Fig. 5D).
3.8. phiPA1-3 genome – general features
We chose third-generation sequencing for the DNA sequence of phage phiPA1-3, manually curated using primers 19F and 19R (corresponding to the ORF19 region), 75F (corresponding to the ORF75 region), and 81F (corresponding to the ORF81 region), to finally obtain the complete sequence. The genome size of phiPA1-3 was 73,402 bp and G+C content was 54.7%, which was lower than that of the host P. aeruginosa (ca. 66%). This discrepancy in G+C content between P. aeruginosa and its phages has also been reported (Yang et al., 2015). ARAGORN analysis of the phage genome revealed that it did not contain tRNA or tmRNA, suggesting that the phage adapted to the host without requiring tRNA molecules. VirulenceFinder 2.0 and ResFinder revealed that phiPA1-3 does not carry any antibiotic resistance or virulence factor genes. PhageLead analysis revealed no genes related to temperate phages, thereby suggesting that phiPA1-3 is a lytic phage. Genome BLASTN comparison revealed that phiPA1-3 had the highest sequence similarity (96.3%) to Pseudomonas phage YH6 (accession number: KM974184), with 98% coverage.
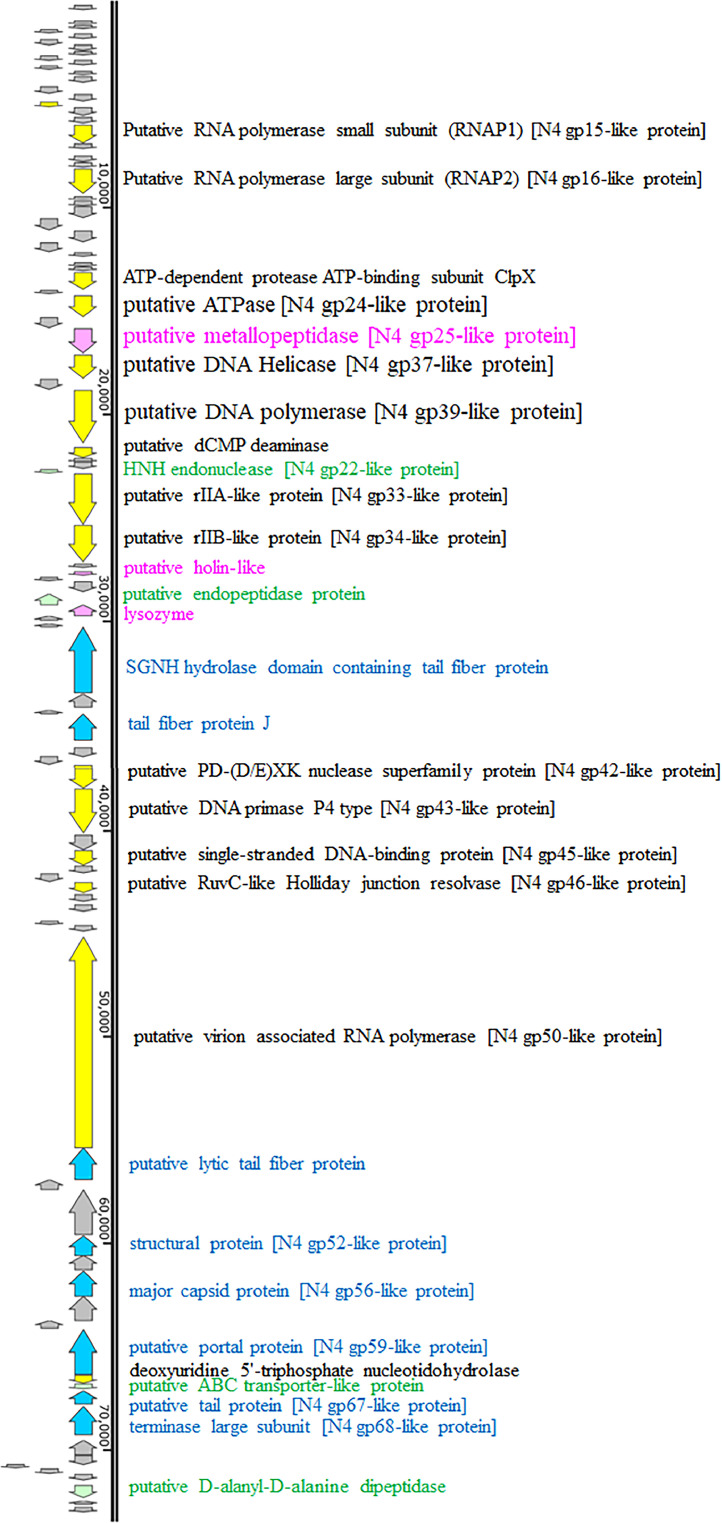
RAST analysis of the phiPA1-3 genome predicted that it contains 93 ORFs. The sequence displayed repeat regions at both ends, spanning 605 and 606 bp, with a 99.5% sequence similarity between the two regions. Of the 93 ORFs, 62 were annotated as hypothetical proteins, whereas the remaining 31 had known functions (Fig. 6 and Table S2). Proteins with known functions were categorized into the following four groups: DNA/RNA processing and metabolism, structural and assembly proteins, lysis proteins, and unclassified proteins. Proteins involved in DNA/RNA processing and metabolism were recognized by their domain signature, as shown in Table S2; these included ORF16 (transcription regulator), ORF19 and ORF23 (putative RNA polymerase subunits), ORF33 (ATP-dependent protease subunit), ORF35 (putative ATPase), ORF37 (putative metallopeptidase), ORF38 (putative helicase), ORF40 (putative DNA polymerase), ORF41 (putative dCMP deaminase), ORF44 (HNH endonuclease), ORF45 and ORF46 (putative rIIA- and rIIB-like proteins, respectively), ORF61 (putative nuclease), ORF62 (putative DNA primase), ORF64 (putative single-stranded DNA-binding protein), ORF72 (putative virion-associated RNA polymerase), and ORF82 (deoxyuridine 5′-triphosphate nucleotidohydrolase).
Fig. 6.
A schematic genome map of phiPA1-3 generated by SnapGene Viewer. The ORFs with known functions are shown as colored blocks and annotations. The gray-colored blocks represent hypothetical proteins. The genome map was created using SnapGene Viewer (www.snapgene.com) and subsequently modified manually.
Eight ORFs belonged to the structure and assembly protein categories; these included ORF55 (tail fiber protein), ORF58 (tail fiber protein J), ORF73 (lytic tail fiber protein), ORF78 (major capsid protein), ORF81 (portal protein), ORF84 (tail protein), and ORF85 (terminase large subunit). Their classification as structural and assembled proteins was based on the specific domain blocks (Table S2). Additionally, 3 ORFs were classified as lysis proteins, including ORF48 (holin-like protein), ORF51 (endopeptidase protein), and ORF52 (lysozyme). The remaining three ORFs had unclassified functions: ORF67 (RuvC-like Holliday junction resolvase), ORF83 (ABC transporter-like protein), and ORF91 (d-alanyl-d-alanine dipeptidase).
Based on the similarity of the ORFs (Table S2), it can be concluded that phage phiPA1-3 belongs to the N4-like phage family since the genome contains several protein products that are similar to those found in N4-like phages. It carries three types of RNA polymerases, with small and large subunits responsible for transcribing middle mRNAs, and the virion-associated RNA polymerase responsible for transcribing early mRNAs. Once injected into the host DNA, N4-like phages inhibit the host RNA polymerase, thereby providing the necessary RNA polymerase for transcription (Lenneman and Rothman-Denes, 2015).
3.9. Comparative genomics and phylogenetic analysis
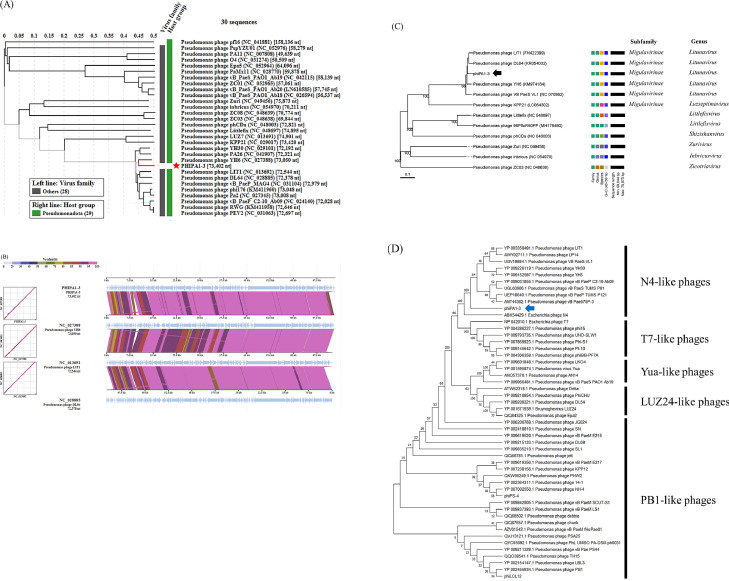
BLASTN showed that the genome of Pseudomonas phage YH6 was most closely related to that of phiPA1-3. Interestingly, although this phage was isolated from sewage, it infected minks, resulting in murine hemorrhagic pneumonia (Yang et al., 2015). Notably, YH6 can also infect the human clinical reference strain ATCC27853. To trace the evolutionary relationship between phiPA1-3 and YH6, as well as, other related Pseudomonas phages, we performed a VIPTree analysis (Nishimura et al., 2017) based on the whole-proteome and constructed an evolutionary tree. We first compared phiPA1-3 with all reference phages in the database, and found that phiPA1-3 clustered with YH6 and several Pseudomonas phages belonging to Schitoviridae (Fig. 7A), all of which displayed similar genome arrangements, except that both ends showed a mild degree of dissimilarity and small ORFs arrangement. The representative phages are shown in Fig. 7B.
Fig. 7.
Genome comparison and phylogenetic tree analysis of phiPA1-3 with related phages. (A) The tree was constructed using VIPTree, by uploading phiPA1-3 genome sequence, and compared with all references in the database, phiPA1-3 was clustered with related Pseudomonas phages, the selection filter in Host group set with “Pseudomonadota”. (B) Alignment of selected genomes similar to phiPA1-3. Colored blocks show% identity of sequence. The comparison was carried out using VIPTree (https://www.genome.jp/viptree/). (C) The phylogenetic tree generated by VICTOR using the complete genomic sequence selected from various members of subfamilies and genera of Pseudomonas N4-like phages. (D) Phylogenetic tree of phage-encoded large terminase subunit (ORF85) of phiPA1-3. The amino acid sequences of the large terminase subunits were compared using MEGA11; the tree was generated using the neighbor-joining method and 1000 bootstrap replicates.
Genome annotation revealed that phiPA1-3 harbors N4-like homologs (Fig. 6), while comparative genomics demonstrated that it constantly clustered with Pseudomonas phages belonging to Schitoviridae. Various subfamilies and genera are included in the classification of N4-like phages (Wittmann et al., 2015). To determine the phylogenetic relationship between phiPA1-3 and other N4-like phages, we conducted a whole-proteome analysis using VICTOR (Meier-Kolthoff and Göker, 2017), which revealed that phiPA1-3 clustered under the Migulavirinae and Litunavirus genera of N4-like phages (Fig. 7C). The terminal large subunit is the main protein involved in phage assembly and an excellent marker for tracing the evolutionary relationship between phages (Casjens et al., 2005; Fouts et al., 2013). We used MEGA11 to analyze the amino acid sequences of the terminal large subunit of various P. aeruginosa, including N4-like, T7-like, LUZ24-like, and PB1-like phages, to reveal the phylogenetic relationships between phiPA1-3 and other phages. These results indicated that the assembly system of phiPA1-3 belongs to N4-like phages (Fig. 7D). Genomic analysis and assembly classification confirmed that phiPA1-3 is a novel N4-like phage.
3.10. Phage rescue potential in zebrafish model
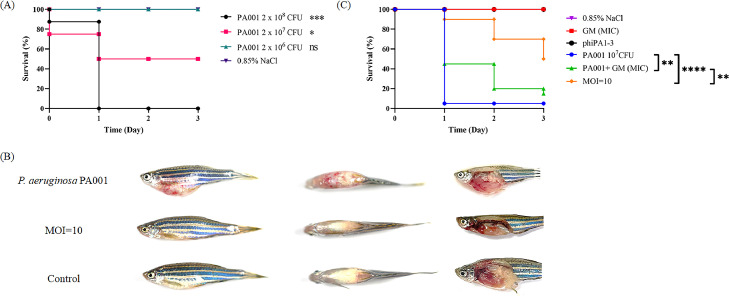
The therapeutic potential of phiPA1-3 was evaluated in the P. aeruginosa PA001-infected zebrafish model. In the bacterial infection group, when each fish was injected with 2 × 108 CFU of P. aeruginosa PA001 (n=8), all died within 24 h, and 2 × 107 CFU per fish reached the Lethal Dose 50 (LD50) after 24 h (Fig. 8A). In addition, fish that died in both the groups showed petechial hemorrhages on the ventral surface, with the postmortem results showing that the infected fish had typical signs of sepsis, such as hepatic congestion, an enlarged spleen, and accumulation of blood plasma in the abdominal cavity (Fig. 8B). However, after injecting 106 CFU of PA001 per fish, no fish died within 24 h, but tiny petechial hemorrhage symptoms persisted in the abdomen.
Fig. 8.
PhiPA1-3 rescued the P. aeruginosa PA001-infected zebrafish. (A) Assessment of lethal dosage; three groups were considered, each containing eight fish. These were injected with PA001 cells (2 × 108, black line; 2 × 107, red line; 2 × 106, green line; CFU/20 µL, respectively); (B) The disease symptoms of zebrafish infected with PA001, as compared to those of the phage-rescued group (MOI=10) and control (0.85% NaCl). P. aeruginosa PA001 was introduced first into the fish, followed by administration of phiPA1-3 (MOI=10) after 30 min. Left: side view; middle: top view; right: abdomen anatomy; (C) The survival rates of PA001-infected zebrafish were measured under different treatments: MIC-gentamicin (4 µg/mL, indicated by the green line), MIC-gentamicin alone (red line), phiPA1-3 treatment (MOI=10) after a 30 min interval following bacterial challenge (orange line), PA001 treatment alone (2 × 107 CFU/20 µL, blue line), phiPA1-3 injection alone (black line), and 0.85% NaCl buffer as the negative control (purple line) The X-axis represents the time post-infection, while the Y-axis represents the survival percentage of the zebrafish. The significance of the differences between groups was assessed by means of the log-rank and Gehan–Breslow–Wilcoxon tests using GraphPad Prism 9. “****” indicates p<0.0001, “**” indicates p<0.001, and ns means no significance. MOI, multiplicity of infection.
We conducted further experiments utilizing MIC-gentamicin (gentamicin at the minimal inhibitory concentration) and phage phiPA1-3 for the treatment of bacterial-infected zebrafish. Notably, zebrafish treated with MIC-gentamicin (Fig. 8C, red line) exhibited results similar to the control group treated with 0.85% NaCl (Fig. 8C, purple line). However, when zebrafish were infected with PA001 and subsequently treated with gentamicin (Fig. 8C, green line), there was an initial increase in survival at day 1. Unfortunately, all zebrafish succumbed to the infection by day 3, aligning with PA001 infection alone (Fig. 8C, blue line). In contrast, treatment of PA001-infected zebrafish with phage phiPA1-3 at an MOI of 10 (Fig. 8C, orange line) significantly improved efficacy. The survival rate increased from day 1 (90%) to day 3 (50%) (Fig. 8C), demonstrating the efficacy of phage treatment in promoting survival without any signs of infection (Fig. 8B). No effect was observed when the phage suspension (1.75 × 108 PFU/fish) was administered to uninfected fish (Fig. 8C, black line).
4. Discussion
P. aeruginosa, one of the six bacterial pathogens in the ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species), is commonly found in diverse environments. Most of these are MDR isolates, which pose greatest challenge in clinical practice. In our surveillance study, 45 clinical isolates were collected from different clinical sources, of which 32 were CRPA (Table S1). RAPD combined with phage sensitivity demonstrated genetic diversity (Fig. S1). Drug resistance in P. aeruginosa involves sophisticated systems (Bitar et al., 2022; Lepe and Martínez-Martínez, 2022; Lorusso et al., 2022) and regulations (Liu et al., 2022a, 2022b; Sikdar and Elias, 2022). However, exploring this detailed mechanism is time-consuming and difficult. Bacteriophages or phages combined with antibiotics have recently gained attention as substitutes for treating drug-resistant bacteria (Qin et al., 2022; Racenis et al., 2022), compared to global Pseudomonas phages, phages isolated from Taiwan against local P. aeruginosa isolates are relatively limited. Therefore, we isolated and characterized phiPA1-3 for further applications. Phage phiPA1-3 forms large plaques with an expanding halo zone surrounding the plaques. The halo zone has been proven to be a depolymerase in many studies (Cornelissen et al., 2012; Li et al., 2022; Wu et al., 2019) and its diversity and potential applications have been reviewed elsewhere (Knecht et al., 2019; Pires et al., 2016). Considering the safety of phage usage, phage-based therapeutics (e.g., depolymerases) have also been evaluated (Tan et al., 2022). Therefore, gene identification and expression analyses may be useful. In the phiPA1-3 genome, we found a tail fiber product containing the SGNH hydrolase domain, which is a good candidate for further study. phiPA1-3 has a larger burst size (619 PFU/infected cell) than those of YH6 (approximately 100 phages per cell) and VL1 (approximately 404 phages per cell) (Lerdsittikul et al., 2022), a newly published Pseudomonas phage similar to YH6. A large burst size has the potential to increase the efficiency of phage therapy and lower the possibility of emergence of phage-resistant bacteria (Lerdsittikul et al., 2022). Apart from the large burst size of phiPA1-3, its widely adapted stability and bacterial killing (Fig. 4) also reveal its effectiveness as an alternative therapeutic agent. Although the host range is limited, the plating efficiency is higher than that of the propagating host PA001. This approach appears to have the potential to be applied to more susceptible strains. Interestingly, when phiPA1-3 infected different P. aeruginosa, it displayed different plaque morphologies (Fig. S2). It is known that plaque morphology is affected by burse size and media (Pan et al., 2021; Ramesh et al., 2019); however, whether these factors contribute to the plaque morphology requires further investigation.
Genome analysis reveals that phiPA1-3 contains 93 ORFs. Of the 93 annotated ORFs, 62 belong to unknown functions. Fortunately, ResFinder (Bortolaia et al., 2020) and VirulenceFinder (Kleinheinz et al., 2014) did not predict any harmful or antibiotic resistance genes, which increases the possibility of their application. Comparative genomic analysis showed different arrangements at both ends of the phiPA1-3-related phages (Fig. 7B). These areas contain many small ORFs, which are mostly annotated as hypothetical proteins. Phage-based small peptides are usually unknown proteins that have been shown to inhibit the growth of bacterial hosts (Glover, 2020; Liu et al., 2004; Shibayama and Dabbs, 2011). At the right end of phiPA1-3, some small ORFs differed from those of YH6 and related phages. A whole-genome approach has been used to explore the functionality of the unknown small proteins. This is a promising direction for an alternative antibacterial approach (Mohanraj et al., 2019).
Our study on anti-biofilm and zebrafish rescue from bacterial infections demonstrated that the functionality of phiPA1-3. P. aeruginosa has the potential to form biofilms with diverse textures and surfaces (Coffey and Anderson, 2014; Lee and Yoon, 2017; Wei and Ma, 2013), which helps initiate the early steps of infection (Chegini et al., 2020). Our results demonstrated that phiPA1-3 prevented the attached bacteria in the co-culture assay (Fig. 5A) and helped eradicate the attached biofilm (Fig. 5B). Notably, the prevention and eradication functions were observed in glass (Fig. 5A and B) or plastic materials (Fig. 5C and D). This suggests that phiPA1-3 has broad-spectrum anti-biofilm activity and can effectively inhibit biofilm-formation on different surfaces.
In addition, in vivo experiments using zebrafish infection models showed that treatment with phiPA1-3 significantly reduced the bacterial burden and increased survival rates (Fig. 8). These results suggest that phiPA1-3 has potential to serve as a therapeutic agent for treating P. aeruginosa infections. However, there is a need for further studies to optimize these conditions and evaluate their safety and efficacy in clinical trials.
5. Conclusions
In summary, in this study, we isolated a novel lytic phage targeting CRPA and determined its biological properties. The ability of phiPA1-3 to prevent/eradicate biofilms and rescue bacterial infections in zebrafish highlights its potential as an alternative agent in clinical settings. Using genome analysis, we successfully classified it as an N4-like member and placed it under the Schitoviridae, Migulavirinae subfamily, and Litunavirus genus. This study provides essential information for further understanding of the intricate interactions between phages and their hosts, as well as, for potential clinical applications.
CRediT authorship contribution statement
Yu-Chuan Tsai: Formal analysis, Data curation, Validation, Writing – review & editing. Yi-Pang Lee: Formal analysis, Data curation, Validation, Funding acquisition, Conceptualization, Project administration. Nien-Tsung Lin: Formal analysis, Data curation, Validation. Hsueh-Hui Yang: . Soon-Hian Teh: Funding acquisition, Conceptualization, Writing – review & editing, Project administration. Ling-Chun Lin: Funding acquisition, Supervision, Writing – review & editing, Project administration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
We thank the Electron Microscopy Laboratory of Tzu Chi University for their technical assistance, and the mass spectrometry provided by Advanced Instrumentation Center of Department of Medical Research, Hualien Tzu Chi Hospital, Buddhist Tzu Chi Medical Foundation and Bioinnovation Center, Buddhist Tzu Chi Medical Foundation, Hualien, Taiwan.
Funding sources
This work was supported by the Hualien Tzu Chi Hospital, Buddhist Tzu Chi Medical Foundation (grant nos. TCRD111-060 and TCRD111-037) and TCMRC-P-111005 from Tzu Chi University.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2023.199178.
Contributor Information
Soon-Hian Teh, Email: jimmyteh2000@gmail.com.
Ling-Chun Lin, Email: aphrodite@gms.tcu.edu.tw.
Appendix. Supplementary materials
Data availability
Data will be made available on request.
References
- Adriaenssens E., Brister J.R. How to name and classify your phage: an informal guide. Viruses. 2017;9(4):70. doi: 10.3390/v9040070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitar I., Salloum T., Merhi G., Hrabak J., Araj G.F., Tokajian S. Genomic characterization of mutli-drug resistant pseudomonas aeruginosa clinical isolates: evaluation and determination of ceftolozane/tazobactam activity and resistance mechanisms. Front. Cell Infect. Microbiol. 2022;12 doi: 10.3389/fcimb.2022.922976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette C.M., Noone J.M., Stone G., Zacherle E., Patel R.P., Howden R., Mapel D. Healthcare cost and utilization before and after diagnosis of pseudomonas aeruginosa among patients with non-cystic fibrosis bronchiectasis in the U.S. Med. Sci. (Basel) 2017;5(4):20. doi: 10.3390/medsci5040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolaia V., Kaas R.S., Ruppe E., Roberts M.C., Schwarz S., Cattoir V., Philippon A., Allesoe R.L., Rebelo A.R., Florensa A.F., Fagelhauer L., Chakraborty T., Neumann B., Werner G., Bender J.K., Stingl K., Nguyen M., Coppens J., Xavier B.B., Malhotra-Kumar S., Westh H., Pinholt M., Anjum M.F., Duggett N.A., Kempf I., Nykäsenoja S., Olkkola S., Wieczorek K., Amaro A., Clemente L., Mossong J., Losch S., Ragimbeau C., Lund O., Aarestrup F.M. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020;75(12):3491–3500. doi: 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S.R., Gilcrease E.B., Winn-Stapley D.A., Schicklmaier P., Schmieger H., Pedulla M.L., Ford M.E., Houtz J.M., Hatfull G.F., Hendrix R.W. The generalized transducing Salmonella bacteriophage ES18: complete genome sequence and DNA packaging strategy. J. Bacteriol. 2005;187(3):1091–1104. doi: 10.1128/JB.187.3.1091-1104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P.P., Lowe T.M. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol. Biol. 2019;1962:1–14. doi: 10.1007/978-1-4939-9173-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chegini Z., Khoshbayan A., Taati Moghadam M., Farahani I., Jazireian P., Shariati A. Bacteriophage therapy against Pseudomonas aeruginosa biofilms: a review. Ann. Clin. Microbiol. Antimicrob. 2020;19(1):45. doi: 10.1186/s12941-020-00389-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey B.M., Anderson G.G. Biofilm formation in the 96-well microtiter plate. Methods Mol. Biol. 2014;1149:631–641. doi: 10.1007/978-1-4939-0473-0_48. [DOI] [PubMed] [Google Scholar]
- Cornelissen A., Ceyssens P.J., Krylov V.N., Noben J.P., Volckaert G., Lavigne R. Identification of EPS-degrading activity within the tail spikes of the novel Pseudomonas putida phage AF. Virology. 2012;434(2):251–256. doi: 10.1016/j.virol.2012.09.030. [DOI] [PubMed] [Google Scholar]
- De Coster W., D'Hert S., Schultz D.T., Cruts M., Van Broeckhoven C. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics. 2018;34(15):2666–2669. doi: 10.1093/bioinformatics/bty149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts D.E., Klumpp J., Bishop-Lilly K.A., Rajavel M., Willner K.M., Butani A., Henry M., Biswas B., Li M., Albert M.J., Loessner M.J., Calendar R., Sozhamannan S. Whole genome sequencing and comparative genomic analyses of two Vibrio cholerae O139 Bengal-specific Podoviruses to other N4-like phages reveal extensive genetic diversity. Virol. J. 2013;10:165. doi: 10.1186/1743-422X-10-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover L. mSphere of influence: expanding the CRISPR sphere with single-locus proteomics. mSphere. 2020;5(1):e00001–20. doi: 10.1128/mSphere.00001-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29(8):1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth M., Kovács T., Koderivalappil S., Ábrahám H., Rákhely G., Schneider G. Identification of a newly isolated lytic bacteriophage against K24 capsular type, carbapenem resistant Klebsiella pneumoniae isolates. Sci. Rep. 2020;10(1):5891. doi: 10.1038/s41598-020-62691-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.T., Liu P.Y., Shih P.W. Homopolish: a method for the removal of systematic errors in nanopore sequencing by homologous polishing. Genome Biol. 2021;22(1):95. doi: 10.1186/s13059-021-02282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jault P., Leclerc T., Jennes S., Pirnay J.P., Que Y.A., Resch G., Rousseau A.F., Ravat F., Carsin H., Le Floch R., Schaal J.V., Soler C., Fevre C., Arnaud I., Bretaudeau L., Gabard J. Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): a randomised, controlled, double-blind phase 1/2 trial. Lancet Infect. Dis. 2019;19(1):35–45. doi: 10.1016/S1473-3099(18)30482-1. [DOI] [PubMed] [Google Scholar]
- Jung S.H., Lee H.Y., Chow S.C. Statistical methods for conditional survival analysis. J. Biopharm. Stat. 2018;28(5):927–938. doi: 10.1080/10543406.2017.1405012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinheinz K.A., Joensen K.G., Larsen M.V. Applying the ResFinder and VirulenceFinder web-services for easy identification of acquired antibiotic resistance and E. coli virulence genes in bacteriophage and prophage nucleotide sequences. Bacteriophage. 2014;4(1):e27943. doi: 10.4161/bact.27943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knecht L.E., Veljkovic M., Fieseler L. Diversity and function of phage encoded depolymerases. Front. Microbiol. 2019;10:2949. doi: 10.3389/fmicb.2019.02949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S., Walenz B.P., Berlin K., Miller J.R., Bergman N.H., Phillippy A.M. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27(5):722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropinski A.M., Mazzocco A., Waddell T.E., Lingohr E., Johnson R.P. Enumeration of bacteriophages by double agar overlay plaque assay. Methods Mol. Biol. 2009;501:69–76. doi: 10.1007/978-1-60327-164-6_7. [DOI] [PubMed] [Google Scholar]
- Laslett D., Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32(1):11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Yoon S.S. Pseudomonas aeruginosa biofilm, a programmed bacterial life for fitness. J. Microbiol. Biotechnol. 2017;27(6):1053–1064. doi: 10.4014/jmb.1611.11056. [DOI] [PubMed] [Google Scholar]
- Lenneman B.R., Rothman-Denes L.B. Structural and biochemical investigation of bacteriophage N4-encoded RNA polymerases. Biomolecules. 2015;5(2):647–667. doi: 10.3390/biom5020647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepe J.A., Martínez-Martínez L. Resistance mechanisms in Gram-negative bacteria. Med. Intensiva (Engl. Ed.) 2022;46(7):392–402. doi: 10.1016/j.medine.2022.05.004. [DOI] [PubMed] [Google Scholar]
- Lerdsittikul V., Thongdee M., Chaiwattanarungruengpaisan S., Atithep T., Apiratwarrasakul S., Withatanung P., Clokie M.R.J., Korbsrisate S. A novel virulent Litunavirus phage possesses therapeutic value against multidrug resistant Pseudomonas aeruginosa. Sci. Rep. 2022;12(1):21193. doi: 10.1038/s41598-022-25576-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Chang R.Y.K., Lin Y., Morales S., Kutter E., Chan H.K. Phage cocktail powder for Pseudomonas aeruginosa respiratory infections. Int. J. Pharm. 2021;596 doi: 10.1016/j.ijpharm.2021.120200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Wang H., Chen L., Guo G., Li P., Ma J., Chen R., Du H., Liu Y., Zhang W. Identification of a phage-derived depolymerase specific for KL47 capsule of Klebsiella pneumoniae and its therapeutic potential in mice. Virol. Sin. 2022;37(4):538–546. doi: 10.1016/j.virs.2022.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenberg M., Jakobsen T.H., Kühl M., Kolpen M., Jensen P., Bjarnsholt T. The structure-function relationship of Pseudomonas aeruginosa in infections and its influence on the microenvironment. FEMS Microbiol. Rev. 2022;46(5) doi: 10.1093/femsre/fuac018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin N.T., Chiou P.Y., Chang K.C., Chen L.K., Lai M.J. Isolation and characterization of phi AB2: a novel bacteriophage of Acinetobacter baumannii. Res. Microbiol. 2010;161(4):308–314. doi: 10.1016/j.resmic.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Liu J., Dehbi M., Moeck G., Arhin F., Bauda P., Bergeron D., Callejo M., Ferretti V., Ha N., Kwan T., McCarty J., Srikumar R., Williams D., Wu J.J., Gros P., Pelletier J., DuBow M. Antimicrobial drug discovery through bacteriophage genomics. Nat. Biotechnol. 2004;22(2):185–191. doi: 10.1038/nbt932. [DOI] [PubMed] [Google Scholar]
- Liu P., Yue C., Liu L., Gao C., Lyu Y., Deng S., Tian H., Jia X. The function of small RNA in Pseudomonas aeruginosa. PeerJ. 2022;10:e13738. doi: 10.7717/peerj.13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Sun W., Ma L., Xu R., Yang C., Xu P., Ma C., Gao C. Metabolic mechanism and physiological role of glycerol 3-phosphate in Pseudomonas aeruginosa PAO1. MBio. 2022;13(6) doi: 10.1128/mbio.02624-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorusso A.B., Carrara J.A., Barroso C.D.N., Tuon F.F., Faoro H. Role of efflux pumps on antimicrobial resistance in Pseudomonas aeruginosa. Int. J. Mol. Sci. 2022;23(24):15779. doi: 10.3390/ijms232415779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manni M., Berkeley M.R., Seppey M., Zdobnov E.M. BUSCO: assessing genomic data quality and beyond. Curr. Protoc. 2021;1(12):e323. doi: 10.1002/cpz1.323. [DOI] [PubMed] [Google Scholar]
- Meier-Kolthoff J.P., Göker M. VICTOR: genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics. 2017;33(21):3396–3404. doi: 10.1093/bioinformatics/btx440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaei M.K., Nilsson A.S. Isolation of phages for phage therapy: a comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLOS One. 2015;10(3) doi: 10.1371/journal.pone.0118557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanraj U., Wan X., Spruit C.M., Skurnik M., Pajunen M.I. A toxicity screening approach to identify bacteriophage-encoded anti-microbial proteins. Viruses. 2019;11(11):1057. doi: 10.3390/v11111057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales E., Cots F., Sala M., Comas M., Belvis F., Riu M., Salvadó M., Grau S., Horcajada J.P., Montero M.M., Castells X. Hospital costs of nosocomial multi-drug resistant Pseudomonas aeruginosa acquisition. BMC Health Serv. Res. 2012;12:122. doi: 10.1186/1472-6963-12-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y., Yoshida T., Kuronishi M., Uehara H., Ogata H., Goto S. ViPTree: the viral proteomic tree server. Bioinformatics. 2017;33(15):2379–2380. doi: 10.1093/bioinformatics/btx157. [DOI] [PubMed] [Google Scholar]
- Pan L., Li D., Sun Z., Lin W., Hong B., Qin W., Xu L., Liu W., Zhou Q., Wang F., Cai R., Qian M., Tong Y. First characterization of a hafnia phage reveals extraordinarily large burst size and unusual plaque polymorphism. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.754331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires D.P., Oliveira H., Melo L.D., Sillankorva S., Azeredo J. Bacteriophage-encoded depolymerases: their diversity and biotechnological applications. Appl. Microbiol. Biotechnol. 2016;100(5):2141–2151. doi: 10.1007/s00253-015-7247-0. [DOI] [PubMed] [Google Scholar]
- Qin S., Xiao W., Zhou C., Pu Q., Deng X., Lan L., Liang H., Song X., Wu M. Pseudomonas aeruginosa: pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal Transduct. Target Ther. 2022;7(1):199. doi: 10.1038/s41392-022-01056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racenis K., Rezevska D., Madelane M., Lavrinovics E., Djebara S., Petersons A., Kroica J. Use of phage cocktail bfc 1.10 in combination with Ceftazidime-Avibactam in the treatment of multidrug-resistant Pseudomonas aeruginosa femur osteomyelitis-a case report. Front. Med. (Lausanne) 2022;9 doi: 10.3389/fmed.2022.851310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh N., Archana L., Madurantakam Royam M., Manohar P., Eniyan K. Effect of various bacteriological media on the plaque morphology of Staphylococcus and Vibrio phages. Access Microbiol. 2019;1(4) doi: 10.1099/acmi.0.000036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K., Stoodley P., Goeres D.M., Hall-Stoodley L., Burmølle M., Stewart P.S., Bjarnsholt T. The biofilm life cycle: expanding the conceptual model of biofilm formation. Nat. Rev. Microbiol. 2022;20(10):608–620. doi: 10.1038/s41579-022-00767-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- Shahed-Al-Mahmud M., Roy R., Sugiokto F.G., Islam M.N., Lin M.D., Lin L.C., Lin N.T. Phage φAB6-borne depolymerase combats Acinetobacter baumannii biofilm formation and infection. Antibiotics (Basel) 2021;10(3):279. doi: 10.3390/antibiotics10030279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibayama Y., Dabbs E.R. Phage as a source of antibacterial genes: multiple inhibitory products encoded by Rhodococcus phage YF1. Bacteriophage. 2011;1(4):195–197. doi: 10.4161/bact.1.4.17746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikdar R., Elias M.H. Evidence for complex interplay between quorum sensing and antibiotic resistance in Pseudomonas aeruginosa. Microbiol. Spectr. 2022;10(6) doi: 10.1128/spectrum.01269-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M.J., Petty N.K., Beatson S.A. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27(7):1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D.L., Pulcini C., Kahlmeter G., Kluytmans J., Carmeli Y., Ouellette M., Outterson K., Patel J., Cavaleri M., Cox E.M., Houchens C.R., Grayson M.L., Hansen P., Singh N., Theuretzbacher U., Magrini N. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018;18(3):318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Kumar S. MEGA11: molecular evolutionary genetics analysis Version 11. Mol. Biol. Evol. 2021;38(7):3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., Su J., Fu M., Zhang H., Zeng H. Recent advances in phage-based therapeutics for multi-drug resistant Acinetobacter baumannii. Bioeng. (Basel) 2022;10(1):35. doi: 10.3390/bioengineering10010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolker-Nielsen T. Pseudomonas aeruginosa biofilm infections: from molecular biofilm biology to new treatment possibilities. APMIS Suppl. 2014;(138):1–51. doi: 10.1111/apm.12335. [DOI] [PubMed] [Google Scholar]
- Turner D., Shkoporov A.N., Lood C., Millard A.D., Dutilh B.E., Alfenas-Zerbini P., van Zyl L.J., Aziz R.K., Oksanen H.M., Poranen M.M., Kropinski A.M., Barylski J., Brister J.R., Chanisvili N., Edwards R.A., Enault F., Gillis A., Knezevic P., Krupovic M., Kurtböke I., Kushkina A., Lavigne R., Lehman S., Lobocka M., Moraru C., Moreno Switt A., Morozova V., Nakavuma J., Reyes Muñoz A., Rūmnieks J., Sarkar B.L., Sullivan M.B., Uchiyama J., Wittmann J., Yigang T., Adriaenssens E.M. Abolishment of morphology-based taxa and change to binomial species names: 2022 taxonomy update of the ICTV bacterial viruses subcommittee. Arch. Virol. 2023;168(2):74. doi: 10.1007/s00705-022-05694-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaitekenas A., Tai A.S., Ramsay J.P., Stick S.M., Kicic A. Pseudomonas aeruginosa resistance to bacteriophages and its prevention by strategic therapeutic cocktail formulation. Antibiotics (Basel) 2021;10(2):145. doi: 10.3390/antibiotics10020145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Twest R., Kropinski A.M. Bacteriophage enrichment from water and soil. Methods Mol. Biol. 2009;501:15–21. doi: 10.1007/978-1-60327-164-6_2. [DOI] [PubMed] [Google Scholar]
- Vaser R., Sović I., Nagarajan N., Šikić M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 2017;27(5):737–746. doi: 10.1101/gr.214270.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q., Ma L.Z. Biofilm matrix and its regulation in Pseudomonas aeruginosa. Int. J. Mol. Sci. 2013;14(10):20983–21005. doi: 10.3390/ijms141020983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick R.R., Judd L.M., Holt K.E. Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol. 2019;20(1):129. doi: 10.1186/s13059-019-1727-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann J., Klumpp J., Moreno Switt A.I., Yagubi A., Ackermann H.W., Wiedmann M., Svircev A., Nash J.H., Kropinski A.M. Taxonomic reassessment of N4-like viruses using comparative genomics and proteomics suggests a new subfamily - "Enquartavirinae". Arch. Virol. 2015;160(12):3053–3062. doi: 10.1007/s00705-015-2609-6. [DOI] [PubMed] [Google Scholar]
- Wu Y., Wang R., Xu M., Liu Y., Zhu X., Qiu J., Liu Q., He P., Li Q. A novel polysaccharide depolymerase encoded by the phage SH-KP152226 confers specific activity against multidrug-resistant Klebsiella pneumoniae via biofilm degradation. Front. Microbiol. 2019;10:2768. doi: 10.3389/fmicb.2019.02768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Du C., Gong P., Xia F., Sun C., Feng X., Lei L., Song J., Zhang L., Wang B., Xiao F., Yan X., Cui Z., Li X., Gu J., Han W. Therapeutic effect of the YH6 phage in a murine hemorrhagic pneumonia model. Res. Microbiol. 2015;166(8):633–643. doi: 10.1016/j.resmic.2015.07.008. [DOI] [PubMed] [Google Scholar]
- Yin R., Cheng J., Wang J., Li P., Lin J. Treatment of Pseudomonas aeruginosa infectious biofilms: challenges and strategies. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.955286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.