Abstract
Background:
Heart failure with preserved ejection fraction (HFpEF) is increasingly prevalent and has few treatments. The molecular mechanisms and resultant signaling pathways that underlie the development of HFpEF are poorly defined. It has been proposed that activation of pro-inflammatory pathways plays a role in the development of cardiac fibrosis. The signature of gene expression (transcriptome) of previously validated left ventricular (LV) biopsies obtained from patients with HFpEF and matched referent controls allows for an unbiased assessment of pro-inflammatory and pro-fibrotic signaling pathways and genes.
Methods:
Epicardial LV biopsies from stringently selected HFpEF patients (HFpEF, n=16) and referent control patients (CTR, n=14) were obtained during aortocoronary bypass surgery. The sub-epicardial myocardium was flash-frozen to build a repository that was parallel-processed for RNA sequencing to allow for an unsupervised in-depth comparison of the LV transcriptome.
Results:
The average patient age was 67 ± 10 years. When compared to controls, HFpEF patients were hypertensive with a higher body mass index (kg/m2: 30 ± 5 vs. 37 ± 6, p<0.01) and elevated NTproBNP levels (pg/mL: 155(89–328) vs. 1554(888–2178), p<0.001). The transcriptome analysis revealed differential expression of 477 genes many of which were involved in pro-fibrotic pathways including extracellular matrix production and post-translational modification but no pro-inflammatory signature.
Conclusions:
The transcriptome analysis of left ventricular myocardial samples from patients with HFpEF confirms an overabundant extracellular matrix gene expression, the basis of myocardial fibrosis, without a signature of activated pro-inflammatory pathways or genes.
Keywords: diastolic dysfunction, heart failure with preserved ejection fraction, cardiac fibrosis, inflammation
INTRODUCTION
About half of heart failure patients have a preserved ejection fraction (HFpEF) and more than a quarter of the adult population is at risk.1,2 Despite an increasing prevalence and socioeconomic burden, only few therapies are available.3 Clinical heterogeneity, a high comorbidity-burden combined with a limited understanding of the myocardial substrate have been put forward to explain why HFpEF is difficult to treat.4
While tissue-level research in heart failure with reduced ejection fraction has greatly benefited from the availability of explanted failing hearts, HFpEF patient studies are limited by available myocardium. Autopsy studies have revealed left ventricular (LV) microvascular rarefication and fibrosis.5 Freshly obtained endo- and epimyocardial biopsies are the only source of HFpEF tissues that allow for comparative analyses.6–9 These studies suggested that collagen overabundance, sarcomere hypo-phosphorylation and cellular calcium retention play a pathophysiological role.6,7,9–12 It was proposed that an obesity associated cardiac pro-inflammatory state may drive several abnormalities chief among them fibrosis and hypophosporylation.13–15 A recent transcriptome analysis of patient HFpEF biopsies suggested involvement of pro-inflammatory pathways while another more limited analysis did not.11,16
We procured a repository of epicardial biopsies of patients with HFpEF and referent controls that has been validated for the presence of several key abnormalities in HFpEF such as increased collagen levels, titin modifications and an abnormal cellular calcium handling.9,12 Using strict inclusion and exclusion criteria epicardial LV biopsies were obtained during aortocoronary bypass surgery of HFpEF patients with an ejection fraction of 50% or higher and referent control patients with the same degree of coronary artery disease but no hypertension or heart failure. Here we provide an unbiased transcriptome (RNA-sequencing) analysis with a focus on pathway analyses and the differential regulation of known or suspected genes to play a role in HFpEF.17,18
METHODS
Data Availability
All raw RNA-sequencing data are deposited at National Center for Biotechnology Information (NCBI’s) sequence read archive (SRA) under Bioproject accession number PRJNA879763. Parallel processed gene array data (Affymetrix ClariomD) are also available upon a reasonable request to the corresponding author.
General Approach
The study cohort consisted of 30 patients from August 2015 to March 2018 recruited to undergo an intraoperative left ventricular (LV) myocardial biopsy from among those scheduled for coronary artery bypass grafting at the University of Vermont (UVM) Medical Center in Burlington, Vermont. All patients signed consent forms approved by the UVM Institutional Review Board. LV anterior wall myocardial biopsies were obtained from the epicardial surface as described previously.9,12
Clinical Measurements
Demographics, medications, laboratory data, medical history, echocardiographic measurements, and cardiac catheterization results were tabulated. The severity of coronary artery disease was graded based on the number of major vessels (left main, left anterior descending, left circumflex, and right coronary artery) with a stenosis of more than 70%. All patients underwent an echocardiographic evaluation to assess LV structure and function.
Patient Enrollment and Clinical Data
Inclusion Criteria, Classification and Enrollment:
Adult patients scheduled to undergo coronary artery bypass grafting with preserved left ventricular ejection fraction (≥50% by echocardiography), normal wall motion, and end-diastolic volume index (≤75 mL/m2) were eligible. The patients were categorized into 2 groups: (1) referent controls without hypertension or heart failure (control patients [CTR]), (2) subjects with hypertension, evidence of hypertensive heart disease and HFpEF. CTR patients fulfilled the general inclusion criteria. HFpEF patients fulfilled the general inclusion criteria and had a documented and confirmed history of hypertension plus echocardiographic evidence of concentric remodeling or overt hypertrophy (relative wall thickness ≥0.42 or left ventricular mass >115g/m2 in men, >95g/m2 in women) and evidence of HFpEF based on the European Society of Cardiology and Heart Failure Society of America criteria: (1) EF≥50%, (2) LVEDVI≤75 mL/m2, (3) presence of at least 1 symptom (dyspnea at rest or with minimal exertion or orthopnea) or 1 sign of heart failure (pulmonary crackles, elevated jugular venous pressure, or peripheral edema), (4) LV end-diastolic pressure >16mm Hg) or elevated NT-proBNP (N-terminal pro-B-type natriuretic peptide) levels (>1000ng/mL), and (5) exclusion of non-cardiac diseases that could result in symptoms commonly present in heart failure.19,20 We have demonstrated that this choice is reasonable, since we also require objective evidence of elevated filling pressures in the absence of ongoing ischemia.9,12
Exclusion criteria:
LV wall motion abnormality or resting ischemia, left ventricular ejection fraction <50%, LV end-diastolic volume index >75mL/m2, more than moderate valvular stenosis or regurgitation, severe chronic pulmonary disease, any non-cardiac disease or condition known to affect myocardial function, anemia (Hbg<11g/dL in men and <10g/dL in women), creatinine >2.0mg/dL, off-pump or emergency cardiac surgery, substance abuse, inability to provide informed consent, active malignancy, severe connective tissue disease, severe liver disease, hypertrophic or restrictive cardiomyopathies, and constrictive pericarditis.
After the informed consent, a blood sample for NT-proBNP was obtained and a history and physical examination directed at signs and symptoms of HF was performed. De-identified demographics and clinical data, e.g., medications and co-morbidities, were tabulated for future analyses.
Experimental Procedures
Biopsy processing:
LV myocardial biopsies were obtained from the non-ischemic viable LV anterior free wall myocardium at a site near its minor axis while patients were on cardiopulmonary bypass. The entire biopsy was immediately placed in ice-cold protective Tyrode solution. After removal of the epicardium and fat tissue, the myocardial tissue was micro-dissected into 2–3 strips and snap frozen in liquid nitrogen for future transcriptome analyses or used for an immediate contractile and functional assessment of excitable strip preparations as reported within 15 minutes.12 The presence of amyloidosis was not assessed. All biopsies were identically procured, processed and stored.
RNA extraction:
Total RNA was extracted from samples using the RNeasy Plus Micro kit (Qiagen) according to the manufactures protocol. Briefly, tissue was transferred to a 1.5ml tube and homogenized with a motorized pestle in RLT buffer containing β-mercaptoethanol (RLT-BME). The homogenate was adjusted to 250μL with RLT-BME, to which 250 μl of 70% ethanol was added before transferring to a spin column. The column was centrifuged at 10,000g. Thereafter, the sample was DNase treated and washed twice with RPE according to the protocol. The final RNA was then eluted in 14μl of RNase-free water and stabilized by adding 0.5μl of an RNase Inhibitor (Ribolock Therm Fisher). The RNA was quantified with the Qubit spectrofluorometer and assayed for quality using the Bioanalyzer with a RNA Pico chip (Agilent Technologies).
Library preparation and next generation RNA sequencing:
Total RNA from each heart sample was converted to Illumina-compatible libraries using the SMARTer® Stranded Total RNA-Seq Kit v2 (Takara) according to the manufacturer’s protocol. The final RNA-Seq libraries were checked for quality using Qubit DNA high sensitivity reagents and the Bioanalyzer DNA HS chip. Libraries were pooled at equimolar concentrations and sequenced with the Illumina HiSeq 1500/2500 system. Depths of 40–44 million paired-end 80bp reads were generated for each sample. The Collection of Hierarchical UMII/RIS Pipelines (PURR), developed by the Research Informatics Solutions (RIS) group at the Minnesota Supercomputing Institute and funded by the University of Minnesota Informatics Institute (UMII), was employed to process raw sequencing data and analyze differentially expressed genes (DEGs). Briefly, 2 × 80bp FastQ paired-end reads for 30 samples were filtered by trimming adaptors and inadequate quality bases using Trimmomatic (v 0.33) enabled with the optional “-q” option. Quality control on raw data for each sample was performed with FastQC. Only paired reads with an average quality higher than 30 were kept and further aligned to the human reference genome sequence (GRCh38) using the Hisat2 (v2.1.0). RNA quantification was done via Feature Counts for raw read counts. Principal component analysis (PCA) was performed using the PCAtools package in R. Only genes meeting the inclusion criteria of 0.5 counts per million (CPM) in at least one sample were used in the PCA. Differentially expressed genes were identified using the edgeR (negative binomial) feature in CLC Genomics Workbench (Qiagen, Redwood City, CA) using raw read counts based on a false discovery rate (FDR) corrected p value of less than 0.1 using the Benjamini-Hochberg (BH) method. RNA-seq analysis was based on uniquely aligned reads. The average gene expression levels in control and HFpEF groups were normalized and presented as average counts per million (CPM) or logarithmically transformed CPM. Reads per kilobase of transcript per million mapped reads (RPKM) was applied to evaluate the transcript-level abundance of different genes in the same heart biopsy sample.
Gene ontology (GO) analysis refers to determining and describing the biological characteristics of genome or transcriptome data in different GO term databases under three domains including biological process (BP), cellular component (CC) and molecular function (MF). The REACTOME pathway (https://reactome.org/) database compiles genomic, chemical, and systematic functional information. In this investigation, GO terms were analyzed using the GEne SeT AnaLysis Toolkit (WebGestalt, http://www.webgestalt.org), a web-based bioinformatics resources with tools for the functional interpretation of large-scale gene datasets.21 REACTOME pathways were analyzed and visualized with ReactomeGSA, a pathway analysis tool integrated into the Reactome bionetwork for analyzing and anticipating functional profiles of gene and gene clusters.22 An FDR corrected p value of 0.05 was applied to determine whether a pathway is significantly impacted.
Definition of Pro-fibrotic and Pro-Inflammatory Markers:
In addition, we tabulated the expression of known or suspected pro-inflammatory and pro-fibrotic genes considered to play a role in HFpEF.18 These include markers of (a) systemic inflammation (i.e. CRP and sST2), (b) cell based pro-inflammatory and pro-fibrotic tissue response that involve monocytes, macrophages, fibroblasts and endothelial cells to include cytokines and chemokines (i.e. TNFα, TGFB1, IL6) and (c) structural and extracellular matrix modifying proteins that have been implicated in the pathogenesis of HFpEF (i.e. collagens, TIMP1).
Statistical Plan
Continuous data are generally reported as mean and standard deviations (SD) or median with interquartile range were indicated. Categorical data are generally reported as number (%). The RNA sequencing data was subjected to log2 transformation of counts per million values prior to performing principal component analysis in the R environment. Gene expression analysis between the CTR and HFpEF groups was performed using the quasi-likelihood test implemented in edgeR. Statistical differences were defined as FDR corrected p value of less than 0.1, using the Benjamini-Hochberg method for multiple hypothesis testing. Gene pathway enrichment analyses were performed in WebGestalt and REACTOME with an FDR corrected p value of less than 0.05.
For the clinical data, student’s unpaired t-tests (parametric) or Mann-Whitney U tests (non-parametric) were used. The Fisher’s exact test was employed for categorical variables. Pearson correlation analyses and ANOVA models were used to determine the association of gene clusters with clinical characteristics, e.g. obesity (BMI) and diabetes.
RESULTS
Patient Characteristics and Diagnostic Parameters
The mean age was 67±10 years (Table 1). On average HFpEF patients were obese with BMI’s ranging from 27 to 46 kg/m2 and diabetes was prevalent. In the HFpEF cohort, systolic blood pressures were higher and antihypertensive and diuretic medication use was common. In addition, LVEDP and NT-proBNP levels were higher with echocardiographic evidence of left ventricular hypertrophy and left atrial dilation. Coronary artery disease was equally severe in the CTR and HFpEF group.
Table 1.
Patient Baseline Characteristics
| Patient Data | Control (n=14) | HFpEF (n=16) | p value |
|---|---|---|---|
| Age (years) | 68 ± 12 | 66 ± 7 | 0.70 |
| Female (%) | 29 | 25 | 0.83 |
| Heart Rate (bpm) | 67 ± 11 | 72 ± 10 | 0.28 |
| BMI (kg/m2) | 30 ± 5 | 37 ± 6 | 0.002 |
| BSA (m2) | 2.0 ± 0.2 | 2.2 ± 0.3 | 0.01 |
| SBP (mmHg) | 123 ± 12 | 138 ± 11 | 0.002 |
| DBP (mmHg) | 75 ± 9 | 73 ± 20 | 0.68 |
| Creatinine (mg/dL) | 0.87 ± 0.19 | 1.03±0.19 | 0.19 |
| NT-proBNP (pg/ml)* | 155 (89–328) | 1554 (888–2178) | <0.001 |
| LVEDP (mmHg) | 13 ± 5 | 23 ± 7 | 0.003 |
| Medications, Medical History and H2FPEF Score | |||
| ACE/ARB (%) | 14 | 75 | <0.001 |
| CCB (%) | 7 | 36 | <0.001 |
| Diuretic Agent (%) | 0 | 71 | <0.001 |
| Beta-blocker (%) | 54 | 81 | 0.12 |
| Statin (%) | 46 | 75 | 0.12 |
| Diabetes (%) | 14 | 69 | 0.03 |
| Atrial Fibrillation (%) | 0 | 31 | 0.05 |
| CAD Severity (# vessels) | 3.0 ± 0.6 | 3.2 ± 0.8 | 0.55 |
| H2FPEF Score | 2.0 ± 1.2 | 5.1 ± 1.7 | <0.001 |
| Echocardiography | |||
| EF (%) | 60 ± 5 | 59 ± 7 | 0.96 |
| Septal Wall (mm) | 10.0 ± 1.3 | 13.6 ± 1.9 | <0.001 |
| Posterior Wall (mm) | 8.4 ± 0.6 | 12.1 ± 1.4 | <0.001 |
| LVEDD (mm) | 49 ± 5 | 48 ± 5 | 0.49 |
| LVESD (mm) | 32 ± 5 | 32 ± 7 | 0.95 |
| RWT | 0.35 ± 0.03 | 0.52 ± 0.10 | <0.001 |
| LVmass (gr) | 163 ± 33 | 244 ± 45 | <0.001 |
| LVmass / BSA (gr/m2) | 82 ± 14 | 111 ± 22 | <0.001 |
| LVmass / H (gr/m) | 96 ± 17 | 144 ± 25 | <0.001 |
| LVmass / H2.7 | 40 ± 7 | 60 ± 13 | <0.001 |
| LA volume (ml) | 44 ± 9 | 79 ± 36 | 0.002 |
| LA volume / BSA | 22 ± 5 | 36 ± 15 | 0.004 |
| E (cm/sec) | 66 ± 12 | 88 ± 26 | 0.01 |
| A (cm/sec) | 79 ± 20 | 82 ± 29 | 0.76 |
| E/A ratio | 0.9 ± 0.4 | 1.1 ± 0.3 | 0.21 |
| E/E’ med | 8.6 ± 4.1 | 17.6 ± 8.0 | 0.002 |
| E/E’ lat | 8.0 ± 2.2 | 14.2 ± 8.8 | 0.02 |
BMI, body mass index; BSA, body surface area; SBP, systolic blood pressure; DBP, diastolic blood pressure; ACE/ARB, ACE-inhibitor/Angiotensin Receptor Blocker; CCB, Calcium Channel Blocker; LVEDP, left ventricular end-diastolic pressure; CAD, coronary artery disease; EF, ejection fraction; LVEDD, left ventricular end-diastolic diameter; LVESD, left ventricular end-systolic diameter; RWT, relative wall thickness; LVmass, left ventricular mass; LVmass/H, left ventricular mass to height ratio; LVmass/H2.7, allometric left ventricular mass index; LA, left atrial; E, E-wave peak velocity; A, A-wave peak velocity; E/E’med, ratio of mitral peak velocity of early filling (E) to early diastolic septal mitral annular velocity (E’), E/E’lat, ratio of mitral peak velocity of early filling (E) to early diastolic lateral mitral annular velocity;
Median (Interquartile Range) Mann Whitney U test.
Left Ventricular Myocardial Transcriptome
Quality Metrics
The transcriptome sequencing resulted in an average of 34 million paired-end reads per biopsy sample. After trimming and filtering, over 97% reads with a quality score of 30 or higher remained. Uniquely mapped reads per sample ranged from 18 to 33 million with an average of 25 million (Figure. S1A) that covered more than 76% of the sequenced reads with 93% total mapped reads. The clean RNAseq read-set identified 42994 genes. To ascertain tissue specificity we compared absolute transcript abundances of three established gene markers for each of the 11 major cardiac cell types in adult human heart tissues as recently described.23,24 This confirmed a signature of ventricular myocardium across all samples with consistently high expression levels of canonical ventricular cardiac myocyte genes, such as beta-myosin heavy chain (MYH7) and myosin light chain 2 (MYL2) and low expression levels of marker genes of atrial myocytes, mesothelial cells, adipocytes, neuronal cells, lymphoid cells and myeloid cells (Figure. S1B). The relative proportion of different cell types was consistent and comparable among all samples.
Differential expression
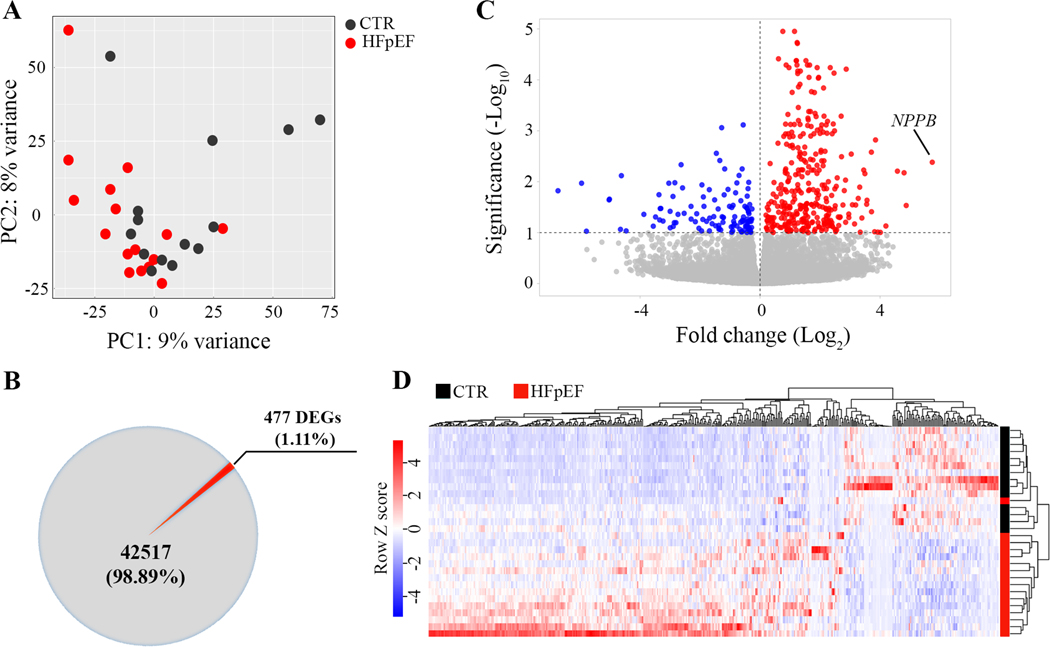
The gene expression signature of the HFpEF and CTR suggested a substantial overlap and some differences as reflected in the unsupervised principal component analysis (PCA) which reduces complex high-dimensional data to retain trends and patterns as shown in Figure 1A. In total, 477 differentially expressed genes (DEG) were identified between HFpEF and CTR, including 353 upregulated (74%) and 124 downregulated (26%) genes (Figure 1B and C). Expression of B-type natriuretic peptide gene (NPPB), which encodes brain natriuretic peptide (BNP) was upregulated 53-fold in HFpEF (Figure 1C). The heatmap of variances in the hierarchical clustering analysis (Pearson correlation) of all differentially expressed genes in individual control and HFpEF samples is provided in Figure 1D.
Figure 1. Differential gene expression analysis.
A, Principal component analysis of all identified genes shows the overlaps and differences between the RNA-Seq transcriptome profiles between left ventricular myocardium from referent controls (CTR, ●) and HFpEF (●). First principal component (PC1) is shown on x-axis while the second principal component (PC2) is shown on y-axis. B, Pie chart for differentially expressed genes (DEGs) identified between CTR and HFpEF. C, Volcano plot with the log2 fold change in gene expression in HFpEF on the x-axis. The red and blue circles represent significantly up- and down-regulated genes on the y-axis. The B-type natriuretic peptide (NPPB) gene encoding BNP is markedly upregulated in HFpEF. D, Heat map of RNA-seq expression data showing the hierarchical clustering of genes that are differentially expressed between control and HFpEF. Z-scores for each of the 477 DEGs were calculated and used as input for complete linkage clustering analysis.
Gene Ontology and Pathway Analysis
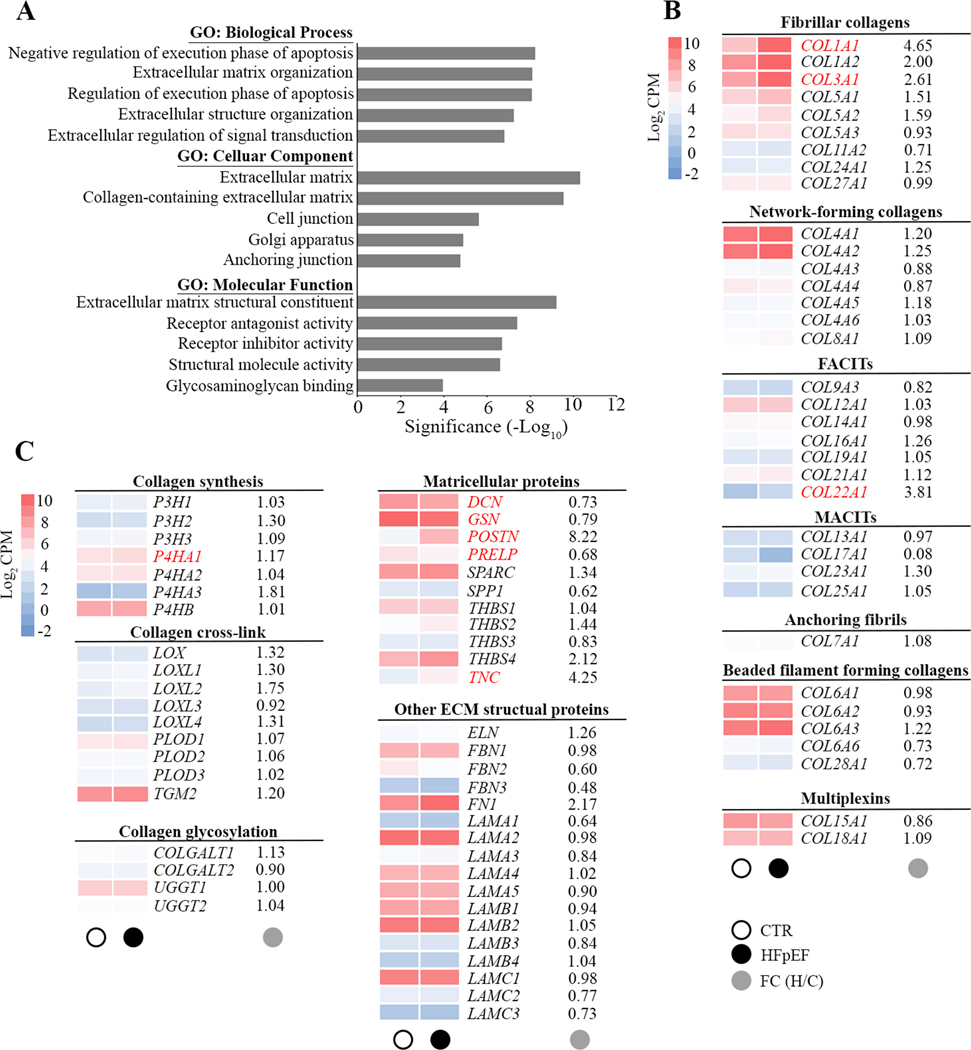
Further gene ontology (GO) analysis of cellular components, biological processes, and molecular function was performed in order to obtain a GO classification for the 477 DEGs in HFpEF patients. The GO analysis of these DEGs revealed an involvement in extracellular matrix, cellular anchoring and interaction. Receptor ligands, intracellular hemostasis regulators of apoptosis and genes relevant to the function of the Golgi apparatus were also differentially expressed (Figure 2A). Changes were seen in three domains of extracellular matrix homeostasis: synthesis, processing (matricellular) and post-translational modification. Increased expression of extracellular matrix genes was associated with HFpEF, including fibrillar type I and type III collagen (COL1A1 and COL3A1), and nonfibrillar type XXII collagen (COL22A1). Prolyl 4-hydroxylase (P4HA1), a key enzyme in collagen synthesis, was also upregulated in HFpEF. Furthermore, nonstructural matrix proteins, known as matricellular proteins (MCPs) including periostin (POSTN), proline arginine-rich end leucine-rich repeat protein (PRELP) and tenascin C (TNC) were associated with HFpEF as was the proteoglycan decorin (DCN). Overrepresented genes were not modified by the degree of obesity (body mass index) or the presence of diabetes.
Figure 2. Gene Ontology enrichment analysis of differentially expressed genes.
A, Bar chart showing the top 5 gene ontology (GO) terms for biological process, cellular component and molecular function ranked by fold enrichment following analysis of all differentially expressed HFpEF genes. Significant enriched GO terms are provided. B and C, Heat maps depict the average expression levels (average log2 CPM) of individual genes in the extracellular matrix in CTR (○) and HFpEF(●). Main targets in subfamilies of fibrillar collagen, nonfibrillar collagen, collagen synthesis and modification, matricellular proteins and other extracellular matrix (ECM) structural proteins are shown as average gene expression levels of CTR (n=14) and HFpEF (n=16) hearts. The fold changes (●) in gene expression in HFpEF is provided and significant changes in gene expression are highlighted in red. FACITs, fibril-associated collagens with interrupted triple helices; MACITs, membrane-associated collagens with interrupted triple helices.
Notably, both GO terms and REACTOME pathway analysis did not reveal any differential expression of inflammation-associated signaling pathways or gene sets in HFpEF. The top 30 pathways ranked by p value obtained from the ReactomeGSA analysis are listed in Table S1.
Expression of Genes implicated in HFpEF
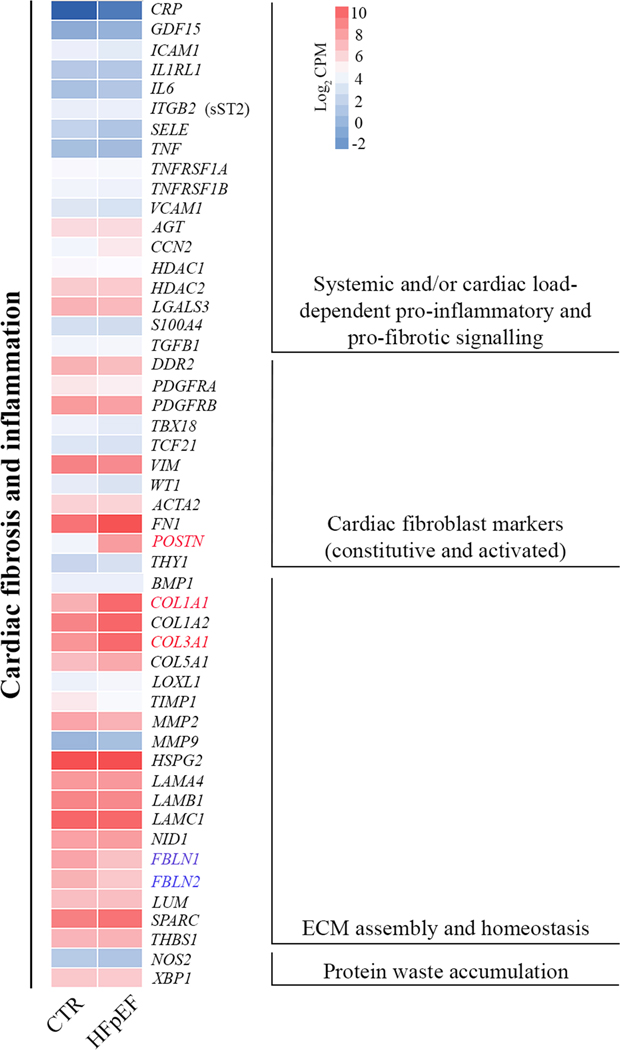
Figure 3 depicts heat maps of key regulators and targets implicated in cardiac pro-fibrotic and pro-inflammatory signaling. mRNA level of interleukins (ILs), C-C motif chemokine ligands (CCLs), C-X-C motif chemokine ligands (CXCLs) and colony stimulating factors (CSFs) were not altered in HFpEF (Table 2). An extended list of low abundance fibrillar and nonfibrillar collagens, matrix metalloproteinases (MMPs) and inhibitors of metalloproteinases (TIMPs) are provided in the supplement (Tables S2 and S3). Genes encoding cytoskeletal and myofilament proteins are listed in the supplement Table S4.
Figure 3. Expression of genes implicated in the pathophysiology of HFpEF.
A heat map of the average log2 CPM is provided to include genes involved in pro-fibrotic and pro-inflammatory pathways. Identifiers of significant down-regulated (blue) and up-regulated (red) genes are provided.
Table 2.
Pro-inflammatory mediators in Control and HFpEF.
| Gene ID | Mean CTR (CPM) | Mean HFpEF (CPM) | FDR p value |
|---|---|---|---|
| IL1B | 0.3851 | 0.5376 | 0.7868 |
| IL6 | 0.6835 | 0.9751 | 0.6745 |
| IL7 | 0.3129 | 0.2797 | 0.9760 |
| IL10 | 0.2665 | 0.1869 | 0.8322 |
| IL12A | 0.2578 | 0.2131 | 0.8643 |
| IL15 | 4.9894 | 4.9001 | 0.9947 |
| IL16 | 15.0293 | 12.0188 | 0.5825 |
| IL17D | 0.3983 | 0.3874 | 0.9851 |
| IL18 | 2.5176 | 1.2784 | 0.4690 |
| IL23A | 0.0706 | 0.0929 | 0.8647 |
| IL32 | 14.0823 | 13.5219 | 0.9748 |
| IL33 | 8.5082 | 7.8487 | 0.8757 |
| IL34 | 1.0356 | 1.3530 | 0.5625 |
| CCL2 | 4.9154 | 4.5330 | 0.9051 |
| CCL3 | 0.3286 | 0.3006 | 0.9587 |
| CCL3L1 | 0.1334 | 0.1286 | 0.9845 |
| CCL4 | 0.1920 | 0.1893 | 0.9941 |
| CCL4L2 | 0.1179 | 0.1471 | 0.9150 |
| CCL5 | 1.2513 | 1.4761 | 0.7694 |
| CCL8 | 0.2299 | 0.3275 | 0.7048 |
| CCL11 | 0.8289 | 1.1145 | 0.6579 |
| CCL13 | 0.2318 | 0.0882 | 0.6244 |
| CCL14 | 0.2098 | 0.3059 | 0.7627 |
| CCL16 | 0.0691 | 0.0713 | 0.9922 |
| CCL17 | 0.0374 | 0.0244 | 0.8693 |
| CCL18 | 0.2356 | 0.1688 | 0.9228 |
| CCL19 | 0.2033 | 0.1634 | 0.9648 |
| CCL20 | 0.0276 | 0.0071 | 0.8555 |
| CCL21 | 6.7012 | 5.1999 | 0.8132 |
| CCL22 | 0.0659 | 0.0643 | 0.9964 |
| CCL23 | 0.0285 | 0.0178 | 0.9785 |
| CCL24 | 0.2568 | 0.0618 | 0.4358 |
| CCL26 | 0.0090 | 0.0514 | 0.5650 |
| CCL28 | 0.4620 | 0.3277 | 0.7164 |
| CXCL1 | 1.2774 | 0.9655 | 0.7194 |
| CXCL2 | 21.7835 | 19.2811 | 0.9046 |
| CXCL3 | 0.7977 | 0.5684 | 0.6745 |
| CXCL5 | 0.0679 | 0.0868 | 0.9156 |
| CXCL6 | 0.0953 | 0.0938 | 0.9845 |
| CXCL8 | 0.2114 | 0.4075 | 0.6745 |
| CXCL9 | 0.9959 | 1.3412 | 0.6745 |
| CXCL10 | 0.2631 | 0.5891 | 0.2150 |
| CXCL11 | 0.0286 | 0.1430 | 0.3613 |
| CXCL12 | 70.0774 | 70.5659 | 0.8887 |
| CXCL14 | 2.0714 | 1.1787 | 0.5671 |
| CXCL16 | 2.1066 | 2.0347 | 0.9845 |
| CXCL17 | 0.0146 | 0.0109 | 0.9852 |
| CSF1 | 15.0473 | 12.7733 | 0.5671 |
| CSF3 | 0.0881 | 0.0261 | 0.6745 |
Table 2. List of pro-inflammatory mediators in control and HFpEF myocardium. The mean CPM values for control and HFpEF samples and p values are provided. CPM, counts per million; IL, interleukin; CCL, C-C motif chemokine ligand; CXCL, C-X-C motif chemokine ligand; CSF, colony stimulating factor.
DISCUSSION
Heart failure with a preserved ejection fraction (HFpEF) is prevalent and has few treatments that reduce symptoms and heart failure events but none that reduces mortality.3 A better understanding of the myocardial substrate may aid the development of HFpEF specific therapies. It has been suggested that activation of cardiac pro-inflammatory pathways may contribute to HFpEF by increasing fibrosis and hypophosphorylation of sarcomeric proteins.11,14,25 This is a field of extensive investigation and prioritized research investments.26
We have performed an unbiased gene analysis of previously validated left ventricular HFpEF biopsies and matched referent control biopsies to reveal HFpEF-specific differences. Overexpression of genes involved in tissue fibrosis, e.g. collagen I and III, with overexpression of matrix-modifying and stabilizing proteins and enzymes was present confirming our previously reported protein findings.9 The analysis of this cohort of HFpEF patients does not support a tissue level activation of genes involved in pro-inflammatory pathways. The raw RNA sequencing data is available through the National Center for Biotechnology Information (NCBI’s) sequence read archive (SRA) biorepository to allow for inquiries of all detected transcripts.
In the following we will provide an in-depth discussion of patient background, differential expression and some of the pitfalls of transcriptome studies.
Patient Background
Our prospective patient cohort consisted of patients scheduled to undergo aortocoronary bypass surgery for severe coronary artery disease (CAD). Patients with HFpEF were required to have a history of hypertension with clinical and objective evidence of HFpEF. Subjects in the HFpEF cohort were typically obese with a high prevalence of diabetes requiring multiple antihypertensive medications and diuretic agents, which recapitulates the prevalent cardiometabolic HFpEF phenotype. Although the duration of heart failure was not determined, the enrollment criteria and clinical characteristics suggest that our patients had advanced HFpEF, e.g. atrial dilation, NT-proBNP levels and medications, associated with an increased mortality.27 Referent control myocardium was obtained from age and gender matched normotensive patients, with the same degree of severe coronary artery disease and echocardiographic findings reflective of the general population.28
In comparison to a published cohort that provided right ventricular expression signatures of HFpEF, our patients were older and had more pronounced left ventricular hypertrophy.11 The prevalence of obesity and hypertension was similar. While the right ventricular analysis used samples from rejected donor hearts as a control, all our biopsies were identically procured and processed, similar to a previous smaller analysis of epicardial left ventricular biopsies from 5 HFpEF and 11 control patients.16
HFpEF Expression Signature
Changes were seen in the four domains of extracellular matrix homeostasis - synthesis, processing, post-translational modification and potentially degradation. The genes included collagen I and III. Also, genes encoding collagen chaperones, such as prolyl 4-hydroxylase, which catalyzes the conversion from proline to hydroxyproline, which is essential in the three-dimensional folding of newly synthesized procollagen were upregulated to suggest excessive extracellular matrix synthesis and maintenance as a predominant theme. Importantly, extracellular matrix overexpression was present in the absence of activated pro-inflammatory pathways or genes to suggest that hemodynamic load and circulating systemic mediators of fibrosis, e.g. aldosterone, sST2 and galectin-3 may be sufficient to maintain a pro-fibrotic expression signature in more advanced stages of HFpEF.18
Pro-inflammatory Signaling Pathways and Genes
It has been proposed that a systemic and myocardial pro-inflammatory state may play a role in HFpEF.11,13–15,18 Similar to a smaller and less stringent comparison of LV biopsies, that did not focus on these pathways and genes, our results do not suggest that pro-inflammatory signaling pathway and gene activation is a prerequisite for extracellular matrix overexpression in this patient cohort with stable HFpEF.16
The lack of activation of general or vascular pro-inflammatory pathways in our analysis could have several reasons. Pro-inflammatory mediators and their respective genes are either not overexpressed or detectable. This includes the possibility of a spatially restricted increase of inflammation-associated cells, such as perivascular macrophages, that may not be readily detected. Although an overabundance of pro-inflammatory factors was reported in HFpEF myocardium, i.e. TGFB1 and VCAM1, low-level lineage specific overexpression could still be missed.13 Also, inflammation may not be present in compensated HFpEF patients able to undergo aortocoronary bypass surgery. However, BNP expression as a marker of wall stress, was markedly elevated in HFpEF myocardium to suggest that key pathophysiological stimuli were still at play. It is also possible that the presence of coronary artery disease which was equally severe in both groups (Table 1) neutralized potential differences in vascular inflammation, i.e. the NLRP3 inflammasome interleukin-1 pathway.29
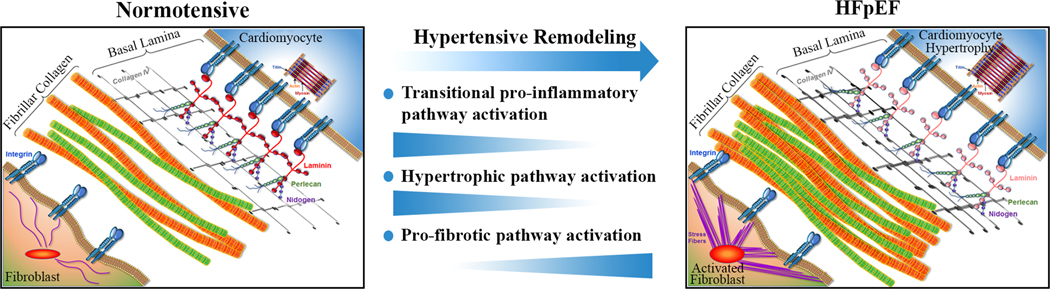
However, the most likely explanation for our results is the recent finding that pro-inflammatory signaling is predominantly active during the initial phase of pressure overload remodeling, i.e. development of concentric hypertrophy, and not required to maintain a more advanced HFpEF substrate.30,31 These findings are in line with a recent report by Kaye et al. that did not detect excess cardiac inflammatory cytokine release in HFpEF patients.32 Combined with the current results this would refine the cardiac pro-inflammatory HFpEF paradigm as being temporally restricted to active concentric remodeling as shown in Figure 4. The innate program that facilitates the remodeling, in skeletal muscle often referred to as myokine signaling, involves many of the same cytokines and inflammation mediators reported in HFpEF.33 An absence of pro-inflammatory cardiac pathway activation in compensated HFpEF patients has clinical implications as it reduces the likelihood that anti-inflammatory therapies will benefit patients with more advanced HFpEF. This may explain why interleukin-1 blockade with anakinra, has yielded inconclusive results.34,35 Studies that test the efficacy of colchicine (NCT04857931, NCT05637398), a IL-6 ligand monoclonal antibody (NCT05636176), or a myeloperoxidase inhibitor (NCT03611153, NCT04986202) are ongoing. In contrast, medications with anti-fibrotic efficacy such as sacubitril/valsartan and spironolactone have become part of the HFpEF treatment algorithm.36,37
Figure 4. Revised model of the hemodynamic load–mediated activation of pro-inflammatory and pro-fibrotic pathways highlighting the time-varying expression patterns in the development of HFpEF.
Once the pro-inflammatory pathway dependent hypertensive remodeling is complete the pro-inflammatory and hypertrophic pathway activation subside while extracellular matrix overproduction continues. This is most prominently reflected in an excess collagen I and III synthesis.
Although spatial factors (e.g. right vs. left and subendocardial vs. subepicardial myocardium) and disease activity (compensated vs. acute heart failure) can be put forward to explain the observed differences in gene expression between the studies divergent findings can also be introduced by exogenous factors such as methodological differences in tissue sourcing, which can introduce systematic errors.
Limitations and Pitfalls
The vast number of comparisons, almost 43000 in our transcriptome analysis, inevitably produces numerous incorrect rejections of the null hypothesis, which leads to false discoveries. In experimental studies, the large number of false positive findings is reduced by only accepting results with a two-fold or higher difference in expression levels between the groups. Such thresholds are unrealistic in the analysis of human myocardium, which commonly displays much more subtle changes in gene or protein expression.9,38
Identical tissue sourcing and sample processing are of critical importance in omics-based tissue comparisons, i.e. surgery and cardioplegia-induced ischemia alters myocardial gene expression.39 Endomyocardial biopsies obtained from contracting hearts may therefore have an advantage as long as the comparator group is equally sourced and processed.11,13 However, if control myocardium from rejected cardioplegia-arrested donor hearts is compared to freshly obtained RV biopsies, source confounding increases the risk for spurious results, e.g. marked upregulation of early transcription factors that play a role in inflammation (FOS, MYC and JUNB) or higher expression of IL6 and BNP in control samples.11
As discussed our cross-sectional comparison of LV myocardium obtained from HFpEF patients with hypertensive heart disease and control myocardium from normotensive patients with equally severe coronary artery disease will not capture time-varying gene expression signatures at different HFpEF stages and phenotypes. However, our analysis confirms an overabundant extracellular matrix expression in HFpEF and substantially extends the list of contributing matrix genes in HFpEF.
Conclusions
Our findings suggest that overexpression of extracellular matrix, which is the source of fibrosis, is a leading abnormality in HFpEF. Our analysis does not support a general or specific tissue level activation of pro-inflammatory pathways in advanced compensated HFpEF.
Supplementary Material
WHAT IS NEW ?
It has been proposed that some of the key abnormalities in the myocardial substrate of patients with heart failure with preserved ejection fraction (HFpEF) are driven by a local activation of cardiac pro-inflammatory pathways or genes.
This analysis revealed an overabundant pro-fibrotic gene expression signature in HFpEF without a general or specific activation of pro-inflammatory pathways or genes.
WHAT ARE THE CLINICAL IMPLICATIONS ?
The gene expression profile of patients with heart failure with a preserved ejection fraction does not suggest the activation of cardiac pro-inflammatory pathways or genes while confirming activation of pro-fibrotic pathways. These results reduce the prospect for anti-inflammatory treatment approaches in HFpEF.
ACKNOWLEGEMENTS
The authors would like to express their profound appreciation to all patients and surgeons, for their willingness to donate and procure myocardial biopsies. The next-generation sequencing was performed in the Vermont Integrative Genomics Resource Massively Parallel Sequencing Facility and was supported by the University of Vermont Cancer Center, Lake Champlain Cancer Research Organization, UVM College of Agriculture and Life Sciences, and the UVM Larner College of Medicine.
SOURCES OF FUNDING
Part of this research was in part supported by grant R01 HL-122744 from the National Institutes of Health and the Engdahl Family Foundation, Minneapolis, Minnesota (Dr. Meyer).
ABBREVIATIONS:
- HF
heart failure
- HFpEF
heart failure with preserved ejection fraction
- EF
ejection fraction
- NT-proBNP
N-terminal pro brain natriuretic peptide
- BMI
Body mass index
Footnotes
DISCLOSURES
None relevant to this work.
REFERENCES
- 1.Drazner MH. The progression of hypertensive heart disease. Circulation. 2011;123:327–334. doi: 10.1161/CIRCULATIONAHA.108.845792 [DOI] [PubMed] [Google Scholar]
- 2.Heidenreich PA, Albert NM, Allen LA, Bluemke DA, Butler J, Fonarow GC, Ikonomidis JS, Khavjou O, Konstam MA, Maddox TM, et al. Forecasting the impact of heart failure in the United States: a policy statement from the American Heart Association. Circ Heart Fail. 2013;6:606–619. doi: 10.1161/HHF.0b013e318291329a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heidenreich PA, Bozkurt B, Aguilar D, Allen LA, Byun JJ, Colvin MM, Deswal A, Drazner MH, Dunlay SM, Evers LR, et al. 2022 AHA/ACC/HFSA Guideline for the Management of Heart Failure: Executive Summary: A Report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. Circulation. 2022;145:e876–e894. doi: 10.1161/CIR.0000000000001062 [DOI] [PubMed] [Google Scholar]
- 4.Shah SJ, Katz DH, Selvaraj S, Burke MA, Yancy CW, Gheorghiade M, Bonow RO, Huang CC, Deo RC. Phenomapping for novel classification of heart failure with preserved ejection fraction. Circulation. 2015;131:269–279. doi: 10.1161/CIRCULATIONAHA.114.010637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohammed SF, Hussain S, Mirzoyev SA, Edwards WD, Maleszewski JJ, Redfield MM. Coronary microvascular rarefaction and myocardial fibrosis in heart failure with preserved ejection fraction. Circulation. 2015;131:550–559. doi: 10.1161/CIRCULATIONAHA.114.009625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borbély A, Falcao-Pires I, van Heerebeek L, Hamdani N, Édes In, Gavina C, Leite-Moreira AF, Bronzwaer JGF, Papp Zn, van der Velden J, et al. Hypophosphorylation of the Stiff N2B Titin Isoform Raises Cardiomyocyte Resting Tension in Failing Human Myocardium. Circulation Research. 2009;104:780–786. doi: 10.1161/circresaha.108.193326 [DOI] [PubMed] [Google Scholar]
- 7.van Heerebeek L, Borbély A, Niessen HW, Bronzwaer JG, van der Velden J, Stienen GJ, Linke WA, Laarman GJ, Paulus WJ. Myocardial structure and function differ in systolic and diastolic heart failure. Circulation. 2006;113:1966–1973. doi: 10.1161/CIRCULATIONAHA.105.587519 [DOI] [PubMed] [Google Scholar]
- 8.Selby DE, Palmer BM, LeWinter MM, Meyer M. Tachycardia-induced diastolic dysfunction and resting tone in myocardium from patients with a normal ejection fraction. J Am Coll Cardiol. 2011;58:147–154. doi: 10.1016/j.jacc.2010.10.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zile MR, Baicu CF, Ikonomidis JS, Stroud RE, Nietert PJ, Bradshaw AD, Slater R, Palmer BM, Van Buren P, Meyer M, et al. Myocardial stiffness in patients with heart failure and a preserved ejection fraction: contributions of collagen and titin. Circulation. 2015;131:1247–1259. doi: 10.1161/CIRCULATIONAHA.114.013215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aslam MI, Hahn VS, Jani V, Hsu S, Sharma K, Kass DA. Reduced Right Ventricular Sarcomere Contractility in Heart Failure With Preserved Ejection Fraction and Severe Obesity. Circulation. 2021;143:965–967. doi: 10.1161/CIRCULATIONAHA.120.052414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hahn VS, Knutsdottir H, Luo X, Bedi K, Margulies KB, Haldar SM, Stolina M, Yin J, Khakoo AY, Vaishnav J, et al. Myocardial Gene Expression Signatures in Human Heart Failure With Preserved Ejection Fraction. Circulation. 2021;143:120–134. doi: 10.1161/CIRCULATIONAHA.120.050498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Runte KE, Bell SP, Selby DE, Häußler TN, Ashikaga T, LeWinter MM, Palmer BM, Meyer M. Relaxation and the Role of Calcium in Isolated Contracting Myocardium From Patients With Hypertensive Heart Disease and Heart Failure With Preserved Ejection Fraction. Circulation: Heart Failure. 2017;10:e004311. doi: 10.1161/CIRCHEARTFAILURE.117.004311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Westermann D, Lindner D, Kasner M, Zietsch C, Savvatis K, Escher F, von Schlippenbach J, Skurk C, Steendijk P, Riad A, et al. Cardiac inflammation contributes to changes in the extracellular matrix in patients with heart failure and normal ejection fraction. Circ Heart Fail. 2011;4:44–52. doi: 10.1161/CIRCHEARTFAILURE.109.931451 [DOI] [PubMed] [Google Scholar]
- 14.Paulus WJ, Tschöpe C. A novel paradigm for heart failure with preserved ejection fraction: comorbidities drive myocardial dysfunction and remodeling through coronary microvascular endothelial inflammation. J Am Coll Cardiol. 2013;62:263–271. doi: 10.1016/j.jacc.2013.02.092 [DOI] [PubMed] [Google Scholar]
- 15.Franssen C, Chen S, Unger A, Korkmaz HI, De Keulenaer GW, Tschöpe C, Leite-Moreira AF, Musters R, Niessen HW, Linke WA, et al. Myocardial Microvascular Inflammatory Endothelial Activation in Heart Failure With Preserved Ejection Fraction. JACC Heart Fail. 2016;4:312–324. doi: 10.1016/j.jchf.2015.10.007 [DOI] [PubMed] [Google Scholar]
- 16.Das S, Frisk C, Eriksson MJ, Walentinsson A, Corbascio M, Hage C, Kumar C, Asp M, Lundeberg J, Maret E, et al. Transcriptomics of cardiac biopsies reveals differences in patients with or without diagnostic parameters for heart failure with preserved ejection fraction. Sci Rep. 2019;9:3179. doi: 10.1038/s41598-019-39445-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfeffer MA, Shah AM, Borlaug BA. Heart Failure With Preserved Ejection Fraction In Perspective. Circ Res. 2019;124:1598–1617. doi: 10.1161/CIRCRESAHA.119.313572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paulus WJ, Zile MR. From Systemic Inflammation to Myocardial Fibrosis: The Heart Failure With Preserved Ejection Fraction Paradigm Revisited. Circ Res. 2021;128:1451–1467. doi: 10.1161/CIRCRESAHA.121.318159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paulus WJ, Tschöpe C, Sanderson JE, Rusconi C, Flachskampf FA, Rademakers FE, Marino P, Smiseth OA, De Keulenaer G, Leite-Moreira AF, et al. How to diagnose diastolic heart failure: a consensus statement on the diagnosis of heart failure with normal left ventricular ejection fraction by the Heart Failure and Echocardiography Associations of the European Society of Cardiology. Eur Heart J. 2007;28:2539–2550. doi: 10.1093/eurheartj/ehm037 [DOI] [PubMed] [Google Scholar]
- 20.Lindenfeld J, Albert NM, Boehmer JP, Collins SP, Ezekowitz JA, Givertz MM, Katz SD, Klapholz M, Moser DK, Rogers JG, et al. HFSA 2010 Comprehensive Heart Failure Practice Guideline. J Card Fail. 2010;16:e1–194. doi: 10.1016/j.cardfail.2010.04.004 [DOI] [PubMed] [Google Scholar]
- 21.Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47:W199–W205. doi: 10.1093/nar/gkz401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griss J, Viteri G, Sidiropoulos K, Nguyen V, Fabregat A, Hermjakob H. ReactomeGSA - Efficient Multi-Omics Comparative Pathway Analysis. Mol Cell Proteomics. 2020;19:2115–2125. doi: 10.1074/mcp.TIR120.002155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Litviňuková M, Talavera-López C, Maatz H, Reichart D, Worth CL, Lindberg EL, Kanda M, Polanski K, Heinig M, Lee M, et al. Cells of the adult human heart. Nature. 2020;588:466–472. doi: 10.1038/s41586-020-2797-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tucker NR, Chaffin M, Fleming SJ, Hall AW, Parsons VA, Bedi KC, Akkad AD, Herndon CN, Arduini A, Papangeli I, et al. Transcriptional and Cellular Diversity of the Human Heart. Circulation. 2020;142:466–482. doi: 10.1161/CIRCULATIONAHA.119.045401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donaldson C, Taatjes DJ, Zile M, Palmer B, VanBuren P, Spinale F, Maughan D, Von Turkovich M, Bishop N, LeWinter MM. Combined immunoelectron microscopic and computer-assisted image analyses to detect advanced glycation end-products in human myocardium. Histochem Cell Biol. 2010;134:23–30. doi: 10.1007/s00418-010-0706-x [DOI] [PubMed] [Google Scholar]
- 26.Shah SJ, Borlaug BA, Kitzman DW, McCulloch AD, Blaxall BC, Agarwal R, Chirinos JA, Collins S, Deo RC, Gladwin MT, et al. Research Priorities for Heart Failure With Preserved Ejection Fraction: National Heart, Lung, and Blood Institute Working Group Summary. Circulation. 2020;141:1001–1026. doi: 10.1161/CIRCULATIONAHA.119.041886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Senni M, Caravita S, Paulus WJ. Do Existing Definitions Identify Subgroup Phenotypes or Reflect the Natural History of Heart Failure With Preserved Ejection Fraction? Circulation. 2019;140:366–369. doi: 10.1161/CIRCULATIONAHA.119.041657 [DOI] [PubMed] [Google Scholar]
- 28.Lang RM, Badano LP, Mor-Avi V, Afilalo J, Armstrong A, Ernande L, Flachskampf FA, Foster E, Goldstein SA, Kuznetsova T, et al. Recommendations for cardiac chamber quantification by echocardiography in adults: an update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. J Am Soc Echocardiogr. 2015;28:1–39.e14. doi: 10.1016/j.echo.2014.10.003 [DOI] [PubMed] [Google Scholar]
- 29.Grebe A, Hoss F, Latz E. NLRP3 Inflammasome and the IL-1 Pathway in Atherosclerosis. Circ Res. 2018;122:1722–1740. doi: 10.1161/CIRCRESAHA.118.311362 [DOI] [PubMed] [Google Scholar]
- 30.McDonald LT, Zile MR, Zhang Y, Van Laer AO, Baicu CF, Stroud RE, Jones JA, LaRue AC, Bradshaw AD. Increased macrophage-derived SPARC precedes collagen deposition in myocardial fibrosis. Am J Physiol Heart Circ Physiol. 2018;315:H92–H100. doi: 10.1152/ajpheart.00719.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Brien M, Baicu CF, Van Laer AO, Zhang Y, McDonald LT, LaRue AC, Zile MR, Bradshaw AD. Pressure overload generates a cardiac-specific profile of inflammatory mediators. Am J Physiol Heart Circ Physiol. 2020;319:H331–H340. doi: 10.1152/ajpheart.00274.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaye DM, Nanayakkara S, Wang B, Shihata W, Marques FZ, Esler M, Lambert G, Mariani J. Characterization of Cardiac Sympathetic Nervous System and Inflammatory Activation in HFpEF Patients. JACC Basic Transl Sci. 2022;7:116–127. doi: 10.1016/j.jacbts.2021.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee JH, Jun HS. Role of Myokines in Regulating Skeletal Muscle Mass and Function. Front Physiol. 2019;10:42. doi: 10.3389/fphys.2019.00042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Tassell BW, Trankle CR, Canada JM, Carbone S, Buckley L, Kadariya D, Del Buono MG, Billingsley H, Wohlford G, Viscusi M, et al. IL-1 Blockade in Patients With Heart Failure With Preserved Ejection Fraction. Circ Heart Fail. 2018;11:e005036. doi: 10.1161/CIRCHEARTFAILURE.118.005036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Everett BM, Cornel JH, Lainscak M, Anker SD, Abbate A, Thuren T, Libby P, Glynn RJ, Ridker PM. Anti-Inflammatory Therapy With Canakinumab for the Prevention of Hospitalization for Heart Failure. Circulation. 2019;139:1289–1299. doi: 10.1161/CIRCULATIONAHA.118.038010 [DOI] [PubMed] [Google Scholar]
- 36.Cunningham JW, Claggett BL, O’Meara E, Prescott MF, Pfeffer MA, Shah SJ, Redfield MM, Zannad F, Chiang LM, Rizkala AR, et al. Effect of Sacubitril/Valsartan on Biomarkers of Extracellular Matrix Regulation in Patients With HFpEF. J Am Coll Cardiol. 2020;76:503–514. doi: 10.1016/j.jacc.2020.05.072 [DOI] [PubMed] [Google Scholar]
- 37.De Marco C, Claggett BL, de Denus S, Zile MR, Huynh T, Desai AS, Sirois MG, Solomon SD, Pitt B, Rouleau JL, et al. Impact of diabetes on serum biomarkers in heart failure with preserved ejection fraction: insights from the TOPCAT trial. ESC Heart Fail. 2021;8:1130–1138. doi: 10.1002/ehf2.13153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyer M, Schillinger W, Pieske B, Holubarsch C, Heilmann C, Posival H, Kuwajima G, Mikoshiba K, Just Hr, Hasenfuss G. Alterations of Sarcoplasmic Reticulum Proteins in Failing Human Dilated Cardiomyopathy. Circulation. 1995;92:778–784. doi: 10.1161/01.cir.92.4.778 [DOI] [PubMed] [Google Scholar]
- 39.Ruel M, Bianchi C, Khan TA, Xu S, Liddicoat JR, Voisine P, Araujo E, Lyon H, Kohane IS, Libermann TA, et al. Gene expression profile after cardiopulmonary bypass and cardioplegic arrest. J Thorac Cardiovasc Surg. 2003;126:1521–1530. doi: 10.1016/s0022-5223(03)00969-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw RNA-sequencing data are deposited at National Center for Biotechnology Information (NCBI’s) sequence read archive (SRA) under Bioproject accession number PRJNA879763. Parallel processed gene array data (Affymetrix ClariomD) are also available upon a reasonable request to the corresponding author.