Abstract
Antiserum prepared against an amino-terminal fragment of rubella virus (RUB) nonstructural polyprotein was used to study RUB-infected Vero cells. Replicase protein P150 was associated with vesicles and vacuoles of endolysosomal origin and later with large, convoluted, tubular membrane structures. Newly incorporated bromouridine was associated with the same structures and specifically with small membrane invaginations, spherules, indicating that these structures may be the sites of viral RNA synthesis.
Rubella virus (RUB) is an enveloped positive-strand RNA virus, the only member of the Rubivirus genus of the family Togaviridae (28). The RUB genome (9,756 nucleotides [nt]) contains two long open reading frames (ORFs) organized in a way similar to the genome of alphaviruses, the other genus in the family Togaviridae. The 3′ ORF (3,189 nt) codes for a capsid protein (C) and two envelope glycoproteins (E1 and E2) (for a review, see reference 7). The larger 5′-proximal ORF (6,645 nt) encodes viral nonstructural proteins P150 and P90, which are processed from a P200 (237-kDa) polyprotein by a single cis cleavage mediated by a papain-like cysteine protease (4, 6, 18, 20). Amino acid sequence comparisons have shown that RUB belongs to a large superfamily consisting of alphaviruses, hepatitis E virus, and a number of plant viruses, the replicase proteins of which harbor the same methyltransferase, helicase, and polymerase motifs (15).
In RUB-infected Vero cells, a gradual virus titer increase starts after 12 h postinfection (p.i.), leveling off at 36 to 48 h p.i. The synthesis rate of viral genome 40S RNA and the subgenomic mRNA of structural proteins peaks at about 24 h p.i. (7, 14). Immunoelectron microscopy using antibodies against double-stranded RNA has suggested that RUB RNA synthesis takes place in cytoplasmic structures resembling type I cytopathic vacuoles (CPVIs) (17, 19), which have previously been described for alphaviruses (1, 9, 10). Less is known about the functions and intracellular localization of RUB P150 and P90. Here we have prepared a potent antiserum against P150 and studied the intracellular localization of this replicase protein in RUB-infected Vero cells.
RNA was isolated from purified RUB strain Therian virions (21) with the RNeasy kit (Qiagen), and cDNA was synthesized with reverse transcriptase (Gibco BRL) by using an oligo(dT) primer. This was used in a PCR with primers 5′ CGGAATTCCCATGGAGAAACTCCTAGATGAGG 3′ and 5′ TCACAAGCTTATTCGCGCGGGACGTCGCAGCGGGGA 3′. The product was cloned into vector pCR2.1 (Invitrogen), and the insert was sequenced. For expression in Escherichia coli, the insert was transferred to vector pHAT (25), giving pHATRUB (encoding amino acids 1 to 505 of P150, here called p55).
Plasmid pHATRUB was transformed into E. coli BL21, and expression was induced by incubation with 300 μM isopropyl-β-d-thiogalactopyranoside for 4 h. Cells were pelleted, resuspended in 50 mM Tris-HCl (pH 8.0)–50 mM NaCl–0.1% Tween 20–1 mM phenylmethylsulfonyl fluoride (buffer A), and broken with a French press. The cell lysate was centrifuged at 15,000 × g for 15 min, and the pellet fraction was washed twice with buffer A supplemented with 20% glycerol and with 2 M urea. Inclusions consisting of p55 protein were placed in 0.1% sodium dodecyl sulfate (SDS) and mixed with complete Freund’s adjuvant. Antigen (20 μg) was injected into the popliteal lymph nodes of each of two rabbits. Two weeks later, subcutaneous injections of 50 μg of antigen per rabbit in incomplete Freund’s adjuvant were given at a total of four different sites. Identical booster injections were given 6, 10, and 14 weeks after the first injection. Blood was collected at day 10 after the fifth injection. Antiserum was absorbed with HeLa or Vero cells fixed with 2% paraformaldehyde and permeabilized with 0.1% Triton X-100 for 60 min at room temperature (RT).
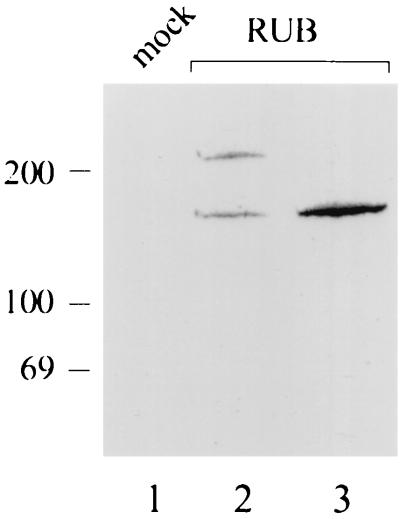
The reactivity of the immune serum was studied by immunoprecipitation using RUB-infected Vero cells (5 PFU/cell) which had been labeled for 60 min with [35S]methionine (200 μCi/60-mm-diameter dish) at 44 h p.i. and then chased with excess unlabeled methionine. Proteins were denatured with SDS and immunoprecipitated by using protein A-Sepharose as previously described (26). After a 15-min chase, 220- and 150-kDa proteins were precipitated (Fig. 1, lane 2). The 220-kDa protein disappeared, and the intensity of the 150-kDa protein increased after a chase period of 90 min (Fig. 1, lane 3). No proteins were immunoprecipitated from similarly labeled mock-infected cells (lane 1). In accordance with earlier studies (6, 20), the larger, unstable protein was designated RUB-specific nonstructural polyprotein P200 and the smaller protein was designated its N-terminal cleavage product P150.
FIG. 1.
Detection of RUB nonstructural proteins. Vero cells were infected with RUB, labeled with [35S]methionine for 1 h at 44 h p.i., and chased with excess unlabeled methionine. Cell lysates were immunoprecipitated with anti-p55 antibodies and subjected to analysis by SDS–10% polyacrylamide gel electrophoresis, followed by fluorography. Molecular mass markers (kilodaltons) are shown on the left. Lanes: 1, precipitate from labeled mock-infected cells; 2, RUB-infected cells after a 15-min chase; 3, RUB-infected cells after a 90-min chase.
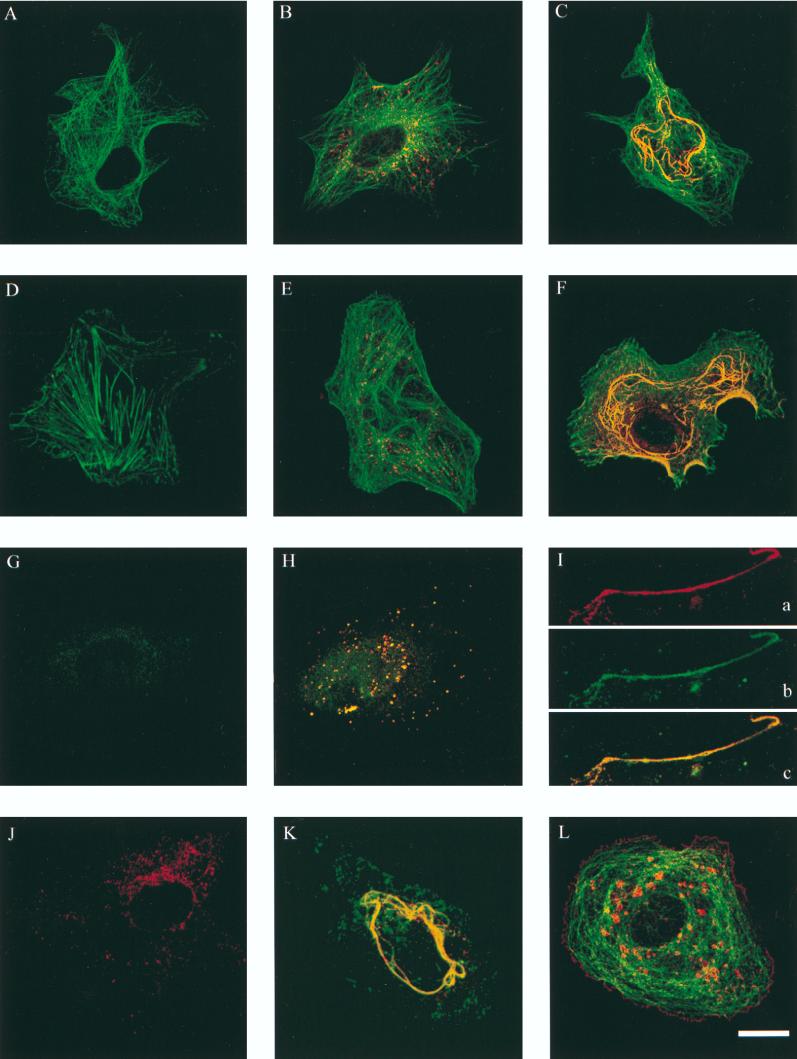
Localization of P150 in RUB-infected cells by confocal microscopy.
Indirect immunofluorescence microscopy (24) was carried out for RUB-infected Vero cells (multiplicity of infection of 50) at various time points between 18 and 72 h p.i. by using a Bio-Rad MRC-1024 confocal microscope. In mock-infected cells double labeled with tetramethyl rhodamine isocyanate (TRITC)-stained anti-tubulin (B-5-1-2; Sigma) and fluorscein isothiocyanate (FITC)-stained anti-p55 antibodies, only the tubulin network was visible (Fig. 2A). In RUB-infected cells at 24 (Fig. 2B) and 33 (data not shown) h p.i., bright spotted fluorescent staining (red) was seen in addition to the microtubulin network (green). At 48 h p.i., convoluted tubular structures were visualized by anti-p55 antibody staining (Fig. 2C). Oregon green-conjugated phalloidin (Molecular Probes Europe BV) revealed F-actin stress fibers in the mock-infected control cells (green fluorescence in Fig. 2D). In RUB-infected cells at 24 h p.i., stress fibers were still visible (Fig. 2E), whereas in cells infected with RUB for 48 h, they had disappeared (Fig. 2F). Double staining with anti-p55 antibodies again showed the spotted (red) fluorescence at 24 h p.i. and the tubular structures at 48 h p.i. (Fig. 2E and F, respectively). Disruption of F-actin stress fibers was regularly seen in RUB-infected cells 48 to 72 h p.i. This phenomenon has been described previously for both Vero and BHK cells infected with RUB (2, 3). We have recently shown that NSP1 of Semliki Forest virus (SFV) and Sindbis virus is responsible for the disappearance of stress fibers during alphavirus infection (16).
FIG. 2.
Confocal fluorescence images of RUB-infected Vero cells. TRITC-stained RUB P150 (red) is shown double labeled with FITC-stained (green) microtubules (A, B, and C), actin filaments (D, E, and F), metabolically incorporated bromouridine (G, H, and I), and RUB capsid protein (K). In mock-infected cells (A, D, and E), no P150-specific staining is seen, while at 24 h post-RUB infection (B, E, and H), P150 appears in small vacuolar structures, which are extended to a large, convoluted, tubular network at 48 h p.i. (C, F, I, and K). Colocalization (yellow) of P150 with bromouridine is evident early in infection as bright dots and also later at tubular structures (H and I, respectively). A three-dimensional reconstruction image of P150 and the capsid protein (K) shows colocalization of these antigens in the virus-induced tubular structures late in infection. In contrast to the capsid protein, TRITC-stained RUB structural protein E2 (J; red) does not localize into tubular structures but shows staining in the perinuclear region. For comparison, a Vero cell infected with SFV for 4 h and stained with antisera against SFV NSP1 (TRITC; red) and microtubules (FITC; green) is shown in panel L. Bar, 10 μm.
Localization of RNA synthesis sites.
To visualize the sites of cytoplasmic RNA synthesis, RUB-infected Vero cells were exposed to 20 mM bromo-UTP (Sigma) between 23 and 24 or 47 and 48 h p.i. using Lipofectin (GIBCO BRL) as a carrier. Dactinomycin (5 μg/ml) was added 30 min before exposure to bromo-UTP to shut off host RNA synthesis. The bromouridine incorporated into RNA was visualized by rat monoclonal antibody (BU1/75 [ICR1]; Harlan Sera-Lab Ltd., Loughborough, United Kingdom) against bromodeoxyuridine (BrdU) plus FITC–anti-rat immunoglobulin G (green fluorescence). In the presence of actinomycin D, negligible fluorescence was detected in mock-infected cells (Fig. 2G). Double labeling with anti-p55 antibody (red revealed colocalization (yellow) of both labels in vesicular-vacuolar structures at 24 h p.i. (Fig. 2H), suggesting that the newly synthesized RNA and P150 were localized in the same structures. At 48 h p.i., the long, tubular structures were labeled with both anti-p55 (Fig. 2Ia) and anti-BrdU (Fig. 2Ib) antibodies and the labels colocalized at the light microscopic level, as shown by the yellow staining in Fig. 2Ic.
The distribution of RUB envelope glycoprotein E2 was visualized with a rabbit polyclonal antiserum (22). E2 was concentrated in the perinuclear area at 24 h p.i. (Fig. 2J) and also at the plasma membrane at 48 h p.i. (not shown). In contrast, anti-capsid antibodies (31) at 48 h p.i. decorated structures similar to those seen by anti-p55 antibodies at the same time point. Both stains colocalized in the convoluted tubule-like structures in a three-dimensional reconstruction (Fig. 2K), indicating close proximity of P150 and the capsid protein in unique virus-specific structures. The tubular structures found in RUB-infected cells could not be seen in SFV-infected Vero cells with antibodies against NSP1, -2, -3, or -4 (24). As an example, we show anti-NSP1 labeling of plasma membrane and CPVIs in SFV-infected Vero cells double stained with anti-tubulin (green) antibodies (Fig. 2L).
Ultrastructure of RUB-infected cells.
RUB-infected (multiplicity of infection; 50) and mock-infected Vero cells were exposed at the time of infection to colloidal gold particles (27) (5-nm diameter), 250 μl/ml, and coated with bovine serum albumin (BSA) (29). After 1 h of adsorption at 37°C, the inoculum was removed and replaced with culture medium containing 2% fetal calf serum. At the indicated times, the cells on coverslips were fixed with 2.5% glutaraldehyde in 0.2 M cacodylate, pH 7.2, for 20 min at RT and postfixed in 1% OsO4 in 0.2 M cacodylate for 30 min at RT and then left overnight in 1% uranyl acetate in 0.3 M sucrose at 4°C before ethanol dehydration and embedding Epon resin.
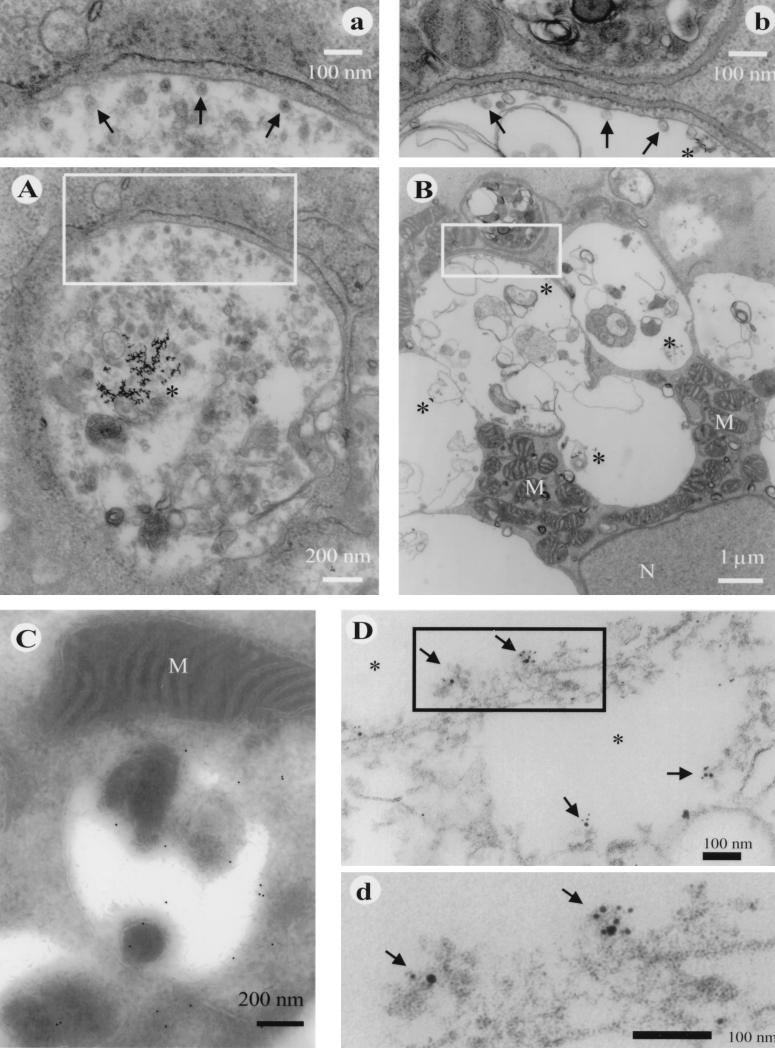
Electron micrographs of thin sections at 18 to 72 h p.i. revealed vacuolar structures of various sizes (0.5 to 2 μm in diameter), some of which were filled with membranous material (Fig. 3). Gold particles were regularly seen in filled (Fig. 3A and insert) and empty vacuoles (Fig. 3B). The inner surface of the vacuoles had small vesicles or spherules (Fig. 3A and B) which, at a higher magnification, turned out to be invaginations with a connecting channel to the overlying smooth membrane (inserts a and b). The spherules, 30 to 60 nm in diameter, were morphologically similar to those described for SFV- and Sindbis virus-infected cells (9, 10, 12, 17), but their number on the membrane surface was clearly less than in the typical alphavirus CPVIs. The vacuolar membrane was often surrounded by a rim of rough endoplasmic reticulum (ER) within a distance of 50 to 200 nm (Fig. 3A and B, inserts). Although similar-size vacuoles containing BSA-gold were also seen in the mock-infected cells, these vacuoles contained no spherules and had no rims of ER membranes in their close vicinity (not shown).
FIG. 3.
Ultrastructure of cytopathic vacuoles in RUB-infected Vero cells. (A) CPVI at 18 h after RUB infection showing endocytosed BSA-gold in the lumen (asterisks) and small spherules at the inner aspect of the vacuolar membrane (enlarged in panel a). (B) General view of a large vacuolar structure at 24 h p.i. containing BSA-gold (asterisks) with numerous mitochondria (M) near the nucleus (N). The enlargement in panel b demonstrates spherules (arrows), BSA-gold (asterisks), and the close proximity of rough ER membranes aligning with the vacuole membrane. (C) Localization of P150 by the cryoimmuno technique to the membrane of a vacuole. Anti-p55 antibody was detected by 10-nm gold–protein A particles. (D) Pre-embedding immunoelectron microscopic image of Triton X-100-treated Vero cells at 48 h p.i. Shown is the intracellular localization of RUB P150 together with metabolically incorporated bromouridine. P150 is labeled with 5-nm gold particles, the bromouridine-RNA is labeled with 10-nm gold particles, and both are visualized best in the enlargement (d). Arrows point to labeled spherule structures. The vacuolar lumen is marked with asterisks, and M stands for mitochondria.
By cryoimmunoelectron microscopy, for which ultrathin frozen sections were prepared as described by Tokuyasu (30), P150 was localized by protein A-conjugated colloidal gold particles (J. Slot laboratory). These were found mostly in close proximity to the inner smooth membranes of the vacuolar structures and the surrounding rough ER membranes (Fig. 3C). The localization of P150 was also studied by treating the fixed and permeabilized cells as for immunofluorescence staining. The pre-embedded samples were postfixed with 3% glutaraldehyde in 0.2 M piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES, pH 7.2) before osmium treatment and treated otherwise as described above. The double labeling with anti-p55 and anti-BrdU antibodies was carried out by using 5-nm anti-rabbit and 10-nm anti-rat gold particles (Sigma), respectively. At 48 h p.i., spherule-like structures on the inner surface of vacuolar remnants were costained with both 5- and 10-nm gold particles (Fig. 3D), suggesting that the spherules are the sites of the replication complexes in RUB-infected cells.
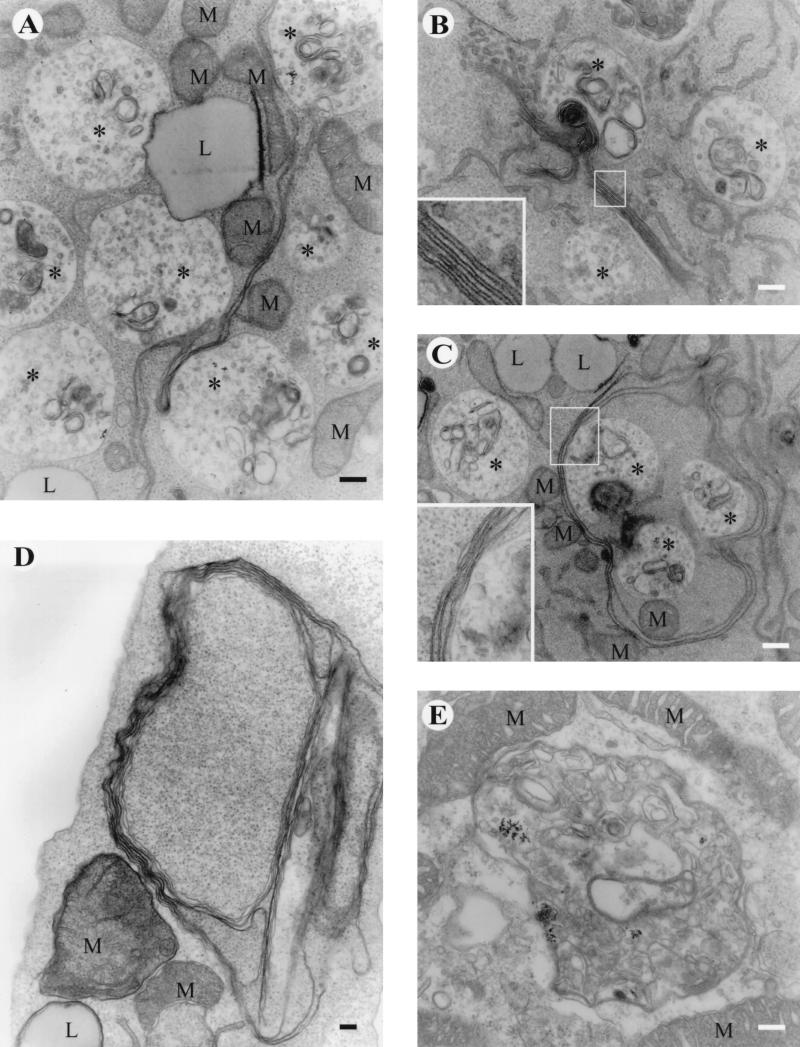
From 48 to 72 h p.i., conventional electron micrographs of thin sections of RUB-infected cells showed similar vacuoles, which contained BSA-gold (Fig. 4A). The association with ER membranes was not as evident as earlier. Instead, long, hairy, tubular structures seemed to connect the vacuoles to each other (Fig. 4A and C). Sometimes these large proliferated membrane structures consisted of several stacked lamina-like membranes (Fig. 4B, C, and D). Similar structures were not seen in mock-infected Vero cells maintained under similar conditions for 72 h (Fig. 4E).
FIG. 4.
Electron microscopy of BSA-gold-labeled Vero cells at 48 (A, B, and C) or 72 (D) h after RUB infection. A mock-infected Vero cell at 72 h is shown in panel E. Panels A to C show long, virus-induced, tubular membrane structures connecting several endocytosed gold-containing CPVIs (asterisks). These large, proliferated membrane structures often consisted of several stacked lamina-like membranes close to CPVIs (B, C, and the enlarged inserts). In panel D, a large, convoluted membrane structure surrounds part of the cell cytoplasm. M stands for mitochondria, and L stands for membrane-bound lipid vesicles. Bars, 200 nm.
Here we have shown by confocal microscopy and immunoelectron microscopy that starting at 24 h p.i., RUB-specific P150 in Vero cells is localized in vacuolar structures which costained with newly synthesized BrdU-labeled RNA (Fig. 2 and 3). The vacuoles must be of endosomal and lysosomal origin, since they contained BSA-coated gold particles which had been endocytosed from the medium during the virus adsorption period (Fig. 3). Like late endosomes and lysosomes, they also contained membranous material, which was evidently derived by an autophagocytosis type of process taking place upon the maturation of endosomes to lysosomes 5, 13, 29. The luminal surface of the vacuoles had vesicular invaginations or spherules similar to those described in the inner surface of alphavirus CPVIs (1, 11). These structures have been described for RUB-infected cells by Lee et al. (17), and their lysosomal origin was suggested by costaining with anti-double-stranded RNA serum and anti-human lamp1 antibodies (19). An interesting progression in the endosomal-lysosomal compartment took place later in RUB infection, resulting in large, tubular, membranous structures which stained intensively with anti-p55 and anti-capsid protein antibodies (Fig. 2 and 4).
Information concerning the origin and biogenesis of the CPVI-type structures in togavirus-infected cells has accumulated slowly since they were first described in 1967 (1, 8, 12). Grimley et al. (12) showed that some of the CPVIs stained with Gomoris stain, indicating that they contained acid phosphatase. Later, CPVIs were stained with both endosomal and lysosomal markers in both alphavirus- and RUB-infected cells (10, 19, 23). Our finding that SFV NSP1, when expressed alone, has affinity for endosomes and lysosomes suggested a possible targeting mechanism of the replication complex to the endosomal apparatus (24). The mechanism by which the invaginations or spherules arise is still unknown. If P150 of RUB is responsible for the recognition of lysosomal membranes, alphavirus NSP1 and P150 may have some features in common. The localization of the site of RNA synthesis has been difficult to define within the CPVI structure. Electron microscopic autoradiography with incorporated tritiated uridine and adenine suggested that the source of radiation was indeed CPVI (9, 17). Lee et al. (17) showed that double-stranded RNA in RUB- and SFV-infected cells was localized in the lumen of the CPVI in permeabilized cells. Our present finding that pulse-labeled BrdU-RNA seemed to localize to the spherules, together with P150 (Fig. 3D), suggests that these structures are the actual RNA replication sites. The invaginations are only about 30 to 60 nm in diameter, but they ought to be able to accommodate the 40S RNA minus strand of approximately 4 μm. Further studies are needed to determine whether and how the template is confined to such a small space.
Acknowledgments
We thank Airi Sinkko, Arja Strandell, and Tarja Välimäki for excellent technical assistance, and Marja Makarow and Eeva-Liisa Punnonen for critical reading of the manuscript.
This work was supported by the Technology Development Center (TEKES) and the Academy of Finland (grant 8397). L.K. is a Biocentrum Helsinki fellow.
REFERENCES
- 1.Acheson N H, Tamm I. Replication of Semliki Forest virus: an electron microscopic study. Virology. 1967;32:128–143. doi: 10.1016/0042-6822(67)90261-9. [DOI] [PubMed] [Google Scholar]
- 2.Bowden D S, Westaway E G. Rubella virus products and their distribution in infected cells. Subcell Biochem. 1989;15:203–231. doi: 10.1007/978-1-4899-1675-4_7. [DOI] [PubMed] [Google Scholar]
- 3.Bowden D S, Pedersen J S, Toh B H, Westaway E G. Distribution by immunofluorescence of viral products and actin-containing cytoskeletal filaments in rubella virus-infected cells. Arch Virol. 1987;92:211–219. doi: 10.1007/BF01317478. [DOI] [PubMed] [Google Scholar]
- 4.Chen J, Strauss J H, Strauss E G, Frey T K. Characterization of the rubella virus nonstructural protease domain and its cleavage site. J Virol. 1996;70:4707–4713. doi: 10.1128/jvi.70.7.4707-4713.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clague M J. Molecular aspects of the endocytic pathway. Biochem J. 1998;336:271–282. doi: 10.1042/bj3360271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forng R-Y, Frey T K. Identification of rubella virus nonstructural proteins. Virology. 1995;206:843–853. doi: 10.1006/viro.1995.1007. [DOI] [PubMed] [Google Scholar]
- 7.Frey T K. Molecular biology of rubella virus. Adv Virus Res. 1994;44:69–161. doi: 10.1016/S0065-3527(08)60328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedman R M, Berezesky I K. Cytoplasmic fractions associated with Semliki Forest virus ribonucleic acid replication. J Virol. 1967;1:374–383. doi: 10.1128/jvi.1.2.374-383.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedman R M, Levin J G, Grimley P M, Berezesky I K. Membrane-associated replication complex in arbovirus infection. J Virol. 1972;10:504–515. doi: 10.1128/jvi.10.3.504-515.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Froshauer S, Kartenbeck J, Helenius A. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J Cell Biol. 1988;107:2075–2086. doi: 10.1083/jcb.107.6.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grimley P M, Berezesky I K, Friedman R M. Cytoplasmic structures associated with an arbovirus infection: loci of viral ribonucleic acid synthesis. J Virol. 1968;2:1326–1338. doi: 10.1128/jvi.2.11.1326-1338.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grimley P M, Levin J G, Berezesky I K, Friedman R M. Specific membranous structures associated with the replication of group A arboviruses. J Virol. 1972;10:492–503. doi: 10.1128/jvi.10.3.492-503.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gruenberg J, Maxfield F R. Membrane transport in the endocytic pathway. Curr Opin Cell Biol. 1995;7:552–563. doi: 10.1016/0955-0674(95)80013-1. [DOI] [PubMed] [Google Scholar]
- 14.Hemphill M L, Forng R, Abernathy E S, Frey T K. Time course of virus-specific macromolecular synthesis during rubella virus infection in Vero cells. Virology. 1988;162:65–75. doi: 10.1016/0042-6822(88)90395-9. [DOI] [PubMed] [Google Scholar]
- 15.Koonin E V, Dolja V V. Evolution and taxonomy of positive-strand RNA-viruses: implications of comparative analysis of amino acid sequences. Crit Rev Biochem Mol Biol. 1993;28:375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- 16.Laakkonen P, Auvinen P, Kujala P, Kääriäinen L. Alphavirus replicase protein NSP1 induces filopodia and rearrangement of actin filaments. J Virol. 1998;72:10265–10269. doi: 10.1128/jvi.72.12.10265-10269.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee J-Y, Marshall J A, Bowden D S. Characterization of rubella virus replication complexes using antibodies to double-stranded RNA. Virology. 1994;200:307–312. doi: 10.1006/viro.1994.1192. [DOI] [PubMed] [Google Scholar]
- 18.Liu X, Ropp S L, Jackson R J, Frey T K. The rubella virus nonstructural protease requires divalent cations for activity and functions in trans. J Virol. 1998;72:4463–4466. doi: 10.1128/jvi.72.5.4463-4466.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magliano D, Marshall J A, Bowden D S, Vardaxis N, Meanger J, Lee J. Rubella virus replication complexes are virus-modified lysosomes. Virology. 1998;240:57–63. doi: 10.1006/viro.1997.8906. [DOI] [PubMed] [Google Scholar]
- 20.Marr L D, Wang C-Y, Frey T K. Expression of the rubella virus nonstructural protein ORF and demonstration of proteolytic processing. Virology. 1994;198:586–592. doi: 10.1006/viro.1994.1070. [DOI] [PubMed] [Google Scholar]
- 21.Oker-Blom C, Kalkkinen N, Kääriäinen L, Pettersson R F. Rubella virus contains one capsid protein and three envelope proteins, E1, E2a, and E2b. J Virol. 1983;46:964–973. doi: 10.1128/jvi.46.3.964-973.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oker-Blom C. The gene order for rubella virus structural proteins is NH2-C-E2-E1-COOH. J Virol. 1984;51:354–358. doi: 10.1128/jvi.51.2.354-358.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peränen J, Kääriäinen L. Biogenesis of type I cytopathic vacuoles in Semliki Forest virus-infected BHK cells. J Virol. 1991;65:1623–1627. doi: 10.1128/jvi.65.3.1623-1627.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peränen J, Laakkonen P, Hyvönen M, Kääriäinen L. The alphavirus replicase protein nsP1 is membrane-associated and has affinity to endocytic organelles. Virology. 1995;208:610–620. doi: 10.1006/viro.1995.1192. [DOI] [PubMed] [Google Scholar]
- 25.Peränen J, Rikkonen M, Hyvönen M, Kääriäinen L. T7 vectors with a modified T7lac promoter for expression of proteins in Escherichia coli. Anal Biochem. 1996;236:371–373. doi: 10.1006/abio.1996.0187. [DOI] [PubMed] [Google Scholar]
- 26.Peränen J, Takkinen K, Kalkkinen N, Kääriäinen L. Semliki Forest virus-specific non-structural protein nsP3 is a phosphoprotein. J Gen Virol. 1988;69:2165–2178. doi: 10.1099/0022-1317-69-9-2165. [DOI] [PubMed] [Google Scholar]
- 27.Slot J W, Geuze H J. A new method of preparing gold probes for multiple-labeling cytochemistry. Eur J Cell Biol. 1985;38:87–93. [PubMed] [Google Scholar]
- 28.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tjelle T E, Brech A, Juvet L K, Griffiths G, Berg T. Isolation and characterization of early endosomes, late endosomes and terminal lysosomes: their role in protein degradation. J Cell Sci. 1996;109:2905–2914. doi: 10.1242/jcs.109.12.2905. [DOI] [PubMed] [Google Scholar]
- 30.Tokuyasu K T. Use of polyvinylpyrrolidone and polyvinyl alcohol for cryoultratomy. Histochem J. 1989;21:163–171. doi: 10.1007/BF01007491. [DOI] [PubMed] [Google Scholar]
- 31.Wolinsky J S, McCarthy M, Allen-Cannady O, Moore W T, Jin R, Cao S, Lovett A, Simmons D. Monoclonal antibody-defined epitope map of expressed rubella virus protein domains. J Virol. 1991;65:3986–3994. doi: 10.1128/jvi.65.8.3986-3994.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]