Abstract
The loss of IL-10R function leads to severe early onset colitis and, in murine models, is associated with the accumulation of immature inflammatory colonic macrophages. We have shown that IL-10R-deficient colonic macrophages exhibit increased STAT1-dependent gene expression, suggesting that IL-10R-mediated inhibition of STAT1 signaling in newly recruited colonic macrophages might interfere with the development of an inflammatory phenotype. Indeed, STAT1−/− mice exhibit defects in colonic macrophage accumulation after Helicobacter hepaticus infection and IL-10R blockade, and this was phenocopied in mice lacking IFNγR, an inducer of STAT1 activation. Radiation chimeras demonstrated that reduced accumulation of STAT1-deficient macrophages was based on a cell-intrinsic defect. Unexpectedly, mixed radiation chimeras generated with both wild-type and IL-10R-deficient bone marrow indicated that rather than directly interfering with STAT1 function, IL-10R inhibits the generation of cell extrinsic signals that promote the accumulation of immature macrophages. These results define the essential mechanisms controlling the inflammatory macrophage accumulation in inflammatory bowel diseases.
INTRODUCTION
Rare homozygous loss-of-function mutations in interleukin (IL)-10 or its receptor are among the most common monogenic disorders associated with severe forms of infantile colitis1–3. Affected patients often fail to respond to conventional therapies, and although bone marrow transplantation may be curative, there is still a significant unmet need for new and bridging therapies. Despite significant study, a detailed mechanistic understanding of how IL-10 signaling interferes with the development of intestinal inflammation remains elusive. Increasing evidence implicates macrophages as central regulators of intestinal inflammation4,5. Gut macrophages reside in the lamina propria (LP) beneath the epithelial monolayer and help maintain mucosal homeostasis under steady state condition. Macrophages within the intestinal LP are phenotypically diverse. At birth, the tissue resident macrophages in the colon are primarily embryonically derived5. These macrophages are gradually replaced and continuously replenished by the recruitment of lymphocyte antigen 6 complex (Ly6C)high major histocompatibility complex (MHC)IIlow blood-derived monocytes, which differentiate into mature Ly6C− and MHCIIhigh macrophages with a tissue resident-like phenotype. Under homeostasis, the recruitment of these Ly6Chigh monocytes is largely driven by the microbiota and depends on the expression of C-C chemokine receptor type 2 (CCR2)6–10.
Mice lacking either IL-10 itself or either of the two chains of the IL-10 receptor (IL-10R) develop microbiota-dependent colitis, which is associated with a marked increase in the accumulation of inflammatory macrophages in the colon7,11–13. Remarkably, we and others have shown that mice specifically lacking the IL-10R in myeloid cells (Lyz2-Cre) also develop colitis characterized by the accumulation of immature Ly6C+ macrophages within the colonic LP6,7, indicating that IL-10R signaling within the myeloid compartment inhibits the accumulation of immature macrophages. Although both newly recruited Ly6C+ immature macrophages and Ly6C− mature macrophages are capable of producing pro-inflammatory cytokines after stimuli, it has been argued that newly recruited monocytes are the most significant source of the observable pro-inflammatory cytokine signature, including IL-12, IL-23, and interferon-stimulated genes6,14, suggesting that the ability of IL-10 to prevent the accumulation of these cells and/or their ability to produce inflammatory cytokines could be an important function involved in inhibiting the development of colitis.
We have recently shown that the loss of CCR2 in mice lacking IL-10R interferes with the accumulation of immature Ly6C+ colonic macrophages, likely secondary to the loss of CCR2-mediated monocyte recruitment10. However, the identification of intrinsic cytokine response pathways that are necessary for monocyte/macrophage accumulation within the colon of IL-10R-deficient mice is incompletely understood. We have demonstrated that intestinal macrophages isolated from Il10ra−/− or Il10rb−/− mice exhibit an increased expression of interferon (IFN)-γ-inducible genes, including Cxcl9, Cxcl11, and Iigp17,10.Increased IFN-γ production is a hallmark of disease in IL-10R-deficient mouse models, likely as a result of failure to control IL-12-mediated T helper (Th)1 responses15,16. IFN-γ is also identified as a macrophage-activating cytokine that turns off gene expression associated with M2 type macrophage polarization17,18. In addition, patients with defects in IL-10R signaling present with strong IFN-γ expression profiles in colonic tissue19.
Signal transducer and activator of transcription 1 (STAT1) is the primary transducer of IFN-γ signals20,21; however, the role of STAT1 in driving the development of colitis in IL-10R-deficient mice has yet to be defined. The deletion of STAT1 interferes with the development of dextran sodium sulfate (DSS)-induced colitis and is associated with decreased accumulation of inflammatory macrophages within the colonic LP22, a phenotype also observed in mice lacking Ifngr1 but not Ifnar1. STAT1 has previously been implicated in the development of human inflammatory bowel disease (IBD)23–25, and a robust induction of a STAT1-regulated gene network was identified within the terminal ileum of pediatric patients with newly diagnosed Crohn’s disease26. It has previously been suggested that IL-10 may directly inhibit STAT1 function and interfere with STAT1-induced gene expression27, but to the best of our knowledge, the direct evaluation of STAT1 function in regulating monocyte/macrophage accumulation within the colon of IL-10R-deficient mice has not been previously determined. Given the potential promise of JAK/STAT inhibitors in treating inflammatory conditions, including IBD, a detailed understanding of STAT1 function in a physiologic model of microbiota-driven colitis could lead to important insights with the potential to guide therapeutic approaches28–30.
Here, we show that a cell autonomous function of STAT1 is necessary for the accumulation of immature macrophages within the intestine of mice lacking IL-10R signaling and is Phenocopied by the absence of IFN-γR, suggesting that an intrinsic IFN-γR/STAT1 pathway drives the accumulation of immature macrophages in mice with defects in IL-10R signaling. Interestingly, IL-10R signaling does not appear to directly interfere with STAT1 function in macrophages but rather prevents the generation of environmental signals necessary for the accumulation of immature macrophages within the inflammatory microenvironment. These mechanistic studies shed further light on the function of key pathways targeted for therapeutic intervention in IBD.
RESULTS
Macrophage accumulation within the colon after the blockade of IL-10R signaling requires STAT1
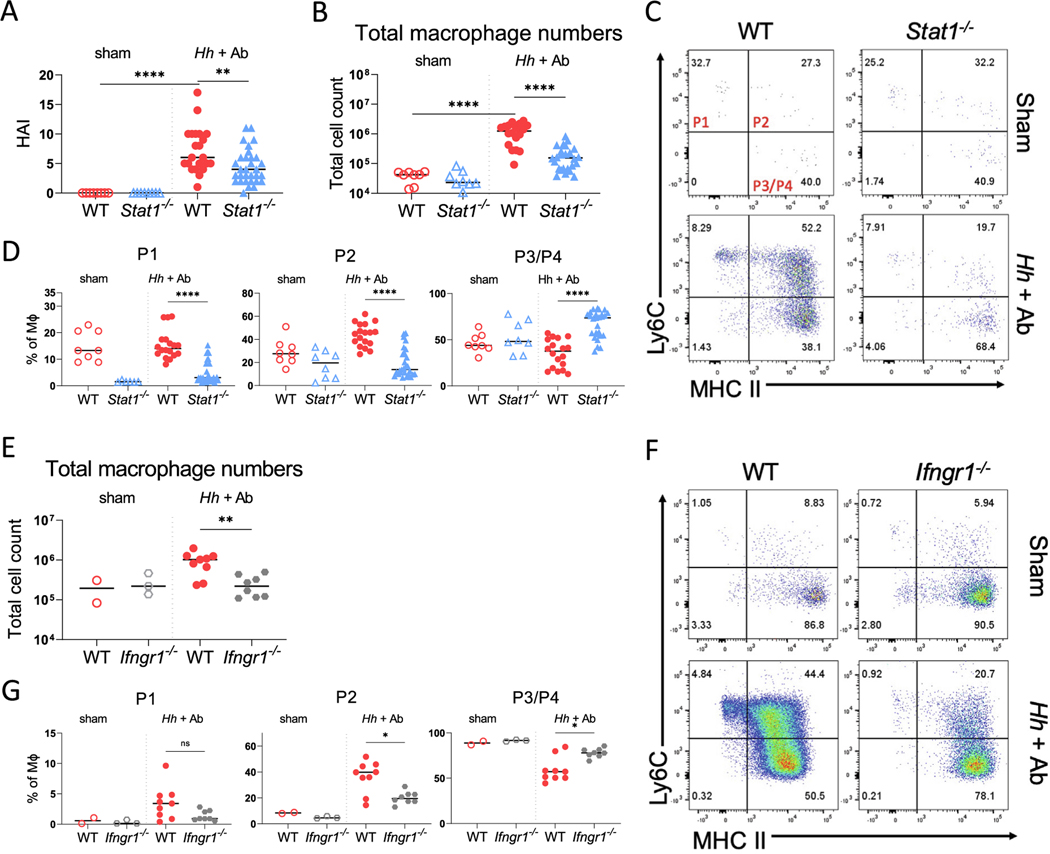
To determine whether STAT1 is necessary for the accumulation of inflammatory macrophages within the colon of mice after the blockade of IL-10R signaling, we gavaged parallel groups of age-matched wild-type (WT) and Stat1−/− mice with Helicobacter hepaticus (Hh) and simultaneously treated them with anti-IL-10RA antibody (Ab) to induce colitis (Supplementary Fig. 1A). Mice were euthanized 16–18 days later. As expected, the Hh-infected WT mice treated with anti-IL-10RA Ab exhibited histopathological signs of colitis, including crypt hyperplasia, abscesses, mononuclear inflammation, and epithelial injury, which resulted in a significantly higher histologic activity index (HAI) than in untreated mice (Fig. 1A and Supplementary Fig. 1B). Infection with Hh and treatment with IL-10RA Ab in WT mice was also accompanied by an increased expression of pro-inflammatory genes (Il1b, Cxcl2, Il12b), STAT1-inducible genes (Cxcl9 and Cxcl11), and the antimicrobial peptide Reg3g within the total RNA extracted from combined samples of the cecum and colon (Supplementary Fig. 1C). Hh-infected WT mice treated with anti-IL-10RA Ab exhibited a dramatic increase in the absolute number of colonic macrophages (CD45+ Ly6G− CD103− CD64+ CD11b+) compared with sham-treated mice (Fig. 1B and Supplementary Fig. 1D). In contrast to WT mice, colitis was significantly less severe within the colons of Hh-infected STAT1-deficient mice treated with anti-IL-10RA Ab (Fig. 1A and Supplementary Fig. 1B). This was accompanied by significantly lower expression of STAT1-dependent genes, as well as Il12b and Il1b within the colon of Stat1−/− mice than WT mice; although, we were unable to detect significant differences in the expression of Cxcl2 and indeed found higher expression of Reg3g (Supplementary Fig. S1C). Notably, there was a significant reduction in the absolute number of macrophages within the LP (Fig. 1B), and this was accompanied by a significant reduction in the frequency of immature Ly6C+ P1 and P2 macrophages and increases in the frequency of mature Ly6C− P3/P4 macrophages within the total LP macrophage population of STAT1-deficient mice compared with their frequencies in WT mice (Figs. 1C and 1D). These results indicate that STAT1 is necessary for the accumulation of immature macrophages observed within the colon of Hh-infected mice after the blockade of the IL-10R.
Fig. 1.
STAT1 and IFN-γR are required for colonic macrophage recruitment following infection with H. hepaticus and treatment with anti-IL-10RA Ab. (A) Histopathologic scores of WT and STAT1-deficient mice infected with Hh and treated with anti-IL-10RA Ab (Hh + Ab), or sham-treated with PBS; (B) total macrophage numbers from LP of WT or Stat1−/− mice with or without Hh + anti-IL-10RA Ab treatment; (C) representative flow cytometry of MHC II and Ly6C staining of LP macrophages gated on CD45+ CD103− Ly6G− CD11b+ CD64+ cells; (D) percentages of Ly6C+ MHC II− (P1), Ly6C+ MHC II+ (P2) Ly6C− MHC II+ (P3/P4) macrophages (MΦ) were determined for mice of indicated genotypes treated with Hh and anti-IL-10R Ab or sham-treated; (E) total macrophage numbers from the colon of WT or Ifngr1−/− mice with or without Hh and anti-IL-10RA Ab treatment; (F) representative staining; and (G) percentages of LP macrophage (MΦ) subpopulations in WT and Ifngr1−/− mice treated with Hh and anti-IL-10RA Ab (Hh + Ab) or sham-treated with PBS. Results are pooled from minimum three independent experiments for Stat1−/− mice and two independent experiments for Ifngr−/− mice, and median values are shown. Statistical significance was determined by nonparametric Mann-Whitney U test. * p < 0.05 ** p < 0.01 **** p < 0.0001. Ab = antibody; HAI = histologic activity index; Hh = Helicobacter hepaticus; IFN-γR = interferon gamma receptor; IL = interleukin; LP = lamina propria; PBS = phosphate-buffered saline; STAT1 = signal transducer and activator of transcription 1; WT = wild-type.
To determine whether the defects in the accumulation of immature macrophages observed in STAT1-deficient mice could be caused by decreased numbers of circulating monocytes, we compared the frequency of circulating CD45+ Ly6G− CD11b+ Ly6Chigh monocytes in WT and Stat1−/− mice either before or after infection with Hh and treatment with anti-IL-10RA Ab (Supplementary Fig. 1E). Regardless of inflammatory state of the animals, we were unable to detect significant differences in the frequency of circulating monocytes between WT and STAT1-deficient mice. This result indicates that the reduced accumulation of immature macrophages observed within the colons of Hh and anti-IL-10RA treated Stat1−/− mice is not due to reduced frequency of circulating monocytes.
STAT1-dependent accumulation of immature macrophages within the colon after the blockade of IL-10R signaling depends on IFN-γ
Elevated production of IFN-γ is a hallmark of microbiota-dependent colitis that develops in mice lacking IL-10R signaling, and STAT1 is the primary transducer of IFN-γR signaling. This raises the possibility that IFN-γ-induced activation of STAT1 could be required for the accumulation of immature intestinal macrophages observed after the blockade of the IL-10R. To address this possibility, we infected Ifngr1−/− and WT mice with Hh, followed by anti-IL-10RA Ab treatment, as described previously (See Supplementary Fig. 1A). Similar to STAT1-deficient mice, we found a significant overall decrease in the absolute number of colonic macrophages in IFN-γR1-deficient mice compared with WT mice (Fig. 1E), as well as a decrease in the proportion of the P2 LP macrophage population, and this was accompanied by a reciprocal increase in the proportion of P3/P4 macrophages in IFN-γR1-deficient mice (Figs. 1F and 1G). The median disease severity score was also lower in Ifngr1−/− mice than in control mice, although this did not reach statistical significance (p = 0.12) (Supplementary Fig. 2A). There were significant decreases in the expression of Il12b, Cxcl9, and Cxcl11 within total colonic RNA isolated from IFN-γR1-deficient mice compared with WT mice (Supplementary Fig. 2B). Consistent with results from STAT1-deficient mice, we did not detect a difference in expression of Cxcl2 between WT and IFN-γR1-deficient mice and expression of Reg3g was significantly higher in IFN-γR1-deficient mice (Supplementary Fig. 2B). The similar phenotypes observed in mice lacking STAT1 or IFN-γR1 suggest that IFN-γ-induced STAT1 signaling is essential for the Helicobacter-driven accumulation of immature macrophages in the intestine of mice lacking IL-10R signaling.
Loss of STAT1 within the hematopoietic compartment interferes with the accumulation of immature colonic macrophages
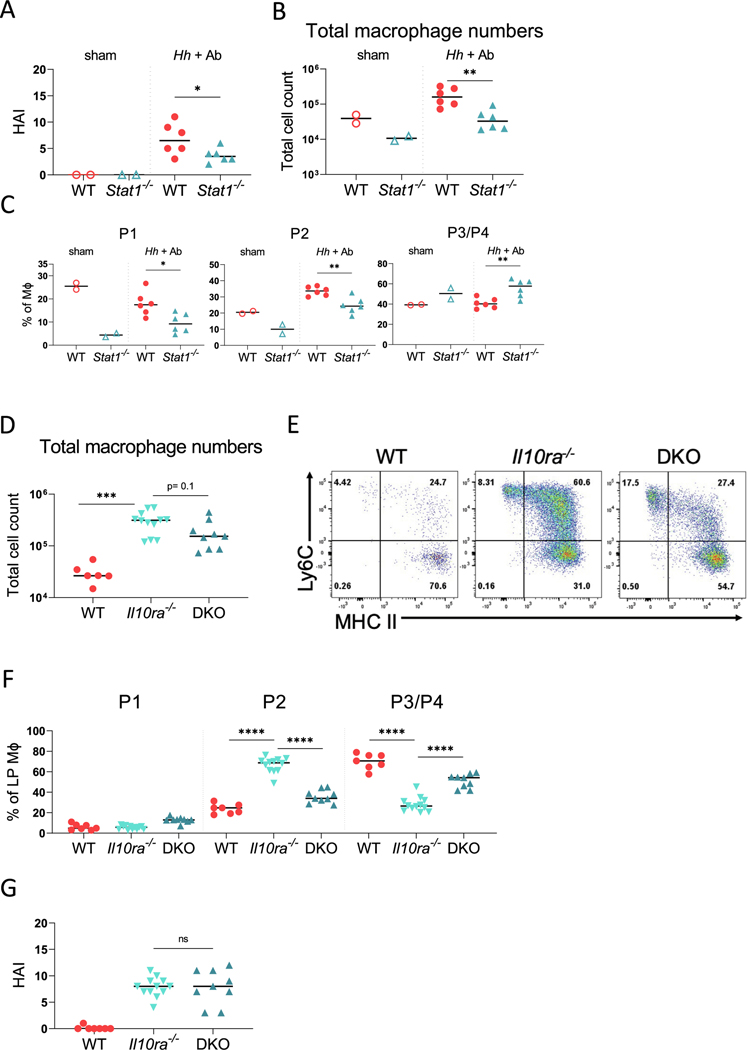
To determine whether the function of STAT1 within the hematopoietic compartment is necessary for the accumulation of immature macrophages after the blockade of the IL-10R, we generated a series of bone marrow chimeras. In the first set of experiments, we reconstituted lethally irradiated WT (CD45.1) hosts with bone marrow-derived from either WT (CD45.1) mice or Stat1−/− (CD45.2) mice (Supplementary Fig. 3A). After an 8 week reconstitution period, the mice were infected with Hh by oral gavage and treated with anti-IL-10RA Ab, as described previously. The mice were euthanized 16–18 days later, and the colitis severity and LP macrophages were analyzed, as described previously. Similar to the total Stat1−/− mice, mice that received STAT1-deficient bone marrow exhibited significantly less severe colitis than the mice that received WT bone marrow (Fig. 2A). The absolute number of macrophages were significantly lower in mice that were reconstituted with STAT1-deficient bone marrow than those that were reconstituted with WT bone marrow cells (Fig. 2B). Furthermore, the frequency of both P1 and P2 macrophages within the donor-derived LP macrophage population was significantly lower in mice that were reconstituted with STAT1-deficient bone marrow cells than those reconstituted with WT bone marrow cells, whereas the frequency of P3/P4 macrophages was significantly higher (Fig. 2C). Consistent with our previous results, the mice that received STAT1-deficient bone marrow cells demonstrated significantly lower expression of Il12b, Il1b, and Cxcl9 within total colonic RNA than mice that received WT bone marrow after infection with Hh and treatment with IL-10RA Ab; although, as observed in complete knockouts, the Cxcl2 levels remained similar and Reg3g expression was significantly higher in mice that received STAT1-deficient bone marrow than those that received WT bone marrow (Supplementary Fig. 3B). These results indicated that the loss of STAT1 within the hematopoietic compartment interferes with the microbiota-dependent accumulation of inflammatory macrophages after the IL-10R blockade.
Fig. 2.
Loss of STAT1 within the hematopoietic compartment interferes with the accumulation of LP macrophages. (A-C) WT hosts were reconstituted with either WT or STAT1-deficient bone marrow: (A) histopathologic scores of generated chimeric mice; (B) total macrophage numbers from colon and cecum of chimeric mice with or without Hh and anti-IL-10RA Ab treatment; (C) frequencies of colonic macrophage (MΦ) subsets. Figs. A-C show the results from one of two experiments with similar results. (D-G) Cdcs1 (CD45.1) (WT) host were reconstituted with either Cdcs1 (CD45.1) (WT), Cdcs1 Il10ra−/− (CD45.2) (Il10ra−/−), or Cdcs1 Il10ra−/− Stat1−/− (CD45.2) (DKO) bone marrow cells: (D) total numbers of donor colonic macrophages isolated from chimeric mice; (E) representative flow cytometry showing phenotype of donor-derived colonic macrophages; and (F) percentages of macrophage (MΦ) subsets for each group; (G) histopathological scores for each group. Results in D–G are pooled from three independent experiments and median values are shown. * p < 0.05 ** p < 0.01 *** p < 0.001 **** p < 0.0001 Mann-Whitney U test. Ab = antibody; HAI = histologic activity index; Hh = Helicobacter hepaticus; IL = interleukin; LP = lamina propria; STAT1 = signal transducer and activator of transcription 1; WT = wild-type.
Deletion of STAT1 interferes with the development of spontaneous colitis observed in IL-10R-deficient mice
We and others have previously shown that the loss of IL-10RA in mice carrying the Cdcs1 colitis susceptibility locus results in the development of spontaneous colitis initiating as early as 3 weeks of age7. Therefore, to complement the bone marrow chimera experiments described previously, we generated an additional set of radiation chimeras in which Cdcs1 (CD45.1) host mice were reconstituted with bone marrow isolated from either Cdcs1 Il10ra+/− (CD45.2) (WT) mice, Cdcs1 Il10ra−/− (CD45.2) (Il10ra−/−) mice, or Cdcs1 Il10ra−/− Stat1−/− (CD45.2) (DKO) mice (Supplementary Fig. 4A). After reconstitution, the mice were housed in conventional specific-pathogen-free (SPF) conditions for 8 weeks to allow the development of colitis and then analyzed. We observed a marked increase in the absolute numbers of donor-derived macrophages within the LP of mice reconstituted with Il10ra−/− bone marrow cells compared with those reconstituted with control bone marrow cells (Fig. 2D). Furthermore, the frequency of P2 macrophages within the donor-derived macrophage population isolated from the LP was significantly higher in mice that were reconstituted with Il10ra−/− bone marrow cells than those reconstituted with WT bone marrow cells, and we observed a reciprocal decrease in the frequency of P3/P4 macrophages (Figs. 2E and 2F). In contrast to mice reconstituted with Il10ra−/− bone marrow, the median number of donor-derived LP macrophages were reduced in mice that were reconstituted with DKO bone marrow cells, and this difference approached statistical significance (p = 0.1) (Fig. 2D). This was accompanied by a significantly lower frequency of donor-derived P2 macrophages and increased frequency of P3/P4 macrophages in the colon of DKO reconstituted mice (Figs. 2E and 2F). As expected, the histologic scores were quite low in mice that received WT bone marrow and moderately severe in mice reconstituted with Il10ra−/− bone marrow (Fig. 2G and Supplementary Fig. 4B). We did not observe a significant difference in the histologic scores between mice reconstituted with Il10ra−/− and DKO bone marrow cells (Fig. 2G). These results confirm that the STAT1 function within the hematopoietic compartment is necessary for the accumulation of immature macrophages observed within the colon of Il10ra−/− mice.
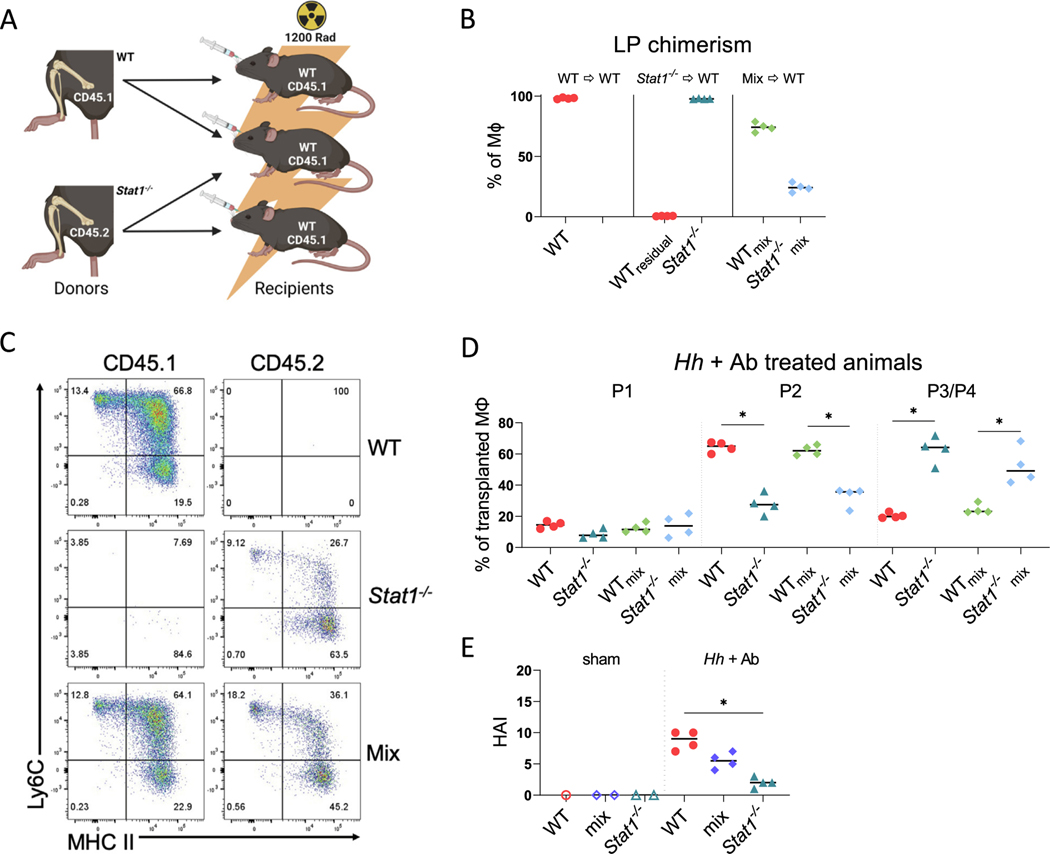
A macrophage-autonomous function of STAT1 is necessary for the accumulation of immature LP macrophages following IL-10RA blockade
As described previously, our results indicate that the presence of STAT1 within the hematopoietic compartment is necessary for the accumulation of immature LP macrophages after the blockade of IL-10R, suggesting that STAT1 may have a cell autonomous function that is necessary for macrophage accumulation in the LP. To directly test this hypothesis, we generated mixed radiation chimeras in which irradiated WT (CD45.1) hosts were reconstituted with either WT bone marrow cells, Stat1−/− (CD45.2) bone marrow cells, or a 1:1 mixture of both (Fig. 3A). A total of 8 weeks later, the reconstituted mice were infected with Hh and treated with anti-IL-10RA Ab to induce colitis. After tissue harvest, reconstitution was verified by identifying macrophages of the expected genotypes in the LP of these mice (Supplementary Fig. 5). As expected, virtually all LP macrophages in the mice reconstituted with WT bone marrow cells were CD45.1+ and all macrophages in mice reconstituted with Stat1−/− bone marrow cells were CD45.2+, whereas in mice reconstituted with a mixture of the two, we observed both CD45.1+ and CD45.2+ macrophages in the LP, indicating reconstitution with both WT- and Stat1−/−-derived bone marrow cells (Fig. 3B). We did note that the proportion in this mixture was skewed toward WT macrophages, reflecting either unequal proportions in the initial mixture or perhaps differential reconstitution efficiency.
Fig. 3.
Cell autonomous defect in the accumulation of Stat1−/− immature colonic macrophages following blockade of IL-10R signaling. (A) Scheme of generated bone marrow chimeras. WT (CD45.1) hosts were reconstituted with either WT or Stat1−/− (CD45.2) bone marrow or a 1:1 mixture of both. Image created using BioRender; (B) percentages of LP macrophages of indicated genotypes in chimeric mice 2 weeks after the induction of colitis; (C) representative flow cytometry of colonic macrophage subsets CD45.1 (WT) and CD45.2 (Stat1−/−) in chimeric mice; (D) percentages of each macrophage (MΦ) subset within total macrophage populations of each genotype; (E) histopathologic scores from reconstituted mice. Results are from one experiment and median values are shown. * p < 0.05 Mann-Whitney U test. Ab = antibody; HAI = histologic activity index; Hh = Helicobacter hepaticus; IL = interleukin; LP = lamina propria; STAT1 = signal transducer and activator of transcription 1; WT = wild-type.
We next analyzed the frequency of immature and mature LP macrophages derived from WT or Stat1−/−bone marrow cells in these chimeras. As expected, based on the previously mentioned experiments, the host mice reconstituted with Stat1−/− bone marrow alone displayed significant reduction in the frequency of donor-derived P2 Ly6C+ colonic macrophages compared with hosts that received WT bone marrow cells alone (Figs. 3C and 3D). Remarkably, in mixed chimeras, the frequency of Stat1−/− P2 macrophages among all Stat1−/− macrophages in the LP was significantly lower than the frequency of WT P2 macrophages among all WT macrophages, and reciprocally, the proportion of P3/P4 macrophages was significantly higher (Fig. 3D). Parallel to our previous results from Stat1−/− mice infected with Hh and treated with anti-IL-10RA Ab, the mice reconstituted with Stat1−/− bone marrow developed significantly less severe colitis than mice reconstituted with WT bone marrow (Fig. 3E); although, the severity of colitis in mice that received mixtures of both was intermediate and not significantly different than the severity in mice that received either WT- or STAT1-deficient bone marrow cells alone. These results indicate that the defect in P2 macrophage accumulation observed in the absence of STAT1 is indeed cell autonomous, demonstrating that cell autonomous STAT1 signaling is necessary for the accumulation of immature colonic macrophages observed after the blockade of IL-10R signaling.
IL-10R-deficient myeloid cells confer an inflammatory phenotype on WT macrophages
We have previously demonstrated that the specific deletion of IL-10RA in macrophages results in the development of colitis and the accumulation of immature P2 macrophages within the colon7. Therefore, our observations that STAT1 has a cell autonomous function in macrophages that is necessary for the accumulation of immature macrophages suggest that a critical function of IL-10R signaling could be to directly inhibit STAT1 function.
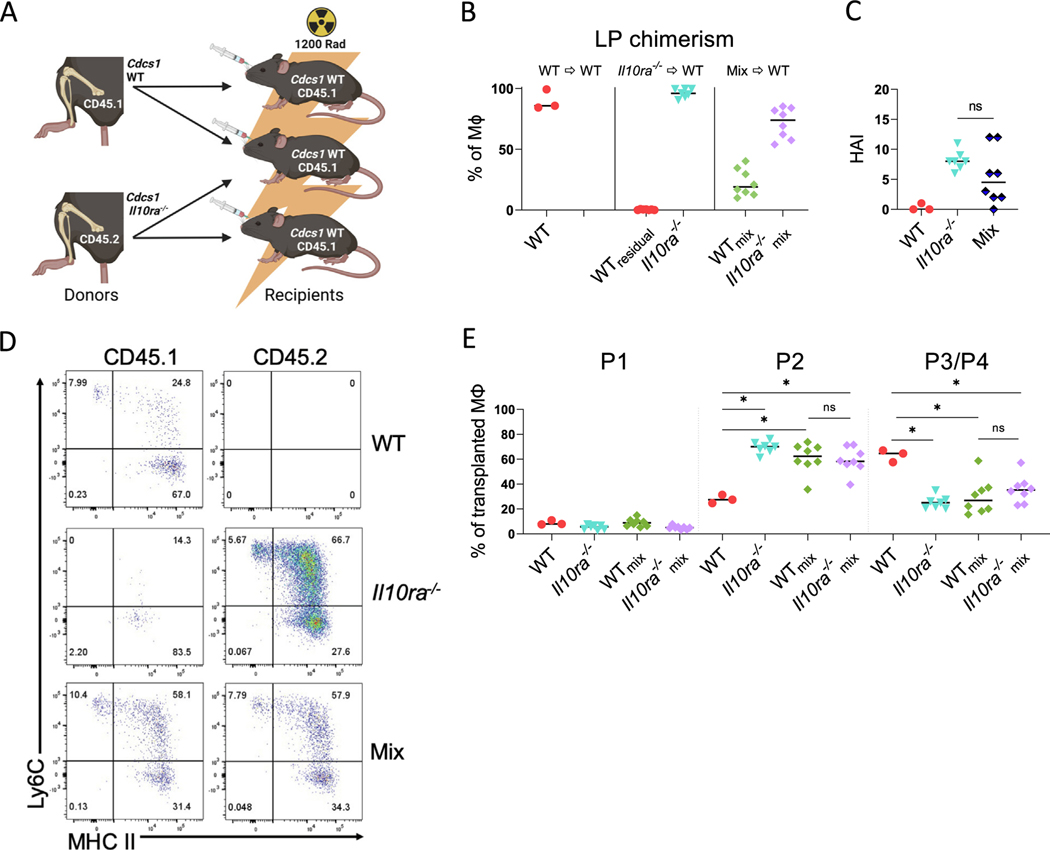
To directly examine this possibility, we produced a series of radiation chimeras in which Cdcs1 (CD45.1) (WT) mice were lethally irradiated and reconstituted with either WT bone marrow cells, Cdcs1 Il10ra−/− (CD45.2) (Il10ra−/−), or a 1:1 mixture of both (Mix) (Fig. 4A). A total of 8 weeks after bone marrow transplantation, the mice that received mixtures of WT and Il10ra−/− bone marrow exhibited the presence of both WT and Il10ra−/− monocytes within the LP, although the proportion was skewed toward Il10ra−/−, suggesting, as previously mentioned, the possibility of either unequal proportions in the initial mixture or differential reconstitution efficiency (Fig. 4B). As anticipated, the mice reconstituted with WT bone marrow alone did not develop colitis. Interestingly, the mice that received Il10ra−/− bone marrow cells alone showed clear histologic signs of colitis as did those that received a mixture of WT and Il10ra−/− bone marrow (Fig. 4C). Intestinal LP macrophages in hosts reconstituted with WT bone marrow primarily consisted of P3/P4 macrophages and the frequency of immature P2 macrophage was low, whereas in hosts reconstituted with Il10ra−/− bone marrow cells, the frequency of donor P2 macrophages was significantly higher (Figs. 4D and 4E). Remarkably, the frequency of P2 macrophages derived from either WT or Il10ra−/− donors mice reconstituted with mixtures of WT and Il10ra−/− bone marrow cells was virtually identical to the frequency observed in mice reconstituted with Il10ra−/− bone marrow cells alone, and significantly higher than the frequency of P2 macrophages in hosts reconstituted with WT bone marrow cells alone (Figs. 4D and 4E). Because in this system WT macrophages accumulate in the P2 stage even though they have intact IL-10R function, these results imply that IL-10R is unlikely to directly inhibit the accumulation of immature colonic macrophages or the function of STAT1. Rather these data suggest that IL-10R-deficient hematopoietic cells generate a cell extrinsic signal that promotes the accumulation of immature macrophages.
Fig. 4.
IL-10R-deficient hematopoietic cells confer an inflammatory phenotype on WT colonic macrophages. (A) Experiment scheme of generated bone marrow chimeras. Cdcs1 (CD45.1) (WT) hosts were reconstituted with either WT or Cdcs1 Il10ra−/− (Il10ra−/−) bone marrow or a 1:1 mixture of both (Mix); (B) percentages of LP macrophages of indicated genotypes in chimeric mice after the induction of colitis (WTmix and Il10ra−/−mix indicates that cells were isolated from mixed chimeras); (C) histopathologic scores; (D) representative flow cytometry showing macrophage subsets in the colon. Cdcs1 WT (CD45.1) and Cdcs1 Il10ra−/− (CD45.2); (E) percentages of each macrophage (MΦ) subset as a proportion of total macrophages of the indicated genotype. Results are pooled from two independent experiments and median values are shown. * p < 0.05 Mann-Whitney U test. IL = interleukin; LP = lamina propria; WT = wild-type.
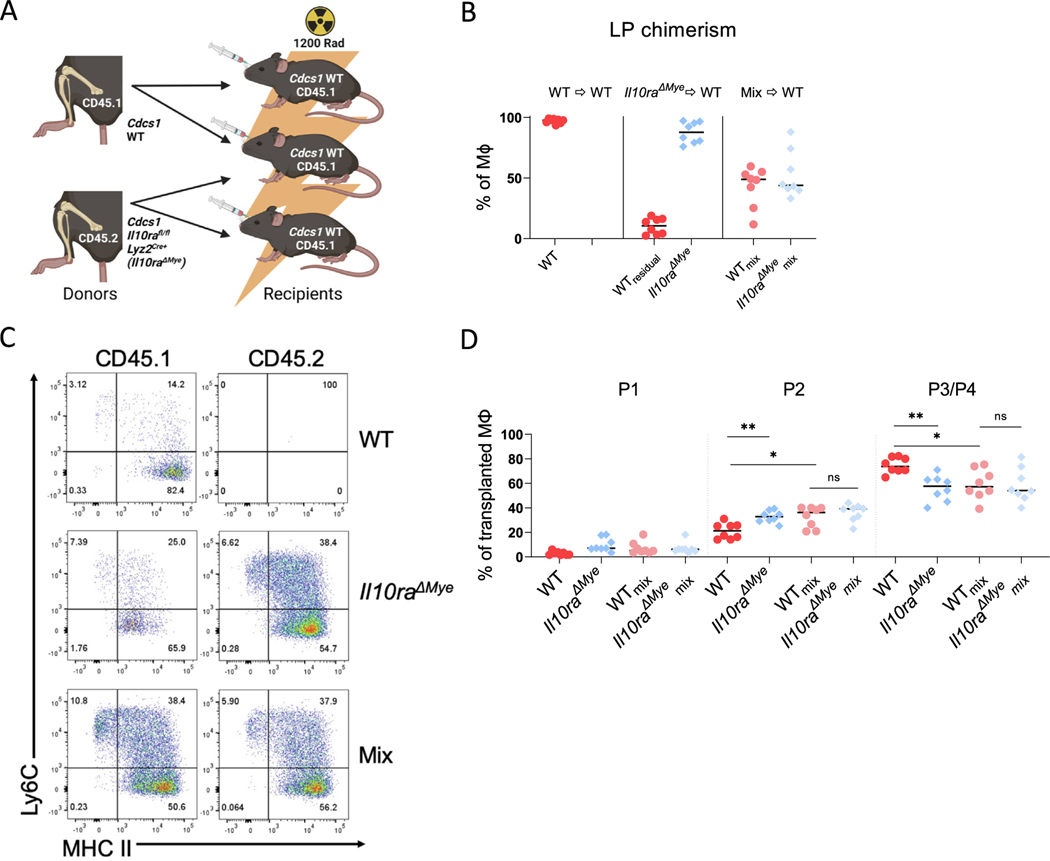
IL-10R is expressed within both the myeloid and lymphoid compartments, but our previous results using mice with a myeloid-specific deletion of IL-10RA (Cdcs1 Il10rafl/fl Lyz2Cre+) suggested that IL-10RA signaling is specifically required in myeloid cells to prevent the development of colitis7. To determine whether IL-10R signaling is also specifically required in myeloid cells to block the signal that promotes the accumulation of immature macrophages, we produced 1:1 mixed radiation chimeras reconstituted with both Cdcs1 (CD45.1) (WT) mice and Cdcs1 Il10rafl/fl Lyz2Cre+ (CD45.2) (Il10raΔmye) mice (Fig. 5A). Reconstitution was confirmed by analyzing the macrophage subsets from the LP of irradiated mice 8 weeks after irradiation. WT and Il10raΔmye LP macrophages were present in near equal proportions in animals that received 1:1 mixtures of bone marrow cells, indicating successful reconstitution of both genotypes (Fig. 5B). As expected, mice reconstituted with bone marrow cells isolated from Il10raΔmye mice alone exhibited significantly higher percentages of donor P2 macrophages and lower percentages of P3/P4 macrophages within the LP than irradiated host mice reconstituted with bone marrow cells isolated from WT mice (Figs. 5C and 5D). Consistent with the hypothesis that IL-10R signaling in myeloid cells interferes with a signal that promotes the accumulation of immature macrophages, the percentage of WT P2 macrophages in mice reconstituted with mixtures of bone marrow cells isolated from both WT mice and Il10raΔmye was significantly higher than the percentages of P2 macrophages observed in mice reconstituted with WT bone marrow cells alone. Conversely, the percentage of WT P3/P4 macrophages was significantly lower in mice reconstituted with mixtures of bone marrow cells isolated from both WT and Il10raΔmye mice than the percentages of P3/P4 macrophages observed in mice reconstituted with WT bone marrow cells alone (Figs. 5C and 5D). We believe that these results suggest that intrinsic IL-10R signaling on macrophages themselves is necessary to prevent the elaboration of extrinsic signal that drives the accumulation of immature macrophages.
Fig. 5.
IL-10R-deficient myeloid cells confer an inflammatory phenotype on WT colonic macrophages. (A) Experiment scheme of generated bone marrow chimeras. Cdcs1 (CD45.1) (WT) hosts were reconstituted with either WT or Cdcs1 Il10rafl/fl Lyz2Cre+ (CD45.2) (Il10raΔmye) bone marrow or a 1:1 mixture of both (Mix); (B) percentages of LP macrophages of indicated genotype following the induction of colitis (WTmix and Il10raΔmyemix indicates that cells were gated from mixed chimeras); (C) representative flow cytometry showing macrophage subsets in the colon; (D) percentages of each macrophage (MΦ) subset as a proportion of total macrophages of the indicated genotype. Results are pooled from two independent experiments and median values are shown. * p < 0.05 ** p < 0.01 Mann-Whitney U test. IL = interleukin; LP = lamina propria; WT = wild-type.
Distinguishing cell autonomous and non-autonomous functions of IL-10R signaling in LP macrophages
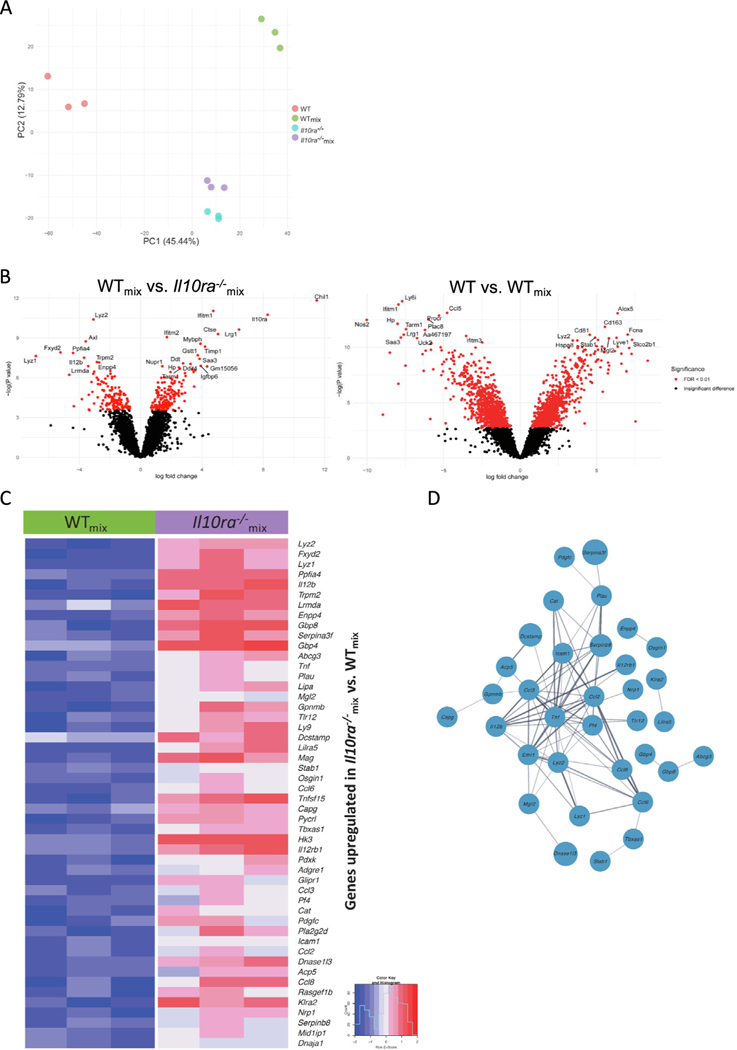
To further explore the hypothesis that Il10ra−/− macrophages drive the accumulation of immature colonic macrophages through the generation of a cell extrinsic signal, we isolated the LP macrophages from host mice reconstituted with WT bone marrow cells (WT) from those reconstituted with Il10ra−/− bone marrow cells (Il10ra−/−) and from those reconstituted with a 1:1 mixture of both (WTmix and Il10ra−/−mix, respectively), using fluorescence-activated cell sorting. RNA was isolated from these macrophage populations and gene expression was analyzed by bulk RNA sequencing. Gene expression profiles were then compared by principal component analysis. WT and Il10ra−/− LP macrophages clustered independently along both PC1 and PC2. Interestingly, Il10ra−/− and Il10ra−/−mix macrophages clustered quite closely together, indicating that the presence of WT macrophages had very little influence on the gene expression characteristics of Il10ra−/− macrophages. In contrast, WTmix macrophages clustered independently from WT, Il10ra−/−, or Il10ra−/−mix macrophages, suggesting that the presence of Il10ra−/− macrophages strongly influence the gene expression profile of WT macrophages. The differences between WT and WTmix macrophages are most distinguished along PC1, whereas the differences between WTmix and Il10ra−/−mix, are largely along PC2 (Fig. 6A). These results suggest that the differences along PC1 are largely driven by cell extrinsic microenvironmental signals generated by the presence of Il10ra−/− hematopoietic cells, whereas we suggest that differences along PC2 are determined by genotype (rather than microenvironment) and are therefore cell autonomous.
Fig. 6.
Bulk RNA sequencing from sorted colonic macrophages from mixed bone marrow chimeric mice. (A) Principal component analysis of sorted LP macrophages identifies cell autonomous and non-cell autonomous differences in gene expression caused by the absence of the IL-10R; (B) Left panel: comparison of WTmix versus Il10ra−/− identifies genes regulated in a cell autonomous fashion. Genes shown on the left arm of the volcano are expressed at lower levels in WTmix macrophages and those shown on the right arm of the volcano are expressed at higher levels in WTmix macrophages. Right panel: Comparison of WT versus WTmix reveals genes that are regulated in non-cell autonomous fashion by the presence of Il10ra−/− macrophages. Genes shown on the left arm of the volcano are expressed at lower levels in WT macrophages and those shown on the right arm of the volcano are expressed at higher levels in WT macrophages; (C) a heatmap depicts the expression of the top 50 upregulated genes in Il10ra−/− as compared with WTmix mice; (D) a network of physical and functional relations between the protein products of the top 50 upregulated genes in Il10ra−/−mix compared with WTmix mice, as determined by STRING v11.551, and visualized in Cytoscape 3.9.154. IL = interleukin; WT = wild-type.
To identify individual genes that are regulated in a cell-autonomous and non-cell autonomous fashion by IL-10RA signaling, we generated a volcano plot of genes differentially expressed between Il10ra−/−mix and WTmix macrophages, and WTmix and WT macrophages respectively (Fig. 6B). We observed that Il12b is expressed at lower levels in WTmix macrophages than in Il10ra−/−mix macrophages, whereas the converse is true for Ddit4, indicating cell autonomous regulation by IL-10, consistent with our previous observations of rapid inhibition of Il12b and induction of Ddit4 after IL-10 treatment of Lipopolysaccharide (LPS) stimulated bone marrow-derived macrophages31,32. Additional exploration of genes that were expressed at higher levels in Il10ra−/−mix macrophages that WTmix macrophages revealed significant enrichment of several CCL family chemokines, including Ccl2, Ccl3, Ccl6, and Ccl8 (Fig. 6C), and that these genes can be found in a functional network with Il12b and TNF, well-described targets of IL-10 mediated suppression33 (Fig. 6D).
In contrast to the intrinsic differences observed in gene expression between WTmix and Il10ra−/−mix LP macrophages, we observed that Nos2 (a marker of inflammatory macrophages) expression is lower in WT than in WTmix macrophages, whereas Cd163 and Lyve1 (previously defined markers of mature M2-like colonic macrophages) expression is higher in WT macrophages than in WTmix macrophages (Fig. 6B)9. This suggests that the ability of IL-10R to prevent the development of inflammatory macrophages and promote the development of anti-inflammatory macrophages in the colon is not a cell autonomous effect of IL-10R (because IL-10R is present on both of these populations) but rather is the result of the differences in the intestinal microenvironment caused by the presence (or absence) of IL-10RA-deficient macrophages.
IL-10R signaling does not directly inhibit IFN-γ induced STAT1 function
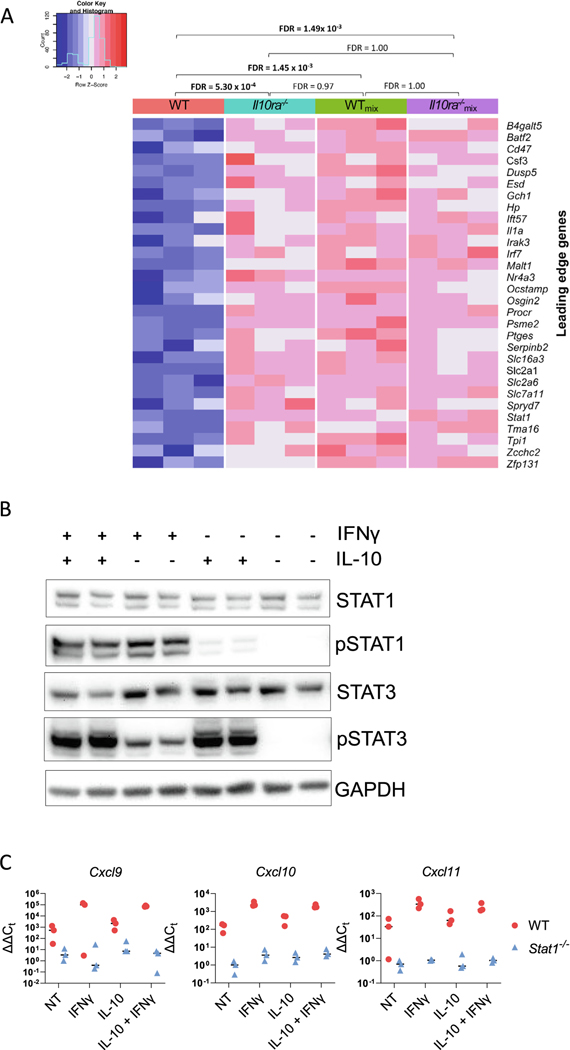
To further evaluate our hypothesis that IL-10R does not directly regulate IFN-γ-induced STAT1-dependent gene expression in LP macrophages, we performed a gene set enrichment analysis focusing on a previously published gene set that is induced in macrophages by treatment with IFN-γ (GSE35825). Our analysis demonstrates that genes in this set are significantly enriched in comparison between WT and Il10ra−/− macrophages, WT and WTmix macrophages, and WT and Il10ra−/−mix macrophages, whereas no significant enrichment was detected for all other comparisons (Il10ra−/− vs. WTmix, Il10ra−/− vs. Il10ra−/−mix, or WTmix vs. Il10ra−/−mix) (Fig. 7A). This confirms that macrophages with intact IL-10R function (WTmix) exhibit elevated IFN-γ/STAT1-dependent gene expression in the presence of Il10ra−/− macrophages, consistent with our hypothesis that IL-10R signaling does not directly inhibit STAT1-dependent gene expression but rather inhibits STAT1-dependent gene expression and the accumulation of immature P2 macrophages through a non-cell autonomous mechanism.
Fig. 7.
IL-10R signaling does not directly inhibit IFN-γ-induced STAT1 function. (A) A heatmap shows the Z-normalized expression of each of 30 leading edge genes from a previously defined gene set of IFN-γ-induced genes in bone marrow-derived macrophages55 that was found by GSEA analysis to be significantly enriched in comparisons between WT and Il10ra−/−, WT and WTmix, and WT and Il10ra−/−mix colonic macrophages. Gene set enrichment false discovery rate Q values are shown for the comparison of all experimental groups; (B) bone marrow-derived macrophages from duplicate C57BL/6 mice were stimulated with 1 ng/ml IFN-γ for 30 minutes, with or without addition of 5 ng/ml IL-10. Cell extracts were prepared and assayed for STAT1, pSTAT1 (Y701), STAT3, and pSTAT3 (Y705). This experiment was performed twice and the figure shows the results of one representative experiment; (C) bone marrow-derived macrophages from three individual C57BL/6 mice were isolated and stimulated with 1 ng/ml IFN-γ and/or 5 ng/ml IL-10 for 2 hours. Cells were washed with phosphate-buffered saline and harvested into TRIzol reagent. RNA was isolated and the expression of Cxcl9, Cxcl10, and Cxcl11 were measured by quantitative reverse transcription-polymerase chain reaction. The figure shows the results from one representative experiment. GADPH = glyceraldehyde 3-phosphate dehydrogenase; GSEA = gene set enrichment analysis; IFN = interferon; IL = interleukin; STAT1 = signal transducer and activator of transcription 1; WT = wild-type.
To test the hypothesis that IL-10R signaling does not directly inhibit STAT1 activation, we prepared bone marrow-derived macrophages from both WT and Stat1−/− mice. These macrophages were then stimulated with IFN-γ alone, IL-10 alone, or both, or left unstimulated. Cell extracts were prepared and the induction of pSTAT1(Y701) was analyzed by western blotting, and the induction of the STAT1-dependent genes Cxcl9, Cxcl10, and Cxcl11 were evaluated by quantitative reverse transcription-polymerase chain reaction. As expected, IFN-γ alone induced a robust activation of pSTAT1 but, including IL-10 had minimal influence on STAT1 activation (Fig. 7B). Likewise, although IFN-γ but not IL-10 induced robust STAT1-dependent induction of Cxcl9, Cxcl10, and Cxcl11, the addition of IL-10 had quite modest which only attained borderline significance for Cxcl9 (Fig. 7C). These experiments demonstrate that IL-10 does not inhibit IFN-γ-induced STAT1 activation and has quite modest inhibitory effects on IFN-γ-induced gene expression. We suggest that these results are therefore consistent with our hypothesis that IL-10R signaling inhibits the induction of STAT1-dependent gene expression in colonic LP macrophages through a non-cell autonomous mechanism.
DISCUSSION
Based on previous data from our laboratory and others, we hypothesized that the suppression of STAT1 signaling in macrophages was an essential function of IL-10 necessary to inhibit the accumulation of immature macrophages within the LP. Here, we have demonstrated that the deletion of STAT1 results in a marked decrease in the accumulation of immature macrophages within the LP of Hh-infected mice after IL-10R blockade and a significant reduction in the histologic signs of colitis. A similar phenotype was identified in mice lacking IFN-γR1. The production of bone marrow radiation chimeras demonstrated that the absence of STAT1 within hematopoietic cells was sufficient to cause a defect in immature macrophage accumulation. Furthermore, using a strategy in which irradiated hosts were reconstituted with mixtures of both WT- and STAT1-deficient bone marrow cells, we demonstrated that reduced accumulation of STAT1-deficient macrophages within the LP is the result of a cell autonomous phenotype. Finally, although these results supported the hypothesis that suppression of STAT1 function by IL-10 is necessary to inhibit immature macrophage accumulation, the evaluation of mixed chimeras reconstituted with mixtures of WT- and IL-10R-deficient bone marrow cells demonstrated enhanced accumulation of immature WT macrophages that displayed elevated expression of STAT1-dependent genes. Further, IL-10 treatment of bone marrow-derived macrophages did not interfere with IFN-γ-induced STAT1 activation nor the induction of STAT-1-dependent gene expression. These results indicate that IL-10R does not directly suppress immature macrophage accumulation and STAT1-dependent gene expression but rather suppresses these functions in a non-cell autonomous fashion. These observations significantly expand our understanding of the mechanisms through which IL-10 signaling inhibits intestinal inflammation.
Our observation that cell autonomous function of STAT1 is required for the accumulation of immature macrophages within the LP of mice that lack effective IL-10R signaling is consistent with previous results showing reduced accumulation of immature macrophages in STAT1-deficient mice compared with WT mice after treatment with DSS22. The observation that STAT1 is required for macrophage accumulation in mice lacking IL-10R signaling suggests that the dependence of immature macrophage recruitment on STAT1 extends to microbiota-driven models of disease and further is not bypassed even when IL-10R signaling is compromised. This could have important implications for therapy of human disease in situations where macrophage accumulation is a prominent feature. A key unanswered question is how the intrinsic STAT1 function supports the accumulation of immature intestinal macrophages. We have not formally investigated the dynamics of STAT1-dependent accumulation of immature macrophage observed in mice after IL-10R blockade, but the accumulation could potentially be the result of increased recruitment as a previous study suggested that IFN-γ-primed macrophages exhibit enhanced migration in response to CCL2, likely a STAT1-dependent function34. It is also possible that in situ expansion, decreased cell death, or reduced differentiation into mature macrophage subsets are responsible for enhanced accumulation of immature macrophages. Although we observed an increase in the proportion of Ly6C+ MHCII+ LP P2 macrophages compared with other subsets, the absolute number of LP macrophages is markedly elevated after IL-10R blockade, and this extends to all subsets (P1-P3/P4) suggesting that a STAT1-mediated block in differentiation cannot fully explain the macrophage accumulation observed in these animals. A caveat of these studies is that our mouse models did not include the CX3CR1-GFP transgene that is often used to distinguish between maturing P3 and fully mature P4 macrophages6, thus we do not know with certainty whether the increase in total macrophages observed in mice after IL- 10R blockade extends to fully mature resident-like macrophages, although a previous study using similar methodology did find an increase in the absolute number of P4 LP macrophage after anti-IL-10R Ab treatment of Hh-infected mice14. An increase in the percentage of circulating monocytes also does not explain our observations because we have not observed differences in the percentage of circulating monocytes after IL-10R blockade. We also believe that STAT1-dependent alterations in the microbiome are unlikely to explain increased accumulation of immature LP macrophages because we have previously demonstrated very similar microbiota in co-housed WT-and IL10R-deficient mice7. Thus, additional studies that include a detailed evaluation of LP macrophage dynamics will be required to fully understand the basis for STAT1-dependent macrophage accumulation in the LP of mice lacking IL-10R signaling.
STAT1 is the primary signal transducer of IFN-γ-mediated signaling, and indeed, mice lacking IFN-γR1 essentially phenocopied the STAT1-deficient mice after anti-IL-10R Ab treatment and Hh infection. This observation coupled with previous observations regarding the role of IFN-γR in macrophage accumulation during DSS colitis strongly suggests that IFN-γ-induced STAT1 signaling plays a central role in macrophage accumulation observed in murine colitis15,22. Nonetheless, we were quite surprised that despite the marked reduction in macrophage accumulation observed in both STAT1- and IFN-γR1-deficient mice after IL-10R blockade, we observed levels of residual histologic signs of colitis that remained elevated over untreated control mice. This is not inconsistent with previous studies showing quite variable effects of IFN-γR blockade or deletion on the development of colitis in mice lacking IL-10R signaling; although, to the best of our knowledge, previous studies have not simultaneously evaluated LP macrophage accumulation in parallel with colitis15. We suggest that there are several potential explanations for this observation. It is well documented that IFN-γ/ STAT1 signaling inhibits expansion of Th17 cells, and it is possible that Th17-mediated pathology could compensate for reduced macrophage induced inflammation observed in the absence of STAT135–37. Indeed, the macrophage-specific deletion of IL-10RA in recombination activating gene (RAG)-deficient mice resulted in enhanced IL-17 responses38, and we have also observed increased Th17 responses both in RAG1−/−Il10rb−/− mice after the adoptive transfer of unfractionated T cells13, as well as in humans with IL-10R-deficiency39. Interestingly, disease in IL-10R-deficient RAG mice after T-cell adoptive transfer is abrogated when T cells lack the IL-1R38,40, and IL-10R signaling interferes with both transcription of pro-IL-1b and inflammasome-dependent production of mature IL-1b in mouse and human macrophages38,40. These results raise the possibility that failure to suppress the IL-1b production in macrophages may, in part, be responsible for enhanced IL-17 responses observed in mice and humans with defects in IL-10 receptor signaling. Our data also show an elevated expression of the IL-22-induced antimicrobial peptide Reg3g in the colon of STAT1-deficient mice, suggesting an increased activation of the Th17 pathway. However, previous experiments demonstrate that the depletion of IL-17 in WT mice treated with anti-IL-10R Ab and infected with Hh actually exacerbated disease severity41. Moreover, Hh colonization and colitis were amplified in Il17a−/− mice compared with WT mice42. Both of these observations are inconsistent with a compensatory effect of IL-17 in STAT1-deficient mice but does not rule-out compensation from another pro-inflammatory pathway.
Another possibility to explain the residual colitis observed in STAT1-deficient mice after IL-10R blockade is that the accumulation of immature macrophages is not required for the development of histological signs of colitis in this model. This is consistent with a recent publication from our group demonstrating that despite the marked reductions in LP macrophage accumulation observed in mice lacking both CCR2 and IL-10R compared with mice lacking IL-10R alone, the mice lacking both CCR2 and IL-10R still exhibited significantly elevated histologic colitis scores compared with WT mice, indicating that interfering with the accumulation of immature macrophages in the intestine only has a limited role in regulating the histologic manifestations of colitis10. This potentially limited role for intestinal macrophages in regulating the severity of colitis seems at odds with strong evidence that macrophage-specific loss of IL-10R signaling is sufficient to drive the development of disease. One potential possibility to explain this phenomenon is that although the loss of IL-10R signaling on resident LP macrophages is sufficient to activate inflammatory pathways that drive disease, the accumulation of newly recruited immature macrophages into the colon observed in the absence of IL-10R signaling is not absolutely required for disease development.
Regardless of their exact role in driving histologic signs of disease, the observation of marked STAT1-dependent accumulation of immature macrophages in the absence IL-10R signaling suggested that the direct inhibition of STAT1 function by IL-10R could be necessary to prevent immature macrophage accumulation. It has previously been suggested that STAT3, the primary transducer of IL-10R signaling can directly interfere with STAT1-induced gene expression in macrophages by sequestering STAT1 and preventing formation of DNA binding STAT1 homodimers43; although, in contrast, we have previously argued that IL-10 does not have direct inhibitory effects on IFN-β-induced transcription32. In fact, data presented here strongly suggest that the ability of IL-10R to inhibit both macrophage accumulation in the colon and the expression of STAT1-dependent genes in colonic macrophages is not cell autonomous because WT macrophages accumulate and express STAT1-dependent genes at similar levels as IL-10R-deficient macrophages in mixed chimeras between WT- and IL-10R-deficient bone marrow cells. Further, despite previous suggestions that activated STAT3 might directly interfere with the function of STAT143, we demonstrated that addition of IL-10 did not interfere with the ability of IFN-γ to induce STAT1 activation or IFN-γ induced STAT1-dependent genes in bone marrow-derived macrophages. These observations strongly suggest that suppression of macrophage accumulation and inhibition of STAT1-dependent gene expression is not a cell autonomous function of IL-10R but rather more likely an indirect effect caused by suppression of inflammatory mediators within the intestinal environment with the ability to secondarily induce STAT1 activation. Indeed, the previous observation that IFN-γ and STAT1-dependent colonic macrophage recruitment is also observed in DSS colitis22 suggests that this pathway may be engaged more broadly in inflammatory states characterized by the accumulation of colonic macrophages. Thus, further exploration of this pathway in patients with increased accumulation of inflammatory monocytes in the colon could have therapeutic implications.
The recognition that IL-10R has potent cell autonomous and non-cell autonomous functions can provide important clues regarding the pathways that are directly and indirectly inhibited by IL-10R signaling, and we propose that understanding these different functions is essential to developing effective therapeutics for children with IL-10R-deficiency and potentially IBD in general. As an example, we and others have proposed that IL-10R signaling may play an important role in generating anti-inflammatory resident macrophages within the intestine7,13,44,45. However, our observations that in irradiated host mice reconstituted with mixtures of WT and IL-10R-deficient bone marrow cells, the expression of key markers of resident macrophages, including Cd163 and Lyve1, are markedly reduced in WTmix intestinal macrophages compared with WT macrophages isolated from mice reconstituted with WT bone marrow cells alone strongly argues that the induction of anti-inflammatory resident macrophages is not due to an autonomous inhibitory effect of IL-10R signaling but rather the ability of IL-10R signaling to suppress the secondary mediators that interfere with the development of anti-inflammatory resident macrophages in the intestine.
In contrast to non-cell autonomous functions of the IL10R, we have identified a set of genes that appear to be regulated in a cell autonomous fashion by IL-10R signaling, and these include known targets of IL-10R-mediated suppression, such as Il12b. The identification of these cell autonomous factors could provide important clues regarding the key drivers of intestinal inflammation in children lacking IL10R function and possibly in IBD more broadly, with potential therapeutic implications. It has previously been shown that IL-12 is a potent inducer of IFN-γ production, and the neutralization of IL-12 p40 (a subunit of both IL-12 and IL-23) interferes with the development of Hh-induced colitis in IL-10-deficient mice15. Further, IL-12 and IL-23 are highly relevant therapeutic targets. Therefore, it is tempting to speculate that the ability of IL-10R signaling to inhibit Il12b expression in a cell autonomous fashion interferes with IFN-γ production, ultimately resulting in the secondary suppression of STAT1 activation and STAT1-dependent macrophage accumulation in the colon. Other genes regulated in a cell autonomous fashion by IL10R include a number of CCL family chemokines including Ccl2, Ccl3, Ccl6, and Ccl8. Because we have shown that CCR2, one of the receptors for CCL family chemokines is intimately involved in the accumulation of immature macrophages into the colon of Il10ra−/− mice with colitis10, increased expression of these factors by Il10ra−/− macrophages could play a central role in the recruitment of immature and inflammatory macrophages into the colon of these mice. However, the exact pathways through which IL-10 suppresses these secondary mediators and their relevance to IBD in humans remains to be completely defined.
Based on these observations presented here, we hypothesize that the genes regulated in a cell autonomous fashion by IL-10R within intestinal macrophages are essential to prevent the development of colitis, and that further study of this gene set could yield essential clues regarding disease pathogenesis. We suggest that approaches including single-cell RNA sequencing in mice and comparison of findings using this technique between mice and patients lacking the IL-10R are essential to unravel the key cellular aspects of IL-10R signaling necessary to prevent the development of microbiota-driven colonic inflammation.
MATERIALS AND METHODS
Mouse strains
All transgenic and WT mice presented here have a C57BL/6 background. C57BL/6J, Stat1−/− [B6.129S(Cg)-Stat1tm1Dlv/J], Ifngr1−/− (B6.129S7-Ifngr1tm1Agt/J), and CD45.1 (B6.SJL-Ptprca Pepcb/ BoyJ) mice were purchased from Jackson Laboratories (Bar Harbor, Maine, USA) and housed under specific pathogen- and viral antibody-free conditions. All other strains used in this study were maintained under specific pathogen-free conditions. Cdcs1 Il10ra−/− and Cdcs1 Il10rafl/fl Lyz2Cre+ were generated as we have previously described7. Cdcs1 (CD45.1) mice were generated by crossing Cdcs1 (B6.C3Bir-Cdcs1 (BC-R))46 and CD45.1 mice. Cdcs1 Il10ra−/− Stat1−/− mice were generated by crossing Cdcs1 Il10ra−/− and Stat1−/− [B6.129S(Cg)-Stat1tm1Dlv/J] mice. Both male and female mice were used throughout the experiments, and wherever it was possible, we used littermate controls. At least 2 weeks before the initiation of an experiment bedding was mixed to standardize microbiota. The protocols for breeding, housing, and experiments were approved by the Animal Resources at Children’s Hospital, according to the Institutional Animal Care and Use Committees (Assurance number: A3303-01). Schematic diagrams describing experimental study designs were created with BioRender.com.
Colitis induction by Hh and anti-IL-10RA antibody
Eight-week-old Helicobacter-free mice were gavaged with 200 μl (optical density (OD) 1.5) Hh inoculums [ATCC 51449 (re-isolated from mice with colitis)] on days 1, 3, and 5. In parallel, 0.5 mg anti-IL-10RA mAb (1B1.3A, Bio X Cell, Lebanon, New Hampshire, USA) was administered through intraperitoneal injection in 150 μl phosphate-buffered saline (PBS) on days 1, 5, and 12. Age-matched littermate controls were included and sham-treated with PBS. Mice were euthanized and tissue was collected for further analysis.
LP cell isolation
Cecal and colonic LP cell preparation was carried out as previously described7,10. Briefly, colon and cecum were harvested and stripped from epithelial cells by agitating tissue in Hank's Balanced Salt Solution (HBSS) media (Gibco, Life Technologies, Grand Island, New York, USA) containing 10 mM EDTA and 4.3 mg/ml dithiothreitol (DTT) for 30 minutes at 37°C. This was followed by collagenase VIII (MilliporeSigma, Burlington, Massachusetts, USA) digestion for 35–45 minutes at 37°C. The remaining undigested tissue was homogenized by repeatedly passing through a 10-ml syringe. Cell suspensions were filtered, washed with PBS, and kept on ice for further use.
Histology and histopathological analysis
Disease severity was assessed using a HAI score evaluating hematoxylin- and eosin-stained sections obtained from the cecum and proximal-, mid-, and distal colon. Scoring was based on the severity of mononuclear inflammation (0–4), crypt hyperplasia (0–4), epithelial injury (0–4), and neutrophilic inflammation/crypt abscesses (0–4). Each tissue sample was scored separately for all four metrics by one of the authors (J. G.). The reported HAI score is the sum of the individual component scores.
Microscopic image acquisition
Hematoxylin- and eosin-stained proximal colonic tissue slides were imaged using a Zeiss Axio Imager Z2 upright microscope at a 10× magnification, and the files were processed by Zen 3.1 (blue edition) (Zeiss, Dublin, California, USA).
Flow cytometry
All cells were run using a BD LSRFortess Cell Analyzer (BD Biosciences, San Jose, California, USA) and analyzed using FlowJo v10.8 (Treestar, Ashland, OR, USA). For blocking non-specific binding, we used TruStain FcX (anti-mouse CD16/32) antibody (BioLegend, San Diego, California, USA). Representative examples for the gating strategies are provided in Supplementary Figs. 1D and 5. For LP macrophage analysis, cells were gated as follows: SSC-A/FSC-A, FSC-A/FSC-H, FSC-A/FSC-W, Live, CD45+ or separated to CD45.2+ and CD45.1+, CD103−, Ly6G−, CD64+, CD11b+, Ly6C/MHC II.
RNA isolation and quantitative reverse transcription-polymerase chain reaction
Tissue from the cecum and distal, mid-, and rectal colon were collected and preserved in TRIzol reagent (Gibco, Life Technologies, Grand Island, New York, USA). Total RNA was extracted following the manufacturer’s protocol, and cDNA generation was carried out using 1 μg total RNA and TaqMan Reverse Transcription Reagents (Applied Biosystems, Carlsbad, California, USA) following the manufacturer’s protocols. Quantitative reverse transcription-polymerase chain reaction was performed using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, Hercules, California, USA) or TaqMan Universal Master Mix II, with UNG (Applied Biosystems) on a QuantStudio Flex 6 System (Thermo Fisher Scientific, Waltham, Massachusetts, USA) and custom primers. Primer sequences are available upon request. Expression was normalized to Gapdh or β-actin expression, and the differences between samples were calculated using the 2−ΔΔ cycle threshold method. Fold change is reported relative to a sham or uninflamed control.
Bone marrow chimeras
Recipient mice were irradiated (Gamma Cell 40, 137Cs) with a split dose of 1200 Rads 4 hours apart. The following day, the bone marrow was harvested from the hind leg bones of donor mice by flushing with sterile PBS. Bone marrow cells were counted, and 4 × 106 cells/mouse were injected into irradiated recipients through retro-orbital injections in 150 μl sterile PBS. Drinking water was supplemented with trimethoprim/sulfamethoxazole (Aurobindo Pharmaceuticals, East Windsor, New Jersey, USA) for 3 weeks (240 mg/250 ml water). Eight weeks after transfer, blood was collected through retro-orbital sampling under anesthesia, stained with CD45.1 and/or CD45.2, and analyzed by flow cytometry to evaluate for successful reconstitution.
Bulk RNA sequencing and analysis
A total of 5000-25,000 P3/P4 LP macrophages from bone marrow chimeras 8 weeks after reconstitution were FACS-sorted (gated on: CD103−, Ly6G−, CD11b+, CD11cint, CD64+, Ly6C−, CD45.1+ or CD45.2+) directly into RLT buffer (Qiagen, Valencia, California, USA) on a fluorescence-activated cell sorting (FACS) Aria II (BD Biosciences, San Jose, California, USA) and mRNA was extracted using the RNeasy Micro kit (Qiagen, Valencia, California, USA). Libraries were prepared using Kapa mRNA Hyper-Prep strand specific sample preparation kits (Roche, Wilmington, Massachusetts) from 200 ng of purified total RNA according to the manufacturer’s protocol on a Biomek i7 (Beck-man Coulter, Indianapolis, Indiana, USA). The finished dsDNA libraries were quantified by Qubit fluorometer (Thermo Fisher Scientific, Waltham, Massachusetts, USA) and 4200 TapeStation System (Agilent, Santa Clara, California, USA). Uniquely dual-indexed libraries were pooled in an equimolar ratio and shal-lowly sequenced on a MiSeq (Illumina, San Diego, California, USA) to further evaluate the library quality and pool balance. The final pool was sequenced on a NovaSeq 6000 (Illumina, San Diego, California, USA) targeting 40 million 50 bp read pairs per library at the Dana-Farber Cancer Institute Molecular Biology Core Facilities.
Sequenced reads were aligned to the UCSC mm10 reference genome assembly and gene counts were quantified using STAR (v2.7.3a)47. Gene counts were then normalized using the trimmed mean of M-values approach in edgeR (v 3.15)48. Differential gene expression testing was performed using limma (v3.15)49, blocking on individual mice.
Because all mice in this study were Cdcs1 knockouts, in the differential gene expression analysis, we excluded all Cdcs1 proximal genes, which might have escaped the targeted knockout. Specifically, we ignored all genes found within 1Mb up and downstream of the expanded Cdcs1 locus, as previously defined46. Moreover, although WT mice in the differential gene expression studies were male, and mice of all other groups (Il10ra−/−, WTmix, and Il10ra−/−mix) were female, we excluded sex dimorphic genes in all comparisons involving WT macrophages. Specifically, we ignored the genes found to be sexually dimorphic in at least three murine cell types, as previously reported50. Bulk RNA-seq data generated as part of this study are deposited in the National Center for Biotechnology Information Gene Expression Omnibus under the accession number GSE211841.
We used STRING (v11.5)51 to identify functional relationships between genes expressed at higher levels in Il10ra−/−mix than WTmix LP macrophages and further used gene set enrichment analysis (GSEA; v4.2.352 to examine the concordant dysregulation of immunologic signatures (C7) of the Molecular Signatures Database (MSigDB) v7.5.1 between groups. In these analyses, we removed Il10ra and its proximal genes (within 1Mb of its 9q locus) to ensure that Il10ra per se and potentially unequal cross-over around its locus do not bias the set and pathway level convergence.
Reagents and antibodies
Distributors and origin of reagents, equipment, and materials used in this study are indicated in the Methods section.
Reagents and antibodies used for flow cytometry and cell sorting: Zombie Violet fixable dye (BioLegend), CD45 (clone 30-F11, BioLegend), CD45.1 (clone A20, BioLegend), CD45.2 (clone 104, BioLegend), CD103 (clone 2E7, BioLegend), Ly-6G (1A8, BioLegend), CD64 (clone X54-5/7.1, BioLegend), CD11b (clone M1/70 BioLegend), CD11c (clone N418, BioLegend) Ly-6C (clone HK1.4, BioLegend), MHC II (clone M5/114.15.2, BioLegend).
Bone marrow-derived macrophage (BMDM) preparation and stimulation
BMDMs were isolated and grown as previously described32. BMDMs were split and plated onto 24-well plates a day before stimulation and were cultured in 500 μl of DMEM supplemented with 10% FBS, penicillin/streptomycin, HEPES, and GlutaMAX at a density of 5 × 105 cells per well for RNA extraction. For stimulation, media was replaced with fresh media and cells were stimulated with the addition of IL-10 (PeproTech, Cranbury, New Jersey, USA) or IFN-γ (eBioscience, San Diego, California, USA) or both.
Immunoblotting
BMDMs were cultured as described previously in 6-well plates and 3 × 106 cells per well were plated for western blotting. After stimulation, BMDMs were washed with 1× PBS (Gibco) and lysed in RIPA buffer completed with phenylmethylsulfonyl fluoride and PhosSTOP (all from Roche, San Francisco, California, USA). 10–20 ug protein were loaded onto Novex 4%–12% Bis-Tris gels (Thermo Fisher Scientific, Waltham, Massachusetts, USA). Proteins were transferred to polyvinylidene difluoride membranes and blotted with anti-STAT1 (D1K9Y), anti-pSTAT1 (58D6; Y701), anti-STAT3 (D372G), anti-pSTAT3 (D3A7; Y705), and anti-GAPDH (14C10; all from Cell Signaling Technology).
Statistical analysis
Statistical analyses were performed using GraphPad Prism 9 unless stated otherwise. A p value < 0.05 was considered statistically significant and asterisks were used to indicate significance as follows: *p < 0.05, **p < 0.01, *** p < 0.001, **** p < 0.0001. Multiple testing correction in the analysis of bulk RNA sequencing data was achieved using the Benjamini-Hochberg procedure, ensuring that the overall false discovery rate of this study was below 0.0553.
Supplementary Material
ACKNOWLEDGMENTS
The authors are grateful to Jared Barends, Zach Herbert, and Sandra M. Frei for technical assistance. The authors also thank the IDDRC Cellular Imaging Core, funded by NIH P50 HD105351. The authors would also like to thank the Beth Israel Deaconess Medical Center Histology Core Facility for processing of histology samples.
FUNDING
This research was supported by a Crohn’s and Colitis Foundation Senior Research Award to B. H. H. and by grants from NIHP30-ES002109, P01CA28842 to J. G. F.. S. B. S. is supported by the National Institute of Diabetes and Digestive Kidney Diseases of the National Institutes of Health under (award number P30DK03485 and RC2DK122532), the Wolpow Family Chair in IBD Treatment and Research, the Translational Investigator Service at Boston Children’s Hospital, and the Children’s Rare Disease Cohort Study.
S. B. S. declares the following interests: scientific advisory board participation for Pfizer, BMS, Hoffman La Roche, Lilly, IFM Therapeutics, Merck, and Pandion and grant support from Pfizer, Novartis, Takeda, Amgen; consulting for Hoffman La Roche, Merck, Takeda, and Amgen.
Footnotes
DECLARATIONS OF COMPETING INTEREST
The other authors have no competing interests to declare.
APPENDIX A. SUPPLEMENTARY DATA
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mucimm.2023.02.006.
DATA AVAILABILITY
The datasets generated during and/or analyzed during the current study are available in the National Center for Biotechnology Information Gene Expression Omnibus, under the accession number GSE211841.
REFERENCES
- 1.Glocker EO et al. Infant colitis-it’s in the genes. Lancet 376, 1272 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Glocker EO et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N. Engl. J. Med 361, 2033–2045 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glocker EO, Kotlarz D, Klein C, Shah N. & Grimbacher B. IL-10 and IL-10 receptor defects in humans. Ann. N. Y. Acad. Sci 1246, 102–107 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Na YR, Stakenborg M, Seok SH & Matteoli G. Macrophages in intestinal inflammation and resolution: a potential therapeutic target in IBD. Nat. Rev. Gastroenterol. Hepatol 16, 531–543 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Bain CC & Mowat AM Macrophages in intestinal homeostasis and inflammation. Immunol. Rev 260, 102–117 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zigmond E. et al. Ly6C hi monocytes in the inflamed colon give rise to proinflammatory effector cells and migratory antigen-presenting cells. Immunity 37, 1076–1090 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Redhu NS et al. Macrophage dysfunction initiates colitis during weaning of infant mice lacking the interleukin-10 receptor. eLife 6, 1–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caruso R, Lo BC & Núñez G. Host-microbiota interactions in inflammatory bowel disease. Nat. Rev. Immunol 20, 411–426 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Kang B. et al. Commensal microbiota drive the functional diversification of colon macrophages. Mucosal Immunol. 13, 216–229 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El Sayed S. et al. CCR2 promotes monocyte recruitment and intestinal inflammation in mice lacking the interleukin–10 receptor. Sci. Rep 12, 452 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kühn R, Löhler J, Rennick D, Rajewsky K. & Müller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell 75, 263–274 (1993). [DOI] [PubMed] [Google Scholar]
- 12.Sellon RK et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect. Immun 66, 5224–5231 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shouval DS et al. Interleukin-10 receptor signaling in innate immune cells regulates mucosal immune tolerance and anti-inflammatory macrophage function. Immunity 40, 706–719 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bain CC, Oliphant CJ, Thomson CA, Kullberg MC & Mowat AM Proinflammatory role of monocyte-derived CX3CR1int macrophages in Helicobacter hepaticus-induced colitis. Infect. Immun 86, e00579–e617 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kullberg MC et al. Helicobacter hepaticus triggers colitis in specific-pathogen-free interleukin-10 (IL-10)-deficient mice through an IL-12-and gamma interferon-dependent mechanism. Infect. Immun 66, 5157–5166 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryzhakov G. et al. Alpha kinase 1 controls intestinal inflammation by suppressing the IL-12/Th1 axis. Nat. Commun 9, 3797 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang K. et al. Interferon-γ represses M2 gene expression in human macrophages by disassembling enhancers bound by the transcription factor MAF. Immunity 47, 235–250.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu X. & Ivashkiv LB Cross-regulation of signaling pathways by interferon-γ: implications for immune responses and autoimmune diseases. Immunity 31, 539–550 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Begue B. et al. Defective IL10 signaling defining a subgroup of patients with inflammatory bowel disease. Am. J. Gastroenterol 106, 1544–1555 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Ramana CV, Chatterjee-Kishore M, Nguyen H. & Stark GR Complex roles of Stat1 in regulating gene expression. Oncogene 19, 2619–2627 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Rauch I, Müller M. & Decker T. The regulation of inflammation by interferons and their STATs. JAKSTAT 2, 1–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakanishi Y, Sato T, Takahashi K. & Ohteki T. IFN-γ-dependent epigenetic regulation instructs colitogenic monocyte/macrophage lineage differentiation in vivo. Mucosal Immunol. 11, 871–880 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Wu F. et al. Genome-wide gene expression differences in Crohn’s disease and ulcerative colitis from endoscopic pinch biopsies: insights into distinctive pathogenesis. Inflamm. Bowel Dis 13, 807–821 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Mudter J. et al. Activation pattern of signal transducers and activators of transcription (STAT) factors in inflammatory bowel diseases. Am. J. Gastroenterol 100, 64–72 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Schreiber S. et al. Activation of signal transducer and activator of transcription (STAT) 1 in human chronic inflammatory bowel disease. Gut 51, 379–385 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haberman Y. et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J. Clin. Invest 124, 3617–3633 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito S. et al. Interleukin-10 inhibits expression of both interferon α- and interferon γ-induced genes by suppressing tyrosine phosphorylation of STAT1. Blood 93, 1456–1463 (1999). [PubMed] [Google Scholar]
- 28.Rogler G. Efficacy of JAK inhibitors in Crohn’s disease. J. Crohns Colitis 14, S746–S754 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cordes F, Foell D, Ding JN, Varga G. & Bettenworth D. Differential regulation of JAK/STAT-signaling in patients with ulcerative colitis and Crohn’s disease. World J. Gastroenterol 26, 4055–4075 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salas A. et al. JAK–STAT pathway targeting for the treatment of inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol 17, 323–337 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Ip WKE, Hoshi N, Shouval DS, Snapper S. & Medzhitov R. Anti-inflammatory effect of IL-10 mediated by metabolic reprogramming of macrophages. Science 356, 513–519 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conaway EA, de Oliveira DC, McInnis CM, Snapper SB & Horwitz BH Inhibition of inflammatory gene transcription by IL-10 is associated with rapid suppression of lipopolysaccharide-induced enhancer activation. J. Immunol 198, 2906–2915 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lang R, Patel D, Morris JJ, Rutschman RL & Murray PJ Shaping gene expression in activated and resting primary macrophages by IL-10. J. Immunol 169, 2253–2263 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Hu X, Park-Min KH, Ho HH & Ivashkiv LB IFN-γ-primed macrophages exhibit increased CCR2-dependent migration and altered IFN-γ responses mediated by Stat1. J. Immunol 175, 3637–3647 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Cypowyj S, Picard C, Maródi L, Casanova JL & Puel A. Immunity to infection in IL-17-deficient mice and humans. Eur. J. Immunol 42, 2246–2254 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernshtein B. et al. IL-23 – producing IL-10Ra – deficient gut macrophages elicit an IL-22 – driven proinflammatory epithelial cell response. Sci. Immunol 6571, 1–15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu L. et al. Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. J. Exp. Med 208, 1635–1648 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li B. et al. IL-10 engages macrophages to shift Th17 cytokine dependency and pathogenicity during T-cell-mediated colitis. Nat. Commun 6, 6131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shouval DS et al. Enhanced TH17 responses in patients with IL10 receptor deficiency and infantile-onset IBD. Inflamm. Bowel Dis 23, 1950–1961 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Shouval DS et al. Interleukin 1β Mediates intestinal Inflammation in Mice and Patients with interleukin 10 Receptor Deficiency. Gastroenterology 151, 1100–1104 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morrison PJ et al. Differential requirements for IL-17A and IL-22 in cecal versus colonic inflammation induced by Helicobacter hepaticus. Am. J. Pathol 185, 3290–3303 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu L. et al. The deletion of IL-17A enhances Helicobacter hepaticus colonization and triggers colitis. J. Inflamm. Res 15, 2761–2773 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ho HH & Ivashkiv LB Role of STAT3 in type I interferon responses. Negative regulation of STAT1-dependent inflammatory gene activation. J. Biol. Chem 281, 14111–14118 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Schridde A. et al. Tissue-specific differentiation of colonic macrophages requires TGFβ receptor-mediated signaling. Mucosal Immunol. 10, 1387–1399 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Girard-Madoux MJH et al. IL-10 control of CD11c+ myeloid cells is essential to maintain immune homeostasis in the small and large intestine. Oncotarget 7, 32015–32030 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bleich A. et al. Cdcsl a major colitis susceptibility locus in mice; subcongenic analysis reveals genetic complexity. Inflamm. Bowel Dis 16, 765–775 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dobin A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson MD & Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ritchie ME et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu T. & Mar JC Investigating transcriptome-wide sex dimorphism by multi-level analysis of single-cell RNA sequencing data in ten mouse cell types. Biol. Sex Differ 11, 61 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Szklarczyk D. et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/ measurement sets. Nucleic Acids Res. 49, D605–D612 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Subramanian A. et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. U. S. A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benjamini Y. & Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995). [Google Scholar]
- 54.Shannon P. et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu SY, Sanchez DJ, Aliyari R, Lu S. & Cheng G. Systematic identification of type I and type II interferon-induced antiviral factors. Proc. Natl Acad. Sci. U. S. A 109, 4239–4244 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available in the National Center for Biotechnology Information Gene Expression Omnibus, under the accession number GSE211841.