Abstract
Tomato bushy stunt virus (TBSV) cDNA, positioned between a modified cauliflower mosaic virus 35S promoter and the hepatitis delta virus antigenomic ribozyme with a downstream nopaline synthase gene polyadenylation signal, established infections upon rub-inoculation of plants with intact plasmids. Application of this methodology produced a TBSV DNA-based gene vector which yielded readily detectable levels of localized foreign gene expression in inoculated leaves. This is the first demonstration of an infectious DNA from a member of the Tombusviridae which permits rapid TBSV-mediated foreign-gene expression upon direct rub-inoculation of miniprep DNA onto a variety of plant species.
In the past decade, molecular genetic studies of plant RNA viruses have benefited tremendously from the methodology introduced by Ahlquist et al. (1), which permits the generation of infectious RNA upon in vitro transcription of full-length cDNA clones. This technique has since been applied to many different RNA plant viruses (4), including tomato bushy stunt virus (TBSV) (12). Infectious cDNA clones have expedited studies expanding our knowledge of molecular virus genetics and have stimulated the use of RNA plant viruses as foreign gene vectors for research and commercial purposes (13, 14, 26). Gene delivery systems based on TBSV have several advantages (26), which include the broad experimental host range of the virus and rapidly occurring, high levels of replication and gene expression. These features have contributed to the successful use of TBSV gene vectors to quickly assess the biological activity of heterologous foreign proteins and/or RNA in protoplasts or inoculated leaves (3, 25, 26, 32).
While the reactions for in vitro transcription are generally straightforward, the costs associated with these procedures may be considerable if large numbers of constructs are tested, for example, when virus vectors are used to screen cDNA expression libraries in plants (15). The objective of this study was to optimize the usefulness of the TBSV vector system by alleviating the necessity of in vitro linearization or transcription of cDNA and to allow direct delivery without the need for agroinoculation or biolistics. For this purpose, plant and animal virus transcription and processing signals were tested to optimize the infectivity of intact, untreated plasmid DNA on several plant species.
Constructs were generated with full-length TBSV cDNA positioned downstream of a cauliflower mosaic virus (CaMV) 35S promoter for the initiation of transcription at the authentic 5′ end of the viral RNA in the plant nucleus (8). The nopaline synthase poly(A) signal [nos-poly(A)] (28) was used to permit the in vivo termination of transcription through polyadenylation, and the hepatitis delta virus antigenomic ribozyme (HDVagrz) (21) was incorporated to generate 3′ termini resembling those of the native TBSV RNA. These modifications permitted the simple rub-inoculation of untreated plasmid DNA onto leaves to infect different plants. Application of this technology to a TBSV construct in which a multiple-cloning region (MCR) replaces the coat protein (CP) gene permitted the convenient introduction of foreign genes which were rapidly expressed to yield detectable levels of proteins in the inoculated leaves of various plants.
Infectivity of TBSV DNA.
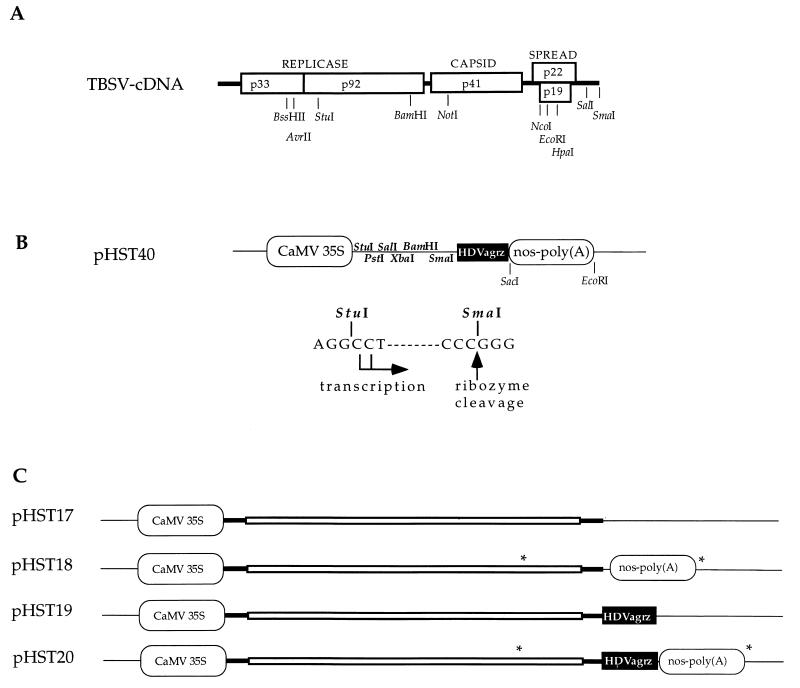
To obtain clones with the 5′ end of TBSV cDNA inserted immediately downstream of the CaMV 35S promoter sequence, a PCR product was obtained with a 5′ primer identical to the 5′ terminus of TBSV (12) and a 3′ primer covering the TBSV StuI site (Fig. 1A) (29). Standard molecular biology protocols were used, as described by Sambrook et al. (23) or as provided by the suppliers of the reagents. The PCR product was cleaved at the internal AvrII site (Fig. 1A), which yielded a fragment with a blunt 5′ end and a 3′ AvrII terminus, which was ligated into the pUC-35S (8) vector between the StuI and XbaI (compatible with AvrII) sites in the MCR to yield pHST7. Subsequently, a TBSV cDNA segment from the BssHII site to the BamHI site (Fig. 1) was inserted between the BssHII and BamHI sites of pHST7. Lastly, this intermediate was digested with BamHI and SmaI, to introduce the BamHI to SmaI fragment of pHST2-14 in which an MCR replaces the CP gene (26). The resulting plasmid, pHST8, was digested with NotI (present inside the MCR) and HpaI (Fig. 1), treated with DNA polymerase Klenow fragment to create blunt ends, and religated. This created pHST9, in which the internal SacI and EcoRI sites were removed to permit the following cloning steps. The nos-poly(A) element of p3′NT (28) was released with SacI and EcoRI, and the resulting ca. 270-bp nos-poly(A) fragment was inserted between the SacI and EcoRI sites at the 3′ end of the TBSV cDNA insert of pHST9 to give pHST10. Subsequently, the ca. 90-bp HDVagrz cDNA fragment was removed with SmaI and SacI from plasmid 2.0 (a generous gift from A. Ball) and inserted into pHST10, which was opened with the same restriction enzymes, to generate pHST11. To obtain a cloning intermediate without the nos-poly(A) signal, pHST11 was digested with SacI and EcoRI, treated with Klenow fragment, and religated to generate pHST15. Sequence analyses were performed at all intermediate steps to ensure the proper positioning and sequence composition of the cloned fragments.
FIG. 1.
Plasmids and regulatory sequences. (A) Infectious cDNA of TBSV (12). The open reading frames are indicated by boxes, and encoded proteins (numbers show molecular masses, in kilodaltons) are provided with their function; thick lines represent untranslated sequences. Selected restriction enzyme sites that are relevant for this study are shown. (B) The cDNA cloning vector (pHST40) for in vivo transcription. Two alternative transcriptional start sites are indicated, and the HDVagrz RNA cleavage site is shown at the SmaI site on the DNA. Any cDNA can be cloned between the StuI and SmaI sites, which will facilitate in vivo transcription of RNAs with authentic 5′ and 3′ termini. Thin lines represent pUC18 sequences. (C) The plasmids pHST17 through pHST20 show the DNA-based TBSV constructs with the CaMV 35S promoter and the combinations of nos-poly(A) and/or HDVagrz at the 3′ end. The asterisks denote the positions of primers utilized to amplify the PCR products used in the assays described for Fig. 3.
The EcoRI-to-SacI fragment from pHST11, comprising the HDVagrz and nos-poly(A), was transferred into pUC-35S to yield pHST40. This potential universal cloning vector has been constructed to permit the insertion of plant viral cDNAs between the StuI and SmaI sites (Fig. 1B). To obtain pHST17 through pHST20 (Fig. 1C), the BamHI-to-SalI fragment from the kanamycin-resistant plasmid pHS24, which harbors a TBSV cDNA fragment with the intact CP gene (27), was used to replace the BamHI-to-SalI fragments from pHST9, pHST10, pHST15, and pHST11, respectively.
As reported for cucumoviruses (8, 9, 30) and cowpea mosaic virus (8), the infectivity of DNA-based systems may benefit strongly from the linearization of plasmids. To test this possibility for TBSV, pHST17 (Fig. 1C) was inoculated onto plants either as untreated supercoiled DNA or after linearization at the 3′ end of the TBSV cDNA insert. The results in Fig. 2A show that infectivity was substantially improved by linearization prior to inoculation. Because the treatment of DNA with a restriction enzyme affected infectivity, this experiment provided indirect evidence that the infections were initiated by DNA rather than contaminant RNA. This was further confirmed by the observation that incubation of plasmid DNA with DNase I destroyed infectivity, whereas RNase A had no effect (Fig. 2B). The enzymatic treatments were followed by phenol-chloroform extractions, ethanol precipitation, and resuspension in water or Tris-EDTA buffer prior to inoculation.
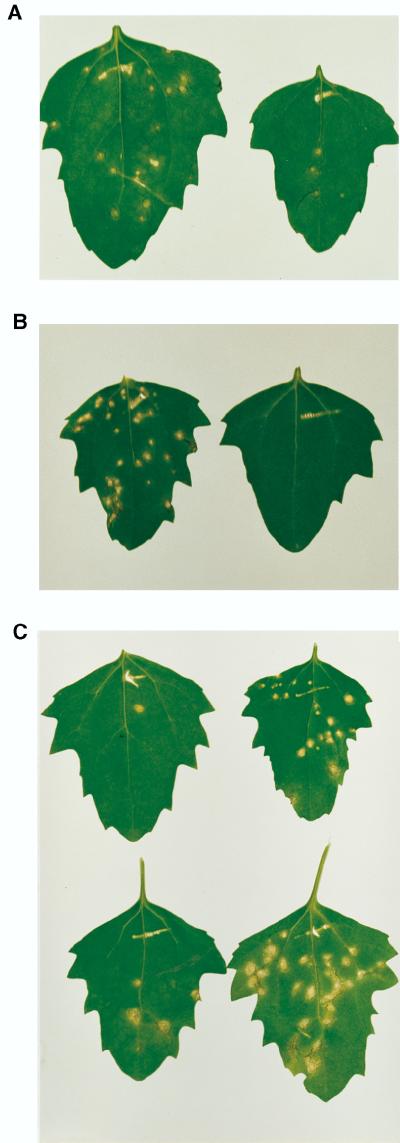
FIG. 2.
Infectivity assays of plasmids. (A) C. quinoa leaves 5 days after inoculation with pHST17 DNA that was SmaI linearized (left) or supercoiled (right). (B) C. quinoa leaves 6 days after inoculation with pHST20 DNA, treated with RNase A (left) or DNase I (right). (C) C. quinoa leaves 9 days after inoculation with supercoiled DNA of pHST17 (top left), pHST18 (top right), pHST19 (bottom left), or pHST20 (bottom right).
To examine if 3′ processing signals enhanced infectivity and thereby eliminated the necessity for in vitro linearization, pHST18, pHST19, and pHST20 DNA were rub-inoculated onto Chenopodium quinoa (Fig. 2C). Approximately 10 μg of CsCl gradient-purified supercoiled plasmid DNA was mixed with RNA inoculation buffer (25) to a total volume of 50 to 100 μl for the inoculation of two to four leaves per plant. Irrespective of the construct, local lesions became visible ca. 3 to 4 days postinoculation. Although the total number of local lesions varied between inoculation experiments, the trend shown in Fig. 2C was consistent. The results of four experiments in which the number of lesions per leaf was tabulated yielded a range of 1 to 10 lesions for pHST17 and pHST19, 10 to 15 lesions for pHST18, and 30 to 40 lesions for pHST20. The same trend was supported by comparative tests on cowpea (Vigna unguiculata), another sensitive local-lesion host. Combined, these results illustrated that the combination of the HDVagrz and nos-poly(A) signals provided the highest level of infectivity.
Hosts which are very susceptible to inoculations with in vitro-generated transcripts, e.g., Nicotiana benthamiana, Nicotiana clevelandii, C. quinoa, cowpea, and Spinacia oleracea (spinach), were also susceptible to direct DNA inoculation. Five days prior to DNA inoculation, N. benthamiana and N. clevelandii plants were transferred from the greenhouse or growth chambers to the laboratory, where presumably the low light conditioned the leaves, resulting in improved inoculation efficiency (unpublished results). Compared to inoculations with transcripts, inoculation with pHST20 DNA would result in a 1- to 2-day delay in the appearance of symptoms. The substitution of virus inoculation buffer (1% Celite, 50 mM KH2PO4 [pH 7.0]) for RNA inoculation buffer (pH 9.3) had no obvious effect on infectivity.
The CaMV 35S promoter has been used by itself and in combination with a 3′ poly(A) signal to generate infectious cDNA plasmids of several plant virus RNAs (5, 6, 8–10, 17–20, 30, 31, 35). The results in this study demonstrate that the modified CaMV 35S promoter (8) is very effective in promoting the transcription of infectious TBSV RNA in vivo. Furthermore, the polyadenylation of TBSV RNA from pHST18, which is predicted to add ca. 300 extra bases to the 3′ end of the viral RNA, does not abolish infectivity (Fig. 1 and 2). This is in agreement with observations by Dalmay et al. (7), who also observed that cymbidium ringspot tombusvirus containing extra sequences at the 3′ end of the RNA maintained the ability to replicate. This phenomenon may also explain the infectivity of intact circular pHST17, which lacks any 3′ processing signals (Fig. 1 and 2).
Ribozyme activity.
The bioassays showed that the presence of the HDVagrz at the 3′ end of TBSV cDNA, upstream of the nos-poly(A) signal, improved infectivity. To confirm the activity of the ribozyme within the TBSV and nos-poly(A) context, transcripts were generated directly from PCR products containing the 3′ proximal 950 bp of the TBSV cDNA and the nos-poly(A) sequence either alone or in combination with the HDVagrz. PCR fragments were amplified with TaqI polymerase (Promega, Madison, Wis.) or Vent DNA polymerase (New England Biolabs, Beverly, Mass.), with either pHST18 or pHST20 as a template (Fig. 1C). For this purpose, a 5′ primer was used which contained the T7 promoter sequence attached to the transcriptional start site sequence of subgenomic RNA2 (sgRNA2) on the cDNA (Fig. 1C). The 3′ universal reverse-sequencing primer annealed immediately downstream of the nos-poly(A) signal (Fig. 1C).
Ribozyme activity was routinely analyzed with ca. 0.2 μg of in vitro-generated transcripts in reaction buffer with a final concentration of 1 mM MgCl2, 5 M urea (optional), and 1.2% sodium dodecyl sulfate to prevent RNase activity. The mixture was incubated for 2 h at room temperature, followed by 30 min of incubation on ice and centrifugation at 9,000 × g for ca. 1 min. The entire sample was electrophoresed through a 2% agarose gel in Tris-borate-EDTA buffer (23). After electrophoresis, the gels were incubated in water to remove excess urea, followed by standard Northern blotting and hybridization procedures.
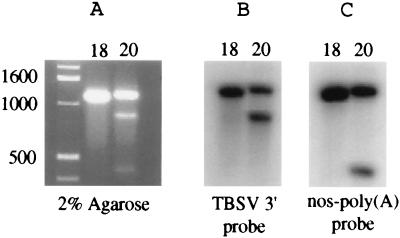
The results in Fig. 3 demonstrate that no specific cleavage product was obtained when the ca. 1,280-nucleotide (nt) transcript was derived from pHST18 which lacks the HDVagrz. However, the ca. 1,370-nt transcript from pHST20, containing the HDVagrz, was processed into two RNA products of ca. 430 and 940 nt. As predicted, the larger cleavage product was the same size as the transcripts that were obtained when cDNA templates were digested with SmaI, which cleaves DNA at the position where the ribozyme cleaves RNA (Fig. 1 and data not shown). The larger RNA product hybridized with TBSV but not with the nos-poly(A) segment (Fig. 3B), whereas the smaller RNA fragment hybridized only with the nos-poly(A) DNA (Fig. 3C), which is in agreement with the anticipated position of the ribozyme cleavage site.
FIG. 3.
In vitro HDVagrz activity. (A) Two percent agarose gel with RNA transcribed from ca. 1,300- and 1,400-bp PCR products obtained from the 3′ proximal fragments of pHST18 (lane 18) or pHST20 (lane 20), respectively (see Fig. 1 for the locations of primer annealing sites). The upper band represents uncleaved RNA, and the lower bands in the lane with RNA from pHST20 (lane 20) represent the HDVagrz cleavage products. The positions of double-stranded DNA size markers are indicated in basepairs. (B) RNA hybridization assay with pHS49 (24) as a specific probe for detection of 3′ proximal TBSV RNA sequences. (C) RNA hybridization of a similar Northern blot with p3′NT (28) as a probe for detection of nos-poly(A) sequences.
Since the proper parameters for HDVagrz activity are not predictable (2), it was determined if ribozyme activity was maintained under varying conditions. For this purpose, RNA was incubated by using the following combinations: room temperature or 55°C; 0, 1, or 10 mM MgCl2; and 0 or 5 M urea. Although complete cleavage was not observed under any of these conditions, compared to the results shown in Fig. 3, ca. threefold less ribozyme activity was obtained upon incubation of ca. 0.2 μg of RNA and 1 mM MgCl2 for 2 h at room temperature. However, in vitro ribozyme activity was again improved twofold by denaturation of the RNA at 55°C with or without 1 mM MgCl2. Regardless of the slight quantitative influences, the expected ratio and sizes of cleavage products were readily obtained under a variety of incubation conditions. These results strongly suggest that the improved infectivity on plants upon inclusion of the HDVagrz in pHST20 results from ribozyme activity.
A positive influence of ribozymes on the infectivity of in vivo-transcribed RNAs was previously demonstrated for tobacco mosaic virus (TMV) (6, 34). Compared with these reports, an ancillary finding with the TBSV DNA system is that the bioassays on local-lesion hosts revealed that the highest levels of infectivity were obtained with plasmid DNA containing the HDVagrz in addition to a poly(A) signal. Omission of the nos-poly(A) signal resulted in a reduced number of lesions, suggesting that the termination of transcription by polyadenylation either increases the efficiency of the HDVagrz activity or properly positions the transcript for nuclear export.
Comparison of TBSV DNA inoculations with those of other DNA-based plant RNA viruses.
CaMV 35S promoter-mediated in vivo-transcribed cDNAs of TMV (35) and brome mosaic virus (19) were infectious only on Chenopodium species, and TMV DNA was not infectious on its natural host, Nicotiana tabacum (31). Although cucumber mosaic virus cDNA was infectious on a variety of plant species, for high levels of infectivity the DNA cassette needed to be released with restriction enzymes prior to inoculation (9, 30). Subsequent studies have shown that the infectivity of cDNA plasmids of two potyviruses (10, 11), two luteoviruses (16, 22), or TMV on N. tabacum (6) requires either biolistic delivery or agroinoculation. Within this context, the novel or attractive features of the new TBSV DNA system include the following: (i) linearization of plasmid DNA is not required; (ii) infectivity is obtained upon simple rub-inoculation; (iii) DNA-mediated infections are established on a wide variety of plants; and (iv) although inoculations with TBSV DNA were initially performed with CsCl-purified DNA, inoculation of less-pure miniprep DNA also yields very efficient infections, which provides a practical level of convenience.
The reasons for the ability of the DNA-based TBSV constructs to efficiently establish infections after simple rub-inoculation of many plant species are unknown. This capacity is probably not due to a more infectious nature of the DNA, since the ca. 10 μg of TBSV DNA used for our inoculations (final concentration of 50 to 125 ng/μl) is within the range reported for other systems (6, 30). However, it is conceivable that the combination of the high infectivity of TBSV transcripts and the nonselective invasion of different tissues contributes to the effectiveness of the DNA constructs.
Rub-inoculation of TBSV vector DNA for expression of foreign genes.
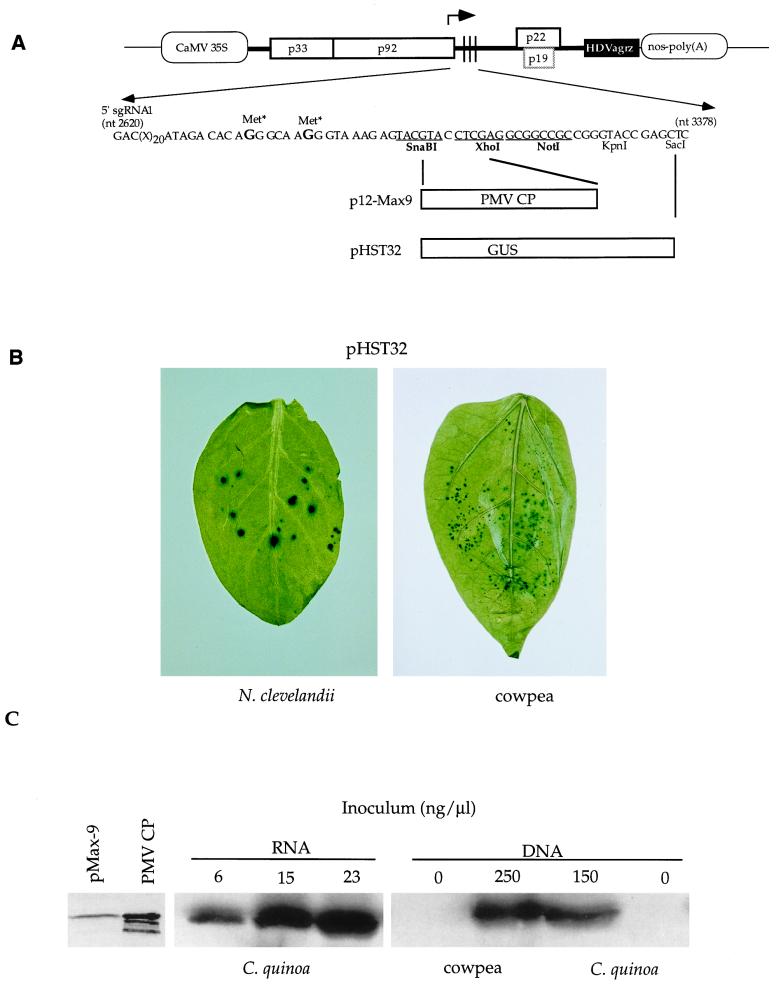
Previously, it was shown that TBSV-mediated β-glucuronidase (GUS) expression could be readily detected in inoculated leaves following inoculation with in vitro-generated transcripts (25, 27). In those experiments, the GUS gene was fused to the 5′ end of the CP gene, which resulted in translational initiation from the authentic CP start codon and the production of an enzymatically active fusion protein. Plasmid pHST12 (Fig. 4A) was designed to allow the transcription of an sgRNA1 with an elongated leader sequence, on which translational initiation occurs from the start codon of the introduced foreign gene. To generate the pHST12 gene vector (Fig. 4A), pHST11 was digested with SnaBI and SalI (Fig. 1), and the released fragment was replaced with the compatible fragment of pHST2-14 (26). To obtain pHST34 (Fig. 4A), a DNA-based vector with an intact p19 gene, the BamHI-to-NcoI fragment of pHST20 was replaced with that of pHST2-14 (26).
FIG. 4.
Design and implementation of TBSV-based DNA gene vectors. (A) Diagram of pHST12, p12-Max9, and pHST32 (not to scale). The stippled box for p19 indicates that this gene is active in pHST32 but inactive in pHST12 and p12-Max9. Nucleotide sequence details are provided for the region containing the MCR, starting at the 5′ end for sgRNA1, which is transcribed from the position indicated by the arrow. The mutated methionine codons (Met*) are indicated, and selected restriction enzyme sites are provided (sites displayed in bold and underlined are unique in the whole plasmid); those sites used for cloning of PMV CP and GUS genes are indicated. (B) Histochemical visualization of GUS expression in N. clevelandii and cowpea leaves inoculated with pHST32 DNA 5 or 4 days previously, respectively. (C) Immunodetection of PMV CP with alkaline phosphatase (left panel) or by horseradish peroxidase chemiluminescence (middle and right panels). The panel on the left shows that PMV CP expressed upon inoculation of C. quinoa with pMax9 transcripts migrates at the same position as 35 ng of purified PMV CP (the lower bands presumably represent CP breakdown products). The two panels on the right show results obtained with C. quinoa and cowpea, inoculated with different concentrations of pMax9 transcripts or p12-Max9 miniprep DNA. Leaves inoculated with RNA or DNA were harvested 4 or 7 days after inoculation, respectively, and samples containing ca. 10 μg of leaf protein were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by Western blot analysis. Protein concentrations were determined with a Micro BCA protein assay reagent kit (Pierce, Rockford, Ill.). The results for the middle and right-hand panels can be compared because these images originate from the same immunoblot.
Although the TBSV DNA-based vectors pHST12 and pHST34 can be directly used for the insertion of foreign genes (Fig. 4A), the GUS insertion vectors were obtained via different intermediates, as is discussed briefly. The GUS donor in these experiments was pHS23, which is a kanamycin-resistant plasmid analogous to pHS24 (27), but instead of containing sequences from the wild type (pTBSV-100), it harbors the StuI-to-SalI fragment from pHS45 (25), which includes the GUS gene. The GUS gene was removed from pHS23 with SmaI and SacI and ligated between the SnaBI to SacI sites in the MCR of pHST2-14 (26) to give pHST6. To permit GUS expression from a TBSV DNA vector with the p19 gene inactivated, an intermediate clone (pHST16) was generated by replacing the BamHI-to-NcoI fragment of pHST12 with the corresponding fragment of pHS24 (27). The GUS gene was subsequently transferred to the DNA-based vectors by exchanging the BamHI-to-NcoI fragments of pHST20 (wild type) and pHST16 (p19 mutant) with the BamHI-to-NcoI fragment of pHST6. These transfers yielded pHST32 (Fig. 4) and pHST33, respectively.
The inoculation of pHST33 or pHST32 DNA onto C. quinoa, N. benthamiana, and spinach, followed by standard histochemical GUS assays (25), resulted in readily detectable levels of GUS expression (data not shown), as was previously demonstrated for the RNA-mediated delivery of TBSV-GUS constructs on these hosts (25, 27). To illustrate that additional hosts are available for TBSV DNA-mediated foreign gene expression, GUS expression for inoculated leaves of N. clevelandii and cowpea is shown in Fig. 4B. The results are provided for pHST32, rather than for pHST33, because in cowpea the p19 gene assists in effective local spread (unpublished data).
To further illustrate the usefulness of the TBSV DNA-based vector system, p12-Max9 was used to express the panicum mosaic virus (PMV) CP, and its accumulation was compared to the levels obtained with the RNA-based analog pMax9. Plasmids pMax9 and p12-Max9 (a gift from M. Turina and K.-B. G. Scholthof) contain the PMV CP gene (33) cloned between the SnaBI and XhoI sites of pHST2-14 (26) and pHST12 (Fig. 4A), respectively. The results in the left-hand panel of Fig. 4C show that the TBSV vector expresses PMV CP that is the same size as purified PMV CP. The inoculation of plants with miniprep p12-Max9 DNA resulted in detectable levels of PMV CP expression in N. benthamiana (data not shown), which supports a systemic infection with TBSV, as well as in the local-lesion hosts C. quinoa and cowpea (Fig. 4C). These results imply that inoculation of C. quinoa with 15 ng of RNA per μl yields severalfold higher levels of PMV CP than inoculations with 150 ng of DNA per μl. This difference in accumulation correlates with the observation that 15 to 20 local lesions were obtained with a concentration of 100 to 150 ng of p12-Max9 DNA per μl, whereas about twice the number of lesions (20 to 50) appeared following inoculation with 3 to 23 ng of pMax9 transcripts per μl.
These experiments demonstrated that foreign proteins can be expressed from modified TBSV sgRNA1 that contains an elongated leader promoting translational initiation at the authentic start codon on the foreign gene. The histochemical GUS assays did not reveal an obvious difference between the intensity of blue color obtained with RNA versus DNA-based vectors (data not shown). However, the yield of PMV CP obtained with TBSV DNA was inferior to that obtained with the analogous RNA-based system, based on micrograms of input nucleic acid. Nevertheless, it is debatable whether there is any relevance to a comparison between results obtained upon inoculation with single-stranded plus-sense transcripts that initiate infections in the cytoplasm and those derived from experiments with double-stranded plasmid DNA, which needs to enter the nucleus. Irrespective of the intrinsic and quantitative differences between RNA-based vectors and DNA-based analogues, the advantage is that sufficient amounts of TBSV DNA inoculum are easily obtained with standard miniprep procedures for the immediate inoculation of plants.
As reviewed previously, RNA-based tobamo-, potex-, and potyvirus gene vector systems offer the advantage of systemic expression of the foreign gene (26). Thus far, instability features restrict the use of TBSV vectors to inoculated leaves (25), but the broad host range and rapid and relatively high levels of gene expression permit the convenient and rapid TBSV-mediated transient introduction of foreign genes in plants. The usefulness of the TBSV RNA-based vector has been documented through studies on the behavior of foreign proteins or RNA in protoplasts or inoculated leaves (3, 25, 32). The present results illustrate the potential for cost-effective, high-throughput screening using TBSV DNA-based vectors that are suitable for the direct rub-inoculation of many different plant species with miniprep plasmid DNA. The application of this easy and rapid alternative expression system may substantially expedite gene (or cDNA library) screening schemes or be used to rapidly evaluate the biochemical behavior of foreign RNA segments. This vector system may also be applied to investigate signaling in gene silencing by inducing or suppressing this phenomenon through the overexpression of particular genes in inoculated leaves.
In summary, this report constitutes the first example of an in vivo-transcribed cDNA of a member of the economically important and diverse family Tombusviridae. Another novel aspect of the present study is that the presence of the HDVagrz in combination with a poly(A) signal improves infectivity upon inoculation of plants with untreated plasmid DNA. These features have been linked to generate a new versatile and robust TBSV-mediated gene vector system that permits the rapid, transient expression of foreign genes after simple rub-inoculation of miniprep DNA onto plant species of different families.
Acknowledgments
I am grateful to Steve Garcia, Joan Kuecker, and Beth Whitehead for excellent technical assistance. I thank Karen-Beth G. Scholthof for the many valuable suggestions during the experimentation and preparation of the manuscript and, together with Massimo Turina, for the plasmids pMax9 and p12-Max9. I also thank Bénédicte Desvoyes for providing purified PMV CP and valuable contributions to the experimental analyses and Mike Hughes for his assistance in optimizing the DNA inoculation conditions. I thank George Lomonossoff for pUC-35S and Andy Ball for plasmid 2.0 and for helpful suggestions regarding in vitro ribozyme activity assays.
This work was funded by grants from USDA/CSREES-NRI-CGP (95-37303-2289) and the Texas Higher Education Coordinating Board Advanced Research Program (999902-056).
REFERENCES
- 1.Ahlquist P, French R, Janda M, Loesch-Fries L S. Multicomponent RNA plant virus infection derived from cloned viral cDNA. Proc Natl Acad Sci USA. 1984;81:7066–7070. doi: 10.1073/pnas.81.22.7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Been M D. Cis- and trans-acting ribozymes from a human pathogen, hepatitis delta virus. Trends Biochem Sci. 1994;19:251–256. doi: 10.1016/0968-0004(94)90151-1. [DOI] [PubMed] [Google Scholar]
- 3.Blanc S, Schmidt I, Vantard M, Scholthof H B, Kuhl G, Esperandieu P, Cerutti M, Louis C. The aphid transmission factor of cauliflower mosaic virus forms a stable complex with microtubules in both insect and plant cells. Proc Natl Acad Sci USA. 1996;93:15158–15163. doi: 10.1073/pnas.93.26.15158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyer J-C, Haenni A-L. Infectious transcripts and cDNA clones of RNA viruses. Virology. 1994;198:415–426. doi: 10.1006/viro.1994.1053. [DOI] [PubMed] [Google Scholar]
- 5.Commandeur U, Jarausch W, Li Y, Koenig R, Burgermeister W. cDNAs of beet necrotic yellow vein virus RNAs 3 and 4 are rendered biologically active in a plasmid containing the cauliflower mosaic virus 35S promoter. Virology. 1991;185:493–495. doi: 10.1016/0042-6822(91)90806-m. [DOI] [PubMed] [Google Scholar]
- 6.Dagless E M, Shintaku M H, Nelson R S, Foster G D. A CaMV 35S promoter driven cDNA clone of tobacco mosaic virus can infect host plant tissue despite being uninfectious when manually inoculated onto leaves. Arch Virol. 1997;142:183–191. doi: 10.1007/s007050050069. [DOI] [PubMed] [Google Scholar]
- 7.Dalmay T, Russo M, Burgyan J. Repair in vivo of altered 3′ terminus of cymbidium ringspot tombusvirus RNA. Virology. 1993;192:551–555. doi: 10.1006/viro.1993.1071. [DOI] [PubMed] [Google Scholar]
- 8.Dessens J T, Lomonossoff G P. Cauliflower mosaic virus 35S promoter-controlled DNA copies of cowpea mosaic virus RNAs are infectious on plants. J Gen Virol. 1993;74:889–892. doi: 10.1099/0022-1317-74-5-889. [DOI] [PubMed] [Google Scholar]
- 9.Ding S-W, Rahtjen J P, Li W-X, Swanson R, Healy H, Symons R H. Efficient infection from cDNA clones of cucumber mosaic cucumovirus RNAs in a new plasmid vector. J Gen Virol. 1995;76:459–464. doi: 10.1099/0022-1317-76-2-459. [DOI] [PubMed] [Google Scholar]
- 10.Fakhfakh H, Vilaine F, Makni M, Robaglia C. Cell-free cloning and biolistic inoculation of an infectious cDNA of potato virus Y. J Gen Virol. 1996;77:519–523. doi: 10.1099/0022-1317-77-3-519. [DOI] [PubMed] [Google Scholar]
- 11.Gal-On A, Meiri E, Huet H, Hua W J, Raccah B, Gaba V. Particle bombardment drastically increases the infectivity of cloned DNA of zucchini yellow mosaic potyvirus. J Gen Virol. 1995;76:3223–3227. doi: 10.1099/0022-1317-76-12-3223. [DOI] [PubMed] [Google Scholar]
- 12.Hearne P Q, Knorr D A, Hillman B I, Morris T J. The complete genome structure and synthesis of infectious RNA from clones of tomato bushy stunt virus. Virology. 1990;177:141–151. doi: 10.1016/0042-6822(90)90468-7. [DOI] [PubMed] [Google Scholar]
- 13.Hohn T, Goldbach R. Vectors: plant viruses. In: Webster R G, Granoff A, editors. Encyclopedia of virology. Vol. 3. San Diego, Calif: Academic Press; 1994. pp. 1536–1543. [Google Scholar]
- 14.Johnson J E, Lin T, Lomonossoff G. Presentation of heterologous peptides on plant viruses: genetics, structure, and function. Annu Rev Phytopathol. 1997;35:67–86. doi: 10.1146/annurev.phyto.35.1.67. [DOI] [PubMed] [Google Scholar]
- 15.Karrer E E, Beachy R N, Holt C A. Cloning of tobacco genes that elicit the hypersensitive response. Plant Mol Biol. 1998;36:681–690. doi: 10.1023/a:1005949304445. [DOI] [PubMed] [Google Scholar]
- 16.Leiser R-M, Ziegler-Graff V, Reutenauer A, Herrbach E, Lemaire O, Guilley H, Richards K, Jonard G. Agroinfection as an alternative to insects for infecting plants with beet western yellows luteovirus. Proc Natl Acad Sci USA. 1992;89:9136–9140. doi: 10.1073/pnas.89.19.9136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacFarlane S A, Gilmer D, Davies J W. Efficient inoculation with CaMV 35S promoter-driven DNA clones of the tobravirus PEBV. Virology. 1992;187:829–831. doi: 10.1016/0042-6822(92)90488-b. [DOI] [PubMed] [Google Scholar]
- 18.Maiss E, Timpe U, Brisske-Rode A, Lesemann D-E, Casper R. Infectious in vivo transcripts of a plum pox full-length cDNA clone containing the cauliflower mosaic virus 35 S promoter. J Gen Virol. 1992;73:709–713. doi: 10.1099/0022-1317-73-3-709. [DOI] [PubMed] [Google Scholar]
- 19.Mori M, Mise K, Kobayashi K, Okuno T, Furusawa I. Infectivity of plasmids containing brome mosaic virus cDNA linked to the cauliflower mosaic virus 35S RNA promoter. J Gen Virol. 1991;72:243–246. doi: 10.1099/0022-1317-72-2-243. [DOI] [PubMed] [Google Scholar]
- 20.Neeleman L, van der Vossen E A G, Bol J F. Infection of tobacco with alfalfa mosaic virus cDNAs sheds light on the early function of the coat protein. Virology. 1993;196:883–887. doi: 10.1006/viro.1993.1551. [DOI] [PubMed] [Google Scholar]
- 21.Pattnaik A K, Ball L A, LeGrone A W, Wertz G W. Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell. 1992;69:1011–1020. doi: 10.1016/0092-8674(92)90619-n. [DOI] [PubMed] [Google Scholar]
- 22.Prufer D, Wipf Scheibel C, Richards K, Guilley H, Lecoq H, Jonard G. Synthesis of a full-length infectious cDNA clone of cucurbit aphid-borne yellows virus and its use in gene exchange experiments with structural proteins from other luteoviruses. Virology. 1995;214:150–158. doi: 10.1006/viro.1995.9945. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Scholthof H B, Jackson A O. The enigma of pX: a host dependent cis-acting element with variable effects on tombusvirus RNA accumulation. Virology. 1997;237:56–65. doi: 10.1006/viro.1997.8754. [DOI] [PubMed] [Google Scholar]
- 25.Scholthof H B, Morris T J, Jackson A O. The capsid protein gene of tomato bushy stunt virus is dispensable for systemic movement and can be replaced for localized expression of foreign genes. Mol Plant-Microbe Interact. 1993;6:309–322. [Google Scholar]
- 26.Scholthof H B, Scholthof K-B G, Jackson A O. Plant virus gene vectors for transient expression of foreign proteins in plants. Annu Rev Phytopathol. 1996;34:299–323. doi: 10.1146/annurev.phyto.34.1.299. [DOI] [PubMed] [Google Scholar]
- 27.Scholthof H B, Scholthof K-B G, Kikkert M, Jackson A O. Tomato bushy stunt virus spread is regulated by two nested genes that function in cell-to-cell movement and host-dependent systemic invasion. Virology. 1995;213:425–438. doi: 10.1006/viro.1995.0015. [DOI] [PubMed] [Google Scholar]
- 28.Scholthof H B, Wu F C, Gowda S, Shepherd R J. Regulation of caulimovirus gene expression and the involvement of cis-acting elements on both viral transcripts. Virology. 1992;190:403–412. doi: 10.1016/0042-6822(92)91226-k. [DOI] [PubMed] [Google Scholar]
- 29.Scholthof K-B G, Scholthof H B, Jackson A O. The tomato bushy stunt virus replicase proteins are coordinately expressed and membrane associated. Virology. 1995;208:365–369. doi: 10.1006/viro.1995.1162. [DOI] [PubMed] [Google Scholar]
- 30.Shi B-J, Ding S-W, Symons R H. Plasmid vector for cloning infectious cDNAs from plant RNA viruses: high infectivity of cDNA clones of tomato aspermy virus. J Gen Virol. 1997;78:1181–1185. doi: 10.1099/0022-1317-78-5-1181. [DOI] [PubMed] [Google Scholar]
- 31.Shintaku M H, Carter S A, Bao Y, Nelson R S. Mapping nucleotides in the 126-kDa protein gene that control the differential symptoms induced by two strains of tobacco mosaic virus. Virology. 1996;221:218–225. doi: 10.1006/viro.1996.0368. [DOI] [PubMed] [Google Scholar]
- 32.Sit T L, Vaewhongs A, Lommel S A. RNA-mediated transactivation of transcription from a viral RNA. Science. 1998;281:829–832. doi: 10.1126/science.281.5378.829. [DOI] [PubMed] [Google Scholar]
- 33.Turina M, Maruoka M, Monis J, Jackson A O, Scholthof K-B G. Nucleotide sequence and infectivity of a full-length cDNA clone of panicum mosaic virus. Virology. 1998;241:141–155. doi: 10.1006/viro.1997.8939. [DOI] [PubMed] [Google Scholar]
- 34.Turpen T H, Turpen A M, Weinzettl N, Kumagai M H, Dawson W O. Transfection of whole plants from wounds inoculated with Agrobacterium tumefaciens containing cDNA of tobacco mosaic virus. J Virol Methods. 1993;42:227–240. doi: 10.1016/0166-0934(93)90035-p. [DOI] [PubMed] [Google Scholar]
- 35.Weber H, Haeckel P, Pfitzner A J P. A cDNA clone of tomato mosaic virus is infectious in plants. J Virol. 1992;66:3909–3912. doi: 10.1128/jvi.66.6.3909-3912.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]