Abstract
Production of gutted, or helper-dependent, adenovirus vectors by current methods is inefficient. Typically, a plasmid form of the gutted genome is transfected with helper viral DNA into 293 cells; the resulting lysate is serially passaged to increase the titer of gutted virions. Inefficient production of gutted virus particles after cotransfection is likely due to suboptimal association of replication factors with the abnormal origins found in these plasmid substrates. To test this hypothesis, we explored whether gutted virus production would be facilitated by transfection into cells expressing various viral replication factors. We observed that C7 cells, coexpressing adenoviral DNA polymerase and preterminal protein, converted plasmid DNA into replicating virus approximately 50 times more efficiently than did 293 cells. This property of C7 cells can be used to greatly increase the efficiency of gutted virus production after cotransfection of gutted and helper viral DNA. These cells should also be useful for generation of recombinant adenovirus from any plasmid-based precursor.
Conventional adenovirus (Ad) gene delivery vectors are based on replacement of early regions of the viral genome with an expression cassette coding for a gene of interest. Unfortunately, Ad vectors have drawbacks that limit their usefulness for many applications. First, the cloning capacity of these vectors is limited to 8 to 10 kb. Second, despite deletion of the E1 region, leaky expression of immunogenic viral proteins occurs in vivo, which leads to a host immune response and elimination of gene expression from transduced tissues (9, 10, 12, 27, 40, 42–45). Gutted, or helper-dependent, Ad vectors may overcome these drawbacks (11, 20, 21). Gutted vectors contain cis-acting DNA sequences necessary for viral replication and packaging but no viral coding sequences. These vectors can accommodate up to 36 kb of exogenous DNA and are unable to express viral proteins. Gutted vectors are produced by replication in the presence of a helper virus, which provides all necessary viral proteins in trans. Since the viral proteins act to replicate both gutted and helper genomes, gutted Ad particles are prepared as a mixture with helper virions, though selection against helper virus packaging can reduce this contamination (16, 28). Particles containing gutted viral genomes, rather than helper genomes, must subsequently be purified on the basis of their lower density (11, 20, 21).
The starting point for production of a gutted virus is plasmid DNA (11, 20, 21). The plasmid contains the viral inverted terminal repeats (ITRs), the viral packaging signal, and exogenous DNA to be carried by the gutted virus. To increase production of gutted virus, most investigators linearize the gutted viral plasmid; some systems require the ligation of viral ITRs after linearization (11, 20). The plasmid DNA is cointroduced with helper sequences into a cell line that can replicate the helper virus, normally 293 cells. Replication of the helper virus eventually causes lysis of the cells; the lysate contains a large number of helper virions and a comparatively small number of gutted virions. The number and proportion of gutted virions is small because plasmid DNA, whether circular (with fused ITRs) or linear, is a poor substrate for initiation of adenoviral DNA replication (15, 39). As a result, replication of the helper virus occurs in many cells without concomitant production of gutted virus, despite the presence of gutted viral plasmid substrate. To increase the number and proportion of gutted virions in the lysate, the initial mixture must be serially passaged (21).
The production of gutted virus particles from plasmid DNA in the first step of gutted vector production is so inefficient that titers of less than 100 particles per milliliter are often obtained (reference 28 and unpublished observations). In some cases no gutted virions can be detected until at least one serial passage has been performed. We hypothesized that the basis for this problem is the low efficiency of initiation of adenoviral DNA replication on plasmid DNA substrates, probably due to suboptimal association of replication factors with the abnormal origins found in these substrates. The normal substrate for initiation of adenoviral DNA replication is terminal protein-DNA complex (6, 33, 34); plasmid-based substrates propagated in Escherichia coli obviously lack terminal protein. The presence of terminal protein bound to the template confers higher affinity for incoming Ad polymerase-preterminal protein complex, a critical viral replication factor (29). Affinity for the cellular replication factor NF-I, by contrast, is not affected by the presence of terminal protein on the initiation substrate (29). NF-I, like terminal protein, enhances the specific binding of pTP-Pol complex to the Ad origin (25).
We hypothesized that provision of higher levels of replication factors, especially Ad polymerase-preterminal protein complex, might facilitate initiation of DNA replication at origins lacking terminal protein, lead to higher infectivity of viral DNA containing such origins, and allow production of higher levels of gutted virus after cointroduction of gutted viral plasmid and helper sequences. We found that cells expressing both Ad DNA polymerase and preterminal protein more efficiently convert plasmid DNA to replicating virus. We show that such cells can be used to increase greatly the efficiency and reproducibility of gutted virus production after cotransfection of gutted and helper viral DNA. Use of these cells to increase production of gutted virus, together with strategies that select against helper virus contamination during serial passage (16, 28), should facilitate more general use of gutted Ad vectors. C7 cells should also facilitate generation of conventional recombinant Ad by using precursor genomes manipulated in E. coli.
Increased infectivity of circular Ad genomes in C7 cells.
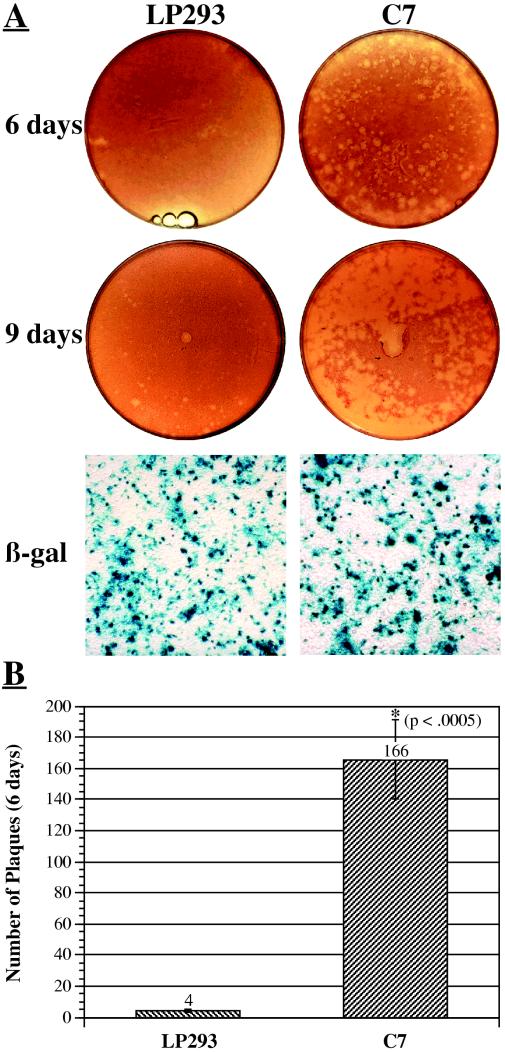
C7 cells are modified 293 cells that stably express both preterminal protein and Ad DNA polymerase (3). Since these proteins are important for the initiation of Ad DNA replication, we hypothesized that their presence might allow for more efficient conversion of suboptimal Ad templates to replicating virus. To test this possibility, we transfected 293 cells or C7 cells with pFG140, a plasmid that contains a complete Ad genome and fused ITRs (13). C7 cells, compared with 293 cells, produced dramatically higher numbers of plaques at all time points (Fig. 1A). To estimate the magnitude of this effect, we reduced the amount of pFG140 transfected so that plates of both cell types displayed well-separated plaques. One such experiment revealed a 40-fold stimulation of plaquing efficiency in C7 cells (Fig. 1B). Our average across all such experiments was 49-fold stimulation (Fig. 2). The transfection efficiencies of the two cell lines, as assessed by transfection of a 27-kb plasmid containing a β-galactosidase expression cassette, were indistinguishable (Fig. 1A).
FIG. 1.
Infectivity of circular Ad genomes in low-passage (LP) 293 and C7 cells. (A) Qualitative comparison. Plates of low-passage 293 (left) or C7 (right) cells were transfected with 8.8 μg of pFG140 or pAd5βdys. The top set of two plates shows neutral red staining of pFG140-transfected plates 6 days after overlay. The middle set of two plates shows neutral red staining of pFG140-transfected plates 9 days after overlay. The bottom set of two fields shows X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining of pAd5βdys-transfected plates 3 days after transfection. The numbers of blue cells per unit of area are not significantly different in either cell line. (B) Quantitative comparison. Plates of low-passage 293 (n = 8) or C7 (n = 8) cells were transfected with 2.5 μg of pFG140 and stained 6 days after overlay. The graph shows the average total number of visible plaques per plate at 6 days, error bars represent the standard error of the mean, and the asterisk represents statistical significance according to the paired-samples t test.
FIG. 2.
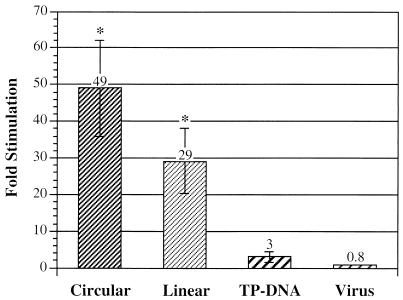
Fold stimulation of plaquing efficiency in C7 cells by physical state of the Ad genome. Plates of low-passage 293 and C7 cells were transfected with the indicated form of viral DNA or infected with intact virus, overlaid, and stained with neutral red after formation of plaques. Fold stimulation in C7 cells was then calculated as the number of plaques observed on a plate of C7 cells divided by the number of plaques observed on an equivalently treated plate of 293 cells. From left to right, the bars represent transfection with pFG140; transfection with linear, deproteinized hpAP DNA extracted from virions; transfection with terminal protein (TP)-DNA complex; and infection with hpAP virus. Bars represent average fold stimulation observed over all experiments (n ≥ 3), error bars represent the standard error of the mean, and asterisks indicate that mean fold stimulation is significantly different from 1. In typical experiments, we observed an average of 8 versus 227 plaques with linear viral DNA, 13 versus 45 plaques with TP-DNA, and 8 versus 7 plaques with intact virus.
Stimulation of infectivity by C7 cells depends on the physical state of the Ad origins of replication (ITRs).
pFG140 is a suboptimal template for initiation of adenoviral DNA replication because it contains fused ITRs, whereas normal Ad origins are found at the ends of linear molecules and are bound covalently to terminal protein (37, 39). We hypothesized that the improved plaquing efficiency of pFG140 in C7 cells was due to the interaction of high levels of Ad DNA polymerase and preterminal protein with the fused ITRs in the plasmid. If so, then the magnitude of the effect should vary according to the physical state of the ITRs, since this would alter the character of the interaction.
293 or C7 cells were transfected with various forms of adenoviral DNA or infected with intact virus. In the case of DNA transfections, the amount of DNA was adjusted so that plates of both cell types displayed well-separated plaques. Typical transfections used 3 μg of circular pFG140 DNA, 100 ng of deproteinized DNA extracted from virions, or 5 ng of intact terminal protein-DNA complex extracted from virions. C7 cells displayed statistically significant increases in the specific activities of the two suboptimal replication templates, pFG140 and deproteinized virion DNA (49- and 29-fold increases in plaque-forming efficiency, respectively, compared with 293 cells [Fig. 2]). By contrast, the specific activity of a normal replication template, terminal protein-DNA complex, was not significantly different. The plaquing efficiency of an intact virus was also not significantly different. These data indicate that the improved plaquing efficiency of pFG140 in C7 cells results from an interaction that depends on the physical state of the viral ITRs.
Production of gutted viral particles and DNA after cotransfection into C7 or low-passage 293 cells.
The production of gutted, or helper-dependent, adenoviral vectors requires initiation of Ad replication on a plasmid-based gutted viral genome, an inefficient process (11, 17, 20, 21). We hypothesized that the production of packaged, gutted virus after cotransfection with helper viral DNA would be more efficient in C7 than in 293 cells, since C7 cells more efficiently convert suboptimal forms of viral DNA to replicating virus. To test this idea, we transfected 293 and C7 cells with 8 μg of circular gutted viral plasmid and 40 ng, 200 ng, or 1 μg of deproteinized linear helper viral DNA extracted from virions. To avoid the problem of pseudotransduction (1), which causes overestimation of vector titers, we chose a gutted viral plasmid that expresses β-galactosidase from an inducible ecdysone promoter. Use of this reporter prevents expression in 293 cells but allows titering in EcR-293 cells (Invitrogen Corporation, Carlsbad, Calif.). The helper viral genome used constitutively expresses alkaline phosphatase. Transfected cells were allowed to lyse, and cells and supernatant were harvested together as a mixture. A portion of this lysate was used for DNA extraction; the remainder was titered for gutted and helper virions.
Plates of C7 cells always lysed before plates of low-passage 293 cells that had been transfected with the same amount of viral DNA, reflecting the higher proportion of transfected cells that initiated replication of the helper. For example, transfection of 293 cells with 40 ng of helper virus DNA failed to produce lysis after 2 weeks, whereas C7 cells lysed within 1 week (Fig. 3B and data not shown).
FIG. 3.
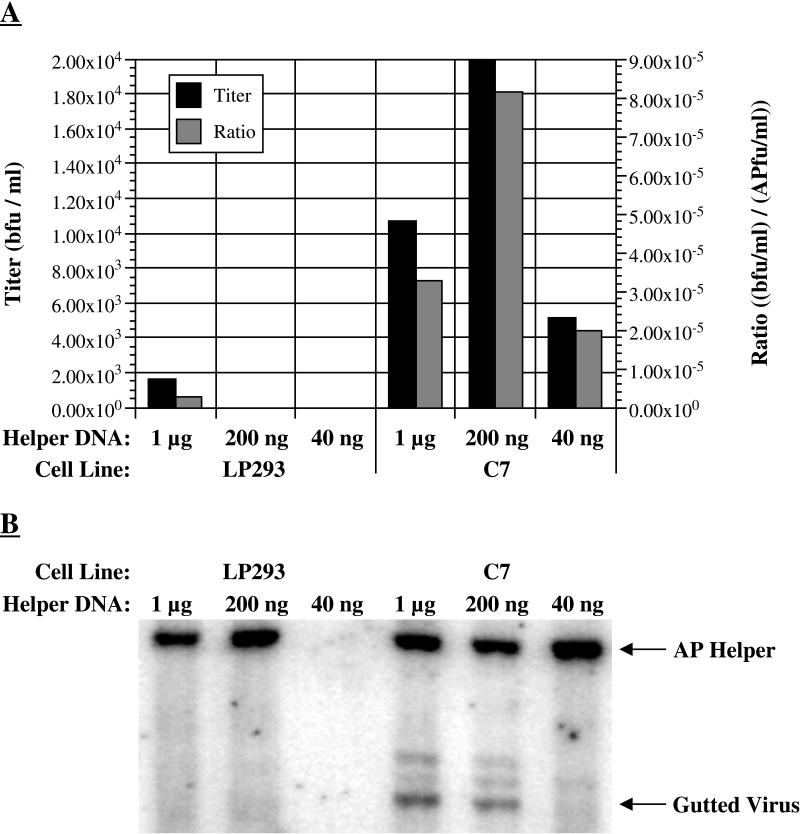
Production of gutted virus particles and DNA after cotransfection into low-passage (LP) 293 or C7 cells. Cells were cotransfected with 8 μg of pGEβdys gutted viral plasmid and 1 μg, 200 ng, or 40 ng of hpAP helper viral DNA. Resulting virus was harvested after lysis or, in the case of one unlysed sample, after 2 weeks. (A) Titer of gutted virus produced after lysis of cotransfected plates. Black bars show the titer of gutted virus, in blue-forming units per milliliter (bfu/ml). These results may be converted to blue-forming units per infected cell by dividing by about 2 × 106, the approximate number of cells in a 6-cm plate. Grey bars show the ratio of gutted titer to helper titer, where the latter is determined by titering lysates for alkaline phosphatase transducing units. (B) Southern analysis of viral DNA produced after cotransfection. DNA was extracted from a small amount of lysate, digested with HindIII, and analyzed by Southern blotting with a probe that overlaps the Ad packaging signal. The left end of the helper virus is the dark 6.6-kb band near the top of the panel shown; the left end of the gutted virus is the 3.4-kb band near the bottom of the panel.
The titer of gutted viral particles produced after cotransfection was invariably higher with C7 cells than with 293 cells (Fig. 3A). Using 40 or 200 ng of helper virus DNA, the titer of gutted viral particles produced in 293 cells was below the limit of detection (about 100 particles per ml); however, C7 cells produced 5 × 103 and 2 × 104 particles per ml with 40 or 200 ng of helper, respectively. Using 1 μg of helper DNA, 293 cells produced 1.6 × 103 particles per ml; C7 cells produced 1.1 × 104 particles per ml, a sevenfold increase. In addition, we found that helper virus titers after lysis were comparable in the two cell types. The ratio of gutted to helper virions was therefore also improved by the use of C7 cells (Fig. 3A).
To verify that the higher titer of gutted virus produced in C7 cells would be maintained during serial passage, lysates produced with 1 μg of helper viral DNA were diluted at a ratio of 1:10 onto fresh plates of 293 cells. These cells were allowed to lyse and were titered for the presence of gutted and helper virions. We found that the secondary lysate derived from a transfected plate of C7 cells maintained at least a sixfold advantage in both the number and proportion of gutted virions compared to the secondary lysate derived from a transfected plate of 293 cells (data not shown).
The accumulation of linear viral DNA in transfected plates was also examined after lysis (Fig. 3B). DNA extracted from 50 μl of lysate was examined by Southern analysis by using as probe a fragment of the Ad packaging signal at the left end of the genome. In lysed 293 cells, the amount of gutted viral DNA was below the level of detection. In contrast, linear gutted viral DNA was easily detected after cotransfection of C7 cells with 200 ng or 1 μg of helper DNA. These data indicate that C7 cells had more efficiently converted the transfected gutted viral substrate to a linear replicating form. Note that the ratio of gutted to helper viral DNA is much higher than the ratio of gutted to helper virions; the reason for this finding is not clear.
Cotransfection of pTP or Ad polymerase expression plasmids.
Preterminal protein and Ad DNA polymerase bind to each other and function as a complex in the initiation of Ad DNA replication (22, 38). We hypothesized that the increased infectivity of pFG140 in C7 cells was due to the presence of both polymerase and preterminal protein in the cells, probably acting as a complex. To test this idea, we first tested the B6 cell line to see whether these cells, stably expressing Ad polymerase but not pTP, would plaque pFG140 with increased efficiency (2). We observed a modest, twofold increase in plaquing efficiency that was not statistically significant (data not shown). This result suggested that Ad DNA polymerase alone was not sufficient to increase the efficiency of plaque formation from supercoiled pFG140. The reciprocal experiment could not be performed for pTP since we were not able to isolate a cell line that stably expressed only preterminal protein; the deleterious effect of pTP on cell growth has been noted previously (36).
We next performed cotransfections of pFG140 with Ad DNA polymerase, pTP, or both into 293, B6, and C7 cells (Fig. 4 and data not shown). Cotransfection of a plasmid expressing Ad DNA polymerase with pFG140 resulted in a striking inhibition of plaque formation in all cell lines. The fact that this phenomenon was observed in B6 and C7 cells, which both express some Ad DNA polymerase, suggests that high levels of Ad DNA polymerase, in the absence of correspondingly high levels of pTP, are detrimental to the conversion of transfected plasmid into replicating DNA.
FIG. 4.
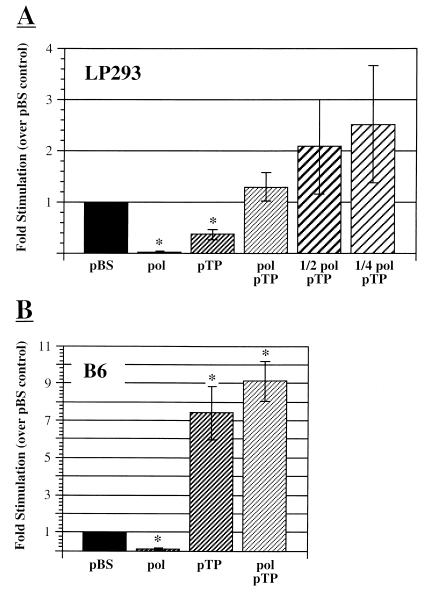
Cotransfection of pTP- or Ad polymerase-expressing plasmids showing the effect on infectivity of pFG140 in various cell lines. Plates of low-passage (LP) 293 (A) or B6 (B) cells were cotransfected with 3 μg of pFG140 and various amounts of pBSKSII+ (pBS), pRSV-Pol, or pRSV-pTP. Plates were then overlaid with agarose and, after formation of plaques, stained with neutral red. Fold stimulation was then calculated as the number of plaques observed after cotransfection of pFG140 with a test plasmid divided by the number of plaques observed after cotransfection of pFG140 with pBS into the same cell line. The lefthand scale indicates average fold stimulation. The righthand scale indicates a calculated estimate of the total increase in infectivity obtained by the combined effect of the cotransfected test plasmid and use of the indicated cell line. The first bar in each set represents the control value of 1. The second bar in each set represents the average effect of adding 3 μg of pRSV-Pol, instead of 3 μg of pBS, to the transfection mixture. The third bar represents the average effect of 3 μg of pRSV-pTP. The fourth bar in each set shows the effect of cotransfecting 3 μg of pRSV-Pol and 3 μg of pRSV-pTP with 3 μg of pFG140. In panel A, the fifth and sixth bars show the effect of cotransfecting 1.5 or 0.75 μg of pRSV-Pol, respectively, together with 3 μg each of pFG140 and pRSV-pTP. Note that in all cases a total of 9 μg of DNA was transfected into a 6-cm plate. Error bars represent the standard error of the mean, and asterisks indicate that mean fold stimulation is significantly different from 1.
Cotransfection of a plasmid expressing preterminal protein resulted in a 2.8-fold inhibition of plaque formation in 293 cells (Fig. 4A). This result suggested that pTP alone is not sufficient to cause the enhancement of plaquing efficiency observed in C7 cells. Cotransfection of pFG140 with a plasmid expressing pTP into the B6 cell line, by contrast, produced a statistically significant, sevenfold increase in the number of plaques observed (Fig. 4B). The effect of pTP cotransfection is therefore quite different depending on the presence or absence of stably expressed Ad DNA polymerase. This observation suggests that there is direct or indirect interaction between the two proteins and that they may cooperate to produce the effects observed in C7 cells.
Cotransfection of pFG140 with both pTP and Ad polymerase expression plasmids also yielded evidence for interaction between the two proteins. Cotransfection of equal weights of pFG140, pTP, and Ad-Pol plasmids into 293 cells resulted in numbers of plaques that were not significantly different from those observed after transfection with pFG140 and pBluescript (Fig. 4A). This result stands in contrast to cotransfection of pFG140 with either the pTP or the Ad polymerase expression plasmids individually, where a significant inhibition of plaquing efficiency was observed. If the two proteins acted independently of each other, then one would expect an additive decrease in plaquing efficiency when both plasmids were cotransfected; instead, no decrease was observed. Thus, the inhibitory effect of either protein is relieved by cotransfection with the other. While cotransfection of the polymerase and pTP plasmids into 293 cells was able to overcome the inhibitory effect of either alone, a stimulation of plaque formation similar to that in C7 cells was not observed (Fig. 4). Given the potent inhibitory activity of excess Ad polymerase, we hypothesized that a slight excess of Ad polymerase over pTP might mask the potent stimulatory effect of pTP-Pol complex. To test this possibility, we repeated the experiment with smaller ratios of polymerase to pTP plasmids. Under these conditions, a modest increase in the infectivity of pFG140 was observed, though the result was not statistically significant (Fig. 4A).
At low concentrations of pTP-Pol complex, the NF-I DNA binding domain has also been shown to enhance initiation of Ad replication by up to 50-fold (25). To test whether the addition of higher levels of NF-I DNA binding domain to cells might stimulate the replication of suboptimal Ad templates, we created stable cell lines expressing the NF-I DNA binding domain and tested them for improved plaquing efficiency of pFG140. No large effect was observed in any of these cell lines (data not shown).
The low infectivity of naked Ad DNA, relative to that of other well-studied DNA viruses, was first recognized over 20 years ago (37). One reason for the reduced infectivity was identified with the observation that terminal protein-DNA complex is 100-fold more infective than protease-treated DNA (37). Other determinants of the efficiency of initiation of DNA replication, including DNA sequences and cellular replication factors, have been studied in cell-free systems (7, 14, 15, 18, 19, 23, 25, 26, 35, 39, 41). It is now clear that at least part of the reason for the low infectivity of naked Ad DNA is inefficient initiation of DNA replication. This problem also occurs with cloned Ad DNA, which not only lacks terminal protein but also suffers from imperfections in origin sequence and/or structure (13, 15, 39).
The low infectivity of cloned Ad DNA limits the efficiency of some artificial systems for manipulation of Ad. For example, creation of recombinant Ad from plasmids by cotransfection and recombination in 293 cells, while simple in principle, is frustratingly inefficient (4, 24). In this scheme an unpackageable parent virus, in the form of plasmid DNA with fused ITRs, is cotransfected with a left end shuttle plasmid. If, as suggested by McGrory et al. (24), recombination occurs after a few rounds of replication of the parent virus, then recombination efficiency could be improved by increasing the number of cells in which replication of the parent occurs. Even when Ad DNA fragments, incapable of independent replication, are used to produce recombinant virus, C7 cells should facilitate replication of successfully recombined genomes. We have in fact observed that production of recombinant Ad is facilitated by use of C7 cells (unpublished results).
Production of gutted Ad from cloned DNA is similarly inefficient. Even when many micrograms of plasmid-derived gutted vector DNA are introduced with a few hundred nanograms of helper virus DNA, very few gutted viral particles are produced (reference 21 and unpublished observations). This low yield presumably occurs because initiation of replication on plasmid-derived gutted viral genomes does not always occur efficiently after infection of the cell with a helper virus particle. Some evidence for this effect may be discerned in the observation that linearization of the gutted viral plasmid increases the yield of gutted virus after cotransfection with helper DNA (11). Linearization likely increases the efficiency of replication initiation on gutted viral templates, which leads to replication of gutted viral genomes in a higher proportion of cells infected by helper.
Despite our understanding of the mechanism of initiation of Ad DNA replication, techniques to facilitate vector manipulation by increasing replication have not been described. Here we report that C7 cells, which stably coexpress Ad DNA polymerase and preterminal protein, plaque circular or deproteinized Ad genomes with higher efficiency than do low-passage 293 cells. These results confirm that such templates exhibit reduced infectivity at least in part due to reduced affinity for replication factors and show that the deficiency can be partially overcome by provision of higher levels of these factors. Addition of NF-I, a cellular factor known to facilitate the interaction of pTP-Pol with the Ad origin, was not similarly effective.
We found that increased infectivity of suboptimal Ad genomes was sensitive to the ratio between preterminal protein and DNA polymerase, as might be expected if the active factor is a complex between the two proteins. Provision of excess Ad DNA polymerase alone was strongly inhibitory to plaque formation in all cell lines tested. We hypothesize that excess polymerase blocks the recruitment of pTP-Pol complex to the Ad origin. Since NF-I is known to assist in the recruitment of pTP-Pol complex through an interaction with Ad polymerase, rather than preterminal protein, available NF-I binding sites for pTP-Pol complex could be occupied by solitary polymerase molecules (5, 8). These molecules are unable to initiate DNA replication in the absence of the appropriate primer, pTP. Alternatively, large amounts of newly synthesized polymerase might be improperly phosphorylated and incapable of properly initiating DNA replication (30–32). Provision of excess pTP alone also slightly decreased the plaquing efficiency of pFG140. Addition of both molecules together reversed these negative effects and provided a small increase in infectivity, which was not statistically significant. The dramatic increase in plaquing efficiency observed in C7 cells could not be reproduced by transient transfection of both pTP and Ad polymerase expression plasmids into 293 cells. Large increases in plaquing efficiency were observed only in cells that stably expressed Ad-Pol and that were either transiently or stably transfected with pTP.
We also used the C7 cell line to test whether inefficient replication of gutted viral templates contributes to the poor yield of gutted virus obtained after cotransfection of gutted viral and helper viral DNA. Production of gutted virus was improved about 10-fold by use of C7 cells for cotransfection (Fig. 3). We find that the use of C7 cells for cotransfection invariably provides increased yields and more consistent results relative to low-passage 293 cells (unpublished results). We have not observed any dramatic advantage in the use of C7 cells for subsequent passage and expansion of gutted virus preparations. This result is consistent with our observation that C7 cells do not provide greatly increased plaquing efficiency for adenoviral templates covalently bound to pTP, such as those delivered to cells by infection during serial passage.
Finally, we observed higher levels of replicating, linear gutted viral DNA after cotransfection into C7 cells. This result indicates that the higher infectivity of pFG140 translates into more efficient initiation of DNA replication on gutted viral templates with similar conformations at the origin. Although all of the studies reported here were performed by using circular gutted plasmids with fused ITRs, we presume that the higher infectivity of deproteinized viral DNA demonstrated in Fig. 2 and 3B should also translate into higher yields of linearized gutted viruses derived from plasmids. Since linearized gutted plasmids have been shown to give higher titers of gutted virus than do circular plasmids, transfection of linearized gutted viruses into C7 cells should allow further improvement in the starting titer (11).
The use of C7 or similar cells, together with strategies that select against packaging of the helper virus, should allow routine production of high-titer gutted virus stocks and more widespread adoption of the technology.
Acknowledgments
This work was supported by a grant from the Muscular Dystrophy Association (United States) and by NIH grant AG015434 (to J.S.C.).
We thank Giovanni Salvatori for helpful discussions.
REFERENCES
- 1.Alexander I E, Russell D W, Miller A D. Transfer of contaminants in adeno-associated virus vector stocks can mimic transduction and lead to artifactual results. Hum Gene Ther. 1997;8:1911–1920. doi: 10.1089/hum.1997.8.16-1911. [DOI] [PubMed] [Google Scholar]
- 2.Amalfitano A, Begy C R, Chamberlain J S. Improved adenovirus packaging cell lines to support the growth of replication-defective gene-delivery vectors. Proc Natl Acad Sci USA. 1996;93:3352–3356. doi: 10.1073/pnas.93.8.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amalfitano A, Chamberlain J S. Isolation and characterization of packaging cell lines that coexpress the adenovirus E1, DNA polymerase, and preterminal proteins: implications for gene therapy. Gene Ther. 1997;4:258–263. doi: 10.1038/sj.gt.3300378. [DOI] [PubMed] [Google Scholar]
- 4.Bett A J, Haddara W, Prevec L, Graham F L. An efficient and flexible system for construction of adenovirus vectors with insertions or deletions in early regions 1 and 3. Proc Natl Acad Sci USA. 1994;91:8802–8806. doi: 10.1073/pnas.91.19.8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosher J, Robinson E C, Hay R T. Interactions between the adenovirus type 2 DNA polymerase and the DNA binding domain of nuclear factor I. New Biol. 1990;2:1083–1090. [PubMed] [Google Scholar]
- 6.Carusi E A. Evidence for blocked 5′-termini in human adenovirus DNA. Virology. 1977;76:380–394. doi: 10.1016/0042-6822(77)90310-5. [DOI] [PubMed] [Google Scholar]
- 7.Challberg M D, Rawlins D R. Template requirements for the initiation of adenovirus DNA replication. Proc Natl Acad Sci USA. 1984;81:100–104. doi: 10.1073/pnas.81.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen M, Mermod N, Horwitz M S. Protein-protein interactions between adenovirus DNA polymerase and nuclear factor I mediate formation of the DNA replication preinitiation complex. J Biol Chem. 1990;265:18634–18642. [PubMed] [Google Scholar]
- 9.Dai Y, Schwarz E M, Gu D, Zhang W W, Sarvetnick N, Verma I M. Cellular and humoral immune responses to adenoviral vectors containing factor IX gene: tolerization of factor IX and vector antigens allows for long-term expression. Proc Natl Acad Sci USA. 1995;92:1401–1405. doi: 10.1073/pnas.92.5.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong J Y, Wang D, Van Ginkel F W, Pascual D W, Frizzell R A. Systematic analysis of repeated gene delivery into animal lungs with a recombinant adenovirus vector. Hum Gene Ther. 1996;7:319–331. doi: 10.1089/hum.1996.7.3-319. [DOI] [PubMed] [Google Scholar]
- 11.Fisher K J, Choi H, Burda J, Chen S J, Wilson J M. Recombinant adenovirus deleted of all viral genes for gene therapy of cystic fibrosis. Virology. 1996;217:11–22. doi: 10.1006/viro.1996.0088. [DOI] [PubMed] [Google Scholar]
- 12.Gaynor R B, Berk A J. Cis-acting induction of adenovirus transcription. Cell. 1983;33:683–693. doi: 10.1016/0092-8674(83)90011-9. [DOI] [PubMed] [Google Scholar]
- 13.Graham F L. Covalently closed circles of human adenovirus DNA are infectious. EMBO J. 1984;3:2917–2922. doi: 10.1002/j.1460-2075.1984.tb02232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guggenheimer R A, Nagata K, Kenny M, Hurwitz J. Protein-primed replication of plasmids containing the terminus of the adenovirus genome. II. Purification and characterization of a host protein required for the replication of DNA templates devoid of the terminal protein. J Biol Chem. 1984;259:7815–7825. [PubMed] [Google Scholar]
- 15.Guggenheimer R A, Nagata K, Lindenbaum J, Hurwitz J. Protein-primed replication of plasmids containing the terminus of the adenovirus genome. I. Characterization of an in vitro DNA replication system dependent on adenoviral DNA sequences. J Biol Chem. 1984;259:7807–7814. [PubMed] [Google Scholar]
- 16.Hardy S, Kitamura M, Harris-Stansil T, Dai Y, Phipps M L. Construction of adenovirus vectors through Cre-lox recombination. J Virol. 1997;71:1842–1849. doi: 10.1128/jvi.71.3.1842-1849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hay R T, Stow N D, McDougall I M. Replication of adenovirus mini-chromosomes. J Mol Biol. 1984;175:493–510. doi: 10.1016/0022-2836(84)90181-5. [DOI] [PubMed] [Google Scholar]
- 18.Kenny M K, Balogh L A, Hurwitz J. Initiation of adenovirus DNA replication. I. Mechanism of action of a host protein required for replication of adenovirus DNA templates devoid of the terminal protein. J Biol Chem. 1988;263:9801–9808. [PubMed] [Google Scholar]
- 19.Kenny M K, Hurwitz J. Initiation of adenovirus DNA replication. II. Structural requirements using synthetic oligonucleotide adenovirus templates. J Biol Chem. 1988;263:9809–9817. [PubMed] [Google Scholar]
- 20.Kochanek S, Clemens P R, Mitani K, Chen H H, Chan S, Caskey C T. A new adenoviral vector: replacement of all viral coding sequences with 28 kb of DNA independently expressing both full-length dystrophin and beta-galactosidase. Proc Natl Acad Sci USA. 1996;93:5731–5736. doi: 10.1073/pnas.93.12.5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar-Singh R, Chamberlain J S. Encapsidated adenovirus minichromosomes allow delivery and expression of a 14 kb dystrophin cDNA to muscle cells. Hum Mol Genet. 1996;5:913–921. doi: 10.1093/hmg/5.7.913. [DOI] [PubMed] [Google Scholar]
- 22.Lichy J H, Field J, Horwitz M S, Hurwitz J. Separation of the adenovirus terminal protein precursor from its associated DNA polymerase: role of both proteins in the initiation of adenovirus DNA replication. Proc Natl Acad Sci USA. 1982;79:5225–5229. doi: 10.1073/pnas.79.17.5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lichy J H, Nagata K, Friefeld B R, Enomoto T, Field J, Guggenheimer R A, Ikeda J E, Horwitz M S, Hurwitz J. Isolation of proteins involved in the replication of adenoviral DNA in vitro. Cold Spring Harbor Symp Quant Biol. 1983;47:731–740. doi: 10.1101/sqb.1983.047.01.084. [DOI] [PubMed] [Google Scholar]
- 24.McGrory W J, Bautista D S, Graham F L. A simple technique for the rescue of early region I mutations into infectious human adenovirus type 5. Virology. 1988;163:614–617. doi: 10.1016/0042-6822(88)90302-9. [DOI] [PubMed] [Google Scholar]
- 25.Mul Y M, Van der Vliet P C. Nuclear factor I enhances adenovirus DNA replication by increasing the stability of a preinitiation complex. EMBO J. 1992;11:751–760. doi: 10.1002/j.1460-2075.1992.tb05108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mul Y M, Verrijzer C P, van der Vliet P C. Transcription factors NFI and NFIII/oct-1 function independently, employing different mechanisms to enhance adenovirus DNA replication. J Virol. 1990;64:5510–5518. doi: 10.1128/jvi.64.11.5510-5518.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nevins J R. Mechanism of activation of early viral transcription by the adenovirus E1A gene product. Cell. 1981;26:213–220. doi: 10.1016/0092-8674(81)90304-4. [DOI] [PubMed] [Google Scholar]
- 28.Parks R J, Chen L, Anton M, Sankar U, Rudnicki M A, Graham F L. A helper-dependent adenovirus vector system: removal of helper virus by Cre-mediated excision of the viral packaging signal. Proc Natl Acad Sci USA. 1996;93:13565–13570. doi: 10.1073/pnas.93.24.13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pronk R, van der Vliet P C. The adenovirus terminal protein influences binding of replication proteins and changes the origin structure. Nucleic Acids Res. 1993;21:2293–2300. doi: 10.1093/nar/21.10.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramachandra M, Nakano R, Mohan P M, Rawitch A B, Padmanabhan R. Adenovirus DNA polymerase is a phosphoprotein. J Biol Chem. 1993;268:442–448. [PubMed] [Google Scholar]
- 31.Ramachandra M, Padmanabhan R. Adenovirus DNA polymerase is phosphorylated by a stably associated histone H1 kinase. J Biol Chem. 1993;268:17448–17456. . (Erratum, 272:11670, 1997.) [PubMed] [Google Scholar]
- 32.Ramachandra M, Padmanabhan R. Expression, nuclear transport, and phosphorylation of adenovirus DNA replication proteins. Curr Top Microbiol Immunol. 1995;199:50–88. [PubMed] [Google Scholar]
- 33.Rekosh D M, Russell W C, Bellet A J, Robinson A J. Identification of a protein linked to the ends of adenovirus DNA. Cell. 1977;11:283–295. doi: 10.1016/0092-8674(77)90045-9. [DOI] [PubMed] [Google Scholar]
- 34.Robinson A J, Younghusband H B, Bellett A J. A circular DNA-protein complex from adenoviruses. Virology. 1973;56:54–69. doi: 10.1016/0042-6822(73)90287-0. [DOI] [PubMed] [Google Scholar]
- 35.Rosenfeld P J, O’Neill E A, Wides R J, Kelly T J. Sequence-specific interactions between cellular DNA-binding proteins and the adenovirus origin of DNA replication. Mol Cell Biol. 1987;7:875–886. doi: 10.1128/mcb.7.2.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaack J, Guo X, Ho W Y, Karlok M, Chen C, Ornelles D. Adenovirus type 5 precursor terminal protein-expressing 293 and HeLa cell lines. J Virol. 1995;69:4079–4085. doi: 10.1128/jvi.69.7.4079-4085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharp P A, Moore C, Haverty J L. The infectivity of adenovirus 5 DNA-protein complex. Virology. 1976;75:442–456. doi: 10.1016/0042-6822(76)90042-8. [DOI] [PubMed] [Google Scholar]
- 38.Stillman B W, Tamanoi F, Mathews M B. Purification of an adenovirus-coded DNA polymerase that is required for initiation of DNA replication. Cell. 1982;31:613–623. doi: 10.1016/0092-8674(82)90317-8. [DOI] [PubMed] [Google Scholar]
- 39.van Bergen B G, van der Ley P A, van Driel W, van Mansfeld A D, van der Vliet P C. Replication of origin containing adenovirus DNA fragments that do not carry the terminal protein. Nucleic Acids Res. 1983;11:1975–1989. doi: 10.1093/nar/11.7.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Ginkel F W, Liu C, Simecka J W, Dong J Y, Greenway T, Frizzell R A, Kiyono H, McGhee J R, Pascual D W. Intratracheal gene delivery with adenoviral vector induces elevated systemic IgG and mucosal IgA antibodies to adenovirus and beta-galactosidase. Hum Gene Ther. 1995;6:895–903. doi: 10.1089/hum.1995.6.7-895. [DOI] [PubMed] [Google Scholar]
- 41.Wides R J, Challberg M D, Rawlins D R, Kelly T J. Adenovirus origin of DNA replication: sequence requirements for replication in vitro. Mol Cell Biol. 1987;7:864–874. doi: 10.1128/mcb.7.2.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Y, Nunes F A, Berencsi K, Furth E E, Gonczol E, Wilson J M. Cellular immunity to viral antigens limits E1-deleted adenoviruses for gene therapy. Proc Natl Acad Sci USA. 1994;91:4407–4411. doi: 10.1073/pnas.91.10.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Y, Su Q, Wilson J M. Role of viral antigens in destructive cellular immune responses to adenovirus vector-transduced cells in mouse lungs. J Virol. 1996;70:7209–7212. doi: 10.1128/jvi.70.10.7209-7212.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Y, Wilson J M. Clearance of adenovirus-infected hepatocytes by MHC class I-restricted CD4+ CTLs in vivo. J Immunol. 1995;155:2564–2570. [PubMed] [Google Scholar]
- 45.Zsengeller Z K, Wert S E, Hull W M, Hu X, Yei S, Trapnell B C, Whitsett J A. Persistence of replication-deficient adenovirus-mediated gene transfer in lungs of immune-deficient (nu/nu) mice. Hum Gene Ther. 1995;6:457–467. doi: 10.1089/hum.1995.6.4-457. [DOI] [PubMed] [Google Scholar]