Abstract
Human immunodeficiency virus type 1 (HIV-1) non-syncytium-inducing (NSI) strains predominantly use the chemokine receptor CCR5, while syncytium-inducing (SI) strains use CXCR4. In vitro, SI isolates infect and replicate in a range of CD4+ CXCR4+ T-cell lines, whereas NSI isolates usually do not. Here we describe three NSI strains that are able to infect two CD4+ T-cell lines, Molt4 and SupT1. For one strain, a variant of JRCSF selected in vitro, replication on Molt4 was previously shown to be conferred by a single amino-acid change in the V1 loop (M.T. Boyd et al., J. Virol. 67:3649–3652, 1993). On CD4+ cell lines expressing different coreceptors, these strains use CCR5 predominantly and do not replicate in CCR5-negative peripheral blood mononuclear cells derived from individuals homozygous for Δ32 CCR5. Furthermore, infection of Molt4 and SupT1 by each of these three strains is potently inhibited by ligands for CCR5, including 2D7, a monoclonal antibody specific for CCR5. CCR5 mRNA was present in both Molt4 and SupT1 by reverse transcription-PCR, although CCR5 protein could not be detected either on the cell surface or in intracellular vesicles. The expanded tropism of the three strains shown here is therefore not due to adaptation to a new coreceptor but due to the capacity to exploit extremely low levels of CCR5 on Molt4 and SupT1 cells. This novel tropism observed for a subset of primary HIV-1 isolates may represent an extended tropism to new CD4+ cell types in vivo.
CD4+ T cells and macrophages are important cell targets of human immunodeficiency virus (HIV) infection. HIV strains have been classified into two main types: (i) syncytium-inducing (SI), T-cell line tropic (T-tropic), rapid/high strains and (ii) non-syncytium-inducing (NSI), macrophage-tropic (M-tropic), slow/low strains. In vitro, NSI viruses infect both macrophages and T-cell cultures but rarely T-cell lines. SI strains, however, replicate in a range of transformed CD4+ T-cell lines (31), while their capacity to infect macrophages is controversial (29, 38, 60, 66, 67, 81, 82). During primary acute infection, the majority of HIV type 1 (HIV-1) isolates are NSI (84), while SI strains emerge during disease progression in about 50% of AIDS patients (70). This emergence often precedes or coincides with a rapid decline in CD4+ cells in blood (41).
Two receptors are required on the surface of the target cell to trigger HIV entry: the CD4 receptor and a coreceptor (22, 28, 30). Coreceptors have seven transmembrane domains (7TM) and are either members of or related to the chemokine receptor family. More than 10 7TM receptors have been shown to act as coreceptors for entry of different HIV-1 strains in vitro (reviewed in references 4, 19, 26, and 47). All HIV-1 strains studied so far use either CCR5 or CXCR4 or both (67, 83). The discovery of HIV coreceptors has mainly explained the NSI/M-tropic versus SI/T-tropic phenotype, by showing that the former strains use CCR5 (1, 17, 22, 27, 28), a receptor for CC chemokines RANTES, MIP-1α, MIP-1β, and MCP-2 (21, 33, 54, 58), while the latter use CXCR4 (30), a receptor for the CXC chemokine stromal cell-derived factor 1 (10, 50). A new nomenclature for HIV strains has been adopted, so that isolates that use CCR5 are termed R5 viruses, those using CXCR4 are designated X4 viruses, and viruses able to use both coreceptors are called R5X4 viruses (5). CCR5 is predominantly expressed on macrophages (53, 73, 79, 82), dendritic cells (3, 9, 34, 82), brain microglial cells (32, 36, 62), and memory T cells (11) but absent on most T-cell lines, while CXCR4 is more widely expressed and present on both naive and memory T cells (11, 46). Thus, the cellular tropism of different strains of HIV-1 is largely determined by differential usage of chemokine receptors. However, this simple picture has several exceptions. Hence, some CCR5-dependent HIV-1 strains do not infect macrophages, although they express high levels of CCR5 (14, 23). Moreover, particular primary CXCR4-using strains do not replicate in several cell lines that express high levels of CXCR4 (43).
In this study, we show that while the majority of CCR5-using viruses do not infect T-cell lines, some strains (called Molt4/SupT1 strains) can infect the T-cell lines Molt4 and SupT1 (12). These strains include a molecularly cloned variant virus (C3) that was adapted in vitro for Molt4 replication and derived from JRCSF. A single amino acid change in the V1 loop accounts for C3’s extended tropism for both Molt4 and SupT1 cells. The V3 loop on gp120 is a major determinant of both cell tropism (7, 15, 16, 37, 45, 61, 64, 72, 74) and more recently of coreceptor usage (8, 56, 68, 80). However, other envelope elements are also involved (39, 55, 56, 71), and several reports have implicated the V1 and V2 loops of gp120 (2, 12, 35, 40, 57, 69), which in addition to the required V3 domain influence the efficiency of replication of HIV-1 in primary macrophages (40, 63, 75) and in Jurkat T cells (13). Groenink et al. (35) described the configuration of a hypervariable locus in the V2 domain that appeared to be predictive for a switch from an NSI to an SI phenotype. V1 and V2 sequences act in conjunction with a CCR5-tropic V3 loop to confer CCR3 usage to some NSI strains (57). Kwong et al. (44) have recently reported the crystal structure of gp120 complexed with CD4 and a neutralizing antibody. This structure shows that the stems of the V1 and V2 loops and the V3 loops are located, respectively, on inner and outer domains of gp120 and on either side of a bridging sheet that spans these two domains. The coreceptor binding site is thought to contain amino acids in this bridging sheet and probably residues in the V3 loop. In some circumstances, the V1 and V2 loops are dispensible for high-affinity binding to coreceptors (77) and viral replication (13), yet when present on gp120 they can have a profound influence on tropism and coreceptor use.
In our study, we aimed to assess the coreceptor(s) used by the C3 variant of JRCSF that differs by a single amino acid in the V1 loop yet can infect the T-cell lines Molt4 and SupT1. We also assessed if the tropism of C3 for Molt4 and SupT1 cells reflected the phenotype of any unselected primary HIV-1 strains and may therefore represent tropism with in vivo relevance.
Replication of R5 viruses in Molt4 and SupT1 T-cell lines.
We assessed if primary HIV-1 R5 strains passaged only in peripheral blood mononuclear cells could infect Molt4 or SupT1 cells. JRCSF and JRFL (42), ADA (74), and E80 (67) are previously described R5 strains. BR49, BR53, BR90, BR92, SL2, SL3, and SL4 are primary R5 isolates provided by St. Mary’s Hospital, London, England. BR49, BR53, BR90, and BR92 were obtained from Brazilian patients (Infectious Disease Service, Porto Alegre, Brazil), while SL2, SL3, and SL4 were from asymptomatic patients from Thailand (Siriraj Hospital, Bangkok) (24, 67).
Of 10 NSI viruses tested, 2 strains (E80 and BR92) consistently replicated in Molt4 or SupT1; 8 other isolates failed to yield supernatant reverse transcriptase activity during 38 days culture. For one of these isolates, ADA, we prepared pseudotype virus that carried the vesicular stomatitis virus envelope glycoprotein G. This pseudotype efficiently infected both Molt4 and SupT1, thus confirming that the block to infection occurred early in the replication cycle and could be bypassed by virions carrying a foreign envelope glycoprotein.
The efficiency of E80 and BR92 as well as the C3 variant of JRCSF to infect SupT1 or Molt4 cells was assessed by estimating endpoint infectivity titers (expressed as 50% tissue culture infective dose [TCID50] per milliliter) (Table 1). These were lower than titers for U87/CD4/CCR5 cells, which express high cell surface concentrations of CCR5. When stocks of E80, BR92, or the C3 variant were prepared from and retitrated back on SupT1 or Molt4 cells, slightly higher titers were noted. For instance, over an endpoint titer of 103 TCID50/ml was observed for BR92 passaged through SupT1 cells. However, these viruses also had higher infectivity titers for U87/CD4/CCR5 cells; therefore, there was no convincing evidence of further adaptation for SupT1 and Molt4 replication (Table 1).
TABLE 1.
Molt4 and SupT1 infection and coreceptor use by R5 strainsa
| HIV-1 strains | Infectivity
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| TCID50/ml
|
FFU/ml
|
||||||||
| On SupT1 | On Molt4 | On U87/CD4 cells
|
On CCC/CD4 cells
|
||||||
| CXCR4, CCR1, CCR2b | CCR3 | CCR5 | CCR8 | GPR-15 | STRL-33 | GPR-1, CX3CR1, D6 | |||
| C3 | 1.7 | 1 | —b | — | 3.5 × 104 | — | — | — | — |
| C3/SupT1 | 1.7 × 101 | 1.7 × 101 | — | — | 7 × 105 | — | — | 20 | — |
| JRCSF | — | — | — | — | 4 × 103 | — | — | — | — |
| E80 | 1 | 1 | — | 3 × 102 | 2.4 × 104 | — | — | — | — |
| E80/SupT1 | 1.7 × 101 | 1.7 × 101 | — | 6 × 102 | 5 × 105 | — | — | — | — |
| BR92 | 1 × 101 | 1.7 | — | — | 2.4 × 105 | — | 3.5 × 102 | 10 | — |
| BR92/SupT1 | 1.7 × 103 | 1.7 × 102 | — | — | 2 × 106 | — | 5 × 102 | 102 | — |
| BR53 | — | — | — | — | 5 × 103 | — | — | — | — |
| BR90 | — | — | — | — | 1.2 × 105 | — | — | — | — |
| SL2 | — | — | — | 4 × 102 | 3 × 105 | — | 3 × 102 | 10 | — |
| SL3 | — | — | — | — | 2.2 × 104 | — | — | — | — |
| SL4 | — | — | — | — | 2 × 103 | — | — | — | — |
| JRFL | — | — | — | 2 × 101 | 7 × 103 | — | — | — | — |
| ADA | — | — | — | 6 × 102 | 7 × 104 | 102 | 2 × 102 | — | — |
U87 cells stably expressing human CD4 and either human CCR1, CCR2b, CCR3, CCR5, or CXCR4 as well as CCC/CD4 cells transfected with expression vectors encoding either human CCR8, GPR-15, STRL-33, GPR-1, CX3CR1, or D6 (49) were seeded into 48-well trays (Costar) at 6 × 104 cells per well. After 24 h, the cells were challenged with 100 μl of serial dilutions of NSI HIV-1 strains for 3 h at 37°C. After 4 days, the wells were fixed and immunostained as previously described (18). The number of positively stained foci was estimated by light microscopy, and the average number of FFU per milliliter was calculated from duplicate wells.
—, no infection detected on SupT1 or Molt4 cells, and <10 FFU/ml on U87/CD4 or CCC/CD4 cells.
Coreceptor use of Molt4/SupT1 R5 strains.
It was possible that Molt4 and SupT1 infection was due to the capacity of these strains to use a novel Molt4/SupT1 coreceptor. We therefore tested the coreceptors that were used by each strain. To determine the coreceptor usage of the isolates studied, we challenged U87 cells stably expressing human CD4 and either human CCR1, CCR2b, CCR3, CCR5, or CXCR4 with HIV-1 strains and monitored infection after 4 to 5 days by immunostaining with an anti-p24 antibody as the primary antibody, followed by incubation with a secondary antibody conjugated to β-galactosidase as described previously (18). For other coreceptors, CCC/CD4 cells were transiently transfected with either CCR8, GPR-15, STRL-33, GPR-1, CX3CR1, or D6 as previously described (67). Table 1 shows infectivity titers in focus-forming units (FFU) per milliliter. JRCSF and C3 infected only cell lines expressing CCR5. All the other M-tropic viruses efficiently infected CCR5+ cells but additionally utilized one or more of the following coreceptors, albeit less efficiently: CCR3 (E80, SL2, ADA, and JRFL), GPR-15 (BR92, SL2, and ADA), STRL-33 (BR92 and SL2), and CCR8 (ADA). None of the viruses used CXCR4 on either U87/CD4 or GHOST/CD4 cells (data not shown). Thus, there was no correlation between Molt4 or SupT1 infection and the use of a particular coreceptor, except for CCR5. Moreover, none of these strains were able to replicate in peripheral blood mononuclear cells from patients homozygous for the CCR5 Δ32 deletion (data not shown).
Inhibition of Molt4 and SupT1 infection by using coreceptor ligands.
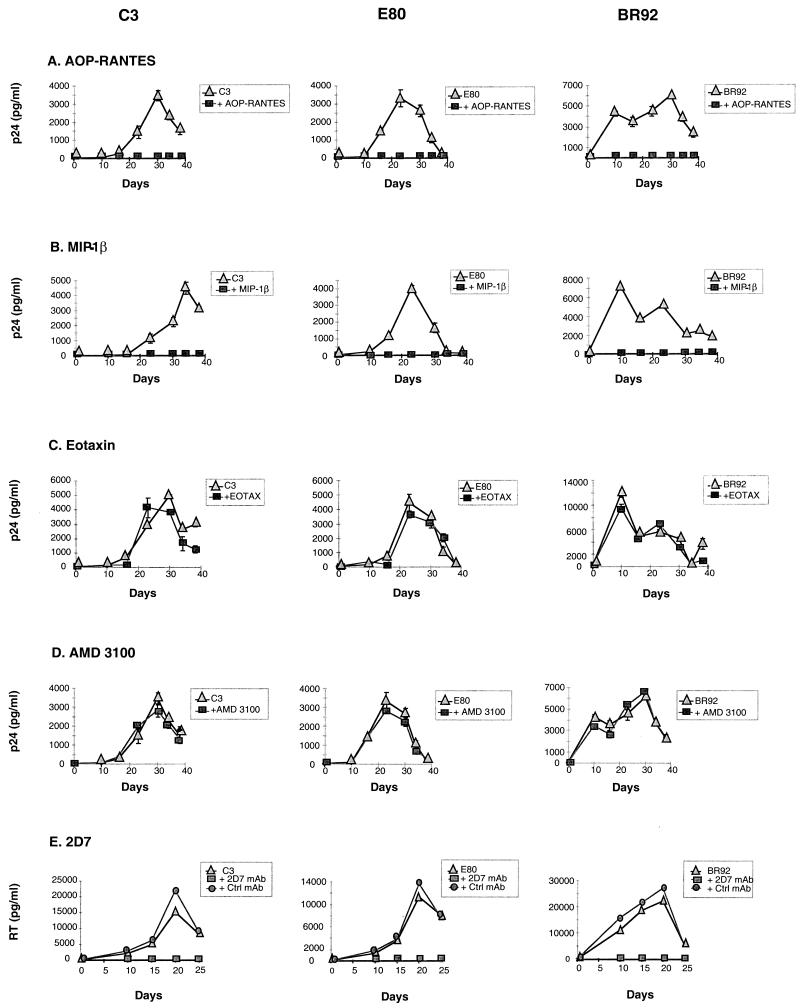
To assess the coreceptors used by C3, E80, and BR92 for SupT1 or Molt4 infection, we tried to inhibit infection by using various coreceptor ligands, including chemokines, modified chemokines, monoclonal antibodies (MAbs), and small organic molecules (reviewed in reference 52). AOP-RANTES is a chemically modified form of RANTES and a potent inhibitor of infection via CCR5 (65). We also tested RANTES itself as well as MIP-1β (20) and eotaxin, a chemokine specific for CCR3 (76). To assess if CXCR4 was involved, we tested inhibition by AMD3100, a bicyclam that is a potent and specific inhibitor of CXCR4 infection (25, 59). SupT1 and Molt4 cells were seeded at 105 cells per well in 96-well trays; 50 μl of medium containing an appropriate chemokine receptor ligand was added at twice the final concentration and incubated at 37°C for 30 min; 50 μl of virus was then added, and the medium was incubated for 3 h at 37°C. The cells were washed four times, and fresh medium containing the relevant chemokine at the required concentration (500, 1,500, or 3,000 ng/ml) was added. For the time course experiment, supernatants were harvested every 3 to 4 days from days 0 to 40, and fresh medium containing appropriate ligands was added.
The results for chemokine receptor ligands used on SupT1 are shown in Fig. 1A to D. Similar results were obtained on Molt4 (data not shown). AOP-RANTES (Fig. 1A) and MIP-1β (Fig. 1B) completely inhibited the replication of C3, E80, and BR92, as did RANTES (data not shown), while eotaxin (Fig. 1C) and AMD3100 (Fig. 1D) had no effect. Although RANTES and MIP-1β bind CCR5, they also bind other 7TM receptors that are potential coreceptors. For instance, MIP-1β binds CCR8 (6) and D6 (49) as well as CCR5. However, C3, E80, and BR92 do not use either as coreceptor (Table 1).
FIG. 1.
Inhibition of C3, E80, and BR92 infection by chemokine receptor ligands. SupT1 and Molt4 cells were treated with virus alone (triangles) or virus plus chemokine receptor ligand (AOP-RANTES, MIP-1β, eotaxin, AMD3100, or the CCR5-specific MAb 2D7, as indicated) (squares). p24 antigen or RT activity (E) in supernatants was measured every 3 to 4 days as previously described (65, 67). For 2D7 inhibition, a control anti-CXCR4 antibody was also tested (circles).
To confirm that C3, E80, and BR92 used CCR5 to infect Molt4 and SupT1 cells, we tested inhibition by using 2D7, a MAb specific for CCR5 that was previously used to block R5 entry or infection in other studies (73, 78). Complete inhibition of replication was observed by 2D7 used at 20 μg/ml but not by a control MAb, used at the same concentration, that recognized CXCR4 (Fig. 1E). Thus, data shown in Fig. 1A, B, and D indicate that CCR5 is the coreceptor used for infectivity of Molt4 and SupT1 cells, even though it could not be detected on the cell surface or internally by immunofluorescence (data not shown).
A single amino acid change in the V1 loop of JRCSF allows the C3 variant to enter and replicate in Molt4 and SupT1 but not several other T-cell lines (12). Two primary R5 isolates out of ten also infected Molt4 and SupT1, providing evidence that viruses like the C3 variant do exist in vivo. In this study, we aimed to investigate whether Molt4/SupT1 tropism was conferred by the use of a specific coreceptor. Our results show that several CCR5 ligands including 2D7 (a MAb specific for CCR5) blocked infection of Molt4 and SupT1 cells, indicating that these strains were able to exploit undetectable levels of CCR5 on these cell lines. Most of the M-tropic CCR5-using strains tested could not infect Molt4 or SupT1 cells, indicating that the use of CCR5 as a coreceptor does not accurately predict the cell tropism of any particular HIV-1 strain. In other systems, receptor expression level has been shown to influence virus entry (48, 79). For instance, the concentrations of CD4 and CCR5 required for efficient infection by R5 viruses are interdependent and the requirement for either is increased when the other is limiting (51). CCR5 expression is variable in vivo (46). Moreover, JRCSF was unable to produce infection in culture when less than 2% of the cells expressed CCR5 (79).
A striking point is that although CCR5 mRNA was present in both Molt4 and SupT1 cells, the protein was not clearly identified, either on the cell surface or internally in permeabilized cells (data not shown). This presumably reflects a very low level of expression, although we cannot rule out that a different conformation of CCR5 or yet unidentified factors impair detection by interfering with the binding of the CCR5-specific MAbs. The CCR5 cDNAs obtained from mRNA extracted from Molt4 and SupT1 were sequenced and found to be 100% homologous to the GenBank sequence. Furthermore, neither of these cell lines produced significant amounts of β-chemokines in the cell supernatant (data not shown). Thus, C3, E80, and BR92 are able to exploit apparently undetectable levels of CCR5 on Molt4 and SupT1 to trigger entry into cells whereas other strains cannot.
To conclude, our results show that a small subset of primary HIV-1 R5 strains are able to infect CD4+ T-cell lines, Molt4 and SupT1. These strains do not use an alternative coreceptor but are able to exploit low concentrations of CCR5 for infection. Molt4/SupT1 tropism therefore identifies primary HIV R5 strains that are likely to have an expanded or altered tropism for CD4+ cells in vivo.
Acknowledgments
We thank Robin Weiss for constructive criticism, as well as Sam Hibbitts, Jackie Reeves, and Áine McKnight for help and suggestions. Thanks go to Mark Marsh and Nathalie Signoret for help with the intracellular detection of CCR5. Rob Nibbs (The Beatson Institute, Glasgow, Scotland) provided the D6 expression vector. AMD3100 was provided by Dominique Schols and Erik de Clercq. Thanks go also to Anna Vyakarnam for testing β-chemokines in supernatants of T-cell lines.
This work was supported by an MRC program grant and partly by an EU Biomed II grant. Nathalie Dejucq is a recipient of an INSERM fellowship.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Andeweg A C, Leeflang P, Osterhaus A D, Bosch M L. Both the V2 and V3 regions of the human immunodeficiency virus type 1 surface glycoprotein functionally interact with other envelope regions in syncytium formation. J Virol. 1993;67:3232–3239. doi: 10.1128/jvi.67.6.3232-3239.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayehunie S, Garcia Zepeda E A, Hoxie J A, Horuk R, Kupper T S, Luster A D, Ruprecht R M. Human immunodeficiency virus-1 entry into purified blood dendritic cells through CC and CXC chemokine coreceptors. Blood. 1997;90:1379–1386. [PubMed] [Google Scholar]
- 4.Berger E A. HIV entry and tropism: the chemokine receptor connection. AIDS. 1997;11:S3–S16. [PubMed] [Google Scholar]
- 5.Berger E A, Doms R W, Fenyo E M, Korber B T, Littman D R, Moore J P, Sattentau Q J, Schuitemaker H, Sodroski J, Weiss R A. A new classification for HIV-1. Nature. 1998;391:240. doi: 10.1038/34571. [DOI] [PubMed] [Google Scholar]
- 6.Bernardini G, Hedrick J, Sozzani S, Luini W, Spinetti G, Weiss M, Menon S, Zlotnik A, Mantovani A, Santoni A, Napolitano M. Identification of the CC chemokines TARC and macrophage inflammatory protein-1 beta as novel functional ligands for the CCR8 receptor. Eur J Immunol. 1998;28:582–588. doi: 10.1002/(SICI)1521-4141(199802)28:02<582::AID-IMMU582>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 7.Bhattacharyya D, Brooks B R, Callahan L. Positioning of positively charged residues in the V3 loop correlates with HIV type 1 syncytium-inducing phenotype. AIDS Res Hum Retroviruses. 1996;12:83–90. doi: 10.1089/aid.1996.12.83. [DOI] [PubMed] [Google Scholar]
- 8.Bieniasz P D, Fridell R A, Aramori I, Ferguson S S G, Caron M G, Cullen B R. HIV-1 induced cell fusion is mediated by multiple regions within both the viral envelope and the CCR-5 co-receptor. EMBO J. 1997;16:2599–2609. doi: 10.1093/emboj/16.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blauvelt A, Asada H, Saville M W, Klaus Kovtun V, Altman D J, Yarchoan R, Katz S I. Productive infection of dendritic cells by HIV-1 and their ability to capture virus are mediated through separate pathways. J Clin Investig. 1997;100:2043–2053. doi: 10.1172/JCI119737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bleul C C, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer T A. The lymphocyte chemoattractant SDF-1 is a ligand for LESTR/fusin and blocks HIV-1 entry. Nature. 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 11.Bleul C C, Wu L, Hoxie J A, Springer T A, Mackay C R. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc Natl Acad Sci USA. 1997;94:1925–1930. doi: 10.1073/pnas.94.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyd M T, Simpson G R, Cann A J, Johnson M A, Weiss R A. A single amino acid substitution in the V1 loop of human immunodeficiency virus type 1 gp120 alters cellular tropism. J Virol. 1993;67:3649–3652. doi: 10.1128/jvi.67.6.3649-3652.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao J, Sullivan N, Desjardin E, Parolin C, Robinson J, Wyatt R, Sodroski J. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J Virol. 1997;71:9808–9812. doi: 10.1128/jvi.71.12.9808-9812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng Mayer C, Liu R, Landau N R, Stamatatos L. Macrophage tropism of human immunodeficiency virus type 1 and utilization of the CC-CKR5 coreceptor. J Virol. 1997;71:1657–1661. doi: 10.1128/jvi.71.2.1657-1661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chesebro B, Nishio J, Perryman S, Cann A, O’Brien W A, Koyanagi Y, Chen I S Y, Wehrly K. Identification of human immunodeficiency virus envelope gene sequences influencing viral entry into CD4-positive HeLa cells, T-leukemia cells, and macrophages. J Virol. 1991;65:5782–5789. doi: 10.1128/jvi.65.11.5782-5789.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chesebro B, Wehrly K, Nishio J, Perryman S. Mapping of independent V3 envelope determinants of human immunodeficiency virus type 1 macrophage tropism and syncytium formation in lymphocytes. J Virol. 1996;70:9055–9059. doi: 10.1128/jvi.70.12.9055-9059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 18.Clapham P R, McKnight Á, Weiss R A. Human immunodeficiency virus type 2 infection and fusion of CD4-negative human cell lines: induction and enhancement by soluble CD4. J Virol. 1992;66:3531–3537. doi: 10.1128/jvi.66.6.3531-3537.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clapham P R, Weiss R A. Immunodeficiency viruses. Spoilt for choice of co-receptors. Nature. 1997;388:230–231. doi: 10.1038/40758. [DOI] [PubMed] [Google Scholar]
- 20.Cocchi F, DeVico A L, Garzino Demo A, Arya S K, Gallo R C, Lusso P. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 21.Combadiere C, Ahuja S K, Tiffany H L, Murphy P M. Cloning and functional expression of CC CKR5, a human monocyte CC chemokine receptor selective for MIP-1(alpha), MIP-1(beta), and RANTES. J Leukoc Biol. 1996;60:147–152. doi: 10.1002/jlb.60.1.147. [DOI] [PubMed] [Google Scholar]
- 22.Deng H, Liu R, Elmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 23.Dittmar M T, McKnight Á, Simmons G, Clapham P R, Weiss R A, Simmonds P. HIV-1 tropism and co-receptor usage. Nature. 1997;385:495–496. doi: 10.1038/385495a0. [DOI] [PubMed] [Google Scholar]
- 24.Dittmar M T, Simmons G, Hibbitts S, O’Hare M, Louisirirotchanakul S, Beddows S, Weber J, Clapham P R, Weiss R A. Langerhans cell tropism of human immunodeficiency virus type 1 subtype A through F isolates derived from different transmission groups. J Virol. 1997;71:8008–8013. doi: 10.1128/jvi.71.10.8008-8013.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donzella G A, Schols D, Lin S W, Este J A, Nagashima K A, Maddon P J, Allaway G P, Sakmar T P, Henson G, De Clercq E, Moore J P. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat Med. 1998;4:72–77. doi: 10.1038/nm0198-072. [DOI] [PubMed] [Google Scholar]
- 26.Doranz B J, Berson J F, Rucker J, Doms R W. Chemokine receptors as fusion cofactors for human immunodeficiency virus type 1 (HIV-1) Immunol Res. 1997;16:15–28. doi: 10.1007/BF02786321. [DOI] [PubMed] [Google Scholar]
- 27.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 28.Dragic T, Litwin V, Allaway G, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 29.Fear W R, Kesson A M, Naif H, Lynch G W, Cunningham A L. Differential tropism and chemokine receptor expression of human immunodeficiency virus type 1 in neonatal monocytes, monocyte-derived macrophages, and placental macrophages. J Virol. 1998;72:1334–1344. doi: 10.1128/jvi.72.2.1334-1344.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane G-protein coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 31.Fenyo E M, Morfeldt Manson L, Chiodi F, Lind B, von Gegerfelt A, Albert J, Olausson E, Asjo B. Distinct replicative and cytopathic characteristics of human immunodeficiency virus isolates. J Virol. 1988;62:4414–4419. doi: 10.1128/jvi.62.11.4414-4419.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghorpade A, Xia M Q, Hyman B T, Persidsky Y, Nukuna A, Bock P, Che M, Limoges J, Gendelman H E, Mackay C R. Role of the beta-chemokine receptors CCR3 and CCR5 in human immunodeficiency virus type 1 infection of monocytes and microglia. J Virol. 1998;72:3351–3361. doi: 10.1128/jvi.72.4.3351-3361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gong W, Howard O M, Turpin J A, Grimm M C, Ueda H, Gray P W, Raport C J, Oppenheim J J, Wang J M. Monocyte chemotactic protein-2 activates CCR5 and blocks CD4/CCR5-mediated HIV-1 entry/replication. J Biol Chem. 1998;273:4289–4292. doi: 10.1074/jbc.273.8.4289. [DOI] [PubMed] [Google Scholar]
- 34.Granelli Piperno A, Moser B, Pope M, Chen D, Wei Y, Isdell F, O’Doherty U, Paxton W, Koup R, Mojsov S, Bhardwaj N, Clark Lewis I, Baggiolini M, Steinman R M. Efficient interaction of HIV-1 with purified dendritic cells via multiple chemokine coreceptors. J Exp Med. 1996;184:2433–2438. doi: 10.1084/jem.184.6.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Groenink M, Fouchier R A M, Broersen S, Baker C H, Koot M, van’t Wout A B, Huisman H G, Miedema F, Tersmette M, Schuitemaker H. Relation of phenotype evolution of HIV-1 to envelope V2 configuration. Science. 1993;260:1513–1515. doi: 10.1126/science.8502996. [DOI] [PubMed] [Google Scholar]
- 36.He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hofmann W, Newman W, Mackay C R, Sodroski J, Gabuzda D. CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature. 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 37.Hwang S R, Boyle T J, Lyerly K, Cullen B R. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science. 1992;253:71–74. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
- 38.Karita E, Nkengasong J N, Willems B, Vanham G, Fransen K, Heyndrickx L, Janssens W, Piot P, van der Groen G. Macrophage-tropism of HIV-1 isolates of different genetic subtypes. AIDS. 1997;11:1303–1304. doi: 10.1097/00002030-199710001-00010. [DOI] [PubMed] [Google Scholar]
- 39.Kim F M, Kolson D L, Balliet J W, Srinivasan A, Collman R G. V3-independent determinants of macrophage tropism in a primary human immunodeficiency virus type 1 isolate. J Virol. 1995;69:1755–1761. doi: 10.1128/jvi.69.3.1755-1761.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koito A, Harrowe G, Levy J A, Cheng-Mayer C. Functional role of the V1/V2 region of human immunodeficiency virus type 1 envelope glycoprotein gp120 in infection of primary macrophages and soluble CD4 neutralization. J Virol. 1994;68:2253–2259. doi: 10.1128/jvi.68.4.2253-2259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koot M, Keet I P, Vos A H, de Goede R E, Roos M T, Coutinho R A, Miedema F, Schellekens P T, Tersmette M. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann Intern Med. 1993;118:681–688. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- 42.Koyanagi Y, Miles S, Mitsuyasu R T, Merrill J E, Vinters H V, Chen I S. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science. 1987;236:819–822. doi: 10.1126/science.3646751. [DOI] [PubMed] [Google Scholar]
- 43.Kozak S L, Platt E J, Madani N, Ferro F E, Jr, Peden K, Kabat D. CD4, CXCR-4, and CCR-5 dependencies for infections by primary patient and laboratory-adapted isolates of human immunodeficiency virus type 1. J Virol. 1997;71:873–882. doi: 10.1128/jvi.71.2.873-882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mammano F, Salvatori F, Ometto L, Panozzo M, Chieco Bianchi L, De Rossi A. Relationship between the V3 loop and the phenotypes of human immunodeficiency virus type 1 (HIV-1) isolates from children perinatally infected with HIV-1. J Virol. 1995;69:82–92. doi: 10.1128/jvi.69.1.82-92.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mo H, Monard S, Pollack H, Ip J, Rochford G, Wu L, Hoxie J, Borkowsky W, Ho D D, Moore J P. Expression patterns of the HIV type 1 coreceptors CCR5 and CXCR4 on CD4+ T cells and monocytes from cord and adult blood. AIDS Res Hum Retroviruses. 1998;14:607–617. doi: 10.1089/aid.1998.14.607. [DOI] [PubMed] [Google Scholar]
- 47.Moore J P. Coreceptors: implications for HIV pathogenesis and therapy. Science. 1997;276:51–52. doi: 10.1126/science.276.5309.51. [DOI] [PubMed] [Google Scholar]
- 48.Naif H M, Li S, Alali M, Sloane A, Wu L, Kelly M, Lynch G, Lloyd A, Cunningham A L. CCR5 expression correlates with susceptibility of maturing monocytes to human immunodeficiency virus type 1 infection. J Virol. 1998;72:830–836. doi: 10.1128/jvi.72.1.830-836.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nibbs R J B, Wylie S M, Pragnell I B, Graham G J. Cloning and characterization of a novel murine beta chemokine receptor, D6. Comparison to three other related macrophage inflammatory protein-1alpha receptors, CCR-1, CCR-3, and CCR-5. J Biol Chem. 1997;272:12495–12504. doi: 10.1074/jbc.272.19.12495. [DOI] [PubMed] [Google Scholar]
- 50.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J-L, Arenzana-Seisdedos F, Schwartz O, Heard J-M, Clark-Lewis I, Legler D F, Loetscher M, Baggiolini M, Moser B. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 51.Platt E J, Wehrly K, Kuhmann S E, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Proudfoot A E I, Wells T N C, Clapham P R. Chemokine receptors—future therapeutic targets for HIV? Biochem Pharmacol. 1998;57:451–463. doi: 10.1016/s0006-2952(98)00339-6. [DOI] [PubMed] [Google Scholar]
- 53.Rana S, Besson G, Cook D G, Rucker J, Smyth R J, Yi Y, Turner J D, Guo H H, Du J G, Peiper S C, Lavi E, Samson M, Libert F, Liesnard C, Vassart G, Doms R W, Parmentier M, Collman R G. Role of CCR5 in infection of primary macrophages and lymphocytes by macrophage-tropic strains of human immunodeficiency virus: resistance to patient-derived and prototype isolates resulting from the Δccr5 mutation. J Virol. 1997;71:3219–3227. doi: 10.1128/jvi.71.4.3219-3227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raport C J, Gosling J, Schweickart V L, Gray P W, Charo I F. Molecular cloning and functional characterization of a novel human CC chemokine receptor (CCR5) for RANTES, MIP-1β, and MIP-1α. J Biol Chem. 1996;271:17161–17166. doi: 10.1074/jbc.271.29.17161. [DOI] [PubMed] [Google Scholar]
- 55.Reeves J D, Schulz T F. The CD4-independent tropism of human immunodeficiency virus type 2 involves several regions of the envelope protein and correlates with a reduced activation threshold for envelope-mediated fusion. J Virol. 1997;71:1453–1465. doi: 10.1128/jvi.71.2.1453-1465.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rizzuto C D, Wyatt R, Hernandez Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 57.Ross T M, Cullen B R. The ability of HIV type 1 to use CCR-3 as a coreceptor is controlled by envelope V1/V2 sequences acting in conjunction with a CCR-5 tropic V3 loop. Proc Natl Acad Sci USA. 1998;95:7682–7686. doi: 10.1073/pnas.95.13.7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Samson M, Labbe O, Mollereau C, Vassart G, Parmentier M. Molecular cloning and functional expression of a new human CC-chemokine receptor gene. Biochemistry. 1996;11:3362–3367. doi: 10.1021/bi952950g. [DOI] [PubMed] [Google Scholar]
- 59.Schols D, Este J A, Henson G, De Clercq E. Bicyclams, a class of potent anti-HIV agents, are targeted at the HIV coreceptor fusin/CXCR-4. Antivir Res. 1997;35:147–156. doi: 10.1016/s0166-3542(97)00025-9. [DOI] [PubMed] [Google Scholar]
- 60.Schuitemaker H, Kootstra N A, Groenink M, De Goede R E, Miedema F, Tersmette M. Differential tropism of clinical HIV-1 isolates for primary monocytes and promonocytic cell lines. AIDS Res Hum Retroviruses. 1992;8:1679–1682. doi: 10.1089/aid.1992.8.1679. [DOI] [PubMed] [Google Scholar]
- 61.Sharpless N E, O’Brien W A, Verdin E, Kufta C V, Chen I S Y, Dubois-Dalcq M. Human immunodeficiency virus type 1 tropism for brain microglial cells is determined by a region of the env glycoprotein that also controls macrophage tropism. J Virol. 1992;66:2588–2593. doi: 10.1128/jvi.66.4.2588-2593.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shieh J T, Albright A V, Sharron M, Gartner S, Strizki J, Doms R W, Gonzalez Scarano F. Chemokine receptor utilization by human immunodeficiency virus type 1 isolates that replicate in microglia. J Virol. 1998;72:4243–4249. doi: 10.1128/jvi.72.5.4243-4249.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shioda T, Levy J A, Cheng Mayer C. Small amino acid changes in the V3 hypervariable region of gp120 can affect the T-cell-line and macrophage tropism of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1992;89:9434–9438. doi: 10.1073/pnas.89.20.9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shioda T, Levy J A, Cheng-Mayer C. Macrophage and T-cell line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature. 1991;349:167–169. doi: 10.1038/349167a0. [DOI] [PubMed] [Google Scholar]
- 65.Simmons G, Clapham P R, Picard L, Offord R E, Rosenkilde M M, Schwartz T W, Buser R, Wells T N C, Proudfoot A E. Potent inhibition of HIV-1 infectivity in macrophages and lymphocytes by a novel CCR5 antagonist. Science. 1997;276:276–279. doi: 10.1126/science.276.5310.276. [DOI] [PubMed] [Google Scholar]
- 66.Simmons G, Reeves J, McKnight A, Dejucq N, Hibbitts S, Power C, Aarons E, Schols D, de Clercq E, Proudfoot A, Clapham P. CXCR4 as a functional co-receptor for human immunodeficiency virus type 1 infection of primary macrophages. J Virol. 1998;72:8453–8457. doi: 10.1128/jvi.72.10.8453-8457.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simmons G, Wilkinson D, Reeves J D, Dittmar M T, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, Clapham P R. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Speck R F, Wehrly K, Platt E J, Atchison R E, Charo I F, Kabat D, Chesebro B, Goldsmith M A. Selective employment of chemokine receptors as human immunodeficiency virus type 1 coreceptors determined by individual amino acids within the envelope V3 loop. J Virol. 1997;71:7136–7139. doi: 10.1128/jvi.71.9.7136-7139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sullivan N, Thali M, Furman C, Ho D D, Sodroski J. Effects of amino acid changes in the V2 region of the human immunodeficiency virus type 1 gp120 glycoprotein on subunit association, syncytium formation, and recognition by neutralizing antibody. J Virol. 1993;67:3674–3679. doi: 10.1128/jvi.67.6.3674-3679.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tersmette M, de Goede R E, Al B J, Winkel I N, Gruters R A, Cuypers H T, Huisman H G, Miedema F. Differential syncytium-inducing capacity of human immunodeficiency virus isolates: frequent detection of syncytium-inducing isolates in patients with acquired immunodeficiency syndrome (AIDS) and AIDS-related complex. J Virol. 1988;62:2026–2032. doi: 10.1128/jvi.62.6.2026-2032.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Meyer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 72.Trujillo J R, Wang W K, Lee T H, Essex M. Identification of the envelope V3 loop as a determinant of a CD4-negative neuronal cell tropism for HIV-1. Virology. 1996;217:613–617. doi: 10.1006/viro.1996.0158. [DOI] [PubMed] [Google Scholar]
- 73.Tuttle D L, Harrison J K, Anders C, Sleasman J W, Goodenow M M. Expression of CCR5 increases during monocyte differentiation and directly mediates macrophage susceptibility to infection by human immunodeficiency virus type 1. J Virol. 1998;72:4962–4969. doi: 10.1128/jvi.72.6.4962-4969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Westervelt P, Gendelman H E, Ratner L. Identification of a determinant within the human immunodeficiency virus 1 surface envelope glycoprotein critical for productive infection of primary monocytes. Proc Natl Acad Sci USA. 1991;88:3097–3101. doi: 10.1073/pnas.88.8.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Westervelt P, Trowbridge D B, Epstein L G, Blumberg B M, Li Y, Hahn B H, Shaw G M, Price R W, Ratner L. Macrophage tropism determinants of human immunodeficiency virus type 1 in vivo. J Virol. 1992;66:2577–2582. doi: 10.1128/jvi.66.4.2577-2582.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Williams T J, Griffiths-Johnson D A, Jose P J, Collins P D. Eosinophil chemoattractants generated in vivo. Agents Actions Suppl. 1995;46:1–9. doi: 10.1007/978-3-0348-7276-8_1. [DOI] [PubMed] [Google Scholar]
- 77.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 78.Wu L, LaRosa G, Kassam N, Gordon C J, Heath H, Ruffing N, Chen H, Humblias J, Samson M, Parmentier M, Moore J P, Mackay C R. Interaction of chemokine receptor CCR5 with its ligands: multiple domains for HIV-1 gp120 binding and a single domain for chemokine binding. J Exp Med. 1997;186:1373–1381. doi: 10.1084/jem.186.8.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu L, Paxton W A, Kassam N, Ruffing N, Rottman J B, Sullivan N, Choe H, Sodroski J, Newman W, Koup R A, Mackay C R. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J Exp Med. 1997;185:1681–1691. doi: 10.1084/jem.185.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xiao L, Owen S M, Goldman I, Lal A A, deJong J J, Goudsmit J, Lal R B. CCR5 coreceptor usage of non-syncytium-inducing primary HIV-1 is independent of phylogenetically distinct global HIV-1 isolates: delineation of consensus motif in the V3 domain that predicts CCR-5 usage. Virology. 1998;240:83–92. doi: 10.1006/viro.1997.8924. [DOI] [PubMed] [Google Scholar]
- 81.Yi Y, Rana S, Turner J D, Gaddis N, Collman R G. CXCR-4 is expressed by primary macrophages and supports CCR5-independent infection by dual-tropic but not T-tropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:772–777. doi: 10.1128/jvi.72.1.772-777.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zaitseva M, Blauvelt A, Lee S, Lapham C K, Klaus Kovtun V, Mostowski H, Manischewitz J, Golding H. Expression and function of CCR5 and CXCR4 on human Langerhans cells and macrophages: implications for HIV primary infection. Nat Med. 1997;3:1369–1375. doi: 10.1038/nm1297-1369. [DOI] [PubMed] [Google Scholar]
- 83.Zhang L, Huang Y, He T, Cao Y, Ho D D. HIV-1 subtype and second-receptor use. Nature. 1996;383:768. doi: 10.1038/383768a0. [DOI] [PubMed] [Google Scholar]
- 84.Zhu T, Mo H, Wang N, Nam D S, Cao Y, Koup R A, Ho D D. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]