Abstract
Background:
Previous research demonstrated that a homozygous mutation of g.136372044G>A (S12N) in caspase recruitment domain family member 9 (CARD9) is critical for producing Aspergillus fumigatus-induced (Af-induced) T helper 2 (TH2)-mediated responses in allergic bronchopulmonary aspergillosis (ABPA). However, it remains unclear whether the CARD9S12N mutation, especially the heterozygous occurrence, predisposes the host to ABPA.
Methods:
A total of 61 ABPA patients and 264 controls (including 156 healthy controls and 108 asthma patients) were recruited for sequencing the CARD9 locus to clarify whether patients with this heterozygous single-nucleotide polymorphisms are predisposed to the development of ABPA. A series of in vivo and in vitro experiments, such as quantitative real-time polymerase chain reaction, flow cytometry, and RNA isolation and quantification, were used to illuminate the involved mechanism of the disease.
Results:
The presence of the p.S12N mutation was associated with a significant risk of ABPA in ABPA patients when compared with healthy controls and asthma patients, regardless of Aspergillus sensitivity. Relative to healthy controls without relevant allergies, the mutation of p.S12N was associated with a significant risk of ABPA (OR: 2.69 and 4.17 for GA and AA genotypes, P = 0.003 and 0.029, respectively). Compared with patients with asthma, ABPA patients had a significantly higher heterozygous mutation (GA genotype), indicating that p.S12N might be a significant ABPA-susceptibility locus (aspergillus sensitized asthma: OR: 3.02, P = 0.009; aspergillus unsensitized asthma: OR: 2.94, P = 0.005). The mutant allele was preferentially expressed in ABPA patients with heterozygous CARD9S12N, which contributes to its functional alterations to facilitate Af-induced TH2-mediated ABPA development. In terms of mechanism, Card9 wild-type (Card9WT) expression levels decreased significantly due to Af-induced decay of its messenger RNA compared to the heterozygous Card9S12N. In addition, ABPA patients with heterozygous CARD9S12N had increased Af-induced interleukin-5 production.
Conclusion:
Our study provides the genetic evidence showing that the heterozygous mutation of CARD9S12N, followed by allele expression imbalance of CARD9S12N, facilitates the development of ABPA.
Keywords: Allergic bronchopulmonary aspergillosis, CARD9 polymorphism, Allelic expression imbalance, Asthma
Introduction
Allergic bronchopulmonary aspergillosis (ABPA) is a hypersensitivity lung disease attributed to bronchial colonization by the mold Aspergillus fumigatus (Af) in susceptible patients with asthma or cystic fibrosis (CF). ABPA affects approximately 12% of patients with severe asthma and 7–9% of those with CF,[1,2] and has low but significant prevalence in European, Hispanic, African American, and Asian people. ABPA affects approximately 30,000 individuals in the United States and a further 70,000 people worldwide.[3] The inflammatory response in ABPA is characterized by T helper 2 (TH2) responses to Af allergens that stimulate immunoglobulin E (IgE) synthesis, eosinophil activation, and production of the cytokines interleukin-4 (IL-4) and IL-5.[4,5]
Clinically, ABPA is characterized by wheezing, pulmonary infiltrates, bronchiectasis, and parenchymal fibrosis. Both genetic and environmental influences drive the development of ABPA. To date, several genetic risk factors for the development of ABPA have been identified, including human leukocyte antigen DR (HLA-DR) and human leukocyte antigen DQ (HLA-DQ) restriction, the Ile75Val polymorphism in the IL-4 receptor alpha chain (IL-4Rα), IL-10 promoter polymorphisms, the Ala91Pro variant of the surfactant protein A2 (SP-A2), and heterozygous mutations of the CF transmembrane conductance regulator (CFTR) gene.[6–11] ABPA patients are genetically at risk to develop heightened TH2 responses to Af antigens, given the prevalence of Af colonization.
Caspase recruitment domain family member 9 (CARD9) is a key adaptor protein involved in orchestrating C-type lectin receptor (CLR)-mediated canonical nuclear factor kappa B (NF-κB [p65]) signaling to initiate IL-12, IL-6, and IL-1β production, which drives TH1 and TH17 responses.[12,13] As a critical adaptor protein, CARD9 is mainly expressed on myeloid cells, downstream of pattern recognition receptors (PRRs) involved in innate and adaptive immunity, especially for macrophages and dendritic cells (DCs).[14] Clinically, CARD9 single-nucleotide polymorphisms (SNP) have been found to be involved in autoimmune diseases, such as Crohn's disease, ulcerative colitis (UC), ankylosing spondylitis, immune globulin A (IgA) nephropathy, and rheumatoid arthritis.[15]
CARD9 null or loss-of-function mutations are responsible for defective innate and adaptive immunity, leading to life-threatening fungal infections in patients.[16,17] Inheritance of the CARD9S12N mutation is reported to confer susceptibility to Crohn's disease and UC (Genome Wide Association Study [GWAS] analyses with an odds ratio of 1.2 for both diseases).[18] In addition, CARD9S12N is also associated with primary sclerosing cholangitis in patients with UC.[19] Our previous study demonstrated that the homozygous Card9S12N is critical for Af-induced TH2 responses.[20] In terms of mechanism, Card9S12N facilitates Af-induced activation of non-canonical NF-κB (RelB) in alveolar macrophages, which regulates IL-5 expression to induce TH2 responses. However, we did not clarify whether and how patients with this heterozygous SNP are predisposed to the development of ABPA.
Methods
Ethical approval
The study design, which conforms to the ethical guidelines of the 1975 Declaration of Helsinki, was approved by the Human Research Committee of Tongji University School of Medicine (No. 2015YXY95) and other hospitals where an individual committee review was required. Written informed consents were obtained from all participants. All animal experiments were performed in compliance with institutional guidelines and the protocol, which was approved by the Animal Ethics Committee of Tongji University School of Medicine (No. 2015YXY95).
Human subject enrollment, sample collection, and antigen detection
A case-control study was conducted involving 61 ABPA patients, 108 asthma patients, and 156 healthy controls (HCs) from Shanghai Pulmonary Hospital, Ruijin Hospital, and Shanghai Putuo District People's Hospital from 2015 to 2019. Diagnosis of ABPA was based on the 2013 International Society for Human and Animal Mycology (ISHAM) criteria.[21] All patients with asthma fulfilled the definition given by the Global Initiative for Asthma (update 2015).[22] All asthma patients were proved to be sensitive to special allergens and divided by the presence of a positive specific immunoglobulin E (sIgE) in their serum into asthma patients with Aspergillus sensitivity, as well as those with other sensitivities. Healthy controls without clinical symptoms and a clinical history of asthma or allergic diseases were recruited from medical examination population. The level of total serum IgE and the levels of sIgE specific to allergens such as Aspergillus, house dust, mixed pollens, and other fungi were determined for ABPA patients, asthma patients, and healthy controls using the Phadia ImmunoCAP 1000 instrument system (Pharmacia Diagnostic, Uppsala, Sweden). Peripheral blood mononuclear cells (PBMCs) were obtained from patients and controls with informed consent.
Af exposure and sensitization animal model
Wild-type (WT),homozygous Card9S12N knock-in mice, and heterozygous Card9S12N knock-in mice (Het), on a C57BL/6J background, described previously,[20] were housed in the specific pathogen-free animal facilities at Tongji University. We established chronic asthma model (five animals per group) in homozygous and heterozygous Card9S12N knock-in mice and Card9WT, and high dose exposure model (six animals per group) in heterozygous Card9S12N knock-in mice and CardWT mice. The chronic asthma model was established as described in the previous study.[25] Briefly, eight-week-old female mice were first administered Af (5 × 106) intratracheally in 50 μL of phosphate buffer saline (PBS). Seven days after the initial exposure, mice were intratracheally re-administered Af conidia (1 × 106) daily for 5 days; after resting for 2 days, all mice were re-challenged with Af conidia (1 × 106) for another 3 days, and were sacrificed 24 h after the final challenge (day 18). For high dose exposure model, mice were sacrificed at day 8 after a single exposure to Af conidia (5 × 107).
Cell culture
Primary cultures of bone marrow-derived macrophages (BMDMs) from CardWT mice, and heterozygous and homozygous Card9S12N knock-in mice were prepared as previously described.[24] After 6–7 days, flow cytometry analysis indicated that the cell population contained >97% CD11b+F4/80+ cells, which were subsequently prepared for use. PBMCs from both patients with ABPA and healthy donors were isolated from heparinized blood using Lymphoprep (MP Biomedicals, Irvine, CA, USA), and then cultured for use. To detect messenger RNA (mRNA) decay of CARD9, the cells were treated with actinomycin D (5 μg/mL), and then challenged with Af conidia for indicated time.
RNA isolation and quantification of CARD9 expression
Complete RNA was extracted from PBMCs using the RNeasy Minikit (Qiagen, Duesseldorf, Germany) and subsequently reversely transcribed into complementary deoxyribonucleic acid (cDNA) using a PrimeScript™ II 1st Strand cDNA Synthesis Kit (TaKaRa, Tokyo, Japan), according to the manufacturer's instructions. Then, the cDNA samples were analyzed by pyrosequencing at Sangon Biotech in Shanghai to determine allelic expression levels according to the Card9S12N polymorphism. The fundamental basis of pyrosequencing is that pyrophosphate is released when a deoxyribonucleotide triphosphate is added to the end of a nascent strand of DNA. As the deoxyribonucleotide triphosphates are sequentially added to the reaction and the pyrophosphate concentration is continuously monitored, the DNA sequence can be determined.[23]
Flow cytometry
Antibodies to mouse integrin αX subunit (CD11c), integrin αM (CD11b), lymphocyte antigen 6 complex locus G6D (Ly6G), SiglecF, and antibodies to human IL-5, CD14, and CD11b were used in flow cytometry [Supplementary Table 1, http://links.lww.com/CM9/B644]. For IL-5 intracellular staining, PBMCs were cultured for 2 h, and non-adherent cells were gently removed. After stimulation with heat-killed swollen conidia (SC) for 2 h, cells were cultured and collected for CD14 and CD11b staining, followed by intracellular IL-5 staining as previously described.[20] Cells were examined using a BD LSRFortessa cell analyzer (BD Immunocytometry Systems,San Jose, CA, USA), and data were analyzed with FlowJo (Tree Star, Ashland, OR, USA).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was isolated using TRIzol (Invitrogen, Carlsbad, CA, USA) and then RNA was reverse-transcribed using PrimeScript RT Master Mix (Takara, Tokyo, Japan). qRT-PCR was performed using Power SYBR Green PCR Master Mix (TaKaRa, Tokyo, Japan). The amounts of transcript were normalized to glyceraldehyde-phosphate dehydrogenase (GAPDH). CARD9 mRNA expression was tested by qRT-PCR. The primers used in this study are shown in Supplementary Table 2, http://links.lww.com/CM9/B644.
Cytokine measurement
BMDM culture supernatants and mouse serum were collected. IL-4, IL-5 and IgE concentrations were measured by SET-Ready-GO ELISA kits (eBioscience, San Diego, CA, USA) according to the manufacturer's protocol.
Statistical analysis
For clinical data, values are presented as mean ± standard deviation, median (Q1, Q3), or number (percentage). For animal experimental data, all values in the paper are presented as the mean ± standard error of the mean (SEM), unless stated otherwise. To analyze the in vitro cell stimulation and in vivo measurements, the experimental data were analyzed using Student's t test, Mann-Whitney U test, or one-way analysis of variance with GraphPad Prism 6.0 (La Jolla, CA, USA). For the analysis of single nucleotide polymorphisms (SNPs), Pearson's chi-squared test was used for exploring the differences in categorical variables among groups, and Hardy-Weinberg equilibrium was evaluated by testing for deviation from the expected values. Logistic regression analysis was used to evaluate the relationship between CARD9 genotype and ABPA. Statistical analyses were performed using SAS software (version 9.4, SAS Institute Inc., Raleigh, NC, USA). All tests were two-sided, and a P <0.05 was considered statistically significant.
Results
Detailed information on recruited ABPA patients, asthma patients, and healthy controls
All recruited Han Chinese ABPA patients (n = 61) in this study met the ISHAM criteria for ABPA diagnosis [Table 1 and Supplementary Table 3, http://links.lww.com/CM9/B644]. The detailed information on enrolled asthma patients (n = 108) and healthy controls (n = 156) is listed in Supplementary Tables 4 and 5, http://links.lww.com/CM9/B644, respectively. The serum aspergillus-specific IgE, total serum IgE levels, and median eosinophil count of ABPA patients were 1.09 (0.57–3.78) kUA/L, 2435 (1694–2500) IU/mL, and 1.06 (0.81–1.65) × 109 cells/L, respectively, and were significantly higher than those in asthma patients and healthy controls (all P <0.001) [Table 2]. The classic radiological features in chest CT scans of ABPA patients are showed in Supplementary Table 3, http://links.lww.com/CM9/B644.
Table 1.
Diagnostic data of ABPA patients (n = 61) .
| Diagnostic items | Number (%) |
|---|---|
| Predisposing conditions | |
| Only asthma | 1 (2) |
| Only bronchiectasis | 0 (0) |
| Both asthma and bronchiectasis | 60 (98) |
| Obligatory criteria | |
| Total IgE >1000 IU/mL | 61 (100) |
| Af specific IgE >0.35 kUA/L | 60 (98) |
| Type I Af skin test (+) | 52 (85) |
| Other criteria | |
| Peripheral EOS >0.5 cells/μL | 61 (100) |
| ABPA chest CT features exist | 61 (100) |
Diagnosis of ABPA was based on the 2013 ISHAM criteria. ABPA: Allergic bronchopulmonary aspergillosis; Af: Aspergillus fumigatus; CT: Computed tomography; EOS: Eosinophils; IgE: Immunoglobulin E; ISHAM: International Society for Human and Animal Mycology.
Table 2.
Demographic information of the ABPA patients, asthma patients and healthy controls.
| Variables |
ABPA (n = 61) |
Asthma with positive allergens (n = 108) | Healthy control (n = 156) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Aspergillus sensitized (n = 43) |
P values (vs. ABPA) |
Aspergillus unsensitized (n = 65) |
P values (vs. ABPA) |
With allergy (n = 34) |
P values (vs. ABPA) |
Without allergy (n = 122) |
P values (vs. ABPA) |
|||
| Age (years) | 48 (38, 59) | 56 (43, 63) | 0.033 | 54 (34, 62) | 0.209 | 35 (28, 40) | <0.001 | 33 (27, 39) | <0.001 | |
| Gender | 0.144 | 0.445 | 0.060 | 0.007 | ||||||
| Male | 24 (39.3) | 24 (55.8) | 30 (46.2) | 21 (61.8) | 75 (61.5) | |||||
| Female | 37 (60.7) | 19 (44.2) | 35 (53.8) | 13 (38.2) | 47 (38.5) | |||||
| Af-specific IgE (kUA/L) | 1.09 (0.57, 3.78) | 0.26 (0.18, 0.37) | <0.001 | 0.01 (0, 0.02) | <0.001 | 0.02 (0, 0.06) | <0.001 | 0 (0, 0) | <0.001 | |
| Total IgE (IU/mL) | 2435 (1694, 2500) | 154 (125, 306) | <0.001 | 183 (82, 392) | <0.001 | 179 (41, 327) | <0.001 | 33 (15, 80) | <0.001 | |
| EOS counts (×109/L) | 1.06 (0.81, 1.65) | 0.24 (0.10, 0.38) | <0.001 | 0.29 (0.13, 0.56) | <0.001 | 0.09 (0.06, 0.18) | <0.001 | 0.08 (0.05, 0.14) | <0.001 | |
| Genotype | 0.002 | 0.009 | 0.036 | 0.003 | ||||||
| G/G | 21 (34.4) | 28 (65.1) | 40 (61.5) | 21 (61.8) | 73 (59.8) | |||||
| G/A | 34 (55.7) | 15 (34.9) | 22 (33.8) | 11 (32.4) | 44 (36.1) | |||||
| A/A | 6 (9.8) | 0 (0) | 3 (4.6) | 2 (5.9) | 5 (4.1) | |||||
Data are expressed as number (percentage) or median (Q1, Q3). ABPA: Allergic bronchopulmonary aspergillosis; Af: Aspergillus fumigatus; EOS: Eosinophils; IgE: Immune globulin E.
The p.S12N mutation in CARD9 was associated with ABPA development
We examined whether CARD9S12N mutations occur in ABPA patients by sequencing the CARD9 locus in the genomic DNA from 61 confirmed ABPA patients, 108 asthma patients, and 156 healthy controls. Notably, 55.7% (34/61) of ABPA patients had the GA genotype of p.S12N in CARD9 and 9.8% (6/61) had AA genotype [Table 2]. As the frequency of the A allele of p.S12N was higher in ABPA patients than that in asthmatic or healthy individuals, we analyzed their associations with ABPA [Table 3]. Relative to healthy controls without relevant allergies, ABPA patients had a significantly higher frequency of mutation, which means that CARD9S12N mutations are associated with the risk of ABPA (OR: 2.69 and 4.17 for GA and AA genotypes, P = 0.003 and 0.029, respectively), and relative to healthy controls with allergies, the association of ABPA and CARD9S12N mutations was significant (OR: 3.09 for GA genotypes, P = 0.015, respectively). It is worth noting that when comparing with patients with asthma, regardless of Aspergillus sensitivity, the association of ABPA risk and the heterozygote (GA genotype) was both significant, indicating that p.S12N is a significant ABPA-susceptibility locus (Aspergillus sensitized asthma: OR: 3.02, P = 0.009; Aspergillus unsensitized asthma: OR: 2.94, P = 0.005).
Table 3.
Association of p.S12N polymorphism of CARD9 and ABPA risk.
| Items | Odds ratio | 95% Confidence interval | P values |
|---|---|---|---|
| ABPA vs. healthy control without allergy | |||
| GG (reference) | 1.00 | ||
| GA | 2.69 | (1.39–5.20) | 0.003 |
| AA | 4.17 | (1.16–15.04) | 0.029 |
| ABPA vs. healthy control with allergy | |||
| GG (reference) | 1.00 | ||
| GA | 3.09 | (1.24–7.68) | 0.015 |
| AA | 3.00 | (0.54–16.60) | 0.208 |
| ABPA vs. Aspergillus sensitized asthma | |||
| GG (reference) | 1.00 | ||
| GA | 3.02 | (1.32–6.93) | 0.009 |
| AA* | – | – | – |
| ABPA vs. Aspergillus unsensitized asthma | |||
| GG (reference) | 1.00 | ||
| GA | 2.94 | (1.39–6.25) | 0.005 |
| AA | 3.81 | (0.86–16.79) | 0.077 |
*Participants with aspergillus sensitized asthma had no AA genotype. Logistic models were used to estimate the odds ratios (ORs) and 95% confidence intervals (95% CIs) for ABPA compared with controls respectively. All statistical tests were 2-sided, and probabilities of less than 0.05 were considered to be statistically significant. ABPA: Allergic bronchopulmonary aspergillosis; –: Not avaiable.
The heterozygous S12N mutation can facilitate Af-induced type-2 responses
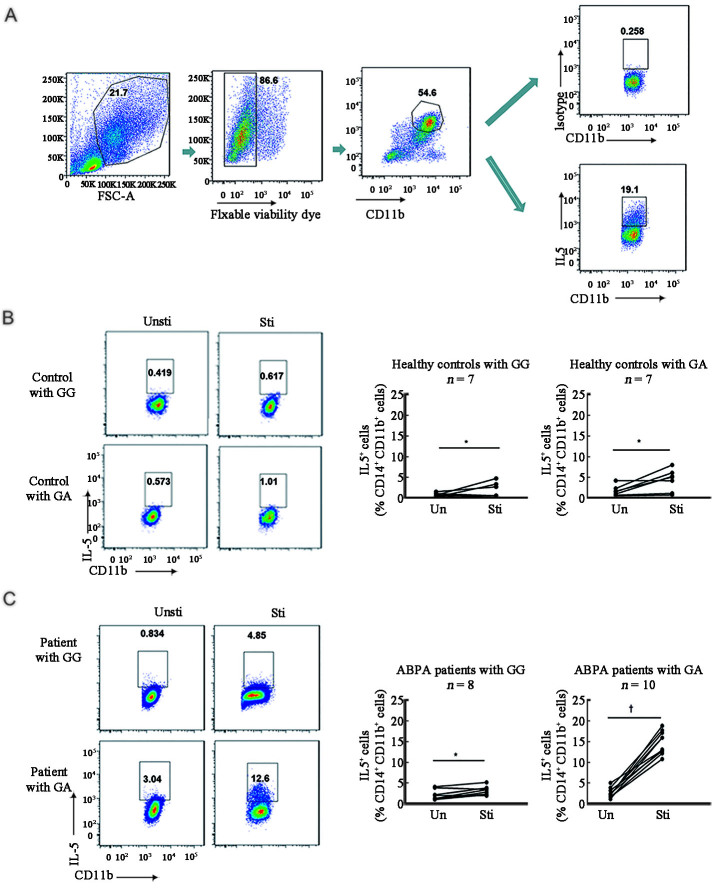
As both GA and AA genotypes of the non-synonymous SNP of the CARD9 gene were significantly associated with ABPA, it seems likely, therefore, that carrying the A allele has a dominant effect in conferring susceptibility to ABPA. To explore this, we determined whether the heterozygous CARD9 GA genotype of this SNP affected CARD9 function in patients with ABPA and healthy controls by intracellular IL-5 cytokine staining in PBMCs [Figure 1A]. There were few Af-induced IL-5-producing PBMCs (defined as live CD11b+CD14+) in samples from healthy controls (GG or GA genotypes) [Figure 1B and Supplementary Figure 1A, http://links.lww.com/CM9/B644]. By contrast, numerous Af-induced IL-5-producing PBMCs were observed in samples from ABPA patients with GA genotypes but not GG genotypes [Figure 1C and Supplementary Figure 1B, http://links.lww.com/CM9/B644]. These data imply that in ABPA patients, the A allele of S12N in CARD9 dominantly confers host susceptibility to ABPA.
Figure 1.
Heterozygous CARD9S12N facilitated Af-induced Type-2 responses. (A) Gating strategy of immunocytochemical staining of IL-5 in PBMCs by flow cytometry. (B) Intracellular levels of IL-5 in PBMCs from healthy controls with WT or heterozygous CARD9S12N mutation were determined before and after stimulation with Af by flow cytometry. (C) Intracellular levels of IL-5 in PBMCs from ABPA patients with WT or heterozygous CARD9S12N mutation were determined before and after stimulation with Af by flow cytometry. *P >0.05, †P <0.001; two-tailed paired t test was used for comparison. ABPA: Allergic bronchopulmonary aspergillosis; Af: Aspergillus fumigatus; CARD9: Caspase recruitment domain family member 9; CD: Cluster of differentiation. GA: Heterozygous CARD9S12N; GG: Wild type CARD9S12N; IL-5: Interleukin 5; n.s: No statistical significance; PBMCs: Peripheral blood mononuclear cells; Sti: Stimulation; Un/Unsti: Unstimulation; WT: Wild type.
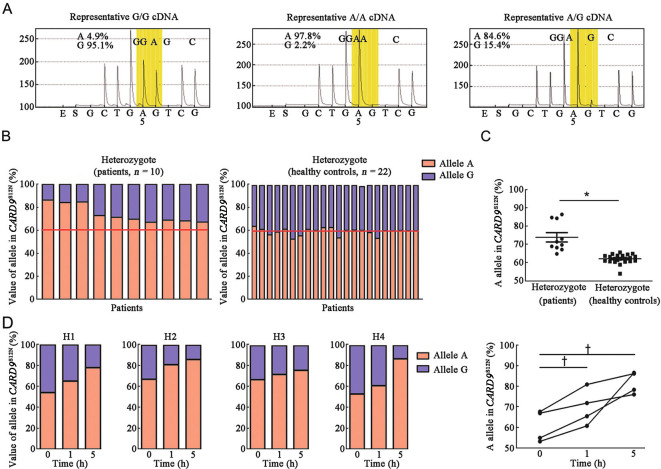
Heterozygous p.S12N mutation with allelic expression imbalance (AEI) facilitates host susceptibility to ABPA
We further sequenced cDNA in PBMCs randomly selected from 10 ABPA patients and 22 healthy controls to examine the AEI of heterozygous S12N of CARD9 [Figure 2A and Supplementary Figure 2A, http://links.lww.com/CM9/B644]. Pyrosequencing showed that healthy control subjects expressed the S12N allele and the WT allele, whereas ABPA patients showed allelic imbalance, prominently expressing the CARD9 allele with S12N mutation but not the WT allele [Figures 2B,2C and Supplementary Figure 2B, http://links.lww.com/CM9/B644]. Next, we determined whether AEI of CARD9 was dependent on Af stimulation. Our data showed that Af stimulation significantly increased the relative percentage of allele A expression from an average of 60% to 80% in four healthy controls with the GA genotype [Figure 2D and Supplementary Figure 2C, http://links.lww.com/CM9/B644], indicating that AEI of CARD9 is promoted by Af stimulation.
Figure 2.
Allelic expression imbalance of CARD9S12N confers host susceptibility to ABPA. (A) Allelic expression imbalance of CARD9S12N polymorphism was determined by pyrosequencing analysis. (B and C) Quantifications of allelle of CARD9S12N polymorphism by pyrosequencing in PBMCs from 10 ABPA patient and 22 healthy control samples. (D) PBMCs from 4 healthy control samples were isolated and challenged with Af swollen conidia. Allelic expression imbalance of CARD9S12N polymorphism was determined. *P <0.001, †P <0.05. Mann–Whitney U-test (C) or one-way ANOVA (D) was used for comparison. ABPA: Allergic bronchopulmonary aspergillosis; Af: Aspergillus fumigatus; cDNA: Complementary deoxyribonucleic acid; H: Healthy control; PBMCs: Peripheral blood mononuclear cells.
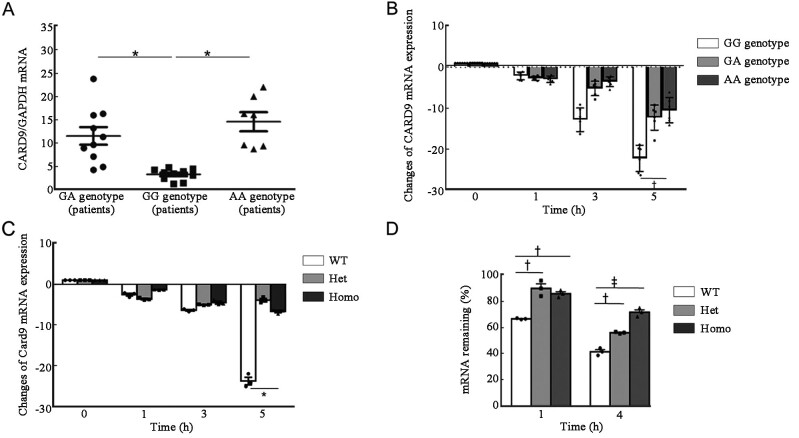
The p.S12N mutation decreased mRNA decay of CARD9 induced by Af conidia
To further assess whether the A allele of p.S12N mutation is associated with CARD9 gene expression levels, patients with ABPA were classified according to their genotypes, GG, GA, and AA, and CARD9 expression levels in their PBMCs were determined by qRT-PCR [Figure 3A]. ABPA patients with GA or AA genotypes exhibited significantly higher levels of CARD9 expression than those with the GG genotype. To further explore this phenomenon, we stimulated PBMCs from ABPA patients with GG, GA, or AA genotypes with Af conidia, and then detected CARD9 expression levels. The results showed that Af conidia stimulation dramatically decreased the mRNA abundance of CARD9 in PBMCs with the GG genotype, compared with GA or AA genotypes [Figure 3B]. We further stimulated BMDMs isolated from wild-type, heterozygous, and homozygous mice with Af conidia, and found that CardWT mRNA levels decreased more rapidly than heterozygous Card9S12N and homozygous Card9S12N [Figure 3C].
Figure 3.
CARD9S12N decreased the mRNA decay of CARD9 induced by Af. (A) qRT-PCR results for CARD9 expression, normalized to that of GAPDH, in PBMCs from ABPA patients expressing wild-type, or heterozygous or homozygous S12N variant of CARD9. (B) CARD9 expression in Af-challenged PBMCs from patients with ABPA who were wild-type, heterozygous, or homozygous for the S12N variant was determined by qRT-PCR. (C) qRT-PCR results for Af-induced Card9 expression in BMDMs isolated from wild-type, heterozygous and homozygous mice. (D) Remaining mRNA was determined by qRT-PCR 1 h and 4 h after treatment with actinomycin D (5 μg/mL) in BMDMs isolated from WT, heterozygous, and homozygous Card9S12N knock-in mice. Data are mean ± s.e.m. *P <0.001, †P <0.05, ‡P <0.01; Mann–Whitney U test (A) or two-tailed unpaired t-test (B–D) was used for comparison. ABPA: Allergic bronchopulmonary aspergillosis; Af: Aspergillus fumigatus; BMDMs: Bone marrow-derived macrophages; CARD9: Caspase recruitment domain family member 9; GAPDH: Glyceraldehyde-phosphate dehydrogenase; Het: Heterozygous; Homo: Homozygous; mRNA: Messenger RNA; PBMCs: Peripheral blood mononuclear cells; qRT-PCR: Quantitative reverse transcription-polymerase chain reaction (PCR); s.e.m: Standard error of mean; WT: Wild type.
Next, we performed Card9 mRNA stability assays in BMDMs from mice with different genotypes and found that Af stimulation significantly induced Card9 mRNA decay in BMDMs from wild-type mice relative to those from either heterozygous or homozygous Card9S12N knock-in animals [Figure 3D]. Together, these data confirm that Af stimulation can induce more mRNA decay in CardWT mice than that in heterozygous and homozygous Card9S12N mice.
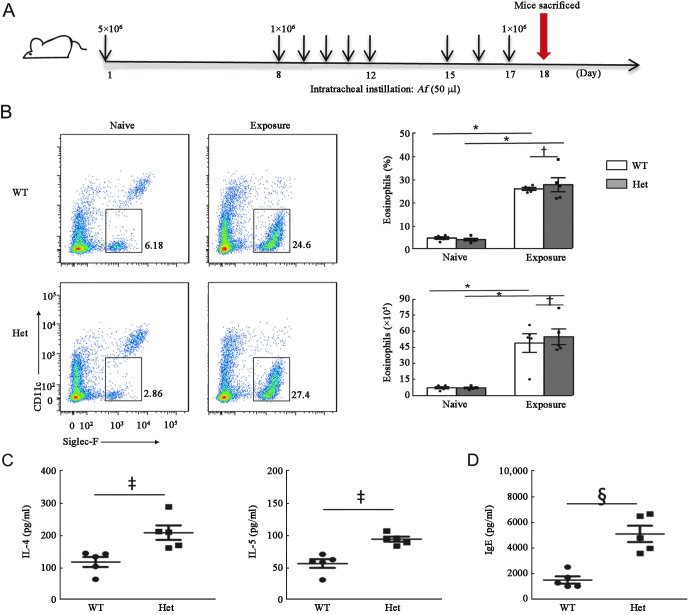
The p.S12N mutation aggravated Af-induced type-2 responses in heterozygous Card9S12N mice
The IL-4 and IL-5 levels are below the detection limit and there was no difference in IgE level in serum between naive WT and heterozygous mice [Supplementary Figure 3A, http://links.lww.com/CM9/B644]. These results showed that the S12N mutation had no substantial influence on physiological responses before fungal exposure. To determine the function of Card9S12N in the development of ABPA, we proposed repeated challenges with low doses of Af conidia to mimic persistent local antigenic stimulation [Figure 4A]. We found that no statistically significant differences in fungal burden were observed between wild-type and heterozygous Card9S12N knock-in mice [Supplementary Figure 3B, http://links.lww.com/CM9/B644]. The results also showed that Af conidia stimulation significantly induced lung eosinophilia in wild-type and heterozygous Card9S12N knock-in mice [Figure 4B]. However, protein levels of pulmonary IL-4, IL-5, and serum IgE were higher in heterozygous Card9S12N knock-in mice than those in WT mice [Figures 4C and 4D]. There was no significant difference in neutrophil accumulation between Card9WT and heterozygous Card9S12N knock-in mice [Supplementary Figure 3C, http://links.lww.com/CM9/B644]. Our data confirmed that heterozygous Card9S12N could facilitate Af-induced TH2-mediated development of ABPA symptoms.
Figure 4.
Heterozygous Card9S12N can facilitate Af-induced TH2-mediated responses. (A) Strategy for development of the chronic murine asthma model, in which WT and heterozygous Card9S12N knock-in mice were sensitized with 5 × 106 Af conidia, challenged with 1 × 106 Af conidia for eight times, and sacrificed for subsequent assay at day 18. (B) Eosinophil (Siglec-F+) counts in lungs of the mice after multiple exposures to Af conidia. (C and D) ELISA results for lung IL-4 and IL-5 (C), and serum IgE (D), in the mice described above. Data are shown as mean ± s.e.m. *P <0.001, †P >0.05, ‡P <0.05, §P <0.01; two-tailed unpaired t-test was used for comparison (B–D). In B–D, n = 5 mice per group. Af: Aspergillus fumigatus; CARD9: Ccaspase recruitment domain family member 9; CD: Cluster of differentiation; ELISA: Enzyme linked immunosorbent assay; Het: Heterozygote; IgE: Immunoglobulin E; IL: Interleukin; s.e.m.: Standard error of mean; TH2: T helper 2: WT: Wild type.
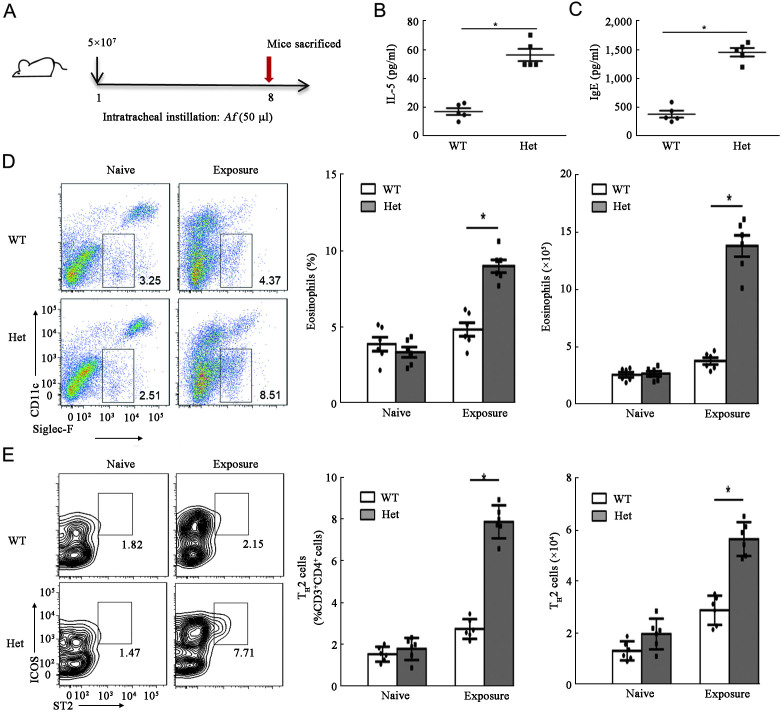
High dose exposure of Af induced TH2-mediated responses in heterozygous Card9S12N mice
Furthermore, our recent study demonstrated that a challenge with a low dose of Af conidia failed to induce TH2-mediated responses in wild-type and heterozygous Card9S12N knock-in mice, whereas a high dose of Aspergillus infection always caused an acute lung inflammatory response.[20,26] Therefore, in this study, we administered intratracheal injections with high doses of Af conidia to determine whether heterozygous Card9S12N is critical for directing Af-induced TH2 polarization [Figure 5A]. We observed that the levels of pulmonary IL-5 and serum IgE were significantly higher in the heterozygous Card9S12N knock-in mice compared with those in wild-type controls [Figures 5B and 5C]; the number and percentage of eosinophils were increased more in the heterozygous Card9S12N knock-in mice than in Card9WT [Figure 5D]. However, there was no significant difference in Af-induced neutrophil accumulation in the comparison between Card9WT and heterozygous Card9S12N mice [Supplementary Figure 4A, http://links.lww.com/CM9/B644]. Quantification of lung TH2 lymphocytes, which are defined here as live SSClowCD3+CD4+ICOS+ST2+ cells[27] [Supplementary Figure 4B, http://links.lww.com/CM9/B644], revealed significantly higher levels of these immune cells in the lungs of heterozygous Card9S12N knock-in mice than those of wild-type mice, after challenged with Af conidia [Figure 5E]. These results indicate that heterozygous Card9S12N can facilitate Af-induced TH2 differentiation.
Figure 5.
TH2-mediated responses can be induced by high-dose aspergillus conidia in heterozygous Card9S12N mice. (A) Eosinophil (SiglecF+) counts in lungs from Card9WT and heterozygous Card9S12N knock-in mice after a single exposure to Af conidia (5 × 107) assayed at day 8 by flow cytometry. (B and C) ELISA results for IL-5 (B) in extracts of lung homogenates and serum IgE (C) levels from Card9WT and heterozygous Card9S12N knock-in mice. (D and E): Eosinophils (D) and TH2 (E) cell counts (CD3+CD4+ICOS+ST2+) in the lungs of Card9WT and heterozygous Card9S12N knock-in mice. Data are shown as means ± SEM. Two-tailed unpaired t-test was used for comparison, *P <0.001 (B–E). In B–C, n = 5 (one sacrificed during infection); In D–E, n = 6 mice per group. Af: Aspergillus fumigatus; CARD9: Caspase recruitment domain family member 9; CD: Cluster of differentiation; ELISA: Enzyme linked immunosorbent assay; Het: Heterozygote; ICOS: Inducible costimulator; IgE: Immunoglobulin E; IL: Interleukin; SEM: Standard error of mean; ST2: Growth stimulation expressed gene 2; WT: Wild type.
Discussion
The inflammatory response of ABPA is characterized by TH2 responses to Aspergillus allergens that stimulate immunoglobulin E (IgE) synthesis, eosinophil activation and the production of IL-4, IL-5, and IL-13 cytokines rather than TH1 molecules such as IL-12 and interferon-γ (IFN-γ). Recent studies indicate that ABPA patients carry genetic risk factors that contribute to skewed and heightened TH2 responses to Af antigens.[28] Our results demonstrate that the non-synonymous SNP in CARD9, rs4077515, encoding the S12N amino acid substitution, favors ABPA development in patients with heterozygous and homozygous genotypes by orchestrating CARD9S12N-mediated signaling to induce TH2 responses. Our studies provide the genetic and functional evidence that indicates the role of S12N polymorphism in CARD9 in regulating immune responses to the development of ABPA. As the frequency of the rs4077515 AA genotype in CARD9 was significantly lower in the Chinese Han population with Aspergillus unsensitized asthma, the prediction of this polymorphism for ABPA did not have enough power, and the differences between ABPA patients and Aspergillus unsensitized asthma patients need a larger number of samples to identify.
To date, more than 24 mutations in CARD9 have been reported to be associated with severe fungal infections.[29] Among these, two loss-of-function homozygous mutations, Q289X (c.865C >T) and Q295X (c.883C >T), in CARD9 are associated with an elevated risk of candidiasis and dermatophytosis, respectively.[16,17] Besides deleterious mutations predisposing to fungal infection, our previous and present studies show that a genetic variant in CARD9 is critical for the development of Af-induced TH2 responses.[20] Our data show that ABPA patients with heterozygous or homozygous genetype of CARD9S12N showed significantly higher CARD9 expression levels than those with wild-type CARD9, hinting that the S12N variant induces the occurrence of AEI in CARD9.
AEI is an indicator of the effects of regulatory polymorphisms residing within a gene locus and is defined by the percentage levels of RNA alleles expressed, and determined by the measurement of cDNA. Epigenetic mechanisms are fundamental players that determine the allele expression. DNA methylation and histone modifications are two mediators of the epigenetic phenomena. Non-coding RNAs (ncRNAs) are also players in regulating gene expression.[30,31] For example, allelic imbalance of variant rs1047643 and B-cell-specific super-enhancer was associated with systemic lupus erythematosus mediated by signal transducer and activator of transcription 3 (STAT3).[32] Besides epigenetic events, AEI exists in autosomal non-imprinted genes and about 4.6% of heterozygous SNP-sample pairs show AEI.[33] However, in most cases, functional significance resulting from AEI and the causes of AEI phenomenon are still unknown. We observed that 10 samples from ABPA patients with heterozygous CARD9S12N showed AEI, with dominant expression of the minor A allele. Furthermore, we showed that AEI of CARD9 was dependent on Af stimulation. Heterozygous Y91H (c.439T >C) mutation of CARD9 has also been reported to be associated with AEI, resulting in CARD9 deficiency in two patients with spontaneous central nervous system candidiasis.[34] Taken together, our results identify the S12N SNP as a major regulatory variant in CARD9, accounting for the AEI observed in this study.
Our study has some limitations. First, we did not collect bone marrow-derived dendritic cells (BMDC) from asthma patients but this doesn't affect our conclusion. Then, the samples of ABPA patients are limited because of the low morbidity. Third, in most cases, functional significance resulting from AEI and the causes of AEI phenomenon are still unknown.
In summary, we reported that a SNP of CARD9 gene encoding S12N is a genetic risk factor for human ABPA disease. Further, we demonstrated that Af-induced AEI of the minor CARD9S12N allele can contribute to autosomal dominant functional alterations, favoring TH2-mediated ABPA development. This study provides genetic evidence to show that the heterozygous mutation of CARD9S12N, followed by AEI of CARD9S12N, favors the development of ABPA. It could be a good screening factor in determining patient susceptibility to ABPA.
Acknowledgments
The authors acknowledge the patients who kindly participated in this study.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Nos. 81925001, 81970036, and 31970889), the Innovation Program of Shanghai Municipal Education Commission (Nos. 202101070007-E00097 and 201901070007E00022), the Program of Shanghai Municipal Science and Technology Commission (No. 21DZ2201800), the Shanghai Municipal Health Commission (Nos. 201740019 and ZY2018-2020 FWTX3022), and Innovative Research Ream of High-Level Local Universities in Shanghai.
Conflicts of interest
None.
Supplementary Material
Footnotes
Xia Xu, Haiwen Lu, and Jianxiong Li contributed equally to this study.
How to cite this article: Xu X, Lu HW, Li JX, Duan JL, Wang ZW, Yang JW, Gu SY, Luo RG, Liang S, Tang W, Zhang FY, Hang JQ, Ge J, Lin X, Qu JM, Jia XM, Xu JF. Heterozygous CARD9 mutation favors the development of allergic bronchopulmonary aspergillosis. Chin Med J 2023;136:1949–1958. doi: 10.1097/CM9.0000000000002786
References
- 1.Greenberger PA. Allergic bronchopulmonary aspergillosis. J Allergy Clin Immunol 2002; 110: 685–692. doi: 10.1067/mai.2002.130179. [DOI] [PubMed] [Google Scholar]
- 2.Knutsen AP, Slavin RG. Allergic bronchopulmonary aspergillosis in asthma and cystic fibrosis. Clin Dev Immunol 2011; 2011: 843763. doi: 10.1155/2011/843763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denning DW, Pleuvry A, Cole DC. Global burden of allergic bronchopulmonary aspergillosis with asthma and its complication chronic pulmonary aspergillosis in adults. Med Mycol 2013; 51: 361–370. doi: 10.3109/13693786.2012.738312. [DOI] [PubMed] [Google Scholar]
- 4.Kauffman HF. Immunopathogenesis of allergic bronchopulmonary aspergillosis and airway remodeling. Front Biosci 2003; 8: e190–196. doi: 10.2741/990. [DOI] [PubMed] [Google Scholar]
- 5.Knutsen AP, Hutchinson PS, Albers GM, Consolino J, Smick J, Kurup VP. Increased sensitivity to IL-4 in cystic fibrosis patients with allergic bronchopulmonary aspergillosis. Allergy 2004; 59: 81–87. doi: 10.1046/j.1398-9995.2003.00129.x. [DOI] [PubMed] [Google Scholar]
- 6.Chauhan B, Ap K, Hutcheson PS, Slavin RG, Bellone CJ. T cell subsets, epitope mapping, and HLA-restriction in patients with allergic bronchopulmonary aspergillosis. J Clin Invest 1996; 97: 2324–2331. doi: 10.1172/JCI118675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chauhan B Santiago L Kirschmann DA Hauptfeld V Knutsen AP Hutcheson PS, et al. The association of HLA-DR alleles and T cell activation with allergic bronchopulmonary aspergillosis. J Immunol 1997; 159: 4072–4076. [PubMed] [Google Scholar]
- 8.Risma KA Wang N Andrews RP Cunningham CM Ericksen MB Bernstein JA, et al. V75R576 IL-4 receptor alpha is associated with allergic asthma and enhanced IL-4 receptor function. J Immunol 2002; 169: 1604–1610. doi: 10.4049/jimmunol.169.3.1604. [DOI] [PubMed] [Google Scholar]
- 9.Brouard J Knauer N Boelle PY Corvol H Henrion-Caude A Flamant C, et al. Influence of interleukin-10 on Aspergillus fumigatus infection in patients with cystic fibrosis. J Infect Dis 2005; 191: 1988–1991. doi: 10.1086/429964. [DOI] [PubMed] [Google Scholar]
- 10.Saxena S, Madan T, Shah A, Muralidhar K, Sarma PU. Association of polymorphisms in the collagen region of SP-A2 with increased levels of total IgE antibodies and eosinophilia in patients with allergic bronchopulmonary aspergillosis. J Allergy Clin Immunol 2003; 111: 1001–1007. doi: 10.1067/mai.2003.1395. [DOI] [PubMed] [Google Scholar]
- 11.Miller PW Hamosh A Macek M Jr Greenberger PA MacLean J Walden SM, et al. Cystic fibrosis transmembrane conductance regulator (CFTR) gene mutations in allergic bronchopulmonary aspergillosis. Am J Hum Genet 1996; 59: 45–51. [PMC free article] [PubMed] [Google Scholar]
- 12.Gross O Gewies A Finger K Schäfer M Sparwasser T Peschel C, et al. Card9 controls a non-TLR signalling pathway for innate anti-fungal immunity. Nature 2006; 442: 651–656. doi: 10.1038/nature04926. [DOI] [PubMed] [Google Scholar]
- 13.Jia XM Tang B Zhu LL Liu YH Zhao XQ Gorjestani S, et al. CARD9 mediates Dectin-1-induced ERK activation by linking Ras-GRF1 to H-Ras for antifungal immunity. J Exp Med 2014; 211: 2307–2321. doi: 10.1084/jem.20132349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu YM Zhang Y You Y Wang D Li H Duramad O, et al. The adaptor protein CARD9 is required for innate immune responses to intracellular pathogens. Nat Immunol 2007; 8: 198–205. doi: 10.1038/ni1426. [DOI] [PubMed] [Google Scholar]
- 15.Vornholz L, Ruland J. Physiological and Pathological Functions of CARD9 Signaling in the Innate Immune System. Curr Top Microbiol Immunol. 2020; 429: 177–203. doi: 10.1007/82_2020_211. [DOI] [PubMed] [Google Scholar]
- 16.Glocker EO Hennigs A Nabavi M Schäffer AA Woellner C Salzer U, et al. A homozygous CARD9 mutation in a family with susceptibility to fungal infections. N Engl J Med 2009; 361: 1727–1735. doi: 10.1056/NEJMoa0810719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lanternier F Pathan S Vincent QB Liu L Cypowyj S Prando C, et al. Deep dermatophytosis and inherited CARD9 deficiency. N Engl J Med 2013; 369: 1704–1714. doi: 10.1056/NEJMoa1208487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhernakova A Festen EM Franke L Trynka G van Diemen CC Monsuur AJ, et al. Genetic analysis of innate immunity in Crohn's disease and ulcerative colitis identifies two susceptibility loci harboring CARD9 and IL18RAP. Am J Hum Genet 2008; 82: 1202–1210. doi: 10.1016/j.ajhg.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janse M Lamberts LE Franke L Raychaudhuri S Ellinghaus E Muri Boberg K, et al. Three ulcerative colitis susceptibility loci are associated with primary sclerosing cholangitis and indicate a role for IL2, REL, and CARD9. Hepatology 2011; 53: 1977–1985. doi: 10.1002/hep.24307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu X Xu JF Zheng G Lu HW Duan JL Rui W, et al. CARD9(S12N) facilitates the production of IL-5 by alveolar macrophages for the induction of type 2 immune responses. Nat Immunol 2018; 19: 547–560. doi: 10.1038/s41590-018-0112-4. [DOI] [PubMed] [Google Scholar]
- 21.Agarwal R Chakrabarti A Shah A Gupta D Meis JF Guleria R, et al. Allergic bronchopulmonary aspergillosis: review of literature and proposal of new diagnostic and classification criteria. Clin Exp Allergy 2013; 43: 850–873. doi: 10.1111/cea.12141. [DOI] [PubMed] [Google Scholar]
- 22.FitzGerald JM Reddel HK Hurd S Bateman ED Boulet LP Becker A, et al. Global Initiative for Asthma. Global Strategy for asthma management and prevention–Update 2015. Available from: www.ginasthma.com. [Last accessed on April 15, 2015].
- 23.Harrington CT, Lin EI, Olson MT, Eshleman JR. Fundamentals of pyrosequencing. Arch Pathol Lab Med 2013; 137: 1296–1303. doi: 10.5858/arpa.2012-0463-RA. [DOI] [PubMed] [Google Scholar]
- 24.Zhu LL Zhao XQ Jiang C You Y Chen XP Jiang YY, et al. C-type lectin receptors Dectin-3 and Dectin-2 form a heterodimeric pattern-recognition receptor for host defense against fungal infection. Immunity 2013; 39: 324–334. doi: 10.1016/j.immuni.2013.05.017. [DOI] [PubMed] [Google Scholar]
- 25.Fei M Bhatia S Oriss TB Yarlagadda M Khare A Akira S, et al. TNF-alpha from inflammatory dendritic cells (DCs) regulates lung IL-17A/IL-5 levels and neutrophilia versus eosinophilia during persistent fungal infection. Proc Natl Acad Sci U S A 2011; 108: 5360–5365. doi: 10.1073/pnas.1015476108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo XL Li JX Huang HR Duan JL Dai RX Tao RJ, et al. LL37 Inhibits Aspergillus fumigatus Infection via Directly Binding to the Fungus and Preventing Excessive Inflammation. Front Immunol 2019; 10: 283. doi: 10.3389/fimmu.2019.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toussaint M Jackson DJ Swieboda D Guedán A Tsourouktsoglou TD Ching YM, et al. Host DNA released by NETosis promotes rhinovirus-induced type-2 allergic asthma exacerbation. Nat Med 2017; 23: 681–691. doi: 10.1038/nm.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poore TS, Hong G, Zemanick ET. Fungal Infection and Inflammation in Cystic Fibrosis. Pathogens 2021; 10: 618. doi: 10.3390/pathogens10050618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaezi A Fakhim H Abtahian Z Khodavaisy S Geramishoar M Alizadeh A, et al. Frequency and Geographic Distribution of CARD9 Mutations in Patients With Severe Fungal Infections. Front Microbiol 2018; 9: 2434. doi: 10.3389/fmicb.2018.02434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Massah S, Beischlag TV, Prefontaine GG. Epigenetic events regulating monoallelic gene expression. Crit Rev Biochem Mol Biol 2015; 50: 337–358. doi: 10.3109/10409238.2015.1064350. [DOI] [PubMed] [Google Scholar]
- 31.Marion-Poll L Forêt B Zielinski D Massip F Attia M Carter AC, et al. Locus specific epigenetic modalities of random allelic expression imbalance. Nat Commun 2021; 12: 5330. doi: 10.1038/s41467-021-25630-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang YF, Day K, Absher DM. STAT3-mediated allelic imbalance of novel genetic variant Rs1047643 and B-cell-specific super-enhancer in association with systemic lupus erythematosus. eLife. 2022; 11: e72837. doi: 10.7554/eLife.72837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heap GA Yang JH Downes K Healy BC Hunt KA Bockett N, et al. Genome-wide analysis of allelic expression imbalance in human primary cells by high-throughput transcriptome resequencing. Hum Mol Genet 2010; 19: 122–134. doi: 10.1093/hmg/ddp473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gavino C Hamel N Zeng JB Legault C Guiot MC Chankowsky J, et al. Impaired RASGRF1/ERK-mediated GM-CSF response characterizes CARD9 deficiency in French-Canadians. J Allergy Clin Immunol 2016; 137: 1178–1188.e7. doi: 10.1016/j.jaci.2015.09.016. [DOI] [PubMed] [Google Scholar]