Abstract
A high prevalence (42.6%) of hepatitis B virus (HBV) infection was suspected in 195 formerly captive orangutans due to a large number of serum samples which cross-reacted with human HBV antigens. It was assumed that such viral infections were contracted from humans during captivity. However, two wild orangutans were identified which were HBV surface antigen positive, indicating that HBV or related viruses may be occurring naturally in the orangutan populations. Sequence analyses of seven isolates revealed that orangutans were infected with hepadnaviruses but that these were clearly divergent from the six known human HBV genotypes and those of other nonhuman hepadnaviruses reported. Phylogenetic analyses revealed geographic clustering with Southeast Asian genotype C viruses and gibbon ape HBV. This implies a common origin of infection within this geographic region, with cross-species transmission of hepadnaviruses among hominoids.
Orangutans (Pongo pygmaeus) are the only great apes found outside Africa. The wild populations are restricted to the islands of Borneo and Sumatra. Orangutans are highly endangered as a result of poaching and the widespread destruction of their habitats. In an approach to save this great ape from extinction, orphaned juveniles and confiscated pet animals are housed at centers established to rehabilitate orangutans. Increasing human encroachment on rainforest habitat and fragmentation of declining populations increases the interactions and consequently the risks of disease transmission between wild primates and human populations (16). The accumulation of relatively solitary orangutans at reintroduction centers also increases the potential of transmission of viral pathogens, either of orangutan or human origin. Due to a high incidence of morbidity and mortality in these captive animals, concern arose that certain infectious diseases not found in orangutans had been acquired from humans.
We recently undertook a serological survey of orangutans to determine if certain human pathogens may have been transmitted to orangutans. Of the human viruses tested, by far the most orangutan sera cross-reacted with human hepatitis B virus (HBV) antigens (15). The distribution of HBV is worldwide, and there are six different genotypic groups characterized to date, designated A to F. Each genomic group has a characteristic geographic distribution in human populations (7, 12). Group A genomes are found predominantly in northern Europe and sub-Saharan Africa. Group B and C genomes are confined to human populations in the Far East. In the Mediterranean and the Near and Middle East, group D genomes are found, while group E is indigenous to West Africa. Group F has its origins in aboriginal populations of the New World (10). Because HBV is highly endemic in Southeast Asia (6) and because of the close contact between humans and captive orangutans, it was assumed that these animals were infected with human HBV. This study aimed to characterize this virus infection of orangutans in order to determine its origin.
Serum samples were obtained from orangutans (P. pygmaeus) that were housed at the Wanariset Orangutan Reintroduction Center in East Kalimantan, Indonesia. Samples were assayed for antibodies to HBV core antibody (HBcAb) and surface antibody (HBsAb), using commercial enzyme-linked immunosorbent assays (ELISA) (Wellcozyme anti-HBs and Wellcozyme anti-HBc IgM and IgG kits; Murex Diagnostic Ltd., Dartford, United Kingdom). HBV surface antigen (HBsAg) was detected by using the Murex HBsAg kit (Murex Diagnostic). All assays were performed according to the manufacturer’s instructions. All serum samples were assayed at least twice to ensure consistent results.
Serum samples from 195 orangutans were examined for evidence of HBV infection, and multiple samples from different time points were studied from most individuals. The results indicated that 42.6% (n = 83) of the individuals had evidence of exposure to HBV or a related virus (Table 1). Twenty-eight animals were surface antibody positive upon arrival, suggesting a past infection with HBV. Fifty-five individuals showed evidence of active virus infection, as these were positive in the HBsAg assay at one or more sampling dates. While housed at the center, 40 animals converted from HBsAg positive to HBsAb positive; 15 animals remained HBsAg positive for longer than 1 year. These orangutans were considered chronically infected. Of these, four individuals had been antigen positive for more than 4 years and were defined as chronic carriers. New infections at the reintroduction center were documented in only two instances (14a). Two of three orangutans which were caught directly from the wild were found to be HBsAb positive, suggesting infection with an indigenous virus.
TABLE 1.
ELISA and diagnostic PCR screening of orangutan sera
| HBV status | No. of individuals |
|---|---|
| Exposure to HBV (positive in HBsAb and/or HBsAg ELISA) | 83 |
| HBsAg-positive individuals | 55 |
| HBsAb-positive individuals upon arrival | 28 |
| Infected with HBsAg >1 year (chronic carriers) | 15 |
| HBsAg positive for >4 years | 4 |
| PCR-confirmed viremia | 32 |
Confirmation of infection with an HBV-like virus was obtained by amplification of HBV core gene sequences from viral DNA isolated from sera of HBsAg-positive animals. DNA was isolated from 100 μl of serum by a modified RNA isolation procedure (13). HBV-related sequences were detected using a diagnostic PCR assay (detection limit, 250 genomes/ml) essentially as described by Zaaijer et al. (17), which amplified a 521-bp fragment from the HBV core protein gene. The PCR products were analyzed on a 1.8% agarose gel. Samples from each individual were examined at least twice in this assay. Of the 55 HBsAg-positive individuals screened by this assay, 32 were positive by this PCR, confirming infection of these great apes with an HBV-like virus.
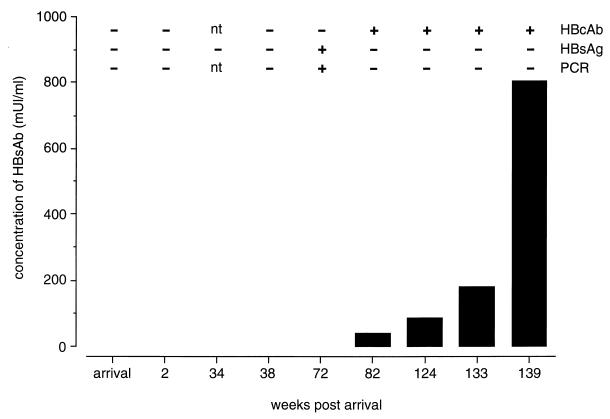
Since sequential serum samples from the animals were not available, a comprehensive picture of the kinetics of this viral infection is difficult to piece together. Furthermore, the exact time point of infection cannot be determined in most cases. However, the fragmented data available from a number of animals suggested that this infection and the hosts’ responses follow trends similar to those observed in HBV-infected humans (3). Such was the case for one orangutan, Abau, which acquired infection while at the center. During his stay, four samples tested negative before the first detection of HBsAg in serum. There was an 8-month period between the last negative sample and the first HBsAg- and PCR-positive serum samples. Two months later this animal developed HBcAb and HBsAb, which coincided with the time the serum became HBsAg and PCR negative (Fig. 1).
FIG. 1.
Antibody responses to HBV and detection of viral surface antigen and genomic DNA in sequential serum samples of an orangutan, Abau. Levels of HBsAb were quantitated and calculated as milliunits per milliliter. Negative and positive results and time points not tested (nt) in HBcAb, HBsAg, and diagnostic PCR assays are indicated.
To determine if liver pathology was associated with this infection, serum alanine-aminotransferase (ALT) levels were examined in the sera of 18 individuals, including 8 chronic carriers, 4 antibody-positive individuals, and 6 individuals that were seronegative for HBV. Two to three time points were evaluated for all chronic carriers, and for two individuals 7 and 9 successive time points were available for testing, respectively. In all cases, the ALT levels from infected individuals were within normal limits by comparison with unexposed animals (10 to 44 U/liter), with one exception (83 U/liter). In the latter case, this animal was HBV PCR positive and had a record of being HBsAb negative followed by seroconversion 4 months later. Furthermore, a liver section obtained from a chronic carrier (Mojo) that had died of a cause unrelated to HBV revealed no histological evidence of HBV-related pathology. Our findings do not suggest evidence of an overt hepatitis caused by this infection in these animals. However, only a limited number of orangutans were tested. Future investigation using more animals and using samples taken just after infection are needed to draw a more definitive conclusion.
To determine if the virus that was detected by serology and PCR was of human origin, we amplified and sequenced the S gene, which is known to be characteristic for the different HBV genotypes. Oligonucleotide primers hepB-SF1 and BR were designed to bind to sequences conserved among HBV strains. Primers hepB-SF1 and BR were used for the amplification of a 1,359-bp DNA fragment encompassing the entire S gene. PCR was performed in a 50-μl volume, using 10 μl of DNA, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 0.01% bovine serum albumin, 50 pmol of each primer, 0.2 mM (each) deoxynucleoside triphosphate, 2.5 mM MgCl2, and 2 U of AmpliTaqGold (Perkin Elmer). Samples were preheated for 15 min at 95°C to activate the enzyme and then cycled for 15 s at 95°C, 15 s at 55°C, and 2 min at 72°C for 30 rounds of amplification. The PCR products were isolated from agarose gel, using a QIAquick gel extraction kit (Qiagen, Hilden, Germany), and cloned in the pGEM-T Easy vector (Promega Corporation, Madison, Wis.). Sequencing of the cloned insert was performed with pUC/M13 forward and reverse sequencing primers which bind on either side of the cloned insert and with HBV-specific oligonucleotides. Analysis was done on an ABI PRISM 310 Genetic Analyzer (Perkin Elmer). Primers used for PCR and sequence analysis and their positions in the chimpanzee HBV genome are given in Table 2.
TABLE 2.
Oligonucleotide primers used for amplification and sequence analysis of the OHV S gene sequence
| Primer | Sequence (5′ to 3′) | Positiona |
|---|---|---|
| hepB-SF1 | TGYGGGTCACCWTATTCTTGGG | 912–932 |
| hepB-SRin | CCTGAGCCTGAGGGCTC | 1145–1161 |
| hepB-SRout | CACTGTTCCTGAACTGGAGC | 1348–1367 |
| BF | CTTCTTRTTGGTTCTTCTGG | 1712–1731 |
| BR | CAYACTTTCCARTCWATAGG | 2252–2271 |
| AF | CGAVGAYTGGGGACCCTG | 1411–1428 |
| AR | CCTGGAAKTAGAGGACAAACG | 1751–1771 |
| preR | AACCCCGCCTGTAACACGAG | 1473–1492 |
Position on chimpanzee HBV isolate LSH (GenBank accession no. D00220).
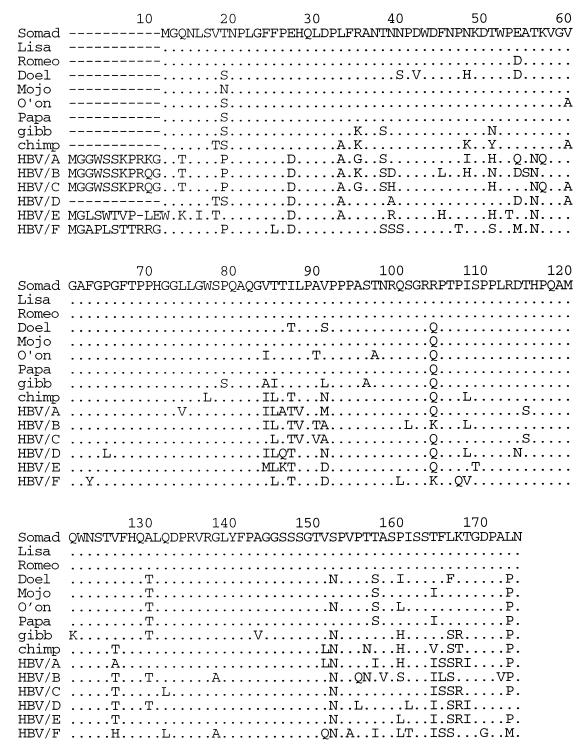
We first determined the sequence of the entire S gene, including the pre-S region, of the virus infecting the chronic carrier Somad (SO). Sequence analysis of the pre-S and S gene sequences was performed using MacVector 6.0 and AssemblyLIGN software packages (Oxford Molecular Ltd.). The nucleotide sequence of the SO isolate showed similarities ranging from 84.1 to 92.6% with published HBV S gene sequences (Table 3). The highest degree of similarity was found with viruses isolated from gibbon (9), chimpanzee (14), and human HBV genotype D (92.6, 90.4, and 89.8% similarity, respectively). This is primarily due to a 33-bp deletion in the pre-S region that is characteristic of these HBV strains. This feature is illustrated in Fig. 2 in which the deduced pre-S amino acid sequence of the SO isolate and six other isolates are aligned with the same regions of the HBV isolates from chimpanzee, gibbon, and representatives of all known human genotypes (A to F). In Fig. 3, the small S proteins of these HBV strains are compared with those of the orangutan isolates. From Fig. 2 and 3 it is evident that the viruses infecting orangutans are related to HBV but that they have multiple amino acid residues which distinguish them from HBV. In the pre-S region they have three unique amino acid residues (Thr85, Val/Ser91, and Leu/Phe166), while the small S protein contains six unique residues (Ser/Leu5, Leu56, Val118, Ser127, Pro129, and Ala224). Division into genotypes and serotypes is based on specific amino acid residues in the small S protein (1, 5, 7, 8, 12). The hepadnaviruses infecting orangutans thus belong to the ayw or ayr serotypes, as an arginine (R) residue can be found at position 122 and a lysine (K) or arginine (R) is at position 160. However, genotype w1 to w4 subtyping was not possible as the S proteins contain a unique serine (S) residue at position 127 (Fig. 3). The analysis of nucleotide and amino acid sequences consistently points to the possibility that orangutans are infected with a hepadnavirus distinct from the known human HBV types.
TABLE 3.
Percentage sequence similarity of HBV SO S gene with other HBV strains
| SO | GIBBa | CHb | Ac | Bd | Ce | Df | Eg | Fh | |
|---|---|---|---|---|---|---|---|---|---|
| SO | 100 | 92.6 | 90.4 | 88.4 | 86.5 | 88 | 89.8 | 87.6 | 84.1 |
| Gibbon | 100 | 91.8 | 88.2 | 86.6 | 88.6 | 89.9 | 88.3 | 84.6 | |
| Chimpanzee | 100 | 89.0 | 88.2 | 88.3 | 92.5 | 90.6 | 85.4 | ||
| A | 100 | 90.9 | 92.8 | 88.0 | 89.8 | 86.0 | |||
| B | 100 | 91.0 | 88.0 | 88.5 | 85.4 | ||||
| C | 100 | 88.0 | 89.3 | 86.2 | |||||
| D | 100 | 90.7 | 84.4 | ||||||
| E | 100 | 85.3 | |||||||
| F | 100 |
Gibbon HBV (GenBank no. U46935).
Chimpanzee HBV (GenBank no. D00220).
Human HBV genotype A (GenBank no. M57663).
Human HBV genotype B (GenBank no. M54923).
Human HBV genotype C (GenBank no. X01587).
Human HBV genotype D (GenBank no. V01460).
Human HBV genotype E (GenBank no. X75664).
Human HBV genotype F (GenBank no. X75663).
FIG. 2.
Alignment of the pre-S protein sequence from seven orangutan isolates with sequences from viruses isolated from gibbon (gibb), chimpanzee (chimp), and representatives of human HBV genotypes A to F (HBV/A to HBV/F). Identical amino acid residues are marked by a dot, and deletions are indicated by a dash.
FIG. 3.
Alignment of the small S protein of seven orangutan isolates with those from gibbon HBV (gibb), chimpanzee HBV (chimp), and representatives of human HBV genotypes A to F (HBV/A to HBV/F). Identical amino acid residues are marked by a dot.
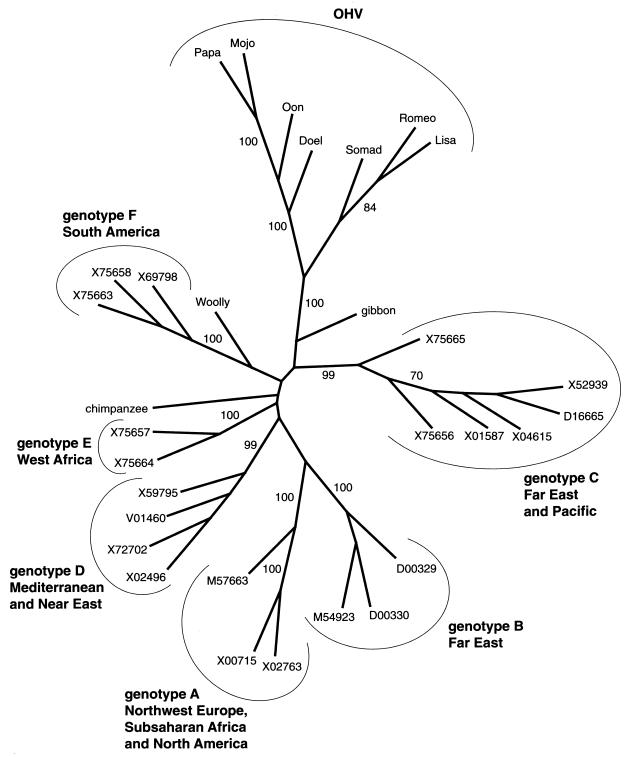
Phylogenetic analysis of S gene sequences was performed using the PHYLIP package, version 3.572 (2). The PHYLIP program SEQBOOT was used to bootstrap data in which 100 data sets were analyzed. DNAPARS (maximum parsimony) and NEIGHBOR (neighbor joining) were used to create dendrograms. CONSENSE was used to create consensus trees which were visualized using DRAWTREE. In Fig. 4 we have depicted the results of the neighbor-joining analysis. In this phylogenetic tree, the orangutan viruses form a cluster separated from the six known human HBV genotype clades and from the chimpanzee and woolly monkey (4) hepadnaviruses. Formation of a separate orangutan hepadnavirus (OHV) cluster is supported by a high bootstrap value of 100%. Consistent results were also obtained with maximum parsimony analysis (data not shown). Closest to the orangutan cluster is the HBV isolated from gibbons, a primate species that shares habitat with orangutans. The human genotype C viruses, which also originate from Southeast Asia, are the human HBV most closely related to the OHVs. This suggests a common ancestor of hepadnavirus in Southeast Asia that by cross-species transmission has spread through humans and apes in this geographic region. The time of divergence between the orangutan viruses and the human genotype C viruses remains unclear as the rate of variation of hepadnaviruses in different hominoids is not exactly defined. However, a recent zoonotic event causing this hepadnavirus infection in orangutans is highly unlikely. Ogata et al. (11) report on the transmission of HBV to another great ape, the chimpanzee. In this comparable situation, no mutations were found in viruses recovered from the chimpanzees after several rounds of replication in this new host. In addition, orangutans are solitary animals with limited contact with other individuals in the wild. This situation may preclude a rapid spread of viruses among the wild orangutan populations. The combination of both of these factors argues against a rapid accumulation of the mutational changes observed in these orangutan viruses within their recent natural history.
FIG. 4.
Relationship between OHV with HBV isolated from humans and hepadnaviruses isolated from chimpanzees, gibbons, and woolly monkeys (Woolly). The tree represents an unrooted consensus tree obtained by the neighbor-joining method based on the nucleotide sequence of the small S protein. Bootstrap analysis was applied using 100 values. Values on the branches represent the percentage of trees for which the sequences at one end of the branch are a monophyletic group. The OHV clade as well as the different HBV genotypes and their geographic origins are indicated. The human HBV isolates are identified by their GenBank database accession numbers. Accession numbers for chimpanzee and gibbon HBV are described elsewhere. The accession number of the woolly monkey hepadnavirus is AF046996. Sequence data of OHV isolates described in this article are deposited in the EMBL and GenBank data libraries.
The data compiled to date reveal that orangutans are infected with a novel hepadnavirus that is distinct but related to human HBV. In those cases possible to observe, laboratory test have not revealed evidence of actual hepatitis in animals infected with OHV. However, the impact of OHV on the survival of existing wild orangutan populations remains to be determined.
Nucleotide sequence accession numbers.
Sequence data of OHV isolates described in this article are deposited in the EMBL and GenBank data libraries. The accession numbers are Y17559, Y17560, Y17561, Y17562, Y17563, Y17564, and Y17565.
Acknowledgments
We thank the technicians at the Wanariset Orangutan Reintroduction Project for assistance in sample collection and the staff of the Wanariset I Samboja Research Station, especially A. Susilo, W. Smits, and T. de Kam. Thanks also to the Indonesian Institute for Scientific Research for enabling research at the center. The support and technical assistance of Henk Niphuis, Nel Otting, Natasja de Groot, and colleagues at the Biomedical Primate Research Centre and the staff at the Division of Veterinary and Biomedical Sciences, Murdoch University, are greatly appreciated.
We gratefully acknowledge financial support provided by BHP Minerals, Melbourne, PT Kaltim Prima Coal, CRA Foundation, PT Kelian Equatorial Mining, and the Merck Foundation.
REFERENCES
- 1.Blitz L, Pujol F H, Swenson P D, Porto L, Atencio R, Araujo M, Costa L, Callejas-Monsalve D, Torres J R, Fields H A, Lambert S, Van Geyt C, Norder H, Magnius L O, Echevarria J M, Stuyver L. Antigenic diversity of hepatitis B virus strains of genotype F in Amerindians and other population groups from Venezuela. J Clin Microbiol. 1998;36:648–651. doi: 10.1128/jcm.36.3.648-651.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Felsenstein J. PHYLIP: phylogeny inference package, version 3.57. Seattle, Wash: University of Washington; 1995. [Google Scholar]
- 3.Hollinger F B. Hepatitis B virus. In: Fields B N, Knipe D M, editors. Virology. Vol. 2. New York, N.Y: Raven Press Ltd; 1990. pp. 2171–2236. [Google Scholar]
- 4.Lanford R E, Chavez D, Brasky K M, Burns R B, Rico-Hesse R. Isolation of a hepadnavirus from the woolly monkey, a New World primate. Proc Natl Acad Sci USA. 1998;95:5757–5761. doi: 10.1073/pnas.95.10.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Magnius L O, Norder H. Subtypes, genotypes and molecular epidemiology of the hepatitis B virus as reflected by sequence variability of the S-gene. Intervirology. 1995;38:24–34. doi: 10.1159/000150411. [DOI] [PubMed] [Google Scholar]
- 6.Margolis H. Prevention of acute and chronic liver disease through immunisation: hepatitis B and beyond. J Infect Dis. 1993;168:9–14. doi: 10.1093/infdis/168.1.9. [DOI] [PubMed] [Google Scholar]
- 7.Norder H, Courouce A M, Magnius L O. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology. 1994;198:489–503. doi: 10.1006/viro.1994.1060. [DOI] [PubMed] [Google Scholar]
- 8.Norder H, Courouce A M, Magnius L O. Molecular basis of hepatitis B virus serotype variations within the four major subtypes. J Gen Virol. 1992;73:3141–3145. doi: 10.1099/0022-1317-73-12-3141. [DOI] [PubMed] [Google Scholar]
- 9.Norder H, Ebert J W, Fields H A, Mushahwar I K, Magnius L O. Complete sequencing of a gibbon hepatitis B virus genome reveals a unique genotype distantly related to the chimpanzee hepatitis B virus. Virology. 1996;218:214–223. doi: 10.1006/viro.1996.0181. [DOI] [PubMed] [Google Scholar]
- 10.Norder H, Hammas B, Lee S, Bile K, Courouce A M, Mushahwar I K, Magnius L O. Genetic relatedness of hepatitis B viral strains of diverse geographical origin and natural variations in the primary structure of the surface antigen. J Gen Virol. 1993;74:1341–1348. doi: 10.1099/0022-1317-74-7-1341. [DOI] [PubMed] [Google Scholar]
- 11.Ogata N, Miller R H, Ishak K G, Purcell R H. The complete nucleotide sequence of a pre-core mutant of hepatitis B virus implicated in fulminant hepatitis and its biological characterization in chimpanzees. Virology. 1993;194:263–276. doi: 10.1006/viro.1993.1257. [DOI] [PubMed] [Google Scholar]
- 12.Okamoto H, Tsuda F, Sakugawa H, Sastrosoewignjo R I, Imai M, Miyakawa Y, Mayumi M. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J Gen Virol. 1988;69:2575–2583. doi: 10.1099/0022-1317-69-10-2575. [DOI] [PubMed] [Google Scholar]
- 13.Ten Haaft P, Verstreppen B, Uberla K, Rosenwirth B, Heeney J. A pathogenic threshold of virus load defined in simian immunodeficiency virus- or simian-human immunodeficiency virus-infected macaques. J Virol. 1998;72:10281–10285. doi: 10.1128/jvi.72.12.10281-10285.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaudin M, Wolstenholme A J, Tsiquaye K N, Zuckerman A J, Harrison T J. The complete nucleotide sequence of the genome of a hepatitis B virus isolated from a naturally infected chimpanzee. J Gen Virol. 1988;69:1383–1389. doi: 10.1099/0022-1317-69-6-1383. [DOI] [PubMed] [Google Scholar]
- 14a.Warren, K. S. Unpublished observations.
- 15.Warren K S, Niphuis H, Heriyanto, Verschoor E J, Swan R A, Heeney J L. Seroprevalence of specific viral infections in confiscated orangutans (Pongo pygmaeus) J Med Primatol. 1998;27:33–37. doi: 10.1111/j.1600-0684.1998.tb00066.x. [DOI] [PubMed] [Google Scholar]
- 16.Wolfe N D, Escalant A A, Karesh W B, Kilbourn A, Spielman A, Lal A A. Wild primate populations in emerging infectious disease research: the missing link? Emerging Infect Dis. 1998;4:149–158. doi: 10.3201/eid0402.980202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zaaijer H L, Ter Borg F, Cuypers H T M, Hermus M C A H, Lelie P N. Comparison of methods for detection of hepatitis B virus DNA. J Clin Microbiol. 1994;32:2088–2091. doi: 10.1128/jcm.32.9.2088-2091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]