ABSTRACT
Fucoxanthin is a carotenoid that possesses various beneficial medicinal properties for human well-being. However, the current extraction technologies and quantification techniques are still lacking in terms of cost validation, high energy consumption, long extraction time, and low yield production. To date, artificial intelligence (AI) models can assist and improvise the bottleneck of fucoxanthin extraction and quantification process by establishing new technologies and processes which involve big data, digitalization, and automation for efficiency fucoxanthin production. This review highlights the application of AI models such as artificial neural network (ANN) and adaptive neuro fuzzy inference system (ANFIS), capable of learning patterns and relationships from large datasets, capturing non-linearity, and predicting optimal conditions that significantly impact the fucoxanthin extraction yield. On top of that, combining metaheuristic algorithm such as genetic algorithm (GA) can further improve the parameter space and discovery of optimal conditions of ANN and ANFIS models, which results in high R2 accuracy ranging from 98.28% to 99.60% after optimization. Besides, AI models such as support vector machine (SVM), convolutional neural networks (CNNs), and ANN have been leveraged for the quantification of fucoxanthin, either computer vision based on color space of images or regression analysis based on statistical data. The findings are reliable when modeling for the concentration of pigments with high R2 accuracy ranging from 66.0% − 99.2%. This review paper has reviewed the feasibility and potential of AI for the extraction and quantification purposes, which can reduce the cost, accelerate the fucoxanthin yields, and development of fucoxanthin-based products.
Keywords: Fucoxanthin, extraction, machine learning, quantification, artificial intelligence, microalgae, response surface methodology, digital images, identification, statistical data
1. Introduction
In recent years, carotenoids such as β-carotene, astaxanthin, lutein, canthaxanthin and especially fucoxanthin have always been considered as one of the important bioactive ingredients due to their major roles in human health (e.g. cancer, muscular dystrophy, cardiovascular, and neurological disorders) [1]. All the while, carotenoids are pigments synthesized from most non-photosynthetic organisms such as fungi and bacteria, fruits, and vegetables [2]. Nowadays, tremendous research efforts have been devoted into investigating microalgae due to their simple cell structure, high lipid accumulation than conventional crops, rapid reproduction and growth rate, nontoxic, biodegradable, and carbon dioxide as carbon source for growth [3–5].
Fucoxanthin is an organic pigment, marine carotenoid that can be found in the photosynthetic cells of edible brown seaweeds, macro-microalgae, diatoms algae [6]. Among all carotenoids, fucoxanthin are present abundantly and account for more than 10% of the predicted total natural production of carotenoids [7]. Fucoxanthin, a xanthophyll derivative that can be described as an orange-colored pigment found in heterokont algae (monophyletic group of photosynthetic species with tripartite tubular hairs and non-photosynthetic with reduced or lost tripartite hairs), including brown seaweeds (Phaeophyceae), diatoms (Bacillariophyceae), and chrysophytes [8,9]. Moreover, fucoxanthin, is available in some brown seaweeds such as Hijika fusiformis [10], Laminaria japonica [11], Sargassum siliquastrum [12], Padina tetrastromatica [13], and Undaria pinnatifida [14]. On the other hand, diatoms representing Phaeodactylum tricornutum, Chaetoceros calcitrans, and Skeletonema costatum are well-known for their sustainable bioactive sources of essential fatty acids, phenolic compounds, and carotenoids [15].
Up to this moment, there is an increasing demand to incorporate efficient downstream processing techniques as plenty effort and progression in the upstream production have led to advancement in the biomanufacturing industry. The most common approaches to extract fucoxanthin would be the solvent, maceration, and Soxhlet extraction along with the aid of organic solvents such as acetone, ethanol, hexane, and ethyl acetate. However, the toxicity of the solvents used, long extraction time, poor extraction performance, and high temperature exposure are the drawbacks of conventional approach [16]. Moving forward, the emerging green extraction technologies, such as using ultrasound-assisted extraction (UAE), microwave-assisted extraction (MAE), enzyme-assisted extraction (EAE) and pressurized liquid extraction (PLE), are of current trend in various research work due to rapid extraction, low-cost, reduced used of solvents, and high extraction efficiency [17]. As promising as it sounds, the recent extraction procedures however are still lacking in determining both qualitatively and quantitatively analysis for the extracted bioactive ingredient through the identification of retention time and absorbance, respectively [18]. High-performance liquid chromatography (HPLC) is considered as one of the well-known techniques in quantitative and qualitative analysis of bioactive ingredients, yet still uses large amount of organic solvents as well as time- and cost-consuming [19]. Nonetheless, conventional extraction techniques often face challenges in terms of slow extraction time, extensive use of toxic solvents, laborious and energy-intensive procedures that influenced the reproducibility and up-scaling of solvent-extraction methods. Hence, some of the green and environmental friendly extraction techniques such as the UAE, MAE, PLE, and EAE have shown varying degrees of success, but still require further refinement and adjustment to overcome the bottlenecks such as costly validation, high energy consumption, prolonged extraction times, and low yields [20].
The application of AI has been successfully adopted in different studies and industries such as prediction of coal wettability using ML for the application of CO2 sequestration [21], application of ML to predict the recovery of multistage hydraulically fractured wells [22], and prediction of in-situ stresses from logging data [23]. The uprising of AI digital computation techniques has developed as novel, reliable, and future chemometric approach in different extraction studies. SVM, ANFIS, and ANN are implemented for the quantification, identification, and prediction of fucoxanthin and its antioxidant activity due to less analysis time, less toxic solvent usage, high accuracy, good prediction, and low analysis cost. Moreover, the accuracy of ML models depends on the number of training dataset and suitable input variables to provide desired output value.
This present review highlights the research gap of AI digital computation techniques for the quantification of fucoxanthin recovery by incorporating various AI strategies including machine learning (ML) and deep learning (DL) models to study the feasibility of extraction process along with the quantification of fucoxanthin from microalgae. It is worth mentioning that the novelty of the review can be summarized as follows: a) to provide a critical analysis on the variability of fucoxanthin content, quality among different microalgae species, influence of environment, cultivation factors on fucoxanthin production stability; b) evaluating the comparison between conventional and non-conventional extraction techniques in terms of solvent consumption, extraction time, purity, temperature, and extraction efficiency; c) implementing a constructive research methodology on the development of AI models that allow researchers to understand each of the process involved; d) constructing an insightful bibliometric analysis for accurate and reliable model selection; e) evaluating the integration of AI in the extraction techniques of fucoxanthin by comparing RSM, ANN, and hybrid models; f) leveraging various ML and DL models for the quantification of fucoxanthin; and g) discovering current challenges of AI models while providing future research opinions for the betterment of AI toward microalgae research.
2. Overview of fucoxanthin
Surprisingly, fucoxanthin from microalgae have sparked many attentions due to several merits such as antioxidant, anticancer, antiangiogenic, antidiabetic, anti-obesity, antimalaria, anti-inflammatory, and photo-protection [2]. Based on Miyashita et al. [24], marine products indeed promote a healthy lifestyle which results in lower rate of cardiovascular diseases, cancer, diabetes, and stroke. However, the detailed processes and mechanisms of their physiological activities are still unclear. As discussed by Leong et al. [25], there is a need to consider appropriate upstream microalgal production (typically culture conditions and pilot-scale studies) and downstream processing (typically advanced extraction technologies and pre-treatment process). In the upstream section, there are various factors affecting the cultivation of microalgae in order to achieve maximum fucoxanthin bioaccumulation. For instance, temperature, light intensity, carbon and air source, photoperiod, types of nutrient medium, pH value, type of cultivation mode, and period of cultivation. Among these factors, pH and temperature do not significantly affect the production of fucoxanthin, instead increase the biomass production. Most microalgae strains such as P. trinorcutum, T. lutea, Isochrysis spp., and Nitzschia spp. are best grown under mesophilic condition at a moderate temperature from 20–30°C, and some for instance, O. aurita best grown under low temperature from −1.5–6°C [25]. Table 1 shows a list of several micro-macroalgae capable for the production of fucoxanthin that emphasis on the variability of fucoxanthin content, quality among different algae species, influence of the environment, cultivation factors on the fucoxanthin production, and the stability.
Table 1.
Fucoxanthin content from various macro-microalgae species.
| Fucoxanthin-producing microalgae species | Cultivation factors on fucoxanthin production | Stability and optimised condition | Growth environment | Yield of fucoxanthin composition | References |
|---|---|---|---|---|---|
| Chaetoceros gracilis (diatom from microalgae) | Temperature = 30 ± 1°C, CO2 gas = 2% at flow rate of .2 L/min, light intensity = 300 µmol.m2/s, photoperiod = 12 L:12 D, fresh medium supplied for 100, 200, and 400 mL at dilution rates of .1, .2, and .4/d, respectively | High silicon uptake at dilution rate of .4/d under 300 µmol.m2/s to reduce cell size with sufficient nutrients | Photobioreactor based on semi-continuous culture, using modified Conway medium | 17.1 mg/g DW | [26] |
| Isochrysis galbana (diatom from microalgae) | Temperature = 20°C, air supply = 5 L/min, 60 W fluorescent light intensity = 2500 lux, continuous culture for 4–5 days | Two-solvent system of n-hexane-ethanol-water with volume ratio = 10:9:1 | 30 L plastic cylinder, using f/2-Si medium | 18.23 mg/g DW | [27] |
| Chaetoceros sp., Skeletonema sp., Thalassiosira sp. (diatom from microalgae) | Incubation temperature = 30°C, photoperiod = 12:12 h light diurnal cycle, light intensity = 100 µmol.m2/s, continuous culture for 21 days | Highest biomass concentration and cell count observed during day 14 | Flasks using f/2-Si medium made with artificial seawater | .403 ± .036 mg/L, .097 ± .007 mg/L, .241 ± .016 mg/L | [28] |
| Chaetoceros calcitrans (diatom from microalgae) | Temperature = 23–25°C, pH = 8–8.5, photoperiod = 150 µmol.m2/s under light/dark 12:12 cycle, continuous culture for 14 days | Optimized extraction condition via crude methanolic extract (CME) with liquid-liquid partitioning of dichloromethane-water solvent system | 120 L capacity annular photobioreactor using Conway medium | 5.25 ± .03 mg/g DW | [29] |
| Odontella aurita (diatom from microalgae) | Temperature = 25°C, irradiance = 150 µmol.m2/s (first 2 days) and 300 µmol.m2/s (remaining duration of cultivation), CO2 gas = 1% at flow rate of .2 vvm, continuous culture for 11 days | Among three nitrogen supply (initial low (ILN), initial high (IHN), and initial high + supplementary nitrogen (SN)), SN conditions maximized the fucoxanthin production | 1.2 L cylindrical glass photobioreactor using modified L1 medium | 6.01 ± .36 mg/L | [30] |
| Phaeodactylum tricornutum (diatom from microalgae) | Temperature = 25°C, Air and CO2 gas = 5 L/min, irradiation using 1,2, or 3 light strips = 100, 150, and 210 µmol.m2/s, respectively, photoperiod of 12h light:12 h dark cycle, continuous culture for 7 days | Optimzed fucoxanthin production at day 7 at low light conditions (100 µmol.m2/s) along with addition of nitrate nutrient. Effect of CO2 gas addition was not significant for fucoxanthin production | 5 L flat panel photobioreactor using modified f/2 medium (without addition of vitamins) | 42.8 ± 19.5 mg/g | [31] |
| Sargassum fusiforme (brown seaweed from macroalgae) | Combination of three temperature (10, 15, and 20°C) and four salinities (10, 20, 30, and 40 psu), photoperiod of 12:12 h light/dark cycle, irradiance of 150 µmol.m2/s, | Optimized temperature = 15°C and salinity = 10 psu. Increase in temperature and decrease in salinity significantly improved the fucoxanthin production | 36 side-arm flasks, each flask containing 500 mL seawater enriched with 25% PESI medium | 2.62 ± .04 mg/g | [32] |
| Laminaria japonica (brown seaweed from macroalgae) | Incubation temperature = 40°C for 4 h, pH 7.0 for 4 h, different hydrolysate (LPH) dosages (.75–3.0 ml/L), continuous culture for 10 days | Optimized with low addition proportion of 1.5 ml/L LPH achieved the highest fucoxanthin production after 10 days cultivation | 500 mL flasks, each containing 300 mL of f/2 medium | 17.55 mg/g | [33] |
Nevertheless, there are numerous challenges to be addressed such as identifying and isolating the best diatom strains for fucoxanthin production, optimal nutrient conditions in a photobioreactor-based production system, and standardization of protocols to acquire pure cultures. Other than that, there is a need to reduce the input cost during extraction process for purchasing high grade and quantity of fucoxanthin at a much reasonable cost [34]. Furthermore, an optimized selection is required to ensure the consistent production of fucoxanthin and biomass under various conditions, either grown in laboratory or outdoor situation. The commercial viability in fucoxanthin production was limited to only a few microalgae strains [35]. Therefore, there is a need of rapid and accurate methods for screening and selecting of optimal microalgae sources, quantification, and the extraction conditions for fucoxanthin recovery.
3. AI key strategies into extraction and quantification process
There are few main steps to be carried out to develop a good AI model such as: (i) data collection, (ii) data pre-processing, (iii) sensitive analysis test and feature selection, (iv) model selection (v) model development and optimization, and (iv) model validation. A schematic flow diagram of methodology for the extraction and quantification of fucoxanthin is illustrated in Figures 2 and 3, respectively. According to Maleki et al. [36], the rules in developing successful machine learning (ML) models are such: (i) representation, (ii) evaluation, and (iii) optimization. First, representation means choosing the type of ML model to generate output value of interest by inserting input variables. Next, evaluation defines the capability of that respective model producing qualitative results between the inputs and outputs. Lastly, optimization evaluates the model accuracy, mean squared error, and precision.
Figure 2.
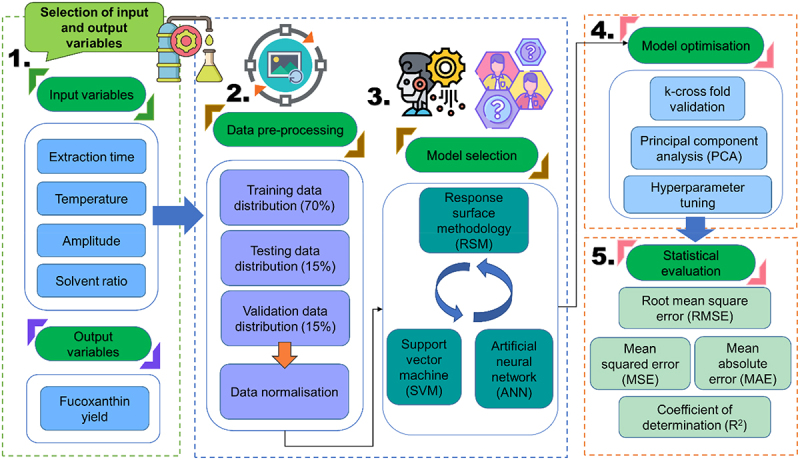
An overview methodology pipeline for the configuration of AI models to be incorporated into the extraction of fucoxanthin from microalgae. The first key step involves selection of input variables and output variable, followed by data pre-processing to divide the data into the desired proportion of training, testing, and validation data prior data normalization. The third key step involves the appropriate model selection and optimization to improve the accuracy and precision of the respective model. lastly, model evaluation to determine the robustness and accuracy of the model based on testing or unseen dataset.
Figure 3.
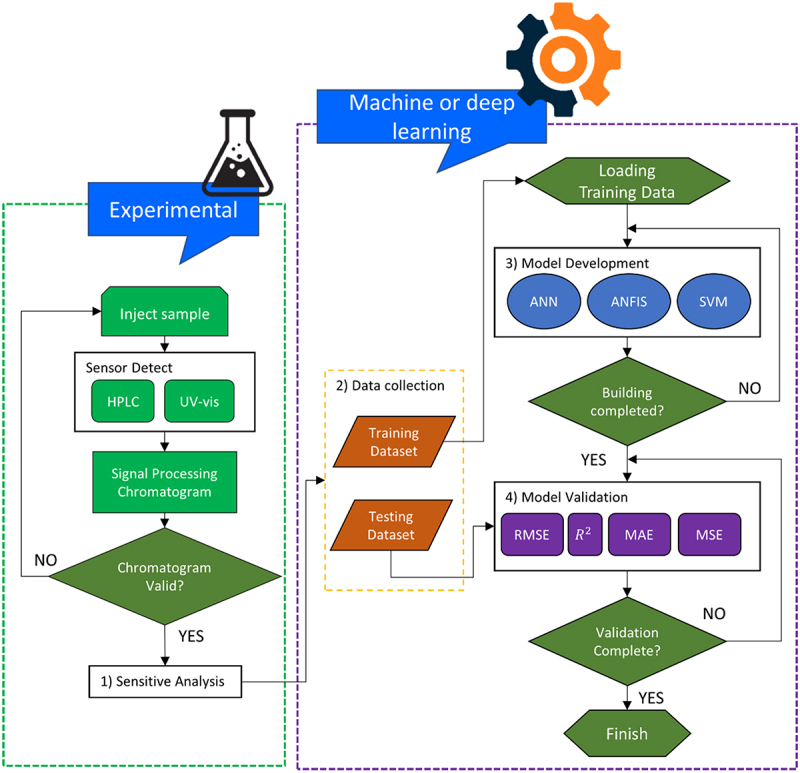
Methodology for incorporating machine learning and deep learning into quantification of fucoxanthin. The first step begins with data acquisition by injecting sample into either HPLC or UV-vis spectrophotometry for the signal processing of chromatogram. If valid, then proceed for sensitive analysis or feature selection to select the desired parameters or otherwise, return to the first step. The data acquired will be divided into training and testing dataset, in which training dataset will be used for model development, while testing dataset for model validation. If training of dataset is completed, proceed for model validation or otherwise, return to model development. If model validation is successful, the model is completely built, otherwise return to model validation.
3.1. Data collection
Data collection is crucial for the development of AI model, which refers to the process of extracting relevant and representative data that will be utilized to train, validate, and test the AI model. The quality and preparation of dataset can affect the accuracy and reliability of the trained models [37]. It involves systematically collecting and organizing data from various sources, such as data collected over time and statistical analysis of that information [38]. For instance, Sarkar et al. [39] collected a total of 69 values of input data for each output parameter from 23 experiment cases. A total of seven most critical extraction variables such as homogenization time, speed, temperature of solvent, solid: solvent ratio, boiling temperature, and microwave time were considered for the modeling of ANN for the optimization of fucoxanthin concentration. As for the quantification of fucoxanthin from algae blooms (consists of green algae, diatoms, and cyanobacteria), Pyo et al. [40] collected both reflectance and absorption coefficient data from a total of 126 sampling points. The image data was taken with a drone setup equipped with hyperspectral imaging sensor ranging from 400–1000 nm and spectral resolution of 4 nm.
3.2. Data pre-processing
As mentioned by Ramírez-Gallego et al. [41], data pre-processing in general required more effort (>50% of total effort) and time to process the entire data analysis prior distributing data into training, testing, and validation. Commonly, the outcome of raw data will generate various flaws such as missing values, noise/redundancies, and inconsistencies. If adhered to appropriate pre-processing steps, low quality data can be avoided and thus, subsequent learning algorithm will not be undermined. Moreover, there are different phases to data pre-processing which includes data collection, data cleaning, session identification, user identification, and path completion [42]. Statistical and image data are two different types of data that can be fed into AI models. Statistical data refers to numerical and categorical data that is typically obtained through experiments, surveys, and observations. This data includes variables such as extraction and quantification parameters of fucoxanthin that are most essential. Data pre-processing for image data involves a series of steps to prepare the images prior analyzing or training AI models. Generally, handling image data includes image acquisition, image resizing (to standardized all input images) followed by optional pre-processing techniques such as gray-scaling, denoising, thresholding, and segmentation [43]. On the other hand, data pre-processing for statistical data includes data normalization between a specific range (0–1), prior training the model [39]. According to Sarkar et al. [39], a total of 138 dataset was split into 70% for model training, 15% for model testing, and the remaining 15% for data cross-validation to prevent overfitting. Depending on the amount of dataset available, the acquired dataset can be categorized into two parts. For instance in most common practices, 70% and 30% of the data will be used for training and testing of AI models, respectively [44].
3.3. Sensitive analysis test and feature selection
The purpose of feature selection or sensitive analysis assists in the reduction of dataset size by removing any falsification or redundant dataset that would affect the performance of AI models. Reducing the size of dataset can be advantageous in scenario where the number of features in the dataset is equal to or greater than the number of samples. Excessive and unknowingly large dataset can often lead to overfitting, where the model becomes too specific to the training data and performs poorly on the validation dataset [45]. Feature selection techniques such as principal component analysis (PCA), linear discriminant analysis (LDA), and multidimensional scaling are applied for correlation analysis, mutual information, and identifying relevant features while removing irrelevant or redundant features to improve model’s efficiency and interpretability [43]. The sensitive analysis methods such as Monte Carlo [46], correlation [47], and non-linear global sensitive analysis: PAWN [48] are performed prior input into the AI models. Such act of performing sensitive analysis is to identify important parameter by reducing redundant big dataset and load of the model. The current framework will lead to some information losses throughout the machine learning process. However, the major problem that correlate to the substantial amount of redundant parameter input into the respective model will be significantly reduced which eventually increased the accuracy of desired output.
3.4. Bibliometric analysis for AI model selection: review methodology
Scopus database has been utilized to collect publication data to study the linkage of AI techniques for the possible application and improvement in the extraction and quantification of fucoxanthin. The keywords used for the data extraction are ‘artificial intelligence’ OR ‘machine learning’ OR ‘deep learning’ OR ‘artificial neural network,’ ‘carotenoid’ OR ‘fucoxanthin’ OR ‘antioxidant’ OR ‘microalgae’ OR ‘bioactive compound’ AND ‘extraction’ OR ‘quantification’ in the last five years, from 2018 to 2023. The Scopus database generated a total of 247 research articles for the selected keywords. Next, the collected database of research articles is interpreted and clustered by leveraging the VOS viewer software (version 1.6.19). The keywords co-occurrence tool in the VOS viewer interface was selected in order to visualize the network between the most recurrent keywords of AI, extraction, and quantification of fucoxanthin. A threshold of three occurrences was set as the minimum requirement for keywords to be included in the analysis to avoid over-crowded during mapping of visualization. The co-occurrence network map is illustrated in Figure 1. The size of each circle in the visualization corresponds to the frequency of a particular keyword appearing in the article title, abstract, and keywords. Consequently, a larger circle indicates a higher occurrence of that keyword. As shown in Figure 1, the keywords ‘artificial neural network,’ ‘machine learning,’ and ‘antioxidant activity,’ and ‘antioxidant’ have dominated the central position with total link strength of 64, 18, 29, and 18, respectively. In addition, AI models such as ‘support vector machine’ (SVM), ‘artificial neural networks’ (ANN), and ‘anfis’ (adaptive neuro fuzzy inference system or ANFIS) have interconnections with the extraction optimization and quantification of antioxidant activity. Meanwhile, the ‘microalgae’ node has connections with ‘machine learning,’ ‘artificial intelligence,’ and ‘carotenoids.’ It is noteworthy to mention that the application of AI models can be seen in various microalgae related research studies such as image classification, biomass prediction, system optimization, smart cultivation, genetic modification, bioactive compound quantification, and extraction optimization [49,50]. This summarizes the potential of these AI models to be incorporated or applied in the extraction and quantification studies of fucoxanthin from microalgae which will be explained further in subsequent sections.
Figure 1.
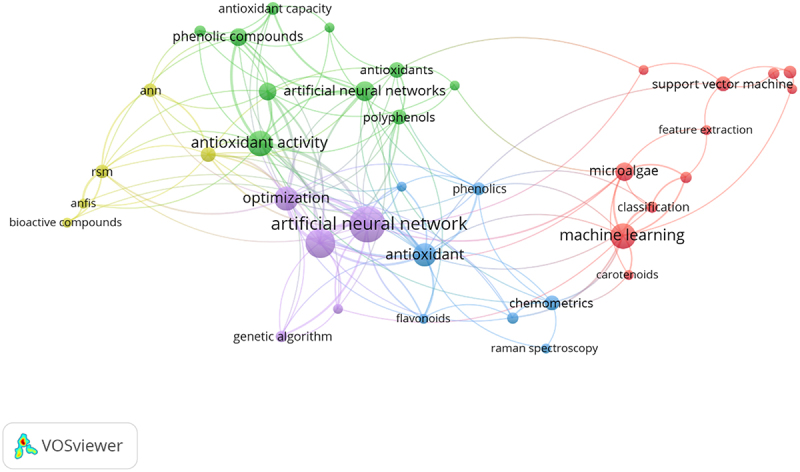
Keyword co-occurrence network map of AI application in the extraction and quantification of fucoxanthin from microalgae. source link: https://www.vosviewer.com/.
3.4.1. Artificial neural networks
The attractiveness of using ANN as a modeling tool arises from their excellent information-processing abilities, which can primarily be attributed to their non-linear nature, high parallelism, capability to handle errors and noise, and their adaptability in learning and generalization. Unlike traditional computation tools, ANN provide a model-free approach that is adaptive, capable of parallel processing, and robust toward error and failure tolerance. Besides, the algorithm of ANN was designed with remarkable learning capabilities to handle inaccurate and fuzzy information, as well as the ability to generalize patterns that have not been previously observed [51]. The rapid progress of algorithms and information technology has emerged as a motivation for the extensive application of ANN algorithm in research and development. While ANN offer significant advantages, there are still limitation that need to be addressed to further improve the efficiency of the model. For example, large amounts of data are required for training, which can be a constraint in situations where data availability is limited. The complexity of ANN architectures also presents a challenge, requires determining optimum factors that influence the model development such as data division, data pre-processing, hyperparameter tuning, and model validation [52].
3.4.2. Support vector machines
Support Vector Machine (SVM) is a very powerful and flexible ML model, which is capable of solving problems that include linear or nonlinear classification, prediction, pattern recognition, and regression [53]. The general concept of SVM adopted the Structural Risk Minimisation (SRM) induction principle, thus, capable of generating better generalized solutions that are sparse and unique, with simple geometric interpretation. Due to its flexibility to adapt sparse training data, SVM can avoid the risk of data overfitting. This is achieved through the maximizing of margin between two classes of vectors while minimizing the training data set error. Similar to ANN model, SVM is also robust toward the ambiguous data and able to solve non-linear relationships between input and output data [54]. Based on Wang et al. [55], SVM performs slightly better as compared to ANN model due to its regularization mechanism and less frequent prone to overfitting, thus reduced the structural risk in training process. The main limitation of SVM is during the hyperparameter tuning, especially the selection of optimum kernel function and regularization parameter. In addition, long computational duration in handling huge dataset, inappropriate data collection and pre-processing such as noisy input and overlapping classes of data can affect the model performance [54].
3.4.3. Adaptive neuro fuzzy inference system
Adaptive Neuro Fuzzy Inference System (ANFIS) is an AI model developed by Jang in 1993, also known as a universal estimator to solve complicated problems [56]. ANFIS represents a hybrid computational model that combines both adaptive learning capability of ANN with fuzzy logic principles. The implementation of fuzzy logic is able to resolve the time consuming and complex process when designing the operation point and linearization of mathematical models based on human intelligence assisted IF-THEN rules. The combination of ANN and ANFIS is much preferred, since ANN only deals with datasets rather than linguistic expressions [57]. Along with ANN and SVM, ANFIS model is also a universal approximator representing highly non-linear functions with high level computational structure and works by perform reasoning and make decisions based on available information. The fuzzy system is limited in terms of its inability to learn and subsequently adjust the components, thus integrating with ANN with the ability of learning and adjusting their internal components based on input and output of the available data can be a beneficial approach [58]. Other than that, Kushwaha et al. [57] mentioned ANFIS has better smoothness as compared to ANN and able to solve complicated engineering tasks. However, low convergence rate, learning rate, high risk in being caught in local extreme and suggested other potential optimization algorithm such as genetic algorithm for the hybrid of ANN instead, for better optimization efficiency.
3.5. Model development, optimization, and validation
Hyperparameters are known as tuning parameters of machine learning models, which will be predetermined prior model training. In model development, hyperparameter optimization or tuning is a crucial step to obtain optimal performance from machine learning models [59]. The objective is to provide enough information into the respective machine learning model to make good prediction. If the error existing in predicted value is high, several alternations can be considered such as substitution of models, training of algorithm model with better features, and reducing the constraints [44]. Additionally, overfitting is another frequent issue of machine learning that occurred due to several potential problems such as data, overloaded test set, inappropriate activation function, and missing normalization [60]. Although, overfitting models perform well on training set but fail to generalize new dataset that interferes with predicting the true pattern of the specific data [61]. A comprehensive discussion regarding the control parameters for each AI model associated with their advantages and limitations for the extraction and quantification of pigments is shown in Table 2.
Table 2.
Development and optimization of model for the quantification and extraction process of organic pigment.
| Objective | Model selection | Control parameters | Advantages & limitations | References |
|---|---|---|---|---|
| Optimization for the extraction of flavonoids from Juglans mandshurica Maxim | Hybrid RSM-ANN-GA |
ANN backpropagation was adopted, 11 samples for training, 2 samples for validation, and remaining 2 samples for testing, trained using Levenberg-Marquardt backpropagation, degree of approximation (Da) was used to determine the number of hidden nodes GA parameters optimization (population type, double vector; population size, 20; initial population, given randomly; selection function, stochastic uniform; elite count, 2; crossover fraction, 0.8; crossover function, scattered; migration fraction, 0.2; migration interval, 20), evaluation of GA after 100 epochs |
-ANN algorithm and genetic algorithm (GA) can compute faster, more cost effective when solving optimisation problems, reduce experiment runs and reagent consumption as compared to traditional RSM model | [62] |
| Optimization of polyphenols from sunflower & corn oils | ANN & MLR (multiple linear regression) |
ANN backpropagation was adopted, among the number of neurons (5, 10, 15, 20), 10 hidden neurons were selected which best determine the best results |
-ANN can solve various engineering problems owning to its non-linearity, learning ability, and capacity to complete missing data. –Both ANN and MLR contributes to positive results for modelling, optimizing, and predicting phenolic contents in wide range of materials | [63] |
| Optimisation of phenolic compound from pomegranate | ANN-GA |
ANN feed-forward backpropagation was adopted, a tangent sigmoid transfer function (transig) was applied at a hidden layer while linear transfer function (pureline) applied at the output layer, 70% data for training, 15% data for testing and remaining 15% for validation, 5 hidden neurons were selected, best performance achieved at 3 epochs GA parameters optimization (51 generations, population size of 20, and scattered crossover function of 0.8) |
-RSM is limited in terms of solving quadratic non-linear problems, while ANN and GA can perform otherwise. | [64] |
| Optimization of total antioxidants from cashew apple bagasse | ANN-GA |
ANN multilayer perceptron (MLP) based on feed-forward backpropagation was adopted, 70% data for training, 15% data for testing and remaining 15% for validation, 10 hidden neurons were selected which best determine the best results, trained using Levenberg-Marquardt backpropagation RSM parameters optimization (population type of double vector, size of 200, crossover function of 0.8, migration of forward migration, creation function of feasible population, fitness scaling function of rank, selection function of roulette wheel function, crossover function of scattered and mutation function of adaptive feasibility |
-Traditional “one-factor-at-a-time” approach is laborious and time consuming for the optimization process. -ANN model can solve non-linear multivariate tasks with better computational and mathematical techniques along with GA for the non-linear optimization formalism that further optimise the input variables of ANN models |
[65] |
| Optimisation of total phenolic content from jujube | ANN |
ANN MLP based on feed-forward backpropagation was adopted, 70% data for training, 15% data for testing and remaining 15% for validation, trained using Levenberg-Marquardt backpropagation, 5-fold cross-validation |
-ANN outperformed RSM in terms of superior properties and increased the workable suitability of the dataset | [66] |
| Spectrophotometric quantification of lutein, violaxanthin, and zeaxanthin from Chlorella vulgaris & Scenedesmus almeriensis | ML model based on particle swarm optimiser-assisted partial least square regression (PSO-assisted PLS) |
PSO with swarm social parameter of 0.6, particle cognitive parameter of 0.6, inertia of the best value was modelled using random chaotic function, 80% data for training and remaining 20% for validation, best performance achieved after 50 iterations |
-PSO improved the flexibility for high dimensionality configurations, limiting cost, significantly reduce the delay in obtaining samples of carotenoids concentrations compared to liquid chromatography while maintaining adequate accuracy | [67] |
| Smartphone-based quantification of chlorophyll & carotenoids contents in olive and avocado oils | MLR & LS-SVM (least squares support vector machine) | MLR & LS-SVM where dataset divided into calibration set (75%, 104 samples) and test set (25%, 34 samples) using Kennard-Stone algorithm, performed cross validation using leave-one-out method LS-SVM Leveraged estimation type and radial basis function kernel, fit parameters were adjusted automatically, and additional parameters were standard |
-Conventional methods for determining pigments based on spectroscopic and chromatographic requires sample preparation, usage of expensive solvents, and equipment. -Smartphone assisted with computer vision models are simple, low-cost, and promising techniques for monitoring quality of pigments |
[68] |
| Non-destructive quantification of carotenoid in carrots | PCR (principal component regression), PLS, & LS-SVM | PCR, PLS, & LS-SVM were adopted among 114 carrot samples collected, 76 carrot samples were calibrated set, and the remaining 38 carrot samples were prediction setLS-SVMLeveraged radial basis function kernel Among 114 carrot samples collected, 76 carrot samples were calibrated set, and the remaining 38 carrot samples were prediction set LS-SVM leveraged radial basis function kernel |
-Conventional methods requires complicated pre-processing, expensive validation, skilled operators, and destructive to samples -These models reduce the modelling time while improving the prediction accuracy |
[69] |
| Drone-borne sensing for the prediction of microalgae pigments | 1D-CNN (One dimensional convolutional neural network) | 1D-CNN was adopted for batch normalisation to promote stable model, 3 convolutional layers with increasing number of filters by 32, 64, and 256 with filter size of 1x3, learning rate of .001 and batch size of 16, max pooling and dropout layers were used after 3rd convolutional layer, 70% (88 data for training) and 30% (38 data for validation) |
-Superior performance while dealing with complex dataset, widely applied to spectral type data, capable of providing promising information regarding important features of input data -Limited in terms of trace concentration levels and similar spectral properties of auxiliary pigments which affect the estimation performance |
[40] |
| Smartphone-based quantification of chlorophyll content in microalgae | Linear regression & ANN |
ANN MLP based on feedforward was adopted, among 1000 samples, 70% of data for training and remaining 30% of data for testing, at least one hidden layer and a maximum of 3 hidden layers were set, training type in batch mode, scaled conjugate gradient was used as optimization algorithm, training options were initial lambda, initial sigma, interval centre, and interval offset |
-Conventional spectrophotometric quantification method proven to be inaccurate due to overlapping of absorption spectra of other pigments such as chlorophyll and carotenoids, unclear selection of numerous formulated simultaneous equations, and standard curve can be inaccurate if pigment integrity is subjected by the changes in temperature, pH, and other factors - ANN can act as universal approximator with the ability to capture linear and non-linear relationships between input and output data, ability to calculate an error based on target output and actual input - Setup of linear regression is simple and easy to implement in the case of forecasting, prediction, and observing data patterns |
[49] |
| Prediction of total phenolic compound from Spirulina platensis | ANFIS, MLP, & step-wise-linear regression (SWLR) | ANFIS was adopted using adaptive multi-layer and feed forward networks using instructions of the Takagi-Sugeno type MLP was trained using Levenberg-Marquardt algorithm, the range of hidden neurons were 3–27 |
-ANFIS is benefit in terms of a universal, wide ranging, and multipurpose model for the estimation of various tasks -MLR also be considered as universal estimator to solve both linear and non-linear problems similar to ANN - SWLR excels in finding the best set of parameters between single and numerous variables |
[70] |
3.6. Statistical evaluation
The testing dataset is used for the evaluation of models for its performance error [44]. According to Witek-Krowiak et al. [71] and Tao et al. [72], the validation process can be calculated by several continuous error matric such as root mean square error (RMSE), mean absolute error (MAE), mean squared error (MSE), coefficient of determination (R2), regression analysis. Evaluation of AI models is vital to produce solid and reasonable results that can fulfil real-world problems.
4. Potential AI integration in extraction techniques: AI techniques based on various optimizer algorithms
The integration of AI has revolutionized various fields of wastewater treatment, food processing, analytical chemistry, and its potential extends even to the prediction of extraction efficiencies of valuable bioactive compounds from microalgae [73]. The extraction of fucoxanthin from microalgae is a process that holds promising values for various applications ranging from health and food industries to cosmetics and bio-energy production [74].
4.1. Connecting AI and extraction techniques of biomolecules
The many considering factors such as solvent selection, proportion of solvents, temperature, extraction techniques, extraction time, and other variables are considered as the initial and most crucial step in achieving the maximum fucoxanthin yield. Yet, these approaches often face limitations such as lengthy extraction times and low extraction efficiency [75]. Optimum values of the factors influencing the extraction process must be known in order to achieve the maximum yield of the intracellular compound of interest. Figure 2 illustrates the concept and feasibility of leveraging AI models for the optimization of fucoxanthin extraction process.
4.1.1. Connecting response surface methodology with extraction methods
First and foremost, statistical methods such as response surface methodology (RSM) is a widely used conventional technique for modeling and optimizing the process for the extraction of bioactive compounds [76]. RSM optimization tool has been extensively used among researchers to analyze the effects of independent variables, their interactions on the response, and determine the optimum conditions for the variables to maximize the extraction of bioactive compounds from microalgae. A comprehensive application of RSM for the optimization of fucoxanthin from microalgae are shown in Table 3.
Table 3.
Opportunities and potential application of AI models in the optimization of fucoxanthin extraction process.
| Model | Carotenoid source | Extraction method | Variable range | Optimum conditions | Optimised response conditions | Statistical validation & results | References |
|---|---|---|---|---|---|---|---|
| RSM with CCD | Fucoxanthin from Irish seaweed (Fucus vesiculosus) | SE using acetone | Time (30–70 min), temperature (30–70°C), solvent pH (5.0–9.0), acetone percentage (30–70%) | Incubating temperature at 40°C for 40 min, pH 6.0, 60% acetone | Incubating temperature at 30°C for 36.51 min, pH 5.7, 62.15% acetone | Before optimized yield = 0.696 ± 0.02 mg/g (dm) After optimized R2 = 93.3%, yield = 0.745 mg/g (dm) |
[77] |
| RSM | Fucoxanthin from Irish brown seaweeds | EAE | Seaweed: water ratio (0.1–50.0%), enzyme: water ratio (0.1–50.0), enzyme incubation time (0.1–24.0 h) | - | Seaweed: water ratio of 5.37%, enzyme: water ratio of .52%, enzyme incubation time of 3.05 h | Before optimized yield = 0.657 mg/g DW After optimized R2 = 91.16%, yield = 0.706 mg/g DW |
[78] |
| RSM with CCD | Fucoxanthin from brown macroalgae (Sargassum muticum) | UAE | Ethanol concentration (S, 35–100%), time (t, 5–55 min), power (p, 100–500 W) | t-value of 45.00 ± 3.35 min, s-value of 37.50 ± 3.06%, p-value of 409.46 ± 1.12 W | t-value of 45.00 ± 3.35 min, s-value of 84.22 ± 4.9%, p-value of 339.73 ± 9.22 W, | Before optimized R2 = 96.5%, yield = 29.98 ± 1.03 g/100 g DW After optimized R2 = 81.99%, yield = 93.00 ± 0.10 g/100 g DW |
[79] |
| RSM with CCD | Fucoxanthin from brown algae (Sargassum fusiforme) | UAE | Liquid/solid ratio (10–60 mL/g), time (10–35 min), temperature (55–75°C), amplitude (20–70%) |
Liquid/solid ratio of 40 mL/g, time of 25 min, temperature of 75°C, amplitude of 5.00% | Liquid/solid ratio of40 mL/g, time of 27 min, temperature of 75°C, amplitude of 53.00% 40 mL/g, time of 27 min, temperature of 75°C, amplitude of 53.00% |
Before optimized yield = 662.51 µg/g After optimized R2 = 99.08%, yield = 696.85 ± 2.84 µg/g |
[80] |
| RSM with BBD | Fucoxanthin from brown, red, and green macroalgae (Fucus virsoides, Amphiroa rigida, Codium bursa) | UAE | Temperature (T, 30–70°C), extraction time (t, 15–45 min), solvent: solid ratio (S, 10–30 mL/g) | - | A.rigida with (T-value of 70°C, t-value of 29.88 min, S-value of 19.63 mL/g), F.virsoides with (T-value of 31.13°C, t-value of 15.06 min, S-value of 1.05 mL/g), C. bursa with (T-value of 3.29°C, t-value of 15.06 min, S-value of 15.17 mL/g), | Before optimized yield (A. rigida) = 0.66 mg/g, yield (F. virsoides) = 1.00 mg/g, yield (C. bursa) = 1.00 mg/g After optimized Standard deviation = ± 10% yield (A. rigida) = 0.69 mg/g, yield (F. virsoides) = 1.90 mg/g, yield (C. bursa) = 0.45 mg/g |
[81] |
| Hybrid RSM-ANN-GA | Flavonoids from Juglans mandshurica Maxim. | UAE | Ethanol concentration (E, 30–80%), ultrasonic power (P, 125–250 W), extraction temperature (T, 30–80°C), solid: liquid ratio (S, 1–35 g/mL), extraction time (t, 20–70 min), extraction times (Et, 1–5) | E-value of 60%, P-value of 225 W, T-value of 60°C, S-value of 1:20 g/mL, t-value of 40 min, Et-value of 1 | E-value of 62%, P-value of 228 W, T-value of 60°C, S-value of 1:20 g/mL, t-value of 40 min, Et-value of 1 | Before optimized R2 = 97.00%, yield = 6.16 mg/g After optimized R2 = 98.28%, yield = 6.28 mg/g |
[62] |
| RSM, ANN & MLR | Polyphenols from sunflower & corn oils | UAE | Olive oil content (C, 2000–6000 ppm), time (t, 15–45 min), amplitude (P, 20–30%) | RSM sunflower oil with (C-value of 6000 ppm, t-value of 15 min, P-value of 30%), RSMcorn oil with (C-value of 6000 ppm, t-value of 15 min, P-value of 20) | - | Before optimized yieldsunflower oil (RSM) = 43.33 ppm, yieldcorn oil (RSM) = 30.02 After optimized R2sunflower oil (ANN) = 85.00%, R2corn oil (ANN) = 88.00%, R2sunflower oil (MLR) = 51.00%, R2corn oil (MLR) = 66.00%, yieldsunflower oil (ANN) = 42.44 ppm, yieldsunflower oil (MLR) = 45.38 ppm, yieldcorn oil (ANN) = 28.45 ppm, yieldcorn oil (MLR) = 27.13 ppm |
[63] |
| RSM & ANN-GA | Phenolic compound (punicalagin) from pomegranate (Punica granatum) | UAE | Solvent volume (S, 20–60 mL), Amplitude (P, 20–70), time (t, 5–25 min), duty cycle (d, 60–100%) | RSM with (S-value of 30 mL, P-value of 50%, t-value of 15 min, d-value of 80%) | ANN-GA with (S-value of 30 mL, P-value of 35%, t-value of 23 min, d-value of 100%), | Before optimized yield (RSM)= 168.70 mg/g After optimized R2 = 99.60%, Yield (ANN-GA) = 179.89 mg/g |
[64] |
| RSM & ANN-GA | Total antioxidants from cashew apple bagasse (CAB) | UAE | Time of extraction (t, 5–15 min), ultrasonic amplitude (P, 30–60%), CAB: solvent ratio (S, 30–50 g/mL) | RSM with (t-value of 15 min, P-value of 58%, S-value of 1:45 g/mL) | ANN-GA with (t-value of 15 min, P-value of 53%, S-value of 1:38 g/mL) | Before optimized R2 = 93.20%, yield (RSM)= 90.98% After optimized R2 = 99.60%, Yield (ANN-GA) = 94.97% |
[65] |
| RSM & ANN | Total phenolic content (TPC) from jujube (Ziziphus jujuba Mill.) | UAE | Time of extraction (t, 2–10 min), ultrasonic amplitude (P, 40–80%) | RSM with (t-value of 6.0 min, P-value of 60%) | ANN with (t-value of 6.2 min, P-value of 6.6%) | Before optimized R2 = 98.10%, yield (RSM)= 58.94 mg GAE/100 mL After optimized R2 = 98.90%, Yield (ANN) = 58.99 mg GAE/100 mL |
[66] |
Although, the usage of RSM for the modeling and optimization of bioactive compound extraction is favored, numerous limitations and challenges have been monitored. In several optimization studies, the inability to identify the true optimum points is due to the failure to properly select the factor ranges. Such failure will lead to the possibility of optimal conditions to fall outside the experiment region. As a consequence, the maximum or minimum responses will be mistakenly assigned as the optimal conditions, leading to unclear and unreliable optimization process. This is true since RSM models only work within a narrow experimental range and limited toward the applicability of extrapolation function [71]. Thus, preliminary experiments are recommended to first determine the correct range of independent variables prior experimental design. Furthermore, certain studies lack experiment repetition, and considering that experimental design relies on the minimum number of experiments, any inaccuracies in the data can significantly impact the results and overall optimization process. Another notable challenge arises in the accuracy of RSM model in predicting the extraction processes. The mathematical strategy of RSM relies on fitting all experimental data to a second-order polynomial equation, which restricts the number of curves available for data fitting. Subsequently, the RSM model may provide an estimation of the process response within a narrow range but not necessarily be the most accurate model for describing the response, leading to relatively low accurate predictions [76]. In addition, the constraint of RSM model becomes evident when it comes to handling non-linearity and accommodating inaccurate experimental data.
4.1.2. Connecting artificial neural network with extraction methods
As digitalization and computational capabilities began to advance, ANN models emerged as a ground-breaking approach for various scientific and technological applications. In the domain of bioactive compound extraction, ANN has gained much attention as an alternative mathematical algorithm to traditional methods such as RSM. ANN harnesses the ability of ML and neural networks (NNs) to model and optimize complex and non-linear extraction data, offering unparallel advantages in flexibility, accuracy, and adaptability. The primary inspiration for the ANN model as a prediction tool came from the structure and functioning of biological NNs, specifically the human brain [82]. The execution of ANN network involves training the input data from its surrounding to acquire the knowledge, and the connections between neurons are utilized to store the acquired knowledge. The breakthrough of ANN model outperforms RSM in terms of flexibility, effectiveness, and precision in fitting experimental data, modeling non-linear correlation, and making predictions [83].
A study was performed by Sarkar et al. [39], using ANN for the optimization and modeling of the extraction process of chlorophylls and carotenoids from Chlorella thermophilia. The architecture of ANN comprises six total number of input parameters (homogenization time, homogenization speed, solvent’s initial temperature, solid-solvent ration, boiling time, and microwave time) and 9 number of neurons in a single hidden layer, mapped onto two outputs (yield of carotenoids and chlorophylls) which results an overall R value of 0.98302. A comparison between RSM and ANN model was conducted by Ousaadi et al. [84], to evaluate the prediction accuracy and reliability for the optimization case study of culture conditions to maximize the yield of microbial enzymes. A total of 4 independent variables such as substrate concentration, inoculum size, sodium chloride powder, and pH were optimized based on RSM-CCD and ANN (with ten-fold cross-validation), which results in prediction accuracy of R2RSM = 0.725 and R2ANN = 0.884, respectively. A continuation of ANN studies for the optimization of fucoxanthin from microalgae is shown in Table 3.
4.1.3. Connecting hybrid models with extraction methods
Leveraging hybrid models with extraction methods presents an innovative and promising approach to optimize and enhance the efficiency of bioactive compounds extraction processes. As mentioned by Alshammari et al. [85], the downside of ANN model lies within the ‘black box’ character cannot be exploited to correlate the input factors and output variables and no clear definition on how or why the proposed ANN arrived at a particular result. Thus, the issue can be solved by incorporate additional RSM model to specially analyze the interaction between both input and response variables. Moreover, there are no definite and strict rules when constructing the ANN network structure, specifically the number of hidden layers and neurons in the hidden layer, as the network is performed by trail-and-error experience. As compared to RSM, ANN perform well typically on large dataset and prone to overfitting [83]. In order to strengthen the ANN model, GA can be applied to evaluate the optimized conditions from the predicted parameters. GA can provide a powerful optimization framework by mimicking the process of natural selection and genetics principle. They employ genetic operators such as selection, crossover, and mutation to generate new candidate solutions [86]. In the perspective of extraction, GA can be used to search for optimal combination of extraction parameters and by iteratively evaluating while refining these parameters, the performance of extraction process can be enhanced toward higher yields [87]. Combining both ANN (data processing and analysis) and GA (parameter optimizing) algorithm can help to further improve the prediction and optimization process [88]. On the other hand, adaptive neuro-fuzzy inference system (ANFIS) is defined as a hybrid soft computing approaches of ANN and fuzzy inference system (FIS) [89]. The architecture of ANFIS model can input and output both linear and non-linear relationship variables. Starting by training the data based on the least squares and back propagation of ANN, then output of ANN will be used to fuzzy logic membership functions for the desired input variables. The advantage of ANFIS model lies on the ability of FIS to increase the correctness of the optimization of these models [90].
An optimization case study by leveraging RSM-GA and ANN-GA hybrids were adapted by Joshi and Singhal [91], for the optimization of zeaxanthin from Paracoccus zeaxanthinifaciens ATCC 21,588. Initially, Taguchi design was considered to identify the most influential input parameters, followed by developing RSM and ANN models (unoptimized condition) for process optimization. It was found that ANN model outperformed RSM model for its mapping abilities based on statistical evaluation of standard error of prediction (SEP), RMSE, and R2. Lastly, both input variables of RSM and ANN model were optimized using GA. The final optimization was conducted by integrating GA model with ANN and RSM model, which results in prediction error of 1.55% and 5.09% for ANN-GA and RSM-GA, respectively. The overall optimization study was enhanced by 21% after incorporating GA as compared to unoptimized condition. Next, Aung et al. [92] coupled RSM-ANN-GA and compared it to RSM to study the predictability and accuracy for the optimization of infusion extraction (IE) and UAE of laver extract. The modeling of hybrid model was carried out by inputting extraction condition of ANN to be optimized by GA and the data generated from RSM experimental responses were mapped to the output layer. Between both RSM and hybrid model, RSM-ANN-GA hybrid model shown to provide higher reliability and accuracy in terms of higher R2 value. Baskararaj et al. [90] focused on the optimization parameters for maximum yield of β-carotene, biomass, chlorophyll, and the antioxidants from Kappaphycus alvarezii by comparing ANFIS and RSM model. The statistical analysis based on both RSM with CCD and ANFIS modeling was successful applied for the optimization of MAE extraction process. Similarly, no significant difference was observed between the prediction of RSM and ANFIS for the optimization of bioactive compound extraction, since both models were able to deliver relatively well consistent results with the predicted values [93]. More case studies related to the optimization of fucoxanthin extraction using hybrid models will be listed in Table 3.
As observed from Table 3, it can be summarized that the majority of studies focused on optimizing the extraction of fucoxanthin from microalgae have predominantly employed conventional mathematical tools like RSM. However, there is a growing interest in exploring metaheuristic algorithm such as GA, and neural networks inspired from the human brain such as ANN and ANFIS to further enhance the optimization process and proved to surpass the results obtained from using RSM model. The RSM approach has proven to be effective in certain cases but have limitations in handling non-linear relationships and complex interactions between extraction variables. On the contrary, ANN with the ability to learn patterns and relationship from large datasets, capturing non-linearity, predicting optimal conditions and provide information into the factor that significantly impact fucoxanthin yield. On top of that, GA can handle both continuous and discrete variables, allowing more comprehensive exploration of the parameter space and discovery of optimal conditions. ANFIS model can effectively model complex relationships between input factors and output variables due to its combined strengths of fuzzy logic and neural networks to handle uncertainties and imprecise information. By harnessing the power of AI, full potential of extraction process can be achieved in terms of higher extraction yields, reduced costs, and accelerate the development of fucoxanthin-based products.
5. Digitalised perspectives on the quantification of organic pigment content: AI techniques based on color and statistical features
An emerging approach is to uphold the development of an efficient method to quantify the changes in microalgae pigment, specifically fucoxanthin based on a nondestructive approach to maintain the perception on environmental and sustainability. A study by Ho Thanh Lam et al. [94] had identified the properties of antioxidant proteins with a wide range of different biochemical tests and rapid aid-kits, however the process has high cost and is time consuming. Furthermore, HPLC is among the useful technique in quantitative and qualitative analysis of bioactive compound but utilizes large amount of solvents, long analysis time, and high analysis cost [18]. Generally, chromatography techniques are complex which limits one’s ability to generate one approach that includes necessary parameters to foresee the quantitative and qualitative of desired bioactive compound and optimizing its efficiency [18]. Recently, researchers have been brainstorming on new approaches to produce reliable results rapidly by reinforcing multiple chromatography techniques that are capable of forecasting the behavior of chromatographic system [18]. Thus, the evolving AI models have become reality acting as future tools and dependable chemometric methodologies in many research areas. Figure 3 shows the key steps for the incorporation of ML and DL techniques into the quantification of fucoxanthin.
5.1. Machine learning approaches for smart organic pigment quantification
In quantification task, ML has the ability to learn from datasets that contain known measurements of the target parameter through recognizing patterns, relationships, and correlation within the data. The effectiveness of ML can capture intricate relationships between input variables (i.e. spectral data, image data, color features) and the target variable (i.e. concentration of fucoxanthin) that may not easily discernible through conventional methods [95].
Relevant studies, for instance Yew et al. [96] leverage the k-nearest neighbor (k-NN) algorithm through RGB (Red, Green, Blue) model pixel raster in the images to predict the nitrogen concentration, biomass concentration, and pH of microalgae Chlorella vulgaris FSP-E. This smart approach simplifies the process by capturing images of the microalgae cultivation using a smartphone and results shown to be successful with optimized conditions at k-value of 4, where the average RSME between predicted and actual was the lowest. An ML workflow was incorporated to develop spectrophotometric equations for the quantification of lutein, violaxanthin, zeaxanthin, and chlorophyll a, b, simultaneously from Chlorella vulgaris and Scenedesmus almeriensis. The data of various pigment concentrations along with their associated visible spectra obtained from HPLC were collected and were used to train the ML model based on particle swarm optimizer-assisted partial least square regression (PSO-assisted PLS). In total, seven feature models including one absorbance and six absorbance derivatives were obtained which leads to less time consumption for the quantification of carotenoid concentrations while retaining adequate accuracy [67]. In addition, de Carvalho and Nunes [68] proposed a calibration transfer approach based on digital images taken by smartphones to predict the levels of chlorophyll and carotenoids in olive and avocado oils. ML models such as MLR and LS-SVM were trained on the color space of images which includes RGB, Y (luma), HSV (hue, saturation, value), CMYK (cyan, magenta, yellow, black), and L*a*b (ranging from black (0) to white (100), ranging from green if -ve to red if +ve, ranging from yellow if +ve to blue if -ve), and XYZ (red, green, blue) under different type of camera and lightning conditions. The best model for predicting both chlorophyll and carotenoid contents were XYZ/LS-SVM showing R2 of 0.96 and Y/LS-SVM showing R2 of 0.83, respectively. A hybrid model of competition adaptive reweighted sampling (CARS)-SVM model was proposed by Yang et al. [97] to predict the mannitol, polysaccharide, fucosterol, and fucoxanthin from Sargassum fusiforme. The near-infrared (NIR) spectroscopy was utilized to obtain spectra data in the range of 833–2500 nm at approximate 0.8 nm sampling interval to be fed into the model. The CARS algorithm was implemented for the proper selection of valuable wavelengths prior SVM model training, which results in R2mannitol = 0.81, R2polysaccharide = 0.86, R2fucosterol = 0.84, and R2fucoxanthin = 0.78. Besides, total carotene was successfully predicted based on PCR, PLS, and LS-SVM by integrating HPLC for the data acquisition of Raman spectrum ranging from 800–1800 cm−1 (74 data as calibration set & 38 data as prediction set). The best fitted model was found to be PLS followed by PCR and SVM with R2PLS (predicted) = 0.950, R2PCR (predicted) = 0.942,, R2SVM (predicted) = 0.919 [69]. Another nondestructive method for the quantification of carotenoid content in Manihot esculenta (Cassava plant) using SVM Kernel algorithm associated with CIELAB color measurement as input variable [98]. The comparison between analytical methods such as HPLC and UV-vis spectrophotometry and colorimetric such as CIELAB color measurement is associated with SVM model to determine the output error and accuracy. The final result showed that chromatographic method has higher R2 and lower RMSE values due to the differences in their physicochemical bases, however CIELAB method produces acceptable error margin. Therefore, CIELAB method associated with SVM can produce rapid, low-cost, nondestructive, good accuracy, and low error as training progresses in near future.
5.2. Deep learning embedded into digital organic pigment quantification
DL is a subset of ML that implements deep neural algorithms such as ANN and convolutional neural networks (CNNs), have the ability to solve non-linear problems and automatically extract relevant features from spectral data or digital images. On the contrary, ML models often rely on manual feature engineering, where domain experts manually select and design features to be fed into the learning algorithms [45]. Based on Shishodia et al. [73], they stressed that ANN model was successful in modeling complex non-linear input-output relationships in some extremely interdisciplinary field. ANN model was widely applied in wastewater treatment [99], food processing [100], and analytical chemistry [101] to predict the extraction and quantification efficiencies.
The application of CNN model, specifically 1D-CNN model has been used for the prediction of microalgae pigments such as chlorophyll-a, phycocyanin, lutein, fucoxanthin, and zeaxanthin from green algae, diatoms, and cyanobacteria to gain better understanding on the occurrence of algae blooms. The experiment data was obtained from water samples of interest based on reflectance and absorption coefficient spectral inputs measured from field monitoring and drone hyperspectral image sensing devices. The 1D-CNN model managed to predict the concentrations of pigments with R2 ranging from 0.74–0.70 [40]. Furthermore, Tang et al. [49] investigated the comparison between linear regression and ANN model in predicting the chlorophyll content from mixed microalgae species (Desmodesmus sp. and Scenedesmus sp.) based on RBG, CYMK, and HSV color models. Multilayer perceptron was chosen for the training of ANN algorithm with a total of 1000 samples and 1–3 hidden layers. The experiment shown that ANN model surpassed the linear regression model with R2 of 0.66 and R2 of 0.58, respectively. In short, providing potential alternative for the estimation of chlorophyll concentration when using ANN model with rapid, low cost, and more efficient analysis. The development of high-throughput quantification tools is important for screening hundreds of cassava genotypes in a short period of time for the estimation of total carotenoids content (TCC). Therefore, de Carvalho et al. [102] aimed to extract color features from digital images taken from pulp color of cassava roots based on RGB, L*a*b, hue, and chroma color spaces by leveraging 12 different models associated with PCA (principal component analysis) clustering. Among 12 different models, ANN presented the best predictive ability showing R2 of 0.94 as compared to other models with R2 ranging from 0.81–0.94, demonstrating economical, rapid, and effective option for the development of TCC tools. An AI-based approach was conducted by Asnake Metekia et al. [70], to study the effects of Spirulina platensis growth mediums on total phenolic compounds by comparing ANFIS, MLP, and SWLR algorithms. These algorithms were trained on several input variables such as algae productivity (P), extraction yield (EY), total flavonoids (TF), percentage flavonoid (%F), and percentage phenols (%P) to predict the concentration of total phenolic compounds. Overall, both ANFIS and SWLR models were superior in the testing phase with increased in 2% higher accuracy as compared to MLP model. Similarly, Taghadomi-Saberi et al. [103] integrated fixed-grid wavelet network (FGWN) and image processing techniques for the quantification of chlorophyll and carotenoid pigments from orange peel. The methodology included image acquisition from smartphone followed by feature extraction based on RGB and CIE color spaces, then applying PCA for effective feature selection and finally leveraging ANN and ANFIS models for prediction. The findings were reliable when modeling both chlorophyll and carotenoids using ANN and ANFIS, showing R2chlorophyll-ANN = 0.975, R2carotenoid-ANN = 0.926, R2chlorophyll-ANFIS = 0.992, and R2carotenoid-ANFIS = 0.984.
In summary, these AI models have different approaches for the quantification of fucoxanthin which can be categorized into computer vision based on color space of images and regression analysis based on statistical data. For instance, collecting input data of visible spectra or absorbance data from HPLC along with the associated output data of various carotenoid concentrations prior training the ML model was conducted. Likewise, collecting input of spectra data in the range of 833–2500 at every 0.8 nm sampling interval, a total of 112 data of Raman spectrum in the range of 800–1800 cm−1, reflectance, and absorption coefficient spectral input data are considered prior subjecting into AI models for regression analysis [40]. On the other hand, collecting images data followed by extraction of color measurements from various color spaces such as RGB, HSV, XYZ, and CIELAB are considered prior fed into AI models for training [49]. Although the study of these techniques is limited toward the application of fucoxanthin, these findings have proven the feasibility and potential of AI for quantification purposes, which can be applied in future fucoxanthin studies.
6. Current challenges of AI in the downstream processing of fucoxanthin
In this review, an insightful perception of incorporating several AI models such as ANN, SVM, and ANFIS to further improve the extraction, quantification, and identification of fucoxanthin from microalgae is provided. Analytical detection tools such as HPLC, UV-vis spectrophotometer, NMR, FTIR, LC, and combination of these are still widely employed without any automation strategy which results in long experimental time, high analysis cost, exploitation of organic solvents, and restriction in high yield due to lack of knowledge in optimization. These limitations can be solved with the help of AI, however there are some challenges that required to take into consideration.
The aforementioned AI models for the quantification and optimization of carotenoids along with antioxidant activity have shown significant progression in terms of high-level accuracy, good predictions, nondestructive, and good optimization. However, recent research is still lacking in terms of research knowledge, data, capability for up-scaling, and the scarcity of integrating AI studies in the field of microalgae, especially related to the extraction and quantification of biomolecules. Recently, ML has been widely applied in the pharmaceutical and biotechnological companies such as Amgen, Bayer, Eli Lilly, Johnson & Johnson, and Merck & Co, Pfizer for the discovery and biomarker identification, discovery alongside manufacturing of drugs, diagnosis with identification of diseases, developing personalized treatments, and clinical trial research [104]. In addition, companies such as Roche, Takeda, Janssen, and Novartis have utilized ML models for drug discovery, identification of drug molecules for central nervous systems disorders, development of drug to treat patients with Parkinson’s disease, and optimization of cell & gene therapies, respectively [105]. Moreover, ML models have been intensively used in cancer research which aims to decode cancer cell images to differentiate normal cells from tumor cells [106]. In the healthcare sector, AI models are used to detect patterns in data automatically to predict data trends or allow decision making under uncertain conditions. Adding on, predicting clinical possibilities, simplifying process management, providing active guidance to physicians when making clinical decisions, and detecting diabetic retinopathy which will greatly improve the quality of patients’ lives and reducing complexity along with cost expenditure [107].
According to Kalyane et al. [108], pharmaceutical industries usually requires to screen through about 10,000 chemical and biological component and most likely just to produce a single potent molecule, however if taking into account the probability of molecule failure rate during experimentation, this could lead to considerably high investment in research and development (R&D). In 2018, 92% of companies invested around $2.6 billion US and managed to development just a single drug molecule and thus, incorporation of AI models with computational algorithm have designed to overcome associated failures, reducing the cost for R&D, less time consumption, high molecules success rate, providing superior platform for drug design, reducing number of experiments required, and avoiding redundant information in both biotechnological and pharmaceutical sectors [108,109]. The fast and reliable nature of AI models can promote higher chances of ML models to be incorporated into the extraction, identification, and quantification of fucoxanthin to produce satisfying and potential results in industrial settings. In the pursuit of integrating AI into fucoxanthin quantification and recovery, however there are several challenges that need to be considered and addressed in terms of data availability and quality, model interpretability and transparency, model generalization, and transferability. Such cases where large dataset is unavailable, mainly due to high cost, complex measurements, limited resources, confidentiality, ethical problems, or just simply non-existent [36]. A simple question one doubted ‘How much data should be collected, and what defines the quality of data required to train a model to achieve better generalization results … ?.’ As suggested by Chong et al. [43], there are no definite rule of the size of data, instead lacking of desired information when selecting optimal input variables for desired output results. In order to maximize the potential of AI integrated into the extraction and quantification of fucoxanthin from microalgae, suggestions such as more lenient approach toward platform transparency, sharing research data, and data transferability to make data easily accessible and usable across multiple systems and platforms ought to be taken into action.
7. Research gap and recommendations
The advent of ChatGPT has marked a significant milestone in the domain of AI. The advancement and breakthrough of AI due to the popular successes of ChatGPT have encouraged and engaged researchers to integrate the advances of biotechnology with AI to create and solving new potential solutions efficiently and effectively. As mentioned by Holzinger et al. [110], digital transformation in the context of biotechnology can establish new technologies and processes to improve the speed, accuracy, and efficiency of research. Concurrently, to accelerate the development of completely new, disruptive products and services by providing access to big data and automating certain tasks. Recently, digitalization approaches such as AI algorithms workflow and IoTs (Internet of Things) sensors on microalgae biotechnology have been studied for system design, economic, process optimization, and environmental impact improvement [111]. Moreover, Teng et al. [50] provide comprehensive review on the utilization of AI algorithms to improve microalgae cultivation, identification, genome editing, and system optimization. While, Wang et al. [112] emphasized more toward how IoT technology can assist in real-time monitoring, automation, low-cost, efficient, smart prediction and decision of microalgae biorefinery. To the best of author’s knowledge, there is a lack and scarcity of studies concerning the digitalized perspective on the extraction and quantification of bioactive compounds, particularly fucoxanthin from microalgae.
Therefore, similar studies related to the application and contribution of AI to improve the extraction and quantification of bioactive compounds should be further studied. Decision tree (DT) and random forest (RF) are other supervised ML models that can be considered for making predictions in microalgae cultivation and bioproduct extraction. Similar to the case of ANN, SVM, and ANFIS, DT has the advantage of simple interpretation, fast to train, and can solve non-linear relationship between the input and output variables. On the other side, RF is an ensemble learning model that can successfully prevent the chance of data overfitting [113]. For instance, Zhang et al. [114] leverage the RF model to study the optimization process of bio-oil yield from microalgae hydrothermal liquefaction, while Singh and Mishra [115] leverage the DT model to identify optimized key variables, including microalgal class, cultivation factors, and operating parameters. These variables will then have the potential to impact both biomass productivity and wastewater treatment efficiency. As previously mentioned, ANFIS model has the high chance of being trap in local extreme, in which GA algorithm is much preferable for better performance. In addition to GA algorithm, there are many other optimization algorithms or heuristic algorithms such as tabu search (TS), simulated annealing (SA), PSO, differential evolution (DE), biogeography-based optimization (BBO), and social network (SNO). However, lack computational power and flexibility to consistently generate optimal designs over time. Moving on, swarm intelligence (SI) algorithms such as starling murmuration optimizer (SMO), golden jackal optimization (GJO), white shark optimizer (WSO), dandelion optimizer (DO), search in forest optimizer (SIFO), snake optimizer (SO), and beluga whale optimization (BWO) are broadly applied in various sectors and proven to perform better as compared to conventional optimization methods. Out of these optimizers, BWO is a trending nature inspired system that mimics the attacking and feeding behaviors of beluga whales [116]. The advantages of BWO can provide higher stability, search ability, convergence rate and speed, but lack in terms of premature convergence and risk of being trapped in local optimum. Recently, Horng and Lin [116] came out with a improved BWO or IBWO, with improvement in the learning approach, acceleration of searching process, and variety and consistency of chosen candidates in order to provide a much reliable optimization process.
The potential of incorporating AI models for the quantification of fucoxanthin have known to reduce experimental time, reduce in solvent usage, accurate, and cost-efficient. However, choosing the most suitable machine learning model for a specific purpose can be difficult for pioneers. To our best knowledge, there have been scarce reports on the quantification of fucoxanthin related to AI. The concept of quantification and identification of bioactive compounds (e.g. antioxidants, carotenoids, pigments) using AI models are more developed toward food products [117] (e.g. antioxidants from banana, carotenoids from tomato), plants such as antioxidant from green tea leaves [73], and carotenoids from Manihot esculenta [98]. Fucoxanthin is defined as carotenoid with orange colored pigmentation along with the presence of antioxidant properties. Hence, a comprehensive review on the application of machine and deep learning techniques for the quantification process of pigments that closely resembled the characteristics of fucoxanthin from microalgae should be of focus. An exploration into deep learning models such as CNNs, self-attention mechanism in transformers (i.e. Vision Transformer (ViT), DEtection Transformer (DETR)), and image segmentation (i.e. Segment anything model) can be applicable for the quantification task for microalgae due to their potential ability to deal with image data in computer vision tasks [45,118].
7.1. Conclusions
The downstream processing of fucoxanthin which follows the conventional trend leads to long analysis time, toxic solvent utilization, and high-cost analysis. Several robust and effective AI models such as ANN, SVM, and ANFIS can promote rapid, precise, and accurate analytical results without consuming large amount of toxic solvents. In order to incorporate these AI models effectively, a systematic methodology approach has been considered. This approach involves several key steps to ensure a structured and reliable implementation which includes data collection, data pre-processing, sensitive analysis test and feature selection, model selection, model development, optimization, validation, and lastly statistical evaluation. Furthermore, evaluating the feasibility of AI integration in the extraction techniques of fucoxanthin by comparing the advantages and limitations of conventional RSM model with ANN and hybrid models. In addition, a comprehensive study between different ML and DL models for the quantification of fucoxanthin due to their ability to deliver rapid and accurate results in a nondestructive manner. Despite the effectiveness of these AI models, there is a lack and scarcity of studies concerning the digitalized perspective on the extraction and quantification of bioactive compounds. Nonetheless, collaboration among multidisciplinary researchers can foster knowledge sharing to help bridge the gap between microalgae research and AI integration, leading to improved model, more comprehensive data, better understanding of the quantification and extraction process. There is also a need to consider computational resources, scalability, and efficiency for developing large-scale AI models that could handle industrial applications in real-world scenarios.
Nomenclature
- Artificial intelligence
AI
- Artificial neural network
ANN
- Support vector machine
SVM
- Adaptive neuro fuzzy inference system
ANFIS
- Ultrasound-assisted extraction
UAE
- Microwave-assisted extraction
MAE
- Enzyme-assisted extraction
EAE
- Pressurized liquid extraction
PLE
- High performance liquid chromatography
HPLC
- Machine learning
ML
- Deep learning
DL
- Crude methanolic extract
CME
- Initial low nitrogen
ILN
- Initial high nitrogen
IHN
- Supplementary nitrogen
SN
- Laminaria japonica hydrolysate
LPH
- Structural Risk Minimisation
SRM
- Root mean square error
RMSE
- Mean absolute error
MAE
- Mean squared error
MSE
- Coefficient of determination
R2
- Degree of approximation
Da
- Genetic algorithm
GA
- Multiple linear regression
MLR
- Multilayer perceptron
MLP
- Particle swarm optimiser
PSO
- Partial least square regression
PLS
- Least squares support vector machine
LS-SVM
- Principal component regression
PCR
- One dimensional convolutional neural network
1D-CNN
- Step-wise-linear regression
SWLR
- Central composite design
CCD
- Box-Behnken design
BBD
- Supercritical carbon dioxide
SC-CO2
- Neural networks
NNs
- Fuzzy inference system
FIS
- Standard error of prediction
SEP
- Infusion extraction
IE
- Cashew apple bagasse
CAB
- Total phenolic content
TPC
- k-nearest neighbour
k-NN
- RGB
Red, Green, Blue
- Y
luma
- HSV
hue, saturation, value
- CMYK
cyan, magenta, yellow, black
- L*a*b
ranging from black (0) to white (100), ranging from green if -ve to red if +ve, ranging from yellow if +ve to blue if -ve
- XYZ
red, green, blue
- Competition adaptive reweighted sampling
CARS
- Near-infrared spectroscopy
NIR
- Convolutional neural networks
CNNs
- Total carotenoids content
TCC
- Productivity
(P)
- Extraction yield
EY
- Total flavonoids
TF
- Percentage flavonoid
%F
- Percentage phenols
%P
- Fixed-grid wavelet network
FGWN
- Ultraviolet – visible spectroscopy
UV-vis spectrophotometer
- Nuclear magnetic resonance
NMR
- Fourier-transform infrared spectroscopy
FTIR
- Liquid chromatography
LC
- IoTs
Internet of Things
- Decision tree
DT
- Random forest
RF
- Tabu search
TS
- Simulated annealing
SA
- Differential evolution
DE
- Biogeography-based optimisation
BBO
- Social network
SNO
- Swarm intelligence
SI
- Starling murmuration optimiser
SMO
- Golden jackal optimisation
GJO
- White shark optimiser
WSO
- Dandelion optimiser
DO
- Search in forest optimiser
SIFO
- Snake optimiser
SO
- Beluga whale optimisation
BWO
- Improved beluga whale optimisation
IBWO
- Vision Transformer
ViT
- DEtection Transformer
DETR
Acknowledgments
The authors would like to thank Nanyang Technological University (Singapore) for the facilities and resources provided for the completion of this work. The authors would also like to acknowledge the support of the recent algae initiative in gathering peers of algae researchers by launching an Algal Biotechnology Consortium (ABC), which is a platform for algae researchers to share, communicate and outreach their algae research with the intention to create sustainable solutions and greener future.
Funding Statement
The author(s) reported there is no funding associated with the work featured in this article.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Data not available
Credit author statement
Jun Wei Chong Roy: Writing – original draft, Writing – review & editing, Kit Wayne Chew: Conceptualization, Formal analysis, Data curation, Funding acquisition, Investigation, Doris Ying Ying Tang: Supervision, Validation, Writing – review & editing, Hui Yi Leong: Data curation, Formal analysis, Funding acquisition, Investigation, Visualization, Kuan Shiong Khoo: Conceptualization, Visualization, Pau Loke Show: Supervision, Validation, Writing – review & editing, Kit Wayne Chew: Funding acquisition, Supervision, Validation
References
- [1].Guedes AC, Amaro HM, Malcata FX.. Microalgae as sources of carotenoids. Marine Drugs query. 2011;9(4):625–386. doi: 10.3390/md9040625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Mohamadnia S, Tavakoli O, Faramarzi MA, et al. Production of fucoxanthin by the microalga tisochrysis lutea: A review of recent developments. Aquaculture. 2020;516. doi: 10.1016/j.aquaculture.2019.734637 [DOI] [Google Scholar]
- [3].Chong JWR, Yew GY, Khoo KS, et al. Recent advances on food waste pretreatment technology via microalgae for source of polyhydroxyalkanoates. J Environ Manage. 2021;293:112782. doi: 10.1016/j.jenvman.2021.112782 [DOI] [PubMed] [Google Scholar]
- [4].Roy CJW, Tan X, Khoo KS, et al. Microalgae-based bioplastics: future solution towards mitigation of plastic wastes. Environ Res. 2021;206:112620. doi: 10.1016/j.envres.2021.112620 [DOI] [PubMed] [Google Scholar]
- [5].Suganya T, Varman M, Masjuki HH, et al. Macroalgae and microalgae as a potential source for commercial applications along with biofuels production: A biorefinery approach. Renewable Sustainable Energy Rev. 2016;55:909–941. doi: 10.1016/j.rser.2015.11.026 [DOI] [Google Scholar]
- [6].Foo SC, Khoo KS, Ooi CW, et al. Meeting sustainable development goals: alternative extraction processes for fucoxanthin in algae. Front Bioeng Biotechnol. 2020;8:546067. doi: 10.3389/fbioe.2020.546067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Miyashita K, Hosokawa M. Carotenoids as a nutraceutical therapy for visceral obesity. in: nutrition in the prevention and treatment of abdominal obesity. 2014. pp. 329–340. 10.1016/B978-0-12-407869-7.00030-1 [DOI] [Google Scholar]
- [8].Andersen RA. Biology and systematics of heterokont and haptophyte algae. Am J Bot. 2004;91(10):1508–22. doi: 10.3732/ajb.91.10.1508 [DOI] [PubMed] [Google Scholar]
- [9].Beppu F, Niwano Y, Tsukui T, et al. Single and repeated oral dose toxicity study of fucoxanthin (FX), a marine carotenoid, in mice. J Toxicol Sci query. 2009;34(5):501. doi: 10.2131/jts.34.501 [DOI] [PubMed] [Google Scholar]
- [10].Yan X, Chuda Y, Suzuki M, et al. Fucoxanthin as the major antioxidant in hijikia fusiformis, a common edible seaweed. Bioscience, Biotechnology, And Biochemistry query. 1999;63(3):605–607. doi: 10.1271/bbb.63.605 [DOI] [PubMed] [Google Scholar]
- [11].Ming JX, Wang ZC, Huang Y, et al. Fucoxanthin extracted from laminaria japonica inhibits metastasis and enhances the sensitivity of lung cancer to gefitinib. J Ethnopharmacol. 2021;265:113302. doi: 10.1016/j.jep.2020.113302 [DOI] [PubMed] [Google Scholar]
- [12].Heo S-J, Yoon W-J, Kim K-N, et al. Anti-inflammatory effect of fucoxanthin derivatives isolated from sargassum siliquastrum in lipopolysaccharide-stimulated RAW 264.7 macrophage. Food Chem Toxicol. 2012;50(9):3336–3342. doi: 10.1016/j.fct.2012.06.025 [DOI] [PubMed] [Google Scholar]
- [13].Raguraman V, SA L, M D, et al. Unraveling rapid extraction of fucoxanthin from padina tetrastromatica: purification, characterization and biomedical application. Process Biochem. 2018;73:211–219. doi: 10.1016/j.procbio.2018.08.006 [DOI] [Google Scholar]
- [14].Maeda H, Hosokawa M, Sashima T, et al. Fucoxanthin from edible seaweed, undaria pinnatifida, shows antiobesity effect through UCP1 expression in white adipose tissues. Biochemical And Biophysical Research Communications query. 2005;332(2):392–397. doi: 10.1016/j.bbrc.2005.05.002 [DOI] [PubMed] [Google Scholar]
- [15].Foo SC, Yusoff FM, Imam MU, et al. Increased fucoxanthin in chaetoceros calcitrans extract exacerbates apoptosis in liver cancer cells via multiple targeted cellular pathways. Biotechnol Report (Amst). 2019;21:e00296. doi: 10.1016/j.btre.2018.e00296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Siahaan EA, Chun BS. Innovative alternative technology for fucoxanthin recovery. In: Encyclopedia of Marine Biotechnology. 2020. pp. 3213–3227. 10.1002/9781119143802.ch143 [DOI] [Google Scholar]
- [17].Mena-García A, Ruiz-Matute AI, Soria AC, et al. Green techniques for extraction of bioactive carbohydrates. TrAC - Trends Anal Chem. 2019;119:115612. DOI: 10.1016/j.trac.2019.07.023 [DOI] [Google Scholar]
- [18].Usman AG, S IS, Abba SI, et al. Artificial intelligence-based models for the qualitative and quantitative prediction of a phytochemical compound using HPLC method. Turk J Chem. 2020;44(5):1339–1351. doi: 10.3906/kim-2003-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Tüzen M, Özdemir M. Chromatographic determination of phenolic acids in the snowdrop by HPLC. Turk J Chem. 2003;27:49–54. [Google Scholar]
- [20].Pocha CKR, Chia WY, Chew KW, et al. Current advances in recovery and biorefinery of fucoxanthin from phaeodactylum tricornutum. Algal Res. 2022;65:102735. doi: 10.1016/j.algal.2022.102735 [DOI] [Google Scholar]
- [21].Ibrahim AF. Prediction of coal wettability using machine learning for the application of CO2 sequestration. Int J Greenhouse Gas Control. 2022;118:103670. doi: 10.1016/j.ijggc.2022.103670 [DOI] [Google Scholar]
- [22].Ibrahim AF, Alarifi SA, Elkatatny S. Application of machine learning to predict estimated ultimate recovery for multistage hydraulically fractured wells in niobrara shale formation. Computer Intellgent Neuroscience. 2022;2022. doi: 10.1155/2022/7084514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ibrahim AF, Gowida A, Ali A, et al. Machine learning application to predict in-situ stresses from logging data. Sci Rep. 2021;11(1):23445. doi: 10.1038/s41598-021-02959-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Miyashita K, Nishikawa S, Beppu F, et al. The allenic carotenoid fucoxanthin, a novel marine nutraceutical from brown seaweeds. J Sci Food Agric query. 2011;91(7):1166–74. doi: 10.1002/jsfa.4353 [DOI] [PubMed] [Google Scholar]
- [25].Leong YK, Chen C-Y, Varjani S, et al. Producing fucoxanthin from algae–recent advances in cultivation strategies and downstream processing. Bioresour Technol. 2022;344:126170. DOI: 10.1016/j.biortech.2021.126170 [DOI] [PubMed] [Google Scholar]
- [26].Tachihana S, Nagao N, Katayama T, et al. High productivity of eicosapentaenoic acid and fucoxanthin by a marine diatom chaetoceros gracilis in a semi-continuous culture. Front Bioeng Biotechnol. 2020;8(1435): doi: 10.3389/fbioe.2020.602721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kim SM, Kang S-W, Kwon ON, et al. Fucoxanthin as a major carotenoid in isochrysis aff. galbana: characterization of extraction for commercial application. J Korean Society Applied Biology Chemical. 2012b;55(4):477–483. doi: 10.1007/s13765-012-2108-3 [DOI] [Google Scholar]
- [28].Bhattacharjya R, Kiran Marella T, Tiwari A, et al. Bioprospecting of marine diatoms thalassiosira, skeletonema and chaetoceros for lipids and other value-added products. Bioresour Technol. 2020;318:124073. doi: 10.1016/j.biortech.2020.124073 [DOI] [PubMed] [Google Scholar]
- [29].Foo SC, Yusoff FM, Ismail M, et al. Production of fucoxanthin-rich fraction (FxRF) from a diatom, chaetoceros calcitrans (paulsen) takano 1968. Algal Res. 2015a;12:26–32. doi: 10.1016/j.algal.2015.08.004 [DOI] [Google Scholar]
- [30].Xia S, Gao B, Fu J, et al. Production of fucoxanthin, chrysolaminarin, and eicosapentaenoic acid by odontella aurita under different nitrogen supply regimes. J Biosci Bioeng. 2018;126(6):723–729. doi: 10.1016/j.jbiosc.2018.06.002 [DOI] [PubMed] [Google Scholar]
- [31].McClure DD, Luiz A, Gerber B, et al. An investigation into the effect of culture conditions on fucoxanthin production using the marine microalgae phaeodactylum tricornutum. Algal Res. 2018;29:41–48. doi: 10.1016/j.algal.2017.11.015 [DOI] [Google Scholar]
- [32].Li J, Liu Y, Liu Y, et al. Effects of temperature and salinity on the growth and biochemical composition of the brown alga sargassum fusiforme (fucales, phaeophyceae). J Appl Phycol. 2019;31(5):3061–3068. doi: 10.1007/s10811-019-01795-9 [DOI] [Google Scholar]
- [33].Wang Z-P, Wang P-K, Ma Y, et al. Laminaria japonica hydrolysate promotes fucoxanthin accumulation inphaeodactylum tricornutum. Bioresour Technol. 2022b;344:126117. doi: 10.1016/j.biortech.2021.126117 [DOI] [PubMed] [Google Scholar]
- [34].Seth K, Kumar A, Rastogi RP, et al. Bioprospecting of fucoxanthin from diatoms—challenges and perspectives. Algal Res. 2021;60:102475. DOI: 10.1016/j.algal.2021.102475 [DOI] [Google Scholar]
- [35].Khaw YS, Yusoff FM, Tan HT, et al. The critical studies of fucoxanthin research trends from 1928 to june 2021: A bibliometric review. Mar Drugs. 2021;19(11):606. doi: 10.3390/md19110606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Maleki F, Muthukrishnan N, Ovens K, et al. Machine learning algorithm validation: from essentials to advanced applications and implications for regulatory certification and deployment. Neuroimaging Clinical N Am. 2020;30(4):433–445. doi: 10.1016/j.nic.2020.08.004 [DOI] [PubMed] [Google Scholar]
- [37].Willemink MJ, Koszek WA, Hardell C, et al. Preparing medical imaging data for machine learning. Radiology. 2020;295(1):4–15. doi: 10.1148/radiol.2020192224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Jha K, Doshi A, Patel P, et al. A comprehensive review on automation in agriculture using artificial intelligence. Artifical Intellgent Agriculture. 2019;2:1–12. doi: 10.1016/j.aiia.2019.05.004 [DOI] [Google Scholar]
- [39].Sarkar S, Manna MS, Bhowmick TK, et al. Extraction of chlorophylls and carotenoids from dry and wet biomass of isolated chlorella thermophila: optimization of process parameters and modelling by artificial neural network. Process Biochem. 2020;96:58–72. DOI: 10.1016/j.procbio.2020.05.025 [DOI] [Google Scholar]
- [40].Pyo J, Hong SM, Jang J, et al. Drone-borne sensing of major and accessory pigments in algae using deep learning modeling. GIScience & Remote Sensing. 2022;59(1):310–332. doi: 10.1080/15481603.2022.2027120 [DOI] [Google Scholar]
- [41].Ramírez-Gallego S, Krawczyk B, García S, et al. A survey on data preprocessing for data stream mining: current status and future directions. Neurocomputing. 2017;239:39–57. DOI: 10.1016/j.neucom.2017.01.078 [DOI] [Google Scholar]
- [42].Dwivedi SK, Rawat B. 2015. A review paper on data preprocessing: A critical phase in web usage mining process. 2015 International Conference on Green Computing and Internet of Things (ICGCIoT), Greater Noida, India, 8-10 Oct. 8-10. pp. 506–510. [Google Scholar]
- [43].Chong JWR, Khoo KS, Chew KW, et al. Trends in digital image processing of isolated microalgae by incorporating classification algorithm. Biotechnol Adv. 2023a;63:108095. [DOI] [PubMed] [Google Scholar]
- [44].Douglass MJJ. Book review: hands-on machine learning with scikit-learn, keras, and tensorflow, 2nd edition by aurélien géron. Physics Engineering Science Medical. 2020;43(3):1135–1136. doi: 10.1007/s13246-020-00913-z [DOI] [Google Scholar]
- [45].Chong JWR, Khoo KS, Chew KW, et al. Microalgae identification: future of image processing and digital algorithm. Bioresour Technol. 2023b;369:128418. doi: 10.1016/j.biortech.2022.128418 [DOI] [PubMed] [Google Scholar]
- [46].Pham BT, Nguyen MD, Dao DV, et al. Development of artificial intelligence models for the prediction of compression coefficient of soil: an application of monte carlo sensitivity analysis. Sci Total Environ. 2019;679:172–184. doi: 10.1016/j.scitotenv.2019.05.061 [DOI] [PubMed] [Google Scholar]
- [47].Haghbakhsh R, Hayer H, Saidi M, et al. Density estimation of pure carbon dioxide at supercritical region and estimation solubility of solid compounds in supercritical carbon dioxide: correlation approach based on sensitivity analysis. Fluid Ph Equilibria. 2013;342:31–41. doi: 10.1016/j.fluid.2012.12.029 [DOI] [Google Scholar]
- [48].Puy A, Lo Piano S, Saltelli A. A sensitivity analysis of the PAWN sensitivity index. Environ Model Softw. 2020;127:104679. doi: 10.1016/j.envsoft.2020.104679 [DOI] [Google Scholar]
- [49].Tang DYY, Chew KW, Ting H-Y, et al. Application of regression and artificial neural network analysis of Red-Green-Blue image components in prediction of chlorophyll content in microalgae. Application Of Regression And Artificial Neural Network Analysis Of Red-Green-Blue Image Components In Prediction Of Chlorophyll Content In Microalgae Bioresour Technologies. 2022;370:128503. doi: 10.1016/j.biortech.2022.128503 [DOI] [PubMed] [Google Scholar]
- [50].Teng SY, Yew GY, Sukačová K, et al. Microalgae with artificial intelligence: A digitalized perspective on genetics, systems and products. Biotechnol Adv. 2020;44:107631. doi: 10.1016/j.biotechadv.2020.107631 [DOI] [PubMed] [Google Scholar]
- [51].Thakur A, Konde A. Fundamentals of neural networks. International Journal Research Applied Science Engineering Technologies. 2021;9(VIII):407–426. doi: 10.22214/ijraset.2021.37362 [DOI] [Google Scholar]
- [52].Sewsynker-Sukai Y, Faloye F, Kana EBG. Artificial neural networks: an efficient tool for modelling and optimization of biofuel production (a mini review). Biotechnology & Biotechnological Equipment query. 2017;31(2):221–235. doi: 10.1080/13102818.2016.1269616 [DOI] [Google Scholar]
- [53].Elkiran G, Nourani V, Abba SI. Multi-step ahead modelling of river water quality parameters using ensemble artificial intelligence-based approach. J Hydrol. 2019;577:123962. doi: 10.1016/j.jhydrol.2019.123962 [DOI] [Google Scholar]
- [54].Otchere DA, Arbi Ganat TO, Gholami R, et al. Application of supervised machine learning paradigms in the prediction of petroleum reservoir properties: comparative analysis of ANN and SVM models. J Pet Sci Eng. 2021;200:108182. doi: 10.1016/j.petrol.2020.108182 [DOI] [Google Scholar]
- [55].Wang Z, Peng X, Xia A, et al. The role of machine learning to boost the bioenergy and biofuels conversion. Bioresour Technol. 2022c;343:126099. doi: 10.1016/j.biortech.2021.126099 [DOI] [PubMed] [Google Scholar]
- [56].Jang J-SR. ANFIS: adaptive-network-based fuzzy inference system. IEEE transactions on systems, man, and cybernetics. 1993;23:665–685. [Google Scholar]
- [57].Kushwaha OS, Uthayakumar H, Kumaresan K. Modeling of carbon dioxide fixation by microalgae using hybrid artificial intelligence (AI) and fuzzy logic (FL) methods and optimization by genetic algorithm (GA). Environ Sci Pollut Res. 2023;30(10):24927–24948. doi: 10.1007/s11356-022-19683-0 [DOI] [PubMed] [Google Scholar]
- [58].Kunjuraman S, Velusamy B. Performance evaluation of shell and tube heat exchanger through ANN and ANFIS model for dye recovery from textile effluents. Energy Sources A. 2021;43(13):1600–1619. doi: 10.1080/15567036.2020.1832627 [DOI] [Google Scholar]
- [59].Hertel L, Collado J, Sadowski P, et al. Sherpa:robust hyperparameter optimization for machine learning. SoftwareX. 2020;12. doi: 10.1016/j.softx.2020.100591 [DOI] [Google Scholar]
- [60].Lopes BT, Eliasy A, Ambrosio R. Artificial intelligence in corneal diagnosis: where are we? Current Ophthalmol Reports. 2019;7(3):204–211. doi: 10.1007/s40135-019-00218-9 [DOI] [Google Scholar]
- [61].Shouval R, Fein JA, Savani B, et al. Machine learning and artificial intelligence in haematology. Br J Haematol. 2021;192(2):239–250. doi: 10.1111/bjh.16915 [DOI] [PubMed] [Google Scholar]
- [62].Chu G, Liang R, Wan C, et al. Ultrasonic-assisted extraction of flavonoids from juglans mandshurica maxim.: artificial intelligence-based optimization, kinetics estimation, and antioxidant potential. Molecules. 2022;27(15):4837. doi: 10.3390/molecules27154837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Samli R, Aydin ZBG, Şahin S. Computer modelling of the enrichment process of sunflower and corn oils with olive leaves through ultrasound treatment. Biomass Convers Biorefin. 2022;12(12):5571–5581. doi: 10.1007/s13399-020-00974-w [DOI] [Google Scholar]
- [64].Rakshit M, Srivastav P. Optimization of pulsed ultrasonic‐assisted extraction of punicalagin from pomegranate (punica granatum) peel: A comparison between response surface methodology and artificial neural network‐multiobjective genetic algorithm. J Food Process Preserv. 2021;45(1):e15078. doi: 10.1111/jfpp.15078 [DOI] [Google Scholar]
- [65].Patra A, Abdullah S, Pradhan RC. Optimization of ultrasound-assisted extraction of ascorbic acid, protein and total antioxidants from cashew apple bagasse using artificial neural network-genetic algorithm and response surface methodology. J Food Process Preserv. 2022;46(3):e16317. doi: 10.1111/jfpp.16317 [DOI] [Google Scholar]
- [66].Yıkmış S, Altıner DD, Ozer H, et al. Modeling and optimization of bioactive compounds from jujube (ziziphus jujuba mill.) vinegar using response surface methodology and artificial neural network: comparison of ultrasound processing and thermal pasteurization. J Food Process Preserv. 2022;46(11):e17102. doi: 10.1111/jfpp.17102 [DOI] [Google Scholar]
- [67].Victor P, Camarena-Bernard C. Lutein, violaxanthin, and zeaxanthin spectrophotometric quantification: A machine learning approach. J Appl Phycol. 2023;35(1):73–84. doi: 10.1007/s10811-022-02855-3 [DOI] [Google Scholar]
- [68].de Carvalho TCL, Nunes CA. Smartphone-based method for the determination of chlorophyll and carotenoid contents in olive and avocado oils: an approach with calibration transfer. J Food Compos Anal. 2021;104:104164. doi: 10.1016/j.jfca.2021.104164 [DOI] [Google Scholar]
- [69].Wang X, Zhang X, Hong H, et al. Non-destructive quantitative analysis of carotene content in carrots using raman spectroscopy. European Food Res And Tech. 2021b;247(9):2299–2307. doi: 10.1007/s00217-021-03788-w [DOI] [Google Scholar]
- [70].Asnake Metekia W, Garba Usman A, Hatice Ulusoy B, et al. Artificial intelligence-based approaches for modeling the effects of spirulina growth mediums on total phenolic compounds. Saudi J Biol Sci. 2022;29(2):1111–1117. doi: 10.1016/j.sjbs.2021.09.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Witek-Krowiak A, Chojnacka K, Podstawczyk D, et al. Application of response surface methodology and artificial neural network methods in modelling and optimization of biosorption process. Bioresour Technol. 2014;160:150–160. doi: 10.1016/j.biortech.2014.01.021 [DOI] [PubMed] [Google Scholar]
- [72].Tao Y, Wu D, Zhang Q-A, et al. Ultrasound-assisted extraction of phenolics from wine lees: modeling, optimization and stability of extracts during storage. Ultrason Sonochem. 2014;21(2):706–715. doi: 10.1016/j.ultsonch.2013.09.005 [DOI] [PubMed] [Google Scholar]
- [73].Shishodia A, Kumar K, Manna MS. Modeling for the efficient separation of bio-active catechins from green tea leaves. sep. Sci Technol. 2017;52(4):671–678. doi: 10.1080/01496395.2016.1252777 [DOI] [Google Scholar]
- [74].Priyadarshani I, Rath B. Commercial and industrial applications of micro algae–A review. Journal Of Algal Biomass Utilization. 2012;3(4):89–100. [Google Scholar]
- [75].Wang S, Wu S, Yang G, et al. A review on the progress, challenges and prospects in commercializing microalgal fucoxanthin. Biotechnol Adv. 2021a;53:107865. doi: 10.1016/j.biotechadv.2021.107865 [DOI] [PubMed] [Google Scholar]
- [76].Weremfo A, Abassah‐Oppong S, Adulley F, et al. Response surface methodology as a tool to optimize the extraction of bioactive compounds from plant sources. J Sci Food Agric. 2023;103(1):26–36. doi: 10.1002/jsfa.12121 [DOI] [PubMed] [Google Scholar]
- [77].Shannon E, Abu-Ghannam N. Optimisation of fucoxanthin extraction from irish seaweeds by response surface methodology. J Appl Phycol. 2017;29(2):1027–1036. doi: 10.1007/s10811-016-0983-4 [DOI] [Google Scholar]
- [78].Shannon E, Abu-Ghannam N. Enzymatic extraction of fucoxanthin from brown seaweeds. International Journal Food Science Technologies. 2018;53(9):2195–2204. doi: 10.1111/ijfs.13808 [DOI] [Google Scholar]
- [79].Carreira-Casais A, Cassani L, Soria-López A, et al. Green extraction of fucoxanthin with promising nutraceutical applications. Biology And Life Sciences Forum. 2022;12(1):34. [Google Scholar]
- [80].Nie J, Chen D, Ye J, et al. Optimization and kinetic modeling of ultrasonic-assisted extraction of fucoxanthin from edible brown algae sargassum fusiforme using green solvents. Ultrason Sonochem. 2021;77:105671. doi: 10.1016/j.ultsonch.2021.105671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Cikoš A-M, Aladić K, Velić D, et al. Evaluation of ultrasound-assisted extraction of fucoxanthin and total pigments from three croatian macroalgal species. Chem Papers. 2023;77(3):1545–1559. doi: 10.1007/s11696-022-02524-2 [DOI] [Google Scholar]
- [82].Okwu MO, Samuel OD, Ewim DRE, et al. Estimation of biogas yields produced from combination of waste by implementing response surface methodology (RSM) and adaptive neuro-fuzzy inference system (ANFIS). IntJ Energy Environ Eng. 2021;12(2):353–363. doi: 10.1007/s40095-021-00381-5 [DOI] [Google Scholar]
- [83].Boateng ID, Kuehnel L, Daubert CR, et al. Updating the status quo on the extraction of bioactive compounds in agro-products using a two-pot multivariate design. A comprehensive review. Food Funct. 2023;14(2):569–601. doi: 10.1039/D2FO02520E [DOI] [PubMed] [Google Scholar]
- [84].Ousaadi MI, Merouane F, Berkani M, et al. Valorization and optimization of agro-industrial orange waste for the production of enzyme by halophilic streptomyces sp. Environ Res. 2021;201:111494. doi: 10.1016/j.envres.2021.111494 [DOI] [PubMed] [Google Scholar]
- [85].Alshammari F, Badrul Alam M, Naznin M, et al. Optimization of Portulaca oleracea L. extract using response surface methodology and artificial neural network and characterization of bioactive compound by high-resolution mass spectroscopy. Arab J Chem. 2023;16(2):104425. doi: 10.1016/j.arabjc.2022.104425 [DOI] [Google Scholar]
- [86].Hamdia KM, Zhuang X, Rabczuk T. An efficient optimization approach for designing machine learning models based on genetic algorithm. Neural Comput Appl. 2021;33(6):1923–1933. doi: 10.1007/s00521-020-05035-x [DOI] [Google Scholar]
- [87].Shekhar S, Prakash P, Singha P, et al. Modeling And Optimization Of Ultrasound-Assisted Extraction Of Bioactive Compounds From Allium Sativum Leaves Using Response Surface Methodology And Artificial Neural Network Coupled With Genetic Algorithm Foods. 2023;12(9):1925. doi: 10.3390/foods12091925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Zhang Q, Wang C. 2008. Using genetic algorithm to optimize artificial neural network: a case study on earthquake prediction. 2008 Second International Conference on Genetic and Evolutionary Computing, Jinzhou, China, 25-26 Sept. 2008. pp. 128–131. [Google Scholar]
- [89].Loganathan C, Girija K. Hybrid learning for adaptive neuro fuzzy inference system. Int J Eng Sci. 2013;2(11):6–13. [Google Scholar]
- [90].Baskararaj S, Theivendren P, Palanisamy P, et al. Optimization of bioactive compounds extraction assisted by microwave parameters from kappaphycus alvarezii using RSM and ANFIS modeling. J Food Meas Charact. 2019;13(4):2773–2789. doi: 10.1007/s11694-019-00198-1 [DOI] [Google Scholar]
- [91].Joshi C, Singhal RS. Modelling and optimization of zeaxanthin production by Paracoccus zeaxanthinifaciens ATCC 21588 using hybrid genetic algorithm techniques. Biocatal Agric Biotechnol. 2016;8:228–235. doi: 10.1016/j.bcab.2016.10.004 [DOI] [Google Scholar]
- [92].Aung T, Kim S-J, Eun J-B. A hybrid RSM-ANN-GA approach on optimisation of extraction conditions for bioactive component-rich laver (porphyra dentata) extract. Food Chem. 2022;366:130689. doi: 10.1016/j.foodchem.2021.130689 [DOI] [PubMed] [Google Scholar]
- [93].Kumar V, Sharma HK. Process optimization for extraction of bioactive compounds from taro (colocasia esculenta), using RSM and ANFIS modeling. J Food Meas Charact. 2017;11(2):704–718. doi: 10.1007/s11694-016-9440-y [DOI] [Google Scholar]
- [94].Ho Thanh Lam L, Le NH, Van Tuan L, et al. Machine learning model for identifying antioxidant proteins using features calculated from primary sequences. Biology (Basel). 2020;9(10):325. doi: 10.3390/biology9100325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Solovchenko A. Seeing good and bad: optical sensing of microalgal culture condition. Algal Res. 2023;71:103071. doi: 10.1016/j.algal.2023.103071 [DOI] [Google Scholar]
- [96].Yew GY, Puah BK, Chew KW, et al. Chlorella vulgaris FSP-E cultivation in waste molasses: photo-to-property estimation by artificial intelligence. Chem Eng J. 2020;402:126230. doi: 10.1016/j.cej.2020.126230 [DOI] [Google Scholar]
- [97].Yang Y, Tong H, Yang L, et al. Application of near-infrared spectroscopy and chemometrics for the rapid quality assessment of sargassum fusiforme. Postharvest Biol Technol. 2021;173:111431. doi: 10.1016/j.postharvbio.2020.111431 [DOI] [Google Scholar]
- [98].Moresco R, Afonso T, Uarrota VG, et al. Classification Tools for carotenoid content estimation in manihot esculenta via metabolomics and machine learning. Cham: Springer International Publishing; 2017. pp. 280–288. doi: 10.1007/978-3-319-60816-7_34 [DOI] [Google Scholar]
- [99].Ranade NV, Nagarajan S, Sarvothaman V, et al. ANN based modelling of hydrodynamic cavitation processes: biomass pre-treatment and wastewater treatment. Ultrason Sonochem. 2021;72:105428. doi: 10.1016/j.ultsonch.2020.105428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Amini G, Salehi F, Rasouli M. Drying kinetics of basil seed mucilage in an infrared dryer: application of GA-ANN and ANFIS for the prediction of drying time and moisture ratio. J Food Process Preserv. 2021;45(3):e15258. doi: 10.1111/jfpp.15258 [DOI] [Google Scholar]
- [101].Rácz A, Bajusz D, Héberger K. Chemometrics in analytical chemistry. In: Applied Chemoinformatics. 2018. pp. 471–499. 10.1002/9783527806539.ch9 [DOI] [Google Scholar]
- [102].de Carvalho RRB, Marmolejo Cortes DF, Bandeira e Sousa M, et al. Image-based phenotyping of cassava roots for diversity studies and carotenoids prediction. Plos One. 2022;17(1):e0263326. doi: 10.1371/journal.pone.0263326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Taghadomi-Saberi S, Masoumi AA, Sadeghi M, et al. Integration of wavelet network and image processing for determination of total pigments in bitter orange (Citrus aurantium L.) peel during ripening. J Food Proc Eng. 2019;42(5):e13120. doi: 10.1111/jfpe.13120 [DOI] [Google Scholar]
- [104].Lamberti MJ, Wilkinson M, Donzanti BA, et al. A study on the application and use of artificial intelligence to support drug development. Clin Ther. 2019;41(8):1414–1426. doi: 10.1016/j.clinthera.2019.05.018 [DOI] [PubMed] [Google Scholar]
- [105].Paul D, Sanap G, Shenoy S, et al. Artificial intelligence in drug discovery and development. Drug Discov Today. 2021;26(1):80–93. doi: 10.1016/j.drudis.2020.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Simm J, Klambauer G, Arany A, et al. Repurposing high-throughput image assays enables biological activity prediction for drug discovery. Cell Chem Biol. 2018;25(5):611–618.e3. doi: 10.1016/j.chembiol.2018.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Guan J. Artificial intelligence in healthcare and medicine: promises, ethical challenges and governance. Chin Med J. 2019;34(2):76–83. doi: 10.24920/003611 [DOI] [PubMed] [Google Scholar]
- [108].Kalyane D, Sanap G, Paul D, et al. Artificial intelligence in the pharmaceutical sector: current scene and future prospect. In: The future of pharmaceutical product development and research. 2020. pp. 73–107. 10.1016/B978-0-12-814455-8.00003-7 [DOI] [Google Scholar]
- [109].Narayanan H, Dingfelder F, Butté A, et al. Machine learning for biologics: opportunities for protein engineering, developability, and formulation. Trends Pharmacol Sci. 2021;42(3):151–165. doi: 10.1016/j.tips.2020.12.004 [DOI] [PubMed] [Google Scholar]
- [110].Holzinger A, Keiblinger K, Holub P, et al. AI for life: trends in artificial intelligence for biotechnology. N Biotechnol. 2023;74:16–24. doi: 10.1016/j.nbt.2023.02.001 [DOI] [PubMed] [Google Scholar]
- [111].Goswami RK, Agrawal K, Upadhyaya HM, et al. Microalgae conversion to alternative energy, operating environment and economic footprint: an influential approach towards energy conversion, and management. Energy Convers Manag. 2022;269:116118. doi: 10.1016/j.enconman.2022.116118 [DOI] [Google Scholar]
- [112].Wang K, Khoo KS, Leong HY, et al. How does the internet of things (IoT) help in microalgae biorefinery? Biotechnol Adv. 2022a;54:107819. doi: 10.1016/j.biotechadv.2021.107819 [DOI] [PubMed] [Google Scholar]
- [113].Oruganti RK, Biji AP, Lanuyanger T, et al. Artificial intelligence and machine learning tools for high-performance microalgal wastewater treatment and algal biorefinery: A critical review. Sci Total Environ. 2023;876:162797. doi: 10.1016/j.scitotenv.2023.162797 [DOI] [PubMed] [Google Scholar]
- [114].Zhang W, Li J, Liu T, et al. Machine learning prediction and optimization of bio-oil production from hydrothermal liquefaction of algae. Bioresour Technol. 2021;342:126011. DOI: 10.1016/j.biortech.2021.126011 [DOI] [PubMed] [Google Scholar]
- [115].Singh V, Mishra V. Exploring the effects of different combinations of predictor variables for the treatment of wastewater by microalgae and biomass production. Biochem Eng J. 2021;174:108129. doi: 10.1016/j.bej.2021.108129 [DOI] [Google Scholar]
- [116].Horng S-C, Lin S-S. Improved beluga whale optimization for solving the simulation optimization problems with stochastic constraints. Mathematics. 2023;11(8):1854. doi: 10.3390/math11081854 [DOI] [Google Scholar]
- [117].Nayak J, Vakula K, Dinesh P, et al. Intelligent food processing: journey from artificial neural network to deep learning. Comput Sci Rev. 2020;38:100297. doi: 10.1016/j.cosrev.2020.100297 [DOI] [Google Scholar]
- [118].Kirillov A, Mintun E, Ravi N, et al. 2023. Segment anything. arXiv preprint arXiv:2304.02643
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data not available


