ABSTRACT
The bottom-up assembly of biological components in synthetic biology has contributed to a better understanding of natural phenomena and the development of new technologies for practical applications. Over the past few decades, basic RNA research has unveiled the regulatory roles of RNAs underlying gene regulatory networks; while advances in RNA biology, in turn, have highlighted the potential of a wide variety of RNA elements as building blocks to construct artificial systems. In particular, synthetic mRNA-based translational regulators, which respond to signals in cells and regulate the production of encoded output proteins, are gaining attention with the recent rise of mRNA therapeutics. In this Review, we discuss recent progress in RNA synthetic biology, mainly focusing on emerging technologies for sensing intracellular protein and RNA molecules and controlling translation.
KEYWORDS: Synthetic biology, Translational regulation, miRNA, RNA-binding protein
Introduction to RNA synthetic biology
Synthetic biology is a constructive approach to studying biological phenomena, built upon a broad range of scientific disciplines, including biology, physics, chemistry, and engineering. Synthetic biologists seek to (1) synthesize artificial living matters and (2) redesign and assemble natural, pre-existing biological components for the generation of fabricated systems [1]. Not only does synthetic biology help us achieve a greater understanding of biological systems in a manner distinct from classical analytical biology, but it also has enormous potential for practical applications beneficial to humankind [2], ranging from healthcare [3–5], agriculture [6], and energy supply [7] to biomanufacturing [8].
Two trailblazing studies published in 2000 mark the rise of synthetic biology research [9,10]. In these papers, biological components (transcription repressors and corresponding repressible promoters) were combined to create two artificial systems using E. coli as a chassis: (1) an oscillating function that periodically expresses genes [9] and (2) a genetic toggle switch that can generate a bi-stable pattern of gene expression between ON and OFF states [10]. Following these reports, early studies have constructed various synthetic gene circuits using transcriptional regulators to program gene expression [11–13], inspired by Boolean algebra, electronic engineering, and computer science.
The central dogma of molecular biology – the flow of genetic information from DNA to protein – illustrates RNA as a template for protein synthesis, but it is crystal clear from our current knowledge that its role goes far beyond functioning as mere intermediate messages. There are myriads of RNA elements in nature, and recent studies have shown many of them to play important roles in gene expression control [14,15]. RNA sequences fold into secondary and further as tertiary structures. Similar to proteins, structured RNAs can bind to specific ligands [16], catalyse various chemical reactions [17], and regulate transcription and translation through interaction with ligands [18]. In addition, the functional properties of RNA molecules as both storage of genetic information and for catalysis suggest the possibility that an RNA-centric primitive life-like system had existed during evolutionary history [19]. It is therefore not surprising that pioneering synthetic biologists recognized the potential of engineering these functional RNA elements to create artificial systems [20]. By utilizing such RNA elements as building blocks, it is possible, for example, to construct gene switches that tweak gene expression on and off in a ligand-dependent manner.
Therapeutic applications of mRNA unexpectedly attracted tremendous public attention following the outbreak of SARS-CoV-2 at the end of 2019. In recent years, mRNA vaccines and therapeutics have made remarkable progress, and several bio-ventures and pharmaceutical companies, including Moderna and BioNTech, are conducting clinical trials of mRNA-based therapies [21,22]. Important characteristics of mRNA therapeutics, composed of in vitro synthesized mRNA administered to the body, are: (1) transgenes are delivered into cells virtually without any risk of harmful genomic integration; (2) transgenes are rapidly expressed because they are delivered to the cytosol, where translation primarily occurs, without ever entering the nucleus; and (3) mRNAs are translated in both non-dividing and actively dividing cells [23]. However, since simple (over)expression of transgenes in off-target cells might cause unexpected adverse effects, researchers are currently taking advantage of synthetic biology approaches for sophisticated gene expression control to develop ‘smarter’ mRNA therapeutics.
In this Review, we aim to introduce recent progress in RNA synthetic biology, especially focusing on emerging mRNA-based technologies to post-transcriptionally regulate gene expressions by sensing cellular proteins and RNA molecules. We also discuss recent advances in related technologies which may contribute to the further development of RNA synthetic biology.
Protein sensors
Proteins are responsible for a wide range of biological phenomena within cells. Variations in protein expression underlie cellular states and identities. Therefore, sensors that detect proteins and generate measurable outputs are helpful tools for cellular engineering applications. mRNA-based technologies that convert the information of cellular proteins or other inputs (e.g. small molecules, and light) to transgene expression through RNA – protein interaction are summarized in Table 1.
Table 1.
Summary of mRNA-based protein sensors that work in mammalian cells.
| Sensor | Expression system | Target molecule/signature | Regulated gene | Cell type | Reference |
|---|---|---|---|---|---|
| Protein-responsive mRNA switch | Plasmid DNA | L7Ae | Fluorescent protein, antiapoptotic protein (Bcl-xL), and proapoptotic protein (Bim) | HeLa | Saito et al., 2010 [24]; Saito et al., 2011 [25] |
| Plasmid DNA | L7Ae, MS2CP, Bacillus ribosomal protein (S15) | Fluorescent protein | HeLa | Endo et al., 2013 [26] | |
| Plasmid DNA, invitro transcribed mRNA | U1A, LIN28A | Fluorescent protein | HEK293FT, HeLa, and human iPS cell | Kawasaki et al., 2017 [28] | |
| Plasmid DNA | TetR, doxycycline | Fluorescent protein | HeLa and HEK293 | Mol et al., 2019 [56] | |
| In vitro transcribed mRNA | L7Ae, MS2CP | Fluorescent protein, MS2CP, and hBax | HEK293FT and HeLa | Matsuura et al., 2018 [30] | |
| Plasmid DNA and in vitro transcribed mRNA | L7Ae, MS2CP, PP7CP, U1A, LIN28A | Fluorescent protein | HEK293FT | Ono et al., 2020 [29] | |
| Plasmid DNA and in vitro transcribed mRNA | CRISPR-Cas proteins | Fluorescent proteins, CRISPR-Cas proteins, and hBax | HEK293FT, HeLa, A549 and human iPS cell | Kawasaki et al., 2023 [37] | |
| Csy4-mediated regulation of transgene expression | Plasmid DNA | CRISPR-associated endoribonuclease (Csy4) | Fluorescent protein and luciferase | HEK293 | Borchardt et al., 2015 [35] |
| PERSIST | Plasmid DNA and genome-integrated | CRISPR-associated endoribonucleases (Csy4, CasE, Cse3, Cas6, LwaCas13a, PspCas13b, RanCas13b, PguCas13b, RfxCas13d) | Fluorescent proteins and endoribonucleases (Csy4, CasE, and Cse3) tagged with a degradation domain (PEST) | HEK293FT and CHO-K1 | DiAndreth et al., 2022 [36] |
| L7Ae tagged with a destabilization domain | In vitro transcribed mRNA, self-replicating RNA (replicon) | Trimethoprim (TMP) | Fluorescent proteins, luciferase, and TetR (with or without fusion with DDX6) | BHK-21 and C2C12 | Wagner et al., 2018 [34] |
| TEV protease-responsive L7Ae | Plasmid DNA | TEV protease (TEVp), viral protein NS3 | Fluorescent proteins | HEK293FT and HeLa | Cella et al., 2018 [38] |
| Protein-responsive ribozyme | Plasmid DNA | U1Ap, N-peptide | SEAP | HEK293, CHO-K1, HeLa and HT-1080 | Ausländer et al., 2014 [44] |
| Plasmid DNA | MS2CP | Fluorescent proteins | Flp-In T-REx HEK293 | Kennedy et al., 2014 [45] | |
| cis-acting switch inverter module utilizing NMD pathway | Plasmid DNA | L7Ae, Bacillus ribosomal protein (S15) | Fluorescent proteins and proapoptotic protein (Bim-EL) | HeLa | Endo et al., 2013 [46] |
| CaVT | In vitro transcribed mRNA | CaVT (a fusion protein of MS2CP and VPg), small molecule (A/C heterodimerizer), photocaged TMP-HL, UV light, eDHFR, EGFP | Fluorescent proteins and luciferase | HeLa | Nakanishi et al., 2020 [48]; Nakanishi et al., 2021 [50]; Nakanishi et al., 2022 [51] |
| TetR Splicing Device (TSD) | Plasmid DNA and genome-integrated | TetR, doxycycline (Dox) | Fluorescent protein and luciferase | HEK293, HeLa and HF1–3 | Mol et al., 2019 [56] Mol et al., 2021 [57] |
| PAL-mediated translational control | Plasmid DNA | Light-oxygen-voltage receptor (PAL), blue light | Luciferase | HeLa | Weber et al., 2019 [58] |
One classic example of an RNA-based protein sensor is an mRNA that contains an aptamer in the untranslated region (UTR) (Figure 1A). An aptamer is an RNA molecule that specifically binds to a ligand. Our group has previously developed a translational control system, called protein-responsive mRNA switches, based on the interaction between RNA aptamers and RNA-binding proteins (RBPs) [24–30]. In the absence of a specific target protein, the protein-responsive mRNA switch is translated; while in its presence, a complex of the target protein and the aptamer strategically inserted into the 5’-UTR of the mRNA probably serves as a roadblock for scanning ribosomes, thereby repressing translation. There are several mRNA switches that are designed to sense specific RBPs. For example, the archaeal RBP L7Ae [24,25], RNA-binding capsid proteins derived from bacteriophages MS2 [30,31] and PP7 [29], tetracycline-responsive repressors (TetR) [32–34], the mammalian spliceosomal protein U1A [28], an RBP highly expressed in pluripotent stem cells (LIN28A) [28], and clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) proteins [35–37].
Figure 1.
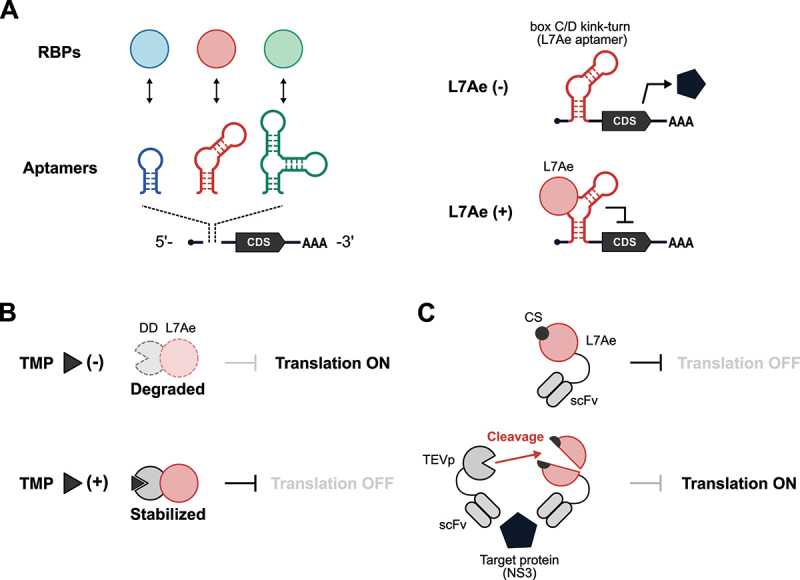
Protein-responsive OFF-type translational regulators.
(A) Aptamers are RNA elements that can interact with specific ligands. In protein-responsive mRNA switches, aptamers that interact with RNA-binding proteins (RBPs) are inserted into the 5’-UTR of mRNAs (left). In the absence of the target RBP, L7Ae, an mRNA harbouring the BoxC/D motif (L7Ae aptamer) is translated, while the L7Ae−BoxC/D interaction represses the translation (right). CDS: coding sequence. (B) A drug-dependent translation regulator using the drug-inducible stabilization of L7Ae. TMP: trimethoprim. DD: destabilization domain. (C) An L7Ae-based protein sensor. TEVp: TEV protease. CS: cleavage site of TEVp. scFv: single-chain variable fragment.
L7Ae is an RBP and one of the most frequently used triggers for protein-responsive mRNA switches because of its strong translational repression ability. Aimed at conditional regulation of translation, Ron Weiss and his group engineered L7Ae to control its stability in a small molecule-dependent manner (Figure 1B). They fused L7Ae with a destabilization domain (DD) [34], thus creating a fusion protein that undergoes proteasomal degradation unless a stabilizing ligand (Trimethoprim; TMP) is added. Another example of L7Ae engineering is to regulate translation in a target protein-dependent manner (Figure 1C) [38]. In this system, a cleavage site (CS) of tobacco etch virus protease (TEVp) is inserted into L7Ae. TEVp disrupts L7Ae function by cleaving at the CS. Next, TEVp and L7Ae are fused to two single-chain variable fragments (scFv162 and scFv35, respectively). Both scFvs recognize the same viral protein NS3 but via distinct epitopes. In the presence of NS3, it serves as a scaffold for the assembly of TEVp-scFv162:NS3:scFv35-L7Ae and enhances TEVp cleavage to de-repress translation.
Several mRNA switches can also sense endogenous proteins to distinguish specific cell types. For example, LIN28A is an RBP involved in stem cell-related gene regulatory networks [39]. The LIN28A-responsive switch has been proven to distinguish differentiated cells and human induced pluripotent stem cells (iPSCs) based on differences in LIN28A expression levels between the two cell types [28]. Recently, several groups have reported translational regulators with CRISPR/Cas proteins by leveraging their RNA-binding abilities [35–37]. Cas proteins are recently being discovered not only in microbial genomes [40,41] but also in bacteriophages [42,43], and are thus promising RBPs to significantly expand the repertoire of translational regulators.
Protein-responsive mRNA switches described above are generally OFF-type systems in which gene expression is suppressed in the presence of a target protein. In contrast, ON-type systems that induce gene expression in the presence of a target are also possible to design and could have vast applications. One strategy to activate translation is to utilize a chimera of an aptamer and a self-cleaving ribozyme (Figure 2A) [44,45]. In two independent studies by Ausländer et al. and Kennedy et al. in 2014, self-cleaving ribozymes were engineered to switch between active and inactive conformations in response to an RNA-binding N-peptide from λ bacteriophage and an RNA-recognition domain derived from U1A (U1Ap) [44], or a bacteriophage-derived RBP (coat protein of bacteriophage MS2, MS2CP) [45], respectively. As an alternative strategy to making ON-type systems, a study utilized the nonsense-mediated mRNA decay (NMD) pathway [46], a natural mRNA surveillance pathway for degrading mRNAs with premature termination codons [47]. This ON-type system employs a bicistronic mRNA that encodes two open reading frames (ORFs): (1) the one upstream contains premature termination codons (PTCs) to trigger translation termination and NMD to degrade the mRNAs and (2) the one downstream operates under the control of an internal ribosome entry site (IRES) to produce an output signal. An aptamer is placed within the 5’-UTR, and the RBP – aptamer interaction represses the pioneer round of translation of the upstream ORF, thereby preventing mRNA degradation by NMD and allowing IRES-dependent output expression.
Figure 2.
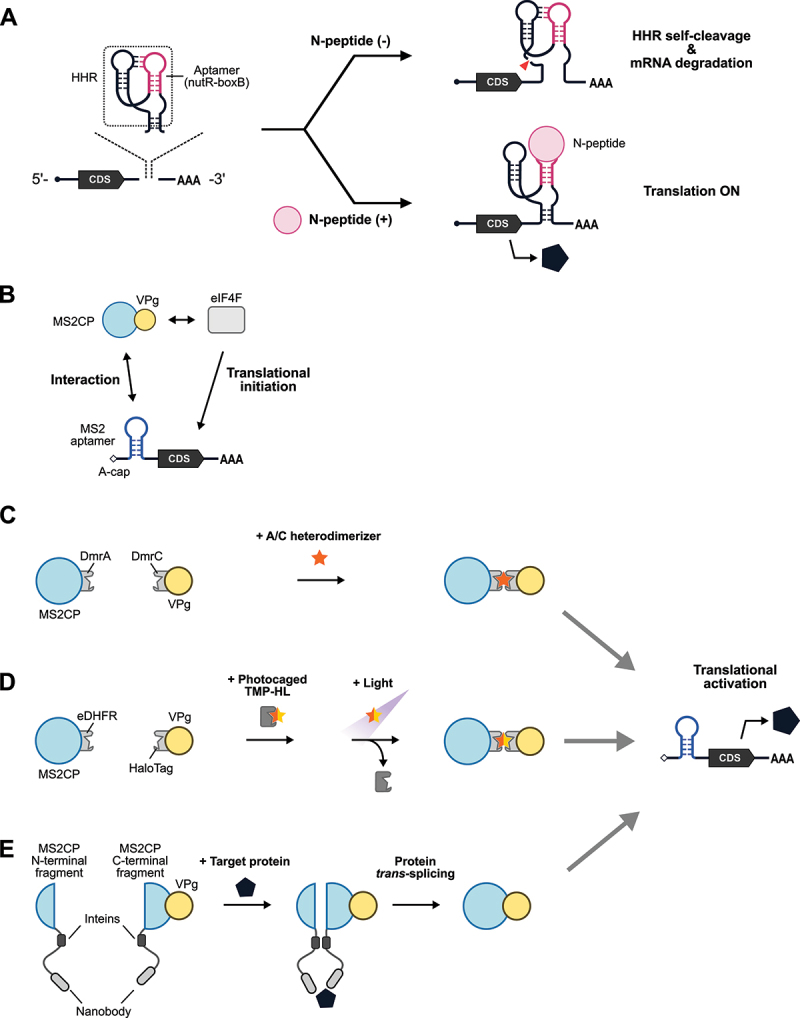
Protein-responsive ON-type translational regulators.
(A) Schematic illustration of protein-responsive riboswitches. A chimera of an aptamer (nutR-boxB hairpin) and a hammerhead ribozyme (HHR) is inserted into the 3’-UTR of mRNA. In the absence of the trigger RNA-binding protein, N-peptide, HHR undergoes self-cleavage, and the mRNA is degraded (translation OFF, top right). In the presence of N-peptide, the binding to nutR-boxB prevents the self-cleavage activity of HHR, resulting in stabilization and translation of the mRNA (translation ON, bottom right). CDS: coding sequence. (B) Schematic illustration of CaVT (Caliciviral VPg-based Translational activator). VPg is capable of interacting with eIF4F to initiate translation. MS2CP fused with VPg interacts with MS2 aptamer inserted into the 5’-UTR of an A-capped target mRNA, recruiting eIF4F proximal to the target mRNA to initiate translation. (C) Schematic illustration of drug-controllable CaVT. A/C heterodimerizer induces the heterodimerization of DmrA and DmrC to tether VPg to the target mRNA for translational initiation. (D) Schematic illustration of light-inducible CaVT. Before light irradiation, photocaged TMP-HL cannot bind to MS2CP-eDHFR. Light irradiation removes the photocage, and a ternary complex of MS2CP-eDHFR, TMP-HL, and HaloTag-VPg activates target mRNA translation. (E) CaVT-based protein sensor. Target proteins act as a molecular hub for MS2CP (N-terminal fragment)-intein nanobody and nanobody-intein-MS2CP (C-terminal fragment)-VPg and enhance intein-mediated protein trans-splicing between these proximal proteins, resulting in reconstitution of full-length MS2CP-VPg.
In a more recent study, a virus-derived protein, VPg (viral protein genome-linked), was used to activate translation (Figure 2B) [48]. VPg is a protein that binds to translation initiation factors and acts as a substitute for the 5’-cap structure [49], which is necessary for efficient initiation of translation. This system, named CaVT (Caliciviral VPg-based Translational activator), consists of two components: (1) a fusion protein of VPg and an RBP (MS2CP); and (2) an mRNA with a 5’-UTR that contains an MS2CP aptamer and a noncanonical 5’-cap structure (A-cap) that is incompetent for efficient translational initiation. When VPg is recruited to the 5’-UTR of the mRNA via MS2CP-aptamer interaction, VPg interacts with translation initiation factors to activate mRNA translation.
The CaVT-based approach has been expanded to drug-dependent translational activation. In this system, drug-inducible heterodimerization domains (DmrA and DmrC) and an inducer A/C Heterodimerizer ligand are used [48]. DmrC and DmrA are fused with VPg and MS2CP, respectively (Figure 2C). In the presence of the A/C Heterodimerizer ligand, the DmrA – DmrC heterodimerization occurs and tethers VPg to the mRNA, resulting in VPg-mediated translational activation. Furthermore, light-activated translation was also demonstrated recently by using a light-responsive small molecule to trigger the interaction between VPg and MS2CP (Figure 2D) [50]. Another application of CaVT is to sense a target protein [51], similar to the L7Ae-based system described above (Figure 2E). For the CaVT-based intracellular protein sensor, MS2CP is split into two fragments, with two nanobodies against distinct epitopes of a single target protein fused to each fragment. In addition, split inteins, which mediate protein trans-splicing events, are also fused to each MS2CP fragment. VPg is fused with the fusion protein of the nanobody, the C-terminal intein (C-intein), and the C-terminal MS2CP fragment. Only when both CaVT component proteins and the target protein are present does the target protein act as a hub to bring the separated CaVT components into proximity to facilitate protein trans-splicing and reconstitute full-length CaVT. The reconstituted CaVT protein, in turn activates target mRNA translation. Such control systems induced by drugs, light, and intracellular proteins are useful tools for regulating translation in a spatiotemporal or cell type-specific manner.
Next, we will discuss the development of technologies that paved the way to further expand the toolbox of mRNA-based protein sensors. In 1990, two pioneering studies reported methods to obtain RNA aptamers with high specificity and affinity to ligands [52,53], now widely recognized as in vitro selection (evolution) or ‘systematic evolution of ligands by exponential enrichment’ (SELEX) (Figure 3A) [52]. SELEX mimics natural processes of evolution: variation, selection, and replication. A pool of stretches of random RNA bases, some of which contain specific sequences that can bind to a specific ligand, is subjected to selection for binding to a ligand of interest by using ligand-immobilized columns, membranes, or microbeads, for example. Selected RNAs are reverse-transcribed, PCR amplified, and in vitro transcribed. The recovered RNA pool is subjected to consecutive rounds of selection, gradually enriching high-affinity RNAs in the pool, and aptamers are finally identified through Sanger sequencing or massively parallel sequencing.
Figure 3.
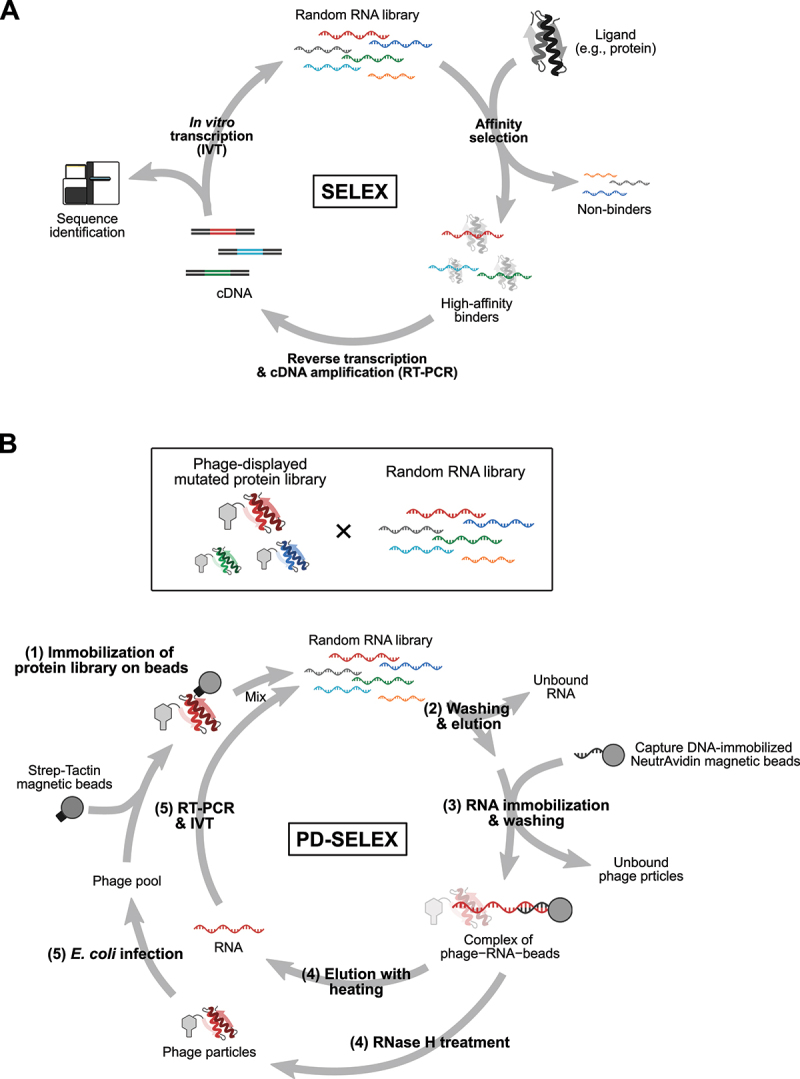
Methods to obtain aptamers and expand the number of RNA – RBP pairs.
(A) Overview of SELEX (systematic evolution of ligands by exponential enrichment) to obtain aptamers. (B) Overview of PD-SELEX (phage display coupled with SELEX) for co-evolution of RNA – RBP pairs by screening through protein and RNA libraries.
One example of a novel RNA – protein pair identified via SELEX for the engineering of RNA-based devices is an RNA aptamer specific for TetR [54]. TetR is a well-characterized transcriptional factor that binds to cognate tetO operator DNA and represses transcription in the presence of ligand, tetracycline (Tc), or its derivatives (e.g. doxycycline: Dox). Upon the addition of Tc, TetR is unbound from tetO, and gene expression is activated. In 2009, Suess and colleagues identified TetR-binding RNA aptamers and demonstrated that the aptamers, instead of Tc, can release TetR from tetO to induce gene expression in E. coli [54]. In a subsequent study, the TetR aptamer was utilized for conditional control of miRNA biogenesis in mammalian cells [55]. To regulate TetR-dependent miRNA biogenesis, the TetR aptamer was inserted into the terminal loop of precursor miRNAs (pre-miRNAs). If TetR is absent or both TetR and Tc are present, miRNA-processing enzyme Dicer cleaves the pre-miRNAs to generate mature miRNA. If TetR is present and Tc is absent, TetR binds to the aptamer and impairs cleavage by Dicer. Another example of TetR aptamer applications is the TetR-dependent regulation of alternative mRNA splicing, named TetR Splicing Device (TSD) [56]. In TSD, the TetR aptamer was inserted into an intron close to a 5’ splice site (5’SS) such that TetR-aptamer binding inhibits spliceosome assembly, consequently leading to intron retention. A more recent study employed the TetR aptamer to control switching between two alternative splice isoforms depending on the binding of TetR to the aptamer [57]. In this system, an intron harbours two 3’ splices sites (3’SSs), and TetR binding to the aptamer in the intron alters 3’SSs selection.
Another interesting work is a study in which Möglich and colleagues employed SELEX to discover aptamers that interact with a bacterial light-oxygen-voltage (LOV) receptor, PAL, for light-activated RNA-protein interaction [58]. PAL switches between light- and dark-adapted states depending on the presence or absence of blue light, respectively. To select aptamers that bind to light-adapted PAL but not dark-adapted PAL, an RNA library was first subjected to PAL binding in the presence of blue light and eluted the RNA in the dark during the selection. They successfully identified aptamers with high affinity for PAL (KD ~20 nM) under blue light and low affinity (KD >1 µM) without. Finally, they demonstrated light-dependent control of translation in mammalian cells by placing the PAL aptamer in the 5’-UTR of mRNA. Collectively, these studies highlight the power of SELEX to engineer RNA-based devices.
Further expansion of the repertoire of orthogonal RNA – RBP pairs – RBPs specifically binding to cognate RNAs – is a helpful tool for various applications (e.g. construction of synthetic gene circuits based on protein-responsive mRNA switches [30,59,60], transcriptional regulation with CRISPR/Cas systems coupled with RNPs [61,62], and imaging of RNAs with multiple RNPs [63–65], however, the limited number of RNA – RBP pairs hinders multiplexing with RNPs. To overcome this limitation, creating novel RNA – RBP pairs by redesigning existing pairs was thought to be a reasonable strategy. One recently established method by Fukunaga and Yokobayashi, PD-SELEX, combined SELEX with phage display (PD), an in vitro screening method to identify ligands for proteins displayed on phage particles [66–68], for the systematic co-evolution of RNA – RBP pairs (Figure 3B) [69]. In addition to a random RNA library, they generated a library of randomly mutated L7Ae (Figure 1). A single cycle of PD-SELEX consists of five steps: (1) immobilizing a library of phage particles displaying L7Ae mutants onto magnetic beads and allowing the RNA pool to bind to the displayed protein library; (2) removing unbound RNA and releasing the phage particles from the beads; (3) capturing the phage-bound RNAs with DNA oligos, which is complementary to a part of the RNA library and immobilized on beads to enrich complexes of RNAs and proteins; (4) eluting RNAs and phage-displayed proteins from the collected beads through heating and RNase H treatment, respectively; and (5) building the RNA and protein libraries via reverse transcription and cDNA amplification by PCR, and in vitro transcription and amplification of the phage pools in E. coli, respectively. Through steps (1)-(3), the RNA and protein libraries are subjected to selective pressure, and interacting RNA – RBP pairs are refined. From an astronomical number of combinations of RNAs and proteins – the theoretical diversity of the RNA library is 1.1 × 1012 molecules, while the practical diversity of the protein library was estimated to be 4 × 108 molecules – favourable RNA – RBP pairs (CS1 RNA – LS4 protein and CS2 RNA – LS12 protein) were selected after six rounds of PD-SELEX and deconvolution of RNA – protein pairs. Importantly, the dissociation constant of CS1 RNA and LS4 protein was 4,030- and 218-fold lower than those of CS1 RNA – LS12 and CS1–L7Ae, indicating the ultra-selective binding of CS1 to LS4 over LS12 and the parental L7Ae. Applying PD-SELEX to redesign other RNA – RBP pairs (e.g. a stem-loop RNA motif of bacteriophage MS2 and its binding partner, MS2CP) might be beneficial because MS2CP, aside from L7Ae, is one of most frequently used RBPs in synthetic biology applications in mammalian cells and may therefore be suitable as a parental protein for generating novel RNA – RBP pairs to work in mammalian cells. One potential application of orthogonal RNA – RBP pairs is constructing synthetic gene circuits by assembling multiple protein-responsive mRNA switches [30,59]. Because of the strong translational repression ability of L7Ae and MS2CP, the current generation of circuits heavily relies on these proteins. However, if PD-SELEX could expand the repertoire of strong and orthogonal translational repressors, it would be possible to further expand the complexity of synthetic gene circuits. Collectively, PD-SELEX has the potential to generate novel orthogonal RNA – RBP pairs derived from existing pairs for expanding the toolbox available for various synthetic biology applications.
RNA sensors
As recent advances in single-cell transcriptome analysis clearly illustrate, the diversity of transcripts underlies cellular identity. In addition to protein sensors, RNA sensing systems would not only help to identify specific cell types but enable us to manipulate cell fate in a cell type-specific manner. Table 2 summarizes mRNA-based technologies to sense cellular transcripts, including microRNAs (miRNAs) and mRNAs.
Table 2.
Summary of mRNA-based RNA sensors that work in mammalian cells.
| Sensor | Expression system | Target molecule/signature | Regulated gene | Cell type | Reference |
|---|---|---|---|---|---|
| miRNA-responsive OFF-type switch | In vitro transcribed mRNA | miRNAs | Fluorescent proteins, proapoptotic protein (Bim), puromycin-resistance gene, membrane protein (CD4), CRISPR-Cas protein, and L7Ae | HEK293FT,HeLa, human pluripotent stem cells (PSCs), PSC-derived cardiomyocytes, PSC-derived hepatocytes, PSC-derived insulin producing cells, PSC-derived endothelial cells, Normal human dermal fibroblast cells (NHDF), and PSC-derived midbrain dopaminergic-like neuronal cells | Miki et al., 2015 [84];Parr et al., 2016 [85];Hirosawa et al., 2017 [87];Tsujisaka et al., 2022 [86] |
| miRNA-responsive ON-type switch | In vitro transcribed mRNA | miRNAs | Fluorescent proteins, lethal ribonuclease (Barnase), and inhibitor of Barnase (Barstar) | HEK293FT,HeLa, and human iPS cell, iPSC-derived cardiomyocytes | Fujita et al., 2022 [88] |
| eToehold | Plasmid DNA and lentiviral vector | Exogenously expressed mRNAs of fluorescent proteins, ySUMO, mouse metalloprotease 9, and mouse tyrosinase, viral RNA, human endogenous transcripts of hsp40 and hsp70 | Fluorescent proteins and luciferase | HEK293T, HeLa, Vero E6, B16-F10 melanoma cell, and D1 marrow stromal cell | Zhao et al., 2022 [105] |
| CellREADR/RADAR/RADARS | Plasmid DNA, and AAV | Exogenously expressed mRNAs of fluorescent protein and ChETA (a variant of light-gated channelrhodopsin-2) Human endogenous transcripts of EEF1A1, ACTB, PCNA, TP53, XIST, HER2, ARC, FOXP2, and VGAT Mouse endogenous transcripts of Fezf2, Ctip2, Plxnd1, Satb2, Rorb, and Vgat Rat endogenous transcripts of Vgat and Tle4 |
Fluorescent proteins and tTA2 | HeLa, HEK293T, KPC1242, and Mouse Neuroblastoma, Neuro-2A, Mouse brain (in vivo) | Qian et al., 2022 [106] |
| Plasmid DNA | Exogenously expressed transcripts Human endogenous transcripts of GAPDH and DNAJB1 (hsp40) |
Fluorescent proteins and Cre recombinase | Flp-In T-Rex HEK293 | Kaseniit et al., 2022 [107] | |
| Plasmid DNA and in vitro transcribed mRNA | Exogenously expressed transcripts of IL6, EGFP, and NPY Endogenous transcripts of RPL41, GAPDH, ACTB, HSP90AA1, PPIB, KRAS, RPS5, UBC, PABPC1, TOP2A, HSP70, and SERPINA1 |
Fluorescent proteins, luciferase, Cre recombinase, and iCaspase | HEK293FT, A549, HepG2, and HeLa | Jiang et al., 2022 [108] |
miRNA sensors
miRNA is a group of short (around 22-nt) non-coding RNAs responsible for the post-transcriptional regulation of gene expression [70]. After processing from primary transcripts and precursor miRNAs, mature miRNAs are loaded onto the protein Argonaute (Ago) and guide the resulting miRNA – protein complex to target mRNAs for translational repression and degradation. More than 2,600 miRNAs are encoded in the human genome [71,72], and miRNA isoforms further diversify the regulatory network of miRNAs [73]. Since different types of tissues and cells have different miRNA signatures [74–77], miRNAs can also be used as markers to identify different cell types [78–83]. Although target sequences of miRNAs are usually observed in the 3’-UTR of mRNAs in nature, we have previously demonstrated that inserting miRNA target sites into the 5’-UTR can greatly improve the performance of miRNA-regulated translation (Figure 4A) [84]. By controlling drug resistance genes, suicide genes, and a membrane protein in a miRNA-dependent manner, we have reported miRNA-based systems to specifically select target cells from heterogeneous cell populations [84–86]. In addition, miRNA-controlled expression of CRISPR/Cas proteins enables cell type-specific genome editing [87]. Such systems will likely be vital to future medical applications such as regenerative medicine and cell therapy.
Figure 4.
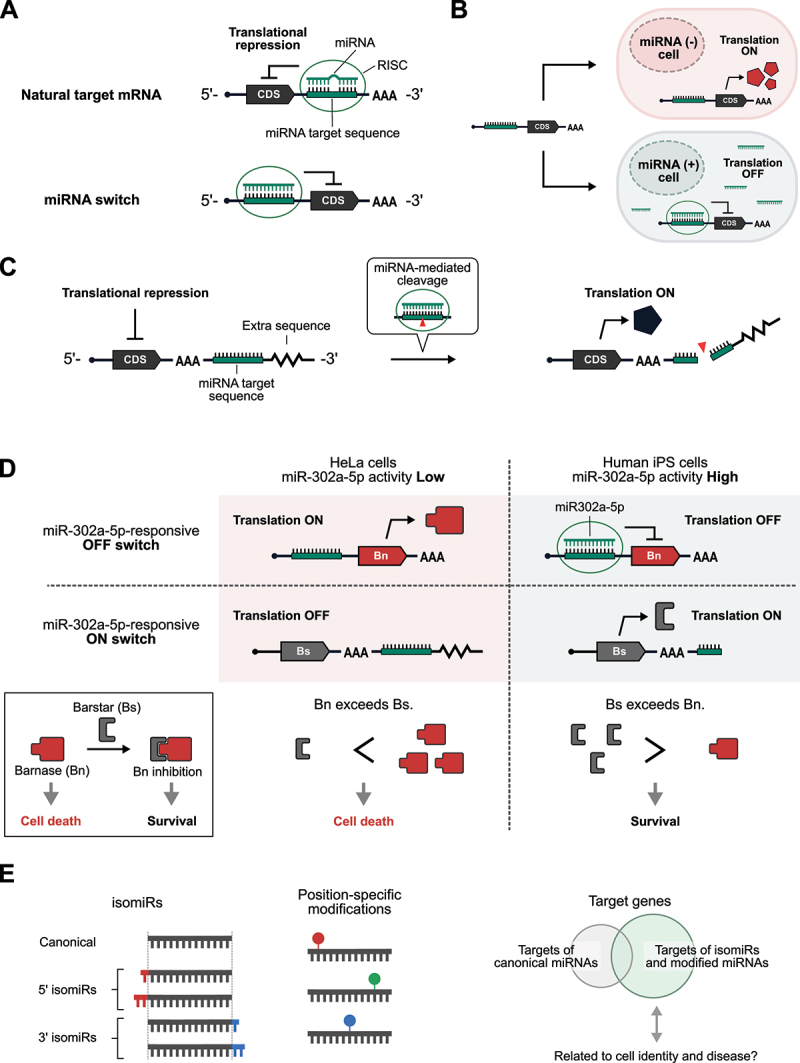
mRNA-based miRNA sensors.
(A) MicroRNA (miRNA) usually binds to target sequences in the 3’-UTR of target mRNAs with imperfect miRNA – mRNA complementarity (top). miRNA-loaded RISC (RNA-induced silencing complex) recognizes target mRNAs and represses their translation. For miRNA switches, target sequences complementary to miRNAs are inserted into the 5’-UTR (bottom). CDS: coding sequence.
(B) Distinguishing different cell types based on miRNA activities. (C) Schematic illustration of miRNA-responsive ON-type switches. (D) Combinatorial use of miRNA-responsive OFF-type switch and ON-type switch for efficient elimination of undesired cells. (E) IsomiRs and modifications in miRNA diversify the regulatory roles of miRNAs. Further studies are needed to elucidate the relationship between these alterations, cellular identity, and diseases.
Since miRNAs usually repress the translation of target mRNAs, the translation of artificial mRNAs containing miRNA target sites is generally inhibited by the presence of target miRNAs (Figure 4B). Although these miRNA-controlled translational repressors (OFF switches) have proven to be useful for cell type-specific control of gene expression and cell fate, miRNA-responsive translational activators (ON switches) would also have great utility. A recent study reported a miRNA-responsive ON switch in which mRNA translation is activated by the presence of target miRNAs (Figure 4C) [88]. In this system, an additional sequence following the miRNA target site was added downstream of the poly(A) tail to repress translation in the absence of trigger miRNAs. Whereas the 3’ ends of naturally-occurring mRNAs usually end with poly(A) tails, it is possible to add extra sequences downstream of the poly(A) tails of artificial mRNAs synthesized in vitro. Surprisingly, the added downstream sequence inhibited translation, which could be derepressed by target miRNAs, probably due to miRNA-mediated cleavage and removal of the extra downstream sequence. With this miRNA-responsive ON switch, the expression level of reporter proteins is ~ 3-fold higher in HeLa cells compared with cells with inhibited miRNA activity. Although further studies are required to elucidate the molecular mechanism of translational repression by such extra sequences following poly(A) tails, it may provide new clues to better our understanding of translational regulation in nature.
Turning on the expression of suicide genes in specific subfractions of a heterogeneous cell population is a promising strategy for mitigating contamination by unwanted cells for cell therapy and eliminating tumour cells while keeping normal cells alive [25,59,89]. To trigger cell death, pro-apoptotic proteins in the Bcl-2 family (e.g. hBax and Bim) have been frequently used in synthetic biology applications [25,59,89]. Although miRNAs can repress the translation of a suicide gene to prevent cell death when there is sufficient miRNA activity, the application of miRNA-responsive OFF switches alone to eliminate specific cell types often suffers from the leaky expression of suicide genes caused by insufficient translational repression by miRNAs, resulting in a loss of the desired cells. In addition, the efficiency in mRNA delivery to cells is not 100%, and the expression levels of mRNA-encoded proteins vary from cell to cell, together making it difficult to eliminate non-transfected, low output-expressing, undesired cells. To overcome these issues, it has been shown that the combined use of both miRNA-responsive ON and OFF switches could increase the purity of desired cells by fine-tuning the balance between activators and inhibitors of cell death [88]. Figure 4D illustrates the elimination of HeLa cells from a mixed culture of HeLa and iPS cells using this system. It consists of two miR-302a-5p-responsive switches and an mRNA for the elimination of non-transfected cells: (1) an OFF switch that encodes a ribonuclease lethal to cells, Barnase (Bn) [90]; (2) an ON switch encoding a protein inhibitor of Bn, Barstar (Bs); and (3) an mRNA that encodes a drug-resistance gene (e.g. Blasticidin-resistance gene) for positive selection. When these three mRNAs are introduced into a co-culture of HeLa (low miR-302a-5p activity) and iPS cells (high miR-302a-5p activity), the translation of Bn is inhibited such that if it leaks, the activated expression of Bs shuts off the activity of Bn in iPS cells, resulting in strong inhibition of cell death. In contrast, there is inadequate miR-302a-5p activity in HeLa cells to suppress Bn expression nor activate Bs expression. Thus, cell death is induced because Bn expression far exceeds the amount of Bs. In addition, non-transfected cells are eliminated by blasticidin treatment. By utilizing this system, it is now possible to purify cells without needing expensive equipment such as fluorescence-activated cell sorters (FACSs). Indeed, it has been shown that cardiomyocytes induced from human iPS cells could be enriched to near-purity. This RNA-based strategy could be adopted widely as an easy and scalable cell purification technology for regenerative medicine applications.
When considering the purification of a specific cell type of interest from a heterogeneous cell population using a miRNA-based system, a critical question is how miRNA markers should be selected. In many studies, medium- and high-throughput techniques like RT-qPCR array, microarray, and massively parallel sequencing have greatly contributed to the identification of cell type-specific miRNA signatures and the elucidation of the roles of miRNAs in complex gene regulatory networks. However, selecting miRNAs solely based on their abundance might overlook suitable miRNAs as markers because of the low correlation between miRNA abundance and their translational repression efficiencies, as measured by miRNA-sensing exogenous reporters [91]. Although the reasons for this low correlation have not been fully elucidated, it may be partly explained by the competitive binding of intracellular miRNAs to their endogenous targets and the exogenous miRNA-sensing constructs [92–94]. In addition, growing evidence from recent studies indicates that isoforms of miRNAs (isomiRs) and position-specific modifications of miRNAs vastly diversify the modes of target recognition and regulation by miRNAs (Figure 4E) [95–98]. IsomiRs vary from canonical miRNA sequences, which are catalogued in miRBase 22 [71,72], by base alterations or nucleotide additions or deletions at the 5’ and/or 3’ ends. Recent advances in deep sequencing of small RNAs have enabled in-depth characterization of sequence variations in miRNAs together with quantitative measurements. According to the current model, nucleotides 2 to 8 on the 5’ end of miRNA are mainly responsible for target recognition by miRNAs [70]. As such, the addition or deletion of nucleotides at the 5’ end of miRNAs shifts the seed regions, thereby altering target recognition [95,96]. One example of position-specific modifications of miRNA is oxidized guanine by reactive oxygen species (ROS). ROS causes oxidative modifications in protein, DNA, and RNA, with RNA being notably more vulnerable to oxidative modifications than protein and DNA [99,100]. Since oxidized guanine can pair with either cytosine or adenine, oxidized guanine in DNA is mutagenic. Interestingly, oxidized guanine in miRNA has been reported to alter the mode of target recognition through mispairing between oxidized guanine and adenine [92,93]. In a study reported in 2015, oxidized miR-184 (but not its unmodified native form) promotes apoptosis by reducing the levels of anti-apoptotic proteins, Bcl-xL and Bcl-w, due to base pairing between oxidized guanine in the miR-184 seed region and adenine in the targets. In addition, oxidized miR-139-3p and miR-204-3p were suggested to switch their targets [97]. A more recent study also reported target alteration in miR-1 in a rat model of cardiac hypertrophy [98]. In this study, the authors developed o8 G-miSeq, in which small RNAs are immunoprecipitated with an antibody that recognizes 8-oxoguanine (o8G) and are deep-sequenced to identify oxidized bases in miRNAs. Collectively, an important implication from these studies is that there is a need to search for miRNA variants related to cellular states and diseases, not simply rely on differences in the abundance of canonical miRNAs because of the aforementioned low correlation between (canonical) miRNA expression levels and their measured activities. This may be vital to dissecting cellular heterogeneity using miRNA-sensing RNA technology. The development of novel technologies for high-throughput analysis of base modifications on miRNAs and RNA-based technologies to sense them will be instrumental in attaining further insights into the roles of miRNAs.
Sensors for longer RNAs
The ability to sense RNA molecules, regardless of whether they appear to have specific regulatory functions, is of great importance. A milestone study in 2014 reported the ‘Toehold switch’ system in bacterial cells to control translational initiation based on strand displacement (Figure 5A) [101]. The toehold switch consists of two components: (1) a ‘switch RNA’ that encodes an arbitrary output protein and (2) a ‘trigger RNA’ that is required for activating switch RNA translation. In the switch RNA, a stem-loop motif is located upstream of the protein-coding sequence. A ribosome binding site (RBS), which facilitates translational initiation, is inserted into the single-stranded loop region, and the start codon is embedded in the partially single-stranded region of the RNA duplex. When the trigger RNA is absent, the region surrounding the RBS and start codon are sequestered, so ribosomes cannot initiate translation. The trigger RNA uses the single-stranded region upstream of the stem-loop sequence as a ‘toehold’ to unwind the duplex and expose the RBS to initiate translation. The toehold switch works not only in bacterial cells but also in cell-free systems on paper [102–104], which relies on bacterial components for translation.
Figure 5.
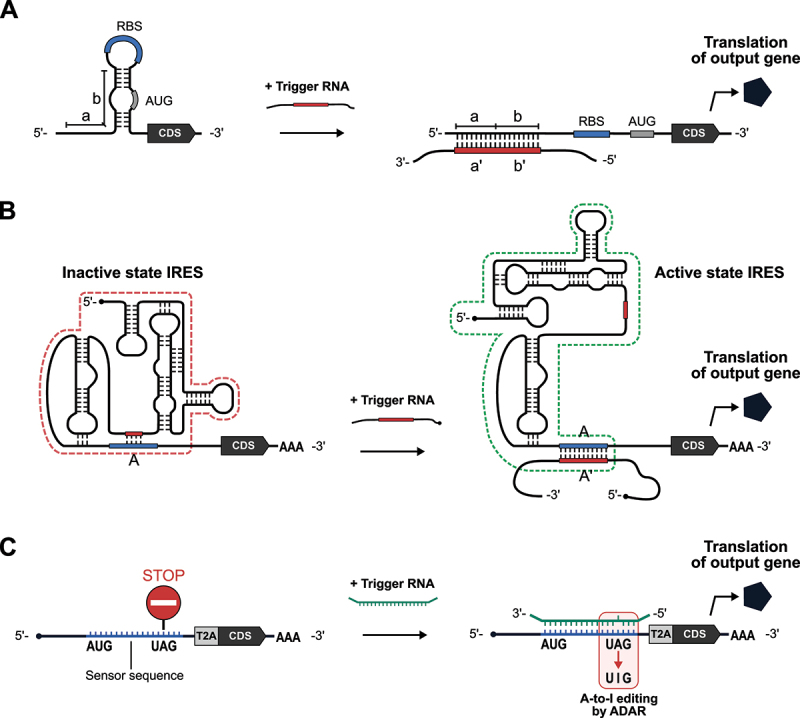
mRNA-based longer RNA sensors.
(A) Schematic illustration of bacterial toehold switches. In the absence of trigger RNA, ribosome binding site (RBS) and AUG initiation codon are sequestered. Interaction between a’ on the trigger RNA and a toehold sequence (a) on the toehold switch initiates RNA – RNA strand displacement interactions. The extended single-stranded sequence on the trigger RNA unwinds the hairpin structure through the b – b’ interaction, thereby initiating translation. (B) Schematic illustration of the eukaryotic version of toehold switch, eToehold. Short (red) and long (blue) RNA segments inserted into IRES (internal ribosome entry site) disrupt the functional structure of IRES. The trigger RNA – eToehold interaction (A – A’) restores the functional IRES structure and activates translation. (C) Schematic illustration of ADAR-mediated RNA sensors. In the absence of trigger RNA, translation terminates at the stop codon upstream of the CDS of an output gene. In the presence of trigger RNA, the RNA duplex surrounding the UAG stop codon recruits ADAR. ADAR-mediated A-to-I editing converts the UAG stop codon to the UIG codon for tryptophan, resulting in the translation of the downstream output CDS. The T2A peptide induces ribosome skipping to translate the output protein and the upstream CDS separately.
However, implementing such a system in eukaryotic cells has lagged until very recently because of large differences in the mechanisms of translational initiation between eukaryotes and prokaryotes. In eukaryotic cells, it has been a challenge to artificially regulate translational initiation using a short sequence like RBS. In 2021, James Collins and his group reported a eukaryotic version of the toehold switch, ‘eToehold’, that works in mammalian cells (Figure 5B) [105]. eToehold uses an IRES, an RNA element derived from viruses that can initiate translation independently of the 5’ cap structure of mRNAs. The translational control activity of IRESs depends on the higher-order structure. In eToehold, the structure required for translational initiation is disrupted by the insertion of long (40–50-nt-long) and short (6–15-nt-long) complementary RNA segments to form a new intramolecular interaction, which is displaced via sense – antisense RNA interactions in the presence of a trigger RNA, thereby restoring IRES functionality. After optimizing the design to reduce leaky expression, eToehold switches were shown to sense Zika virus infection and endogenous heat shock protein mRNAs in mammalian cells.
In 2022, three groups leveraged an RNA-editing enzyme, ADAR (Adenosine Deaminase Acting on RNA), as a new RNA-sensing technology: CellREADR (Cell access through RNA sensing by Endogenous ADAR) [106], RADAR (RNA sensing using ADAR) [107], and RADARS (Reprogrammable ADAR Sensors) (Figure 5C) [108]. ADAR is an A-to-I RNA editing enzyme that converts adenosine in the double-stranded region of RNA – through a hydrolytic deamination reaction – to inosine [109–111], which is then recognized as guanosine. A common mechanism of action for these three sensors is that: (1) in the sensor RNA, UAG stop codons are placed upstream of the coding region of an output gene (e.g. fluorescent protein) to block its translation in the absence of a trigger RNA; (2) sequences flanking the stop codon in the sensor RNA (sensor sequences) are reverse complementary to the trigger RNA, but designed to have a mismatched C to create a bubble at the UAG stop codon that enhances ADAR-mediated adenosine deamination (e.g. 5’- UAG −3’ in the sensor RNA and 3’- ACC −5’ in the trigger); and (3) ADAR recognizes the trigger-sensor duplex and converts adenosine to inosine to generate a UIG (recognized as a UGG and codes for tryptophan) upon binding between trigger and sensor RNAs, thereby allowing the translation of the downstream sequence. The length of sensor sequences required to efficiently turn on output gene translation is different for each system, probably due to differences in sensor and trigger RNA designs, but > 50-bp double-stranded RNA seems to be capable of activating the translation of ADAR-based RNA sensors. All three systems succeeded in detecting endogenous RNA transcripts. In addition, by placing sensor RNA sequences for different triggers in tandem, 2-input AND-like behaviour, where the output gene is only expressed in the presence of both input triggers, was achieved. Future work may focus on further improvement of the specificity, sensitivity, and reliability of ADAR-based RNA sensors by optimizing the design of the sensor RNAs (e.g. the length and sequences of sensors, position of the stop codon) and choice of the trigger RNA sequences, as well as engineering of the RNA-editing enzymes themselves.
An important property of eToehold and ADAR-based sensors is that they can detect RNAs, such as mRNAs, through simple base pairing but do not require specific regulatory functions (e.g. translational repression ability of miRNAs) from the target transcripts. The transcriptome is a critical piece of information that defines the type and state of the cell. With the emergence of deep sequencing, transcriptome analysis at the single-cell resolution is becoming more accessible. RNA-sensing systems like eToehold and ADAR-based sensors are compatible with transcriptome analysis and will likely be further developed in the future to meet the increasing demand for specific control of gene expression and cell fate using synthetic mRNAs.
Conclusion and perspective
In this review, we highlighted emerging technologies in RNA-based sensors for cellular RNA and protein. RNA engineering and its medical application are gaining momentum with the rise of mRNA therapeutics. RNA-based sensors will contribute significantly to precision diagnostics and medicine in the future. For example, mRNA-based medicine, in theory, can sense cancer-related signatures to eliminate tumour cells specifically.
As discussed above, various RNA-based sensors in development leverage a wide range of molecular mechanisms to detect intracellular molecules and regulate translation. Nonetheless, there remains room to significantly improve the performance of such systems (e.g. sensitivity and specificity) before they are ready for practical applications. As deep learning has recently made remarkable progress in designing new proteins [112] and predicting protein structure [113], advances in RNA engineering will be similarly aided using machine learning techniques. To achieve this, accumulating large collections of open-source data on RNA – experimentally validated secondary and tertiary structures and functional properties, preferably with the cellular contexts – will be necessary. Another potential challenge is the use of non-human proteins in some sensors (e.g. L7Ae, MS2CP, VPg, and CRISPR-Cas proteins) because these non-self-proteins might trigger immune responses once expressed in the hosts. Indeed, it has been documented that there is a pre-existing humoral and cellular immunity against Cas9 nucleases in humans [114,115]. The immunogenicity of these proteins in humans remains largely unknown, but this should be a matter requiring careful consideration when utilizing them in vivo . Perhaps there is a need to develop sensors that utilize improved or novel proteins with reduced immunogenicity in humans.
In the past two decades, synthetic biology and RNA science have made momentous progress. We hope that scientists from diverse backgrounds will join this field and boost the development of novel technologies for practical applications in the next two decades.
Acknowledgments
We thank Dr. Kelvin Hui and Chen Jung (Kyoto University) for the critical reading of this manuscript. We also thank Hiromi Takemoto and Yoshiko Ogawa (Kyoto University) for their administrative support.
Funding Statement
The work was supported by the Japan Society for the Promotion of Science [JP20H05626]; Japan Society for the Promotion of Science [JP22K20628].
Author contributions
H.O. and H.S wrote and edited the manuscript.
Disclosure statement
H.S. owns shares of aceRNA Technologies Ltd. and is an outside director of aceRNA Technologies Ltd.
Data availability statement
Data sharing is not applicable to this article as no new data were created or analysed for this review article.
References
- [1].Benner SA, Sismour AM.. Synthetic biology. Nat Rev Genet. 2005;6(7):533–543. doi: 10.1038/nrg1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Voigt CA. Synthetic biology 2020–2030: six commercially-available products that are changing our world. Nat Commun. 2020;11(1):10–15. doi: 10.1038/s41467-020-20122-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ruder WC, Lu T, Collins JJ. Synthetic biology moving into the clinic. Science. 2011;333(6047):1248–1252. doi: 10.1126/science.1206843 [DOI] [PubMed] [Google Scholar]
- [4].Weber W, Fussenegger M. Emerging biomedical applications of synthetic biology. Nat Rev Genet. 2012;13(1):21–35. doi: 10.1038/nrg3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kitada T, DiAndreth B, Teague B, et al. Programming gene and engineered-cell therapies with synthetic biology. Science. 2018;359(6376). doi: 10.1126/science.aad1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wurtzel ET, Vickers CE, Hanson AD, et al. Revolutionizing agriculture with synthetic biology. Nat Plants. 2019;5(12):1207–1210. doi: 10.1038/s41477-019-0539-0 [DOI] [PubMed] [Google Scholar]
- [7].Jagadevan S, Banerjee A, Banerjee C, et al. Recent developments in synthetic biology and metabolic engineering in microalgae towards biofuel production. Biotechnol Biofuels. 2018;11(1):1–21. doi: 10.1186/s13068-018-1181-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kelwick RJR, Webb AJ, Freemont PS. Biological materials: the next frontier for cell-free synthetic biology. Front Bioeng Biotechnol. 2020;8: doi: 10.3389/fbioe.2020.00399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403(6767):335–338. doi: 10.1038/35002125 [DOI] [PubMed] [Google Scholar]
- [10].Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403(6767):339–342. doi: 10.1038/35002131 [DOI] [PubMed] [Google Scholar]
- [11].Guet CC, Elowitz MB, Hsing W, et al. Combinatorial synthesis of genetic networks. Science. 2002;296(5572):1466–1470. doi: 10.1126/science.1067407 [DOI] [PubMed] [Google Scholar]
- [12].Yokobayashi Y, Weiss R, Arnold FH. Directed evolution of a genetic circuit. Proc Natl Acad Sci U S A. 2002;99(26):16587–16591. doi: 10.1073/pnas.252535999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hasty J, McMillen D, Collins JJ. Engineered gene circuits. Nature. 2002;420(6912):224–230. doi: 10.1038/nature01257 [DOI] [PubMed] [Google Scholar]
- [14].Serganov A, Patel DJ. Ribozymes, riboswitches and beyond: Regulation of gene expression without proteins. Nat Rev Genet. 2007;8(10):776–790. doi: 10.1038/nrg2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Statello L, Guo CJ, Chen LL, et al. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22(2):96–118. doi: 10.1038/s41580-020-00315-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhou J, Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat Rev Drug Discov. 2017;16(3):181–202. doi: 10.1038/nrd.2016.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Doherty EA, Doudna JA. Ribozyme structures and mechanisms. Annu Rev Biochem. 2000;69(1):597–615. doi: 10.1146/annurev.biochem.69.1.597 [DOI] [PubMed] [Google Scholar]
- [18].Mandal M, Breaker RR. Gene regulation by riboswitches. Nat Rev Mol Cell Biol. 2004;5(6):451–463. doi: 10.1038/nrm1403 [DOI] [PubMed] [Google Scholar]
- [19].Higgs PG, Lehman N. The RNA world: molecular cooperation at the origins of life. Nat Rev Genet. 2015;16(1):7–17. doi: 10.1038/nrg3841 [DOI] [PubMed] [Google Scholar]
- [20].Isaacs FJ, Dwyer DJ, Collins JJ. RNA synthetic biology. Nat Biotechnol. 2006;24(5):545–554. doi: 10.1038/nbt1208 [DOI] [PubMed] [Google Scholar]
- [21].Sahin U, Karikó K, Türeci Ö. mRNA-based therapeutics — developing a new class of drugs. Nat Rev Drug Discov. 2014;13(10):759–780. doi: 10.1038/nrd4278 [DOI] [PubMed] [Google Scholar]
- [22].Qin S, Tang X, Chen Y, et al. mRNA-based therapeutics: powerful and versatile tools to combat diseases. Signal Transduct Target Ther. 2022;7(1). doi: 10.1038/s41392-022-01007-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Andries O, De Filette M, Rejman J, et al. Comparison of the gene transfer efficiency of mRNA/GL67 and pDNA/GL67 complexes in respiratory cells. Mol Pharm. 2012;9(8):2136–2145. doi: 10.1021/mp200604h [DOI] [PubMed] [Google Scholar]
- [24].Saito H, Kobayashi T, Hara T, et al. Synthetic translational regulation by an L7Ae–kink-turn RNP switch. Nat Chem Biol. 2010;6(1):71–78. doi: 10.1038/nchembio.273 [DOI] [PubMed] [Google Scholar]
- [25].Saito H, Fujita Y, Kashida S, et al. Synthetic human cell fate regulation by protein-driven RNA switches. Nat Commun. 2011;2(1):160–168. doi: 10.1038/ncomms1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Endo K, Stapleton JA, Hayashi K, et al. Quantitative and simultaneous translational control of distinct mammalian mRnas. Nucleic Acids Res. 2013;41(13):e135–e135. doi: 10.1093/nar/gkt347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hara T, Saito H, Inoue T. Directed evolution of a synthetic RNA–protein module to create a new translational switch. Chem Commun. 2013;49(37):3833–3835. doi: 10.1039/c3cc38688k [DOI] [PubMed] [Google Scholar]
- [28].Kawasaki S, Fujita Y, Nagaike T, et al. Synthetic mRNA devices that detect endogenous proteins and distinguish mammalian cells. Nucleic Acids Res. 2017;45(12):e117. doi: 10.1093/nar/gkx298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ono H, Kawasaki S, Saito H. Orthogonal protein-responsive mRNA switches for mammalian synthetic Biology. ACS Synth Biol. 2020;9(1):169–174. doi: 10.1021/acssynbio.9b00343 [DOI] [PubMed] [Google Scholar]
- [30].Matsuura S, Ono H, Kawasaki S, et al. Synthetic RNA-based logic computation in mammalian cells. Nat Commun. 2018;9(1):4847. doi: 10.1038/s41467-018-07181-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Parr CJC, Wada S, Kotake K, et al. N 1-methylpseudouridine substitution enhances the performance of synthetic mRNA switches in cells. Nucleic Acids Res. 2020;48(6):e35–e35. doi: 10.1093/nar/gkaa070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A. 1992;89(12):5547–5551. doi: 10.1073/pnas.89.12.5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Belmont BJ, Niles JC. Engineering a direct and inducible protein−RNA interaction to regulate RNA Biology. ACS Chem Biol. 2010;5(9):851–861. doi: 10.1021/cb100070j [DOI] [PubMed] [Google Scholar]
- [34].Wagner TE, Becraft JR, Bodner K, et al. Small-molecule-based regulation of RNA-delivered circuits in mammalian cells. Nat Chem Biol. 2018;14(11):1043–1050. doi: 10.1038/s41589-018-0146-9 [DOI] [PubMed] [Google Scholar]
- [35].Borchardt EK, Vandoros LA, Huang M, et al. Controlling mRNA stability and translation with the CRISPR endoribonuclease Csy4. RNA. 2015;21(11):1921–1930. doi: 10.1261/rna.051227.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].DiAndreth B, Wauford N, Hu E, et al. PERSIST platform provides programmable RNA regulation using CRISPR endoRnases. Nat Commun. 2022;13(1). doi: 10.1038/s41467-022-30172-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kawasaki S, Ono H, Hirosawa M, et al. Programmable mammalian translational modulators by CRISPR-associated proteins. Nat Commun. 2023;14(1):2243. doi: 10.1038/s41467-023-37540-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Cella F, Wroblewska L, Weiss R, et al. Engineering protein-protein devices for multilayered regulation of mRNA translation using orthogonal proteases in mammalian cells. Nat Commun. 2018;9(1). doi: 10.1038/s41467-018-06825-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Shyh-Chang N, Daley GQ. Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell. 2013;12(4):395–406. doi: 10.1016/j.stem.2013.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Mojica FJM, Diez-Villasenor C, Soria E, et al. Biological significance of a family of regularly spaced repeats in the genomes of archaea, bacteria and mitochondria. Mol Microbiol. 2000;36(1):244–246. doi: 10.1046/j.1365-2958.2000.01838.x [DOI] [PubMed] [Google Scholar]
- [41].Jansen R, Van Embden JDA, Gaastra W, et al. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43(6):1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x [DOI] [PubMed] [Google Scholar]
- [42].Al-Shayeb B, Sachdeva R, Chen L-X, et al. Clades of huge phages from across Earth’s ecosystems. Nature. 2020;578(7795):425–431. doi: 10.1038/s41586-020-2007-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Pausch P, Al-Shayeb B, Bisom-Rapp E, et al. CRISPR-Casφ from huge phages is a hypercompact genome editor. Science. 2020;369(6501):333–337. doi: 10.1126/science.abb1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ausländer S, Stücheli P, Rehm C, et al. A general design strategy for protein-responsive riboswitches in mammalian cells. Nat Methods. 2014;11(11):1154–1160. doi: 10.1038/nmeth.3136 [DOI] [PubMed] [Google Scholar]
- [45].Kennedy AB, Vowles JV, D’Espaux L, et al. Protein-responsive ribozyme switches in eukaryotic cells. Nucleic Acids Res. 2014;42(19):12306–12321. doi: 10.1093/nar/gku875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Endo K, Hayashi K, Inoue T, et al. A versatile cis-acting inverter module for synthetic translational switches. Nat Commun. 2013;4(1):1–9. doi: 10.1038/ncomms3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lykke-Andersen S, Jensen TH. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol. 2015;16(11):665–677. doi: 10.1038/nrm4063 [DOI] [PubMed] [Google Scholar]
- [48].Nakanishi H, Saito H. Caliciviral protein-based artificial translational activator for mammalian gene circuits with RNA-only delivery. Nat Commun. 2020;11(1):1297. doi: 10.1038/s41467-020-15061-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Royall E, Locker N. Translational control during calicivirus infection. Viruses. 2016;8(4):104–113. doi: 10.3390/v8040104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Nakanishi H, Yoshii T, Kawasaki S, et al. Light-controllable RNA-protein devices for translational regulation of synthetic mRnas in mammalian cells. Cell Chem Biol. 2021;28(5):662–674.e5. doi: 10.1016/j.chembiol.2021.01.002 [DOI] [PubMed] [Google Scholar]
- [51].Nakanishi H, Saito H, Itaka K. Versatile design of intracellular protein-responsive translational regulation system for synthetic mRNA. ACS Synth Biol. 2022;11(3):1077–1085. doi: 10.1021/acssynbio.1c00567 [DOI] [PubMed] [Google Scholar]
- [52].Tuerk C, Gold L. Systematic Evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DBA polymerase. Science. 1990;249(4968):505–510. doi: 10.1126/science.2200121 [DOI] [PubMed] [Google Scholar]
- [53].Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346(6287):818–822. doi: 10.1038/346818a0 [DOI] [PubMed] [Google Scholar]
- [54].Hunsicker A, Steber M, Mayer G, et al. An RNA aptamer that induces transcription. Chem Biol. 2009;16(2):173–180. doi: 10.1016/j.chembiol.2008.12.008 [DOI] [PubMed] [Google Scholar]
- [55].Atanasov J, Groher F, Weigand JE, et al. Design and implementation of a synthetic pre-miR switch for controlling miRNA biogenesis in mammals. Nucleic Acids Res. 2017;45(22):e181–e181. doi: 10.1093/nar/gkx858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Mol AA, Groher F, Schreiber B, et al. Robust gene expression control in human cells with a novel universal TetR aptamer splicing module. Nucleic Acids Res. 2019;47(20):e132–e132. doi: 10.1093/nar/gkz753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].MOL AA, VOGEL M, SUESS B. Inducible nuclear import by TetR aptamer-controlled 3′ splice site selection. RNA. 2021;27(2):234–241. doi: 10.1261/rna.077453.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Weber AM, Kaiser J, Ziegler T, et al. A blue light receptor that mediates RNA binding and translational regulation. Nat Chem Biol. 2019;15(11):1085–1092. doi: 10.1038/s41589-019-0346-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Wroblewska L, Kitada T, Endo K, et al. Mammalian synthetic circuits with RNA binding proteins for RNA-only delivery. Nat Biotechnol. 2015;33(8):839–841. doi: 10.1038/nbt.3301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Ausländer S, Ausländer D, Müller M, et al. Programmable single-cell mammalian biocomputers. Nature. 2012;487(7405):123–127. doi: 10.1038/nature11149 [DOI] [PubMed] [Google Scholar]
- [61].Zalatan JG, Lee M, Almeida R, et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 2015;160(1–2):339–350. doi: 10.1016/j.cell.2014.11.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Shechner DM, Hacisuleyman E, Younger ST, et al. Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat Methods. 2015;12(7):664–670. doi: 10.1038/nmeth.3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Hocine S, Raymond P, Zenklusen D, et al. Single-molecule analysis of gene expression using two-color RNA labeling in live yeast. Nat Methods. 2013;10(2):119–121. doi: 10.1038/nmeth.2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Wu B, Chen J, Singer RH. Background free imaging of single mRnas in live cells using split fluorescent proteins. Sci Rep. 2014;4(1):11–13. doi: 10.1038/srep03615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Tutucci E, Vera M, Biswas J, et al. An improved MS2 system for accurate reporting of the mRNA life cycle. Nat Methods. 2018;15(1):81–89. doi: 10.1038/nmeth.4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Smith GP. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228(4705):1315–1317. doi: 10.1126/science.4001944 [DOI] [PubMed] [Google Scholar]
- [67].Mccafferty J, Griffiths AD, Winter G, et al. Phage antibodies: filamentous phage displaying antibody variable domains. Nature. 1990;348(6301):552–554. doi: 10.1038/348552a0 [DOI] [PubMed] [Google Scholar]
- [68].Danner S, Belasco JG. T7 phage display: A novel genetic selection system for cloning RNA-binding proteins from cDNA libraries. Proc Natl Acad Sci U S A. 2001;98(23):12954–12959. doi: 10.1073/pnas.211439598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Fukunaga K, Yokobayashi Y. Directed evolution of orthogonal RNA–RBP pairs through library-vs-library in vitro selection. Nucleic Acids Res. 2022;50(2):601–616. doi: 10.1093/nar/gkab527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Griffiths-Jones S, Grocock RJ, van Dongen S, et al. miRbase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(90001):D140–D144. doi: 10.1093/nar/gkj112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kozomara A, Birgaoanu M, Griffiths-Jones S. MiRBase: From microRNA sequences to function. Nucleic Acids Res. 2019;47(D1):D155–D162. doi: 10.1093/nar/gky1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14(8):475–488. doi: 10.1038/nrm3611 [DOI] [PubMed] [Google Scholar]
- [74].Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- [75].Kosik KS. MicroRNAs and cellular phenotypy. Cell. 2010;143(1):21–26. doi: 10.1016/j.cell.2010.09.008 [DOI] [PubMed] [Google Scholar]
- [76].Lagos-Quintana M, Rauhut R, Yalcin A, et al. Identification of tissue-specific MicroRNAs from mouse. Curr Biol. 2002;12(9):735–739. doi: 10.1016/S0960-9822(02)00809-6 [DOI] [PubMed] [Google Scholar]
- [77].Chen CZ, Li L, Lodish HF, et al. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83–86. doi: 10.1126/science.1091903 [DOI] [PubMed] [Google Scholar]
- [78].Mansfield JH, Harfe BD, Nissen R, et al. MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet. 2004;36(10):1079–1083. doi: 10.1038/ng1421 [DOI] [PubMed] [Google Scholar]
- [79].Brown BD, Venneri MA, Zingale A, et al. Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nat Med. 2006;12(5):585–591. doi: 10.1038/nm1398 [DOI] [PubMed] [Google Scholar]
- [80].Gentner B, Visigalli I, Hiramatsu H, et al. Identification of hematopoietic stem cell–specific miRNAs enables gene therapy of globoid cell leukodystrophy. Sci Transl Med. 2010;2(58):ra5884–ra5884. doi: 10.1126/scitranslmed.3001522 [DOI] [PubMed] [Google Scholar]
- [81].Endo K, Hayashi K, Saito H. High-resolution identification and separation of living cell types by multiple microRNA-responsive synthetic mRnas. Sci Rep. 2016;6(1):1–8. doi: 10.1038/srep21991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Nakanishi H, Miki K, Komatsu KR, et al. Monitoring and visualizing microRNA dynamics during live cell differentiation using microRNA-responsive non-viral reporter vectors. Biomaterials. 2017;128:121–135. doi: 10.1016/j.biomaterials.2017.02.033 [DOI] [PubMed] [Google Scholar]
- [83].Endo K, Hayashi K, Saito H. Numerical operations in living cells by programmable RNA devices. Sci Adv. 2019;5(8):eaax0835. doi: 10.1126/sciadv.aax0835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Miki K, Endo K, Takahashi S, et al. Efficient detection and purification of cell populations using synthetic microRNA switches. Cell Stem Cell. 2015;16(6):699–711. doi: 10.1016/j.stem.2015.04.005 [DOI] [PubMed] [Google Scholar]
- [85].Parr CJC, Katayama S, Miki K, et al. MicroRNA-302 switch to identify and eliminate undifferentiated human pluripotent stem cells. Sci Rep. 2016;6(1):1–14. doi: 10.1038/srep32532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Tsujisaka Y, Hatani T, Okubo C, et al. Purification of human Ipsc-derived cells at large scale using microRNA switch and magnetic-activated cell sorting. Stem Cell Rep. 2022;17(7):1772–1785. doi: 10.1016/j.stemcr.2022.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Hirosawa M, Fujita Y, Parr C, et al. Cell-type-specific genome editing with a microRNA-responsive CRISPR–Cas9 switch. Nucleic Acids Res. 2017;45(13):e118–e118. doi: 10.1093/nar/gkx309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Fujita Y, Hirosawa M, Hayashi K, et al. A versatile and robust cell purification system with an RNA-only circuit composed of microRNA-responsive on and off switches. Sci Adv. 2022;8(1):1–16. doi: 10.1126/sciadv.abj1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Xie Z, Wroblewska L, Prochazka L, et al. Multi-Input RNAi-based logic circuit for identification of specific cancer cells. Science. 2011;333(6047):1307–1311. doi: 10.1126/science.1205527 [DOI] [PubMed] [Google Scholar]
- [90].Hartley RW. Barnase and barstar: two small proteins to fold and fit together. Trends Biochem Sci. 1989;14(11):450–454. doi: 10.1016/0968-0004(89)90104-7 [DOI] [PubMed] [Google Scholar]
- [91].Mullokandov G, Baccarini A, Ruzo A, et al. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat Methods. 2012;9(8):840–846. doi: 10.1038/nmeth.2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Poliseno L, Salmena L, Zhang J, et al. A coding-independent function of gene and pseudogene mRnas regulates tumour biology. Nature. 2010;465(7301):1033–1038. doi: 10.1038/nature09144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Cesana M, Cacchiarelli D, Legnini I, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–369. doi: 10.1016/j.cell.2011.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Tan GC, Chan E, Molnar A, et al. 5′ isomiR variation is of functional and evolutionary importance. Nucleic Acids Res. 2014;42(14):9424–9435. doi: 10.1093/nar/gku656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Bofill-De Ros X, Kasprzak WK, Bhandari Y, et al. Structural differences between pri-miRNA paralogs promote alternative drosha cleavage and expand target repertoires. Cell Rep. 2019;26(2):447–459.e4. doi: 10.1016/j.celrep.2018.12.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Wang J-X, Gao J, Ding S-L, et al. Oxidative modification of mir-184 enables it to target Bcl-Xl and Bcl-w. Mol Cell. 2015;59(1):50–61. doi: 10.1016/j.molcel.2015.05.003 [DOI] [PubMed] [Google Scholar]
- [98].Seok H, Lee H, Lee S, et al. Position-specific oxidation of miR-1 encodes cardiac hypertrophy. Nature. 2020;584(7820):279–285. doi: 10.1038/s41586-020-2586-0 [DOI] [PubMed] [Google Scholar]
- [99].Tanaka M, Chock PB, Stadtman ER. Oxidized messenger RNA induces translation errors. Proc Natl Acad Sci U S A. 2007;104(1):66–71. doi: 10.1073/pnas.0609737104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Kong Q, Lin CG. Oxidative damage to RNA: mechanisms, consequences, and diseases. Cell Mol Life Sci. 2010;67(11):1817–1829. doi: 10.1007/s00018-010-0277-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Green AA, Silver PA, Collins JJ, et al. Toehold switches: De-novo-designed regulators of gene expression. Cell. 2014;159(4):925–939. doi: 10.1016/j.cell.2014.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Pardee K, Green A, Ferrante T, et al. Paper-based synthetic gene networks. Cell. 2014;159(4):940–954. doi: 10.1016/j.cell.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Pardee K, Green AA, Takahashi MK, et al. Rapid, low-cost detection of zika virus using programmable biomolecular components. Cell. 2016;165(5):1255–1266. doi: 10.1016/j.cell.2016.04.059 [DOI] [PubMed] [Google Scholar]
- [104].Takahashi MK, Tan X, Dy AJ, et al. A low-cost paper-based synthetic biology platform for analyzing gut microbiota and host biomarkers. Nat Commun. 2018;9(1):1–12. doi: 10.1038/s41467-018-05864-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Zhao EM, Mao AS, de Puig H, et al. RNA-responsive elements for eukaryotic translational control. Nat Biotechnol. 2022;40(4):539–545. doi: 10.1038/s41587-021-01068-2 [DOI] [PubMed] [Google Scholar]
- [106].Qian Y, Li J, Zhao S, et al. Programmable RNA sensing for cell monitoring and manipulation. Nature. 2022;610(7933):713–721. doi: 10.1038/s41586-022-05280-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Kaseniit KE, Katz N, Kolber NS, et al. Modular, programmable RNA sensing using ADAR editing in living cells. Nat Biotechnol. 2023;41(4):482–487. doi: 10.1038/s41587-022-01493-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Jiang K, Koob J, Chen XD, et al. Programmable eukaryotic protein synthesis with RNA sensors by harnessing ADAR. Nat Biotechnol. 2023;41(5):698–707. doi: 10.1038/s41587-022-01534-5 [DOI] [PubMed] [Google Scholar]
- [109].Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71(1):817–846. doi: 10.1146/annurev.biochem.71.110601.135501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79(1):321–349. doi: 10.1146/annurev-biochem-060208-105251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Savva YA, Rieder LE, Reenan RA. The ADAR protein family. Genome Biol. 2012;13(12):252. doi: 10.1186/gb-2012-13-12-252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Anishchenko I, Pellock SJ, Chidyausiku TM, et al. De Novo protein design by deep network hallucination. Nature. 2021;600(7889):547–552. doi: 10.1038/s41586-021-04184-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Jumper J, Evans R, Pritzel A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Charlesworth CT, Deshpande PS, Dever DP, et al. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat Med. 2019;25(2):249–254. doi: 10.1038/s41591-018-0326-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Wagner DL, Amini L, Wendering DJ, et al. High prevalence of Streptococcus pyogenes Cas9-reactive T cells within the adult human population. Nat Med. 2019;25(2):242–248. doi: 10.1038/s41591-018-0204-6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analysed for this review article.


