Summary
Sexually dimorphic traits in morphologies are widely studied1–4, but those in essential molecular pathways remain largely unexplored. Previous work showed substantial sex differences in Drosophila gonadal piRNAs5, which guide PIWI proteins to silence selfish genetic elements thereby safeguarding fertility6–8. However, the genetic control mechanisms of piRNA sexual dimorphism remain unknown. Here, we showed that most sex differences in the piRNA program originate from the germline rather than gonadal somatic cells. Building on this, we dissected the contribution of sex chromosome and cellular sexual identity towards the sex-specific germline piRNA program. We found that the presence of the Y chromosome is sufficient to recapitulate some aspects of the male piRNA program in a female cellular environment. Meanwhile, sexual identity controls the sexually divergent piRNA production from X-linked and autosomal loci, revealing a crucial input from sex determination into piRNA biogenesis. Sexual identity regulates piRNA biogenesis through Sxl and this effect is mediated in part through chromatin proteins Phf7 and Kipferl. Together, our work delineated the genetic control of a sex-specific piRNA program, where sex chromosome and sexual identity collectively sculpt an essential molecular trait.
Keywords: piRNA, sexual dimorphism, Y chromosome, sex determination, Sxl, Phf7, Kipferl, transposon, Drosophila, germline
Graphical Abstract
eTOC blurb / In Brief
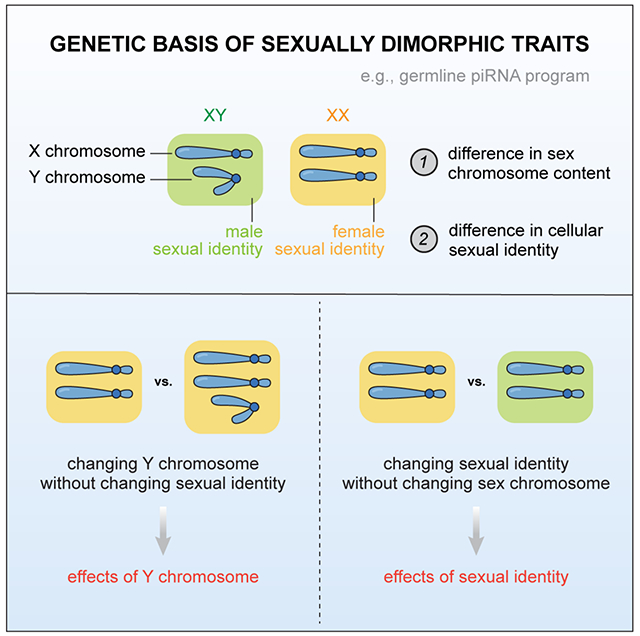
The genetic basis of sexually dimorphic traits is often unknown. Chen and Aravin genetically separated the actions of sex chromosomes and the sex determination pathway to dissect their contributions towards the sex-specific piRNA program in D. melanogaster germline.
Results
Sexual dimorphism, where a trait is modified by the biological sex to manifest in distinct ways between males and females, is pervasive in nature. While sexually dimorphic traits in morphologies have been widely studied1–4, those in essential molecular pathways remain largely unexplored. In Drosophila melanogaster gonads, the piRNA program executes a critical function by guiding the PIWI-clade Argonaute proteins to silence selfish genetic elements such as transposons6–8, thereby safeguarding fertility. To pass on the transgenerational memory of proper piRNA targets, mothers deposit piRNAs to the embryo, instructing the zygotic genome to mount a homologous piRNA program in the next generation that reflects the maternal response to genomic parasites9–12. However, males implement a piRNA program distinct from their female siblings5, the underlying mechanism of which is elusive. We previously found evidence for both differential transcription of piRNA loci in the nucleus13 and differential processing of piRNA precursor transcripts in the cytoplasm5 between the two sexes, but the upstream control of these sexually dimorphic molecular events is unknown. In this work, we sought to decipher the genetic control of piRNA sexual dimorphism, in order to gain insights into the mechanisms by which sexual dimorphism in essential molecular traits is sculpted.
Prior work compared the male and female piRNA profiles from two different D. melanogaster lab strains5, where distinct genetic backgrounds confounded the characterization of piRNA sexual dimorphism. In addition, the sex of D. melanogaster is determined independently of the presence of the Y chromosome (both XY and XO flies are phenotypic males, while XX and XXY flies are phenotypic females)14–16, so the morphology-based identification of males and females does not directly translate to an interpretation of Y chromosome status. Given that several piRNA-producing loci reside on the Y5, the inability to infer Y chromosome content from the phenotypic sex complicates the characterization of piRNA sexual dimorphism. To circumvent these issues, we introduced a Y chromosome marked by y+ and w+ genes (hereafter y+w+Y) into an inbred yw stock and backcrossed it to yw for multiple consecutive generations (see STAR Methods). This line allowed us to unequivocally identify XY males (red-eyed flies with black body color and male genitalia) and XX females (white-eyed flies with yellow body color and female genitalia) (Figure 1A), from which we profiled the gonadal piRNAs in each of the two sexes. Analysis of the piRNA libraries showed substantial intersexual differences in the abundance of piRNAs targeting different transposon families (Figure 1B) and expression levels of individual major piRNA loci in the genome (Figure 1C), largely in agreement with our previous study5. Having excluded the possible confounding effects of genetic backgrounds and Y chromosome status, we confirmed that the piRNA program in D. melanogaster gonads is sexually dimorphic.
Figure 1 |. Germline is the major cell type origin of piRNA sexual dimorphism in D. mel gonads.
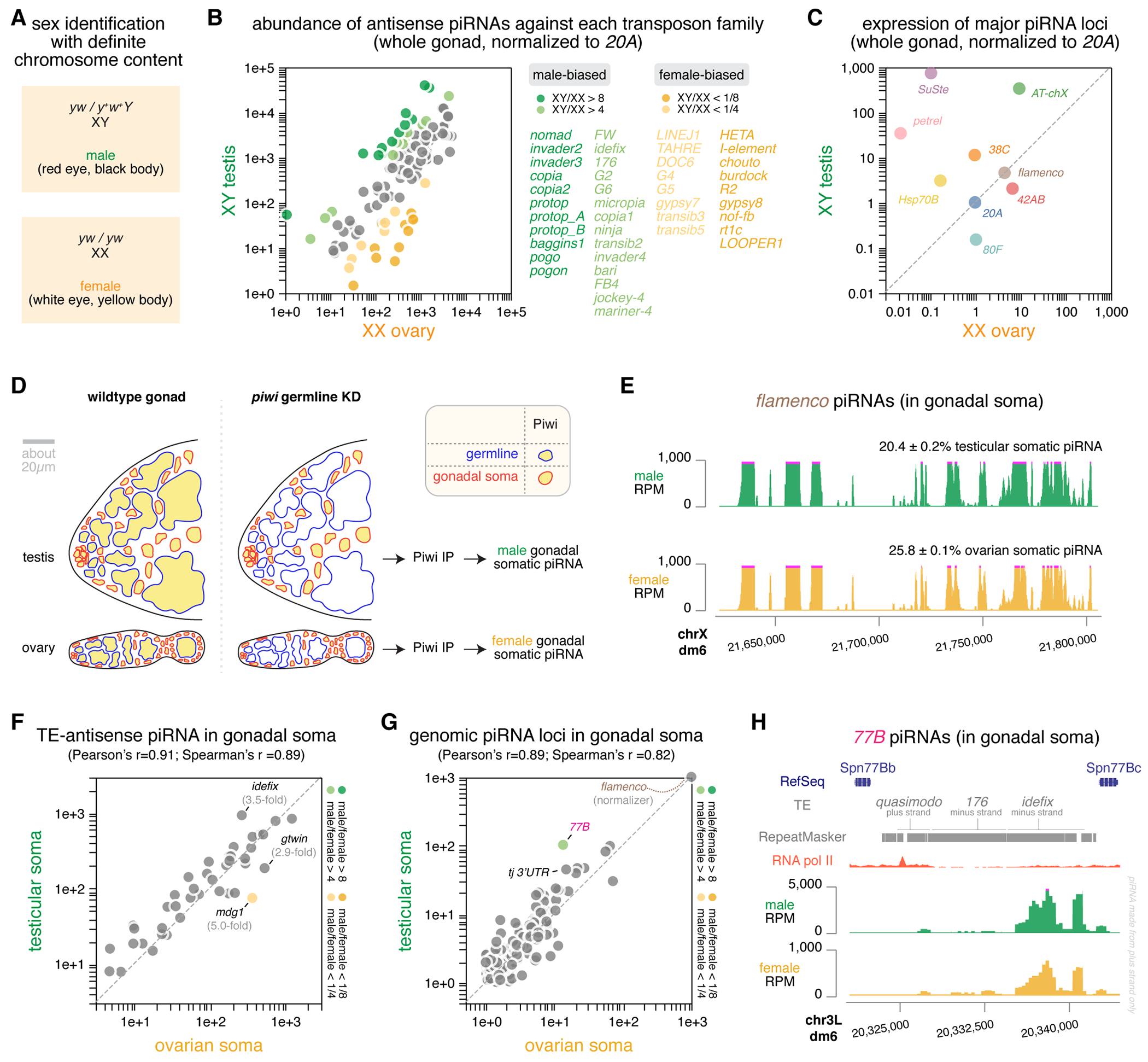
(A) Genotype and phenotype of males and females that can be identified with definite chromosome content, employing an X chromosome lacking y and w genes as well as a Y chromosome that carries the wildtype y+ and w+ genes.
(B) Comparison of the abundance of piRNAs targeting individual transposon families in XY testis versus XX ovary, normalized to the expression of 20A piRNAs. Sex-biased transposon-targeting piRNAs are color coded and listed on the right.
(C) Comparison of the expression of major piRNA loci in the genome5 in XY testis versus XX ovary, normalized to the expression of 20A piRNAs. Each locus is marked by a different color.
(D) Illustration of the experimental strategy to isolate somatic piRNAs in the gonad. Left: cartoon showing the cell type composition of testis and ovary, with germline having a blue outline and gonadal somatic cells having a red outline. Both germline and gonadal somatic cells express Piwi, which is marked by yellow. Right: cartoon showing Piwi expression in testis and ovary upon efficient, germline-specific knock-down of piwi that completely depletes Piwi in the germline, leaving the somatic cells as the only source of Piwi in the gonad. Gonadal somatic piRNAs are isolated by immunoprecipitating Piwi from these gonads that lose germline Piwi.
(E) UCSC genome browser view of the flamenco piRNA locus showing flamenco piRNAs take up similar fractions of gonadal somatic piRNAs in testis and ovary with similar coverage profiles.
(F) Comparison of the abundance of transposon-antisense piRNAs in testicular and ovarian soma, normalized to the expression of flamenco piRNAs. Sex-biased piRNAs are color coded in the same way as in (B) and the correlation coefficients are reported.
(G) Comparison of the expression of different piRNA loci in testicular and ovarian soma, normalized to the expression of flamenco piRNAs. Sex-biased piRNAs are color coded in the same way as in (B) and the correlation coefficients are reported.
(H) UCSC genome browser view of the 77B piRNA locus, showing its flanking protein-coding genes, its transposon contents and piRNA coverage profiles in two sexes. Note the difference in y-axis scales that reflects a higher relative activity of 77B in the testicular soma than the female counterpart. A putative promoter marked by an RNA pol II peak likely drives the expression of piRNAs from the plus strand that are antisense to two transposons, 176 and idefix.
See also Figure S1.
Germline is the major cell type origin of piRNA sexual dimorphism.
In D. melanogaster gonads, the piRNA program operates in both the germline and gonadal somatic cells, but piRNA biogenesis and targets differ between the two cell types10,17. Thus, the male-female differences seen in gonad-wide piRNA quantification could reflect sexual dimorphism in either germline or gonadal somatic cells, or both cell types, which could be further skewed by distinct germline-soma ratios in testis and ovary. To distinguish these possibilities, we isolated the somatic piRNAs in the gonad, by immunoprecipitating Piwi upon germline-specific piwi knock-down (see STAR Methods) and then sequencing the small RNAs associated with Piwi in gonadal somatic cells (Figure 1D). This allowed us to profile the gonadal somatic piRNA program in each of the two sexes.
Experimentally isolated gonadal somatic piRNAs from testes and ovaries (Figure 1D) allowed us to directly compare the piRNA program in gonadal soma between sexes. We found that the flamenco piRNA locus shows a similar piRNA coverage profile and produces piRNAs that take up comparable fractions of total piRNAs in testicular and ovarian soma (Figure 1E). Most of the highly expressed piRNAs in gonadal soma are antisense to transposons in both males and females, and piRNAs targeting different transposon families display a strong positive correlation between the two sexes (Pearson’s r = 0.91, p < 0.0001; Spearman’s r = 0.89, p < 0.0001; Figure 1F). When normalized to flamenco, a few transposons are targeted more in either males (e.g., idefix) or females (e.g., gtwin and mdg1), but these biases are relatively mild (Figure 1F). To examine piRNA production across the genome, we defined piRNA-producing loci in gonadal soma and measured their expression levels (see STAR Methods). Akin to piRNA quantification based on their transposon targets, quantifying piRNAs based on their genomic origins also revealed a strong positive correlation between the two sexes (Pearson’s r = 0.89, p < 0.0001; Spearman’s r = 0.82, p < 0.0001; Figure 1G). We did, however, note an exception: a novel piRNA locus we identified in the gonadal soma, 77B (Figure 1H), produces more piRNAs in males than females (Figure 1G). This locus resembles flamenco, as it makes piRNAs from one genomic strand downstream of a prominent RNA pol II peak that is indicative of a promoter, producing antisense piRNAs against transposons active in the gonadal soma (e.g., idefix; Figure 1H). Nevertheless, the genome-wide view of the piRNA production in gonadal soma highly correlates between sexes. The 3’ UTR of some genes (e.g., tj) is known to produce piRNAs in ovarian soma18, and the same holds in the male counterpart (Figure 1G). Overall, the piRNA program operating in gonadal soma shows very few sex differences.
Taking advantage of the fact that the flamenco piRNA locus is active exclusively in the gonadal soma but not in the germline of both testis (Figure S1) and ovary7,19, we inferred that germline piRNAs make up about 97% and 79% of the gonadal piRNAs in testis and ovary, respectively (see STAR Methods). Because germline piRNAs dominate the whole gonad piRNA pool to comparable extents in both sexes (97% / 79% = 1.2-fold difference), gonad-wide piRNA quantification is a close approximation of the germline piRNA program when studying male-female differences. Consistent with this, almost all sex-biased piRNAs have been previously annotated as germline-specific5,10,17,20. These results suggest that the piRNA sexual dimorphism we observed in the whole gonads originates from the germline rather than gonadal soma. For the rest of this work, we used the gonad-wide piRNA sexual dimorphism to approximate germline piRNA sexual dimorphism.
Y chromosome is necessary and sufficient to recapitulate aspects of male piRNA program.
Having found that germline is the major cell type origin of piRNA sexual dimorphism, we aimed to dissect its underlying genetic control mechanisms (Figure 2A). Distinct sex chromosome contents between sexes, specifically, the presence of Y chromosome in males, could in theory explain some sex differences in piRNAs. On the other hand, distinct sexual identities could lead to differential piRNA production even from identical piRNA loci located outside the Y. Importantly, the sex determination in D. melanogaster does not involve the Y14, which provides us with a unique opportunity to manipulate the Y chromosome without perturbing sexual identities.
Figure 2 |. Y chromosome produces piRNAs in both males and females.
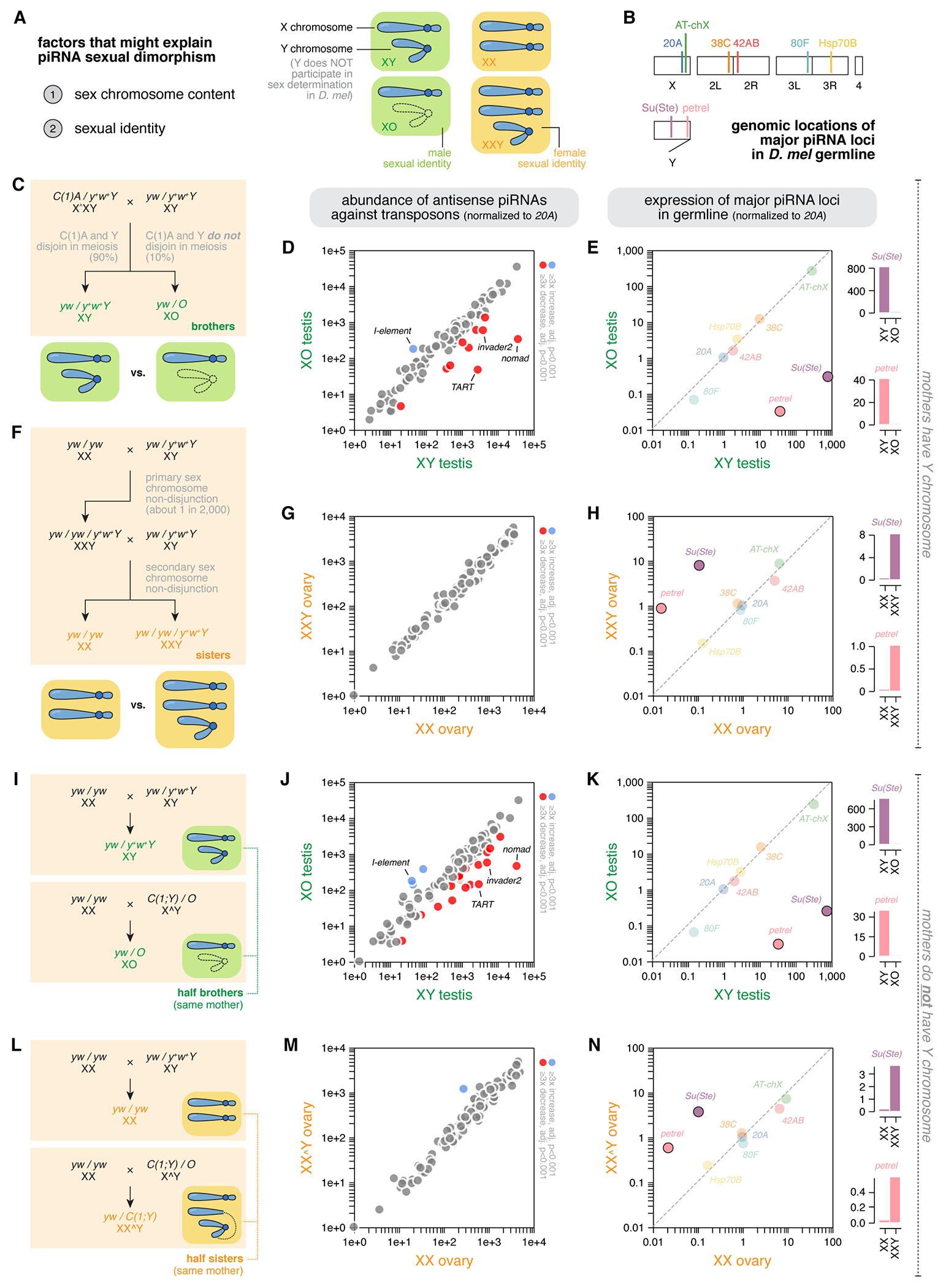
(A) Left: listed are factors that might explain piRNA sexual dimorphism. Right: cartoon showing different sex chromosome contents and respective sexual identities. X and Y chromosomes are depicted in different ways, and sexual identities are color coded with the male identity being green and the female one being orange. Note that the Y chromosome of D. melanogaster does not participate in sex determination, and the sex instead depends on the number of X. Hence, XY and XO are both males, and XX and XXY are both females.
(B) Illustration showing the karyotype of D. melanogaster with five chromosomes – X, Y, 2, 3, and 4, as well as the rough genomic locations of major piRNA loci in the germline.
(C) Cross scheme of the generation of XY and XO brothers.
(D) The abundance of transposon-targeting piRNAs in XO males compared to their XY brothers, showing the loss of piRNAs targeting several transposon families.
(E) The expression of major germline piRNA loci in XO males compared to their XY brothers, showing a specific loss of piRNAs from two Y-linked loci, Su(Ste) and petrel.
(F) Cross scheme of the generation of XX and XXY sisters.
(G) The abundance of transposon-targeting piRNAs in XXY females compared to their XX sisters, showing very limited differences.
(H) The expression of major germline piRNA loci in XXY females compared to their XX sisters, showing piRNA production from two Y-linked loci, Su(Ste) and petrel.
(I) Cross scheme of the generation of XY and XO half-brothers, with the same XX mothers.
(J) The abundance of transposon-targeting piRNAs in XO males compared to their XY half-brothers, both of which are sired by XX mothers, showing similar loss of piRNAs targeting several transposon families as seen in (D).
(K) The expression of major germline piRNA loci in XO males compared to their XY half-brothers, both of which are sired by XX mothers, showing a similar loss of piRNAs from two Y-linked loci, Su(Ste) and petrel, as seen in (E).
(L) Cross scheme of the generation of XX and XXY half-sisters, with the same XX mothers.
(M) The abundance of transposon-targeting piRNAs in XXY females compared to their XX half-sisters, both of which are sired by XX mothers, showing very few differences similar to (G) .
(N) The expression of major germline piRNA loci in XXY females compared to their XX half-sisters, both of which are sired by XX mothers, showing piRNA production from two Y-linked loci, Su(Ste) and petrel, as seen in (H).
To pinpoint the contribution of the Y chromosome to sex differences in the piRNA program, we first generated XY and XO male sibling flies that only differ in the Y chromosome content but are otherwise genetically identical. This is done by using spontaneous sex chromosome nondisjunction that occurs at about 10% frequency in X^XY females carrying the compound X chromosome, C(1)A (Figure 2C). When compared to their XY brothers, XO males lose piRNAs targeting several transposon families (Figure 2D), suggesting that the Y chromosome is likely a source of transposon-targeting piRNAs in the male. For example, the absence of the Y chromosome causes decreases in piRNAs against nomad and invader2 (Figure 2D) – two transposons that are normally targeted by more piRNAs in males than females (Figure 1B), suggesting that these sex differences could be explained by males having the Y chromosome. We also note that, piRNAs targeting I-element appear upregulated in males lacking the Y chromosome (Figure 2D), which warrants future investigation. Removing the Y also led to a specific loss of piRNAs from two Y-linked loci – Su(Ste) and petrel (Figure 2B) – while leaving the piRNA production from other loci on X and autosomes unperturbed (Figures 2B and 2E). Therefore, Y chromosome contributes to the male piRNA program via production of piRNAs from two loci on the Y, Su(Ste) and petrel, as well as piRNAs targeting a select group of transposons. While it is possible that Y could upregulate the expression of transposon-targeting piRNAs from other chromosomes, its specific effect on a few transposons led us to favor the possibility that Y chromosome encodes these transposon-targeting piRNAs.
Complementing the male experiment, we generated XX and XXY female sibling flies that only differ in their Y chromosome contents but are otherwise genetically identical. This is achieved by first obtaining an exceptional XXY female from primary sex chromosome nondisjunction that occurs naturally about 1 in 2,000 wildtype flies14, and then crossing this XXY female with XY males to sire XX and XXY females through secondary sex chromosome nondisjunction (Figure 2F). The extra Y chromosome barely altered the overall transposon-targeting piRNA program in females (Figure 2G). Nonetheless, the presence of the Y chromosome in females triggers piRNA biogenesis from Su(Ste) and petrel (Figure 2H), two loci that reside on the Y chromosome. Even though these two Y-linked piRNA loci appear to be less active in females than males, they have expression levels comparable to other top piRNA loci, including 42AB, 38C, and 80F (Figure 2H), suggesting that the Y is an active and productive piRNA source in a female cellular environment.
We generated genetically identical male and female siblings that only differ in their Y chromosome contents, however, these crosses necessitated the employment of mothers carrying a Y chromosome (Figures 2C and 2F). Given that maternally deposited piRNAs instruct piRNA biogenesis in the progeny9,11,12, Y-bearing mothers might create a permissive environment to produce Y piRNAs in the offspring by depositing Y-derived piRNAs to the embryo. Consequently, it is unclear whether the effects of Y chromosome on male and female piRNA production we observed (Figures 2D, 2E, 2G, and 2H) depends on mothers carrying a Y chromosome. To empirically test the role of Y chromosome in piRNA production without mothers bearing a Y, we devised a strategy to generate half siblings of both sexes that share similar, albeit not identical, genetic backgrounds with and without Y chromosome from XX mothers (Figures 2I and 2L). We observed similar effects of the Y chromosome on piRNA production when mothers do not have a Y: Y chromosome seems to be an important source of transposon-targeting piRNAs in males but not in females (Figures 2J and 2M), and the two Y-linked piRNA loci, Su(Ste) and petrel, produce piRNAs in both sexes, irrespective of the cellular sexual identity (Figures 2K and 2N). We noticed that the Y exerts a slightly greater effect on the transposon-targeting piRNA program in this latter cross scheme (Figures 2I and 2L) compared to the former one involving Y-bearing mothers (Figures 2C and 2F), which likely results from having different fathers and thus distinct paternally inherited haploid genome. Nevertheless, the results between mothers with and without a Y chromosome are qualitatively very similar. We conclude that the presence of the Y chromosome alters the piRNA profiles independently of its presence in the mothers.
Y chromosome in D. melanogaster is known to exhibit imprinting effects21–23, that is, Y can behave differently when inherited from the mother or the father. To test if Y-linked piRNA loci show parent-of-origin effects, we designed crosses that allow females to inherit a Y chromosome from either parent. In the case of paternally inheriting the Y, we also designed crosses either with or without mothers bearing a Y (Figure 3A top). In all cases, we detected nascent transcripts from Su(Ste) and petrel piRNA loci located on the Y chromosome (Figure 3A middle), indicating that piRNA loci on the Y are transcriptionally active in the female germline when inherited from either parent. We also observed similar behaviors of the Y-linked piRNA loci in the male counterpart (Figure 3B) – when inherited from either parent, with or without mothers carrying a Y, Y chromosome activates both Su(Ste) and petrel loci in the male germline. Thus, there are no obvious imprinting effects of the two Y-linked piRNA loci, and the mere presence of the Y can translate to an effect on the germline piRNA program in both sexes. Interestingly, in our cross scheme of passing the Y from mothers to daughters, Su(Ste) piRNA precursor transcription is also activated in the follicle cells (a gonadal somatic cell type), an unexpected finding that calls for future studies.
Figure 3 |. Y-linked piRNA loci are active and functional in both sexes, when inherited from either parent, regardless of whether mothers carry a Y chromosome.
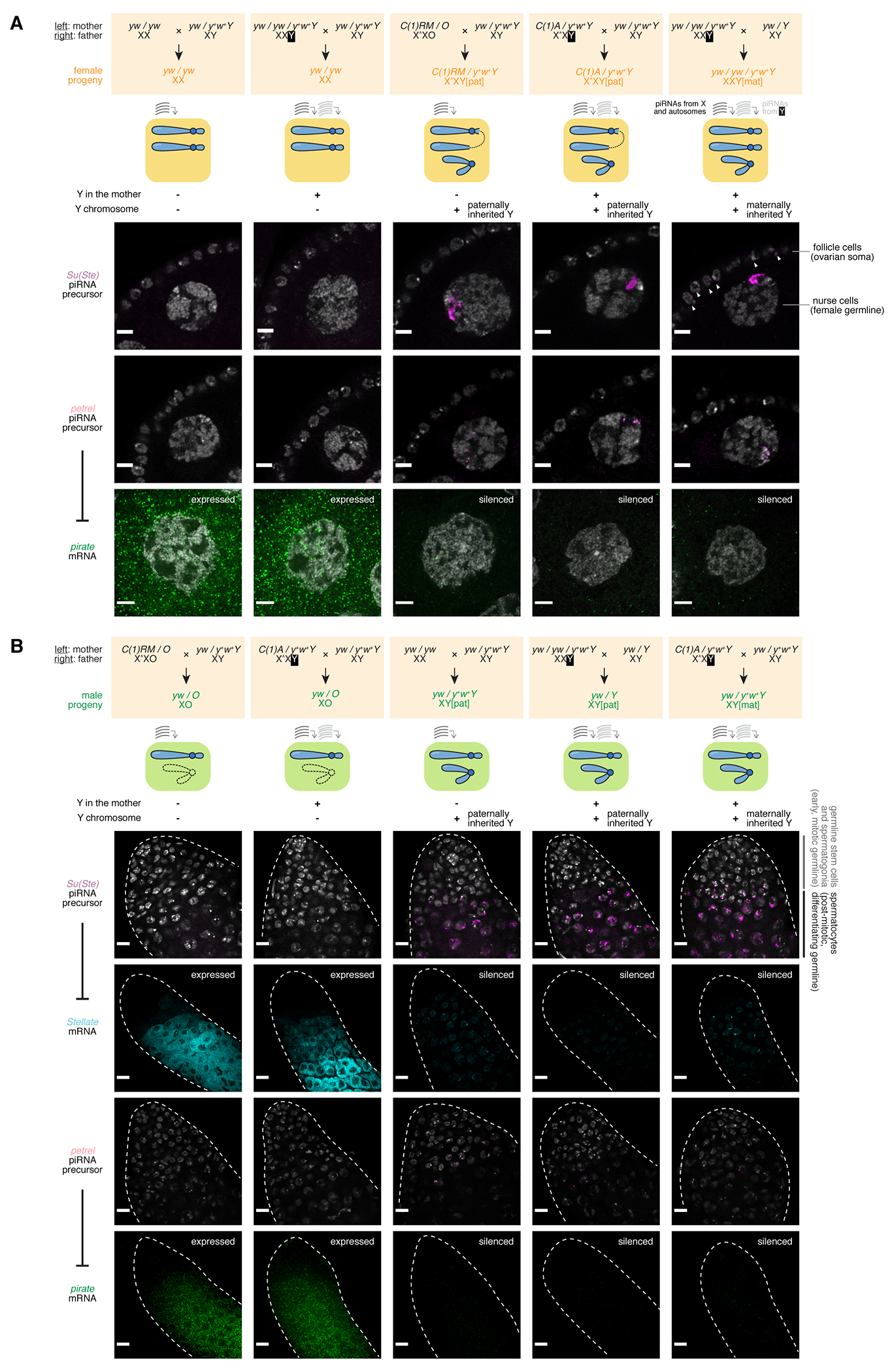
(A) Top: Cross schemes that generate: XX females without mothers bearing a Y (column 1), XX females with Y-bearing mothers (column 2), XXY females without mothers bearing a Y (column 3), XXY females with Y-bearing mothers but inhering the Y from the father (column 4), and XXY females with Y-bearing mothers and inheriting the Y from the mother (column 5). Middle: cartoon showing the genotype of each kind of females generated and whether their mothers carry a Y is reflected by whether they receive maternally deposited Y-derived piRNAs. Bottom: RNA in situ HCR detecting Su(Ste) piRNA precursors (row 1), petrel piRNA precursors (row 2), and pirate mRNAs (row 3) in stage 6-7 egg chambers. Scale bar: 5μm.
(B) Top: Cross schemes that generate: XO males without mothers bearing a Y (column 1), XO males with Y-bearing mothers (column 2), XY males without mothers bearing a Y (column 3), XY males with Y-bearing mothers but inhering the Y from the father (column 4), and XY males with Y-bearing mothers and inheriting the Y from the mother (column 5). Middle: cartoon showing the genotype of each kind of males generated and whether their mothers carry a Y is reflected by whether they receive maternally deposited Y-derived piRNAs. Bottom: RNA in situ HCR detecting Su(Ste) piRNA precursors (row 1), Stellate mRNAs (row 2), petrel piRNA precursors (row 3), and pirate mRNAs (row 4) and at the apical tips of the testes. Scale bar: 10μm.
Whereas Su(Ste) piRNAs silence the Stellate genes that are only active in the male germline, petrel piRNAs silence the pirate gene that is ubiquitously expressed in all tissues including female germline, which allows us to explore if activating petrel piRNA biogenesis in the female germline leads to pirate silencing. In wildtype XX female germline, the pirate gene is active, and its transcripts can be readily detected by RNA in situ HCR (Figure 3A bottom). However, introducing a Y into the female germline from either parent led to a marked silencing of the pirate gene (Figure 3A bottom), suggesting that making petrel piRNAs in female germline has a direct functional outcome. Meanwhile, the presence of Y chromosome in mothers was neither necessary nor sufficient for the silencing of pirate in the female progeny. Thus, pirate silencing in the female germline requires the presence of the Y chromosome, regardless of the parental origin of the Y. Similarly, in the male germline, having a Y-bearing mother is neither sufficient nor required for the male germline to tame Stellate and pirate, and the presence of Y chromosome triggers silencing of both Stellate and pirate genes regardless of the Y’s inheritance path (Figure 3B). Taken together, the differences in piRNA profiles caused by the presence of Y chromosome directly translates to differential silencing of several targets, without obvious parent-of-origin effects.
Cellular sexual identity provides a key input into piRNA biogenesis.
Though necessary, the presence of the Y chromosome in males is not sufficient to explain sex differences in the piRNA program, as piRNA loci outside the Y are also differentially expressed in two sexes (Figure 1C). What underlies piRNA sexual dimorphism outside the Y? In D. melanogaster, a cascade of molecular switches takes place after counting the number of X, culminating in either male or female cellular sexual identity14–16,24–29 (Figure 4A). To examine the contribution of sexual identities to germline piRNA sexual dimorphism without confounding impacts of the Y chromosome, we sought to masculinize XX female germline and compare it to XO male germline that lacks a Y (Figure 4B). Unlike the Drosophila soma, where sex determination occurs cell-autonomously, Drosophila germline receives an additional input from the soma on top of its own chromosomal content to determine the germline sex25,26. When the germline sex does not match the somatic sex, germline either dies or becomes tumorous24–26,30–33, so a productive germline sex reversal requires perturbing both the germline and somatic sex. Given that there is very little sexual dimorphism in the gonadal somatic piRNAs (Figures 1E, 1F, 1G, and 1H) and that gonad-wide sex-biased piRNAs are predominantly expressed in the germline5,10,17 20, reversing the somatic sex should not significantly confound our study of germline piRNA sexual dimorphism. Hence, our germline sex reversal was done in sex-reversed soma, which allowed us to interrogate the effect of sexual identities on germline piRNA sexual dimorphism. With the caveat that somatic perturbations could still influence gonad-wide piRNA profiles to some extent, we focused our analysis on piRNAs that are active in the germline.
Figure 4 |. Sexual identity provides a key input into piRNA biogenesis and is a major determinant of the piRNA program.
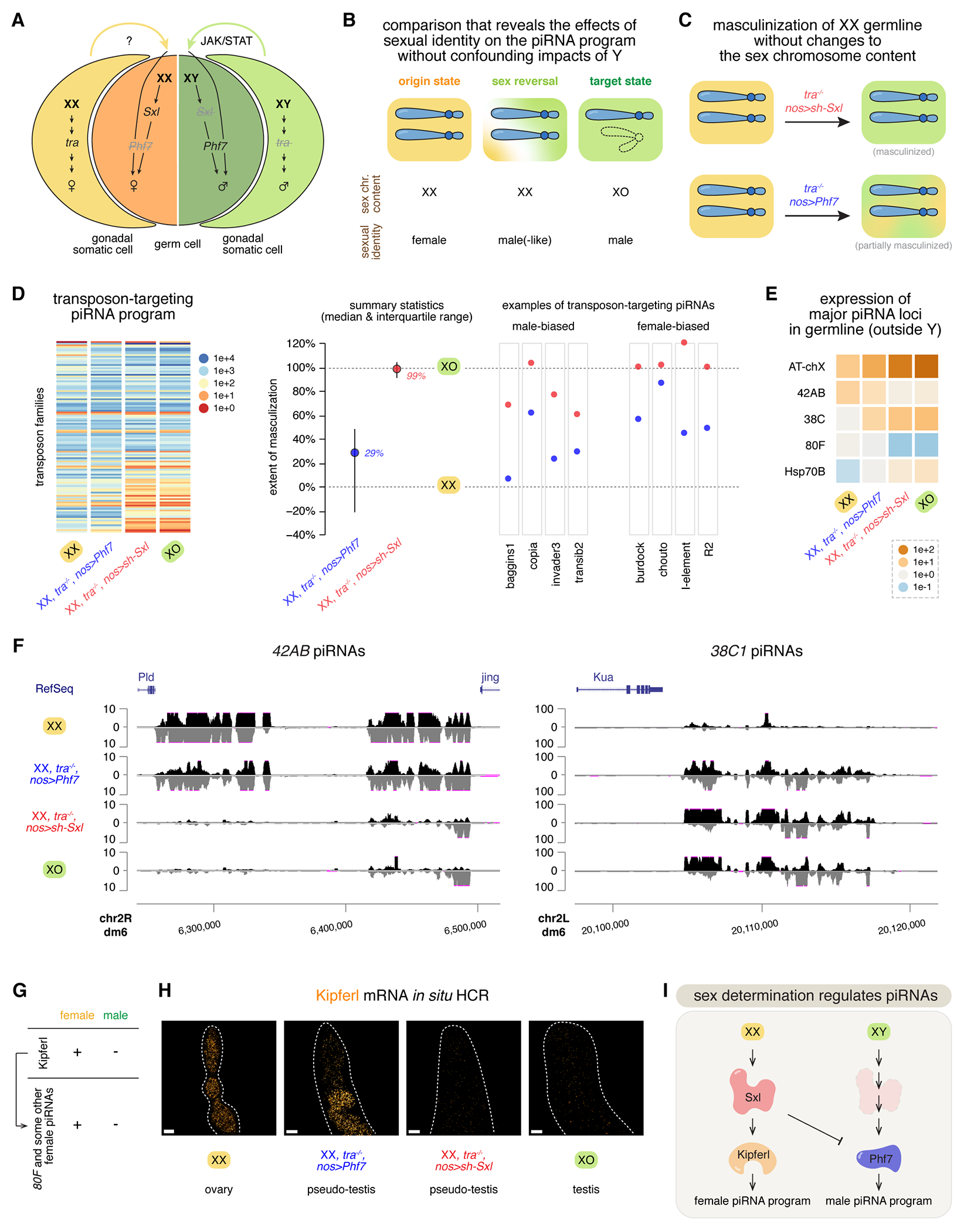
(A) A simplified model of the sex determination pathway in germline and soma. On top of its own chromosome content, germline receives an additional input from the soma to determine its sex.
(B) A comparison scheme that uses sex reversal to examine the effects of sexual identities on the piRNA program. XX female germline is masculinized and compared to both wildtype XX females (the origin state) and XO males (the target state). Thus, any differences observed would reveal the effects of sexual identities on piRNAs, without confounding impacts of Y chromosome.
(C) Cartoon showing the masculinization of the XX female germline by genetic perturbations, without any changes to the sex chromosome content. To facilitate germline masculinization, the soma is masculinized by mutating tra. In addition, germline-specific knock-down of Sxl near completely masculinizes the female germline, while ectopic expression of Phf7 in the germline led to partial masculinization.
(D) Left: a heatmap showing the abundance of piRNAs targeting different transposons in XX female, XO male, or XX masculinized by perturbing either Phf7 or Sxl expression (in the tra mutant background). Each row represents piRNAs that target a different transposon, and their expression levels are color coded. Expression was normalized to the 20A piRNAs. Middle: Quantification of the extent of masculinization for piRNAs targeting individual transposon families. For each transposon family, the abundance of corresponding antisense piRNAs in XX female is scaled to 0% and that in XO male is scaled to 100%, so the levels of transposon-targeting piRNAs in masculinized XX can be normalized to reflect the extent of masculinization. Shown are the summary statistics (median and interquartile range) of the antisense piRNAs targeting different transposon families. Right: piRNA abundance upon XX masculinization for four examples of male-biased and female-biased transposon-targeting piRNAs, respectively. In these examples as well as the overall summary statistics, perturbing Sxl led to a stronger masculinization of the piRNA program than perturbing Phf7.
(E) A heatmap showing the expression of major germline piRNA loci (located outside Y) in XX female, XO male, or XX masculinized by perturbing either Phf7 or Sxl expression (in the tra mutant background), normalized to the expression of 20A piRNAs.
(F) UCSC genome browser view of the piRNA coverage profiles over the locus 42AB (left) and the locus 38C1 (right) in XX female, XO male, or XX masculinized by perturbing either Phf7 or Sxl expression (in the tra mutant background).
(G) Female-specific expression of Kipferl and Kipferl-dependent piRNAs.
(H) RNA in situ HCR detecting Kipferl mRNA in XX female, XO male, or XX masculinized by perturbing either Phf7 or Sxl expression (in the tra mutant background).
(I) A genetic circuit that connects the sex determination pathway and piRNA biogenesis.
To explore if and how sexual identities impact germline piRNA profiles, we masculinized XX female germline by germline-specific knock-down of Sex lethal (Sxl), the major factor that governs the female identity in germline24–27, in the transformer (tra) mutant background that has a masculinized soma31 (Figure 4C). Strikingly, masculinizing the XX female germline converted its transposon-targeting piRNA program to a state that closely resembles the XO male germline (Figure 4D left and S2). When quantified, the median extent of masculinization for piRNAs targeting different transposon families is 99% (see STAR Methods; Figure 4D right). Similarly, for major piRNA loci outside the Y, their expression levels and piRNA coverage profiles also switched from an XX female state to an XO male state upon masculinization of the XX germline (Figures 4E and 4F). For example, abundant piRNAs are made from both proximal and distal ends of the 42AB piRNA locus in XX females, but masculinized XX germline only make some piRNAs from the distal end of 42AB and barely any from the proximal side, reminiscent of the XO male germline (Figure 4F left). In sum, reversing the germline sexual identity is sufficient to switch the germline piRNA program from one sex to the other, suggesting that the cellular sexual identity provides a key input into piRNA biogenesis.
How does the germline interpret its sexual identity to elicit a sex-specific piRNA program? We showed that this sexual-identity effect on piRNA biogenesis is governed by the major switch protein Sxl (Figures 4D, 4E, and 4F), which is active in the female, but not male, germline. Next, we looked into how Sxl orchestrates a female-specific piRNA program in the germline. Sxl is known to regulate two target genes that exhibit sex-specific expression patterns in the germline34: Tdrd5l29, a cytoplasmic protein that forms granules distinct from the piRNA processing sites, and Phf735, a chromatin reader protein that binds H3K4me2/3. Both Tdrd5l and Phf7 promote a male identity, and Sxl represses these two factors to bolster a female identity. Since Tdrd5l and Phf7 act genetically redundantly to support a male identity29, we focused on Phf7 for this study and asked whether and, if so, to what extent Phf7 mediates the sexual-identity effect on piRNA biogenesis. Expressing Phf7 in the female germline accompanied by somatic masculinization partially masculinized the XX female germline, leading to a 29% median extent of masculinization of transposon-targeting piRNAs (Figure 4D). For many transposon-targeting piRNAs (e.g., those targeting copia and burdock), the ectopic expression of Phf7 in XX germline shifted the piRNA profile from a female state towards a male state, but not as completely as losing Sxl did (Figure 4D right and S2). This partial reversal of the piRNA program from one sex to the other by Phf7 activation is also obvious when examining the expression of major piRNA loci in the genome. Each of the major piRNA loci in Phf7-expressing XX female germline resumed an activity somewhere in between the wildtype XX female and XO male, for both male- and female-biased loci (Figures 4E and 4F). For instance, Phf7 dampened the activity of 42AB, a female-biased piRNA locus, and enhanced the activity of 38C, a male-biased piRNA locus (Figures 4E and 4F). These observations indicate that Phf7 promotes a male piRNA program, and Sxl supports a female piRNA program in part through repressing Phf7. Thus, Phf7 mediates part of the sexual-identity effect on piRNA biogenesis, acting downstream of Sxl.
Recently, a female-specific piRNA biogenesis factor, the zinc-finger protein Kipferl, was described to drive a subset of piRNA production in the female germline36. In particular, piRNA production from 80F, a sex-specific piRNA locus only active in the female germline (Figure 4E), depends on Kipferl36 (Figure 4G), indicating that Kipferl is directly responsible for some of the germline piRNA sexual dimorphism. As Kipferl appears dedicated to the piRNA pathway and is absent in the male germline, we hypothesized that Kipferl is an effector protein that acts downstream of the sex determination pathway to elaborate a female piRNA program. Indeed, knocking-down Sxl in the female germline is sufficient to abrogate the expression of Kipferl in XX germline, suggesting that Kipferl expression depends on Sxl (Figure 4H). On the other hand, expressing Phf7 in the XX female germline did not perturb Kipferl expression (Figure 4H), suggesting that Phf7 and Kipferl act in parallel, both downstream of Sxl, to promote male and female piRNA programs, respectively (Figure 4I).
Taken together, we elucidated a genetic circuit that connects the sex determination pathway to germline piRNA sexual dimorphism (Figure 4I). XX germline activates Sxl, which positively regulates Kipferl to produce female-specific piRNAs and negatively regulates Phf7 to suppress a male piRNA program. On the contrary, XY germline lacks functional Sxl to activate Kipferl and instead expresses Phf7 to elaborate a male piRNA program. We conclude that male and female sexual identities enable divergent piRNA production programs, sculpting a sexually dimorphic molecular trait alongside the male-specific Y chromosome.
Discussion
In this work, we identified the germline as the source of piRNA sexual dimorphism in fly gonads. Building on this, we genetically separated the actions of sex chromosomes and the sex determination pathway to dissect their distinct roles towards the sex-specific piRNA program. We characterized the contribution of the Y chromosome to the male piRNA program, and we showed that the presence of the Y is sufficient to recapitulate some aspects of the male piRNA program in a female cellular environment. In fact, the effect of the Y is independent of its parental origin and mothers’ sex chromosome contents. The ability of Y-linked piRNA loci to act in both male and female cellular environments independently of its inheritance path implies unique regulatory mechanisms37 employed by the Y and distinctive evolutionary forces acting on the Y. What could be the forces that drove the evolution of Y-linked piRNAs? We envisage that Y-linked piRNAs might have evolved to facilitate male-specific transposon control and/or to suppress sex-chromosome meiotic drive. Meanwhile, we showed that sexual identity is another major determinant of the piRNA program that regulates piRNA biogenesis outside the Y. Specifically, sexual identity shapes piRNA sexual dimorphism under the control of Sxl, which relays the sexual identity of a cell to piRNA biogenesis through the histone reader protein Phf7 and the zinc-finger protein Kipferl. Even though we cannot completely exclude the effect of somatic perturbation in our germline sex reversal, our work unequivocally demonstrated an input from sex determination into piRNA biogenesis. We speculate that the sex determination pathway has been hijacked by transposons to facilitate their sex-biased germline invasion5, so integrating the information of germline sexual identities into piRNA biogenesis provides a means to directly couple the sex-specific piRNA defense program with sex-specific transposon threats. Together, our work revealed that sex chromosome and sexual identity control different facets of piRNA sexual dimorphism, and it is their collective action that sculpts the sex-specific piRNA program in fly germline. It is very likely that other sexually dimorphic traits are under the control of both sex chromosome and sexual identity, and disentangling the effects of the two promises to offer new insights into how a molecular pathway can be modified by each of the two sexes to execute essential functions.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Alexei Aravin (aaa@caltech.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Sequencing data generated in this study have been deposited at NCBI SRA and are publicly available as of the date of publication. Accession number is listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| HCR hybridization buffer | Molecular Instruments | N/A |
| HCR amplification buffer | Molecular Instruments | N/A |
| HCR wash buffer | Molecular Instruments | N/A |
| GFP-Trap Magnetic Agarose | ChromoTek | gtma-20 |
| Critical commercial assays | ||
| TraPR Small RNA Isolation Kit | Lexogen | N/A |
| NEBNext Multiplex Small RNA Sample Prep Set for Illumina | NEB | E7330 |
| Deposited data | ||
| Small RNA-seq | This paper | PRJNA933224 |
| Experimental models: Organisms/strains | ||
| yw | Bloomington Drosophila Stock Center | BDSC 6599 |
| y+w+Y | Bloomington Drosophila Stock Center | BDSC 7060 |
| C(1)A | Bloomington Drosophila Stock Center | BDSC 2950 |
| C(1;Y) | Bloomington Drosophila Stock Center | BDSC 9460 |
| nos-Gal4 | Bloomington Drosophila Stock Center | BDSC 4937 |
| bam-Gal4 | Bloomington Drosophila Stock Center | BDSC 80579 |
| sh-piwi | Bloomington Drosophila Stock Center | BSDC 33724 |
| sh-Sxl | Bloomington Drosophila Stock Center | BDSC 38195 |
| UAS-Phf7 | Bloomington Drosophila Stock Center | BDSC 15894 |
| tra 1 | Bloomington Drosophila Stock Center | BDSC 675 |
| Df(tra) | Bloomington Drosophila Stock Center | BDSC 5416 |
| Tj-GFP | Vienna Drosophila Resource Center | VDRC 318066 |
| GFP-Piwi | Le Thomas et al.50 | N/A |
| mCherry-Vasa | Lerit et al.51 | N/A |
| Oligonucleotides | ||
| Probes against flam piRNA precursors | Molecular Instruments | 3893/E046 |
| Probes against petrel piRNA precursors | Molecular Instruments | 3872/E024 |
| Probes against pirate mRNAs | Molecular Instruments | 3916/E064 |
| Probes against Stellate mRNAs | Molecular Instruments | 4537/E832 |
| Probes against Kipferl mRNAs | Molecular Instruments | 4708/E1062 |
| Probes against Su(Ste) piRNA precursors | IDT | N/A |
| Software and algorithms | ||
| Fiji | Schindelin et al.39 | https://imagej.net/software/fiji/ |
| numpy 1.20.3 | Oliphant42 | https://numpy.org/ |
| pandas 1.3.3 | McKinney43 | https://pandas.pydata.org/ |
| altair 4.1.0 | VanderPlas et al.44 | https://altair-viz.github.io/ |
| UCSC Genome Browser | Kent et al.45 | https://genome.ucsc.edu/cgi-bin/hgGateway |
| IGV | Robinson et al.46, Thorvaldsdóttir et al.47 | https://software.broadinstitute.org/software/igv/ |
| DESeq2 | Love et al.49 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Other | ||
| RNA pol II ChIP-seq data (D. mel ovary) | Andersen et al.48 | GSM2576144 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
We maintained Drosophila melanogaster stocks and crosses in plastic vials containing standard cornmeal food, in a room with temperature and humidity control (25 °C and 70% relative humidity). Young flies (≤ 5-day old) were used for all experiments. Fly stocks used in this study are listed in the key resources table.
METHOD DETAILS
Fly crosses
To minimize genetic background differences, yw / y+w+Y was backcrossed to the inbred yw line for six consecutive generations, via a single male at every generation. Similarly, after generating C(1)A / y+w+Y females, we backcrossed them to yw / y+w+Y males for six consecutive generations, via 2-3 females at every generation. To obtain an XXY exceptional female, we looked for a female carrying the marked Y chromosome (y+w+Y) in the y+w+Y stock, which typically took no more than two months. To deplete germline Piwi, we expressed sh-piwi using both nos-Gal4 and bam-Gal4, which led to efficient knock-down of Piwi in the germline as evidenced by the loss of germline GFP-Piwi expression in both testis and ovary. For sex reversal experiments, a Y chromosome marked by Bs (present in the Df(tra) stock) that alters the eye shape was employed, such that the sex chromosome content could be inferred independently of the morphological sex. Upon sex reversal, germline remains prevalent but shows phenotypes characteristic of either male or female germline development in Sxl and Phf7 perturbations, respectively (Figure S3).
RNA in situ hybridization and RNA in situ hybridization chain reaction (HCR)
For RNA in situ HCR, probes, amplifiers and buffers were purchased from Molecular Instruments (molecularinstruments.org) for flam (3893/E046), petrel (3872/E024), pirate (3916/E064), Stellate (4537/E832) and Kipferl (4708/E1062) transcripts. RNA in situ HCR v3.038 was done according to manufacturer’s recommendations for generic samples in solution. To detect Su(Ste) transcripts, we did conventional RNA in situ hybridization using DNA probes (75bp, position 994-1068 of Su(Ste): CR42424, sense direction) directly conjugated with fluorophore purchased from IDT.
Image acquisition and processing
Confocal images were acquired with Zeiss LSM 800 or LSM 980 using a 63x oil immersion objective (NA=1.4) and processed using Fiji39. Single focal planes were shown in all images, where dotted outlines were manually drawn for illustration purposes.
Small RNA sequencing
Argonaute-associated small RNAs were isolated from ovaries (20 pairs per sample) or testes (30 pairs per sample) using TraPR columns40. Purified small RNA was subject to library prep using NEBNext Multiplex Small RNA Sample Prep Set for Illumina (NEB E7330). Adaptor-ligated, reverse-transcribed, PCR-amplified samples were purified again by PAGE (6% polyacrylamide gel), from which we cut out the band within the desired size range. This additional size selection by PAGE eliminated other, longer RNAs (>30 nt) captured by TraPR columns. To isolate Piwi-associated small RNAs in gonadal soma, we first immunoprecipitated GFP-Piwi from gonads lacking germline Piwi (see above for fly crosses) using GFP-Trap (ChromoTek) magnetic agarose beads, as described before41. Small RNAs associated with gonadal somatic Piwi are then purified by TraPR columns and library-prepared, as described above for all Argonaute-associated small RNAs. Libraries were sequenced on Illumina HiSeq 2500 or NextSeq 2000.
Analysis of transposon-targeting piRNAs
To computationally extract piRNAs from all Argonaute-associated small RNAs, adaptor-trimmed small RNAs were size-selected for 23-29nt (cutadapt 2.5) and those mapped to rRNA, tRNA, snRNA, snoRNA, miRNA, hpRNA and 7SL RNA were discarded (bowtie 1.2.2 with -v 3). piRNAs were then mapped to RepBase25.08 (manually curated) and those antisense to transposon consensus sequences with ≤3 mismatches are designated as transposon-targeting piRNAs. For LTR transposons, reads mapping to the LTR and internal sequences of a given transposon family were merged for quantification, given their well-correlated behaviors. All quantification was normalized to the expression of 20A piRNAs (unless otherwise noted) and shown as the mean of two biological replicates.
Analysis of the expression of major piRNA loci
piRNAs were computationally extracted as described above. piRNAs were mapped to the dm6 genome using a previously described algorithm5 that considers both piRNAs that map to single unique positions in the genome as well as those that map to “local repeats” (defined as repeats that are contained within a genomic window <2Mb in length in dm6 reference genome). Major piRNA loci, their coordinates and quantification method were described before5. All quantification was normalized to 20A piRNAs. The average of two biological replicates was shown in all figures.
Definition of piRNA-producing loci in gonadal soma
piRNA-producing loci in gonadal soma were defined as previously described for piRNA-producing loci (also known as “piRNA clusters”) in whole gonads5. Briefly, piRNAs isolated from gonadal soma were mapped to the genome and those that map uniquely or to local repeats were kept and quantified over 1Kb windows that tile the entire genome. Neighboring 1Kb widows within 3Kb were merged. If merged windows were ≥5Kb, they were merged again within 15Kb, and this process was repeated twice. This de novo method recapitulated the flamenco locus and the 3’UTR of the tj gene – two loci that are known to make abundant piRNAs in ovarian soma – confirming its utility.
Inference of germline contribution to whole gonad piRNAs
Given that flamenco is only active in the gonadal soma but not in the germline, flamenco piRNAs found in whole gonad piRNAs must come from somatic cells in the gonad. Experimentally isolated gonadal somatic piRNAs revealed the contribution of flamenco piRNAs to total piRNAs in the gonadal soma (e.g., 25%), so if flamenco takes up 5% of whole gonad piRNAs, gonadal soma will contribute to 20% (5% / 25% = 20%) of whole gonad piRNAs. Then, the germline contribution to whole gonad piRNAs is 100% - 20% = 80%. When calculating actual contributions of flamenco piRNAs to gonadal soma and whole gonads of both sexes, the mean of two replicates was used.
Data visualization
All data visualization were done in Python 3 via JupyterLab with the following software packages: numpy42, pandas43 and altair44. The UCSC Genome Brower45 and IGV46,47 were used to explore sequencing data and to prepare genome browser track panels shown. UCSC genome browser tracks of piRNA coverages shown in the figures were normalized to CPM (counts per million 23-29nt reads that map uniquely to dm6 genome). Two biological replicates showed similar coverage profiles on the genome browser, so one of the two replicates was randomly selected to be shown in the figures. RNA pol II ChIP-seq data from wildtype fly ovary (w1118) was visualized using IGV using the publicly available bigwig file from GEO under the accession number GSM257614448.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis was done in Python 3 via JuputerLab. All sequencing experiments were done with two biological replicates. Differential expression analysis was done using DESeq249.
Supplementary Material
Highlights.
Most sexual dimorphism in gonadal piRNA expression originates from the germline.
Sex chromosomes and sex determination collectively control piRNA sexual dimorphism.
The presence of Y chromosome triggers expression of some male piRNAs in females.
Sex determination pathway regulates piRNA biogenesis through Sxl, Phf7 and Kipferl.
Acknowledgement
We thank Grace YC Lee, Felipe Karam Teixeira, Justin Blumenstiel, Katalin Fejes Toth, and members of the Aravin Laboratory for discussion, Jim Kennison for advice on sex chromosome nondisjunction, Mark Van Doren and Helen Salz for advice on sex determination, and Yukiko Yamashita for sharing unpublished results. We thank Angela Stathopoulos, Ellen Rothenberg, and Henry Chung for comments on the manuscript draft. We are grateful to Liz Gavis, Bloomington Drosophila Stock Center and Vienna Drosophila Resource Center for fly lines. We appreciate the help of Igor Antoshechkin (Millard and Muriel Jacobs Genetics and Genomics Laboratory, Caltech) with sequencing, the help of Fan Gao (Bioinformatics Resource Center, Caltech) with bioinformatic analysis, the help of Grace Shin (Molecular Technologies, Caltech) with in situ HCR, and the help of Giada Spigolon and Andres Collazo (Biological Imaging Facility, Caltech) with microscopy. This work was supported by grants from the National Institutes of Health (R01 GM097363) and by the HHMI Faculty Scholar Award to A.A.A.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare no competing interests.
References
- 1.Kopp A, Duncan I, Godt D, and Carroll SB (2000). Genetic control and evolution of sexually dimorphic characters in Drosophila. Nature 408, 553–559. 10.1038/35046017. [DOI] [PubMed] [Google Scholar]
- 2.Williams TM, Selegue JE, Werner T, Gompel N, Kopp A, and Carroll SB (2008). The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell 134, 610–623. 10.1016/j.cell.2008.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox RM, and Calsbeek R (2009). Sexually antagonistic selection, sexual dimorphism, and the resolution of intralocus sexual conflict. Am Nat 173, 176–187. 10.1086/595841. [DOI] [PubMed] [Google Scholar]
- 4.Galouzis CC, and Prud’homme B (2021). Transvection regulates the sex-biased expression of a fly X-linked gene. Science 371, 396–400. 10.1126/science.abc2745. [DOI] [PubMed] [Google Scholar]
- 5.Chen P, Kotov AA, Godneeva BK, Bazylev SS, Olenina LV, and Aravin AA (2021). piRNA-mediated gene regulation and adaptation to sex-specific transposon expression in D. melanogaster male germline. Genes Dev 35, 914–935. 10.1101/gad.345041.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, and Zamore PD (2006). A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313, 320–324. 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 7.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, and Hannon GJ (2007). Discrete Small RNA-Generating Loci as Master Regulators of Transposon Activity in Drosophila. Cell 128, 1089–1103. 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 8.Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, and Siomi MC (2006). Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 20, 2214–2222. 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, and Hannon GJ (2008). An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322, 1387–1392. 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, and Hannon GJ (2009). Specialized piRNA Pathways Act in Germline and Somatic Tissues of the Drosophila Ovary. Cell 137, 522–535. 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Vanssay A, Bougé A-L, Boivin A, Hermant C, Teysset L, Delmarre V, Antoniewski C, and Ronsseray S (2012). Paramutation in Drosophila linked to emergence of a piRNA-producing locus. Nature 490, 112–115. 10.1038/nature11416. [DOI] [PubMed] [Google Scholar]
- 12.Le Thomas A, Stuwe E, Li S, Du J, Marinov G, Rozhkov N, Chen Y-CA, Luo Y, Sachidanandam R, Toth KF, et al. (2014). Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing. Genes Dev. 28, 1667–1680. 10.1101/gad.245514.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen P, Luo Y, and Aravin AA (2021). RDC complex executes a dynamic piRNA program during Drosophila spermatogenesis to safeguard male fertility. PLoS Genet 17, e1009591. 10.1371/journal.pgen.1009591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bridges CB (1916). Non-Disjunction as Proof of the Chromosome Theory of Heredity. Genetics 1, 1–52. 10.1093/genetics/1.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bridges CB (1921). Triploid Intersexes in Drosophila melanogaster. Science 54, 252–254. 10.1126/science.54.1394.252. [DOI] [PubMed] [Google Scholar]
- 16.Erickson JW, and Quintero JJ (2007). Indirect effects of ploidy suggest X chromosome dose, not the X:A ratio, signals sex in Drosophila. PLoS Biol 5, e332. 10.1371/journal.pbio.0050332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, et al. (2009). Collapse of Germline piRNAs in the Absence of Argonaute3 Reveals Somatic piRNAs in Flies. Cell 137, 509–521. 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robine N, Lau NC, Balla S, Jin Z, Okamura K, Kuramochi-Miyagawa S, Blower MD, and Lai EC (2009). A broadly conserved pathway generates 3’UTR-directed primary piRNAs. Curr Biol 19, 2066–2076. 10.1016/j.cub.2009.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pélisson A, Song SU, Prud’homme N, Smith PA, Bucheton A, and Corces VG (1994). Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. EMBO J 13, 4401–4411. 10.1002/j.1460-2075.1994.tb06760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohn F, Sienski G, Handler D, and Brennecke J (2014). The Rhino-Deadlock-Cutoff Complex Licenses Noncanonical Transcription of Dual-Strand piRNA Clusters in Drosophila. Cell 157, 1364–1379. 10.1016/j.cell.2014.04.031. [DOI] [PubMed] [Google Scholar]
- 21.Maggert KA, and Golic KG (2002). The Y chromosome of Drosophila melanogaster exhibits chromosome-wide imprinting. Genetics 162, 1245–1258. 10.1093/genetics/162.3.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menon DU, and Meller VH (2009). Imprinting of the Y chromosome influences dosage compensation in roX1 roX2 Drosophila melanogaster. Genetics 183, 811–820. 10.1534/genetics.109.107219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemos B, Branco AT, Jiang P-P, Hartl DL, and Meiklejohn CD (2014). Genome-wide gene expression effects of sex chromosome imprinting in Drosophila. G3 (Bethesda) 4, 1–10. 10.1534/g3.113.008029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schüpbach T (1985). Normal female germ cell differentiation requires the female X chromosome to autosome ratio and expression of sex-lethal in Drosophila melanogaster. Genetics 109, 529–548. 10.1093/genetics/109.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steinmann-Zwicky M, Schmid H, and Nöthiger R (1989). Cell-autonomous and inductive signals can determine the sex of the germ line of drosophila by regulating the gene Sxl. Cell 57, 157–166. 10.1016/0092-8674(89)90181-5. [DOI] [PubMed] [Google Scholar]
- 26.Nöthiger R, Jonglez M, Leuthold M, Meier-Gerschwiler P, and Weber T (1989). Sex determination in the germ line of Drosophila depends on genetic signals and inductive somatic factors. Development 107, 505–518. 10.1242/dev.107.3.505. [DOI] [PubMed] [Google Scholar]
- 27.Hashiyama K, Hayashi Y, and Kobayashi S (2011). Drosophila Sex lethal gene initiates female development in germline progenitors. Science 333, 885–888. 10.1126/science.1208146. [DOI] [PubMed] [Google Scholar]
- 28.Shapiro-Kulnane L, Smolko AE, and Salz HK (2015). Maintenance of Drosophila germline stem cell sexual identity in oogenesis and tumorigenesis. Development 142, 1073–1082. 10.1242/dev.116590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Primus S, Pozmanter C, Baxter K, and Van Doren M (2019). Tudor-domain containing protein 5-like promotes male sexual identity in the Drosophila germline and is repressed in females by Sex lethal. PLoS Genet 15, e1007617. 10.1371/journal.pgen.1007617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown EH, and King RC (1961). Studies on the Expression of the Transformer Gene of Drosophila Melanogaster. Genetics 46, 143–156. 10.1093/genetics/46.2.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sturtevant AH (1945). A Gene in Drosophila Melanogaster That Transforms Females into Males. Genetics 30, 297–299. 10.1093/genetics/30.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steinmann-Zwicky M (1993). Sex determination in Drosophila: sis-b, a major numerator element of the X:A ratio in the soma, does not contribute to the X:A ratio in the germ line. Development 117, 763–767. 10.1242/dev.117.2.763. [DOI] [PubMed] [Google Scholar]
- 33.Casper A, and Van Doren M (2006). The control of sexual identity in the Drosophila germline. Development 133, 2783–2791. 10.1242/dev.02415. [DOI] [PubMed] [Google Scholar]
- 34.Grmai L, Pozmanter C, and Van Doren M (2022). The Regulation of Germline Sex Determination in Drosophila by Sex lethal. Sex Dev, 1–6. 10.1159/000521235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang SY, Baxter EM, and Van Doren M (2012). Phf7 controls male sex determination in the Drosophila germline. Dev Cell 22, 1041–1051. 10.1016/j.devcel.2012.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baumgartner L, Handler D, Platzer SW, Yu C, Duchek P, and Brennecke J (2022). The Drosophila ZAD zinc finger protein Kipferl guides Rhino to piRNA clusters. Elife 11, e80067. 10.7554/eLife.80067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Venkei ZG, Gainetdinov I, Starostik MR, Choi CP, Chen P, Balsara C, Whitfield TW, Bell GW, Feng S, Jacobsen SE, et al. (2022). Drosophila Males Use 5’-to-3’ Phased Biogenesis to Make Stellate-silencing piRNAs that Lack Homology to Maternally Deposited piRNA Guides (Developmental Biology) 10.1101/2022.09.12.507655. [DOI] [Google Scholar]
- 38.Choi HMT, Schwarzkopf M, Fornace ME, Acharya A, Artavanis G, Stegmaier J, Cunha A, and Pierce NA (2018). Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development 145. 10.1242/dev.165753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grentzinger T, Oberlin S, Schott G, Handler D, Svozil J, Barragan-Borrero V, Humbert A, Duharcourt S, Brennecke J, and Voinnet O (2020). A universal method for the rapid isolation of all known classes of functional silencing small RNAs. Nucleic Acids Res 48, e79. 10.1093/nar/gkaa472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ninova M, Chen Y-CA, Godneeva B, Rogers AK, Luo Y, Fejes Tóth K, and Aravin AA (2020). Su(var)2-10 and the SUMO Pathway Link piRNA-Guided Target Recognition to Chromatin Silencing. Mol Cell 77, 556–570.e6. 10.1016/j.molcel.2019.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oliphant TE (2015). Guide to NumPy (Continuum Press; ). [Google Scholar]
- 43.McKinney W (2010). Data Structures for Statistical Computing in Python. In, pp. 56–61. 10.25080/Majora-92bf1922-00a. [DOI] [Google Scholar]
- 44.VanderPlas J, Granger B, Heer J, Moritz D, Wongsuphasawat K, Satyanarayan A, Lees E, Timofeev I, Welsh B, and Sievert S (2018). Altair: Interactive Statistical Visualizations for Python. JOSS 3, 1057. 10.21105/joss.01057. [DOI] [Google Scholar]
- 45.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, and Haussler D (2002). The human genome browser at UCSC. Genome Res. 72, 996–1006. 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nat Biotechnol 29, 24–26. 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thorvaldsdóttir H, Robinson JT, and Mesirov JP (2013). Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinformatics 14, 178–192. 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andersen PR, Tirian L, Vunjak M, and Brennecke J (2017). A heterochromatin-dependent transcription machinery drives piRNA expression. Nature 549, 54–59. 10.1038/nature23482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, and Tóth KF (2013). Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 27, 390–399. 10.1101/gad.209841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lerit DA, and Gavis ER (2011). Transport of germ plasm on astral microtubules directs germ cell development in Drosophila. Curr Biol 27, 439–448. 10.1016/j.cub.2011.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data generated in this study have been deposited at NCBI SRA and are publicly available as of the date of publication. Accession number is listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| HCR hybridization buffer | Molecular Instruments | N/A |
| HCR amplification buffer | Molecular Instruments | N/A |
| HCR wash buffer | Molecular Instruments | N/A |
| GFP-Trap Magnetic Agarose | ChromoTek | gtma-20 |
| Critical commercial assays | ||
| TraPR Small RNA Isolation Kit | Lexogen | N/A |
| NEBNext Multiplex Small RNA Sample Prep Set for Illumina | NEB | E7330 |
| Deposited data | ||
| Small RNA-seq | This paper | PRJNA933224 |
| Experimental models: Organisms/strains | ||
| yw | Bloomington Drosophila Stock Center | BDSC 6599 |
| y+w+Y | Bloomington Drosophila Stock Center | BDSC 7060 |
| C(1)A | Bloomington Drosophila Stock Center | BDSC 2950 |
| C(1;Y) | Bloomington Drosophila Stock Center | BDSC 9460 |
| nos-Gal4 | Bloomington Drosophila Stock Center | BDSC 4937 |
| bam-Gal4 | Bloomington Drosophila Stock Center | BDSC 80579 |
| sh-piwi | Bloomington Drosophila Stock Center | BSDC 33724 |
| sh-Sxl | Bloomington Drosophila Stock Center | BDSC 38195 |
| UAS-Phf7 | Bloomington Drosophila Stock Center | BDSC 15894 |
| tra 1 | Bloomington Drosophila Stock Center | BDSC 675 |
| Df(tra) | Bloomington Drosophila Stock Center | BDSC 5416 |
| Tj-GFP | Vienna Drosophila Resource Center | VDRC 318066 |
| GFP-Piwi | Le Thomas et al.50 | N/A |
| mCherry-Vasa | Lerit et al.51 | N/A |
| Oligonucleotides | ||
| Probes against flam piRNA precursors | Molecular Instruments | 3893/E046 |
| Probes against petrel piRNA precursors | Molecular Instruments | 3872/E024 |
| Probes against pirate mRNAs | Molecular Instruments | 3916/E064 |
| Probes against Stellate mRNAs | Molecular Instruments | 4537/E832 |
| Probes against Kipferl mRNAs | Molecular Instruments | 4708/E1062 |
| Probes against Su(Ste) piRNA precursors | IDT | N/A |
| Software and algorithms | ||
| Fiji | Schindelin et al.39 | https://imagej.net/software/fiji/ |
| numpy 1.20.3 | Oliphant42 | https://numpy.org/ |
| pandas 1.3.3 | McKinney43 | https://pandas.pydata.org/ |
| altair 4.1.0 | VanderPlas et al.44 | https://altair-viz.github.io/ |
| UCSC Genome Browser | Kent et al.45 | https://genome.ucsc.edu/cgi-bin/hgGateway |
| IGV | Robinson et al.46, Thorvaldsdóttir et al.47 | https://software.broadinstitute.org/software/igv/ |
| DESeq2 | Love et al.49 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Other | ||
| RNA pol II ChIP-seq data (D. mel ovary) | Andersen et al.48 | GSM2576144 |


