Summary
The Healthy Oregon Project (HOP) is a statewide effort that aims to build a large research repository and influence the health of Oregonians through providing no-cost genetic screening to participants for a next-generation sequencing 32-gene panel comprising genes related to inherited cancers and familial hypercholesterolemia. This type of unbiased population screening can detect at-risk individuals who may otherwise be missed by conventional medical approaches. However, challenges exist for this type of high-throughput testing in an academic setting, including developing a low-cost high-efficiency test and scaling up the clinical laboratory for processing large numbers of samples. Modifications to our academic clinical laboratory including efficient test design, robotics, and a streamlined analysis approach increased our ability to test more than 1,000 samples per month for HOP using only one dedicated HOP laboratory technologist. Additionally, enrollment using a HIPAA-compliant smartphone app and sample collection using mouthwash increased efficiency and reduced cost. Here, we present our experience three years into HOP and discuss the lessons learned, including our successes, challenges, opportunities, and future directions, as well as the genetic screening results for the first 13,670 participants tested. Overall, we have identified 730 pathogenic/likely pathogenic variants in 710 participants in 24 of the 32 genes on the panel. The carrier rate for pathogenic/likely pathogenic variants in the inherited cancer genes on the panel for an unselected population was 5.0% and for familial hypercholesterolemia was 0.3%. Our laboratory experience described here may provide a useful model for population screening projects in other states.
Keywords: clinical genetic testing, population screening, laboratory genetics, inherited cancer, familial hypercholesterolemia
This report describes our design, experience, and initial findings in a large population screening project, the Healthy Oregon Project. Details include the steps taken to scale up for a large-scale population screening program and an in-depth discussion of the overall lessons learned.
Introduction
Early detection of inherited genetic diseases through increased screening in susceptible individuals can potentially increase survival rates and is predicted to decrease healthcare costs for affected individuals.1,2,3 However, without a strong family history of disease or other risk factors, an individual is unlikely to meet testing guidelines.4 Therefore, it may be difficult to identify those individuals at increased risk who may require surveillance due to an inherited genetic variant that leads to increased likelihood of disease.3 Even with a positive family history, or other risk factors, there may be barriers to the testing required to determine an individual’s risk due to costs associated with genetic testing or obtaining a correct referral.5,6 Cost-free population-based screening can help alleviate these barriers and has successfully been used to generate this information in the past.7
Several population screening studies have been implemented from nationwide studies like the All of Us Research Program,8 eMERGE,9,10 and the Cancer Moonshot11 to smaller statewide programs such as the Healthy Nevada Project and the Alabama Genomic Health Initiative.7,12 These smaller statewide initiatives have demonstrated the utility of population screening to identify at-risk individuals who would otherwise not be identified with standard clinical practice.7,12
The Healthy Oregon Project (HOP) is an Oregon Health & Science University (OHSU) IRB-approved (18473) population-based study that aims to build a large research repository and impact the health of Oregonians. HOP is supported by OHSU’s CEDAR (Cancer Early Detection Advanced Research) Center. A subsequent federally funded National Cancer Institute Beau Biden Cancer Moonshot initiative clinical trial that utilizes the HOP infrastructure also funds a portion of this work. Goals of this research partnership with Oregonians include the use of data and results provided by participants to create a long-term data repository including samples, surveys, and genetic testing results that will help researchers working on early disease detection research. Participants who enroll in HOP are given the option to have no-cost genetic testing for inherited disorders (initially for inherited cancer and later also familial hypercholesterolemia) with an option to receive results on additional disorders going forward as part of the enrollment process using a Health Insurance Portability and Accountability Act (HIPAA)-compliant app.
Here, we present our clinical laboratory experience and the genetic screening results for the first three years of HOP. We describe the necessary changes to our academic clinical diagnostic laboratory to adapt to the influx of cases. Finally, we present the screening results that demonstrate the genetic risk of inherited cancer and familial hypercholesterolemia in the state of Oregon.
Subjects and methods
Recruitment of participants
The initial recruiting and pilot were done at OHSU starting in December 2018 through events around the OHSU campus. The first participant samples were accessioned in January 2019. Expanded recruitment occurred at local partner businesses and at the county level. This was followed by an additional county rollout with recruitment at partner healthcare centers and through clinics at OHSU. Advertising was done using diverse strategies including social media ads, tabling events at local community functions, and vending machines. Recruitment extended to the entire state and became fully virtual with mailed genetic testing kits in October of 2020 as a result of the COVID-19 pandemic. Participation was open to any individual with an Oregon address aged 18 years or older. The participant did not have to be within the OHSU system to participate. The general population was enrolled from the onset of HOP. This general screening was meant to be unbiased and was not targeted for any one group including underrepresented minorities to better capture the population demographics across the state of Oregon. However, approximately 2.5 years into the study, recruitment of individuals with a prior cancer diagnosis was done through a targeted MyChart message and paper mail from a partner healthcare system from July 19, 2021 onward for patients interested in research studies. The OHSU IRB reviewed this specific work and granted IRB approval for publication (STUDY00024025).
Consent by the HOP app and submission of samples for DNA sequencing
Individuals interested in joining HOP were instructed to download the HIPAA- and 21CFR11-compliant smartphone app for consent. This app was the primary means of contact between HOP and study participants with all consenting done through the app. During enrollment, participants provided their personal information including contact details within the app. Participants could opt into the genetic screening portion after electing to participate in optional surveys (e.g., surveys about cancer history, behaviors, lifestyle, and stress), administered through Let’s Get Healthy!13 (OHSU; Portland, OR) linked through the HOP app. Surveys and saliva samples were fully optional, and participants could decline any components of the study. One section of the enrollment pertained to consenting to DNA testing for inherited cancer and familial hypercholesterolemia. This testing was offered at no cost to the participant. During the consent process in the app, participants learned about how they would receive their DNA testing results. They were informed that if their result was negative, they would receive an e-mail notification alerting them to check their HOP account for their test result; however, if it was positive, they would be contacted by an OHSU genetic counselor by phone to discuss their results. They were also informed that their positive results would be uploaded into an OHSU medical record. Importantly, this information also included text to let the participant know that if the OHSU genetic counselor could not reach them by phone or e-mail, they would not receive a result. Other specific information about the genetic counselor conversation and what to expect was also provided (Note S1). Participants who elected to consent to DNA testing received a HOP kit containing a bottle of mouthwash (Scope original mint),13 a collection tube with a sealable bag, collection instructions, and pre-paid shipping for return by mail. After collection of the mouthwash sample, the kits were sealed and dropped in the mail (or vending machines at OHSU locations prior to March 2020) by participants to send to our testing laboratory. Although participants provided personal information when enrolling in HOP through the app, due to limited accessibility and the need to efficiently test samples, this information was not used in the genetic testing process. These samples came to our clinical diagnostic laboratory coded with an internal ID lacking any details such as demographics, contact information, or personal or family history and were analyzed without this information. This coded ID could later be used to link back to the participant’s detailed enrollment information for the purpose of recontact.
Scaling up for low-cost population screening
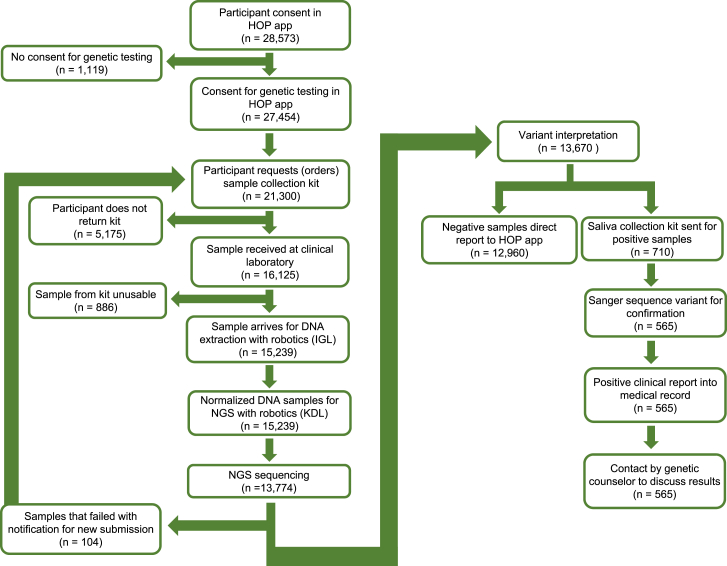
Our laboratory section of the OHSU Knight Diagnostic Laboratories, an academic clinical laboratory, performs diagnostic testing for inherited genetic disorders including all HOP testing. The infrastructure for next-generation sequencing (NGS) testing and analysis were already in place including CLIA/CAP certification, robust internal bioinformatics support with our own database and lab-developed pipeline, multiple NGS instruments, and expertise in generation and analysis of NGS data and variant interpretation. HOP provided the challenge of scaling up testing for larger numbers of samples. Thus, the subsequent methods presented here can be followed with the caveat that clinical laboratories should at a minimum have resources and experience in NGS testing and analysis prior to considering launching a population screening project. A major change to our testing workflow that was not already in place before HOP was the addition of an institutional core laboratory with the necessary equipment and experience for large-scale DNA extractions. Prior to the initiation of HOP, this core lab obtained CAP accreditation for DNA extraction. Here, we summarize the other changes that were required for our laboratory to design a low-cost test and to scale up for HOP that can be followed by other clinical laboratories. These changes allowed the test to be performed at approximately $50 per sample, including the genetic testing components only. Additional factors such as genetic counseling are not included in this number. Our workflow to accommodate the influx of samples for HOP, which was predicted to reach many thousand per year, is illustrated in Figure 1.
-
1.
Robotics: The introduction of automation into the DNA extraction process as well as the NGS library preparation was essential to scale testing up with only one dedicated laboratory technologist for sequencing and one for DNA extractions. A QIAsymphony SP robot (Qiagen) and Eppendorf epMotion Liquid Handling Workstation (Eppendorf) were used for DNA extraction, while two Janus G3 Workstations (PerkinElmer) were used in the NGS setup. For the NGS setup, the two robots were split into one for pre-PCR steps and the other for post-PCR steps to maximize efficiency. This implementation of robotics in the NGS library preparation process was crucial to accommodate the large sample influx. Additional details on the specific uses of the robots is presented in the supplemental methods.
-
2.
NGS assay design: The HOP test was designed as a low-cost test using an efficient testing platform to detect the most commonly occurring variants (SNVs) in inherited cancer and familial hypercholesterolemia. Limiting the number of genes on the panel, along with sequencing at a depth required for germline rather than somatic mutation detection, allowed us to save costs for the overall NGS panel. These trade-offs affected our ability to detect all types of variants such as CNVs, so this was a limitation of the test design that saved on time and costs.
-
3.
Pre-filtering of NGS results before analysis: Several filters were used to obtain a set of variants most likely to be disease-causing (details in supplemental methods). This step helped streamline our analysis.
-
4.
Expedited initial review of variants post filtering: The use of ClinVar14 as a preliminary review process of variants before formal classification helped decrease the analysis time for HOP samples.
-
5.
Types of variants returned to participants: Not returning variants of uncertain significance (VUSs) allowed our laboratory to focus analysis time on variants that have strong evidence of pathogenicity (pathogenic and likely pathogenic).
Figure 1.
Pipeline for the enrollment of HOP participants and sample workflow
HOP participants sequenced as of April 15, 2022. Differences in numbers are reflective of the fluid pipeline and time it takes samples to process from consent in the HOP app through next-generation sequencing (NGS) and analysis. IGL, Integrated Genomics Laboratory, core laboratory; KDL, Knight Diagnostic Laboratories, clinical laboratory.
Further information about HOP processes and how to contact us can be obtained from the HOP website at https://healthyoregonproject.com/.
Next generation sequencing and variant calling
DNA was extracted in the College of American Pathologists (CAP)-accredited Integrated Genomics Lab (IGL) using a QIAsymphony SP robot (Qiagen) quantitated and normalized to a 10 ng/μL DNA concentration (acceptable range at 5–45 ng/μL) and plated out in 96-well PCR plates. Following DNA extraction and normalization, an amplicon-based library preparation was performed using the QIAseq Targeted DNA Custom Panel (Qiagen) targeting coding regions including the reportable HOP genes (for details see supplemental methods). Samples were sequenced in batches of 4× 96-well plates at our laboratory using an Illumina NextSeq 500 or 550 (Illumina). FASTQ files from the sequencer were checked for quality metrics and then run through our internal bioinformatics pipeline. FASTQs that passed QC metrics using FastQC were trimmed of adapter and linker sequences using Cutadapt.15 UMI-tools was then used to isolate 12-bp unique molecular indices (UMIs) for use in downstream deduplication.16 Resulting processed FASTQs were then aligned using BWA-mem17 against the GRCh37 reference genome, assigned read groups, and sorted before deduplication with the UMI-tools “directional” grouping approach. Primer sequences were removed from reads to reduce primer-specific error modes using in-house tools. Genome Analysis Tool Kit (GATK) HaplotypeCaller performed indel and single-nucleotide polymorphism (SNP) variant calling in gVCF mode, restricted by a target region BED file with a genomic region buffer of 100 bases. After all samples on a run had produced a variant call format (VCF) file, they were combined using GATK4 CombineGVCFs, then genotyped using GenotypeGVCFs. Variants from the resultant VCF were split at multi-allelic sites using in-house tools, yielding another VCF. This VCF was then normalized utilizing VT normalize. Annotations from ClinVar, HGMD,18 SnpEff,19 and gnomAD were supplied to the VCF INFO field for use in variant filtering. Further variant annotations were provided by SeattleSeqAnnotation 138.20 Further quality metrics were provided via GATK CollectAlignmentSummaryMetrics and QC Coverage Metrics. Lab relevant metrics were collected utilizing the in-house Sample Level Metrics tool. Sample-level metrics included Q30%, average depth, average coverage at 250×, 100×, 50×, 20×, and 10×, percentage of on-target reads, and GC%. Samples were considered as passing QC with average read depth >100×, depth at 20× > 95%, and Q30% > 70.
Test design
The main genetic focus of HOP was to test genes in which pathogenic variants are associated with the common inherited cancer syndromes hereditary breast and ovarian cancer (HBOC) and Lynch syndrome, while also including genes in which pathogenic variants are associated with rarer types of inherited cancer. The clinical team selected 31 medically actionable inherited cancer-related genes (APC [MIM: 611731], ATM [MIM: 607585], BAP1 [MIM: 603089], BMPR1A [MIM: 601299], BRCA1 [MIM: 113705], BRCA2 [MIM: 600185], BRIP1 [MIM: 605882], CDH1 [MIM: 192090], CDK4 [MIM: 123829], CDKN2A [MIM: 600160], CHEK2 [MIM: 604373], MEN1 [MIM: 613733], MITF [MIM: 156845], MLH1 [MIM: 120436], MSH2 [MIM: 609309], MSH6 [MIM: 600678], MUTYH [MIM: 604933], NBN [MIM: 602667], PALB2 [MIM: 610355], PMS2 [MIM: 600259], POLD1 [MIM: 174761], PTEN [MIM: 601728], RAD51C [MIM: 602774], RAD51D [MIM: 602954], RB1 [MIM: 614041], RET [MIM: 164761], SMAD4 [MIM: 600993], STK11 [MIM: 602216], TP53 [MIM: 191170], TSC1 [MIM: 605284], and TSC2 [MIM: 191092]) for the HOP panel. LDLR (MIM: 606945) was also included to determine the risk of familial hypercholesterolemia (FH), leading to 32 genes in the final HOP panel (Table 1). As a way to maximize panel space while capturing the vast majority of risk, we did not interrogate the other two genes in which pathogenic variants are associated with FH (APOB [MIM: 107730] and PCSK9 [MIM: 607786]) since >90% of reported FH-causing variants occur in LDLR.21 20 of the 32 genes on the HOP panel overlapped with the newest American College of Medical Genetics and Genomics (ACMG) Secondary Findings list22 (SF list), while 12 genes were unique to the HOP panel (Figure S1).
Table 1.
List of genes and their associated disorders for the Healthy Oregon Project
| Gene | OMIM non-somatic inherited cancer disorder(s) or familial hypercholesterolemia | Prevalence of selected disorders or cancer typesa |
|---|---|---|
| APC | adenomatous polyposis coli (MIM: 175100); brain tumor-polyposis syndrome 2 (MIM: 175100); desmoid disease, hereditary (MIM: 135290); Gardner syndrome (MIM: 175100) | familial adenomatous polyposis: 1:6,850 to 1:31,250 |
| ATM | ataxia-telangiectasia (MIM: 208900)b; susceptibility to breast cancer (MIM: 114480) | ataxia-telangiectasia: 1:40,000 to 1:100,000 |
| BAP1 | tumor predisposition syndrome (MIM: 614327) | BAP1-tumor predisposition syndrome: prevalence unknown |
| BMPR1A | polyposis syndrome, hereditary mixed, 2 (MIM: 610069); polyposis, juvenile intestinal (MIM: 174900) | juvenile polyposis syndrome: 1:16,000 to 1:100,000 |
| BRCA1 | Fanconi anemia, complementation group S (MIM: 617883)b; breast-ovarian cancer, familial, 1 (MIM: 604370); pancreatic cancer, susceptibility to, 4 (MIM: 614320) | BRCA1 and BRCA2-associated hereditary breast and ovarian cancer: 1:400 to 1:500 (higher in specific population groups) |
| BRCA2 | Fanconi anemia, complementation group D1 (MIM: 605724)b; Wilms tumor (MIM: 194070); breast cancer, male, susceptibility to (MIM: 114480); breast-ovarian cancer, familial, 2 (MIM: 612555); glioblastoma 3 (MIM: 613029)b; medulloblastoma (MIM: 155255); pancreatic cancer 2 (MIM: 613347); prostate cancer (MIM: 176807) | BRCA1 and BRCA2-associated hereditary breast and ovarian cancer: 1:400 to 1:500 (higher in specific population groups) |
| BRIP1 | Fanconi anemia, complementation group J (MIM: 609054)b; breast cancer, early-onset, susceptibility to (MIM: 114480) | ovarian cancer due to BRIP1: estimated at ∼1:11223 |
| CDH1 | blepharocheilodontic syndrome 1 (MIM: 119580); gastric cancer, hereditary diffuse, with or without cleft lip and/or palate (MIM: 137215); breast cancer, lobular (MIM: 114480); prostate cancer, susceptibility to (MIM: 176807) | hereditary diffuse gastric cancer: prevalence unknown; 1% to 3% of cases due to CDH1 pathogenic variant |
| CDK4 | melanoma, cutaneous malignant, 3 (MIM: 609048) | melanoma, cutaneous malignant due to CDK4: prevalence unknown |
| CDKN2A | melanoma and neural system tumor syndrome (MIM: 155755); pancreatic cancer/melanoma syndrome (MIM: 606719); melanoma, cutaneous malignant, 2 (MIM: 155601) | familial atypical multiple mole melanoma (FAMMM): prevalence unknown |
| CHEK2 | Li-Fraumeni syndrome (MIM: 609265); colorectal cancer, susceptibility to (MIM: 114500); breast cancer, susceptibility to (MIM: 114480); prostate cancer, familial, susceptibility to (MIM: 176807) | colon cancer due to CHEK2: 2:10024 |
| MEN1 | multiple endocrine neoplasia 1 (MIM: 131100) | multiple endocrine neoplasia 1: 1:10,000 to 1:100,000 |
| MITF | melanoma, cutaneous malignant, susceptibility to, 8 (MIM: 614456) | MITF-related melanoma and renal cell carcinoma predisposition syndrome: <1:1,000,00025 |
| MLH1 | colorectal cancer, hereditary nonpolyposis, type 2 (MIM: 609310); mismatch repair cancer syndrome 1 (MIM: 276300)b; Muir-Torre syndrome (MIM: 158320) | Lynch syndrome: 1:279 |
| MSH2 | colorectal cancer, hereditary nonpolyposis, type 1 (MIM: 120435); mismatch repair cancer syndrome 2 (MIM: 619096)b; Muir-Torre syndrome (MIM: 158320) | Lynch syndrome: 1:279 |
| MSH6 | colorectal cancer, hereditary nonpolyposis, type 5 (MIM: 614350); mismatch repair cancer syndrome 3 (MIM: 619097)b; endometrial cancer, familial (MIM: 608089) | Lynch syndrome: 1:279 |
| MUTYH | adenomas, multiple colorectal (MIM: 608456)b | MUTYH-associated polyposis: 1:20,00 to 1:60,000 |
| NBN | aplastic anemia (MIM: 609135); leukemia, acute lymphoblastic (MIM: 613065); Nijmegen breakage syndrome (MIM: 251260)b | Nijmegen breakage syndrome: ∼1:100,000 |
| PALB2 | Fanconi anemia, complementation group N (MIM: 610832)b; breast cancer, susceptibility to (MIM: 114480); pancreatic cancer, susceptibility to, 3 (MIM: 613348) | breast, pancreas, and ovarian cancer due to PALB2: prevalence unknown |
| PMS2 | colorectal cancer, hereditary nonpolyposis, type 4 (MIM: 614337); mismatch repair cancer syndrome 4 (MIM: 619101)b | Lynch syndrome: 1:279 |
| POLD1 | colorectal cancer, susceptibility to, 10 (MIM: 612591) | polymerase proofreading-associated polyposis (PPAP): prevalence unknown |
| PTEN | Cowden syndrome 1 (MIM: 158350); Lhermitte-Duclos syndrome (MIM: 158350); glioma susceptibility 2 (MIM: 613028); meningioma (MIM: 607174) | Cowden syndrome: 1:200,000 |
| RAD51C | Fanconi anemia, complementation group O (MIM: 613390)b; breast-ovarian cancer, familial, susceptibility to, 3 (MIM: 613399) | breast-ovarian cancer, familial due to RAD51C: prevalence unknown |
| RAD51D | breast-ovarian cancer, familial, susceptibility to, 4 (MIM: 614291) | breast-ovarian cancer, familial due to RAD51D: prevalence unknown |
| RB1 | retinoblastoma (MIM: 180200); retinoblastoma, trilateral (MIM: 180200) | retinoblastoma: estimated at 1.5:100,00026 |
| RET | medullary thyroid carcinoma (MIM: 155240); multiple endocrine neoplasia IIA (MIM: 171400); multiple endocrine neoplasia IIB (MIM: 162300); pheochromocytoma (MIM: 171300); | multiple endocrine neoplasia type 2: 1:35,000 |
| SMAD4 | juvenile polyposis/hereditary hemorrhagic telangiectasia syndrome (MIM: 175050); Myhre syndrome (MIM: 139210); polyposis, juvenile intestinal (MIM: 174900) | juvenile polyposis syndrome: 1:16,000 to 1:100,000 |
| STK11 | Peutz-Jeghers syndrome (MIM: 175200) | Peutz-Jeghers syndrome: 1:25,000 to 1:280,000 |
| TP53 | bone marrow failure syndrome 5 (MIM: 618165); Li-Fraumeni syndrome (MIM: 151623); adrenocortical carcinoma, pediatric (MIM: 202300); basal cell carcinoma 7 (MIM: 614740); choroid plexus papilloma (MIM: 260500); colorectal cancer (MIM: 114500); glioma susceptibility 1 (MIM: 137800); osteosarcoma (MIM: 259500) | Li-Fraumeni syndrome: 1:3,555 to 1:5,476 |
| TSC1 | lymphangioleiomyomatosis (MIM: 606690); tuberous sclerosis-1 (MIM: 191100) | tuberous sclerosis complex: 1:10,000 to 1:100,00025 |
| TSC2 | tuberous sclerosis-2 (MIM: 613254) | tuberous sclerosis complex: 1:10,000 to 1:100,00025 |
| LDLR | hypercholesterolemia, familial, 1 (MIM: 143890); LDL cholesterol level QTL2 (MIM: 143890) | familial hypercholesterolemia: 1:500 |
All disorders listed act as autosomal dominant unless otherwise indicated.
All prevalence values listed are from GeneReviews (https://www.ncbi.nlm.nih.gov/books/NBK1116/) unless otherwise indicated
Autosomal recessive
A positive test result was defined as the detection of a pathogenic or likely pathogenic (P/LP) heterozygous variant in 1 of 30 of the 32 genes (excluding MUTYH and NBN). A positive result for MUTYH was defined as the detection of a homozygous P/LP variant or two heterozygous P/LP variants. A positive result for NBN was modified during the course of the project following changes to National Comprehensive Cancer Network (NCCN) cancer risk management recommendations. The current protocol for NBN is to report any homozygous P/LP variant or two heterozygous P/LP variants. For all of the genes on the HOP panel including NBN, no variants of uncertain significance (VUSs) were reported. A negative test result was defined as no P/LP variants detected (or detection of only single heterozygous P/LP variants in MUTYH or NBN). If the classification of a variant that was detected in previous HOP participants but not reported is upgraded to P/LP, a protocol is in place to recontact these participants to ask for a confirmatory sample after IRB review. This process also covers changes in reporting of variants due to updated research and recommendations. The negative report and an example positive report are available in the Notes S2 and S3, respectively. All sequencing occurred in a CLIA/CAP-certified laboratory at OHSU.
Several key differences exist in the way the results from HOP were analyzed versus our normal laboratory diagnostic tests (Table S1). Mosaic and somatic variants (imbalanced allele frequencies) were not reported. Copy number variants were not reported. Due to the high-throughput nature of the study, we developed an expedited variant review process for initially determining if variants likely had enough evidence to reach P/LP. Since most of the genes on the HOP testing panel have been well studied and characterized, we first used ClinVar,14 referencing previous laboratories’ entries of the variant to quickly determine what variants may have appropriate evidence for pathogenicity. Prior to reporting, a thorough literature review and analysis was completed by our laboratory using ACMG-recommended guidelines.27 Variants that were not curated in ClinVar and of the type likely to meet criteria for pathogenic (e.g., loss-of-function variants) were fully classified during the initial review process. Variants deemed possibly P/LP after this initial review were interpreted using our standard approach for diagnostic samples (see variant interpretation below). Variants were considered unreported if at the time of testing there was no ClinVar entry.
Variant filtering
The full list of filtering criteria is described in the supplemental methods. To summarize, after the samples were run through our initial bioinformatics pipeline to call genetic variants, numerous filters were used on the sequence results before analysis. These parameters included filtering on clinical significance in ClinVar,14 classification in Human Gene Mutation Database (HGMD),18 variant type, and population frequency.
Receiving genetic testing results and secondary confirmation
Participants with a negative genetic testing result received that information entirely through the HOP app. Participants with an initial positive result from the NGS screening test were contacted by one of our clinical laboratory staff members using the internal coded ID to extract participant contact information. Participants were not given any information about the initial finding or any indication that they had a preliminary positive result. They were provided a secondary confirmation kit consisting of two Oragene OG-500 saliva collection kits (DNA Genotek) that are used with our standard clinical diagnostic tests with instructions for DNA collection in order to complete the test. The confirmation sample we received at our laboratory included identifiable information as would normally be included for a normal clinical sample received into our lab. This included name, sex at birth and an optional checkbox for legal sex, date of birth, and access to their medical record if one existed at OHSU. This secondary sample was extracted using a Nucleospin Tissue kit (Takara Bio USA) and Sanger sequenced in our CLIA/CAP clinical lab. Upon confirmation of the variant in this secondary sample by targeted capillary Sanger sequencing, a clinical report was uploaded to the participant’s OHSU medical record. If the participant was outside of the OHSU network, then an OHSU medical record was generated for this individual with their positive report. The participant was then contacted by an OHSU genetic counselor by telephone to discuss the results of the testing. The genetic counselors would discuss personal and family history of related disease for participants with a positive result during this call to help inform any recommendations. If the participant wanted to have this report sent to their primary care team outside of OHSU, they could sign a release form to get this information sent. If no release was signed, then their care team could access some of this information through EPIC Care Everywhere (https://www.epic.com/careeverywhere/) if their institute participates in this program, or they could use EPIC Share Everywhere (https://shareeverywhere.epic.com/) to give their primary care team temporary access to some information about their results. One month after genetic counseling, a HOP participant navigator would reach out to offer any assistance the participant may need regarding their result such as resources to pay for recommended changes to timing or type of standard cancer screenings. These recommended resources were external to OHSU and HOP since there were no specific HOP funds allocated for any downstream medical management. Participants were also given an e-mail address to contact HOP with any additional questions. If these emails included testing results or other protected health information (PHI) excluding contact name, date of birth, phone number, and address, the discussion would be migrated to an OHSU-encrypted e-mail system. Further contact with the participant through the encrypted e-mail was done by a HOP team member outside of our clinical laboratory. If the secondary sample was not received within two weeks, a follow-up e-mail was sent, and if the sample was still not received after another two weeks, the participant was contacted by phone. No additional attempts were made to obtain a confirmation sample, and the participant was not issued a report. However, participants can still submit their secondary sample and receive a result.
Variant interpretation
Variants were classified according to the American College of Medical Genetics and Genomics (ACMG)-recommended guidelines.27 Unreported variants within the canonical splice sites or those strongly predicted to affect splicing that have not been previously reported to show an RNA impact were analyzed experimentally with a blood sample from the participant. RNA was extracted using the Qiagen RNeasy Mini Kit (Qiagen) and converted to cDNA using Invitrogen’s SuperScript II Reverse Transcriptase (Thermo Fisher). Sanger sequencing was performed on the resulting cDNA, and traces were analyzed using Mutation Surveyor (Softgenetics) and PolyPeak.28 The final determination of pathogenicity was made by a board-certified clinical molecular geneticist.
Initial validation of the HOP sequencing panel with manual workflow
The HOP screening test is not designed to detect noncoding variants, variants within pseudogene regions, and/or copy number variation (CNV). Thus, the clinical sensitivity is estimated to be approximately 90% based on known contribution of noncoding variants, variants within pseudogenes, and CNVs to inherited cancer in the targeted genes.
The HOP assay was validated according to standard CLIA/CAP guidelines. To summarize, initial validation of the cancer associated genes included 304 Scope mouthwash saliva samples that consisted of 71 unique participants and included 30 positive controls with 99 known variants (82 of the variants were from replicate samples) and 20 no template controls. Positive control libraries were constructed and then sequenced by the same operator in duplicate sample sequencing runs. The same control samples were used to construct libraries and sequence in two additional sample sequencing runs by two additional operators to assess repeatability. Thus, libraries were constructed for 384 samples by three different operators and sequenced on four flow cells. With the addition of LDLR, the assay was revalidated under similar experimental design with the addition of 19 new positive controls containing 21 known cardiomyopathy variants. The results of both validations indicated that the positive predictive value (PPV) is 99% and repeatability, reproducibility, sensitivity, and specificity are all 99% for the HOP assay.
Statistical analysis
Statistical analyses were performed in R software v.4.1.2. Statistically significant was considered as a p value < 0.05.
Results
Participant demographics
The mean age of HOP participants was 47 (range 18–97), and for those who provided their sex at birth, approximately 76% were female and 24% male. The majority of this population self-identified as White (84%), reflecting the primary composition of the Oregon population, although other population groups such as Asian, Hispanic or Latino/a/x, and Black or African American also participated. Detailed demographics of these HOP participants are presented in Table S2. Approximately 96% of HOP participants who enrolled in the study requested genetic testing, suggesting that testing was a major driver in the enrollment process.
DNA sequencing results
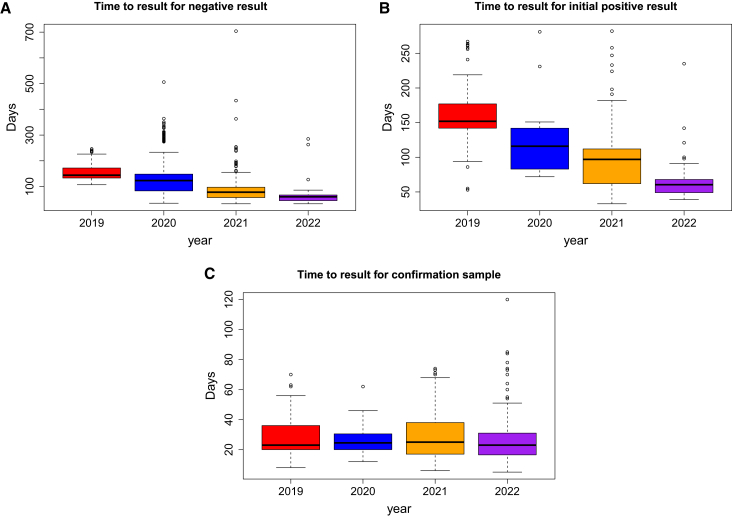
While we have now screened more than 20,000 HOP participants, the results presented here reflect the first 13,774 participants sequenced with the initial version of the test. Because testing was performed to detect germline rather than somatic variants, the test was designed to achieve a minimum of 100× depth of coverage, and in fact, the average depth of sequencing for all samples tested was 764× (range 0–6,665). The failure rate for all samples tested by NGS was 0.8% (104 of 13,774), which was considered an acceptable level. The likely reason for a failed sample was the self-collection process of saliva which can result in lower DNA yields and higher contamination. The mean time to result from receipt of initial mouthwash sample to negative report in the app was 88 days (Figure 2A), while the mean time to result from receipt of initial mouthwash sample to sending the request for a secondary confirmation sample for positive cases was 95 days (Figure 2B). Our time to result decreased as the study continued since the initial recruiting and sample collection occurred before sequencing was initiated in our laboratory and with the addition of robotics to the sequencing pipeline (for details see subjects and methods). After receiving a secondary sample for confirmation, our mean turnaround time to issue a positive report in the participant’s medical record was 28 days (Figure 2C).
Figure 2.
Time to result for samples in HOP
(A) The time to result from when we received a mouthwash sample from a HOP participant to the return of a negative report in the HOP app.
(B) The time to result from when we received a mouthwash sample from a HOP participant to when we shipped out a secondary confirmation kit.
(C) The time to result from when the lab received a secondary saliva sample to the posting of a positive clinical diagnostic report to the participant’s medical record. We fully validated and implemented the use of robotic automation in January 2020.
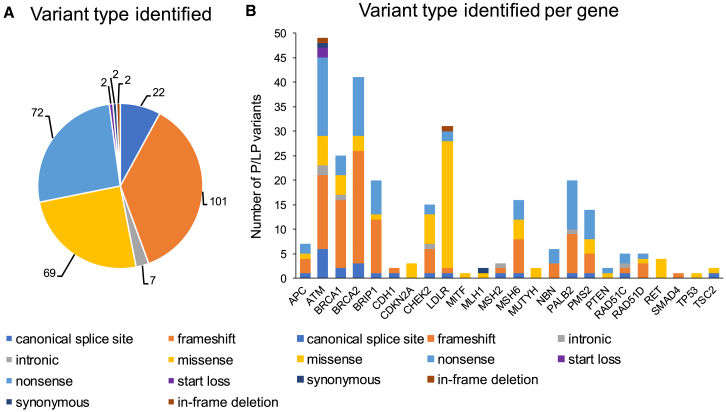
Overall, 730 pathogenic/likely pathogenic (P/LP) variants were identified in 710 participants, representing 75% of the genes on the panel (24/32) (Table S3). While all participants with positive results were contacted to request a confirmatory sample, only 80% (565 of 710) of them actually returned a second sample and received a report. All of the second samples provided from participants for Sanger testing of P/LP variants previously identified by NGS confirmed the initial findings (565 of 565). These results were reassuring that there had been no sample mix-ups of clinical relevance. In viewing the dataset for all HOP participants, there were 12 genes where at least 10 different HOP participants carried a P/LP variant (Table 2). We also found that 98% of the P/LP variants had previously been reported in ClinVar (719 of 730), which is not surprising in this list of well-studied and clinically tested genes. While all variants were reviewed using the ACMG guidelines, the few (11) that were not in the ClinVar database were extensively reviewed. The most common type of P/LP variants detected were frameshifts caused by small deletions or duplications, while the most infrequently detected were in-frame deletions, start loss, and synonymous variants (Figure 3A). However, this differed by specific gene (e.g., genes with missense variants as their primary or only variant; Figure 3B).
Table 2.
Number of P/LP variants detected per gene in HOP
| Gene | Number of P/LP variants |
|---|---|
| CHEK2 | 221 |
| ATM | 78 |
| MITF | 70 |
| BRCA2 | 55 |
| APC | 52 |
| BRCA1 | 44 |
| BRIP1 | 33 |
| PMS2 | 28 |
| PALB2 | 26 |
| MSH6 | 20 |
| RET | 11 |
| MUTYH | 8 |
| NBN | 8 |
| RAD51C | 8 |
| RAD51D | 5 |
| CDKN2A | 4 |
| MSH2 | 3 |
| CDH1 | 2 |
| MLH1 | 2 |
| PTEN | 2 |
| TSC2 | 2 |
| SMAD4 | 1 |
| TP53 | 1 |
| BAP1 | 0 |
| BMPR1A | 0 |
| CDK4 | 0 |
| MEN1 | 0 |
| POLD1 | 0 |
| RB1 | 0 |
| STK11 | 0 |
| TSC1 | 0 |
| LDLR | 46 |
P/LP, pathogenic or likely pathogenic
Figure 3.
Types of P/LP variants identified in HOP
(A) The type of variant identified for all 277 unique P/LP variants found in HOP.
(B) The type of P/LP variant identified per gene in HOP. Deletions, duplications, or insertions leading to a direct nonsense variant are categorized as nonsense. Frameshift variants contain deletions, duplications, and insertions that lead to a frameshift. Intronic variants contain deletions or substitutions that occur outside of the canonical splice sites.
Inherited cancer results
The overall rate of P/LP variants in genes in which P/LP variants are associated with inherited cancer in HOP was approximately 5.0%. P/LP variants (246 unique variants) were identified in 23 out of the 31 inherited cancer genes on the panel, with the most identified P/LP variants occurring in CHEK2 with 221 (30% of total identified inherited cancer P/LP variants), followed by ATM with 78 and MITF with 70 (Table 2). These three genes were responsible for approximately 50% of all detected P/LP variants, while 12 genes (MUTYH, NBN, RAD51C, RAD51D, CDKN2A, MSH2, CDH1, MLH1, PTEN, TSC2, SMAD4, TP53) contained fewer than 10 P/LP variants.
Our results indicate that 0.7% (99 of 13,670) of HOP participants tested carried a P/LP variant in BRCA1/BRCA2, which is associated with HBOC. In total, 14% of P/LP variants associated with inherited cancer were identified in BRCA1/BRCA2. Most of the unique variants that were identified in these two genes were truncating (80%), similar to results of other screening studies.29 To further interrogate the results associated with breast cancer more broadly, we included eight additional genes tested by the HOP panel that are established breast cancer-predisposition genes (ATM, CDH1, CHEK2, PALB2, PTEN, RAD51C, RAD51D, TP53)30 to the BRCA1/BRCA2 HBOC results. Adding these additional genes increased the rate of P/LP variants associated with breast cancer susceptibility to 3.2% in our cohort.
The rate of P/LP variants responsible for Lynch syndrome was 0.4% (53 of 13,670) based on testing MLH1, MSH2, MSH6, and PMS2. Although not as high as the percentage of the variants found in BRCA1 and BRCA2, the majority (63%) of the unique variants identified in these four genes were truncating. The gene where most P/LP variants were detected for Lynch syndrome was PMS2, with 28 P/LP variants, while we identified only two likely pathogenic variants in MLH1.
Familial hypercholesterolemia results
There were 46 participants who harbored a P/LP variant in LDLR, corresponding to a 0.3% rate of P/LP variants associated with familial hypercholesterolemia in our screening population. In contrast to the major variant types found in inherited cancer genes, missense variants comprised 84% of the unique variants found in LDLR. The other two genes in which P/LP variants are associated with FH (APOB, PCSK9) were not tested by HOP.
Recurrent variants and participants with more than one P/LP variant
Approximately 30% (82/277) of the P/LP variants found in HOP participants were detected in more than one individual, with several being known founder variants. Specifically, five of these variants were found in more than 10 participants, with three of the five occurring in CHEK2 (Figure S2). Indeed, the most commonly detected P/LP variant was the c.470T>C (p.Ile157Thr [GenBank: NM_007194.4]) missense variant in CHEK2 and comprised 13% of total identified P/LP variants. Additional commonly detected P/LP variants included founder mutations with high frequency in specific populations such as the c.3920T>A (p.Ile1307Lys [GenBank: NM_000038.6]) Ashkenazi Jewish APC missense variant31 and the c.1100del (p.Thr367Metfs∗15 [GenBank: NM_007194.4]) CHEK2 founder frameshift variant found at high frequency in individuals of Northern European descent.32,33
We also found that 2.8% (20 of 710) of participants harbored more than one P/LP variant (Table 3), including five participants with P/LP variants associated with an inherited cancer syndrome and familial hypercholesterolemia. In total, nine participants with two P/LP variants harbored one P/LP variant in CHEK2, including one participant with two common founder CHEK2 P/LP variants (c.1100del [p.Thr367Metfs∗15] and c.470T>C [p.Ile157Thr]) further highlighting the commonality of P/LP variants in this gene.
Table 3.
List of participants with more than one P/LP variant
| Participant | Variants identified | Genes | Classifications | Variant types |
|---|---|---|---|---|
| 1 | GenBank: NM_000038.6; c.3920T>A (p.Ile1307Lys) | APC | population-specific risk factor | missense |
| GenBank: NM_007194.4; c.1100del (p.Thr367Metfs∗15) | CHEK2 | pathogenic | frameshift | |
| 2 | GenBank: NM_007194.4; c.433C>T (p.Arg145Trp) | CHEK2 | likely pathogenic | missense |
| GenBank: NM_000248.4; c.952G>A (p.Glu318Lys) | MITF | pathogenic | missense | |
| 3 | GenBank: NM_000038.6; c.3920T>A (p.Ile1307Lys) | APC | population-specific risk factor | missense |
| GenBank: NM_007194.4; c.1283C>T (p.Ser428Phe) | CHEK2 | pathogenica | missense | |
| 4 | GenBank: NM_000051.4; c.2T>C (p.Met1?) | ATM | pathogenic | start loss |
| GenBank: NM_000179.3; c.2731C>T (p.Arg911∗) | MSH6 | pathogenic | nonsense | |
| 5 | GenBank: NM_000051.4; c.15dup (p.Asn6∗) | ATM | pathogenic | nonsense |
| GenBank: NM_024675.4; c.2032del (p.Leu678Tyrfs∗31) | PALB2 | pathogenic | frameshift | |
| 6 | GenBank: NM_000051.4; c.15dup (p.Asn6∗) | ATM | pathogenic | nonsense |
| GenBank: NM_024675.4; c.2032del (p.Leu678Tyrfs∗31) | PALB2 | pathogenic | frameshift | |
| 7 | GenBank: NM_007194.4; c.1100del (p.Thr367Metfs∗15) | CHEK2 | pathogenic | frameshift |
| GenBank: NM_007194.4; c.470T>C (p.Ile157Thr) | CHEK2 | likely pathogenic risk factora | missense | |
| 8 | GenBank: NM_000051.4; c.3G>A (p.Met1?) | ATM | pathogenic | start loss |
| GenBank: NM_000535.7; c.2249G>A (p.Gly750Asp) | PMS2 | likely pathogenic | missense | |
| 9 | GenBank: NM_007194.4; c.1263del (p.Ser422Valfs∗15) | CHEK2 | pathogenic | frameshift |
| GenBank: NM_000248.4; c.952G>A (p.Glu318Lys) | MITF | pathogenic | missense | |
| 10 | GenBank: NM_007194.4; c.1100del (p.Thr367Metfs∗15) | CHEK2 | pathogenic | frameshift |
| GenBank: NM_000248.4; c.952G>A (p.Glu318Lys) | MITF | pathogenic | missense | |
| 11 | GenBank: NM_007194.4; c.470T>C (p.Ile157Thr) | CHEK2 | likely pathogenic risk factora | missense |
| GenBank: NM_032043.3; c.1372G>T (p.Glu458∗) | BRIP1 | pathogenic | nonsense | |
| 12 | GenBank: NM_007194.4; c.470T>C (p.Ile157Thr) | CHEK2 | likely pathogenic risk factora | missense |
| GenBank: NM_000249.4; c.1517T>C (p.Val506Ala) | MLH1 | likely pathogenic | missense | |
| 13 | GenBank: NM_001048174.2; c.452A>G (p.Tyr151Cys) | MUTYH | pathogenic | missense |
| GenBank: NM_001048174.2; c.1103G>A (p.Gly368Asp) | MUTYH | pathogenic | missense | |
| 14 | GenBank: NM_001048174.2; c.452A>G (p.Tyr151Cys) | MUTYH | pathogenic | missense |
| GenBank: NM_001048174.2; c.1103G>A (p.Gly368Asp) | MUTYH | pathogenic | missense | |
| 15 | GenBank: NM_001048174.2; c.452A>G (p.Tyr151Cys) | MUTYH | pathogenic | missense |
| GenBank: NM_001048174.2; c.1103G>A (p.Gly368Asp) | MUTYH | pathogenic | missense | |
| 16 | GenBank: NM_007194.4; c.470T>C (p.Ile157Thr) | CHEK2 | likely pathogenic risk factora | missense |
| GenBank: NM_000527.5; c.1775G>A (p.Gly592Glu) | LDLR | pathogenic | missense | |
| 17 | GenBank: NM_000535.7; c.1927C>T (p.Gln643∗) | PMS2 | pathogenic | nonsense |
| GenBank: NM_000527.5; c.798T>A (p.Asp266Glu) | LDLR | pathogenic | missense | |
| 18 | GenBank: NM_007294.4; c.2681_2682del (p.Lys894Thrfs∗8) | BRCA1 | pathogenic | frameshift |
| GenBank: NM_000527.5; c.1027G>A (p.Gly343Ser) | LDLR | pathogenic | missense | |
| 19 | GenBank: NM_000059.4; c.6275_6276del (p.Leu2092Profs∗7) | BRCA2 | pathogenic | frameshift |
| GenBank: NM_000527.5; c.2054C>T (p.Pro685Leu) | LDLR | pathogenic | missense | |
| 20 | GenBank: NM_000059.4; c.4449del (p.Asp1484Thrfs∗2) | BRCA2 | pathogenic | frameshift |
| GenBank: NM_000527.5; c.301G>A (p.Glu101Lys) | LDLR | pathogenic | missense |
P/LP, pathogenic or likely pathogenic
Low penetrance
Results based on filtering by the ACMG secondary findings gene list
The ACMG recommended list of secondary findings22 is composed of genes curated by experts as a minimum set recommended to analyze and report for individuals undergoing unrelated clinical whole-genome or whole-exome diagnostic testing. These genes were selected for multiple reasons, but foremost they must be medically actionable to appear on this gene list. HOP also aims to return actionable results, so we compared these gene lists. If we sequenced only genes on the ACMG’s recommended secondary findings list22 that were also included on the HOP panel, our results would have only been 41% (301/730) of what we identified in participants using the full HOP gene panel. Thus, limiting HOP to this list of genes would have resulted in us missing P/LP variants in more than 50% of HOP participants. Interestingly, 5 of the 8 genes (63%) where we did not identify a single P/LP variant were also on the ACMG secondary findings gene list (BMPR1A, MEN1, RB1, STK11, TSC1), suggesting that some of the disorders associated with genes on the secondary findings list are less common.
Variants requiring functional studies
Throughout the project, we identified several variants that were either outside canonical splice sites and computationally predicted to affect RNA splicing or at canonical sites without any published functional studies to validate altered splicing. To functionally test these computational predictions, blood was requested for RNA studies from nine HOP participants, and seven of these participants consented to this additional testing (78% return rate). These analyses resulted in one computational prediction that was incorrect leading to a negative report for the participant, while another variant was classified and reported as a VUS due to ambiguous RNA results. The full results of the RNA studies are listed in Table S4.
Discussion
The results presented here represent the first three years of HOP and help provide a roadmap that may be useful for other laboratories already performing clinical NGS testing to engage in low-cost population screening (for details see subjects and methods). Major factors that can be emulated to scale up testing include the introduction of robotics into the NGS library preparation protocols as well as the development of expedited initial variant review processes. Costs can be kept low by choice of platform for the test design with a limited set of genes, by reporting only P and LP variants with confirmations, and by the use of a HIPAA-compliant app for all consent and project enrollment as well as choice of sample collection kits. Limiting the return of positive results to a phone call with a genetic counselor rather than in-person contact also contributes to a successful screening project by maximizing counseling resources. Although the changes we have implemented in our clinical laboratory allowed us to scale up to the level needed for this project, it was critical that we already had the basic infrastructure and expertise in place prior to HOP.
A major goal of the overall project was to help Oregonians through providing no-cost genetic screening for major inherited cancer disorders and familial hypercholesterolemia. We have accomplished this goal by empowering people with the knowledge of their genetic risk for inherited cancer and familial hypercholesterolemia so they can make informed decisions about their healthcare. 96% of the participants who enrolled in HOP gave consent for genetic testing, suggesting no-cost genetic testing may have been a primary motivator for people to enroll in the study. Similar to other studies, females were more interested in learning about predisposition to disease.7,12 However, unlike other studies that used on-site sample collection,7,34 only 75% of those who consented and requested a kit sent it back, and 95% of those requests for a kit actually resulted in a usable sample in the laboratory (56% of total participants who consented to testing sent in a viable sample), suggesting that the logistics of self-collection and submission may have been a barrier to some participants. The lower use of the collection kit may also have been influenced by the pause and modifications to HOP due to the COVID-19 pandemic (subjects and methods).
Some positive outcomes from this study include participants with positive results obtaining clinical follow-up with healthcare providers to discuss risks and getting increased screening that was applicable to the variant/gene reported. Participants have also discovered the genetic cause of their family history of disease, while others who would otherwise not have undergone traditional genetic testing for inherited cancer or familial hypercholesterolemia have discovered predisposition to disease. Further details of participant outcomes and the outcomes of trial NCT04494945 will be described at the close of the trial, currently expected in early 2025 with at least 22,000 sequenced participants with results returned.
Are RNA studies useful in population screening?
Throughout this project, we identified several variants that were predicted to affect RNA splicing without published functional studies. As past work has demonstrated the usefulness of RNA analysis in reclassification of splice variants in hereditary cancer,35,36 we collected a blood sample and sequenced the RNA of participants with these variants to determine the functional impact. Although this additional work required updates to our IRB protocol and coordinating a blood draw, it was worthwhile since we were able to provide a definitive result to several participants. However, these studies came at additional time and cost, and thus large screening studies incorporating RNA analyses in the future may want to evaluate these factors.
Possible future updates in variant classification
We identified VUSs that may change classification in the future due to recently published studies. One such variant is c.146T>C (p.Ile49Thr [GenBank: NM_000077.5]) in CDKN2A. This variant is found in high frequency specifically in the Latino population at 0.5% in gnomAD.37 Classifications for this variant are mixed in ClinVar (ClinVar: 127523). This variant was considered a VUS due to conflicting data and our high threshold for a P/LP variant in this project. However, recent work suggests this may be a damaging mutation.38 Thus, we have begun a watch list of variants such as this one that are trending toward P/LP. As more information and research is published, more variants like this one may change classification, and so we developed a protocol to return this updated result (and any future ones) to participants. These changes to interpretation demonstrate that a wide consent covering changes in variant interpretation would be useful in genetic health screening projects going forward. Another option would have been to return VUSs to HOP participants with the option to update that classification in the future if they are upgraded. While returning VUSs would allow us to update an existing clinical report rather than having to issue a new positive one after a participant received a negative report, it could also lead to (1) stress to the participant due to the ambiguity and guidelines for VUSs, (2) a dramatic increase in validation and counseling costs, and (3) the likelihood that more than 90%, and perhaps more than 99%, of these VUSs would be later reclassified as benign.
High rate of P/LP variants associated with inherited cancer
Our rate for P/LP variants associated with inherited cancer in HOP participants was 5.0%. This value is higher than what we would have expected based upon previously reported findings.12,30,39 There are several reasons why our rate may be higher than previous reports. First, although we did not separate participants upon enrollment in the study by previous history of cancer, we did specifically target the enrollment of individuals with a prior cancer diagnosis several years into the study. This may have inflated our positive rate compared to the general population. We also recruited HOP participants with a message about cancer risk, and past research has demonstrated that family history of disease increases the likelihood of participation.40 Thus, there is also a possibility that we could have enrolled a large proportion of participants with a strong family history of inherited cancer. A recent study that recruited previous cancer patients or those with a family history while also including the general population in Vietnam found that their rate of P/LP variants in genes associated with inherited cancer was 3.2%. However, they found a carrier frequency of 4.2% in participants with a personal or family history of cancer versus 2.6% without a history of cancer.29 Given that our rate was 5.0%, this might suggest we may have enrolled participants with greater risk than the general population. However, the most important factor for our higher rate is most likely due to the genes we tested and variants we considered reporting. Comparing our results to this Vietnamese study but limiting them to only the 15 genes that overlap both studies (BRCA1, BRCA2, PALB2, PTEN, TP53, CDH1, MLH1, MSH2, MSH6, PMS2, APC, MUTYH, STK11, RB1, RET) reduced our rate to 1.9%. One limitation of this comparison is the fact that the population types differ since HOP is overwhelmingly of White participants. The Alabama Genomic Health Initiative (AGHI) is a screening study in the United States with a population type closer to HOP whose results were also much lower than we reported.12,34 However, if we limit our gene list to ones that overlap AGHI (APC, BMPR1A, BRCA1, BRCA2, MEN1, MLH1, MSH2, MSH6, MUTYH, PMS2, PTEN, RB1, RET, SMAD4, STK11, TP53, TSC1, TSC2), our rate of P/LP variants decreases to 1.7%. There are also limitations to this comparison since AGHI used a genotyping platform of specific variants rather than NGS.34 Since most of our detected P/LP variants were already reported, the impact of this comparison may not be as much as if we identified primarily unreported variants. Additionally, another population study in the United States found a much lower rate of P/LP variants in inherited cancer genes than HOP, but that study focused only on ACMG SFs.39 Importantly, these other studies did not include ATM, CHEK2, or MITF which were the top three genes with identified P/LP variants in HOP, confirming that our high carrier rate is mostly due to gene choice on the HOP panel. Although these aforementioned studies did not include these three genes, a recent screening study included both ATM and CHEK2 on their gene panel,41 demonstrating that at least including these two genes was not unique to HOP.
Although our overall rate for P/LP variants in genes associated with inherited cancer was higher than expected, the rates in specific subtypes were similar to reported results of another population study in the United States. This comparable study performing unbiased population screening, the Healthy Nevada Project (HNP), found an overall rate of P/LP variants in BRCA1/BRCA2 of approximately 1:150 (0.67%), while our overall rate was approximately 1:138 (0.72%). The HNP also screened for Lynch syndrome using the same set of genes as HOP and found an overall rate of P/LP variants in Lynch syndrome genes of approximately 1:340 (0.29%), while our overall rate was approximately 1:258 (0.39%). These differences in rates between our studies were not statistically significant (p = 0.51, p = 0.16, 2 Sample Z test of proportions, respectively). These results demonstrate a consistent prevalence of P/LP variants in the genes associated with BRCA1/BRCA2 HBOC and Lynch syndrome in different geographic locations in the United States.
Interestingly, we found that the gene with the most P/LP variants in Lynch syndrome for HOP was PMS2, while the one with the fewest was MLH1. Although past studies have shown that P/LP variants in MLH1 contribute most to Lynch syndrome,42,43 more recent studies have found that P/LP variants in PMS2 were the most common, while P/LP variants in MLH1 were one of the least common, mirroring our results.7,29 One explanation of these newer findings could be due to the lower penetrance and older age of onset of Lynch syndrome caused by P/LP variants in PMS2 and MSH6,44 suggesting that individuals with P/LP variants in these genes may not have been selected for targeted testing of Lynch syndrome in the past.
Carrier rate for P/LP variants in genes associated with familial hypercholesterolemia
The rate of P/LP variants in LDLR associated with familial hypercholesterolemia in our cohort of Oregonians in HOP was 0.3%. Although there has been some debate on what the most accurate prevalence is for familial hypercholesterolemia,45 the prevalence of P/LP variants in LDLR found in HOP is similar to overall prevalence of familial hypercholesterolemia observed from a recent large systematic review and meta-analysis of 1:311 (0.32%)46 but a bit lower than other published prevalence values of ∼0.5%.47 Other estimates place the prevalence of familial hypercholesterolemia at 1:200–25047 to 1:300–500.48 Even though P/LP variants in LDLR are responsible for the majority of cases of familial hypercholesterolemia with an identified genetic cause,47 we did not test for the two other genes found in familial hypercholesterolemia (APOB and PCSK9). Thus, the prevalence of familial hypercholesterolemia due to P/LP variants in any of these three genes is probably more common in the Oregon population than our genetic testing results indicate. Importantly, some of these studies did not always specifically determine the rate of P/LP variants in genes that cause familial hypercholesterolemia and instead determined overall disease prevalence using several different metrics. Since penetrance of familial hypercholesterolemia caused by genetic variants is not 100%,47 the rate of familial hypercholesterolemia in HOP is most likely not concordant with a participant harboring a P/LP variant in LDLR. Thus, the most accurate comparison would be to a study focused on the carrier rate of P/LP variants that predispose to familial hypercholesterolemia. The HNP studied three genes for familial hypercholesterolemia and found a rate of P/LP variants that cause familial hypercholesterolemia of approximately 1:260. However, limiting their results to only LDLR gives a positive rate of 0.3%, which is statistically similar to our observed results (p = 0.56, 2 Sample Z test of proportions).
ACMG secondary findings
If the HOP NGS panel had contained only genes on the ACMG secondary findings (SF) list, we would have missed identifying 59% of P/LP variants. Additionally, the three genes with the most identified P/LP variants (ATM, CHEK2, MITF) are not part of the ACMG SF list.22 Many of these variants we detected at higher frequency are considered lower/moderate penetrance variants, risk factors, or founder variants.49,50 Including this expanded gene list allowed us to capture a larger percentage of results than typically returned in other studies that only follow the ACMG SF list.39 Even though some of the genes on the HOP panel are not on the ACMG SF list,51 there are National Comprehensive Cancer Network (NCCN) guidelines for management used in the counseling of HOP participants for the majority of these genes (with the exceptions of BAP1, CDK4, MITF, and NBN).
Contact by HOP participants
Several participants contacted us with questions or concerns when they received unexpected results. For example, several participants with negative HOP results had previous clinical diagnostic testing that had reported a variant in a cancer predisposition gene tested by HOP. In almost all of these instances, the reason for the discrepancy was a test limitation such as their previously reported variant being a P/LP CNV or a VUS. However, in some instances their variant was classified as P/LP by another testing laboratory where we classified it as a VUS in HOP due to the high threshold our clinical team set for reporting. While we detected these variants, they were not reported. This conflict led to some discrepancies between participant results and what they obtained from a provider-ordered clinical diagnostic sequencing test. These testing and reporting limitations are reflected in the negative HOP report to participants that states only pathogenic and likely pathogenic SNVs are reported. However, since we have received participant contact regarding these limitations on more than one occasion, this highlights the opportunity to improve communication with participants.
Limitations
One limitation to our study design is the need for a secondary sample for confirmation of the originally identified P/LP variant. The study was designed this way to assure there was no sample mix-up for participants as well as to confirm the NGS finding by an orthogonal method. To date, our confirmation rate has been 100%, indicating that we have never had a clinically relevant sample mix-up with more than 20,000 HOP specimens sequenced in our lab. However, 20% of HOP participants who did not return a secondary sample for confirmation may never know they have increased risk for disease. This is a concern that is currently undergoing discussion to maximize the benefits to HOP participants while following our study protocol. One possible explanation for why participants did not return a second sample is that they were not informed of their initial positive result. There are several reasons why we did not inform them. One factor was our process of recontact, which was performed by a lab staff member who would not have been able to address questions a participant may have about a positive result. Also, early discussions with participant advocacy groups advised against informing the participants of an initial positive result as this could lead to undue stress while waiting for the results of the confirmation. Additionally, since many HOP participants are outside of the OHSU healthcare system, our genetic counselors would not have access to their medical record to add notes and to detail the conversation.
Another limitation was that we did not report variants with skewed variant allele frequencies outside of the range considered germline.52 There are several possibilities in what these variants could be, including germline, mosaic, somatic, and clonal hematopoiesis of indeterminate potential (CHIP), among others.53 Not reporting these types of variants is not unusual for a large screening study; however, this gives the opportunity for follow-up work that will study these as a separate research project. A final test limitation is that we did not report CNVs. This limitation meant that we would not detect a portion of inherited cancer risk alleles resulting from these types of variants. This was a tradeoff in developing a low-cost test that could efficiently identify most single-nucleotide variants (SNVs) and small indels. This limitation may have an impact on the healthcare of HOP participants and their health outcomes since this test will not return these types of variants. Thus, this screening test would not be the choice for individuals at high risk of inherited cancer, but instead for the general population with average risk.
Another current limitation of HOP is the lack of diversity. Although our mostly White population is similar to the breakdown of the population type in the state of Oregon (US Census Bureau, 2016, https://data.census.gov/table?g=040XX00US41&tid=ACSDP1Y2016.DP05), we are working to increase diversity in the study. The use of a smartphone app may have affected diversity, especially among elderly individuals or those without technical knowledge or possessing a smartphone. We tried to make the app widely available on both Android and Apple iOS to reach as many interested individuals as possible. We are also discussing the possible use of a web-based system in the future to further increase our enrollment efforts across all population groups.
Successful low-cost screening test for inherited cancer and familial hypercholesterolemia
Our results from the implementation of a statewide low-cost population screening test in an academic clinical diagnostic laboratory suggest that it can be a successful model for other states with similar resources. As the only academic medical research institute in Oregon with clinical genetic testing expertise and facilities, it was ideal for OHSU to provide Oregonians the opportunity to learn more about their genetic risks. However, the section of our clinical laboratory that provides testing for inherited disorders is relatively small (5 FTEs) and thus had to undergo significant reorganization to provide high volume testing as required in a population screening project. Part of the success of this study was due to the implementation of automation and robotics that has been previously highlighted,54,55 which resulted in strong benefits including increased efficiency of processing large numbers of samples, increased precision and accuracy, decreased costs per sample, and reduced manual workload with increased ability to multi-task. Robotic automation for library preparation saved us 4–5 manual hours per 96-well plate and allowed one technologist to set up and run four 96-well plates per week by offloading several steps of the library preparation to the Janus workstation robots (Figure S3). Robotic automation for DNA extraction saved additional technician hours per week and was essential for our lab to process large volumes of samples. These changes allowed our laboratory to increase our testing capacity by 10× to more than 1,000 samples per month. Additionally, with a low failure rate of <1%, we can easily expand this project beyond our current capacity.
In summary, HOP is now in its fifth year, and we have learned much about our study design, what has worked well, and what can be improved going forward. The use of a HIPAA-compliant app, combined with an easily accessible sample collection approach (mouthwash), has allowed us to impact the health of more than 20,000 Oregonians. Other factors influencing the success of HOP included the introduction of robotics within our clinical laboratory, the use of the OHSU core laboratory to perform DNA extraction, efficient assay design and modifications to our NGS pipeline that allowed expedited review, and the decision not to report VUSs. However, these successes have come with challenges. The greatest challenge in the study design of HOP is the requirement of a second sample for confirmation of results. It is concerning that 20% of participants with a positive result did not submit a secondary sample, and therefore did not receive a result. This suggests a need for improved education and communication with participants that might result in a better rate of return for the secondary sample. There are a number of subtleties that participants do not understand and which lead to opportunities for improved education in areas such as genes and variant types that are being sequenced and reported for the HOP panel as well as the clinical sensitivity of the test and what it can and cannot detect. Despite these challenges, Oregonians (more than 500 within the first three years of HOP) have directly benefited by learning about their increased risk of disease. HOP is continuing to enroll and expand; the next iteration of the current project includes four additional genes (SDHB [MIM: 185470], SDHC [MIM: 602413], SDHD [MIM: 602690], and VHL [MIM: 608537]) that will enable the project to continue to be a strong component in the overall health and wellbeing of individuals in the state of Oregon.
Acknowledgments
We thank HOP participants. We also thank members of the clinical laboratory including Travis Hayes and Shelbie Haefliger, support staff for our genomic database including William Moore, Charles Dahl and Jay Pleyte, and HOP members Madeleine Matheis, Kami Chiotti, Brittany Daughtry, Aaron Caughey, and Julia Bainbridge. This work was funded by a Cancer Early Detection Advanced Research Center, Knight Cancer Institute (CEDAR) award (Full 2023-1729) and an NIH Moonshot grant (1U01CA232819-01A1) to Jackilen Shannon and Paul Spellman. The Let’s Get Healthy! platform used for survey data collection within the HOP app was developed with the Oregon Clinical and Translational Research Institute (OCTRI; 1UL1TR002369) through funding from the National Institutes of Health (NIH), including Science Education Partnership Awards (R25OD01496, R25GM129840) and infrastructure developed by NIH grants R25RR020443-05S1, UL1RR024140-04S3, RR026008, 3P30CA-69553-13S9, and UL1TR002369.
Declaration of interests
C.S.R. is a Director on the American Board of Genetics and Genomics. P.T.S. has the following conflicts: employee of OHSU, consultant to Natera, Twinstrand, Foresight Diagnostics, and Lab Corp, and equity position in Convergent Genomics. T.D.O., A.B.P., C.C.D., G.G., J.H.L., S. McCabe, A.K., R.W., J.T., and C.S.R. are employees of the Knight Diagnostic Laboratories (KDL).
Published: July 27, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2023.06.014.
Supplemental information
Data and code availability
All P/LP variants identified have been provided in Table S3. Additional individual sequencing level data are not available due to privacy of HOP participants.
References
- 1.Zhang L., Bao Y., Riaz M., Tiller J., Liew D., Zhuang X., Amor D.J., Huq A., Petelin L., Nelson M., et al. Population genomic screening of all young adults in a health-care system: a cost-effectiveness analysis. Genet. Med. 2019;21:1958–1968. doi: 10.1038/s41436-019-0457-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manchanda R., Patel S., Gordeev V.S., Antoniou A.C., Smith S., Lee A., Hopper J.L., MacInnis R.J., Turnbull C., Ramus S.J., et al. Cost-effectiveness of Population-Based BRCA1, BRCA2, RAD51C, RAD51D, BRIP1, PALB2 Mutation Testing in Unselected General Population Women. J. Natl. Cancer Inst. 2018;110:714–725. doi: 10.1093/jnci/djx265. [DOI] [PubMed] [Google Scholar]
- 3.Buchanan A.H., Manickam K., Meyer M.N., Wagner J.K., Hallquist M.L.G., Williams J.L., Rahm A.K., Williams M.S., Chen Z.M.E., Shah C.K., et al. Early cancer diagnoses through BRCA1/2 screening of unselected adult biobank participants. Genet. Med. 2018;20:554–558. doi: 10.1038/gim.2017.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daly M.B., Pilarski R., Yurgelun M.B., Berry M.P., Buys S.S., Dickson P., Domchek S.M., Elkhanany A., Friedman S., Garber J.E., et al. NCCN Guidelines Insights: Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic, Version 1.2020. J. Natl. Compr. Canc. Netw. 2020;18:380–391. doi: 10.6004/jnccn.2020.0017. [DOI] [PubMed] [Google Scholar]
- 5.Germain D.P., Moiseev S., Suárez-Obando F., Al Ismaili F., Al Khawaja H., Altarescu G., Barreto F.C., Haddoum F., Hadipour F., Maksimova I., et al. The benefits and challenges of family genetic testing in rare genetic diseases-lessons from Fabry disease. Mol. Genet. Genomic Med. 2021;9:e1666. doi: 10.1002/mgg3.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swink A., Nair A., Hoof P., Matthews A., Burden C., Johnson K., Blum J.L. Barriers to the utilization of genetic testing and genetic counseling in patients with suspected hereditary breast and ovarian cancers. SAVE Proc. 2019;32:340–344. doi: 10.1080/08998280.2019.1612702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grzymski J.J., Elhanan G., Morales Rosado J.A., Smith E., Schlauch K.A., Read R., Rowan C., Slotnick N., Dabe S., Metcalf W.J., et al. Population genetic screening efficiently identifies carriers of autosomal dominant diseases. Nat. Med. 2020;26:1235–1239. doi: 10.1038/s41591-020-0982-5. [DOI] [PubMed] [Google Scholar]
- 8.The All of Us Research Program Investigators. Denny J.C., Rutter J.L., Goldstein D.B., Philippakis A., Smoller J.W., Jenkins G., Dishman E. The "All of Us" Research Program. N. Engl. J. Med. 2019;381:668–676. doi: 10.1056/NEJMsr1809937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wiesner G.L., Kulchak Rahm A., Appelbaum P., Aufox S., Bland S.T., Blout C.L., Christensen K.D., Chung W.K., Clayton E.W., Green R.C., et al. Returning Results in the Genomic Era: Initial Experiences of the eMERGE Network. J. Pers. Med. 2020;10 doi: 10.3390/jpm10020030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.eMERGE Consortium Harmonizing Clinical Sequencing and Interpretation for the eMERGE III Network. Am. J. Hum. Genet. 2019;105:588–605. doi: 10.1016/j.ajhg.2019.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agus D.B., Jaffee E.M., Van Dang C. Cancer Moonshot 2.0. Lancet Oncol. 2021;22:164–165. doi: 10.1016/S1470-2045(21)00003-6. [DOI] [PubMed] [Google Scholar]
- 12.East K.M., Kelley W.V., Cannon A., Cochran M.E., Moss I.P., May T., Nakano-Okuno M., Sodeke S.O., Edberg J.C., Cimino J.J., et al. A state-based approach to genomics for rare disease and population screening. Genet. Med. 2021;23:777–781. doi: 10.1038/s41436-020-01034-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marriott L.K., Cameron W.E., Purnell J.Q., Cetola S., Ito M.K., Williams C.D., Newcomb K.C., Randall J.A., Messenger W.B., Lipus A.C., Shannon J. Let's Get Healthy! Health awareness through public participation in an education and research exhibit. Prog. Community Health Partnersh. 2012;6:331–337. doi: 10.1353/cpr.2012.0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W., et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. j. 2011;17:10–11. [Google Scholar]
- 16.Smith T., Heger A., Sudbery I. UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 2017;27:491–499. doi: 10.1101/gr.209601.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stenson P.D., Ball E.V., Mort M., Phillips A.D., Shiel J.A., Thomas N.S.T., Abeysinghe S., Krawczak M., Cooper D.N. Human Gene Mutation Database (HGMD): 2003 update. Hum. Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 19.Cingolani P., Platts A., Wang L.L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng S.B., Turner E.H., Robertson P.D., Flygare S.D., Bigham A.W., Lee C., Shaffer T., Wong M., Bhattacharjee A., Eichler E.E., et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sturm A.C., Knowles J.W., Gidding S.S., Ahmad Z.S., Ahmed C.D., Ballantyne C.M., Baum S.J., Bourbon M., Carrié A., Cuchel M., et al. Clinical Genetic Testing for Familial Hypercholesterolemia: JACC Scientific Expert Panel. J. Am. Coll. Cardiol. 2018;72:662–680. doi: 10.1016/j.jacc.2018.05.044. [DOI] [PubMed] [Google Scholar]
- 22.Miller D.T., Lee K., Chung W.K., Gordon A.S., Herman G.E., Klein T.E., Stewart D.R., Amendola L.M., Adelman K., Bale S.J., et al. ACMG SF v3.0 list for reporting of secondary findings in clinical exome and genome sequencing: a policy statement of the American College of Medical Genetics and Genomics (ACMG) Genet. Med. 2021;23:1381–1390. doi: 10.1038/s41436-021-01172-3. [DOI] [PubMed] [Google Scholar]
- 23.Suszynska M., Ratajska M., Kozlowski P. BRIP1, RAD51C, and RAD51D mutations are associated with high susceptibility to ovarian cancer: mutation prevalence and precise risk estimates based on a pooled analysis of ∼30,000 cases. J. Ovarian Res. 2020;13:50. doi: 10.1186/s13048-020-00654-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoffel E.M., Murphy C.C. Epidemiology and Mechanisms of the Increasing Incidence of Colon and Rectal Cancers in Young Adults. Gastroenterology. 2020;158:341–353. doi: 10.1053/j.gastro.2019.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orphanet: An Online Database of Rare Diseases and Orphan Drugs. www.orpha.net
- 26.Yun J., Li Y., Xu C.T., Pan B.R. Epidemiology and Rb1 gene of retinoblastoma. Int. J. Ophthalmol. 2011;4:103–109. doi: 10.3980/j.issn.2222-3959.2011.01.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hill J.T., Demarest B.L., Bisgrove B.W., Su Y.C., Smith M., Yost H.J. Poly peak parser: Method and software for identification of unknown indels using sanger sequencing of polymerase chain reaction products. Dev. Dyn. 2014;243:1632–1636. doi: 10.1002/dvdy.24183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tran V.T., Nguyen S.T., Pham X.D., Phan T.H., Nguyen V.C., Nguyen H.T., Nguyen H.P., Doan P.T.T., Le T.A., Nguyen B.T., et al. Pathogenic Variant Profile of Hereditary Cancer Syndromes in a Vietnamese Cohort. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.789659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu C., Hart S.N., Gnanaolivu R., Huang H., Lee K.Y., Na J., Gao C., Lilyquist J., Yadav S., Boddicker N.J., et al. A Population-Based Study of Genes Previously Implicated in Breast Cancer. N. Engl. J. Med. 2021;384:440–451. doi: 10.1056/NEJMoa2005936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boursi B., Sella T., Liberman E., Shapira S., David M., Kazanov D., Arber N., Kraus S. The APC p.I1307K polymorphism is a significant risk factor for CRC in average risk Ashkenazi Jews. Eur. J. Cancer. 2013;49:3680–3685. doi: 10.1016/j.ejca.2013.06.040. [DOI] [PubMed] [Google Scholar]
- 32.CHEK2 Breast Cancer Case-Control Consortium CHEK2∗1100delC and susceptibility to breast cancer: a collaborative analysis involving 10,860 breast cancer cases and 9,065 controls from 10 studies. Am. J. Hum. Genet. 2004;74:1175–1182. doi: 10.1086/421251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muranen T.A., Greco D., Blomqvist C., Aittomäki K., Khan S., Hogervorst F., Verhoef S., Pharoah P.D.P., Dunning A.M., Shah M., et al. Genetic modifiers of CHEK2∗1100delC-associated breast cancer risk. Genet. Med. 2017;19:599–603. doi: 10.1038/gim.2016.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bowling K.M., Thompson M.L., Gray D.E., Lawlor J.M.J., Williams K., East K.M., Kelley W.V., Moss I.P., Absher D.M., Partridge E.C., et al. Identifying rare, medically relevant variation via population-based genomic screening in Alabama: opportunities and pitfalls. Genet. Med. 2021;23:280–288. doi: 10.1038/s41436-020-00976-z. [DOI] [PubMed] [Google Scholar]
- 35.Agiannitopoulos K., Pepe G., Papadopoulou E., Tsaousis G.N., Kampouri S., Maravelaki S., Fassas A., Christodoulou C., Iosifidou R., Karageorgopoulou S., et al. Clinical Utility of Functional RNA Analysis for the Reclassification of Splicing Gene Variants in Hereditary Cancer. Cancer Genomics Proteomics. 2021;18:285–294. doi: 10.21873/cgp.20259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Truty R., Ouyang K., Rojahn S., Garcia S., Colavin A., Hamlington B., Freivogel M., Nussbaum R.L., Nykamp K., Aradhya S. Spectrum of splicing variants in disease genes and the ability of RNA analysis to reduce uncertainty in clinical interpretation. Am. J. Hum. Genet. 2021;108:696–708. doi: 10.1016/j.ajhg.2021.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kimura H., Paranal R.M., Nanda N., Wood L.D., Eshleman J.R., Hruban R.H., Goggins M.G., Klein A.P., Familial Pancreatic Cancer Genome Sequencing P., Roberts N.J. Functional CDKN2A assay identifies frequent deleterious alleles misclassified as variants of uncertain significance. Elife. 2022;11:e71137. doi: 10.7554/eLife.71137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Natarajan P., Gold N.B., Bick A.G., McLaughlin H., Kraft P., Rehm H.L., Peloso G.M., Wilson J.G., Correa A., Seidman J.G., et al. Aggregate penetrance of genomic variants for actionable disorders in European and African Americans. Sci. Transl. Med. 2016;8:364ra151. doi: 10.1126/scitranslmed.aag2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts L.W., Kim J.P. Receptiveness to participation in genetic research: A pilot study comparing views of people with depression, diabetes, or no illness. J. Psychiatr. Res. 2017;94:156–162. doi: 10.1016/j.jpsychires.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rao N.D., Kaganovsky J., Malouf E.A., Coe S., Huey J., Tsinajinne D., Hassan S., King K.M., Fullerton S.M., Chen A.T., Shirts B.H. Diagnostic yield of genetic screening in a diverse, community-ascertained cohort. Genome Med. 2023;15:26. doi: 10.1186/s13073-023-01174-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jasperson K.W., Tuohy T.M., Neklason D.W., Burt R.W. Hereditary and familial colon cancer. Gastroenterology. 2010;138:2044–2058. doi: 10.1053/j.gastro.2010.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hegde M., Ferber M., Mao R., Samowitz W., Ganguly A., Working Group of the American College of Medical Genetics and Genomics ACMG Laboratory Quality Assurance Committee ACMG technical standards and guidelines for genetic testing for inherited colorectal cancer (Lynch syndrome, familial adenomatous polyposis, and MYH-associated polyposis) Genet. Med. 2014;16:101–116. doi: 10.1038/gim.2013.166. [DOI] [PubMed] [Google Scholar]
- 44.Idos G., Valle L. In: GeneReviews. R)), Adam M.P., Everman D.B., Mirzaa G.M., Pagon R.A., Wallace S.E., Bean L.J.H., Gripp K.W., Amemiya A., editors. 1993. Lynch Syndrome. [Google Scholar]
- 45.Brunham L.R., Hegele R.A. What Is the Prevalence of Familial Hypercholesterolemia? Arterioscler. Thromb. Vasc. Biol. 2021;41:2629–2631. doi: 10.1161/ATVBAHA.121.316862. [DOI] [PubMed] [Google Scholar]
- 46.Hu P., Dharmayat K.I., Stevens C.A.T., Sharabiani M.T.A., Jones R.S., Watts G.F., Genest J., Ray K.K., Vallejo-Vaz A.J. Prevalence of Familial Hypercholesterolemia Among the General Population and Patients With Atherosclerotic Cardiovascular Disease: A Systematic Review and Meta-Analysis. Circulation. 2020;141:1742–1759. doi: 10.1161/CIRCULATIONAHA.119.044795. [DOI] [PubMed] [Google Scholar]
- 47.Youngblom E., Pariani M., Knowles J.W. In: Familial Hypercholesterolemia. GeneReviews((R)), Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J.H., Gripp K.W., Mirzaa G.M., Amemiya A., editors. 1993. [Google Scholar]
- 48.Hopkins P.N., Toth P.P., Ballantyne C.M., Rader D.J., National Lipid Association Expert Panel on Familial Hypercholesterolemia Familial hypercholesterolemias: prevalence, genetics, diagnosis and screening recommendations from the National Lipid Association Expert Panel on Familial Hypercholesterolemia. J. Clin. Lipidol. 2011;5:S9–S17. doi: 10.1016/j.jacl.2011.03.452. [DOI] [PubMed] [Google Scholar]
- 49.Bychkovsky B.L., Agaoglu N.B., Horton C., Zhou J., Yussuf A., Hemyari P., Richardson M.E., Young C., LaDuca H., McGuinness D.L., et al. Differences in Cancer Phenotypes Among Frequent CHEK2 Variants and Implications for Clinical Care-Checking CHEK2. JAMA Oncol. 2022;8:1598–1606. doi: 10.1001/jamaoncol.2022.4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yokoyama S., Woods S.L., Boyle G.M., Aoude L.G., MacGregor S., Zismann V., Gartside M., Cust A.E., Haq R., Harland M., et al. A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma. Nature. 2011;480:99–103. doi: 10.1038/nature10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller D.T., Lee K., Gordon A.S., Amendola L.M., Adelman K., Bale S.J., Chung W.K., Gollob M.H., Harrison S.M., Herman G.E., et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2021 update: a policy statement of the American College of Medical Genetics and Genomics (ACMG) Genet. Med. 2021;23:1391–1398. doi: 10.1038/s41436-021-01171-4. [DOI] [PubMed] [Google Scholar]
- 52.DeLeonardis K., Hogan L., Cannistra S.A., Rangachari D., Tung N. When Should Tumor Genomic Profiling Prompt Consideration of Germline Testing? J. Oncol. Pract. 2019;15:465–473. doi: 10.1200/JOP.19.00201. [DOI] [PubMed] [Google Scholar]
- 53.Maani N., Panabaker K., McCuaig J.M., Buckley K., Semotiuk K., Farncombe K.M., Ainsworth P., Panchal S., Sadikovic B., Armel S.R., et al. Incidental findings from cancer next generation sequencing panels. NPJ Genom. Med. 2021;6:63. doi: 10.1038/s41525-021-00224-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hess J.F., Kohl T.A., Kotrová M., Rönsch K., Paprotka T., Mohr V., Hutzenlaub T., Brüggemann M., Zengerle R., Niemann S., Paust N. Library preparation for next generation sequencing: A review of automation strategies. Biotechnol. Adv. 2020;41 doi: 10.1016/j.biotechadv.2020.107537. [DOI] [PubMed] [Google Scholar]
- 55.Tegally H., San J.E., Giandhari J., de Oliveira T. Unlocking the efficiency of genomics laboratories with robotic liquid-handling. BMC Genom. 2020;21:729. doi: 10.1186/s12864-020-07137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All P/LP variants identified have been provided in Table S3. Additional individual sequencing level data are not available due to privacy of HOP participants.