Abstract
Bdellovibrio bacteriovorus is a predatory bacterium preying upon Gram-negative bacteria. As such, B. bacteriovorus has the potential to control antibiotic-resistant pathogens and biofilm populations. To survive and reproduce, B. bacteriovorus must locate and infect a host cell. However, in the temporary absence of prey, it is largely unknown how B. bacteriovorus modulate their motility patterns in response to physical or chemical environmental cues to optimize their energy expenditure. To investigate B. bacteriovorus’ predation strategy, we track and quantify their motion by measuring speed distributions as a function of starvation time. While an initial unimodal speed distribution relaxing to one for pure diffusion at long times may be expected, instead we observe a bimodal speed distribution with one mode centered around that expected from diffusion and the other centered at higher speeds. What is more, for an increasing amount of time over which B. bacteriovorus is starved, we observe a progressive reweighting from the active swimming state to an apparent diffusive state in the speed distribution. Distributions of trajectory-averaged speeds for B. bacteriovorus are largely unimodal, indicating switching between a faster swim speed and an apparent diffusive state within individual observed trajectories rather than there being distinct active swimming and apparent diffusive populations. We also find that B. bacteriovorus’ apparent diffusive state is not merely caused by the diffusion of inviable bacteria as subsequent spiking experiments show that bacteria can be resuscitated and bimodality restored. Indeed, starved B. bacteriovorus may modulate the frequency and duration of active swimming as a means of balancing energy consumption and procurement. Our results thus point to a reweighting of the swimming frequency on a trajectory basis rather than a population level basis.
Significance
Bdellovibrio bacteriovorus is a predatory bacterium that may help control Gram-negative bacterial populations in environmental and clinical settings. To locate its prey in solution, B. bacteriovorus must expend energy to fight hydrodynamic drag. This raises the question as to how B. bacteriovorus should expend its energy reserves in the absence of chemical cues from its prey. Here, we show that B. bacteriovorus adapts its motility to minimize energy expenditure (due to fighting drag in swimming) upon prolonged starvation by exploiting two motility states.
Introduction
Bacterial cues drawn from the environment—mediated through a number of factors such as hydrodynamic interactions (1,2), external flows (3,4,5), or other direct chemical signaling and quorum sensing (6,7,8,9,10,11,12)—are critical in understanding cell-cell interaction and emergent collective behaviors of biomedical interest (7,13,14) such as biofilm formation (15) and bacterial swarming (16,17).
Here, our focus is in understanding the hunting strategy of predatory bacteria, such as Bdellovibrio and like organisms (sometimes called BALOs), driven by external resources. Understanding such strategies is helpful in exploiting Bdellovibrio and like organisms for a number of tasks including the degradation of hazardous microbial biofilms on surfaces (15,18,19,20,21), purifying water and soil (22,23,24), and serving as a “living antibiotic” (14,25,26) by reducing pulmonary bacterial infections in rats (27), gut infections in poultry (28), corneal infections in rabbits (29), and infections in other animal models such as zebrafish (30). Indeed, the bacterial predator, Bdellovibrio bacteriovorus, has been shown to significantly decrease populations of many other species of Gram-negative bacteria (31,32) across habitats (14,15,18,19,22,23,24,28,29,30,33,34).
The biphasic life cycle of B. bacteriovorus involves an attack phase in which predatory B. bacteriovorus locate their bacterial prey and enter the periplasm where they complete the replicative growth phase, ending with the progeny lysing from the host (35,36,37,38,39,40,41). Although B. bacteriovorus was discovered half a century ago (42,43,44) and is now studied as a model predator, it is largely unknown how B. bacteriovorus locate individual prey. For instance, previous studies suggested that B. bacteriovorus bumps into its prey at random (19,22,34,38,45,46,47,48).
In previous work, we revisited this hypothesis (2) and investigated the role hydrodynamics plays in B. bacteriovorus’ hunting strategy. Our own previous efforts built upon recent literature suggesting that bacteria sense and respond to environmental hydrodynamic flows (3,4,5) and self-generated flows (1,49,50,51,52,53,54). In particular, we found that, on account of B. bacteriovorus’ unusually high speed, passive hydrodynamic forces could drive it toward surfaces where prey tend to be present in larger numbers (2), thereby improving the odds of a chance collision.
This finding raises the following question: given that hydrodynamic forces induced by B. bacteriovorus’ unusually high speeds are critical in bringing it toward prey, how might B. bacteriovorus, in the absence of prey in its environment, allocate its energy reserves to overcome hydrodynamic drag as it propels itself through solution in search of prey? Naively, one may expect a unimodal speed distribution at high speeds across a trajectory to shift to a unimodal speed distribution at longer times to coincide with a peak around that expected for pure diffusion. On the other hand, prey bacteria such as E. coli are known to modulate swimming behavior based on environmental cues (55,56) (mainly the presence of predator bacteria) so it does not seem unreasonable to expect the predator to modulate its swimming behavior in the presence of prey.
A naive search strategy where active motility persists until the available energy—used to maintain metabolic homeostasis, as well as overcoming both translational and rotational hydrodynamic drag—is expended may suggest that B. bacteriovorus would not be revivable once energy resources are depleted and, possibly, that its proteome might be catabolized in an effort to locate prey by active swimming. On the other hand, an alternative use of energy may involve the active modulation of motility as a function of both internal energy reserves (i.e., age) and environmental cues (i.e., potential sources of energy and nutrients) before reaching a point of no return, at which point B. bacteriovorus would no longer have the reserves needed to fight hydrodynamic drag in search of prey despite external cues suggesting its availability. We partly address this question by monitoring changes in large numbers of B. bacteriovorus trajectories under starvation conditions as a function of age and buffer conditions. We found that key dynamical features—instantaneous and trajectory-averaged speed distributions—monitored over the course of 40 h may suggest behavior consistent with what can described as an exploitative search strategy.
We corroborated these results by subsequently performing spiking experiments in which starved bacteria are resuspended in rich media, showing that B. bacteriovorus can be resuscitated and their active motility restored. Our results help shed light on the compromises B. bacteriovorus must make when hunting in the absence of environmental cues and how switching between an active state and an apparent diffusive state can be used to cope with unfavorable environmental conditions.
Materials and methods
Bacterial strains and growth conditions
To avoid any possible variations in the motile behavior of B. bacteriovorus due to expression of a fluorescent protein, the wild-type strain 109 (Bdellovibrio bacteriovorus Stolp and Starr, ATTC no. 15143; American Type Culture Collection, Manassas, VA) was used to carry out these studies. Escherichia coli strain OP50 which was grown in lysogeny broth (LB) (10 g/L yeast extract, 20 g/L tryptone, 20 g/L sodium chloride [pH 7.5]) at 37°C on a 300-rpm shaker was used as B. bacteriovorus’ prey. After multiple washes with HEPES medium, the OD (600 nm) was taken of the E. coli and the bacteria were stored at 4°C until use for synchronization cultures as described below.
Synchronization of B. bacteriovorus
Modifications to the synchronization protocols outlined in the literature (7,57) allow for the synchronized observation of many B. bacteriovorus of the same age. This was achieved by growing B. bacteriovorus in nutrient-poor medium (HEPES medium [HM]) (25 mM HEPES [pH 7.4], 3 mM CaCl2, 2 mM MgCl2) containing approximately 108 (OD 1) E. coli cells overnight at 28°C on a 300-rpm shaker. The next day, B. bacteriovorus was isolated by centrifugation at 7900 rpm for 30 min at 4°C. The predators were then resuspended in fresh HM buffer containing E. coli cells at 0.7 OD. An hour after introduction to fresh E. coli, the formed bdelloplasts were isolated and resuspended in fresh medium several times via centrifugation (2000 rpm for 10 min at room temperature). This eliminated the majority of leftover predators. After an additional 4 h, the lysed progeny were collected by passing the culture through a 0.45-μm Millex HP syringe filter twice. The isolated B. bacteriovorus remained in nutrient-poor medium to begin starvation conditions.
The effectiveness of the isolation method was tested by passing an E. coli OP50 culture of OD 1 through the filter and adding nutrient-rich medium. The culture was monitored for several days, with no bacteria growing, showing that E. coli was effectively removed by filtration.
Starvation of B. bacteriovorus experiments
The synchronized B. bacteriovorus was obtained as described above. The culture remained shaking at 28°C on a 300-rpm shaker during the total duration of the experiment (approximately 48 h). A small aliquot was taken from the culture every hour for sample preparation and data acquisition described below.
Spiking of B. bacteriovorus with rich medium experiments
The B. bacteriovorus was synchronized as described above. Four hours after the culture was filtered, the culture was divided into three 2-mL total volumes: a control, and two cultures that were spiked with rich medium. The B. bacteriovorus was spiked with nutrient-rich medium, LB (10 g/L yeast extract, 20 g/L tryptone, 20 g/L NaCl [pH 7.5]) at hour 4 and hour 20 after filtration. The cultures that were spiked had 1.75 mL of B. bacteriovorus added and 0.25 mL of rich medium. The cultures remained shaking at 28°C on a 300-rpm shaker during the total duration of the experiment. Small aliquots were taken from the three cultures for measurements.
Memory of B. bacteriovorus experiments
The B. bacteriovorus was synchronized according to synchronization of B. bacteriovorus. Six hours after filtration, the culture was divided into three 2-mL total volumes: a control, and two cultures that were spiked with rich medium at hour 6. The cultures that were spiked had 1.75 mL of B. bacteriovorus added and 0.25 mL of rich medium. The cultures were observed 15 min after exposure. To initiate starvation conditions again, all three cultures were centrifuged (30 min after initial exposure time) at 13,000 rpm for 4 min at room temperature. The control culture and one of the spiked cultures were resuspended in 2 mL fresh HM. The other spiked culture was resuspended in 1.75 mL of HM and 0.25 mL of LB. (58). The cultures remained shaking at 28°C on a 300-rpm shaker during the total duration of the experiment. Small aliquots were taken from the three cultures for measurements. The effects of centrifugation can be seen in Fig. S2.
Sample preparation
A small volume (10 μL) of the culture was aliquoted onto a microscope coverslip containing vacuum grease along the outer edge and secured with a slide. Focusing on the midplane (approximately 70 μm from the surface) allowed for the observation of the bacteria without restricting their movement due to a confined vertical depth (2).
Data acquisition
Using an inverted phase contrast microscope (Nikon, Melville, NY), the bacteria were imaged with a 1024 × 1024 pixel ROI using a 60× (1.4 NA) oil immersion objective. Each hour data were collected by recording six 20-s videos at 8 ms exposure using a Hamamatsu (Shizuoka, Japan) ORCA-Flash4.0 3V sCMOS camera. These videos were then processed to obtain velocity distributions over thousands of bacterial trajectories as described in data analysis.
Data analysis
The bacteria in the images are clearly visible by eye and could in principle be tracked by hand (Fig. S3). However, due to the volume of data, we automated tracking. The motility of the bacteria was tracked by using a package for feature detection, TrackPy (59), which provides the ability to locate features (e.g., bacteria) in raw image data and link those features across frames into trajectories. Two of the main parameters used for locating features were the diameter and minmass. The diameter is determined based on the size of the object tracked and the minmass is the minimum integrated brightness used to eliminate spurious features. As B. bacteriovorus is about 1 μm long (i.e., 11 pixels), we conservatively chose an all-inclusive 15-pixel diameter. The minmass varies between runs given the overall brightness of the video. Changes in this value may affect the overall number of bacteria tracked within the focal plane, but would not influence conclusions drawn on the overall motility of individual bacteria; i.e., it only reduces the statistics available. To link bacteria across frames, the search range was consistently set to 20 pixels as the bacteria are unlikely to travel further than this number of pixels between consecutive frames (19). As bacteria move in and out of focus, there is a sampling bias in speed measurements at the start and end of trajectories toward slow speeds (i.e., a bias toward a larger velocity component orthogonal to the focal plane). Removing the first and last two frames of all trajectories was sufficient to mitigate this bias (Fig. S4; Table S1). Also for this reason, we ignored any traces shorter than 10 consecutive frames.
Window averaging of the bacterium’s position was calculated to more accurately track its movement. The effect of different windows can be seen in Fig. S5. Based on these results, a window of three was used for all bacteria located. As windowing essentially decreases the number of original data points, we also tested downsampling (decimating) the data to assure ourselves that the shape of the distributions did not greatly vary (Fig. S6).
Once each trajectory is recorded as a time series of two-dimensional coordinates, the distance the feature moves between each consecutive frame and the exposure time of the camera are used to calculate the instantaneous speeds. A central difference of the positions with respect to time was used to get a more accurate speed estimate. The average speed for each trajectory was obtained by taking the average of the instantaneous speeds of each bacterium’s trajectory along the whole trajectory.
For robustness, we compared the above tracking method with a method leveraging neural networks to localize the bacteria, and parameter-free Gaussian process regression to obtain velocities at each frame (see supporting material). In brief, key results uncovered in the speed histograms (namely bimodality, discussed shortly) (Figs. 1 and 2), were recovered using the parameter-free analysis, further suggesting that the bimodality is a feature of the data and not a tracking artifact.
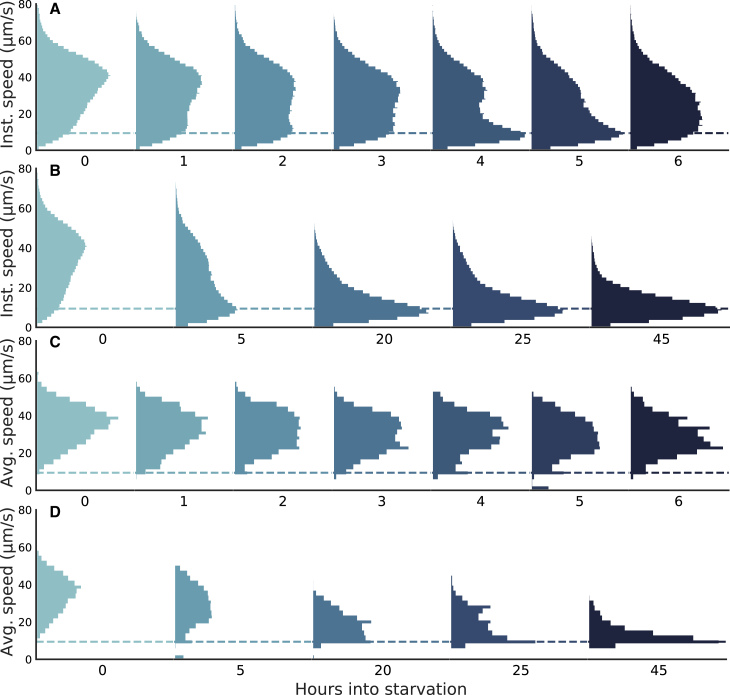
Figure 1.
Under starvation conditions, the instantaneous and average speed distributions of B. bacteriovorus shift across time. (A) Instantaneous speed distributions are bimodal with a substantial shift toward an apparent diffusive state over the first 6 h. (B) Beyond hour 6 during starvation, the instantaneous speed distribution relaxes to a unimodal distribution about the expected diffusive speed for a Brownian particle of B. bacteriovorus’ size (9.40 μm s−1, dashed lines). (C) Distributions of speeds averaged over individual trajectories are largely unimodal over the first 6 h. (D) Average speed distributions over 45 h eventually relax to the expected diffusive speed. The sampling statistics for each histogram can be found in supporting material, sampling statistics for main text figures.
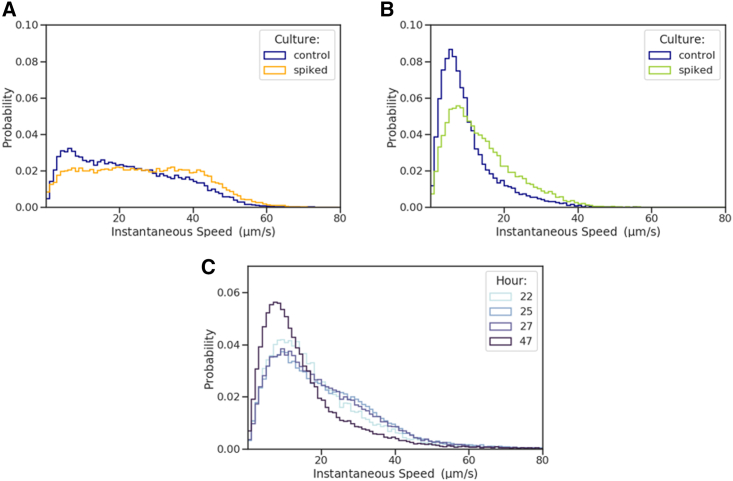
Figure 2.
After addition of LB, B. bacteriovorus begins to swim more frequently even after long starvation times. We compare the instantaneous speed distributions of our control, starving B. bacteriovorus (blue), to that of cultures spiked with LB at hour 4 (orange) and hour 20 (green). Both spiked cultures were observed after 2 h of exposure. (A) After 2 h of exposure, the culture spiked at hour 4 (orange) exhibits a small repopulation of the active swimming speed peak. (B) After 2 h of exposure, the culture spiked at hour 20 (green) has faster speeds than the control (blue). (C) The culture spiked at hour 20 can be seen after several hours of exposure with negligible difference (21,22,23,24,25,26), indicating that, after several hours of exposure, the nutrients are in excess. The sampling statistics for each histogram can be found in supporting material, sampling statistics for main text figures.
Results
To see how B. bacteriovorus adapts to environmental cues, we first observed the swimming behavior of synchronized B. bacteriovorus in the absence of prey and nutrients. The bacterial trajectories after different periods of starvation can be seen in Fig. 3, in which the normalized speeds are plotted in blue. Under such starvation conditions, we monitored the instantaneous speed of each bacterium at each frame as well as the average speed of each bacterium along its trajectory (Fig. 1). The instantaneous speed is obtained using the smoothing algorithm described in the materials and methods. The average speed of each bacterium is simply the average of the instantaneous speeds for the bacterium. We uncover a reproducible bimodal instantaneous speed distribution with peaks at roughly 10 and 40 μm s−1 with the relative weight of each peak being sensitive to B. bacteriovorus’ age and buffer conditions.
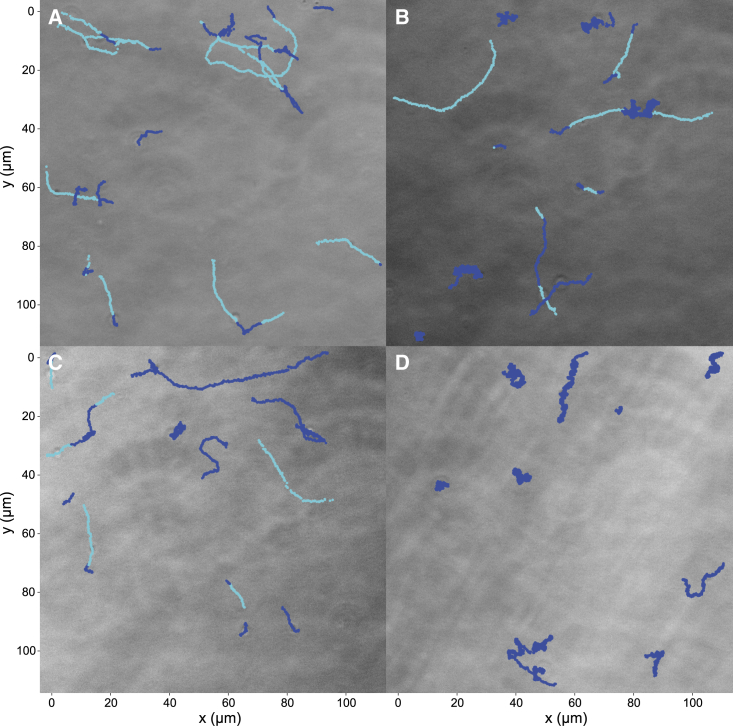
Figure 3.
Representative B. bacteriovorus trajectories depicting instantaneous velocities. Bacterial trajectories are obtained for the first 500 frames at hour 0 (A), hour 4 (B), hour 6 (C), and hour 20 (D). Trajectories are colored according to the state at that time level (active swimming is bright and apparent diffusive is dark) determined by thresholding above and below 30 m/s. This threshold is selected based on the midpoint of the center of both peaks in the bimodal distribution in Fig. 1.
We then introduced B. bacteriovorus to nutrient-rich medium (LB)—effectively providing them with an external energy source—to determine whether the bacteria were merely inviable or swimming less frequently at later times. We monitored the changes in the speed distributions (Fig. 2) compared with a starving control of equal age.
To further investigate the nature of B. bacteriovorus’ search strategy bimodal speed distribution, we monitored B. bacteriovorus’ behavior with a cycle of starvation, rich medium exposure, and then restarvation. This resulted in bacteria repopulating the active swimming state and then the apparent diffusive state (Fig. S7).
Diffusive speed calculations
As mentioned before, Fig. 1 contains a bimodal instantaneous speed distribution with peaks at 10 and 40 μm s−1. We interpret these peaks as a diffusive state and a swimming state, respectively. To justify our interpretation, we compare our inferred diffusive speed (10 μm s−1) with the expected diffusive speed calculated using the Stokes-Einstein equation. Assuming the bacterium is a sphere, we can calculate the diffusion coefficient as given by the Stokes-Einstein equation:
| (1) |
where is the dynamic viscosity and is the radius of the sphere. The dynamic viscosity of water at 28°C (the temperature of the incubator) is approximately 8.318 10−4 kg m s−1, and the radius of the bacterium is approximately 0.375 μm. By substituting these values and Boltzmann’s constant, , the diffusion coefficient can be estimated as 0.707 μm2 s−1. The exposure time, , between two frames is 0.008 s. Therefore, the diffusive speed is approximately 9.40 μm s−1, as given by:
| (2) |
whose outcome is consistent with our measurement.
As an aside, we can compare this to the diffusive speed of an E. coli. A rough calculation for E. coli’s diffusive speed can be obtained by approximating the cell as a sphere and taking its volume to be 1 μm3, giving a radius of 0.6 μm. From Eq. 2 above, we get 7.4 μm s−1. Considering the approximations made in this calculation, E. coli’s diffusive speed of 7.4 μm s−1 is actually similar to the diffusive speed of B. bacteriovorus, which is approximately 9.4 μm s−1.
Starvation of B. bacteriovorus
Representative traces, exhibiting both active swimming states (light blue) and apparent diffusive states (dark blue) visited within individual trajectories, are shown in Fig. 3. As described in the materials and methods, hour 0 is observed 10 min after filtering.
In Fig. 1, we show the instantaneous and average speed distributions as a function of age. Starting from hour 0, the instantaneous speeds exhibit a bimodal distribution, with peaks around 10 and 40 μm s−1. The former peak is centered around the expected diffusive speed of a spherical particle the B. bacteriovorus’ size, as predicted by the Stokes-Einstein relation (Fig. 1, dashed line) (see the previous section for calculation).
As bacteria age over the first 6 h under starvation conditions, we observe a decrease in the higher-speed population and an increase in the apparent diffusive population of our bimodal distribution. The observed shifts suggest the possibility that the bacteria are actively adjusting the amount of time spent swimming in response to persisting starvation conditions. The evolution of the speed distribution, from hour 0 up to hour 45, is shown in Fig. 1 B (with bimodality shifting to unimodality around the 20-h mark). In the materials and methods, we describe tracking details.
We highlight that, by contrast to B. bacteriovorus which continues swimming for hours poststarvation, E. coli stops swimming as soon as we can image it when under starvation as shown in Fig. S1.
To determine whether there are two bacterial populations or two motility states for each bacterium contributing to the bimodal instantaneous distributions (Fig. 1, A and B), the average velocity over each bacterium’s trajectory was calculated (Fig. 1 C). The speed distributions have a similar average velocity over the first 6 h. This indicates that the speed of each bacterium is more similar to that of other bacteria than the speeds sampled within its own trajectory. Indeed the instantaneous speed distribution at hour 6 appears to shift slightly toward higher values compared with hour 5 (see Fig. 1 A). We attribute this shift to noise, for two reasons 1) the width of the distribution at hour 6 is generally consistent with that of previous hours and 2) the number of live bacteria decreases at later hours, resulting in poorer statistical accuracy. However, just as before, the average velocity approaches the expected diffusive speed after hour 20 (Fig. 1 D, dashed line), suggesting that B. bacteriovorus are apparently diffusing. The absence of a well-defined unimodal speed distribution is corroborated by direct visual inspection of bacterial trajectories; representative traces, exhibiting both active swimming and apparent diffusive states within individual trajectories, are shown in Fig. 3. Additional runs can be found in the supporting material (Figs. S8–S10).
To demonstrate that the observations depicted in Fig. 1 are not an artifact of the localization method employed, we also performed deep learning localization on the same data sets and obtained consistent results. Furthermore, we conducted Gaussian mixture fitting and provide a list of all fitted parameters. These results are presented in the supporting material, comparison to another analysis method.
Spiking of B. bacteriovorus
Following up on the preanalysis, we exposed previously starving bacteria to nutrient-rich medium (LB), as described in the materials and methods. One of the main ingredients, yeast extract, has been shown to induce an active chemotactic response in B. bacteriovorus (11). In Fig. 2, we compare the motile response to the addition of LB with a control where B. bacteriovorus is kept in nutrient-poor medium (HM), identical to those in Fig. 1.
To determine if B. bacteriovorus is revivable, we took a B. bacteriovorus culture and separated it into three. One of these cultures was spiked with LB at hour 4, another was spiked with LB at hour 20, and the third was never spiked. The spiked cultures remained in the LB solution for the duration of the remainder of the experiment. For both spiked cultures, we then looked at their instantaneous speed distributions after 2 h of exposure, thus at hour 6 for Fig. 2 A and at hour 22 for Fig. 2 B. We found that bacteria move more frequently after spiking. Additional runs are provided in Figs. S11 and S12.
As seen in Fig. 2 C, after leaving the bacteria in the spiked rich medium, their speed distributions did not greatly vary, indicating that over several hours there is an excess of nutrients. To further determine if the concentration of LB used to spike the cultures at hour 4 and hour 20 had an overall effect on the speed distributions, we spiked cultures at half the concentration of LB used to spike the cultures as well as at 10× the concentration (see Fig. S13). The half LB spiked cultures exhibited similar results to the initial whole concentration seen in Fig. 2. At 10× LB, the speed of B. bacteriovorus was observed to be slower. This can be attributed to the higher osmolarity of the solution, consistent with previous reports in the literature (60). Although high osmolarity is known to generally lower fitness (61), the exact mechanism by which higher osmolarities lead to slower B. bacteriovorus speeds is yet to be fully understood (60). One recent explanation suggests that higher osmolarities can impede the metabolism of B. bacteriovorus and hinder the regeneration of NADH pools, without necessarily causing cell death (62).
Spiking B. bacteriovorus at hour 40 resulted in no change compared with the starving control (Figs. S14 and S15). This indicates that, although the speed distribution in Fig. 1 looks similar for hours 20 and 45, the bacteria cannot be revived after hour 45.
These findings then naturally beg the question as to whether B. bacteriovorus recovers its bimodal behavior after reinitialization of starvation conditions (resuspension in HM) after spiking. Here, we follow up on the spiking experiments (Fig. 2), except that after spiking we centrifuged B. bacteriovorus and resuspended them in nutrient-poor medium. We then ask how the fraction of swimming versus apparently diffusing B. bacteriovorus differ from our control (Fig. S7). Qualitatively, we find that the bacteria continue to spend more time swimming compared with the control—regardless of the tendency of centrifugation to slow down B. bacteriovorus (see Fig. S2)—suggesting that there is some degree of memorylessness. In other words, these results suggest that B. bacteriovorus behave as if they were exposed to rich medium and starved for the very first time.
Discussion
Bimodality in the instantaneous speed distributions over the first 6 h under starvation (Fig. 1, A and B), along with the absence of bimodality in the trajectory-averaged speed distributions (Fig. 1, C and D), suggests that B. bacteriovorus samples both active swimming and apparent diffusive states of motility within the course of a single trajectory. At any given time point (after the initiation of starvation), the motile behavior of a given B. bacteriovorus appears to be representative of other cells within the population. Indeed, B. bacteriovorus cells sample speeds from a bimodal distribution, with active swimming and apparent diffusive states, from the onset of starvation.
B. bacteriovorus remains active over hours and our spiking experiments not only show that B. bacteriovorus can be resuscitated after even 20 h of starvation, but that those bacteria that do stop swimming (and revert to apparent diffusion) can be stimulated back into sampling the high-speed state of its velocity distribution. Indeed, shortly after spiking, B. bacteriovorus increases the fraction of time it spends swimming. As the reinitiation of starvation conditions (Fig. S7) produced behavior similar to initial starvation, it is likely that B. bacteriovorus allocates energy reserves for active motion in direct response to its environment, regardless of previous conditions, and that the B. bacteriovorus population eventually enters an energy-conserving apparent diffusive state if starved for long enough. Previous studies in which bimodal speed distributions are observed are normally associated with the bundling of flagella for peritrichously flagellated species (e.g., the run-and-tumble paradigm (56,63)), which is not a feasible mechanism for the uniflagellated B. bacteriovorus and occurs on second timescales far shorter than the hours and days timescales involved in reweighting motility states.
The subtle way in which B. bacteriovorus modulates which of the states of its speed distribution it samples is worthy, in and of itself, of future molecular attention. This is especially meaningful in light of the stark contrast in its ability to keep swimming under starvation compared with its prey, E. coli. To wit, it is already known that B. bacteriovorus dramatically varies its gene expression levels between attack and growth phases (57,64,65) with, most recently, work exploring B. bacteriovorus’ motility reduction under starvation linked to cyclic-di-GMP effectors playing the role of “motility brakes” (66).
These findings, in turn, beg broader questions associated with the cost, both hydrodynamic and transcriptional, tied to motility reinitiation by B. bacteriovorus. Our work here provides a piece of the puzzle toward assessing the energetic cost associated with motility with time from the onset of starvation. It remains to be seen whether, in the end, active swimming state modulation with age is tied to transcriptional activity and part of a global strategy to jointly minimize hydrodynamic and transcriptional energy expenditure to maximize the probability of locating prey.
Author contributions
L.W.Q.X. and J.S.B. performed all of the analysis. S.P. oversaw all aspects of the research.
Acknowledgments
S.P. acknowledges the support of NIH (NIGMS R01GM130745, NIGMS R01GM134426, NIGMS R35GM148237) and NSF (CAREER Grant MCB-1719537, PHY-2310610). We would like to thank Doug Shepherd for helpful feedback on all experimental aspects of this study. We would also like to give a special thanks to Edouard Jurkevitch, Rajesh Sathyamoorthy, and Daniel Kadouri for their help with B. bacteriovorus growth protocols and Yixin Shi for generously providing E. coli strains.
Declaration of interests
The authors have no conflicts of interest to declare.
Editor: Pablo Iglesias.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2023.06.008.
Supporting material
Data availability
B. bacteriovorus localization data are available at statphysbio.physics.asu.edu.
References
- 1.Drescher K., Dunkel J., et al. Goldstein R.E. Fluid dynamics and noise in bacterial cell-cell and cell-surface scattering. Proc. Natl. Acad. Sci. USA. 2011;108:10940–10945. doi: 10.1073/pnas.1019079108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jashnsaz H., Al Juboori M., et al. Pressé S. Hydrodynamic Hunters. Biophys. J. 2017;112:1282–1289. doi: 10.1016/j.bpj.2017.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meng Y., Li Y., et al. Hoch H.C. Upstream migration of Xylella fastidiosa via pilus-driven twitching motility. J. Bacteriol. 2005;187:5560–5567. doi: 10.1128/JB.187.16.5560-5567.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen Y., Siryaporn A., et al. Stone H.A. Flow directs surface-attached bacteria to twitch upstream. Biophys. J. 2012;103:146–151. doi: 10.1016/j.bpj.2012.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaya T., Koser H. Direct upstream motility in Escherichia coli”. Biophys. J. 2012;102:1514–1523. doi: 10.1016/j.bpj.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker M.D., Wolanin P.M., Stock J.B. Signal transduction in bacterial chemotaxis. Bioessays. 2006;28:9–22. doi: 10.1002/bies.20343. [DOI] [PubMed] [Google Scholar]
- 7.Dori-Bachash M., Dassa B., Jurkevitch E., et al. Proteome-based comparative analyses of growth stages reveal new cell cycle-dependent functions in the predatory bacterium Bdellovibrio bacteriovorus. Appl. Environ. Microbiol. 2008;74:7152–7162. doi: 10.1128/AEM.01736-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waters C.M., Bassler B.L. Quorum Sensing: Cell-to-Cell Communication in Bacteria. Annu. Rev. Cell Dev. Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 9.Mukherjee S., Seok S.C., et al. Das J. Data-driven quantification of the robustness and sensitivity of cell signaling networks. Phys. Biol. 2013;10 doi: 10.1088/1478-3975/10/6/066002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang L., Ouyang Q., Tu Y. Quantitative Modeling of Escherichia coli Chemotactic Motion in Environments Varying in Space and Time. PLoS Comput. Biol. 2010;6 doi: 10.1371/journal.pcbi.1000735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LaMarre A.G., Straley S.C., Conti S.F. Chemotaxis toward amino acids by Bdellovibrio bacteriovorus. J. Bacteriol. 1977;131:201–207. doi: 10.1128/jb.131.1.201-207.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hespell R.B., Thomashow M.F., Rittenberg S.C. Changes in cell composition and viability of Bdellovibrio bacteriovorus during starvation. Arch. Microbiol. 1974;97:313–327. doi: 10.1007/bf00403070. [DOI] [PubMed] [Google Scholar]
- 13.Dashiff A., Junka R.A., et al. Kadouri D.E. Predation of human pathogens by the predatory bacteria Micavibrio aeruginosavorus and Bdellovibrio bacteriovorus”. J. Appl. Microbiol. 2011;110:431–444. doi: 10.1111/j.1365-2672.2010.04900.x. [DOI] [PubMed] [Google Scholar]
- 14.Sockett R.E., Lambert C. Bdellovibrio as therapeutic agents: a predatory renaissance? Nat. Rev. Microbiol. 2004;2:669–675. doi: 10.1038/nrmicro959. [DOI] [PubMed] [Google Scholar]
- 15.Kadouri D., O’Toole G.A. Susceptibility of biofilms to Bdellovibrio bacteriovorus attack. Appl. Environ. Microbiol. 2005;71:4044–4051. doi: 10.1128/AEM.71.7.4044-4051.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Passino K.M. Biomimicry of bacterial foraging for distributed optimization and control. Control Syst. Mag. 2002;22:52–67. [Google Scholar]
- 17.Astling D.P., Lee J.Y., Zusman D.R. Differential effects of chemoreceptor methylation-domain mutations on swarming and development in the social bacterium Myxococcus xanthus. Mol. Microbiol. 2006;59:45–55. doi: 10.1111/j.1365-2958.2005.04926.x. [DOI] [PubMed] [Google Scholar]
- 18.Williams H.N., Kelley J.I., et al. Turng B.F. The association of Bdellovibrios with surfaces in the aquatic environment. Can. J. Microbiol. 1995;41:1142–1147. [Google Scholar]
- 19.Lambert C., Fenton A.K., et al. Sockett R.E. Predatory Bdellovibrio Bacteria Use Gliding Motility To Scout for Prey on Surfaces. J. Bacteriol. 2011;193:3139–3141. doi: 10.1128/JB.00224-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harini K., Ajila V., Hegde S. Bdellovibrio Bacteriovorus: A Future Antimicrobial Agent? 2013. [DOI] [PMC free article] [PubMed]
- 21.Chanyi R.M., Koval S.F. Role of Type IV Pili in Predation by Bdellovibrio bacteriovorus”. PLoS One. 2014;9:e113404. doi: 10.1371/journal.pone.0113404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Straley S.C., Conti S.F. Chemotaxis by Bdellovibrio bacteriovorus toward prey. J. Bacteriol. 1977;132:628–640. doi: 10.1128/jb.132.2.628-640.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oyedara O.O., De Luna-Santillana E.d.J., et al. Rodriguez-Perez M.A. Isolation of Bdellovibrio sp. from soil samples in Mexico and their potential applications in control of pathogens. MicrobiologyOpen. 2016;5:992–1002. doi: 10.1002/mbo3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu R., Zhang S., et al. Li C. Isolation and application of predatory Bdellovibrio-and-like organisms for municipal waste sludge biolysis and dewaterability enhancement. Front. Environ. Sci. Eng. 2017;11:10. [Google Scholar]
- 25.Sun Y., Ye J., et al. Zhou T. Predation Efficacy of Bdellovibrio bacteriovorus on Multidrug-Resistant Clinical Pathogens and Their Corresponding Biofilms. Jpn. J. Infect. Dis. 2017;70:485–489. doi: 10.7883/yoken.JJID.2016.405. [DOI] [PubMed] [Google Scholar]
- 26.Iebba V., Totino V., et al. Artini M. Bdellovibrio bacteriovorus directly attacks Pseudomonas aeruginosa and Staphylococcus aureus cystic fibrosis isolates. Front. Microbiol. 2014;5 doi: 10.3389/fmicb.2014.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shatzkes K., Singleton E., et al. Kadouri D.E. Predatory Bacteria Attenuate Klebsiella pneumoniae Burden in Rat Lungs. mBio. 2016;7 doi: 10.1128/mBio.01847-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atterbury R.J., Hobley L., et al. Sockett R.E. Effects of orally administered Bdellovibrio bacteriovorus on thewell-being and Salmonella colonization of young chicks. Appl. Environ. Microbiol. 2011;77:5794–5803. doi: 10.1128/AEM.00426-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Romanowski E.G., Stella N.A., et al. Shanks R.M.Q. Predatory bacteria are nontoxic to the rabbit ocular surface. Sci. Rep. 2016;6 doi: 10.1038/srep30987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willis A.R., Moore C., et al. Sockett R.E. Injections of Predatory Bacteria Work Alongside Host Immune Cells to Treat Shigella Infection in Zebrafish Larvae. Curr. Biol. 2016;26:3343–3351. doi: 10.1016/j.cub.2016.09.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park S., Kim D., et al. Kim T. A microfluidic concentrator array for quantitative predation assays of predatory microbes. Lab Chip. 2011;11:2916–2923. doi: 10.1039/c1lc20230h. [DOI] [PubMed] [Google Scholar]
- 32.Van Essche M., Quirynen M., et al. Teughels W. Bdellovibrio bacteriovorus attacks Aggregatibacter actinomycetemcomitans”. J. Dent. Res. 2009;88:182–186. doi: 10.1177/0022034508329693. [DOI] [PubMed] [Google Scholar]
- 33.Mitchell R., Yankfsky S., Jannasch H.W. Lysis of Escherichia coli by marine micro-organisms. Nature. 1967;215:891–893. doi: 10.1038/215891a0. [DOI] [PubMed] [Google Scholar]
- 34.Lambert C., Smith M.C.M., Sockett R.E. A novel assay to monitor predator-prey interactions for Bdellovibrio bacteriovorus 109 J reveals a role for methyl-accepting chemotaxis proteins in predation. Environ. Microbiol. 2003;5:127–132. doi: 10.1046/j.1462-2920.2003.00385.x. [DOI] [PubMed] [Google Scholar]
- 35.Rendulic S., Jagtap P., et al. Schuster S.C. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science. 2004;303:689–692. doi: 10.1126/science.1093027. [DOI] [PubMed] [Google Scholar]
- 36.Steyert S.R., Pineiro S.A. Development of a novel genetic system to create markerless deletion mutants of Bdellovibrio bacteriovorus”. Appl. Environ. Microbiol. 2007;73:4717–4724. doi: 10.1128/AEM.00640-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lambert C., Evans K.J., et al. Sockett R.E. Characterizing the flagellar filament and the role of motility in bacterial prey-penetration by Bdellovibrio bacteriovorus”. Mol. Microbiol. 2006;60:274–286. doi: 10.1111/j.1365-2958.2006.05081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lambert C., Morehouse K.A., et al. Sockett R.E. Bdellovibrio: growth and development during the predatory cycle. Curr. Opin. Microbiol. 2006;9:639–644. doi: 10.1016/j.mib.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 39.Thomashow M.F., Cotter T.W. Bdellovibrio host dependence: the search for signal molecules and genes that regulate the intraperiplasmic growth cycle. J. Bacteriol. 1992;174:5767–5771. doi: 10.1128/jb.174.18.5767-5771.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fratamico P.M., Whiting R.C. Ability of Bdellovibrio bacteriovorus 109J to Lyse Gram-Negative Food-Borne Pathogenic and Spoilage Bacteria. J. Food Prot. 1995;58:160–164. doi: 10.4315/0362-028X-58.2.160. [DOI] [PubMed] [Google Scholar]
- 41.Seidler R.J., Starr M.P. Factors affecting the intracellular parasitic growth of Bdellovibrio bacteriovorus developing within Escherichia coli”. J. Bacteriol. 1969;97:912–923. doi: 10.1128/jb.97.2.912-923.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stolp H., Petzold H. Untersuchungen ueber einen obligat parasitishen Mikroorganismus mit lyticher Aktivitaet fuer Pseudomonas-bakterien. Phytopathology. 1962;45:364–390. [Google Scholar]
- 43.Stolp H., Starr M.P. Bdellovibrio bacteriovorus gen. et sp. n., a predatory, ectoparasitic, and bacteriolytic microorganism. Antonie Leeuwenhoek. 1963;29:217–248. doi: 10.1007/BF02046064. [DOI] [PubMed] [Google Scholar]
- 44.Seidler R.J., Starr M.P. Isolation and Characterization of Host-Independent Bdellovibrios”. J. Bacteriol. 1969;100:769–785. doi: 10.1128/jb.100.2.769-785.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Varon M., Zeigler B.P. Bacterial predator-prey interaction at low prey density. Appl. Environ. Microbiol. 1978;36:11–17. doi: 10.1128/aem.36.1.11-17.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lambert C., Hobley L., et al. Sockett L. A predatory patchwork: membrane and surface structures of Bdellovibrio bacteriovorus”. Adv. Microb. Physiol. 2009;54:313–361. doi: 10.1016/S0065-2911(08)00005-2. [DOI] [PubMed] [Google Scholar]
- 47.Straley S.C., Conti S.F. Chemotaxis by Bdellovibrio bacteriovorus”. J. Bacteriol. 1974;120:549–551. doi: 10.1128/jb.120.1.549-551.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strauch E., Schwudke D., Linscheid M. Predatory mechanisms of Bdellovibrio and like organisms. Future Microbiol. 2007;2:63–73. doi: 10.2217/17460913.2.1.63. [DOI] [PubMed] [Google Scholar]
- 49.Drescher K., Goldstein R.E., et al. Tuval I. Direct Measurement of the Flow Field around Swimming Microorganisms. Phys. Rev. Lett. 2010;105 doi: 10.1103/PhysRevLett.105.168101. [DOI] [PubMed] [Google Scholar]
- 50.Lushi E., Wioland H., Goldstein R.E. Fluid flows created by swimming bacteria drive self-organization in confined suspensions. Proc. Natl. Acad. Sci. USA. 2014;111:9733–9738. doi: 10.1073/pnas.1405698111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frymier P.D., Ford R.M., et al. Cummings P.T. Three-dimensional tracking of motile bacteria near a solid planar surface. Proc. Natl. Acad. Sci. USA. 1995;92:6195–6199. doi: 10.1073/pnas.92.13.6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lauga E., DiLuzio W.R., et al. Stone H.A. Swimming in Circles: Motion of Bacteria near Solid Boundaries. Biophys. J. 2006;90:400–412. doi: 10.1529/biophysj.105.069401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Di Leonardo R., Dell'Arciprete D., et al. Iebba V. Swimming with an Image. Phys. Rev. Lett. 2011;106 doi: 10.1103/PhysRevLett.106.038101. [DOI] [PubMed] [Google Scholar]
- 54.Hu J., Wysocki A., et al. Gompper G. Physical sensing of surface properties by microswimmers: directing bacterial motion via wall slip. Sci. Rep. 2015;5 doi: 10.1038/srep09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matz C., Kjelleberg S. Off the hook–how bacteria survive protozoan grazing. Trends Microbiol. 2005;13:302–307. doi: 10.1016/j.tim.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 56.Matz C., Jürgens K. High motility reduces grazing mortality of planktonic bacteria. Appl. Environ. Microbiol. 2005;71:921–929. doi: 10.1128/AEM.71.2.921-929.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karunker I., Rotem O., et al. Sorek R. A global transcriptional switch between the attack and growth forms of Bdellovibrio bacteriovorus. PLoS One. 2013;8:e61850. doi: 10.1371/journal.pone.0061850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lambert C., Lerner T.R., et al. Sockett R.E. Interrupting peptidoglycan deacetylation during Bdellovibrio predator-prey interaction prevents ultimate destruction of prey wall, liberating bacterial-ghosts. Sci. Rep. 2016;6:26010. doi: 10.1038/srep26010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Allan D., Caswell T., et al. van der Wel C.M. soft-matter/trackpy: Trackpy v0.4.2. Version v0.4.2. Zendo. 2019 doi: 10.5281/zenodo.3492186. Preprint at. [DOI] [Google Scholar]
- 60.Hansol I., Son S., et al. Ghim C.M. Serum albumin and osmolality inhibit Bdellovibrio bacteriovorus predation in human serum. Sci. Rep. 2017;7:2045–2322. doi: 10.1038/s41598-017-06272-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ghosh K., de Graff A.M.R., et al. Dill K.A. Role of proteome physical chemistry in cell behavior. J. Phys. Chem. B. 2016;120:9549–9563. doi: 10.1021/acs.jpcb.6b04886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jang H., Mun W., et al. Mitchell R.J. Use of Resazurin To Rapidly Enumerate Bdellovibrio and Like Organisms and Evaluate Their Activities. Microbiol. Spectr. 2022;10:e0082522. doi: 10.1128/spectrum.00825-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Toley B.J., Forbes N.S. Motility is critical for effective distribution and accumulation of bacteria in tumor tissue. Integr. Biol. 2012;4:165–176. doi: 10.1039/c2ib00091a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waso M., Khan S., et al. Khan W. Expression of attack and growth phase genes of Bdellovibrio bacteriovorus in the presence of Gram-negative and Gram-positive prey. Microbiol. Res. 2020;235:126437. doi: 10.1016/j.micres.2020.126437. [DOI] [PubMed] [Google Scholar]
- 65.Herencias C., Salgado-Briegas S., et al. Nogales J. Providing new insights on the byphasic lifestyle of the predatory bacterium Bdellovibrio bacteriovorus through genome-scale metabolic modeling. PLoS Comput. Biol. 2020;16 doi: 10.1371/journal.pcbi.1007646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sathyamoorthy R., Kushmaro Y., et al. Jurkevitch E. To hunt or to rest: prey depletion induces a novel starvation survival strategy in bacterial predators. ISME J. 2020;15:109–123. doi: 10.1038/s41396-020-00764-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
B. bacteriovorus localization data are available at statphysbio.physics.asu.edu.