Abstract
Atomically detailed simulations of RNA folding have proven very challenging in view of the difficulties of developing realistic force fields and the intrinsic computational complexity of sampling rare conformational transitions. As a step forward in tackling these issues, we extend to RNA an enhanced path-sampling method previously successfully applied to proteins. In this scheme, the information about the RNA’s native structure is harnessed by a soft history-dependent biasing force promoting the generation of productive folding trajectories in an all-atom force field with explicit solvent. A rigorous variational principle is then applied to minimize the effect of the bias. Here, we report on an application of this method to RNA molecules from 20 to 47 nucleotides long and increasing topological complexity. By comparison with analog simulations performed on small proteins with similar size and architecture, we show that the RNA folding landscape is significantly more frustrated, even for relatively small chains with a simple topology. The predicted RNA folding mechanisms are found to be consistent with the available experiments and some of the existing coarse-grained models. Due to its computational performance, this scheme provides a promising platform to efficiently gather atomistic RNA folding trajectories, thus retain the information about the chemical composition of the sequence.
Significance
Identifying the conformational changes that RNA molecules undergo during their folding process is a fundamental problem in molecular biology. In principle, atomistic molecular dynamics (MD) simulations could provide this information. In practice, conventional MD is currently limited in both efficiency and accuracy because RNA systems are complex and intrinsically frustrated. In this work, we overcome these limitations using an advanced simulation scheme and show that the folding of RNAs of known native structures can be accurately reconstructed with atomistic resolution. Our results are validated against experiments and highlight important differences between the RNA and the protein folding processes. This method paves the way to investigating longer and more complex RNAs, including many involved in gene expression regulation.
Introduction
The importance of noncoding RNA molecules has become more and more evident in recent years with the discovery of the central role of these systems in regulating gene expression (1,2) and other vital cellular processes (3). Moreover, many viruses rely on RNA systems to hijack the host cellular machinery and spread the infection (4,5). Just like for most proteins, to properly function, these molecules need to adopt a well-defined three-dimensional structure. Understanding their folding can shed light on their function and may inspire new therapeutic strategies. However, the limited chemical alphabet of nucleic acid bases and their ability to form both canonical and noncanonical pairings (6) make RNA folding prone to frustration, as alternative overall architectures can be adopted by the same sequence. This is what happens for riboswitches, where the presence of a ligand triggers a full reorganization of the structure (7), but also for other systems, now known to adopt alternative structures (8,9). Moreover, even single-point mutations and posttranscriptional modifications such as methylations can trigger global structural rearrangements (10,11,12).
RNA folding has been tackled before by computational means in various models (13). Plain molecular dynamics (MD) simulations at atomistic resolution have been performed for specific RNAs of limited size (14,15,16). However, most RNA molecules of biological interest, comprising from a few dozen to a few hundred nucleotides, remain out of reach. Larger systems can be studied mostly in the proximity of known conformations and by simulations largely guided by experimental evidence. Notable examples are the recent studies of SARS-CoV-2 frame-shifting elements addressing the existence of alternative conformations (17,18,19,20,21).
Similarly, the existence of alternative RNA structures and the determination of their low-energy interconversion pathways have been tackled using atomistic path-sampling methods (10,22,23). These appealing, although computationally demanding methods require an implicit solvent description because they are based on a geometrical optimization scheme rather than on integrating the equations of motion. Different coarse-grained models (24,25,26,27), including native-centric ones, have been adopted to overcome computational limitations and complement the atomistic approaches. These schemes are used to capture the main features of RNA folding of biologically relevant systems. However, these models do not provide detailed physicochemical insight, limiting the potential for translational applications.
In response to these challenges, here we report on the first application to RNA of an enhanced path-sampling technique called the bias functional (BF) approach, which was originally developed for protein folding simulations. We show that this method enables the simulation of RNA folding within state-of-the-art all-atom force fields in explicit solvent.
The BF scheme relies on a specific type of biased dynamics called ratchet-and-pawl MD (rMD) (28,29) to efficiently generate a statistically significant number of productive trajectories. In rMD, an auxiliary history-dependent potential depending on a suitable collective variable (CV) is introduced to prevent the chain from backtracking toward the initial state (unfolded, reactant). Conversely, the biasing force is inactive when the system spontaneously progresses toward the final state (folded, product). In the ideal case in which the CV is the committor function (30), rMD simulations yield the correct Boltzmann sampling in the region explored by the transition path ensemble (31,32). In our simulation of RNA folding, we use a CV that measures the distance in contact map space of the instantaneous configurations from the known native structure. Since this CV is only a proxy of the ideal reaction coordinate, rMD yields an approximate sampling of the equilibrium distribution in the transition region. However, it is still possible to keep systematic errors to a minimum by applying the so-called BF filtering (33): a variational principle derived from Langevin dynamics is used for scoring the folding trajectories generated by rMD to identify those with the highest probability of occurring in the absence of any biasing force.
In this study, we use BF simulations to highlight key differences between folding of RNAs and proteins of comparable size and topological structure. Then, we apply the same scheme on the folding of a more challenging RNA molecule, previously addressed in experiments and with coarse-grained models, and we present the first fully atomistic reconstruction of its folding mechanism.
Materials and methods
RNA folding simulation
RNA folding simulations were carried out in explicit TIP3P water using the Amber RNA force field with OL3 refinement (ff99bsc0χOL3 combination) (34) implemented in GROMACS 2020-dev (35). After imposing charge neutrality on the system by introducing monovalent sodium ions, we further added a buffer of 0.15 M sodium chloride. The size of the cubic periodic simulation box was chosen large enough to accommodate the fully unfolded conformations. The total number of atoms in our systems ranged from tens of thousands (for the two smallest RNAs) to several hundreds of thousands (for the largest RNA).
For each of these three nucleic acids, we sampled their native states with 100 ns of plain MD at body temperature (C) and standard pressure starting from the energy-minimized PDB configuration and discarding the first 10 ns. To keep temperature and pressure fixed, we employed a stochastic velocity rescaling procedure (36). We used this local equilibrium sampling to compute the native contact maps used to define the CV employed in rMD simulations (see further below).
To obtain 10 initial unfolded configurations for the hairpin and tmRNA pseudoknot, we performed 5 ns MD runs at the nominal temperature of 800 K, starting from configurations sampled in the native state. For hTR, which is more compact, we instead relied on UnityMol (37), a software enabling the interactive visual manipulation of coarse-grained RNAs, to generate efficiently three fully unfolded structures. We relaxed all the unfolded RNA structures with 5 ns of MD at standard temperature and pressure conditions. The pairwise relative root mean-square deviation (RMSD) among the unfolded and the native structure is above 10 Å; they are different enough to sample a vast/significant portion of the unfolded state (Fig. 4).
Figure 4.
Initial unfolded structures for the three considered RNA molecules. The corresponding .pdb files are available for download as Supporting material. To see this figure in color, go online.
Then, we initiated a set of rMD simulations from each unfolded configuration obtained at the end of this relaxation run. In particular, 10 independent rMD trajectories for each configuration of the hairpin and the tmRNA pseudoknot, and 20 for each configuration of the hTR pseudoknot. The protocol has been employed previously in protein folding simulations, as detailed, e.g., in (38).
In ideal rMD simulations, the history-dependent biasing force is defined as:
| (1) |
where is the Heaviside step function, denotes the set of atomic coordinates in the nucleic acid, is the committor function, while denotes the maximum value attained by the committor function up to time t, i.e.
| (2) |
In this ideal limit, rMD samples the Boltzmann distribution in the transition region. Unfortunately, it also loses its computational advantage: indeed, the gradient of the committor (and, therefore, the biasing force) is exponentially suppressed within metastable basins. In practice, rMD simulations are based on a proxy of the committor function—hereby denoted with —which typically remains finite in metastable states, thus accelerating the reaction kinetics. In particular, in our simulations we adopted an ansatz that measures the instantaneous overlap with the native contact map:
| (3) |
In this equation, is a switching function that approaches 1 when atom i and j are in contact and vanishes when they are far apart. In particular, we use:
| (4) |
Here, and Å is a threshold reference distance and are entries of the contact map of the native state, which are obtained from an average of over the configurations that have been generated by 100 ns of MD, starting from the energy-minimized crystal native structure. Finally, the symbol denotes a summation that excludes atoms that belong to neighboring amino acids or nucleotides, to weaken the effect of the rMD bias in forming local contacts. Specifically, in protein folding simulations denotes summation prescription , while in RNA folding simulations it refers to a summation over pairs of atoms that are at least four nucleotides apart. These restrictions are introduced to avoid the ratchet force acting on atoms that are subject to strong correlations determined by the local chemical structure of the chain.
We note that the biasing force (1) indirectly promotes the formation of the native contacts encoded in by counteracting any conformational change that decreases the overlap of with . The native structures and the Python code we have used to compute the reference contact maps for all RNA and protein molecules are available for download as Supporting material.
The constant was set to kJ/mol for the hairpin and the PK1 pseudoknot, and kJ/mol for the hTR pseudoknot. These values are typical for rMD protein folding simulations, where they allow for efficiently generating many folding trajectories while applying a gentle total biasing force. Indeed, the biasing force provided only a small correction to the physical force acting on each atom. In RNA folding simulations, we noticed that the same choice of leads to larger biasing forces, which can become comparable with the physical ones. This is in line with the fact that our choice of CV is inspired by energy landscape theory arguments, which have been introduced in the context of protein folding and may be less fit for RNAs, which are more frustrated systems.
To reduce the effect of the bias, we resorted to the so-called BF method (33). According to this scheme, rigorously derived for Langevin dynamics, the rMD trajectories that have the largest probability to occur in an unbiased simulation are those with the least value of the BF
| (5) |
where denote the mass and viscosity coefficient of each atom, respectively, and is the time duration of the folding transition. Our rMD trajectories could last up to 2 nominal ns (for the hairpin and PK1 pseudoknot) and 5 nominal ns (for the hTR pseudoknot). However, the trajectories were terminated as soon as the chain entered the native state—defined by an RMSD to native Å and fraction of native contacts . This was done to avoid the total value of the BF being largely affected by the value of the biasing force in the native state. Indeed, the instantaneous contact map becomes very close to , but does not coincide with it, so the biasing force is always switched on, leading to large contributions T. We used the BF criterion to identify and discard atypical transitions, i.e., outliers characterized by a very large value of T.
We stress that, in rMD simulations, the history-dependent biasing force alters all the timescales, so the resulting trajectories cannot be used to directly infer the kinetics of the system. On the other hand, the individual trajectories do carry qualitative information about the reaction mechanism. In particular, the comparative analysis reported in (38) showed that the order of formation of native contacts along rMD folding trajectories for a selected representative set of fast-folding proteins is statistically indistinguishable from the order found by plain MD simulations, performed with the same force field on the Anton supercomputer. The rMD simulations were performed with a custom modification of the GROMACS2020 simulation engine, the setup files being provided as Supporting material. A summary of the salient parameters of the simulations, and the typical durations, are given in Table 1.
Table 1.
Details about the RNA folding simulations
| Molecule | RNA atoms | Total atoms | (kJ/mol) | Number of trajectories | Duration (ns) | Run time (ns/day) |
|---|---|---|---|---|---|---|
| HHR | 644 | 81,696 | 2 | 146 | ||
| PK1 | 708 | 100,344 | 2 | 127 | ||
| hTR | 1497 | 1,484,247 | 5 | 15 |
The parameter was chosen to keep the biasing force to a minimum while yielding productive folding trajectories in a few ns of nominal simulation time. Because of the folding acceleration provided by the biassing term, the rMD folding times are lower bounds to the actual folding times. The notation in the sixth column indicates that x different initial conditions were used, collecting y folding trajectories for each of them. The structure files of the different initial conditions are provided in the Supporting material, along with the rMD setup files. The real-time run time (last column) refers to simulations performed on a workstation equipped with an i9-9900KF CPU and an RTX2080 GPU.
Simulation analysis
Folding trajectories have been analyzed through software specifically designed to detect key interactions in RNA structures. In particular, we monitored the RMSD to native, the number of heavy-atom contacts (native and nonnative), and the number of basepairs. The latter CV was used to monitor the formation of the secondary structures. RMSD to native and contacts were computed using the MDtraj python package (39), with a contact distance cutoff of 4.5 Å.
For each structure in the trajectory, we computed basepairing based on geometric criteria using the DSSR (40) software. Since the detection of hydrogen bonds can depend on the specific cutoffs employed by the algorithm, for a sample of structures we computed basepairing also using Barnaba (41) to verify the robustness of the results with an independent algorithm, and found a substantial agreement between the two methods. We also monitored the formation of multiplets, since they appear in the native structure of hTR and in the triple helix. The formation of the various secondary structural elements is computed as the ratio between the native basepairs of the element present in the structure under scrutiny and the number of basepairs in the native structure. Folded trajectories are identified as those for which all structural elements are formed at the end of the simulation. They are then grouped together in folding pathways according to the order of formation of the elements. For trajectories belonging to the same folding pathway, we compute average properties combining the information of the formation of the secondary structural elements versus native overlap.
Results
rMD can be used to map the overall features of free-energy landscapes by projecting the generated folding trajectories onto suitable CVs. For example, in proteins, despite the fact that free-energy barriers may be underestimated, this approach could predict protein folding intermediates that have been later experimentally confirmed (42,43) and exploited for drug discovery purposes (44). BF filtering subsequently enables removing the most biased (thus unphysical) trajectories from the generated transition path ensemble (see materials and methods).
First, we use this scheme to characterize the main relative differences in the landscapes of RNAs and proteins of comparable size and topological architectures. This comparison is especially aimed at identifying landscape features that are shared by unrelated RNA molecules and yet have no analog in proteins.
For this comparative endeavor, which to our knowledge has not been pursued before, we selected two RNAs of comparable length and markedly different secondary and tertiary organizations. Specifically, we considered 1) the domain II of the CCHMVD hammerhead ribozyme (PDB: 2RPK, hereby referred to as HHR) (45) and 2) the PK1 pseudoknot of the Aquifex aeolicus tmRNA (PDB: 2G1W) (46). The former consists of 20 nucleotides and has a typical hairpin structure, while the latter is 22 nucleotides long and adopts an H pseudoknot configuration where two stems of 4 and 3 basepairs, respectively, are interlaced. For an equal footing comparison with protein counterparts, we considered the TRP-CAGE miniprotein (PDB: 2JOF) and a WW domain (PDB: 2F21), which have about the same number of monomeric units and similar pattern of local versus nonlocal contacts as the two representative RNAs: the TRP-CAGE consists of 20 amino acids and has a helix structure, while the WW domain comprises 36 amino acids in a β sheet motif. For each system, we generated independent folding trajectories.
Salient features of the folding landscape
First, we analyzed the trajectories in terms of two order parameters commonly used in folding contexts: the RMSD from the experimental native structure and the fraction of established native contacts, Q. For each molecule, we cumulated data from all productive trajectories obtaining the Q and RMSD histograms shown in Fig. 1. Notice that the plots are truncated just short of reaching the native state basin to discount the contribution due to dwelling time in the native state, which can be made indefinitely large in an rMD setup.
Figure 1.
Probability distributions of the fraction of native contacts Q and the RMSD to the crystal native structures, estimated from a frequency histogram of the productive rMD folding trajectories for four different macromolecules: (A) HHR, (B) protein TRP-CGE, (C) tmRNA fragment, and (D) protein WW domain. In all cases, trajectories were terminated immediately after entering the native region (gray areas). To see this figure in color, go online.
The probability distributions estimated from RNA folding paths are riddled with pronounced minima, indicative of trapping in multiple metastable basins. Direct inspection of the trajectories confirms that these basins correspond to states with an increasing fraction of formed pairs. The metastability of these configurations is due to the entropy cost of bringing together a new pair. It is important to emphasize that BF simulations systematically underestimate the height of the probability lumps associated with metastable regions, thus smoothing out the energy landscapes. Indeed, the rMD biasing force promotes the escape rate from metastable basins, thus leading to a lower free energy barrier.
Despite that rMD simulations tend to underestimate the roughness of the landscape, some differences between RNA and protein emerge. The density distributions of both protein counterparts are significantly smoother than those of RNA molecules, indicating a smaller degree of frustration. On the one hand, this finding is in line with the established notion that the folding of small globular proteins is highly cooperative (47,48,49,50). On the other hand, the higher frustration observed in RNA is consistent with the observed larger heterogeneity of RNA conformers (51,52,53,54,55,56,57).
Folding mechanism
The analysis of the ensemble of folding trajectories provides insight into the folding mechanism of these chains. In particular, we consider the progress of folding in time and increasing values of the fraction of native contacts.
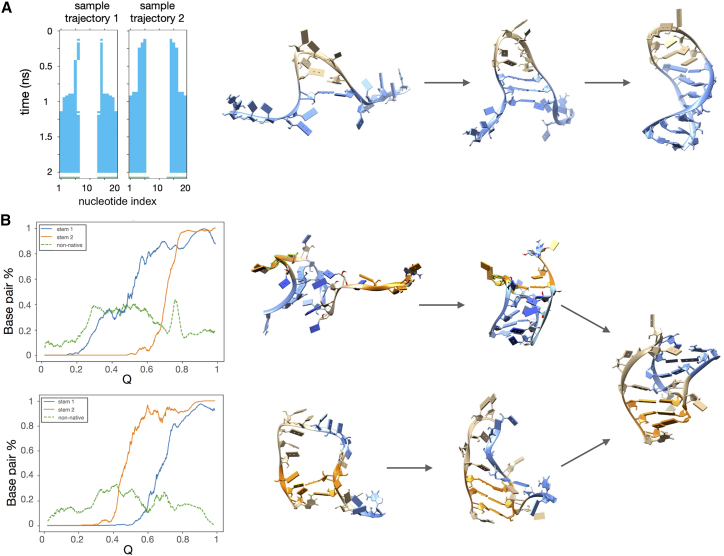
We note that the formation of the stems is a key step of the folding process for both representative RNAs, albeit with important differences informed by their architectures, see Fig. 2. For HHR, the most explored folding pathway is one in which the transition is nucleated by the formation of key contacts at the turn of the loop, following a “zipping mechanism.” This is exemplified by the two folding trajectories of Fig. 2 A. Zipping from the loop toward the termini can proceed more or less rapidly and is by far the dominant mechanism: among all trajectories that reached the native structure (49 out of 100) only one featured a reverse zipping mechanism, i.e., with the ladder of pairings starting from the termini. (Movies displaying representative events after the zipping (Videos S1 and S2) and reversed zipping (Video S3) mechanisms are available for download as Supporting material.) Our results show that reverse zipping is possible but, as expected, its occurrence is statistically marginal, even for medium-sized hairpins such as HHR. The limited statistical incidence is consistent with the expected higher entropic cost of forming the hairpin from the termini.
Figure 2.
Folding pathways of (A) the HHR hairpin and (B) the tmRNA pseudoknot. (A) Left: typical mechanism of hairpin formation, from two independent trajectories. The color bands indicate the time order of formation of contacts between nucleotides in the hairpin. The hairpin zipping initiates at the hairpin tip and proceeds toward the termini. Representative snapshots are shown on the right. (B) Left: the progressive formation of tmRNA structural elements is shown as a function of the fraction of native contacts, Q, for two typical productive trajectories. The curves indicate the percentage of established native contacts in stems 1 and 2. Canonical and noncanonical nonnative contacts are shown too, normalized by 10 to have a comparable scale with native contacts. In the first example (top), stem 1 (blue) forms first, followed by stem 2 (orange). In the second example (bottom), the order is reversed. Representative snapshots of the two folding pathways are shown on the right, adopting the same color code convention as in the plots on the left. To see this figure in color, go online.
For the tmRNA pseudoknot, folding can proceed by two distinct pathways: either forming stem 1 first (G1-G4:C10-C13) followed by stem 2 (G6-C8:G19-C21) or the other way around (Fig. 2, B and C). In our set of productive folding trajectories, the first option is slightly more represented (28 vs. 22 out of 50 folding events). This is coherent with the fact that stem 1 is more stable than stem 2 thanks to the additional C-G pair, in line with the correlation between folding order and thermodynamics stability reported in (58,59). In the plots, we report also the number of nonnative basepairs, canonical or noncanonical. For illustration purposes, we arbitrarily normalized these curves by dividing the number of nonnative pairs by 10, a typical number observed in nonproductive trajectories. In productive trajectories, the incidence of nonnative pairs is limited. However, in all cases, the folding begins with the formation of a couple of nonnative pairs. This reflects the fact that the chain needs to form at least a minimum number of initial contacts to collapse and start folding the stems.
Comparison with experiment and coarse-grained models
To compare the predictions of our method against experimental results, we simulated the folding of the P2B-P3 pseudoknot from human telomerase (delta U177 variant, PDB: 2K96, referred to as hTR) (60), which has been extensively characterized by NMR, calorimetry, and FRET experiments (61,62), and also studied using different coarse-grained models (59,63,64,65). This molecule consists of 47 nucleotides and in its native state adopts the conformation of an H pseudoknot with two stems and two loops. The longer loop (loop 1) wraps around one of the stems (stem 2) forming a series of successive U-A-U triplets and a triple helix. This gives the molecule a straight and tight shape.
As before, we analyzed of the formation of the secondary structure elements as a function of native overlap Q. This highlights the presence of three folding pathways, one of which is largely dominant. Nineteen out of 25 productive trajectories fold by first forming stem 2 (Fig. 3 A). This is followed by the formation of the triplets of the triple helix and the stem 1, closing the pseudoknot. Noncanonical interactions of loop 2 build up all along the folding process. They begin to form during the formation of stem 2 and they complete during the folding of stem 1. In the meantime, other nonnative pairs form, mainly involving nucleotides of loop 2. This results in the early formation of a proto-loop involving the same nucleotides as loop 2, but with an out-of-register pairing with respect to the native structure. This process is followed by a successive readjustment of the basepairs, progressively matching the native contact pattern.
Figure 3.
Formation of native secondary structural elements as a function of the fraction of native contacts (Q) for hTR. Three separate productive folding pathways emerge. In (A) stem 2 (yellow) forms first, followed by the formation of part of loop 2 (green), of the triplets (red), and, finally, the formation of stem 1 (blue). In (B) and (C) stem 1 forms before stem 2, but in (B) the first elements that forms are the triplets, while in (C) the first element is stem 1 directly. The right panel shows representative structures from each pathway toward the experimental structure, which is the rightmost structure on the right in the middle row (magnified). To see this figure in color, go online.
In the remaining six productive trajectories, stem 2 forms last. In half of them, the triplets form first, followed by the simultaneous formation of stem 1 and loop 2 (Fig. 3 B). In this case, a fraction of the basepairs of stem 2 forms early on, before stem 1 begins to form, but the rest of the stem forms later. In the other three productive events, stem 1 forms first followed by the simultaneous formation of stem 2 and the triplets. Loop 2 is, however, partially formed when stem 1 begins to fold (Fig. 3 C).
As we observed for the small pseudoknot, also in this case the folding reaction is seeded by transient nonnative pairs that induce the collapse of the chain.
The picture emerging from these folding mechanisms is in qualitative agreement with the results of thermal denaturation experiments (61). Following previous simulation studies of the same system (59), from the observation of the hierarchical folding of RNA molecules (66) it is plausible to draw a heuristic connection between thermal stability and temporal order of events in folding. Thermal denaturation of the mutant of hTR occurs at a melting temperature of around C for both stems. However, the melting of the loops differs significantly, with C for loop 1, which contains the triplets, and C for loop 2. This hints at a faster folding of loop 2 than loop 1 and agrees with our observation that loop 2 starts to form consistently before the triplets, which are the main structural feature of loop 1. Moreover, the interactions in loop 2 are more short-ranged than those of the two stems, which have comparable enthalpy. Therefore, this argument suggests that the formation of loop 2 is likely the initial folding step.
Our landscape agrees with this picture, with loop 2 being the first structure to form, followed by stem 2, and then stem 1 together with the triplets.
This preferred pathway has also been observed in previous coarse-grained simulations of the same system, both with (59) and without (65) native bias. The biased simulations were based on the TIS model and reported the same main folding steps detailed in this study (59). Two folding pathways were found with stem1 forming before stem 2 and vice versa, but it was not possible to assess whether one was more likely than the other. The unbiased simulations, carried out with replica-exchange (HiRE-RNA) (65), found a plurality of metastable states, including partially folded ones that we also observe here, with stem 2 formed but not the pseudoknot, but also included traps with significantly different arrangements than the folded state. In both these earlier studies, the pathways are consistent with the observation from our rMD simulations about the roughness of the RNA folding landscape. Our current results can go one step further by providing the atomistic detail that is missing in coarse-grained models and by explicitly accounting for solvation effects. Moreover, the rMD scheme allows us to gather better statistics on folding trajectories. This makes it possible for us to estimate the relative statistical weight of the competing folding mechanisms.
Discussion
The comparison of the frequency histograms of RNAs and proteins of similar length and topology highlights key differences between the folding landscapes of these two classes of biopolymers. In particular, our results indicate that the landscape of RNAs is significantly more frustrated, displaying a larger number of local minima. This is suggestive of a larger conformational heterogeneity of the RNA equilibrium ensembles. These results, along with those of other recent path-sampling simulations (10,22,23,67,68), structure prediction (63,65,69,70,71,72) and experiments (53,73,74) coherently point toward the conclusion that RNAs are moderately frustrated systems, with energy landscapes displaying several alternative competing minima.
The computational cost of generating many folding pathways for the hTR pseudoknot in explicit solvent was relatively low, enabling the completion of our simulations in a just few days on a small GPU cluster. Our calculations are already in good agreement with the mechanism inferred by thermodynamic experiments and with previous simulations performed in coarse-grained models.
An important direction for future improvement would be to obtain more precise estimates of free energy barriers, which are generally underestimated when the biasing CV is only a proxy of the committor function. In this first application to RNAs, we adopted a CV based on the measure of the instantaneous overlap with the native contact map. This is known to be a good reaction coordinate for protein folding, but may not be as fit for more frustrated systems. In the future, this possible limitation may be overcome by employing more computationally demanding approaches in which the biasing CV is optimized using self-consistent refinement iterations (75,76) or by machine learning (77,78,79,80).
In the context of protein folding, the BF scheme has been successfully validated against available plain MD folding trajectories (33,38) and several experiments (42,43,81,82). The accuracy of the predictions allowed for rationalizing the effect of point mutations related to protein misfolding (81) and even provided an innovative scheme to identify small molecules that can interfere with the folding process, leading to a new class of protein degraders (44). Further extending the scope of such folding simulation methods to RNA would open numerous perspectives. For instance, it would allow for detecting bifurcation points of the folding process, determining how the sequence folds on a specific state over the possible alternatives, or identifying metastable states that can become dominant upon changes in environmental conditions. This would provide invaluable insights to better understand the folding mechanisms of these systems as well as to envision strategies for drug design down the road.
Author contributions
G.L. performed research and analyzed the data. C.M., S.P., and P.F. designed the research, supervised the work, analyzed the data, and wrote the manuscript.
Acknowledgments
We thank Elisa Marchioro for providing scripts for data analysis and free energy landscape calculations. This work was supported by an STSM Grant from COST Action CA17139 (eutopia.unitn.eu) funded by COST (www.cost.eu).
Declaration of interests
P.F. is cofounder of Sibylla Biotech (www.sibyllabiotech.it), a company involved in early-stage drug discovery.
Editor: Filip Lankas.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2023.06.012.
Contributor Information
Cristian Micheletti, Email: cristian.micheletti@sissa.it.
Samuela Pasquali, Email: samuela.pasquali@u-paris.fr.
Pietro Faccioli, Email: pietro.faccioli@unimib.it.
Supporting material
References
- 1.Sharp P.A. The Centrality of RNA. Cell. 2009;136:577–580. doi: 10.1016/j.cell.2009.02.007. https://www.cell.com/cell/abstract/S0092-8674(09)00143-3 [DOI] [PubMed] [Google Scholar]
- 2.Jiao A.L., Slack F.J. RNA-mediated gene activation. Epigenetics. 2014;9:27–36. doi: 10.4161/epi.26942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michelini F., Jalihal A.P., et al. Fagagna F.d. d. From “Cellular” RNA to “Smart” RNA: Multiple Roles of RNA in genome stability and beyond. Chem. Rev. 2018;118:4365–4403. doi: 10.1021/acs.chemrev.7b00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balvay L., Lopez Lastra M., et al. Ohlmann T. Translational control of retroviruses. Nat. Rev. Microbiol. 2007;5:128–140. doi: 10.1038/nrmicro1599. https://www.nature.com/articles/nrmicro1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaafar Z.A., Kieft J.S. Viral RNA structure-based strategies to manipulate translation. Nat. Rev. Microbiol. 2019;17:110–123. doi: 10.1038/s41579-018-0117-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leontis N.B., Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garst A.D., Edwards A.L., Batey R.T. Riboswitches: structures and mechanisms. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saldi T., Riemondy K., et al. Bentley D.L. Alternative RNA structures formed during transcription depend on elongation rate and modify RNA processing. Mol. Cell. 2021;81:1789–1801.e5. doi: 10.1016/j.molcel.2021.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinez-Zapien D., Legrand P., et al. Dock-Bregeon A.-C. The crystal structure of the 5' functional domain of the transcription riboregulator 7SK. Nucleic Acids Res. 2017;45:3568–3579. doi: 10.1093/nar/gkw1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Röder K., Stirnemann G., et al. Pasquali S. Structural transitions in the RNA 7SK 5’ hairpin and their effect on HEXIM binding. Nucleic Acids Res. 2020;48:373–389. doi: 10.1093/nar/gkz1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X., Lu Z., et al. He C. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manners O., Baquero-Perez B., Whitehouse A. m6A: Widespread regulatory control in virus replication. Biochimica et Biophysica Acta. Gene Regulatory Mechanisms. 2019;1862:370–381. doi: 10.1016/j.bbagrm.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Šponer J., Bussi G., et al. Otyepka M. RNA Structural Dynamics As Captured by Molecular Simulations: A Comprehensive Overview. Chem. Rev. 2018;118:4177–4338. doi: 10.1021/acs.chemrev.7b00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miner J.C., Chen A.A., García A.E. Free-energy landscape of a hyperstable RNA tetraloop. Proc. Natl. Acad. Sci. USA. 2016;113:6665–6670. doi: 10.1073/pnas.1603154113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kührová P., Best R.B., et al. Banáš P. Computer folding of RNA tetraloops: identification of key force field deficiencies. J. Chem. Theory Comput. 2016;12:4534–4548. doi: 10.1021/acs.jctc.6b00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mlýnský V., Janeček M., et al. Šponer J. Toward Convergence in Folding Simulations of RNA Tetraloops: Comparison of Enhanced Sampling Techniques and Effects of Force Field Modifications. J. Chem. Theory Comput. 2022;18:2642–2656. doi: 10.1021/acs.jctc.1c01222. [DOI] [PubMed] [Google Scholar]
- 17.Bottaro S., Bussi G., Lindorff-Larsen K. Conformational Ensembles of Noncoding Elements in the SARS-CoV-2 Genome from Molecular Dynamics Simulations. J. Am. Chem. Soc. 2021;143:8333–8343. doi: 10.1021/jacs.1c01094. [DOI] [PubMed] [Google Scholar]
- 18.Omar S.I., Zhao M., et al. Woodside M.T. Modeling the structure of the frameshift-stimulatory pseudoknot in SARS-CoV-2 reveals multiple possible conformers. PLoS Comput. Biol. 2021;17 doi: 10.1371/journal.pcbi.1008603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schlick T., Zhu Q., et al. Yan S. Structure-altering mutations of the SARS-CoV-2 frameshifting RNA element. Biophys. J. 2021;120:1040–1053. doi: 10.1016/j.bpj.2020.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlick T., Zhu Q., et al. Laederach A. To Knot or Not to Knot: Multiple Conformations of the SARS-CoV-2 Frameshifting RNA Element. J. Am. Chem. Soc. 2021;143:11404–11422. doi: 10.1021/jacs.1c03003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan S., Zhu Q., et al. Schlick T. Length-dependent motions of SARS-CoV-2 frameshifting RNA pseudoknot and alternative conformations suggest avenues for frameshifting suppression. Research Square. 2022 doi: 10.1038/s41467-022-31353-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joseph J.A., Röder K., et al. Wales D.J. Exploring biomolecular energy landscapes. Chem. Commun. 2017;53:6974–6988. doi: 10.1039/c7cc02413d. [DOI] [PubMed] [Google Scholar]
- 23.Röder K., Barker A.M., et al. Pasquali S. Investigating the structural changes due to adenosine methylation of the Kaposi’s sarcoma-associated herpes virus ORF50 transcript. bioRxiv. 2021 doi: 10.1101/2021.11.16.468829. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cragnolini T., Derreumaux P., Pasquali S. Ab initio RNA folding. J. Phys. Condens. Matter. 2015;27 doi: 10.1088/0953-8984/27/23/233102. [DOI] [PubMed] [Google Scholar]
- 25.Dawson W.K., Maciejczyk M., et al. Bujnicki J.M. Coarse-grained modeling of RNA 3D structure. Methods. 2016;103:138–156. doi: 10.1016/j.ymeth.2016.04.026. [DOI] [PubMed] [Google Scholar]
- 26.Li J., Chen S.-J. RNA 3D structure prediction using coarse-grained models. Front. Mol. Biosci. 2021;8:e720937. doi: 10.3389/fmolb.2021.720937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poblete S., Bottaro S., Bussi G. A nucleobase-centered coarse-grained representation for structure prediction of RNA motifs. Nucleic Acids Res. 2018;46:1674–1683. doi: 10.1093/nar/gkx1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paci E., Karplus M. Forced unfolding of fibronectin type 3 modules: an analysis by biased molecular dynamics simulations. J. Mol. Biol. 1999;288:441–459. doi: 10.1006/jmbi.1999.2670. [DOI] [PubMed] [Google Scholar]
- 29.Camilloni C., Broglia R.A., Tiana G. Hierarchy of folding and unfolding events of protein G, CI2, and ACBP from explicit-solvent simulations. J. Chem. Phys. 2011;134 doi: 10.1063/1.3523345. [DOI] [PubMed] [Google Scholar]
- 30.E W., Vanden-Eijnden E. Transition-Path Theory and Path-Finding Algorithms for the Study of Rare Events. Annu. Rev. Phys. Chem. 2010;61:391–420. doi: 10.1146/annurev.physchem.040808.090412. [DOI] [PubMed] [Google Scholar]
- 31.Bartolucci G., Orioli S., Faccioli P. Transition path theory from biased simulations. J. Chem. Phys. 2018;149 doi: 10.1063/1.5027253. [DOI] [PubMed] [Google Scholar]
- 32.Cameron M., Vanden-Eijnden E. Flows in Complex Networks: Theory, Algorithms, and Application to Lennard-Jones Cluster Rearrangement. J. Stat. Phys. 2014;156:427–454. [Google Scholar]
- 33.A Beccara S., Fant L., Faccioli P. Variational scheme to compute protein reaction pathways using atomistic force fields with explicit solvent. Phys. Rev. Lett. 2015;114 doi: 10.1103/PhysRevLett.114.098103. [DOI] [PubMed] [Google Scholar]
- 34.Zgarbová M., Otyepka M., et al. Jurečka P. Refinement of the Cornell et al. nucleic acids force field based on reference quantum chemical calculations of glycosidic torsion profiles. J. Chem. Theory Comput. 2011;7:2886–2902. doi: 10.1021/ct200162x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abraham M.J., Murtola T., et al. Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1-2:19–25. [Google Scholar]
- 36.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126 doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 37.Doutreligne S., Gageat C., et al. Baaden M. Virtual and Augmented Reality for Molecular Science (VARMS@IEEEVR), 2015 IEEE 1st International Workshop on. 2015. UnityMol: interactive and ludic visual manipulation of coarse-grained RNA and other biomolecules. [Google Scholar]
- 38.Terruzzi L., Spagnolli G., et al. Faccioli P. All-atom simulation of the HET-s prion replication. PLoS Comput. Biol. 2020;16 doi: 10.1371/journal.pcbi.1007922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGibbon R.T., Beauchamp K.A., et al. Pande V.S. MDTraj: A Modern Open Library for the Analysis of Molecular Dynamics Trajectories. Biophys. J. 2015;109:1528–1532. doi: 10.1016/j.bpj.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu X.-J., Bussemaker H.J., Olson W.K. DSSR: an integrated software tool for dissecting the spatial structure of RNA. Nucleic Acids Res. 2015;43:e142. doi: 10.1093/nar/gkv716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bottaro S., Bussi G., et al. Lindorff-Larsen K. Barnaba: software for analysis of nucleic acid structures and trajectories. RNA. 2019;25:219–231. doi: 10.1261/rna.067678.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ianeselli A., Orioli S., et al. Mennucci B. Atomic Detail of Protein Folding Revealed by an Ab Initio Reappraisal of Circular Dichroism. J. Am. Chem. Soc. 2018;140:3674–3682. doi: 10.1021/jacs.7b12399. [DOI] [PubMed] [Google Scholar]
- 43.Dingfelder F., Macocco I., et al. Schuler B. Slow Escape from a Helical Misfolded State of the Pore-Forming Toxin Cytolysin A. JACS Au. 2021;1:1217–1230. doi: 10.1021/jacsau.1c00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spagnolli G., Massignan T., et al. Biasini E. Pharmacological inactivation of the prion protein by targeting a folding intermediate. Commun. Biol. 2021;4:62. doi: 10.1038/s42003-020-01585-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dufour D., de la Peña M., et al. Gallego J. Structure-function analysis of the ribozymes of chrysanthemum chlorotic mottle viroid: a loop-loop interaction motif conserved in most natural hammerheads. Nucleic Acids Res. 2009;37:368–381. doi: 10.1093/nar/gkn918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nonin-Lecomte S., Felden B., Dardel F. NMR structure of the Aquifex aeolicus tmRNA pseudoknot PK1: new insights into the recoding event of the ribosomal trans-translation. Nucleic Acids Res. 2006;34:1847–1853. doi: 10.1093/nar/gkl111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dill K.A., Chan H.S. From Levinthal to pathways to funnels. Nat. Struct. Biol. 1997;4:10–19. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 48.Wolynes P.G., Onuchic J.N., Thirumalai D. Navigating the folding routes. Science. 1995;267:1619–1620. doi: 10.1126/science.7886447. [DOI] [PubMed] [Google Scholar]
- 49.Baldwin R.L. The nature of protein folding pathways: The classical versus the new view. J. Biomol. NMR. 1995;5:103–109. doi: 10.1007/BF00208801. [DOI] [PubMed] [Google Scholar]
- 50.Onuchic J.N., Luthey-Schulten Z., Wolynes P.G. Theory of protein folding: the energy landscape perspective. Annu. Rev. Phys. Chem. 1997;48:545–600. doi: 10.1146/annurev.physchem.48.1.545. [DOI] [PubMed] [Google Scholar]
- 51.Zhao L., Xia T. Direct Revelation of Multiple Conformations in RNA by Femtosecond Dynamics. J. Am. Chem. Soc. 2007;129:4118–4119. doi: 10.1021/ja068391q. [DOI] [PubMed] [Google Scholar]
- 52.Gupta P., Khadake R.M., et al. Rode A.B. Alternative RNA Conformations: Companion or Combatant. Genes. 2022;13:1930. doi: 10.3390/genes13111930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu M.T.-P., D’Souza V. Alternate RNA Structures. Cold Spring Harb. Perspect. Biol. 2020;12:a032425. doi: 10.1101/cshperspect.a032425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kalinina M., Skvortsov D., et al. Pervouchine D.D. Multiple competing RNA structures dynamically control alternative splicing in the human ATE1 gene. Nucleic Acids Res. 2021;49:479–490. doi: 10.1093/nar/gkaa1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tomezsko P.J., Corbin V.D.A., et al. Rouskin S. Determination of RNA structural diversity and its role in HIV-1 RNA splicing regulation. Nature. 2020;582:438–442. doi: 10.1038/s41586-020-2253-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vicens Q., Kieft J.S. Thoughts on how to think (and talk) about RNA structure. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2112677119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woodson S.A. Compact Intermediates in RNA Folding. Annu. Rev. Biophys. 2010;39:61–77. doi: 10.1146/annurev.biophys.093008.131334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tinoco I., Bustamante C. The preferential route. J. Mol. Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 59.Cho S.S., Pincus D.L., Thirumalai D. Assembly mechanisms of RNA pseudoknots are determined by the stabilities of constituent secondary structures. Proc. Natl. Acad. Sci. USA. 2009;106:17349–17354. doi: 10.1073/pnas.0906625106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim N.-K., Zhang Q., et al. Feigon J. Solution Structure and Dynamics of the Wild-type Pseudoknot of Human Telomerase RNA. J. Mol. Biol. 2008;384:1249–1261. doi: 10.1016/j.jmb.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Theimer C.A., Blois C.A., Feigon J. Structure of the human telomerase rna pseudoknot reveals conserved tertiary interactions essential for function. Mol. Cell. 2005;17:671–682. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 62.Gavory G., Symmons M.F., et al. Balasubramanian S. Structural Analysis of the Catalytic Core of Human Telomerase RNA by FRET and Molecular Modeling. Biochemistry. 2006;45:13304–13311. doi: 10.1021/bi061150a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Denesyuk N.A., Thirumalai D. Crowding Promotes the Switch from Hairpin to Pseudoknot Conformation in Human Telomerase RNA. J. Am. Chem. Soc. 2011;133:11858–11861. doi: 10.1021/ja2035128. [DOI] [PubMed] [Google Scholar]
- 64.Biyun S., Cho S.S., Thirumalai D. Folding of Human Telomerase RNA Pseudoknot Using Ion-Jump and Temperature-Quench Simulations. J. Am. Chem. Soc. 2011;133:20634–20643. doi: 10.1021/ja2092823. [DOI] [PubMed] [Google Scholar]
- 65.Cragnolini T., Laurin Y., et al. Pasquali S. Coarse-Grained HiRE-RNA Model for ab Initio RNA Folding beyond Simple Molecules, Including Noncanonical and Multiple Base Pairings. J. Chem. Theory Comput. 2015;11:3510–3522. doi: 10.1021/acs.jctc.5b00200. [DOI] [PubMed] [Google Scholar]
- 66.Li P.T.X., Vieregg J., Tinoco I. How RNA unfolds and refolds. Annu. Rev. Biochem. 2008;77:77–100. doi: 10.1146/annurev.biochem.77.061206.174353. [DOI] [PubMed] [Google Scholar]
- 67.de Souza V.K., Stevenson J.D., et al. Wales D.J. Defining and quantifying frustration in the energy landscape: Applications to atomic and molecular clusters, biomolecules, jammed and glassy systems. J. Chem. Phys. 2017;146 doi: 10.1063/1.4977794. [DOI] [PubMed] [Google Scholar]
- 68.Cragnolini T., Chakraborty D., et al. Wales D.J. Multifunctional energy landscape for a DNA G-quadruplex: An evolved molecular switch. J. Chem. Phys. 2017;147 doi: 10.1063/1.4997377. [DOI] [PubMed] [Google Scholar]
- 69.Schroeder S.J. Challenges and approaches to predicting RNA with multiple functional structures. RNA. 2018;24:1615–1624. doi: 10.1261/rna.067827.118. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6239171/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cruz J.A., Blanchet M.-F., et al. Westhof E. RNA-Puzzles: a CASP-like evaluation of RNA three-dimensional structure prediction. RNA (New York, N.Y.) 2012;18:610–625. doi: 10.1261/rna.031054.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miao Z., Adamiak R.W., et al. Westhof E. RNA-Puzzles Round II: assessment of RNA structure prediction programs applied to three large RNA structures. RNA. 2015;21:1066–1084. doi: 10.1261/rna.049502.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Miao Z., Adamiak R.W., et al. Westhof E. RNA-Puzzles Round III: 3D RNA structure prediction of five riboswitches and one ribozyme. RNA. 2017;23:655–672. doi: 10.1261/rna.060368.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ritz J., Martin J.S., Laederach A. Evolutionary Evidence for Alternative Structure in RNA Sequence Co-variation. PLoS Comput. Biol. 2013;9 doi: 10.1371/journal.pcbi.1003152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu A.M., Gasper P.M., et al. Lucks J.B. Computationally reconstructing cotranscriptional RNA folding from experimental data reveals rearrangement of non-native folding intermediates. Mol. Cell. 2021;81:870–883.e10. doi: 10.1016/j.molcel.2020.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Orioli S., a Beccara S., Faccioli P. Self-consistent calculation of protein folding pathways. J. Chem. Phys. 2017;147 doi: 10.1063/1.4997197. [DOI] [PubMed] [Google Scholar]
- 76.Pérez de Alba Ortíz A., Tiwari A., et al. Ensing B. Advances in enhanced sampling along adaptive paths of collective variables. J. Chem. Phys. 2018;149 doi: 10.1063/1.5027392. [DOI] [PubMed] [Google Scholar]
- 77.Rogal J., Schneider E., Tuckerman M.E. Neural-network-based path collective variables for enhanced sampling of phase transformations. Phys. Rev. Lett. 2019;123 doi: 10.1103/PhysRevLett.123.245701. [DOI] [PubMed] [Google Scholar]
- 78.Jung H., Covino R., et al. Hummer G. Autonomous artificial intelligence discovers mechanisms of molecular self-organization in virtual experiments. arXiv. 2021 doi: 10.48550/arXiv.2105.06673. Preprint at. [DOI] [Google Scholar]
- 79.Bolhuis P.G., Brotzakis Z.F., Vendruscolo M. A maximum caliber approach for continuum path ensembles. Eur. Phys. J. B. 2021;94:188–221. [Google Scholar]
- 80.Bonati L., Rizzi V., Parrinello M. Data-driven collective variables for enhanced sampling. J. Phys. Chem. Lett. 2020;11:2998–3004. doi: 10.1021/acs.jpclett.0c00535. [DOI] [PubMed] [Google Scholar]
- 81.Wang F., Orioli S., et al. Wintrode P.L. All-Atom simulations reveal how single-point mutations promote serpin misfolding. Biophys. J. 2018;114:2083–2094. doi: 10.1016/j.bpj.2018.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang F., Cazzolli G., et al. Faccioli P. Folding mechanism of proteins Im7 and Im9: insight from all-atom simulations in implicit and explicit solvent. J. Phys. Chem. B. 2016;120:9297–9307. doi: 10.1021/acs.jpcb.6b05819. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.