Abstract
Animals rely on chemosensory cues to survive in pathogen-rich environments. In Caenorhabditis elegans, pathogenic bacteria trigger aversive behaviors through neuronal perception and activate molecular defenses throughout the animal. This suggests that neurons can coordinate the activation of organism-wide defensive responses upon pathogen perception. In this study, we found that exposure to volatile pathogen-associated compounds induces activation of the endoplasmic reticulum unfolded protein response (UPRER) in peripheral tissues after xbp-1 splicing in neurons. This odorant-induced UPRER activation is dependent upon DAF-7/transforming growth factor beta (TGF-β) signaling and leads to extended lifespan and enhanced clearance of toxic proteins. Notably, rescue of the DAF-1 TGF-β receptor in RIM/RIC interneurons is sufficient to significantly recover UPRER activation upon 1-undecene exposure. Our data suggest that the cell non-autonomous UPRER rewires organismal proteostasis in response to pathogen detection, pre-empting proteotoxic stress. Thus, chemosensation of particular odors may be a route to manipulation of stress responses and longevity.
Subject terms: Stress signalling, Gene regulation, Ageing
De-Souza, Thompson and Taylor demonstrate that pathogen-associated odorants can activate the UPR cell non-autonomously in C. elegans via neuronal TGF-β signaling, leading to extended lifespan and enhanced clearance of toxic proteins.
Main
To adapt and survive, organisms must be able to detect and respond to environmental changes. In animals, this is mediated by the sensory nervous system, which activates defensive responses upon identification of hazards, such as reduced oxygen availability, temperature increase or food shortage1. In addition, the detection of stress within cells activates cellular stress responses, such as the unfolded protein response of the endoplasmic reticulum (UPRER), which respond to homeostatic imbalance by activating mechanisms that restore homeostasis2. As animals age, they lose this ability to recognize and respond to stress, resulting in increased mortality and age-related disease1,3–5. In particular, reduced activity of the IRE-1/XBP-1 signaling branch of the UPRER has been linked to brain aging and neurodegeneration, whereas genetic activation of XBP-1 can protect animals against proteotoxic insults5,6.
Recent evidence suggests that neurons can trigger the cell non-autonomous activation of cellular stress responses in peripheral tissues, leading to coordinated increases in organismal resilience and lifespan. Consistent with this, genetic activation of the UPRER in a subset of neuronal or glial cells can extend lifespan in Caenorhabditis elegans via neuronal signaling mechanisms that result in UPRER activation in distal tissues7,8. However, whether specific environmental situations or exogenous molecules can trigger the activation of the cell non-autonomous UPRER in wild-type animals remains unknown. We therefore decided to identify physiologically relevant cues that drive cell non-autonomous UPRER activation in C. elegans.
Results
Pathogen-associated odorants can activate the UPRER
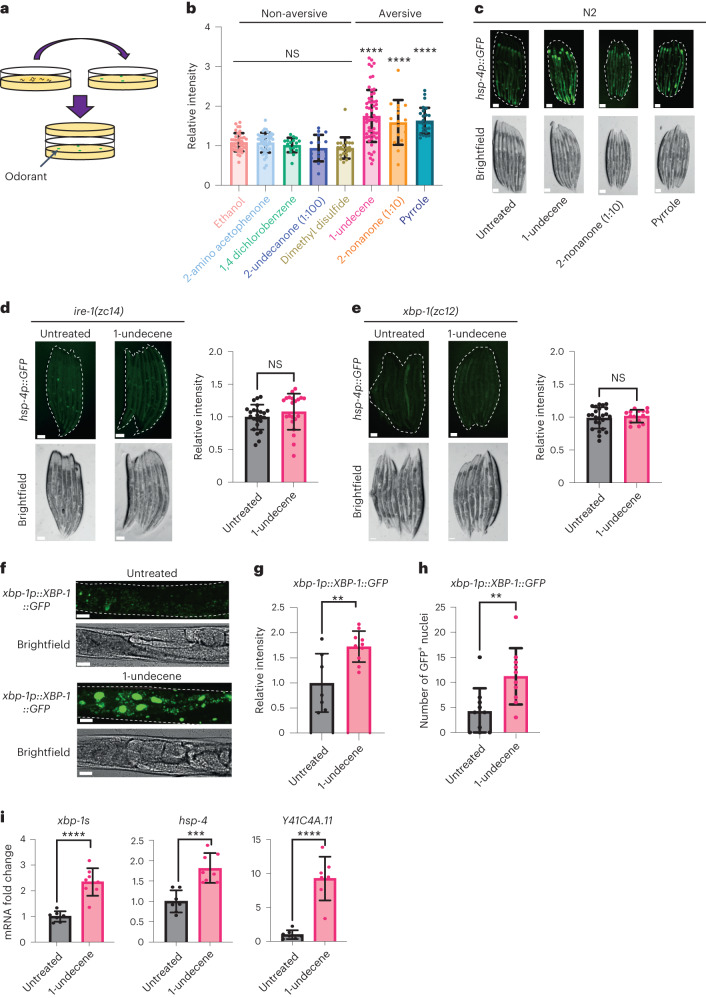
Olfactory perception of bacteria alters gene expression in invertebrates9, and the immune response to Pseudomonas spp is associated with UPRER activation in C. elegans3,10. The smell of pathogenic bacteria can also sensitize the heat shock response in worms11, suggesting a possible link between olfaction and proteostasis. We therefore asked whether pathogen-associated odor could activate the cell non-autonomous UPRER in C. elegans. We exposed animals to a variety of odorant molecules secreted by pathogenic bacteria, including Pseudomonas aeruginosa and Staphylococcus aureus12, and monitored the expression of hsp-4p::GFP, a transcriptional reporter of UPRER activation. Notably, because the volatile molecules and the worms were placed on different plates, there was no direct contact between them (Fig. 1a). We observed that the UPRER could be activated in the intestine by exposure to three odorant molecules: 1-undecene, pyrrole and 2-nonanone (Fig. 1b,c and Extended Data Fig. 1a). Curiously, all three compounds have previously been linked to aversive behavioral responses in worms13,14 (Extended Data Fig. 1b). We decided to focus on 1-undecene in subsequent experiments. As previously observed by others15, exposure to higher concentrations of 2-nonanone caused lethal toxicity in a majority of the population; however, exposure to 1-undecene did not cause any overt alteration in the physiology of exposed animals, such as changes to brood size (Extended Data Fig. 1c).
Fig. 1. Pathogen-associated odor activates the IRE-1/XBP-1 branch of the UPRER.
a, Schematic showing the experimental setup for the odorant exposure assay. In brief, young adult worms were sealed for 12 h in NGM plates together with another NGM plate containing four spots of 3 μl of odorant. b, Fluorescence intensity of hsp-4p::GFP after odorant exposure. Quantification of hsp-4p::GFP expression was performed in ImageJ, and data were normalized to untreated hsp-4p::GFP animals. This assay was independently performed three times (n = 39, 42, 17, 15, 21, 60, 16 and 27 animals). Graphs show mean ± s.d. ****P < 0.0001 (one-way ANOVA with Dunnett’s multiple comparison test). c, Representative fluorescence microscopy images of worms untreated or exposed to 1-undecene, 2-nonanone (diluted 10×) and pyrrole for 12 h. This assay was independently performed three times. Scale bars, 200 μm. d,e, Representative fluorescence microscopy images and quantification of hsp-4p::GFP fluorescence in ire-1(zc14) (d) and xbp-1(zc12) (e) worms with or without exposure to 1-undecene odor for 12 h. These experiments were repeated four times (n = 26 and 21 animals for d and n = 25 and 16 animals for e). Scale bars, 200 μm. Graphs show mean ± s.d. NS, not significant (two-tailed unpaired Student’s t-test). f, Representative image. g, Quantification of fluorescence. h, Number of GFP+ nuclei in the intestine of worms expressing an xbp-1p::xbp-1::GFP transgene with or without exposure to 1-undecene for 8 h. This experiment was repeated three times (n = 7 and 10 animals for g and n = 10 and 10 animals for h). Scale bars, 200 μm. Graphs show mean ± s.d. ****P < 0.0001 and **P < 0.01 (two-tailed unpaired Student’s t-test). i, mRNA levels of xbp-1s, hsp-4 and Y41C4A.11 were measured by qRT–PCR in animals exposed to 1-undecene for 8 h relative to untreated worms (n = 7 and 8 biological replicates). Graphs show mean ± s.d. ****P < 0.0001 and ***P < 0.001 (two-tailed unpaired Student’s t-test). Precise P values are provided in Source Data.
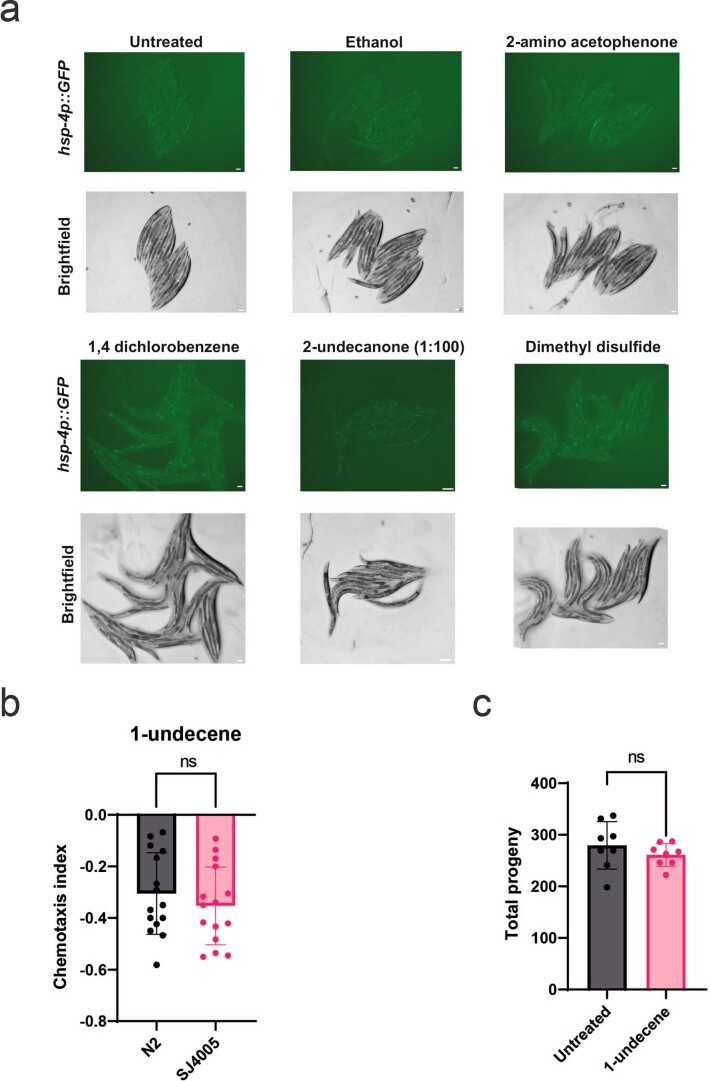
Extended Data Fig. 1. Non-aversive odorants do not induce UPRER activation and 1-undecene does not affect brood size.
a, Representative images from the hsp-4p::GFP odorant screen. Experiments were repeated three times with at least 10 worms per group. Scale bars, 200 μm. b, Chemotaxis index (CI) of N2 and SJ4005 (zcIs4 [hsp-4p::GFP]) worms exposed to 1-undecene. N = 15 biological repeats per group, 60 animals per replicate. Graph represents mean CI ± s.d. Ns, not significant (two-tailed unpaired Student’s t test). c, Number of eggs laid per worm in animals exposed to 1-undecene odor for 12 hours at day 1 of adulthood. n = 8 worms per group. Graphs show mean ± s.d. Ns, not significant (two-tailed unpaired Student’s t test). Source Data contains precise P values.
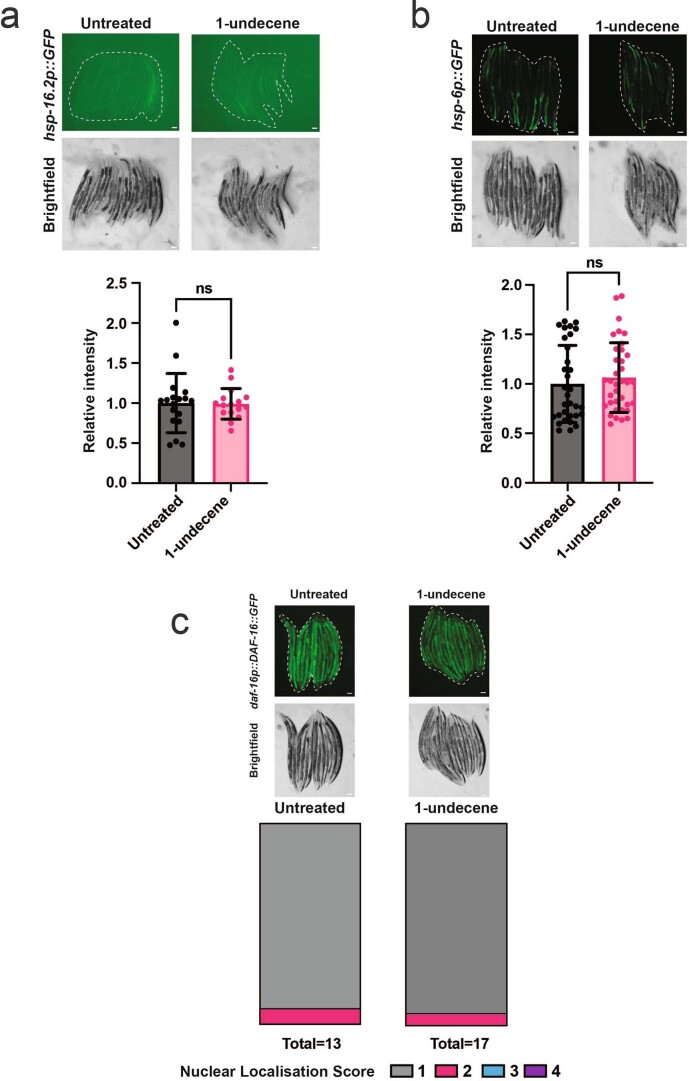
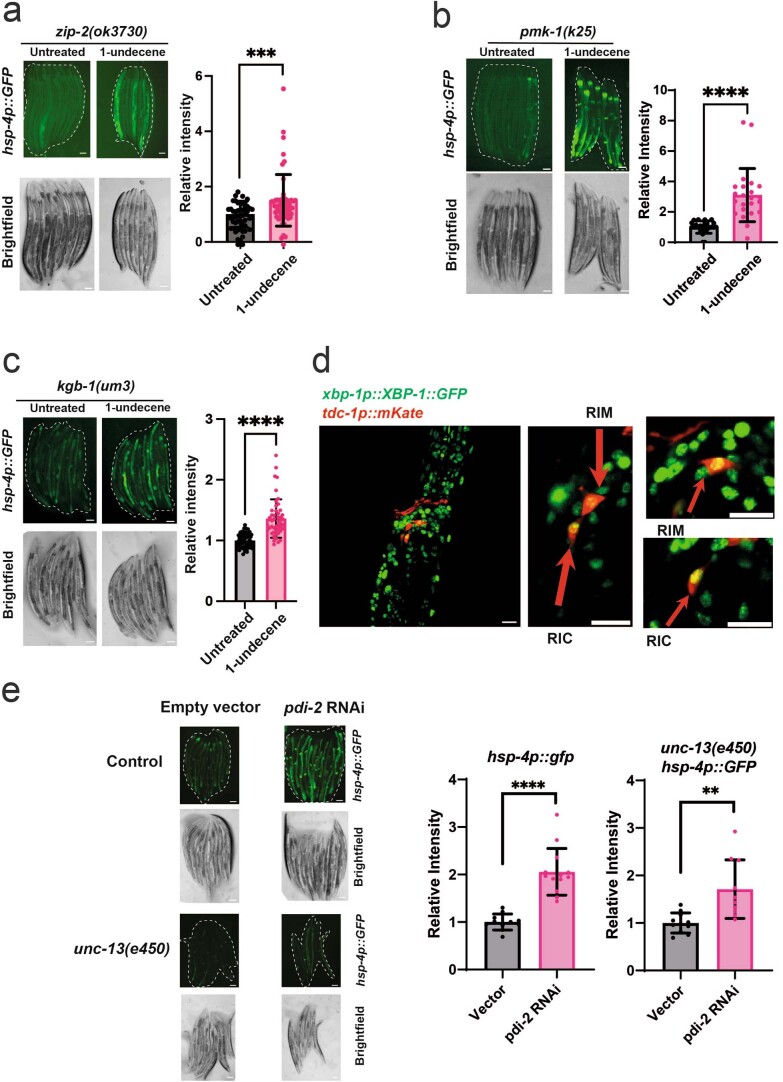
We found that mutation of the UPRER regulators ire-1 or xbp-1 abolished UPRER activation by 1-undecene odor, indicating that the IRE-1/XBP-1 signaling pathway is essential for activation of the UPRER by this compound (Fig. 1d,e). Consistent with this, an XBP-1s::GFP splicing reporter that expresses XBP-1s::GFP from an xbp-1p::xbp-1::GFP transgene only when xbp-1 mRNA is spliced by IRE-1 (ref. 8) revealed an increase in XBP-1s::GFP within the intestinal cells of animals exposed to 1-undecene (Fig. 1f–h). Furthermore, we observed a significant increase in transcript levels of spliced xbp-1 and two XBP-1s target genes (hsp-4 and Y41C4A.11), confirming activation of the IRE-1/XBP-1 pathway by 1-undecene (Fig. 1i). Interestingly, we were unable to detect activation of other cellular stress response pathways, including nuclear DAF-16 localization and hsp-16.2 (heat shock response) or hsp-6 (mitochondrial UPR) upregulation, suggesting that the UPRER is specifically activated by pathogen-associated odor (Extended Data Fig. 2a–c). Finally, a recent study found that the C. elegans immune system can also be activated by olfactory perception of 1-undecene16. However, odor-induced UPRER activation is unlikely to be a downstream consequence of immune response activation, as animals with mutations in the key immunity transcription factor zip-2, or in the immunity-associated kinases pmk-1 and kgb-1, still showed UPRER activation in response to 1-undecene (Extended Data Fig. 3a–c).
Extended Data Fig. 2. 1-undecene exposure does not activate other stress response pathways.
a,b, Representative fluorescence microscopy images and quantification of a, hsp-16.2p::GFP and b, hsp-6p::GFP fluorescence. These experiments were repeated three times. n = 18, 17 animals in a and n = 32, 36 animals in b. Scale bars, 200 μm. Graphs show mean ± s.d. Ns, not significant (two-tailed unpaired Student’s t test). c, Representative fluorescence microscopy images and quantification of the subcellular localization of DAF-16::GFP in worms expressing a daf-16p::DAF-16::GFP transgene. Worms were scored based on the number of intestinal cells that presented nuclear GFP localization, 1 = 0 cells (cytosolic GFP only), 2 = 2-4 cells, 3 = 5-8 cells, 4 = more than 8 cells. This experiment was repeated three times. n = 13, 17 animals. Scale bars, 200 μm. Source Data contains precise P values.
Extended Data Fig. 3. Activation of the UPRER in by 1-undecene odor does not require immune response regulators and occurs in RIM/RIC interneurons.
a, b, c, Representative fluorescence microscopy images and quantification of hsp-4p::GFP fluorescence in a, zip-2(ok3730), b, pmk-1(k25) and c, kgb-1(um3) animals with or without exposure to 1-undecene odor for 12 hours. Experiments were repeated three times. n = 50, 58 animals in a, n = 23, 18 animals in b and n = 54, 48 animals in c. Scale bars, 200 μm. Graphs show mean ± s.d. ***P < 0.001 in a and ****P < 0.0001 in b and c (two-tailed unpaired Student’s t test). d, Representative image of worms expressing a tdc-1p::mKate; xbp-1p::xbp-1::GFP transgene exposed to 8 hours of 1-undecene odor. Scale bars, 10 μm. Experiment repeated one time. Scale bars, 10 μm. e, Representative fluorescence microscopy images and quantification of hsp-4p::GFP fluorescence in hsp-4p::GFP animals with or without an unc-13(e450) mutant background grown from L1 larval stage on NGM plates containing bacteria harboring empty vector (L4440) or pdi-2 RNAi. Data were normalized by samples treated with vector only. The experiment was repeated twice. n = 11, 13, 11, 10. Scale bars, 200 μm. Graphs show mean ± s.d. **P < 0.01, ****P < 0.0001 (two-tailed unpaired Student’s t test). Source Data contains precise P values.
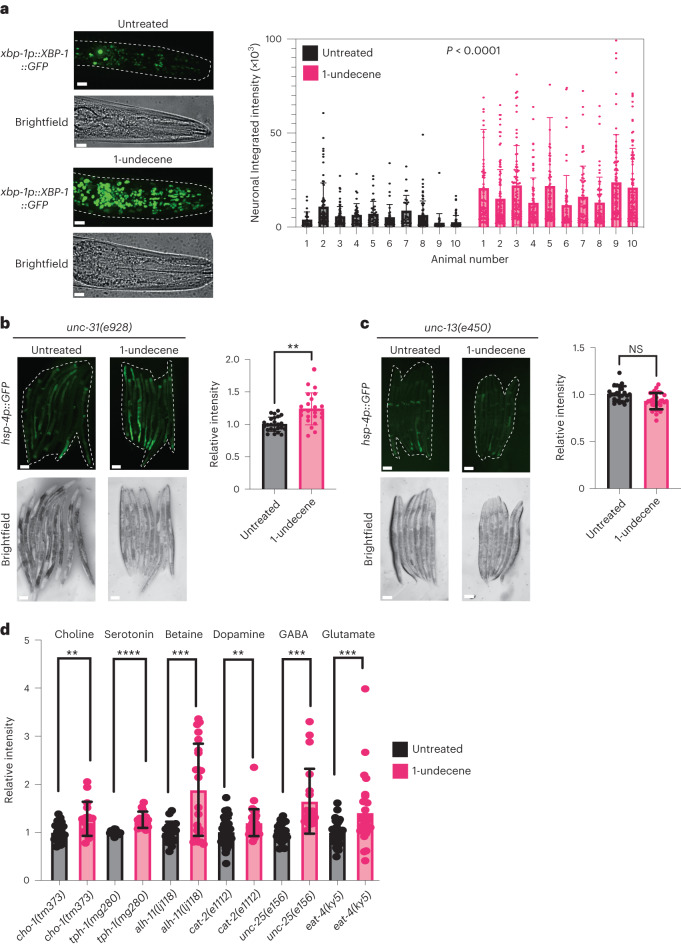
Previous work from our group and others demonstrated that neuronal signaling can activate the UPRER in peripheral tissues, such as the intestine3,17. We wondered whether signals produced by the nervous system were also responsible for odor-induced UPRER activation. We observed that animals exposed to pathogen-associated odor showed a significant increase in the number and fluorescence intensity of XBP-1s::GFP+ cells surrounding the pharynx (Fig. 2a), including neurons such as RIM and RIC (Extended Data Fig. 3d). To establish whether UPRER activation arising from 1-undecene exposure was cell non-autonomous in nature, we tested the dependency of this effect on the neuronal signaling regulators unc-31 and unc-13—mutations in the former blocking release of neuropeptides from dense core vesicles and in the latter preventing the release of a range of signaling molecules, including small-molecule neurotransmitters3,7. We observed that the hsp-4p::GFP reporter was activated in the intestine of unc-31(e928) mutant animals (Fig. 2b), whereas the unc-13(e450) mutation entirely inhibited activation of the UPRER in the periphery (Fig. 2c), suggesting that a non-neuropeptide neuronal signal is involved in cell non-autonomous UPRER activation by exposure to 1-undecene. Notably, mutation of unc-13 does not prevent animals from responding to cell-autonomous ER stress, as hsp-4p::GFP is still activated in animals exposed to RNA interference (RNAi) against pdi-2 (Extended Data Fig. 3e)18.
Fig. 2. Neuronal signaling is required for downstream UPRER activation by 1-undecene exposure.
a, Representative image and quantification of GFP+ cells in the head of worms expressing an xbp-1p::xbp-1::GFP transgene with or without exposure to 1-undecene for 8 h. This experiment was repeated three times with 10 worms per group. Scale bars, 10 μm. Graphs show mean ± s.d. Significance was calculated by unpaired Student’s t-test. b,c, Representative fluorescence microscopy images and quantification of hsp-4p::GFP fluorescence in unc-31(e928) (b) and unc-13(e450) (c) with or without exposure to 1-undecene odor for 12 h. These experiments were repeated three times (n = 24 and 23 animals for b and n = 23 and 30 animals for c). Scale bars, 200 μm. Graphs show mean ± s.d. NS, not significant and **P < 0.01 (two-tailed unpaired Student’s t-test) for b and c. d, Quantification of hsp-4p::GFP fluorescence in cho-1(tm373), tph-1(mg280), alh-1(ij118), cat-2(e1112), unc-25(e156) and eat-4(ky5) mutants. Intensity was normalized to untreated animals for each mutant strain. These experiments were repeated three times (n = 20, 16, 12, 15, 18, 22, 44, 39, 21, 20, 36 and 28). Graphs show mean ± s.d. **P < 0.01, ***P < 0.01 and ****P < 0.001 (two-tailed unpaired Student’s t-test comparison between untreated and 1-undecene). Precise P values are provided in Source Data.
TGF-β signaling is required for odorant-induced UPRER activation
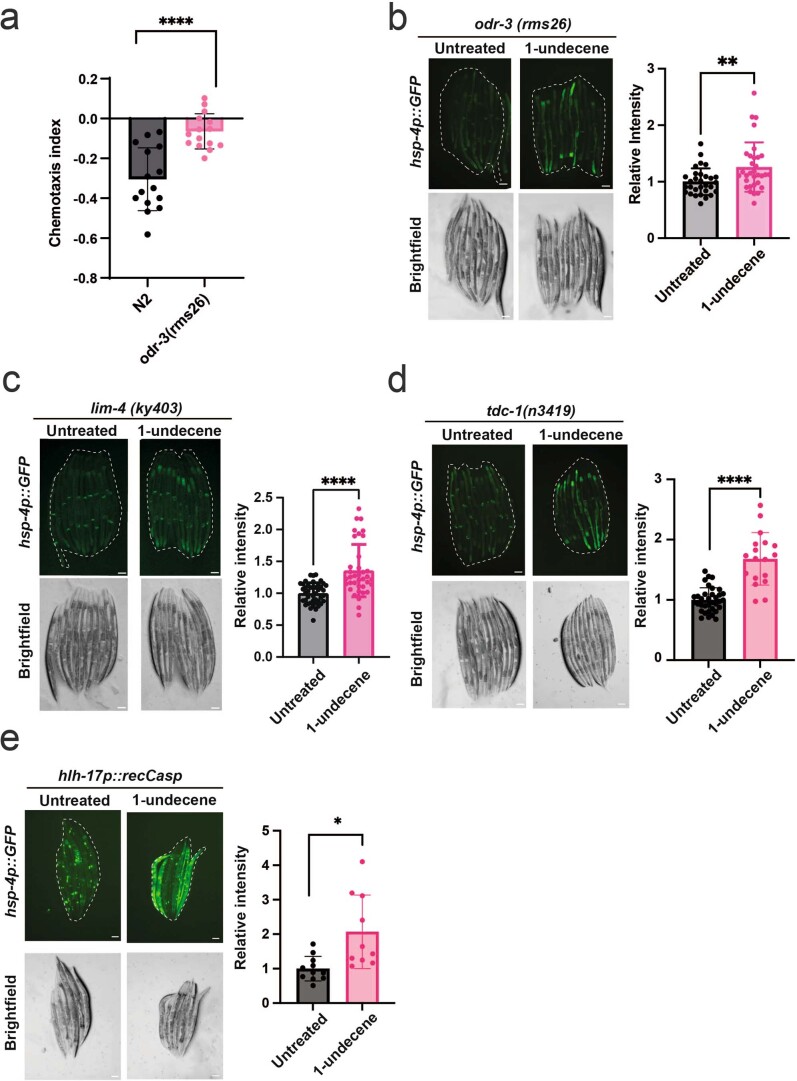
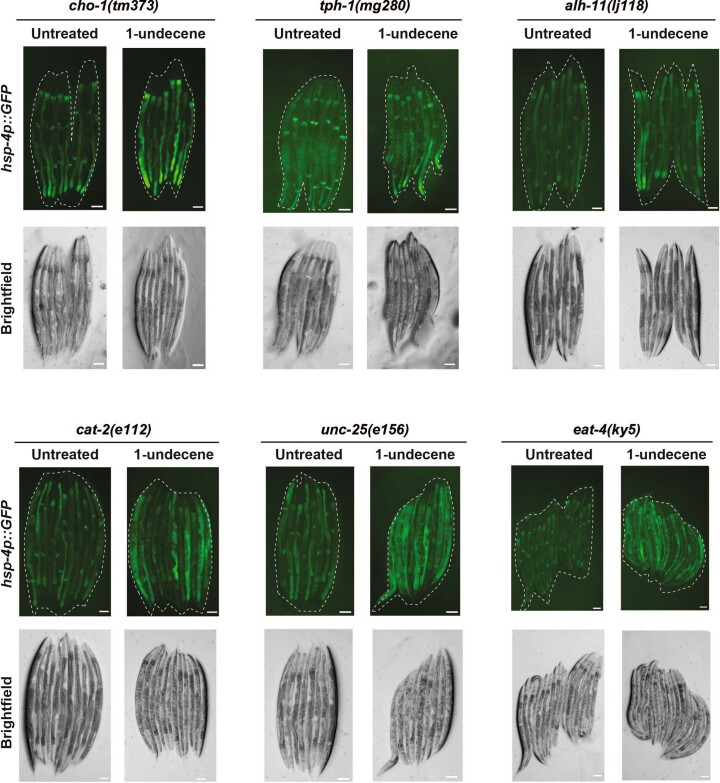
The Gα protein ODR-3 was previously shown to be required for activation of the immune response by 1-undecene16. We therefore asked whether ODR-3 is also required for 1-undecene-induced UPRER activation. To do this, we used CRISPR to generate an odr-3 deletion mutation in the hsp-4p::GFP background and confirmed that this mutation abolished the aversive behavioral response to 1-undecene (Extended Data Fig. 4a). However, we observed a full hsp-4p::GFP response in this odr-3 null background, suggesting that this gene is not required for UPRER activation by 1-undecene (Extended Data Fig. 4b). The immune response to 1-undecene also requires the AWB sensory neurons16. However, a lim-4 mutation, which results in dysfunction of AWB neurons, also failed to abolish 1-undecene-induced UPRER activation (Extended Data Fig. 4c). This suggests that the neuronal circuitry involved in the immune and UPRER activation responses to 1-undecene are different. In addition, tyramine synthesis is necessary for cell non-autonomous UPRER activation in strains constitutively expressing neuronal xbp-1s8. Unexpectedly, we found that tdc-1, a gene essential for the synthesis of tyramine, was not required for activation of hsp-4p::GFP in strains exposed to 1-undecene (Extended Data Fig. 4d). We also ruled out the possibility that the CEPsh glia are involved in this response, as animals in which these cells were genetically ablated still displayed increased hsp-4p::GFP levels after 1-undecene exposure (Extended Data Fig. 4e)7. We then tested mutants that fail to synthesize a variety of neurotransmitters, including dopamine, serotonin, GABA, glutamate, choline and betaine, for their ability to activate the UPRER in response to 1-undecene exposure, but we did not identify a role for any of these molecules (Fig. 2d and Extended Data Fig. 5).
Extended Data Fig. 4. ODR-3, LIM-4, tyramine synthesis, and CEPsh glia are not required for UPRER activation by 1-undecene odor.
a, Chemotaxis index of N2 and odr-3(rms31) animals following exposure to 1-undecene. n = 15 biological repeats per group, 60 animals per replicate (N2 data displayed is the same as Extended Data Fig. S1b as they were performed at the same time). Graphs show mean CI ± s.d. ****P < 0.0001 (two-tailed unpaired Student’s t test). b, c, d, e Representative fluorescence microscopy images and quantification of hsp-4p::GFP fluorescence in animals with b, odr-3(rms31), c, lim-4(ky403) d, tdc-1(n3419), and e, nsIs180[hlh-17p::recCaspase-3, unc-122p::GFP] backgrounds with or without exposure to 1-undecene for 12 hours. Experiments were repeated three (b, c, e) or four (d) times. n = 30, 32 animals in b, n = 42, 37 animals in c, n = 32, 18 animals in d, n = 11, 10 animals in e. Scale bars, 200 μm. Graphs show mean ± s.d. *P < 0.05, **P < 0.01, ****P < 0.0001 (two-tailed unpaired Student’s t test). Source Data contains precise P values.
Extended Data Fig. 5. A range of neurotransmitters are dispensable for activation of the hsp-4p::GFP reporter transgene by 1-undecene exposure.
Representative fluorescence microscopy images of hsp-4p::GFP fluorescence in cho-1(tm373), tph-1(mg280), alh-11(lj118), cat-2(e1112), unc-25(e156), and eat-4(ky5) backgrounds after exposure or no exposure to 1-undecene for 12 hours. Experiments were repeated three times with at least 8 worms per group. Scale bars, 200 μm.
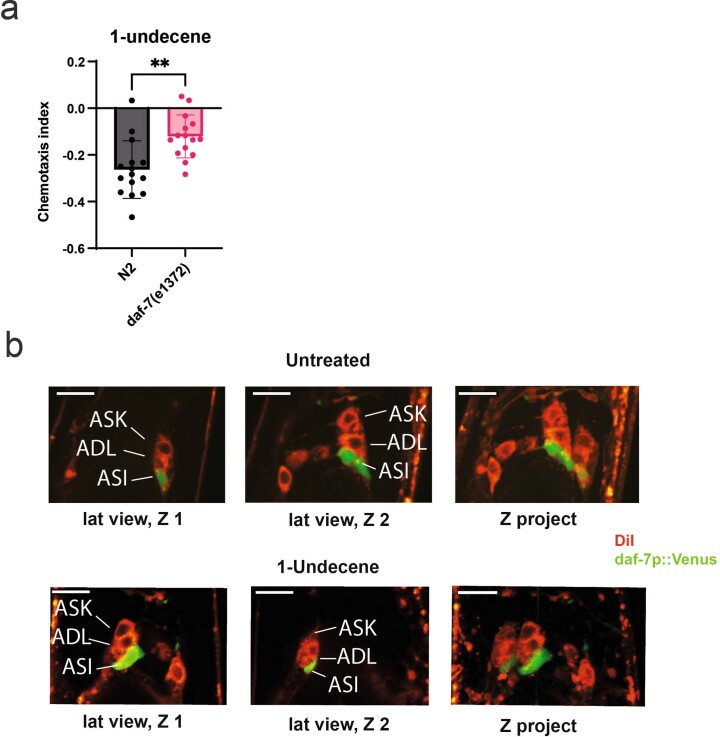
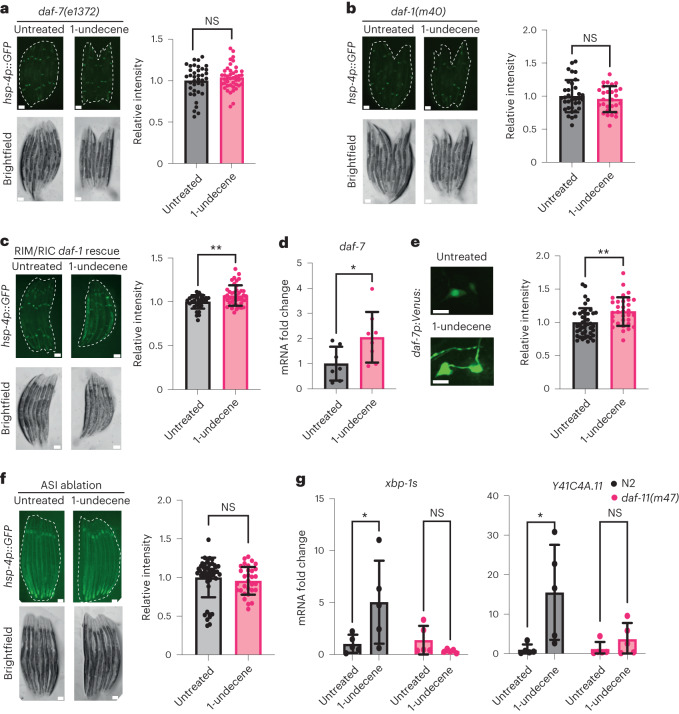
Worms avoid food containing pathogenic bacteria through aversive olfactory learning19. The same aversive behavior is seen in animals exposed to pathogen-associated molecules20,21. One signaling molecule that plays a key role in the neuronal circuits that govern these behaviors is transforming growth factor-beta (TGF-β)20,22. DAF-7, a worm homolog of TGF-β, is necessary for the avoidance of 2-nonanone23, a molecule whose odor induced UPRER activation in our initial odorant screen (Fig. 1b). We observed that DAF-7 is also necessary for behavioral avoidance of 1-undecene (Extended Data Fig. 6a). We therefore asked whether DAF-7/TGF-β is required for UPRER activation by 1-undecene. Strikingly, we found that daf-7 was indeed necessary for UPRER activation after 1-undecene exposure (Fig. 3a). In addition, a mutation in a specific DAF-7 receptor, daf-1, completely inhibited 1-undecene-induced UPRER activation (Fig. 3b). Notably, DAF-1 is expressed in the RIM/RIC interneurons, and our previous work showed that UPRER activation in these neurons is sufficient to drive inter-tissue intestinal UPRER activation8,20. We found that rescue of DAF-1 in these interneurons alone was sufficient to partially restore UPRER activation in daf-1(m40) mutants (Fig. 3c). DAF-7 is primarily expressed in the ASI chemosensory neurons, and animals exposed to P. aeruginosa exhibit increased expression of daf-7 (ref. 20). We therefore asked whether daf-7 expression was elevated by chemosensation of 1-undecene. Indeed, daf-7 mRNA levels were upregulated upon 1-undecene exposure (Fig. 3d). To confirm this, we also employed a daf-7p::Venus fluorescent reporter transgene and observed an increase in expression of daf-7 only in the ASI neurons upon treatment with 1-undecene (Fig. 3e and Extended Data Fig. 6b). In addition, genetic ablation of the ASI neurons prevented UPRER activation by 1-undecene exposure (Fig. 3f). This suggests that an ASI-RIM/RIC neuronal circuit plays a role in the regulation of UPRER activation after odorant exposure.
Extended Data Fig. 6. The daf-7 pathway is activated by 1-undecene exposure and is involved in 1-undecene avoidance.
a, Chemotaxis index of N2 and daf-7(e1372) strains exposed to 1-undecene. N = 15 biological repeats per group, 60 animals per replicate. Graphs show mean ± s.d. **P < 0.01 (two-tailed unpaired Student’s t test). Source Data contains precise P values. b, Confocal images of worms exposed to 1-undecene, expressing daf-7p::Venus and stained with DiI to label ciliated sensory neurons. The experiment was repeated 3 times. Scale bars, 20 μm.
Fig. 3. DAF-7/TGF-β signaling is required for odor-induced UPRER activation.
a–c, Representative fluorescence microscopy images and quantification of hsp-4p::GFP fluorescence in daf-7(e1372) (a), daf-1(m40) (b) and daf-1(m40);ftEx205[tdc-1p::daf-1:gfp] (c) strains with or without exposure to 1-undecene for 12 h. Each experiment was repeated four times (n = 38 and 42 animals in a, n = 34 and 28 animals in b and n = 34 and 42 animals in c). Scale bars, 200 μm. Graphs show mean ± s.d. NS, not significant (two-tailed unpaired Student’s t-test) or **P < 0.01 (two-tailed unpaired Student’s t-test with Welch’s correction). d, mRNA levels of daf-7 were measured by qRT–PCR in animals exposed to 1-undecene for 8 h relative to untreated worms (n = 7 and 8 biological replicates). Graph shows mean ± s.d. *P < 0.05 (two-way ANOVA with Tukey’s multiple comparisons test). e, Representative fluorescence microscopy images and quantification of daf-7p::Venus fluorescence in ASI neurons after worms were exposed or not exposed to 1-undecene odor for 12 h. This experiment was repeated three times (n = 42 and 32 animals). Scale bars, 7 μm. Graph shows mean ± s.d. **P < 0.01 (two-tailed unpaired Student’s t-test). f, Representative fluorescence microscopy images and quantification of hsp-4p::GFP fluorescence in an ASI-ablated strain (oyIs84[gpa-4p::TU#813 + gcy-27p::TU#814 + gcy-27p::GFP + unc-122p::DsRed]). This experiment was repeated three times (n = 48 and 33 animals). Graph shows mean ± s.d. NS, not significant (two-tailed unpaired Student’s t-test). Scale bars, 200 μm g, mRNA levels of xbp-1s and Y41C4A.11 were measured by qRT–PCR in animals exposed to 1-undecene for 8 h relative to untreated worms (n = 5 biological replicates). Graphs show mean ± s.d. NS, not significant and *P < 0.05 (two-way ANOVA with Tukey’s multiple comparison test). Precise P values are provided in Source Data.
Expression levels of daf-7 have been linked to activation of the guanylate cyclase DAF-11 in ASI neurons during starvation24. We therefore asked whether DAF-11 is also required for UPRER activation upon 1-undecene exposure and observed that DAF-11 was indeed necessary for transcriptional upregulation of xbp-1s and its target gene Y41C4A.11 (Fig. 3g). This suggests that DAF-11 is involved in the neuronal perception of 1-undecene odor and subsequent UPRER activation. Thus, our data implicate a TGF-β signaling circuit in connecting the recognition of pathogen-related odorants to inter-tissue regulation of the UPRER.
Chemosensory perception can extend lifespan and enhance proteostasis
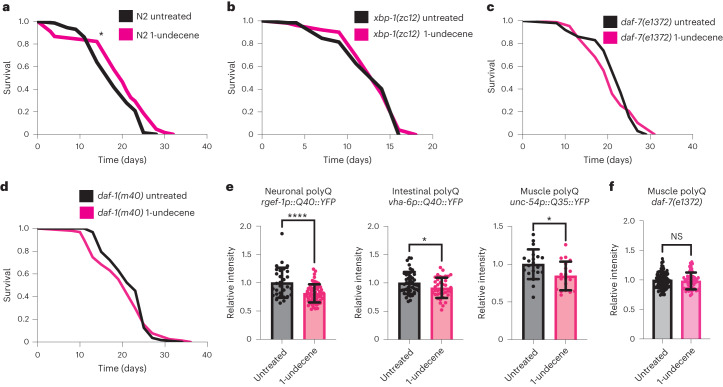
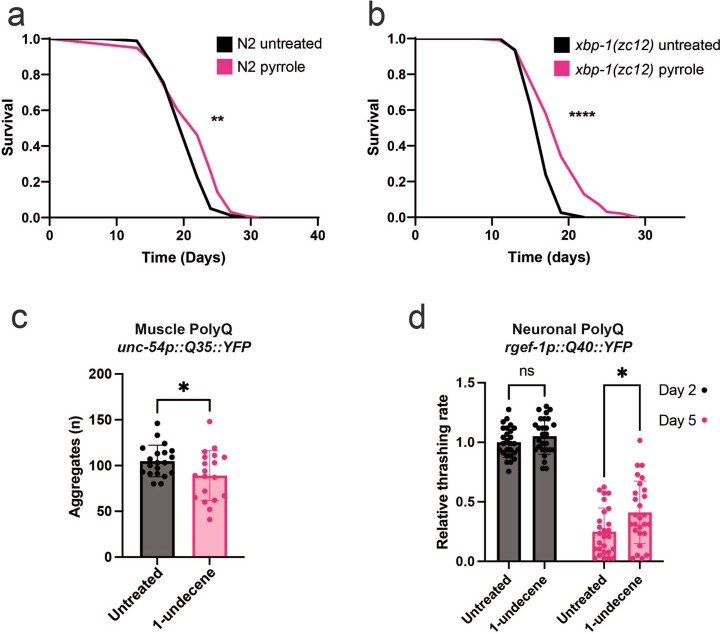
Activation of cellular stress responses is associated with increased lifespan and improved resistance to disease-associated toxic proteins6,25,26. This prompted us to ask whether 1-undecene exposure on the first day of adulthood could impact organismal lifespan and proteostasis. Excitingly, 1-undecene-exposed animals consistently had significantly longer lifespans than untreated animals (Fig. 4a and Supplementary Table 1). This increase in survival was dependent upon xbp-1 (Fig. 4b and Supplementary Table 1), suggesting that 1-undecene odor extends lifespan through the activation of the UPRER. Furthermore, 1-undecene-induced lifespan extension was also dependent upon daf-7 and daf-1, confirming the importance of UPRER activation via TGF-β signaling downstream of 1-undecene exposure (Fig. 4c,d and Supplementary Table 1). Treatment with the UPRER-inducing odorant pyrrole (Fig. 1b) also extended lifespan (Extended Data Fig. 7a and Supplementary Table 1). However, this extension of longevity was less consistent and was not dependent upon xbp-1 (Extended Data Fig. 7b and Supplementary Table 1), suggesting the involvement of additional mechanisms.
Fig. 4. 1-undecene odor increases C. elegans lifespan and reduces polyQ accumulation.
a–d, Lifespan analysis of N2 wild-type (a), xbp-1(zc12) (b), daf-7(e1372) (c) and daf-1(m40) (d) animals with or without exposure to 1-undecene for 24 h at day 1 of adulthood. Graphs are plotted as Kaplan–Meier survival curves. n = 100–120 animals in each group in each of three biological replicates (a,b) and n = 50–100 in two biological replicates (c,d). *P < 0.05 (Mantel–Cox log-rank test). e, Animals expressing polyQ::YFP repeats in neurons, intestine or body wall muscle exposed to 1-undecene for 12 h at day 1 of adulthood and imaged 72 h after treatment. YFP levels were quantified using ImageJ and normalized to untreated animals. This experiment was repeated three times (n = 31 and 65 animals (neuronal); n = 21 and 20 animals (muscle); n = 44 and 39 animals (intestine)). Graphs show mean ± s.d. ****P < 0.0001 and *P < 0.05 (two-tailed unpaired Student’s t-test). f, daf-7(e1372) animals expressing polyQ::YFP repeats in body wall muscle exposed to 1-undecene for 12 h at day 1 of adulthood and imaged 72 h after treatment, as in e. This experiment was repeated three times with at least 15 worms per group. Graphs show mean ± s.d. NS, not significant (two-tailed unpaired Student’s t-test). Precise P values are provided in Source Data.
Extended Data Fig. 7. Pathogen-associated odor regulates longevity and proteostasis in C. elegans.
a, b, Lifespan of a, N2 or b, xbp-1(zc12) strains exposed or not to pyrrole for 24 hours on day 1 of adulthood. Graphs are plotted as Kaplan-Meier survival curves. n = 90-100 animals in each group in 2-3 biological replicates. ****P < 0.0001, **P < 0.01 (Mantel-Cox log-rank test). c, The number of polyQ aggregates quantified in muscle cells on day 4 of adulthood in animals that were exposed or not for 12 hours to 1-undecene on day 1 of adulthood. n = 20, 19 worms per group in 3 biological replicates. Graphs show mean ± s.d. *P < 0.05 (two-tailed unpaired Student’s t test). d, Number of body bends quantified in worms expressing polyQ in neurons at day 2 or 5 of adulthood. Worms were exposed to 1-undecene or not for 16 hours on day 1 of adulthood. n = 30 worms per group in 3 biological replicates. Graphs show mean ± s.d. *P < 0.05, ns, not significant (two-tailed unpaired Student’s t test). Source Data contains precise P values.
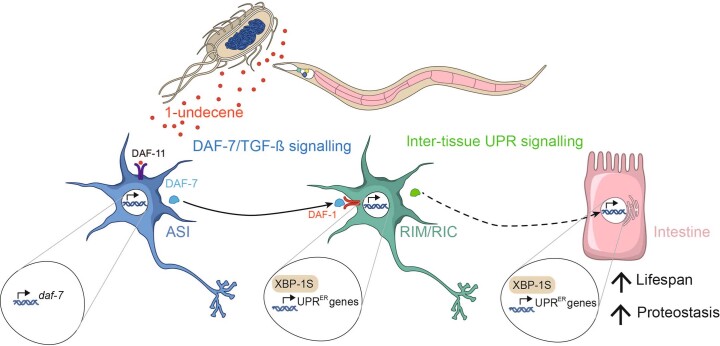
To examine the impact of pathogen-related odor on a C. elegans model of neurodegeneration-associated proteotoxicity, we measured levels of YFP-tagged polyglutamine (polyQ) repeats in different tissues of the animal after 1-undecene exposure at day 1 of adulthood. Remarkably, 1-undecene induced a consistent decrease in levels of polyQ in all tissues examined (intestine, muscle and neurons) (Fig. 4e). It also decreased the number of polyQ aggregates observed in muscle cells (Extended Data Fig. 7c) and ameliorated motility decline in worms expressing neuronal polyQ (Extended Data Fig. 7d). This suggests that 1-undecene-induced UPRER activation enhances clearance of toxic proteins across the animal. Decreased polyQ levels upon 1-undecene exposure were also dependent upon daf-7 (Fig. 4f). These results therefore suggest a model in which the neuronal perception of a pathogen-associated odorant molecule enhances organismal proteostasis and lifespan through TGF-β signaling and UPRER activation (Extended Data Fig. 8).
Extended Data Fig. 8.
Model showing that olfactory stimulation, through exposure to 1-undecene, can activate the cell non-autonomous UPRER in C. elegans via TGF-β signaling.
Discussion
Previous studies reported the cell non-autonomous activation of the UPRER by signals from neurons and glia. In each case, however, transgenes driving xbp-1s were used to achieve this activation, and the evolutionary logic for the development of such systems has been unclear. Here we demonstrate that C. elegans can trigger a cell non-autonomous UPRER without such transgenes, in response to pathogen-associated odorant molecules that induce an aversive behavioral response. We reason that the cell non-autonomous UPRER may have evolved to enable animals to enhance defensive mechanisms in anticipation of the increased translation associated with an immune response or the direct proteostatic challenge of the pathogen itself. Animals that constitutively activate a PMK-1-driven immune response require xbp-1 to survive the demands imposed by an active immune system10, suggesting that UPRER capacity is of critical importance in conditions that require an immune response.
Although the action of 1-undecene on C. elegans is likely a specific interaction informed by the complex evolutionary relationship between pathogen and host, existing evidence supports the idea that the broader principle underlying cell non-autonomous UPRER activation may be conserved. In mice, sensory perception of food activates pro-opiomelanocortin (POMC)-expressing neurons, resulting in hepatic xbp-1 splicing as a predictive physiological response in anticipation of food consumption27. In addition, driving xbp-1s genetically in murine POMC neurons is sufficient to increase hepatic xbp-1s levels via a cell non-autonomous mechanism17. There are significant similarities between the roles of ASI neurons in the worm and the hypothalamus and POMC neurons in mice28. ASI neurons regulate food intake and food-seeking behavior through the action of DAF-7/TGF-β29. Similarly, POMC is expressed in subsets of cells, including neurons in the arcuate nucleus of the hypothalamus30, and POMC neurons also regulate food intake and energy expenditure via locomotion in some contexts31. Furthermore, expression of the TGF-β antagonist Smad7 in POMC neurons regulates peripheral glucose metabolism, suggesting that TGF-β signaling is also important for POMC neurons to achieve anticipatory, cell non-autonomous effects in the periphery32. These mammalian studies suggest that major interactions in the pathway we describe here are likely to be conserved in mammals.
Although earlier work showed that food-associated odor can prevent lifespan extension induced by caloric restriction33,34, the present study is, to our knowledge, the first demonstration that the perception of a specific odorant molecule can increase the lifespan of an animal. It was noted recently35 that a great many mechanisms that regulate aging in model organisms include cell non-autonomous protective pathways that are subject to neuronal control, often by sensory neurons. Dietary restriction-mediated longevity requires the UPRER (refs. 36,37) as well as functional ASI neurons expressing daf-7 (refs. 38,39) and is regulated by olfactory perception40. Furthermore, cell non-autonomous regulation of the mitochondrial UPR41, heat shock response42, AMP-activated protein kinase (AMPK)43 and target of rapamycin complex 1 (TORC1)44, as well as lifespan regulation by temperature45 and the hypoxia response46, are all similarly orchestrated, with signals originating in sensory neurons leading via cell non-autonomous routes to regulation of pro-longevity pathways. Here we show that direct stimulation of chemosensory neurons can extend lifespan. We therefore speculate that directly manipulating the activity of sensory neurons via their sensory inputs and/or corresponding receptors may be a way to activate these pro-longevity pathways.
Finally, mounting evidence suggests that Ire1/Xbp1 activity is highly correlated with the pathophysiology observed in neurodegenerative disorders in animal models, including Alzheimer’s, Parkinson’s and Huntington’s diseases, and age-associated decline in the activation of this pathway may be associated with disease progression47–49. Activation of the UPRER through stimulation of sensory pathways by olfactory compounds may therefore represent a promising strategy to prevent the disease-related proteostasis collapse associated with aging.
Methods
C. elegans strains and maintenance
Strains were made in the course of this study, provided by the Caenorhabditis Genetics Center (CGC) or kindly gifted by other laboratories. A list of strains used in this work can be found in Supplementary Table 2. The CGC Bristol N2 hermaphrodite stock was used as wild-type. Worms were maintained at 20 °C on nematode growth medium (NGM) plates seeded with Escherichia coli OP50 using standard techniques50. For RNAi by feeding51, NGM plates were supplemented with 1 mM IPTG and 100 μg ml−1 carbenicillin and then seeded with HT115 bacteria harboring L4440 empty vector or the RNAi of interest. All RNAi used are from the Ahringer RNAi library (Source BioScience) and were confirmed by sequencing.
Transgenic strain construction
The odr-3(rms31) mutant was generated by CRISPR using a dual crRNA dpy-10 co-CRISPR strategy and a custom protocol based on previous methods52,53 and optimization for our laboratory. In brief, 1 µl of 320 µM solution of each CRISPR RNA (crRNA) and 0.5 µl of dpy-10 crRNA (50 µM) was annealed to 0.4 µl of 100 µM trans-acting CRISPR RNA (tracrRNA) (Integrated DNA Technologies) by heating to 95 °C in a PCR machine and cooling to 4 °C at 0.1 °C s−1. Then, 0.5 µl of Cas9 protein (Invitrogen) was added, and the mixture was incubated for 10 min at 37 °C. Next, 0.5 µl of 100 µM stock of each repair template (target and dpy-10) was added, and the solution was made up to 20 µl with DPEC water. This mix was centrifuged for 30 min at 20,000g at 4 °C before injection. Oligonucleotides used in this study are provided in Supplementary Tables 3 and 4.
Brood size measurement
Brood size was determined by counting the number of eggs laid per worm during their fertile period (from day 1 to day 4 of adulthood).
Treatment with 1-undecene
To expose animals to the odor of 1-undecene (Sigma-Aldrich, 242527), worms were placed on NGM plates with a diameter of 60 mm, which were sealed with another 60 mm NGM plate on which was pipetted 4 × 3-μl drops of 1-undecene.
Epifluorescence microscopy
To investigate the effect of 1-undecene on reporter transgene expression (for example, hsp-4p::gfp), worms were exposed to 1-undecene odor for 12 h in plates sealed with Parafilm M and then immobilized with 20 mM sodium azide (Sigma-Aldrich) and imaged using a Leica M205 FA microscope. To image worms expressing polyQ::YFP, animals were exposed to 1-undecene for 12 h on day 1 of adulthood and imaged on day 4 of adulthood. For DAF-16::GFP analysis, worms were scored based on the subcellular localization of GFP in intestinal cells, as described previously24. Worms were randomly selected from a synchronized population before imaging. Fluorescence values (mean intensity) were obtained by analyzing microscope images on ImageJ or Fiji.
Confocal microscopy
Worms expressing daf-7p::Venus or xbp-1p::xbp-1::GFP transgenes were treated with 1-undecene for 8 h. They were then immobilized with 20 mM sodium azide (Sigma-Aldrich) and mounted on a 2% agarose pad. Animals were imaged on an LSM 710 confocal microscope using the ×40 and ×63 oil immersion objectives and on an Andor Revolution spinning disk microscope using the ×20 and ×60 water immersion objectives. All images were acquired using Leica LAS X (version 5.1.0) and analyzed using ImageJ (version 1.53e).
RNA extraction and qRT–PCR
Approximately 300 young adult animals were collected with M9 after being exposed or not to 1-undecene for 8 h. TRIzol was added to samples, which were immediately frozen in liquid nitrogen. RNA isolation was carried out using the Direct-zol RNA MiniPrep Kit (Zymo Research) following the manufacturer’s instructions. RNA was quantified by NanoDrop. One microgram of RNA was used for cDNA synthesis with the QuantiTect Reverse Transcription Kit (Qiagen). Samples were diluted 2.5× after cDNA synthesis, and SYBR Select Master Mix (Applied Biosystems) was used for qRT–PCR on a Vii7 Real-Time PCR machine (Thermo Fisher Scientific) to quantify alterations in the transcript level of genes of interest. Data were analyzed using the comparative 2−ΔΔCt method. A list of primers used in this work is provided in Supplementary Table 3.
Chemotaxis assay
For odorant chemotaxis assays, chemotaxis was performed in a two-plate setup. On the lower plate, 1-undecene (test) or water (control) was placed approximately 2 cm from the center of the plate. Worms at day 1 of adulthood were placed on the center of the upper plate, and a test zone and a control zone were designated opposite the odorant, with the remaining space scored as the center zone14. Water mixed with sodium azide (1:1) was placed in the center of each the control and test zone. After 30 min, the number of worms in each zone was quantified, and the chemotaxis index was calculated by the formula: CI = (number of wormstest − number of wormscontrol) / (total number of worms).
Thrashing assay
Animals expressing neuronal polyQ were exposed to 1-undecene on day 1 of adulthood for 16 h. At day 2 or day 5 of adulthood, these animals were transferred to M9 solution, and the number of body bends per 30 s was quantified.
Survival assays
Approximately 100 worms were exposed or not exposed to 1-undecene odor for 24 h at day 1 of adulthood. Worms were then placed on NGM plates containing 100 μg ml−1 FUDR and seeded with E. coli OP50 and were kept at 20 °C. Animals were monitored as alive or dead every second day by a blinded investigator, and data were analyzed on GraphPad Prism 8.4.2 software.
Statistics and reproducibility
Statistical analysis was performed using GraphPad Prism 8.4.2 software. All bar graphs show the mean with error bars representing s.d. Appropriate tests for each experiment were chosen and are described (including tests for multiple comparisons) in the figure legends. With the exception of lifespan assays, data collection and analysis were not performed blinded to the conditions of the experiments. Unless specified otherwise in the figure legend, a minimum of three individual experiments were conducted for each assay. All replication efforts consistently yielded similar results. No animals were excluded from the analysis; however, for qRT–PCR experiments, samples that did not meet the predetermined quality control standards were excluded. Where used, n is immediately defined. Information regarding the number of repeats, number of animals per repeat and the results of the statistical tests performed are given in the figure legends. No statistical methods were used to pre-determine sample sizes, but our sample sizes are similar to those reported in previous publications from our group6. Data distribution was assumed to be normal, but this was not formally tested. Animals were randomly selected based upon developmental stage and not screened in any way before analysis.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We are grateful to the Medical Research Council Laboratory of Molecular Biology (MRC-LMB) Visual Aids department for assistance with figures. Some C. elegans strains were provided by A. Dillin (UC Berkeley), W. Schafer (MRC-LMB), J. Tullet (University of Kent), D. Kim (Harvard Medical School) and the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). This work was supported by the MRC (R.C.T.) and by the European Union’s Horizon 2020 Research and Innovation Programme under Marie Skłodowska‐Curie grant agreement number 894039 (E.A.D.-S.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Extended data
Source data
Statistical Source Data.
Statistical Source Data.
Statistical Source Data.
Statistical Source Data.
Statistical Source Data.
Statistical Source Data.
Statistical Source Data.
Statistical Source Data.
Statistical Source Data.
Statistical Source Data.
Author contributions
Conceptualization: E.A.D.-S., M.A.T. and R.C.T. Methodology: E.A.D.-S., M.A.T. and R.C.T. Investigation: E.A.D.-S., M.A.T. and R.C.T. Visualization: E.A.D.-S., M.A.T. and R.C.T. Funding acquisition: E.A.D.-S. and R.C.T. Project administration: R.C.T. Supervision: R.C.T. Writing—original draft, review and editing: E.A.D.-S., M.A.T. and R.C.T.
Peer review
Peer review information
Nature Aging thanks Thorsten Hoppe, Konstantinos Palikaras and the other anonymous reviewer for their contribution to the peer review of this work.
Data availability
All data reported in this paper will be shared by the lead contact upon reasonable request. Any additional information required to re-analyze the data reported in this paper is available from the lead contact upon reasonable request. This paper does not report original code. Source data are provided with this paper.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Evandro A. De-Souza, Maximillian A. Thompson.
Contributor Information
Evandro A. De-Souza, Email: desouza.evs92@gmail.com
Maximillian A. Thompson, Email: max.thompson@babraham.ac.uk
Rebecca C. Taylor, Email: rebecca.c.taylor@uea.ac.uk
Extended data
is available for this paper at 10.1038/s43587-023-00467-1.
Supplementary information
The online version contains supplementary material available at 10.1038/s43587-023-00467-1.
References
- 1.Linford, N. J., Kuo, T.-H., Chan, T. P. & Pletcher, S. D. Sensory perception and aging in model systems: from the outside in. Ann. Rev. Cell Dev. Biol.27, 759–785 (2011). [DOI] [PMC free article] [PubMed]
- 2.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 3.Taylor RC, Dillin A. XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell. 2013;153:1435–1447. doi: 10.1016/j.cell.2013.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Labbadia J, Morimoto RI. Repression of the heat shock response is a programmed event at the onset of reproduction. Mol. Cell. 2015;59:639–650. doi: 10.1016/j.molcel.2015.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cabral-Miranda, F. et al. Unfolded protein response IRE1/XBP1 signaling is required for healthy mammalian brain aging. EMBO J.41, e111952 (2022). [DOI] [PMC free article] [PubMed]
- 6.Imanikia S, Özbey NP, Krueger C, Casanueva MO, Taylor RC. Neuronal XBP-1 activates intestinal lysosomes to improve proteostasis in C. elegans. Curr. Biol. 2019;29:2322–2338. doi: 10.1016/j.cub.2019.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frakes AE, et al. Four glial cells regulate ER stress resistance and longevity via neuropeptide signaling in C. elegans. Science. 2020;367:436–440. doi: 10.1126/science.aaz6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Özbey NP, et al. Tyramine acts downstream of neuronal XBP-1s to coordinate inter-tissue UPRER activation and behavior in C. elegans. Dev. Cell. 2020;55:754–770. doi: 10.1016/j.devcel.2020.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masuzzo A, Montanari M, Kurz L, Royet J. How bacteria impact host nervous system and behaviors: lessons from flies and worms. Trends Neurosci. 2020;43:998–1010. doi: 10.1016/j.tins.2020.09.007. [DOI] [PubMed] [Google Scholar]
- 10.Richardson CE, Kooistra T, Kim DH. An essential role for XBP-1 in host protection against immune activation in C. elegans. Nature. 2010;463:1092–1095. doi: 10.1038/nature08762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ooi, F. K. & Prahlad, V. Olfactory experience primes the heat shock transcription factor HSF-1 to enhance the expression of molecular chaperones in C. elegans. Sci. Signal.10, eaan4893 (2017). [DOI] [PMC free article] [PubMed]
- 12.Filipiak W, et al. Molecular analysis of volatile metabolites released specifically by Staphylococcus aureus and Pseudomonas aeruginosa. BMC Microbiol. 2012;12:113. doi: 10.1186/1471-2180-12-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.L’Etoile ND, Bargmann CI. Olfaction and odor discrimination are mediated by the C. elegans guanylyl cyclase ODR-1. Neuron. 2000;25:575–586. doi: 10.1016/S0896-6273(00)81061-2. [DOI] [PubMed] [Google Scholar]
- 14.Prakash, D., Siddiqui, R., Chalasani, S. H. & Singh, V. Pyrrole produced by Pseudomonas aeruginosa influences olfactory food choice of Caenorhabditis elegans. Preprint at bioRxiv10.1101/2022.01.27.477966 (2022).
- 15.Kimura KD, Fujita K, Katsura I. Enhancement of odor avoidance regulated by dopamine signaling in Caenorhabditis elegans. J. Neurosci. 2010;30:16365–16375. doi: 10.1523/JNEUROSCI.6023-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prakash D, et al. 1‐Undecene from Pseudomonas aeruginosa is an olfactory signal for flight‐or‐fight response in Caenorhabditis elegans. EMBO J. 2021;40:e106938. doi: 10.15252/embj.2020106938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams KW, et al. Xbp1s in Pomc neurons connects ER stress with energy balance and glucose homeostasis. Cell Metab. 2014;20:471–482. doi: 10.1016/j.cmet.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eletto D, Eletto D, Dersh D, Gidalevitz T, Argon Y. Protein disulfide isomerase A6 controls the decay of IRE1α signaling via disulfide-dependent association. Mol. Cell. 2014;53:562–576. doi: 10.1016/j.molcel.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Lu H, Bargmann CI. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature. 2005;438:179–184. doi: 10.1038/nature04216. [DOI] [PubMed] [Google Scholar]
- 20.Meisel JD, Panda O, Mahanti P, Schroeder FC, Kim DH. Chemosensation of bacterial secondary metabolites modulates neuroendocrine signaling and behavior of C. elegans. Cell. 2014;159:267–280. doi: 10.1016/j.cell.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beale E, Li G, Tan M-W, Rumbaugh KP. Caenorhabditis elegans senses bacterial autoinducers. Appl. Environ. Microbiol. 2006;72:5135–5137. doi: 10.1128/AEM.00611-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore RS, Kaletsky R, Murphy CT. Piwi/PRG-1 argonaute and TGF-β mediate transgenerational learned pathogenic avoidance. Cell. 2019;177:1827–1841. doi: 10.1016/j.cell.2019.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris G, et al. Molecular and cellular modulators for multisensory integration in C. elegans. PLoS Genet. 2019;15:e1007706. doi: 10.1371/journal.pgen.1007706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tataridas-Pallas N, et al. Neuronal SKN-1B modulates nutritional signalling pathways and mitochondrial networks to control satiety. PLoS Genet. 2021;17:e1009358. doi: 10.1371/journal.pgen.1009358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 26.Klaips CL, Jayaraj GG, Hartl FU. Pathways of cellular proteostasis in aging and disease. J. Cell Biol. 2018;217:51–63. doi: 10.1083/jcb.201709072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brandt C, et al. Food perception primes hepatic ER homeostasis via melanocortin-dependent control of mTOR activation. Cell. 2018;175:1321–1335. doi: 10.1016/j.cell.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tullet JMA, et al. Direct inhibition of the longevity promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gallagher T, Kim J, Oldenbroek M, Kerr R, You Y-J. ASI regulates satiety quiescence in C. elegans. J. Neurosci. 2013;33:9716–9724. doi: 10.1523/JNEUROSCI.4493-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y, Atasoy D, Su HH, Sternson SM. Hunger states switch a flip-flop memory circuit via a synaptic AMPK-dependent positive feedback loop. Cell. 2011;146:992–1003. doi: 10.1016/j.cell.2011.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhan C, et al. Acute and long-term suppression of feeding behavior by POMC neurons in the brainstem and hypothalamus, respectively. J. Neurosci. 2013;33:3624–3632. doi: 10.1523/JNEUROSCI.2742-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan F, et al. Overexpression of Smad7 in hypothalamic POMC neurons disrupts glucose balance by attenuating central insulin signaling. Mol. Metab. 2020;42:101084. doi: 10.1016/j.molmet.2020.101084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Libert S, et al. Regulation of Drosophila life span by olfaction and food-derived odors. Science. 2007;315:1133–1137. doi: 10.1126/science.1136610. [DOI] [PubMed] [Google Scholar]
- 34.Park S, et al. Diacetyl odor shortens longevity conferred by food deprivation in C. elegans via downregulation of DAF-16/FOXO. Aging Cell. 2021;20:e13300. doi: 10.1111/acel.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller HA, Dean ES, Pletcher SD, Leiser SF. Cell non-autonomous regulation of health and longevity. eLife. 2020;9:e62659. doi: 10.7554/eLife.62659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, P. W., Karpac, J., Jasper, H. & Kapahi, P. Intestinal IRE1 is required for increased triglyceride metabolism and longer lifespan under dietary restriction. Cell Rep.17, 1207–1216 (2016). [DOI] [PMC free article] [PubMed]
- 37.Matai L, et al. Dietary restriction improves proteostasis and increases life span through endoplasmic reticulum hormesis. Proc. Natl Acad. Sci. USA. 2019;116:17383–17392. doi: 10.1073/pnas.1900055116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- 39.Fletcher, M. & Kim, D. H. Age-dependent neuroendocrine signaling from sensory neurons modulates the effect of dietary restriction on longevity of Caenorhabditis elegans. PLoS Genet.13, e1006544 (2017). [DOI] [PMC free article] [PubMed]
- 40.Zhang B, Jun H, Wu J, Liu J, Xu XZS. Olfactory perception of food abundance regulates dietary restriction-mediated longevity via a brain-to-gut signal. Nat. Aging. 2021;1:255–268. doi: 10.1038/s43587-021-00039-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prahlad V, Cornelius T, Morimoto RI. Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science. 2008;320:811–814. doi: 10.1126/science.1156093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burkewitz K, et al. Neuronal CRTC-1 governs systemic mitochondrial metabolism and lifespan via a catecholamine signal. Cell. 2015;160:842–855. doi: 10.1016/j.cell.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, et al. Neuronal TORC1 modulates longevity via AMPK and cell nonautonomous regulation of mitochondrial dynamics in C. elegans. eLife. 2019;8:e49158. doi: 10.7554/eLife.49158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee S-J, Kenyon C. Regulation of the longevity response to temperature by thermosensory neurons in Caenorhabditis elegans. Curr. Biol. 2009;19:715–722. doi: 10.1016/j.cub.2009.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leiser SF, Begun A, Kaeberlein M. HIF-1 modulates longevity and healthspan in a temperature-dependent manner. Aging Cell. 2011;10:318–326. doi: 10.1111/j.1474-9726.2011.00672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sado M, et al. Protective effect against Parkinson’s disease-related insults through the activation of XBP1. Brain Res. 2009;1257:16–24. doi: 10.1016/j.brainres.2008.11.104. [DOI] [PubMed] [Google Scholar]
- 48.Casas-Tinto S, et al. The ER stress factor XBP1s prevents amyloid-β neurotoxicity. Hum. Mol. Genet. 2011;20:2144–2160. doi: 10.1093/hmg/ddr100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vidal RL, et al. Targeting the UPR transcription factor XBP1 protects against Huntington’s disease through the regulation of FoxO1 and autophagy. Hum. Mol. Genet. 2012;21:2245–2262. doi: 10.1093/hmg/dds040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/S0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 52.Vicencio J, Martínez-Fernández C, Serrat X, Cerón J. Efficient generation of endogenous fluorescent reporters by nested CRISPR in Caenorhabditis elegans. Genetics. 2019;211:1143–1154. doi: 10.1534/genetics.119.301965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paix A, et al. Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 sites in Caenorhabditis elegans. Genetics. 2014;198:1347–1356. doi: 10.1534/genetics.114.170423. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon reasonable request. Any additional information required to re-analyze the data reported in this paper is available from the lead contact upon reasonable request. This paper does not report original code. Source data are provided with this paper.