Abstract
Background
Currently, surgical resection is the mainstay for colorectal liver metastases (CRLM) management and the only potentially curative treatment modality. Prognostication tools can support patient selection for surgical resection to maximize therapeutic benefit. This study aimed to develop a survival prediction model using machine learning based on a multicenter patient sample in Hong Kong.
Methods
Patients who underwent hepatectomy for CRLM between 1 January 2009 and 31 December 2018 in four hospitals in Hong Kong were included in the study. Survival analysis was performed using Cox proportional hazards (CPH). A stepwise selection on Cox multivariable models with Least Absolute Shrinkage and Selection Operator (LASSO) regression was applied to a multiply-imputed dataset to build a prediction model. The model was validated in the validation set, and its performance was compared with that of Fong Clinical Risk Score (CRS) using concordance index.
Results
A total of 572 patients were included with a median follow-up of 3.6 years. The full models for overall survival (OS) and recurrence-free survival (RFS) consist of the same 8 established and novel variables, namely colorectal cancer nodal stage, CRLM neoadjuvant treatment, Charlson Comorbidity Score, pre-hepatectomy bilirubin and carcinoembryonic antigen (CEA) levels, CRLM largest tumor diameter, extrahepatic metastasis detected on positron emission-tomography (PET)-scan as well as KRAS status. Our CRLM Machine-learning Algorithm Prognostication model (CMAP) demonstrated better ability to predict OS (C-index =0.651), compared with the Fong CRS for 1-year (C-index =0.571) and 5-year OS (C-index =0.574). It also achieved a C-index of 0.651 for RFS.
Conclusions
We present a promising machine learning algorithm to individualize prognostications for patients following resection of CRLM with good discriminative ability.
Keywords: Machine-learning, colorectal liver metastasis (CRLM), prognostic model, survival analysis, hepatectomy outcome
Introduction
Colorectal cancer (CRC) has the third highest incidence and second highest mortality among all malignancies globally (1). In Hong Kong, CRC is the most common malignancy and second highest cause of cancer-related mortality, with over 20% of patients having metastasis at initial presentation (2). The liver is the most common site, accounting for approximately 50% of colorectal cancer metastases (3).
Despite contemporary advances in chemotherapy, hepatectomy remains the only curative treatment option for resectable CRLM (4). However, recurrence occurs in nearly three-quarters of patients within 16 months post-resection (5). As such, prognostication tools have been developed for the selection of patients to receive surgical resection, to maximize therapeutic benefit (Table S1).
Of these, the most commonly cited is the Fong Clinical Risk Score (CRS) for Colorectal Cancer Recurrence (6), which considers overall survival (OS) based on 5 variables. As all factors were equally weighted in a multivariable analysis, this may have oversimplified the prognostic impact of each. Since it was based on a single institution with a limited number of patients, its external validity and applicability in diverse population groups remain controversial (7,8). Aside from CRS, other older but well-established models include those from Nordlinger et al. (9) and Iwatsuki et al. (10).
More recent scoring systems such as the Genetic And Morphological Evaluation (GAME) score (11) and modified clinical score (m-CS) (12) have adapted to the modern age of genomic and chemotherapeutic advancements by including KRAS mutational status. However, as scores, they still suffer from the same limitations as CRS and are not as widely validated. There have been fewer studies on Asia-Pacific populations, with existing literature based on single-institution samples in Korea and China respectively (13,14) and a multi-center cohort in Japan (15).
This study aimed to develop a survival prediction model using machine learning on a multicenter patient sample in Hong Kong incorporating established and novel demographic, and clinicopathological variables. We present this article in accordance with the TRIPOD reporting checklist (available at https://hbsn.amegroups.com/article/view/10.21037/hbsn-21-453/rc).
Methods
Ethical approval
All research processes were conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Institutional Review Board of The University of Hong Kong and the Hong Kong West Cluster of Hospital Authority, Hong Kong (Reference number UW 21-471).
Data source and variables
Data were anonymously extracted from electronic health records from the Clinical Management System (CMS) and Clinical Data Analysis and Reporting System (CDARS), an electronic healthcare database managed by the Hong Kong Hospital Authority. Patient-related variables were extracted from patient records. This included dates of CRC and CRLM diagnoses, dates of colorectal resection and hepatectomy, sex, age, Charlson Comorbidity Score (16), pre-hepatectomy bilirubin and carcinoembryonic antigen (CEA) levels, and the use of systemic treatments including neoadjuvant and adjuvant chemotherapy/radiotherapy for CRC and CRLM. Post-hepatectomy variables including follow-up status at the conclusion of the study, date of last follow-up, date of death, postoperative mortality and cause of death were also recorded if applicable.
Surgical variables for CRC [e.g., primary tumor location, operation category (emergency or elective)], hepatectomy approach (laparoscopic or open), hepatectomy extent [major involving 3 or more segments, or minor operation involving less than 3 segments (17)] and whether patients underwent combined radiofrequency ablation (RFA) treatment were recorded from operation reports. Pathological variables for CRC, namely the largest tumor diameter, lymphovascular invasion, venous infiltration, resection margin, tumor differentiation, number of positive lymph nodes, and KRAS status; for CRLM, namely number of tumor nodules, largest tumor diameter, lymphovascular invasion, resection margin, lobar involvement, and the new Edmondson grading for differentiation were extracted from pathology reports.
Findings from the positron emission-tomography (PET) imaging reports for CRLM were also noted, including tumor location, number of tumors, largest tumor diameter, and the presence of extrahepatic metastasis along with the number and location of extrahepatic lesions. Additionally, whether extrahepatic lesions were resected and the timing of resection were also noted when applicable.
The manuscript adheres to the Transparent Reporting of multivariable prediction models for Individual Prognosis or Diagnosis (TRIPOD) statement (18). Investigators were not blinded from the outcome or predictor variables.
Study population
Patients who underwent hepatectomy for CRLM between 1 January 2009 and 31 December 2018 in four hospitals in Hong Kong (two teaching hospitals and two peripheral hospitals) were included in this study. Patients were excluded for the following: (I) unresected primary colorectal malignancy before/during the entire study period, (II) first liver metastasis treated by RFA only, (III) postoperative mortality, and (IV) multiple synchronous primary malignancies.
Statistical analysis
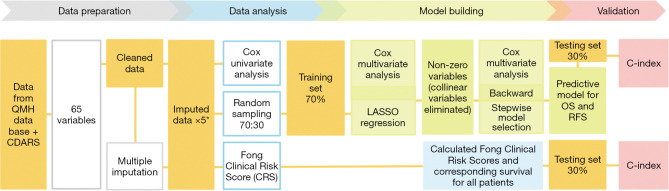
Statistical analysis and model building were performed using R 4.0.6. (Figure 1).
Figure 1.
The research pipeline for data preparation, analysis as well as model building and validation. All results from analyses on imputed datasets were pooled. QMH, Queen Mary Hospital; CDARS, Clinical Data Analysis and Reporting System; LASSO, Least Absolute Shrinkage and Selection Operator; OS, overall survival; RFS, recurrence-free survival.
Data preparation
Using the mice (Multivariate Imputation by Chained Equations) statistical package (19), multiple imputation was performed to handle missing data and to increase statistical power and reduce bias. Missing values were replaced by five plausible data values, predicted upon other baseline values and/or outcomes. For all imputations, the results generated from analyses of the multiply imputed data were pooled, i.e., the results were averaged and the total variance was computed over the repeated analyses by Rubin’s rules (20). Multiple imputation has been shown to provide more validity than simple ad-hoc approaches such as excluding entries with missing data or mean imputation (21,22). Certain continuous variables were made categorical using cutoffs derived from existing scoring systems including preoperative CEA level and bilirubin levels, whereas and the number of colorectal cancer lymph nodes affected was grouped according to TNM nodal status staging. OS (time between date of hepatectomy and the date of death), and recurrence-free survival (RFS) (time between the date of hepatectomy and the date of recurrence) were calculated.
Data analysis
Univariate Cox proportional hazards analysis was performed on the data using the survival package (23), investigating the association between all variables and the OS and RFS, as well as their statistical significance.
Model building and selection
Data was randomized into training and validation sets in a 70:30 ratio. To address multicollinearity, arising from the inevitable correlations between clinical predictors, Least Absolute Shrinkage and Selection Operator (LASSO) regression was performed using the glmnet package (24) to shrink the estimates of collinear predictors to zero and improve stability of predictions (25). With Cox multivariable analysis as the foundation of the model, backward model selection was applied on the non-zero variables using the mice package (19). Each variable was removed in turn, and the pooled likelihood ratio P value was calculated and compared between the Cox multivariable models with and without the variable. If the P value of the comparison between the two models was >0.05, the corresponding variable would be removed. The procedure was repeated on the smaller model until all significant variables were removed.
The predictive model built on the training set predicts both OS and RFS, and was internally validated on the validation set, using Harrell’s Concordance index (C-index), which handles censored data (26). The C-indices of our model were compared with those of CRS. Comparison with other more recent scoring systems such as GAME and m-CS could not be done since the predicted survival probabilities at different time points were not accessible.
Results
Baseline characteristics
Six hundred and fifty-two patients were screened, and a total of 572 patients were included with a median follow-up of 3.6 years [interquartile range (IQR): 2.5–5.7]. During the 9-year study period, 329 patients died. The important clinical and pathologic characteristics of the population are summarized in Table 1, and further detailed in Table S2. 62.9% of patients were male, and the median age was 62 (range, 24–94). Overall, left-sided CRC was the most common primary tumor location (38.5%). 61.4% patients presented with synchronous liver metastasis, but only 30.4% received synchronous resection. The median Charlson Comorbidity Index was 10 (IQR: 9.0–11.0). The median pre-CRC resection CEA was 9.7 ng/mL (IQR: 3.9–42.0), and the median pre-hepatectomy CEA was 12.0 ng/mL (IQR: 4.6–45.4). 33.3% of patients received 6 cycles of 5-FU-based neoadjuvant chemotherapy prior to hepatectomy.
Table 1. Baseline characteristics of the population.
| Variables | Total (n=572) |
|---|---|
| Sex, n (%) | |
| Male | 360 (62.9) |
| Female | 212 (37.1) |
| Age (years), median (Q1, Q3); range (min–max) | 62 (56.8, 69.0); (24–94) |
| Charlson Comorbidity Score, median (Q1, Q3); range (min–max) | 10 (9.0, 11.0); (8–16) |
| Presence of co-existing cancers (Hong Kong Top 10), n (%) | |
| No | 557 (97.4) |
| Yes | 15 (2.6) |
| Number of lymph nodes positive in primary CRC, median (Q1, Q3) | 1 (0, 4.0) |
| Colorectal liver metastasis | |
| Pre-treatment PET scan findingsª | |
| Number of tumor nodules, median (Q1, Q3); range (min–max) | 1 (1.0, 3.0); (0–12) |
| Tumor location, n (%) | |
| Unilobar | 343 (70.6) |
| Bilobar | 143 (29.4) |
| Largest tumor diameter (cm), median (Q1, Q3); range (min–max) | 2.5 (1.6, 4.0); (0–16.8) |
| Extrahepatic metastasis, n (%) | |
| No | 423 (89.4) |
| Yes | 50 (10.6) |
| Extrahepatic metastasis site, n (%) | |
| Lung metastasis | 28 (56.0) |
| Bone metastasis | 6 (12.0) |
| Other metastasis | 16 (32.0) |
| Extrahepatic metastasis number, n (%) | |
| Single extrahepatic metastasis | 47 (94.0) |
| Multiple extrahepatic metastases | 3 (7.0) |
| Extrahepatic metastasis resection status, n (%) | |
| Not resected | 28 (56.0) |
| Resected before hepatic resection | 10 (20.0) |
| Resected during hepatic resection | 6 (12.0) |
| Resected after hepatic resection | 6 (12.0) |
| Pre-operative investigations | |
| Albumin (g/L), median (Q1, Q3) | 40.0 (36.0, 43.0) |
| Bilirubin (µmol/L), median (Q1, Q3); range (min–max) | 8.2 (6.0, 12.9); (2.0–51.0) |
| Liver CEA (ng/mL), median (Q1, Q3); range (min–max) | 12.0 (4.6, 45.4); (0.7–4,040.0) |
| Liver pathology report findings | |
| Largest diameter of liver metastasis (cm), median (Q1, Q3); range (min–max) | 3.0 (2.0, 4.0); (0.4–18.0) |
| Number of tumor nodules, median (Q1, Q3); range (min–max) | 2 (1.0,3.0); (0–8) |
| Lymphovascular invasion, n (%) | |
| No | 84 (52.5) |
| Yes | 76 (47.5) |
| Tumor lobar involvement, n (%) | |
| Unilobar | 395 (69.3) |
| Bilobar | 175 (30.7) |
| KRAS, n (%) | |
| No mutation | 250 (58.8) |
| Mutation | 175 (41.2) |
| Treatment received | |
| Neoadjuvant treatment, n (%) | |
| No | 378 (66.7) |
| Yes | 189 (33.3) |
| Adjuvant chemotherapy, n (%) | |
| No | 161 (28.1) |
| Yes | 411 (71.9) |
| Patients who received only neoadjuvant therapy, n (%) | |
| No | 523 (91.6) |
| Yes | 48 (8.4) |
| Patients who received only adjuvant therapy, n (%) | |
| No | 302 (53.2) |
| Yes | 266 (46.8) |
| Patients who received both neoadjuvant and adjuvant chemotherapy, n (%) | |
| No | 427 (75.2) |
| Yes | 141 (24.8) |
| Follow-up | |
| Status, n (%) | |
| Censored (alive at the end of the study or was lost to follow up) | 243 (42.5) |
| Dead | 329 (57.5) |
| RFS, median (Q1, Q3), days | 419.0 (186.5, 1,159.8) |
| OS, median (Q1, Q3), days | 1,169.0 (771.5, 1,893.0) |
| DFI, median (Q1, Q3), days | 0 (0, 218.0) |
This table summarizes patient data on key clinically significant variables only. Additional data on all the 65 variables extracted from patient records is summarized in Table S2. ª, For all patients, data was extracted from their pretreatment/baseline PET scan, where available; for patients who received neoadjuvant chemotherapy, data was extracted from their initial PET scan prior to commencing any neoadjuvant treatment; for the 24 patients who only had PET scan data available after commencing treatment, this was not considered to be representative of their baseline characteristics and thus their PET scan data was not included in analysis. CRC, colorectal cancer; CEA, carcinoembryonic antigen; PET, positron emission-tomography; RFS, recurrence-free survival; OS, overall survival; DFI, disease free interval.
Regarding pathology, the median largest CRC and CRLM diameters were 4.0 cm (IQR: 3.0–5.0) and 3.0 cm (IQR: 2.0–4.0) respectively. 41.2% of patients had KRAS mutations. More than two thirds of the patients (69.3%) had unilobar liver metastasis, and the median number of CRLM nodules was 2 (IQR: 1.0–3.0; range, 0–8). This result was consistent with the preoperative PET scan, which also showed 10.6% of patients having extrahepatic metastasis.
There were no statistically significant differences between the data distributions of the training and validation sets (Table S2).
Univariate cox proportional hazard analysis
Univariate analysis revealed that OS and RFS were significantly associated with 27 and 22 variables respectively (P<0.05) (Table S3). Significant variables for both OS and RFS included CRLM clinical risk scores and their parameters, CRC and CRLM pathological factors, liver PET scan parameters, and treatment factors. Patient demographic factors were not associated with either OS or RFS.
Final model results
LASSO regression returned 17 non-zero variables for both OS and RFS (Table 2). Nine additional variables were eliminated using backward model selection. The final models for OS and RFS were conducted with the same list of shortlisted 8 predictors (Table 2).
Table 2. Predictors considered and selected for the final models.
| Overall survival | Recurrence-free survival | |||
|---|---|---|---|---|
| Final model | Global P value: 2.583e-08 | Global P value: 6.005e-06 | ||
| Survival probability is 1 at baseline, which is the date of surgery | Survival probability is 1 at baseline, which is the date of surgery | |||
| Predictor variables | Pooled coefficient | Predictor variables | Pooled coefficient | |
| 1. Bilirubin (>35 μmol/L) pre-hepatectomy | 2.31 | 1. Bilirubin (>35 μmol/L) pre-hepatectomy | 1.46 | |
| 2. Extrahepatic Metastasis detected on PET-scan | 0.41 | 2. CEA pre-hepatectomy (>200 ng/mL) | 0.51 | |
| 3. CEA pre-hepatectomy (>200 ng/mL) | 0.25 | 3. Neoadjuvant treatment for CRLM | 0.43 | |
| 4. Neoadjuvant treatment for CRLM | 0.28 | 4. Extrahepatic Metastasis detected on PET-scan | 0.30 | |
| 5. CRC N stage | 0.25 | 5. CRC N stage | 0.30 | |
| 6. KRAS mutation status | 0.18 | 6. KRAS mutation status | 0.18 | |
| 7. Charlson Comorbidity Score | 0.17 | 7. Charlson Comorbidity Score | 0.12 | |
| 8. CRLM largest tumor diameter | 0.06 | 8. CRLM largest tumor diameter | 0.06 | |
| Other non-0 predictors from LASSO regression excluded from the final model | 1. Adjuvant Chemotherapy following CRLM resection | 1. Adjuvant Chemotherapy following CRLM resection | ||
| 2. Number of CRLM | 2. Number of CRLM | |||
| 3. CRLM lymphovascular invasion | 3. Number CRLM >3 (Nordlinger) | |||
| 4. Largest CRLM tumor diameter on PET-scan | 4. CRLM lymphovascular invasion | |||
| 5. CRC # of lymph nodes | 5. Bilobar CRLM on PET-scan | |||
| 6. CRC lymphovascular invasion | 6. CRC lymphovascular invasion | |||
| 7. CRC resection margin | 7. CRC resection margin | |||
| 8. Disease free interval >1 year (Fong Score) | 8. Disease Free interval >1 year (Fong Score) | |||
| 9. Laparoscopic hepatectomy | 9. Laparoscopic hepatectomy | |||
LASSO, Least Absolute Shrinkage and Selection Operator; PET, positron emission-tomography; CEA, carcinoembryonic antigen; CRLM, colorectal liver metastases; CRC, colorectal cancer.
Kaplan-Meier curves plotted on the model’s predictions on the imputed validation cohort demonstrated good accuracy compared with the actual survival in the same dataset, for both OS and RFS (Figure 2).
Figure 2.
Kaplan-Meier curves plotted on the model’s predictions on the imputed validation cohort, compared with the actual survival and the OS predicted by CRS. CRS, Clinical Risk Score; OS, overall survival.
Our model gives OS and RFS predictions at continuous time points with a global C-index for OS and RFS respectively. Since CRS only gives 1-year and 5-year OS predictions, our C-indices could only be compared with those of CRS at these two timepoints. Validated in the imputed validation cohort, our model demonstrated better ability to predict OS, with a C-index of 0.651, compared with the CRS for 1-year and 5-year OS, with a relative improvement of 13.9% and 13.5% respectively. Our model achieved a C-index of 0.651 for RFS (Table 3).
Table 3. The performance of our model compared with CRS.
| Our model | Fong CRS | Relative improvement | |
|---|---|---|---|
| Overall survival | |||
| 1-year survival | 0.651 (95% CI: 0.537–0.765) | 0.571 (95% CI: 0.514–0.628) | 13.9% |
| 5-year survival | 0.574 (95% CI: 0.517–0.630) | 13.5% | |
| Recurrence-free survival | 0.651 (95% CI: 0.597–0.705) |
CRS, Clinical Risk Score; CI, confidence interval.
Discussion
Given that long-term outcomes post-hepatectomy may vary widely among patients, prognostic tools are crucial in facilitating individualized patient care and multidisciplinary discussion and in identifying patients who may benefit from specific therapeutic modalities. While a few traditional scoring systems (6,9,10) have been validated in international cohorts, they largely rely on histopathological variables and do not consider newer treatment regimens or molecular variables. More recent scores (11,12) which address KRAS mutation status and neoadjuvant chemotherapy are not yet widely validated in external cohorts, and their integer risk scores inherently limit their accuracy in reflecting nonlinear and complex interaction effects between predictors, especially in the modern era of computed recording and prediction.
We developed and internally validated a model that predicts survival of patients with CRLM post-hepatectomy for colorectal liver metastasis using data from 572 patients across 4 major public hospitals in Hong Kong. This is the first prognostication tool for CRLM patients in this locality. When validated in an internal validation set, our model CMAP demonstrated better ability to predict OS (C-index =0.651), compared with the Fong CRS for 1-year (C-index =0.571) and 5-year OS (C-index =0.574). It also achieved a C-index of 0.651 for RFS.
In addition to tailoring to the local population, CMAP accounts for recent advances in CRLM treatment by integrating holistic prognostic information about a patient’s premorbid status, genetic profile, imaging and blood test findings as well as management in predicting patients’ recurrence and survival post-hepatectomy. The need for individualized prognostication is being increasingly emphasized in the modern era of precision medicine (27). Instead of giving generalized estimates of survival by categorizing patients into subgroups, our model is able to account for 8 unique variables that are clinically important in the current standard of care and provide individualized predictions for recurrence and survival at continuous points of time. This can facilitate more discussions tailored to each patient’s unique situation depending on their priorities and expectations of the disease course.
Furthermore, each variable of our model is independently weighed in our machine learning model to enable more accurate prediction of both OS and RFS. The a-CS model developed by Paredes et al. (28), like ours, incorporated machine learning to improve their robustness. Although they reported higher discriminatory ability than the CRS and m-CS, this comparison’s significance is debatable as their model calculates RFS, whereas CRS and m-CS predict OS (29). In our model-building process, predictors for RFS and OS were analyzed separately and while the final model consists of the same list of variables, each model accounts for different weights and interactions between the variables. To ensure a fair comparison, only the OS model’s performance was compared with that of CRS, which only predicts OS.
Variables included in our model which are supported in existing scoring systems include preoperative CEA (6,11,14,15,28,30) size of liver metastases (6,9,10,12,14, 15,28,30), the presence of extrahepatic metastasis (11,14,15,28,30), and positive nodal status of primary CRC (6,11,12,30). Preoperative CEA has been found in studies to impact OS and RFS as well as response to systemic therapy (6,31). The largest tumor diameter of CRLM was used as a continuous rather than a dichotomized variable as studies have shown that binary cut-off values for this parameter may not accurately represent its prognostic significance (32). Existing literature reports that positive primary tumor nodal status in patients with resectable CRLM predicts poor RFS (33). The presence of extrahepatic metastasis was previously considered an absolute contraindication to hepatectomy; however, this notion has been challenged with the advent of novel systemic therapeutic agents which have expanded the criteria for surgical resectability (34). Our model joins recent scoring systems which have similarly included extrahepatic disease as a poor prognostic factor for both OS and RFS (11,14,28,30). Further investigation into specific factors impacting survival outcomes for surgically fit patients with extrahepatic disease should be conducted with the aim of identifying clearer determinants of surgical futility.
In addition to these variables, the current model also includes newer prognostic variables including the use of neoadjuvant chemotherapy and KRAS mutation status, similar to the recent m-CS and GAME score (11,12). Patients with KRAS mutant status have poorer prognostic outcomes given their associated resistance to Epidermal Growth Factor Receptor (EGFR)-targeted therapies as well as more aggressive tumor behavior (31); this has been echoed in existing studies (35,36).
Neoadjuvant chemotherapy pre-hepatectomy was found to be associated with worse OS and RFS, a result also reported to be of interest in other studies (11,28,37,38). This association may be due to the heterogeneity of chemotherapy regimens used, interactions between underlying factors such as tumor molecular status (11), or the fact that patients receiving neoadjuvant chemotherapy often have worse disease status and poorer surgical prognosis to begin with (28). Upon additional retrospective analysis of our study population, patients who received neoadjuvant therapy were more likely to have had bilobar liver metastasis (45.2% vs. 23.3%, P<0.001) and multiple liver lesions (64.9% vs. 42.3%, P<0.001), when compared to those directly undergoing hepatectomy. Future research should assess the prognostic impact of patients’ baseline disease status as well as specific therapeutic regimens including chemotherapeutic agents used, number of cycles, and objective parameters to measure treatment response.
Our inclusion of Charlson Comorbidity Score and pre-hepatectomy bilirubin level reflects our effort to include clinically available and practical predictors, especially in this era where there is increasing evidence for the use of parenchymal-sparing approaches for patient tolerability (39). A high Charlson comorbidity score was associated with higher short-term and long-term mortality in patients by Robertson et al. (40), and was also deemed useful in predicting lower OS in CRC patients over 75 years of age in Japan (41). Yang et al. (42) reported that pre-treatment direct bilirubin was an independent prognostic factor for OS in stage IV CRC patients. Ma et al. (43) found that a bilirubin level of >35 mmol/L was a significant predictor of poor post-hepatectomy outcomes for primary and secondary liver tumors. Additionally, we utilized preoperative PET-CT results to detect the presence of extrahepatic metastasis. Although some studies suggest that preoperative PET-CT is not related to improvements in OS or DFS in CRLM patients (44), PET-CT has demonstrated high sensitivity for detecting distant metastases (45). With PET-CT becoming a more popular preoperative staging tool, future studies may fare better should they incorporate the value of PET findings in prognostic models.
However, certain variables included in previous scoring systems were not found to be significant within this model, including the number of metastatic hepatic lesions (6,9,10,15,28,30) and a shorter disease-free interval (DFI) (6,9,10,28). For the number of hepatic lesions, Jang et al. (46) also reported no statistically significant difference in OS between patients with 1−2 CRLM nodules, compared to those with 3−8 nodules. The advent of modern treatment modalities may explain similar survival outcomes regardless of the number of liver metastases (12). Similarly, other newer scores have also excluded DFI as a variable including the m-CS (12,47) which did not find DFI to significantly predict OS. Conversely, there is expanding literature regarding the worse prognosis of metastases detected after surgical resection (i.e., metachronous lesions) and adjuvant therapy for the primary tumor compared to those synchronously detected, potentially due to unfavorable biological characteristics such as chemotherapy-resistance (12,48).
Our tool takes a holistic view of the patient, integrating prognostic information about their premorbid status, genetic profile, imaging and blood test findings as well as management, allowing for more individualized risk estimates that can be applied to complex clinical scenarios. It will be deployed as a web-based or phone-based clinical decision tool for clinicians to easily input 8 clinical data points. Since our tool supports the predictions of OS and RFS at continuous points in time, this can facilitate more discussions tailored to each patient’s unique situation depending on their priorities and expectations of the disease course.
Limitations
Retrospective data extraction meant that potentially important investigation results and reports, especially those from in the private sector, were not always accessible through electronic health records. Variables with a high proportion of missing data included BRAF status (98.4%), New Edmondson grading for colorectal cancer histology (79.7%), and the presence of CRLM lymphovascular invasion (72.0%). Additionally, certain molecular variables such as NRAS status were not available on patient records. Although the study included consideration of potential confounders such as age and sex, others, like socioeconomic status were not closely examined; this may warrant further analysis. Furthermore, the retrospective nature of this study may have introduced potential selection bias with respect to the variables examined. This was minimized by a priori inclusion of 68 clinically available raw variables for analysis to avoid selective reporting or inclusion.
Data was collected on several other prognostic variables that have been reported in existing literature such as BRAF mutation status and PET findings, however there was insufficient information from our database to draw meaningful conclusions. BRAF mutation status was found by recent studies and a meta-analysis to be strongly associated with worse prognosis (49,50). However, information on BRAF status was only available for 1.6% of patients in our database, thus precluding further conclusions from being drawn. Tumor pathological response to chemotherapy has also been associated with improved survival (51,52). While we collected data on the use of neoadjuvant chemotherapy, data unavailability prevented measurement of tumor response by either radiological (e.g., RECIST) or circulating tumor cells (e.g., CyCAR) criteria, both found to be independent prognostic factors for OS (53).
One of the variables in our model, the largest liver tumor diameter, was determined post-operatively using pathology reports. This is a common limitation faced by other prognostication tools (6,11,32,54). It does not necessarily affect the utility of the tool since CT imaging, as the mainstay for CRLM diagnosis, can be readily substituted as an accurate tool for determining the size of metastases (55). However, future studies should attempt to build models solely with pre-operative findings and compare them with those built with post-operative findings to account for CT’s limitations as well as the differences between the initial CT scan and operation.
The statistical methodology’s weaknesses should also be considered when interpreting the results. Every statistical model comes with certain assumptions in which the model’s validity can be threatened if they are not met (56). A key assumption in the Cox model is the proportional hazard assumption, meaning the ratio of hazards (the output of a Cox model) is constant over time. Future research should include external validation in diverse cohorts as well as prospective studies both locally and internationally to assess accuracy. In addition, Margonis et al. (29) pointed out the inherent limitation of building “One size fits all” prediction models in heterogeneous populations. To account for effect modification by factors that potentially affect CRLM prognosis, patients should be stratified into more homogenous groups according to variables such as comorbid status, extrahepatic metastases, and biological markers to increase accuracy and applicability. Future research can use machine learning to better develop personalized predictions for these sub-stratified populations as identified through the preliminary findings of clinically significant variables in this study. Additionally, given the rapid progress in the field of newer oncological therapies for late-stage cancers, we believe our model provides a backbone for future analyses in this locality, allowing for the incorporation of newer therapeutic regimens in future models.
Conclusions
We presented a promising machine learning algorithm for survival prediction for patients following resection of CRLM. Our model was built on multicenter patient data in Hong Kong, considered new variables and addressed missing data and multicollinearity. Our model demonstrated superior performance than CRS for OS prediction, and an excellent concordance index for both RFS and OS predictions.
In the future, other machine learning algorithms should also be considered to further improve the model performance. The deployment of the model, its evaluation and external validation is reserved for a future publication as it merits more discussion than can be included here.
Supplementary
The article’s supplementary files as
Acknowledgments
We would like to thank Ms. Chrissie Chiu and Mr. Kim Yuen (Department of Surgery, QMH) for their kind support and guidance in data collection.
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All research processes were conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Institutional Review Board of The University of Hong Kong and the Hong Kong West Cluster of Hospital Authority, Hong Kong (Reference number UW 21-471).
Footnotes
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://hbsn.amegroups.com/article/view/10.21037/hbsn-21-453/rc
Data Sharing Statement: Available at https://hbsn.amegroups.com/article/view/10.21037/hbsn-21-453/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://hbsn.amegroups.com/article/view/10.21037/hbsn-21-453/coif). CML serves as the unpaid editorial board member of Hepatobiliary Surgery and Nutrition. The other authors have no conflicts of interest to declare.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Overview of Hong Kong cancer statistics of 2018. Hong Kong Cancer Registry. 2020. Available online: http://www3.ha.org.hk/cancereg
- 3.Filip S, Vymetalkova V, Petera J, et al. Distant Metastasis in Colorectal Cancer Patients—Do We Have New Predicting Clinicopathological and Molecular Biomarkers? A Comprehensive Review. Int J Mol Sci 2020;21:5255. 10.3390/ijms21155255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu F, Tang B, Jin TQ, et al. Current status of surgical treatment of colorectal liver metastases. World J Clin Cases 2018;6:716-34. 10.12998/wjcc.v6.i14.716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tabchouri N, Gayet B, Okumura S, et al. Recurrence patterns after laparoscopic resection of colorectal liver metastases. Surg Endosc 2018;32:4788-97. 10.1007/s00464-018-6229-6 [DOI] [PubMed] [Google Scholar]
- 6.Fong Y, Fortner J, Sun RL, et al. Clinical score for predicting recurrence after hepatic resection for metastatic colorectal cancer: analysis of 1001 consecutive cases. Ann Surg 1999;230:309-18; discussion 318-21. 10.1097/00000658-199909000-00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beamish P, Lemke M, Li J, et al. Validation of clinical risk score for colorectal liver metastases resected in a contemporary multicenter cohort. HPB (Oxford) 2017;19:675-81. 10.1016/j.hpb.2017.03.010 [DOI] [PubMed] [Google Scholar]
- 8.Mann CD, Metcalfe MS, Leopardi LN, et al. The clinical risk score: emerging as a reliable preoperative prognostic index in hepatectomy for colorectal metastases. Arch Surg 2004;139:1168-72. 10.1001/archsurg.139.11.1168 [DOI] [PubMed] [Google Scholar]
- 9.Nordlinger B, Guiguet M, Vaillant JC, et al. Surgical resection of colorectal carcinoma metastases to the liver: a prognostic scoring system to improve case selection, based on 1568 patients. Association Française de Chirurgie. Cancer 1996;77:1254-62. [DOI] [PubMed] [Google Scholar]
- 10.Iwatsuki S, Dvorchik I, Madariaga JR, et al. Hepatic resection for metastatic colorectal adenocarcinoma: a proposal of a prognostic scoring system. J Am Coll Surg 1999;189:291-9. 10.1016/S1072-7515(99)00089-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Margonis GA, Sasaki K, Gholami S, et al. Genetic And Morphological Evaluation (GAME) score for patients with colorectal liver metastases. Br J Surg 2018;105:1210-20. 10.1002/bjs.10838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brudvik KW, Jones RP, Giuliante F, et al. RAS Mutation Clinical Risk Score to Predict Survival After Resection of Colorectal Liver Metastases. Ann Surg 2019;269:120-6. 10.1097/SLA.0000000000002319 [DOI] [PubMed] [Google Scholar]
- 13.Kim WJ, Lim TW, Kang SH, et al. Development and validation of novel scoring system for the prediction of disease recurrence following resection of colorectal liver metastasis. Asian J Surg 2020;43:438-46. 10.1016/j.asjsur.2019.06.001 [DOI] [PubMed] [Google Scholar]
- 14.Liang JY, Lin HC, Liu J, et al. A novel prognostic nomogram for colorectal cancer liver metastasis patients with recurrence after hepatectomy. Cancer Med 2021;10:1535-44. 10.1002/cam4.3697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beppu T, Sakamoto Y, Hasegawa K, et al. A nomogram predicting disease-free survival in patients with colorectal liver metastases treated with hepatic resection: multicenter data collection as a Project Study for Hepatic Surgery of the Japanese Society of Hepato-Biliary-Pancreatic Surgery. J Hepatobiliary Pancreat Sci 2012;19:72-84. 10.1007/s00534-011-0460-z [DOI] [PubMed] [Google Scholar]
- 16.Charlson ME, Pompei P, Ales KL, et al. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis 1987;40:373-83. 10.1016/0021-9681(87)90171-8 [DOI] [PubMed] [Google Scholar]
- 17.Dahiya D, Wu TJ, Lee CF, et al. Minor versus major hepatic resection for small hepatocellular carcinoma (HCC) in cirrhotic patients: a 20-year experience. Surgery 2010;147:676-85. 10.1016/j.surg.2009.10.043 [DOI] [PubMed] [Google Scholar]
- 18.Collins GS, Reitsma JB, Altman DG, et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ 2015;350:g7594. 10.1136/bmj.g7594 [DOI] [PubMed] [Google Scholar]
- 19.van Buuren S, Groothuis-Oudshoorn K, Robitzsch A. Package ‘mice’: Multivariate Imputation by Chained Equations. CRAN Repos. 2019. Available online: https://cran.r-project.org/web/packages/mice/mice.pdf
- 20.Rubin DB. Multiple imputation for nonresponse in surveys: John Wiley & Sons; 2004. [Google Scholar]
- 21.Laird NM. Missing data in longitudinal studies. Stat Med 1988;7:305-15. 10.1002/sim.4780070131 [DOI] [PubMed] [Google Scholar]
- 22.McCleary L. Using multiple imputation for analysis of incomplete data in clinical research. Nurs Res 2002;51:339-43. 10.1097/00006199-200209000-00012 [DOI] [PubMed] [Google Scholar]
- 23.Therneau TM, Lumley T, Elizabeth A, Cynthia C, Therneau MTM. Package ‘survival’. 2020. Available online: https://CRAN.R-project.org/package=survival
- 24.Friedman JH, Hastie T, Tibshirani R. glmnet: lasso and elastic-net regularized generalized linear models, 2010b. 2020:1.-5. Available online: https://cran.r-project.org/web/packages/glmnet/glmnet.pdf
- 25.Dormann CF, Elith J, Bacher S, et al. Collinearity: a review of methods to deal with it and a simulation study evaluating their performance. Ecography 2013;36:27-46. 10.1111/j.1600-0587.2012.07348.x [DOI] [Google Scholar]
- 26.Harrell Jr FE, Harrell Jr MFE. Package ‘hmisc’. CRAN2018. 2019;2019:235-6. Available online: https://cran.r-project.org/web/packages/Hmisc/Hmisc.pdf
- 27.Mahar AL, Compton C, Halabi S, et al. Personalizing prognosis in colorectal cancer: A systematic review of the quality and nature of clinical prognostic tools for survival outcomes. J Surg Oncol 2017;116:969-82. 10.1002/jso.24774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paredes AZ, Hyer JM, Tsilimigras DI, et al. A Novel Machine-Learning Approach to Predict Recurrence After Resection of Colorectal Liver Metastases. Ann Surg Oncol 2020;27:5139-47. 10.1245/s10434-020-08991-9 [DOI] [PubMed] [Google Scholar]
- 29.Margonis GA, Andreatos N, Brennan MF. Predicting Survival in Colorectal Liver Metastasis: Time for New Approaches. Ann Surg Oncol 2020;27:4861-3. 10.1245/s10434-020-09053-w [DOI] [PubMed] [Google Scholar]
- 30.Rees M, Tekkis PP, Welsh FK, et al. Evaluation of long-term survival after hepatic resection for metastatic colorectal cancer: a multifactorial model of 929 patients. Ann Surg 2008;247:125-35. 10.1097/SLA.0b013e31815aa2c2 [DOI] [PubMed] [Google Scholar]
- 31.Spolverato G, Ejaz A, Azad N, et al. Surgery for colorectal liver metastases: The evolution of determining prognosis. World J Gastrointest Oncol 2013;5:207-21. 10.4251/wjgo.v5.i12.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasaki K, Morioka D, Conci S, et al. The tumor burden score: a new “metro-ticket” prognostic tool for colorectal liver metastases based on tumor size and number of tumors. Ann Surg 2018;267:132-41. 10.1097/SLA.0000000000002064 [DOI] [PubMed] [Google Scholar]
- 33.Seeberg LT, Brunborg C, Waage A, et al. Survival Impact of Primary Tumor Lymph Node Status and Circulating Tumor Cells in Patients with Colorectal Liver Metastases. Ann Surg Oncol 2017;24:2113-21. 10.1245/s10434-017-5818-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pawlik TM, Schulick RD, Choti MA. Expanding criteria for resectability of colorectal liver metastases. Oncologist 2008;13:51-64. 10.1634/theoncologist.2007-0142 [DOI] [PubMed] [Google Scholar]
- 35.Karagkounis G, Torbenson MS, Daniel HD, et al. Incidence and prognostic impact of KRAS and BRAF mutation in patients undergoing liver surgery for colorectal metastases. Cancer 2013;119:4137-44. 10.1002/cncr.28347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nash GM, Gimbel M, Shia J, et al. KRAS mutation correlates with accelerated metastatic progression in patients with colorectal liver metastases. Ann Surg Oncol 2010;17:572-8. 10.1245/s10434-009-0605-3 [DOI] [PubMed] [Google Scholar]
- 37.Passot G, Denbo JW, Yamashita S, et al. Is hepatectomy justified for patients with RAS mutant colorectal liver metastases? An analysis of 524 patients undergoing curative liver resection. Surgery 2017;161:332-40. 10.1016/j.surg.2016.07.032 [DOI] [PubMed] [Google Scholar]
- 38.Ito H, Are C, Gonen M, et al. Effect of postoperative morbidity on long-term survival after hepatic resection for metastatic colorectal cancer. Ann Surg 2008;247:994-1002. 10.1097/SLA.0b013e31816c405f [DOI] [PubMed] [Google Scholar]
- 39.Evrard S, Poston G, Kissmeyer-Nielsen P, et al. Combined ablation and resection (CARe) as an effective parenchymal sparing treatment for extensive colorectal liver metastases. PLoS One 2014;9:e114404. 10.1371/journal.pone.0114404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robertson DJ, Stukel TA, Gottlieb DJ, et al. Survival after hepatic resection of colorectal cancer metastases: a national experience. Cancer 2009;115:752-9. 10.1002/cncr.24081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tominaga T, Nonaka T, Takeshita H, et al. The Charlson Comorbidity Index as an Independent Prognostic Factor in Older Colorectal Cancer Patients. Indian J Surg 2018;80:54-60. 10.1007/s12262-016-1544-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang L, Ge LY, Yu T, et al. The prognostic impact of serum bilirubin in stage IV colorectal cancer patients. J Clin Lab Anal 2018;32:e22272. 10.1002/jcla.22272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma KW, Cheung TT, She WH, et al. Risk prediction model for major complication after hepatectomy for malignant tumor - A validated scoring system from a university center. Surg Oncol 2017;26:446-52. 10.1016/j.suronc.2017.08.007 [DOI] [PubMed] [Google Scholar]
- 44.Daza JF, Solis NM, Parpia S, et al. A meta-analysis exploring the role of PET and PET-CT in the management of potentially resectable colorectal cancer liver metastases. Eur J Surg Oncol 2019;45:1341-8. 10.1016/j.ejso.2019.03.025 [DOI] [PubMed] [Google Scholar]
- 45.Grut H, Dueland S, Line PD, et al. The prognostic value of 18F-FDG PET/CT prior to liver transplantation for nonresectable colorectal liver metastases. Eur J Nucl Med Mol Imaging 2018;45:218-25. 10.1007/s00259-017-3843-9 [DOI] [PubMed] [Google Scholar]
- 46.Jang KU, Kim CW, Kim KH, et al. Prognostic Factors in Terms of the Number of Metastatic Nodules in Patients With Colorectal Cancer Liver Metastases. Ann Coloproctol 2016;32:92-100. 10.3393/ac.2016.32.3.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Höppener DJ, Nierop PMH, van Amerongen MJ, et al. The Disease-Free Interval Between Resection of Primary Colorectal Malignancy and the Detection of Hepatic Metastases Predicts Disease Recurrence But Not Overall Survival. Ann Surg Oncol 2019;26:2812-20. 10.1245/s10434-019-07481-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andreou A, Kopetz S, Maru DM, et al. Adjuvant chemotherapy with FOLFOX for primary colorectal cancer is associated with increased somatic gene mutations and inferior survival in patients undergoing hepatectomy for metachronous liver metastases. Ann Surg 2012;256:642-50. 10.1097/SLA.0b013e31826b4dcc [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsilimigras DI, Ntanasis-Stathopoulos I, Bagante F, et al. Clinical significance and prognostic relevance of KRAS, BRAF, PI3K and TP53 genetic mutation analysis for resectable and unresectable colorectal liver metastases: A systematic review of the current evidence. Surg Oncol 2018;27:280-8. 10.1016/j.suronc.2018.05.012 [DOI] [PubMed] [Google Scholar]
- 50.Margonis GA, Buettner S, Andreatos N, et al. Association of BRAF Mutations With Survival and Recurrence in Surgically Treated Patients With Metastatic Colorectal Liver Cancer. JAMA Surg 2018;153:e180996. 10.1001/jamasurg.2018.0996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Primavesi F, Fadinger N, Biggel S, et al. Early response evaluation during preoperative chemotherapy for colorectal liver metastases: Combined size and morphology-based criteria predict pathological response and survival after resection. J Surg Oncol 2020;121:382-91. 10.1002/jso.25796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berardi G, De Man M, Laurent S, et al. Radiologic and pathologic response to neoadjuvant chemotherapy predicts survival in patients undergoing the liver-first approach for synchronous colorectal liver metastases. Eur J Surg Oncol 2018;44:1069-77. 10.1016/j.ejso.2018.03.008 [DOI] [PubMed] [Google Scholar]
- 53.Delgado-Ureña M, Ortega FG, de Miguel-Pérez D, et al. Circulating tumor cells criteria (CyCAR) versus standard RECIST criteria for treatment response assessment in metastatic colorectal cancer patients. J Transl Med 2018;16:251. 10.1186/s12967-018-1624-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Byun SS, Heo TS, Choi JM, et al. Deep learning based prediction of prognosis in nonmetastatic clear cell renal cell carcinoma. Sci Rep 2021;11:1242. 10.1038/s41598-020-80262-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cantisani V, Grazhdani H, Fioravanti C, et al. Liver metastases: Contrast-enhanced ultrasound compared with computed tomography and magnetic resonance. World J Gastroenterol 2014;20:9998-10007. 10.3748/wjg.v20.i29.9998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Freedman DA. Statistical models: theory and practice. Cambridge University press; 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as