Abstract
Experimental studies of microbial evolution have largely focused on monocultures of model organisms, but most microbes live in communities where interactions with other species may impact rates and modes of evolution. Using the cheese rind model microbial community, we determined how species interactions shape the evolution of the widespread food- and animal-associated bacterium Staphylococcus xylosus. We evolved S. xylosus for 450 generations alone or in co-culture with one of three microbes: the yeast Debaryomyces hansenii, the bacterium Brevibacterium aurantiacum, and the mold Penicillium solitum. We used the frequency of colony morphology mutants (pigment and colony texture phenotypes) and whole-genome sequencing of isolates to quantify phenotypic and genomic evolution. The yeast D. hansenii strongly promoted diversification of S. xylosus. By the end of the experiment, all populations co-cultured with the yeast were dominated by pigment and colony morphology mutant phenotypes. Populations of S. xylosus grown alone, with B. aurantiacum, or with P. solitum did not evolve novel phenotypic diversity. Whole-genome sequencing of individual mutant isolates across all four treatments identified numerous unique mutations in the operons for the SigB, Agr, and WalRK global regulators, but only in the D. hansenii treatment. Phenotyping and RNA-seq experiments highlighted altered pigment and biofilm production, spreading, stress tolerance, and metabolism of S. xylosus mutants. Fitness experiments revealed antagonistic pleiotropy, where beneficial mutations that evolved in the presence of the yeast had strong negative fitness effects in other biotic environments. This work demonstrates that bacterial-fungal interactions can have long-term evolutionary consequences within multispecies microbiomes by facilitating the evolution of strain diversity.
Subject terms: Microbial ecology, Evolution
Introduction
Variation in ecological, genetic, physiological, and biochemical traits is commonly observed within microbial species. From metabolic heterogeneity in the bacterium Escherichia coli [1, 2] to divergent stress responses of the yeast Saccharomyces cerevisiae [3, 4], strain-level diversity is a fundamental feature of most microbes. Intraspecific variation can impact microbial ecology [5–7] and provides useful diversity to exploit for industrial purposes [4, 8, 9]. Despite its biological and economic significance, the mechanisms that generate intraspecific phenotypic variation within microbiomes are poorly characterized [10].
Adaptation to the challenges and opportunities presented by other microbial species may contribute to the generation of microbial strain diversity. Due to their large population sizes and fast generation times, microbes can rapidly adapt to a range of selective pressures [11–15]. Microbiologists have used experimental evolution to repeatedly subculture microbial species over many generations and monitor rapid adaptation to novel environments [11, 13, 16]. These experiments typically use monocultures of model species in the lab [17–19], but most microbes in nature live in multispecies communities where they experience strong selective pressures from other microbes [20–25]. Because interspecific microbial interactions have been largely excluded from experimental evolution, our understanding of the dynamics and mechanisms of microbial adaptation in multispecies microbiomes is incomplete and lacking biotic context.
Based on examples from macroorganisms and some preliminary studies in synthetic microbial systems, microbe-microbe interactions can mediate adaptation by reducing population size and subsequent rates of mutations [26], inducing shifts to alternative niches via competition [27, 28], producing novel niches via resource provisioning [29, 30], or by inducing or releasing environmental stresses and impacting selection on stress and defense systems [31]. This is not an exhaustive set of all potential mechanisms underlying microbial adaptation to biotic environments and multiple mechanisms may be operating in the presence of a single neighbor.
A few highly synthetic studies of laboratory isolates have started to identify different mechanisms of microbial adaptation to varying biotic environments [28, 30, 32–37]. These studies begin to add biological complexity to microbial evolution, but they rarely attempt to explain adaptation of microbial populations in naturally occurring communities where ecological environments of ancestral and evolved strains can be clearly defined. These studies also often use selective environments that may not be relevant to the interactions that microbes experience in naturally forming microbial systems. Moreover, this work has been limited to a few target species (mainly Pseudomonas fluorescens) and does not usually identify specific mechanisms underlying microbial adaptation to novel biotic environments [33].
The domestication of wild microbes in fermented food environments provides a unique opportunity to fill major gaps in our understanding of how interspecific interactions impact microbial adaptation. In some fermented foods - such as sourdough, kimchi, and certain cheeses - wild microbial species colonize the food substrate and become part of the food microbiome [38–43]. As continuous batches of food are made in the same location, recirculating microbial populations can adapt to novel abiotic and biotic environments [43, 44]. Hints of adaptive evolution come from comparative studies of domesticated and wild microbes [44–46]. But the mechanisms that drive the adaptation of fermented food microbes are unknown due to a limited number of studies experimentally recreating the domestication process [43, 44, 47].
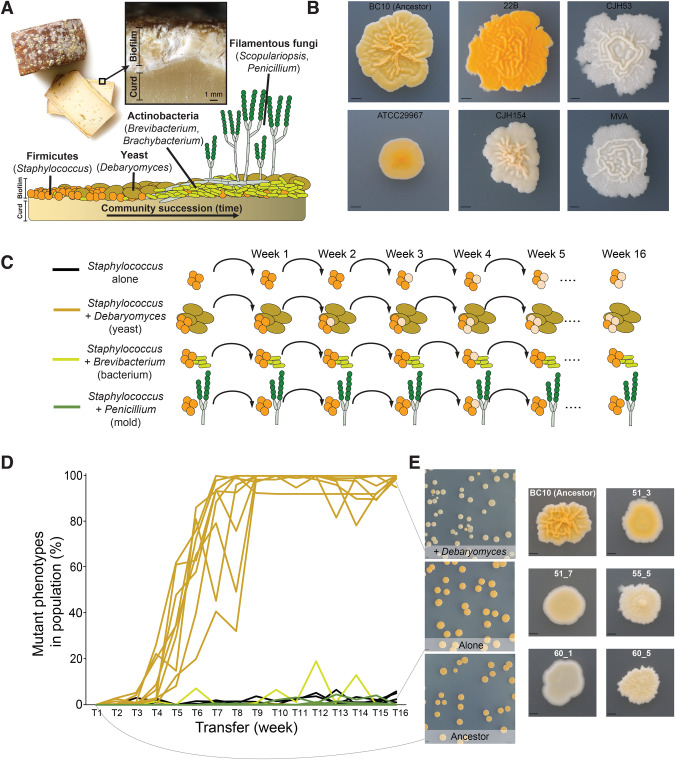
In this work, we use one bacterium that is widespread in cheese rinds - Staphylococcus xylosus - as a target species for evolution in different biotic environments. This Staphylococcus species is often used as a starter culture in the production of cheese and fermented meats [48], but can also be found associated with various mammals [49]. This bacterium is an early colonizer of cheese rinds and interacts with a range of fungal and bacterial species during cheese rind succession ([50], Fig. 1A). We have observed considerable variation in colony morphology and pigmentation of strains isolated from cheese and other fermented food environments (Fig. 1B). The evolutionary processes that shape this remarkable phenotypic diversity in S. xylosus have not been characterized.
Fig. 1. The yeast Debaryomyces hansenii causes phenotypic diversification of S. xylosus BC10 during experimental evolution.
A Overview of the cheese rind microbiome and the microbial species used in this manuscript including Staphylococcus xylosus, Debaryomyces hansenii, Brevibacterium aurantiacum, and Penicillium solitum. B Phenotypic variation across different isolates of Staphylococcus xylosus. All isolates are from cheese except for ATCC29967, the reference isolate for the species. BC10 is the strain used in this study. The scale bar represents 2.5 mm. C Overview of the experimental design used for the evolution study. D Frequency of phenotypic mutants in replicate populations across the four experimental treatments. Line colors correspond with the treatments indicated in C. Each line represents a replicate population, with ten replicates per treatment. One +Debaryomyces replicate became contaminated and had to be removed from the experiment. Mutant frequency was significantly higher in the +Debaryomyces treatment compared to the other three treatments at the final transfer (Kruskal–Wallis p < 0.0001, with Mann–Whitney post-hoc tests). E Examples of typical colony morphotypes within a population of Ancestor (the wild-type BC10), a population evolved Alone (without another microbial species), and a population in the +Debaryomyces treatment. Note the distinct difference in color in +Debaryomyces compared to the other two populations. More detailed examples of colony morphologies are shown on the right. 51_3, 51_7, 55_5, 60_1, and 60_5 were used for additional experiments in Figs. 3–5. The scale bar in the population photos on the left represents 5 mm and the scale bar in the closeup photos represents 1 mm.
To determine if microbial interactions contribute to the phenotypic and genomic diversification of S. xylosus, we evolved an isolate of this bacterium (strain BC10) either alone or co-cultured with each of three different cheese rind neighbor species. We predicted that microbe-microbe interactions might promote diversification in the presence of some neighboring species that provide novel ecological opportunities and inhibit diversification in the presence of other species that are strong competitors. Because the evolution of colony morphology often correlates with clear underlying adaptive mutations in Staphylococcus species [51, 52], we were able to track phenotypic diversity of colonies over time as a proxy for diversification under the different biotic conditions. We also conducted whole-genome sequencing of evolved isolates from the end of the experiment to measure genomic diversity and determine mutations that were driving phenotypic diversity. Competition experiments, phenotypic assays, and RNA sequencing of select isolates revealed underlying mechanisms of adaptation and intriguing trade-offs associated with the adaptation of S. xylosus to different biotic environments.
Results
The yeast Debaryomyces hansenii promotes phenotypic evolution of S. xylosus
We serially transferred populations of S. xylosus grown in four different biotic environments: (1) without a neighbor (hereafter Alone), (2) with the bacterium Brevibacterium aurantiacum (hereafter +Brevibacterium), (3) with the yeast Debaryomyces hansenii (hereafter +Debaryomyces), and (4) with the filamentous fungus Penicillium solitum (hereafter +Penicillium). All of these microbes were isolated from the same natural rind cheese that has been the focus of previous research in this model system, and span a range of life history strategies and temporal dynamics in community succession [50, 53–55]. Both S. xylosus species and yeasts such as D. hansenii are abundant in early stages of cheese rind development, while Penicillium molds and Actinobacteria such as Brevibacterium species thrive in the later stages of rind development [50, 54]. Because representatives of all three neighboring taxa have been shown to both inhibit or promote the growth of S. xylosus and other Staphylococcus species [50, 54], we predicted that the different biotic environments created by these neighbors would impact phenotypic and genomic evolution of S. xylosus.
Ten replicate populations of S. xylosus in each of the four different conditions were transferred weekly for 16 weeks (about 450 generations). At each transfer, populations were plated out to determine the number of wild-type and mutant colonies and to determine the presence of the interacting neighbor species. We did not attempt to distinguish different types of mutant colony morphologies within populations because some of these differences are subtle and difficult to track over time. We simply tracked the proportion of total mutant (not wild-type/ancestor) colonies over time as an approximation of phenotypic evolution within the S. xylosus populations. We acknowledge that this approach misses some important dynamics of strain evolution over time and will not capture genomic diversification that does not cause colony morphology changes. But it is a useful tool to track phenotypic diversification that also pointed to some fascinating patterns of genomic evolution at the end of the experiment.
Five weeks into the experiment, we noticed that many colonies across replicate +Debaryomyces populations had morphotypes that were different in appearance compared to the ancestor (Fig. 1D). These colonies were often reduced in pigmentation, were not as wrinkly, and often had a shiny appearance that was not apparent in the ancestor (Fig. 1E). By the end of the experiment, most of the +Debaryomyces replicate populations were taken over by mutant colony phenotypes (99% mean phenotypic mutant frequency across populations; Fig. 1D). In contrast to the +Debaryomyces treatment, very few phenotypic mutant colony phenotypes were observed in the Alone (0.02% frequency), +Brevibacterium (0% frequency), or +Penicillium (0% frequency) treatments. There were fluctuations at certain transfer points in phenotypic mutant frequencies in these other treatments (sometimes up to nearly 20%), but only in the +Debaryomyces treatment did phenotypic mutant frequencies stabilize at or nearly 100% frequency. Interacting neighbor species were maintained throughout the 16 weeks of the experiment (Fig. S1).
Differences in population sizes caused by both abiotic and biotic factors can impact rates of evolution [26, 56] and may explain the differences in phenotypic evolution across the four treatments. For example, biotic interactions that suppress population size may inhibit evolution [57, 58]. There were significant differences in population size over time across the four treatments, with the +Penicillium treatment having the highest mean population size (ANOVA F3,38 = 58.2; p < 0.001). However, the +Debaryomyces populations where we observed the high phenotypic mutant frequency were not significantly different in population size from the Alone or +Penicillium treatments with low phenotypic mutant frequencies (Fig. S2), suggesting that population size does not explain the vastly different amounts of phenotypic evolution we observed.
Mutations in global regulatory genes explain S. xylosus phenotypic evolution with Debaryomyces hansenii
To better understand genomic changes that drove the phenotypic diversification of S. xylosus, we randomly selected seven colonies from three representative populations from each of the four treatments for whole-genome sequencing. We mapped Illumina sequence reads from each of these evolved isolates to the S. xylosus reference genome to identify SNPs and other genomic changes in each evolved isolate. Corroborating the phenotypic evolution observed in the +Debaryomyces treatment, we observed a higher frequency of non-synonymous mutations in isolates from that treatment (ANOVA F3,11 = 6.4, p < 0.05; Fig. 2A). All isolates from the +Debaryomyces treatment contained at least one non-synonymous mutation, whereas fewer than half of the isolates from the other treatments had a non-synonymous mutation. One intergenic and seven synonymous mutations were observed across the sequenced isolates (Table S1).
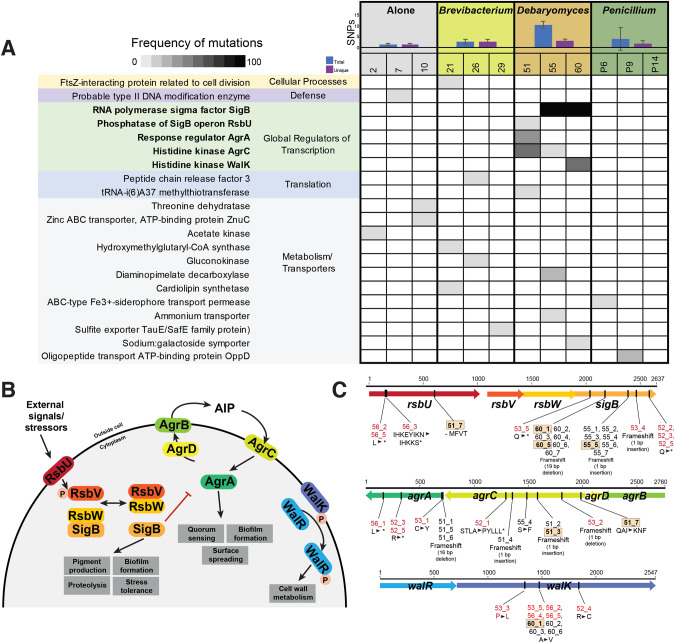
Fig. 2. Whole-genome sequencing identifies mutations in putative global regulators of S. xylosus BC10 in the +Debaryomyces treatment.
A Blue and purple bar graphs show number of SNPs (total in blue, number across unique genes in purple) detected across S. xylosus from three populations within each of the four biotic treatments (Alone, +Brevibacterium, +Debaryomyces, and +Penicillium). Shaded cells show frequency of non-synonymous mutations within a gene within a population (columns) based on sequencing 7 isolates per population. Only genes with a putative function assigned are shown. See Fig. S3 for a full table of all mutations, including those with unknown functions and those in the additional +Debaryomyces treatments that were sequenced. Numbers 1–10, 21–30, 51–60, and 61–70 are used to indicate unique populations. There were other experimental populations included in the initial experiment (31–40, 41–50), but they are excluded from this manuscript because the neighbor treatment went extinct. B Overview of the components of the three global regulatory systems where many mutations were detected in the +Debaryomyces treatment. These systems are poorly characterized in S. xylosus, so the potential structure and function of these systems is inferred from what is known in S. aureus. C Location and type of mutations observed in the sigB, agr, and walRK loci. Strains indicated in red font were from the additional +Debaryomyces treatments that were sequenced and are not included in (A), but are found in Fig. S3.
When considering the types of genes with mutations across all treatments, a very distinct pattern emerged: all isolates from the +Debaryomyces treatment had at least one mutation in a global regulator of transcription, including the alternative sigma factor B (SigB), the accessory gene regulator (Agr), and the WalRK system (Fig. 2A). As discussed below, all three of these systems regulate key cellular processes, including biofilm and pigment formation, and could explain the increase of novel colony morphotypes in the +Debaryomyces treatment. We did not detect mutations in global regulators of transcription in any of the other treatments. Mutations in genes related to metabolism and nutrient transport, translation, defense, and other cellular processes occurred across all four treatments, but there was not a specific enrichment of these mutations in any treatment (Fig. 2A). To confirm that this pattern of mutations in global regulators was consistent beyond the three +Debaryomyces populations that we sampled (51, 55, 60), we sequenced genomes of 5 isolates from an additional three populations (52, 53, 56) from the +Debaryomyces treatment and found an identical pattern of mutations in global regulators of transcription across all three populations (Fig. S3).
SigB is very well-characterized in Staphylococcus aureus and other Gram-positive bacterial species, where it has been shown to regulate the expression of hundreds of genes [59–61] (Fig. 2B). The sigB operon includes the genes sigB, rsbU, rsbV, and rsbW (Fig. 2C). A diverse set of mutations in the sigB operon were detected across populations, with most mutations observed in sigB, a few in rsbU, and no mutations in rsbV or rsbW (Fig. 2C). Mutations in sigB ranged from amino acid substitutions to both small (1 bp insertion) and large (19 bp deletion) frameshift mutations. Some populations contained the same mutation across all isolates (e.g. a major deletion of 19 base pairs in all isolates from population 60; Fig. 2C) whereas other populations had multiple strains with unique sigB mutations (e.g. a predicted truncation in 53_5 and a 1 bp insertion causing a frameshift in 53_4). Three different mutations were detected in rsbU: a 12 base-pair deletion causing a predicted loss of 4 amino acids in an isolate from population 51 (strain 51_7, Fig. 2C), a predicted truncation in two isolates from population 56, and amino acid substitutions and a truncation in another isolate from population 56.
Agr is a quorum sensing regulator that has been well-characterized in S. aureus and other Staphylococcus species [62–64] (Fig. 2B). The agr operon consists of agrA and agrC (encoding a two-component signal transduction system), agrD (encoding a propeptide that becomes the quorum sensing molecule), and agrB (encoding a transmembrane protein). Consistent with patterns of Agr evolution in S. aureus [63], all mutations we observed were in agrA and agrC (Fig. 2C). Most mutations were observed in population 51, with four unique mutations distributed across the 7 isolates, including both large deletions (in agrA) and small insertions/deletions (in agrC) causing frameshifts (Fig. 2C). An amino acid substitution was observed in one isolate from population 55. Additional agrA and agrC mutations were observed in the additional +Debaryomyces populations that were sequenced (Figs. 2C and S3).
The walRK operon (sometimes referred to as walKR, yycGF, or vicKR) is another regulatory region where multiple mutations were observed in +Debaryomyces isolates. WalRK is a two-component system that controls a variety of processes in S. aureus, including cell wall metabolism [65] (Fig. 2B). Although the functions of WalRK have not been characterized in S. xylosus, previous work in S. aureus has demonstrated that single mutations in walK or walR can impact drug resistance and cell wall structure [66]. Three different amino acid substitutions were observed in the predicted walK gene, including one mutation that occurred across three different populations (53, 56, and 60; Fig. 2C). Some strains had mutations in both the walK and either rsbU (strains 56_2, 56_5) or sigB (strains 53_5, 60_1, 60_2, 60_3, 60_6).
Collectively, our resequencing of evolved mutants highlights parallel mutations in global regulators across replicate +Debaryomyces populations. The regulatory systems noted above have not been characterized in S. xylosus, so we cannot know for sure that they regulate the same genes and traits as in S. aureus where they have been well-studied. But many components of these systems are conserved across Staphylococcus species or Gram-positive bacteria more generally [67–69], and may operate in a similar manner in S. xylosus.
S. xylosus mutants have altered pigment, biofilm, and stress tolerance phenotypes
Because whole-genome sequence data identified mutations in global regulators known to control phenotypes in Staphylococcus species, we used a series of assays to characterize these traits in a subset of evolved strains of S. xylosus BC10. Our goal with these assays was to better understand how the mutations impacted the biology of S. xylosus and whether the mutations in the global regulators had similar effects in S. xylosus as they do in other Staphylococcus species. We selected mutants from different replicate populations of the +Debaryomyces treatment that spanned a range of genes and different mutations within those genes (strains 51_3, 51_7, 55_5, 60_1, and 60_5). These mutants did not have other mutations in predicted coding regions, so changes in phenotypes observed in these strains could be attributed to mutations in the global regulators.
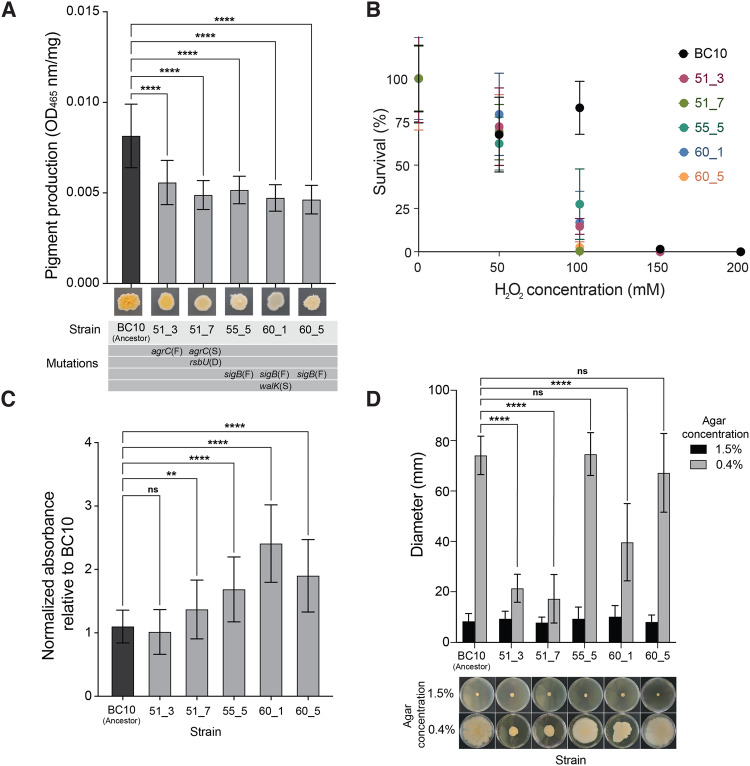
Colonies of S. xylosus evolved in the +Debaryomyces treatment had noticeably lighter colonies compared to other populations (Fig. 3A). We predicted that these differences could be due to changes in production of the carotenoid staphyloxanthin. This pigment gives S. aureus and other Staphylococcus species a golden or orange appearance and is encoded by the crtOPQMN operon that is controlled by SigB [70]. Mutations in the SigB operon can lead to reduced pigmentation [52, 71]. Colorimetric assays indicated that the ancestor produced significantly more staphyloxanthin pigment compared to all other evolved strains (one-way ANOVA; p < 0.001). The strains containing a mutation in the sigB system produced the least amount of pigment, with strain 60_5 only producing 43% compared to the ancestor. Strains 51_3 and 51_7 also produced significantly less pigment, 68 and 60% compared to the ancestor, respectively.
Fig. 3. Phenotypic assays reveal biological impacts of global regulator mutations in evolved S. xylosus BC10.
A Pigment production in the ancestor and evolved strains of S. xylosus BC10. Bars represent means and error bars represent standard deviations. **** indicates p < 0.0001 based on ANOVA with Dunnett’s test. Data are from three independent experiments with six replicates of each treatment within each experiment. B Survival of the ancestor and evolved strains of S. xylosus BC10 across a range of H2O2 concentrations. Dots represent means and error bars represent standard deviations. At 100 mM, the S. xylosus BC10 ancestor had a higher survival compared to all evolved strains (two-way ANOVA, p < 0.0001). Data are from two independent experiments with eight replicates of each treatment within each experiment. C Biofilm production of ancestor and evolved strains of S. xylosus BC10. Bars represent means and error bars represent standard deviations. ** indicates p < 0.01 and **** indicates p < 0.0001 based on ANOVA with Dunnett’s test. ns indicates not significant. Data are from three independent experiments with nine replicates of each treatment within each experiment. D Spreading of ancestor and evolved strains of S. xylosus BC10. Photos at the bottom of the graph show representative plates from the 1.5% (low spreading) and 0.4% (high spreading) conditions. Bars represent means and error bars represent standard deviations. **** indicates p < 0.0001 based on ANOVA with Dunnett’s test. ns indicates not significant. Data are from three independent experiments with five replicates of each treatment within each experiment.
Because carotenoids like staphyloxanthin can function as antioxidants in Staphylococcus species [72, 73] and may play a role in how S. xylosus interacts with the cheese rind environment, we next tested tolerance of the S. xylosus strains to oxidative stress in the form of hydrogen peroxide. All strains were equally susceptible to 50 mM of H2O2, but the ancestor BC10 had a higher tolerance to 100 mM than the evolved strains (Fig. 3B). At 150 mM, there were no viable cells in any population. These data suggest that S. xylosus carotenoid levels can provide similar oxidative protection as in S. aureus and other Staphylococcus species.
Biofilm formation is a key ecological trait that determines how microbes interact with both abiotic and biotic elements of their environment [74–76]. To understand if the observed mutations in the evolved S. xylosus affected biofilm production, we used a crystal violet staining assay to quantify biofilm production. In previous studies of S. aureus, expression of sigB has been linked with the ability to produce biofilms and mutations in sigB have resulted in biofilm-negative phenotypes [77, 78]. Mutations in the agr system can cause increased biofilm production in S. aureus because a functional agr inhibits biofilm production [79]. Based on this, we predicted that S. xylosus strains that had mutations in sigB would result in decreased biofilm formation compared to the ancestor, while the strains with agr mutations would have increased or similar levels of production. In contrast to these predictions, strains 55_5, 60_1, and 60_5 (all with sigB frameshift mutations) produced significantly more biofilm at 1.53, 2.18, and 1.78 times more than the BC10 ancestor (Fig. 3C). Aligning with our predictions, the two strains that had agrC mutations (51_3 and 51_7) had increased or equal biofilm formation compared to the ancestor (1.2 times more for 51_7 produced and equal production for 51_3, Fig. 3C).
The ability to spread across surfaces is an important trait for the virulence of S. aureus [80, 81], and may also be important for how S. xylosus spreads across cheese surfaces [82, 83]. Previous studies have shown that the agr system is involved in the production of a biosurfactant that allows Staphylococcus species to spread, and that disruption of the agr system can cause reduced spreading [51, 52, 84]. Both agr mutants were found to be deficient in colony spreading; strain 51_3 and 51_7 had a 71.1% and 76.7% reduction in spreading compared to the BC10 ancestor, respectively (Fig. 3D). Strain 60_1 had an average of 46.5% reduction in spreading with irregular colony edges (Fig. 3D). Strains 55_5 and 60_5 were not statistically different from the ancestor (Fig. 3D).
These targeted phenotypic data demonstrate that S. xylosus mutations that evolved in the presence of D. hansenii can have major impacts on the biology of this bacterium. Our data also suggest that some of the functions of these global regulators from S. aureus may be conserved in S. xylosus; predictions about how mutations in the global regulators might alter phenotypes based on S. aureus biology were often correct.
RNA-seq reveals decreased expression of key metabolic pathways in evolved S. xylosus strains
To better understand other underlying changes in the biology of the evolved strains that were not captured with the phenotypic assays, we used RNA-seq to quantify global transcriptional differences between the ancestor BC10 and three mutants: 51_3, 55_5, and 60_1. We selected these three strains for RNA-seq because they ranged in sigB, agr, and walRK mutations and had no or few other genetic changes in other regions of their genome. We compared global gene expression at 3 days of growth on cheese curd agar because the bacterium is in late exponential phase and RNA can be easily extracted at this time point.
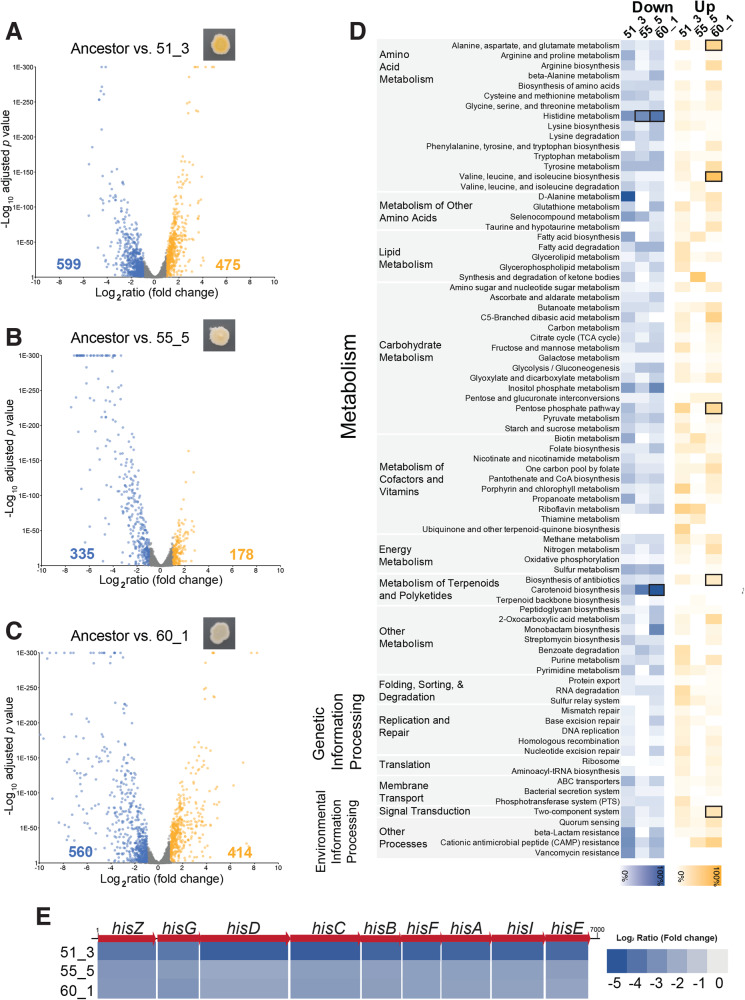
Each of the three evolved strains had major shifts in global transcription compared to the ancestor BC10 (Fig. 4A–C; Tables S2–S5). With a differentially expressed gene (DEG) cutoff of a log2 ratio of 1/−1 and a false discovery rate (FDR) corrected p value of 0.05, many genes across the genome of the bacterium had lower expression levels in the mutants: 23% of predicted genes across the genome in 51_3, 13% in 55_5, and 22% in 60_1. A lower, but still substantial number of genes were also upregulated across the three strains: 18% of genes in 51_3, 7% in 55_5, and 16% in 60_1. These broad patterns of DEGs support previous studies showing that SigB, Agr, and WalRK can have broad transcriptional control in Staphylococcus species and that mutations in these global regulators can alter transcriptional networks within these species [52, 85–89].
Fig. 4. RNA sequencing highlights transcriptomic impacts of global regulator mutations in S. xylosus BC10.
Volcano plots showing changes in gene expression in mutant strain 51_3 (A), 55_5 (B), and 60_1 (C) compared to the ancestor BC10. Each dot represents a gene in the BC10 genome. Yellow dots are genes with significant increases in expression in the mutants. Blue dots are genes with significant decreases in expression compared to the ancestor. Numbers on the left and right of the x-axes indicate the number of genes that were significantly higher (yellow) or lower (blue) in expression for each mutant. D KOBAS pathway analysis of differentially expressed genes for each mutant. Blue or yellow shading indicates percent of genes in a pathway that were differentially expressed. Bold boxes indicate significant enrichment of a pathway based on a Fisher’s exact test with FDR correction. E Fold-change in expression of the nine genes in the his operon for histidine biosynthesis.
Across the three mutants, most of the differentially expressed genes were associated with metabolism, including the biosynthesis and degradation of amino acids, lipids, carbohydrates, and secondary metabolites (Fig. 4D; Tables S6–S9). When we used pathway enrichment analysis to identify what DEGs were significantly associated with all three mutants, two patterns of decreased pathway expression emerged. One pattern involves the production of carotenoids. Most of the genes in the staphyloxanthin biosynthesis operon were downregulated in all three mutants compared to the ancestor (Fig. 4D). This was especially pronounced in strains 55_5 and 60_1 and corroborates our observations and pigment assays above where 55_5 and 60_1 were much lighter in color compared to the ancestor. This also aligns with previous studies demonstrating that disruption of normal SigB functioning can alter pigment production in S. aureus [52].
The other strong pattern of decreased pathway expression across all three mutants was histidine metabolism (Fig. 4D). Specifically, all genes in a putative histidine biosynthesis operon were strongly downregulated in all three mutants compared to the ancestor (Fig. 4E). In a previous study of another S. xylosus strain, this histidine operon was upregulated in growth in a dairy matrix due to the low availability of histidine and other free amino acids [90]. Downregulation of histidine biosynthesis and other amino acid metabolism in the evolved S. xylosus BC10 mutants suggests a parallel shift in nutrient requirements in all three strains, despite different mutations that caused the downregulation.
Competition experiments reveal fitness trade-offs of evolved S. xylosus strains in different biotic environments
To better understand how the mutations and altered phenotypes observed above impact the fitness of evolved S. xylosus BC10 strains, we conducted a series of assays to measure growth and competitive abilities of all strains in different biotic environments. First, we measured the growth of the ancestor and evolved strains growing individually on cheese curd agar for 7 days. All strains demonstrated typical growth curves on cheese, reaching stationary phase around 3 days of growth. There were significant differences in growth over time across all strains (F5,84 = 6.9, p < 0.001; Table S10), which was driven by two major patterns. First, early in growth at day 1, all strains except 60_5 had lower growth compared to the ancestor (Fig. 5A), suggesting a lag in log phase for most mutant strains. Second, at the final time point of 7 days, two strains (55_5, 60_6) had significantly higher abundance compared to the ancestor (Fig. 5A). These data demonstrate that the genomic changes in the evolved strains had minor and inconsistent effects on their growth compared to the ancestor.
Fig. 5. Growth and competition experiments reveal fitness of evolved S. xylosus mutants in different biotic environments.
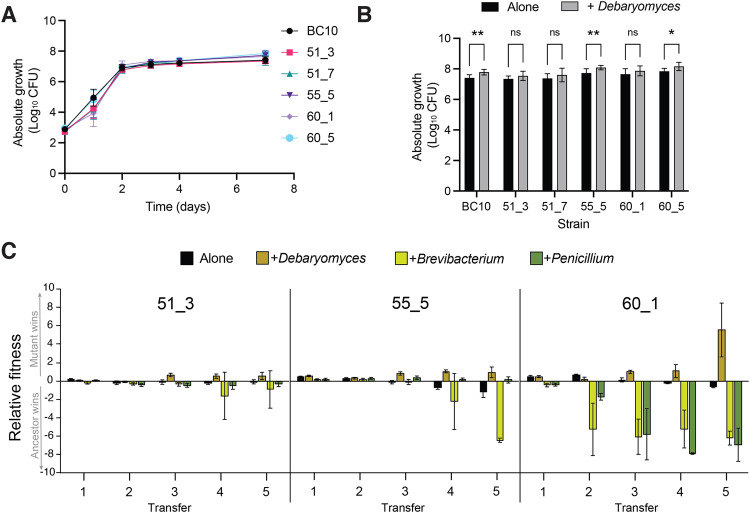
A Growth of ancestor and evolved strains on cheese curd agar alone. Data represent mean CFUs at each time point and error bars are standard errors of the mean. Data are from three independent experiments with five replicates of each treatment within each experiment. B Growth of ancestor and evolved strains on cheese curd agar with and without the yeast D. hansenii after 7 days of growth. Data represent mean CFUs and error bars are one standard error of the mean. Data are from three independent experiments with five replicates of each treatment within each experiment. D. hansenii increased the growth of the isolates compared to growth alone (F5,161 = 3.4; p < 0.001). For post-hoc comparisons, ** = p < 0.005; * = p < 0.05. C Fitness of ancestor and mutant strains of S. xylosus when competed in initially identical ratios in different biotic environments. The ancestor:mutant mix was grown in four treatments and passaged five times to mimic the repeated cycles of growth in the evolution experiment. Relative fitness is expressed as log10((CFUs of mutant strain +1) ÷ (CFUs of ancestor strain +1)). A positive relative fitness means that the mutant strain reached a higher proportion of the total number of CFUs when competing with the ancestor. A negative fitness means the ancestor strain reached a higher proportion. Error bars represent one standard deviation of the mean with eight replicates of each treatment.
Given the high frequency of global regulator mutations in the +Debaryomyces treatment, we next tested how the presence of D. hansenii impacted the growth of each of the ancestor and evolved strains. We predicted that D. hansenii may increase the growth of evolved strains relative to the ancestor because they should have a fitness benefit from living with the yeast. Surprisingly, all strains were slightly stimulated by the presence of D. hansenii, with significant increases in growth observed for the Ancestor, 55_5, and 60_5 (Fig. 5B).
Growth as single isolates does not fully capture mutant fitness in competitive environments; evolved strains need to compete with ancestor strains during evolution to become dominant lineages at the end of the experiment. Additionally, very small fitness differences that could accumulate to have major impacts over time may not be apparent in the short time scales used in the previous experiments. Therefore, we next conducted fitness experiments where we grew each evolved strain in a 50:50 ratio with the ancestor BC10 strain. Because the evolved strains had unique colony morphologies, we were able to directly compete the strains and distinguish them on output plates. The ancestor:mutant pairing was grown either alone (without any other neighboring microbes) or in the presence of the biotic treatments used in the evolution experiment (+Debaryomyces, +Brevibacterium, and +Penicillium). We transferred the competition experiments five times after 7 days of growth to mimic the passaging that occurred in the original evolution experiment. We predicted that evolved strains would have a higher fitness relative to ancestor strains in the +Debaryomyces given how they dominated those populations in the evolution experiment. We only did these experiments with the three strains used in the RNA-seq analysis (51_3, 55_5, and 60_1) because they spanned the range of phenotypes and mutations observed in the evolution experiment.
As predicted, strains 51_3, 55_5, and 60_1 all had a significantly higher relative fitness in the +Debaryomyces treatment after multiple rounds of passaging (ANOVA 51_3 F3,29 = 7.9, p < 0.001; 55_5 F3,29 = 8.5 p < 0.001; 60_1 F3,29 = 10.2 p < 0.001; Fig. 5C). In the Alone treatment with no neighbors, all evolved strains had slight negative fitness compared to the ancestor, which corroborates the lack of these mutants in the Alone evolution populations. We repeated this experiment again with the Alone and +Debaryomyces treatment and observed the same pattern of increased fitness of the evolved strains in the +Debaryomyces treatment and lower fitness in the Alone treatment (Fig. S4).
Some evolved strains had strong fitness defects in other biotic environments, especially 60_1 in the +Brevibacterium and +Penicillium environments (Fig. 5C). This suggests that the altered SigB, Agr, and WalRK regulation in these evolved strains impairs their growth relative to the ancestor strain and explains why they did not evolve in the Alone, +Brevibacterium, or +Penicillium treatments. We do not know the exact impact of the mutations on WalRK functions in 60_1, but this regulator is known to play key roles in cell wall metabolism [89], so altered functions of WalRK may make evolved strains unable to grow well in the presence of +Brevibacterium or +Penicillium. The severe fitness trade-offs observed for the evolved strains in different biotic environments is an example of antagonistic pleiotropy [91], where beneficial mutations in the presence of the yeast D. hansenii can have strong negative fitness effects in other biotic environments.
Discussion
One major theme from this work is that interspecific microbe-microbe interactions can shape the evolutionary trajectories of bacteria. While in the Alone, +Brevibacterium, and +Penicillium treatments we did not see considerable phenotypic evolution or consistent mutations in populations, the yeast D. hansenii promoted genomic and phenotypic diversification of S. xylosus. In a short period of time, many different mutant colony types emerged that were distinct from the ancestral colony morphology, with unique pigmentation and colony structure. This pattern emerged across replicate populations and involved distinct mutations in the same global regulator loci, providing strong evidence of parallel adaptation in this biotic environment.
There are several key nuances and limitations of our work that should be considered when contextualizing it within the cheese rind microbiome and other microbial systems. First, we did not attempt to prevent or control the evolution of the neighbors in our experiment, so the selection pressures experienced by S. xylosus throughout time may have shifted if ecologically relevant traits of the neighbors evolved. This was a conscious decision as we were trying to mimic population dynamics in cheese aging facilities where both target microbes and neighboring microbes might evolve. We did not observe any obvious growth or colony morphology changes in the neighbors and previous research in our lab has demonstrated that one neighbor (Penicillium) does not evolve new phenotypes if bacteria are present [47]. Second, our studies were confined to a controlled laboratory environment, so we do not know how often mutants we have observed here would evolve in cheese production and aging environments. However, we believe there is considerable potential for similar evolutionary processes to unfold in cheese aging facilities. In these environments, S. xylosus populations experience varying biotic environments due to patchiness of cheese rind communities across the surfaces of cheese and variation in community composition across wheels of cheese [50, 54, 92]. Third, we only considered how one neighboring species at a time impacts the evolution of S. xylosus and did not consider how combinations of species drive phenotypic and genomic evolution of this bacterium. For example, combining D. hansenii with a neighbor that decreased the fitness of evolved strains might inhibit the global regulator evolution observed when only D. hansenii was present.
A major question that remains unanswered is why does D. hansenii promote the diversification of S. xylosus on cheese? We did not identify specific mechanisms underlying Staphylococcus-Debaryomyces interactions, but several lines of evidence from our data and the cheese rind system provide some helpful clues. One ecological explanation is that the degree of niche overlap between these two species may promote diversification of S. xylosus. A common theme in the study of adaptation to novel environments is that interspecific interactions can promote diversification when there is strong niche overlap [32]. Staphylococcus species and yeasts such as D. hansenii share similar resource and temporal niches in cheese rind microbiomes. At the early stages of cheese ripening, the pH of cheese is low, salt concentrations are high, and resources such as free amino acids are locked up in the casein of cheese. Previous work in our lab and others has shown that both S. xylosus and D. hansenii grow early in cheese succession [50, 54, 93]. These microbes are both tolerant of high salt concentrations and have proteolytic abilities that help them access free amino acids in cheese. In contrast, the other two biotic neighbors (Brevibacterium and Penicillium) grow later in cheese rind succession and occupy different niches compared to Staphylococcus. During our evolution experiment, competition for resources between S. xylosus and D. hansenii may have promoted the adaptation of new lineages of S. xylosus with different metabolic and ecological traits.
Another potential explanation for the abundance of evolved strains with altered global regulation is that these strains are only able to persist in the presence of D. hansenii due to facilitative interactions between the yeast and evolved strains. Both the ancestor and evolved S. xylosus strains had slightly higher growth in the presence of D. hansenii, suggesting that the yeast does provide some benefits for the bacterium. RNA-seq demonstrated strong metabolic downregulation of many pathways in evolved strains, including biosynthesis of histidine and other amino acids. D. hansenii may have promoted the rapid and reproducible rise of these strains through unknown mechanisms of supplementing their altered metabolism. For example, D. hansenii may break down some cheese curd components and release nutrients that are needed to support the altered metabolism of the S. xylosus with mutations in global regulators. Future work using metabolomics will help pinpoint the chemical mediators of sigB, agr, and walRK evolution in the D. hansenii environment.
Our evolution experiment demonstrates how interspecific interactions might facilitate the generation of strain diversity within microbiomes. These data also contribute to an emerging body of work demonstrating that microbes have very different evolutionary outcomes when biotic complexity is incorporated into experimental evolution [30, 33, 35, 36, 94, 95]. Because most medically and industrially important microbes often live with other microbial species that may impose strong biotic selection [23, 25, 96, 97], the management and engineering of microbiomes may need to incorporate how microbe-microbe interactions can shape evolution. As additional studies use a broader range of target microbes and more diverse biotic environments, we can begin to create a predictive framework for when and how interspecific interactions can impact microbial evolution.
Methods
Experimental evolution
All yeast and bacterial cultures were maintained as frozen stocks in 15% glycerol in a −80 °C freezer. The main target for experimental evolution, S. xylosus strain BC10, was originally isolated from the surface of a natural rind cheese made in Vermont, USA and has been previously described [50]. Three different microbial strains were used as neighbor treatments: Brevibacterium aurantiacum strain JB5 (as previously described in [53]), Penicillium solitum strain #12 (as previously described in [47]), and the yeast D. hansenii strain 135B (as previously described in [50]). All of these strains were isolated from one natural rind cheese made in the same aging facility and commonly co-occur in natural rind cheeses made in different regions of the world [50, 54, 98–100].
To initiate the evolution experiments, cultures were inoculated into 1.5 mL microcentrifuge tubes filled with a 150 µL aliquot of cheese curd agar (CCA) containing 3% salt (w/w) [101]. Inoculum for each culture came from frozen glycerol stocks where a known CFU/µL concentration of the microbial culture had been previously determined [101]. In the alone and +neighbor treatments, 50 CFUs of S. xylosus were inoculated into each replicate tube by adding 10 µL of inoculum suspended in 1X phosphate buffered saline (PBS) to the CCA surface. In each of the neighbor treatments, 50 CFUs of a neighbor were added to the S. xylosus inoculum. Each treatment was replicated 10 times. One replicate from the +Debaryomyces treatment was lost during the experiment due to contamination. Tubes were covered with sterile AeraSeal films and then placed in microcentrifuge tube racks containing a snack Ziplock bag with a paper towel moistened with 5 mL of sterile PBS to maintain high humidity. A lid was placed on the tube rack to contain the moisture and tubes were incubated in the dark for 7 days at 24 °C.
After 7 days of growth, each population was transferred by adding 300 µL of 15% glycerol to the existing population, homogenizing the sample with a micropestle, and pipetting 10 µL of the homogenate using a wide orifice pipette tip to a fresh 1.5 mL microcentrifuge tube containing 150 µL of CCA. Each homogenized sample was then frozen at −80 °C to serve as fossil stocks for isolating colonies. This was repeated over a period of 15 weeks.
To determine the frequency of mutant phenotypes within the population at each transfer, 20 µL of homogenate was serially diluted and plated onto plate count agar with milk and salt (PCAMS, [101]) plates with 50 mg/L of cycloheximide to inhibit the growth of fungi. Plates were incubated at 24 °C with natural room light to allow for the development of colony pigmentation. Plates were scored for mutant frequency by counting colonies and noting the presence of colonies with altered phenotypes. Wild-type S. xylosus BC10 is intensely orange/yellow in color, wrinkly, and shiny. Colonies deemed phenotypic mutants had abnormal pigmentation (white, light yellow), colony texture (less wrinkly to completely smooth), and colony surface appearance (matte or partially shiny). Abundances of neighbors were also determined by plating onto selective media for fungi (PCAMS + 50 mg/L of chloramphenicol to inhibit bacteria) or by counting colonies of Brevibacterium that grew alongside the S. xylosus colonies.
Whole-genome sequencing and analysis
To identify genomic changes in evolved isolates, whole-genome sequencing was conducted on seven randomly selected colonies from the final time point from three populations across the four treatments (84 genomes in total). DNA was extracted from streaks of pure cultures growing on PCAMS plates using a MoBio PowerSoil DNA Isolation kit. Sequencing libraries for each genome were constructed using the NEBNext Ultra DNA Library Prep Kit for Illumina (New England Biolabs) using the manufacturer’s protocol with DNA input concentrations of 5–15 ng/µL, a 20-min fragmentase incubation for DNA fragmentation, and 5 rounds of PCR enrichment. Equimolar concentrations of each library representing isolate genomes were pooled and sequenced in one HiSeq 2500 run at the Harvard FAS Center for Systems Biology Core Facility. We aimed to have at least ~20–30X coverage of each genome for variant detection. Mean coverage across all libraries was 39.4.
Reads were mapped to a reference genome of S. xylosus BC10 (NCBI WGS Project LNPU00000000) using end-to-end alignment in Bowtie2 [102]. We also mapped reads of the ancestral isolate to the reference genome to identify any errors in the original genome assembly that could cause erroneous variant calling. Variant calling was conducted using FreeBayes in pooled continuous mode with the following settings: ploidy = 1, minimum alternate count = 10 (a coverage of at least 10 reads needed to support variant calls), minimum alternate fraction = 0.9 (at least 90% of mapped reads needed to contain the variant in order for it to be called), and minimum probability = 0.1. Any SNPs or other variants identified from mapping raw reads of the ancestor were excluded from variant calls of the evolved isolates. When read mapping indicated potential deletion or other structural variants, we re-assembled the genomes of those isolates using SPAdes and annotated the genome using RASTk.
Phenotypic assays
To measure staphyloxanthin production, bacteria were streaked across a PCAMS plate and were left to grow for one week to develop pigment. Each plate was laid out singly in a plastic bag to ensure each plate received even exposure to light. After one week of growth at 24 °C, 20 mg of cells were removed from the plate with a wooden dowel. Cells were resuspended in 800 µL of methanol and thoroughly mixed via vortexing. The suspensions were left overnight at 55 °C with shaking at 525 rpm on a benchtop shaking incubator to extract the pigment from cells. After extractions, the suspensions were centrifuged at 5000 g for 10 min. The pellets appeared colorless (white) with the pigment in the supernatant. The absorbance of the supernatant was read at 465 nm using methanol as a blank. The absorbance readings were normalized to cell weight.
To quantify biofilm production, bacteria were grown in 5 mL brain heart infusion (BHI) broth overnight at 24 °C. The cells were diluted 100-fold in BHI, and 200 µL of inoculum was added to eight wells of a flat-bottom 96-well plate (Falcon). After 16 h of growth at 24 °C, the OD600 was recorded to account for differential growth of each strain. Wells were washed three times by manually flipping plates to dispose of liquid and adding 200 µL of 1X PBS to the wells to remove dislodged cells. The cells were heat fixed to the plate by incubating for 1 h at 60 °C. Fifty microliters of a solution of 0.06% crystal violet stain was added to each sample. These were incubated for 5 min at room temperature and rinsed three times with 200 µL of 1× PBS. Biofilm formation was estimated by solubilization of crystal violet by adding 200 µL of 30% acetic acid and determining the OD600. For each of three independent experiments, 8–12 replicate wells in three replicate plates were used.
The effect of mutations on bacterial spreading was assessed on soft BHI agar (0.4% w/v). After autoclaving, the medium was cooled down to 50 °C and 20 mL was poured into Petri dishes. The plates were kept at room temperature for 3 h prior to spreading assay. Bacterial strains were grown in 5 mL BHI broth overnight at room temperature. The cells were adjusted to OD600 of 1, and 2 µL of the cell suspension was spotted in the center of a 0.4% and 1.5% BHI agar plate. The plates were incubated for 5 days at 24 °C. Spreading diameter was calculated by measuring the largest diameter of the colony for each of five replicates.
To measure oxidative stress tolerance, inocula of BC10, 51_3, 51_7, 55_5, 60_1, 60_5 grown for 16 h in BHI broth were standardized to OD600 of 0.5 and incubated in 200 µL of different concentrations of H2O2 (0, 50, 100, 150, 200 mM) in the dark at 24 °C. After 45 min, the reaction was stopped with the addition of 2 U/mL catalase and incubation for 20 min. Cells were serially diluted and plated on PCAMS to quantify CFUs. Values are expressed as a percentage of the CFU in the control lacking H2O2 and are the averages of eight replicates in two independent experiments.
RNA sequencing
To characterize the transcriptomes of the ancestor and three evolved strains with mutant colony phenotypes (51_3, 55_5, and 60_1), we constructed and analyzed RNA-seq libraries. Lawns of each strain were grown on 100 mm Petri dishes containing 10% CCA media with 3% salt. Bacteria were inoculated at 50,000 CFUs and plates were incubated at 24 °C for 3 days. Bacterial cells were harvested from the surface of the CCA using a sterile razor blade and were then immediately placed into 3 mL of RNAProtect Bacteria Reagent (Qiagen) and frozen at −80 °C until RNA extraction. Total RNA was extracted as previously detailed using a 125:24:1 (v/v/v) phenol/chloroform/isoamyl alcohol extraction [55, 101]. Bacterial rRNA was depleted using a NEBNext Bacteria rRNA Depletion Kit, and depleted RNA was used to construct libraries using the NEBNext Ultra RNA Library Prep Kit for Illumina. Libraries were sequenced using a NextSeq 550 with single-end 75 base-pair reads. Reads were mapped to the S. xylosus BC10 draft genome (GenBank Assembly Accession: GCA_001747745.1) using Bowtie2, and differentially expressed genes were identified using DESeq2 with a log2 ratio of 1/−1 and FDR corrected p value of <0.05. To identify pathways that were enriched in the differentially expressed genes, we used the Gene-List Enrichment tool of KOBAS [103] with our BC10 draft genome as the background genes to test for enrichment.
Growth and competition experiments
To determine the ability of the ancestor and evolved strains to grow on cheese, each was inoculated at a concentration of 667 CFUs onto the surface of 150 µL of CCA in a 1.5 mL Eppendorf tube. Experimental units were incubated as described for the evolution experiment above. Growth of cells over time was determined at 1, 2, 3, 4 and 7 days by resuspending the cells into 300 µL of 30% glycerol. The cells and CCA were homogenized via pestling, serially diluted, and plated onto PCAMS to determine CFUs. To determine how the yeast D. hansenii strain 135B impacted the short-term growth of the ancestor and evolved S. xylosus strains, the same experimental approach was used with 667 CFUs of D. hansenii added to the +Debaryomyces treatment.
To determine the fitness of mutant strains, we competed three evolved strains (51_3, 55_5, and 60_1) against the ancestor strain in 50:50 initial ratios either alone (with no neighbor) or in the three biotic treatments (+Debaryomyces, +Brevibacterium, +Penicillium). Each strain (S. xylosus BC10 ancestor, S. xylosus BC10 evolved strain, and Debaryomyces/Brevibacterium/Penicillium if present) was inoculated at a concentration of 667 CFUs into 1.5 mL Eppendorf tubes containing 150 µL of CCA. After one week of incubation at 24 °C under aerobic conditions, cells were harvested by resuspending the wells into 300 µL of 30% glycerol and homogenizing via pestling. Ten microliters of the homogenized communities were transferred to a new tube and were incubated for another week to mimic the multiple rounds of growth that are experienced in the experimental evolution design and to amplify small fitness differences that might be observed in only one round of growth. This was repeated for a total of five transfers. We acknowledge that some mutations may occur in the ancestor strains during the five transfers and that this could increase the observance of mutant colony phenotypes. However, this should happen relatively rarely based on the frequency of mutant colonies we observed in our original evolution experiment (Fig. 1D). To determine the abundance of ancestor and mutant colonies at each transfer, 20 µL of the homogenized sample was serially diluted and plated onto PCAMS. Mutant colonies could be distinguished from ancestor colonies due to their reduced pigmentation and altered colony morphology. This experiment continued for a total of five weeks. Relative fitness of the mutant strains was calculated as log10((CFUs of mutant strain +1) ÷ (CFUs of ancestor strain +1)). A second competition experiment was conducted to confirm the observed fitness patterns observed in the first experiment. It was executed using the same setup, transfer, and plating methods as above, but with only the Alone and +Debaryomyces treatments. This second experiment was conducted for only three weeks.
Statistics
To assess whether there were significant differences in the frequencies of phenotypic mutants or mutations across the experimental evolution populations (Figs. 1 and 2), we used ANOVA with Tukey’s post-hoc tests. We also used ANOVAs with Dunnett’s multiple comparison tests or Tukey’s post-hoc tests to assess differences in the phenotypic assays (Fig. 3). To identify differentially expressed genes in the RNA-seq data (Fig. 4), DESeq2 was used with a significance cutoff of a log2 ratio of 1/−1 and a FDR corrected p value of <0.05. For RNA-seq pathway enrichment, we used Fisher’s exact tests with Benjamini and Hochberg false discovery rate correction in the Gene-List Enrichment Tool of KOBAS. To determine if there were differences in growth of the strains over time (Fig. 5A), a repeated-measures ANOVA was used. To assess whether D. hansenii impacted the growth of S. xylosus BC10 ancestor and mutant strains (Fig. 5B), an ANOVA on the day 7 CFU data was used. To determine how biotic environments affected the fitness of ancestor and mutant BC10 strains (Fig. 5C), ANOVAs were used for each mutant strain. Log transformations were applied when appropriate. All statistical analyses were conducted in PRISM 9 or R.
Supplementary information
Acknowledgements
This work was funded by a grant from the United States National Science Foundation (CAREER IOS/BIO 1942063) to BEW. The authors are grateful to members of the Wolfe lab, including Robert May, Nicolas Louw, Kasturi Lele, Dillon Arrigan, Chris Tomo, and Mak Boylan, for providing extensive feedback on previous versions of this manuscript.
Author contributions
BN, CC, and BEW designed the study. BN conducted the evolution experiment. CC and BEW conducted the whole-genome resequencing. CC conducted the fitness experiments. BEW conducted and analyzed the RNA-seq experiment. CC, NK, MD, and MP conducted the phenotypic assays. CC, BN, and BEW conducted statistical analyses, created figures, and wrote the first drafts of the manuscript. All authors read, revised, and approved the final manuscript.
Data availability
All raw fastq read files of re-sequenced evolved isolates of S. xylosus BC10 have been deposited in NCBI in BioProject PRJNA856679. Raw RNA-seq read files from this study have been deposited in NCBI in BioProject PRJNA856810.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Casey Cosetta, Brittany Niccum.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-023-01462-5.
References
- 1.Monk JM, Koza A, Campodonico MA, Machado D, Seoane JM, Palsson BO, et al. Multi-omics quantification of species variation of Escherichia coli links molecular features with strain phenotypes. Cell Syst. 2016;3:238–51. doi: 10.1016/j.cels.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCloskey D, Xu J, Schrübbers L, Christensen HB, Herrgård MJ. RapidRIP quantifies the intracellular metabolome of 7 industrial strains of E. coli. Metab Eng. 2018;47:383–92. doi: 10.1016/j.ymben.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 3.Garay-Arroyo A, Covarrubias AA, Clark I, Niño I, Gosset G, Martinez A. Response to different environmental stress conditions of industrial and laboratory Saccharomyces cerevisiae strains. Appl Microbiol Biotechnol. 2004;63:734–41. doi: 10.1007/s00253-003-1414-4. [DOI] [PubMed] [Google Scholar]
- 4.Gallone B, Steensels J, Prahl T, Soriaga L, Saels V, Herrera-Malaver B, et al. Domestication and divergence of Saccharomyces cerevisiae beer yeasts. Cell. 2016;166:1397–410. doi: 10.1016/j.cell.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Des Roches S, Post DM, Turley NE, Bailey JK, Hendry AP, Kinnison MT, et al. The ecological importance of intraspecific variation. Nat Ecol Evol. 2018;2:57–64. doi: 10.1038/s41559-017-0402-5. [DOI] [PubMed] [Google Scholar]
- 6.Leventhal GE, Boix C, Kuechler U, Enke TN, Sliwerska E, Holliger C, et al. Strain-level diversity drives alternative community types in millimetre-scale granular biofilms. Nat Microbiol. 2018;3:1295–303. doi: 10.1038/s41564-018-0242-3. [DOI] [PubMed] [Google Scholar]
- 7.Ellegaard KM, Engel P. Beyond 16S rRNA community profiling: intra-species diversity in the gut microbiota. Front Microbiol. 2016;7:1475. doi: 10.3389/fmicb.2016.01475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warringer J, Zörgö E, Cubillos FA, Zia A, Gjuvsland A, Simpson JT, et al. Trait variation in yeast is defined by population history. PLoS Genet. 2011;7:e1002111. doi: 10.1371/journal.pgen.1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steensels J, Snoek T, Meersman E, Picca Nicolino M, Voordeckers K, Verstrepen KJ. Improving industrial yeast strains: exploiting natural and artificial diversity. FEMS Microbiol Rev. 2014;38:947–95. doi: 10.1111/1574-6976.12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Rossum T, Ferretti P, Maistrenko OM, Bork P. Diversity within species: interpreting strains in microbiomes. Nat Rev Microbiol. 2020;18:491–506. doi: 10.1038/s41579-020-0368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lenski RE. Experimental evolution and the dynamics of adaptation and genome evolution in microbial populations. ISME J. 2017;11:2181–94. doi: 10.1038/ismej.2017.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elena SF, Lenski RE. Microbial genetics: evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat Rev Genet. 2003;4:457. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- 13.Cooper VS. Experimental evolution as a high-throughput screen for genetic adaptations. mSphere. 2018;3:e00121–18. doi: 10.1128/mSphere.00121-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long A, Liti G, Luptak A, Tenaillon O. Elucidating the molecular architecture of adaptation via evolve and resequence experiments. Nat Rev Genet. 2015;16:567–82. doi: 10.1038/nrg3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kassen R. Experimental evolution of innovation and novelty. Trends Ecol Evol. 2019;34:712–22. [DOI] [PMC free article] [PubMed]
- 16.Kawecki TJ, Lenski RE, Ebert D, Hollis B, Olivieri I, Whitlock MC. Experimental evolution. Trends Ecol Evol. 2012;27:547–60. doi: 10.1016/j.tree.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 17.Kassen R. Experimental evolution and the nature of biodiversity. Greenwood Village, CO: Roberts & Company; 2014.
- 18.Bailey SF, Bataillon T. Can the experimental evolution programme help us elucidate the genetic basis of adaptation in nature? Mol Ecol. 2016;25:203–18. doi: 10.1111/mec.13378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jessup CM, Kassen R, Forde SE, Kerr B, Buckling A, Rainey PB, et al. Big questions, small worlds: microbial model systems in ecology. Trends Ecol Evol. 2004;19:189–97. doi: 10.1016/j.tree.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 20.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Little AEF, Robinson CJ, Peterson SB, Raffa KF, Handelsman J. Rules of engagement: interspecies interactions that regulate microbial communities. Annu Rev Microbiol. 2008;62:375–401. doi: 10.1146/annurev.micro.030608.101423. [DOI] [PubMed] [Google Scholar]
- 22.Frey-Klett P, Burlinson P, Deveau A, Barret M, Tarkka M, Sarniguet A. Bacterial-fungal interactions: hyphens between agricultural, clinical, environmental, and food microbiologists. Microbiol Mol Biol Rev. 2011;75:583–609. doi: 10.1128/MMBR.00020-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deveau A, Bonito G, Uehling J, Paoletti M, Becker M, Bindschedler S, et al. Bacterial–fungal interactions: ecology, mechanisms and challenges. FEMS Microbiol Rev. 2018;42:335–52. doi: 10.1093/femsre/fuy008. [DOI] [PubMed] [Google Scholar]
- 24.Pacheco AR, Segrè D. A multidimensional perspective on microbial interactions. FEMS Microbiol Lett. 2019;366:fnz125. doi: 10.1093/femsle/fnz125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cosetta CM, Wolfe BE. Causes and consequences of biotic interactions within microbiomes. Curr Opin Microbiol. 2019;50:35–41. doi: 10.1016/j.mib.2019.09.004. [DOI] [PubMed] [Google Scholar]
- 26.Johansson J. Evolutionary responses to environmental changes: how does competition affect adaptation? Evolution. 2008;62:421–35. doi: 10.1111/j.1558-5646.2007.00301.x. [DOI] [PubMed] [Google Scholar]
- 27.Stuart YE, Campbell TS, Hohenlohe PA, Reynolds RG, Revell LJ, Losos JB. Rapid evolution of a native species following invasion by a congener. Science. 2014;346:463–6. doi: 10.1126/science.1257008. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Q-G, Ellis RJ, Godfray HCJ. The effect of a competitor on a model adaptive radiation. Evolution. 2012;66:1985–90. doi: 10.1111/j.1558-5646.2011.01559.x. [DOI] [PubMed] [Google Scholar]
- 29.Harcombe W. Novel cooperation experimentally evolved between species. Evolution. 2010;64:2166–72. doi: 10.1111/j.1558-5646.2010.00959.x. [DOI] [PubMed] [Google Scholar]
- 30.Lawrence D, Fiegna F, Behrends V, Bundy JG, Phillimore AB, Bell T, et al. Species interactions alter evolutionary responses to a novel environment. PLoS Biol. 2012;10:e1001330. doi: 10.1371/journal.pbio.1001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lahti DC, Johnson NA, Ajie BC, Otto SP, Hendry AP, Blumstein DT, et al. Relaxed selection in the wild. Trends Ecol Evol. 2009;24:487–96. doi: 10.1016/j.tree.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 32.Bailey SF, Dettman JR, Rainey PB, Kassen R. Competition both drives and impedes diversification in a model adaptive radiation. Proc Biol Sci. 2013;280:20131253. doi: 10.1098/rspb.2013.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hall JPJ, Harrison E, Brockhurst MA. Competitive species interactions constrain abiotic adaptation in a bacterial soil community. Evol Lett. 2018;2:580–9. doi: 10.1002/evl3.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jousset A, Eisenhauer N, Merker M, Mouquet N, Scheu S. High functional diversity stimulates diversification in experimental microbial communities. Sci Adv. 2016;2:e1600124. doi: 10.1126/sciadv.1600124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scheuerl T, Hopkins M, Nowell RW, Rivett DW, Barraclough TG, Bell T. Bacterial adaptation is constrained in complex communities. Nat Commun. 2020;11:754. doi: 10.1038/s41467-020-14570-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Golzar FS, Ferguson GC, Hendrickson HL. Protozoan predation drives adaptive divergence in Pseudomonas fluorescens SBW25; ecology meets experimental evolution. bioRxiv. 2021.10.1101/2021.07.12.452127.
- 37.Chu X-L, Zhang Q-G, Buckling A, Castledine M. Interspecific niche competition increases morphological diversity in multi-species microbial communities. Front Microbiol. 2021;12:2103. doi: 10.3389/fmicb.2021.699190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolfe BE, Dutton RJ. Fermented foods as experimentally tractable microbial ecosystems. Cell. 2015;161:49–55. doi: 10.1016/j.cell.2015.02.034. [DOI] [PubMed] [Google Scholar]
- 39.Di Cagno R, Coda R, De Angelis M, Gobbetti M. Exploitation of vegetables and fruits through lactic acid fermentation. Food Microbiol. 2013;33:1–10. doi: 10.1016/j.fm.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 40.De Vuyst L, Van Kerrebroeck S, Harth H, Huys G, Daniel H-M, Weckx S. Microbial ecology of sourdough fermentations: diverse or uniform? Food Microbiol. 2014;37:11–29. doi: 10.1016/j.fm.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 41.Montel M-C, Buchin S, Mallet A, Delbes-Paus C, Vuitton DA, Desmasures N, et al. Traditional cheeses: rich and diverse microbiota with associated benefits. Int J Food Microbiol. 2014;177:136–54. doi: 10.1016/j.ijfoodmicro.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 42.Macori G, Cotter PD. Novel insights into the microbiology of fermented dairy foods. Curr Opin Biotechnol. 2018;49:172–8. doi: 10.1016/j.copbio.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 43.Steensels J, Gallone B, Voordeckers K, Verstrepen KJ. Domestication of industrial microbes. Curr Biol. 2019;29:R381–R393.. doi: 10.1016/j.cub.2019.04.025. [DOI] [PubMed] [Google Scholar]
- 44.Gibbons JG, Rinker DC. The genomics of microbial domestication in the fermented food environment. Curr Opin Genet Dev. 2015;35:1–8. doi: 10.1016/j.gde.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gibbons JG, Salichos L, Slot JC, Rinker DC, McGary KL, King JG, et al. The evolutionary imprint of domestication on genome variation and function of the filamentous fungus Aspergillus oryzae. Curr Biol. 2012;22:1403–9. doi: 10.1016/j.cub.2012.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ropars J, Rodríguez de la Vega RC, López-Villavicencio M, Gouzy J, Sallet E, Dumas É, et al. Adaptive horizontal gene transfers between multiple cheese-associated fungi. Curr Biol. 2015;25:2562–9. doi: 10.1016/j.cub.2015.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bodinaku I, Shaffer J, Connors AB, Steenwyk JL, Kastman E, Rokas A, et al. Rapid phenotypic and metabolomic domestication of wild Penicillium molds on cheese. mBio. 2019;10:e02445–19. doi: 10.1128/mBio.02445-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heo S, Lee J-H, Jeong D-W. Food-derived coagulase-negative Staphylococcus as starter cultures for fermented foods. Food Sci Biotechnol. 2020;29:1023–35. doi: 10.1007/s10068-020-00789-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nagase N, Sasaki A, Yamashita K, Shimizu A, Wakita Y, Kitai S, et al. Isolation and species distribution of staphylococci from animal and human skin. J Vet Med Sci. 2002;64:245–50. doi: 10.1292/jvms.64.245. [DOI] [PubMed] [Google Scholar]
- 50.Kastman EK, Kamelamela N, Norville JW, Cosetta CM, Dutton RJ, Wolfe BE. Biotic interactions shape the ecological distributions of Staphylococcus Species. mBio. 2016;7:e01157–16.. doi: 10.1128/mBio.01157-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Savage VJ, Chopra I, O’Neill AJ. Population diversification in Staphylococcus aureus biofilms may promote dissemination and persistence. PLoS ONE. 2013;8:e62513. doi: 10.1371/journal.pone.0062513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koch G, Yepes A, Förstner KU, Wermser C, Stengel ST, Modamio J, et al. Evolution of resistance to a last-resort antibiotic in Staphylococcus aureus via bacterial competition. Cell. 2014;158:1060–71. doi: 10.1016/j.cell.2014.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Niccum BA, Kastman EK, Kfoury N, Robbat A, Jr, Wolfe BE. Strain-level diversity impacts cheese rind microbiome assembly and function. mSystems. 2020;5:e00149–20.. doi: 10.1128/mSystems.00149-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolfe BE, Button JE, Santarelli M, Dutton RJ. Cheese rind communities provide tractable systems for in situ and in vitro studies of microbial diversity. Cell. 2014;158:422–33. doi: 10.1016/j.cell.2014.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cosetta CM, Kfoury N, Robbat A, Wolfe BE. Fungal volatiles mediate cheese rind microbiome assembly. Environ Microbiol. 2020;22:4745–60. doi: 10.1111/1462-2920.15223. [DOI] [PubMed] [Google Scholar]
- 56.Hu G, Wang Y, Liu X, Strube ML, Wang B, Kovács ÁT. Species and condition dependent mutational spectrum in experimentally evolved biofilms of Bacilli. bioRxiv. 2022. 10.1101/2022.12.07.519423.
- 57.Lanfear R, Kokko H, Eyre-Walker A. Population size and the rate of evolution. Trends Ecol Evol. 2014;29:33–41. doi: 10.1016/j.tree.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 58.Zhao X-F, Buckling A, Zhang Q-G, Hesse E. Specific adaptation to strong competitors can offset the negative effects of population size reductions. Proc Biol Sci. 2018;285:2018000. doi: 10.1098/rspb.2018.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schaik A. The role of σB in the stress response of Gram-positive bacteria–targets for food preservation and safety. Curr Opin Biotechnol. 2005;16:218–24. doi: 10.1016/j.copbio.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 60.Bischoff M, Entenza JM, Giachino P. Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus. J Bacteriol. 2001;183:5171–9. doi: 10.1128/JB.183.17.5171-5179.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guldimann C, Boor KJ, Wiedmann M, Guariglia-Oropeza V. Resilience in the face of uncertainty: sigma factor B fine-tunes gene expression to support homeostasis in gram-positive bacteria. Appl Environ Microbiol. 2016;82:4456–69. doi: 10.1128/AEM.00714-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson DA, Monk AB, Cooper JE, Feil EJ, Enright MC. Evolutionary genetics of the accessory gene regulator (agr) locus in Staphylococcus aureus. J Bacteriol. 2005;187:8312–21. doi: 10.1128/JB.187.24.8312-8321.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Painter KL, Krishna A, Wigneshweraraj S, Edwards AM. What role does the quorum-sensing accessory gene regulator system play during Staphylococcus aureus bacteremia? Trends Microbiol. 2014;22:676–85. doi: 10.1016/j.tim.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 64.Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42:541–64. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 65.Martin PK, Li T, Sun D, Biek DP, Schmid MB. Role in cell permeability of an essential two-component system in Staphylococcus aureus. J Bacteriol. 1999;181:3666–73. doi: 10.1128/JB.181.12.3666-3673.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ji Q, Chen PJ, Qin G, Deng X, Hao Z, Wawrzak Z, et al. Structure and mechanism of the essential two-component signal-transduction system WalKR in Staphylococcus aureus. Nat Commun. 2016;7:11000. doi: 10.1038/ncomms11000. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Dufour P, Jarraud S, Vandenesch F, Greenland T, Novick RP, Bes M, et al. High genetic variability of the agr locus in Staphylococcus species. J Bacteriol. 2002;184:1180–6. doi: 10.1128/jb.184.4.1180-1186.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wuster A, Babu MM. Conservation and evolutionary dynamics of the agr cell-to-cell communication system across firmicutes. J Bacteriol. 2008;190:743–6. doi: 10.1128/JB.01135-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ferreira A, Gray M, Wiedmann M, Boor KJ. Comparative genomic analysis of the sigB operon in Listeria monocytogenes and in other Gram-positive bacteria. Curr Microbiol. 2004;48:39–46. doi: 10.1007/s00284-003-4020-x. [DOI] [PubMed] [Google Scholar]
- 70.Xue L, Chen YY, Yan Z, Lu W, Wan D, Zhu H. Staphyloxanthin: a potential target for antivirulence therapy. Infect Drug Resist. 2019;12:2151–60. doi: 10.2147/IDR.S193649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marbach H, Mayer K, Vogl C, Lee JYH, Monk IR, Sordelli DO, et al. Within-host evolution of bovine Staphylococcus aureus selects for a SigB-deficient pathotype characterized by reduced virulence but enhanced proteolytic activity and biofilm formation. Sci Rep. 2019;9:13479. doi: 10.1038/s41598-019-49981-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Clauditz A, Resch A, Wieland K-P, Peschel A, Götz F. Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect Immun. 2006;74:4950–3. doi: 10.1128/IAI.00204-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu GY, Essex A, Buchanan JT, Datta V, Hoffman HM, Bastian JF, et al. Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J Exp Med. 2005;202:209–15. doi: 10.1084/jem.20050846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Davey ME, O’toole GA. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64:847–67. doi: 10.1128/MMBR.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Flemming H-C, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016;14:563–75. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 76.Moons P, Michiels CW, Aertsen A. Bacterial interactions in biofilms. Crit Rev Microbiol. 2009;35:157–68. doi: 10.1080/10408410902809431. [DOI] [PubMed] [Google Scholar]
- 77.Bateman BT, Donegan NP, Jarry TM, Palma M, Cheung AL. Evaluation of a tetracycline-inducible promoter in Staphylococcus aureus in vitro and in vivo and its application in demonstrating the role of sigB in microcolony formation. Infect Immun. 2001;69:7851–7. doi: 10.1128/IAI.69.12.7851-7857.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lauderdale KJ, Boles BR, Cheung AL, Horswill AR. Interconnections between Sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect Immun. 2009;77:1623–35. doi: 10.1128/IAI.01036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vuong C, Saenz HL, Götz F, Otto M. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J Infect Dis. 2000;182:1688–93. doi: 10.1086/317606. [DOI] [PubMed] [Google Scholar]
- 80.Pollitt EJG, Crusz SA, Diggle SP. Staphylococcus aureus forms spreading dendrites that have characteristics of active motility. Sci Rep. 2015;5:17698. doi: 10.1038/srep17698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pollitt EJG, Diggle SP. Defining motility in the Staphylococci. Cell Mol Life Sci. 2017;74:2943–58. doi: 10.1007/s00018-017-2507-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kaito C, Sekimizu K. Colony spreading in Staphylococcus aureus. J Bacteriol. 2007;189:2553–7. doi: 10.1128/JB.01635-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dordet-Frisoni E, Gaillard-Martinie B, Talon R, Leroy S. Surface migration of Staphylococcus xylosus on low-agar media. Res Microbiol. 2008;159:263–9. doi: 10.1016/j.resmic.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 84.Tsompanidou E, Denham EL, Becher D, de Jong A, Buist G, van Oosten M, et al. Distinct roles of phenol-soluble modulins in spreading of Staphylococcus aureus on wet surfaces. Appl Environ Microbiol. 2013;79:886–95. doi: 10.1128/AEM.03157-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu T, Wang X-Y, Cui P, Zhang Y-M, Zhang W-H, Zhang Y. The agr quorum sensing system represses persister formation through regulation of phenol soluble modulins in Staphylococcus aureus. Front Microbiol. 2017;8:2189. doi: 10.3389/fmicb.2017.02189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Aubourg M, Pottier M, Léon A, Bernay B, Dhalluin A, Cacaci M, et al. Inactivation of the response regulator AgrA has a pleiotropic effect on biofilm formation, pathogenesis and stress response in Staphylococcus lugdunensis. Microbiol Spectr. 2022;10:e0159821. doi: 10.1128/spectrum.01598-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rao Y, Peng H, Shang W, Hu Z, Yang Y, Tan L, et al. A vancomycin resistance-associated WalK(S221P) mutation attenuates the virulence of vancomycin-intermediate Staphylococcus aureus. J Adv Res. 2021;40:167–78. doi: 10.1016/j.jare.2021.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Altman DR, Sullivan MJ, Chacko KI, Balasubramanian D, Pak TR, Sause WE, et al. Genome plasticity of agr-defective Staphylococcus aureus during clinical infection. Infect Immun. 2018;86:e00331–18.. doi: 10.1128/IAI.00331-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shoji M, Cui L, Iizuka R, Komoto A, Neoh H-M, Watanabe Y, et al. walK and clpP mutations confer reduced vancomycin susceptibility in Staphylococcus aureus. Antimicrob Agents Chemother. 2011;55:3870–81. doi: 10.1128/AAC.01563-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Leroy S, Even S, Micheau P, de La Foye A, Laroute V, Le Loir Y, et al. Transcriptomic analysis of Staphylococcus xylosus in solid dairy matrix reveals an aerobic lifestyle adapted to rind. Microorganisms. 2020;8:1807. doi: 10.3390/microorganisms8111807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen P, Zhang J. Antagonistic pleiotropy conceals molecular adaptations in changing environments. Nat Ecol Evol. 2020;4:461–9. doi: 10.1038/s41559-020-1107-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Quigley L, O’Sullivan O, Beresford TP, Paul Ross R, Fitzgerald GF, Cotter PD. High-throughput sequencing detects subpopulations of bacteria not previously associated with artisanal cheeses. Appl Environ Microbiol. 2012;78:5717–23. doi: 10.1128/AEM.00918-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mounier J, Monnet C, Vallaeys T, Arditi R, Sarthou A-S, Hélias A, et al. Microbial interactions within a cheese microbial community. Appl Environ Microbiol. 2008;74:172–81. doi: 10.1128/AEM.01338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Barber JN, Nicholson LC, Woods LC, Judd LM, Sezmis AL, Hawkey J, et al. Species interactions constrain adaptation and preserve ecological stability in an experimental microbial community. ISME J. 2022;16:1442–52. doi: 10.1038/s41396-022-01191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gómez P, Hall AR, Paterson S, Buckling A. Rapid decline of adaptation of Pseudomonas fluorescens to soil biotic environment. Biol Lett. 2022;18:20210593. doi: 10.1098/rsbl.2021.0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Peleg AY, Hogan DA, Mylonakis E. Medically important bacterial–fungal interactions. Nat Rev Microbiol. 2010;8:340–9. doi: 10.1038/nrmicro2313. [DOI] [PubMed] [Google Scholar]
- 97.Pierce EC, Dutton RJ. Putting microbial interactions back into community contexts. Curr Opin Microbiol. 2022;65:56–63. doi: 10.1016/j.mib.2021.10.008. [DOI] [PubMed] [Google Scholar]
- 98.Addis E, Fleet GH, Cox JM, Kolak D, Leung T. The growth, properties and interactions of yeasts and bacteria associated with the maturation of Camembert and blue-veined cheeses. Int J Food Microbiol. 2001;69:25–36. doi: 10.1016/S0168-1605(01)00569-4. [DOI] [PubMed] [Google Scholar]
- 99.Bertuzzi AS, Walsh AM, Sheehan JJ, Cotter PD, Crispie F, McSweeney PLH, et al. Omics-based insights into flavor development and microbial succession within surface-ripened cheese. mSystems. 2018;3:e00211–17.. doi: 10.1128/mSystems.00211-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Larpin-Laborde S, Imran M, Bonaïti C, Bora N, Gelsomino R, Goerges S, et al. Surface microbial consortia from Livarot, a French smear-ripened cheese. Can J Microbiol. 2011;57:651–60. doi: 10.1139/w11-050. [DOI] [PubMed] [Google Scholar]
- 101.Cosetta CM, Wolfe BE. Deconstructing and reconstructing cheese rind microbiomes for experiments in microbial ecology and evolution. Curr Protoc Microbiol. 2020;56:e95. doi: 10.1002/cpmc.95. [DOI] [PubMed] [Google Scholar]
- 102.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bu D, Luo H, Huo P, Wang Z, Zhang S, He Z, et al. KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021;49:W317–W325.. doi: 10.1093/nar/gkab447. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw fastq read files of re-sequenced evolved isolates of S. xylosus BC10 have been deposited in NCBI in BioProject PRJNA856679. Raw RNA-seq read files from this study have been deposited in NCBI in BioProject PRJNA856810.