Abstract
Ammonia oxidising archaea are among the most abundant living organisms on Earth and key microbial players in the global nitrogen cycle. They carry out oxidation of ammonia to nitrite, and their activity is relevant for both food security and climate change. Since their discovery nearly 20 years ago, major insights have been gained into their nitrogen and carbon metabolism, growth preferences and their mechanisms of adaptation to the environment, as well as their diversity, abundance and activity in the environment. Despite significant strides forward through the cultivation of novel organisms and omics-based approaches, there are still many knowledge gaps on their metabolism and the mechanisms which enable them to adapt to the environment. Ammonia oxidising microorganisms are typically considered metabolically streamlined and highly specialised. Here we review the physiology of ammonia oxidising archaea, with focus on aspects of metabolic versatility and regulation, and discuss these traits in the context of nitrifier ecology.
Subject terms: Environmental microbiology, Biogeochemistry, Metabolism, Archaeal physiology, Biogeochemistry
Introduction
Our understanding of the global nitrogen cycle has been revolutionised in the last few decades thanks to the discovery of novel processes and microbial players. This includes the detection of anaerobic ammonia oxidising (anammox) bacteria in natural and engineered ecosystems [1–3] and, more recently, the characterisation of complete ammonia oxidation (comammox) by comammox Nitrospira, which oxidise both ammonia and nitrite within the same cell to produce nitrate [4, 5]. Perhaps the most unanticipated breakthrough though, was the discovery of aerobic ammonia oxidation within the domain Archaea nearly two decades ago [6, 7]. Ammonia oxidising archaea (AOA) constitute a major fraction of microbial biomass on Earth [8] and play a vital role in the global biogeochemical cycling of nitrogen. Understanding the drivers of ammonia oxidation in the environment is of major importance since it could contribute towards food security and help mitigate the release of the greenhouse gas nitrous oxide (N2O) and nitrate (NO3-), which play roles in climate change and groundwater pollution, respectively [9]. Whilst significant progress has been made into cultivation and physiology of AOA, as well as environmental factors affecting their distribution and activity, many knowledge gaps still remain in our understanding of their metabolism, cellular regulation and adaptation to environmental changes. Ammonia oxidisers, including AOA, are autotrophic and able to fix their own carbon from inorganic CO2 and to generate reductant from ammonia [6, 10]. They are considered specialists in this specific metabolism. Here we review the physiology and metabolism of ammonia oxidising archaea, including whether AOA are as metabolically constrained as often thought. This is an ever-unfolding field of research as more ammonia oxidisers are isolated or highly enriched in culture, and novel insights into their metabolism revealed.
Ammonia oxidation kinetics: the roles of ammonia and oxygen
Ammonia (NH3), rather than ammonium (NH4+), is the substrate oxidised by the key enzyme ammonia monooxygenase (AMO) from the AOB Nitrosomonas europaea [11]. The current consensus is that ammonia is also the preferred substrate for the archaeal AMO [12]. Ammonia oxidisers are widely considered to be specialist microorganisms, for whom ammonia is usually the sole source of reducing power [4–6, 13]. It is therefore not surprising that the affinity for ammonia has been a subject of intensive research [12, 14, 15]. Ammonia concentration is a key eco-physiological factor influencing the abundance and distribution of nitrifiers. Early kinetics studies suggested that AOA and comammox bacteria are adapted to low ammonia concentrations, although it is now known that ammonia oxidisers exhibit a wide range of ammonia affinities. Within AOA, the Km(app) for ammonia ranges across four orders of magnitude from representatives of genus Nitrosocosmicus with the highest Km values (>12 µM), comparable to many characterised soil AOB [16–19], to the lowest values in Nitrosopumilales and ‘Ca. Nitrosotaleales’ genera with Km(app) in the low nM range (<2.8 nM) [12, 14]. In addition, there could be differences in the kinetics between AOA strains, but potentially also depending on growth conditions. For instance, Nitrosocosmicus-affiliated AOA outcompeted other ammonia oxidisers in ammonia-limited soil enrichments, suggesting that some members of this genus are adapted to low ammonia concentrations at least under certain conditions [19]. Although cultured representatives of genus Nitrosocosmicus have a high tolerance to ammonia compared to other AOA [16–19], there is no indication that high ammonia tolerance is necessarily linked to low ammonia affinity.
Ammonia and ammonium exist in pH-dependent equilibrium with a pKa of 9.25, meaning that ammonia decreases exponentially with decreasing pH. Laboratory cultures usually have ample supply of ammonia, but nitrifiers in the environment are frequently exposed to low and fluctuating substrate concentrations [6, 20]. Affinity for ammonia is thus likely to be an important factor for survival in the environment [6, 20]. A high affinity for ammonia would be advantageous to nitrifiers found in environments with low ammonia concentrations or low pH. The ammonia oxidation kinetics are also relevant to agriculture and engineered systems [21–25]. Nitrifiers, denitrifiers and anammox bacteria often integrate themselves into biofilms, where diffusion causes gradients in ammonia concentrations [11, 26, 27]. This means that microorganisms embedded deeper in the biofilm may experience lower ammonia concentrations. Subsequently, ammonia availability can influence competition and cooperation between these communities [26–29].
Whilst more research efforts have focused on the affinity for ammonia, the AMO has two substrates: ammonia and oxygen. Oxygen plays an important role in nitrification both as a substrate for the AMO enzyme and as a terminal electron acceptor for ammonia oxidisers [26]. Even though oxygen is a substrate for AMO, it is often overlooked, despite the potential value of characterising kinetic parameters and linking to niche, as has been done with ammonia. In marine environments, AOA of the family Nitrosopumilaceae, particularly those associated with the marine low ammonia ecotype, are successful under oxygen-limited conditions, with the highest abundances often detected in oxygen minimum zones (OMZs) and deep ocean sediments [30, 31]. Rates of ammonia oxidation in OMZs were measurable at <0.01 μM O2, likely driven exclusively by communities of AOA [31]. The enrichment of AOA from marine sediments was attributed to their ability to outcompete co-occurring bacteria at low O2 [32]. Recently, it was demonstrated Nitrosopumilus maritimus SCM1 was able to generate small amounts of oxygen under anoxic conditions, most likely by nitric oxide disproportionation [33]. Although the potential oxygen production by other AOA strains or in the environment is not yet fully explored, the ability to produce oxygen may provide an explanation for the presence of AOA in oxygen-limited habitats such as OMZs. In N. maritimus, some of the oxygen produced is used by the aerobic metabolism, which includes both the ammonia oxidation pathway and the respiratory chain, and some oxygen is released from the cells [33]. The metabolic pathway of oxygen production in Nitrosopumilus maritimus is not yet fully resolved, but involves nitric oxide and nitrous oxide [33, 34]. Many AOA encode chlorite dismutase-like enzymes [35], however, the function of these in AOA is unknown. Chlorite dismutase from the NOB “Candidatus Nitrospira defluvii” has been shown to reduce chlorite to chloride and O2 [36, 37]. Further studies are required to test the impact of oxygen producing metabolism in the environment. Affinity for oxygen and the ability to produce oxygen could influence niche specialisation and competition between different groups of ammonia oxidisers. For instance, comammox bacteria are well-adapted to the low oxygen concentrations in the oxic-anoxic interface of the biofilms and have been enriched from bioreactors under low dissolved oxygen conditions [3, 4, 38, 39]. AOA from wet tropical soils were resistant to prolonged intervals of anoxia [40] and reacted faster to anoxic/oxic fluctuations compared to AOB [41]. AOA seem to have a somewhat higher affinity for oxygen than AOB, with Nitrosopumilus representatives having the highest affinity (Table 1). However, studying the affinity for oxygen in further AOA, AOB and comammox strains and mixed communities, could provide insights into how oxygen availability shapes nitrifying communities.
Table 1.
Oxygen uptake kinetics by ammonia oxidisers.
| Strain | Km (μM) | Vmax (μmol O2 mg prot−1 h−1) | Reference |
|---|---|---|---|
| AOA | |||
| Nitrosarchaeum koreense MY1 | 10.4 (1.1) | 20 | [37] |
| Nitrosopumilus maritimus SCM1 | 3.9 (0.6) | 36 | [15] |
| Nitrosopumilus AR Enrichment | 2.0 (0.5) | 11 | [30] |
| Ca. Nitrososphaera sp. JG1 | 4.7 (0.2) | 35 | [38] |
| Ca. Nitrosocosmicus franklandus C13 | 11.5 (1.9) | N.D. | Lehtovirta-Morley, unpublished |
| AOB | |||
| Nitrosomonas europaea (ATCC 19178) | 1.3–14.9 | N.D. | [39] |
| Nitrosomonas europaea C-31 (ATCC 25978) | 186 | 129 | [30] |
| Nitrosomonas mobilis Ms1 | 21.7 (4.0) | N.D. | [40] |
| Activated sludge | 15.6 | N.D. | [41] |
N.D. not determined.
Despite intense research on ammonia oxidation kinetics over the past few years, there are still outstanding questions, particularly with regards to mechanisms which underpin the whole-cell kinetics. Structural basis of AMO conferring high or low affinity is not understood, nor is the role of ammonia transport. Furthermore, although AMO is known to be a membrane-bound enzyme [13, 42], the location and orientation of the AMO active site remain open research questions. In addition, the role of oxygen in AOA is understudied and could shed light on the niche adaptation of ammonia oxidisers in the future.
Nitrogen uptake and metabolism
Ammonium transport mechanisms
Much research focus has been on ammonia oxidation, but ammonium is required for both energy metabolism and assimilation in ammonia oxidisers. There must presumably exist reasonably sophisticated regulation for ammonium uptake, assimilation, and oxidation, particularly because ammonia is a relatively poor energy-yielding substrate. An overwhelming majority of ammonia is used for energy metabolism rather than anabolism, indicated by the near stoichiometry of 1:1 for ammonia:nitrite typically observed for ammonia oxidation [6, 13]. It is not known, whether ammonium transport is in any way coupled to oxidation in AOA, or if the transport is solely required for assimilation. Ammonia can cross biological membranes, but many organisms rely on the import of ammonium to meet nitrogen demands. Ammonium transport is mediated by a class of ubiquitous membrane proteins comprising ammonium transporters (Amts), methylammonium permeases (Meps) and rhesus (Rh) proteins [43, 44]. Many previously characterised Amts function as energy-dependent electrogenic ammonium transporters whilst the Rh-type proteins act as low-affinity ammonia channels [43, 45]. Mechanism of ammonium transport has been studied for decades and is still not fully understood. Recently, a two-lane mechanism was demonstrated for electrogenic ammonium transport by Escherichia coli AmtB and Nitrosomonas europaea [46]. Active transport against an ammonium gradient would presumably require energy, although energetics of ammonium transport remain enigmatic and intracellular ammonium concentrations in AOA are not known. Ammonia oxidation provides low energy yield, and active transport would be energetically costly [13]. On the other hand, active transport of ammonium could be advantageous to nitrifiers in acidic habitats and nitrogen-limited environments (Fig. 1), because, despite requiring energy, it could enable these nitrifiers to colonise otherwise inaccessible niches.
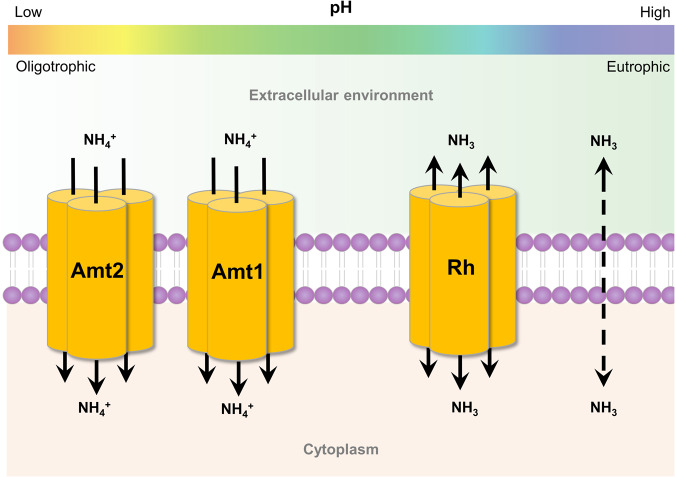
Fig. 1. Amt and Rh-type transporters encoded by ammonia oxidising microorganisms.
Amt transporters are energy-dependent and bind ammonium. Amt2 is a high-affinity Amt and can function at low pH and substrate concentration. Rh proteins facilitate the bidirectional flow of NH3 and therefore function at high pH and substrate concentration. NH3 can also cross the bilipid membrane by diffusion. NH4+ and NH3 exist in equilibrium based on pH. Membrane permeability between AOA and AOB may differ as their membrane compositions are different [60].
Most AOA encode at least two Amt-type transporters, whilst approximately half of the available AOB genomes contain Rh proteins [47]. Other AOB lack recognisable transporters and presumably rely on ammonia diffusion [47]. The two different clades of comammox bacteria appear to utilise distinct ammonium uptake mechanisms. Clade A encode Rh-type transporters with >70% amino acid similarity to those of the β-AOB, whereas clade B encode Amt-type transporters [48]. Anammox bacteria encode both types of transporters [49] (Supplementary Fig. S1). The Rh protein (Rh50) from Nitrosomonas europaea and Amt5 from ‘Ca. Kuenenia stuttgartiensis’ have both been isolated by recombinant expression and structurally characterised [50, 51]. Rh50 from Nitrosomonas europaea has been experimentally demonstrated to function as an ammonia transporter [46, 50, 52]. Electrophysiological analysis of ‘Ca. Kuenenia stuttgartiensis’ Amt5 revealed no transport function and instead this protein acts as an ammonium sensor [51]. Sequence and structural dissimilarities between the ammonium transporters in AOA and bacterial ammonia oxidisers indicate that they are functionally distinct [53]. Additionally, the transcriptional response of the archaeal Amt transporters to different ammonia concentration suggests they operate as high- and low-affinity transporters [54–56]. All sequenced representatives of the Nitrosocosmicus genus only encode one low-affinity Amt [16–18, 57]. Additionally, these strains lack the S-layers, which can function as ammonium concentrating mechanisms [58, 59]. The affinities of archaeal ammonium transporters have been inferred from transcriptomic studies, but not yet tested directly [56]. In addition, the role of the thaumarchaeal cell envelope in concentrating ions remains underexplored, as does the regulation of how ammonia is partitioned for assimilation and oxidation by AOA. Furthermore, archaeal and bacterial membranes have distinctly different compositions, which may affect membrane permeability and the rate at which ammonia can diffuse [60], although this question has not yet been fully explored.
Ammonia assimilation pathways
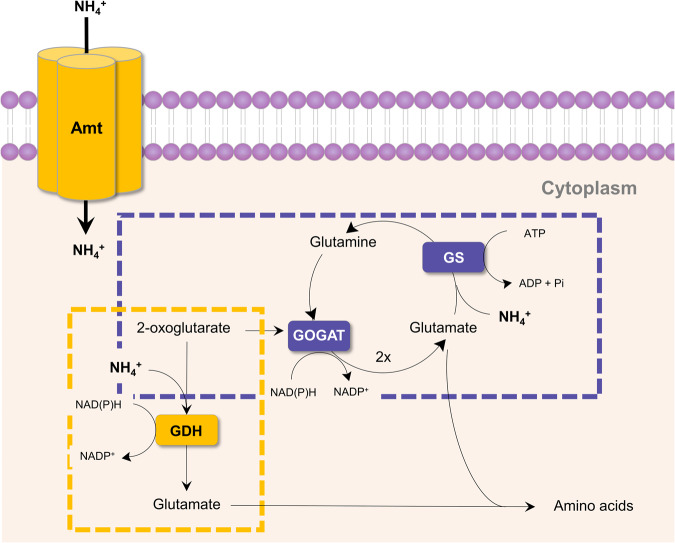
All characterised AOA contain glutamate dehydrogenase (GDH) and glutamine synthetase (GS) [57]. GDH is a key enzyme in ammonia assimilation and catalyses the reversible reductive amination of 2-oxoglutarate to glutamate [57, 61] (Fig. 2). In heterotrophs, this low-affinity pathway is favourable under energy and carbon limiting conditions since no ATP is consumed and less carbon is used per ammonia molecule assimilated [62]. The role of GDH in AOA is understudied. It seems likely that intracellular ammonium concentration and the regulation between ammonia uptake, assimilation and oxidation would be important for the function of this pathway in AOA, although this currently remains untested. GS catalyses an ATP-dependent conversion of ammonia and glutamate into glutamine and is considered to play an important role in central nitrogen metabolism. Nearly all AOA contain PII signal transduction protein homologues which belong to the glnK/B subfamily and regulate nitrogen metabolism [60, 63]. GlnK and GlnB interact directly with Amt transporters and glutamine synthetase (GS), respectively, to regulate ammonium influx into the cell by uridylylation of PII in response to low ammonia concentration, and also GS activity in response to extracellular and intracellular nitrogen concentrations [63]. The external ammonia concentration is likely important for ammonia assimilation, and the transcriptional activity of both GS and GDH in Nitrosocosmicus agrestis was upregulated in response to high ammonia concentrations [64].
Fig. 2. Ammonia assimilation pathways.
The glutamate dehydrogenase (GDH) pathway, represented in yellow, has a low-affinity for NH4+ and is found in all sequenced AOA. The high-affinity glutamine synthetase-glutamate synthase (GS-GOGAT) pathway is represented in purple and is only present in some representatives of the Nitrosocosmicus genera. One ATP is required per NH4+.
Two known AOA, Nitrosocosmicus arcticus and Nitrosocosmicus oleophilus, encode genes for the glutamine synthetase-glutamate synthase (GS-GOGAT) pathway [17, 57]. In this pathway, GS which is conserved in AOA, converts glutamate to glutamine. Glutamine-oxoglutarate amidotransferase (GOGAT) catalyses the NADPH-dependent formation of two glutamate molecules from glutamine and 2-oxoglutarate (Fig. 2). This high affinity, energy consuming pathway for ammonia assimilation functions well at low ammonia concentrations, and when the cell is not energy or carbon limited [61]. The reason for the presence of GS/GOGAT in some AOA is unknown as are many details of ammonia assimilation in AOA. It is possible that most AOA use GDH, as this pathway is energetically less costly than GS/GOGAT, even if it requires high concentrations of ammonia. It was also postulated that some Nitrosocosmicus strains might have as-yet unidentified, auxiliary energy sources, which may explain their use of ATP-dependent GS/GOGAT pathway [57].
The biochemistry of energy metabolism of AOA
The structure, function, and substrate range of the AMO
The AMO is a copper-dependent multimeric transmembrane enzyme belonging to the CuMMO superfamily which comprises of ammonia, methane, and alkane monooxygenases [4–6, 13, 42]. Based on the similarities to bacterial AMO and pMMO, it is assumed that the oxidation of ammonia by the archaeal AMO requires two electrons [13]. These electrons are provided via electron carriers from the downstream pathway, the oxidation of hydroxylamine, and possibly nitric oxide, although the role of nitric oxide in AOA is not fully understood [65, 66]. Assuming that the archaeal AMO functions similarly to its bacterial counterparts, two net electrons per ammonia molecule are generated from the ammonia oxidation pathway and this reductant powers the ATP synthesis and cellular anabolism, including carbon fixation [13]. Nitrous oxide is also produced by AOA during ammonia oxidation, and in 15N-labelling studies with Nitrososphaera viennensis, nitrous oxide was generated through N-nitrosating hybrid formation [67]. A recent on study on Nitrosopumilus maritimus found that, although hybrid formation is a key mechanism of nitrous oxide production, there are multiple pathways through which the constituent atoms of nitrous oxide are derived from ammonia, nitrite, O2 and H2O [68]. In addition, Nitrosopumilus maritimus can produce nitrous oxide through NO dismutation under anoxic conditions [31]. Active AMO is difficult to purify and many predictions about the structure have been based on homology to the better-characterised particulate methane monooxygenase (pMMO) from methanotrophs [69, 70]. The AMO is predicted to exist as a heterotrimeric complex composed of three subunits in bacteria: AmoA, AmoB, and AmoC [71]. The archaeal AMO is very divergent from bacterial AMO and other CuMMOs and appears to have additional subunits including AmoX, AmoY, and AmoZ [42] (Fig. 3). The location and nature of the AMO active site has not been identified. Analysis of the AmoB and AmoC protein structure favours an extracellular active site (outwards facing) [47], which would be logical considering the toxicity of hydroxylamine. The location and nature of the pMMO active site also remain uncertain, although mono- and di-copper sites have been proposed to reside in the soluble region of the PmoB [72] and a newly discovered tri-copper site is found in the PmoC subunit, close to the periplasm [73]. Mutagenesis studies on the hydrocarbon monooxygenase, a member of the CuMMO superfamily, in Mycobacterium NBB4 have demonstrated that the metal-binding residues on the C subunit are essential for activity [74]. Substrates and inhibitors of the AMO are largely non-polar, suggesting the active site is hydrophobic, and consistent with ammonia rather than ammonium as the natural substrate [75].
Fig. 3. Schematic of the archaeal ammonia monooxygenase (AMO).
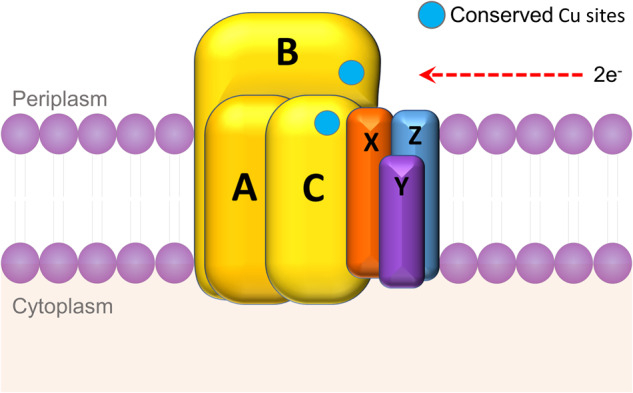
AmoA, AmoB, and AmoC (yellow), are conserved and form trimers (AmoABC). AmoB and AmoC contain copper binding sites, which are conserved in AOA and AOB. AmoX (orange), AmoY (purple), and AmoZ (blue), are putative archaeal AMO accessory proteins.
Copper is thought to be a co-factor in both the archaeal and bacterial AMO. The respiratory chain in AOA is predicted to be copper-based, and they contain numerous small blue copper proteins and multicopper oxidases which may be involved in electron transfer [61, 76]. In contrast, the respiratory chain and key enzymes e.g. hydroxylamine dehydrogenase (HAO) in AOB use proteins which require heme [13]. Nevertheless, it is estimated that both iron and copper may be limiting factors to the growth of AOA in the ocean [77, 78]. There are no known examples of AOA producing chalkophores (copper-binding molecules), and Nitrososphaera viennensis has a higher affinity for copper uptake than Nitrosopumilus maritimus does [78, 79]. Translating culture-based findings to the environmental context is challenging because copper bioavailability in many habitats is affected by factors such as pH and complexation with organic molecules [80].
Due to the difficulty in purifying AMO in its active state, much of what is known about the AMO has been discovered using inhibitors. Acetylene is a well characterised inhibitor of both the AMO and pMMO [81, 82]. With Nitrosomonas europaea, acetylene acts as a suicide substrate and cells require de novo protein synthesis of new AMO to re-establish ammonia-oxidising activity [83]. Incubations with 14[C]-acetylene resulted in the covalent radiolabelling of Nitrosomonas europaea AMO, enabling identification of the genes coding for AMO [84]. A subsequent study found that the ketene product of acetylene activation bound covalently to a histidine residue (H191) on the AmoA subunit of Nitrosomonas europaea, a residue thought to be in close proximity to the putative active site [85]. While acetylene is also an irreversible inhibitor of the archaeal AMO [86], the AMO from archaea lack the histidine residue responsible for binding in Nitrosomonas europaea, suggesting that acetylene must bind at a different position on the enzyme [85].
Insights into the structure of the archaeal AMO active site(s) and its potential substrate range has been provided by characterising the inhibition of archaeal AMOs to linear 1-alkynes [87–89]. The archaeal AMO demonstrates a reduced sensitivity to inhibition by larger 1-alkynes compared to bacterial AMO, suggesting they have a narrower hydrocarbon substrate range [87–89]. In fact, archaeal 1-alkyne inhibition profiles were similar to that of pMMO which can only oxidise linear C1-C5 alkanes and alkenes [88, 89]. Recent reconstitution of the pMMO in a lipid bilayer revealed the PmoC tri-copper binding site, which is adjacent to a hydrophobic cavity capable of accommodating up to C5 linear hydrocarbons [73, 90]. The aromatic alkyne, phenylacetylene, inhibited the archaeal and bacterial AMO at different threshold concentrations and by different mechanisms of inhibition, highlighting functional differences between the archaeal and bacterial AMO [89]. Kinetic analysis of the inhibition of ammonia oxidation by Nitrosomonas europaea demonstrated that unlike acetylene, phenylacetylene does not compete with ammonia for the same binding site and behaved as an uncompetitive inhibitor, suggesting phenylacetylene only had affinity for the AMO-ammonia complex [89]. Phenylacetylene inhibition of ‘Ca. Nitrosocosmicus franklandus’ was found to be non-competitive [89]. The results indicate the presence of secondary, non-ammonia, binding sites on both the archaeal and bacterial AMO, as previously suggested for the AMO from Nitrosomonas europaea and the pMMO [91–93].
It is proposed that the downstream metabolism refines the functional role of microorganisms containing CuMMO [94]. For example, the bacterial ammonia oxidisers Nitrosococcus oceani and Nitrosomonas europaea can oxidise methane but lack necessary downstream enzymes to gain energy from methane oxidation [95]. Likewise, several methanotrophs have been shown to co-oxidise ammonia, but this does not support growth [96]. AMO- and pMMO-expressing microorganisms have received interest for their potential use in bioremediation due to their capability to co-oxidize persistent organic pollutants such as halogenated alkanes and alkenes and chlorinated hydrocarbons [97, 98]. It was shown that ‘Ca. Nitrososphaera gargensis Ga9.2’ was capable of co-metabolising two tertiary amines, mianserin and ranitidine (pharmaceutical drugs), with the initial oxidative reaction possibly carried out by AMO [99]. Co-oxidation of compounds other than ammonia by AOA is an open question and has not been fully explored yet. The work on alkyne inhibitors suggests that the substrate range for archaeal AMO may include hydrocarbons with chain-lengths <C5. If AOA participate in co-oxidation of such compounds e.g. methane, ethane, or propane, or even more complex branched hydrocarbons, this would be of importance for both bioremediation and for understanding global biogeochemical carbon cycling.
Metabolic versatility
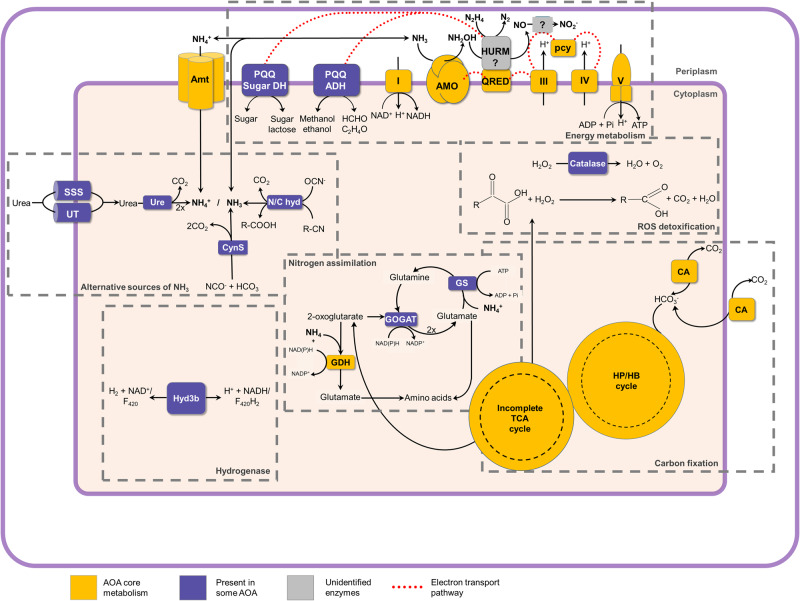
Autotrophic ammonia oxidising microorganisms are generally considered metabolically streamlined, and specialised in using ammonia as their sole source of energy. In contrast, several nitrifying microorganisms demonstrate remarkable metabolic flexibility [100]. Best known examples are nitrite-oxidising bacteria, which can derive energy for growth from formate, hydrogen and sulfide [100]. Some, but not all, ammonia oxidisers can use urea or cyanate as the sole source of energy and reductant as both are enzymatically converted to ammonium [101–104] (Fig. 4). Growth on urea or cyanate is not pH-dependent and therefore may be advantageous in acidic and low ammonium environments [105, 106]. Urea uptake is mediated by specific urea transporters (UTs) and solute/sodium symporters (SSS (DUR3)), after which urea hydrolysed intracellularly by a urease [107]. SSS transporters are phylogenetically divided into several distinct clusters, of which AOA share the closest sequence similarity with the plant urea transporters [107]. Urea uptake systems and urease enzymes have been reported in AOB, AOA and comammox bacteria [61, 101–103]. The hydrolysis of urea can support the growth of AOB such as Nitrosoglobus terrae and Nitrosospira sp. at low pH [105, 108]. However, the use of urea by marine AOA is not directly related to pH unlike often reported for ammonia oxidisers in acidic soils [102]. Urea is commonly present in marine habitats, and being able to use urea in addition to ammonia may give these AOA a competitive advantage [109]. Nitrososphaera gargensis Ga9.2 is currently the only genome-sequenced AOA that encodes a known cyanase, which catalyses conversion of cyanate to ammonium and CO2 [104]. Nitrosopumilus maritimus lacks a canonical cyanase, but also produces ammonia from cyanate, which suggests there must exist an as-yet unknown mechanism, which breaks down cyanate in Nitrosopumilus maritimus [109]. Genes putatively encoding for enzymes of a novel class of nitrilases or cyanide hydratases are found in genomes of AOA from the Nitrosocaldus, Nitrosotenuis and Nitrosopumilus genera [110–112]. Nitrilases catalyse the conversion of nitriles to the corresponding acid and cyanide hydratases convert hydrogen cyanide (HCN) to formamide, both of which can produce ammonia [112]. These homologues might be involved in the conversion of cyanate to ammonia, but their function has not yet been experimentally proven (Fig. 4).
Fig. 4. Schematic of predicted metabolic features from the genomes of AOA.
Yellow represents genomic features encoded by all AOA. Blue indicates metabolisms not shared by all AOA. AMO ammonia monooxygenase, QRED quinone reductase, HURM hydroxylamine ubiquinone redox module, PQQ pyrroloquinoline quinone, DH dehydrogenase, ADH alcohol dehydrogenase, Amt ammonium transporter, UT urea transporter, SSS solute/sodium symporter, Ure urease, N/C hyd nitrile/cyanide hydratases, Cyn cyanase, TCA tricarboxylic acid cycle, HP/HB hydroxypropionate/hydroxybutyrate cycle, CA carbonic anhydrase, Hyd hydrogenase, GDH glutamate dehydrogenase, GS-GOGAT glutamine synthetase-glutamate synthase, ROS reactive oxygen species.
Hydroxylamine is a physiological intermediate in the archaeal ammonia oxidation pathway [113]. No HAO homologue exists in AOA, nor do they have the genetic repertoire to fully synthesise c-type hemes. Therefore, AOA have a novel enzymology for the oxidation of hydroxylamine [113]. Hydrazine, a key intermediate in anammox catabolism and a structural analogue of hydroxylamine [114], was found to be an inhibitor of hydroxylamine oxidation by AOA [115]. In addition, the AOA isolate “Ca. Nitrosocosmicus franklandus” oxidised hydrazine to dinitrogen, with O2 consumption coupled with ATP production [115] (Fig. 4). Hydrazine is also a substrate for the bacterial HAO, and is oxidised to N2 (Eq. (1), [116]).
| 1 |
Electrons derived from this reaction can serve as reducing equivalents for the AMO and hydrazine has been used as an external source of reductant to fuel alternative substrate oxidations by Nitrosomonas europaea [117–119]. Ammonia, urea, cyanate, hydroxylamine, and hydrazine are currently the only experimentally confirmed energy-yielding substrates in AOA. However, there are many other predicted, as-yet unproven types of metabolism in AOA as discussed below.
Ammonia oxidisers are chemolithoautotrophs and able to fix their own carbon. Ammonia oxidising archaea fix their carbon using the 3-hydroxypropionate/4-hydroxybutyrate pathway and reductant generated from ammonia oxidation [10]. Nevertheless, there are some indirect indications of mixotrophic growth in ammonia oxidisers. Early studies of carbon metabolism in marine archaea indicated both autotrophic and heterotrophic modes of carbon assimilation occur, although it is possible that not all archaea in these environmental samples were AOA [120, 121]. A recent study found that in the Pacific Ocean, a proportion of the Marine Group I archaea, which includes AOA, incorporated auxiliary carbon from urea and amino acids [122]. However, it can be challenging to disentangle mixotrophy and autotrophy, because e.g. urea could be metabolised into CO2 and fixed autotrophically, resulting in indirect incorporation. In addition, a discrepancy between a high abundance of AOA marine water column and a low nitrification rate was reported [30]. One possible explanation for this observation would be mixotrophic growth, where AOA would use electron donors other than ammonia or fix carbon from alternative organic carbon sources instead of CO2. However, the only direct evidence of mixotrophy in ammonia oxidisers is known in Nitrosomonas europaea, the growth of which is stimulated by fructose and pyruvate which are assimilated as carbon sources [123]. There have been efforts to investigate the potential of AOA for mixotrophy using organic acids, some of which are tricarboxylic acid cycle intermediates [18, 124, 125]. However, the apparent stimulation of catalase-negative AOA by α-keto acids is due to the alleviation of oxidative stress, rather than mixotrophic growth [126] (Fig. 4). In addition, organic compounds can inhibit some AOA strains, potentially due to their high copper complexation potential or their toxic effect at low pH [80, 125].
The genome of “Ca. Nitrosocosmicus hydrocola” encodes for genes putatively involved in one-carbon (C1) metabolism, including methanol oxidation to formaldehyde and formate oxidation to CO2 [18]. Some AOA from deep seafloor sediments have the genetic repertoire to perform proteolysis and deamination to regenerate ammonia, coupling mixotrophic and autotrophic metabolisms [127]. Intriguingly, the growth of the recently described AOA “Ca. Nitrosocosmicus arcticus” was uncoupled from ammonia oxidation, suggesting that this strain has alternative or supplementing energy metabolism(s) [57]. The genomes of some “Ca. Nitrosocosmicus” strains encode putative periplasmic or membrane-bound pyrroloquinoline quinone (PQQ)-dependent dehydrogenases, which oxidise sugars/alcohols by simultaneously reducing electron acceptors, potentially contributing reducing equivalents to the respiratory chain [57] (Fig. 4). PQQ-dependent dehydrogenases were among the most highly expressed genes by the newly discovered heterotrophic marine thaumarchaea, and therefore are likely to be important for energy metabolism since these Thaumarchaeota lack the ability to oxidise ammonia [128].
Some AOA genomes contain coding sequences related to hydrogenases, although their function has not been verified in any AOA. The genomes of the thermophilic AOA “Ca. Nitrosocaldus cavascurensis” and “Ca. Nitrosocaldus islandicus” both contain genes encoding for the four subunits of a putative cytoplasmic Group 3b [NiFe]-hydrogenase [35, 129] (Fig. 4). Some previously characterised members of Group 3b hydrogenases are oxygen-tolerant and bidirectional, and can couple oxidation of H2 to reduction of NAD(P), or oxidation of NAD(P)H to fermentative production of H2 [130–132]. In addition, Abby and colleagues also speculate that oxidised F420 could be a potential cofactor for the 3b-[NiFe]-hydrogenase in “Ca. Nitrosocaldus cavascurensis”, although this has not yet been tested experimentally [129]. All members of genus ‘Ca. Nitrosocaldus’ are thermophilic and thrive in hot springs at temperatures of ~70 °C, although the link between the growth temperature and presence of Group 3b [NiFe] hydrogenases in AOA remains unproven. Hydrogenases appear to be mainly absent in the genomes of AOB, apart from two representatives from the Nitrosomonas cluster 6a and Nitrosospira multiformis [103, 133], both of which originate from soil ecosystems and contain putative Group 3d hydrogenases. Group 3b hydrogenases are also found in comammox Nitrospira, predominantly in the representatives from clade A [101]. The genomes of other AOA (including some representatives of genera Nitrosotalea, Nitrososphaera and Nitrosocosmicus) contain genes with homology to Group 4a [NiFe]-hydrogenases. These proteins, termed energy conserving hydrogenase-related complexes (Ehr), lack the CxxC motif required for hydrogenase activity and might play another role in electron transfer [134]. Ehr complexes are also found in comammox Nitrospira [135, 136]. It is unknown whether the Ehr complexes confer any advantages in terms of metabolic flexibility and environmental adaptation in ammonia oxidisers.
Outlook
This review aimed to highlight some key knowledge gaps in AOA research, aside from the seemingly elusive enzymology of ammonia oxidation pathway and its intermediates [137, 138]. Ammonia oxidisers are widely regarded as relatively inflexible, but we have covered some possibilities of alternative metabolisms, including predicted pathways and potential co-oxidation, which require further exploration. In contrast to the laboratory, where culture conditions can be carefully controlled, AOA in the environment are exposed to fluctuating conditions in a complex ecosystem where substrates can often be limiting for growth (Table 2). There are knowledge gaps in how AOA respond to these environmental changes and which types of metabolisms are required for them to survive and thrive. Ammonia oxidation is highly specialised and not a widespread trait, and an argument could be made that there is little return on investing in alternative metabolisms. In contrast, ammonia oxidation offers little energetic reward and supplementing autotrophic growth could benefit nitrifiers, especially in oligotrophic environments, in environments where they co-exist with heterotrophs or in environments where ammonia oxidation is difficult, such as low oxygen conditions. For example, utilisation of small nitrogenous compounds such as urea and cyanate by AOA has been reported previously, but there are gaps in our understanding of the function and ecological importance of these pathways (Table 2). Likewise, it is known that compounds other than ammonia can interact with the archaeal AMO enzyme, but little is known about the potential role of alternative substrates and inhibitors of the AMO in the environment. Some of these substrates and inhibitors of the AMO occur naturally, but their influence on biogeochemical cycling is unknown (Table 2). An additional aspect of regulation and energy metabolism in ammonia oxidising microorganisms is that ammonia is both a source of energy as well as being required for constructing biomass. There must therefore presumably exist a system for sensing and regulating how ammonia is allocated within the cell (Table 2), particularly as ammonia oxidisers need to be able to rapidly respond to a changing environment, and to switch from growing to persisting or vice versa.
Table 2.
Future perspectives.
| Ammonia uptake, assimilation, and oxidation. |
| Ammonia has a dual role in ammonia oxidisers, because it is required for both energy and building biomass. |
| ● How is the partitioning between ammonia assimilation and oxidation regulated, and does ammonium transport play a role in both? |
| Co-oxidation of alternative substrates. |
| The archaeal ammonia monooxygenase (AMO) can interact with a range of compounds, including hydrocarbons. |
| ● Can the archaeal AMO also co-oxidise alternative substrates? |
| ● What are the consequences of inhibition of, and co-oxidation by, AMO for biogeochemical cycling of nitrogen and other elements? |
| Alternative energy yielding pathways. |
| Metabolic pathways, including energy conservation coupled to H2 production, have been predicted, but not yet validated. |
| ● Are these pathways functional, and under which environmental conditions do they operate? |
| Some alternative types of metabolism, such utilisation of cyanate and urea, have been confirmed in AOA. |
| ● How important are the cyanate and urea metabolism in the adaptation of AOA to the environment? |
| Linking metabolism and adaptation of AOA to their environments. |
| Environmental conditions, including ammonia and oxygen concentrations, fluctuate and ammonia oxidisers will need to adapt to these potentially sudden changes. |
| ● How do AOA sense and respond to these changes, and which metabolisms contribute to their success and resilience in the environment? |
| ● Are AOA as metabolically inflexible as often thought? |
| ● Do AOA use different strategies to grow and persist, and how important is dormancy in underpinning their cosmopolitanism? |
The cultivation of ammonia oxidising microorganisms is notoriously difficult owing to their low yield and slow growth [139]. Nevertheless, validation of metabolism cannot be done from genetic repertoire alone. The study of isolated or highly enriched cultures is paramount to testing if the predicted metabolic pathways are functional, and to obtaining a mechanistic understanding of how they operate. The development of a genetic system for AOA would be especially valuable for investigating some of the putative metabolisms highlighted in this review and providing information about the regulation of major genes (Table 2). Furthermore, heterologous expression of AOA proteins could provide crucial insights on structure and biochemistry of key enzymes and transporters in vitro. In addition, single-cell and systems biology can deliver new knowledge which is not to easily accessible by other means. There is a promising outlook to further our understanding of nitrogen cycling and AOA by linking culture-based studies and culture-independent experiments on mixed communities from the environment. Novel mechanistic insights on the metabolism and biochemistry, including regulation, structure and function of key enzymes and discovery of new pathways, may help explain and predict how nitrifying communities respond to environmental changes, and how these factors together influence nitrification process rates in the environment.
Supplementary information
Acknowledgements
CLW was supported by an ERC Starting Grant awarded to LLM (UNITY 852993). LLM was supported by an ERC Starting Grant (UNITY 852993) and a Royal Society Dorothy Hodgkin Research Fellowship (DH150187). We thank Prof Colin Murrell and Dr Andrew Crombie for insightful discussions and advice on the manuscript.
Author contributions
CLW and LLM conceived and designed the study. CLW collected and analysed data. CLW and LLM wrote the paper.
Data availability
All data generated or analysed during this study are included in this published article and its supplementary information files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-023-01467-0.
References
- 1.Thamdrup B, Dalsgaard T. Production of N2 through anaerobic ammonium oxidation coupled to nitrate reduction in marine sediments. Appl Environ Microbiol. 2002;68:1312–8. doi: 10.1128/AEM.68.3.1312-1318.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dalsgaard T, Thamdrup B, Canfield DE. Anaerobic ammonium oxidation (anammox) in the marine environment. Res Microbiol. 2005;156:457–64. doi: 10.1016/j.resmic.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 3.Jetten MSM, Cirpus I, Kartal B, van Niftrik LAMP, Van De Pas-Schoonen KT, Sliekers O, et al. 1994–2004: 10 years of research on the anaerobic oxidation of ammonium. Biochem Soc Trans. 2005;33:119–23. doi: 10.1042/BST0330119. [DOI] [PubMed] [Google Scholar]
- 4.Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, et al. Complete nitrification by Nitrospira bacteria. Nature. 2015;528:504–9. doi: 10.1038/nature16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Kessel MA, Speth DR, Albertsen M, Nielsen PH, den Camp HJO, Kartal B, et al. Complete nitrification by a single microorganism. Nature. 2015;528:555–9. doi: 10.1038/nature16459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Könneke M, Bernhard AE, José R, Walker CB, Waterbury JB, Stahl DA. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–6. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 7.Francis CA, Beman JM, Kuypers MM. New processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J. 2007;1:19–27. doi: 10.1038/ismej.2007.8. [DOI] [PubMed] [Google Scholar]
- 8.Offre P, Spang A, Schleper C. Archaea in biogeochemical cycles. Ann Rev Microbiol. 2013;67:437–57. doi: 10.1146/annurev-micro-092412-155614. [DOI] [PubMed] [Google Scholar]
- 9.Prosser JI, Hink L, Gubry‐Rangin C, Nicol GW. Nitrous oxide production by ammonia oxidizers: physiological diversity, niche differentiation and potential mitigation strategies. Glob Chang Biol. 2020;26:103–18. doi: 10.1111/gcb.14877. [DOI] [PubMed] [Google Scholar]
- 10.Könneke M, Schubert DM, Brown PC, Hügler M, Standfest S, Schwander T, et al. Ammonia-oxidizing archaea use the most energy-efficient aerobic pathway for CO2 fixation. Proc Natl Acad Sci USA. 2014;111:8239–44. doi: 10.1073/pnas.1402028111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki I, Dular U, Kwok S. Ammonia or ammonium ion as substrate for oxidation by Nitrosomonas europaea cells and extracts. J Bacteriol. 1974;120:556–8. doi: 10.1128/jb.120.1.556-558.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung MY, Sedlacek CJ, Kits KD, Mueller AJ, Rhee SK, Hink L, et al. Ammonia-oxidizing archaea possess a wide range of cellular ammonia affinities. ISME J. 2022;16:272–83. doi: 10.1038/s41396-021-01064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arp DJ, Sayavedra-Soto LA, Hommes NG. Molecular biology and biochemistry of ammonia oxidation by Nitrosomonas europaea. Arch Microbiol. 2002;178:250–5. doi: 10.1007/s00203-002-0452-0. [DOI] [PubMed] [Google Scholar]
- 14.Martens-Habbena W, Berube PM, Urakawa H, José R, Stahl DA. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature. 2009;461:976–9. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- 15.Kits KD, Sedlacek CJ, Lebedeva EV, Han P, Bulaev A, Pjevac P, et al. Kinetic analysis of a complete nitrifier reveals an oligotrophic lifestyle. Nature. 2017;549:269–72. doi: 10.1038/nature23679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehtovirta-Morley LE, Ross J, Hink L, Weber EB, Gubry-Rangin C, Thion C, et al. Isolation of ‘Candidatus Nitrosocosmicus franklandus’, a novel ureolytic soil archaeal ammonia oxidiser with tolerance to high ammonia concentration. FEMS Microbiol Ecol. 2016;92:fiw057. [DOI] [PMC free article] [PubMed]
- 17.Jung MY, Kim JG, Sinninghe Damsté JS, Rijpstra WIC, Madsen EL, Kim SJ, et al. A hydrophobic ammonia‐oxidizing archaeon of the Nitrosocosmicus clade isolated from coal tar‐contaminated sediment. Environ Microbiol Rep. 2016;8:983–92. doi: 10.1111/1758-2229.12477. [DOI] [PubMed] [Google Scholar]
- 18.Sauder LA, Albertsen M, Engel K, Schwarz J, Nielsen PH, Wagner M, et al. Cultivation and characterization of Candidatus Nitrosocosmicus exaquare, an ammonia-oxidizing archaeon from a municipal wastewater treatment system. ISME J. 2017;11:1142–57. doi: 10.1038/ismej.2016.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodriguez J, Chakrabarti S, Choi E, Shehadeh N, Sierra-Martinez S, Zhao J. et al. Nutrient-Limited enrichments of nitrifiers from soil yield consortia of Nitrosocosmicus-affiliated AOA and Nitrospira-affiliated NOB. Front Microbiol. 2021;12:671480. [DOI] [PMC free article] [PubMed]
- 20.Verhamme D, Prosser J, Nicol G. Ammonia concentration determines differential growth of ammonia-oxidising archaea and bacteria in soil microcosms. ISME J. 2011;5:1067–71. doi: 10.1038/ismej.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jia Z, Conrad R. Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ Microbiol. 2009;11:1658–71. doi: 10.1111/j.1462-2920.2009.01891.x. [DOI] [PubMed] [Google Scholar]
- 22.Sauder LA, Peterse F, Schouten S, Neufeld JD. Low‐ammonia niche of ammonia‐oxidizing archaea in rotating biological contactors of a municipal wastewater treatment plant. Environ Microbiol. 2012;14:2589–2600. doi: 10.1111/j.1462-2920.2012.02786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ouyang Y, Norton JM, Stark JM. Ammonium availability and temperature control contributions of ammonia oxidizing bacteria and archaea to nitrification in an agricultural soil. Soil Biol Biochem. 2017;113:161–72. doi: 10.1016/j.soilbio.2017.06.010. [DOI] [Google Scholar]
- 24.Langone M, Yan J, Haaijer SCM, Op den Camp HJM, Jetten MSM, Andreottola G. Coexistence of nitrifying, anammox and denitrifying bacteria in a sequencing batch reactor. Front Microbiol. 2014;5:28. doi: 10.3389/fmicb.2014.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strous M, Kuenen JG, Jetten MS. Key physiology of anaerobic ammonium oxidation. Appl Environ Microbiol. 1999;65:3248–50. doi: 10.1128/AEM.65.7.3248-3250.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oshiki M, Satoh H, Okabe S. Ecology and physiology of anaerobic ammonium oxidizing (anammox) bacteria. Environ Microbiol. 2016;18:2784–96. doi: 10.1111/1462-2920.13134. [DOI] [PubMed] [Google Scholar]
- 27.Straka LL, Meinhardt KA, Bollmann A, Stahl DA, Winkler MK. Affinity informs environmental cooperation between ammonia-oxidizing archaea (AOA) and anaerobic ammonia-oxidizing (Anammox) bacteria. ISME J. 2019;13:1997–2004. doi: 10.1038/s41396-019-0408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan J, Haaijer SC, Op den Camp HJ, van Niftrik L, Stahl DA, Könneke M, et al. Mimicking the oxygen minimum zones: stimulating interaction of aerobic archaeal and anaerobic bacterial ammonia oxidizers in a laboratory‐scale model system. Environ Microbiol. 2012;14:3146–58. doi: 10.1111/j.1462-2920.2012.02894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bouskill NJ, Eveillard D, Chien D, Jayakumar A, Ward BB. Environmental factors determining ammonia‐oxidizing organism distribution and diversity in marine environments. Environ Microbiol. 2012;14:714–29. doi: 10.1111/j.1462-2920.2011.02623.x. [DOI] [PubMed] [Google Scholar]
- 30.Muck S, De Corte D, Clifford EL, Bayer B, Herndl GJ, et al. Niche differentiation of aerobic and anaerobic ammonia oxidizers in a high latitude deep oxygen minimum zone. Front Microbiol. 2019;10:2141. doi: 10.3389/fmicb.2019.02141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bristow LA, Dalsgaard T, Tiano L, Mills DB, Bertagnolli AD, Wright JJ, et al. Ammonium and nitrite oxidation at nanomolar oxygen concentrations in oxygen minimum zone waters. Proc Natl Acad Sci USA. 2016;113:10601–6. doi: 10.1073/pnas.1600359113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park BJ, Park SJ, Yoon DN, Schouten S, Sinninghe Damsté JS, Rhee SK. Cultivation of autotrophic ammonia-oxidizing archaea from marine sediments in coculture with sulfur-oxidizing bacteria. Appl Environ Microbiol. 2010;76:7575–87. doi: 10.1128/AEM.01478-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kraft B, Jehmlich N, Larsen M, Bristow LA, Könneke M, Thamdrup B, et al. Oxygen and nitrogen production by an ammonia-oxidizing archaeon. Science. 2022;375:97–100. doi: 10.1126/science.abe6733. [DOI] [PubMed] [Google Scholar]
- 34.Martens-Habbena W, Qin W. Archaeal nitrification without oxygen. Science. 2022;375:27–8. doi: 10.1126/science.abn0373. [DOI] [PubMed] [Google Scholar]
- 35.Daebeler A, Herbold CW, Vierheilig J, Sedlacek CJ, Pjevac P, Albertsen M, et al. Cultivation and genomic analysis of “Candidatus Nitrosocaldus islandicus,” an obligately thermophilic, ammonia-oxidizing thaumarchaeon from a hot spring biofilm in Graendalur Valley, Iceland. Front Microbiol. 2018;9:193. doi: 10.3389/fmicb.2018.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kostan J, Sjöblom B, Maixner F, Mlynek G, Furtmüller PG, et al. Structural and functional characterisation of the chlorite dismutase from the nitrite-oxidizing bacterium “Candidatus Nitrospira defluvii”: identification of a catalytically important amino acid residue. J Struct Biol. 2010;172:331–42. doi: 10.1016/j.jsb.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 37.Maixner F, Wagner M, Lücker S, Pelletier E, Schmitz-Esser S, et al. Environmental genomics reveals a functional chlorite dismutase in the nitrite-oxidizing bacterium ‘Candidatus Nitrospira defluvii’. Environ Microbiol. 2008;10:3043–56. doi: 10.1111/j.1462-2920.2008.01646.x. [DOI] [PubMed] [Google Scholar]
- 38.Camejo PY, Santo Domingo J, McMahon KD, Noguera DR. Genome-enabled insights into the ecophysiology of the comammox bacterium “Candidatus Nitrospira nitrosa”. Msystems. 2017;2:e00059–17. doi: 10.1128/mSystems.00059-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roots P, Wang Y, Rosenthal AF, Griffin JS, Sabba F, Petrovich M, et al. Comammox Nitrospira are the dominant ammonia oxidizers in a mainstream low dissolved oxygen nitrification reactor. Water Res. 2019;157:396–405. doi: 10.1016/j.watres.2019.03.060. [DOI] [PubMed] [Google Scholar]
- 40.Pett-Ridge J, Petersen DG, Nuccio E, Firestone MK. Influence of oxic/anoxic fluctuations on ammonia oxidizers and nitrification potential in a wet tropical soil. FEMS Microbiol Ecol. 2013;85:179–94. doi: 10.1111/1574-6941.12111. [DOI] [PubMed] [Google Scholar]
- 41.Chen XP, Zhu YG, Xia Y, Shen JP, He JZ. Ammonia‐oxidizing archaea: important players in paddy rhizosphere soil? Environ Microbiol. 2008;10:1978–87. doi: 10.1111/j.1462-2920.2008.01613.x. [DOI] [PubMed] [Google Scholar]
- 42.Hodgskiss LH, Melcher M, Kerou M, Chen W, Ponce-Toledo RI, Savvides SN, et al. Unexpected complexity of the ammonia monooxygenase in archaea. ISME J. 2023;17:588–99. [DOI] [PMC free article] [PubMed]
- 43.Andrade SL, Dickmanns A, Ficner R, Einsle O. Crystal structure of the archaeal ammonium transporter Amt-1 from Archaeoglobus fulgidus. Proc Natl Acad Sci USA. 2005;102:14994–9. doi: 10.1073/pnas.0506254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ellerbeck M, Schüßler A, Brucker D, Dafinger C, Loos F, Brachmann A. Characterization of three ammonium transporters of the glomeromycotan fungus Geosiphon pyriformis. Eukaryot Cell. 2013;12:1554–62. doi: 10.1128/EC.00139-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wacker T, Garcia-Celma JJ, Lewe P, Andrade SL. Direct observation of electrogenic NH4+ transport in ammonium transport (Amt) proteins. Proc Natl Acad Sci USA. 2014;111:9995–10000. doi: 10.1073/pnas.1406409111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Williamson G, Tamburrino G, Bizior A, Boeckstaens M, Dias Mirandela G, Bage MG, et al. A two-lane mechanism for selective biological ammonium transport. Elife. 2020;9:e57183. doi: 10.7554/eLife.57183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lehtovirta-Morley LE, Sayavedra-Soto LA, Gallois N, Schouten S, Stein LY, Prosser JI, et al. Identifying potential mechanisms enabling acidophily in the ammonia-oxidizing archaeon “Candidatus Nitrosotalea devanaterra”. Appl Environ Microbiol. 2016;82:2608–19. doi: 10.1128/AEM.04031-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palomo A, Pedersen AG, Fowler SJ, Dechesne A, Sicheritz-Pontén T, Smets BF. Comparative genomics sheds light on niche differentiation and the evolutionary history of comammox Nitrospira. ISME J. 2018;12:1779–93. doi: 10.1038/s41396-018-0083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matassi G. Horizontal gene transfer drives the evolution of Rh50 permeases in prokaryotes. BMC Evol Biol. 2017;17:1–14. doi: 10.1186/s12862-016-0850-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lupo D, Li XD, Durand A, Tomizaki T, Cherif-Zahar B, Matassi G, et al. The 1.3-Å resolution structure of Nitrosomonas europaea Rh50 and mechanistic implications for NH3 transport by Rhesus family proteins. Proc Nat Acad Sci USA. 2007;104:19303–8. doi: 10.1073/pnas.0706563104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pflüger T, Hernández CF, Lewe P, Frank F, Mertens H, Svergun D, et al. Signaling ammonium across membranes through an ammonium sensor histidine kinase. Nat Commun. 2018;9:164. doi: 10.1038/s41467-017-02637-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cherif-Zahar B, Durand A, Schmidt I, Hamdaoui N, Matic I, Merrick M, et al. Evolution and functional characterization of the RH50 gene from the ammonia-oxidizing bacterium Nitrosomonas europaea. J Bacteriol. 2007;189:9090–100. doi: 10.1128/JB.01089-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Offre P, Kerou M, Spang A, Schleper C. Variability of the transporter gene complement in ammonia-oxidizing archaea. Trends Microbiol. 2014;22:665–75. doi: 10.1016/j.tim.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 54.Qin W, Amin SA, Lundeen RA, Heal KR, Martens-Habbena W, Turkarslan S, et al. Stress response of a marine ammonia-oxidizing archaeon informs physiological status of environmental populations. ISME J. 2018;12:508–19. doi: 10.1038/ismej.2017.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Santoro AE, Casciotti KL. Enrichment and characterization of ammonia-oxidizing archaea from the open ocean: phylogeny, physiology and stable isotope fractionation. ISME J. 2011;5:1796–808. doi: 10.1038/ismej.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakagawa T, Stahl DA. Transcriptional response of the archaeal ammonia oxidizer Nitrosopumilus maritimus to low and environmentally relevant ammonia concentrations. Appl Environ Microbiol. 2013;79:6911–6. doi: 10.1128/AEM.02028-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alves RJE, Kerou M, Zappe A, Bittner R, Abby SS, Schmidt HA, et al. Ammonia oxidation by the arctic terrestrial thaumarchaeote “Candidatus Nitrosocosmicus arcticus” is stimulated by increasing temperatures. Front Microbiol. 2019;10:1571. doi: 10.3389/fmicb.2019.01571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li PN, Herrmann J, Tolar BB, Poitevin F, Ramdasi R, Bargar JR, et al. Nutrient transport suggests an evolutionary basis for charged archaeal surface layer proteins. ISME J. 2018;12:2389–402. doi: 10.1038/s41396-018-0191-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nicol GW, Hink L, Gubry-Rangin C, Prosser JI, Lehtovirta-Morley LE. Genome Sequence of “Candidatus Nitrosocosmicus franklandus” C13, a terrestrial ammonia-oxidizing archaeon. Microbiol Resour Announc. 2019;8:e00435–19. doi: 10.1128/MRA.00435-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Valentine DL. Adaptations to energy stress dictate the ecology and evolution of the Archaea. Nat Rev Microbiol. 2007;5:316–23. doi: 10.1038/nrmicro1619. [DOI] [PubMed] [Google Scholar]
- 61.Kerou M, Offre P, Valledor L, Abby SS, Melcher M, Nagler M, et al. Proteomics and comparative genomics of Nitrososphaera viennensis reveal the core genome and adaptations of archaeal ammonia oxidizers. Proc Natl Acad Sci USA. 2016;113:7937–46. doi: 10.1073/pnas.1601212113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Heeswijk WC, Westerhoff HV, Boogerd FC. Nitrogen assimilation in Escherichia coli: putting molecular data into a systems perspective. Microbiol Mol Biol Rev. 2013;77:628–95. doi: 10.1128/MMBR.00025-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Forchhammer K. PII signal transducers: novel functional and structural insights. Trends Microbiol. 2008;16:65–72. doi: 10.1016/j.tim.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 64.Liu L, Liu M, Jiang Y, Lin W, Luo J. Production and excretion of polyamines to tolerate high ammonia, a case study on soil ammonia-oxidizing archaeon “Candidatus Nitrosocosmicus agrestis”. Msystems. 2021;6:e01003–20. doi: 10.1128/mSystems.01003-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martens-Habbena W, Qin W, Horak RE, Urakawa H, Schauer AJ, Moffett JW, et al. The production of nitric oxide by marine ammonia-oxidizing archaea and inhibition of archaeal ammonia oxidation by a nitric oxide scavenger. Environ Microbiol. 2015;17:2261–74. doi: 10.1111/1462-2920.12677. [DOI] [PubMed] [Google Scholar]
- 66.Caranto JD, Lancaster KM. Nitric oxide is an obligate bacterial nitrification intermediate produced by hydroxylamine oxidoreductase. Proc Natl Acad Sci USA. 2017;114:8217–22. doi: 10.1073/pnas.1704504114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stieglmeier M, Mooshammer M, Kitzler B, Wanek W, Zechmeister-Boltenstern S, Richter A, et al. Aerobic nitrous oxide production through N-nitrosating hybrid formation in ammonia-oxidizing archaea. ISME J. 2014;8:1135–46. doi: 10.1038/ismej.2013.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wan X, Lei H, Kao S, Zhang Y, Sheng H, et al. Pathways of N2O production by marine ammonia-oxidizing archaea determined from dual-isotope labelling. Proc Natl Acad Sci USA. 2013;120:e2220697120. doi: 10.1073/pnas.2220697120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Holmes AJ, Costello A, Lidstrom ME, Murrell JC. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol Lett. 1995;132:203–8. doi: 10.1111/j.1574-6968.1995.tb07834.x. [DOI] [PubMed] [Google Scholar]
- 70.Lawton TJ, Ham J, Sun T, Rosenzweig AC. Structural conservation of the B subunit in the ammonia monooxygenase/particulate methane monooxygenase superfamily. Proteins. 2014;82:2263–7. doi: 10.1002/prot.24535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tolar BB, Herrmann J, Bargar JR, van den Bedem H, Wakatsuki S, Francis CA. Integrated structural biology and molecular ecology of N‐cycling enzymes from ammonia‐oxidizing archaea. Environ Microbiol Rep. 2017;9:484–91. doi: 10.1111/1758-2229.12567. [DOI] [PubMed] [Google Scholar]
- 72.Ross MO, MacMillan F, Wang J, Nisthal A, Lawton TJ, Olafson BD, et al. Particulate methane monooxygenase contains only mononuclear copper centers. Science. 2019;364:566–70. doi: 10.1126/science.aav2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Koo CW, Tucci FJ, He Y, Rosenzweig AC. Recovery of particulate methane monooxygenase structure and activity in a lipid bilayer. Science. 2022;375:1287–91. doi: 10.1126/science.abm3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liew EF, Tong D, Coleman NV, Holmes AJ. Mutagenesis of the hydrocarbon monooxygenase indicates a metal centre in subunit-C, and not subunit-B, is essential for copper-containing membrane monooxygenase activity. Microbiol. 2014;160:1267–77. doi: 10.1099/mic.0.078584-0. [DOI] [PubMed] [Google Scholar]
- 75.Lancaster KM, Caranto JD, Majer SH, Smith MA. Alternative bioenergy: updates to and challenges in nitrification metalloenzymology. Joule. 2018;2:421–41. doi: 10.1016/j.joule.2018.01.018. [DOI] [Google Scholar]
- 76.Reyes C, Hodgskiss LH, Kerou M, Pribasnig T, Abby SS, Bayer B, et al. Genome wide transcriptomic analysis of the soil ammonia oxidizing archaeon Nitrososphaera viennensis upon exposure to copper limitation. ISME J. 2020;14:2659–74. doi: 10.1038/s41396-020-0715-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shafiee RT, Snow JT, Zhang Q, Rickaby RE. Iron requirements and uptake strategies of the globally abundant marine ammonia-oxidising archaeon, Nitrosopumilus maritimus SCM1. ISME J. 2019;13:2295–305. doi: 10.1038/s41396-019-0434-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Amin SA, Moffett JW, Martens-Habbena W, Jacquot JE, Han Y, Devol A, et al. Copper requirements of the ammonia‐oxidizing archaeon Nitrosopumilus maritimus SCM1 and implications for nitrification in the marine environment. Limnol Oceanogr. 2013;58:2037–45. doi: 10.4319/lo.2013.58.6.2037. [DOI] [Google Scholar]
- 79.Reyes C, Hodgskiss LH, Baars O, Kerou M, Bayer B, Schleper C, et al. Copper limiting threshold in the terrestrial ammonia oxidizing archaeon Nitrososphaera viennensis. Res Microbiol. 2020;171:134–42. doi: 10.1016/j.resmic.2020.01.003. [DOI] [PubMed] [Google Scholar]
- 80.Gwak JH, Jung MY, Hong H, Kim JG, Quan ZX, Reinfelder JR, et al. Archaeal nitrification is constrained by copper complexation with organic matter in municipal wastewater treatment plants. ISME J. 2020;14:335–46. doi: 10.1038/s41396-019-0538-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Prior SD, Dalton H. Acetylene as a suicide substrate and active site probe for methane monooxygenase from Methylococcus capsulatus (Bath) FEMS Microbiol Lett. 1985;29:105–9. doi: 10.1111/j.1574-6968.1985.tb00843.x. [DOI] [Google Scholar]
- 82.Hyman MR, Wood PM. Suicidal inactivation and labelling of ammonia monooxygenase by acetylene. Biochem J. 1985;227:719–25. doi: 10.1042/bj2270719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hyman MR, Arp DJ. 14C2H2-and 14CO2-labeling studies of the de novo synthesis of polypeptides by Nitrosomonas europaea during recovery from acetylene and light inactivation of ammonia monooxygenase. J Biol Chem. 1992;267:1534–45. doi: 10.1016/S0021-9258(18)45979-0. [DOI] [PubMed] [Google Scholar]
- 84.McTavish HJAF, Fuchs JA, Hooper AB. Sequence of the gene coding for ammonia monooxygenase in Nitrosomonas europaea. J Bacteriol. 1993;175:2436–44. doi: 10.1128/jb.175.8.2436-2444.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gilch S, Vogel M, Lorenz MW, Meyer O, Schmidt I. Interaction of the mechanism-based inactivator acetylene with ammonia monooxygenase of Nitrosomonas europaea. Microbiol. 2009;155:279–84. doi: 10.1099/mic.0.023721-0. [DOI] [PubMed] [Google Scholar]
- 86.Vajrala N, Bottomley PJ, Stahl DA, Arp DJ, Sayavedra-Soto LA. Cycloheximide prevents the de novo polypeptide synthesis required to recover from acetylene inhibition in Nitrosopumilus maritimus. FEMS Microbiol Ecol. 2014;88:495–502. doi: 10.1111/1574-6941.12316. [DOI] [PubMed] [Google Scholar]
- 87.Taylor AE, Vajrala N, Giguere AT, Gitelman AI, Arp DJ, Myrold DD, et al. Use of aliphatic n-alkynes to discriminate soil nitrification activities of ammonia-oxidizing thaumarchaea and bacteria. Appl Environ Microbiol. 2013;79:6544–51. doi: 10.1128/AEM.01928-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Taylor AE, Taylor K, Tennigkeit B, Palatinszky M, Stieglmeier M, Myrold DD, et al. Inhibitory effects of C2 to C10 1-alkynes on ammonia oxidation in two Nitrososphaera species. Appl Environ Microbiol. 2015;81:1942–8. doi: 10.1128/AEM.03688-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wright CL, Schatteman A, Crombie AT, Murrell JC, Lehtovirta-Morley LE. Inhibition of ammonia monooxygenase from ammonia-oxidizing archaea by linear and aromatic alkynes. Appl Environ Microbiol. 2020;86:e02388–19. doi: 10.1128/AEM.02388-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chan SI, Yu SSF. Controlled oxidation of hydrocarbons by the membrane-bound methane monooxygenase: The case for a tricopper cluster. Acc Chem Res. 2008;41:969–79. doi: 10.1021/ar700277n. [DOI] [PubMed] [Google Scholar]
- 91.Lontoh S, DiSpirito AA, Semrau JD. Dichloromethane and trichloroethylene inhibition of methane oxidation by the membrane-associated methane monooxygenase of Methylosinus trichosporium OB3b. Arch Microbiol. 1999;171:301–8. doi: 10.1007/s002030050714. [DOI] [Google Scholar]
- 92.Keener WK, Arp DJ. Kinetic studies of ammonia monooxygenase inhibition in Nitrosomonas europaea by hydrocarbons and halogenated hydrocarbons in an optimized whole-cell assay. Appl Environ Microbiol. 1993;59:2501–10. doi: 10.1128/aem.59.8.2501-2510.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Keener WK, Russell SA, Arp DJ. Kinetic characterization of the inactivation of ammonia monooxygenase in Nitrosomonas europaea by alkyne, aniline and cyclopropane derivatives. Biochim Biophys Acta Protein Struct Mol Enzymol. 1998;1388:373–85. doi: 10.1016/S0167-4838(98)00188-5. [DOI] [PubMed] [Google Scholar]
- 94.Pester M, Schleper C, Wagner M. The Thaumarchaeota: an emerging view of their phylogeny and ecophysiology. Curr Opin Microbiol. 2011;14:300–6. doi: 10.1016/j.mib.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hyman MR, Wood PM. Methane oxidation by Nitrosomonas europaea. Biochem J. 1983;212:31–37. doi: 10.1042/bj2120031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nyerges G, Stein LY. Ammonia cometabolism and product inhibition vary considerably among species of methanotrophic bacteria. FEMS Microbiol Lett. 2009;297:131–6. doi: 10.1111/j.1574-6968.2009.01674.x. [DOI] [PubMed] [Google Scholar]
- 97.Sayavedra-Soto LA, Gvakharia B, Bottomley PJ, Arp DJ, Dolan ME. Nitrification and degradation of halogenated hydrocarbons—a tenuous balance for ammonia-oxidizing bacteria. Appl Microbiol Biotechnol. 2010;86:435–44. doi: 10.1007/s00253-010-2454-1. [DOI] [PubMed] [Google Scholar]
- 98.Semrau J. Bioremediation via methanotrophy: overview of recent findings and suggestions for future research. Front Microbiol. 2011;2:209. doi: 10.3389/fmicb.2011.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Men Y, Han P, Helbling DE, Jehmlich N, Herbold C, Gulde R, et al. Biotransformation of two pharmaceuticals by the ammonia-oxidizing archaeon Nitrososphaera gargensis. Environ Sci Technol. 2016;50:4682–92. doi: 10.1021/acs.est.5b06016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Daims H, Lücker S, Wagner M. A new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends Microbiol. 2016;24:699–712. doi: 10.1016/j.tim.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Koch H, van Kessel MAHJ, Lücker S. Complete nitrification: insights into the ecophysiology of comammox Nitrospira. Appl Microbiol Biotechnol. 2019;103:177–89. doi: 10.1007/s00253-018-9486-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tourna M, Stieglmeier M, Spang A, Könneke M, Schintlmeister A, Urich T, et al. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci Usa. 2011;108:8420–5. doi: 10.1073/pnas.1013488108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Norton JM, Klotz MG, Stein LY, Arp DJ, Bottomley PJ, Chain PS, et al. Complete genome sequence of Nitrosospira multiformis, an ammonia-oxidizing bacterium from the soil environment. Appl Environ Microbiol. 2008;74:3559–72. doi: 10.1128/AEM.02722-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Palatinszky M, Herbold C, Jehmlich N, Pogoda M, Han P, von Bergen M, et al. Cyanate as an energy source for nitrifiers. Nature. 2015;524:105–8. doi: 10.1038/nature14856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Burton SA, Prosser JI. Autotrophic ammonia oxidation at low pH through urea hydrolysis. Appl Environ Microbiol. 2001;67:2952–7. doi: 10.1128/AEM.67.7.2952-2957.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lu L, Han W, Zhang J, Wu Y, Wang B, Lin X, et al. Nitrification of archaeal ammonia oxidizers in acid soils is supported by hydrolysis of urea. ISME J. 2012;6:1978–84. doi: 10.1038/ismej.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang WH, Kohler B, Cao FQ, Liu LH. Molecular and physiological aspects of urea transport in higher plants. Plant Sci. 2008;175:467–77. doi: 10.1016/j.plantsci.2008.05.018. [DOI] [Google Scholar]
- 108.Hayatsu M, Tago K, Uchiyama I, Toyoda A, Wang Y, Shimomura Y, et al. An acid-tolerant ammonia-oxidizing γ-proteobacterium from soil. ISME J. 2017;11:1130–41. doi: 10.1038/ismej.2016.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kitzinger K, Padilla CC, Marchant HK, Hach PF, Herbold CW, Kidane AT, et al. Cyanate and urea are substrates for nitrification by Thaumarchaeota in the marine environment. Nat Microbiol. 2019;4:234–43. doi: 10.1038/s41564-018-0316-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinel N, Arp DJ, et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci Usa. 2010;107:8818–882. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bayer B, Vojvoda J, Offre P, Alves RJ, Elisabeth NH, Garcia JA, et al. Physiological and genomic characterization of two novel marine thaumarchaeal strains indicates niche differentiation. ISME J. 2016;10:1051–63. doi: 10.1038/ismej.2015.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Luo ZH, Narsing Rao MP, Chen H, Hua ZS, Li Q, Hedlund BP, et al. Genomic insights of “Candidatus Nitrosocaldaceae” based on nine new metagenome-assembled genomes, including “Candidatus Nitrosothermus” gen nov. and two new species of “Candidatus Nitrosocaldus”. Front Microbiol. 2021;11:3412. doi: 10.3389/fmicb.2020.608832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vajrala N, Martens-Habbena W, Sayavedra-Soto LA, Schauer A, Bottomley PJ, Stahl DA, et al. Hydroxylamine as an intermediate in ammonia oxidation by globally abundant marine archaea. Proc Natl Acad Sci USA. 2013;110:1006–11. doi: 10.1073/pnas.1214272110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kartal B, de Almeida NM, Maalcke WJ, Op den Camp HJM, Jetten MSM, et al. How to make a living from anaerobic ammonium oxidation. FEMS Microbiol Rev. 2013;37:428–61. doi: 10.1111/1574-6976.12014. [DOI] [PubMed] [Google Scholar]
- 115.Schatteman A, Wright CL, Crombie AT, Murrell JC, Lehtovirta-Morley LE. Hydrazines as Substrates and Inhibitors of the Archaeal Ammonia Oxidation Pathway. Appl Environ Microbiol. 2022;88:e02470–21. doi: 10.1128/aem.02470-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Logan MS, Hooper AB. Suicide inactivation of hydroxylamine oxidoreductase of Nitrosomonas europaea by organohydrazines. Biochem. 1995;34:9257–64. doi: 10.1021/bi00028a039. [DOI] [PubMed] [Google Scholar]
- 117.Hyman MR, Murton IB, Arp DJ. Interaction of ammonia monooxygenase from Nitrosomonas europaea with alkanes, alkenes, and alkynes. Appl Environ Microbiol. 1988;54:3187–90. doi: 10.1128/aem.54.12.3187-3190.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rasche ME, Hyman MR, Arp DJ. Factors limiting aliphatic chlorocarbon degradation by Nitrosomonas europaea: cometabolic inactivation of ammonia monooxygenase and substrate specificity. Appl Environ Microbiol. 1991;57:2986–94. doi: 10.1128/aem.57.10.2986-2994.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Keener WK, Arp DJ. Transformations of aromatic compounds by Nitrosomonas europaea. Appl Environ Microbiol. 1994;60:1914–20. doi: 10.1128/aem.60.6.1914-1920.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ouverney CC, Fuhrman JA. Marine planktonic archaea take up amino acids. Appl Environ Microbiol. 2000;66:4829–33. doi: 10.1128/AEM.66.11.4829-4833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ingalls AE, Shah SR, Hansman RL, Aluwihare LI, Santos GM, Druffel ER, et al. Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. Proc Natl Acad Sci USA. 2006;103:6442–7. doi: 10.1073/pnas.0510157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Parada AE, Mayali X, Weber PK, Wollard J, Santoro AE, Fuhrman JA, et al. Constraining the composition and quantity of organic matter used by abundant marine Thaumarchaeota. Environ Microbiol. 2022. [DOI] [PubMed]
- 123.Hommes NG, Sayavedra-Soto LA, Arp DJ. Chemolithoorganotrophic growth of Nitrosomonas europaea on fructose. J Bacteriol. 2003;185:6809–14. doi: 10.1128/JB.185.23.6809-6814.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Spang A, Poehlein A, Offre P, Zumbrägel S, Haider S, Rychlik N, et al. The genome of the ammonia‐oxidizing “Candidatus Nitrososphaera gargensis”: insights into metabolic versatility and environmental adaptations. Environ Microbiol. 2012;14:3122–45. doi: 10.1111/j.1462-2920.2012.02893.x. [DOI] [PubMed] [Google Scholar]
- 125.Lehtovirta-Morley LE, Ge C, Ross J, Yao H, Nicol GW, Prosser JI. Characterisation of terrestrial acidophilic archaeal ammonia oxidisers and their inhibition and stimulation by organic compounds. FEMS Microbiol Ecol. 2014;89:542–52. doi: 10.1111/1574-6941.12353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kim JG, Park SJ, Damsté JSS, Schouten S, Rijpstra WIC, Jung MY, et al. Hydrogen peroxide detoxification is a key mechanism for growth of ammonia-oxidizing archaea. Proc Natl Acad Sci USA. 2016;113:7888–93. doi: 10.1073/pnas.1605501113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Vuillemin A, Wankel SD, Coskun ÖK, Magritsch T, Vargas S, Estes ER, et al. Archaea dominate oxic subseafloor communities over multimillion-year time scales. Sci Adv. 2019;5:eaaw4108. doi: 10.1126/sciadv.aaw4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Aylward FO, Santoro AE. Heterotrophic Thaumarchaea with small genomes are widespread in the dark ocean. mSystems. 2020;5:e00415–20. doi: 10.1128/mSystems.00415-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Abby SS, Melcher M, Kerou M, Krupovic M, Stieglmeier M, Rossel C, et al. “Candidatus Nitrosocaldus cavascurensis”, an ammonia oxidizing, extremely thermophilic archaeon with a highly mobile genome. Front Microbiol. 2018;9:28. doi: 10.3389/fmicb.2018.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Greening C, Biswas A, Carere CR, Jackson CJ, Taylor MC, Stott MB, et al. Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J. 2016;10:761–77. doi: 10.1038/ismej.2015.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Greening C, Islam ZF, Bay SK. Hydrogen is a major lifeline for aerobic bacteria. Trends Microbiol. 2022;30:330–7. doi: 10.1016/j.tim.2021.08.004. [DOI] [PubMed] [Google Scholar]
- 132.Berney M, Greening C, Conrad R, Jacobs WR, Cook GM. An obligately aerobic soil bacterium activates fermentative hydrogen production to survive reductive stress during hypoxia. Proc Natl Acad Sci Usa. 2014;111:11479–84. doi: 10.1073/pnas.1407034111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sedlacek CJ, McGowan B, Suwa Y, Sayavedra-Soto L, Laanbroek HJ, Stein LY, et al. A physiological and genomic comparison of Nitrosomonas cluster 6a and 7 ammonia-oxidizing bacteria. Micro Ecol. 2019;78:985–94. doi: 10.1007/s00248-019-01378-8. [DOI] [PubMed] [Google Scholar]
- 134.Batista AP, Marreiros BC, Pereira MM. The antiporter-like subunit constituent of the universal adaptor of complex I, group 4 membrane-bound [NiFe]-hydrogenases and related complexes. Biol Chem. 2013;394:659–66. doi: 10.1515/hsz-2012-0342. [DOI] [PubMed] [Google Scholar]
- 135.Lücker S, Wagner M, Mzixnera F, et al. Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc Natl Acad Sci USA. 2010;107:13479–84. doi: 10.1073/pnas.1003860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Koch H, Galushko A, Albertsen M, et al. Growth of nitrite-oxidizing bacteria by aerobic hydrogen oxidation. Science. 2014;345:1052–4. doi: 10.1126/science.1256985. [DOI] [PubMed] [Google Scholar]
- 137.Lehtovirta-Morley LE. Ammonia oxidation: Ecology, physiology, biochemistry and why they must all come together. FEMS Microbiol Lett. 2018;365:fny058. doi: 10.1093/femsle/fny058. [DOI] [PubMed] [Google Scholar]
- 138.Stein LY. Insights into the physiology of ammonia-oxidizing microorganisms. Curr Opin Chem Biol. 2019;49:9–15. doi: 10.1016/j.cbpa.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 139.Prosser JI, Nicol GW. Archaeal and bacterial ammonia-oxidisers in soil: the quest for niche specialisation and differentiation. Trends Microbiol. 2012;20:523–31. doi: 10.1016/j.tim.2012.08.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files.