Abstract
Spumaviruses, or foamy viruses, express Gag proteins that are incompletely processed by the viral protease in cell cultures. To delineate the proteolytic cleavage sites between potential Gag subdomains, recombinant human spumaretrovirus (HSRV) Gag proteins of different lengths were expressed, purified by affinity chromatography, and subjected to HSRV protease assays. HSRV-specific proteolytic cleavage products were isolated and characterized by Western blotting. Peptides spanning potential cleavage sites, as deduced from the sizes of the proteolytic cleavage products, were chemically synthesized and assayed with HSRV protease. The cleaved peptides were then subjected to mass spectrometry. In control experiments, HSRV protease-deficient mutant proteins were used to rule out unspecific processing by nonviral proteases. The cleavage site junctions identified and the calculated sizes of the cleavage products were in agreement with those of the authentic cleavage products of the HSRV Gag proteins detectable in viral proteins from purified HSRV particles and in virus-infected cells. The biological significance of the data was confirmed by mutational analysis of the cleavage sites in a recombinant Gag protein and in the context of the infectious HSRV DNA provirus.
The prototypic human spumaretrovirus (HSRV) has unique features of gene expression different from those of other known retroviruses. For a recent review that stresses the unconventional aspects of foamy viruses (FV), see reference 9. FV as complex retroviruses express genes from two different promoters (10, 11), and unlike onco- and lentiviruses, HSRV and nonprimate FV such as feline spumaretrovirus express subgenomic pol-specific transcripts (3, 4, 23). Genetic analysis showed that the HSRV protease (PR) is absolutely required for infectivity and processing, since the PR D/A mutant, in which the Asp residue of the catalytic center was replaced by Ala, resulted in a noninfectious HSRV provirus (8). The HSRV PR starts at the first Met of the residues encoded by the pol reading frame (12). Proteolytic processing of the HSRV Gag has been described as inefficient and incomplete (1, 2, 5–7, 11, 23). To date, a single cleavage site that is located between the p68Gag and p3Gag, close to the C terminus of the precursor pr71Gag, has been molecularly defined (5, 15). Nevertheless, reports by several groups on the existence of smaller cleavage products reactive with HSRV Gag-specific antibodies indicate that further proteolytic processing of HSRV Gag apparently occurs (1, 2, 6, 7, 12, 13, 20), as is also suggested by the electron microscopic morphology of pentameric or doughnut-shaped HSRV capsid-like structures (13, 25).
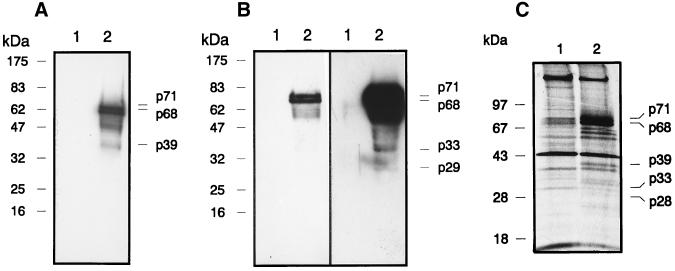
To obtain the patterns of HSRV Gag proteins from virus particles, supernatants of HSRV-infected HEL cell cultures three days postinfection were purified by sucrose density centrifugation as reported recently (25). Immunoblotting of the disrupted virions with two different polyclonal antibodies, Gagn and Gagc, revealed the patterns of viral polypeptides as shown in Fig. 1A and B. Antiserum Gagn (α-Gagn) is directed against the N-terminal region and α-Gagc is directed against the C-terminal part of HSRV Gag, as outlined in Fig. 2. The immunoblot analysis with α-Gagn showed a band at 39 kDa in addition to the characteristic doublet of p68 and p71 (Fig. 1A) reported previously (1, 2). It is noteworthy that after reaction with α-Gagc, polypeptide bands of 29 and 33 kDa were detectable upon longer exposure (Fig. 1B). The data suggest that the p39 cleavage product is derived from the N terminus of HSRV Gag; in contrast, the p29 and p33 bands have to be mapped to the C-terminal region of Gag.
FIG. 1.
Detection of HSRV proteins in purified virus particles by immunoblot analysis. (A) Purified virions were disrupted and reacted with polyclonal Gagn antiserum directed against the N-terminal region of HSRV Gag and detected by enhanced chemiluminescence (lane 2). Supernatants from mock-infected HEL cells were analyzed in parallel as a control (lane 1). Molecular size markers are shown on the left. (B) Immunoblot analysis as in panel A with the Gagc antiserum. The blot on the left was exposed for 15 s, and that on the right was exposed for 4 min. (C) Detection of HSRV proteins in virus-infected HEL cells by immunoprecipitation. Shown is an autoradiogram of metabolically labeled and immunoprecipitated proteins from mock-infected cells (lane 1) and virus-infected cells 6 days postinfection (lane 2). Labeling of HEL cells with l-[35S]methionine and [35S]cysteine (14.3 mCi/ml; Promix; Amersham, Little Chalfont, Buckinghamshire, United Kingdom) was carried out as reported elsewhere (2). For labeling, 50 μCi/ml of medium was used. Cell lysates were reacted with the Gagm antiserum from the central part of the HSRV Gag. The characteristic double band of the HSRV pr71Gag and p68Gag and other Gag-specific proteins are marked. Sizes of marker proteins are shown on the left.
FIG. 2.
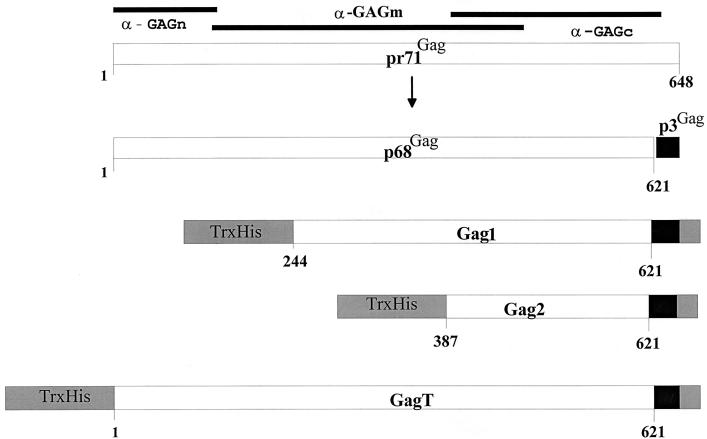
Schematic diagrams of HSRV Gag proteins. The large open rectangles in the top two diagrams represent the pr71Gag and p68Gag proteins, while the rest represent Gag1, Gag2, and GagT, expressed as His-tagged Trx fusion proteins. Small solid boxes, p3Gag domain (15); shaded boxes, the Trx-His part of the recombinant proteins and the coding regions derived from the pET32b vector backbone. The solid bars on top delineate the Gag regions against which the three polyclonal antisera Gagn, Gagm, and Gagc were raised. Numbers correspond to the positions of the amino acid residues of the HSRV pr71Gag. Cloning, expression, and purification of Gag proteins were carried out as described previously (15, 16).
To gain an overview of the patterns of HSRV Gag protein expression and processing in virus-infected cells, HEL cells were metabolically labeled with [35S]methionine and [35S]cysteine (2). Cell extracts were immunoprecipitated with polyclonal antibodies raised against the centrally located Gagm domain (Fig. 2), followed by sodium dodecyl sulfate-gel electrophoresis. The resulting pattern of polypeptides was compared to that of uninfected HEL cells as shown in Fig. 1C. Distinct bands of about 71, 68, 39, 33, and 28 kDa and some additional bands of smaller molecular sizes that were not detectable in mock-infected HEL cells (Fig. 1C, lane 1) were observed. The prominent double band of about 68 and 71 kDa observed by independent immunodetection methods corresponds to the HSRV pr71Gag and p68Gag proteins (Fig. 2). Taken together, the HSRV protein patterns of purified virions and infected cells showed a distinct set of defined viral polypeptides formed by proteolytic processing. Remarkably, more processed bands were detectable in infected cells than in virions (Fig. 1C). However, detection of smaller proteins and peptides requires even more sensitive techniques.
In order to define precisely the peptide sequences that span the suspected proteolytic cleavage sites, synthetic peptides were subjected to digestion with recombinant HSRV PR followed by matrix-assisted laser desorption ionization (MALDI) mass spectrometry of the cleaved substrates. The peptides were selected by combining several guidelines: (i) determination of the molecular sizes of the HSRV Gag proteins in infected cells and virions (see above), (ii) secondary-structure predictions by two different algorithms, and (iii) comparison to known cleavage sites within HSRV Pol and Gag (16). By using this approach various synthetic HSRV Gag-derived peptides (Table 1) were subjected to HSRV PR treatment, using the truncated recombinant Pol protein PR-ΔRT-His (16). This form of HSRV PR contains the N-terminally located 159 residues encoded by the HSRV pol reading frame with 20 additional residues derived from the vector backbone, including the C-terminal six-His tag (16).
TABLE 1.
Synthetic peptides derived from the HSRV Gag sequence and analyzed for susceptibility to cleavage by the HSRV PR
| No. of peptide | Peptide sequencea | Position within Gag | PR assay result | Turnover (nmol/60 min [%]) |
|---|---|---|---|---|
| 1 | PRAVNPHTMFMIS | 76–87 | Not cleaved | |
| 2 | DELEDVLNTQSEI | 141–153 | Not cleaved | |
| 3 | RSFSGLPSLPSIPGRb | 199–211 | Not cleaved | |
| 4 | PIQHIR↓SVTGE | 306–316 | Cleaved | 0.25 (15) |
| 5 | PAIDGVF↓PVTTPDLRCRIIN↓AILGGNI | 333–359 | Cleaved twice | NDc |
| 6 | PDGVF↓PVTTPDLRb | 336–347 | Cleaved | ND |
| 7 | PRCRIIN↓AILGG | 347–357 | Cleaved | 0.60 (36) |
| 8 | PGNIGLSLTPGDCLTWDS | 357–372 | Not cleaved | |
| 9 | PMHQLGNVIKG | 387–397 | Not cleaved | |
| 10 | PRAVN↓TVTQRb | 618–625 | Cleaved | 1.65 (100) |
N termini were modified by adding a Pro residue for fluorogenic assays. Residues in boldface flank the cleavage sites (arrows).
C termini were modified by adding an Arg residue for higher solubility.
ND, not done.
Analysis of peptide 4 illustrates the approach taken (Table 1). In the MALDI mass spectrum of the 11-mer peptide 4, a molecular ion at m/z 1,236.3 was observed in the control digestion with the enzymatically inactive HSRV PR D/A mutant (data not shown). The two signals observed after HSRV PR-ΔRT-His treatment correspond to the N- and C-terminal fragments PIQHIR and SVTGE, respectively. Proteolytic cleavage took place at the peptide bond between Arg311 and Ser312 (Table 1). Surprisingly, peptide 5 was cleaved twice at sites shown in Table 1. The results of the peptide analyses are compiled in Table 1, which also contains relative turnover numbers of the essential Gag cleavages. Taken together, the data obtained reveal three potential cleavage junctions within HSRV Gag besides p68/p3. Table 2 lists the processing sites and compares those experimentally identified in HSRV with those at analogous sites in related FV.
TABLE 2.
Locations of proteolytic cleavage sites of spumaviral Gag proteins
| Virusa | p33–p5 | p3.5–p1.5 | p1.5–p29 | p68–p3b |
|---|---|---|---|---|
| HFV | QHIR↓SVTG | DGVF↓PVTT | RIIN↓AILG | RAVN↓TVTQ |
| SFV-1 | QHIR↓AVTG | EGVF↓PIPT | RVIN↓ALLG | RSVN↓TVTA |
| SFV-3 | QHIR↓AVTG | EGVF↓MTTP | RVVN↓ALIG | RNVD↓TVTA |
| BFV | THIR↓AVIG | QGVY↓PVQD | RTVN↓ALTV | SAVH↓SVRL |
| FeFV | NHLR↓SVIG | EGVF↓PIVD | RRVN↓ALVA | AAVH↓TVKA |
| Consensus | XH(I/L)R↓(A/S)V(T/I)G | XGV(F/Y)↓(P/M)XXX | RX(I/V)N↓A(I/L)XX | XXVX↓(T/S)VXX |
HFV, human FV; SFV, simian FV; BFV, bovine FV; FeFV, feline FV.
The p68/p3 cleavage has been reported previously (15). Conserved residues are boldfaced.
To show that the cleavage sites identified are not used within peptide substrates only, different recombinant HSRV Gag proteins were expressed and subjected to HSRV PR assays. Plasmid clones expressing defined regions of the HSRV Gag proteins were constructed as shown in Fig. 2 and as described previously (15). The recombinant HSRV Gag His-tagged proteins were purified by Ni2+-chelate column affinity chromatography and reacted with the Gagm antiserum (Fig. 2). The resulting Western blot analysis revealed that the main recombinant Gag1 and GagT proteins had molecular sizes of about 65 and 90 kDa (Fig. 3), in agreement with the calculated values of 65.8 and 92.8 kDa, respectively, for the thioredoxin (Trx)-His-Gag fusion proteins. HSRV PR-ΔRT-His assays of the Gag1 protein showed that it was specifically cleaved, yielding the distinct products marked by arrows in Fig. 3A. The largest proteolytic cleavage product migrated as an intense band of approximately 59 kDa and corresponds to the efficient processing site of p68Gag/p3Gag at position 621 retained in the recombinant Trx-His-tagged fusion protein (Fig. 2). The size difference of about 7 kDa between this and the substrate Gag1 as starting material is in close agreement with the calculated value, since the p3Gag domain is extended by 38 residues of the vector backbone, as shown in Fig. 2 (15).
FIG. 3.
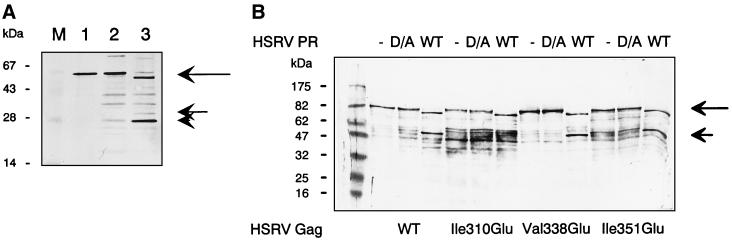
Proteolytic processing of affinity-purified recombinant HSRV Gag assayed with HSRV PR-ΔRT-His and subsequent detection by immunoblot analysis. (A) Shown are Gag1 proteins without HSRV PR treatment (lane 1), after treatment with an HSRV PR-deficient (D/A) mutant (lane 2), and after HSRV PR treatment (lane 3), and marker proteins (lane M). The long arrow marks the processed Gag1 from which the p3Gag was cleaved, the short arrow marks the specific cleavage product of 33 kDa, and the arrowhead marks the specific cleavage product of 28 kDa. (B) Proteolytic cleavage of wild-type (WT) and mutant GagT proteins Ile310Glu, Val338Glu, and Ile351Glu. Western blot analysis of GagT protein without HSRV PR treatment (lanes marked by hyphens), after treatment with an HSRV PR-deficient (D/A) mutant (lanes D/A), and after treatment with active PR (lanes WT) was performed. The long arrow marks the processed GagT cleavage products from which the p3Gag plus vector-derived part was cleaved, and the short arrow indicates the specific cleavage product of 56 kDa, corresponding to the N-terminal half of GagT plus vector-derived sequences. The Gagm antiserum was used. Left lane, marker proteins.
The second specifically cleaved intense protein band (Fig. 3A) was formed by proteolysis from p59 and migrated with a size of about 28 kDa.
The third cleavage product, of relatively low intensity, migrated at about 33 kDa (Fig. 3A) and was formed by processing at cleavage site 311/312. This result was confirmed by N-terminal amino acid sequencing of the purified 33-kDa proteolytic cleavage product (Fig. 3A) of the recombinant Gag1 protein (Fig. 2) (see below). Edman degradation revealed the N-terminal residues (NH2)-(X)VTGE (data not shown), corresponding to the sequence SVTGE of peptide 4. This sequence occurs only once in HSRV Gag, at positions 312 to 315. The intensity of the p33Gag band was low, since this cleavage event is of low efficiency in Gag1, analogous to that in the corresponding peptide, which is also poor compared to the complete cleavage of the peptide at Gag site 621; the low intensity might also be due to the relatively low titer of the Gagm antiserum used for detection, since it covers only a small part of the protein sequence of the p33 product (Fig. 2). Importantly, C-terminal cleavage products that still contained the unprocessed p3 site were not detectable, indicating that C-terminal p68Gag/p3Gag processing is a prerequisite for further proteolysis.
To confirm that additional cleavage sites did not remain undetected, purified GagT protein (Fig. 2) that comprised the complete HSRV Gag sequence was also assayed with HSRV PR. After incubation of the 90-kDa GagT protein with HSRV PR-ΔRT-His and the PR-deficient mutant, two cleavage products were observed by immunodetection with the Gagm antiserum (Fig. 3B). In analogy to the Gag1 protein cleavage described above, the reaction product of about 82 kDa was formed by cleaving off the C-terminal part, thereby removing approximately 7 kDa from GagT (Fig. 3B). The second cleavage product migrated with a size of about 56 kDa and was due to the same cleavage event at site 311/312 that was described above for Gag1. To rule out unspecific proteolytic cleavages, the PR-defective D/A mutant was used (lanes D/A).
To further confirm the biological significance of the three Gag cleavage sites, site-directed mutagenesis was used. We chose to mutate the P2 position of the substrate GagT protein in each case (for definition of P sites, see reference 19) and performed immunoblot analysis after treatment with recombinant HSRV PR. The result shown in Fig. 3B indicates that when P2, i.e., residue 310, was changed from Ile to Glu, site 311/312 was resistant to proteolytic cleavage with wild-type PR. Cryptic processing events occurred less efficiently at both downstream sites but were barely detectable (data not shown). When either residue Val338 or residue Ile351 was mutated to Glu, the mutant GagT protein showed almost the same cleavage patterns as wild-type GagT. Peptides with the corresponding mutations (data not shown) were completely resistant to proteolysis, indicating that the 311/312 site is the major central cleavage site and that once the 311/312 site was mutated, cryptic and inefficient cleavages at the remaining sites occurred.
The three Gag cleavage site mutations were introduced into the infectious pHSRV13 provirus DNA clone (9a) and analyzed after transfection into BHK cells. Upon titration of extracellular virions on FAB cells (23), the three HSRV proviruses with mutations at the P2 positions of cleavage sites 311/312, 339/340, and 352/353 consistently showed no infectivity in contrast to wild-type pHSRV13.
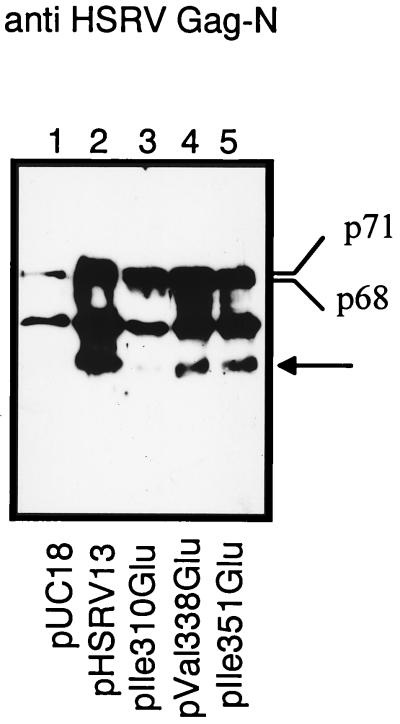
Western blot analysis of proteins from the supernatant virus particles after centrifugation through 20% sucrose showed virus release, as pr71Gag and p68Gag were detectable (Fig. 4). In addition, the p33 proteins were clearly detectable in virions from pHSRV13-, pVal338Glu-, and pIle351Glu-transfected cells. This band was virtually absent in virus particles from pIle310Glu, indicating that cleavage at the 311/312 site was suppressed. These results show that assembly and release of virions were not abrogated in the proviral Gag mutants, and they are in agreement with the cleavage data of the recombinant GagT. Taken together, the data indicate the biological relevance of specific proteolytic processing at the three Gag processing sites and point to a prominent role of the 311/312 cleavage site.
FIG. 4.
Proteolytic processing of HSRV Gag proviral mutants and subsequent detection by immunoblot analysis. Plasmids pHSRV13 (lane 2), pIle310Glu (lane 3), pVal338Glu (lane 4), pIle351Glu (lane 5), and pUC18 (lane 1) were electrotransfected into BHK-21 cells. Virions from cell supernatants were centrifuged through 20% sucrose 3 days after transfection and analyzed by immunoblotting with antiserum Gagn. Antigens from pHSRV13-derived virions were loaded at only 1/20 the amount of those used for mutant viruses to compensate for the reduced amounts of mutant viruses. The arrow marks the position of the p33Gag proteins.
In this study we have carried out analyses to define the cleavage junctions of HSRV Gag proteins. The HSRV pr71Gag is efficiently but not completely cleaved at the p68Gag/p3 cleavage site. When wild-type and mutant recombinant Gag were subjected to HSRV PR, a secondary cleavage site at position 311/312 was identified. This site was found to be resistant to the viral PR upon mutagenesis and could not be functionally substituted for by the two other cleavage sites further downstream. The data of another group showed that the p68Gag/p3 cleavage site is required for viral replication (5). This primary cleavage is followed by secondary processing events described here that might be required for maturation and/or disassembly.
Site-directed mutagenesis of cleavage sites at positions 311/312, 339/340, and 352/353 in the context of the infectious HSRV provirus led to noninfectious viral genomes, indicating an important role of these sites in the virus life cycle, although formally other functions of the mutated residues cannot be ruled out. The predicted sizes of the resulting cleavage products are in agreement with reports on the cleavage patterns of HSRV Gag of lysates from virus-infected BHK, COS, and other cells (1, 2, 6, 7, 11–14). Combining the data on processed HSRV proteins of virus particles and virus-infected cells reported here with those of the cleavage sites of peptides, mutant recombinant Gag proteins, and mutant proviruses, the following processed HSRV Gag proteins were detectable: p39/p29 and p34/p33, besides p68Gag/p3Gag. We assume that the 311/312 cleavage site may be utilized during disassembly.
The data on mutant Gag proteins seem to indicate two cleavage sites; processing close to the C terminus is a primary cleavage, whereas the remaining central cleavages in Gag are secondary events; in vivo they might require viral and/or cellular cofactors. The reaction products of slightly different molecular sizes detectable in virus particles and in virus-infected cells might be due to posttranslational modification(s). The mechanism of regulation of proteolytic processing during infection remains unknown. A mechanism of controlling proteolytic processing that also includes viral inhibitors cannot be ruled out and requires more experiments.
Judging from the relative degrees of cleavage efficiency of the peptides analyzed, which seem to reflect that of the wild-type Gag proteins, some processing sites are apparently preferred. In most cases hydrophobic residues are the preferred amino acids at the P2 and P2′ sites. When one takes the cleavage junctions of the HSRV Pol polyprotein into account (16), this preference for branched hydrophobic residues at the two P2 positions is pronounced. In particular, Val and Ile residues are by far the most conserved amino acids. In general, these observations are not inconsistent with the rules established for the specificity of other retroviral cleavage sites (16–19, 21, 24).
Acknowledgments
We thank David Baldwin and Maxine Linial for providing Gag antisera, Hajo Delius for sequencing, Andrea Wagner for providing cell cultures, Helmut Bannert for excellent technical assistance, Jennifer Reed for critical reading of the manuscript, and Harald zur Hausen for support.
REFERENCES
- 1.Aguzzi A, Wagner E F, Netzer K O, Bothe K, Anhauser I, Rethwilm A. Human foamy virus proteins accumulate in neurons and induce multinucleated giant cells in the brains of transgenic mice. Am J Pathol. 1992;142:1061–1072. [PMC free article] [PubMed] [Google Scholar]
- 2.Bartholomä A, Muranyi W, Flügel R M. Bacterial expression of the capsid antigen domain and identification of native Gag proteins from spumavirus-infected cells. Virus Res. 1992;23:27–38. doi: 10.1016/0168-1702(92)90065-h. [DOI] [PubMed] [Google Scholar]
- 3.Bodem J, Löchelt M, Winkler I, Flower R T, Delius H, Flügel R M. Characterization of the spliced pol transcript of feline foamy virus: the splice acceptor of the pol transcript is located in gag of foamy viruses. J Virol. 1996;70:9024–9027. doi: 10.1128/jvi.70.12.9024-9027.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Enssle J, Jordan I, Mauer B, Rethwilm A. Foamy virus reverse transcriptase is expressed independently from the Gag protein. Proc Natl Acad Sci USA. 1996;93:4137–4141. doi: 10.1073/pnas.93.9.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enssle J, Fischer N, Moebes A, Mauer B, Smola U, Rethwilm A. Carboxy-terminal cleavage of the human foamy virus Gag precursor molecule is an essential step in the viral life cycle. J Virol. 1997;71:7312–7317. doi: 10.1128/jvi.71.10.7312-7317.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giron M-L, Colas S, Wybier J, Rozain F, Emanoil-Ravier R. Expression and maturation of human foamy virus Gag precursor polypeptides. J Virol. 1997;71:1635–1639. doi: 10.1128/jvi.71.2.1635-1639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hahn H, Baunach G, Bräutigam S, Mergia A, Neumann-Haefelin D, Daniel M D, McClure M O, Rethwilm A. Reactivity of primate sera to foamy virus Gag and Bet proteins. J Gen Virol. 1994;75:2635–2644. doi: 10.1099/0022-1317-75-10-2635. [DOI] [PubMed] [Google Scholar]
- 8.Konvalinka J, Löchelt M, Zentgraf H, Flügel R M, Kräusslich H-G. Active spumavirus proteinase is essential for virus infectivity but not for formation of the Pol polyprotein. J Virol. 1995;69:7264–7268. doi: 10.1128/jvi.69.11.7264-7268.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Linial M L. Foamy viruses are unconventional retroviruses. J Virol. 1999;73:1747–1755. doi: 10.1128/jvi.73.3.1747-1755.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Löchelt M, Zentgraf H, Flügel R M. Construction of an infectious DNA clone of the full-length human spumaretrovirus genome and mutagenesis of the bel1 gene. Virology. 1991;184:43–54. doi: 10.1016/0042-6822(91)90820-2. [DOI] [PubMed] [Google Scholar]
- 10.Löchelt M, Muranyi W, Flügel R M. Human foamy virus genome possesses an internal, Bel 1-dependent and functional promoter. Proc Natl Acad Sci USA. 1993;90:7317–7321. doi: 10.1073/pnas.90.15.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Löchelt M, Flügel R M. The molecular biology of primate spumaviruses. In: Levy J A, editor. The Retroviridae. Vol. 4. New York, N.Y: Plenum Press; 1995. pp. 239–292. [Google Scholar]
- 12.Löchelt M, Flügel R M. The human foamy virus pol gene is expressed as a Pro-Pol polyprotein and not as a Gag-Pol fusion protein. J Virol. 1996;70:1033–1040. doi: 10.1128/jvi.70.2.1033-1040.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morozov V A, Copeland T D, Nagashima K, Gonda M A, Oroszlan S. Protein composition and morphology of human foamy virus intracellular cores and extracellular particles. Virology. 1997;228:307–317. doi: 10.1006/viro.1996.8379. [DOI] [PubMed] [Google Scholar]
- 14.Netzer K-O, Rethwilm A, Maurer B, ter Meulen V. Identification of the major immunological structural proteins of human foamy virus. J Gen Virol. 1990;71:1237–1241. doi: 10.1099/0022-1317-71-5-1237. [DOI] [PubMed] [Google Scholar]
- 15.Pfrepper K-I, Löchelt M, Schnölzer M, Flügel R M. Expression and molecular characterization of an enzymatically active recombinant human spumaretrovirus protease. Biochem Biophys Res Commun. 1997;237:548–553. doi: 10.1006/bbrc.1997.7187. [DOI] [PubMed] [Google Scholar]
- 16.Pfrepper K-I, Rackwitz H-R, Schnölzer M, Heid H, Löchelt M, Flügel R M. Molecular characterization of proteolytic processing of the Pol proteins of human foamy virus reveals novel features of the viral protease. J Virol. 1998;72:7648–7652. doi: 10.1128/jvi.72.9.7648-7652.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rao J K M, Erickson J W, Wlodawer A. Structural and evolutionary relationships between retroviral and eukaryotic aspartic proteinases. Biochemistry. 1991;30:4663–4671. doi: 10.1021/bi00233a005. [DOI] [PubMed] [Google Scholar]
- 18.Swanstrom R, Wills J W. Synthesis, assembly and processing of viral proteins. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 19.Vogt V. Proteolytic processing and particle maturation. Curr Top Microbiol Immunol. 1997;214:95–131. doi: 10.1007/978-3-642-80145-7_4. [DOI] [PubMed] [Google Scholar]
- 20.Winkler I, Bodem J, Haas L, Zemba M, Delius H, Flower R, Flügel R M, Löchelt M. Characterization of the genome of feline foamy virus and its proteins shows distinct features different from those of primate spumaviruses. J Virol. 1997;71:6727–6741. doi: 10.1128/jvi.71.9.6727-6741.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wlodawer A, Erickson J. Structure-based inhibitors of HIV-1. Annu Rev Biochem. 1993;62:543–585. doi: 10.1146/annurev.bi.62.070193.002551. [DOI] [PubMed] [Google Scholar]
- 22.Yu S F, Baldwin D N, Gwynn S R, Yendapalli S, Linial M L. Human foamy virus replication—a pathway distinct from that of retroviruses and hepadnaviruses. Science. 1996;271:1579–1582. doi: 10.1126/science.271.5255.1579. [DOI] [PubMed] [Google Scholar]
- 23.Yu S F, Linial M L. Analysis of the role of the bel and bet open reading frames of human foamy virus by using a new quantitative assay. J Virol. 1993;67:6618–6624. doi: 10.1128/jvi.67.11.6618-6624.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zabransky A, Andreansky M, Hruskova-Heidingsfeldova O, Havlicek V, Hunter E, Rum T, Pichova I. Three active forms of aspartic proteinase from Mason-Pfizer monkey virus. Virology. 1998;245:250–256. doi: 10.1006/viro.1998.9173. [DOI] [PubMed] [Google Scholar]
- 25.Zemba M, Wilk T, Rutten T, Wagner A, Flügel R M, Löchelt M. The carboxy-terminal p3Gag domain of the human foamy virus Gag precursor is required for efficient virus infectivity. Virology. 1998;247:7–13. doi: 10.1006/viro.1998.9234. [DOI] [PubMed] [Google Scholar]