Abstract
Respiratory infections, whether chronic or acute, are frequent in both children and adults and result in an economic burden in health care systems. In particular, for an immunocompromised patient, respiratory infection leads to acute hypoxemic respiratory failure, a leading cause of intensive care unit (ICU) admission. Most respiratory infections are caused by bacteria, viruses, parasites, smoking, or air pollution. Over the last two decades, considerable improvements have been made in understanding and identifying respiratory infections. Various biosensing techniques have been developed with a range of targets to identify the infection at earlier stages. Recently, nanomaterials have been effectively applied to improve biosensors and their analytical performances. This review discusses recent biosensor developments for identifying respiratory infections caused by viruses and bacteria assisted by different types of nanomaterials and target molecules.
Keywords: Respiratory infections, Biosensor: viruses, Bacteria, Nanomaterial
1. Introduction
Respiratory infection is a complication in the breathing system involving the throat, sinuses, lungs, and airways. Respiratory infections include upper and lower infections of the respiratory tract. Upper respiratory infection generally infects the throat and sinuses and causes the common cold, laryngitis, epiglottitis, sinus infection, and sore throat. Lower respiratory infection affects the lungs and airways. In most cases, lower respiratory infection has a longer duration and causes more serious health issues, such as bronchitis (lung infection causing fever and coughing), bronchiolitis, pneumonia, and chest infection. Respiratory infections are generally caused by viruses, bacteria, and fungi. Viruses are the primary causes of bronchiolitis, whereas Streptococcus pneumoniae (bacteria) is the common cause of pneumonia. Viruses or bacteria enter the respiratory tract through inhaled droplets and invade the mucosa. Respiratory viruses may cause severe health issues, and they can induce acute respiratory failure that could lead to acute respiratory distress syndrome. In particular, respiratory infection is more serious in immunocompromised patients. Acute hypoxemic respiratory failure (ARF) is most common in immunocompromised patients and a major cause of intensive care unit (ICU) admission [1]. This increases the economic burden on the health care system and elevates mortality and morbidity. Over the last two decades, considerable improvement has been made in understanding disease infection mechanisms, strategies for infection control, and identification of diseases, which could help to reduce the burden in the health care system. At the same time, early identification of respiratory infections helps to choose the right treatment and control the spread of diseases. Methods such as sputum tests, tuberculin skin tests, spirometry, chest X-ray, pulse oximetry, computed tomography, bronchoscopy, and pleural fluid culture are commonly used methods to identify respiratory infections and their conditions (Fig. 1). However, these methods are expensive and inconvenient for patients, and it is necessary to develop an easy identification system for respiratory infections.
Fig. 1.
Respiratory diseases. Different diseases are displayed. Healthy lungs and images of the infection are shown.
Clinical evaluation, medical history, physical examination, and in certain cases, further testing is used to diagnose respiratory infections. Notably, the specific diagnostic tests employed may differ depending on the suspected respiratory infection and the available resources. A physical examination is performed by a health care practitioner to assess patient symptoms, using a stethoscope to look for indicators of respiratory infection such as abnormal lung sounds, increased respiratory rate, or nasal congestion. To test for the presence of certain viral or bacterial infections, a swab sample from the nose or throat is obtained. This can aid in identifying the organism responsible for the infection. A complete blood count (CBC) or C-reactive protein (CRP) test can also reveal the presence and severity of illness. These tests look for inflammatory indicators or the body's immunological response. On the other hand, a chest X-ray can aid in the diagnosis of lung abnormalities such as pneumonia or lung infiltrates. A computed tomography (CT) scan produces detailed cross-sectional images of the lungs, allowing for a more exact assessment of lung abnormalities and infection severity. Recently, detection of the genetic material or particular proteins of viruses by viral antigen tests and polymerase chain reaction (PCR) have been developed. These tests are often used to identify respiratory viral diseases such as influenza, respiratory syncytial virus (RSV), and COVID-19.
Despite many demonstrations, these sensing strategies have different hurdles, such as lower sensitivity, time-consuming processes, and the necessity of training to handle the measurements. To overcome these issues, various biosensors and biosensing strategies have been developed by researchers to identify respiratory infections at their earlier stages [[2], [3], [4]]. The generated biosensors fulfill the current demands, including label-free, accurate detection, faster diagnosis, and lower cost, and complement the current detection systems. In addition, researchers have used nanomaterials to enhance the analytical performance of biosensors. This review discusses recent biosensor developments for diagnosing respiratory infections assisted by nanomaterial-mediated biosensing.
2. Respiratory viruses
Viral pathogens are the common cause of lower and upper respiratory infections. Viruses such as influenza virus, mumps virus, rhinoviruses, parainfluenza virus, measles virus, coronavirus, and adenovirus are common viruses that cause respiratory infections [5,6]. These viruses exhibit varying severity at different levels of infection in patients, with seasonal variations and locations. Respiratory viruses are causally associated with 16–49% of patients with acute respiratory failure requiring ICU care [7,8]. Respiratory viral infections are clinically classified by the causative virus, such as influenza, and they are classified according to the resulting syndrome, which includes the common cold, croup, bronchiolitis, and pneumonia. At the same time, specific pathogens cause characteristic clinical manifestations. For example, respiratory syncytial virus (RSV) commonly causes bronchiolitis, and rhinovirus (RV) typically causes the common cold. RSV is the most common cause of respiratory tract infection among infants [9], and RV causes colds and approximately two-thirds of asthma exacerbations [10]. Influenza is a common cause of pneumonia-related death in developed countries [11]. In most cases, influenza, RSV and RV infections lead to hospitalization and ICU admission. The severity of respiratory infection widely varies; severe disease is more likely in elderly patients and infants. In particular, premature infants aged less than six months, individuals with lung disease or chronic heart disease, immunocompromised individuals, and elderly adults older than 65 are at high risk of respiratory viral infection. The severity of the symptoms can markedly vary from mild cold to serious wheezing, pneumonia, or bronchiolitis [12]. Morbidity may occur directly due to the virus or indirectly due to the exacerbation of underlying cardiopulmonary conditions.
2.1. Spectrum of infection from asymptomatic to mild disease
The range of clinical manifestations exhibited in persons infected with a specific pathogen is referred to as the infection spectrum, which ranges from asymptomatic to mild infectious illness. Some people who are infected with a pathogen may exhibit no symptoms or only extremely mild symptoms of which they are unaware. They can, however, still spread the virus to others. Asymptomatic carriers can significantly contribute to the transmission of infectious diseases. Individuals with subclinical illness have minor symptoms that may go undiagnosed or be attributed to other causes. These symptoms are usually not severe enough to interfere with everyday activities or necessitate medical intervention. Mildly infected people often have recognizable but not severe symptoms. A low-grade fever, slight weariness, cough, sore throat, nasal congestion, headache, or body aches are all possible symptoms. These symptoms may resemble those of a normal cold or flu and are frequently self-limiting, resolving without the need for medical attention. It is important to highlight that the infection spectrum does not end with mild disease; the disease can also be moderate, severe, or critical, depending on the pathogen and individual variables. Furthermore, the intensity of symptoms and the clinical course of different viral diseases might vary greatly.
3. Basic classes of respiratory diseases
Agent-based respiratory disorders are infectious diseases spread predominantly through respiratory droplets, aerosols, or close contact with infected people. Pathogens that cause these disorders include bacteria, viruses, and fungi. Influenza viruses cause influenza, sometimes known as the flu. When an infected individual coughs, sneezes, or speaks, the virus spreads through respiratory droplets. Influenza symptoms can range from mild to severe, and complications might occur, particularly in high-risk individuals. The new coronavirus SARS-CoV-2 causes COVID-19, and the virus spreads when an infected individual coughs, sneezes, talks, or breathes heavily. COVID-19 symptoms might range from asymptomatic to severe respiratory disease, multiorgan failure, or death. The bacterium Mycobacterium tuberculosis causes tuberculosis. It primarily affects the lungs, although it can affect other organs as well. Symptoms include a chronic cough, fever, weight loss, and exhaustion. The measles virus causes the disease, which is highly contagious. Measles symptoms include a high fever, cough, runny nose, and a distinctive rash. Pertussis is a highly contagious disease caused by Bordetella pertussis. Pertussis can cause violent coughing fits, difficulties breathing, and a distinctive “whooping” sound when inhaling.
3.1. Respiratory infections caused by bacteria
Bacterial pathogens are common causes of respiratory infections. The species Mycoplasma pneumoniae, Streptococcus pneumoniae, Chlamydophila pneumoniae, and Haemophilus influenzae are common bacteria causing upper respiratory tract infections [13,14]. In addition, Legionella pneumophila and Coxiella burnetiid can cause outbreaks and sporadic cases of respiratory-related issues. To cause an infection, bacteria first need to colonize the nasopharyngeal niche. The process of colonization includes the elimination and acquisition of species, interactions between the host and the microbes, and other environmental factors that also cause the dynamic and complex interplay of microbes. Imbalances in the ecosystem may result in the invasion and overgrowth of bacterial species and cause invasion of the respiratory system, particularly in children with weaker immune systems. Bacteria can cause upper respiratory infections, such as the common cold, sinusitis, pharyngitis, epiglottitis, and laryngotracheitis. The symptoms of upper respiratory infections are sneezing, cough, fever, headache, chill, sore throat, and shortness of breath. Infections of the lower respiratory tract cause bronchitis, bronchiolitis, and pneumonia, with similar symptoms to upper respiratory infections. However, tests such as throat swabbing, chest X-ray, CT scan, and lateral neck X-ray can identify the precise infected area and help to provide the appropriate treatment; however, a more accurate sensing system is necessary.
4. Potential sensing systems for diagnosing respiratory infections
Electrochemical impedance spectroscopy (EIS) is an effective method for investigating the electrical behavior and processes in electrochemical systems. It contributes to the creation and optimization of diverse electrochemical devices and systems by providing significant insights into the underlying characteristics of materials and surfaces. EIS is commonly utilized in corrosion science, battery research, fuel cells, sensors, and materials science. EIS provides useful information about the processes that take place at the electrode–electrolyte interface. In EIS, a low-amplitude alternating current signal is supplied to the electrochemical system over a wide frequency range. The system's response is then measured, often in terms of impedance. Impedance is a complicated quantity made up of two parts: resistance and reactance. The two types of reactance are capacitive reactance and inductive reactance. EIS impedance data are often shown as a Nyquist plot or a Bode plot, where the x-axis of a Nyquist plot indicates the real part of the impedance (Z′) and the y-axis shows the imaginary part of the impedance (Z″). A Nyquist plot can be used to extract parameters such as charge transfer resistance, double-layer capacitance, and diffusion processes by providing information about processes occurring at different frequencies. A Bode plot depicts the impedance magnitude and phase angle as a function of frequency. It provides information on the frequency response of the system and can aid in understanding the dynamics of various electrochemical processes. EIS can be used to investigate the rate of electrochemical reactions at the electrode surface. The charge transfer resistance derived from impedance measurements can provide information about reaction kinetics and catalyst performance.
Cyclic voltammetry (CV) is widely utilized in domains such as electrochemistry, analytical chemistry, materials science, and biochemistry. It enables the identification and study of redox-active species, as well as the determination of electrode kinetics and the examination of electrochemical mechanisms. CV is an electrochemical technique used to evaluate the electrochemical processes occurring at an electrode surface and to study the redox behavior of electroactive species. It describes analytes' oxidation and reduction potentials, as well as their reaction rates and electron transfer processes. A voltage is applied to an electrochemical cell, and the resulting current is measured. Between the starting potential and the ending potential, the potential varies linearly with time, typically in a triangle waveform or staircase waveform. The current response is recorded while the potential is scanned, resulting in a voltammogram. A working electrode, a reference electrode, and a counter electrode are immersed in an electrolyte solution in the basic cyclic voltammetry setup. The working electrode is usually where the electrochemical reaction of interest takes place. The reference electrode offers a stable potential against which the working electrode's potential is measured. By permitting electricity to flow, the counter electrode completes the electrical circuit. The potential is initially swept in the positive direction (forward scan) from the starting potential to the ending potential. This causes electroactive species at the working electrode to undergo oxidation or reduction processes, resulting in a current response. The current direction is determined by the type of reaction (oxidation or reduction) and the possible sweep direction. The potential sweep is reversed (reverse scan) back to the starting potential once it reaches the finishing potential. This allows the electroactive species to undergo the redox reaction in the opposite direction. During the reverse scan, the ensuing current response is captured. A cyclic voltammogram is created by plotting the recorded current values against the applied voltage. The voltammogram shape and characteristics reveal important information about electrochemical processes.
4.1. Nanomaterial-mediated biosensors for diagnosing respiratory infections
Medical societies and research scientists have recently paid more attention to cost-effective biosensors to test water and food contaminants and identify diseases and human biological processes. A rapidly expanding sector of technology is biosensing, which is used in areas such as food, environmental monitoring, and medical diagnostics. Especially in the medical field, it is important to identify biomarkers at earlier stages of diseases, which helps to prevent spreading and treat the diseases. Biosensors are analytical devices used to identify biomolecular interactions by using biological components and a physiochemical detector (Fig. 2). Biosensors are used to screen biological molecules, which helps to customize medicine. An improved sensor is mainly determined by biomolecular affinity, surface modification, and sensor equipment. Researchers are working toward improving surface functionalization with biomolecules to enhance the analytical performance of biosensors. In addition, nanoscale nanomaterials are effectively applied in various sensors for biomolecular immobilization and for fabricating sensing surfaces. Various nanomaterials, including gold, silver, silica, zeolite copper, and graphene, are commonly used for biosensor development. Among them, gold is one of the major materials that improves biosensor performance due to its unique characteristics, such as biocompatibility, water dispersal, and easy surface functionalization. In addition, gold is used to fabricate sensing surfaces, and various biomarkers are detected on gold-coated sensor surfaces with a different sensor. Lakshmipriya et al. used a gold-coated sensor surface to identify the clotting protein factor IX (FIX) with the assistance of a surface plasmon resonance (SPR) sensor. The authors conducted an aptamer-antibody-based assay to identify the target protein. The thiolated aptamer was attached to the gold surface, and the target molecule (FIX) was sandwiched with an anti-FIX antibody, reaching a lower level of FIX identification [15]. In addition, gold is a very popular material in various kinds of lateral flow assays (LFAs). LFAs have been used extensively for point-of-care diagnosis of various diseases, particularly for virus detection. LFAs have many advantages, such as low cost, user friendliness, and rapid detection. Recently, researchers have focused on LFAs for COVID-19 infection [16]. Similar to gold, carbon-derived materials such as graphene, single-walled carbon nanotubes, multiwalled carbon nanotubes, and graphene oxide are used for various surface functionalizations to enhance the detection of targets [17]. Carbon-based materials show good conductivity of electricity and are mainly used in electrochemical sensors to fabricate sensor surfaces. Furthermore, silica is a common material for biosensor development due to its stability and easy surface functionalization. The APTES linker is generally used to attach biomolecules to silica surfaces. Zeolite is a composition of silica and alumina, which is an established material in electrochemical sensors due to its excellent conductivity and stability with biomolecules. Similarly, the loop-mediated isothermal amplification (LAMP) method has been utilized for the detection of microbial pathogens and viral infection [18]. Nanomaterial-utilizing LAMP methods enhance the identification of viruses and bacteria [19]. In addition, the combination of two materials can also be used to improve the biomolecular interactions on the sensing surfaces. Similar to other diseases, nanomaterial-based sensors are widely used to identify various respiratory infections and their conditions. The following sections describe some of the respiratory infections detected on nanomaterial-modified sensing surfaces.
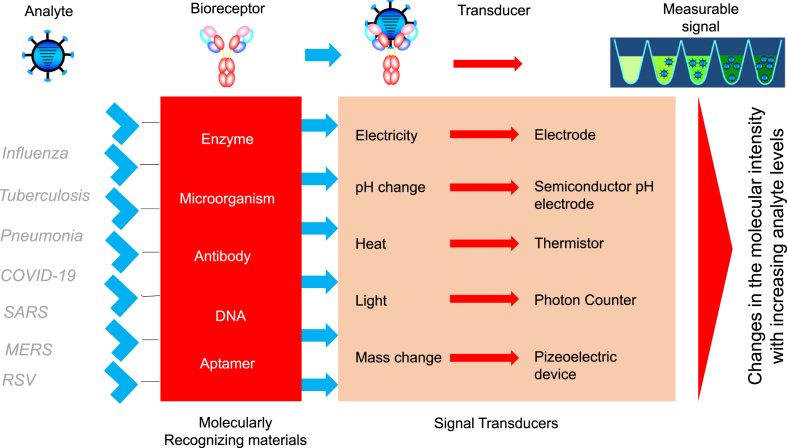
Fig. 2.
Biosensing principle. The analyte, receptor, transducer, and signal output are displayed. Biomolecular complex formation is shown.
4.2. Detection of tuberculosis
Mycobacterium tuberculosis is a threatening bacterium that causes tuberculosis (TB), one of the most devastating respiratory diseases [20]. At present, approximately one-third of humans are affected by TB worldwide. Infection by TB is a serious issue because it causes complications with acquired immune deficiency syndrome infection. Most people affected by TB live in low- and middle-income countries. Until now, TB has mainly been diagnosed with smear microscopy, which is not readily available in low-income areas. In addition, traditional culture-based tests are also used to identify M. tuberculosis, but the test involves many steps, such as cell culture, counts, and enrichments. These steps are time-consuming, less sensitive, and expensive [21]. Therefore, it is necessary to create a sensing strategy to identify TB in its earlier stages with a suitable biomarker. Various sensing strategies have been developed to identify TB biomarkers at earlier stages. ESAT-6 is an early secretory protein that is an important biomarker for TB identification. An antibody-based colorimetric assay was developed by researchers to identify the ESAT-6 protein at lower levels. First, ESAT-6 and its antibody are mixed, and then, this mixture is complexed with gold nanoparticles (GNPs). If ESAT-6 is present, it interacts with the antibodies, and then when the GNPs are added, no antibody remains to attach to the GNPs. Under this condition, NaCl is added to conduct the salt-induced colorimetric assay, and it aggregates the GNPs. In the presence of ESAT-6, the color of the solution changes to purple, while it remains red if there is no ESAT-6 in the solution (Fig. 3). This easy and simple colorimetric assay lowered the limit of detection of ESAT-6 to 1.25 pM [22]. In another study, a GNP-based ELISA was developed to quantify the level of ESAT-6. For this assay, ESAT-6 was attached to the ELISA well, and then the GNP-conjugated ESAT-6 antibody was added. Next, biotinylated secondary antibodies followed by streptavidin-conjugated HRP were added. Finally, signal amplification was monitored with a substrate for HRP. This ELISA method improved the detection of ESAT-6 and lowered the detection limit to 1 nM [23]. Similar to ESAT-6, 16 kDa heat shock protein (HSP) plays a major role in developing TB. A simple impedimetric sensor was developed by researchers to quantify the level of HSP. Anti-HSP was attached to the sensing electrode through the amine linker on the nanogapped dielectric sensor, and the interaction of HSV with its antibody was monitored by the impedimetric sensor. The detection limit of HSV was lowered to 100 fM with a detection range of 100 fM to 1 nM [24]. In addition, the clustered regularly interspaced short palindromic repeats (CRISPR) technique was used to identify viruses and bacteria. This system was used to identify M. tuberculosis, L. monocytogenes, E. coli, and S. aureus. Higher sensitivity with CRISPR techniques was achieved by isothermal nucleic acid amplification and polymerase chain reaction techniques such as SDA, RCA, LAMP, and EXPAR [25].
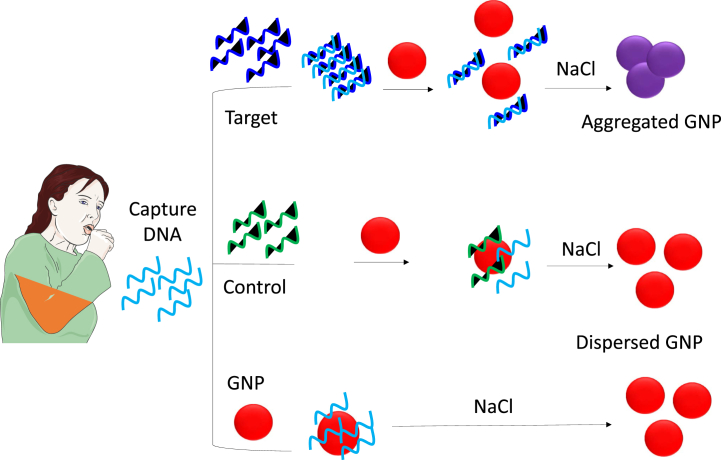
Fig. 3.
Colorimetry detection of tuberculosis. Detection of the biomarker ESAT-6 with controlled assembly and disassembly of gold nanoparticles is described. If the target is present, gold nanoparticles aggregate and display a purple colored solution, whereas if no target is present, the solution remains red in the presence of NaCl.
4.3. Detection of pneumonia
Lower respiratory infections, such as nosocomial pneumonia and acquired pneumonia, are another major cause of death worldwide. More specifically, viral respiratory infection is the major cause of mortality and morbidity among critically ill patients. Acute respiratory infections, pneumonia in particular, cause death in both children and adults, and a sharp peak of mortality due to respiratory infection is observed in infancy and later adulthood. Approximately 1.4–1.8 million fatal cases per year are registered, and pneumonia causes more fatalities than malaria, AIDS, and measles combined. Therefore, identifying the earlier stages of pneumonia helps to provide the necessary treatment and improve the condition of the patient. Various biosensors have been developed by researchers to identify pneumonia biomarkers [26]. Mycoplasma pneumoniae (M.pne) is a dangerous pathogen that affects the respiratory tract and causes pneumonia, bronchiolitis, and tracheobronchitis. A gold nanoparticle-based colorimetric assay was developed by researchers to detect the M. pne target gene. Captured DNA was duplexed with target DNA, and then GNPs were added. In the presence of the target, duplex formation occurred, and it could not attach to the GNPs. Under high salt conditions, the solution color changes to purple due to GNP aggregation. In the absence of a target, captured DNA can attach to the surface of the GNPs and prevent aggregation under high salt conditions. This easy colorimetric assay helps to identify M. pne and diagnose respiratory infections [27] (Fig. 4). Procalcitonin is a peptide that is widely used as a biomarker for pneumonia identification. Research has proven that procalcitonin levels in serum are elevated during bacterial infection but not during viral infection, which helps to distinguish common pneumonia from viral pathogens [28,29]. Zhu et al. (2022) constructed a boronated affinity recognition-enhanced dynamic light scattering (DLS) sensor with an antibody-modified magnetic nanoparticle to detect procalcitonin. The serum samples generate the DLS signal upon binding of procalcitonin with its antibody, and the limit of detection was 0.03 pg/mL [30]. Staphylococcus aureus (S. aureus) is a gram-positive bacterium and is responsible for hospital-acquired pneumonia. Identifying bacterial infections in the earlier stages helps control them and avoid hospitalization [31]. Researchers developed a sensing mechanism for the proteolytic activity of S. aureus proteases on a peptide substrate sandwiched between the gold surface and magnetic beads on top of a paper substrate. An external magnet was attached to the sensor back to accelerate the cleavage of the peptide from the sensor upon the introduction of test samples. The color changes from the dissociation of the bead moieties were identified by naked-eye detection at as low as 7 CFU/mL in pure broth culture [32].
Fig. 4.
Colorimetric detection of oligonucleotides as targets. Detection by complementation with controlled assembly and disassembly of gold nanoparticles is described. In principle, double-stranded oligonucleotides do not attach to gold nanoparticles, whereas single-stranded oligonucleotides do.
4.4. Detection of influenza
Influenza, called the flu, is caused by the influenza virus, which infects part of the respiratory tract, including the throat, nose, and lungs. According to the World Health Organization, more than 2 million people are affected by influenza viruses every year. Unless other viral infections occur, influenza causes more severe illness and possibly leads to life-threatening conditions. The influenza virus spreads through respiratory droplets by sneezing and coughing and also through touching contaminated surfaces and entering the body through the nose, eyes, or mouth. Influenza is categorized into three types, influenza A, influenza B, and influenza C. Among them, influenza A and B are common to humans, while influenza C is not. The genome of the influenza virus encodes 11 proteins, of which hemagglutinin, neuraminidase, matrix protein, and nucleoprotein are the major proteins [33]. Within influenza A and B, there are various types of influenza categorized by their surface proteins hemagglutinin (H) and neuraminidase (N). For example, the swine flu, H1NI, is a more dangerous type that has killed many people. Rapid diagnosis of influenza tests is available on the market, but these tests can only differentiate between influenza A and B, and they cannot differentiate the subtypes within influenza viruses. Researchers have worked toward identifying the seasonal variations of influenza viruses with developed biosensors. In general, antibodies were used as the probes to develop the influenza biosensor. Researchers developed a novel influenza sensor for the matrix protein M1. An anti-M1-modified diamond electrode was used to identify the M1 protein on electrochemical impedance spectra with a limit of detection of 1 fg/mL [34]. Antibody-based sensors can identify the influenza virus; however, they can only detect the types of influenza virus. Antibodies display difficulty in differentiating the subtypes of influenza viruses. Therefore, researchers have recently focused on aptamer-based biosensors for diagnosing influenza. The aptamer is a nucleic acid, and either DNA or DNA can be selected from the randomized library of molecules by using a method called SELEX. The aptamer is often called ‘an artificial antibody’; it can work like an antibody and be used in various applications instead of antibodies. Since aptamers can only bind with the smaller region of the target molecule, they can differentiate closely related molecules. At the same time, multiple aptamers can be used for a particular target, and many aptamers with different binding sites have been generated [[35], [36], [37]]. This will help to develop a sandwich aptamer-aptamer biosensor and lower the detection limit. Many aptamers against influenza A and B have been selected and used for sensor development. Lakshmipriya et al. produced an aptamer for influenza B Tokio intact virus and used it to identify the selected aptamers with fluorescent-based surface plasmon resonance spectroscopy. A silica surface was used as the sensing substrate, which was modified with influenza virus through the amine-aldehyde linker, and then aptamer was added on the surface. The interaction of aptamer and influenza virus was identified by the fluorescence of cy5. The authors found that the aptamers showed a greater affinity, and the detection limit of influenza with aptamer was found to be 8 ng/mL [38]. In another study, an aptamer was selected against influenza A H3N2, and detection analysis was conducted using SPR. The selected aptamers showed a 15-fold higher affinity than anti-HA antibodies and could differentiate closely related strains such as A/Wuhan/359/95 and A/Sydney/05/97 [39]. Similarly, a highly selective RNA aptamer was generated against influenza A HA, and its binding affinity was identified on an SPR-based Biacore system. The HA protein is needed for membrane fusion with host cells to mediate the early stages of influenza infection. The selected aptamers against HA not only help to distinguish the influenza viruses but also help to inhibit the HHA-mediated membrane protein [40]. In another study, an anti-A H3N2 antibody was attached to GNPs and used as the detection probe to identify influenza A on wave in the guide mode sensor. Gold conjugate complexed with the viruses and formed a ring of nanodots on the membrane of the viruses, and it had a limit of detection for influenza viruses of 8 × 103 PFU/mL [41]. Not only aptamers but also some antibodies can potentially discriminate closely related influenza viruses. Anti-influenza A H3N2 was used as a probe to distinguish other influenza viruses. The results proved that it can discriminate the other influenza A and B viruses, which helps in the accurate identification of influenza viruses [42] (Fig. 5).
Fig. 5.
Waveguide mode detection of influenza virus. The detection with the waveguide setup is shown. It is an optical detection system based on surface plasmons; however, it is a guiding mode of light reflection on the prism.
4.5. Detection of SARS-CoV-2
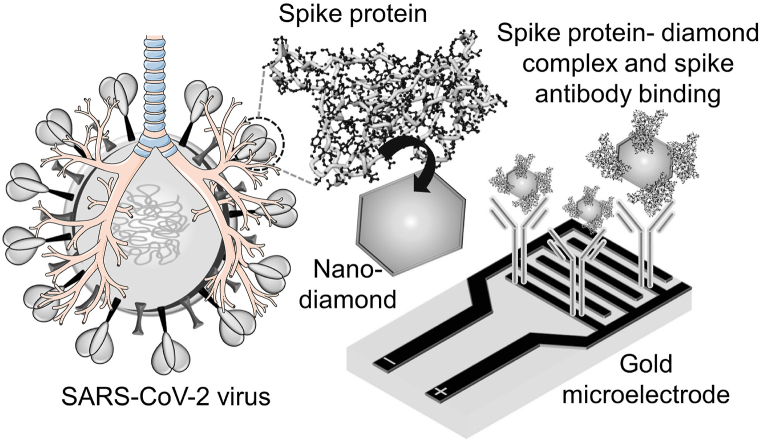
In February 2020, the WHO announced the outbreak situation due to severe acute syndrome coronavirus (SARS), which causes a respiratory disease named COVID-19 [[43], [44], [45], [46]]. COVID-19 led to a panic situation due to the lack of suitable medication [47,48]. COVID-19 belongs to the β coronavirus family and is the 7th known coronavirus that affects humans. Among these, SARS-CoV, SARS-CoV-2, and MERS-CoV can cause severe symptoms, and the fatality rates are 10, 5, and 37%, respectively. Identification of COVID-19 at its earlier stages helps to avoid spreading the virus and to save patient lives. At the same time, new virus strains are emerging, and it is difficult to develop a suitable sensor for diagnosing COVID-19. In this crucial situation, most sensors rely on polymerase chain reaction (PCR)-based testing, but it is expensive and time-consuming, and researchers are working toward developing a highly sensitive COVID-19 biosensor with various COVID-19 biomarkers. Ramanathan et al. (2022) used antibody-based electrochemical impedance spectroscopy to detect the surface spike protein of coronavirus. The anti-spike protein antibody was attached to the electrode through the GOPTS chemical linker. The electrode was modified with GOPTS to attach the anti-spike protein antibody. At the same time, spike protein was immobilized on the surface of the diamond through the CDI linker. The modified diamond-spike protein was allowed to interact with the antibody-modified surface, and current responses were monitored by impedance spectroscopy. The limit of detection of the spike protein was calculated to be 189 fM [49] (Fig. 6). In another study, a carbon nanodot-modified interdigitated electrode was used to detect SARS-COVID-19-specific miRNA. The capture miRNA sequence was attached to the electrode through the APTES amine linker and then detected by its target DNA [43]. Furthermore, researchers developed a field effect transistor (FET)-based biosensor for diagnosing COVID-19 in clinical samples. The sensing surface was developed by immobilizing a graphene sheet with an anti-COVID-19 spike protein for detecting the antigen spike protein in the cultured virus and nasopharyngeal swab specimens from COVID patients. The FET sensor identified the spike protein at 100 fg/mL in clinical transport medium and 1 fg/mL in phosphate-buffered saline. The limit of detection of the spike protein was calculated as 1.6 × 101 PFU/mL in culture medium and 2.42 × 102 copies/mL in clinical samples [50]. In another study, a smartphone-based portable system was developed by researchers for the rapid identification of pathogens such as bacteria and viruses via the application of colorimetric loop-mediated isothermal amplification (LAMP). This method of detection involves purifying the viral samples with specific reagents, which are preconjugated with magnetic beads; lysing the virus; executing isothermal nucleic acid amplification; and quantifying the result with a colorimetric assay. The limit of detection of the viral antigen was 3.2 × 10−3 units per reaction, which helps to identify the virus at its earlier stages [51]. It should be noted that the development and commercialization of electrochemical sensors for COVID-19 detection are ongoing research and development projects. While certain electrochemical sensors have been proposed and are in the works. These sensors identify specific biomarkers or antigens linked with COVID viruses and the detected signal is analysed and interpreted to determine the existence or concentration of SARS-CoV-2-associated biomarkers [[52], [53], [54], [55]].
Fig. 6.
Detection of SARS-CoV-2 (COVID). The detection of spike protein in the interdigitated electrode sensor is clearly shown. The interdigitated electrode has the desired number of fingers with proper gaps, and the output signal changes with the alteration of the surface ions.
4.6. Detection of respiratory syncytial virus (RSV)
Respiratory syncytial virus (RSV) is a single standard RNA virus that belongs to the family Paramyxoviridae. There are two antigenic subtypes, RSV-A and RSV-B. The genome of RSV encodes various structural and nonstructural proteins, including small hydrophobic proteins, protein G, fusion protein, and glycoproteins. Viral entry is mediated by the interaction of the F protein with the host cell nucleolin [56]. RSV is one of the most common causes of respiratory infections in infants, young children, and elderly people [57,58]. RSV infection leads to croup, pneumonia, and bronchiolitis. In addition, RSV causes nosocomial diseases and significantly impacts hospitalization [59]. Researchers have developed various biosensing methods for diagnosing RSV infection. A label-free electrochemical sensor with screen-printed gold and carbon disks was developed for rapid identification of RSV. F protein was used as the target, and anti-F protein was used as the detection probe. The interaction of the target and antibody was monitored by electrochemical impedance spectroscopy (EIS) and cyclic voltammetry (CV). The limit of detection of F protein on glossy carbon was 2.85 × 106 PFU/mL, and it was lowered to 1.1 × 103 PFU/mL with a gold electrode [60]. In another study, specific and sensitive detection and differentiation of subgroups of RSA, such as A and B, was performed by using colorimetric toehold switch biosensors. The authors used an RNA-based riboswitch to regulate the translation activity of the β-galactosidase gene. In the presence of the target RNA, the sensor was activated, and the expression of β-galactosidase-hydrolyzed chromogenic substrates produced a colorimetric result that could be observed directly with the naked eye [61]. Furthermore, research was focused on the DNA hairpin structure to identify RSV. The DNA hairpin was covalently attached to the gold filament, and the gold filament served to create fluorescence-based detection. The filament with probes was dipped in a capillary tube containing viral RNA, moved to the capillary tube for washing, and scanned for fluorescence. This method lowered the detection limit of RSV to 11.9 PFU [62]. A highly selective immunosensor was developed by accompanying catalyzed reporter deposition with a gold nanoparticle-deposited sensing surface, which amplified the anti-RSV antibody. The immunosensor could detect RSV as low as 0.01 pg/mL with a detection range of 0.05–30 pg/mL [63]. In another study, carbon-printed sensors and monoclonal RSV antibodies were coated on a polystyrene microarray plate for the rapid identification of RSV. The signal was amplified by horseradish peroxidase on a thin layer-based amperometric enzyme immunoassay, and this method can be compared with the commercially available PCR assay [64].
5. Validations in clinical settings
Clinical validation is a complete process that includes clinical evaluation, patient history, physical examination, diagnostic testing, and medical assessment. These multifaceted methods assist health care workers in making accurate diagnoses, determining the severity of respiratory infections, and determining the best treatment strategies. The process of establishing the presence of a respiratory infection and measuring its severity and influence on an individual's health by clinical examination, diagnostic testing, and medical assessment is referred to as clinical validation of respiratory infections. To learn more about respiratory infection, health care experts examine the patient's symptoms, medical history, and physical examination findings. Cough, fever, shortness of breath, nasal congestion, sore throat, lethargy, or body aches are all possible symptoms. The severity and duration of symptoms are critical in determining the type of illness. Potential causes of respiratory symptoms, such as allergies, asthma, chronic obstructive pulmonary disease (COPD), or other noninfectious respiratory disorders, must be considered during clinical validation. It is critical to distinguish between viral and bacterial infections to make proper treatment decisions. Several diagnostic tests can help with the clinical validation of respiratory infections. Among these tests are molecular tests, serological tests, imaging studies and laboratory diagnostics.
A health care practitioner assesses the patient's general health, vital signs, and laboratory test findings. Blood tests such as complete blood count, C-reactive protein, and procalcitonin levels can reveal information regarding inflammation, bacterial infection, or systemic response to infection. The severity of respiratory distress and oxygenation status must be assessed to determine the necessary amount of medical intervention. At the molecular level, PCR tests are routinely used to determine the presence of certain viral or bacterial genetic material. Pathogens such as influenza viruses, respiratory syncytial virus (RSV), SARS-CoV-2 (the virus that causes COVID-19), and Streptococcus pneumoniae can be identified with these assays. Serological tests look for antibodies that are created by the body's immunological response to a specific illness. They can be used to determine previous or current exposure to specific respiratory infections. In respiratory infections, chest X-rays or computed tomography scans of the chest may be used to determine the existence and amount of lung involvement. These imaging examinations can aid in the diagnosis of pneumonia, lung inflammation, and other abnormalities. Health care professionals can establish an appropriate treatment plan based on the clinical examination and diagnostic test results. This could include antiviral or antibiotic drugs, supportive care to manage symptoms, and precautions to prevent illness from spreading to others. In addition, the diagnostic methods discussed above play encouraging roles in complementing routine clinical diagnosis in a high-performance manner with the involvement of different nanomaterials (Table 1).
Table 1.
List of biosensors for identifying respiratory infection biomarkers.
| Disease | Biosensor | Target | Probe | Material | Limit of detection | References |
|---|---|---|---|---|---|---|
| Pneumonia | Colorimetric assay | DNA | DNA | Gold | 1 pM | [27] |
| Pneumonia | DLS | Procalcitonin | Antibody | Magnetic | 0.03 pg/mL | [30] |
| Pneumonia | Colorimetric assay | S. aureus | Peptide | Gold and magnets | 7 CFU/mL | [32] |
| Influenza | LFA | Influenza virus | Aptamer and antibody | Gold | 2 × 106 virus particles | [65] |
| Influenza | Impedance sensor | Matrix protein | Antibody | Diamond | 1 pg/mL | [34] |
| Influenza A | Waveguide mode sensor | Influenza A virus | Aptamer | Gold | 8 × 103 PFU/mL | [41] |
| Influenza | LFA | Capsid protein | Antibody | Gold | 1:10,000 dilution | [66] |
| Influenza A | LOOP | cDNA | DNA | – | – | [67] |
| Influenza B | SPR | Influenza B virus | Aptamer | Silica | 7 ng/mL | [38] |
| SARS CoV-19 | Electrochemical sensor | Spike protein | Antibody | Gold | 1 pg/mL | [68] |
| SARS-CoV-2 | FET sensor | Spike protein | Antibody | Graphene | 1.6 × 101 PFU/mL | [50] |
| COVID-19 | Impedance spectroscopy | Spike protein | Antibody | Diamond | 189 fM | [49] |
| RSV | Cyclic voltammetry | F protein | Antibody | Gold | 1.1 × 103 PFU/mL | [60] |
| RSV | Cyclic voltammetry | F protein | Antibody | Glossy carbon | 2.85 × 106 PFU/mL | [60] |
| RSV | Fluorescence | DNA | DNA | Gold | 11.9 PFU | [62] |
| RSV | CARD | RSV | Antibody | Gold | 0.01 pg/mL | [63] |
| Salmonella | CRISPR | DNA | DNA | Gold | 1 CFU/mL | [25] |
| S. typhimurium | CRISPR | DNA | DNA | Dyna beads | 20 CFU/mL | [69] |
| TB | Colorimetric assay | ESAT-6 | Gold | 1.25 pM | [22] | |
| TB | Impedance | 16 kDA HSP | Antibody | Gold | 100 fM | [18] |
| TB | Electrochemical sensor | Aptamer | DNA | Gold | 100 CFU/mL | [70] |
| Staphylococcus aureus | CRISPR | MecA gene | Guide RNA | Silver | 10 fM | [71] |
6. Conclusion
Respiratory viruses cause infection in parts of the respiratory system, such as the throat, sinuses, lungs, and airways, and lead to various health complications, including difficulty breathing. Severe respiratory infection is the leading cause of intensive care unit admission in infants, children, and elderly individuals who are immunocompromised. Most of the time, viruses, bacteria, and fungi cause respiratory infections. Early identification of the infection helps to avoid ICU admission and speed recovery. Various biosensing methods have been used for the early identification of respiratory infections. In addition, nanomaterial-modified sensors show greater efficiency in biomarker identification. In this review, we discussed respiratory infections and recent biosensor development with nanomaterial-based systems. The implementation of nanoparticles has had several successes and yielded good surveillance systems. The current developments are continuing with attempts to generate multiplex detection of different subtypes or among different respiratory viruses.
Author contribution statement
All authors listed have significantly contributed to the development and the writing of this article.
Data availability statement
No data was used for the research described in the article.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Azoulay E., Mokart D., Kouatchet A., Demoule A., Lemiale V. Acute respiratory failure in immunocompromised adults. Lancet Respir. Med. 2019;7:173–186. doi: 10.1016/S2213-2600(18)30345-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang Z., Feng B., Xu J., Qing T., Zhang P., Qing Z. Graphene biosensors for bacterial and viral pathogens. Biosens. Bioelectron. 2020;166 doi: 10.1016/j.bios.2020.112471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qureshi A., Niazi J.H. Biosensors for detecting viral and bacterial infections using host biomarkers: a review. Analyst. 2020;145:7825–7848. doi: 10.1039/d0an00896f. [DOI] [PubMed] [Google Scholar]
- 4.Ribeiro B.V., Cordeiro T.A.R., Oliveira e Freitas G.R., Ferreira L.F., Franco D.L. Biosensors for the detection of respiratory viruses: a review. Talanta Open. 2020;2 doi: 10.1016/j.talo.2020.100007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cameron R.J., De Wit D., Welsh T.N., Ferguson J., Grissell T.V., Rye P.J. Virus infection in exacerbations of chronic obstructive pulmonary disease requiring ventilation. Intensive Care Med. 2006;32:1022–1029. doi: 10.1007/s00134-006-0202-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luyt C.E., Combes A., Nieszkowska A., Trouillet J.L., Chastre J. Viral infections in the ICU. Curr. Opin. Crit. Care. 2008;14:605–608. doi: 10.1097/MCC.0b013e32830f1e12. [DOI] [PubMed] [Google Scholar]
- 7.Schnell D., Gits-Muselli M., Canet E., Lemiale V., Schlemmer B., Simon F., Azoulay E., Legoff J. Burden of respiratory viruses in patients with acute respiratory failure. J. Med. Virol. 2014;86:1198–1202. doi: 10.1002/jmv.23760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daubin C., Vincent S., Vabret A., Du Cheyron D., Parienti J.J., Ramakers M., Freymuth F., Charbonneau P. Nosocomial viral ventilator-associated pneumonia in the intensive care unit: a prospective cohort study. Intensive Care Med. 2005;31:1116–1122. doi: 10.1007/s00134-005-2706-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kusel M.M.H., De Klerk N.H., Holt P.G., Kebadze T., Johnston S.L., Sly P.D. Role of respiratory viruses in acute upper and lower respiratory tract illness in the first year of life: a birth cohort study. Pediatr. Infect. Dis. J. 2006;25:680–686. doi: 10.1097/01.inf.0000226912.88900.a3. [DOI] [PubMed] [Google Scholar]
- 10.Gern J.E. How rhinovirus infections cause exacerbations of asthma. Clin. Exp. Allergy. 2015;45:32–42. doi: 10.1111/cea.12428. [DOI] [PubMed] [Google Scholar]
- 11.Newton A.H., Cardani A., Braciale T.J. The host immune response in respiratory virus infection: balancing virus clearance and immunopathology. Semin. Immunopathol. 2016;38:471–482. doi: 10.1007/s00281-016-0558-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Troy N.M., Bosco A. Respiratory viral infections and host responses; insights from genomics. Respir. Res. 2016;17:156. doi: 10.1186/s12931-016-0474-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cappelletty D. Microbiology of bacterial respiratory infections. Pediatr. Infect. Dis. J. 1998;17:S55–S61. doi: 10.1097/00006454-199808001-00002. [DOI] [PubMed] [Google Scholar]
- 14.Watson K., Carville K., Bowman J., Jacoby P., Riley T.V., Leach A.J., Lehmann D. Upper respiratory tract bacterial carriage in aboriginal and non-aboriginal children in a semi-arid area of Western Australia. Pediatr. Infect. Dis. J. 2006;25:782–790. doi: 10.1097/01.inf.0000232705.49634.68. [DOI] [PubMed] [Google Scholar]
- 15.Lakshmipriya T., Horiguchi Y., Nagasaki Y. Co-immobilized poly(ethylene glycol)-block-polyamines promote sensitivity and restrict biofouling on gold sensor surface for detecting factor IX in human plasma. Analyst. 2014;139:3977–3985. doi: 10.1039/c4an00168k. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Y., Wu Y., Ding L., Huang X., Xiong Y. Point-of-care COVID-19 diagnostics powered by lateral flow assay. TrAC, Trends Anal. Chem. 2021;145 doi: 10.1016/j.trac.2021.116452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mao X., Rutledge G.C., Hatton T.A. Nanocarbon-based electrochemical systems for sensing, electrocatalysis, and energy storage. Nano Today. 2014:405–432. doi: 10.1016/j.nantod.2014.06.011. [DOI] [Google Scholar]
- 18.Garg N., Ahmad F.J., Kar S. Recent advances in loop-mediated isothermal amplification (LAMP) for rapid and efficient detection of pathogens. Curr. Res. Microb. Sci. 2022;3 doi: 10.1016/j.crmicr.2022.100120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu H.H., Li Y., Wu L.X., Wang K.S., Zhang Y., Fan Q.Y., Ming Z.Z., Chen W.Q., Liu W.W. Internal heating method of loop-mediated isothermal amplification for detection of HPV-6 DNA. Microchim. Acta. 2022;189:212. doi: 10.1007/s00604-022-05283-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diouani M.F., Ouerghi O., Refai A., Belgacem K., Tlili C., Laouini D., Essafi M. Detection of ESAT-6 by a label free miniature immuno-electrochemical biosensor as a diagnostic tool for tuberculosis. Mater. Sci. Eng. C. 2017;74:465–470. doi: 10.1016/j.msec.2016.12.051. [DOI] [PubMed] [Google Scholar]
- 21.He X., Zhou L., He D., Wang K., Qin D. Biosensing technologies for mycobacterium tuberculosis detection: status and new developments. Clin. Dev. Immunol. 2011 doi: 10.1155/2011/193963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang F., Lakshmipriya T., Gopinath S.C.B. Red spectral shift in sensitive colorimetric detection of tuberculosis by ESAT-6 antigen-antibody complex : a new strategy with gold nanoparticle. Nanoscale Res. Lett. 2018;13:331. doi: 10.1186/s11671-018-2753-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hashim U. Signal enhancement in ELISA : biotin- streptavidin technology against gold nanoparticles. J. Taibah Univ. Med. Sci. 2016;11:432–438. doi: 10.1016/j.jtumed.2016.05.010. [DOI] [Google Scholar]
- 24.Gopinath S.C.B., Perumal V., Kumaresan R., Lakshmipriya T., Rajintraprasad H., Rao B.S., Arshad M.K.M., Chen Y., Kotani N., Hashim U. Nanogapped impedimetric immunosensor for the detection of 16Â kDa heat shock protein against. Mycobacterium tuberculosis, Microchim. Acta. 2016;183:2697–2703. doi: 10.1007/s00604-016-1911-7. [DOI] [Google Scholar]
- 25.Ma L., Peng L., Yin L., Liu G., Man S. CRISPR-Cas12a-Powered dual-mode biosensor for ultrasensitive and cross-validating detection of pathogenic bacteria. ACS Sens. 2021;6 doi: 10.1021/acssensors.1c00686. [DOI] [PubMed] [Google Scholar]
- 26.Black R.E., Cousens S., Johnson H.L., Lawn J.E., Rudan I., Bassani D.G., Jha P., Campbell H., Walker C.F., Cibulskis R., Eisele T., Liu L., Mathers C. Global, regional, and national causes of child mortality in 2008: a systematic analysis. Lancet. 2010;375:1969–1987. doi: 10.1016/S0140-6736(10)60549-1. [DOI] [PubMed] [Google Scholar]
- 27.Dapeng Qin Z.Y., Gong Qiuping, Li Xin, Gao Yanping, Subash C., Gopinath B., Chen Yeng. Identification of Mycoplasma pneumoniae by DNA-modified gold nanomaterials in a colorimetric assay. Biotechnol. Appl. Biochem. 2022 doi: 10.1002/bab.2377. [DOI] [PubMed] [Google Scholar]
- 28.Kamat I.S., Ramachandran V., Eswaran H., Guffey D., Musher D.M. Procalcitonin to distinguish viral from bacterial pneumonia: a systematic review and meta-analysis. Clin. Infect. Dis. 2020;70:538–542. doi: 10.1093/cid/ciz545. [DOI] [PubMed] [Google Scholar]
- 29.Müller B., Harbarth S., Stolz D., Bingisser R., Mueller C., Leuppi J., Nusbaumer C., Tamm M., Christ-Crain M. Diagnostic and prognostic accuracy of clinical and laboratory parameters in community-acquired pneumonia. BMC Infect. Dis. 2007;7 doi: 10.1186/1471-2334-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu K., Chen J., Hu J., Xiong S., Zeng L., Huang X., Xiong Y. Low-sample-consumption and ultrasensitive detection of procalcitonin by boronate affinity recognition-enhanced dynamic light scattering biosensor. Biosens. Bioelectron. 2022;200 doi: 10.1016/j.bios.2021.113914. [DOI] [PubMed] [Google Scholar]
- 31.Jiang L., Li X., Gu R., Mu D. Nanoparticles-based biosensor coupled with multiplex loop-mediated isothermal amplification for detection of staphylococcus aureus and identification of methicillin-resistant S. Aureus. Infect. Drug Resist. 2020;13:1251–1263. doi: 10.2147/IDR.S243881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suaifan G.A.R.Y., Alhogail S., Zourob M. Rapid and low-cost biosensor for the detection of Staphylococcus aureus. Biosens. Bioelectron. 2017;90:230–237. doi: 10.1016/j.bios.2016.11.047. [DOI] [PubMed] [Google Scholar]
- 33.Shinya K., Ebina M., Yamada S., Ono M., Kasai N., Kawaoka Y. Avian flu: influenza virus receptors in the human airway. Supplementary Info, Nature. 2006;440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 34.Nidzworski D., Siuzdak K., Niedziałkowski P., Bogdanowicz R., Sobaszek M., Ryl J., Weiher P., Sawczak M., Wnuk E., Goddard W.A., Jaramillo-Botero A., Ossowski T. A rapid-response ultrasensitive biosensor for influenza virus detection using antibody modified boron-doped diamond. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-15806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gopinath S.C.B., Kumar P.K.R. Aptamers that bind to the hemagglutinin of the recent pandemic influenza virus H1N1 and efficiently inhibit agglutination. Acta Biomater. 2013;9:8932–8941. doi: 10.1016/j.actbio.2013.06.016. [DOI] [PubMed] [Google Scholar]
- 36.Gopinath S.C.B., Awazu K., Fujimaki M., Shimizu K. Neu5Acα2,6Gal and Neu5Acα2,3Gal receptor specificities on influenza viruses determined by a waveguide-mode sensor. Acta Biomater. 2013;9:5080–5087. doi: 10.1016/j.actbio.2012.09.027. [DOI] [PubMed] [Google Scholar]
- 37.Gopinath S.C.B., Awazu K., Fons P., Tominaga J., Kumar P.K.R. A sensitive multilayered structure suitable for biosensing on the BioDVD platform. Anal. Chem. 2009;81:4963–4970. doi: 10.1021/ac802757z. [DOI] [PubMed] [Google Scholar]
- 38.Lakshmipriya T., Fujimaki M., Gopinath S.C.B., Awazu K. Generation of anti-influenza aptamers using the systematic evolution of ligands by exponential enrichment for sensing applications. Langmuir. 2013;29:15107–15115. doi: 10.1021/la4027283. [DOI] [PubMed] [Google Scholar]
- 39.Gopinath S.C.B., Misono T.S., Kawasaki K., Mizuno T., Imai M., Odagiri T., Kumar P.K.R. An RNA aptamer that distinguishes between closely related human influenza viruses and inhibits haemagglutinin-mediated membrane fusion. J. Gen. Virol. 2006;87:479–487. doi: 10.1099/vir.0.81508-0. [DOI] [PubMed] [Google Scholar]
- 40.Gopinath S.C.B. An efficient RNA aptamer against human influenza B virus hemagglutinin. J. Biochem. 2006;139:837–846. doi: 10.1093/jb/mvj095. [DOI] [PubMed] [Google Scholar]
- 41.Gopinath S.C.B., Awazu K., Fujimaki M., Shimizu K., Shima T. Observations of immuno-gold conjugates on influenza viruses using waveguide-mode sensors. PLoS One. 2013;8:1–10. doi: 10.1371/journal.pone.0069121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gopinath S.C.B., Awazu K., Fujimaki M., Shimizu K. Evaluation of anti-A/Udorn/307/1972 antibody specificity to influenza A/H3N2 viruses using an evanescent-field coupled waveguide-mode sensor. PLoS One. 2013;8 doi: 10.1371/journal.pone.0081396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun P., Lu X., Xu C., Sun W., Pan B. Understanding of COVID-19 based on current evidence. J. Med. Virol. 2020;92:548–551. doi: 10.1002/jmv.25722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/nejmoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Velusamy P., Kiruba K., Su C.H., Arun V., Anbu P., Gopinath S.C.B., Vaseeharan B. SARS-CoV-2 spike protein: site-specific breakpoints for the development of COVID-19 vaccines. J. King Saud Univ. Sci. 2021;33 doi: 10.1016/j.jksus.2021.101648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu C., Zhou Q., Li Y., Garner L.V., Watkins S.P., Carter L.J., Smoot J., Gregg A.C., Daniels A.D., Jervey S., Albaiu D. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent. Sci. 2020;6:315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gopinath N. Artificial intelligence: potential tool to subside SARS-CoV-2 pandemic. Process Biochem. 2021;110:94–99. doi: 10.1016/j.procbio.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santheraleka Ramanathan S.S., Gopinath Subash C.B. Zool Hilmi Ismail, No Title Nanodiamond conjugated SARS-CoV-2 spike protein: electrochemical impedance immunosensing on a gold microelectrode. Microchim. Acta. 2022;189:226. doi: 10.1007/s00604-022-05320-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seo G., Lee G., Kim M.J., Baek S.H., Choi M., Ku K.B., Lee C.S., Jun S., Park D., Kim H.G., Kim S.J., Lee J.O., Kim B.T., Park E.C., Il Kim S. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 51.Ma Y.D., Li K.H., Chen Y.H., Lee Y.M., Chou S.T., Lai Y.Y., Huang P.C., Ma H.P., Bin Lee G. A sample-to-answer, portable platform for rapid detection of pathogens with a smartphone interface. Lab Chip. 2019;19:3804–3814. doi: 10.1039/c9lc00797k. [DOI] [PubMed] [Google Scholar]
- 52.Liv L., Kayabay H. An electrochemical biosensing platform for the SARS-CoV-2 spike antibody detection based on the functionalised SARS-CoV-2 spike antigen modified electrode. ChemistrySelect. 2022;7 doi: 10.1002/slct.202200256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liv L., Yener M., Çoban G., Can Ş.A. Electrochemical biosensing platform based on hydrogen bonding for detection of the SARS-CoV-2 spike antibody. Anal. Bioanal. Chem. 2022;414:1313–1322. doi: 10.1007/s00216-021-03752-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liv L., Baş A. Discriminative electrochemical biosensing of wildtype and omicron variant of SARS-CoV-2 nucleocapsid protein with single platform. Anal. Biochem. 2022;657 doi: 10.1016/j.ab.2022.114898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Olgaç N., Şahin Y., Liv L. Development and characterisation of cysteine-based gold electrodes for the electrochemical biosensing of the SARS-CoV-2 spike antigen. Analyst. 2022;147:4462–4472. doi: 10.1039/d2an01225a. [DOI] [PubMed] [Google Scholar]
- 56.Il Goh K., Cusick M.E., Valle D., Childs B., Vidal M., Barabási A.L. The human disease network. Proc. Natl. Acad. Sci. U.S.A. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hall C.B., Weinberg G.A., Iwane M.K., Blumkin A.K., Edwards K.M., Staat M.A., Auinger P., Griffin M.R., Poehling K.A., Erdman D., Grijalva C.G., Zhu Y., Szilagyi P. The burden of respiratory syncytial virus infection in young children. N. Engl. J. Med. 2009;360:588–598. doi: 10.1056/nejmoa0804877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haber N. Respiratory syncytial virus infection in elderly adults. Med. Maladies Infect. 2018;48:577–587. doi: 10.1016/j.medmal.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 59.French C.E., McKenzie B.C., Coope C., Rajanaidu S., Paranthaman K., Pebody R., Nguyen-Van-Tam J.S., Higgins J.P.T., Beck C.R. Risk of nosocomial respiratory syncytial virus infection and effectiveness of control measures to prevent transmission events: a systematic review. Influenza Other Respi. Viruses. 2016;10:268–290. doi: 10.1111/irv.12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Białobrzeska W., Firganek D., Czerkies M., Lipniacki T., Skwarecka M., Dziąbowska K., Cebula Z., Malinowska N., Bigus D., Bięga E., Pyrć K., Pala K., Żołędowska S., Nidzworski D. Electrochemical immunosensors based on screen-printed gold and glassy carbon electrodes: comparison of performance for respiratory syncytial virus detection. Biosensors. 2020;10:175. doi: 10.3390/BIOS10110175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cao M., Sun Q., Zhang X., Ma Y., Wang J. Detection and differentiation of respiratory syncytial virus subgroups A and B with colorimetric toehold switch sensors in a paper-based cell-free system. Biosens. Bioelectron. 2021;182 doi: 10.1016/j.bios.2021.113173. [DOI] [PubMed] [Google Scholar]
- 62.Perez J.W., Haselton F.R., Wright D.W. Viral detection using DNA functionalized gold filaments. Analyst. 2009;134:1548–1553. doi: 10.1039/b904191e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhan L., Yang T., Li C.M., Wu W.B., Huang C.Z. Sensitive immunosensor for respiratory syncytial virus based on dual signal amplification of gold nanopaticle layer-modified plates and catalyzed reporter deposition. Sensor. Actuator. B Chem. 2018;255:1291–1297. doi: 10.1016/j.snb.2017.08.110. [DOI] [Google Scholar]
- 64.Rochelet M., Solanas S., Grossiord C., Maréchal P., Résa C., Vienney F., Barranger C., Joannes M. A thin layer-based amperometric enzyme immunoassay for the rapid and sensitive diagnosis of respiratory syncytial virus infections. Talanta. 2012;100:139–144. doi: 10.1016/j.talanta.2012.07.088. [DOI] [PubMed] [Google Scholar]
- 65.Le T.T., Chang P., Benton D.J., McCauley J.W., Iqbal M., Cass A.E.G. Dual recognition element lateral flow assay toward multiplex strain specific influenza virus detection. Anal. Chem. 2017;89:6781–6786. doi: 10.1021/acs.analchem.7b01149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu W., Meng K., Zhang Y., Bu Z., Zhao D., Meng G. Lateral flow assay for the detection of african swine fever virus antibodies using gold nanoparticle-labeled acid-treated p72. Front. Chem. 2022;9 doi: 10.3389/fchem.2021.804981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Poon L.L.M., Leung C.S.W., Chan K.H., Lee J.H.C., Yuen K.Y., Guan Y., Peiris J.S.M. Detection of human influenza A viruses by loop-mediated isothermal amplification. J. Clin. Microbiol. 2005;43:427–430. doi: 10.1128/JCM.43.1.427-430.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yousefi H., Mahmud A., Chang D., Das J., Gomis S., Chen J.B., Wang H., Been T., Yip L., Coomes E., Li Z., Mubareka S., Mcgeer A., Christie N., Gray-Owen S., Cochrane A., Rini J.M., Sargent E.H., Kelley S.O. Detection of SARS-CoV-2 viral particles using direct, reagent-free electrochemical sensing. J. Am. Chem. Soc. 2021;143:1722–1727. doi: 10.1021/jacs.0c10810. [DOI] [PubMed] [Google Scholar]
- 69.Liu X., Bu S., Feng J., Wei H., Wang Z., Li X., Zhou H., He X., Wan J. Electrochemical biosensor for detecting pathogenic bacteria based on a hybridization chain reaction and CRISPR-Cas12a. Anal. Bioanal. Chem. 2022;414:1073–1080. doi: 10.1007/s00216-021-03733-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang X., Feng Y., Duan S., Su L., Zhang J., He F. Mycobacterium tuberculosis strain H37Rv electrochemical sensor mediated by aptamer and AuNPs-DNA. ACS Sens. 2019;4:849–855. doi: 10.1021/acssensors.8b01230. [DOI] [PubMed] [Google Scholar]
- 71.Suea-Ngam A., Howes P.D., Demello A.J. An amplification-free ultra-sensitive electrochemical CRISPR/Cas biosensor for drug-resistant bacteria detection. Chem. Sci. 2021;12:12733–12743. doi: 10.1039/d1sc02197d. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.