Summary
In endangered species, low-genetic variation and inbreeding result from recent population declines. Genetic screenings in endangered populations help to assess their vulnerability to extinction and to create informed management actions toward their conservation efforts. The leopard, Panthera pardus, is a highly generalist predator with currently eight different subspecies. Yet, genomic data are still lacking for the Critically Endangered Arabian leopard (P. p. nimr). Here, we sequenced the whole genome of two Arabian leopards and assembled the most complete genomic dataset for leopards to date. Our phylogenomic analyses show that leopards are divided into two deeply divergent clades: the African and the Asian. Conservation genomic analyses indicate a prolonged population decline, which has led to an increase in inbreeding and runs of homozygosity, with consequent purging of deleterious mutations in both Arabian individuals. Our study represents the first attempt to genetically inform captive breeding programmes for this Critically Endangered subspecies.
Subject areas: Genomics, Zoology
Graphical abstract

Highlights
-
•
Two new whole genomes are generated for the Critically Endangered Arabian leopard
-
•
Phylogenomics support Arabian leopards as a distinct lineage sister to Asian leopards
-
•
Arabian leopard has suffered a prolonged bottleneck
-
•
High inbreeding and ROH but purging of deleterious mutations in Arabian leopards
Genomics; Zoology; Leopards
Introduction
In endangered species, low-genetic variation and high levels of inbreeding are usually a consequence of population decline, which may have negative effects on their adaptive potential and rates of reproduction, and thus increase their extinction risk.1 From a conservation point of view, exploring the fine-scale population structure, genetic diversity, and intraspecific demographic dynamics in an endangered species is of crucial importance to correctly design plans for its conservation.1,2 For instance, populations within a species may have different evolutionary histories, substructure, or genetic adaptations to their local environment and, if so, they should be considered as different conservation units.3 Genetic data play a key role in detecting all these factors and inferring their effect on demographic changes and inbreeding. In particular, the use of genome-wide approaches is highly recommended in captive breeding programmes, as such datasets can help to identify deleterious mutations and guide the management of endangered species.2,4,5,6,7,8
In the field of conservation genetics, a few nuclear and mitochondrial markers on a high number of individuals have been used as the standard methodology to infer population parameters.9 These techniques, although very useful, lack the power and precision to reflect all the genomic information from both individuals and populations.10 With the rise and wide implementation of next-generation sequencing (NGS) technologies, the field’s paradigm is shifting toward conservation genomics.11 This emerging discipline takes advantage of genome-wide data, such as whole-genome sequencing (WGS), to assess with unprecedented resolution and accuracy both the taxonomy of a focal species, as well as population dynamics such as hybridization events, demographic changes, disease outbreaks, and local genetic adaptation.12,13,14,15,16,17,18 More importantly, endangered species are elusive and usually found at low-population densities, making the task of finding a high number of individuals for multi-locus approaches nearly impossible.2 Sequencing the genomes of a few individuals has yielded results comparable to those obtained by genotyping a high number of individuals with traditional markers,4 making WGS a powerful tool for the conservation of endangered species.8
The leopard (Panthera pardus, Linnaeus 1758) is a highly generalist species, present in a wide range of ecological conditions such as semi-desert, savanna, rainforest, and montane habitats, from sea level to high-mountain ranges.19,20 Its diverse diet and capacity to adapt to different environments21 has allowed this species to expand across Africa and Asia,19 and even across Europe during the Early Pleistocene,22 thus having the largest distributional range within the genus Panthera. Currently, leopards are still present across much of the African continent and from the Middle East to the Pacific Ocean in Asia,19,20,23 although they are only occupying around 25–37% of their historical range.24 A pioneer study with mitochondrial markers by Uphyrkina et al.20 found two main monophyletic groups: the African and the Asian leopards. Fossil evidence and high levels of genetic diversity pointed to an Eastern African origin for the species around 2 million years ago (Mya).20,24,25 However, paleontological data suggested that more than one out-of-Africa event occurred and that the Javan leopard (P. p. melas) could represent an isolated population resulting from one of those events.20,25,26 Using mitogenomes from historical and ancient samples, a single out-of-Africa dispersion was proposed and dated around 710 thousand years ago (Kya).25 Studies with WGS data supported a similar scenario, with a split between African and Asian leopards around 500–600 Kya, and confirmed the monophyly of both groups.23 Nevertheless, the origin of the Asian colonization is still uncertain. Paijmans et al.23 suggested a colonization by north-western African leopards. Nonetheless, none of these studies incorporated samples from the Arabian leopard (P. p. nimr), a subspecies that due to its geographical distribution is key to resolve the uncertainty on how leopards colonized Eurasia.
The Arabian leopard (P. p. nimr) is the Arabian flagship predator and has been listed as Critically Endangered by the IUCN’s Red List of Threatened Species.27 This subspecies faces a significant reduction in population size and is on the brink of extinction, with current estimates of fewer than 250 individuals in the wild.28,29 Moreover, the Arabian leopard has lost as much as 98% of its historical range,24 with populations highly isolated and fragmented. This has prevented acquisition of knowledge on the current status of wild populations. Previous evaluations estimated 50 individuals in Saudi Arabia, 25–30 individuals in Oman, and a captive stock of 82 individuals, mostly in United Arab Emirates (UAE).27,28,30,31 Populations within Yemen have not been evaluated. Furthermore, just 9% of its current distribution is within protected areas.24 Nowadays, several wildlife research centers in the Arabian Peninsula are focused on captive breeding programmes for this species.30 However, conservation management efforts for these populations are being formulated without a complete understanding of population genomic patterns, which could result in sub-optimal conservation outcomes.18 Thus, an extensive study of the genomic diversity and population structure of the Arabian leopard and its relationships with other leopard subspecies will strongly benefit its conservation, as a comprehensive knowledge of a species’ history is essential for both evolutionary research and conservation management.18
Here, we sequence for the first time two whole genomes for the Critically Endangered Arabian leopard at medium coverage (10.31x and 7.52x, respectively) in order to explore its past evolutionary history. Together with the recently released genomes of all currently accepted leopard subspecies (a combined dataset from Paijmans et al.23 and Pečnerová et al; 32), we investigate the phylogenomic position of the Arabian leopard, a highly discussed topic in the past.20,23 Moreover, we explore introgression between the different subspecies of leopards with special focus on the Arabian leopard’s potential introgression with both the African (P. p. pardus) and the Anatolian (P. p. tulliana) leopards. Finally, with the high resolution provided by WGS data, we assess the current levels of genomic diversity and mutational load, setting the first step for a genomic-informed conservation strategy for the Critically Endangered Arabian leopard.
Results
Population structure
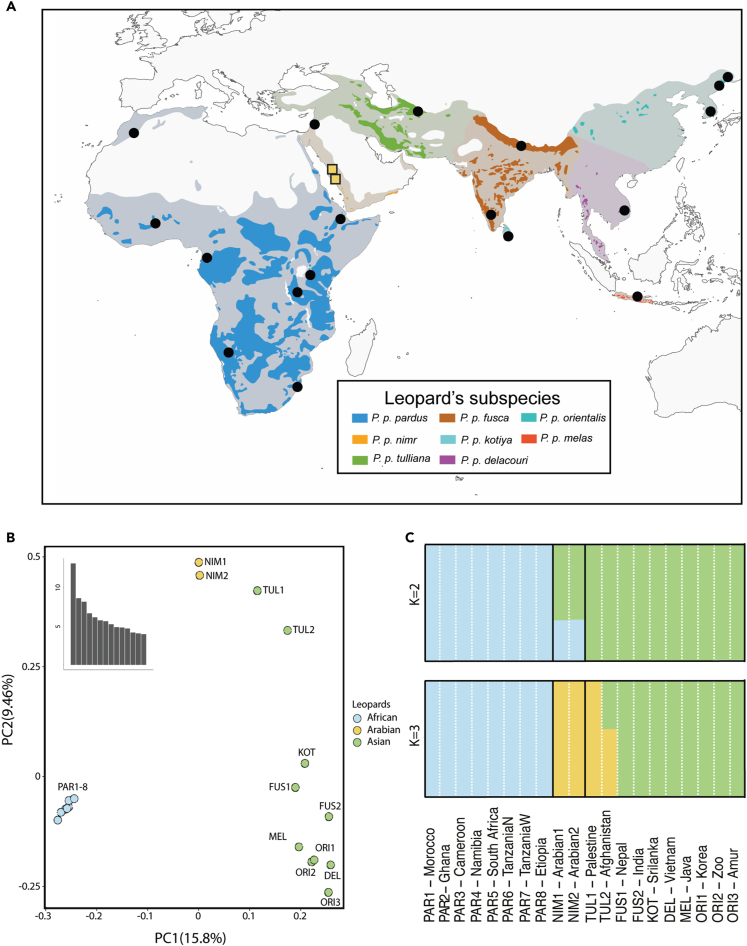
We generated the whole genome from two Critically Endangered Arabian leopards at an average coverage of 10.32x and 7.53x (Table 1). Together with the African and Asian samples gathered from previous studies,16,23,32 we assembled a genomic dataset for the leopard lineage that, for the first time, includes representatives from all currently recognized subspecies33 (Figure 1A; Table 1). With this comprehensive dataset, we explored the population structure of leopards with a principal component analysis (PCA) of 1.35 million polymorphic positions, after LD pruning. We found a segregation of African and Asian populations along PC1 (Figure 1B) while PC2 informed about the variability within the Asian specimens, recovering a gradient following geographical locations. The Arabian leopards clustered next to their geographically closest populations of Anatolian leopards. Similarly, admixture analyses reported k = 2 as the most likely number of ancestral populations (Table S1), dividing African and Asian populations (Figure 1C). Arabian leopards grouped within the Asian cluster for k = 2, but showed up to 37% of African component. The Arabian and the two Anatolian leopards formed a unique cluster for k = 3 (the latter with 41% of ancestral component from the Asian clade).
Table 1.
Information of all samples used in this study, including coverage for all the specimens and heterozygosity for the two Arabian leopards and all other samples with coverage higher than 15x
| Id | Species | Coverage | Heterozygosity | Publication |
|---|---|---|---|---|
| PAR1- Morocco | P. p. pardus | 7 | – | Paijmans et al. 202123 |
| PAR2 – Ghana | P. p. pardus | 18.82 | 0.0021 | Pečnerová et al. 202132 |
| PAR3 – Cameroon | P. p. pardus | 8.75 | – | Paijmans et al. 202123 |
| PAR4 – Namibia | P. p. pardus | 20.84 | 0.0022 | Pečnerová et al. 202132 |
| PAR5 – South Africa | P. p. pardus | 12.64 | – | Paijmans et al. 202123 |
| PAR6 – TanzaniaN | P. p. pardus | 20.7 | 0.0021 | Pečnerová et al. 202132 |
| PAR7 – TanzaniaW | P. p. pardus | 20.4 | 0.0024 | Pečnerová et al. 202132 |
| PAR8 – Etiopia | P. p. pardus | 14.22 | – | Paijmans et al. 202123 |
| NIM1-Arabian1 | P. p. nimr | 10.31 | 0.00054 | This study |
| NIM2-Arabian2 | P. p. nimr | 7.52 | 0.0004 | This study |
| TUL1 – Palestine | P. p. tulliana | 5.6 | – | Paijmans et al. 202123 |
| TUL2 – Afghanistan | P. p. tulliana | 9.8 | – | Paijmans et al. 202123 |
| FUS1 – Nepal | P. p. fusca | 41 | 0.001 | Paijmans et al. 202123 |
| FUS2 – India | P. p. fusca | 12 | – | Paijmans et al. 202123 |
| KOT – Sri Lanka | P. p. kotya | 11 | – | Paijmans et al. 202123 |
| DEL – Vietnam | P. p. delacouri | 7.4 | – | Paijmans et al. 202123 |
| MEL – Java | P. p. melas | 17 | 0.00067 | Paijmans et al. 202123 |
| ORI1 – Korea | P. p. orientalis | 13 | – | Paijmans et al. 202123 |
| ORI2 – Zoo | P. p. orientalis | 17 | 0.00057 | SRA SRR5382750 |
| ORI3 - Amur | P. p. orientalis | 35 | 0.00098 | Kim et al., 201616 |
| Botswana lion | P. leo | 7.09 | – | de Manuel et al., 202034 |
| Central African lion | P. leo | 9.53 | – | de Manuel et al., 202034 |
| Tanzania lion | P. leo | 5.75 | – | de Manuel et al., 202034 |
Figure 1.
Sampling and population genetic structure
(A) Distribution for all non-extinct leopard subspecies, including historical distribution for all the species (in lighter color). Yellow squares show new individuals sequenced for this study while circles show data already available and included in this work.
(B) PCA of genetic variation over 1.35 million SNPs for all subspecies of leopards.
(C) Admixture analysis with k = 2 and k = 3. Abbreviations are as follows: PAR, P. p. pardus; NIM, P. p. nimr; TUL, P. p. tulliana; FUS, P. p. fusca; KOT, P. p. kotiya; DEL, P. p. delacouri; ORI, P. p. orientalis and MEL, P. p. melas.
Evolutionary relationships
To explore in more detail the evolutionary relationships of all leopard subspecies and in particular to shed light on the phylogenetic position of the Arabian leopards, we inferred their phylogenetic relationships using 2,330 non-overlapping 1 Mbp windows that covered around 96% of the whole genome. The resulting phylogenomic tree shows two well-supported main clades, the African and the Asian (Figure 2A). Although the internal phylogenomic relationships within the African clade were not well supported (a result previously discussed by Paijmans et al.23), the internal phylogenomic relationships within the Asian clade support the Arabian leopard as sister taxon to all the remaining subspecies of Asian leopards. All the other phylogenomic relationships within the Asian clade were not fully supported, and the analysis did not recover monophyly for some of the subspecies of Asian leopards (Figure 2A). Phylogenomic trees with sliding windows of reduced size (i.e., 500 Kbp and 100 Kbp) showed similar topologies with overall lower support (although some subspecies within the Asian clade were not recovered as monophyletic) (Figure S1). In both cases, Arabian leopards were found within the Asian clade. Interestingly, the phylogenomic tree with a sliding window of 100 Kbp showed one individual of the Anatolian leopard (P. p. tulliana) clustering together with the Arabian leopards, a result in line with introgression analyses (see the following text).
Figure 2.
Phylogenomic trees for the nuclear and mitochondrial data
(A) Phylogenomic consensus tree from 2,330 maximum likelihood trees of 1 Mbp non-overlapping sliding windows along the reference genome for all currently accepted subspecies of leopards, and with the lion as outgroup. Node values indicate clade frequency.
(B) Maximum likelihood mitogenome phylogeny for a subset of the samples, with lion as outgroup. Bootstrap values are shown in each node. Abbreviations are as follows: PAR, P. p. pardus; NIM, P. p. nimr; TUL, P. p. tulliana; FUS, P. p. fusca; KOT, P. p. kotiya; DEL, P. p. delacouri; ORI, P. p. orientalis; MEL, P. p. melas and SPE, P. p. spelaea.
The mitogenomic phylogeny split the samples into two main groups: a well-supported clade comprising the African and Arabian subspecies (contrary to the nuclear genome phylogeny of Figure 2A), and an Eurasian clade comprising European samples (P. p. spelaea) from the late Pleistocene and all other remaining leopard Asian subspecies, showing a topology similar to previous phylogenies20,25 (Figure 2B).
Ancient demographic history
PSMC analysis showed a continuous negative trend in population size for the two Arabian leopards (Figures 3 and S2). Individuals from other subspecies showed results previously observed,23,32 with African samples reporting the highest current effective population size of the species and Javan and Amur samples showing lower population sizes, with the Amur leopard following an ancestral trajectory different from African samples, as reported in Paijmans et al.23 Owing to the lower coverage of both Arabian leopards compared to the other subspecies, we downsampled the high-coverage data at similar coverage levels (∼10x) to those of the Arabian leopards (Figure S3). As expected, lower effective population sizes were observed with the downsampled dataset but general trends persisted (compare Figure 3 with Figure S3), indicating that the pattern observed in the Arabian samples was likely not strongly affected by their coverage levels. Beyond >105 years, PSMC inferred different effective population sizes for the two Arabian leopards, most likely the product of inaccuracy due to their low heterozygosity and lack of coalescent events dating at that point in the past.35
Figure 3.
PSMC analysis for the high-coverage samples plus the two Arabian leopards
Generation time was set to 5 years and substitution rate to 1 × 10−8 per site per year. Abbreviations are as follows: NIM, P. p. nimr; MEL, P. p. melas; PAR, P. p. pardus, and ORI, P. p. orientalis.
Genomic diversity, inbreeding, and ROHs
Genome-wide heterozygosity levels varied across subspecies, with the African lineage harboring the highest diversity levels (Figure 4A; Table S2). Heterozygosity in Asian individuals varied among the subspecies, with the Arabian leopards having some of the lowest heterozygosity values (5.4 × 10−4 bp−1 and 4 × 10−4 bp−1 for Arabian1 and Arabian2, respectively). Individuals within the Amur leopard subspecies (P. p. orientalis), classified as Critically Endangered by IUCN, showed in general low heterozygosity but higher than the Arabian samples, while the Nepalese sample of Indian leopard (P. p. fusca) contained the highest heterozygosity values within the Asian clade. Out-of-ROH heterozygosity recovered higher estimates but showed similar tendencies to genome-wide heterozygosity, with Arabian leopards showing more than double their genome-wide heterozygosity (Figure S4). To test the effect of coverage on the detection of heterozygous sites, we downsampled six high-coverage individuals to similar coverage levels to the Arabian leopards (∼10x), and we did not detect any statistically significant change between non-downsampled and downsampled individuals (t = −0.66, p value = 0.51), although as expected, the resulting heterozygosity levels were slightly lower after downsampling (Figure 4A). After correcting for the coverage, Arabian leopards still showed very low-heterozygosity levels, with significant differences from downsampled African leopards (t = 16.61, p value = 0.007) but not with other downsampled Asian leopards (t = 0.62, p value = 0.57). However, wild and captive samples with higher coverage should be included to explore the real genetic variation of this subspecies. ROH analyses reported high disparity among all the samples tested (Figure 4B). Both captive samples of Arabian leopards contained more than 50% of the genome under ROH (Figure 4B; Table S3), with the highest percentage of short (<500 Kbp) and medium (0.5–1 Mbp) ROH, indicative of a small and common long-term effective population size. Moreover, there was an important difference in the number of long ROH (>1 Mbp) between the two Arabian leopards (Figure 4B; Table S3). This could be explained by different levels of recent inbreeding as both individuals are captive-bred leopards. Arabian2, a fourth-generation captive-bred female, had 10% longer ROH than Arabian1, a third-generation captive-bred male. The Javan leopard had around 30% of short ROH, indicating ancient inbreeding in this island subspecies. Interestingly, one captive P. p. orientalis individual (ORI2-Zoo) accumulated almost 50% of its genome within ROH and 20% of them being long (>1 Mbp). All these results were supported by a correlation between number (NROH) and cumulative sum of RoHs (SROH) (Pearson correlation t = 3.14, correlation value = 0.76 and p value = 0.016; Figure S5; Table S4), with large and well-connected populations having a reduced NROH and SROH and small and fragmented populations showing high values of both NROH and SROH.
Figure 4.
Heterozygosity and Runs of Homozygosity for the high-coverage individuals plus the Arabian leopards
(A) Genome-wide heterozygosity for all samples with coverage levels higher than 15x and the two Arabian leopards. In dark, genome-wide heterozygosity for downsampled individuals, in light heterozygosity with non-downsampled individuals. Arabian leopards were not downsampled. Statistically significant differences exist between downsampled African and Arabian leopards (p value <0.01) but not between downsampled Asian and Arabian (p value = 0.57).
(B) Percentage of the genome in ROH. Different colors within the columns indicate the relative percentage of long (>1 Mbp), medium (>500 Kbp and <1 Mbp) and short (<500 Kbp) ROHs along the genome for the high-coverage samples and the two Arabian leopards; see Table S2 for more detailed information. Abbreviations are as follows: PAR, P. p. pardus; NIM, P. p. nimr; FUS, P. p. fusca; MEL, P. p. melas and ORI, P. p. orientalis.
Introgression
We initially tested introgression between the Arabian leopard and its two geographically closest subspecies, the African (P. p. pardus) and the Anatolian (P. p. tulliana) leopards, and later between all other leopard subspecies. We tested a simple tree-like model where the lion was set as an outgroup following the new phylogenomic relationships to establish the comparisons. Introgression analyses revealed past introgression between Arabian and both African and Anatolian leopards (Figure 5). D-statistic values varied depending on the comparison, but all of them were significant and with absolute Z-scores higher than three. We also found introgression between several other subspecies, highlighting a complex history of gene flow within the species (Table S5).
Figure 5.
Introgression analyses between the Arabian leopard (P. p. nimr) and the two geographically closest subspecies, the African (P. p. pardus) and the Anatolian (P. p. tulliana) leopards, using the lion as outgroup
Introgression between Pop2 and Pop3 will be reported when D-statistic is negative and introgression between Pop1 and Pop3 will happen when the D-statistic is positive. All D-statistics had an absolute Z score value higher than 3 (shown in yellow). Error bars are shown within the dots.
Mutational load
We compared the mutational load of the Arabian leopard (an example of a small and long-term isolated subspecies) to that of the African (a subspecies with large and connected populations) and to other Asian leopards (an intermediate case, with some subspecies with well-connected populations and others with small and isolated populations). As a first approach, we counted the number of high-, moderate-, or low-impact deleterious alleles for each individual (see star methods). Later, we calculated the number of homozygous (multiplied by two as they are represented twice) and heterozygous alleles separately, as a proxy of realized (i.e., homozygous alleles with effects on the current generation) and masked (heterozygous alleles that can be expressed in future generations) genetic load, thus assuming most deleterious alleles are recessive.36 Moreover, we calculated the ratio of derived alleles between African, Arabian, and Asian individuals (although Arabian leopards are within Asia, we analyzed them separately for comparative reasons). Arabian leopards showed the lowest number of high-impact deleterious alleles but similar levels of moderate- and low-impact deleterious alleles compared to African and the rest of Asian leopards (Figures 6A and S6; Table S6). Interestingly, Arabian leopards significantly differed from both African and the rest of Asian leopards in the number of high-impact deleterious alleles (t = 2.93, p value = 0.021 and t = 4.35, p value = 0.002, respectively) and with Asian leopards but not African leopards in the number of moderate-impact deleterious alleles (t = 3.75, p value = 0.005 and t = −0.28, p value = 0.78, respectively) (Figures 6A and S6; Table S6). Interestingly, low-impact deleterious alleles did not show differences between Arabian and African leopards (t = 1.68, p value = 0.13), Arabian and the rest of Asian leopards (t = 1.74, p value = 0.11) and African and Asian leopards (excluding Arabian leopards) (t = 0.39, p value = 0.7). African and Asian (excluding Arabian) leopards did not differ in the number of high-impact deleterious alleles (t = −1.95, p value = 0.07) but did in the number of moderate-impact deleterious alleles (t = −2.66, p value = 0.01). The number of high-impact and moderate-impact derived deleterious alleles in homozygosity (the realized load) showed significantly higher number of alleles for Arabian leopards compared with African leopards (t = −14.24, p value <0.0001 for high-impact deleterious alleles; t = −26.15, p value <0.0001 for moderate-impact deleterious alleles) and only for moderate-impact deleterious alleles when compared with other Asian leopards (t = 0.53, p value = 0.60 for high-impact deleterious alleles; t = −2.68, p value = 0.02 for moderate-impact deleterious alleles) (Figures 6A and S6; Table S6). Conversely, Arabian leopards showed significantly lower numbers of heterozygous alleles (the masked load) compared to African (t = 10.7, p value <0.0001 for high-impact deleterious alleles and t = 13.48, p > 0.0001 for moderate-impact deleterious alleles) and the rest of Asian leopards (t = 3.39 p value = 0.09 for high-impact deleterious alleles and t = 3.99, p value = 0.003 for moderate-impact deleterious alleles) (Figures 6A and S6; Table S6). Ratio of derived alleles (RXY) between Arabian leopards and both African and the rest of Asian leopards were in line with previous results and showed reduced high-impact and moderate-impact deleterious alleles for the former while the ratio between Asian (excluding Arabian) and African leopards showed Asian leopards to have slightly more deleterious alleles for both categories (Figure 6B). Around half of the observed alleles were within ROH for the Arabian leopards and the P. p. orientalis specimen analyzed from a zoo, while all other leopards had at least 75% of the deleterious alleles outside ROH (Figure S7).
Figure 6.
Mutational load in leopards
(A) Number of high-impact deleterious alleles found in total, realized and masked genetic load for leopards from Africa, Arabia and the rest of Asia.
(B) Ratio of high and moderate impact deleterious alleles between Arabian leopards with both African and the rest of Asian leopards and between African and Asian leopards (excluding Arabian) for both high- and moderate-impact deleterious alleles.
Discussion
In this study, we have sequenced for the first time the complete genome of two Arabian leopards (P. p. nimr), a Critically Endangered subspecies with less than 250 wild individuals distributed into continuously declining and severely fragmented populations across Saudi Arabia, Yemen, and south Oman.28,29 Apart from its interest from a conservation point of view, its geographical distribution at the intersection between the two main leopard clades (Africa and Asia) make this subspecies key to understanding the evolutionary history of leopards. Together with genomic data available from all other leopard subspecies (Figure 1A), we have analyzed their evolutionary history and demography. In an attempt to guide future conservation plans in the Arabian leopard and other subspecies, we have applied a conservation genomics approach to assess the genetic consequences of isolation and inbreeding.
Population structure analyses suggest that the Arabian leopards are closer to the Asian than to the African leopards (Figures 1B and 1C). We replicated the PCA results from Paijmans et al.,23 with all African leopards clustering very closely both in PC1 and PC2, while the Asian leopard subspecies appeared separated in PC2 according to their geographical distribution. These contrasting clusterings highlight a lower genetic differentiation within African leopards than within Asian leopards (Figure 1B). Genetic similarity in African leopards can be explained by high mobility, habitat versatility, and weak signatures of dispersal barriers across Africa.24,32,37 Conversely, Asian leopards are characterized by high levels of structuring (Figure 1B). This can be expected as the demographic history of Asian leopards is defined by an initial founder event followed by expansions and bottlenecks, with drift causing population structure at neutral loci.23,38
Owing to the almost complete absence of genetic information for the Arabian leopard to date, its phylogenetic position has been a highly discussed topic for decades.20,23,32 Previous to this study, the only genetic information available was from a study based on mitochondrial markers suggesting that the Arabian leopard was the sister group to the African leopard.20 Our phylogenomic analysis, including thousands of genome-wide autosomal markers of all currently accepted subspecies of leopards, was a key to successfully resolve the evolutionary relationships of leopards and the enigmatic phylogenetic position of the Arabian leopard (Figure 2A). Contrary to the results of the analysis of the mitochondrial dataset by Uphyrkina et al.20 and the results of our mitogenome analyses presented in this study (Figure 2B), the genome-wide autosomal dataset fully supported the position of the Arabian leopard as sister taxon to the rest of Asian leopards (Figure 2A); a result consistent with the population structure analyses (Figure 1) and supporting only one out-of-Africa dispersal event. This is not the first case of mitonuclear discordance in the genus Panthera,39 highlighting the need to use information from the nuclear genome to correctly infer the evolutionary relationships among species.
Although the autosomal phylogenomic tree was based on complete genomes, low resolution within African and Asian clades was observed and monophyly was not recovered for some of the subspecies (Figure 2A). Establishing the phylogenetic position of the Arabian leopard is critical for conservation programmes, since even recently33 the Arabian leopard was considered consubspecific to the African P. p. pardus, as the mitochondrial inference indicates. Our genome-wide data advocates for the genomic distinction of the Arabian leopard, confirming that it needs to be managed as a separate conservation unit, as it has been done so far. Given that the Arabian leopard is the sister group to all other Asian subspecies, our findings suggest Arabia may have served as a stepping stone for the subsequent expansion across the rest of the Asian continent and perhaps Europe. Whole nuclear genomes of European individuals should be sequenced to resolve the complete evolutionary history of the species. All other phylogenomic relationships were found to be in line with previous phylogenomic results.23
When estimating the past evolutionary history, a continuous trend of reduction in effective population size toward the present was observed for both Arabian samples (Figure 3). Low-heterozygosity levels for both Arabian leopards (Figure 4A; Table S2) and the medium coverage for both samples could influence the PSMC analysis, as this software relies on the heterozygosity levels to reconstruct the historical evolutionary history of each sample.35 Wild animals from different locations need to be sequenced at higher coverage to explore their evolutionary history and investigate the potential variability of population trends within the Arabian leopard populations.
As previously discussed by Pečnerová et al.,32 African leopards present high levels of genetic diversity and high continent-wide genetic connectivity, considering their trophic position. Here, we replicated their findings showing that African leopards have the highest genome-wide heterozygosity levels and the lowest percentage of genome in ROH of all leopard populations (Figure 4; Tables S2 and S3). The two Arabian leopards included in our study showed low-heterozygosity levels (Tables 1 and S2; Figure 4A). This heterozygosity could be consistent with a scenario of strong genetic drift acting upon the population, most likely due to the observed low long-term effective population size observed (Figure 3) as well as inbreeding (Figure 4; Table S2). Aridity, anthropogenic habitat degradation and persecution are factors that could have caused this continuous decline,28 together with genetic drift after colonization of Arabia. This genetic decrease might turn into a reduced ability to adapt to any rapid environmental change or the emergence of new diseases such as SARS-CoV-2 in leopards.40,41 More than 50% of both Arabian leopard genomes are under ROH (Figure 4B; Table S3), with different sizes of ROH explaining long-term and recent inbreeding. Short and medium ROH are features of populations that have experienced an old population bottleneck.42 Both captive individuals of Arabian leopard contained almost the same percentage of short and medium ROH, indicating a similar evolutionary history (Figure 4B; Table S3). These short and medium ROH are consistent with the historical effective population reduction trends (Figure 3), suggesting that this subspecies might have suffered a prolonged past bottleneck. Interestingly, the female Arabian2 had about 10% more recent and long ROHs than the male Arabian1 (Table S3). This may be explained by the captive breeding program, as long ROH are caused by recent inbreeding loops.42 This is an unexpected result, as in every breeding event inside the conservation program, there is planned inbreeding avoidance by mating with at least one wild-born individual. However, the founder population (wild-born individuals) could have been close relatives or have come from the same population, contributing to the increase in inbreeding. Due to the low number of individuals studied, wild and captive individuals should be sampled to have a clearer idea about the genomic situation in the wild and how the captive breeding program is affecting the genomic status of this endangered Arabian subspecies. Only the highly inbred P. p. orientalis leopard from a zoo shows similar levels of genome-wide heterozygosity and ROH as the ones observed in the Arabian leopards. We strongly recommend incorporating genomic data to estimate relatedness between individuals in the continuing leopard breeding program, as genomics, together with other disciplines, is an essential tool for the conservation success of target species. All other Asian samples were in line with previous results, with the Javan leopard (P. p. melas) from a small island population also showing low heterozygosity and high percentage of short ROH and the Amur leopard with relatively low-heterozygosity values and low percentage of the genome under ROH (Figures 4 and S5; Tables S2 and S3).
Despite the existence of morphological differences,23 we found introgression signals between Arabian and Anatolian (P. p. tulliana) leopards and between Arabian and African leopards (Figure 5). Gene flow between big cats is a well-known phenomenon.13 Thus, the description of gene flow between geographically close subspecies of leopards is not surprising. On the one hand, the introgression between African and Arabian leopards is also partially supported by the mitochondrial phylogeny, as both groups cluster together, suggesting a possible migration corridor for females between Africa and the Arabian Peninsula. However, we cannot rule out that this topology is due to mitogenomic incomplete lineage sorting, as in leopards, males have higher dispersal than females.43 On the other hand, the introgression between Arabian and Anatolian leopards is supported by the autosomal phylogeny, as the 100 Kbp sliding windows phylogeny clusters Arabian and one sample of Anatolian leopard together (Figure S1B). Their past continuous distribution through Northern Arabia could have helped in promoting gene flow between these two genetically and morphologically distinct subspecies. Finally, we also found several other comparisons between subspecies of leopards to be significant, mainly between Asian subspecies, revealing a complex history of gene flow within the species (Table S5).
Genetic load is described as the resulting reduction in individual and mean population fitness due to deleterious mutations originating from mutation or gene flow and maintained or even increased by genetic drift or reduced efficacy of purifying selection.36 In diploid organisms, genetic load can be separated (assuming recessivity) into the realized load (homozygous, expressed and with effects on the current generation) and the masked load (heterozygous, recessive deleterious mutations that can be expressed in future generations).36 Recently, several studies have reported purging of genetic load, especially in long-term isolated and inbred populations.44,45 However, few studies on big cats have focused on reporting mutational load levels (but see de Manuel et al.34 and Khan et al; 46). In leopards, several genomic studies have recently focused on genomics and landscape ecology, but the incidence of mutational load across subspecies has not been evaluated yet.23,32 Here, we found a reduced total genetic load (in the number of high-impact deleterious alleles) for both Arabian samples compared with African and the rest of Asian leopards (Figure 6A). This result is also supported by the ratio of derived alleles (Figure 6B). Interestingly, when high-impact, moderate-impact, and low-impact deleterious alleles were split in homozygous or heterozygous derived alleles (as a proxy of realized and masked load), we observed a high-realized load (with a large number of both alleles in the homozygous state) and a lower masked load (with less alleles in the heterozygous state) in Arabian leopards (Figures 6A and S6), following the theoretical predictions after a prolonged bottleneck.36 During population decline, the composition of genetic load changes, with many previously masked mutations becoming expressed and, as a consequence, increasing the realized load and decreasing the masked load.36 We also observe an increase of the realized load and a decrease of the masked load for Asian (excluding Arabian) compared to African leopards (Figure 6; Figure S6). However, in contrast to Arabian leopards, the total load seems to have experienced a relaxation of the purging, possibly due to a posterior expansion. The Arabian leopard has been long-term isolated and together with the putative slow increase in inbreeding (and subsequent reduction of masked load), this has possibly allowed the purging of high-impact deleterious alleles, mainly in the heterozygous state. As a consequence, around 40% and 60% of the high-impact deleterious alleles were found within ROH of both Arabian leopards (Figure S7). During bottlenecks, inbreeding and drift increase homozygosity, turning masked load into realized load. Then, purifying selection acts upon the high-impact deleterious mutations but the moderate-impact deleterious mutations escape it, and if the bottleneck is prolonged enough, some of them become fixed.36 Interestingly, the Arabian leopard shows similar levels of moderate-impact and low-impact deleterious alleles compared to African and the rest of Asian leopards, as selection has not been as strong in these alleles. This is consistent with the purging of recessive high-impact deleterious alleles as a consequence of increased inbreeding.36,47 For instance, purging has been reported in some island populations of the Endangered kakapo (Strigops habroptilus), with lower genetic diversity and population effective size and higher inbreeding and longer ROH than other mainland populations.44
Inbreeding depression is defined as the increased homozygosity resulted from inbreeding, causing a reduction in fitness.48 The genome-wide heterozygosity values for the two Arabian individuals were among the lowest ever reported for the species, and more than half of the genomes of the two Arabian individuals were within ROH (Figure 4; Table S2). However, the amount of high-impact deleterious alleles was significantly lower and concordant with purging of deleterious mutations along a prolonged past bottleneck, as reported by evolutionary history analyses (Figures 3 and 6), highlighting the importance of performing a comprehensive genomic study to evaluate the status of a species. If only heterozygosity values of the Arabian leopard were taken into account, the subspecies could be considered to be on the brink of an important inbreeding depression, but until now no reduction in fitness has been reported. Surprisingly, the out-of-ROH heterozygosity for both Arabian leopards is almost twice that of the genome-wide heterozygosity of some Asian individuals, highlighting that the areas without ROH contain higher genetic diversity. Nonetheless, low genome-wide genetic diversity can result in a loss of adaptive potential, leading to health issues and causing a negative impact on reproductive fitness and life quality.4,49 In fact, genetic depletion has been already highlighted as one of the current threats for the Arabian leopard.27
Overall, our results highlight that genomic tools are essential to assess the situation of endangered and elusive species. Genome-wide data successfully resolved the phylogenetic position of the Arabian leopard as sister to the rest of Asian subspecies, a topic highly discussed over the last decades.20,23,32 Moreover, using genomic data we provided accurate estimations of genetic diversity, population structure, and demographic history for the Critically Endangered Arabian leopard, confirming a prolonged past bottleneck with subsequent inbreeding and purging of deleterious mutations. Ultimately, our study stresses the benefits of using genomic tools both from an evolutionary and a conservation perspective and highlights the importance of integrating the field of genomics when managing in-situ and ex situ endangered and elusive species.
Limitations of the study
This study presents the first two complete genomes for the Arabian leopard subspecies. However, these two samples derive from captive-bred individuals, which could affect some genomic analyses performed here. Moreover, the medium coverage and number of individuals sampled in this study could mask the complete picture of genetic variation in this subspecies. Despite this, we hope that this work will positively contribute to increase the general knowledge about the Critically Endangered Arabian leopard as well as its ongoing captive breeding programmes.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Panthera pardus nimr whole blood samples | This study | SRR23089148 |
| Panthera pardus nimr whole blood samples | This study | SRR23089147 |
| Critical commercial assays | ||
| QIAGEN DNeasy Blood&Tissue Kit | Psomagen | N/A |
| Deposited Data | ||
| Panthera pardus pardus | Paijmans et al. 202123 | ERR5671309 |
| Panthera pardus pardus | Pečnerová et al. 202132 | ERR5056141 |
| Panthera pardus pardus | Paijmans et al. 202123 | ERR5671306 |
| Panthera pardus pardus | Pečnerová et al. 202132 | ERR5056152 |
| Panthera pardus pardus | Paijmans et al. 202123 | ERR5671308 |
| Panthera pardus pardus | Pečnerová et al. 202132 | ERR5056108 |
| Panthera pardus pardus | Pečnerová et al. 202132 | ERR5056111 |
| Panthera pardus pardus | Paijmans et al. 202123 | ERR5671300 |
| Panthera pardus nimr | This study | SRR23089148 |
| Panthera pardus nimr | This study | SRR23089147 |
| Panthera pardus tulliana | Paijmans et al. 202123 | ERR5671303 |
| Panthera pardus tulliana | Paijmans et al. 202123 | ERR5671312 |
| Panthera pardus fusca | Paijmans et al. 202123 | ERR5671313 |
| Panthera pardus fusca | Paijmans et al. 202123 | SRR11286171 |
| Panthera pardus kotya | Paijmans et al. 202123 | ERR5671301 |
| Panthera pardus delacouri | Paijmans et al. 202123 | ERR5671302 |
| Panthera pardus melas | Paijmans et al. 202123 | ERR5671317 |
| Panthera pardus orientalis | Paijmans et al. 202123 | ERR5671311 |
| Panthera pardus orientalis | SRA SRR5382750 | SRA SRR5382750 |
| Panthera pardus orientalis | Kim et al., 201616 | SRR3041424 |
| Panthera leo | de Manuel et al., 202034 | SRR11286181 |
| Panthera leo | de Manuel et al., 202034 | ERR5056108 |
| Panthera leo | de Manuel et al., 202034 | SRR836361 |
| Felis catus | Buckley et al., 2020 | GCA_000181335.4 |
| Panthera pardus spelaea | Paijmans et al. 201823 | MH588611 |
| Panthera pardus saxicola | Paijmans et al. 201823 | MH588612 |
| Software and algorithms | ||
| Fastp | Chen et al., 201850 | https://github.com/OpenGene/fastp |
| FastQC | Andrews, 201051 | https://github.com/s-andrews/FastQC |
| bwa-mem v0.7.17 | H. Li, 201352 | https://github.com/lh3/bwa |
| Samtools v1.9 | Li et al., 200953 | https://github.com/samtools/ |
| PicardTools | Broad Institute, 202154 | https://github.com/broadinstitute/picard |
| GATK v.4.1.7 | McKenna et al., 201055 | https://github.com/broadinstitute/gatk |
| bcftools | Danecek et al., 202156 | https://github.com/samtools/bcftools |
| vcftools | Danecek et al., 201157 | https://github.com/vcftools/vcftools |
| Plink v1.9 | Chang et al., 201558 | https://github.com/insilico/plink |
| ADMIXTURE | Alexander et al., 200959 | https://github.com/NovembreLab/admixture |
| R v.3.6.3 | R Core Team, 202160 | https://www.r-project.org/ |
| ggplot2 | Wickham, 201661 | https://ggplot2.tidyverse.org/ |
| ANGSD v.0.933 | Korneliussen et al., 201462 | https://github.com/ANGSD/NgsRelate |
| IQ-TREE2 | Nguyen et al., 201563 | http://www.iqtree.org/ |
| BEAST2 v2.6.6 | Bouckaert et al., 201964 | http://www.beast2.org/ |
| MitoFinder v.1.4.1 | Allio et al., 202065 | https://github.com/RemiAllio/MitoFinder |
| RAxML-NG | Kozlov et al., 201966 | https://github.com/amkozlov/raxml-ng |
| PSMC | Li & Durbin, 201135 | https://github.com/lh3/psmc |
| bedtools | Quinlan & Hall, 201067 | https://github.com/arq5x/bedtools2 |
| Admixtools | Patterson et al., 201268 | https://github.com/uqrmaie1/admixtools |
| SNPeff v.4.3 | Cingolani et al., 201269 | https://github.com/pcingola/SnpEff |
Resource availability
Lead contact
Further information can be requested via the lead contact, Gabriel Mochales Riaño (gabriel.mochales@csic.es).
Materials availability
This study did not generate any new reagents.
Experimental model and subject details
Materials and methods
Data collection
We generated WGS data for two Arabian leopards (Panthera pardus nimr): a male and a female from the captive breeding programme at Prince Saud Al Faisal Wildlife Research Centre, National Center for Wildlife / the Royal Commission for AlUla (RCU). The male (NIM1 – Arabian1) is the third generation of the captive breeding programme whilst the female (NIM2 - Arabian2) is the fourth generation of the same programme. Genomic DNA was extracted from whole blood samples using the MagAttract HMW Kit (Qiagen) following manufacturer’s protocols. Then, we prepared Illumina libraries following the BEST protocol70 with minor modifications. The libraries were sequenced in a 2 × 101 bp HiSeq4000 lane aiming for a 10x depth of coverage. Additionally, we gathered the raw sequencing data (FASTQs) from 18 leopards of all the other subspecies and three lions (Panthera leo) (See Table 1) generated in previous publications.16,23,32,34 Additionally, we obtained two full mitochondrial genomes for two extinct Pleistocene samples from Paijmans et al.25
Method details
Data processing
Raw reads for each of the 23 genomes were filtered, and adapters removed with fastp.50 A minimum base quality score was set to 30, and adapter detection for paired-end sequencing was activated, with a required fragment length of 50bp. Trimming of poly-G/X tails and correction in overlapped regions were specified. All other parameters were set as default. Filtered sequences were visually explored with FastQC51 to ensure data quality and absence of adapters. Filtered reads were mapped against the reference genome of a female domestic cat (Felis_catus_9.0; GenBank assembly accession: GCA_000181335.4)71 with bwa-mem v0.7.17.52 Mapped reads were sorted with Samtools v1.9.53 Duplicated reads were marked and removed with PicardTools54 and reads with mapping quality lower than 30 were discarded. SNP calling was carried out with HaplotypeCaller from GATK,55 with BP_resolution and split by chromosome. For each chromosome, individual genotypes were joined using CombineGVCFs with convert-to-base-pair-resolution and the GenotypeGVCFs tool was then applied to include non-variant sites. Finally, the whole dataset split by chromosome was concatenated with bcftools concat,56 keeping only the autosomes. For some analyses we generated a separate dataset including only leopards. Then, we filtered the raw callset by excluding variants matching at least one of the following criteria: Quality by Depth (QD) < 10, Mapping Quality (MQ) < 50, Fisher Strand test (FS) > 10, StrandOddsRatio (SOR) > 4, MQRankSum < −5 && MQRankSum >5 and ReadPosRankSum < −5 && ReadPosRankSum >5. Later, genotypes were filtered using vcftools,57 with a minimum variant quality of 30, removing indels, keeping only biallelic SNPs, allowing 10% missing data and filtering for minor allele frequency (MAF) of 0.001, filtering out monomorphic sites and keeping only variable positions. Repetitive regions were identified from the cat reference genome and removed. A second dataset was created for population genetic analyses keeping only unlinked SNPs through bcftools56 using a maximum value of r2 = 0.5.
Quantification and statistical analyses
Population structure analyses
We performed a Principal Component Analysis (PCA) with the leopard unlinked filtered dataset using Plink v1.9.58 We applied ADMIXTURE59 to detect ancestral populations from k = 2 to k = 5. A total of 20 replicates for each K were calculated, selecting the best K value after 20 cross validations. Visualization of results from these analyses was performed with R v.3.6.360 and the R package ggplot2.61
Phylogeny
We reconstructed the phylogenomic relationships of all samples (including the three lion genomes) using only autosomal chromosomes selected from bam files with the view function from Samtools v1.9.53 These bam files were used to generate individual pseudohaploid consensus sequences with ANGSD v.0.933,62 taking a consensus-based sampling approach (-doFasta 3) in non-overlapping sliding windows of 1 Mbp. Maximum likelihood trees for each window along the cat reference genome were calculated with IQ-TREE263 applying a GTR+I+G substitution model, with the lion samples as outgroup. Windows where one individual had >50% missing data were removed, leaving a total of 2,330 non-overlapping windows. A maximum clade-credibility tree was created with TreeAnnotator v2.6.4. from BEAST2 v2.6.6.64 We tested the effect of window size by repeating the analyses for smaller window sizes (500 Kbp and 100 Kbp) for the largest chromosome (240 Mbp) and similar topologies were obtained. Additionally, the mitochondrial DNA (mtDNA) was assembled with MitoFinder v.1.4.165 from a subset of the samples and a maximum likelihood phylogeny was reconstructed using RAxML-NG,66 with the GTR+I+G model and performing 1,000 bootstraps.
Demographic history
We inferred the demographic history of leopards with the Pairwise Sequential Markovian Coalescent (PSMC) software,35 for which we used four high-coverage individuals together with the two Arabian leopards. Heterozygous positions were obtained from bam files with Samtools v1.953 and data was filtered for low mapping (<30) and base quality (<30). Minimum and maximum depths were set at half and double the average coverage for each sample, respectively. Only autosomal data was considered. A rate of 1.1 x 10−8 substitutions/site/generation and a generation time of 5 years were used, following.32 Other parameters were set as default following previous knowledge on leopard genomics.16 For the two Arabian leopards, 100 bootstraps were calculated. Final results were plotted with the psmc_plot.pl function from PSMC (https://github.com/lh3/psmc). Finally, high-coverage individuals were downsampled to the coverage level of the Arabian leopards and PSMC was run again for comparative purposes.
Genomic diversity and ROHs
We used the raw dataset to calculate average genome heterozygosity per individual. We generated non-overlapping sliding windows of 100 Kbp for the domestic cat reference genome and took only sites (both variant and invariant) with site quality higher than 30 (QUAL field in a VCF file from GATK). Only windows containing more than 60,000 unfiltered sites were considered. Later, six individuals were downsampled to coverage levels similar to those of the Arabian leopards, for comparative reasons. After obtaining homozygous regions per individual (see below), out-of-ROH heterozygosity was calculated using the intersect function from bedtools.67 Runs of Homozygosity (ROHs) were calculated per individual based on the density of heterozygous sites in the genome using the implemented Hidden Markov Model (HMM) in bcftools roh function72 with –AF-dflt 0.4 (following Armstrong et al.73) on the filtered dataset. We kept ROHs with a Phred Score of at least 70 and with a minimum length of 100 Kbp. Finally, we performed a correlation test between the number and cumulative length of ROHs. Visualisation for all analyses was carried out with ggplot2.61
Introgression
We used the D-Statistics (ABBA-BABA tests) method using the qpDstat function from Admixtools68 to test for gene flow between the different subspecies of leopards and especially between the Arabian leopard and its geographically closest subspecies of leopards (i.e., African and Anatolian leopards). To do so, we used the filtered VCF file (16.32 million SNPs) containing all subspecies of leopards and using the lion as an outgroup. We first explored the possible introgression between African and Arabian leopards, as previous mitochondrial phylogenetic analyses showed African and Arabian leopards clustering together.20 When introgression between African and Arabian leopards was tested, we created a model with (((X, Arabian),African),Lion), where X are all other subspecies of leopards. Posterior comparisons used the same procedure, changing the order of the samples accordingly. Later, and following population genetic analyses, we examined if the Arabian leopards contained past signals of introgression with the Anatolian leopard. Finally, we tested the subsequent possible introgressions between the remaining subspecies.
Mutational load
We estimated the mutational load for coding regions in all leopard individuals using SNPeff v.4.3.69 with the filtered dataset, not allowing missingness (10.35 million SNPs). A database was created using the cat annotation file available in GenBank (Felis_catus_9.0; GenBank assembly accession: GCA_000181335.4).71 We identified putative deleterious variants in the four categories established by the manual: 1) Low: mostly harmless or unlikely to change protein behaviour (i.e., synonymous variants); 2) Moderate: non-disruptive variants that might change protein effectiveness (i.e., missense variants); 3) High: assumed to have a high (disruptive) impact in the protein, probably causing protein truncation, loss of function (LoF) or triggering nonsense mediated decay (i.e., stop codons, splice donor variant and splice acceptor, start codon lost, etc.); 4) Modifier: usually non-coding variants or variants affecting non-coding genes, where predictions are difficult or there is no evidence of impact (i.e., downstream or upstream variants).69 Then, following the author’s recommendations,69 we counted the number of derived alleles based on the cat reference genome with low, moderate and high predicted levels for homozygous (multiplied by two as they are represented twice) and heterozygous alleles, removing observations with warnings. Later, we calculated the amount of high-impact deleterious alleles in and outside of ROH regions using bcftools. Because we only used sites found in all individuals, variants in these two categories were separately counted and no bootstrapping was needed, as discussed in Dussex et al.44 Following Xue et al.,74 we calculated a statistic which compares two populations in relation to the number of derived alleles found at sites in one population rather than the other, for a particular variant category. Basically, for each category of variants we estimated at each site i the observed allele frequency in Population X as fxi = dxi/nxi, where nxi is the total number of alleles called in population X and dxi is the total number of called derived alleles. Similarly, we define fyi for population Y. Then, for each category C of variants, we estimated:
Finally, we calculated the ratio RXY = Freqpop-x/Freqpop-y, where a value of 1 corresponds to no change in frequency, RXY > 1 represents a decrease in frequency in population Y relative to population X and RXY < 1 results from an increase in frequency in population Y relative to population X.
Acknowledgments
This work was supported by funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement No. 864203), “Unidad de Excelencia María de Maeztu”, funded by the AEI (CEX2018-000792-M) to TMB, and grants PGC2018-098290-B-I00 (MCIU/AEI/FEDER, UE) Spain, PID2021-128901NB-I00 (MCIN/AEI/10.13039/501100011033 and by ERDF, A way of making Europe), Spain to SC. We acknowledge both Anders Albrechtsen and Nicolas Dussex for useful comments with introgression and mutational load analyses, respectively, and Núria Hermosilla for interesting comments about phylogenetic analysis. We further acknowledge Johanna Paijmans for helping during the downloading of some of the leopard genomes. We want to thank Prem Aguilar for his useful comments in a previous version of this manuscript as well as two anonymous reviewers. We are deeply thankful to Dr. Mohammed Qurban, chief executive officer of the National Center for Wildlife (NCW), and the former vice president of the formal Saudi Wildlife Authority Dr. Hani Tatwany for their support and encouragement since the beginning of the project. We are also grateful to all personnel involved in the Arabian Leopard programme at the Royal Commission for AlUla and their partners in the Arabian Leopard project for support, with the Approval of Saudi Wildlife Authority under permit SWA/6386. GM-R was funded by an FPI grant from the Ministerio de Ciencia, Innovación y Universidades, Spain (PRE2019-088729), AT is supported by “la Caixa”” doctoral fellowship program (LCF/BQ/DR20/11790007), BB-C was funded by FPU grant from Ministerio de Ciencia, Innovación y Universidades, Spain (FPU18/04742) and HT-C was supported by a "Juan de la Cierva - Formación" postdoctoral fellowship (FJC2021-046832-I) funded by MCIN/AEI/10.13039/501100011033 and by the European Union NextGenerationEU/PRTR.
Author contributions
Conceptualization, G.M.R., C.F., M.M., A.T., B.B.C., H.T.C., M.S., T.M.B., and S.C.; Formal Analyses, G.M.R. and C.F.; Investigation, G.M.R., C.F., R.H.M.A., and M.S.; Writing—Original Draft, G.M.R.; Writing—Review & Editing, all authors have read, edited, and approved the manuscript; Visualization, G.M.R.; Funding Acquisition, T.M.B. and S.C.
Declaration of interests
The authors declare no competing interests.
Published: July 27, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107481.
Contributor Information
Gabriel Mochales-Riaño, Email: gabriel.mochales@csic.es.
Claudia Fontsere, Email: claudia.fontsere@sund.ku.dk.
Tomas Marques-Bonet, Email: tomas.marques@upf.edu.
Salvador Carranza, Email: salvador.carranza@ibe.upf-csic.es.
Supplemental information
Data and code availability
-
•
Genomic data for this study has been deposited in GenBank and it is publicly available as of the date of publication under the GenBank BioProject: PRJNA924233. Accession numbers are listed in the key resources table.
-
•
All the code used in this study has been uploaded to GitHub (https://github.com/gubrins/Arabian-leopard) and it is publicly available as of the date of publication.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact (gabriel.mochales@csic.es) upon request.
References
- 1.Frankham R., Ballou J.D., Briscoe D.A. Camb. Univ. Press Camb.; 2002. Introduction to Conservation Genetics. [Google Scholar]
- 2.Shafer A.B.A., Wolf J.B.W., Alves P.C., Bergström L., Bruford M.W., Brännström I., Colling G., Dalén L., De Meester L., Ekblom R., et al. Genomics and the challenging translation into conservation practice. Trends Ecol. Evol. 2015;30:78–87. doi: 10.1016/J.TREE.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 3.Palsbøll P.J., Bérubé M., Allendorf F.W. Identification of management units using population genetic data. Trends Ecol. Evol. 2007;22:11–16. doi: 10.1016/J.TREE.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Irizarry K.J.L., Bryant D., Kalish J., Eng C., Schmidt P.L., Barrett G., Barr M.C. Integrating Genomic Data Sets for Knowledge Discovery: An Informed Approach to Management of Captive Endangered Species. Int. J. Genomics. 2016;2016:2374610. doi: 10.1155/2016/2374610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson W.E., Onorato D.P., Roelke M.E., Darrell Land E., Cunningham M., Belden R.C., McBride R., Jansen D., Lotz M., Shindle D., et al. Genetic Restoration of the Florida Panther. 31 R Leb . C Tomé Mater Sci Eng A. 2005;102:63–87. doi: 10.1126/science.1192465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohn M.H., Murphy W.J., Ostrander E.A., Wayne R.K. Genomics and conservation genetics. Trends Ecol. Evol. 2006;21:629–637. doi: 10.1016/J.TREE.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 7.Romanov M.N., Koriabine M., Nefedov M., De Jong P.J., Ryder O.A. Construction of a California condor BAC library and first-generation chicken-condor comparative physical map as an endangered species conservation genomics resource. Genomics. 2006;88:711–718. doi: 10.1016/j.ygeno.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 8.Wright B.R., Farquharson K.A., McLennan E.A., Belov K., Hogg C.J., Grueber C.E. A demonstration of conservation genomics for threatened species management. Mol. Ecol. Resour. 2020;20:1526–1541. doi: 10.1111/1755-0998.13211. [DOI] [PubMed] [Google Scholar]
- 9.Luikart G., England P.R., Tallmon D., Jordan S., Taberlet P. The power and promise of population genomics: from genotyping to genome typing. Nat. Rev. Genet. 2003;4:981–994. doi: 10.1038/NRG1226. [DOI] [PubMed] [Google Scholar]
- 10.Camacho-Sanchez M., Velo-Antón G., Hanson J.O., Veríssimo A., Martínez-Solano Í., Marques A., Moritz C., Sílvia C. Comparative assessment of range-wide patterns of genetic diversity and structure with SNPs and microsatellites: A case study with Iberian amphibians. Ecol. Evol. 2020;10:10353–10363. doi: 10.1002/ece3.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khan S., Nabi G., Ullah M.W., Yousaf M., Manan S., Siddique R., Hou H. Overview on the Role of Advance Genomics in Conservation Biology of Endangered Species. Int. J. Genomics. 2016;2016:3460416. doi: 10.1155/2016/3460416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho Y.S., Hu L., Hou H., Lee H., Xu J., Kwon S., Oh S., Kim H.M., Jho S., Kim S., et al. The tiger genome and comparative analysis with lion and snow leopard genomes. Nat. Commun. 2013;4:2433. doi: 10.1038/ncomms3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Figueiró H.V., Li G., Trindade F.J., Assis J., Pais F., Fernandes G., D Santos S.H., Hughes G.M., Komissarov A., Antunes A., et al. Genome-wide signatures of complex introgression and adaptive evolution in the big cats. Sci Adv. 2017;3:e1700299. doi: 10.1126/sciadv.1700299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fontsere C., Kuhlwilm M., Morcillo-Suarez C., Alvarez-Estape M., Lester J.D., Gratton P., Schmidt J.M., Dieguez P., Aebischer T., Álvarez-Varona P., et al. Population dynamics and genetic connectivity in recent chimpanzee history. Cell Genom. 2022;2 doi: 10.1016/j.xgen.2022.100133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frandsen P., Fontsere C., Nielsen S.V., Hanghøj K., Castejon-Fernandez N., Lizano E., Hughes D., Hernandez-Rodriguez J., Korneliussen T.S., Carlsen F., et al. Targeted conservation genetics of the endangered chimpanzee. Hered. 2020;125:15–27. doi: 10.1038/s41437-020-0313-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim S., Cho Y.S., Kim H.M., Chung O., Kim H., Jho S., Seomun H., Kim J., Bang W.Y., Kim C., et al. Comparison of carnivore, omnivore, and herbivore mammalian genomes with a new leopard assembly. Genome Biol. 2016;17:211. doi: 10.1186/s13059-016-1071-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lorenzana G.P., Figueiró H.V., Kaelin C.B., Barsh G.S., Johnson J., Karlsson E., Morato R.G., Sana D.A., Cullen L., May J.A., et al. Whole-genome sequences shed light on the demographic history and contemporary genetic erosion of free-ranging jaguar (Panthera onca) populations. J. Genet. Genomics. 2022;49:77–80. doi: 10.1016/J.JGG.2021.10.006. [DOI] [PubMed] [Google Scholar]
- 18.Supple M.A., Shapiro B. Conservation of biodiversity in the genomics era. Genome Biol. 2018;19:131. doi: 10.1186/S13059-018-1520-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miththapala S., Seidensticker J., O’Brien S.J. Phylogeographic Subspecies Recognition in Leopards {Pantherapardus): Molecular Genetic Variation. Conserv. Biol. 1996;10:1115–1132. [Google Scholar]
- 20.Uphyrkina O., Johnson W.E., Quigley H., Miquelle D., Marker L., Bush M., O’Brien S.J. Phylogenetics, genome diversity and origin of modern leopard, Panthera pardus. Mol. Ecol. 2001;10:2617–2633. doi: 10.1046/j.0962-1083.2001.01350.x. [DOI] [PubMed] [Google Scholar]
- 21.Hayward M.W., Henschel P., O’brien J., Hofmeyr M., Balme G., Kerley G.I.H., Hayward M.W. Prey preferences of the leopard (Panthera pardus) J. Zool. 2006;270:298–313. doi: 10.1111/j.1469-7998.2006.00139.x. [DOI] [Google Scholar]
- 22.Diedrich C.G. Late Pleistocene leopards across Europe – northernmost European German population, highest elevated records in the Swiss Alps, complete skeletons in the Bosnia Herzegowina Dinarids and comparison to the Ice Age cave art. Quat. Sci. Rev. 2013;76:167–193. doi: 10.1016/j.quascirev.2013.05.009. [DOI] [Google Scholar]
- 23.Paijmans J.L.A., Barlow A., Becker M.S., Cahill J.A., Fickel J., Förster D.W.G., Gries K., Hartmann S., Havmøller R.W., Henneberger K., et al. African and Asian leopards are highly differentiated at the genomic level. Curr. Biol. 2021;31:1872–1882.e5. doi: 10.1016/j.cub.2021.03.084. [DOI] [PubMed] [Google Scholar]
- 24.Jacobson A.P., Gerngross P., Lemeris J.R., Schoonover R.F., Anco C., Breitenmoser-Würsten C., Durant S.M., Farhadinia M.S., Henschel P., Kamler J.F., et al. Leopard (Panthera pardus) status, distribution, and the research efforts across its range. PeerJ. 2016;4:e1974. doi: 10.7717/peerj.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paijmans J.L.A., Barlow A., Förster D.W., Henneberger K., Meyer M., Nickel B., Nagel D., Worsøe Havmøller R., Baryshnikov G.F., Joger U., et al. Historical biogeography of the leopard (Panthera pardus) and its extinct Eurasian populations. BMC Evol. Biol. 2018;18:156. doi: 10.1186/s12862-018-1268-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hemmer H. Fossil history of living Felidae. Carnivore. 1979;2:58–61. [Google Scholar]
- 27.Mallon D., Budd K. Regional Red list status of Carnivores in the arabian Peninsula. Camb. UK Gland Switz. IUCN Sharjah UAE Environ. Prot. Areas Auth. 2011 [Google Scholar]
- 28.Islam M., Boug A., Al-Shehri A. PSFWRC SWA Saudi Arab; 2017. National Strategy and Action Plan Arabian Leopard Saudi Arabia. [Google Scholar]
- 29.Jackson R., Islam Z., Boug A., Al Shehri A. 2011. Camera-Trapping Manual for the Arabian Leopard. [Google Scholar]
- 30.Budd J., Leus K. The Arabian Leopard Panthera pardus nimr conservation breeding programme. Zool. Middle East. 2013;54:141–150. doi: 10.1080/09397140.2011.10648905. [DOI] [Google Scholar]
- 31.Perez I., Geffen E., Mokady O. Critically Endangered Arabian leopards Panthera pardus nimr in Israel: Estimating population parameters using molecular scatology. Oryx. 2006;40:295–301. doi: 10.1017/S0030605306000846. [DOI] [Google Scholar]
- 32.Pečnerová P., Garcia-Erill G., Liu X., Nursyifa C., Waples R.K., Santander C.G., Quinn L., Frandsen P., Meisner J., Stæger F.F., et al. High genetic diversity and low differentiation reflect the ecological versatility of the African leopard. Curr. Biol. 2021;31:1862–1871.e5. doi: 10.1016/j.cub.2021.01.064. [DOI] [PubMed] [Google Scholar]
- 33.Kitchener A.C., Breitenmoser-Würsten C., Eizirik E., Gentry A., Werdelin L., Wilting A., Yamaguchi N., Abramov A.V., Christiansen P., Driscoll C., et al. Cat News; 2017. A Revised Taxonomy of the Felidae : The Final Report of the Cat Classification Task Force of the IUCN Cat Specialist Group. [Google Scholar]
- 34.De Manuel M., Barnett R., Sandoval-Velasco M., Yamaguchi N., Garrett Vieira F., Zepeda Mendoza M.L., Liu S., Martin M.D., Sinding M.H.S., Mak S.S.T., et al. The evolutionary history of extinct and living lions. Proc. Natl. Acad. Sci. USA. 2020;117:10927–10934. doi: 10.1073/pnas.1919423117/-/DCSupplemental. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H., Durbin R. Inference of human population history from individual whole-genome sequences. Nat. 2011;475:493–496. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bertorelle G., Raffini F., Bosse M., Bortoluzzi C., Iannucci A., Trucchi E., Morales H.E., van Oosterhout C. Genetic load: genomic estimates and applications in non-model animals. Nat. Rev. Genet. 2022;23:492–503. doi: 10.1038/s41576-022-00448-x. [DOI] [PubMed] [Google Scholar]
- 37.Ray-Brambach R.R., Stommel C., Rödder D. Home ranges, activity patterns and habitat preferences of leopards in Luambe National Park and adjacent Game Management Area in the Luangwa Valley, Zambia. Mamm. Biol. 2018;92:102–110. doi: 10.1016/j.mambio.2017.11.002. [DOI] [Google Scholar]
- 38.Excoffier L., Ray N. Surfing during population expansions promotes genetic revolutions and structuration. Trends Ecol. Evol. 2008;23:347–351. doi: 10.1016/J.TREE.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 39.Li G., Davis B.W., Eizirik E., Murphy W.J. Phylogenomic evidence for ancient hybridization in the genomes of living cats (Felidae) Genome Res. 2016;26:1–11. doi: 10.1101/GR.186668.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahajan S., Karikalan M., Chander V., Pawde A.M., Saikumar G., Semmaran M., Lakshmi P.S., Sharma M., Nandi S., Singh K.P., et al. Detection of SARS-CoV-2 in a free ranging leopard (Panthera pardus fusca) in India. Eur. J. Wildl. Res. 2022;68:59. doi: 10.1007/s10344-022-01608-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ralls K., Sunnucks P., Lacy R.C., Frankham R. Genetic rescue: A critique of the evidence supports maximizing genetic diversity rather than minimizing the introduction of putatively harmful genetic variation. Biol. Conserv. 2020;251 doi: 10.1016/j.biocon.2020.108784. [DOI] [Google Scholar]
- 42.Ceballos F.C., Joshi P.K., Clark D.W., Ramsay M., Wilson J.F. Runs of homozygosity: windows into population history and trait architecture. Nat. Rev. Genet. 2018;19:220–234. doi: 10.1038/nrg.2017.109. [DOI] [PubMed] [Google Scholar]
- 43.Oliveira M.E., Saranholi B.H., Dirzo R., Galetti P.M., Jr. A review of philopatry and dispersal in felids living in an anthropised world. Mamm Rev. 2022;52:208–220. doi: 10.1111/mam.12275. [DOI] [Google Scholar]
- 44.Dussex N., van der Valk T., Morales H.E., Wheat C.W., Díez-Del-Molino D., von Seth J., Foster Y., Kutschera V.E., Guschanski K., Rhie A., et al. Population genomics of the critically endangered kākāpō. Cell Genom. 2021;1 doi: 10.1016/j.xgen.2021.100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kleinman-Ruiz D., Lucena-Perez M., Villanueva B., Fernández J., Saveljev A.P., Ratkiewicz M., Schmidt K., Galtier N., García-Dorado A., Godoy J.A. Purging of deleterious burden in the endangered Iberian lynx. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2110614119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khan A., Patel K., Shukla H., Viswanathan A., van der Valk T., Borthakur U., Nigam P., Zachariah A., Jhala Y.V., Kardos M., Ramakrishnan U. Genomic evidence for inbreeding depression and purging of deleterious genetic variation in Indian tigers. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2023018118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caballero A., Bravo I., Wang J. Inbreeding load and purging: implications for the short-term survival and the conservation management of small populations. Hered. 2017;118:177–185. doi: 10.1038/hdy.2016.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gooley R., Hogg C.J., Belov K., Grueber C.E. No evidence of inbreeding depression in a Tasmanian devil insurance population despite significant variation in inbreeding. Sci. Rep. 2017;7:1830. doi: 10.1038/s41598-017-02000-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Farquharson K.A., Hogg C.J., Grueber C.E. A meta-analysis of birth-origin effects on reproduction in diverse captive environments. Nat. Commun. 2018;9:1055. doi: 10.1038/s41467-018-03500-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen S., Zhou Y., Chen Y., Gu J. Bioinformatics. Oxford University Press; 2018. Fastp: An ultra-fast all-in-one FASTQ preprocessor; pp. i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andrews S. 2010. FastQC: A Quality Control Tool for High Throughput Sequence Data. [Google Scholar]
- 52.Li H. 2013. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. [Google Scholar]
- 53.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Broad Institute . Broad Inst. GitHub Repos; 2021. Picard Tools. [Google Scholar]
- 55.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O., Whitwham A., Keane T., McCarthy S.A., Davies R.M., Li H. Twelve years of SAMtools and BCFtools. GigaScience. 2021;10 doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/GR.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.R Core Team . 2021. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 61.Wickham H. Springer-Verlag; 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- 62.Korneliussen T., Albrechtsen A., Nielsen R. 2014. ANGSD: Analysis of Next Generation Sequencing Data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nguyen L.T., Schmidt H.A., Von Haeseler A., Minh B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bouckaert R., Vaughan T.G., Barido-Sottani J., Duchêne S., Fourment M., Gavryushkina A., Heled J., Jones G., Kühnert D., De Maio N., et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019;15:e1006650. doi: 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Allio R., Schomaker-Bastos A., Romiguier J., Prosdocimi F., Nabholz B., Delsuc F. MitoFinder: Efficient automated large-scale extraction of mitogenomic data in target enrichment phylogenomics. Mol. Ecol. Resour. 2020;20:892–905. doi: 10.1111/1755-0998.13160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kozlov A.M., Darriba D., Flouri T., Morel B., Stamatakis A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35:4453–4455. doi: 10.1093/BIOINFORMATICS/BTZ305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/BIOINFORMATICS/BTQ033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Patterson N., Moorjani P., Luo Y., Mallick S., Rohland N., Zhan Y., Genschoreck T., Webster T., Reich D. Ancient Admixture in Human History. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cingolani P., Platts A., Wang L.L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/FLY.19695/SUPPL_FILE/KFLY_A_10919695_SM0001.ZIP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carøe C., Gopalakrishnan S., Vinner L., Mak S.S.T., Sinding M.H.S., Samaniego J.A., Wales N., Sicheritz-Pontén T., Gilbert M.T.P. Single-tube library preparation for degraded DNA. Methods Ecol. Evol. 2018;9:410–419. doi: 10.1111/2041-210X.12871. [DOI] [Google Scholar]
- 71.Buckley R.M., Davis B.W., Brashear W.A., Farias F.H.G., Kuroki K., Graves T., Hillier L.W., Kremitzki M., Li G., Middleton R.P., et al. A new domestic cat genome assembly based on long sequence reads empowers feline genomic medicine and identifies a novel gene for dwarfism. PLoS Genet. 2020;16:e1008926. doi: 10.1371/JOURNAL.PGEN.1008926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Narasimhan V., Danecek P., Scally A., Xue Y., Tyler-Smith C., Durbin R. BCFtools/RoH: A hidden Markov model approach for detecting autozygosity from next-generation sequencing data. Bioinformatics. 2016;32:1749–1751. doi: 10.1093/bioinformatics/btw044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Armstrong E.E., Taylor R.W., Miller D.E., Kaelin C.B., Barsh G.S., Hadly E.A., Petrov D. Long live the king: Chromosome-level assembly of the lion (Panthera leo) using linked-read, Hi-C, and long-read data. BMC Biol. 2020;18:3. doi: 10.1186/s12915-019-0734-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xue Y., Prado-Martinez J., Sudmant P.H., Narasimhan V., Ayub Q., Szpak M., Frandsen P., Chen Y., Yngvadottir B., Cooper D.N., et al. Mountain gorilla genomes reveal the impact of long-term population decline and inbreeding. Science. 2015;348:242–245. doi: 10.1126/science.aaa3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Genomic data for this study has been deposited in GenBank and it is publicly available as of the date of publication under the GenBank BioProject: PRJNA924233. Accession numbers are listed in the key resources table.
-
•
All the code used in this study has been uploaded to GitHub (https://github.com/gubrins/Arabian-leopard) and it is publicly available as of the date of publication.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact (gabriel.mochales@csic.es) upon request.