Abstract
Adeno-associated virus capsids are composed of three proteins, VP1, VP2, and VP3. Although VP1 is necessary for viral infection, it is not essential for capsid formation. The other capsid proteins, VP2 and VP3, are sufficient for capsid formation, but the functional roles of each protein are still not well understood. By analyzing a series of deletion mutants of VP2, we identified a region necessary for nuclear transfer of VP2 and found that the efficiency of nuclear localization of the capsid proteins and the efficiency of virus-like particle (VLP) formation correlated well. To confirm the importance of the nuclear localization of the capsid proteins, we fused the nuclear localization signal of simian virus 40 large T antigen to VP3 protein. We show that this fusion protein could form VLP, indicating that the VP2-specific region located on the N-terminal side of the protein is not structurally required. This finding suggests that VP3 has sufficient information for VLP formation and that VP2 is necessary only for nuclear transfer of the capsid proteins.
Adeno-associated virus (AAV) type 2, a nonpathogenic human parvovirus, has an icosahedral capsid 20 to 25 nm in diameter (1, 7). The capsid consists of three proteins, VP1, VP2, and VP3, which have molecular masses of 87, 72, and 58 kDa, respectively. They are expressed from the same open reading frame by means of alternative initiation codons (2, 3). The ratio of the three proteins in wild-type AAV-2, which has been shown to be 1:1:10 (5), is reflected in the relative amounts of the proteins expressed in the cell. Studies of mutant viruses have indicated that VP1 is necessary for infection but not for capsid formation (6, 14, 16). On the other hand, VP2 and VP3 are considered necessary for capsid formation (12, 17), but the functional role of each protein is still not well understood.
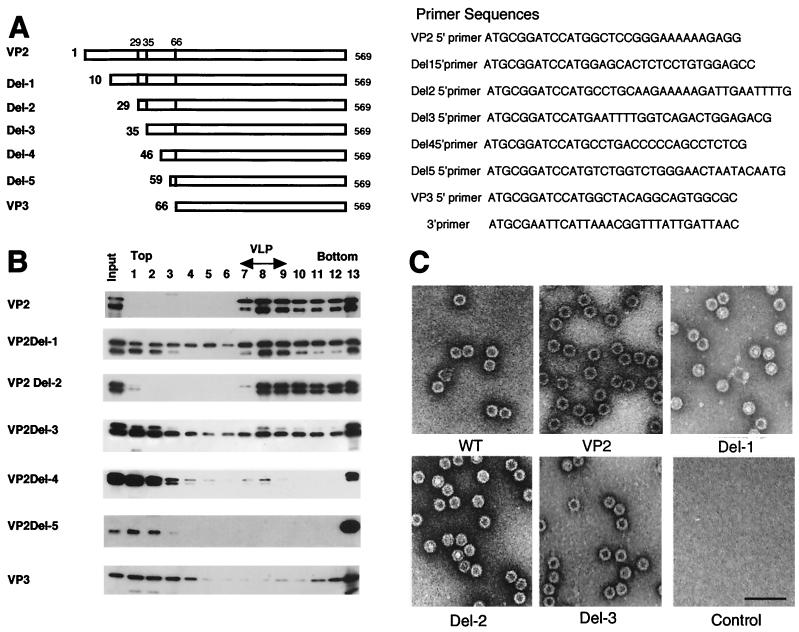
To clarify the functional domain of VP2 essential for capsid formation, we made a series of N-terminal deletion mutants of VP2 and tested their ability to form virus-like particles (VLPs) in the baculovirus protein expression system. DNA fragments encoding VP2 and VP3 and five truncated VP2 proteins (termed Del-1 to Del-5; Fig. 1A) with sequential N-terminal deletions were obtained by PCR using pAV2 as template (11) (a complete clone of AAV-2, kindly provided by B. J. Carter). A common 3′ oligonucleotide and seven different 5′ oligonucleotides containing a BamHI linker and an ATG start codon were used as primers to generate the truncated VP2s as well as VP2 and VP3 (Fig. 1). For construction of the VP2 expression vector, the unusual translation initiation codon, ACG, was mutated to ATG. The amplified DNA fragments were subcloned into pFastbac1 (Invitrogen), and recombinant baculoviruses were generated according to the manufacturer’s instructions (Invitrogen). After purification and amplification of the recombinant viruses, Spodoptera frugiperda Sf9 cells maintained in TC-100 (Gibco BRL) were infected with each of the recombinant viruses and harvested on day 4. The expression of these proteins was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by immunoblot analysis using anti-VP3 antibody raised against a recombinant VP3 (courtesy of M. Ikada and I. Tamai, MBL, Nagoya, Japan).
FIG. 1.
Analysis of VLP formation by N-terminal deletion analysis of VP2. (A) Schematic representation of truncated VP2 proteins. The numbers on the left indicate the first amino acid of each VP2 protein. (B) Sucrose density gradient analysis of N-terminal deletion mutants of VP2. A series of N-terminal deletion mutants were expressed by using recombinant baculoviruses. VLP formation was analyzed by using sucrose density gradient ultracentrifugation. Each fraction was subjected to SDS-PAGE followed by immunoblotting with anti-VP3 serum. Smaller proteins detected in each sample were degradation products of VP2 as determined by amino acid sequence analyses of the smaller proteins. Fraction numbers (from the top to the bottom of the gradient) are shown at the top of the figure. VLP-containing fractions were identified by electron microscopy. (C) Electron microscopy. Purified wild-type empty capsid and VLPs from recombinant baculovirus-infected cells were negatively stained and observed by electron microscopy. Wild-type empty capsid was prepared as described by Ruffing et al. (12). Bar, 100 nm.
To examine the ability of these proteins to form VLPs, each of the cell homogenates infected by each recombinant baculovirus was subjected to sucrose density gradient analysis. A total of 107 infected cells were harvested and suspended in 1 ml of sonication buffer A (20 mM Tris-HCl [pH 8.0]–150 mM NaCl–1 mM phenylmethylsulfonyl fluoride [PMSF]) and sonicated for 5 min at the maximum power of a handheld sonicator (Tomy, Tokyo, Japan). The extract was centrifuged at 15,000 × g for 10 min at 4°C, the pellet was sonicated again, and the supernatants were pooled. An aliquot of the sample was loaded onto a 5-ml sucrose gradient (5 to 20% sucrose in 20 mM Tris-HCl [pH 8.0] and 150 mM NaCl) and centrifuged at 35,000 rpm at 4°C for 2 h using an SW50.1 rotor (Beckman). After the centrifugation, the gradients were fractionated and the fractions were subjected to SDS-PAGE followed by immunoblot analysis (Fig. 1B). For the electron microscopic analysis, 5 × 108 infected cells were used. After centrifugation of the cell extract at 100,000 × g for 1 h, the pellet was resuspended in sonication buffer A and layered onto a CsCl step gradient made up of four densities ranging from 1.15 to 1.44 mg/ml. After centrifugation at 34,000 rpm for 2.5 h at 15°C in an SW50.1 rotor, the VLP-containing fractions were collected and dialyzed against 20 mM Tris-HCl [pH 8.0] and 300 mM NaCl. The dialyzed sample was then treated with DNase and RNase in the presence of 5 mM MgCl2 and again purified by CsCl step gradient centrifugation as described above. The VLP-containing fractions were collected, negatively stained by uranyl acetate, and analyzed by electron microscopy (H-7000; Hitachi, Japan) (Fig. 1C).
Del-1 and Del-2, the deletion mutants which lacked 9 and 28 amino acids, respectively, of the N terminus of VP2, could form VLPs efficiently. In contrast, Del-3, which lacked 34 amino acids of the VP2 N terminus, was much less efficient in forming VLPs. The efficiency of VLP formation further decreased when more amino acids were deleted and Del-5, which lacked 58 amino acids, could not form VLPs at all (Fig. 1). These results indicated that the region between 29 and 34 amino acids from the N terminus is the most important region of VP2 for VLP formation.
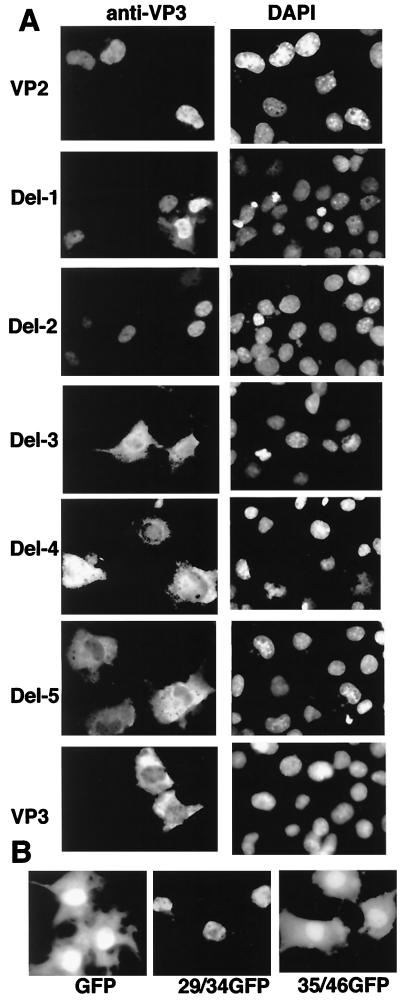
Similarity of the amino acid sequence of the region between 29 and 34, PARKRL, with a typical nuclear localization signal (NLS) raised the possibility that this region functions as an NLS in VP2. To confirm the effect of this region, the deletion mutants were recloned into mammalian expression vectors and the nuclear localization experiment was performed in COS1 cells. DNA fragments encoding VP2 and its truncated versions were subcloned into pcDL-SRα296 (15). COS1 cells were maintained in Dulbecco’s modified Eagle’s medium (Gibco BRL) supplemented with 10% fetal calf serum, and transfection was carried out by lipofection (Gibco BRL). Forty-eight hours after transfection, the locations of the proteins were examined by indirect immunofluorescence staining with antibodies against VP3 as described previously (13).
Del-1 and Del-2 were localized in the nucleus, whereas Del-3, Del-4, and Del-5 were distributed throughout the nucleus and cytoplasm. Except in Del-1 and Del-2, as the number of missing N-terminal residues increased, the nuclear localization efficiency decreased. These results reinforced the conclusion that the main NLS of VP2 is located between residues 29 and 34.
To confirm the NLS function of this region, amino acid residues corresponding to the region were fused to green fluorescent protein (GFP) (Clontech). Oligonucleotides coding for the regions from amino acids 29 to 34 and 35 to 45 in the N terminus of VP2 were synthesized (ATGCGAATTCATGCC TGCAAGAAAAAGAT TGATGG TGAGCAAGGGC and ATGCGAATTCATGAATTTTGGTCAGACTGGAGACGCAGACTCAGTAATGGTGAGCAAGGGC, respectively) and introduced into the GFP vector, pEGFP-N1 (Clontech). These plasmids were transfected into COS1 cells, and the localization of the expressed proteins was examined by fluorescence microscopy 48 h after transfection. When the region between residues 29 to 34 was combined to GFP, the fused protein was detected only in the nucleus whereas GFP alone and the fused protein with the region between residues 35 to 46 were detected both in the cytoplasm and the nucleus (Fig. 2B). These results indicated that the region could function as an NLS even in a heterogeneous protein.
FIG. 2.
NLS activity of the VP2 N-terminal region. (A) Subcellular distribution of the N-terminal deletion mutants of VP2. COS1 cells were transfected with plasmids containing VP2, VP3, and the panel of VP2 deletion mutants. Localization of individual proteins was analyzed by indirect immunofluorescence. Cells were stained with anti-VP3 antibody and subsequently with rhodamine-conjugated rabbit anti-immunoglobulin G. 4′,6-Diamidino-2-phenylindole (DAPI)-stained COS1 cells in the same fields are also shown. (B) NLS activity of the peptide from the VP2 N-terminal region. Fluorescent microscopic images of GFP and GFP-fusion proteins expressed in COS1 cells are shown. 29/34GFP and 35/46GFP indicate the localization of the fusion proteins having the amino acid 29 to 34 and 35 to 46 regions, respectively.
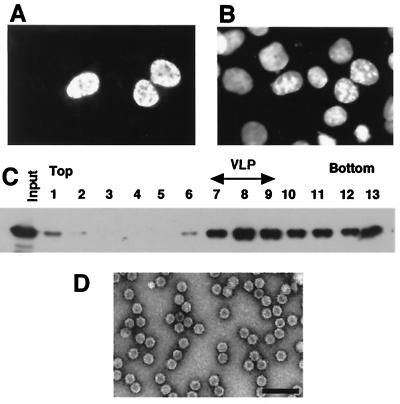
To clarify the relationship between nuclear localization and VLP formation, the NLS of simian virus 40 (SV40) large T antigen was added to the N terminus of VP3 and the resulting NLS-VP3 chimeric protein was examined to determine its subcellular localization and ability to form VLPs. Briefly, an oligonucleotide encoding the NLS of SV40 (ATGCGGATCCATGGCACCACCAAAGAAGAAGCGAAAGG T TATGGC TACAGGCAGTGGCGC) was synthesized and introduced into the VP3 expression vector to allow expression of the SV40 NLS fused VP3 protein. As shown in Fig. 3, NLS-VP3 was localized in the nucleus when it was expressed in COS1 cells, whereas VP3 was found in both the nucleus and the cytoplasm. The DNA fragment encoding NLS-VP3 was recloned into pFastbac1 and analyzed as described above after expression in Sf9 cells. In the sucrose density gradient, NLS-VP3 sedimented to the same position as the wild-type empty particles (Fig. 3C), indicating that NLS-VP3 was proficient for VLP formation. Electron microscopy confirmed the presence of VLPs in the peak fraction and revealed that the NLS-VP3-derived VLPs were morphologically indistinguishable from the wild-type empty particles (Fig. 3D). As it was suggested that capsid formation of AAV is dependent on the level of expression of capsid proteins (17), we expressed the NLS-VP3 protein in COS1 cells and analyzed VLP formation. Sucrose density gradient and electron microscopic analyses showed that it was possible to detect VLP formation in COS1 cells (data not shown), supporting the idea that nuclear transport is crucial for VP3 capsid formation. Thus, the expression level is not the only critical factor for VLP formation.
FIG. 3.
VLP formation of NLS-VP3. (A) Subcellular distribution of NLS-VP3. COS1 cells were infected with plasmids containing the NLS-VP3 construct and after 48 h of transfection, the subcellular distribution of NLS-VP3 was analyzed by immunofluorescence staining with anti-VP3 antibody followed by rhodamine-conjugated rabbit anti-immunoglobulin G. (B) DAPI-stained COS1 cells. The image is the same field as shown in panel A. (C) Sucrose density gradient analysis of NLS-VP3. Sf9 cells were infected with a recombinant baculovirus containing NLS-VP3 and the cell extract was subjected to sucrose density gradient to analyze for VLP formation. Each fraction was subjected to SDS-PAGE followed by immunoblotting with anti-VP3 antibody. (D) Electron microscopy. VLPs were purified by CsCl density gradient ultracentrifugation and analyzed by electron microscopy with negative staining. Bar, 100 nm.
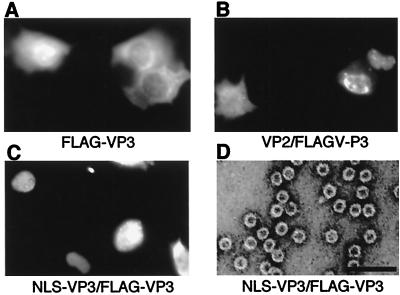
To confirm that the peptide fused to the N terminus of VP3 does not affect VLP formation, we constructed FLAG (8)-fused VP3 (FLAG-VP3) as described above. The oligonucleotide sequence for the fused protein was ATGCGGATCCATGGAC TACAAGGATGACGACAAGATGGC TACAGGCAG TGGCGC. Only when FLAG-VP3 was expressed with NLS-VP3 or VP2 in COS1 cells, nuclear localization of FLAG-VP3 was confirmed by using anti-FLAG antibody (BABCO) (Fig. 4). The DNA fragment encoding FLAG-VP3 was recloned into pFastbac1, the VLP formation of insect-expressed FLAG-VP3 was analyzed, and electron microscopy was performed as described above. VLP formation could be confirmed only when FLAG-VP3 was expressed with NLS-VP3 (Fig. 4D) or VP2 (data not shown). The presence of FLAG-VP3 in VLP was confirmed by immunoblotting using anti-FLAG antibody (data not shown). These results indicated that the peptides fused to the N terminus of VP3 were not essential for VLP formation and that VP3 possesses all the necessary information for VLP formation once it is translocated into the nucleus.
FIG. 4.
Nuclear transport and VLP formation of FLAG-VP3 in the presence of NLS-VP3. (A to C) Subcellular localization of FLAG-VP3 construct alone (A) or together with expression vectors containing VP2 (B) or NLS-VP3 (C). The localization of FLAG-VP3 was examined by indirect immunofluorescence staining. Cells were stained with anti-FLAG monoclonal antibody followed by rhodamine-conjugated mouse anti-immunoglobulin G. (D) Electron microscopy. Sf9 cells were infected with a recombinant baculovirus containing FLAG-VP3 and NLS-VP3. VLP purified by CsCl density gradient analysis was negatively stained and observed. Bar, 100 nm.
Amino acid sequences of the VPs of AAV show significant identity with those of other parvoviruses, such as human B19 and canine parvovirus. In contrast to AAV, it has been shown that the capsids of these parvoviruses can be formed in the presence of the major capsid protein, VP2, alone (4, 9). In previous work, it was shown that the most N-terminal 25-amino-acid residues of VP2 of B19 were not required for capsid assembly (10). Amino acid sequence alignment of VP2 protein of B19 with AAV indicates that the minimal protein capable of B19 capsid formation corresponds to VP3 rather than VP2 of AAV (data not shown). This also supports the notion that VP3 contains the sequences critical for the formation of VLPs.
Our finding that the N-terminal region of VP2 located between amino acid residues 29 and 34 plays an important role in nuclear translocation suggests that the major function of VP2 is translocation of VP3 into the nucleus. This finding should be helpful not only in the development of models for AAV capsid formation but also in the development of VLPs as vaccines.
Acknowledgments
This work was supported by a Research Grant from Core Research for Evolutional Science and Technology (CREST) of Japan Science and Technology Corporation (JST); a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture; and a grant for research and development projects in Cooperation with Academic Institutions from New Energy and Industrial Technology Development Organization (NEDO).
REFERENCES
- 1.Atchison R W, Casto B C, Hammon W M. Electron microscopy of adenovirus-associated virus (AAV) in cell cultures. Virology. 1966;29:353–357. doi: 10.1016/0042-6822(66)90045-6. [DOI] [PubMed] [Google Scholar]
- 2.Becerra S P, Koczot F, Fabisch P, Rose J A. Synthesis of adeno-associated virus structural proteins requires both alternative mRNA splicing and alternative initiations from a single transcript. J Virol. 1988;62:2745–2754. doi: 10.1128/jvi.62.8.2745-2754.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berns K I, Bohenzky R A. Adeno-associated viruses: an update. Adv Virus Res. 1987;32:243–306. doi: 10.1016/s0065-3527(08)60479-0. [DOI] [PubMed] [Google Scholar]
- 4.Brown C S, Van Lent J W M, Vlak J M, Spaan W J M. Assembly of empty capsids by using baculovirus recombinants expressing human parvovirus B19 structural proteins. J Virol. 1991;65:2702–2706. doi: 10.1128/jvi.65.5.2702-2706.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buller R M L, Rose J A. Characterization of adenovirus-associated virus-induced polypeptides in KB cells. J Virol. 1978;25:331–338. doi: 10.1128/jvi.25.1.331-338.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hermonat P L, Labow M A, Wright R, Berns K I, Muzyczka N. Genetics of adeno-associated virus: isolation and preliminary characterization of adeno-associated virus type 2 mutants. J Virol. 1984;51:329–339. doi: 10.1128/jvi.51.2.329-339.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoggan M D. Adenovirus associated viruses. Prog Med Virol. 1970;12:211–239. [PubMed] [Google Scholar]
- 8.Hopp T P, Prickett K S, Price C, Libby R T, March C J, Cerretti P, Urdal D L, Conlon P J. A short marker sequence useful for recombinant protein identification and purification. Bio/Technology. 1988;6:1205–1210. [Google Scholar]
- 9.Kajigaya S, Fujii H, Field A, Anderson S, Rosenfeld S, Anderson L J, Shimada T, Young N S. Self-assembled B19 parvovirus capsids, produced in a baculovirus system, are antigenically and immunogenically similar to native virions. Proc Natl Acad Sci USA. 1991;88:46–50. doi: 10.1073/pnas.88.11.4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawase M, Momoeda M, Young N S, Kajigaya S. Modest truncation of the major capsid protein abrogates B19 parvovirus capsid formation. J Virol. 1995;69:6567–6571. doi: 10.1128/jvi.69.10.6567-6571.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laughlin C A, Tratschin J D, Coon H, Carter B J. Cloning of infectious adeno-associated virus genomes in bacterial plasmids. Gene. 1983;23:65–73. doi: 10.1016/0378-1119(83)90217-2. [DOI] [PubMed] [Google Scholar]
- 12.Ruffing M, Zentgraf H, Kleinschmidt J A. Assembly of virus-like particles by recombinant structural proteins of adeno-associated virus type 2 in insect cells. J Virol. 1992;66:6922–6930. doi: 10.1128/jvi.66.12.6922-6930.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sawa C, Goto M, Suzuki F, Watanabe H, Sawada J-I, Handa H. Functional domains of transcription factor hGABP beta1/E4TF1-53 required for nuclear localization and transcription activation. Nucleic Acids Res. 1996;24:4954–4961. doi: 10.1093/nar/24.24.4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smuda J W, Carter B J. Adeno-associated virus having nonsense mutations in the capsid genes: growth in mammalian cells containing an inducible amber suppressor. Virology. 1991;184:310–318. doi: 10.1016/0042-6822(91)90847-5. [DOI] [PubMed] [Google Scholar]
- 15.Takabe Y, Seiki M, Fujisawa J-I, Hoy P, Yokota K, Arai K-I, Yoshida M, Arai N. SRα promoter: an efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of human T-cell leukemia virus type 1 long terminal repeat. Mol Cell Biol. 1988;8:466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tratschin J D, Miller I L, Carter B J. Genetic analysis of adeno-associated virus: properties of deletion mutants constructed in vitro and evidence for an adeno-associated virus replication function. J Virol. 1984;51:611–619. doi: 10.1128/jvi.51.3.611-619.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wistuba A, Kern A, Weger S, Grimm D, Kleinschmidt J A. Subcellular compartmentalization of adeno-associated virus type 2 assembly. J Virol. 1997;71:1341–1352. doi: 10.1128/jvi.71.2.1341-1352.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]