Summary
Asthma is a global chronic airway disease. The expression and role of RNF125, an E3 ubiquitin ligase, in asthma remain uncertain. In this study, we revealed that RNF125 was downregulated in the bronchial epithelium of mice and patients with asthma. Rnf125 hypermethylation was responsible for the low expression of RNF125 in primary airway epithelial cells of mice treated with OVA. Moreover, we demonstrated that RNF125 could attenuate autophagy, oxidative stress, and protect epithelial barrier in vivo and in vitro. Additionally, we identified HMGB1 as a substrate of RNF125, which interacted with the HMG B-box domain of HMGB1 and induced degradation via the ubiquitin proteasome system, reducing autophagy and oxidative stress. Overall, our findings elucidated that hypermethylation of Rnf125 reduced its expression, which promoted autophagy-induced oxidative stress in asthma by increasing HMGB1 stability. These findings offer a theoretical and experimental basis for the pathogenesis of asthma.
Subject areas: Biological sciences, Molecular biology, Molecular interaction
Graphical abstract

Highlights
-
•
RNF125 is downregulated in bronchial epithelial cells in asthma
-
•
RNF125 overexpression in the bronchial epithelium alleviated asthmatic symptoms
-
•
RNF125 significantly reduced autophagy and oxidative stress
-
•
We identified HMGB1 as a substrate of RNF125
Biological sciences; Molecular biology; Molecular interaction
Introduction
Asthma is a chronic airway disease that involves various cells and cell components. It is a common and frequently occurring disease worldwide, and its incidence continues to increase. Its pathogenesis is very complex, and its specific pathogenesis has not been elucidated. Glucocorticoids are the first-line drugs in the treatment of asthma. However, there are many patients with hormone dependence or hormone resistance, and long-term use of glucocorticoids may cause many adverse reactions, such as osteoporosis.1 Therefore, in order to develop more effective treatment strategies, it is necessary to further understand the underlying pathogenesis of asthma.
The airway epithelium is the first barrier against pathogens and allergens,2 and is one of the main parts of pathological changes in asthma. Airway epithelial cell barrier function is significantly impaired in patients with asthma.3 Autophagy is a highly conserved lysosomal dependent pathway that exists in the body and participates in a variety of biological events. It is generally believed that autophagy is over-activated in the airway epithelium of asthma, which is one of the important factors in the occurrence and development of the disease.4,5 A classic inhibitor of autophagy, 3-MA is reported to reduce airway hyperresponsiveness and airway inflammation in animal models of asthma.6 At present, it is widely accepted that excessive autophagy in asthma aggravates oxidative stress, which is a cardinal characteristic.7 Studies have shown that oxidative stress is also overactivated in asthma. The oxygen free radicals in the breath condensate of asthmatic patients are significantly higher than that of healthy control group.8 The lung tissues of asthmatic mice also show obvious oxidative stress.9 The oxidative stress inhibitor, N-acetyl cysteine, has been reported to reduce asthma symptoms.10 However, the mechanisms underlying bronchial epithelial autophagy and oxidative stress in asthma remain unclear.
High-mobility group box 1 (HMGB1), a highly conserved and abundant protein, plays an important role in the study of autophagy and oxidative stress in airway epithelial cells of asthma. HMGB1 is a star molecule in asthma. The expression of HMGB1 in airway epithelium and lung tissue of asthma is higher than that in the control group.11,12 Studies have shown that HMGB1 promotes autophagy. Knockdown of HMGB1 in airway epithelial cells inhibits autophagy.13 Meanwhile, HMGB1 is also reported to induce oxidative stress. The exposure of bronchial epithelial cells 16HBE to HMGB1 enhances the aggravation of reactive oxygen species (ROS).14 However, the mechanism by which HMGB1 affects autophagy and oxidative stress in asthma remains unclear.
RING finger protein 125 (RNF125) is an E3 ubiquitin ligase belonging to the RING domain family.15 Earlier studies on RNF125 include T cell activation promotion,16 interferon signaling pathway negative regulation,17 and HIV-1 replication inhibition.18 In recent years, RNF125 has been found to be involved in the occurrence and development of inflammatory diseases. Knockdown of Rnf125 activates the NLRP3 inflammasome in mouse bone marrow-derived macrophages.19 In human glioma cells, knockdown of RNF125 can increase the expression of inflammatory factors, such as IL-1β and TNF-α.20 Thus, RNF125 is a protective molecule in many diseases. However, no study has evaluated the role of RNF125 in asthma. Whether RNF125 is also a protective molecule in asthma is unknown. Therefore, we explored the expression, function, and potential mechanism of RNF125 in asthma, which may provide some insights into disease pathogenesis and assist in the development of therapeutics.
DNA methylation is a common chemical modification of DNA associated with gene expression programming. Typically, DNA methylation occurs at CpG islands.21 Hypermethylation of the gene often leads to its low expression, and hypomethylation leads to its high expression. Therefore, exploring the occurrence of DNA methylation is an important way to find the cause of expression alterations. However, up to now, whether there is a CpG island in the promotor of RNF125 and whether it could be methylated remains unknown.
In the present study, we aimed to identify the expression of RNF125 in airway bronchial cells in asthma. Moreover, we sought to investigate the reason for the expression alteration in asthma and the mechanisms underlying the effects of RNF125 on autophagy and oxidative stress in asthma. We revealed the roles of RNF125 in asthma using in vitro, in vivo, and molecular studies. It is helpful to understand the pathogenesis of asthma from a different angle, which may provide clinical and scientific significance.
Results
RNF125 was downregulated in bronchial epithelial cells of OVA-treated mice and patients with asthma
We first evaluated RNF125 expression in asthma to identify its potential role. We established murine models of asthma, including day 3, day 7, and day 14 groups according to the duration of ovalbumin (OVA) inhalation. Detailed asthma model establishment was performed as follows: the mice were first sensitized with a mixture of OVA and aluminum hydroxide by intraperitoneal injection, and then challenged with OVA by aerosol inhalation. Challenge means airway stimulated by OVA and that triggers a cascade of reactions. Immunohistochemistry results demonstrated that RNF125 protein expression was similar in the bronchial epithelium of OVA-treated mice to those in the control group on day 3. However, in day 7 and day 14 groups, the expression of RNF125 in the bronchial epithelium of OVA-treated mice were significantly lower than those in the control group. The expression of RNF125 in day 7 group was similar to that in day 14 group (Figures 1A and 1B). Additionally, western blotting results also showed similar conclusions (Figures 1C and 1D). Therefore, day 7 group was then chosen for further research. Then we tried to collect clinical specimens to prove this problem from another aspect. It is very difficult to recruit subjects without thoracic surgery, so we had to settle for second best. We recruited four asthmatic participants and four control participants with thoracic surgery for the study. To ensure that the data were as comparable as possible, we selected samples that were as similar in age, sex, pathological type, and differentiation degree of tumor as possible. As expected, for spirometry parameters, FEV1, FEC1%, FEV1/FVC, MMEF 75/25 was lower in asthmatic group than those in the control group. Meanwhile, bronchodilation test was all positive in asthmatic patients (Table S1). We collected bronchial samples from asthmatic patients and controls for immunohistochemical experiments to explore the expression of RNF125 in bronchial epithelium. Due to limited bronchial samples, lung tissues of the same asthmatic patients and control groups were collected for WB experiments. Immunohistochemistry results showed that the expression of RNF125 in bronchial epithelium of the group with asthma was lower than in the control group (Figures 1E and 1G). Western blotting results demonstrated that RNF125 was downregulated in the lung tissues of the group with asthma (Figures 1F and 1H). Overall, these results suggested that RNF125 expression was downregulated in bronchial epithelial cells in asthma.
Figure 1.
RNF125 is downregulated in bronchial epithelial cells of OVA-treated mice and patients with asthma
(A and B) RNF125 expression in bronchial epithelial cells of OVA-treated mice and controls on day 3, 7, and 14 detected using immunohistochemistry (n = 6). The length of the scale is 100 μm.
(C and D) Protein expression of RNF125 in lung tissues of OVA-treated mice and controls on day 3, 7, and 14 detected using western blotting (n = 6).
(E and G) RNF125 expression in bronchial epithelial cells of asthma patients and controls detected using immunohistochemistry (n = 4). The length of the scale is 50 μm.
(F and H) The protein expression of RNF125 in lung tissues of asthma patients and controls detected using western blotting (n = 4). Statistical analysis—t test. Values are expressed as mean ± standard deviation (∗p < 0.05, ∗∗∗p < 0.001, compared with the control group).
Hypermethylation of Rnf125 reduced its expression in OVA-treated asthmatic bronchial epithelial cells
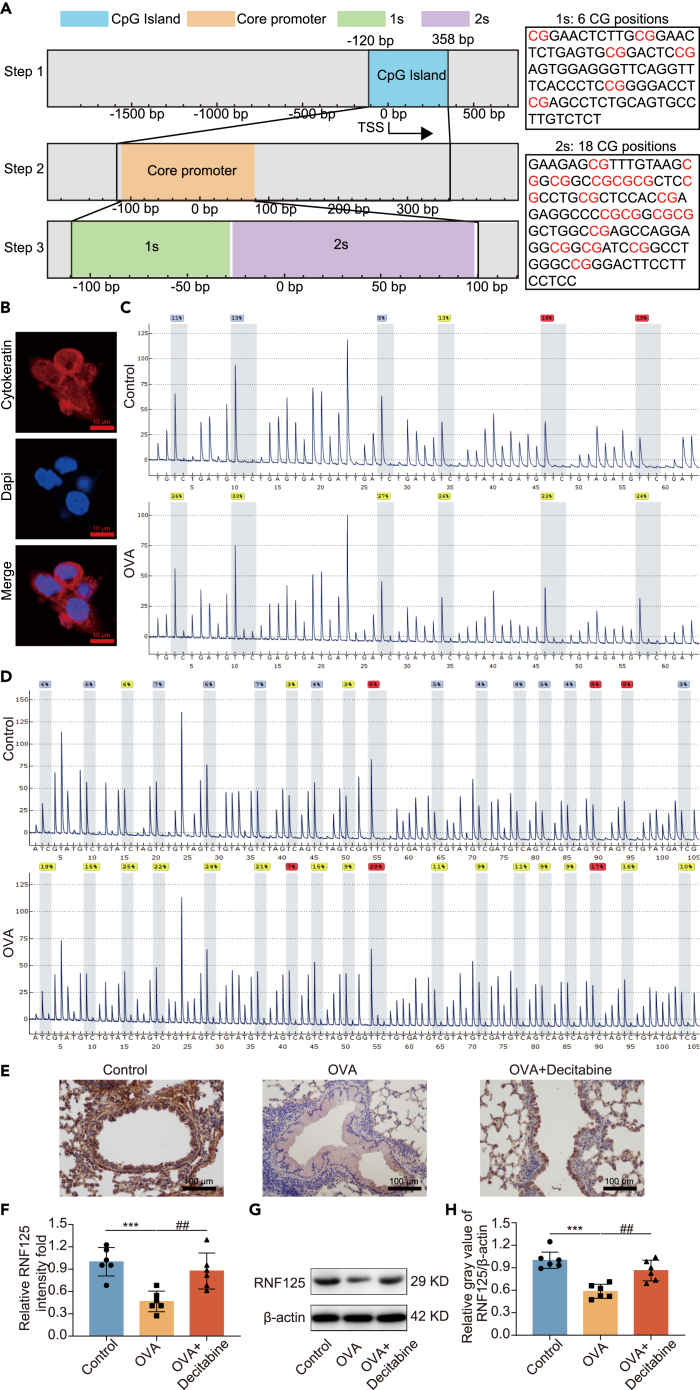
To investigate the reason for the low expression of RNF125 in airway epithelial cells in asthma, Rnf125 gene was searched in UCSC. Firstly, we found there was a CpG island in the promoter region of Rnf125. Secondly, the core promoter region was located exactly at the CpG island. Thirdly, we roughly divided the core promoter region into two regions according to the experimental detection requirements, which were called first sequence (1s) and second sequence (2s). In the 1s, there were 6 CG positions, while in the 2s, there were 18 CG positions (Figure 2A). We isolated primary mice tracheobronchial epithelial cells of the control and OVA-treated mice in vivo. When separated, there were mainly two types of cells, fibroblast, and tracheobronchial epithelial cells. Immunofluorescence showed that the isolated cells were cytokeratin (a marker of epithelial cell) positive cells (> 95%) (Figure 2B). Pyrosequencing showed that the 1s and 2s regions of the Rnf125 gene were hypermethylated in airway epithelial cells of OVA-treated mice compared with those in the control group (Figures 2C and 2D). The percentage of methylation of 1s position 1, 2, 3, 4, 5, and 6 and mean of the 1s in airway epithelial cells of OVA-treated mice were higher than those in the control group were (Figures S1A‒S1F and S1X). The percentage of methylation of 2s position 1, 2, 3, 5, 7, 8, 9, 11, 14, and 18 and mean of the 2s position in airway epithelial cells of OVA-treated mice were higher than those in the control group, while the percentage of methylation of 2s position 4, 6, 12, 13, 15, 16, and 17 in airway epithelial cells of OVA-treated mice were still higher than those in the control group, but not statistically significant (Figures S1G‒S1W and S1Y). The G peak before the 10th position of 2s reaction could not reach the expected peak height, which may be due to the single-nucleotide polymorphism (SNP) site, and the T of the 10th position was also low, so the data of position 10 were not included in the analysis. Next, decitabine, a hypomethylating agent, was used to explore the role of hypermethylation of Rnf125 on its expression. Immunohistochemistry results showed that RNF125 in lung tissues and bronchial epithelium was higher after decitabine treatment (Figures 2E and 2F). Meanwhile, western Blotting results showed that RNF125 in lung tissues was upregulated after decitabine treatment (Figures 2G and 2H). Therefore, these results suggested that hypermethylation of Rnf125 was found in primary airway epithelial cells of OVA-treated mice, which was the reason why RNF125 is downregulated. HMGB1 has been reported to bind to CpG islands to induce methylation.22 We then tried to find the cause of Rnf125 hypermethylation. HMGB1 has been reported to bind to CpG islands to induce methylation.22 Therefore, we explored the effect of HMGB1 on RNF125, and found that there was no change in the expression of RNF125 after overexpression of HMGB1 in 16HBE cells (Figures S2A–S2C). These results suggested that HMGB1 was not the reason affecting RNF125 expression, and there were other factors involved.
Figure 2.
Hypermethylation of Rnf125 reduces its expression in asthmatic bronchial epithelial cells
(A) CpG island (blue box) was identified by the UCSC database, core promotor (orange box) was marked out around about 100 bp above and below the TSS, 1s (green box) contains 6 CG sites, 2s (purple box) contains 18 CG sites.
(B) Cytokeratin protein expression in primary mice epithelial cells detected by immunofluorescence. The length of the scale is 10 μm.
(C) Percentage of methylation of CG sites in 1s in OVA-treated and control mice detected by pyrosequencing.
(D) Percentage of methylation of CG sites in 2s in OVA-treated and control mice detected by pyrosequencing.
(E and F) The expression of RNF125 in bronchial epithelial cells detected using immunohistochemistry in different groups. The length of the scale is 100 μm.
(G and H) RNF125 expression levels in lung tissues after Rnf125 overexpression in bronchial epithelial cells detected using western blotting in different groups. Decitabine group received intraperitoneal injections of 1.5 mg/kg of decitabine 1 h before OVA inhalation.
Detailed methods of OVA-treated asthma mice model were shown in the method. Statistical analysis—one-way ANOVA. Values are expressed as mean ± standard deviation (n = 6, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, compared with the control group; #p < 0.05, ##p < 0.01, ###p < 0.001, compared with the OVA-treated mice).
Overexpression of Rnf125 alleviated lung function, inflammatory cell infiltration, and mucus hypersecretion in vivo
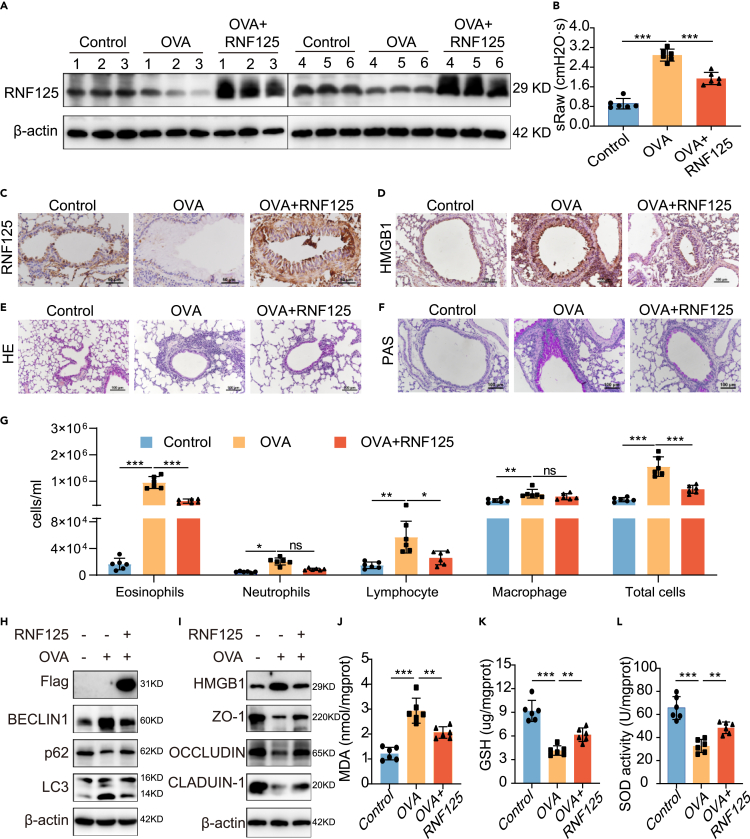
To investigate the function of RNF125 in asthma, we overexpressed Rnf125 in the bronchial epithelium in mice of day 7 group by adeno-associated virus (AAV). OVA-treated asthma mice model was established as follows: the mice were first sensitized with a mixture of OVA and aluminum hydroxide by intraperitoneal injection, and then challenged with OVA by aerosol inhalation. Challenge means airway stimulated by OVA and that triggers a cascade of reactions. The expression of RNF125 in lung tissues was significantly higher after infection with Rnf125 AAV (Figures 3A and S3A). Meanwhile, immunohistochemistry results showed that RNF125 was significantly upregulated in the bronchial epithelium of mice infected with Rnf125 AAV (Figures 3C and S3B). Besides the airway epithelium, overexpression of Rnf125 delivered by adeno-associated Rnf125 increased the expression of airway smooth muscle and alveoli (Figure S3M). These results indicated that we have successfully overexpressed Rnf125 in lung tissues. On this basis, we conducted a series of asthma-related experiments. Lung function analysis revealed that the specific airway resistance (sRaw) of the OVA-treated mice was higher than that of the control group, whereas that of the RNF125 group was significantly lower than that of the OVA-treated mice but higher than that of the control group (Figure 3B). HE staining showed more infiltration of inflammatory cells around the airway in the OVA-treated mice. After Rnf125 overexpression, inflammatory cell infiltration was significantly reduced. The control group showed almost no inflammatory cell infiltration (Figures 3E and S3C). PAS staining of the lungs demonstrated that the epithelial cells of the OVA-treated mice produced more mucus than the control group, whereas the RNF125 group showed less mucus hypersecretion than the OVA-treated mice but was still higher than that in the control group (Figures 3F and S3D).Total and differential leukocyte counts in bronchoalveolar lavage fluid (BALF) showed that the numbers of total leukocytes, eosinophils, neutrophils, lymphocytes, and macrophages in the OVA-treated mice were significantly higher than those in the control group. Rnf125 overexpression reduced the numbers of total leukocytes, eosinophils, and lymphocytes, compared to those in the OVA-treated mice, but the numbers were still higher than those in the control group (Figure 3G). These findings suggested that Rnf125 overexpression in vivo attenuated lung function, airway inflammation, and mucus hypersecretion.
Figure 3.
Overexpression of Rnf125 relieves asthma symptoms and alleviates autophagy, oxidative stress, and protect epithelial barrier in vivo
The 18 mice were divided into 3 groups: control mice (n = 6), OVA-treated mice (n = 6), and Rnf125-treated mice (n = 6). Two weeks before sensitization, mice were intratracheally administered 70 μL of mouse AAV6 (control AAV6 for control mice and OVA-treated mice or Rnf125 overexpression AAV6 for Rnf125-treated mice). On days 15, 22, and 29, mice were sensitized with 200 μL sensitization liquid (for OVA-treated mice and Rnf125-treated mice), containing 50 μg ovalbumin and 1 mg AL(OH)3 intraperitoneally or PBS (for control mice), on days 36–42, the mice were challenged with 2% OVA (for OVA-treated mice and Rnf125-treated mice) or PBS (for control mice) inhalation for 30 min.
(A) RNF125 expression levels in lung tissues after Rnf125 overexpression in bronchial epithelial cells in vivo.
(B) The specific airway resistance (sRaw) in mice after Rnf125 overexpression in bronchial epithelial cells in vivo.
(C) The expression of RNF125 in bronchial epithelial cells detected using immunohistochemistry in different groups (Large magnification). The length of the scale is 50 μm.
(D) The expression of HMGB1 in bronchial epithelial cells detected using immunohistochemistry in different groups. The length of the scale is 100 μm.
(E) Hematoxylin and eosin staining of lung tissues in different groups. The length of the scale is 100 μm.
(F) Periodic acid-Schiff staining of lung tissues in different groups. The length of the scale is 100 μm.
(G) Total and differential leukocyte counts in bronchial alveolar lavage fluid in different groups.
(H) Protein expression levels of Flag, BECLIN1, p62, and LC3 after RNF125 overexpression in vivo.
(I) Protein expression levels of HMGB1, ZO-1, OCCLUDIN, and CLAUDIN 1 after Rnf125 overexpression in vivo.
(J–L) MDA content; (K) GSH content; and (L) SOD activity of lung tissues after overexpression of Rnf125 in vivo.
Statistical analysis—one-way ANOVA. Values are expressed as mean ± standard deviation (n = 6, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, compared with the OVA-treated mice).
Overexpression of Rnf125 alleviated autophagy, oxidative stress, and protected airway epithelial barrier in vivo
Autophagy is a key cellular process, which helps maintain cellular homeostasis. There is growing evidence that autophagy is overactivated in asthma, especially in airway epithelium of asthma patients and asthma mice4,23 and lung tissues in asthma mice,5 which contribute to its pathogenesis. Meanwhile, autophagy inhibitors, 3-methyladenine and chloroquine, have been shown to reduce asthma symptoms.24,25 Therefore, we focused on autophagy. We evaluated the effect of RNF125 on autophagy in vivo. Western blotting results showed that the BECLIN1 and LC3 II/I protein expression levels were increased markedly in the lung tissues of OVA-treated mice, whereas p62 protein expression levels were decreased in the OVA-treated mice compared to those in the control group. These data suggested autophagy was over-activated in the OVA-treated mice, which was widely accepted. Rnf125 overexpression in OVA-treated mice increased the expression of Flag and p62 and reduced the expression of BECLIN1, and LC3 II/I in the lung tissues (Figures 3H and S3E–S3H), demonstrating that RNF125 alleviated autophagy in vivo.
Meanwhile, excessive autophagy induces oxidative stress, which is a cardinal characteristic of asthma.7,26 Oxidative stress refers to the situation in which the capacity of oxidation in the body exceeds that of antioxidation. Oxidative stress is over-activated in the lung tissue of asthmatic mice and asthmatic patients, and inhibition of oxidative stress can help in the treatment of asthma.27 Then we tried to detect changes in oxidative stress. To investigate the role of RNF125 in oxidative stress, we measured MDA, SOD, and GSH levels in the lung tissues. MDA levels were elevated in the OVA-treated mice, whereas SOD and GSH levels were lower in the OVA-treated mice compared to those in the control group. Overexpression of Rnf125 significantly reduced MDA levels and increased SOD and GSH levels compared to those in the OVA-treated mice (Figures 3J‒3L).
Meanwhile, we identified the role of RNF125 on HMGB1. Western blotting and histochemistry results showed that the HMGB1 protein expression levels were increased markedly in the lung tissues of OVA-treated mice and overexpression of Rnf125 in vivo decreased the expression of HMGB1 in lung tissues (Figures 3D, 3I, and S3I). To identify the function of RNF125 in epithelial barrier, we detected the epithelial barrier related protein ZO-1, OCCLUDIN, and CLAUDIN 1 in lung tissues. The expression of ZO-1, OCCLUDIN, and CLAUDIN 1 were decreased in the lung tissues of OVA-treated mice compared to those in the control group. Overexpression of Rnf125 significantly increased all these three proteins compared to those in the OVA-treated mice (Figures 3I and S3J–S3L). Taken together, these data indicated that RNF125 reduced oxidative stress, autophagy, and enhanced epithelial barrier function in vivo.
Overexpression of RNF125 reduced autophagy, oxidative stress, and protected epithelial barrier function in vitro
To further confirm the conclusions obtained from in vivo experiments, we transfected bronchial epithelial cells 16HBE with a Flag-RNF125 overexpression plasmid. After transfection, the expression of RNF125 mRNA and RNF125 protein extremely increased compared to those in the control group (Figures 4A, 4B, and S4A), indicating that RNF125 was successfully overexpressed. We then measured alterations in autophagy after RNF125 overexpression in vitro. Western blotting analysis of autophagy-related proteins showed RNF125 overexpression reduced the autophagy levels. Overexpression of RNF125 decreased the protein expression of BECLIN1, and LC3 II/I and increased the protein expression of p62 (Figures 4D and S4B–S4D). Transmission electron microscopic analysis revealed that RNF125 overexpression reduced autophagosome formation in 16HBE cells (Figure 4C).
Figure 4.
RNF125 inhibits autophagy, oxidative stress, and protect epithelial barrier function in vitro
(A) Relative mRNA expression of RNF125 in 16HBE cells after transfection with RNF125 overexpression plasmid.
(B) Protein expression levels of RNF125 after overexpression of RNF125.
(C) Ultrastructure in 16HBE cells after transfection with RNF125 overexpression plasmid. The black box represents higher magnification. Arrows represent autophagosomes. The length of the scale is 2 μm for lower magnification and 1 μm for higher magnification.
(D) Protein expression levels of BECLIN1, p62, and LC3 after overexpression of RNF125.
(E) Protein expression levels of ZO-1, OCCLUDIN, and CLAUDIN 1 after overexpression of RNF125.
(F and G) ROS detection using fluorescence microscopy and microplate reader after overexpression of RNF125. The length of the scale is 100 μm.
(H) MDA content.
(I and J) GSH content; and (J) SOD activity after overexpression of RNF125 in 16HBE cells.
(K) Relative mRNA expression of RNF125 in 16HBE cells after silencing RNF125.
(L) Protein expression levels of RNF125 after silencing RNF125.
(M) Ultrastructure in 16HBE cells after silencing RNF125. The black box represents higher magnification and arrows represent autophagosomes. The length of the scale is 2 μm for lower magnification and 1 μm for higher magnification.
(N and O) ROS detection using fluorescence microscopy and microplate reader after RNF125 knockdown. The length of the scale is 100 μm.
(P) Protein expression levels of BECLIN1, p62, and LC3 after RNF125 knockdown. (Q) Protein expression levels of ZO-1, OCCLUDIN, and CLAUDIN 1 after RNF125 knockdown.
(R) MDA content.
(S and T) GSH content; and (T) SOD activity after knockdown of RNF125 in 16HBE cells.
Statistical analysis—t test (A, G, H, I, and J), one-way ANOVA (K, O, R, S, and T). Values are expressed as mean ± standard deviation (n = 3, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, compared with the control group).
Moreover, we detected alterations in oxidative stress after RNF125 overexpression in vitro. Fluorescence microscopy and microplate reader analyses demonstrated that RNF125 overexpression reduced ROS levels in 16HBE cells (Figures 4F and 4G). MDA, GSH, and SOD kit analyses revealed that RNF125 overexpression significantly reduced MDA levels but resulted in elevated GSH and SOD levels (Figures 4H‒4J). Then we measured alterations in epithelial barrier related proteins after RNF125 overexpression in vitro. Overexpression of RNF125 increased the protein expression of ZO-1, OCCLUDIN, and CLAUDIN 1 (Figures 4E and S4E–S4G). Overall, these data suggested that RNF125 overexpression decreased autophagy and oxidative stress and increased the epithelial barrier function in 16HBE cells.
Knockdown of RNF125 aggravated autophagy, oxidative stress, and impaired epithelial barrier function in vitro
To further verify the results drawn from RNF125 overexpression, we transfected 16HBE cells with RNF125-siRNA. After transfection, qPCR results showed that the mRNA expression of RNF125 in the RNF125-si1 and RNF125-si2 groups was reduced by more than 80% compared with the control group (Figure 4K). Western blotting results showed that the protein expression of RNF125 decreased in the RNF125-si1 and RNF125-si2 groups compared to that in the control group (Figures 4L and S4H), suggesting that the knockdown of RNF125 was successful. Next, we evaluated alterations in autophagy after RNF125 silencing in vitro. Western blotting results demonstrated that silencing RNF125 increased protein levels of LC3 II/I and decreased protein levels of p62 (Figures 4P and S4I‒S4K). Knockdown of RNF125 in 16HBE slightly increased the expression of BECLIN1 (The increase was small but statistically significant). Compared to the Si-NC group, more autophagosomes were observed in the RNF125-si1 and RNF125-si2 groups, as detected by transmission electron microscopy (Figure 4M). In addition, we measured alterations in oxidative stress after RNF125 silencing in vitro. Knockdown of RNF125 significantly increased ROS levels, as detected by fluorescence microscopy and a microplate reader (Figures 4N and 4O). We also measured MDA, GSH, and SOD levels. Compared to the Si-NC group, there were higher MDA levels and lower GSH and SOD levels in the RNF125-si1 and RNF125-si2 groups (Figures 4R‒4T). Moreover, we measured alterations in epithelial barrier related proteins after RNF125 silencing in vitro. Knockdown of RNF125 significantly decreased ZO-1, OCCLUDIN, and CLAUDIN 1 protein expression (Figures 4Q and S4L–S4N). Taken together, these data indicated that RNF125 silencing increased autophagy, oxidative stress, and decreased epithelial barrier function in vitro.
RNF125 interacted with HMGB1 via its HMG B box domain mainly in nucleus in vitro
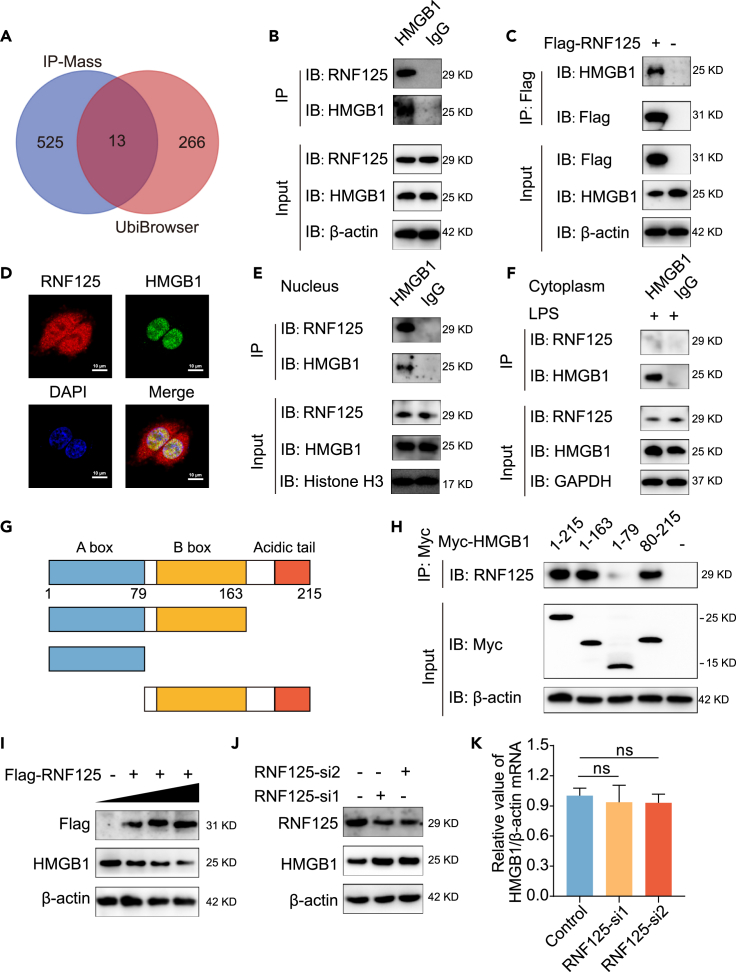
To explore the mechanism of RNF125 reducing autophagy and oxidative stress, we overexpressed RNF125 in bronchial epithelial cells 16HBE, performed immunoprecipitation (IP) experiments, and detected the RNF125 binding protein using mass spectrometry sequencing. Among the interacting protein of RNF125, we found the RNF125 itself was in the detection results of mass spectrometry, which increased the reliability of the results to some extent. Beyond that, however, we did not detect proteins that were known to bind to RNF125. The top 20 biological process, cellular component, and molecular function terms of the RNF125-interacting proteins identified by IP-Mass are shown in Figures S5A‒S5C, which suggests that these proteins were associated with various cellular progresses, biological regulations, and bindings. Meanwhile, the substrate of RNF125 was also analyzed using the bioinformatics database UbiBrowser.28 By intersection of substrates obtained from UbiBrowser and proteins obtained from IP-Mass, a total of 13 proteins were obtained, namely ARHGDIA, DDX17, DDX21, DDX46, DDX3X, DHX9, HMGB1, PCNA, PRKAR2A, SERPINH1, SERPINB12, SNRNP200, SSRP1 (Figure 5A). Of these, only SERPINH1, PCNA, and HMGB1 have been reported to be associated with asthma. Among them, HMGB1 is the most widely studied and has been reported to be highly expressed and play an important role in asthma. Therefore, we chose HMGB1 as the downstream molecule of RNF125 for the study. HMGB1 ranked 138 among all interacting protein of RNF125 (a total 538 proteins) detected by IP-Mass. Therefore, we chose HMGB1 as the downstream molecule of RNF125 for the study, which was not reported before. First, we explored whether RNF125 could bind HMGB1. Co-immunoprecipitation (Co-IP) results demonstrated that RNF125 could interact with HMGB1 (Figures 5B and 5C). Immunofluorescence experiments showed that RNF125 was expressed in both nucleus and cytoplasm, while HMGB1 was expressed in nucleus. The Merge image result showed that RNF125 and HMGB1 co-located in the nucleus, suggesting that the two may bind to each other (Figure 5D). To confirm whether RNF125 binds to HMGB1 in the nucleus or cytoplasm, we extracted nucleoprotein and cytoplasmic protein, respectively, for IP experiment. Co-IP results demonstrated that RNF125 could interact with HMGB1 in the nucleus (Figure 5E). Due to low expression of HMGB1 in the cytoplasm of normal state, we used LPS to stimulate the aggregation of HMGB1 in the cytoplasm. Co-IP results demonstrated that RNF125 could not interact with HMGB1 in cytoplasm (Figure 5F). To identify which domain of HMGB1 interacted with RNF125, HMGB1 was divided into three domains: HMG A box, HMG B box, and acidic tail domains (Figure 5G). Co-IP was performed to analyze the interaction between full-length Myc-tagged HMGB1 or various Myc-HMGB1 truncated mutants and RNF125. The results demonstrated that RNF125 mainly interacted with the HMG B-box domain of HMGB1 (Figure 5H). Thus, these data indicated that RNF125 interacted with HMGB1 through the HMG B-box domain mainly in the nucleus.
Figure 5.
RNF125 interacts with HMGB1 via the HMG B-box domain mainly in nucleus, and reduces HMGB1 expression in vitro
(A) Venn diagram of the intersection of molecules obtained by IP combined with mass spectrometry and RNF125 substrates predicted by UbiBrowser.
(B) HMGB1 co-immunoprecipitated with RNF125.
(C) Flag-RNF125 co-immunoprecipitated with HMGB1.
(D) The immunofluorescence double staining co-localization of RNF125 and HMGB1 in 16HBE cells. Red represents RNF125, green represents HMGB1, blue represents the nucleus. The length of the scale is 50 μm.
(E) HMGB1 co-immunoprecipitated with RNF125 in the nucleus.
(F) HMGB1 co-immunoprecipitated with RNF125 in cytoplasm after LPS (2 μg/mL, 16h) stimulation.
(G) Schematic representation of HMGB1 functional domains.
(H) Full length Myc-tagged HMGB1 or various Myc-HMGB1 truncated mutants co-immunoprecipitated with RNF125.
(I) Protein expression levels of Flag and HMGB1 after transfection with Flag-RNF125 overexpression plasmid in a gradient.
(J) Protein expression levels of RNF125 and HMGB1 after transfection with RNF125-si1 and RNF125-si2.
(K) Relative mRNA expression of HMGB1 in 16HBE cells after silencing RNF125.
Statistical analysis—one-way ANOVA. Values are expressed as mean ± standard deviation (n = 3).
RNF125 reduced HMGB1 expression in vitro
To clarify the regulatory relationship between RNF125 and HMGB1, HMGB1 protein expression was detected in 16HBE cells after overexpression and knockdown of RNF125. First, the cells were transfected with the RNF125 overexpression plasmid in a gradient. As RNF125 expression increased, HMGB1 expression gradually decreased (Figures 5I, S6A, and S6B). Then, cells were transfected with RNF125 siRNA. Compared with those in the Si-NC group, there was a lower expression of RNF125 and higher expression of HMGB1 in the RNF125-si1 and RNF125-si2 groups (Figures 5J, S6C, and S6D). These results suggested that RNF125 reduced the expression of HMGB1 in vitro. Then we detected the difference of mRNA level expression of HMGB1 between the control group and the RNF125 knockdown group in vitro. We found that the mRNA level of HMGB1 did not change after knockdown of RNF125 in 16HBE cells (Figure 5K). These results suggested that RNF125 did not affect the transcription level of HMGB1, and reduced the HMGB1 protein expression, which may due to ubiquitination modification.
RNF125 reduced HMGB1 expression through the ubiquitin proteasome pathway
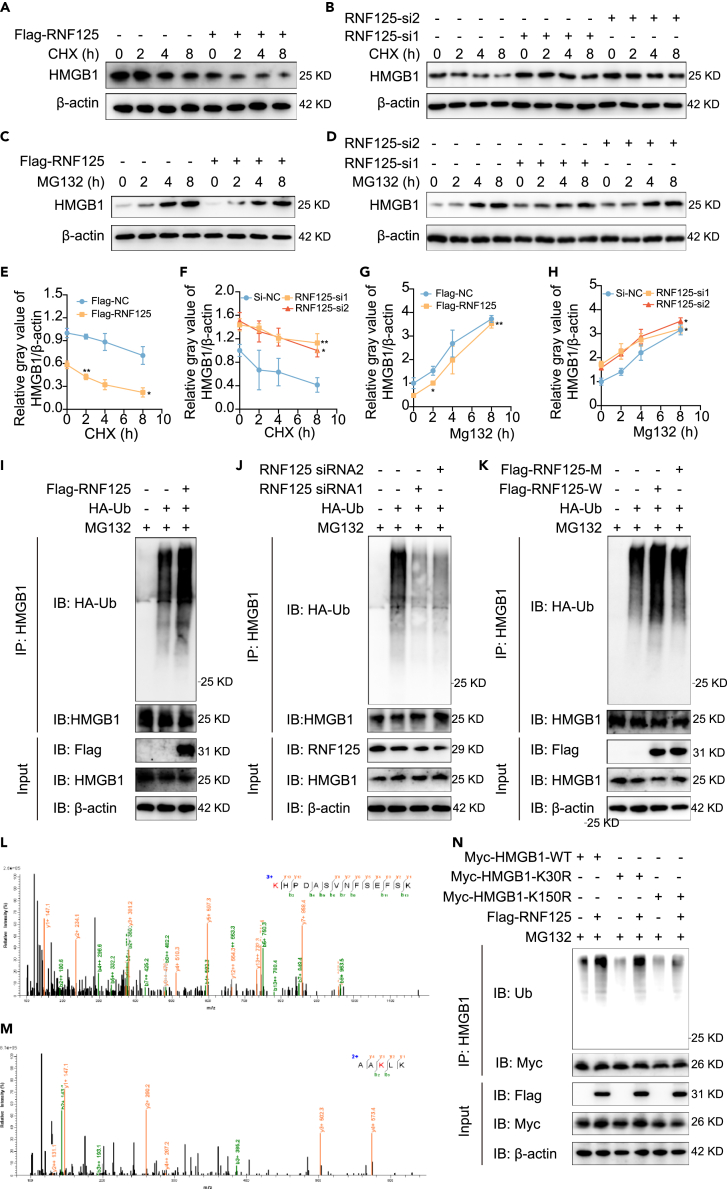
To explore whether RNF125 reduced HMGB1 protein levels by enhancing HMGB1 proteasome degradation, 16HBE cells were treated with the protein synthesis inhibitor cycloheximide (CHX) or a proteasome pathway inhibitor MG132 in a time-gradient manner after overexpression or knockdown of RNF125. First, we measured the protein degradation rate after overexpression and silencing of RNF125. With the extension of CHX use, the expression of HMGB1 in the control group, RNF125 overexpression group, and RNF125 silencing group gradually decreased. Compared to the control group, the expression of HMGB1 decreased more rapidly in the RNF125 overexpression group (Figures 6A and 6E). Expression levels of HMGB1 in the RNF125-si1 and RNF125-si2 groups decreased slower than that in the control group (Figures 6B and 6F). These results indicated that RNF125 increased the rate of HMGB1 protein degradation.
Figure 6.
RNF125 degraded HMGB1 dependent on polyubiquitination partially via the K150 site in vitro
(A and E) RNF125 was overexpressed to evaluate the effect of RNF125 on HMGB1 expression after CHX treatment at various times. The change rate of HMGB1 (compared with 0 h) was compared with the control group.
(B and F) RNF125 was silenced to evaluate the effect of RNF125 on HMGB1 expression after CHX treatment at various times. The change rate of HMGB1 (compared with 0 h) was compared with the control group.
(C and G) RNF125 was overexpressed to evaluate the effect of RNF125 on HMGB1 expression after MG132 treatment at various times. The change rate of HMGB1 (compared with 0 h) was compared with the control group.
(D and H) RNF125 was silenced to evaluate the effect of RNF125 on HMGB1 expression after MG132 treatment at various times. The change rate of HMGB1 (compared with 0 h) was compared with the control group.
(I) HMGB1 was purified using IP, and ubiquitination levels of HMGB1 were detected using anti-HA antibodies after transfection with Flag-RNF125 and HA-Ub and treatment with MG132.
(J) Ubiquitination levels of HMGB1 were detected after knockdown of RNF125.
(K) Ubiquitination levels of HMGB1 were detected after transfection with RNF125 wild type plasmid or RNF125 point mutant plasmid (C72, 75A, lack of ubiquitination function).
(L and M) HMGB1 was purified using IP, and then mass spectrometry was carried out, K30 (L), and K150 (M) may be ubiquitinated.
(N) Flag-RNF125, Myc-HMGB1-WT, Myc-HMGB1-K30R, Myc-HMGB1-K150R were co-expressed and treated with MG132 and Myc-HMGB1 purification was carried out by IP. Ubiquitination levels of HMGB1 were assessed with anti-Ub antibodies.
Statistical analysis—t test (E and G), one-way ANOVA (F and H). Values are expressed as mean ± standard deviation (n = 3, ∗p < 0.05, ∗∗p < 0.01).
Next, we explored the role of the proteasome degradation pathway in RNF125-induced reduction of HMGB1. The expression of HMGB1 in the control, RNF125 overexpression, and RNF125 silencing groups increased gradually in a time-dependent manner. The increased speed and proportion of the expression of HMGB1 in the control group were lower than in the RNF125 overexpression group (Figures 6C and 6G). However, the increased speed and proportion of the expression of HMGB1 in the control group were greater than in the RNF125 siRNA group (Figures 6D and 6H). These data indicated that MG132 could reverse the effects of RNF125 on the expression of HMGB1.
RNF125 degraded HMGB1 dependent on polyubiquitination partially via the K150 site in vitro
To identify whether RNF125-induced degradation of HMGB1 was dependent on polyubiquitination, we detected the ubiquitination level of HMGB1 after transfection with RNF125 overexpression wild-type plasmid, RNF125 siRNA, and RNF125 overexpression point mutant plasmid (C72, 75A). Studies have shown that the C72, 75A mutations of RNF125 lacks its E3 ligase activity.16,17 First, 16HBE cells were transfected with Flag-tagged RNF125 and HA-tagged ubiquitin in the presence of the proteasome pathway inhibitor MG132. After RNF125 overexpression, the ubiquitination level of HMGB1 was significantly increased (Figure 6I). The cells were then transfected with RNF125 siRNA and HA-tagged ubiquitin under MG132. Compared with those of the control group, the ubiquitination levels of HMGB1 were decreased in the RNF125 knockdown group (Figure 6J). Moreover, Flag-tagged point mutant RNF125 was also transfected into cells, and Co-IP results demonstrated that the ubiquitination level in the point mutant RNF125 group, which lose the function of ubiquitination, was remarkably decreased compared to Flag-tagged wild-type RNF125 (Figure 6K). To clarify the specific sites of HMGB1 ubiquitination, we performed IP experiment to enrich HMGB1 protein, and then the sample was electrophoresed using SDS-PAGE, stained by Coomassie Brilliant Blue, and sent to carry out mass spectrometry for ubiquitination sequencing. As shown in Figures 6L and 6M, two sites, K30 and K150, of HMGB1 may be modified by ubiquitination. To identify the sequencing results, we constructed two mutant plasmids, including K30R (K-R) and K150R (K-R). The wild-type plasmid and two mutant plasmids were overexpressed in vitro. Co-IP results showed that overexpression of the K30R mutant plasmid did not affect the ubiquitination of HMGB1 and overexpression of the K150R mutant plasmid significantly decreased the ubiquitination of HMGB1 compared with the wild type group. It is an important site of HMGB1 for its ubiquitination. In addition, overexpression of RNF125 partially increased the ubiquitination of HMGB1 when K150R mutant plasmid overexpression at the same time. This suggests that RNF125 can affect HMGB1 ubiquitination through K150, but there may be other modification sites (Figure 6N). Therefore, these results indicated that RNF125 reduced the expression of HMGB1 in a polyubiquitination-dependent manner partially through the K150 site.
The protection of autophagy-induced oxidative stress by RNF125 overexpression was partially reversed by the overexpression of HMGB1 in vitro
To explore whether the HMGB1 played a role in the mechanism by which RNF125 protection autophagy and oxidative stress, we overexpressed HMGB1 and RNF125 in 16HBE cells simultaneously. Firstly, we measured autophagy alterations in vitro. Transmission electron microscopic results revealed that overexpression of RNF125 reduced the number of autophagosome in 16HBE cells. However, overexpression of HMGB1 during the RNF125 overexpression partially reversed the effect of RNF125 on the autophagosome formation (Figure 7A). Meanwhile, we detected the autophagy-related protein in vitro. As shown in the Figures 7B‒7G, the expression of Myc was significantly increased after Myc-HMGB1 transfected, and the expression of Flag was significantly increased after Flag-RNF125 transfected. Overexpression of RNF125 resulted in decreased BECLIN1 and LC3 II/I expression and increased p62 expression. Overexpression of HMGB1 during overexpression of RNF125 partially reversed the effect of RNF125 on the expression of BECLIN1, LC3 II/I, and p62. Next, we measured the oxidative stress alterations after overexpression of RNF125 and HMGB1 at the same time. RNF125 overexpression significantly reduced ROS and MDA levels and increased GSH and SOD levels. Overexpression of RNF125 and HMGB1 increased the ROS and MDA levels and decreased the GSH and SOD levels compared to those in the RNF125 overexpression group. However, compared to the control group, the ROS and MDA levels were still lower and GSH and SOD levels were still higher in the RNF125 and HMGB1 overexpression group (Figures 7H‒7L). These results suggested that overexpression of RNF125 alleviated autophagy and oxidative stress through HMGB1.
Figure 7.
Overexpression of HMGB1 can partially reverse the inhibition of autophagy-induced oxidative stress caused by RNF125 overexpression in vitro
(A) Ultrastructure in 16HBE cells after overexpression of RNF125 and HMGB1. The black box represents higher magnification, and arrows represent autophagosomes. The length of the scale is 2 μm for lower magnification and 1 μm for higher magnification.
(B–G) Protein expression levels of Myc, Flag, BECLIN1, p62, and LC3 after overexpression of RNF125 and HMGB1.
(H–I) ROS detection using fluorescence microscopy and microplate reader after overexpression of RNF125 and HMGB1 or BECN1. The length of the scale is 50 μm.
(J) MDA content.
(K) GSH content.
(L) SOD activity after overexpression of RNF125 and HMGB1 or BECN1 in 16HBE cells.
Statistical analysis—one-way ANOVA. Values are expressed as mean ± standard deviation (n = 3, ∗∗p < 0.01, ∗∗∗p < 0.001, compared with the control group; #p < 0.05, ##p < 0.01, ###p < 0.001, compared with the Flag-RNF125 group).
Moreover, to explore whether autophagy played a role in the mechanism by which RNF125 protection oxidative stress, we overexpressed BECN1 and RNF125 in 16HBE cells simultaneously. Firstly, BECN1 plasmid was transfected into 16HBE cells, and it was found that BECLIN1 protein expression was significantly increased after BECN1 overexpression (Figures S7A and S7B). Meanwhile, compared with the control group, BECN1 overexpression could increase the autophagy level of cells (Figure S7C). Then, as shown in the Figures 7H‒7L, overexpression of BECN1 and RNF125 increased the ROS and MDA levels and decreased the SOD levels compared to the RNF125 overexpression group. However, the GSH levels were seem to be lower in the overexpression of BECN1 and RNF125 groups compared to the RNF125 overexpression group they were not statistically significant. These results indicated that autophagy could partially reverse the protection of oxidative stress caused by overexpression of RNF125.
Discussion
Asthma is a common chronic airway disease. Currently, approximately 360 million people suffer from asthma.29 The prevalence of adult asthma in China is 4.2%, and the total number of patients is as high as 45.7 million.30 Asthma has become an important public health problem, seriously threatening the physical and mental health of patients, and causing a huge social and economic burden. Therefore, there is an urgent need to explore the possible pathogenesis of asthma to develop more effective treatments.
Ubiquitination is a post-translational modification process in which a substrate molecule covalently binds to one or more ubiquitin molecules via a series of ubiquitination enzymes. E3 ubiquitin ligase is an important enzyme in ubiquitin modification that can specifically recognize substrates and mediate ubiquitin transfer to substrates. Ubiquitin-labeled proteins are recognized and degraded by proteasomes.31 In asthma, ubiquitin modification plays an important role, and several E3 ubiquitin ligases reportedly affect disease progression.32,33,34 RNF125 is a RING-type E3 ubiquitin ligase. In recent years, RNF125 has been found to play an important role in several diseases, including Japanese encephalitis,20 AIDS,35 overgrowth syndrome,36 and cancers.37,38 However, the expression and function of RNF125 in asthma remains unclear. Therefore, we established a classical mouse model of asthma induced by OVA, and collected samples of patients with asthma. Western blotting showed that RNF125 was downregulated in the lung tissues of OVA-treated mice, and lung tissues of patients with asthma. Immunohistochemical localization and semi-quantitative results showed low expression of RNF125 in the bronchial epithelial cells of OVA-treated mice and patients (Figure 1). These results suggested that RNF125 may be involved in the pathogenesis of asthma, and its downregulation in bronchial epithelial cells in asthma may affect its function.
Hypermethylation of DNA core promoter regions is often the cause of low protein expression.39 DNA methylation generally occurs at the CpG sites. Using UCSC database, we found a CpG island in the promoter region of Rnf125. However, up to now, no studies have reported the methylation of Rnf125. We established an asthma mouse model and extracted primary airway epithelial cells of OVA-treated mice. The methylation level of Rnf125 in airway epithelial cells of OVA-treated mice was significantly higher than that in the control group by pyrosequencing. Next, hypomethylating agent, decitabine, treatment in vivo increased the expression of RNF125 in lung tissues and bronchial epithelium (Figure 2). Therefore, hypermethylation of Rnf125 was an important reason for its low expression in the asthmatic airway epithelial cells.
Then we tried to find the mechanisms that lead to RNF125 hypermethylation in asthma. We first focused on the HMGB1 protein. It has been reported that HMGB1 binding to CpG islands of promoter induces methylation.22 However, overexpression of HMGB1 in 16HBE cells did not change the expression of RNF125, indicating that HMGB1 was not a factor affecting the methylation of RNF125. Therefore, we discussed the possible causes of RNF125 hypermethylation to provide some hints for future experiments. Methylation is a reversible enzymatic reaction. DNA methyltransferase and DNA methylation binding proteins are essential for methylation. DNA methyltransferase includes DNMT1, DNMT2, DNMT3a, and DNMT3b, catalyzing the methylation process.40 DNA methylation binding proteins (including MeCP2, MBD1, 2, 3, 4) recognize methylation sites and inhibit transcription.41 The DNA demethylation process, which includes base excision repair and nucleotide excision repair and so on,42 antagonizes the DNA methylation process. Therefore, an imbalance in any of these three aspects (DNA methyltransferase, DNA methylation binding protein, and demethylation pathway) may result in hypermethylation of RNF125.
To investigate the role of RNF125 in asthma, Rnf125 was overexpressed in the lung tissues of mice by adeno-associated viruses. We found that Rnf125 overexpression significantly alleviated airway hyperresponsiveness and reduced airway inflammation and mucus hypersecretion in vivo (Figures 3A–3G). These data indicated that RNF125 was an anti-asthmatic molecule.
Autophagy in bronchial epithelial cells is an important characteristic of asthma. It has been reported that the LC3 II/LC3 I ratio in the lung tissues,43 and the number of autophagosomes in bronchial epithelial cells significantly increase in the asthma group compared to those in the control group.4 After intraperitoneal injection of the autophagy inhibitor 3-MA or bronchial epithelial knockdown of ATG5 in asthma mice, airway inflammation and airway hyperresponsiveness are significantly improved.6 However, the association between RNF125 and autophagy has not yet been reported. Alterations in autophagy were detected in the present study. We found that Rnf125 overexpression in lung tissues inhibited autophagy in vivo (Figure 3H). Similar conclusions were drawn in vitro. Overexpression of RNF125 in 16HBE cells reduced the protein expression levels of BECLIN1, downregulated the LC3 II/LC3 I ratio, increased p62 expression, and reduced the number of autophagosomes (Figures 4C and 4D). Knockdown of RNF125 increased BECLIN1 protein expression levels, upregulated the LC3 II/LC3 I ratio, decreased p62 expression, and increased the number of autophagosomes (Figures 4M and 4P). These results suggested that RNF125 inhibited autophagy in asthma.
Over-activated autophagy in asthma is often accompanied by excessive oxidative stress, which is an important factor influencing the occurrence and development of the disease. Studies have shown that the production of MDA in lung tissues in asthma animal models is significantly higher than that in the control group, and GSH and SOD were lower in lung tissues of the asthma group.44 Natural antioxidants have been reported to alleviate the progression of asthma by inhibiting oxidative stress.45,46 In asthma, autophagy reportedly induces oxidative stress. ATG5 knockdown inhibits oxidative stress in bronchial epithelial cells.26 However, the relationship between RNF125 and oxidative stress remains unclear. Our study found that overexpression of Rnf125 in lung tissues relieved oxidative stress in OVA-treated mice (Figures 3J–3L). Consistent with this, RNF125 overexpression significantly attenuated oxidative stress in vitro, whereas RNF125 knockdown aggravated oxidative stress (Figures 4F–4J, 4N–4O, and 4R–4T). Thus, RNF125 protected against oxidative stress in asthma and may be an important target for asthma treatment.
Airway epithelium is the first barrier of the body to resist the harmful stimulation of the internal and external environment of the airway. The impaired airway epithelial barrier function is an important pathological feature of asthma.2 ZO-1, OCCLUDIN, and CLAUDIN 1 are tight-junction proteins, maintaining the barrier function of the epithelium. These proteins are all decreased in asthmatic lung tissues and airway epithelium.47,48,49,50 Oxidative stress is one of the important causes of airway barrier damage. Hydrogen peroxide, an inducer of oxidative stress, is reported to decrease the expression of ZO-1, OCCLUDIN, and CLAUDIN 1.51 NAC, a ROS scavenger, can block the epithelial barrier dysfunction.52,53 Therefore, oxidative stress is closely related to airway barrier function. We have found that RNF125 inhibited oxidative stress, so we speculated whether RNF125 could therefore affect the barrier function of airway epithelium. We demonstrated that overexpression of Rnf125/RNF125 increases the expression of barrier related proteins in vivo and in vitro studies, while silencing RNF125 decrease their expression in vitro. Thus, RNF125 could protect the barrier of airway epithelium.
Based on the bioinformatics database, IP-Mass results, and related literature, HMGB1 was identified as a potential substrate of RNF125. HMGB1, believed to be a pro-asthmatic molecule, is a highly conserved nuclear protein widely distributed in mammalian cells. Studies have shown that HMGB1 expression in sputum, plasma, bronchial epithelial cells of asthma patients, and bronchial epithelial cells of asthma mice is significantly higher than those in the control groups were.12,54 After anti-HMGB1 treatment in asthma mice, the infiltration of airway inflammatory cells and mucus secretion by epithelial cells are significantly alleviated, and these symptoms are significantly aggravated by the bronchial administration of HMGB1 recombinant protein.55 Therefore, we performed experiments to explore the association between E3 ubiquitin ligase RNF125 and HMGB1. Using Co-IP experiments, we confirmed that RNF125 directly bound to HMGB1 in vitro (Figures 5A–5C). Immunofluorescence and Co-ip experiments showed that RNF125 and HMGB1 interacted mainly in the nucleus (Figures 5D–5H). Next, we explored some details of the binding of RNF125 to HMGB1. Firstly, it was further proved that RNF125 interaction with HMGB1 was dependent on its HMG B-box domain. Studies have shown that HMGB1 can be expressed in both the nucleus and cytoplasm, which is important for its function. Therefore, secondly, we explored whether the binding of RNF125 to HMGB1 occurred in the nucleus or cytoplasm. By extracting nucleoprotein and cytoplasm protein separately, we found that this binding mainly existed in the nucleus. At the same time, we explored the binding of RNF125 to HMGB1 in asthma. Based on the aforementioned, we confirmed that RNF125 bound to HMGB1 through its B box domain mainly in the nucleus. Then, we explored the regulatory relationship between RNF125 and HMGB1. We found that overexpression of RNF125 decreased HMGB1 protein expression, whereas knockdown of RNF125 increased HMGB1 protein expression (Figures 5I and 5J). Thus, these results indicated that RNF125 could interact with HMGB1 mainly in the nucleus and inhibit its expression.
The ubiquitination and deubiquitination of HMGB1 are reportedly involved in the development of various diseases. In esophageal squamous cell carcinoma, E3 ubiquitin ligase SYN1 plays an anticancer role by degrading the HMGB1 protein.56 In multiple myeloma, the deubiquitination enzyme USP12 deubiquitinates HMGB1 and increase its stability, thus promoting autophagy.57 Then, we explored whether RNF125 could ubiquitinate HMGB1. Inhibitors CHX and MG132 were used to demonstrate that RNF125 reduced HMGB1 through the ubiquitination-proteasome pathway. Furthermore, by directly detecting the ubiquitination level of HMGB1, we demonstrated that RNF125 could ubiquitinate HMGB1. By using RNF125 point mutation (ubiquitination enzyme inactivation) plasmid, we further elucidate these results. By IP-Mass and Co-IP, we also demonstrated that a site K150 is important in HMGB1 ubiquitination by RNF125 (Figure 6). Thus, these results suggested that RNF125 reduced HMGB1 expression through the ubiquitin proteasome pathway partially via its K150 site, revealing a signaling pathway (RNF125/HMGB1 axis) that regulates autophagy and oxidative stress in asthma.
Many studies have shown that HMGB1 is an important regulatory factor of autophagy, and that HMGB1 knockdown reduces autophagy in human bronchial epithelial cells.13 Meanwhile, HMGB1 aggravates cellular oxidative stress, while HMGB1 siRNA transfection reduces oxidative stress.58 We demonstrated that overexpression of HMGB1 and RNF125 simultaneously partially reversed the inhibition of autophagy and oxidative stress caused by RNF125 overexpression. Therefore, the HMGB1 appears to be integral to RNF125-related autophagy and oxidative stress. Then we explore whether autophagy is necessary for RNF125 to inhibit oxidative stress. BECLIN1 is one of the most classic autophagy related proteins, and it is widely believed that BECLIN1 can activate autophagy. At the same time, our results showed that RNF125 could inhibit BECLIN1 expression. Thus, overexpression of BECN1 was used to induce autophagy. We demonstrated that overexpression of BECN1 and RNF125 at the same time partially reversed the inhibition of oxidative stress caused by RNF125 overexpression (Figure 7). These results suggested that RNF125 inhibited oxidative stress partially dependent on autophagy. Therefore, based on the aforementioned, we confirmed that RNF125 inhibited autophagy-induced oxidative stress partially dependent on HMGB1.
Overall, this study reports findings not previously reported, such as the expression and effect of RNF125 in asthma; CpG islands in Rnf125; methylation of Rnf125; the association between RNF125 and autophagy or oxidative stress; and the regulation and interaction of RNF125 and HMGB1.
In this study, we found that RNF125 was downregulated in bronchial epithelial cells of OVA-treated mice, and bronchial epithelial cells of patients with asthma, which was due to its core promoter hypermethylation. RNF125 interacted with HMGB1 via the HMG B-box domain mainly in the nucleus partially via its K150 site. The downregulation of RNF125 in asthma inhibited ubiquitination of the HMGB1 protein, resulting in decreased degradation of HMGB1, increased stability, upregulated expression of HMGB1, thereby aggravating autophagy-induced oxidative stress. Meanwhile, low expression of RNF125 impaired bronchial epithelial barrier in asthma (graphical abstract). This study demonstrated the expression, function, and mechanism of RNF125 in asthma. It is an in-depth study of the pathogenesis of asthma, and the results obtained can help us better understand its pathogenesis, find different treatment methods, and lay a theoretical foundation to develop solutions for oxidative stress and autophagy.
Limitations of the study
This study still has some limitations. It was difficult to collect lung tissues and bronchi from patients with asthma, so we could only collect samples from patients undergoing surgery. Therefore, the sample size was small, and it could only explain the problem to a certain extent. Meanwhile, due to the downregulation of RNF125 in lung tissues in asthma group, we overexpressed it in vivo to explore its role. Silencing of RNF125 is also an important pathway to investigate the role of RNF125 in vivo. However, this was not included in our experiments. Furthermore, only one cell line, 16HBE, was used to explore the role and mechanism of RNF125 in vitro, and the use of two or more cell lines could substantiate the conclusion. In this study, we studied oxidative stress by overexpressing RNF125. However, overexpression of protein itself may overload the ribosome and cause stress in cells, so this is one of the limitations in this study. 16HBE cell lines were used to explore the role of RNF125. However, the cell line is immortalized and differs greatly from the primary airway epithelial cells, so the conclusions drawn in this study may not be applicable to the primary airway epithelium, which needs further experimental verification. Moreover, there is no cell specificity in adeno-associated viruses. RNF125 was overexpressed in airway epithelium, airway smooth muscle, and alveolar epithelium. Therefore, in this study, the effect of overexpression of RNF125 on asthma may not depend solely on airway epithelium. Conditional overexpression is a better choice to study the effect of RNF125 of airway epithelium in asthma. Compared with conditional overexpression, adeno-associated virus-induced overexpression and knockdown has its technical disadvantages. In the animal studies, female mice were selected because they are more likely to effectively establish an asthma model. However, as male mice were not considered in the study, the applicability of our findings to them may be limited.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-LC3 | Abcam | Cat#ab192890; RRID: AB_2827794 |
| Anti-BECLIN1 | Abcam | Cat#ab207612; RRID: AB_2692326 |
| Anti-p62 | Abcam | Cat#ab109012; RRID: AB_2810880 |
| Anti-RNF125 | Abclonal | Cat#A15165; RRID: AB_2762054 |
| Anti-HA (rabbit host) | Cell Signaling Technology | Cat#3724; RRID: AB_1549585 |
| Anti-Ubiquitin | Cell Signaling Technology | Cat#58395 |
| Anti-HA (mouse host) | GIN (Japan) | Cat#GNI4110-HA |
| Anti-Myc (mouse host) | GIN (Japan) | Cat#GNI4110-MC |
| Anti-HMGB1 (mouse host) | Novus Biologicals | Cat#NBP2-27396 |
| Anti-HMGB1 (rabbit host) | Proteintech Group | Cat#10829-1-AP; RRID: AB_2232989 |
| Anti-Flag (rabbit host) | Proteintech Group | Cat#20543-1-AP; RRID: AB_11232216 |
| Anti-Myc (rabbit host) | Proteintech Group | Cat#16286-1-AP; RRID: AB_11182162 |
| Anti-β-actin | Proteintech Group | Cat#20536-1-AP; RRID: AB_10700003 |
| Anti-ZO-1 | Proteintech Group | Cat#21773-1-AP; RRID: AB_10733242 |
| Anti-OCCLUDIN | Proteintech Group | Cat#13409-1-AP; RRID: AB_2156308 |
| Anti-CLAUDIN 1 | Proteintech Group | Cat#13050-1-AP; RRID: AB_2079881 |
| Anti-Flag (mouse host) | Sigma-Aldrich | Cat#F1804; RRID: AB_262044 |
| Chemicals, peptides, and recombinant proteins | ||
| LPS | Sigma-Aldrich | Cat#L2880 |
| Ovalbumin | Sigma-Aldrich | Cat#A5503 |
| DCFH-DA | Sigma-Aldrich | Cat#D6883 |
| AL(OH)3 | Thermo Scientific | Cat#77161 |
| RPMI-1640 medium | Procell | Cat#PM150110 |
| Fetal bovine serum | Bioind | Cat#04-001 |
| Pronase | Solarbio | Cat#P8360 |
| DNAseI | Solarbio | Cat#D8071 |
| TRIzol Reagent | Takara | Cat#9108 |
| Phosphatase inhibitor | Meilunbio | Cat#MB12707 |
| Phenylmethanesulfonyl fluoride | Solarbio | Cat#IP0280 |
| Decitabine | Selleckchem | Cat#S1200 |
| MG132 | ApexBio | Cat#a2585 |
| CHX | Aladdin | Cat#C112766 |
| Critical commercial assays | ||
| Wright–Giemsa stain kit | Solarbio | Cat#G1020 |
| Periodic acid-Schiff kit | Beyotime | Cat#C0142 |
| Malondialdehyde kit | Beyotime | Cat#S0131 |
| superoxide dismutase kit | Elabscinece | Cat#E-BC-K020 |
| Glutathione kit | Solarbio | Cat#BC1175 |
| BCA kit | Beyotime | Cat#P0010 |
| cDNA Synthesis Kit | Takara | Cat#RR047A |
| SYBR PrimeScript RT-PCR Kit | Takara | Cat#RR820A |
| Genomic DNA Extraction Kit | Sangon Biotech | Cat#B518264 |
| Deposited data | ||
| Mass data (RNF125 interacting protein) | Beijing Protein Innovation | ProteomeXchange: PXD043937; iProX: IPX0006363000 |
| Mass data (HMGB1 ubiquitination related sites) | Shanghai Applied Protein Technology | ProteomeXchange: PXD043938; iProX: IPX0006367000 |
| Experimental models: Cell lines | ||
| Human bronchial epithelial cell line 16HBE | Xiangya School of Medicine (Changsha, China) | Cat#HTCC23 |
| Experimental models: Organisms/strains | ||
| Female BALB/c mice (6–7 weeks old, 17–18 g) | Beijing Hfk Bioscience Co. Ltd. | Cat#11002A |
| Oligonucleotides | ||
| Primer:human RNF125 Forward | This paper | 5ʹ-GTTCCTGTATTGCTACCAGTCT-3ʹ |
| Primer:human RNF125 Reverse | This paper | 5ʹ-TTCTTTTGGCTACATCAGTTGC-3ʹ |
| Primer:human HMGB1 Forward | This paper | 5ʹ-TCTTCCTCTTCTGCTCTGAGTATCG-3ʹ |
| Primer:human HMGB1 Reverse | This paper | 5ʹ-TTCGCAACATCACCAATGGACAG-3ʹ |
| Primer:human β-actin Forward | This paper | 5ʹ-CCTGGCACCCAGCACAAT-3ʹ |
| Primer:human β-actin Reverse | This paper | 5ʹ-GGGCCGGACTCGTCATAC-3ʹ |
| Recombinant DNA | ||
| Flag-RNF125 | Han bio | N/A |
| Myc-HMGB1 wt | Sino Biological | N/A |
| Myc-HMGB1 (1-163aa) | Sino Biological | N/A |
| Myc-HMGB1 (1-79aa) | Sino Biological | N/A |
| Myc-HMGB1 (80-215aa) | Sino Biological | N/A |
| Software and algorithms | ||
| R 3.6.3 | The R Foundation | https://www.r-project.org/ |
| UCSC | Navarro Gonzalez59 | http://genome.ucsc.edu |
| Gene Ontology | Ashburner et al.60 | http://geneontology.org |
| ClusterProfiler | Yu et al.61 | http://bioconductor.org/packages/release/bioc/html/clusterProfiler.html |
| org.Hs.e.g.,.db | Carlson M et al.62 | http://bioconductor.org/packages/release/data/annotation/html/org.Hs.eg.db.html |
| ImageJ | National Institutes of Health | https://imagej.net/Fiji |
| GraphPad Prism 8.0 | GraphPad | https://www.graphpad.com/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yunxiao Shang (shangyunxiao321@163.com)
Materials availability
This study did not generate new unique reagents.
Experimental model and study participant details
Ethics statement
All clinical specimen collection procedures were approved by ethics committee of Shengjing Hospital of China Medical University (Shenyang, China; approval no. 2022PS546K). All animal procedures were performed in accordance with the guidelines of the Chinese National Regulations for Animal Care and the China Medical University Animal Care and Use Committee (Shenyang, China; approval no. 2022PS187K).
Participants
In the asthma group, three female patients, one male patient with asthma and three female patients, one male control patient without asthma undergoing thoracic surgery for pulmonary masses (lung tumor) were selected. The control group consisted of adults with an average age of 56.25 ± 6.449 years. The asthma group had an average age of 54.75 ± 9.430 years. All participants were adults, and they were recruited from China, representing individuals of Asian ethnicity. The diagnosis of asthma was based on the Global Initiative for Asthma (GINA) guidelines.65 Excluded from this study were patients with a history of other respiratory diseases (such as chronic obstructive pulmonary disease, bronchiectasis, active tuberculosis, etc.); chronic renal diseases (such as glomerulonephritis, nephrotic syndrome, Ig-A nephropathy, and renal failure); autoimmune liver disease, acute and chronic hepatitis, cirrhosis, ulcerative colitis, Crohn disease and other chronic digestive diseases; hyperthyroidism, thyroiditis, Graves’ disease and other endocrine system diseases; various connective tissue diseases; primary hypertension and primary pulmonary hypertension; hemophilia, hereditary spherocytosis and other hereditary diseases; and a variety of systemic diseases associated with autoimmunity. In the control group, three patients without asthma (with normal lung function) undergoing thoracic surgery for pulmonary masses (lung tumor) were selected. In order to ensure the comparability of the two groups, the following criteria was implemented to screen specimens: same gender; similar age; identical pathological tumor type; same differentiation degree of tumor.
Preparation of human lung tissues
Lung tissue specimens were obtained from lung tissue excised during thoracic surgery. Lung tissue specimens were taken as far away from the tumor as possible (at least 5 cm from the lesion site), and appeared as normal tissue. We standardized the location (surrounding tissue extending to exclude the pleural surface) and the number (∼1 cm³) of each sample to ensure a roughly equal number of pulmonary parenchyma, airways, and pulmonary vessels in the sample. The excised lung tissue samples were carefully washed with normal saline to remove blood, and dried with gauze, and immediately placed in liquid nitrogen for preservation. Specimens requiring long-term preservation were kept in a deep freeze freezer at −80° C.
Establishment of asthma mouse model and treatment
Female BALB/c mice (6–7 weeks old, 17–18 g) were supplied by Beijing Hfk Bioscience Co. Ltd. (Beijing, China). Female mice are more prone to asthma, so we chose female mice, which is consistent with the animal selection of most of the literature in asthma. All mice were housed for one week to adapt to the laboratory environment. During the experimental period, the mice were housed in a standard pathogen-free laboratory with water and food ad libitum. On days 1, 8, and 15, mice were sensitized with 200 μL sensitization liquid, containing 50 μg ovalbumin (OVA; Sigma-Aldrich, MO, USA) and 1 mg AL(OH)3 intraperitoneally (i.p.), as previously described.(Wang et al., 2019) On days 22–24 (day 3 group) or days 22–28 (day 7 group), or days 22–35 (day 14 group), the mice were challenged with 2% OVA inhalation for 30 min, delivered using a nebulizer (DeVilbiss, KDU, USA). Decitabine group sensitized with sensitization liquid on days 1, 8, and 15, and received intraperitoneal injections of 1.5 mg/kg of decitabine 1 h before OVA inhalation, and then challenged with 2% OVA inhalation for 30 min on days 22–28. Two weeks before sensitization, mice were intratracheally administered 70 μL of mouse AAV6 (1 × 1012 vg/mL) supplied by Syngenbio (Beijing, China). In this study, AAV6 mainly included RNF125 AAV6 and control AAV6. AAV6 is a type of adeno-associated virus that contains an overexpression plasmid and can infect the cells. And the Rnf125 AAV6 is mouse Rnf125 AAV6. Control AAV is a type of adeno-associated virus just as Rnf125 overexpresses adeno-associated viruses except that Rnf125 cannot be overexpressed. Mice were randomly divided into three groups as follows: mice sensitized with saline water, challenged with saline water, and administered control AAV (control group); mice sensitized with sensitization liquid, challenged with OVA, and administered control AAV (OVA-treated mice); and mice sensitized with sensitization liquid, challenged with OVA, and administered Rnf125 AAV (Rnf125 group).
Method details
Airway hyperresponsiveness
Twenty-four hours after the last challenge, airway hyperresponsiveness was evaluated using a non-invasive lung function detection system (Dsi buxco, MN, USA). Plethysmography was used to measure the sRaw of the mice.
Bronchoalveolar lavage fluid (BALF) collection
Mice were anesthetized with sodium pentobarbital, and BALF was collected. First, we fixed the mouse on the operating table. The abdominal cavity was opened with ophthalmic scissors, a slit in the chest was carefully opened, and t the right main bronchus was ligated with thread. The trachea was carefully isolated, 0.5mL of normal saline was instilled into the trachea using a trocar three times to collect BALF. The fluid was centrifuged (4°C, 188 × g, 8 min) to pellet the cells, and the supernatant was obtained for chemokine and cytokine detection. Cell counts and morphology were evaluated using a cell counting plate and Wright–Giemsa staining (Solarbio, Beijing, China). At least 200 cells were counted independently and typed by two researchers.
Histological evaluation
The lung tissues were fixed with 4% paraformaldehyde for 48 h, dehydrated, and embedded in paraffin. Next, 4 μm thick sections were sliced. Hematoxylin and eosin (HE; Solarbio) staining was performed to evaluate airway inflammation infiltration and semi-quantitative analysis.66 Periodic acid-Schiff (PAS; Beyotime, Shanghai, China) staining was used to detect mucus production.
Cell culture and treatment
The human bronchial epithelial cell line 16HBE, purchased from the Xiangya School of Medicine (Changsha, China), was cultured in 10% fetal bovine serum (Bioind, ISR) in RPMI-1640 medium (Procell, Wuhan, China), as previously described. (Wang et al., 2019) Small interfering RNAs (siRNAs) were used to transfect cells to generate the siRNA-negative control (Si-NC) and RNF125-silencing (RNF125-si) groups. The target sequences were as follows. RNF125 siRNA1: CCGUGUGCCUUGAGGUGUU and RNF125 siRNA2: GACUUGUCAGAAGUACAUA. To form the plasmid-negative control group (Flag-NC group, Flag-tagged) and RNF125 overexpression group (Flag-RNF125 group, Flag-tagged), either RNF125 plasmid encoding full-length RNF125 cDNA or an empty plasmid was used to transfect cells. Cells were transfected using Lipo3000 according to the manufacturer’s instructions. Indicated markers were detected 48 h after transfection. Flag-tagged RNF125 point mutants (C72, 75A) plasmid, HA-tagged ubiquitin, Myc-tagged full-length HMGB1, and three truncated Myc-tagged HMGB1 plasmids (containing:1–163, 1–79, and 80–215) were constructed.
Primary murine airway epithelial cells were isolated using the following procedure. First, we sterilized the operating table with alcohol spray and UV. Then, the animal carcasses were sprayed with alcohol. We removed the skin around the tracheal and exposed the intact trachea. The trachea was placed into a 100 mm sterile Petri dish. The connective tissues around trachea were removed. Next, tracheas were digested with pronase (Solarbio) solution and incubated with DNAseI (Solarbio), and the interference of fibroblasts was eliminated by the differential adherent method.
Oxidative stress analysis
16HBE cells were incubated with 20 μM DCFH-DA (Sigma-Aldrich, USA) for 30 min. Digestion was involved in this experiment. A fluorescence microscope (Olympus, Japan) (Cellular digestion was used in the process) and a multi-mode microplate reader (BioTek Synergy HT, USA) were used to evaluate the expression of ROS. Malondialdehyde (MDA), superoxide dismutase (SOD), and glutathione (GSH) expression were detected using MDA (Beyotime, Shanghai, China), SOD (Elabscinece, Wuhan, China), and GSH (Solarbio, Beijing, China) kits, respectively, according to the manufacturer’s instructions. A multimode microplate reader was used to evaluate the results.
Western blotting, immunoprecipitation, and protein mass spectrometry
Lung samples and cells were lysed in cold immunoprecipitation buffer (Beyotime Biotechnology) containing phenylmethanesulfonyl fluoride (Solarbio) and a phosphatase inhibitor (Meilunbio, Dalian, China). The lysates were incubated on ice for 30 min and centrifuged at 12000 × g to collect the supernatant. The protein concentration was quantified using a BCA kit (Beyotime Biotechnology). For immunoblotting, protein samples (20 μg) were electrophoresed to separate proteins of different molecular weights using 10% or 12.5% sodium dodecyl sulphate-polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes. Membranes were blocked with 5% skim milk for 2 h at room temperature and incubated with primary antibodies overnight at 4°C, followed by incubation with the appropriate secondary antibodies (ZSGB-BIO, Beijing, China). Enhanced chemiluminescent reagent (Wanleibio Group, Inc., Shenyang, China) was used to detect chemiluminescent signals. We used ImageJ software to determine the gray values. For IP, total protein was incubated with 1 μg primary antibody overnight at 4°C, and then incubated with 30 μL of protein A/G beads (Santa Cruz Biotechnology, CA, USA) for 4 h at 4°C. The immunoprecipitants were electrophoresed to separate the proteins using SDS-PAGE. Thereafter, immunoblotting and protein mass spectrometry were performed.
RNA extraction and quantitative real-time PCR (qPCR)
Total RNA was obtained from cells using TRIzol Reagent (Takara, Japan). The RNA and a cDNA Synthesis Kits (Takara) were used to generate complementary DNA. Next, SYBR PrimeScript RT-PCR Kit (Takara) and a LightCycler 480 (Roche Molecular Systems, Inc.) were used to amplify the generated complementary DNA. The sequences of human RNF125 PCR primers were 5ʹ-GTTCCTGTATTGCTACCAGTCT-3ʹ (forward) and 5ʹ-TTCTTTTGGCTACATCAGTTGC-3ʹ (reverse). The sequences of human HMGB1 PCR primers were 5ʹ-TCTTCCTCTTCTGCTCTGAGTATCG-3ʹ (forward) and 5ʹ-TTCGCAACATCACCAATGGACAG-3ʹ (reverse). The sequences of human β-actin PCR primers were 5ʹ-CCTGGCACCCAGCACAAT-3ʹ (forward) and 5ʹ-GGGCCGGACTCGTCATAC-3ʹ (reverse). Relative mRNA fold changes were assessed by the 2−ΔΔCT method.
Gene Ontology enrichment analysis
The gene functions were classified into biological process, cellular component, and molecular function using a biological model by Gene Ontology (http://geneontology.org).60 R software version 3.6.3 was used to conduct the data analysis. Enrichment analysis was performed using clusterProfiler package,61 ID conversion was performed using org.Hs.e.g.,.db package,62 and visualization was performed using ggplot2 package (https://ggplot2.tidyverse.org).67Significant terms were obtained with criteria of p.adj <0.05 and qvalue <0.2.
Immunohistochemistry and immunofluorescence
After 4% paraformaldehyde fixation and paraffin embedding, 3 μm-thick lung tissue sections were stained with an anti-RNF125 antibody. Primary murine airway epithelial cells were seeded in 12-well culture plates with 13 mm cell climbing slices and fixed with 4% paraformaldehyde and methanol. For immunohistochemistry, a biotin-conjugated secondary antibody was used to bind to the primary antibody. For immunofluorescence, 594-conjugated secondary antibodies were used to bind the primary antibody. Tissue sections and cells were photographed using a microscope. ImageJ software (National Institutes of Health, https://imagej.net/Fiji) was used to evaluate protein expression.
Transmission electron microscopic analysis
16HBE cells were harvested after trypsin digestion and fixed with 2.5% glutaraldehyde overnight and 1% OsO4 for 2 h at room temperature. Dehydration, embedding, sectioning, and staining with uranium acetate, lead citrate was then performed. Transmission electron microscopy was used to evaluate autophagy levels.
DNA methylation analysis
Firstly, DNA extraction was performed using a Genomic DNA Extraction Kit (Sangon Biotech, Shanghai, China) according to the manufacturer’s instructions. The CpG island was obtained from “UCSC Genome Browser database: http://genome.ucsc.edu”,59 the criteria were as follows: length over 200 bp, %G + C over 50, observed/expected CpG over 0.6.68 The core promoter was defined TSS +100 bp to −100 bp.69,70 Then DNA methylation around core promoter was analyzed by pyrosequencing at the BGI Company (Beijing Liuhe, Beijing, China).
Quantification and statistical analysis
All experiments were performed at least three times in vitro and six times in vivo, and the data are presented as mean ± standard deviation. Statistical analysis was conducted using GraphPad Prism 8.0 software and SPSS 25. A t test was used to compare the means of two groups, while one-way ANOVA was used to compare the means of a minimum of three groups. Categorical variables were analyzed with Fisher exact test. Statistical significance was set at p < 0.05.
Acknowledgments
This work was supported by the National Natural Science Foundation of China [grant: 82200036], 345 Talent Project of Shengjing Hospital of China Medical University [grant: M1340], and Research Foundation of Shengjing Hospital of China Medical University [grant: M1132].
Author contributions
J.L. and Y.S. conceptualized and supervised this study. J.H. performed most of the experiments and analyzed the data. J.H., R.D., S.L. contributed to the study design. J.H. and R.D. conducted visualization. J.H. and J.W. collected the clinical samples.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: July 27, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107503.
Contributor Information
Jianjun Li, Email: jianjunlicmu@163.com.
Yunxiao Shang, Email: shangyunxiao321@163.com.
Supplemental information
Data and code availability
-
•
The mass spectrometry proteomics data have been deposited to the “ProteomeXchange: http://proteomecentral.proteomexchange.org” via the iProX partner repository63,64 with the dataset identifier PXD043937 (mass data related to RNF125 interacting protein) and PXD043938 (mass data related to HMGB1 ubiquitination related sites). The respective numbers in iProX are IPX0006363000 and IPX0006367000. The mass data are publicly available. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Lane N.E. Glucocorticoid-Induced Osteoporosis: New Insights into the Pathophysiology and Treatments. Curr. Osteoporos. Rep. 2019;17:1–7. doi: 10.1007/s11914-019-00498-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hellings P.W., Steelant B. Epithelial barriers in allergy and asthma. J. Allergy Clin. Immunol. 2020;145:1499–1509. doi: 10.1016/j.jaci.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gon Y., Hashimoto S. Role of airway epithelial barrier dysfunction in pathogenesis of asthma. Allergol. Int. 2018;67:12–17. doi: 10.1016/j.alit.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 4.Liu T., Liu Y., Miller M., Cao L., Zhao J., Wu J., Wang J., Liu L., Li S., Zou M., et al. Autophagy plays a role in FSTL1-induced epithelial mesenchymal transition and airway remodeling in asthma. Am. J. Physiol. Lung Cell Mol. Physiol. 2017;313 doi: 10.1152/ajplung.00510.2016. L27–l40. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y., Tang H., Yuan X., Ran Q., Wang X., Song Q., Zhang L., Qiu Y., Wang X. TGF-β3 Promotes MUC5AC Hyper-Expression by Modulating Autophagy Pathway in Airway Epithelium. EBioMedicine. 2018;33:242–252. doi: 10.1016/j.ebiom.2018.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J.N., Suh D.H., Trinh H.K.T., Chwae Y.J., Park H.S., Shin Y.S. The role of autophagy in allergic inflammation: a new target for severe asthma. Exp. Mol. Med. 2016;48:e243. doi: 10.1038/emm.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang C.Y., Hu R.C., Li J., Chen B.B., Dai A.G. α1-Antitrypsin alleviates inflammation and oxidative stress by suppressing autophagy in asthma. Cytokine. 2021;141:155454. doi: 10.1016/j.cyto.2021.155454. [DOI] [PubMed] [Google Scholar]
- 8.Quimbar M.E., Davis S.Q., Al-Farra S.T., Hayes A., Jovic V., Masuda M., Lippert A.R. Chemiluminescent Measurement of Hydrogen Peroxide in the Exhaled Breath Condensate of Healthy and Asthmatic Adults. Anal. Chem. 2020;92:14594–14600. doi: 10.1021/acs.analchem.0c02929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang W.C., Fang L.W., Liou C.J. Phloretin Attenuates Allergic Airway Inflammation and Oxidative Stress in Asthmatic Mice. Front. Immunol. 2017;8:134. doi: 10.3389/fimmu.2017.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee P.H., Hong J., Jang A.S. N-acetylcysteine decreases airway inflammation and responsiveness in asthma by modulating claudin 18 expression. Korean J. Intern. Med. (Engl. Ed.) 2020;35:1229–1237. doi: 10.3904/kjim.2019.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kianian F., Kadkhodaee M., Sadeghipour H.R., Karimian S.M., Seifi B. An overview of high-mobility group box 1, a potent pro-inflammatory cytokine in asthma. J. Basic Clin. Physiol. Pharmacol. 2020;31 doi: 10.1515/jbcpp-2019-0363. [DOI] [PubMed] [Google Scholar]
- 12.Imbalzano E., Quartuccio S., Di Salvo E., Crea T., Casciaro M., Gangemi S. Association between HMGB1 and asthma: a literature review. Clin. Mol. Allergy. 2017;15:12. doi: 10.1186/s12948-017-0068-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu M., Shan M., Zhang Y., Guo Z. Progranulin Protects Against Airway Remodeling Through the Modulation of Autophagy via HMGB1 Suppression in House Dust Mite-Induced Chronic Asthma. J. Inflamm. Res. 2021;14:3891–3904. doi: 10.2147/jir.s322724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lv Y., Li Y., Zhang D., Zhang A., Guo W., Zhu S. HMGB1-induced asthmatic airway inflammation through GRP75-mediated enhancement of ER-mitochondrial Ca(2+) transfer and ROS increased. J. Cell. Biochem. 2018;119:4205–4215. doi: 10.1002/jcb.26653. [DOI] [PubMed] [Google Scholar]
- 15.Giannini A.L., Gao Y., Bijlmakers M.J. T-cell regulator RNF125/TRAC-1 belongs to a novel family of ubiquitin ligases with zinc fingers and a ubiquitin-binding domain. Biochem. J. 2008;410:101–111. doi: 10.1042/BJ20070995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao H., Li C.C., Pardo J., Chu P.C., Liao C.X., Huang J., Dong J.G., Zhou X., Huang Q., Huang B., et al. A novel E3 ubiquitin ligase TRAC-1 positively regulates T cell activation. J. Immunol. 2005;174:5288–5297. doi: 10.4049/jimmunol.174.9.5288. [DOI] [PubMed] [Google Scholar]
- 17.Arimoto K.i., Takahashi H., Hishiki T., Konishi H., Fujita T., Shimotohno K. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc. Natl. Acad. Sci. USA. 2007;104:7500–7505. doi: 10.1073/pnas.0611551104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shoji-Kawata S., Zhong Q., Kameoka M., Iwabu Y., Sapsutthipas S., Luftig R.B., Ikuta K. The RING finger ubiquitin ligase RNF125/TRAC-1 down-modulates HIV-1 replication in primary human peripheral blood mononuclear cells. Virology. 2007;368:191–204. doi: 10.1016/j.virol.2007.06.028. [DOI] [PubMed] [Google Scholar]
- 19.Tang J., Tu S., Lin G., Guo H., Yan C., Liu Q., Huang L., Tang N., Xiao Y., Pope R.M., et al. Sequential ubiquitination of NLRP3 by RNF125 and Cbl-b limits inflammasome activation and endotoxemia. J. Exp. Med. 2020;217:e20182091. doi: 10.1084/jem.20182091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu B., Ye J., Nie Y., Ashraf U., Zohaib A., Duan X., Fu Z.F., Song Y., Chen H., Cao S. MicroRNA-15b Modulates Japanese Encephalitis Virus-Mediated Inflammation via Targeting RNF125. J. Immunol. 2015;195:2251–2262. doi: 10.4049/jimmunol.1500370. [DOI] [PubMed] [Google Scholar]
- 21.Deaton A.M., Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu R., Yan Y., Ma C., Chen H., Dong Z., Wang Y., Liu Y., Liu M., Yang L. HMGB1 contributes to SASH1 methylation to attenuate astrocyte adhesion. Cell Death Dis. 2019;10:417. doi: 10.1038/s41419-019-1645-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li W., Wu Y., Zhao Y., Li Z., Chen H., Dong L., Liu H., Zhang M., Wu Y., Zhou J., et al. MTOR suppresses autophagy-mediated production of IL25 in allergic airway inflammation. Thorax. 2020;75:1047–1057. doi: 10.1136/thoraxjnl-2019-213771. [DOI] [PubMed] [Google Scholar]
- 24.Silveira J.S., Antunes G.L., Kaiber D.B., da Costa M.S., Ferreira F.S., Marques E.P., Schmitz F., Gassen R.B., Breda R.V., Wyse A.T.S., et al. Autophagy induces eosinophil extracellular traps formation and allergic airway inflammation in a murine asthma model. J. Cell. Physiol. 2020;235:267–280. doi: 10.1002/jcp.28966. [DOI] [PubMed] [Google Scholar]
- 25.McAlinden K.D., Deshpande D.A., Ghavami S., Xenaki D., Sohal S.S., Oliver B.G., Haghi M., Sharma P. Autophagy Activation in Asthma Airways Remodeling. Am. J. Respir. Cell Mol. Biol. 2019;60:541–553. doi: 10.1165/rcmb.2018-0169OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dickinson J.D., Alevy Y., Malvin N.P., Patel K.K., Gunsten S.P., Holtzman M.J., Stappenbeck T.S., Brody S.L. IL13 activates autophagy to regulate secretion in airway epithelial cells. Autophagy. 2016;12:397–409. doi: 10.1080/15548627.2015.1056967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michaeloudes C., Abubakar-Waziri H., Lakhdar R., Raby K., Dixey P., Adcock I.M., Mumby S., Bhavsar P.K., Chung K.F. Molecular mechanisms of oxidative stress in asthma. Mol. Aspect. Med. 2022;85:101026. doi: 10.1016/j.mam.2021.101026. [DOI] [PubMed] [Google Scholar]
- 28.Li Y., Xie P., Lu L., Wang J., Diao L., Liu Z., Guo F., He Y., Liu Y., Huang Q., et al. An integrated bioinformatics platform for investigating the human E3 ubiquitin ligase-substrate interaction network. Nat. Commun. 2017;8:347. doi: 10.1038/s41467-017-00299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.GBD Chronic Respiratory Disease Collaborators Prevalence and attributable health burden of chronic respiratory diseases, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Respir. Med. 2020;8:585–596. doi: 10.1016/S2213-2600(20)30105-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang K., Yang T., Xu J., Yang L., Zhao J., Zhang X., Bai C., Kang J., Ran P., Shen H., et al. Prevalence, risk factors, and management of asthma in China: a national cross-sectional study. Lancet. 2019;394:407–418. doi: 10.1016/S0140-6736(19)31147-X. [DOI] [PubMed] [Google Scholar]
- 31.Zheng N., Shabek N. Ubiquitin Ligases: Structure, Function, and Regulation. Annu. Rev. Biochem. 2017;86:129–157. doi: 10.1146/annurev-biochem-060815-014922. [DOI] [PubMed] [Google Scholar]
- 32.Dimasuay K.G., Schaunaman N., Martin R.J., Pavelka N., Kolakowski C., Gottlieb R.A., Holguin F., Chu H.W. Parkin, an E3 ubiquitin ligase, enhances airway mitochondrial DNA release and inflammation. Thorax. 2020;75:717–724. doi: 10.1136/thoraxjnl-2019-214158. [DOI] [PubMed] [Google Scholar]
- 33.Wu J., Wang Y., Zhou Y., Wang Y., Sun X., Zhao Y., Guan Y., Zhang Y., Wang W. PPARgamma as an E3 Ubiquitin-Ligase Impedes Phosphate-Stat6 Stability and Promotes Prostaglandins E2-Mediated Inhibition of IgE Production in Asthma. Front. Immunol. 2020;11:1224. doi: 10.3389/fimmu.2020.01224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu B., Wang J., Ren Z. SKP2-Promoted Ubiquitination of FOXO3 Promotes the Development of Asthma. J. Immunol. 2021;206:2366–2375. doi: 10.4049/jimmunol.2000387. [DOI] [PubMed] [Google Scholar]
- 35.Britto A.M.A., Amoedo N.D., Pezzuto P., Afonso A.O., Martínez A.M.B., Silveira J., Sion F.S., Machado E.S., Soares M.A., Giannini A.L.M. Expression levels of the innate response gene RIG-I and its regulators RNF125 and TRIM25 in HIV-1-infected adult and pediatric individuals. AIDS. 2013;27:1879–1885. doi: 10.1097/QAD.0b013e328361cfbf. [DOI] [PubMed] [Google Scholar]
- 36.Tenorio J., Mansilla A., Valencia M., Martínez-Glez V., Romanelli V., Arias P., Castrejón N., Poletta F., Guillén-Navarro E., Gordo G., et al. A new overgrowth syndrome is due to mutations in RNF125. Hum. Mutat. 2014;35:1436–1441. doi: 10.1002/humu.22689. [DOI] [PubMed] [Google Scholar]
- 37.Liu Z.Y., Cao J., Zhang J.T., Xu G.L., Li X.P., Wang F.T., Ansari K.H., Mohamed H., Fan Y.Z. Ring finger protein 125, as a potential highly aggressive and unfavorable prognostic biomarker, promotes the invasion and metastasis of human gallbladder cancers via activating the TGF- β1-SMAD3-ID1 signaling pathway. Oncotarget. 2017;8:49897–49914. doi: 10.18632/oncotarget.18180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim H., Frederick D.T., Levesque M.P., Cooper Z.A., Feng Y., Krepler C., Brill L., Samuels Y., Hayward N.K., Perlina A., et al. Downregulation of the Ubiquitin Ligase RNF125 Underlies Resistance of Melanoma Cells to BRAF Inhibitors via JAK1 Deregulation. Cell Rep. 2015;11:1458–1473. doi: 10.1016/j.celrep.2015.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoffman A.R., Hu J.F. Directing DNA methylation to inhibit gene expression. Cell. Mol. Neurobiol. 2006;26:425–438. doi: 10.1007/s10571-006-9057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edwards J.R., Yarychkivska O., Boulard M., Bestor T.H. DNA methylation and DNA methyltransferases. Epigenet. Chromatin. 2017;10:23. doi: 10.1186/s13072-017-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Du Q., Luu P.L., Stirzaker C., Clark S.J. Methyl-CpG-binding domain proteins: readers of the epigenome. Epigenomics. 2015;7:1051–1073. doi: 10.2217/epi.15.39. [DOI] [PubMed] [Google Scholar]
- 42.Gavin D.P., Chase K.A., Sharma R.P. Active DNA demethylation in post-mitotic neurons: a reason for optimism. Neuropharmacology. 2013;75:233–245. doi: 10.1016/j.neuropharm.2013.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y., Tang H., Yuan X., Ran Q., Wang X., Song Q., Zhang L., Qiu Y., Wang X. TGF-beta3 Promotes MUC5AC Hyper-Expression by Modulating Autophagy Pathway in Airway Epithelium. EBioMedicine. 2018;33:242–252. doi: 10.1016/j.ebiom.2018.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ezz-Eldin Y.M., Aboseif A.A., Khalaf M.M. Potential anti-inflammatory and immunomodulatory effects of carvacrol against ovalbumin-induced asthma in rats. Life Sci. 2020;242:117222. doi: 10.1016/j.lfs.2019.117222. [DOI] [PubMed] [Google Scholar]
- 45.Ma Y., Ge A., Zhu W., Liu Y., Ji N., Zha W., Zhang J., Zeng X., Huang M. Morin Attenuates Ovalbumin-Induced Airway Inflammation by Modulating Oxidative Stress-Responsive MAPK Signaling. Oxid. Med. Cell. Longev. 2016;2016:5843672. doi: 10.1155/2016/5843672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang W.C., Fang L.W., Liou C.J. Phloretin Attenuates Allergic Airway Inflammation and Oxidative Stress in Asthmatic Mice. Front. Immunol. 2017;8:134. doi: 10.3389/fimmu.2017.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jia Z., Bao K., Wei P., Yu X., Zhang Y., Wang X., Wang X., Yao L., Li L., Wu P., et al. EGFR activation-induced decreases in claudin1 promote MUC5AC expression and exacerbate asthma in mice. Mucosal Immunol. 2021;14:125–134. doi: 10.1038/s41385-020-0272-z. [DOI] [PubMed] [Google Scholar]
- 48.Looi K., Buckley A.G., Rigby P.J., Garratt L.W., Iosifidis T., Zosky G.R., Larcombe A.N., Lannigan F.J., Ling K.M., Martinovich K.M., et al. Effects of human rhinovirus on epithelial barrier integrity and function in children with asthma. Clin. Exp. Allergy. 2018;48:513–524. doi: 10.1111/cea.13097. [DOI] [PubMed] [Google Scholar]
- 49.Zhou L., Hao M., Fan X., Lao Z., Li M., Shang E. Effects of Houpo Mahuang Decoction on serum metabolism and TRPV1/Ca(2+)/TJs in asthma. J. Ethnopharmacol. 2023;302:115873. doi: 10.1016/j.jep.2022.115873. [DOI] [PubMed] [Google Scholar]
- 50.Sun Y.B., Liu M., Fan X.S., Zhou L.P., Li M.W., Hu F.Y., Yue Q.F., Zhang Y.M. Effects of cigarette smoke on the aggravation of ovalbumin-induced asthma and the expressions of TRPA1 and tight junctions in mice. Mol. Immunol. 2021;135:62–72. doi: 10.1016/j.molimm.2021.04.006. [DOI] [PubMed] [Google Scholar]
- 51.Cao S., Wang C., Yan J., Li X., Wen J., Hu C. Curcumin ameliorates oxidative stress-induced intestinal barrier injury and mitochondrial damage by promoting Parkin dependent mitophagy through AMPK-TFEB signal pathway. Free Radic. Biol. Med. 2020;147:8–22. doi: 10.1016/j.freeradbiomed.2019.12.004. [DOI] [PubMed] [Google Scholar]
- 52.Shukla P.K., Gangwar R., Manda B., Meena A.S., Yadav N., Szabo E., Balogh A., Lee S.C., Tigyi G., Rao R. Rapid disruption of intestinal epithelial tight junction and barrier dysfunction by ionizing radiation in mouse colon in vivo: protection by N-acetyl-l-cysteine. Am. J. Physiol. Gastrointest. Liver Physiol. 2016;310:G705–G715. doi: 10.1152/ajpgi.00314.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang H., Zhai N., Chen Y., Fu C., Huang K. OTA induces intestinal epithelial barrier dysfunction and tight junction disruption in IPEC-J2 cells through ROS/Ca(2+)-mediated MLCK activation. Environ. Pollut. 2018;242:106–112. doi: 10.1016/j.envpol.2018.06.062. [DOI] [PubMed] [Google Scholar]
- 54.Di Candia L., Gomez E., Venereau E., Chachi L., Kaur D., Bianchi M.E., Challiss R.A.J., Brightling C.E., Saunders R.M. HMGB1 is upregulated in the airways in asthma and potentiates airway smooth muscle contraction via TLR4. J. Allergy Clin. Immunol. 2017;140:584–587.e8. doi: 10.1016/j.jaci.2016.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li R., Wang J., Zhu F., Li R., Liu B., Xu W., He G., Cao H., Wang Y., Yang J. HMGB1 regulates T helper 2 and T helper17 cell differentiation both directly and indirectly in asthmatic mice. Mol. Immunol. 2018;97:45–55. doi: 10.1016/j.molimm.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 56.Yao W., Wang J., Zhu L., Jia X., Xu L., Tian X., Hu S., Wu S., Wei L. Epigenetic Regulator KDM4D Restricts Tumorigenesis via Modulating SYVN1/HMGB1 Ubiquitination Axis in Esophageal Squamous Cell Carcinoma. Front. Oncol. 2021;11:761346. doi: 10.3389/fonc.2021.761346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li H., Roy M., Liang L., Cao W., Hu B., Li Y., Xiao X., Wang H., Ye M., Sun S., et al. Deubiquitylase USP12 induces pro-survival autophagy and bortezomib resistance in multiple myeloma by stabilizing HMGB1. Oncogene. 2022;41:1298–1308. doi: 10.1038/s41388-021-02167-9. [DOI] [PubMed] [Google Scholar]
- 58.Pei X., Jiang H., Liu X., Li L., Li C., Xiao X., Li D., Tang S. Targeting HMGB1 inhibits T-2 toxin-induced neurotoxicity via regulation of oxidative stress, neuroinflammation and neuronal apoptosis. Food Chem. Toxicol. 2021;151:112134. doi: 10.1016/j.fct.2021.112134. [DOI] [PubMed] [Google Scholar]
- 59.Navarro Gonzalez J., Zweig A.S., Speir M.L., Schmelter D., Rosenbloom K.R., Raney B.J., Powell C.C., Nassar L.R., Maulding N.D., Lee C.M., et al. The UCSC Genome Browser database: 2021 update. Nucleic Acids Res. 2021;49 doi: 10.1093/nar/gkaa1070. D1046–d1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carlson M.F.S., Pages H., Li N. 2013. org. Hs. eg. db: Genome wide annotation for Human. R package version. [Google Scholar]
- 63.Ma J., Chen T., Wu S., Yang C., Bai M., Shu K., Li K., Zhang G., Jin Z., He F., et al. iProX: an integrated proteome resource. Nucleic Acids Res. 2019;47 doi: 10.1093/nar/gky869. D1211–d1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen T., Ma J., Liu Y., Chen Z., Xiao N., Lu Y., Fu Y., Yang C., Li M., Wu S., et al. iProX in 2021: connecting proteomics data sharing with big data. Nucleic Acids Res. 2022;50 doi: 10.1093/nar/gkab1081. D1522–d1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.GINA r. Global Strategy for Asthma Management and Prevention. https://ginasthma.org/gina-reports/
- 66.Myou S., Leff A.R., Myo S., Boetticher E., Tong J., Meliton A.Y., Liu J., Munoz N.M., Zhu X. Blockade of inflammation and airway hyperresponsiveness in immune-sensitized mice by dominant-negative phosphoinositide 3-kinase-TAT. J. Exp. Med. 2003;198:1573–1582. doi: 10.1084/jem.20030298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.H W. Springer-Verlag; 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- 68.Gardiner-Garden M., Frommer M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 69.Thomson J.P., Hunter J.M., Nestor C.E., Dunican D.S., Terranova R., Moggs J.G., Meehan R.R. Comparative analysis of affinity-based 5-hydroxymethylation enrichment techniques. Nucleic Acids Res. 2013;41:e206. doi: 10.1093/nar/gkt1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kindgren P., Ard R., Ivanov M., Marquardt S. Transcriptional read-through of the long non-coding RNA SVALKA governs plant cold acclimation. Nat. Commun. 2018;9:4561. doi: 10.1038/s41467-018-07010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The mass spectrometry proteomics data have been deposited to the “ProteomeXchange: http://proteomecentral.proteomexchange.org” via the iProX partner repository63,64 with the dataset identifier PXD043937 (mass data related to RNF125 interacting protein) and PXD043938 (mass data related to HMGB1 ubiquitination related sites). The respective numbers in iProX are IPX0006363000 and IPX0006367000. The mass data are publicly available. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.