Summary
There has been an intense focus to uncover the molecular mechanisms by which fasting triggers the adaptive cellular responses in the major organs of the body. Here, we show that in mice, hepatic S-adenosylmethionine (SAMe)—the principal methyl donor—acts as a metabolic sensor of nutrition to fine-tune the catabolic-fasting response by modulating phosphatidylethanolamine N-methyltransferase (PEMT) activity, endoplasmic reticulum-mitochondria contacts, β-oxidation, and ATP production in the liver, together with FGF21-mediated lipolysis and thermogenesis in adipose tissues. Notably, we show that glucagon induces the expression of the hepatic SAMe-synthesizing enzyme methionine adenosyltransferase α1 (MAT1A), which translocates to mitochondria-associated membranes. This leads to the production of this metabolite at these sites, which acts as a brake to prevent excessive β-oxidation and mitochondrial ATP synthesis and thereby endoplasmic reticulum stress and liver injury. This work provides important insights into the previously undescribed function of SAMe as a new arm of the metabolic adaptation to fasting.
Keywords: S-adenosylmethionine, fasting, caloric restriction, phosphatidylethanolamine methyltransferase, β-oxidation, mitochondria-associated-membranes, endoplasmic reticulum stress, liver, adipose tissue, methionine adenosyltransferase a1
Graphical abstract

Highlights
-
•
Decrease of hepatic SAMe levels during fasting acts as a metabolic sensor of nutrition
-
•
During fasting, hepatic SAMe fine-tunes ER-mitochondria contacts and β-oxidation
-
•
Hepatic MAT1A is expressed in MAMs to fuel PEMT activity during fasting
-
•
SAMe synthesis during fasting prevents ER stress and liver damage
Capelo-Diz et al. report that hepatic S-adenosylmethionine (SAMe), the principal methyl donor, acts as a metabolic sensor of nutrition to fine-tune the adaptive cellular responses to fasting by modulating β-oxidation and ATP production in the liver and thus preventing liver damage.
Introduction
Fasting is beneficial to humans by promoting optimal health, retarding the aging process, and protecting against obesity. Fasting triggers the adaptive metabolic response in major organ systems—including the liver, brain, and adipose tissues—which is governed by a complex network of hormones, growth factors, and cytokines.1 In the liver of most mammals, 12–24 h of fasting results in the depletion of hepatic glycogen stores and an increase in the use of fat-derived ketone bodies and free fatty acids as energy sources.1 This adaptation of liver metabolism is remarkably complex, tightly regulated, and encompasses changes in glucose, lipid, and protein metabolism.2,3
It has been proposed that the enzymes in the methionine (Met) pathway may be modulated during fasting to maintain the levels of Met.4 Met is metabolized in the liver by Met adenosyltransferase (MAT) isoenzymes I and III to synthesize S-adenosylmethionine (SAMe).5 There are three main MAT isoenzymes: MATI, MATII, and MATIII. The catalytic subunit of MATI and III (MAT1A) is encoded by the gene Mat1a, which is expressed mainly in the liver, whereas the catalytic subunit of MATII (MAT2A) is encoded by the widely expressed Mat2a gene.5,6,7 In the liver, SAMe contributes to the biosynthesis of polyamines, participates in the synthesis of glutathione (GSH), and is involved in transmethylation.8 Notably, in reactions catalyzed by phosphatidylethanolamine (PE) N-methyltransferase (PEMT), SAMe can also act as methyl donor for the conversion of the lipid PE into phosphatidylcholine (PC).9,10
Several studies have pointed to an important role of the Met cycle in maintaining lipid homeostasis. Rodents fed with a Met restriction diet (MRD), which reduces hepatic Met and SAMe levels, have an extended lifespan, reduced body weight gain, and increased insulin sensitivity, and they do not develop hepatic steatosis.11,12,13 MRD also protects from diet-induced obesity, type 2 diabetes, and hepatic steatosis in mice fed with a high-fat diet (HFD).14 In humans, plasma SAMe levels increase with body mass index,15 and in high-weight gainers,16 SAMe, but not Met, has been found to be independently associated with truncal adiposity and fat mass.15 On the other hand, during fasting, hepatic SAMe levels decrease whereas Met levels remain constant, suggesting an enhanced utilization of hepatic SAMe under starvation.4,13 Taken together, these results open up the possibility that the modulation in SAMe levels, rather than Met per se, could modulate lipid homeostasis during the fed/fasting states. The functional importance of the changes of the essential metabolite SAMe in the metabolic response of the body to fasting however remains largely unexplored.
Here, we show that the physiological decrease of hepatic SAMe levels during fasting acts as a metabolic sensor of nutrition by fine-tuning mitochondrial-associated membranes’ (MAMs) interaction, β-oxidation, and ATP synthesis in the liver. Notably, we observed that chronic decrease of hepatic SAMe levels induced fibroblast growth factor 21 (FGF21) promoter hypomethylation, enhancing its synthesis and secretion to promote lipid catabolism in white and brown adipose tissues (WAT and BAT) and increasing energy expenditure (EE). Fluxomic analyses suggest that during fasting, the Met cycle fluxes toward SAMe and PC synthesis. Strikingly, we also found that despite the global decrease in SAMe levels, there was an increased expression of MAT1A protein at MAMs during fasting, which is required for the localized production of SAMe at these sites. Our data suggest that this localized synthesis of SAMe at MAMs plays a critical role in maintaining PEMT activity and the PC/PE ratio during fasting, which restricts the rate of mitochondrial β-oxidation and ATP production to ultimately prevent endoplasmic reticulum stress (ERS) and liver damage. Altogether, our data unveil a key role of hepatic SAMe in the adaptive metabolic response to fasting.
Results
Nutrient stress increases usage of SAMe in the liver
It was previously reported that hepatic levels of SAMe decrease after 16 h of fasting.4 We performed a detailed time course study of the levels of hepatic Met, SAMe, and S-Adenosyl-L-homocysteine (SAH) after fasting and found that hepatic SAMe levels decreased significantly as early as 6 h. This decrease in SAMe levels was reversible, since 2 h of refeeding was sufficient to partially recover SAMe levels after a 24-h period of fasting. Levels of SAH, the by-product formed after transfer of methyl groups from SAMe, was, on the other hand, increased, resulting in a reduction in the SAMe/SAH ratio (Figure 1A). Notably, we found similar results when mice were maintained under 60% of caloric restriction (CR) over a 5-day period, including markedly decreased hepatic SAMe levels and SAMe/SAH ratio (Figure 1B).
Figure 1.
Nutrient stress leads to an increased usage of SAMe in the liver
(A and B) SAMe, Met, and SAH levels in total liver extracts from male C57BL/6 mice (A) fasted for the indicated times and re-fed for 2 h after fasting (n = 5 per time point) and (B) maintained under 60% of CR over a 5-day period (n = 3–5).
(C–F) qRT-PCR of Mat1a mRNA expression and immunoblotting analyses of MAT1A protein levels in (C) male C57BL/6 mice fasted from 6 to 24 h (n = 5 per time point), (D) fasted for 36 h (n = 3–5), (E) maintained under 60% of CR over a 5-day period (n = 5), and (F) in mouse primary hepatocytes treated with glucagon (n = 3).
(G) RT-qPCR analysis of Mat1a mRNA in livers from 16-h-fasted Gcgr-WT and -KO mice treated with glucagon in vivo for 30 min (n = 9–10 for Gcgr-WT, and n = 6–7 for Gcgr-KO). Results are presented as mean ± SEM; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 by one-way ANOVA in (A), by two-way ANOVA in (C), or Student’s t test in (B) and (D)–(G). f.c., denoted fold change.
See also Figure S1.
Since hepatic SAMe is synthetized mainly by MAT1A, we next analyzed Mat1a mRNA and MAT1A protein levels after fasting. We found that Mat1a mRNA and MAT1A protein expression increased progressively from 12 to 24 h and remained elevated at 36 h (Figures 1C and 1D). Similarly, Mat1a mRNA and MAT1A protein levels were also increased significantly during 60% of CR over a 5-day period (Figure 1E) and in mice fed during 2 months with 40% of CR or with alternate day fasting (ADF) (Figures S1A and S1B).
Glucocorticoids, such as triamcinolone and dexamethasone, have previously been shown to induce expression of Mat1a mRNA.17,18 Here, we examined whether glucagon could also regulate Mat1a mRNA levels. We found an upregulation of Mat1a mRNA and protein levels in primary hepatocyte cultures after glucagon treatment for 12 and 16 h, respectively (Figure 1F). Treatment of overnight-fasted wild-type (WT) mice with glucagon for only 30 min led to a slight increase in hepatic Mat1a mRNA levels in vivo, an effect that was abolished in mice lacking the glucagon receptor (Gcgr-KO [knockout] mice)19 (Figure 1G). This demonstrates that the upregulation of Mat1a mRNA during fasting could be controlled by glucagon.
Altogether, our results show that nutritional restriction induces an upregulation of the enzyme responsible for SAMe synthesis, together with a reduction in hepatic levels of SAMe and enhanced levels of its by-product SAH. This suggests that nutritional restriction could enhance production and concomitant increased usage of SAMe in the liver.
Ablation of hepatic MAT1A increases weight loss and postnatal mortality
Next, we explored the role of hepatic MAT1A in lipid metabolism using the KO mouse model lacking Mat1a (MAT1A-KO mice). These mice are characterized by a marked hypermethioninemia and chronic SAMe deficiency in the liver.20,21 MAT1A-KO mice develop macrovesicular steatosis at 8 months old, and half of the KO mice develop hepatocellular carcinoma at 18 months of age.20,21
We observed that MAT1A-KO pups show enhanced mortality with only 54.8% of the pups surviving until weaning, compared with 76.8% of WT pups (Figure 2A). In addition, we found that there was a consistent reduction in weight of MAT1A-KO mice from 21 to 90 days of age, although these differences were resolved for males at 4 months of age (Figure 2B) and for females around 7 months of age (Figure S1C). Food intake, at night and day periods, was similar in WT and MAT1A-KO mice at 50–60 days of age, indicating that hypophagia or altered feeding patterns were unlikely to be responsible for the lower body weight of MAT1A-KO mice (Figure 2C).
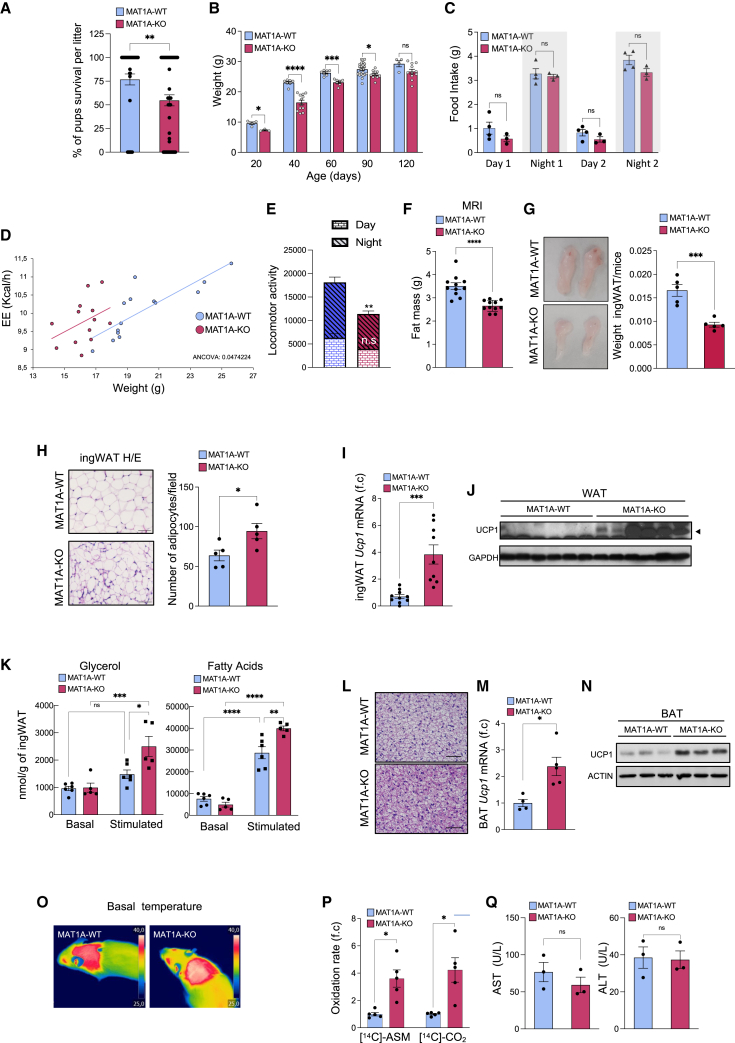
Figure 2.
Chronic MAT1A hepatic ablation increases weight lost, energy expenditure, and postnatal mortality
(A–Q) Male mice were fed ad libitum.
(A and B) (A) Percentage of mice pups surviving at weaning (n = 48–57) and (B) weight of male mice (n = 3–23).
(C–Q) 50- to 60-day-old mice.
(C–H) (C) Food intake (n = 3–4), (D) accumulated energy expenditure (EE) (n = 13–14), (E) locomotor activity (counts of beam breaks) (n = 6), (F) fat mass by MRI (n = 11), (G) macroscopic pictures and ingWAT/mice weights (n = 5), and (H) quantification of density of adipocytes and representative pictures of hematoxylin and eosin (H&E)-stained sections (scale bars, 50 μm) (n = 5).
(I and J) (I) Ucp-1 mRNA (n = 9–10) and (J) protein levels (n = 6–7) in ingWAT.
(K) Basal and stimulated (100 nM isoproterenol) lipolysis, as measured by glycerol (left) and fatty acids (right) released by explants of ingWAT (n = 5–6).
(L) Representative pictures of H&E-stained BAT sections (scale bars, 50 μm).
(M and N) (M) RT-qPCR (n = 4–5) and (N) immunoblot analysis (n = 3–5) of Ucp-1 mRNA and protein levels in BAT.
(O) Termal pictures from BAT interscapular temperatures.
(P and Q) (P) Incomplete and complete fatty acid (palmitate) oxidation, evaluated by measuring the release of [14C]-acid-soluble metabolites (ASMs) and [14C]-CO2 (n = 5), and (Q) serum transaminases (n = 3). Results are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 by two-way ANOVA in (B), (C), and (K), by ANCOVA analysis in (D), or Student’s t test in (A), (E)–(I), (M), (P) and (Q). f.c., denoted fold change.
See also Figure S1.
EE was significantly enhanced in MAT1A-KO mice (Figure 2D), whereas their locomotor activity when fed ad libitum was slightly decreased (Figure 2E), suggesting that the differences in weight gain observed were due to an enhanced EE and not to enhanced locomotor activity.
We observed lower fat content in male MAT1A-KO mice (45–55 days old) as seen by MRI analysis (Figure 2F), inguinal white adipose tissue (ingWAT)/body weight ratio (Figure 2G), and adipocyte size (Figure 2H). The MAT1A-KO mice also showed increased signs of ingWAT browning, as demonstrated by an upregulation of Ucp-1 mRNA expression and protein levels (Figures 2I and 2J), as well as enhanced lipolytic response when stimulated with isoproterenol, as demonstrated by increased secretion of glycerol and free fatty acids in an ex vivo assay (Figure 2K).
BAT from MAT1A-KO mice had similar weights (data not shown) but displayed a distinct morphology with a reduction in size of the multilocular lipid droplets (as seen by hematoxylin and eosin [H&E] staining) (Figure 2L), showed an increased expression of Uncoupling protein 1 (Ucp1) mRNA and protein levels (Figures 2M and 2N), and showed enhanced thermogenesis as seen by a significant increase in the surface temperature of interscapular BAT of MAT1A-KO mice at room temperature or when subjected to acute cold exposure (at 4°C) (Figures 2O and S1D).
In the liver, we observed an increase in incomplete and complete fatty acid oxidation, respectively, from fed WT and MAT1A-KO livers (Figure 2P). This was not accompanied by liver damage, as shown by unaltered serum transaminase levels (Figure 2Q). These observations prompted us to examine whether insulin sensitivity and glucose metabolism was improved in MAT1A-KO mice. MAT1A-KO mice showed enhanced glucose tolerance, lower insulin resistance, and decreased glucose levels in response to pyruvate (Figure S1E).
Collectively, our results suggest that a chronic deficiency of hepatic SAMe, induced by the lack of MAT1A, can induce, even under fed conditions, a hypercatabolic state, which could ultimately lead to the increase in postnatal mortality. This catabolic response is similar to that observed after fasting in WT mice, which is also characterized by a decrease in SAMe levels, suggesting that the sustained decrease in SAMe levels could operate as a metabolic sensor of food restriction to induce the catabolic response.
Chronic MAT1A hepatic ablation exacerbates weight lost and induces hepatic β-oxidation, ATP production, ER stress, and liver damage during fasting
Next, we aimed to further challenge our mouse model of chronic MAT1A deficiency and examined their response to fasting. For this, we used MAT1A-KO mice at 50–60 days old (before the appearance of steatosis) and subjected them to starvation over a 36-h period. We found that both male and female MAT1A-KO mice exhibited a more pronounced loss of body weight during fasting than MAT1A-WT mice (Figures 3A and S2A, respectively).
Figure 3.
Chronic MAT1A hepatic ablation during fasting exacerbates weight loss and induces liver damage
(A–C) 50- to 60-day-old male MAT1A-KO mice were fasted.
(A) Graph showing more pronounced weight loss in mice fasted over a 36-h period (n = 3–8).
(B) Measurement of fat mass by MRI (n = 6) and (C) macroscopic pictures and ingWAT/mice weights (n = 3–5) in mice fasted during 36 h.
(D) Lipolysis measured by glycerol released to the culture medium in explants of ingWAT from WT and MAT1A-KO male mice fasted over 24 h (n = 4).
(E) Accumulated energy expenditure (EE) during the first 8 h of fasting in MAT1A-KO mice (n = 9–10).
(F) Representative pictures of H&E-stained sections of BAT from 24- to 36-h-fasted mice (scale bars, 50 μm).
(G) Immunoblot analysis of UCP1 and MAT1A protein levels in BAT from mice fasted during 24 and 36 h (n = 3–5).
(H) Incomplete and complete fatty acid (palmitate) oxidation in 36-h-fasted mice (n = 4).
(I) ATP levels under basal conditions and after glucagon treatment (10 nM, 24 h) in primary hepatocytes from 36-h-fasted mice (n = 3).
(J and K) Primary MAT1A-KO hepatocytes obtained from 24-h-fasted mice, which were treated with vehicle or 4 mM SAMe for 4 h and compared with untreated MAT1A-WT hepatocytes. (J) Cellular respiration parameters (n = 6–8) and (K) fatty acid oxidation rate (n = 6).
(L) Serum transaminases in 24 h of fasted mice (n = 4–5).
(M–O) Immunoblot analysis of ER stress proteins from liver extracts; (M) fasted for 36 h (n = 4–5 biological replicates for each strain) or (N) fed during 2 months with 40% of CR (n = 5) or (O) ADF (n = 3). Results are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 by two-way ANOVA in (A), (I), and (J), one-way ANOVA in (K), by ANCOVA analysis in (E), or Student’s t test in (B)–(D), (H), and (L). f.c., denoted fold change.
See also Figures S2 and S3.
Fasting led to a reduction in fat content in MAT1A-KO mice as seen by MRI (Figure 3B) and weight of ingWAT (Figures 3C and S2B), which was accompanied by enhanced lipolysis as demonstrated by increased secretion of glycerol in an ex vivo assay (Figure 3D). However, we did not observe an increase in fatty acid secretion in MAT1A-KO ingWAT, which could be due to an exhaustion of the ingWAT after 24 h of fasting or to enhanced fatty acid oxidation (Figure S2C).
EE was significantly enhanced in MAT1A-KO mice (Figure 3E), and BAT from fasted MAT1A-KO mice maintained a reduction in size of the multilocular lipid droplets (Figure 3F) and increased UCP1 protein levels (Figure 3G) at 24 and 36 h after fasting. Importantly, we confirmed as described before,22 that MAT1A could not be detected in BAT and ingWAT (Figures 3G and S2D), and that MAT2A was not differentially expressed in BAT from MAT1A-KO mice (Figure S2E), suggesting that the effects observed in adipose tissues in MAT1A-KO mice were due to a systemic effect triggered by the lack of expression of hepatic MAT1A.
Finally, although we did not observe significant differences in expression of the lipogenic genes Fas and Srebp, and of the lipolytic genes Hsl and Mgl, in BAT and ingWAT from fed or 36-h-fasted WT and MAT1A-KO mice, we found a significant increase in Atgl mRNA expression in ingWAT from MAT1A-KO mice after 36 h of fasting (Figure S2F), in agreement with the enhanced lipolysis observed in these tissues (Figure 3D). We did not find differences in serum glucagon levels between WT and MAT1A-KO mice, indicating that the effects observed in MAT1A-KO mice were not likely due to an increased action of glucagon on the fasting response (Figure S2G).
Livers from fasted MAT1A-KO mice showed a significant increase in incomplete fatty acid palmitate oxidation (acid-soluble metabolite [ASM]) (Figure 3H) that correlated with enhanced serum levels of ketone bodies and lower serum levels of triglycerides (TGs) (Figures S2H and S2I). Importantly, we found that hepatocytes isolated from fasted MAT1A-KO mice produced more ATP in basal and stimulated conditions (glucagon treatment) (Figure 3I).
We next evaluated the impact of SAMe administration on oxygen consumption rate (OCR), ATP production, and β-oxidation in MAT1A-KO hepatocytes during fasting. Hepatocytes isolated from 24-h-fasted MAT1A-KO mice showed enhanced basal OCR, ATP-linked respiration, maximal respiration, and non-mitochondrial oxygen consumption, an effect that was abolished by SAMe supplementation (Figures 3J and S2J). SAMe treatment also prevented the enhanced fatty acid oxidation activity seen in MAT1A-KO hepatocytes (Figure 3K). In summary, these results suggest that low hepatic SAMe levels in MAT1A-KO mice enhance hepatocyte respiration, β-oxidation, and ATP production during fasting.
Interestingly, we observed that 24 and 36 h of fasting induced liver damage in MAT1A-KO mice—as seen by increased alanine aminotransferase (ALT) serum levels (Figures 3L and S3A)—and enhanced p38MAPK and cJun phosphorylation (Figure 3M) and also increased ERS, as seen by increased phosphorylation of eukaryotic initiation factor-2α (eIF2α) and inositol-requiring enzyme 1 (IRE1) (Figure 3M). Similarly, we found enhanced cJun and IRE1 phosphorylation after 5 days of 60% of CR in MAT1A-KO livers (Figure S3B).
SAMe is a physiologic precursor of reduced GSH, which is known to play a protective and antioxidant role in liver.23 To examine whether decreased GSH synthesis was responsible for the observed increase in ERS in fasted MAT1A-KO mice, we treated 36-h-fasted MAT1A-KO mice with the GSH precursor N-acetyl cysteine (NAC). We found that NAC treatment did not prevent hepatic eIF2α phosphorylation. Similar results were obtained in primary hepatocytes isolated from 24-h-fasted MAT1A-KO mice treated during 16 h with NAC (Figure S3C). Altogether, our data suggest that a decrease in GSH levels is not likely to be responsible for the ERS observed in fasted MAT1A-KO mice.
Mammalian target of rapamycin (mTOR) regulates cell growth and coordinates the response to amino acid availability.24 Experiments performed in HEK293T cells have shown that Met starvation can inhibit mTORC1 signaling in a SAMTOR-dependent fashion by reducing SAMe levels and promoting the association of SAMTOR with GATOR1.25 We therefore decided to examine whether decreased levels of hepatic SAMe in MAT1A-KO mice could also inhibit mTORC1 signaling. We did not observe any alteration in mTOR phosphorylation between MAT1A-WT and -KO mice under fed or 36-h-fasted conditions at 50 days of age (Figure S3D). These observations are in agreement with our previous data showing that phosphorylation of mTOR and the mTOR substrate Initiation factor 4E binding protein (4EBP) was strongly inhibited by increasing SAMe levels in the liver or by the treatment of hepatocytes with SAMe.13 Altogether, these data suggest that alterations in mTOR signaling are not likely to be responsible for the observed effects in MAT1A-KO mice.
We next decided to assess whether the increase in liver damage and ERS observed during fasting and 60% of CR during 5 days in MAT1A-KO mice was simply the result of stress during adaptation or if, on the contrary, they were an effect that could be relevant for longer CR regimens. We found that ERS also increased significantly in MAT1A-KO mice fed for 2 months with either ADF or 40% CR (Figures 3N and 3O). Similarly, the percentage of weight loss was constantly higher in MAT1A-KO than in WT mice during 30 consecutive fasting periods of 24 h during ADF, whereas there was a trend to greater weight loss at initial stages of 40% of CR until a plateau of weight loss was reached (Figures S3E and S3F). Notably, two out of the seven MAT1A-KO mice fed with ADF developed liver damage, and we observed a significant increase in ALT in MAT1A-KO mice under 40% CR for 2 months (Figures S3G and S3H).
Altogether, our data show that hepatic ablation of MAT1A during fasting hyperactivates fat usage and mobilization from adipose tissue, as well as hepatic β-oxidation and ATP production, leading to ERS and liver damage. This suggests that the upregulation of MAT1A levels plays a protective role during food restriction to balance the catabolic response.
Decreased hepatic SAMe levels in MAT1A-KO reduces FGF21 promoter methylation in liver, increasing FGF21 circulating levels and EE
FGF21, which is mainly expressed in the liver, is involved in the metabolic response of starvation and protein restriction by playing a central role in the induction of hepatic fat oxidation, ketogenesis and gluconeogenesis, lipolysis, and browning of WAT, and in improving glucose tolerance.26 We observed that hepatic Fgf21 mRNA levels and serum FGF21 were significantly higher in fed and fasted MAT1A-KO mice (Figures 4A, 4B, and S3I). Livers from MAT1A-KO mice fed with ADF or with 40% CR for 2 months had also enhanced Fgf21 mRNA expression (Figures S3J and S3K).
Figure 4.
Chronic MAT1A hepatic ablation reduces FGF21 promoter methylation and FGF21 transcription, enhancing energy expenditure
(A and B) Serum FGF21 and Fgf21 mRNA levels in (A) fed and (B) fasted male MAT1A-KO mice (n = 5–9).
(C and D) Graphs showing percentage of DNA methylation levels of 5 consecutive CGs (CG1 to CG5) of Fgf21 in livers from (C) fed (n = 7) and (D) fasted MAT1A-KO mice (n = 9–11).
(E) FGF21 serum levels (n = 3–5).
(F–L) (F) Fat mass by MRI (n = 7), (G) ingWAT/mice weights and macroscopic pictures, (H) quantification of density of adipocytes (n = 4–7) and representative pictures of H&E-stained sections (scale bars, 50 μm), (I) accumulated energy expenditure (EE) (n = 4–5), (J) incomplete fatty acid (palmitate) oxidation (n = 3–6), (K) p-eIF2α in total liver extracts (n = 5), and (L) serum transaminases (AST, ALT) (n = 6–7) in 36-h-fasted MAT1A-KO mice after silencing of FGF21. Results are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 by one-way ANOVA in (E), (F), and (J), two-way ANOVA in (B)–(D), or Student’s t test in (A), (G)–(I), and (K). f.c., denoted fold change.
See also Figure S3.
It has been shown that reduced DNA methylation ratio of CpG sites located downstream (#10–#21) of the transcription start site (TSS) of Fgf21 is associated with enhanced induction of hepatic FGF21 expression.27 Here, we found a significant decrease in DNA methylation levels at the CpG sites (#10–#15) in livers from fed and fasted MAT1A-KO mice (Figures 4C and 4D). Next, to examine the functional importance of this upregulation of FGF21 in the fasting response of MAT1A-KO mice, we silenced hepatic FGF21 in vivo in MAT1A-KO mice, which effectively reduced the elevated serum levels of FGF21 (Figure 4E) and prevented the enhanced loss of body fat, ingWAT weight, adipocyte size, and EE during fasting in MAT1A-KO mice (Figures 4F–4I). Notably, we did not observe any hepatic phenotype in these mice, including MAT1A-KO-enhanced hepatic β-oxidation rate, ERS, or elevated transaminases (Figures 4J–4L).
Altogether, our results demonstrate that loss of hepatic MAT1A induces FGF21 promoter hypomethylation, likely leading to an increase in hepatic synthesis and secretion of FGF21, which in turn would impact on EE. On the other hand, this increase in FGF21 levels is unlikely to be involved in the increase in β-oxidation and liver damage observed in MAT1A-KO mice after food restriction regimes.
Met cycle fluxes during fasting toward SAMe and PC synthesis, and SAMe-mediated regulation of PEMT modulates hepatic β-oxidation, ER stress, and liver damage
We performed a fluxomic experiment using nuclear magnetic resonance (NMR) spectroscopy to track the incorporation of 13C-labeled Met into other cellular metabolites. For this, we intravenously injected 13C-uniformly labeled Met, 30 min before sacrifice, in 24-h-fasted WT and MAT1A-KO mice. We found, in hepatic lysates, radioactive labeling in several metabolites from the Met cycle (including SAMe, homocysteine, PC, and homoserine) as well as in metabolites from gluconeogenesis (including glycerol 3-phosphate, fructose 1,6-bisphosphate, glucose-6phosphate, and glucose), and in lactate. Notably, we found a decrease in the amount of 13C label in SAMe, homocysteine, homoserine, lactic acid, and fructose 1,6 bis phosphate metabolites, whereas an increase in 13C label was observed in Met, glycerol-3P, glucose 6-P, and glucose in MAT1A-KO mice (Figures 5A and 5B), suggesting a displacement of the metabolic flux of Met from SAMe synthesis toward glucose synthesis by loss of MAT1A during fasting. This displacement could be due to a decrease in the transmethylation and transsufuration pathways (due to the absence of MAT1A) and to an increase in the Met transamination pathway, as often seen in adults with MAT deficiency.28,29
Figure 5.
Methionine cycle fluxes during fasting toward SAMe and PC synthesis
(A and B) (A) Schematic representation of Met flux through one-carbon metabolism, tricarboxylic acid cycle (TCA) cycle, and gluconeogenesis, and (B) relative fold change (log2) in the hepatic content of 13C label of the Met-derived metabolites in 24-h-fasted MAT1A-KO mice, as compared with WT mice (blue and red indicate upregulated and downregulated, respectively) (n = 7).
(C) PC and PE hepatic content and PC/PE ratio in livers from fed and 24-h-fasted WT and MAT1A-KO mice (n = 7).
(D–K) (D) RT-qPCR of hepatic Pemt gene (n = 5), (E) PC hepatic content (n = 5) and PC/PE ratio (n = 5–6), (F) percentage of loss of weight (n = 5), (G) ratios of ingWAT/mice weights (n = 5), (H) hepatic Fgf21 mRNA and serum FGF21 levels (n = 5), (I) hepatic fatty acid (palmitate) oxidation (n = 5), (J) hepatic eIF2α protein phosphorylation (n = 5), and (K) serum transaminases (AST, ALT) (n = 5) in Pemt-silenced mice after 36 h of fasting. Results are presented as the mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 by Student’s t test. f.c., denoted fold change.
See also Figure S4.
Interestingly, these data also showed a trend toward decreased 13C label incorporation into PC during these 30 min of labeling. To confirm this, we measured total PC and PE content by 31P-NMR-based phosphoromics30 and found a clear decrease in PC and the PC/PE ratio in livers of 24-h-starved MAT1A-KO mice (Figure 5C). One of the two pathways for PC synthesis involves trimethylation of PE by hepatic PEMT, a reaction that requires three molecules of SAMe.31 Ablation of PEMT leads to a strong reduction in the PC/PE ratio, which results in an increase in mitochondrial respiration and ATP production.32,33 We previously showed that the decrease in SAMe availability in MAT1A-KO mice fed ad libitum can lead to a decrease in PEMT activity and PC/PE ratio.34 These data suggest that the enhanced β-oxidation, ATP synthesis, ERS, and liver damage observed in fasted MAT1A-KO mice could be due to this decrease in PEMT activity. To demonstrate this, we acutely silenced PEMT in liver prior to a period of 36 h of fasting. We found that hepatic Pemt gene expression was strongly reduced in these mice (Figure 5D), which led to a decrease in the hepatic PC content and PC/PE ratio (Figure 5E). Pemt silencing did not lead to a more pronounced loss of body weight (Figure 5F) and ingWAT mass (Figure 5G) or to enhanced FGF21 gene or protein expression (Figure 5H). Instead, Pemt silencing during fasting induced hepatic β-oxidation, as shown by palmitic acid complete oxidation (Figure 5I) that was accompanied by increased ERS (as seen by enhanced eIF2α phosphorylation) and AST (Figures 5J and 5K). Some of these effects induced by Pemt silencing were also observed after acute silencing of Mat1a, similar to our data above in the MAT1A-KO mice (Figures S4A–S4D). Finally, as expected, FGF21 silencing during fasting did not lead to a recovery of the decrease in PC content in MAT1A-KO livers (Figure S4E), suggesting that elevated levels of hepatic FGF21 in MAT1A-KO mice are unlikely to be involved in regulating PEMT function in MAT1A-KO mice after food restriction regimes.
Altogether, these results suggest that SAMe levels could be essential for regulating PEMT function in hepatic β-oxidation, ER stress, and unfolded protein response (UPR) during fasting. Excessive reduction of SAMe synthesis during fasting thus could lead to a decrease in PC/PE ratio at the ER, inducing ERS, which could ultimately result in liver damage.
Low hepatic SAMe levels enhance mitochondrial interaction with ER
Mitochondrial β-oxidation is fueled by the release of Ca2+ from the ER to the mitochondria. During fasting, certain regions of ER become reversibly tethered to mitochondria at contact points called MAMs to favor this Ca2+ flow. After feeding, glucose reduces the number of MAMs in mouse liver, impairing mitochondrial respiration.35 These observations led us to posit that loss of hepatic MAT1A, and therefore low hepatic SAMe levels, could also modulate mitochondrial interaction with ER. Thus, using transmission electron microscopy (TEM), we examined liver sections from fed and 24-h-fasted WT and MAT1A-KO mice and measured the degree of contact between mitochondria and ER, as previously described.36 In WT mice, as expected, fasting induced an increase in contact between mitochondria and ER. Importantly, we found that MAT1A-KO hepatocytes had a notably enhanced percentage of contact between mitochondria and ER, compared with that of MAT1A-WT mice, both under fed and fasted conditions (Figures 6A and 6B). These data suggest that a decrease in hepatic SAMe levels favors the formation of ER-mitochondria contact points, which could promote Ca2+ transfer to the mitochondria and mitochondrial respiration.
Figure 6.
MAT1A localizes at MAMs during fasting
(A) Representative electron microscopy images, and (B) graph showing the percentage of ER adjacent to mitochondria (n = 309–423), in fixed liver sections from fed and 24-h-fasted MAT1A-WT and -KO mice (from n = 3 biological replicas for each condition). Mitochondrial perimeter is denoted by green lines and ER-mitochondrial contact points by blue lines.
(C) Model of phospholipid synthesis and trafficking at MAMs.
(D and E) Immunoblot analysis showing an increase in MAT1A protein expression in membrane/organelles and cytosolic fractions isolated from n = 4 livers of C57BL/6 mice after (D) 24 h of fasting or acute CR (60% for 5 days) and (E) ADF or chronic CR (40% for 2 months). Controls of purity for membrane/organelles are shown.
(F and G) Homogenates and subcellular fractions from 6- and 24-h-fasted livers of (F) C57BL/6 mice or (G) from WT and MAT1A-KO mice over a 36-h period of fasting (n = 1–2). Controls of purity for homogenates (H), nuclear (N), crude mitochondria (Mc), pure mitochondria (MP), MAMs, and endoplasmic reticulum (ER) extracts are shown.
(H and I) Ultra-performance liquid chromatography (UPLC) analysis of SAMe in (H) total liver extracts (n = 3–5), and (I) crude mitochondrial extracts from WT and MAT1A-KO mice (n = 4) (please see STAR Methods for detailed protocol). Results are presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001 by two-way ANOVA in (B) and (H) or Student’s t test in (I).
See also Figure S4.
MAT1A localizes at MAMs during fasting modulating local SAMe synthesis
As mentioned above, PEMT, which is mainly localized at MAMs, functions during starvation to sustain PC synthesis and the import of phosphatidylserine (PS) into mitochondria (Figure 6C).37,38 Thus, a decrease in SAMe availability in MAT1A-KO mice after fasting can also reduce PEMT activity at MAMs, leading to a decrease in PC/PE32,33 (Figure 6C).
In light of the above findings, we hypothesized that during fasting, the MAT1A protein synthetized could localize close to PEMT at ER/MAM regions for the local production of the SAMe required for its activity (Figure 6C). Indeed, we found that 24-h fasting, ADF, acute CR (60% for 5 days), or chronic CR (40% for 2 months) all led to an increase in MAT1A protein in organelles (Figures 6D and 6E). We then used subcellular fractioning by differential centrifugation to obtain MAMs, ER, and crude and pure mitochondria (MP)-enriched fractions from fresh liver tissue. We found that MAT1A protein was undetectable in MAMs at 6 h after fasting but was highly enriched after 16–24 h of fasting (Figures 6F and S4F). This expression pattern was specific since we found that MAT1A was not detected in MAMs in fasted MAT1A-KO mice and in Mat1a-silenced mice (Figures 6G and S4G).
We next examined whether localization of MAT1A at MAMs during fasting led to compartmentalization of SAMe synthesis at this specific compartment by measuring SAMe and SAH in total liver extracts and crude mitochondria extracts (which include MAMs plus mitochondria) isolated from fed and 24-h-starved WT and MAT1A-KO mice. Nutritional stress (fasting or CR) did not alter SAMe levels in total liver homogenates between WT and MAT1A-KO mice (Figures 6H and S4H–S4J). Strikingly, however, we found that in crude mitochondria extracts, the levels of SAMe were significantly lower in 24-h-fasted MAT1A-KO mice (Figure 6I). We also observed a decrease in SAH levels in crude mitochondria from starved MAT1A-KO mice, suggesting a decrease in methylation reactions during starvation in MAT1A-KO mice (Figure S4K). These data suggest that food deprivation or restriction induces the compartmentalization of SAMe synthesis at specific subcellular locations where methylation reactions are required, which would be an important mechanism to counterbalance the global reduction in SAMe levels caused by lack of nutrients, preventing excessive β-oxidation and ERS and thus protecting against liver damage.
Discussion
In this study, we report that nutritional stress regimes (fasting and CR), which stimulate lipid catabolism, are accompanied by a profound decrease in hepatic levels of the principal biological methyl donor SAMe. Notably, by artificially reducing hepatic SAMe levels in vivo, using mice lacking MAT1A, we were able to mimic the enhanced lipid catabolism seen in nutritional stress, even under a fed state. Furthermore, our data suggest that local synthesis of SAMe during fasting could fine-tune the formation of ER-mitochondria contact points to modulate hepatic β-oxidation and ATP synthesis and to prevent liver damage. This suggests that SAMe levels could play a key physiological role as a metabolic sensor of nutrition, particularly in the hepatic fasting adaptive response.
A major metabolic adaptation from glucose to lipid utilization takes place during the transition from fetal to postnatal life. We have previously shown that Mat1a expression increases in mice from embryonic day 18, increasing 10-fold immediately after birth, and reaching a peak at 10 days of age.17,39 Our results suggest that lack of hepatic MAT1A during postnatal development would interfere with the tuning of this metabolic adaptation by enhancing lipid catabolism, which would prevent body weight gain and increase littermate mortality. In addition, metabolic adaptation plays a critical role in the transition from fasting to fed state, switching from oxidation of fatty acids (fasted state) to glucose oxidation (fed state). Our results suggest that lack of MAT1A (or low hepatic SAMe levels) would impair this metabolic adaptation after a meal, leading to a metabolic inflexibility and decreasing the ability to switch off fatty acid oxidation during the transition from fasting to a fed state. We observed a significant recovery in hepatic SAMe levels 2 h after refeeding in WT mice. The impact of the recovery of SAMe levels on fatty acid and glucose oxidation in the transition from fasting to fed state, however, still remains to be elucidated.
Importantly, we describe a new molecular function of MAT1A in the maintenance of SAMe levels at ER-mitochondria contact sites during fasting. ER-mitochondria contact sites are a reversible contact event that appears to be favored by fasting, leading to increased calcium transfer from ER to mitochondria.40,41,42 Here, we have used standard TEM analyses and biochemical analyses of enriched MAMs to study ER-mitochondria interactions during fasting, in line with other studies,36,42 which could be confirmed using other advanced imaging platforms in future studies, including cryoelectron microscopy (cryo-EM) and serial section electron tomography coupled to three-dimensional reconstruction.41,43 Based on our results, we propose that during fasting, after the hepatic decrease in intracellular SAMe, newly synthetized MAT1A in the liver would translocate to the ER-mitochondria contact sites to participate in the local synthesis of SAMe, fueling the activity of PEMT to modulate the PC/PE ratio. It has been demonstrated that decreases in the PC/PE ratio can profoundly alter mitochondria energy production, enhancing mitochondrial respiration and the activities of proteins of the electron transport chain, and increasing production of ATP in hepatocytes.44 Here, we have observed that Pemt silencing during fasting decreases PC/PE ratio, enhancing β-oxidation and ERS. Thus, we propose that the translocation of MAT1A to the ER-mitochondria during fasting would fuel PEMT activity; maintain the PC/PE ratio in mitochondria; tune ER-mitochondria tethering, lipid β-oxidation, and ATP production; and prevent ERS.
These results are in line with other studies, which have shown that MAT1A could be recruited to subcellular compartments to provide a local source of SAMe. For instance, galactosamine and acetaminophen treatments induce the nuclear accumulation of MAT1A,45 and other studies have found that the nuclear localization of MAT1A correlated with increased histone 3 K27 trimethylation, a gene repression mark.46 Most recently, it was demonstrated that MAT1A is present in the mitochondrial matrix, interacting with mitochondrial proteins, such as cytochrome P450 2E1.47 Altogether, our results suggest that during fasting or CR when the global hepatic levels of SAMe are compromised and when the intracellular levels of SAH rise, inhibiting methylation reactions, MAT1A could be recruited to ER and MAMs to provide a local supply of SAMe required to carry the methylation reactions needed at those specific compartments.
Another important observation is that hepatic MAT1A ablation during fasting induces ER stress, similarly to the one that we have detected in Pemt-silenced fasted mice. We hypothesize that in situations of enhanced lipid mobilization during fasting, the decreased ratio of PC/PE at ER (due to a decrease in availability of SAMe and/or in functionality of PEMT48), would induce ER stress and sensitize the mice to liver injury. Interestingly, PEMT-KO mice fed with HFD are showing a similar phenotype as young MAT1A-KO mice, with both models protected from weight gain and insulin resistance, but showing with time signs of liver damage, hepatic steatosis, and increased ALT.21,49 In MAT1A-KO mice, we propose that this could be due to a disruption in mitochondrial function. Initially, MAT1A deficiency or chronically low hepatic SAMe levels would lead to an excessive lipid mobilization and hepatic β-oxidation, promoting mitochondrial stress and ERS, which, if they persist, would damage mitochondrial functionality, as seen in 8-month-old MAT1A-KO mice, characterized by increased levels of hepatic reactive oxygen species, ERS, reduced mitochondrial potential, and disrupted hepatic very low-density lipoprotein assembly.34,50
Several studies have suggested that restrictions in Met availability can reduce adiposity. But more recent studies have found that it is SAMe, rather than Met, which is independently associated with fat mass and truncal adiposity in individuals with obesity.15,16 Interestingly, Mat1a-silenced mice fed with HFD show protection from weight gain.51 Our own results further corroborate these observations. Here, we show that MAT1A deficiency, which is characterized by reduced hepatic SAMe levels, can reduce adiposity. Importantly, in these mice there is a striking accumulation of hepatic and circulating Met, which supports the notion that it is the decrease in hepatic SAMe levels, rather than Met levels, that is important for controlling body fat mass and also suggests that a decrease in hepatic SAMe levels could act as a metabolic sensor of nutrition to induce hypermetabolism in a scenario where food availability is limited. However, this work also provides important insights into the previously undescribed function of newly synthetized SAMe during fasting as a hepatic protective metabolite, arguing for a potential role of local SAMe production in modulating PC synthesis (by PEMT) and ERS and in fine-tuning ER-mitochondrial contact, β-oxidation, and ATP synthesis during fasting. Finally, our results suggest that lack of hepatic MAT1A and chronic hepatic SAMe deficiency could induce lipolysis and browning in adipose tissue, likely due to epigenetic-mediated synthesis of the lipolytic factor FGF21 by the liver. In summary, our results highlight the importance that intra- and subcellular regulation of SAMe levels in hepatocytes play in the adaptive catabolic lipid response to fasting. On the one hand, the physiological decrease of hepatic SAMe levels during fasting could act as a hepatocyte-metabolic intracellular sensor of nutrition by activating ER-mitochondria tethering and β-oxidation in liver. Remarkably, at the same time, we also show that local synthesis of SAMe at MAMs could act as a brake to prevent excessive mitochondrial respiration to prevent ER stress and liver damage (Figure S6).
Limitations of the study
Although in our study we showed that MAT1A translocate to MAMs during food restriction regimes, we have been unsuccessful to identify the protein responsible for anchoring MAT1A to the MAMs or to find the MAT1A region responsible for its localization at MAMs. This has precluded us from examining the functional significance of this localization.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| β-Actin | Sigma-Aldrich | Cat#A5441; RRID:AB_476744 |
| Calreticulin | Cell Signaling Technology | Cat#12238; RRID:AB_2688013 |
| Calnexin | Enzo Life Sciences | Cat#ADI-SPA-860-F; RRID:AB_11178981 |
| c-Jun | Cell Signaling Technology | Cat#9165; RRID:AB_2130165 |
| Phospho-c-Jun (Ser73) | Cell Signaling Technology | Cat#9164; RRID:AB_330892 |
| Cytocrome C | Cell Signaling Technology | Cat#4272; RRID:AB_2090454 |
| Phospho-eIF2α (Ser51) | Cell Signaling Technology | Cat#9721; RRID:AB_330951 |
| eIF2α | Cell Signaling Technology | Cat#5324; RRID:AB_10692650 |
| FACL4 | Abcam | Cat#ab155282; RRID:AB_2714020 |
| GAPDH | Abcam | Cat#ab8245; RRID:AB_2107448 |
| GRP75 | Cell Signaling Technology | Cat#3593; RRID:AB_2120328 |
| Phospho-Ire1α | Abcam | Cat#Ab48187; RRID:AB_873899 |
| Ire1α | Santa Cruz | Cat#SC-390960; RRID:AB_2927490 |
| IP3R | Abcam | Cat#ab5804; RRID:AB_305124 |
| MATα1 | In-house | N/A |
| MATα2 | Abcam | Cat#ab26174; RRID:AB_881274 |
| PDI | Cell Signaling Technology | Cat#2446; RRID:AB_2298935 |
| PEMT | Sigma-Aldrich | Cat#AV43558; RRID:AB_1855188 |
| Phospho-p38 MAPK | Cell Signaling Technology | Cat#9215; RRID:AB_331762 |
| Phospho-mTOR (Ser 2448) | Cell Signaling Technology | Cat#2971; RRID:AB_330970 |
| Phospho-mTOR (Ser 2481) | Cell Signaling Technology | Cat#2974; RRID:AB_2262884 |
| mTOR | Cell Signaling Technology | Cat#2972; RRID:AB_330978 |
| UCP1 | Fisher | Cat#PA1-24894; RRID:AB_2241459 |
| VDAC | Cell Signaling Technology | Cat#4661; RRID:AB_10557420 |
| Anti-mouse IgG, HRP-linked | Cell Signaling Technology | Cat#7076; RRID:AB_330924 |
| Anti-rabbit IgG, HRP-linked | Cell Signaling Technology | Cat#7074; RRID:AB_2099233 |
| Anti-Chicken-IgY, HRP-linked | Aves Labs | Cat#H-1004; RRID:AB_2313517 |
| Bacterial and virus strains | ||
| pLKO.1 plasmid | Addgene | Cat#8453 |
| Chemicals, peptides, and recombinant proteins | ||
| N-acetyl-L-cysteine (NAC) | EMD Millipore Corporation, Merck | Cat#106425 |
| 3C-uniformly labeled methionine | Tracer Tec, Spain | Cat#LC-1972-C |
| Glucagon | Sigma-Aldrich | Cat#G1774 |
| Invivofectamine™ 3.0 Kit | Life Technologies | Cat#IVF3005 |
| Critical commercial assays | ||
| Mouse/Rat FGF-21 Quantikine ELISA Kit | R&D Systems | Cat#MF2100 |
| ATPlite luminescence assay system | Perkin Elmer | Cat#6016943 |
| Fatty Acid Oxidation (FAO) Kit | Biomedical Research Service | Cat#E-141 |
| NEFA-HR (1) Assay | FUJIFILM Wako Chemicals Europe | Cat#434-91795 |
| NEFA-HR (2) Assay | FUJIFILM Wako Chemicals Europe | Cat#436-91995 |
| Free Glycerol Reagent | Sigma | Cat#F6428 |
| ProteoExtract Subcellular Proteome Extraction Kit | Merck-Millipore | Cat#539790 |
| Deposited data | ||
| Data S1 - Source Data | This paper | This paper |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6 | CEBEGA (USC) | N/A |
| Mouse: Gcgr−/− | B Finan et al.52 | From Prof. Timo D. Müller |
| Mouse: Mat1a-KO | Lu et al.20 | N/A |
| Oligonucleotides | ||
| si-Mat1a (S62313) | Ambion® In Vivo siRNA | Cat#4457308 |
| si-Pemt (S71475) | Ambion® In Vivo siRNA | Cat#4457302 |
| si-Ctrl | Ambion® In Vivo siRNA | Cat#4457289 |
| sh-scrambled | CCTAAGGTTAAGTCGCCCTCG | This paper |
| sh-Fgf21 | TCTATGGATCGCCTCACTTTG | This paper |
| Sequences for Methylation analysis of Fgf21 by pyrosequencing | Sequencing: CTCATCCATTCCATCA, forward: TTAGTTGGGGATTTAATAT AGGAGAAATAG, Reverse: TCCCAA CTCTAAATCTCATCCATTCCA |
This paper |
| qRT-PCR Primer sequences | Table S1 | N/A |
| Software and algorithms | ||
| FLIR-Tools specific software package | Contreras et al.53 | |
| ImageJ software | https://imagej.nih.gov/ij. | https://imagej.nih.gov/ij. |
| PyroMark Assay Design 2.0 | Qiagen, Germany | N/A |
| Image lab software | BIO-Rad Laboratories | N/A |
| QuanLynx software | Waters, Manchester, UK | N/A |
| TopSpin 4.0.7 | Bruker Biospin GmbH | N/A |
| GraphPad Prism 8 software | GraphPad | N/A |
| Graphical abstract and Figures 6C and S5 were created using biorender.com | https://www.biorender.com/ | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Marta Varela-Rey (martavarela.rey@usc.es).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Animals
MAT1A-KO and MAT1A-WT male and female mice (between 40 - 55 days of age for most experiments except for weigt curve analysis, for which mice up to 1 year old were used) were bred at the CIC bioGUNE AAALAC-accredited animal facility, and at the CEBEGA-USC animal facility. C57BL/6J male mice of 45 – 90 days of age were bred and used at our animal facility (CEBEGA/USC). All animal procedures were approved by the CIC bioGUNE Institutional Animal Care and Use Committee, by the Faculty Animal Committee at the University of Santiago de Compostela, the Country Council of Bizkaia and The Xunta de Galicia, and the experiments were performed in agreement with the Rules of Laboratory Animal Care and International Law on Animal Experimentation. Mice were maintained in groups of a maximum of 5 mice per cage, containing woodchip bedding, on a 12 h light (8:00 to 20:00)/12 h dark cycle, under controlled temperature (20 to 22°C) and humidity of 45 ±10%. Mice were maintained with ad libitum access to water and to a rodent maintenance diet (2914 Teklad global 14% protein, or the 2918 Teklad global 18% protein rodent maintenance diets Envigo). An analysis of the influence of sex on the results of the study was performed (Figures S1C, S2A, S2B, S3A, and S3H)
Mouse primary hepatocyte cultures
Mouse primary hepatocytes were obtained by in situ perfusion of the liver with collagenase digestion following the Lefferts method.54 Briefly, mice were anesthetized in an inhalation chamber with 4% IsoFlo (13400264, Proyma), dissolved in 0,9% O2. The abdomen was opened, and intestines were moved to the right side to expose the portal vein. After tying a knot around, the portal vein was punctured with a SurFlash® Polyurethane I.V. Catheters18g x 2' (Terumo) and connected to a peristaltic pump (Econo Pump, BioRad). Inferior cava vein was immediately cut to allow blood drainage. First, 20 ml of warm (37ºC) 1 mM EGTA-Leffert’s buffer (10 mM HEPES, 3 mM KCl, 130 mM NaCl, 1 mM NaH2PO4·H2O, 10 mM D-glucose, pH 7.4) were allowed to flow through the circuit, followed by 20 ml of warm Leffert’s buffer, and ultimately 50 ml of 2.5 mM CaCl2-Leffert’s buffer containing 25 mg of collagenase type I (350 U/mg, LS004196, Worthington). Special care was taken to avoid air bubbles flowing into the liver. After the perfusion, the liver was detached from the animal and placed in a 100 mm tissue culture-treated dish containing ice-cold Minimum Essential Medium (MEM) (31095029, Gibco) supplemented with 10% (v/v) Fetal Bovine Serum (FBS) (10270106, Gibco) and 1% (v/v) Penicillin-Streptomycin-Glutamine (10378016, Gibco). The gallbladder was carefully removed, and the liver was disposed of the Glisson's capsule. With the help of a pair of forceps, the cells were removed from the liver by gently shaking the tissue. The cell solution obtained from the digested livers was filtered through a sterile gauze and resuspended in a final volume of 50 ml of the referred cell culture medium above. Next, mouse primary hepatocytes were isolated by several centrifugation steps. The perfused livers were centrifuged 3 times at 500 rpm for 5 min and the cell pellet was resuspended in 50 ml of fresh cell culture medium. Cell density was estimated in a Neubauer counting chamber and cell viability was assessed by Trypan Blue solution staining (T8154, Sigma-Aldrich). A cell viability superior to 70% was achieved in all experiments. Finally, 0.5x106 mouse primary hepatocytes were deposited on collagen type I (354236, Corning)-coated 6-well culture plates and cultured in Dulbecco’s Modified Eagle’s Medium (DMEM), no glucose (11966025, Gibco) supplemented with 5% (v/v) FBS (10270106, Gibco), 1% (v/v) Antibiotic-Antimycotic (15240062, Gibco), and hepatocytes were allowed to attach to the bottom of the plate during 2h and then culture medium was replaced with FBS-free DMEM, no glucose supplemented with 1% (v/v) Antibiotic-Antimycotic for 1 h. Next, cells were incubated in the absence or presence of 10 nM glucagon (G1774, Sigma-Aldrich) during 12 h and 16h for mRNA and protein analysis respectively.
For total ATP quantification, mitochondrial respiration, and β-oxidation assays in hepatocytes, livers were perfused from mice starved during 24 or 36h, following the procedure described before with slight modifications using Leffert’s buffer without glucose. After the perfusion, the liver was released from the animal and placed into 100 mm TC-treated dishes containing ice-cold containing DMEM, no glucose (11966025, Gibco) supplemented with 5% (v/v) FBS (10270106, Gibco), and 1% (v/v) Antibiotic-Antimycotic (15240062, Gibco). After three washes, the cell pellet was resuspended in 50 ml of fresh cell culture medium and cell density and viability was estimated using Trypan Blue as described before. Finally, 0.5x106 mouse primary hepatocytes were placed on collagen type I (354236, Corning)-coated 6-well culture plates and hepatocytes were allowed to attach to the bottom of the plate during 3h and then culture medium was replaced with medium without FBS.
Method details
Fasting Studies
For the studies done with Mat1a-KO mice, for six, 24 and 36h of fasting, food was removed between six to nine a.m. and mice sacrificed six, 24 or 36h later. Mice had ad libitum access to water. For the course time studies done in C57BL/6 mice, food was removed at 3 p.m. for mice sacrificed after 24h and 6h of fasting, at 4 pm for mice sacrificed after 16 h of fasting, and at 7pm for mice sacrificed after 12h of fasting. When indicated, N-acetyl-L-cysteine (NAC) (400 mg/kg, EMD Millipore Corporation, Merck #106425) was administered intravenously 9h before sacrifice to Mat1a-KO mice fasted during 36h. For the fluxomic studies, 24h-fasted mice were injected intravenously with 13C-uniformly labeled Met (25 mg/kg) (LC-1972-C, Tracer Tec, Spain) 30 min before sacrifice. For the analysis of Mat1a mRNA expression in overnight-fasted Gcgr+/+ and Gcgr-/- mice,19 animals were treated by intraperitoneal injection with 200 μg/kg body weight of glucagon diluted in saline (Sigma-Aldrich, #G1774), 30 min before the sacrifice.55
Alternate day fasting studies
Mice were randomly separated into two groups: one group receive the maintenance diet ad libitum, while the other group was provided with access to food at 11 a.m. every other day.
Caloric Restriction Studies
Five days before the onset of caloric restriction (CR), mice were placed in individual cages and fed with the maintenance diet ad libitum. During these days of acclimatization, food intake was monitored to determine the average amount of food consumed daily by each mouse.
For the 60% of CR for 5 days, mice were randomly separated into two groups: one group continued to receive the maintenance diet ad libitum, while the other group underwent a 60% CR, i.e., each mouse was fed at 6 p.m. every day with an amount of food equivalent to 40% of its own daily food intake during the acclimatization week.
For the 40% of CR for 2 months, mice were randomly separated into two groups: one group continued to receive the maintenance diet ad libitum, while the other group underwent a 40% CR, each mouse was fed at 12 to 2 p.m. every day with an amount of food equivalent to 60% of its own daily food intake during the acclimatization week. Throughout the procedure, mice remained housed individually.
Mat1a and Pemt silencing in vivo
C57BL/6 mice of 45 to 90 days of age were randomly divided into two experimental groups. Tail vein injection was done at 24h before the start of the fasting period and immediately before the start of the fasting period (-24 and 0h) using si-Mat1a, si-Pemt, or si-Ctrl (Ambion® In Vivo siRNA) dissolved in Invivofectamine™ 3.0 Kit (IVF3005, Life Technologies) following manufacturer’s instructions.
Thermal imaging
Mice were placed for 6h in a special room with a stable temperature of 4ºC.56 Interscapular temperature was assessed and visualized using a high-resolution infrared camera (E60bx: Compact-Infrared-Thermal Imaging-Camera; FLIR) and analyzed with a FLIR-Tools specific software package.52
Indirect calorimetric measurements
The metabolic parameters, energy expenditure and locomotor activity were analyzed using an indirect calorimetric system (LabMaster; TSE Systems; Bad Homburg, Germany),57,58 in 12 open circuit sealed chambers. Measurements were performed for the dark (from 8 p.m. to 8 a.m.) and light (from 8 a.m. to 8 p.m.) periods under ad libitum feeding or fasting conditions. Mice were allowed to acclimatize to the individual metabolic cages. Energy expenditure and locomotor activity were calculated using the manufacturer’s system software. Fat Mass was analyzed by ECHO MRI M113 mouse system (Echo Medical Systems).
shRNA Design and Lentivirus Generation
Small hairpin RNAs (shRNAs) and lentiviral production utilized in this study were performed as previously described.53 Briefly, the specific shRNAs for knockdown of Fgf21 and control Luciferase shRNA were designed, synthesized, and subcloned into pLKO.1 vector (Addgene). All shRNA constructs were confirmed by SANGER sequencing and knockdown efficiency was validated by RT-qPCR. The target sequences of the shRNAs used were: sh-scrambled CCTAAGGTTAAGTCGCCCTCG; and shFgf21#2 TCTATGGATCGCCTCACTTTG. For lentiviral production, pLKO.1-shRNA plasmids were co-transfected with packaging vectors into HEK293T cells. Viral supernatants were harvested after 48 and 72h post-transfection, concentrated using centrifugal filter units (Amicon, Ref: UFC903024), and virus titer was determined by QuickTiter Lentivirus Kit (Cell Biolabs, Ref: VPK-107).
GTT, ITT and PTT tolerance tests
Basal blood glucose levels were measured after an overnight fast (12h) for the Glucose (GTT) and Pyruvate (PTT); and after 6h for the Insulin (ITT), with a Glucocard Glucometer (ARKRAY, USA). GTT, ITT, and PTT were done after intraperitoneal injection of D-glucose (2 g/kg, Sigma-Aldrich, G8270), insulin (0.35 U/kg Actrapid, Novo Nordisk), or sodium pyruvate (1.25 g/kg Sigma-Aldrich, P2256) respectively. Area under the curve values were determined.
Hematoxylin and eosin (H&E) staining
Five μm-thick paraffin-embedded sections of formalin-fixed liver, ingWAT and BAT samples were deparaffinised for 20 min using Histo-Clear (HS-200, National Diagnostics) and rehydrated through graded ethanol solutions (100-70%) to distilled water. Sections were incubated with Harris Hematoxylin (Bio-Optica, Milano, Italy) for 15 min, followed by eosin staining (Sigma Aldrich) during 15 min. Next, sections were rinsed in running tap water and differentiated with 0.5% (v/v) HCl. Finally, samples were dehydrated through graded ethanol solutions (70-100%), cleared with Histo-Clear and mounted with DPX mounting medium (06522, Sigma-Aldrich). Five to ten random images per sample were blindly taken using a Leica DM750 microscope. Number of adipocytes per field were counted using ImageJ software https://imagej.nih.gov/ij.
Serum extraction from mice
Anaesthesia was induced by placing each mouse in an inhalation chamber with 4% isoflurane (IsoFlo®, Abbot Laboratories) in 0.8-1% O2. Blood was withdrawn from the retro-orbital plexus with a micro haematocrit capillary (BRAND® micro haematocrit capillary BR749311) and collected in BD Microtainer serum separator gel tubes (Microtainer®, 365967, BD). Blood samples were allowed to clot for 5-15 min and were subsequently centrifuged (6,000 rpm, 10 min, 4ºC). Serum was transferred to a non-autoclaved Eppendorf safe-lock tubes and immediately stored at -80ºC.
Determination of serum transaminases
Aspartate aminotransferase (AST) and alanine aminotransferase (ALT) serum activity was determined by using the Selectra Junior Spinlab 100 automated analyser (Vital Scientific) according to the manufacturer’s instructions. Standard controls were run before each determination, and the values obtained for the different biochemical parameters were always within the expected ranges.
Serum measurements
Mouse serum FGF21 concentration was determined using the Mouse/Rat FGF-21 Quantikine ELISA Kit (MF2100, R&D Systems) according to the manufacturer’s instructions. Glucagon quantification was performed using the Glucagon ELISA kit (10-1271-01, Mercodia) according to the manufacturer’s instructions. Serum triglycerides levels in fasted and fed conditions were measured using 10 μl of serum and commercially available kits (A. Menarini Diagnosis, Spain and Wako Chemicals, USA, respectively) following manufacturer’s instructions. Serum ketone bodies were quantified using a kit from Wako chemicals (Richmond, VA).
FAO determination in frozen tissue
The β-oxidation rate was determined as previously described8 with slight modifications. Liquid N2-frozen tissue was homogenized in ice-cold buffer (25 mM Tris-HCl, 500 nM sucrose, 1 mM EDTA-Na2, pH 7.4) using a Potter homogenizer. The tissue homogenate was subsequently sonicated for 10s and centrifuged (420 x g, 10 min, 4ºC). The supernatant was subjected to protein determination, and 500μg of total protein was used per assay in a final volume o 60μl. The reaction was started by incubating the sample with 340 μl of the assay mixture containing 0.2 mM 0.5 μCi/ml [1-14C]-palmitic acid (NEC075H, Perkin Elmer) for 30 min at 37ºC, in Eppendorf tubes covered with a Whatman paper cap. The reaction was stopped by the injection of 200 μl of 3 M perchloric acid, and the Whatman paper caps were impregnated with 1 M NaOH to allow the capture of released CO2. After leaving the tubes at room temperature for 1h, the filter paper was removed and the acidified medium was centrifuged (21,000 x g, 10 min, 4ºC) to eliminate particulate matter. The radioactivity of the trapped CO2, which corresponds with complete oxidation, and the radioactivity of the acid soluble metabolites (ASM) contained in the supernatant, which is equivalent to incomplete oxidation, were measured in a scintillation counter.
ATP quantification in primary hepatocytes
For total ATP quantification, cells were incubated in the absence or presence of 10 nM glucagon (G1774, Sigma-Aldrich) during 3h. Then, hepatocytes (0.5x106) were harvested in 100 μl of mammalian cell lysis solution from ATPlite luminescence assay system (6016943, Perkin Elmer) diluted in 200 μl of media per well. Five μl of this lysis solution was used for the quantification of total ATP levels in OptiPlate-96 white opaque 96-well microplates (6005290, PerkinElmer), following the ATPlite manufacturer’s instructions. Luminescence was measured in a Veritas microplate luminometer (Turner BioSystems). ATP concentration was calculated by extrapolation to an ATP standard curve and subsequent normalization with the total protein content of each sample determined by the Bradford assay (5000006, Bio-Rad).
FAO determination in primary hepatocytes
Fatty Acid Oxidation (FAO) Kit (E-141Biomedical Research Service) was used as previously described.59,60 Briefly, attached hepatocytes were exposed to glucose-free DMEM supplemented with 5% (v/v) FBS to limit nutrient availability, with or without the addition of 4 mM SAMe. After 4h, cells were washed twice with ice cold PBS and 1x106 cells were lysed in 50 μl of Cell Lysis Solution, incubated at 4ºC for 5 min, followed by centrifugation at 12,000 xg for 5 min. The supernatants were transferred to new tubes, and 50 μl of each was added in 96-well plate, mixed with 50 μl of Reaction Solution, and incubated at 37°C (without CO2) for 60 min. Absorbance was determined at 492 nm, and FAO activity was determined normalizing the results to protein content. All reactions were performed in duplicate.
Mitochondrial respiration in hepatocytes
Mitochondrial respiration was evaluated by monitoring the oxygen consumption rate (OCR) by high-resolution respirometry with the Seahorse Bioscience XF Extracellular Flux Analyzer (Agilent Technologies). Primary hepatocytes (2x104), obtained from Mat1a-KO and Mat1a-WT mice after 24h of fasting, were seeded per well in a XFe96 cell culture microplate (102416–100, Seahorse Bioscience, Agilent Technologies) into DMEM, no glucose (11966025, Gibco) supplemented with 5% (v/v) FBS (10270106, Gibco), and 1% (v/v) Antibiotic-Antimycotic (15240062, Gibco), and maintained for 3h at 37°C (with CO2) to allow cell adhesion. Then, cells were exposed to either 4 mM SAMe or vehicle for 4h. After that, medium was removed, and replaced with pre-warmed assay medium, composed of Seahorse XF DMEM medium (103575-100 Seahorse Bioscience, Agilent Technologies) containing 1 mM sodium pyruvate, 2 mM L-glutamine and 10 mM glucose, with or without SAMe, and cultured at 37°C in room air. Controls were supplemented with the same amount of fatty-acid-free BSA. After equilibration in assay medium for 1h, five basal measurements of OCR were performed. Next, we sequentially added into cells wells the modulators of respiration oligomycin (Oligo, 1.5 μM), carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone (FCCP, 1 μM) and rotenone/antimycin (Rot/AA, 0.5 μM) to evaluate the mitochondrial function (Cell Mito Stress Test Kit, 103015-100, Seahorse Bioscience, Agilent Technologies). The following key parameters of mitochondrial function were calculated according to the manufacturer's user guide: basal respiration, ATP-linked respiration, proton leak, maximal respiration, and spare capacity. All results were normalized to protein content.
Lipolysis measurements in ingWAT
At sacrifice, 20 mg pieces of ingWAT were cut and placed in 1X PBS. The tissues were incubated with 100 μl of basal or stimulated (100 nM isoproterenol) medium [1% DMEM, 4% BSA (Without Fatty Acids)] in 96-well plates for 4h at 37 °C with stirring. After 4h, the pieces of ingWAT were removed and the medium stored at –80ºC. Fatty acids content was determined using 2 μl of the medium using NEFA-HR (1) and HR (2) Assay (434-91795 and 436-91995) from FUJIFILM Wako Chemicals Europe Kit following the manufacturer’s instructions. For glycerol determination, 6 μl of medium was placed in 96-well plates, and 200 μl of the Free Glycerol Reagent (F6428 Sigma) was added to each well. The plates were incubated for 5 min at 37 °C. Standards were prepared using Glycerol Standard. Absorbance was read at 540 nm in a SpectraMax M2/M2e microplate reader (Molecular Devices).
Total protein extraction from tissue
Liquid N2-frozen tissue (25 to 40 mg) was homogenized in 1 ml of RIPA lysis buffer (1.6 mM Na2HPO4, 8.4 mM NaH2PO4, 5 g/L sodium azide, 100 mM NaCl, 0.1% (w/v) SDS, 0.1% (v/v) Triton X-100) supplemented with 5 g/L sodium deoxycholate, 1 mM sodium orthovanadate, 50 mM NaF, protease and phosphatase inhibitor cocktails in a Precellys 24 homogenizer (Bertin Instruments) at 5,000 rpm for 30s. The homogenate was transferred into a clean tube and subjected to one freeze-thaw cycle in liquid N2 to enhance cell disruption. After centrifugation (12,500 rpm, 20 min, 4ºC), total protein concentration from the supernatant was estimated by the Micro BCA Protein Assay Kit (23235, ThermoFisher Scientific) in a SpectraMax M2/M2e microplate reader (Molecular Devices). Samples were stored at -80ºC.
Subcellular protein extraction from tissue
Subcellular fractions from flash-frozen liver tissue were isolated using the ProteoExtract Subcellular Proteome Extraction Kit (539790, Merck-Millipore) with slight modifications to the manufacturer’s instructions. Briefly, 20 mg of tissue was resuspended in 1 ml of ice-cold Extraction Buffer I containing 5 μl of Protease Inhibitor Cocktail, 1 mM sodium orthovanadate and 50 mM NaF and disrupted using borosilacate homogenizers. The mixture was incubated for 20 min at 4ºC in a carousel rotating shaker. The insoluble material was pelleted by centrifugation (1,000 xg, 10 min, 4ºC) and the supernatant (fraction 1, cytosolic proteins) was transferred to a clean tube and stored at -80ºC. The pellet was resuspended again with 1 ml of buffer I for 10 min at 4ºC in a carousel rotating shaker. The insoluble material was pelleted by centrifugation (1,000 xg, 10 min, 4ºC) and the supernatant was discarded. The pellet was resuspended with 1 ml of ice-cold Extraction Buffer II containing 5 μl of Protease Inhibitor Cocktail, 1 mM sodium orthovanadate and 50 mM NaF by gently flicking the tube and incubated for 30 min at 4ºC in a carousel rotating shaker. The insoluble material was pelleted by centrifugation (6,000 xg, 10 min, 4ºC) and the supernatant (fraction 2, membranes and organelles proteins) was transferred to a clean tube and stored at -80ºC. The pellet was resuspended again with 1 ml of buffer II and incubated for 10 min at 4ºC in a carousel rotating shaker. The insoluble material was pelleted by centrifugation (6,000 xg, 10 min, 4ºC) and the supernatant was discarded. The pellet was resuspended with 0,5 ml of Buffer III containing 5 μl of Protease Inhibitor Cocktail, 1,5 μl Benzonase® nuclease, 1 mM sodium orthovanadate and 50 mM NaF by gently flicking the tube and incubated for 20 min at 4ºC in a carousel rotating shaker. The insoluble material was pelleted by centrifugation (7,000 xg, 10 min, 4ºC) and the supernatant (fraction 3, nuclear proteins) was transferred to a clean tube and stored at -80ºC. Protein concentration of each subcellular compartment was determined by the Bradford assay (5000006, Bio-Rad). Controls of purity for membrane/organelles (FACL4, IP3R, CRT, CNX and VDAC), and cytosolic fractions GAPDH were done by immunioblotting.
Mitochondria-associated membranes isolation
Crude and pure mitochondria, endoplasmic reticulum and MAMs were isolated from fresh mouse livers as previously described.61 Protein concentration of each subcellular structure was estimated by the Micro BCA Protein Assay Kit (23235, ThermoFisher Scientific) measured in a SpectraMax M2/M2e microplate reader (Molecular Devices). For the determinations of SAMe, SAH and Met in crude mitochondria the processing time was reduced to 3h, and done at 4ºC or on ice, without adding detergents. This protocol ensures that mitochondria are maintained as close as possible to the in vivo state. Additionally, to ensure lack of cross-contamination with cytoplasmic contents, the pellet of crude mitochondria was washed twice with MRB buffer before metabolite extraction, and the absence of GAPDH on samples was confirmed by western blot. Controls of purity for homogenates (H), nuclear (N), crude mitochondria (Mc), pure mitochondria (Mp), MAMs, and endoplasmic reticulum (ER) extracts, using IP3R, Cytochrome C (Cyt C), Calreticulin (CRT), PDI, Calnexin (CNX) and GRP75 antibodies were done by immunioblotting.
Immunoblotting
Two to 30 ug of protein were combined with 5x sample loading buffer (25% (v/v) Glycerol, 250 mM Tris pH 6.8, 10% (w/v) SDS, 500 mM β-Mercaptoethanol, 0,125% (w/v) bromophenol blue) and boiled at 95ºC for 5 min. Protein samples were separated by SDS-PAGE in 10-15% acrylamide gels using a Mini-PROTEAN Tetra Cell electrophoresis system (Bio-Rad) or 4-12% NuPAGE Bis-Tris (WG1403BOX, ThermoFisher Scientific). The following molecular weight markers were used PiNK Plus Prestained Protein Ladder (GeneDireX) or Blue Star Protein marker from Nippon Genetics. Proteins were transferred from gels into 0.2 μm-pore size nitrocellulose membranes (10600001, GE Healthcare) using a Trans-Blot Cell system electroblotting or using a Bio-Rad Trans-blot SD Semi-dry Transfer Cell (Bio-Rad). The presence of total protein was detected by Ponceau S solution (P7170, Sigma-Aldrich) staining. Non-specific binding was blocked by incubation of the membranes with 0.1% (v/v) Tween 20-TBS, solution containing 5% (w/v) skimmed milk powder or 2,5% (w/v) BSA (A3912, Sigma-Aldrich) for 1 h at room temperature, prior to addition of the primary antibody following the conditions in Table S1. After washing the unbound primary antibody with 0.1% (v/v) Tween 20-TBS three times, membranes were incubated with the corresponding HRP-linked secondary antibody for 1 h at room temperature. Membranes were washed three times to remove the excess of secondary antibody and the Western Lightning Plus-ECL HRP substrate (NEL104001EA, Perkin Elmer) was subsequently added. The chemiluminescent signal from immunoreactive proteins was captured using an iBright™ Imaging System (Model #:FL1500) or by exposition to Super RX Fuji Medical X-Ray films (06-SHRGH1824B, Fujifilm), which were further developed in a Curix 60 processor (AGFA Healthcare). Films were digitalised and transformed into 8-bit images. Bands were quantified by densitometric analysis using the open-source image processing program Image J (http://rsbweb.nih.gov/ij) and normalized to GAPDH or β-actin housekeeping protein expression.
Total RNA isolation
RNA from flash-frozen tissue and primary hepatocytes was extracted with TRIzol reagent (15596026, Invitrogen) as per manufacturer’s instructions. RNA was heated to 65°C for 5 min and quickly chilled on ice. Then concentration was determined using NanoDrop 1000 spectrophotometer (ThermoFisher Scientific).
Reverse transcription (RT)
RNA (1-2 μg) was treated with Amplification Grade DNase I (18068015, Invitrogen) following the manufacturer’s guidelines. cDNA was synthesized with M-MLV Reverse Transcriptase (28025013, Invitrogen) in the presence of Random Primers (48190011, Invitrogen), dNTPs (10297018, Invitrogen), and RNaseOUT Recombinant Ribonuclease Inhibitor (10777019, Invitrogen), by using a Veriti Dx Thermal Cycler (Applied Biosystems). Mix was incubated at 25°C for 10 min, then was incubated 180 min at 37°C, and the reaction was inactivated by heating at 70°C for 15 min. The resulting cDNA was diluted 20-fold in Nuclease-Free Water (W4502, Sigma-Aldrich).
Quantitative PCR (qPCR)
Mus musculus gene primer sequences were designed with the Primer-BLAST tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) and synthesized by Sigma-Aldrich. DNA (1.5 μl) was mixed with specific primers (Table S1) and SYBR Select Master Mix (4472908, Invitrogen) constituting a final volume of 6.5 μl, in MicroAmp Optical 384-Well Reaction Plates (4309849, Applied Biosystems). Each reaction was performed in duplicate using the ViiA 7 Real-Time PCR System (Applied Biosystems). qPCR conditions involved an initial denaturation step (90 s at 95ºC), followed by 40 cycles of annealing (15 s at 95ºC and 1 min at 59ºC), and a final extension phase (15 s at 95ºC, 1 min at 60ºC and 15 s at 95ºC). Ct values were extrapolated from the melting curve and gene expression levels were normalized with GAPDH or HPRT, or 18S ribosomal RNA housekeeping expression by implementing the formula.
Methylation analysis of FGF21
A total of 500 ng of genomic DNA for each sample was used for the bisulfite conversion with the EZ DNA Methylation kit (Zymo Research) following manufacturer’s recommendations. DNA methylation levels of 5 consecutive CGs (CG1 to CG5) of Fgf21 (chr7:45,264,738-45,264,772; GRCm39/mm39) were analyzed by pyrosequencing. Briefly, primer sequences for Fgf21 analysis were designed with PyroMark Assay Design 2.0 (Qiagen, Germany) (Sequencing: CTCATCCATTCCATCA, forward: TTAGTTGGGGATTTAATATAGGAGAAATAG, Reversed: TCCCAACTCTAAATCTCATCCATTCCA). Standard PCR reactions were carried out with ∼10 ng of bisulfite-converted genomic DNA. PCR products were observed in 2% (w/v) agarose gels before pyrosequencing. PyroMark Q24 Vacuum Workstation was used for the immobilization and preparation of PCR products. Pyrosequencing reactions were performed using a PyroMark Gold Q24 Reagent Kit (Qiagen, Germany) following the manufacturer’s instructions. Methylation values were obtained using PyroMark Q24 Software 2.0 (Qiagen, Germany). Assays were conducted including 100% methylated DNA (Universal Methylated Mouse DNA Standard from Zymo Research) as positive control, and water as no template control.
Quantification of Lipids
Livers (30-50 mg) were homogenized in cold PBS in a bead mill homogenizer (OMNI international; USA) at 3m/s for 30 seconds and sonicated with 3 cycles of 30 seconds sonication and 10 seconds rest at a frequency of 23 kHz and with an amplitude ≤ 6 microns. Total protein was measured as described before and lipids were extracted from 1.5 mg of total protein following the Folch method.62 Major lipid species were separated by thin layer chromatography63 and PC and PE levels were quantified using the image lab software (BIO-Rad Laboratories Inc, USA).
Determinations of SAMe, SAH and Methionine
SAMe, SAH and Met levels were determined in serum, liver tissues, and crude mitochondria samples by Ultra-High-Performance Chromatography coupled to Mass Spectrometry (UPLC-MS). Protein concentration of crude mitochondria was estimated by the Micro BCA Protein Assay Kit Briefly, and a pellet from mitochondria samples and liver tissues (50 mg) were homogenized in 500 μL of ice-cold extraction liquid with a tissue homogenizer (FastPrep) in for 40 s at 6000 rpm. The extraction liquid consisted of a mixture of ice-cold methanol/water (50/50 %v/v) with 10 mM acetic acid and 1 μM stable labelled 13CD3-Met (Met-SL) as internal standard. Subsequently 400 μL of the homogenate plus 400 μL of chloroform was transferred to a fresh tube and shaken at 1400 rpm for 60 min at 4 °C. Next the aliquots were centrifuged for 30 min at 13000 rpm at 4 °C. The aqueous phase (150 μL) was transferred to a fresh aliquot? and placed at -80 °C for 20 min. The chilled supernatants were speed-vac evaporated, and the resulting pellets were resuspended in 100 μL water/acetonitrile/formic acid (40/60/0.1 v/v/v) for mitochondria samples and 250 μL for liver samples. Serum samples (50 μL) were mixed with 50 μL of water/0.15% (v/v) formic acid. Subsequently, proteins were precipitated by addition of 150 μL of acetonitrile. To get optimum extraction, after addition of acetonitrile, the samples were sonicated for 10 min at 4ºC and agitated at 1,400 rpm for 30 min at 4ºC. Next, the samples were centrifuged at 14,000 rpm for 60 min at 4ºC. Concentrations of all metabolites were determined with a semi-quantitative method.64 Calibration curves for SAMe, SAH and Met were obtained by measuring serial dilutions of a pooled standard mixture in resuspension solution. The concentrations for all compounds in the dilutions ranged from 10 μM to 0.0025 μM. Samples were measured with a UPLC system (Acquity, Waters Inc., Manchester, UK) coupled to a Time-of-Flight mass spectrometer (ToF MS, SYNAPT G2, Waters Inc.). A 2.1 x 100 mm, 1.7 μm BEH amide column (Waters Inc.), thermostated at 40 °C, was used to separate the analytes before entering the MS. Mobile phase solvent A (aqueous phase) consisted of 99.5% water, 0.5% FA and 20 mM ammonium formate while solvent B (organic phase) consisted of 29.5% water, 70% acetonitrile, 0.5% FA and 1 mM ammonium formate. To obtain a good separation of the analytes the following gradient was used: from 5% A to 50% A in 2.4 min in curved gradient (#8, as defined by Waters), from 50% A to 99.9% A in 0.2 min constant at 99.9% A for 1.2 min, back to 5% A in 0.2 min. The flow rate was 0.250 mL/min and the injection volume was 2 μL. All samples were injected randomly. After every 8 injections QC sample was injected. Analytes were measured in enhanced duty cycle (EDC) mode, optimized for the mass of the analyte in question. Met was measured in scan function 1 (EDC at 152), SAH was measured in scan function 2 (EDC at 385) and SAMe was measured in scan function 3 (EDC at 399). The cone voltage was between 20 and 25 depending on the analyte. A 2 ng/mL leucine-enkephalin solution in water/acetonitrile/formic acid (49.9/50/0.1 v/v/v) was infused at 10 μL/min and used for a lock mass which was measured each 32 s for 0.5 s. Spectral peaks were automatically corrected for deviations in the lock mass. Extracted ion traces are obtained for SAMe (m/z = 399.145), Met (m/z = 150.0589), 13CD3-Met (m/z = 154.0796) and SAH (m/z = 385.1294) in a 20 mDa window for the most abundant isotopes and subsequently smoothed and integrated with QuanLynx software (Waters, Manchester, UK).
Fluxomic analysis by NMR spectroscopy
After 24 h fasting, Mat1a-WT and Mat1a-KO mice were intravenously injected with 13C-uniformly labeled Met (25 mg/kg), 30 min before sacrifice. Aliquots of 300 mg of liver, were divided into three different Eppendorf tubes. Thus, for every 100 mg of liver 774 μL of ice-cold chloroform/methanol mixture (38.8/61.2 v/v) were added. Afterwards, livers were homogenized in a Precellys tissue homogenizer at 4000 g for 2 x 30 s and then, 554 μL of CHCl3:H20 (45.8:54.2 v/v) were added over the homogenate. After vortexing, the solution was centrifugated for 15 min at 9000 xg, 4oC. Next, the two phases were carefully divided into different Eppendorfs before being dried using Speedvac for at least 3h. Finally, the remaining powder was stored at -80oC. Next, for NMR spectroscopy, 300 mg of dry liver was resuspended into 300 μL of solution depending on each extract fraction. In the event of hydrophilic fraction, we used Phosphate buffer in deuterium oxide (200 mM), pD 7, with trimethylsilylpropanoic acid (2.5 mM) as 13C reference compound and gadobutrol (0.5 mM) as paramagnetic relaxation enhancer. Meanwhile, for the lipophilic phase, we used DMSO-d6 with added triphenyl phosphine oxide (4 mM) as 13C and 31P reference compound. Fluxomics experiments were recorded at 298 K on a Bruker 600 MHz (12 T) Avance III spectrometer equipped with a BBO probe head. For each sample, three different experiments were collected: (I) 1D 1H p3919gp with water signals suppression using a binomial 3-9-19 pulse with echo gradient pair, (II) 1D 13C zgpg30 with power-gated decoupling and (III) phase-sensitive gradient-enhanced 2D 1H-13C HSQC (hsqcetgp). However, phosphoromics experiments were recorded at 298 K on a Bruker 400 MHz (9.39 T) Avance Neo spectrometer equipped with a SmartProbe, having done two experiments: (I) a 1D 1H p3919gp with water signals suppression using a binomial 3-9-19 pulse with echo gradient pair and (II) a 1D 31P zgip spectrum with inverse gated 1H decoupling. Chemical shift assignment was done using standard compounds in order to facilitate the identification of metabolites. Then, assigned metabolites were quantified by referencing their 13C or 31P peak integral against the internal reference compound added. In some cases, we used peak deconvolution with the aim of assigning corresponding peak in case of signal overlap. Peak integration and quantification was done using TopSpin 4.0.7 (Bruker Biospin GmbH) and in-house MatLab scripts.
Electron microscopy
For electron microscopy studies, fed and 24 h fasted Mat1a-WT and Mat1a-KO mice were perfused by the inferior vena cava with saline at room temperature with 0.1 mL heparin per mouse. Next, livers were perfused with a mixture of 2% (v/v) paraformaldehyde and 2.5% (v/v) glutaraldehyde in 0.1 M phosphate buffer pH 7.4. After the perfusion, the liver was detached from the animal postfixed with the same fixative solution overnight at 4 ºC. Next, livers were washed four times with phosphate buffer 0.1 M, pH 7.4 for 30 min.To improve the post-fixing and embedding process of the samples, 150 μm sections from the same liver lobe were cut on a vibratome Leica VT-1000 (Leica, Heidelberg, Germany). Sections were post-fixed with 2% osmium, rinsed, dehydrated, and embedded in Durcupan resin (Fluka, Sigma-Aldrich, St. Louis, USA). Ultrathin sections (0.06 -0.08 μm) were cut with an Ultracut UC-6 (Leica microsystems, Wetzlar, Germany) with a diamond knife, stained with lead citrate (Reynold’s solution) and examined under a transmission electron microscope FEI Tecnai G2 Spirit BioTwin (ThermoFisher Scientific company, Oregon, USA). All images were acquired with a Xarosa digital camera (EMSIS GmbH, Münster, Germany) controlled by Radius software (Version 2.1). 30 pictures of each experimental group (from 3 different animals per group) were obtained at 9300x magnification. The images were analyzed using Image J (National Institutes of Health, USA). After calibration of images using the scale bars, the mitochondrial membranes, and the ER in contact with mitochondrial membranes were delineated using the freehand tool. The mitochondrial perimeter in contact with the ER was normalized with the total mitochondrial perimeter.
Quantification and statistical analysis
Statistical analysis was performed using GraphPad Prism 8 software. Data were represented as mean ± SEM for each experimental group. Statistical significance was determined by Student’s t-test, or one-way analysis of variance (one-way ANOVA) and two-way analysis of variance (two-way ANOVA) followed by post hoc Sidak. p < 0.05 was considered as statistically significant.
Acknowledgments
M.V.-R. is supported by Proyecto PID2020-119486RB-100 (funded by MCIN/AEI/10.13039/501100011033), Gilead Sciences International Research Scholars Program in Liver Disease, Acción Estratégica Ciberehd Emergentes 2018 (ISCIII), Fundación BBVA, HORIZON-TMA-MSCA-Doctoral Networks 2021 (101073094), and Redes de Investigación 2022 (RED2022-134485-T). M.L.M.-C. is supported by La CAIXA Foundation (LCF/PR/HP17/52190004), Proyecto PID2020-117116RB-I00 (funded by MCIN/AEI/10.13039/501100011033), Ayudas Fundación BBVA a equipos de investigación científica (Umbrella 2018), and AECC Scientific Foundation (Rare Cancers 2017). A.W. is supported by RTI2018-097503-B-I00 and PID2021-127169OB-I00, (funded by MCIN/AEI/10.13039/501100011033) and by “ERDF A way of making Europe,” Xunta de Galicia (Ayudas PRO-ERC), Fundación Mutua Madrileña, and European Community’s H2020 Framework Programme (ERC Consolidator grant no. 865157 and MSCA Doctoral Networks 2021 no. 101073094). C.M. is supported by CIBERNED. P.A. is supported by Ayudas para apoyar grupos de investigación del sistema Universitario Vasco (IT1476-22), PID2021-124425OB-I00 (funded by MCIN/AEI/10.13039/501100011033 and “ERDF A way of making Europe,” MCI/UE/ISCiii [PMP21/00080], and UPV/EHU [COLAB20/01]). M.F. and M.G.B. are supported by PID2019-105739GB-I00 and PID2020-115472GB-I00, respectively (funded by MCIN/AEI/10.13039/501100011033). M.G.B. is supported by Xunta de Galicia (ED431C 2019/013). C.A., T.L.-D., and J.B.-V. are recipients of pre-doctoral fellowships from Xunta de Galicia (ED481A-2020/046, ED481A-2018/042, and ED481A 2021/244, respectively). T.C.D. is supported by Fundación Científica AECC. A.T.-R. is a recipient of a pre-doctoral fellowship from Fundación Científica AECC. S.V.A. and C.R. are recipients of Margarita Salas postdoc grants under the “Plan de Recuperación Transformación” program funded by the Spanish Ministry of Universities with European Union’s NextGeneration EU funds (2021/PER/00020 and MU-21-UP2021-03071902373A, respectively). T.C.D., A.S.-R., and M.T.-C. are recipients of Ayuda RYC2020-029316-I, PRE2019/088960, and BES-2016/078493, respectively, supported by MCIN/AEI/10.13039/501100011033 and by El FSE invierte en tu futuro. S.L.-O. is a recipient of a pre-doctoral fellowship from the Departamento de Educación del Gobierno Vasco (PRE_2018_1_0372). P.A.-G. is recipient of a FPU pre-doctoral fellowship from the Ministry of Education (FPU19/02704). CIC bioGUNE is supported by Ayuda CEX2021-001136-S financiada por MCIN/AEI/10.13039/501100011033.
Author contributions
Conceptualization, M.L.M.-C. and M.V.-R.; funding acquisition, A.W., M.L.M.-C., and M.V.-R.; investigation, all authors; supervision, M.L.M.-C. and M.V.-R.; writing – original draft, A.W., M.L.M.-C., and M.V.-R.; writing – review & editing, all authors.
Declaration of interests
M.L.M.-C. advises for Mitotherapeutix.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: July 31, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cmet.2023.07.002.
Contributor Information
María Luz Martínez-Chantar, Email: mlmartinez@cicbiogune.es.
Marta Varela-Rey, Email: martavarela.rey@usc.es.
Supplemental information
Data and code availability
-
•
No datasets (e.g. RNA-Seq or proteomics) were generated in this study.
-
•
Data S1: Unprocessed source data underlying all blots and graphs, related to Figures 1, 2, 3, 4, 5, 6, and S1–S4. This file includes: 1) Uncropped scans of all the blots; 2) Excel file containing the values that were used to create all graphs in the paper.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Longo V.D., Mattson M.P. Fasting: molecular mechanisms and clinical applications. Cell Metab. 2014;19:181–192. doi: 10.1016/j.cmet.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geisler C.E., Hepler C., Higgins M.R., Renquist B.J. Hepatic adaptations to maintain metabolic homeostasis in response to fasting and refeeding in mice. Nutr. Metab. (Lond.) 2016;13:62. doi: 10.1186/s12986-016-0122-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rui L. Energy metabolism in the liver. Compr. Physiol. 2014;4:177–197. doi: 10.1002/cphy.c130024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakata S.F., Okumura S., Matsuda K., Horikawa Y., Maeda M., Kawasaki K., Chou J.Y., Tamaki N. Effect of fasting on methionine adenosyltransferase expression and the methionine cycle in the mouse liver. J. Nutr. Sci. Vitaminol. (Tokyo) 2005;51:118–123. doi: 10.3177/jnsv.51.118. [DOI] [PubMed] [Google Scholar]
- 5.Pascale R.M., Feo C.F., Calvisi D.F., Feo F. Deregulation of methionine metabolism as determinant of progression and prognosis of hepatocellular carcinoma. Transl. Gastroenterol. Hepatol. 2018;3:36. doi: 10.21037/tgh.2018.06.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murray B., Barbier-Torres L., Fan W., Mato J.M., Lu S.C. Methionine adenosyltransferases in liver cancer. World J. Gastroenterol. 2019;25:4300–4319. doi: 10.3748/wjg.v25.i31.4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramani K., Mato J.M., Lu S.C. Role of methionine adenosyltransferase genes in hepatocarcinogenesis. Cancers (Basel) 2011;3:1480–1497. doi: 10.3390/cancers3021480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirschey M.D., Shimazu T., Goetzman E., Jing E., Schwer B., Lombard D.B., Grueter C.A., Harris C., Biddinger S., Ilkayeva O.R., et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vance J.E. MAM (mitochondria-associated membranes) in mammalian cells: lipids and beyond. Biochim. Biophys. Acta. 2014;1841:595–609. doi: 10.1016/j.bbalip.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 10.Jacobs R.L., van der Veen J.N., Vance D.E. Finding the balance: the role of S-adenosylmethionine and phosphatidylcholine metabolism in development of nonalcoholic fatty liver disease. Hepatology. 2013;58:1207–1209. doi: 10.1002/hep.26499. [DOI] [PubMed] [Google Scholar]
- 11.Malloy V.L., Krajcik R.A., Bailey S.J., Hristopoulos G., Plummer J.D., Orentreich N. Methionine restriction decreases visceral fat mass and preserves insulin action in aging male Fischer 344 rats independent of energy restriction. Aging Cell. 2006;5:305–314. doi: 10.1111/j.1474-9726.2006.00220.x. [DOI] [PubMed] [Google Scholar]
- 12.Orentreich N., Matias J.R., DeFelice A., Zimmerman J.A. Low methionine ingestion by rats extends life span. J. Nutr. 1993;123:269–274. doi: 10.1093/jn/123.2.269. [DOI] [PubMed] [Google Scholar]
- 13.Zubiete-Franco I., García-Rodríguez J.L., Martínez-Uña M., Martínez-Lopez N., Woodhoo A., Juan V.G., Beraza N., Lage-Medina S., Andrade F., Fernandez M.L., et al. Methionine and S-adenosylmethionine levels are critical regulators of PP2A activity modulating lipophagy during steatosis. J. Hepatol. 2016;64:409–418. doi: 10.1016/j.jhep.2015.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ables G.P., Perrone C.E., Orentreich D., Orentreich N. Methionine-restricted C57BL/6J mice are resistant to diet-induced obesity and insulin resistance but have low bone density. PLoS One. 2012;7 doi: 10.1371/journal.pone.0051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elshorbagy A.K., Nijpels G., Valdivia-Garcia M., Stehouwer C.D., Ocke M., Refsum H., Dekker J.M. S-adenosylmethionine is associated with fat mass and truncal adiposity in older adults. J. Nutr. 2013;143:1982–1988. doi: 10.3945/jn.113.179192. [DOI] [PubMed] [Google Scholar]
- 16.Elshorbagy A.K., Jernerén F., Samocha-Bonet D., Refsum H., Heilbronn L.K. Serum S-adenosylmethionine, but not methionine, increases in response to overfeeding in humans. Nutr. Diabetes. 2016;6:e192. doi: 10.1038/nutd.2015.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gil B., Casado M., Pajares M.A., Boscá L., Mato J.M., Martín-Sanz P., Alvarez L. Differential expression pattern of S-adenosylmethionine synthetase isoenzymes during rat liver development. Hepatology. 1996;24:876–881. doi: 10.1002/hep.510240420. [DOI] [PubMed] [Google Scholar]
- 18.Zeng Z., Huang Z.Z., Chen C., Yang H., Mao Z., Lu S.C. Cloning and functional characterization of the 5′-flanking region of human methionine adenosyltransferase 1A gene. Biochem. J. 2000;346:475–482. [PMC free article] [PubMed] [Google Scholar]
- 19.Finan B., Clemmensen C., Zhu Z., Stemmer K., Gauthier K., Müller L., De Angelis M., Moreth K., Neff F., Perez-Tilve D., et al. Chemical hybridization of glucagon and thyroid hormone optimizes therapeutic impact for metabolic disease. Cell. 2016;167:843–857.e14. doi: 10.1016/j.cell.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 20.Lu S.C., Alvarez L., Huang Z.Z., Chen L., An W., Corrales F.J., Avila M.A., Kanel G., Mato J.M. Methionine adenosyltransferase 1A knockout mice are predisposed to liver injury and exhibit increased expression of genes involved in proliferation. Proc. Natl. Acad. Sci. USA. 2001;98:5560–5565. doi: 10.1073/pnas.091016398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martínez-Chantar M.L., Corrales F.J., Martínez-Cruz L.A., García-Trevijano E.R., Huang Z.Z., Chen L., Kanel G., Avila M.A., Mato J.M., Lu S.C. Spontaneous oxidative stress and liver tumors in mice lacking methionine adenosyltransferase 1A. FASEB J. 2002;16:1292–1294. doi: 10.1096/fj.02-0078fje. [DOI] [PubMed] [Google Scholar]
- 22.Mato J.M., Martínez-Chantar M.L., Lu S.C. S-adenosylmethionine metabolism and liver disease. Ann. Hepatol. 2013;12:183–189. [PMC free article] [PubMed] [Google Scholar]
- 23.Lushchak V.I. Glutathione homeostasis and functions: potential targets for medical interventions. J. Amino Acids. 2012;2012:736837. doi: 10.1155/2012/736837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sancak Y., Bar-Peled L., Zoncu R., Markhard A.L., Nada S., Sabatini D.M. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu X., Orozco J.M., Saxton R.A., Condon K.J., Liu G.Y., Krawczyk P.A., Scaria S.M., Harper J.W., Gygi S.P., Sabatini D.M. SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science. 2017;358:813–818. doi: 10.1126/science.aao3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martínez-Garza Ú., Torres-Oteros D., Yarritu-Gallego A., Marrero P.F., Haro D., Relat J. Fibroblast growth factor 21 and the adaptive response to nutritional challenges. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20194692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan X., Tsujimoto K., Hashimoto K., Kawahori K., Hanzawa N., Hamaguchi M., Seki T., Nawa M., Ehara T., Kitamura Y., et al. Epigenetic modulation of FGF21 in the perinatal mouse liver ameliorates diet-induced obesity in adulthood. Nat. Commun. 2018;9:636. doi: 10.1038/s41467-018-03038-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blom H.J., Boers G.H., van den Elzen J.P., Gahl W.A., Tangerman A. Transamination of methionine in humans. Clin. Sci. (Lond.) 1989;76:43–49. doi: 10.1042/cs0760043. [DOI] [PubMed] [Google Scholar]
- 29.Gahl W.A., Bernardini I., Finkelstein J.D., Tangerman A., Martin J.J., Blom H.J., Mullen K.D., Mudd S.H. Transsulfuration in an adult with hepatic methionine adenosyltransferase deficiency. J. Clin. Invest. 1988;81:390–397. doi: 10.1172/JCI113331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernardo-Seisdedos G., Bilbao J., Fernández-Ramos D., Lopitz-Otsoa F., Gutierrez de Juan V., Bizkarguenaga M., Mateos B., Fondevila M.F., Abril-Fornaguera J., Diercks T., et al. Metabolic landscape of the mouse liver by quantitative (31) Hepatology. 2021;74:148–163. doi: 10.1002/hep.31676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vance D.E. Phospholipid methylation in mammals: from biochemistry to physiological function. Biochim. Biophys. Acta. 2014;1838:1477–1487. doi: 10.1016/j.bbamem.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 32.van der Veen J.N., Kennelly J.P., Wan S., Vance J.E., Vance D.E., Jacobs R.L. The critical role of phosphatidylcholine and phosphatidylethanolamine metabolism in health and disease. Biochim. Biophys. Acta Biomembr. 2017;1859:1558–1572. doi: 10.1016/j.bbamem.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 33.van der Veen J.N., Lingrell S., da Silva R.P., Jacobs R.L., Vance D.E. The concentration of phosphatidylethanolamine in mitochondria can modulate ATP production and glucose metabolism in mice. Diabetes. 2014;63:2620–2630. doi: 10.2337/db13-0993. [DOI] [PubMed] [Google Scholar]
- 34.Cano A., Buqué X., Martínez-Uña M., Aurrekoetxea I., Menor A., García-Rodríguez J.L., Lu S.C., Martínez-Chantar M.L., Mato J.M., Ochoa B., et al. Methionine adenosyltransferase 1A gene deletion disrupts hepatic very low-density lipoprotein assembly in mice. Hepatology. 2011;54:1975–1986. doi: 10.1002/hep.24607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Theurey P., Tubbs E., Vial G., Jacquemetton J., Bendridi N., Chauvin M.A., Alam M.R., Le Romancer M., Vidal H., Rieusset J. Mitochondria-associated endoplasmic reticulum membranes allow adaptation of mitochondrial metabolism to glucose availability in the liver. J. Mol. Cell Biol. 2016;8:129–143. doi: 10.1093/jmcb/mjw004. [DOI] [PubMed] [Google Scholar]
- 36.Arruda A.P., Pers B.M., Parlakgül G., Güney E., Inouye K., Hotamisligil G.S. Chronic enrichment of hepatic endoplasmic reticulum-mitochondria contact leads to mitochondrial dysfunction in obesity. Nat. Med. 2014;20:1427–1435. doi: 10.1038/nm.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cui Z., Vance D.E. Expression of phosphatidylethanolamine N-methyltransferase-2 is markedly enhanced in long term choline-deficient rats. J. Biol. Chem. 1996;271:2839–2843. doi: 10.1074/jbc.271.5.2839. [DOI] [PubMed] [Google Scholar]
- 38.Walkey C.J., Yu L., Agellon L.B., Vance D.E. Biochemical and evolutionary significance of phospholipid methylation. J. Biol. Chem. 1998;273:27043–27046. doi: 10.1074/jbc.273.42.27043. [DOI] [PubMed] [Google Scholar]
- 39.Vázquez-Chantada M., Fernández-Ramos D., Embade N., Martínez-Lopez N., Varela-Rey M., Woodhoo A., Luka Z., Wagner C., Anglim P.P., Finnell R.H., et al. HuR/methyl-HuR and AUF1 regulate the MAT expressed during liver proliferation, differentiation, and carcinogenesis. Gastroenterology. 2010;138:1943–1953. doi: 10.1053/j.gastro.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rowland A.A., Voeltz G.K. Endoplasmic reticulum-mitochondria contacts: function of the junction. Nat. Rev. Mol. Cell Biol. 2012;13:607–625. doi: 10.1038/nrm3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sood A., Jeyaraju D.V., Prudent J., Caron A., Lemieux P., McBride H.M., Laplante M., Tóth K., Pellegrini L. A Mitofusin-2-dependent inactivating cleavage of Opa1 links changes in mitochondria cristae and ER contacts in the postprandial liver. Proc. Natl. Acad. Sci. USA. 2014;111:16017–16022. doi: 10.1073/pnas.1408061111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamasaki M., Furuta N., Matsuda A., Nezu A., Yamamoto A., Fujita N., Oomori H., Noda T., Haraguchi T., Hiraoka Y., et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 43.Ilacqua N., Anastasia I., Raimondi A., Lemieux P., de Aguiar Vallim T.Q., Toth K., Koonin E.V., Pellegrini L. A three-organelle complex made by wrappER contacts with peroxisomes and mitochondria responds to liver lipid flux changes. J. Cell Sci. 2022;135 doi: 10.1242/jcs.259091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tasseva G., Bai H.D., Davidescu M., Haromy A., Michelakis E., Vance J.E. Phosphatidylethanolamine deficiency in Mammalian mitochondria impairs oxidative phosphorylation and alters mitochondrial morphology. J. Biol. Chem. 2013;288:4158–4173. doi: 10.1074/jbc.M112.434183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Delgado M., Garrido F., Pérez-Miguelsanz J., Pacheco M., Partearroyo T., Pérez-Sala D., Pajares M.A. Acute liver injury induces nucleocytoplasmic redistribution of hepatic methionine metabolism enzymes. Antioxid. Redox Signal. 2014;20:2541–2554. doi: 10.1089/ars.2013.5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reytor E., Pérez-Miguelsanz J., Alvarez L., Pérez-Sala D., Pajares M.A. Conformational signals in the C-terminal domain of methionine adenosyltransferase I/III determine its nucleocytoplasmic distribution. FASEB J. 2009;23:3347–3360. doi: 10.1096/fj.09-130187. [DOI] [PubMed] [Google Scholar]
- 47.Murray B., Peng H., Barbier-Torres L., Robinson A.E., Li T.W.H., Fan W., Tomasi M.L., Gottlieb R.A., Van Eyk J., Lu Z., et al. Methionine adenosyltransferase alpha1 is targeted to the mitochondrial matrix and interacts with cytochrome P450 2E1 to lower its expression. Hepatology. 2019;70:2018–2034. doi: 10.1002/hep.30762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao X., van der Veen J.N., Vance J.E., Thiesen A., Vance D.E., Jacobs R.L. Lack of phosphatidylethanolamine N-methyltransferase alters hepatic phospholipid composition and induces endoplasmic reticulum stress. Biochim. Biophys. Acta. 2015;1852:2689–2699. doi: 10.1016/j.bbadis.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 49.Jacobs R.L., Zhao Y., Koonen D.P., Sletten T., Su B., Lingrell S., Cao G., Peake D.A., Kuo M.S., Proctor S.D., et al. Impaired de novo choline synthesis explains why phosphatidylethanolamine N-methyltransferase-deficient mice are protected from diet-induced obesity. J. Biol. Chem. 2010;285:22403–22413. doi: 10.1074/jbc.M110.108514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alonso C., Fernández-Ramos D., Varela-Rey M., Martínez-Arranz I., Navasa N., Van Liempd S.M., Lavín Trueba J.L., Mayo R., Ilisso C.P., de Juan V.G., et al. Metabolomic identification of subtypes of nonalcoholic steatohepatitis. Gastroenterology. 2017;152:1449–1461.e7. doi: 10.1053/j.gastro.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sáenz de Urturi D., Buqué X., Porteiro B., Folgueira C., Mora A., Delgado T.C., Prieto-Fernández E., Olaizola P., Gómez-Santos B., Apodaka-Biguri M., et al. Methionine adenosyltransferase 1a antisense oligonucleotides activate the liver-brown adipose tissue axis preventing obesity and associated hepatosteatosis. Nat. Commun. 2022;13:1096. doi: 10.1038/s41467-022-28749-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Contreras C., González-García I., Martínez-Sánchez N., Seoane-Collazo P., Jacas J., Morgan D.A., Serra D., Gallego R., Gonzalez F., Casals N., et al. Central ceramide-induced hypothalamic lipotoxicity and ER stress regulate energy balance. Cell Rep. 2014;9:366–377. doi: 10.1016/j.celrep.2014.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Covelo-Molares H., Souto Y., Fidalgo M., Guallar D. A simple, rapid, and cost-effective method for loss-of-function assays in pluripotent cells. Methods Mol. Biol. 2022;2520:199–213. doi: 10.1007/7651_2021_434. [DOI] [PubMed] [Google Scholar]
- 54.Leffert H.L., Koch K.S., Moran T., Williams M. Liver cells. Methods Enzymol. 1979;58:536–544. doi: 10.1016/S0076-6879(79)58168-3. [DOI] [PubMed] [Google Scholar]
- 55.Ozcan L., Wong C.C., Li G., Xu T., Pajvani U., Park S.K., Wronska A., Chen B.X., Marks A.R., Fukamizu A., et al. Calcium signaling through CaMKII regulates hepatic glucose production in fasting and obesity. Cell Metab. 2012;15:739–751. doi: 10.1016/j.cmet.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Czyzyk T.A., Romero-Picó A., Pintar J., McKinzie J.H., Tschöp M.H., Statnick M.A., Nogueiras R. Mice lacking delta-opioid receptors resist the development of diet-induced obesity. FASEB J. 2012;26:3483–3492. doi: 10.1096/fj.12-208041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quiñones M., Al-Massadi O., Folgueira C., Bremser S., Gallego R., Torres-Leal L., Haddad-Tóvolli R., García-Caceres C., Hernandez-Bautista R., Lam B.Y.H., et al. p53 in AgRP neurons is required for protection against diet-induced obesity via JNK1. Nat. Commun. 2018;9:3432. doi: 10.1038/s41467-018-05711-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suárez J., Rivera P., Arrabal S., Crespillo A., Serrano A., Baixeras E., Pavón F.J., Cifuentes M., Nogueiras R., Ballesteros J., et al. Oleoylethanolamide enhances beta-adrenergic-mediated thermogenesis and white-to-brown adipocyte phenotype in epididymal white adipose tissue in rat. Dis. Model. Mech. 2014;7:129–141. doi: 10.1242/dmm.013110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheng X., Geng F., Pan M., Wu X., Zhong Y., Wang C., Tian Z., Cheng C., Zhang R., Puduvalli V., et al. Targeting DGAT1 ameliorates glioblastoma by increasing fat catabolism and oxidative stress. Cell Metab. 2020;32:229–242.e8. doi: 10.1016/j.cmet.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kwong S.C., Jamil A.H.A., Rhodes A., Taib N.A., Chung I. Metabolic role of fatty acid binding protein 7 in mediating triple-negative breast cancer cell death via PPAR-alpha signaling. J. Lipid Res. 2019;60:1807–1817. doi: 10.1194/jlr.M092379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wieckowski M.R., Giorgi C., Lebiedzinska M., Duszynski J., Pinton P. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat. Protoc. 2009;4:1582–1590. doi: 10.1038/nprot.2009.151. [DOI] [PubMed] [Google Scholar]
- 62.Folch J., Lees M., Sloane Stanley G.H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 63.Ruiz J.I., Ochoa B. Quantification in the subnanomolar range of phospholipids and neutral lipids by monodimensional thin-layer chromatography and image analysis. J. Lipid Res. 1997;38:1482–1489. [PubMed] [Google Scholar]
- 64.van Liempd S., Cabrera D., Mato J.M., Falcon-Perez J.M. A fast method for the quantitation of key metabolites of the methionine pathway in liver tissue by high-resolution mass spectrometry and hydrophilic interaction ultra-performance liquid chromatography. Anal. Bioanal. Chem. 2013;405:5301–5310. doi: 10.1007/s00216-013-6883-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
No datasets (e.g. RNA-Seq or proteomics) were generated in this study.
-
•
Data S1: Unprocessed source data underlying all blots and graphs, related to Figures 1, 2, 3, 4, 5, 6, and S1–S4. This file includes: 1) Uncropped scans of all the blots; 2) Excel file containing the values that were used to create all graphs in the paper.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.