Abstract
In the past two decades brain connectomics has evolved into a major concept in neuroscience. However, the current perspective on brain connectivity and how it underpins brain function relies mainly on the hemodynamic signal of functional magnetic resonance imaging (MRI). Molecular imaging provides unique information inaccessible to MRI-based and electrophysiological techniques. Thus, positron emission tomography (PET) has been successfully applied to measure neural activity, neurotransmission, and proteinopathies in normal and pathological cognition. Here, we position molecular imaging within the brain connectivity framework from the perspective of timeliness, validity, reproducibility, and resolution. We encourage the neuroscientific community to take an integrative approach whereby MRI-based, electrophysiological techniques, and molecular imaging contribute to our understanding of the brain connectome.
Imaging of brain connectivity
The powerful idea of the brain as a network has gradually gained traction over the past 20 years and has evolved into a major concept in neuroscience. According to this perspective, brain functions in general, and cognitive functions in particular, depend on interactions between distributed brain regions operating in large-scale networks [1]. Accordingly, dysfunction of these networks, or brain disconnectivity, has been observed in numerous cognitive disorders [2]. Brain networks (see Glossary) can be captured at the macroscale using electrophysiological and neuroimaging techniques. Electroencephalography (EEG) was applied first [3], followed by PET [4–6]. In fact, the first formal framework of functional connectivity was proposed based on perfusion PET studies [6]. Only later was the repertoire of techniques expanded to functional MRI (fMRI) [7,8], magnetoencephalography (MEG) [9,10], and, more recently, to functional near-infrared spectroscopy (fNIRS) [11].
The approach of molecular imaging is based on the detection of radioactivity emitted following a small amount of a radiotracer injected into a peripheral vein. Established radiotracers bind to molecular targets at a nano- to picomolar level with high affinity and selectivity [12]. In a broad sense, the term molecular connectivity refers to a statistical dependence between regional measures of molecular imaging [13]. Thus, this term echoes the definition of functional connectivity used by Karl Friston – ‘as statistical dependencies among remote neurophysiological events’ [14]. Although molecular imaging encompasses several techniques such as PET and single-photon emission tomography [15] in vivo, as well as autoradiography ex vivo [16], we explicitly focus here on PET as the most popular in vivo technique used in research settings. In the following, molecular imaging and PET are therefore used as synonyms.
Currently, blood oxygenation level-dependent (BOLD) fMRI is by far the most popular method to study functional connectivity. Compared to PET, fMRI is widely available, cheap, has a higher temporal resolution, and is devoid of ionizing radiation exposure. Even though PET studies on brain connectivity are increasing in absolute terms, their proportion is still very limited: according to a comprehensive PubMed search, fMRI studies make up more than 2/3 of the literature on functional brain connectivity, following a trend of exponential increase over the past 30 years. Thus, the current perspective on how brain connectivity and networks underpin cognitive functions disproportionately relies on the hemodynamic signal of fMRI. Because brain activity emerges from a complex interplay of biochemical and electrical signaling, one method cannot completely characterize the diversity of the inter-regional communication. Thus, there is an increasing need for a multimodal, integrative perspective of the brain connectome. Molecular imaging provides unique information inaccessible to the common MRI-based techniques and electrophysiological tools. This article aims to position molecular imaging within the framework of brain connectivity.
Value of molecular connectivity
In the following we argue that PET represents a valuable imaging tool for brain connectivity research from the perspective of validity, reproducibility, and resolution. Whereas validity and reproducibility are key criteria for any scientific method in the context of good scientific practice [17], resolution is a common crucial characteristic of neuroscientific techniques, from optical microscopy to EEG [18]. By definition, validity, also referred to as accuracy, indicates how close a measurement of the method is to the true biological process of interest, amid other processes and noise. Reproducibility, also referred to as reliability or precision, indicates how close repeated measurements of the method are to each other. In the absence of a ground-truth reference, knowledge of the reproducibility of connectivity measures cannot be overvalued. Replicability, a special case of reproducibility, refers to the ability to obtain the same results in another dataset. Spatial and temporal resolution are measures of the minimum object that can be resolved by the method. These characteristics are important for understanding the scale of inference at which the different forms of brain connectivity operate.
In the following section we focus on PET with 18F-FDG as the most popular and established radiotracer. Where appropriate, we compare 18F-FDG PET-based molecular connectivity to fMRI-based functional connectivity, which is the most popular neuroimaging tool in the field of connectomics. If not noted otherwise, estimates of molecular connectivity are based on intersubject modeling of static PET images (Box 1 for details).
Box 1. Inter- and intrasubject estimation of molecular connectivity.
In the case of intersubject estimation, connectivity is estimated from covariation in measures across subjects, where only one image per subject is available. Although time is commonly used to infer brain connectivity from fMRI and electrophysiological data, it is not mandatory. First, the presence of a temporal correlation does not guarantee that two regions are connected because the temporal dependence might be direct, indirect, or spurious [89]. Second, a full time-series is not necessary to estimate functional connectivity: only a small fraction of frames with the strongest cofluctuation can explain a significant proportion of variance in functional connectivity [90]. Third, as is true for fMRI functional connectivity [91], structural connectivity via white matter fiber tracts is a relevant substrate of intersubject covariance of regional FDG uptake [92]. Finally, the patterns of connectivity in intersubject resting-state 18F-FDG PET and conventional BOLD fMRI data show similar features, such as known RSNs [29] and a stronger homotopic versus heterotopic interhemispheric connectivity [4,92]. Several statistical approaches have been successfully applied to estimate molecular connectivity from intersubject PET data [37,93], similarly to sMRI [94] and fMRI [46,95,96] data. The assumption behind this is that regions with covarying signal intensity across subjects belong to the same network [97]. Of note, fMRI time series and subject series produce similar spatial patterns of functional connectivity [46,95,96,98]. This approach also shares similarities with meta-analytical connectivity mapping [99].
In the case of intrasubject estimation, connectivity is estimated from dynamic PET protocols with a temporal resolution approaching that of fMRI – so-called functional PET [100,101]. Connectivity here is estimated from covariation of measures over time. Functional PET studies apply a constant radiotracer infusion within the same imaging session [100,101]. Hence, resting-state 18F-FDG PET with 1 minute frame durations yielded group-level networks that were similar to those estimated in simultaneously acquired BOLD data, including the default mode network and visual networks [102]. However, functional 18F-FDG PET yielded several unique networks that were not evident in the BOLD data, and vice versa. More recently, molecular connectivity was estimated with a sampling rate of 16 s [27]. A major challenge of functional PET is the inherently low signal-to-noise ratio in comparison to static PET, which hampers meaningful connectivity estimation at the single-subject level. This issue can be partly addressed by longer scan time [101], spatiotemporal filtering [27], and bolus infusion protocols [42]. In the latter case, by providing a portion of the total dose as a bolus, immediately followed by constant infusion, plasma radioactivity shows an early peak that stabilizes during infusion [103].
Validity
Historically, the concept of functional connectivity is rooted in the idea that communication between brain regions can be traced by electrical signaling [6]. However, neural communication also possesses a biochemical component: chemical synapses convey information, where neurons continually convert electrical to chemical signals and then convert chemical to electrical signals [19]. The biochemical transmitters detected by the postsynaptic machinery induce transduction of an electrical signal in the postsynaptic cell; the latter then induces packets (quanta) of biochemical transmitters to be released again in the presynaptic terminal [19]. Because the recording of electrical signaling or its proxies has long been considered to be a reasonable strategy to investigate functional links between brain regions, chemical signaling or its proxies should represent an alternative, equally reasonable means to access patterns of neuronal communication. In fact, because chemical synapses represent by far the predominant mode of signal transduction in the human brain [20], neural communication may be more accurately reflected by biochemical than by electrical signaling. PET with 18F-FDG, a glucose analog, indexes the activity of a glycolytic enzyme called hexokinase [21]. The coupling between glucose metabolism and neural activity via hexokinase is known as neurometabolic coupling [22] (see section on Neural activity for details). By contrast, fMRI measures neural activity via the amount of oxygen in the blood vessels supplied to a given brain region [23]. The neurovascular coupling behind the hemodynamic fMRI response is based on a complex interplay between local cerebral blood flow, volume, and cerebral metabolic rate of oxygen [24]. Thus, 18F-FDG PET and BOLD fMRI record partially different processes operating in parallel. Even though the BOLD signal is dependent upon cerebral blood flow (CBF) and metabolic responses [25], for reasons that are not well understood, neural activity elicits stronger responses in glucose metabolism than in oxygen consumption, with a 10-to-1 proportion of relative increase [26]. The difference in the nature of the two signals is also reflected in brain connectivity, where resting state networks (RSNs) estimated from intersubject 18F-FDG PET and conventional BOLD fMRI data show only moderate spatial similarity [27–29]. A recent fMRI connectivity simulation study found that aberrant connectivity characterizing brain neurovascular disorders not only reflects aberrant neural activity but also brain vessels and hemodynamic/metabolic pathophysiology [30]. Thus, 18F-FDG PET approaches neural activity in a more direct way.
Reproducibility
Whereas structural connectivity estimated with diffusion weighted MRI (dMRI) aims to estimate actual anatomical connectivity, the neural substrates of other putative indices of brain connectivity, which are based on a statistical dependence, are unknown. In this context, we propose the term ‘proxy’ estimates of brain connectivity. Because a gold standard, or ground-truth reference, is missing, the reproducibility of such proxy estimates is of crucial value. The rationale behind this is that a reproducible measure is more likely to reflect a true signal than a spurious one. In fact, test–retest reproducibility is the ‘starting’ condition for the generalizability of any method [31]. PET measures of regional glucose consumption were shown to possess excellent test–retest reproducibility, with an intraclass correlation coefficient (ICC) of ~0.90 [32]. ICC for regional intensity of the BOLD signal, expressed as the average amplitude of low frequency fluctuations (ALFF), was found to be 0.53 [33]. Good reproducibility has also been found for molecular connectivity estimates of neurotransmission [34]. Test–retest reproducibility of fMRI connectivity was shown to be poor [35]. Data on a direct comparison between 18F-FDG PET and BOLD fMRI networks are yet missing.
Replicability of results is related to the amount of variance in the neuroimaging data associated with a cognitive variable of interest. Higher variance concentration enables the construction of such patterns with fewer independent neuroimaging variables, such as principal or independent components. Smaller sets of independent variables generally translate into less statistical noise and better out-of-sample replication [36]. In the extreme case of pure noise (i.e., in the absence of any spatially correlated activity in the data), the number of necessary components will approach the data rank, and, similarly to ordinary regression, there will be over-fitting with no robustness of pattern loadings or imaging–cognition associations. In a real scenario where 18F-FDG PET and fMRI data were simultaneously acquired in the same set of participants, we found a noticeably higher variance concentration in the 18F-FDG PET data [37]. Thus, relative to fMRI, static 18F-FDG PET data apparently suffer from less statistical noise.
Spatial and temporal resolution
Parcellation of the brain in general, and mapping of brain connectivity in particular, depend on the spatial and temporal resolution of the acquired data (e.g., fMRI images or EEG recordings). How does molecular imaging compare to other neuroimaging and electrophysiological tools? Spatial resolution is dependent both on the intrinsic characteristics of a technique and on data post-processing. Assuming a typical resolution of clinical PET scanners of 4.3 mm [38] (Figure 1A) and an average neuronal density of 30 000 neurons per mm3 [39], the minimal spatial ‘unit’ resolved by PET corresponds to 2 385 210 neurons (Figure 1B). These are only threefold more neurons than those within the minimal spatial unit of conventional BOLD fMRI. However, the spatial resolution of each method varies across the brain: by the application of attenuation correction to compensate for signal deterioration around deep brain structures, PET can accurately resolve signals throughout the whole brain. This is not true for most of the other neuroimaging and neurophysiological techniques (Figure 1C,D). As it stands, modern electrophysiological and functional neuroimaging techniques in humans including PET are 5–8 orders of magnitude away from the single neuron. In addition, the spatial resolution of BOLD fMRI and 18F-FDG PET is inherently capped by their spatial specificity [40]. For the former, spatial specificity is limited by, among other things, downstream draining vein effects [40]. A higher spatial specificity of 18F-FDG PET can be achieved using the well-established framework of compartmental modeling that allows researchers to disentangle the metabolic and vascular components of the signal [41].
Figure 1. Spatial and temporal resolution of common neurophysiological techniques.
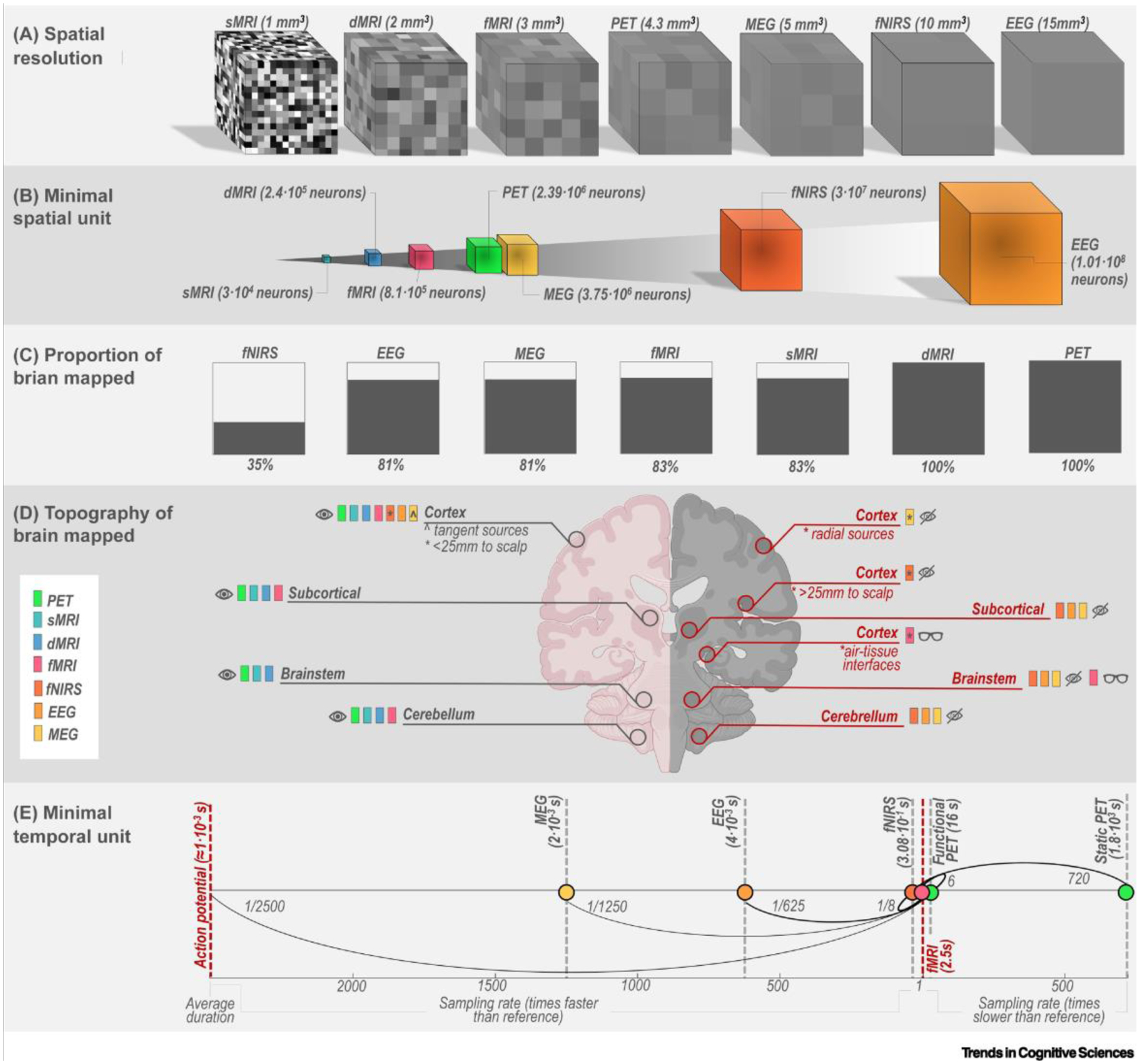
(A) Simulation of how a sample random object is resolved by each method. Spatial resolution is defined for sMRI, dMRI, and fMRI by voxel size, for PET and fNIRS by full width at half maximum (FWHM) [38,109], for EEG and MEG from [110]. For EEG and MEG the reported estimates are likely to be optimistic for functional connectivity studies [111]. Note that the values serve as a rough approximation. (B) The minimal spatial unit – expressed as the number of neurons – that each method can resolve, assuming 30 000 neurons/mm3. (C) Representation of the overall proportion of brain mapped by each method; lack of whole brain coverage by sMRI and fMRI is dependent on the distance factor, here assumed to be 20%.(D) Representation of different portions of the brain where signal is measured accurately (pink side, eye icon), is not measured (gray side, crossed eye icon), or is measured but with a lower resolution and/or susceptibility artifacts (gray side, glasses icon). This part of the figure was created in part with BioRender.com.(E) Average sampling rate of each method, based on an adjusted logarithmic scale centered around an fMRI sampling rate of 2.5 s. The timescale of PET is shown under the case of static PET and functional PET. Note that the values serve as a rough approximation. Action potentials with an assumed duration of 1 ms are reported for reference. Abbreviations: dMRI, diffusion magnetic resonance imaging; EEG, electroencephalography; fNIRS, near-infrared spectroscopy; fMRI, functional magnetic resonance imaging; MEG, magnetoencephalography; PET, positron emission tomography; sMRI, structural magnetic resonance imaging.
Temporal resolution is also dependent on characteristics of the technique itself, acquisition parameters, and post-processing options applied by the user, such as reconstruction parameters, temporal filtering, and smoothing. In the field of 18F-FDG PET, several acquisition and reconstruction protocols have been used so far that provide estimates of molecular connectivity with a sampling rate of minutes to seconds [27,42]. Obviously, even the highest sampling rate in PET studies is several orders of magnitude away from the timescale of neural activity in terms of an action potential (~1 ms), as is the case for fMRI (Figure 1E). Of note, the temporal resolution of these techniques is inherently capped by the underlying transfer functions, namely neurovascular coupling for perfusion PET, hemodynamic response for fMRI and fNIRS, and neurometabolic coupling for 18F-FDG PET [23,43]. These functions have slow dynamics, unfolding over tens of seconds [43,44]. Thus, indefinitely increasing the temporal resolution over inherently relatively slow processes will not necessarily result in additional information on the underlying neural activity [44].
Despite the gap between the spatial and temporal scale of neural activity and sampled measurements of neuroimaging or electrophysiology, functional connectivity seems to be distributed over different spatial scales and temporal frequencies such that no scale is per se uninformative [6,45]. In fact, functional connectivity conveys distinct information at different spatial and temporal scales. In the latter case, longer timescales capture stationary and less-dynamic interactions within the connectome, as determined by for example trophic effects, genetics, and environment [46], whereas shorter timescales are more reflective of the dynamic, instantaneous neural activity [47,48]. Accordingly, at slow timescales (minutes), functional coupling was shown to be a good indicator of an underlying structural link; at intermediate timescales (seconds, closer to the sampling rate of fMRI) correlated fluctuations in time emerged across regions, and these were coordinated in a manner that reveals the existence of anticorrelated clusters; at fast timescales (milliseconds), intermittent synchronization and desynchronizations were observed between regions, producing a large set of metastable states [49]. Similarly, PET measurements obtained at different timescales (16 s vs 60 min) provide distinct patterns of molecular connectivity [27]. Thus, even within the same imaging method, different timescales capture different aspects of inter-regional communication [50]. Whereas it is unclear how events at different timescales interact with each other (or whether they are independent), and whether covariation in mean ‘pooled’ activity at lower timescales influences the emergence of dynamic correlations at faster timescales [6] or vice versa [51], different approaches seem to be equally meaningful and allow different, complementary aspects of brain organization and cognition to be accessed. For example, ‘slow’ temporal scales might be more appropriate for capturing learning-induced neural plasticity [52].
Forms of molecular connectivity
Brain connectivity can be estimated from PET data in different ways. Although a few PET studies have estimated brain connectivity from time series, either in the form of multiple scanning sessions [53] or time-frames [27,47], the majority of studies (>85%, Table S1 in the supplemental information online) have relied on so-called subject series in which a single image per subject is available for analyses. The rationale behind inter- and intrasubject estimation of molecular connectivity is explained in Box 1. Regarding biological targets, molecular imaging is able to target various processes in the living human brain, such as neural activity, neurotransmission, and proteinopathies, resulting in different forms of molecular connectivity. Figure 2 summarizes neurobiological targets that have been and can potentially be approached by using molecular imaging and other neurophysiological techniques. In this section we discuss the neurobiological basis of the different forms of molecular connectivity.
Figure 2. Targets and connectivity estimates of common neurophysiological techniques.
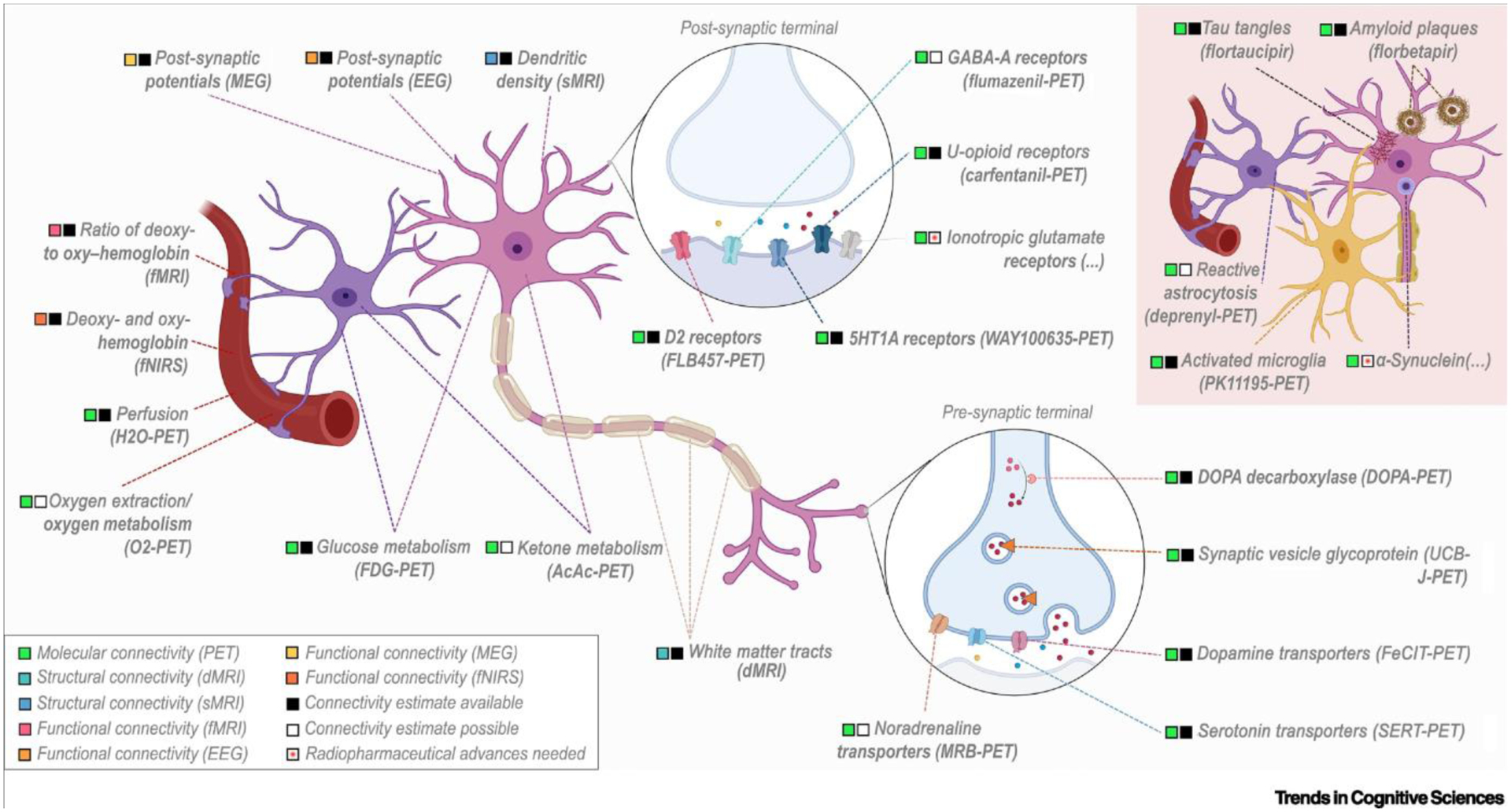
Schematic representation of the main neurobiological targets measured by each neurophysiological technique, and their localization in the brain parenchyma (astrocytes, neurons) or blood vessels. Neurobiological targets found only in pathological conditions are also shown (red panel, top right). The type of connectivity estimate obtained for each neurobiological target is noted, and is color-coded for each technique. Connectivity estimates are considered to be available if at least one study has been published in a peer-reviewed scientific journal, and to be possible if there are no studies but an appropriate positron emission tomography (PET) radiotracer is available; radiopharmaceutical advances are needed if an appropriate PET radiotracer remains under development. This figure was created in part with BioRender.com.
Neural function
Glucose is the obligatory energy substrate for neurons, with synapses representing the major energy consumers [54]. Transferred from blood to the brain via glucose transporters, 18F-FDG is metabolized by hexokinase to FDG-6-phosphate which is then trapped in the cell. Of note, hexokinase represents a ‘gatekeeper’ in glycolysis, regulating the rate at which all subsequent reactions occur [55]. Thus, there is time-dependent accumulation of 18F-FDG in the brain, which is proportional to glucose metabolism [21]. Glucose metabolism is directly coupled to excitatory neural activity mediated via the release and transport of glutamate, the most abundant excitatory neurotransmitter in the brain [54]. Networks of neural activity based on 18F-FDG PET data, so-called metabolic connectivity, have been associated with normal [56–58] and pathological [59] cognition.
The high energy demand of the brain is sustained by continuous CBF which is facilitated by coupling between neuronal activity, CBF, and metabolism. The mechanisms and mediators (e.g., nitric oxide, ion channels, and astrocytes) that regulate CBF–metabolism coupling have been extensively studied ([60] for review). Upon neuronal activation, CBF demand increases to supply glucose and oxygen to the brain parenchyma, thus allowing sustained neuronal activation. CBF is therefore indirectly coupled to neural activity. In the past, CBF PET studies with 15O-H2O as radiotracer have made an essential contribution to the establishment of macro-scale brain connectivity as a discipline of neurosciences [61], together with its substantial role in cognitive neuroscience (Table S1).
Neurotransmission
Neurotransmission is the backbone of signal propagation in the human brain, and underlies all cognitive processes [62]. It can be studied with PET radiotracers that target components of the synaptic function, such as receptors, vesicles, and transporters, or substrates of enzyme activity. Neuronal projections of the major neurotransmission systems (e.g., dopaminergic, serotonergic, and opioidergic) align well with structural and functional connectivity [63]. Thus, mapping the brain-wide distribution of a radiotracer might indirectly provide information on the connectivity within a given neurotransmission system [13,34]. As a practical example, consider the main components of serotonergic transmission – the serotonin transporter and serotonin 1A autoreceptor [64] – that regulate the reuptake and release of serotonin, respectively. Increases in serotonin transporter in the raphe nucleus, the main source of serotonergic projections to the forebrain, result in lower availability of serotonin in the presynaptic terminal, leading to lower activity of the serotonin 1A autoreceptors and, consequently, increased firing rate of the raphe nucleus toward its projections. As a result, more serotonin is available at the raphe projections, which in turn leads to increased expression of serotonin transporter and decreased expression of serotonin 1A autoreceptors at the projection sites. Hence, serotonin transporter expression, as well as serotonin 1A autoreceptor expression, are interdependent (connected) across regions [64]. Thus, spatial patterns of correlations between striatal and extrastriatal dopaminergic D2 receptors were found to be consistent with the known biochemical architecture of the dopaminergic system [65]. Further studies have extended the investigation of molecular connectivity to the serotoninergic [64,66–68] and μ-opioid [66] systems, in relation to cognition [69] and emotions [70]. Recently, replicable networks of synaptic density have been identified in the PET data using the 11C-UCB-J tracer [71].
Proteinopathies
Many neurodegenerative disorders are associated with the aggregation of abnormal misfolded proteins such as TAR DNA-binding protein 43 in frontotemporal dementia, α-synuclein in Parkinson’s disease, and amyloid-β (Aβ) and tau in Alzheimer’s disease. Increasing evidence indicates that the proteinopathies may progress through intercellular transmission trans-synaptically [72]. For example, injection of tau seeds into the mouse brain induced local tau hyperphosphorylation and spread to connected regions through neuronal endocytosis, ampli-fication, transport, and further release of new tau seeds [73]. Thus, multimodal imaging studies have reported the spreading of tau along anatomical connections in humans [74]. If the spread of pathological proteins occurs through connected brain areas, mapping the misfolded proteins with molecular connectivity might provide insights into disease progression in terms of local vulnerability and paths of protein spreading. In the former case, molecular connectivity allows the identi-fication of regions with an elevated number of pathological ‘connections’; these represent pathological hubs in which pathology has a stronger local effect that could then easily propagate to the rest of the network ([75] for a recent example in tau pathology). Regarding protein spreading, molecular connectivity allows the identification of paths of protein propagation, where regions with a covarying amount of pathology can be considered to be stations on the same pathological path. For example, recent PET studies reported that Aβ and tau spread through distinct brain networks [76–78]. The Aβ network was characterized by a spatial pattern that largely overlapped with the default mode network [76,77], whereas the tau networks resembled a wider range of functional networks, in line with the known relationship between this proteinopathy and cognitive deficits [77,78]. Furthermore, Aβ and tau deposits were found to accumulate in cognitively normal individuals across distinct pathways, and the degree of propagation along the tau pathways correlated with general cognitive performance and memory [79]. Instead of relying on connectomes derived exclusively from dMRI [80] and fMRI [81] data, future studies should further explore the possibility of defining networks of tau propagation using molecular connectivity approaches [79].
Connectome mapping
How might connectivity estimates from neurophysiological data in general, and from PET data in particular, contribute to our understanding of brain organization? We introduce here the concept of connectome maps. This concept can be viewed as a special case of interoperable atlases [82] that are specific for brain connectivity. Consider the brain connectome to be a meta-state. Although there is a structural core composed of major neural fibers, units of the connectome continuously interact in space and time at a functional level. The structural skeleton can be captured with dMRI, whereas a functional connectome can be captured with MRI techniques, electrophysiological tools, and molecular imaging. A portion of brain connectivity, both structural and functional, is shared between the majority of healthy individuals. Herewith, functional connectivity is presumed to vary more strongly between individuals than structural connectivity. The so-called invariant part of the connectome is characteristic of a given species and can be referred to as a map. The invariant part, as outlined using for example fMRI, may be referred to as the fMRI map of the (human) connectome. Each technique is presumed to outline a particular aspect of the brain connectome (Figure 3A). To this end, we consider group-level data, both inter- and intrasubject, to be suitable for characterizing the invariant, population-based portion of the connectome (Figure 3A). In our opinion, the integration of the different maps (Figure 3B; Box 2) is the way to advance our understanding of brain organization.
Figure 3. Brain connectomes as estimated with common neurophysiological techniques and scenarios of their integration.
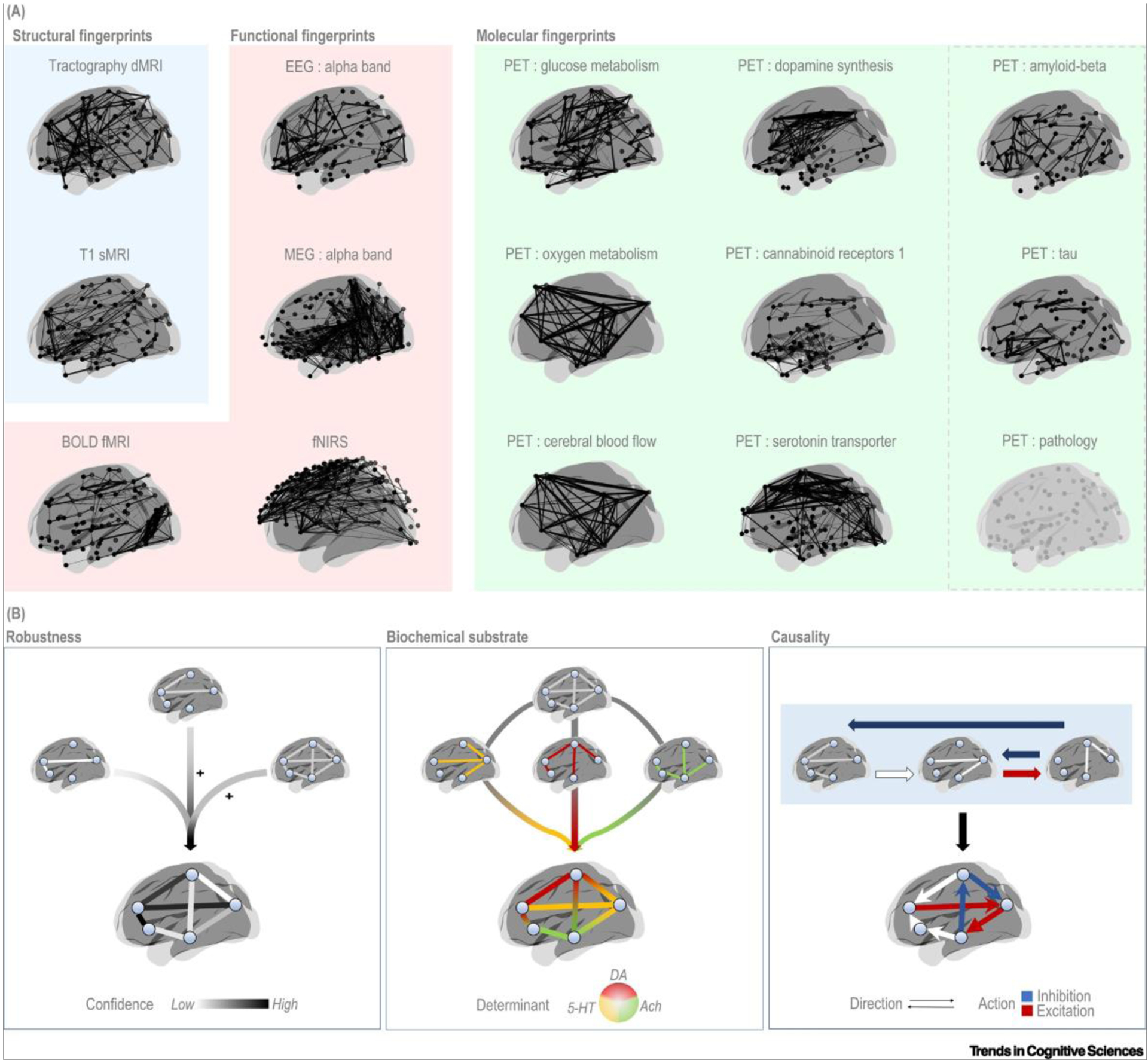
(A) All connectomes refer to group-level maps, with the exception of the EEG and MEG connectomes that are estimated at the single-subject level. All connectomes are based on data from healthy subjects. In each connectome, nodes correspond to brain regions, with the exception of the fNIRS connectome in which nodes correspond to channels on the scalp. The PET pathology render represents an outline of a prototype connectome for any pathological target. (B) Scenarios of integration across connectomes. Integration of molecular connectivity can increase the robustness of connectivity estimates (Left), specify the biochemical determinants underlying a given connection (middle), or clarify the directionality and, eventually, the type of action exerted along each connection (red, blue arrows) (Right). All renders were created using BrainNet [112]. Abbreviations: Ach, acetylcholine; DA, dopamine; 5-HT, serotonin; dMRI, diffusion magnetic resonance imaging; EEG, electroencephalography; fMRI, functional magnetic resonance imaging; fNIRS, near-infrared spectroscopy; MEG, magnetoencephalography; PET, positron emission tomography; sMRI, structural magnetic resonance imaging.
Box 2. Integration of molecular connectivity.
The brain connectome is presumed to provide the scaffold for cognition [104]. So far, MRI and electrophysiological techniques have been widely used to capture the structural and functional connectivity of the brain. Integration of molecular connectivity into a broad framework of brain connectivity would improve the characterization of the connectome in general and as the substrate of cognition in particular. The integration can be achieved in a few generic ways (Figure 3B), depending on the research goal.
Robustness: adding information about the certainty of the estimates of structural and functional connectivity via PET measurements of neural activity; that is, concordant estimates are more likely to be true and not spurious at a given temporal scale.
Biochemical substrate: adding information about the neurotransmitter systems behind the estimates of structural and functional connectivity via PET measurements of specific classes of receptors and transporters.
Causality: adding information about the direction of structural and functional connectivity via PET measurements of excitatory and inhibitory inputs, pre- and postsynaptic signaling, or regional gradients, for example in proteinopathies.
Methodologically, integration of connectivity information from the different techniques can be achieved using different approaches such as those below.
Joint or parallel independent component analyses [105] to quantify correlations (linkage) across brain networks estimated from different techniques.
Multilayer approaches [106] to quantify correlations across brain regions and across different techniques (layers).
Multifactorial brain models [107,108] to model the contribution of different techniques (factors) in generating particular brain states.
Outlook
Although the foundations of molecular connectivity are already in place, several methodological and conceptual issues remain to be addressed. We subdivide this path into two major chronological stages – the stage of validation and the stage of application.
First, validation: (i) consensus agreement on a nomenclature to designate molecular connectivity results and avoid the use of inconsistent terminology across the literature [27,83]. (ii) Definition of best methodological practices that include systematic investigation of the reproducibility of molecular connectivity estimates under varying experimental conditions (sample size, sample heterogeneity, scanner type, and acquisition, reconstruction, and processing pipelines), in comparison to more established MRI-based connectivity estimates; as well as simulation experiments to study how changes in regional intensity impact on patterns of molecular connectivity.
Second, application: (i) developing an atlas of molecular imaging maps of the human connectome based on data with established PET radiotracers; (ii) integration of molecular connectivity into the framework of causal models [84] to enable causal inferences about brain function; and (iii) derivation and validation of molecular connectivity indices at the individual level for clinical applications that are already initiated [85–88] but need further development and replication.
Last but not least, open science practices, including data and code sharing, will hopefully prompt neuroscientists beyond the molecular neuroimaging community to approach the field of molecular connectivity. To facilitate this, we summarized codes and toolboxes for connectivity analyses of PET data (Table S2) as well as readily accessible PET datasets (Table S3). These datasets include PET data of more than 31 000 human subjects. The lists (Tables S1–S3) will soon be available at www.molecularconnectivity.com. We encourage researchers to update these databases.
Concluding remarks
Given that chemical synapses represent the predominant mode of signal transduction in the human brain, targeting the molecular level of neural communication represents a necessary step for advancing brain connectomics. At the macroscale, this can be achieved using molecular imaging. We argue here that molecular imaging represents a useful approach to characterize the brain connectome by providing unique information inaccessible to MRI-based and electrophysiological techniques. Preliminary evidence indicates that molecular imaging may deliver valid and reproducible estimates of brain connectivity with reasonable spatial and temporal resolution. Herewith, both static and functional PET protocols are of value. Weencourage the neuroscientific community to take an integrative perspective on the brain connectome where various methods including MRI-based techniques, electrophysiological tools, and molecular imaging contribute to our understanding of brain organization. To this end, multidisciplinary efforts and rigorous pipelines of data analyses are essential. The road ahead is long (see Outstanding questions), but the path is becoming clearer.
Outstanding questions.
What is the test–retest reproducibility of molecular connectivity estimates compared to more established MR-based connectivity estimates?
What is the impact of different methodological choices (acquisition, quantification, preprocessing, analysis) on estimates of molecular connectivity?
How similar are the connectomes produced using different (e.g., absolute and relative) measures of the same PET method?
How can statistical thresholds be set to isolate molecular connectivity from noise?
How can researchers integrate estimates of brain connectivity obtained at different spatial and temporal scales?
What proportion of molecular connectivity is shared between individuals, and what proportion is unique?
How reproducible and replicable are molecular connectivity estimates at the individual level?
Supplementary Material
Highlights.
The number of molecular imaging studies in the field of brain connectivity is steadily increasing.
Molecular imaging is not yet widely used by the MRI-predominant neuroimaging community as tool of choice for studying brain connectomics.
Because chemical synapses are essential to signal transduction, targeting the molecular level of brain communication is indispensable for our understanding of the brain connectome.
PET as major molecular imaging tool provides various established markers of neural activity, neurotransmitter systems, and proteinopathies.
Integration of connectomes produced with different neurophysiological methods, including molecular imaging, might be key for advancing the field of neuroscience.
Acknowledgments
A.S. is funded by the Belgian National Fund for Scientific Research [grant 40001328, Chargée de Recherches Fonds de la Recherche Scientifique (FRS)/Fonds National de la Recherche Scientifique (FNRS)/Université de Liège/Coma Science Group GIGA]. V.D.C. is supported by the National Institutes of Health (NIH; R01MH118695) and the National Science Foundation (21124550). S.B.E. is funded by the EU Horizon 2020 Research and Innovation Program [grant agreements 945539 (HBP SGA3) and 826421 (VBC)] and the Deutsche Forschungsgemeinschaft (SFB 1451, IRTG 2150). M.V. is funded by the Ministero dell’istruzione, dell’università e della ricerca (MIUR) under the initiative ‘Departments of Excellence’ (Law 232/2016), by a Wellcome Trust Digital Award (215747/Z/19/Z), and by the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley National Health Service Foundation Trust and King’s College London. J.B.P. is funded by the Swedish Research Council. S.D.J. is supported by a National Health and Medical Research Council of Australia Fellowship (APP1174164). We thank Giulia Carli and Hubertus Hautzel for providing data for Table S1. We also thank Rick Adams, Marilyn Albert, Ricardo Allegri, Silvia Alonso-Lana, Andrea Arighi, Murat Bilgel, Alexa Pichet Binette, Gérard N. Bischof, Mercè Boada, Ilaria Bonoldi, Chris Brown, Min S. Byun, Stephen F. Carter, Massimo Castellani, Jaime J. Castrellon, Jiří Cerman, Kewei Chen, Gaël Chetelat, Andrea Chincarini, Patricio Chrem, Lyduine Collij, Anka Cuderman, Tarik Dahoun, Michael D. Devous, Ramon Diaz-Arrastia, Paul Donaghy, Rachelle Doody, Massimo Dottorini, Carole Dufouil, Germán Falasco, Carles Falcon, Sara B. Festini, Christopher Fowle, Peter Fox, Crystal Franklin, Jurgen Fripp, Giovanni Frisoni, Giorgio Fumagalli, Valentin Fuster on behalf of the Progression of Early Subclinical Atherosclerosis (PESA) Study Team, Daniela Galimberti, Melanie Ganz, Valentina Garibotto, Juan D. Gispert, Karl Herholz, Rainer Hinz, Russ Hornbeck, Oliver Howes, Lauren Hudswell, Jordi Huguet, Leonardo Iaccarino, Janne Isojärvi, Takeshi Iwatsubo, Sameer Jahuar, Hyemin Jang, Peter S. Jensen, Sterling Johnson, Kristen Kennedy, David S. Knopman, Pamela LaMontagne, Susan Landau, Jessica Langbaum, Rachael Lawson, Dong Y. Lee, Francisco Lopera, Hanzhang Lu, Emma Luckett, Maura Malpetti, Marta Marquié, Robert McCutcheon, Ian M. McDonough, Richard McIntyre, Patrizia Mecocci, Lefkos Middleton, Yasuhiko Mikari, Valeria Mondelli, Silvia Morbelli, Alice Murphy, Maria A. Nettis, Flavio Nobili, Martin Nørgaard, Matthew Nour, Lauri Nummenmaa, John O’Brien, Grégory Operto, Rik Ossenkoppele, Carmine Pariante, Denise Park, Matej Perovnik, Michela Pievani, Thad Polk, Luca Presotto, Scott A. Przybelski, Giulia Quattrini, Eric Reiman, Susan Resnick, Patricia A. Reuter-Lorenz, Jenny R. Rieck, Silvia Rios-Romenets, Karen Rodrigue, Maria Rogdaki, Melissa Rundle, Luca Sacchi, Gemma Salvadó, Gregory R. Samanez-Larkin, Michela Scamosci, Jolien Schaeverbeke, Christopher G. Schwarz, Michio Senda, Sang W. Seo, Stelvio Sestini, Mahnaz Shekari, Hilmar Sigurdson, Kaycee Sink, Zhuang Song, Oscar Sotolongo-Grau, Yi Su, Pierre Tariot, Szymon Tomczyk, Petra Tomše, Cristina Tranfaglia, Maja Trošt, Federico Turkheimer, Chi Udeh-Momoh, Angela Uecker, Rik Vandenberghe, Sylvia Villeneuve, Pieter J. Visser, Devadas Vivek, Stephanie J.B. Vos, Tyler Ward, Gagan Wig, Chengjie Xiong, Dahyun Yi, David H. Zald, Jingting Zhang, and Shenjun Zhong for providing information on the datasets included in Table S3. The National Alzheimer’s Coordinating Center (NACC) database is funded by National Institute on Aging/NIH (grant U24 AG072122). NACC data are contributed by the NIA-funded Alzheimer’s Disease Research Centers (ADCs): P50 AG005131 (PI James Brewer), P50 AG005133 (PI Oscar Lopez), P50 AG005134 (PI Bradley Hyman), P50 AG005136 (PI Thomas Grabowski), P50 AG005138 (PI Mary Sano), P50 AG005142 (PI Helena Chui), P50 AG005146 (PI Marilyn Albert), P50 AG005681 (PI John Morris), P30 AG008017 (PI Jeffrey Kaye), P30 AG008051 (PI Thomas Wisniewski), P50 AG008702 (PI Scott Small), P30 AG010124 (PI John Trojanowski), P30 AG010129 (PI Charles DeCarli), P30 AG010133 (PI Andrew Saykin), P30 AG010161 (PI David Bennett), P30 AG012300 (PI Roger Rosenberg), P30 AG013846 (PI Neil Kowall), P30 AG013854 (PI Robert Vassar), P50 AG016573 (PI Frank LaFerla), P50 AG016574 (PI Ronald Petersen), P30 AG019610 (PI Eric Reiman), P50 AG023501 (PI Bruce Miller), P50 AG025688 (PI Allan Levey), P30 AG028383 (PI Linda Van Eldik), P50 AG033514 (PI Sanjay Asthana), P30 AG035982 (PI Russell Swerdlow), P50 AG047266 (PI Todd Golde), P50 AG047270 (PI Stephen Strittmatter), P50 AG047366 (PI Victor Henderson), P30 AG049638 (PI Suzanne Craft), P30 AG053760 (PI Henry Paulson), P30 AG066546 (PI Sudha Seshadri), P20 AG068024 (PI Erik Roberson), P20 AG068053 (PI Marwan Sabbagh), P20 AG068077 (PI Gary Rosenberg), P20 AG068082 (PI Angela Jefferson), P30 AG072958 (PI Heather Whitson), P30 AG072959 (PI James Leverenz). We thank Pascal Fries for insightful discussion of Figure 1, Enrico Amico, Jitka Annen, Joe Aoe, Jae Gwan Kim, Hanli Liu, Thien Nguyen, Rajakanat Panda, Tharick Pascoal, Pedro Rosa-Neto, Ekansh Sareen, and Tadashi Watabe for providing data for Figure 3, and Nikita Beliy for coding support for Figures 1 and 3.
Glossary
- Anatomical connectivity
the physical linkage between different elements of the brain, such as synaptic connections between sets of neurons or axon bundles between different gray matter regions.
- Brain network
a set of structurally or functionally interconnected brain regions with a particular function.
- Connectome
the generic sum of all connections in the brain.
- Default mode network
a large-scale brain network that is most active at rest.
- Functional connectivity
statistical dependence between regional measures of neural activity interpreted as inter-regional communication. This umbrella term refers to brain connectivity estimated based on direct measures of neural activity (provided by electroencephalography, EEG; or magnetencephalography, MEG) or proxies of neural activity, such as the ratio of deoxygenated-to-oxygenated hemoglobin (fMRI) or (historically)perfusion PET or glucose metabolism PET.
- Functional PET
dynamic positron emission tomography (PET) combined with constant tracer infusion within the same imaging session, either alone or in combination with an initial bolus injection, where regional fluctuations are quantified along a time–activity curve.
- Interoperable atlases
a collection of brain maps that are organized in such a way that information included in one map can be used in conjunction with the others. Interoperability requires brain maps to be represented in the same reference space.
- Molecular imaging
visualization and quantification of molecules of interest using imaging techniques.
- Neurotransmission
the process by which signaling molecules (neurotransmitters) are released from the presynaptic terminal of a neuron and bind to receptors on the postsynaptic terminal of another neuron, typically with excitatory or inhibitory effects.
- Proteinopathies
conditions characterized by aggregation of abnormal misfolded proteins, such as amyloid-β, tau, and α-synuclein, leading to neurodegeneration.
- Radiotracer
a chemical compound linked to a radioactive isotope whose decay allows its distribution in the living being to be traced. It allows quantification(i.e., tracing) of a specific biological process of interest without altering it.
- Static PET
the acquisition of a short emission scan during equilibrium of the radiotracer; such data are reconstructed as a single PET image that can be analyzed via simplified methods.
- Subject series
concatenation of neurophysiological data of individual subjects to form a vector in which one subject contributes only one value within the vector. Subject series are the basis of intersubject estimation of brain connectivity.
- Synapse
the site where an action potential is transmitted from one neural cell to another. A synapse can be chemical or, rarely, electrical.
Footnotes
Declaration of interests
The authors declare no conflicts of interest.
Supplemental information
Supplemental information associated with this article can be found online at https://doi.org/10.1016/j.tics.2022.11.015.
References
- 1.Wig GS (2017) Segregated systems of human brain networks. Trends Cogn. Sci 21, 981–996 [DOI] [PubMed] [Google Scholar]
- 2.van den Heuvel MP and Sporns O (2019) A cross-disorder connectome landscape of brain dysconnectivity. Nat. Rev. Neurosci 20, 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brazier M and Casby J (1952) Crosscorrelation and autocorrelation studies of electroencephalographic potentials. EEG Clin. Neurophysiol 4, 201–211 [DOI] [PubMed] [Google Scholar]
- 4.Horwitz B et al. (1984) Intercorrelations of glucose metabolic rates between brain regions: application to healthy males in a state of reduced sensory input. J. Cereb. Blood Flow Metab 4, 484–499 [DOI] [PubMed] [Google Scholar]
- 5.Metter EJ et al. (1984) Cerebral metabolic relationships for selected brain regions in Alzheimer’s, Huntington’s, and Parkinson’s diseases. J. Cereb. Blood Flow Metab 4, 500–506 [DOI] [PubMed] [Google Scholar]
- 6.Friston KJ et al. (1993) Functional connectivity: the principal-component analysis of large (PET) data sets. J. Cereb. Blood Flow Metab 13, 5–14 [DOI] [PubMed] [Google Scholar]
- 7.Biswal B et al. (1995) Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn. Reson. Med 34, 537–541 [DOI] [PubMed] [Google Scholar]
- 8.Kleinschmidt A et al. (1994) Correlational imaging of thalamocortical coupling in the primary visual pathway of the human brain. J. Cereb. Blood Flow Metab 14, 952–957 [DOI] [PubMed] [Google Scholar]
- 9.Gross J et al. (2001) Dynamic imaging of coherent sources: studying neural interactions in the human brain. Proc. Natl. Acad. Sci. U. S. A 98, 694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nikouline VV et al. (2001) Interhemispheric phase synchrony and amplitude correlation of spontaneous beta oscillations in human subjects: a magnetoencephalographic study. Neuroreport 12, 2487–2491 [DOI] [PubMed] [Google Scholar]
- 11.Mesquita RC et al. (2010) Resting state functional connectivity of the whole head with near-infrared spectroscopy. Opt. Soc. Am 1, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LongWong W-T, eds (2014) The Chemistry of Molecular Imaging, John Wiley & Sons [Google Scholar]
- 13.Hahn A et al. (2019) Making sense of connectivity. Int.J. Neuropsychopharmacol 22, 194–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friston KJ (2011) Functional and effective connectivity: a review. Brain Connect. 1, 13–36 [DOI] [PubMed] [Google Scholar]
- 15.Sala A et al. (2021) In vivo human molecular neuroimaging of dopaminergic vulnerability along the Alzheimer’s disease phases. Alzheimers Res. Ther 13, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McIntosh AR and Gonzalez-Lima F (1995) Functional network interactions between parallel auditory pathways during Pavlovian conditioned inhibition. Brain Res. 683, 228–241 [DOI] [PubMed] [Google Scholar]
- 17.Zuo XN et al. (2019) Harnessing reliability for neuroscience research. Nat. Hum. Behav 3, 768–771 [DOI] [PubMed] [Google Scholar]
- 18.Munck S et al. (2021) Maximizing content across scales: moving multimodal microscopy and mesoscopy toward molecular imaging. Curr. Opin. Chem. Biol 63, 188–199 [DOI] [PubMed] [Google Scholar]
- 19.Jessell TM and Kandel ER (1993) Synaptic transmission: a bidirectional and self-modifiable form of cell-cell communication. Neuron 10, 1–30 [DOI] [PubMed] [Google Scholar]
- 20.Pereda AE (2014) Electrical synapses and their functional interactions with chemical synapses. Nat. Rev. Neurosci 15, 250–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sokoloff L (1977) Relation between physiological function and energy metabolism in the central nervous system.J. Neurochem 29, 13–26 [DOI] [PubMed] [Google Scholar]
- 22.Magistretti PJ and Allaman I (2015) A cellular perspective on brain energy metabolism and functional imaging. Neuron 86, 883–901 [DOI] [PubMed] [Google Scholar]
- 23.Drew PJ (2019) Vascular and neural basis of the BOLD signal. Curr. Opin. Neurobiol 58, 61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim SG and Ogawa S (2012) Biophysical and physiological origins of blood oxygenation level-dependent fMRI signals.J. Cereb. Blood Flow Metab 32, 1188–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barros LF et al. (2018) Current technical approaches to brain energy metabolism. Glia 66, 1138–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fox PT et al. (1988) Nonoxidative glucose consumption during focal physiologic neural activity. Science 241, 462–464 [DOI] [PubMed] [Google Scholar]
- 27.Jamadar SD et al. (2021) Metabolic and hemodynamic resting-state connectivity of the human brain: a high-temporal resolution simultaneous BOLD-fMRI and FDG-fPET multimodality study. Cereb. Cortex 31, 2855–2867 [DOI] [PubMed] [Google Scholar]
- 28.Di X and Biswal BB (2012) Metabolic brain covariant networks as revealed by FDG-PET with reference to resting-state fMRI networks. Brain Connect. 2, 275–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Savio A et al. (2017) Resting-state networks as simultaneously measured with functional MRI and PET. J. Nucl. Med 58, 1314–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Archila-Meléndez ME et al. (2020) Modeling the impact of neurovascular coupling impairments on BOLD-based functional connectivity at rest. Neuroimage 218, 116871. [DOI] [PubMed] [Google Scholar]
- 31.Nichols TE et al. (2017) Best practices in data analysis and sharing in neuroimaging using MRI. Nat. Neurosci 20, 299–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maquet P et al. (1990) Reproducibility of cerebral glucose utilization measured by PET and the [18F]-2-fluoro-2-deoxy-dglucose method in resting, healthy human subjects. Eur.J. Nucl. Med 16, 267–273 [DOI] [PubMed] [Google Scholar]
- 33.Mao D et al. (2015) Low-frequency fluctuations of the resting brain: high magnitude does not equal high reliability. PLoS One 10, e0128117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Veronese M et al. (2019) Covariance statistics and network analysis of brain PET imaging studies. Sci. Rep 9, 2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noble S et al. (2019) A decade of test-retest reliability of functional connectivity: a systematic review and meta-analysis. Neuroimage 203, 116157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Habeck C and Stern Y (2010) Multivariate data analysis for neuroimaging data: overview and application to Alzheimer’s disease. Cell Biochem. Biophys 58, 53–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yakushev I et al. (2017) Metabolic connectivity: methods and applications. Curr. Opin. Neurol 30, 677–685 [DOI] [PubMed] [Google Scholar]
- 38.Delso G et al. (2011) Performance measurements of the siemens mMR integrated whole-body PET/MR scanner. J. Nucl. Med 52, 1914–1922 [DOI] [PubMed] [Google Scholar]
- 39.Ribeiro PFM et al. (2013) The human cerebral cortex is neither one nor many: neuronal distribution reveals two quantitatively different zones in the gray matter, three in the white matter, and explains local variations in cortical folding. Front. Neuroanat 7, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bollmann S and Barth M (2021) New acquisition techniques and their prospects for the achievable resolution of fMRI. Prog. Neurobiol 207, 101936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bertoldo A et al. (2014) Deriving physiological information from PET images: from SUV to compartmental modelling. Clin. Transl. Imaging 2, 239–251 [Google Scholar]
- 42.Rischka L et al. (2018) Reduced task durations in functional PET imaging with [18F]FDG approaching that of functional MRI. Neuroimage 181, 323–330 [DOI] [PubMed] [Google Scholar]
- 43.Mann K et al. (2021) Causal coupling between neural activity, metabolism, and behavior across the Drosophila brain. Nature 593, 244–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wald LL and Polimeni JR (2015) High-speed, high-resolution acquisitions. In Brain Mapping: An Encyclopedic Reference (Toga AW, ed.), pp. 103–116, Elsevier [Google Scholar]
- 45.Buzsáki Gand Christen Y, eds (2016) Micro-, Meso-and Macro-Dynamics of the Brain, Springer Nature; [PubMed] [Google Scholar]
- 46.Zhang Z et al. (2011) Resting-state brain organization revealed by functional covariance networks. PLoS One 6, e28817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomasi DG et al. (2017) Dynamic brain glucose metabolism identifies anti-correlated cortical-cerebellar networks at rest.J. Cereb. Blood Flow Metab 37, 3659–3670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hellyer PJ et al. (2017) Protein synthesis is associated with high-speed dynamics and broad-band stability of functional hubs in the brain. Neuroimage 155, 209–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Honey CJ et al. (2007) Network structure of cerebral cortex shapes functional connectivity on multiple time scales. Proc. Natl. Acad. Sci. U. S. A 104, 10240–10245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Horwitz B (2003) The elusive concept of brain connectivity. Neuroimage 19, 466–470 [DOI] [PubMed] [Google Scholar]
- 51.Besserve M et al. (2010) Causal relationships between frequency bands of extracellular signals in visual cortex revealed by an information theoretic analysis. J. Comput. Neurosci 29, 547–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang MB and Halassa MM (2022) Thalamocortical contribution to flexible learning in neural systems. Netw. Neurosci 6, 980–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Di X et al. (2019) Interregional causal influences of brain metabolic activity reveal the spread of aging effects during normal aging. Hum. Brain Mapp 40, 4657–4668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stoessl AJ (2017) Glucose utilization: still in the synapse. Nat. Neurosci 20, 382–384 [DOI] [PubMed] [Google Scholar]
- 55.Berg JM et al. , eds (2012) Biochemistry (7th edn), W.H. Freeman & Co [Google Scholar]
- 56.Zou N et al. (2015) Metabolic connectivity as index of verbal working memory. J. Cereb. Blood Flow Metab 35, 1122–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Voigt K et al. (2022) Metabolic and functional connectivity provide unique and complementary insights into cognition-connectome relationships. Cereb. Cortex Published online April 19, 2022. 10.1093/cercor/bhac150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yakushev I et al. (2013) Metabolic and structural connectivity within the default mode network relates to working memory performance in young healthy adults. Neuroimage 79, 184–190 [DOI] [PubMed] [Google Scholar]
- 59.Perani D et al. (2017) The impact of bilingualism on brain reserve and metabolic connectivity in Alzheimer’s dementia. Proc. Natl. Acad. Sci 114, 1690–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peterson EC et al. (2011) Regulation of cerebral blood flow. Int. J. Vasc. Med 2011, 823525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shulman GL et al. (1997) Common blood flow changes across visual tasks II. Decreases in cerebral cortex. J. Cogn. Neurosci 9, 648–663 [DOI] [PubMed] [Google Scholar]
- 62.Shine JM et al. (2019) Human cognition involves the dynamic integration of neural activity and neuromodulatory systems. Nat. Neurosci 22, 289–296 [DOI] [PubMed] [Google Scholar]
- 63.Hansen JY et al. (2022) Mapping neurotransmitter systems to the structural and functional organization of the human neocortex. Nat. Neurosci 25, 1569–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hahn A et al. (2014) Attenuated serotonin transporter association between dorsal raphe and ventral striatum in major depression. Hum. Brain Mapp 35, 3857–3866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cervenka S et al. (2010) PET studies of D2-receptor binding in striatal and extrastriatal brain regions: biochemical support in vivo for separate dopaminergic systems in humans. Synapse 64, 478–485 [DOI] [PubMed] [Google Scholar]
- 66.Tuominen L et al. (2014) Mapping neurotransmitter networks with PET: an example on serotonin and opioid systems. Hum. Brain Mapp 35, 1875–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ashok AH et al. (2019) Reduced mu opioid receptor availability in schizophrenia revealed with [11C]-carfentanil positron emission tomographic Imaging. Nat. Commun 10, 4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pillai RLI et al. (2019) Molecular connectivity disruptions in males with major depressive disorder. J. Cereb. Blood Flow Metab 39, 1623–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wager TD et al. (2007) Placebo effects on human μ-opioid activity during pain. Proc. Natl. Acad. Sci 104, 11056–11061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schneck N et al. (2021) Large-scale network dynamics in neural response to emotionally negative stimuli linked to serotonin 1A binding in major depressive disorder. Mol. Psychiatry 26, 2393–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fang XT et al. (2021) Identifying brain networks in synaptic density PET (11C-UCB-J) with independent component analysis. Neuroimage 237, 118167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peng C et al. (2020) Protein transmission in neurodegenerative disease. Nat. Rev. Neurol 16, 199–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jucker M and Walker LC (2018) Propagation and spread of pathogenic protein assemblies in neurodegenerative diseases. Nat. Neurosci 21, 1341–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vogel JW et al. (2020) Spread of pathological tau proteins through communicating neurons in human Alzheimer’s disease. Nat. Commun 11, 2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee WJ et al. (2022) Dynamic network model reveals distinct tau spreading patterns in early-and late-onset Alzheimer disease. Alzheimers Res. Ther 14, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pereira JB et al. (2018) Amyloid network topology characterizes the progression of Alzheimer’s disease during the predementia stages. Cereb. Cortex 28, 340–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pereira JB et al. (2019) Amyloid and tau accumulate across distinct spatial networks and are differentially associated with brain connectivity. Elife 8, e50830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ossenkoppele R et al. (2019) Tau covariance patterns in Alzheimer’s disease patients match intrinsic connectivity networks in the healthy brain. NeuroImage Clin. 23, 101848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sepulcre J et al. (2018) Neurogenetic contributions to amyloid beta and tau spreading in the human cortex. Nat. Med 24, 1910–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee WJ et al. (2022) Regional Aβ–tau interactions promote onset and acceleration of Alzheimer’s disease tau spreading. Neuron 110, 1932–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Franzmeier N et al. (2020) Patient-centered connectivity-based prediction of tau pathology spread in Alzheimer’s disease. Sci. Adv 6, eabd1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Amunts K et al. (2014) Interoperable atlases of the human brain. Neuroimage 99, 525–532 [DOI] [PubMed] [Google Scholar]
- 83.Sala A et al. (2022) Static versus functional PET: making sense of metabolic connectivity. Cereb. Cortex 32, 1125–1129 [DOI] [PubMed] [Google Scholar]
- 84.Reid AT et al. (2019) Advancing functional connectivity research from association to causation. Nat. Neurosci 22, 1751–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ripp I et al. (2020) Integrity of neurocognitive networks in dementing disorders as measured with simultaneous PET/functional MRI. J. Nucl. Med 61, 1341–1347 [DOI] [PubMed] [Google Scholar]
- 86.Titov D et al. (2017) Metabolic connectivity for differential diagnosis of dementing disorders. J. Cereb. Blood Flow Metab 37, 252–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang M et al. (2020) Individual brain metabolic connectome indicator based on Kullback–Leibler divergence similarity estimation predicts progression from mild cognitive impairment to Alzheimer’s dementia. Eur. J. Nucl. Med. Mol. Imaging 47, 2753–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yao Z et al. (2016) Individual metabolic network for the accurate detection of Alzheimer’s disease based on FDGPET imaging. In 2016 IEEE International Conference of Bioinformatics and Biomedicine (BIBM), pp. 1328–1335, IEEE [Google Scholar]
- 89.Eickhoff SB and Müller VI (2015) Functional connectivity. In Brain Mapping: An Encyclopedic Reference (Toga AW, ed.), pp. 187–201, Elsevier [Google Scholar]
- 90.Esfahlani FZ et al. (2020) High-amplitude cofluctuations in cortical activity drive functional connectivity. Proc. Natl. Acad. Sci. U. S. A 117, 28393–28401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kemmer PB et al. (2018) Evaluating the strength of structural connectivity underlying brain functional networks. Brain Connect. 8, 579–594 [Google Scholar]
- 92.Yakushev I et al. (2022) Mapping covariance in brain FDG up-take to structural connectivity. Eur. J. Nucl. Med. Mol. Imaging 49, 1288–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sala A and Perani D (2019) Brain molecular connectivity in neurodegenerative diseases: recent advances and new perspectives using positron emission tomography. Front. Neurosci 13, 617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Alexander-Bloch A et al. (2013) Imaging structural co-variance between human brain regions. Nat. Rev. Neurosci 14, 322–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Roberts RP et al. (2016) The Simpson’s paradox and fMRI: similarities and differences between functional connectivity measures derived from within-subject and across-subject correlations. Neuroimage 135, 1–15 [DOI] [PubMed] [Google Scholar]
- 96.Taylor PA et al. (2012) Functional covariance networks: obtaining resting-state networks from intersubject variability. Brain Connect. 2, 203–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Seghier ML and Price CJ (2018) Interpreting and utilising intersubject variability in brain function. Trends Cogn. Sci 22, 517–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Calhoun VD and Allen E (2013) Extracting intrinsic functional networks with feature-based group independent component analysis. Psychometrika 78, 243–259 [DOI] [PubMed] [Google Scholar]
- 99.Laird AR et al. (2013) Networks of task co-activations. Neuroimage 80, 505–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Patlak CS and Pettigrew KD (1976) A method to obtain infusion schedules for prescribed blood concentration time courses. J. Appl. Physiol 40, 458–463 [DOI] [PubMed] [Google Scholar]
- 101.Villien M et al. (2014) Dynamic functional imaging of brain glucose utilization using fPET-FDG. Neuroimage 100, 192–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li S et al. (2020) Analysis of continuous infusion functional PET(fPET) in the human brain. Neuroimage 213, 116720. [DOI] [PubMed] [Google Scholar]
- 103.Jamadar SD et al. (2022) Monash DaCRA fPET-fMRI: a dataset for comparison of radiotracer administration for high temporal resolution functional FDG-PET. Gigascience 11, giac031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sporns O (2022) The complex brain: connectivity, dynamics, information. Trends Cogn. Sci 26, 1066–1067 [DOI] [PubMed] [Google Scholar]
- 105.Calhoun VD et al. (2009) A review of group ICA for fMRI data and ICA for joint inference of imaging, genetic, and ERP data. Neuroimage 45, S163–S172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bassett DS and Sporns O (2017) Network neuroscience.Nat. Neurosci 20, 353–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Khan AF et al. (2022) Personalized brain models identify neurotransmitter receptor changes in Alzheimer’s disease. Brain 145, 1785–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kringelbach ML et al. (2020) Dynamic coupling of whole-brain neuronal and neurotransmitter systems. Proc. Natl. Acad. Sci.U. S. A 117, 9566–9576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.White BR and Culver J (2010) Quantitative evaluation of high-density diffuse optical tomography: in vivo resolution and mapping performance. J. Biomed. Opt 15, 026006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wein S et al. (2021) Brain connectivity studies on structure-function relationships: a short survey with an emphasis on machine learning. Comput. Intell. Neurosci 2021, 5573740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Schoffelen JM and Gross J (2009) Source connectivity analysis with MEG and EEG. Hum. Brain Mapp 30, 1857–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xia M et al. (2013) BrainNet Viewer: a network visualization tool for human brain connectomics. PLoS One 8, e68910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


