Abstract
Glioma represents a complex and heterogeneous disease, posing significant challenges to both clinicians and researchers. Despite notable advancements in glioma treatment, the overall survival rate for most glioma patients remains dishearteningly low. Hence, there is an urgent necessity to discover novel biomarkers and therapeutic targets specifically tailored for glioma. In recent years, long non-coding RNAs (lncRNAs) have emerged as pivotal regulators of gene expression and have garnered attention for their involvement in the development and progression of various cancers, including glioma. The dysregulation of lncRNAs plays a critical role in glioma pathogenesis and influences clinical outcomes. Consequently, there is growing interest in exploring the potential of lncRNAs as diagnostic and prognostic biomarkers, as well as therapeutic targets. By understanding the functions and dysregulation of lncRNAs in glioma, researchers aim to unlock new avenues for the development of innovative treatment strategies catered to glioma patients. The identification and thorough characterization of lncRNAs hold the promise of novel therapeutic approaches that could potentially improve patient outcomes and enhance the management of glioma, ultimately striving for better prospects and enhanced quality of life for those affected by this challenging disease. The primary objective of this paper is to comprehensively review the current state of knowledge regarding lncRNA biology and their intricate roles in glioma. It also delves into the potential of lncRNAs as valuable diagnostic and prognostic indicators and explores their feasibility as promising targets for therapeutic interventions.
Keywords: Non-coding RNAs, Long non-coding RNAs, Glioma, Biological significance, Therapeutic targets
1. Introduction
Glioma, the most prevalent and aggressive malignant tumor within the central nervous system (CNS), poses a significant challenge due to its high recurrence rate and mortality [1]. Globally, the number of cancer diagnoses has been on the rise, with approximately 11 million cases reported annually and an estimated increase to 16 million by 2020 [2,3]. Correspondingly, the incidence of glioma is also increasing, with around 200,000 new cases reported each year. In 2016, the World Health Organization (WHO) introduced a novel molecular diagnosis concept for CNS tumors, leading to the reclassification of various tumors, including diffuse glioma, medulloblastoma, and other embryonal tumors, such as IDH-wildtype glioblastoma. Moreover, the classification now includes diffuse midline glioma - H3 K27 M mutant, RELA fusion-positive ependymomas, WNT-activated and SHH-activated medulloblastoma, and C19MC amplified multilayered chrysanthemum embryonal tumor [[4], [5], [6]]. Despite advancements in multimodal treatment approaches encompassing surgical resection, radio- and chemotherapy (Fig. 1), the overall survival rate for glioma patients remains disappointingly low, especially for glioblastoma, where the median survival time is only about 14 months [7].
Fig. 1.
Basic strategies in the treatment of gliomas. One of these is RNA interference (RNAi) between tumor targets and genes, where long non-coding RNAs (lncRNAs) show the potential.
Like other tumors, glioma is considered a genetic disease, with the involvement of two main categories of genes: proto-oncogenes and anti-oncogenes. Among the identified oncogenes are epidermal growth factor receptor (EGFR), basic fibroblast growth factor (bFGF), and platelet-derived growth factor (PDGF) [[8], [9], [10], [11], [12], [13], [14], [15]]. Additionally, tumor suppressor genes such as p53, phosphatase and tensin homolog (PTEN), retinoblastoma protein (Rb), and E2F1 have been found to play crucial roles in glioma development [[16], [17], [18], [19], [20], [21]]. Uncovering novel biomarkers and therapeutic targets is crucial for gaining a better understanding of glioma and developing more effective treatments. Recent research has revealed that only a small fraction (2%) of genes in the human genome encode proteins, while most of the genome consists of non-coding genes [22]. Non-coding RNAs (ncRNAs) are categorized based on their nucleotide length into short ncRNAs and long non-coding RNAs (lncRNAs). While ncRNAs do not directly code for proteins, they exert significant regulatory functions in both transcription and translation processes. Short ncRNAs, such as microRNAs (miRNAs) and transfer RNAs (tRNAs), are typically around 200 nucleotides long. In contrast, lncRNAs, which exceed 200 nucleotides in length, play vital roles in glioma pathogenesis through epigenetic regulation, transcriptional modulation, and post-transcriptional modifications, thereby holding a unique and essential role in glioma's molecular biology [23].
The objective of this review is to explore the molecular biological mechanisms through which lncRNAs contribute to the initiation and progression of glioma. Additionally, the review aims to assess and anticipate the clinical implications of lncRNAs in glioma research.
2. Biological overview of lncRNAs
The complexity of the human gene transcriptome has revolutionized our understanding of RNA's potential, highlighting the significant role of many lncRNAs in transcription [24,25]. Presently, the Coding Project (GENCODE V26) has identified approximately 16,000 human lncRNA genes, generating over 28,000 distinct transcripts. Moreover, protein-coding genes can produce non-coding transcriptional variants, further adding to the diversity of non-coding transcripts in cells. lncRNAs are characterized by their nonprotein-coding nature, with lengths ranging from nearly 200 nucleotides to over 100 kilobases [26]. Based on their genomic locations and backgrounds, lncRNAs can be classified into five categories: sense (transcribed from the sense strand of a protein-coding gene), antisense (transcribed from the opposite mRNA), introns (transcribed from introns of protein-coding genes), intergenic (transcribed from intergenic regions), and two-way (bidirectional transcription at or near the start of sense and antisense transcription) [27].
While many identified lncRNAs are found in both the nucleus and cytoplasm [28], they were previously regarded as transcriptional noise for several decades [29]. However, recent research has demonstrated that lncRNAs are actively involved in various biological processes and may play pivotal roles in the development of diverse diseases, including cancer [30,31]. In glioma cells, lncRNAs primarily regulate gene expression through three mechanisms: epigenetic regulation, transcriptional regulation, and interaction regulation with miRNAs.
2.1. Epigenetic regulation
Upon the discovery of lncRNAs capability to regulate gene expression, researchers recognized their significant role in epigenetic gene regulation, which holds particular importance in the pathogenesis of glioma. Epigenetic mechanisms, including DNA methylation, have been identified as essential factors in glioma development. Numerous studies have highlighted the involvement of lncRNAs in the epigenetic regulation of genes that contribute to glioma pathogenesis. For instance, alterations in the expression of lncRNA- POU Class 3 Homeobox 3 (POU3F3) have been found to modulate the methylation status of POU3F3 gene [32]. Notably, homeobox transcript antisense intergenic RNA (HOTAIR) is a well-known lncRNA epigenetic gene regulator. It indirectly silences homeobox D Cluster (HOXD) genes by facilitating the recruitment of the polycomb repressive complex 2 (PRC2) to the HOXD gene cluster, thereby promoting complex trimethylation of chromatin, leading to the repression of HOXD gene expression [[33], [34], [35]]. LncRNA X-inactive specific transcript (XIST) has been shown to play a role in chromatin regulation by modifying DNA/RNA and histone status [36].
2.2. Regulation of transcription level
LncRNAs have demonstrated their capability to regulate gene transcription activity and modify RNA functions by interacting with transcription factors. One example is the lncRNA tumor suppressor in lung cancer-1 antisense-1 (TSLC1-AS1), which is transcribed from the antisense strand of the tumor suppressor gene TSLC1. This lncRNA functions to silence the expression of TSLC1 by targeting its mRNA. Moreover, lncRNA TSLC1-AS1 has shown a positive correlation with other tumor suppressors, including B-Raf proto-oncogene, serine/threonine kinase (BRAF), while BRAF has displayed a negative correlation with neurofibromatosis type 1 (NF1), Von Hippel-Lindau (VHL), and phosphoinositide-3-kinase regulatory subunit 1 (PIK3R1) [37].
Apart from interacting with transcription factors, lncRNAs may contribute to gliomas through their involvement in various other RNA regulatory processes, such as gene splicing, RNA editing, and even protein translation. For instance, the lncRNA highly upregulated in liver cancer (HULC) has been found to suppress the molecular eukaryotic initiation factor 4E (eIF4E), thereby regulating other proteins that inhibit angiogenesis [38].
2.3. Interaction regulation with miRNAs
Certain lncRNAs possess the ability to interact with miRNAs, effectively sequestering them and preventing their interaction with target mRNAs. For instance, lncRNA nuclear paraspeckle assembly transcript 1 (NEAT1) plays a role in promoting glioma development by regulating miR-449b-5p [39]. This interaction is in line with the competing endogenous RNA (ceRNA) hypothesis, wherein lncRNAs can influence the expression of target genes that are under the control of miRNAs. In both glioma and normal tissues, specific lncRNAs can interact with miRNAs, acting as ceRNAs to modulate the availability of miRNAs for binding to their target mRNAs. For example, knockdown of lncRNA X-inactive specific transcript (XIST) has been shown to suppress cancer stem cells in human glioblastoma by upregulating miR-152 [36]. Another study found that overexpression of the lncRNA glioma tumor suppressor gene cancer susceptibility candidate 2 (CASC2) led to a significant reduction in miR-21 expression, and mutual inhibition between CASC2 and miR-21 was mediated by Argonaute-2 (Ago2) [40,41].
3. Molecular biological role of lncRNAs in glioma
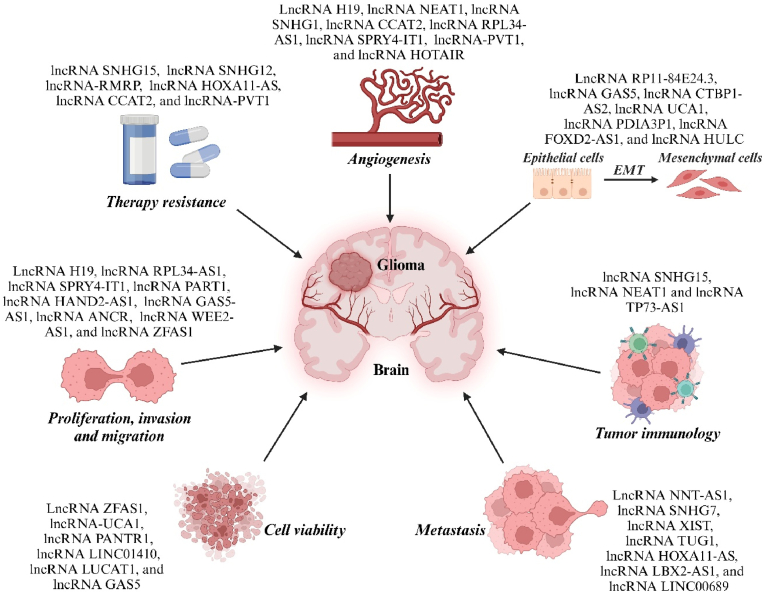
LncRNAs play the role of tumor suppressor genes and oncogenes in the occurrence and development of tumors, thereby affecting tumor phenotype, clinicopathological manifestations and prognosis [42]. Understanding the main biological roles of lncRNAs in glioma is of great significance for fully understanding the occurrence, development, and clinical treatment of glioma (Fig. 2).
Fig. 2.
An example of the most studied long non-coding RNAs (lncRNAs) and the processes of glioma tumorigenesis in which they are involved. The figure shows the role of lncRNAs in glioma by promoting epithelial-mesenchymal transition (EMT), proliferation, metastasis, chemo- and radiotherapy resistance, invasion, immune microenvironment, tumor cell viability, and angiogenesis.
LncRNAs play a crucial role in increasing the incidence, migration, and invasion of malignant glioma. For instance, the proto-oncogene H19 is upregulated, promoting the invasion of glioma cells through the induction of miR-675 expression [43]. Additionally, POU3F3, a highly conserved functional translation regulator, is significantly elevated during glioma development [32]. Two target genes of POU3F3, namely lncRNA ASLNC22381 and ASLNC20819, mediate downstream signaling cascades and insulin-like growth factor 1 (IGF-1) activity, influencing cell proliferation and viability by binding to the IGF-1 receptor correlator. This molecular mechanism is believed to play a critical role in glioma occurrence and development [44].
As already mentioned above, some lncRNAs can interact with miRNAs, hindering miRNAs from binding to their target mRNAs in gliomagenesis. For example, Cao et al. demonstrated that overexpression of lncRNA gastric cancer high expressed transcript 1 (GHET1) led to increased survival, migration, and invasion of glioma U251 cells [45]. The overexpression of lncRNA GHET1 resulted in the upregulation of cell cycle genes (cyclin D1, cyclin-dependent kinase 4 (CDK4), cyclin-dependent kinase 6 (CDK6)) and pre-metastatic genes (matrix metallopeptidase 9 (MMP9) and vimentin). Additionally, lncRNA GHET1 overexpression activated janus kinase 2 (JAK2)/signal transducer and activator of transcription 3 (STAT3) and p53/survivin signaling pathways by downregulating miR-216a, thereby promoting glioma progression. The lncRNA plasmacytoma variant translocation 1 (PVT1) functions as an oncogene and is highly expressed in various tumors, including human gliomas, gastric cancer (GC), and non-small cell lung cancer (NSCLC). When lncRNA PVT1 is downregulated, it negatively regulates miR-424, leading to the inhibition of activity, migration, and invasive capabilities of human glioma cells [46]. On the other hand, increased expression of lncRNA tumor protein P73 antisense RNA 1 (TP73-AS1) is associated with poor patient survival. Downregulation of TP73-AS1 suppresses cell proliferation and invasion, and its interaction with miR-142 plays a crucial role in various diseases, including cancer, with downstream effects on RAS-related C3 botulinum toxin substrate 1 (Rac1) levels, leading to activation of multiple cellular pathways [[47], [48], [49], [50]]. Furthermore, lncRNA ECONEXIN is upregulated in glioma tissues, promoting cell proliferation, and influencing the expression of the topoisomerase 2α (TOP2A) gene through interactions with miR-411–5p [51]. In contrast, certain lncRNAs have been found to slow down or inhibit the malignant progression of glioblastoma. The downregulation of lncRNA maternally expressed gene 3 (MEG3) is associated with the progression of glioblastoma, but its overexpression inhibits the growth and proliferation of glioblastoma cells and promotes cell apoptosis [52,53]. Similarly, overexpression of lncRNA CASC2 can inhibit the proliferation, migration, and invasion of glioblastoma cells and induce tumor cell apoptosis. The relationship between CASC2 and miR-21 is of particular interest, with miR-21 negatively regulating CASC2 [40]. On the other hand, the exact pathway by which lncRNA MDC1-AS induces cell proliferation through mediator of DNA damage checkpoint 1 (MDC1) remains unclear [54]. TSLC1-AS1, the antisense regulatory factor of the tumor suppressor gene TSLC1, also plays a role in inhibiting cell growth, although the exact mechanism is not fully understood [37]. Lastly, ADAMTS9-AS2, which is downregulated in glioblastoma and correlates with glioma grading, has been shown to inhibit cell migration and invasion upon overexpression [55,56]. Numerous studies are currently underway to develop a potential therapeutic approach based on interactions between lncRNAs and miRNAs, which reveal the vital role of this interaction in proliferation, angiogenesis, invasion, metastasis, cell survival and resistance to therapy in glioma (Table 1) [[57], [58], [59], [60], [61], [62], [63], [64], [65], [66]].
Table 1.
Some last studies about long non-coding RNAs (lncRNAs)-microRNAs (miRNAs) interactions in glioma.
| LncRNAs | Expression of lncRNAs | miRNAs | Targets | Function | Ref. |
|---|---|---|---|---|---|
| HOXA11-AS | Up | let-7b-5p | Tpl2-MEK1/2-ERK1/2 axis | Sensitizes glioblastoma cells to ROS, drastically impairing tumor growth and prolonging survival of mouses | [57] |
| MIR210HG | Down | miR-377–3p | LMX1A | Suppresses glioma cell proliferation | [58] |
| PDCD4-AS1 | Down | miR-30b-3p | METTL7B | Inhibits glioma cell proliferation, invasion, migration, and induces cell cycle arrest | [59] |
| GSCAR | Down | miR-6760–5p | SRSF1 | Promotes tumor cell responses to TMZ | [60] |
| SPRY4-IT1 | Down | miR-101–3p | EZH2 and VEGFA | Achieves cell proliferation and angiogenesis | [61] |
| LINC01018 | Up | miR-942–5p | KNG1 | Suppresses the migration, invasion, and proliferation of glioma cells | [62] |
| LINC01018 | Up | miR-182–5p | ADRA2C, RAB6B, RAB27B, RAPGEF5, STEAP2, TAGLN3, and UNC13C | Inhibits cell proliferation and metastasis | [63] |
| TUSC7 | Up | miR-10a-5p | BDNF/ERK axis | Suppresses glioma cell proliferation and migration | [64] |
| LINC01426 | Down | miR-661 | Mdm2 | Inhibits proliferation and induces apoptosis | [65] |
| DANCR | Up | miR-33b | DLX6/ATG7 axis | Promotes intracellular autophagy and proliferation and reduce apoptosis | [66] |
Abbreviations: MIR210HG, MIR210 Host Gene; PDCD4-AS1, PDCD4 Antisense RNA 1; GSCAR, Glioma stem cell associated lncRNA; SPRY4-IT1, Sprouty RTK signaling antagonist 4-intronic transcript 1; LINC01018, Long Intergenic Non-Protein Coding RNA 1018; TUSC7, Tumor Suppressor Candidate 7; LINC01426, Long Intergenic Non-Protein Coding RNA 1426; DANCR, Differentiation Antagonizing Non-Protein Coding RNA; TMZ, Temozolomide; Tpl2, Tumor progression locus 2; MEK1/2, Mitogen-activated protein kinase kinase 1/2; ERK1/2, Extracellular signal-regulated kinase 1/2; LMX1A, LIM Homeobox Transcription Factor 1 Alpha; METTL7B, Methyltransferase-like 7B; SRSF1, Serine/arginine-rich splicing factor 1; EZH2, Enhancer Of Zeste 2 Polycomb Repressive Complex 2 Subunit; VEGFA, Vascular endothelial growth factor A; KNG1, Kininogen 1; ADRA2C, Adrenoceptor Alpha 2C; RAB6B, Ras-related protein Rab-6B; RAB27B, Ras-related protein Rab-27B; RAPGEF5, Rap guanine nucleotide exchange factor 5; STEAP2, Six-transmembrane epithelial antigen of the prostate-2; TAGLN3, Transgelin 3; UNC13C, unc-13 homolog C; BDNF, Brain-derived neurotrophic factor; ERK, Extracellular signal-regulated kinases; Mdm2, Mouse double minute 2 homolog; DLX6, Distal-Less Homeobox 6; ATG7, Autophagy Related 7.
4. Study the clinical significance of lncRNAs in glioma
4.1. LncRNAs serve as targets for the diagnosis and treatment of gliomas
Numerous lncRNAs have been found to influence the expression of specific genes through knockout or overexpression, offering potential as molecular targets for future clinical drug therapy against gliomas. For instance, studies on Chinese glioblastoma patients revealed that overexpression of HOTAIR is negatively correlated with patient prognosis. In animal models, inhibiting HOTAIR was shown to suppress the invasion and metastasis of glioblastoma polymorphic cells [67]. Knocking out HOTAIR in glioma cells hindered their biological development, and miR-326 is believed to play a role in regulating this process [35]. Another potential therapeutic target is the tumor suppressor gene TSLC1, which influences glioma occurrence and development by modulating methylation. Inhibiting or knocking out TSLC1 can accelerate glioma cell proliferation and metastasis, whereas upregulating TSLC1 has the opposite effect, making it a promising clinical molecular target [68].
Moreover, research on lncRNAs in tissue and biofluids samples has identified 80 lncRNAs with different expression levels in tumor tissue samples compared to normal samples. Among them, lncRNA MIR210 Host Gene (miR210HG) showed potential as a diagnostic biomarker for gliomas. Its stable expression was detected in all serum samples from glioma patients, with higher levels in high-risk groups (WHO III or IV). As a glioma biomarker, miR210HG exhibited a sensitivity of 86.21% and a specificity of 72.41% [69]. Table 2 and Table 3 provides a summary of potential diagnostic and prognostic markers of lncRNAs in gliomas [67,[70], [71], [72], [73], [74], [75], [76]].
Table 2.
Long non-coding RNAs (lncRNAs) that hold diagnostic significance in gliomas (tumor tissue and biofluids).
| LncRNAs | Expression | ROC curve | Sensitivity, % | Specificity, % | Ref. |
|---|---|---|---|---|---|
| DLX6-AS1 | Up | 0.736 | – | – | [70] |
| ELF3-AS1 | Up | 0.8073 | 67.23 | 85.22 | [71] |
| ASB16-AS1 | Up | 0.96923 | – | – | [72] |
| NEF | Down | 0.7908 | – | – | [73] |
| ANRIL | Up | 0.860 | 81.62 | 90.83 | [74] |
| HOTAIR | Up | 0.913 | 86.1 | 87.5 | [75] |
| GSCAR | Up | 0.971 | – | – | [76] |
Note: - , not represented; ROC curve, Receiver operating characteristic (ROC) curve analysis was performed to evaluate the diagnostic value of lncRNAs for glioma.
Table 3.
Long non-coding RNAs (lncRNAs) that hold prognostic significance in gliomas (tumor tissue and biofluids).
| LncRNAs | Ensembl ID | Chromosome | Length (bp) | Tumor | Effect on Prognosis | Ref. |
|---|---|---|---|---|---|---|
| HOTAIR | ENSG00000228630 | 12 | 12.649 | glioblastoma | negative | [67] |
| MCM3AP-AS | ENSG00000215424 | 21 | 30.174 | glioblastoma | positive | [77] |
| PART1 | ENSG00000152931 | 5 | 59.945 | glioblastoma | positive | [78] |
| MIAT | ENSG00000225783 | 22 | 30.051 | glioblastoma | positive | [78] |
| GAS5 | ENSG00000234741 | 1 | 4983 | glioblastoma | positive | [78] |
| RP11–838N2.4 | ENSG00000266835 | 18 | 12.729 | glioblastoma | positive | [79] |
| NR_002809 | ENSG00000212694 | 12 | 8640 | astrocytoma | negative | [80] |
| XLOC_010967 | – | – | – | astrocytoma | positive | [80] |
| BC002811 | – | – | – | astrocytoma | positive | [80] |
| TPT1-AS1 | ENSG00000170919 | 13 | 52.069 | glioma | positive | [81] |
| TUSC7 | ENSG00000243197 | 3 | 14.347 | glioma | positive | [82] |
| AGAP2-AS1 | ENSG00000255737 | 12 | 2117 | glioma | negative | [83] |
| LINC01198 | ENSG00000231817 | 13 | 31.372 | glioma | negative | [83] |
| SPRY4-IT1 | ENSG00000281881 | 5 | 703 | glioma | negative | [84] |
| ZEB1-AS1 | ENSG00000237036 | 10 | 114.170 | glioma | negative | [85] |
| KIAA0495 | ENSG00000227372 | 1 | 11.773 | glioblastoma | negative | [73,86] |
| MALAT1 | ENSG00000251562 | 11 | 8755 | glioma | negative | [76,87] |
| HOXA11-AS | ENSG00000240990 | 7 | 4776 | glioma | negative | [77,88] |
Note: - , not represented.
4.2. LncRNAs and chemoresistance in glioma
Chemotherapy is a widely used and effective approach for treating glioblastoma, but its effectiveness is hindered by multidrug resistance (MDR) [89]. Temozolomide (TMZ) is the primary chemotherapeutic drug utilized for glioblastoma treatment due to its alkylating properties [90]. Li et al. discovered that increasing the expression of metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) in U251/TMZ and U87/TMZ cell lines enhanced the sensitivity to TMZ and resulted in reduced expression of zinc finger E-box binding homeobox 1 (ZEB1) protein [91]. The upregulation of MALAT1 was inversely associated with the expression of MDR-related proteins, cell viability, and the epithelial-mesenchymal transition (EMT) status. In in vivo experiments, overexpression of MALAT1 led to enhanced tumor resistance to TMZ.
Additionally, KIAA0495/pipeline defect assessment manual (PDAM) is commonly overexpressed in oligodendroglial tumors, and animal studies have shown that this gene induces resistance to cisplatin, suggesting its significant role in the molecular development of oligodendroglial tumors. Therefore, a molecular targeting therapy could be designed based on this mechanism to target specific points [86].
4.3. The role of lncRNAs in glioma radiotherapy
Radiation therapy is a commonly used treatment for gliomas, and its effectiveness is influenced by the sensitivity of the tumor to radiation. LncRNAs, crucial regulators of various biological processes, play significant roles in glioma development and pathogenesis. Aryankalayil et al. identified several radiation-induced lncRNAs involved in DNA damage response and immune response [92]. These include lncRNAs such as PVT1, taurine upregulated gene 1 (Tug1), tumor protein p53 pathway corepressor 1 (Trp53cor1), damage-induced (DINO), and MEG3, which are affected by radiation and are associated with the tumor suppressor p53. Gm14005 (Morrbid) and Theiler's murine encephalitis virus possible gene1 (TMEVPG1) are also regulated by radiation at different time points and doses, showing potential as blood radiation biomarkers.
Moreover, the oncogene hyaluronan-mediated motility receptor (HMMR) is highly expressed in glioblastoma and promotes tumor growth. Li et al. discovered that the HMMR antisense lncRNA, HMMR-AS1, is highly expressed in glioblastoma cell lines and stabilizes HMMR expression [93]. Deletion of HMMR-AS1 leads to reduced HMMR expression, inhibiting cell migration, invasion, and mesenchymal phenotype, both in vitro and in vivo. Furthermore, HMMR-AS1 expression reduces the radiation sensitivity of glioblastoma cells by reducing the expression of DNA repair proteins like ATM, RAD51, and polycomb group complex 1 (PRC1). Targeting HMMR-AS1 may be a potential strategy for glioblastoma treatment. LncRNAs have a dual role in glioma radiotherapy, and studying the mechanisms of radiotherapy resistance can enhance our understanding of relevant molecular pathways in tumor cells and provide effective countermeasures to improve the clinical efficacy of glioma radiotherapy.
4.4. Correlation between lncRNAs and the prognosis of glioma patients
MALAT1 expression is elevated in glioma patients compared to healthy adults, and it positively correlates with glioma grading, tumor size, and overall survival. Hence, it can serve as a molecular marker to predict the prognosis and survival of glioma patients [87]. Other potential molecular markers for prognosis include HOTAIR, HOXA11-AS, and NEAT1. HOTAIR is linked to glioma's pathological grading, molecular typing, and prognosis [33]. HOXA11-AS, a cell cycle-regulating lncRNA transcribed from the 5′ end of HOXA, can also be used as a molecular marker for glioma progression [88]. NEAT1's expression is significantly upregulated in high-grade gliomas and is associated with patient survival after chemotherapy [94]. Furthermore, lncRNA AB073614 is found to be overexpressed in high-grade gliomas compared to low-grade gliomas and normal brain tissue. Its expression is correlated with overall survival and may serve as a potential negative prognostic biomarker for glioma [95]. Conversely, RP11-838n2.4 downregulation is positively associated with TMZ resistance and poor prognosis [96]. In another study by Zhi et al., 59 differentially expressed lncRNAs were identified when comparing tumor tissue samples (WHO grades II-IV) with normal brain samples [97]. Among these, ENST00000545440 and NR_002809 were linked to the high clinical stage of astrocytoma. High NR_002809 expression and low BC002811 and XLOC-010967 expression were significantly associated with poor prognosis and survival in glioma patients. These lncRNA biomarkers offer accurate predictions of survival rates in low-grade glioma patients without the need for normal brain tissue, which can be difficult to obtain. Larger cohort studies with comprehensive clinical information will be conducted in the future to validate the role of lncRNA signals.
4.5. Correlation between lncRNAs and glioma angiogenesis
One of the primary characteristics of malignant tumors is the continuous generation of blood vessels, known as angiogenesis [98]. Glioblastoma, being the most vascular tumor among solid tumors, exhibits a prominent feature of ongoing vessel generation, which drives its progression. While hypoxia is a common factor promoting angiogenesis, emerging evidence suggests that non-hypoxic mechanisms can also stimulate vessel formation [[99], [100], [101]]. The key regulator of vessel generation under low-oxygen conditions is the vascular endothelial growth factor (VEGF), whose expression is significantly reduced in response to hypoxia. In glioblastoma cells, hypoxic conditions lead to high-level expression of hypoxia-inducible factor (HIF-1) and VEGF, which play critical roles in various processes, including anaerobic glycolysis, metabolism, vessel generation, metastasis, and epithelial-mesenchymal transition (EMT) pathways [102].
Interestingly, under normal oxygen conditions, glioma cells induce vessel generation by activating the phosphoinositide 3-kinases (PI3Ks)/Akt/mammalian target of rapamycin (mTOR) signaling pathway. A serine/arginine-rich protein kinase 1 (SRPK1), abundant in serine/arginine residues, promotes vessel generation in glioma cells under normal oxygen levels by regulating Akt phosphorylation and VEGF splicing [103]. Additionally, lncRNA POU3F3 has been found to be transported via extracellular vesicles to endothelial cells, where it triggers the formation of new blood vessels [32]. Studies have shown that high expression of lncRNA POU3F3 induces vessel generation by promoting the expression of basic fibroblast growth factor (bFGF), vascular endothelial growth factor-A (VEGF-A), and basic fibroblast growth factor receptor (bFGFR). Similarly, lncRNA highly up‐regulated in liver cancer (HULC) regulates the expression of the ESM-1 gene, leading to increased expression of endothelial cell-specific molecule 1 (ESM-1), a molecular marker of vessel generation that is activated by the VEGF/PI3K pathway [38].
4.6. LncRNAs and regulation of blood-tumor barrier in glioma
The efficacy of glioma treatment is affected by the presence of the blood-tumor barrier (BTB), which involves the tight regulation of tight junction proteins such as claudin and occludin, as well as multiprotein complexes containing zonula occludens (ZO) proteins acting as cytoplasmic scaffolds [104,105]. Inhibition of lncRNA MALAT1 expression has been found to increase BTB permeability and decrease the levels of tight junction proteins. Additionally, MALAT1 is targeted by miR-140, which further regulates BTB permeability and the expression of tight junction proteins. The expression levels of MALAT1 and miR-140 are inversely correlated. Furthermore, miR-140 targets the nuclear transcription factor Y subunit alpha (NFYA) gene, acting as a transcription factor to regulate the expression of ZO-1, occludin, and claudin-5 [106].
Another lncRNA, NEAT1, exhibits low expression levels associated with upregulated miR-181–5p, which, in turn, regulates BTB permeability and the expression of ZO-1, occludin, and claudin-5 through SRY-Box transcription factor 5 (SOX5) [107]. Downregulation of lncRNA TUG1 expression leads to increased BTB permeability and decreased expression of BTB-related proteins ZO-1, occludin, and claudin-5. LncRNA TUG1 competes with miR-144 for the expression of tight junction proteins and inhibits the expression of heat shock transcription factor 2 (HSF2), which acts as a transcription factor for tight junction proteins. This relationship has been confirmed in animal models [108]. Similarly, the downregulation of lncRNAs HOTAIR and XIST, along with the upregulation of miR-148b-3p and miR-137, results in increased BTB permeability and decreased expression of ZO-1/2 and Occludin tight junction proteins [109]. The differential expression of lncRNA MEG3 affects BTB permeability through the P-element induced wimpy testis (PIWI)-like protein 1 (PIWI1)/MEG3/miR-330/runt-related transcription factor 3 (RUNX3) axis, influencing the molecular occurrence and development of glioma cells [110]. These findings suggest that lncRNAs hold potential as gene targets for regulating the BTB and should be further explored clinically to provide new approaches for glioma treatment.
5. Conclusion
As research into lncRNAs and their roles in glioma continues to progress, the potential for transformative therapeutic strategies becomes increasingly evident. Harnessing the power of lncRNAs opens up exciting possibilities for targeted therapies that can specifically address the underlying molecular alterations driving glioma development and progression. By leveraging the knowledge gained from high-throughput genomics and transcriptomics research, scientists can identify novel lncRNA candidates with significant functional implications in glioma biology. The advent of advanced technologies, such as RNA interference, provides a powerful tool for exploring the precise functions of lncRNAs in gliomas. This revolutionary approach allows for the selective silencing or modulation of specific lncRNAs, shedding light on their impact on tumorigenesis and potentially paving the way for the development of innovative treatments tailored to individual patients.
Moreover, the integration of lncRNAs into diagnostic and prognostic models holds great promise for precision medicine in glioma patients. By identifying lncRNA signatures associated with distinct glioma subtypes or clinical outcomes, clinicians can make informed decisions regarding treatment approaches, leading to improved patient management and personalized therapeutic strategies. However, it is crucial to acknowledge that the journey towards harnessing the full potential of lncRNAs in glioma therapy is still in its early stages. More extensive research is needed to unravel the complexities of lncRNA biology in the context of glioma pathogenesis fully. Collaborative efforts across various disciplines, including computational biology, functional genomics, and bioinformatics, will be essential to decipher the intricate interactions between lncRNAs and other molecular players in gliomas. In addition to targeted therapy, lncRNAs hold the potential to revolutionize liquid biopsy approaches for glioma detection and monitoring. The stability of lncRNAs in bodily fluids, such as blood and cerebrospinal fluid, presents a valuable opportunity for non-invasive molecular diagnostics, enabling early detection and real-time monitoring of disease progression or treatment response.
Overall, the exploration of lncRNAs in gliomas represents a promising frontier in cancer research, with the potential to transform how we understand, diagnose, and treat these devastating brain tumors. As the field continues to advance, it is hoped that lncRNAs will take center stage as game-changing therapeutic targets, ushering in a new era of precision medicine for glioma patients. The convergence of cutting-edge technologies and collaborative efforts will undoubtedly drive us closer to a future where gliomas can be effectively managed and conquered, offering hope for a brighter outlook for patients and their loved ones.
Funding
This work was supported by the Bashkir State Medical University Strategic Academic Leadership Program (PRIORITY-2030).
CRediT authorship contribution statement
Ilgiz Gareev: made substantial contributions to conception and design, been involved in drafting the manuscript or revising it critically for important intellectual content, has given final approval for the version published. Manuel de Jesus Encarnacion Ramirez: made substantial contributions to acquisition of data. Renat Nurmukhametov: made substantial contributions to acquisition of data. Denis Ivliev: Wang made substantial contributions to analysis and interpretation of data. Alina Shumadalova: Wang made substantial contributions to analysis and interpretation of data. Tatiana Ilyasova: Wang made substantial contributions to analysis and interpretation of data. Aferin Beilerli: Wang made substantial contributions to analysis and interpretation of data. Chunlei Wang: made substantial contributions to analysis and interpretation of data, All authors agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Declaration of competing interest
The authors declare that no conflicts of interest exist.
References
- 1.Khalilian S., Bijanvand A., Abedinlou H., Ghafouri-Fard S. A review on the role of miR-210 in human disorders. Pathol. Res. Pract. 2023 Jan;241 doi: 10.1016/j.prp.2022.154244. [DOI] [PubMed] [Google Scholar]
- 2.Bhatt A.N., Mathur R., Farooque A., et al. Cancer biomarkers-current perspectives. Indian J. Med. Res. 2010;132(8):129–149. [PubMed] [Google Scholar]
- 3.Cho W.C. Contribution of oncoproteomics to cancer biomarker discovery. Mol. Cancer. 2007 Apr 2;6:25. doi: 10.1186/1476-4598-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li M., Deng H., Peng H., et al. Functional nanoparticles in targeting glioma diagnosis and therapies. J. Nanosci. Nanotechnol. 2014 Jan;14(1):415–432. doi: 10.1166/jnn.2014.8757. [DOI] [PubMed] [Google Scholar]
- 5.Louis D.N., Perry A., Wesseling P., Brat D.J., Cree I.A., Figarella-Branger D., Hawkins C., Ng H.K., Pfister S.M., Reifenberger G., Soffietti R., von Deimling A., Ellison D.W. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 2021 Aug 2;23(8):1231–1251. doi: 10.1093/neuonc/noab106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mansouri A., Mansouri S., Hachem L.D., et al. The role of 5-aminolevulinic acid in enhancing surgery for high-grade glioma, its current boundaries, and future perspectives: a systematic review. Cancer. 2016 Aug 15;122(16):2469–2478. doi: 10.1002/cncr.30088. [DOI] [PubMed] [Google Scholar]
- 7.Delgado-López P.D., Corrales-García E.M. Survival in glioblastoma: a review on the treatment modalities. Clin. Transl. Oncol. 2016 Nov;18(11):1062–1071. doi: 10.1007/s12094-016-1497-x. [DOI] [PubMed] [Google Scholar]
- 8.Sun Y., Zhang W., Chen D., et al. A glioma classification scheme based on coexpression modules of EGFR and PDGFRA. Proc. Natl. Acad. Sci. U.S.A. 2014 Mar 4;111(9):3538–3543. doi: 10.1073/pnas.1313814111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Resnier P., David S., Lautram N., et al. EGFR siRNA lipid nanocapsules efficiently transfect glioma cells in vitro. Int. J. Pharm. 2013 Oct 1;454(2):748–755. doi: 10.1016/j.ijpharm.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Paul I., Bhattacharya S., Chatterjee A., et al. Current understanding on EGFR and Wnt/β-catenin signaling in glioma and their possible crosstalk. Genes Cancer. 2013 Nov;4(11–12):427–446. doi: 10.1177/1947601913503341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hua W., Yao Y., Chu Y., et al. The CD133+ tumor stem-like cell-associated antigen may elicit highly intense immune responses against human malignant glioma. J. Neuro Oncol. 2011 Nov;105(2):149–157. doi: 10.1007/s11060-011-0572-y. [DOI] [PubMed] [Google Scholar]
- 12.Liu J., Xu X., Feng X., et al. Adenovirus-mediated delivery of bFGF small interfering RNA reduces STAT3 phosphorylation and induces the depolarization of mitochondria and apoptosis in glioma cells U251. J. Exp. Clin. Cancer Res. 2011 Sep 9;30(1):80. doi: 10.1186/1756-9966-30-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang B., Feng X., Wang J., et al. Combined antitumor effect of ad-bFGF-siRNA and ad-Vpr on the growth of xenograft glioma in nude mouse model. Pathol. Oncol. Res. 2011 Jun;17(2):237–242. doi: 10.1007/s12253-010-9303-5. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Y., Jin G., Mi R., et al. The methylation status of the platelet-derived growth factor-B gene promoter and its regulation of cellular proliferation following folate treatment in human glioma cells. Brain Res. 2014 Mar 27;1556:57–66. doi: 10.1016/j.brainres.2014.01.045. [DOI] [PubMed] [Google Scholar]
- 15.Liu K., Hu B., Cheng S. Platelet-derived growth factor receptor alpha in glioma: a bad seed. Chin. J. Cancer Res. 2011 Sep;30(9):590–602. doi: 10.5732/cjc.011.10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar S. P53 induction accompanying G2/M arrest upon knockdown of tumor suppressor HIC1 in U87MG glioma cells. Mol. Cell. Biochem. 2014 Oct;395(1–2):281–290. doi: 10.1007/s11010-014-2137-9. [DOI] [PubMed] [Google Scholar]
- 17.Wang M., Lin C., Zhang J., et al. Role of PTEN in cholera toxin-induced SWO38 glioma cell differentiation. Mol. Med. Rep. 2013 Jun;7(6):1912–1918. doi: 10.3892/mmr.2013.1434. [DOI] [PubMed] [Google Scholar]
- 18.Errafiy R., Aguado C., Ghislat G., et al. PTEN increases autophagy and inhibits the ubiquitin-proteasome pathway in glioma cells independently of its lipid phosphatase activity. PLoS One. 2013 Dec 13;8(12) doi: 10.1371/journal.pone.0083318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fueyo J., Gomez-Manzano C., Alemany R., et al. A mutant oncolytic adenovirus targeting the Rb pathway produces anti-glioma effect in vivo. Oncogene. 2000 Jan 6;19(1):2–12. doi: 10.1038/sj.onc.1203251. [DOI] [PubMed] [Google Scholar]
- 20.Mitlianga P.G., Gomez-Manzano C., Kyritsis A.P., et al. Overexpression of E2F-1 leads to bax-independent cell death in human glioma cells. Int. J. Oncol. 2002;21(5):1015–1020. [PubMed] [Google Scholar]
- 21.Gomez-Manzano C., Lemoine M.G., Hu M., et al. Adenovirally-mediated transfer of E2F-1 potentiates chemosensitivity of human glioma cells to temozolomide and BCNU. Int. J. Oncol. 2001 Aug;19(2):359–365. doi: 10.3892/ijo.19.2.359. [DOI] [PubMed] [Google Scholar]
- 22.Beylerli O., Gareev I., Sufianov A., Ilyasova T., Guang Y. Long noncoding RNAs as promising biomarkers in cancer. Noncoding RNA Res. 2022 Feb 25;7(2):66–70. doi: 10.1016/j.ncrna.2022.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sufianov A., Begliarzade S., Beilerli A., Liang Y., Ilyasova T., Beylerli O. Circular RNAs as biomarkers for lung cancer. Noncoding RNA Res. 2022 Nov 7;8(1):83–88. doi: 10.1016/j.ncrna.2022.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sufianov A., Begliarzade S., Ilyasova T., Liang Y., Beylerli O. MicroRNAs as prognostic markers and therapeutic targets in gliomas. Noncoding RNA Res. 2022 Jul 6;7(3):171–177. doi: 10.1016/j.ncrna.2022.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iyer M.K., Niknafs Y.S., Malik R., et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015 Mar;47(3):199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sufianov A., Kostin A., Begliarzade S., Kudriashov V., Ilyasova T., Liang Y., Mukhamedzyanov A., Beylerli O. Exosomal non coding RNAs as a novel target for diabetes mellitus and its complications. Noncoding RNA Res. 2023 Feb 7;8(2):192–204. doi: 10.1016/j.ncrna.2023.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gareev I., Kudriashov V., Sufianov A., Begliarzade S., Ilyasova T., Liang Y., Beylerli O. The role of long non-coding RNA ANRIL in the development of atherosclerosis. Noncoding RNA Res. 2022 Sep 6;7(4):212–216. doi: 10.1016/j.ncrna.2022.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ponting C.P., Oliver P.L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009 Feb 20;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 29.Van Bakel H., Hughes T.R. Establishing legitimacy and function in the new transcriptome. Briefings Funct. Genomics Proteomics. 2009 Nov;8(6):424–436. doi: 10.1093/bfgp/elp037. [DOI] [PubMed] [Google Scholar]
- 30.Kapranov P., Cheng J., Dike S., et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007 Jun 8;316(5830):1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 31.Yao Z.T., Yang Y.M., Sun M.M., He Y., Liao L., Chen K.S., Li B. New insights into the interplay between long non-coding RNAs and RNA-binding proteins in cancer. Cancer Commun. 2022 Feb;42(2):117–140. doi: 10.1002/cac2.12254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo H., Wu L., Yang Q., et al. Functional linc-POU3F3 is overexpressed and contributes to tumorigenesis in glioma. Gene. 2015 Jan 1;554(1):114–119. doi: 10.1016/j.gene.2014.10.038. [DOI] [PubMed] [Google Scholar]
- 33.Garbo S., Tripodi M., Battistelli C. lncRNA HOTAIR functions and therapeutic perspectives. Oncoscience. 2022 Sep 13;9:49–51. doi: 10.18632/oncoscience.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pastori C., Kapranov P., Penas C., et al. The Bromodomain protein BRD4 controls HOTAIR, a long noncoding RNA essential for glioblastoma proliferation. Proc. Natl. Acad. Sci. U.S.A. 2015 Jul 7;112(27):8326–8331. doi: 10.1073/pnas.1424220112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J., Zhao J., Hu P., Gao L., Tian S., He Z. Long non-coding RNA HOTAIR in central nervous system disorders: new insights in pathogenesis, diagnosis, and therapeutic potential. Front. Mol. Neurosci. 2022 Jun 23;15 doi: 10.3389/fnmol.2022.949095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao Y., Ma J., Xue Y., et al. Knockdown of long noncoding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. 2015 Apr 1;359(1):75–86. doi: 10.1016/j.canlet.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 37.Qin X., Yao J., Geng P., et al. LncRNA TSLC1-AS1 is a novel tumor suppressor in glioma. Int. J. Clin. Exp. Pathol. 2014;7(6):3065–3072. [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Y., Zhang X., Qi L., et al. HULC long noncoding RNA silencing suppresses angiogenesis by regulating ESM-1 via the PI3K/Akt/mTOR signaling pathway in human gliomas. Oncotarget. 2016 Mar 22;7(12):14429–14440. doi: 10.18632/oncotarget.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhen L., Yun-Hui L., Hong-Yu D., et al. Long noncoding RNA NEAT1 promotes glioma pathogenesis by regulating miR- 449b-5p/c-met axis. Tumour Biol. 2016 Jan;37(1):673–683. doi: 10.1007/s13277-015-3843-y. [DOI] [PubMed] [Google Scholar]
- 40.Wang P., Liu Y.H., Yao Y.L., et al. Long noncoding RNA CASC2 suppresses malignancy in human gliomas by miR-21. Cell. Signal. 2015 Feb;27(2):275–282. doi: 10.1016/j.cellsig.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 41.Ebrahimi A.A., Ashoori H., Vahidian F., Mosleh I.S., Kamian S. Long non-coding RNA panel as a molecular biomarker in glioma. J. Egypt. Natl. Cancer Inst. 2021 Oct 25;33(1):31. doi: 10.1186/s43046-021-00090-4. [DOI] [PubMed] [Google Scholar]
- 42.Sun W., Wu Y., Yu X., et al. Decreased expression of long noncoding RNA AC096655.1-002 in gastric cancer and its clinical significance. Tumour Biol. 2013 Oct;34(5):2697–2701. doi: 10.1007/s13277-013-0821-0. [DOI] [PubMed] [Google Scholar]
- 43.Shi Y., Wang Y., Lung W., et al. Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PLoS One. 2014 Jan 23;9(1) doi: 10.1371/journal.pone.0086295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weroha S.J., Haluska P. The insulin-like growth factor system in cancer. Endocrinol Metab. Clin. N. Am. 2012 Jun;41(2):335–350. doi: 10.1016/j.ecl.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cao W., Liu B., Ma H. Long non-coding RNA GHET1 promotes viability, migration and invasion of glioma cell line U251 by down-regulation of miR-216a. Eur. Rev. Med. Pharmacol. Sci. 2019 Feb;23(4):1591–1599. doi: 10.26355/eurrev_201902_17118. [DOI] [PubMed] [Google Scholar]
- 46.Han Y., Li X., Yan J., et al. Knockdown of LncRNA PVT1 inhibits glioma progression by regulating miR-424 expression. Oncol. Res. 2019 Jun 21;27(6):681–690. doi: 10.3727/096504018X15424939990246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang R., Jin H., Lou F. The long non-coding RNA TP73-AS1 interacted with miR-142 to modulate brain glioma growth through HMGB1/RAGE pathway. J. Cell. Biochem. 2018 Apr;119(4):3007–3016. doi: 10.1002/jcb.26021. [DOI] [PubMed] [Google Scholar]
- 48.Amornsupak K., Thongchot S., Thinyakul C., Box C., Hedayat S., Thuwajit P., Eccles S.A., Thuwajit C. HMGB1 mediates invasion and PD-L1 expression through RAGE-PI3K/AKT signaling pathway in MDA-MB-231 breast cancer cells. BMC Cancer. 2022 May 24;22(1):578. doi: 10.1186/s12885-022-09675-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zang W., Wang T., Wang Y., et al. Knockdown of long noncoding RNA TP73-AS1 inhibits cell proliferation and induces apoptosis in esophageal squamous cell carcinoma. Oncotarget. 2016 Apr 12;7(15):19960–19974. doi: 10.18632/oncotarget.6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bid H.K., Roberts R.D., Manchanda P.K., et al. An emerging therapeutic option for targeting cancer angiogenesis and metastasis. Mol. Cancer Therapeut. 2013 Oct;12(10):1925–1934. doi: 10.1158/1535-7163.MCT-13-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ding F., Tang H., Nie D., et al. Long non-coding RNA Fer- 1-like family member 4 is overexpressed in human glioblastoma and regulates the tumorigenicity of glioma cells. Oncol. Lett. 2017 Aug;14(2):2379–2384. doi: 10.3892/ol.2017.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang X., Zhou Y., Mehta K.R., et al. A pituitary derived MEG3 isoform functions as a growth suppressor in tumor cells. J. Clin. Endocrinol. Metabol. 2003 Nov;88(11):5119–5126. doi: 10.1210/jc.2003-030222. [DOI] [PubMed] [Google Scholar]
- 53.Momtazmanesh S., Rezaei N. Long non-coding RNAs in diagnosis, treatment, prognosis, and progression of glioma: a state-of-the-art review. Front. Oncol. 2021 Jul 12;11 doi: 10.3389/fonc.2021.712786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yue H., Zhu J., Xie S., et al. MDC1-AS, an antisense long noncoding RNA, regulates cell proliferation of glioma. Biomed. Pharmacother. 2016 Jul;81:203–209. doi: 10.1016/j.biopha.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 55.Yao J., Zhou B., Zhang J., et al. A new tumor suppressor LncRNA ADAMTS9-AS2 is regulated by DNMT1 and inhibits migration of glioma cells. Tumour Biol. 2014 Aug;35(8):7935–7944. doi: 10.1007/s13277-014-1949-2. [DOI] [PubMed] [Google Scholar]
- 56.Rynkeviciene R., Simiene J., Strainiene E., et al. Non- coding RNAs in glioma. Cancers. 2018 Dec 22;11(1):17. doi: 10.3390/cancers11010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wei C., Zhang X., Peng D., Zhang X., Guo H., Lu Y., Luo L., Wang B., Li Z., He Y., Du X., Zhang S., Liang H., Li S., Wang S., Han L., Zhang J. LncRNA HOXA11-AS promotes glioma malignant phenotypes and reduces its sensitivity to ROS via Tpl2-MEK1/2-ERK1/2 pathway. Cell Death Dis. 2022 Nov 9;13(11):942. doi: 10.1038/s41419-022-05393-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu Z., Che N., He Y., Zhang B. ceRNA network of lncRNA MIR210HG/miR-377-3p/LMX1A in malignant proliferation of glioma cells. Genes Genomics. 2022 Dec;44(12):1445–1455. doi: 10.1007/s13258-022-01312-2. [DOI] [PubMed] [Google Scholar]
- 59.Li Z., Song Y., Zhang J. lncRNA PDCD4-AS1 promotes the progression of glioma by regulating miR-30b-3p/METTL7B signaling. Oxid. Med. Cell. Longev. 2023 Apr 28;2023 doi: 10.1155/2023/3492480. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 60.Jiang X., Zhang Y., Yuan Y., Jin Z., Zhai H., Liu B., Li Y., Zhang C., Chen M., Shi Y., Yan D., Pu J., Chen Y., Yang C. LncRNA GSCAR promotes glioma stem cell maintenance via stabilizing SOX2 expression. Int. J. Biol. Sci. 2023 Mar 5;19(6):1681–1697. doi: 10.7150/ijbs.80873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang J., Chen Y., Wang Q., Xu H., Jiang Q., Wang M., Li S., Chen Y., Wu C., Yu P., Xiao Z., Chen W., Lan Q. LncRNA SPRY4-IT1 facilitates cell proliferation and angiogenesis of glioma via the miR-101-3p/EZH2/VEGFA signaling axis. Cancer Med. 2023 Mar;12(6):7309–7326. doi: 10.1002/cam4.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu J., Wang J., Zhao M., Li C., Hong S., Zhang J. LncRNA LINC01018/miR-942-5p/KNG1 axis regulates the malignant development of glioma in vitro and in vivo. CNS Neurosci. Ther. 2023 Feb;29(2):691–711. doi: 10.1111/cns.14053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Su H., Hailin Z., Dongdong L., Jiang Y., Shuncheng H., Shun Z., Dan L., Biao P. Long non-coding RNA LINC01018 inhibits human glioma cell proliferation and metastasis by directly targeting miRNA-182-5p. J. Neuro Oncol. 2022 Oct;160(1):67–78. doi: 10.1007/s11060-022-04113-5. [DOI] [PubMed] [Google Scholar]
- 64.Wang R., Wang J., Wang Y., Yang L. lncRNA TUSC7 sponges miR-10a-5p and inhibits BDNF/ERK pathway to suppress glioma cell proliferation and migration. Aging (Albany NY) 2023 Apr 14;15(8):3021–3034. doi: 10.18632/aging.204655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shu B., Gan H., Wang C., Cao C., Tong H., Liang D. LINC01426 aggravates the malignant progression of glioma through miR-661/Mdm2 axis. Brain Res. Bull. 2022 Oct 1;188:110–121. doi: 10.1016/j.brainresbull.2022.06.012. [DOI] [PubMed] [Google Scholar]
- 66.Yu W., Ma L., Li X. DANCR promotes glioma cell autophagy and proliferation via the miR-33b/DLX6/ATG7 axis. Oncol. Rep. 2023 Feb;49(2):39. doi: 10.3892/or.2023.8476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou Q., Liu Z.Z., Wu H., Kuang W.L. LncRNA H19 promotes cell proliferation, migration, and angiogenesis of glioma by regulating Wnt5a/β-catenin pathway via targeting miR-342. Cell. Mol. Neurobiol. 2022 May;42(4):1065–1077. doi: 10.1007/s10571-020-00995-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qin X., Yao J., Geng P., et al. LncRNA TSLC1-AS1 is a novel tumor suppressor in glioma. Int. J. Clin. Exp. Pathol. 2014;7(6):3065–3072. [PMC free article] [PubMed] [Google Scholar]
- 69.Min W., Wang J., Zhang D., et al. Long noncoding RNAmiR210HG as a potential biomarker for the diagnosis of glioma. PLoS One. 2016 Sep 27;11(9) doi: 10.1371/journal.pone.0160451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li X., Zhang H., Wu X. Long noncoding RNA DLX6-AS1 accelerates the glioma carcinogenesis by competing endogenous sponging miR-197-5p to relieve E2F1. Gene. 2019 Feb 20;686:1–7. doi: 10.1016/j.gene.2018.10.065. [DOI] [PubMed] [Google Scholar]
- 71.Mei J.C., Yan G., Mei S.Q. Diagnostic and prognostic potentials of long noncoding RNA ELF3-AS1 in glioma patients. Dis. Markers. 2020 Sep 18;2020 doi: 10.1155/2020/8871746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang D., Zhou H., Liu J., Mao J. Long noncoding RNA ASB16-AS1 promotes proliferation, migration, and invasion in glioma cells. BioMed Res. Int. 2019 Mar 4;2019 doi: 10.1155/2019/5437531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huang Q., Chen H., Zuo B., Cheng C., Yu W., Yang Y. lncRNA NEF inhibits glioma by downregulating TGF-β1. Exp. Ther. Med. 2019 Jul;18(1):692–698. doi: 10.3892/etm.2019.7602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sun Y., Jing Y., Zhang Y. Serum lncRNA-ANRIL and SOX9 expression levels in glioma patients and their relationship with poor prognosis. World J. Surg. Oncol. 2021 Sep 23;19(1):287. doi: 10.1186/s12957-021-02392-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tan S.K., Pastori C., Penas C., Sarkaria J., Komotar R.J., Ayad N. GENE-03. Serum long noncoding RNA HOTAIR as novel diagnostic and prognostic biomarker in glioblastoma multiforme. Neuro Oncol. 2017 Nov;19(Suppl 6):vi92. doi: 10.1093/neuonc/nox168.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jiang X., Zhang Y., Yuan Y., Jin Z., Zhai H., Liu B., Li Y., Zhang C., Chen M., Shi Y., Yan D., Pu J., Chen Y., Yang C. LncRNA GSCAR promotes glioma stem cell maintenance via stabilizing SOX2 expression. Int. J. Biol. Sci. 2023 Mar 5;19(6):1681–1697. doi: 10.7150/ijbs.80873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cao Y., Wang P., Ning S., Xiao W., Xiao B., Li X. Identification of prognostic biomarkers in glioblastoma using a long non-coding RNA-mediated, competitive endogenous RNA network. Oncotarget. 2016;7:41737–41747. doi: 10.18632/oncotarget.9569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang X.Q., Sun S., Lam K.F., Kiang K.M., Pu J.K., Ho A.S., Lui W.M., Fung C.F., Wong T.S., Leung G.K. A long non-coding RNA signature in glioblastoma multiforme predicts survival. Neurobiol. Dis. 2013;58:123–131. doi: 10.1016/j.nbd.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 79.Liu Y., Xu N., Liu B., Huang Y., Zeng H., Yang Z., He Z., Guo H. Long noncoding RNA RP11-838N2.4 enhances the cytotoxic effects of temozolomide by inhibiting the functions of miR-10a in glioblastoma cell lines. Oncotarget. 2016;7:43835–43851. doi: 10.18632/oncotarget.9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhi F., Wang Q., Xue L., Shao N., Wang R., Deng D., Wang S., Xia X., Yang Y. The use of three long non-coding RNAs as potential prognostic indicators of astrocytoma. PLoS One. 2015;10 doi: 10.1371/journal.pone.0135242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gao X, Cao Y, Li J, Wang C, He H. LncRNA TPT1-AS1 Sponges miR-23a-5p in Glioblastoma to Promote Cancer Cell Proliferation. Cancer Biother. Radiopharm. 2021 Sep;36(7):549–555. doi: 10.1089/cbr.2019.3484. [DOI] [PubMed] [Google Scholar]
- 82.Shang C., Guo Y., Hong Y., Xue Y.X. Long non-coding RNA TUSC7, a target of miR-23b, plays tumor-suppressing roles in human gliomas. Front. Cell. Neurosci. 2016;10:235. doi: 10.3389/fncel.2016.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang W., Yang F., Zhang L., Chen J., Zhao Z., Wang H., Wu F., Liang T., Yan X., Li J., et al. LncRNA profile study reveals four-lncRNA signature associated with the prognosis of patients with anaplastic gliomas. Oncotarget. 2016;7:77225–77236. doi: 10.18632/oncotarget.12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhou Y., Wang D.L., Pang Q. Long noncoding RNA SPRY4-IT1 is a prognostic factor for poor overall survival and has an oncogenic role in glioma. Eur. Rev. Med. Pharmacol. Sci. 2016;20:3035–3039. [PubMed] [Google Scholar]
- 85.Lv Q.L., Hu L., Chen S.H., Sun B., Fu M.L., Qin C.Z., Qu Q., Wang G.H., He C.J., Zhou H.H. A long noncoding RNA ZEB1-AS1 promotes tumorigenesis and predicts poor prognosis in glioma. Int. J. Mol. Sci. 2016;17:1431. doi: 10.3390/ijms17091431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pang J., Li K., Lau K., et al. KIAA0495/PDAM is frequently downregulated in oligodendroglial tumors and its knockdown by siRNA induces cisplatin resistance in glioma cells. Brain Pathol. 2010 Nov;20(6):1021–1032. doi: 10.1111/j.1750-3639.2010.00405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ma K., Wang H., Li X., et al. Long noncoding RNA MALAT1 associates with the malignant status and poor prognosis in glioma. Tumour Biol. 2015 May;36(5):3355–3359. doi: 10.1007/s13277-014-2969-7. [DOI] [PubMed] [Google Scholar]
- 88.Wang Q., Zhang J., Liu Y., et al. A novel cell cycle associated lncRNA, HOXA11-AS, is transcribed from the 5-prime end of the HOXA transcript and is a biomarker of progression in glioma. Cancer Lett. 2016 Apr 10;373(2):251–259. doi: 10.1016/j.canlet.2016.01.039. [DOI] [PubMed] [Google Scholar]
- 89.Yi W., Yan D., Wang D., Li Y. Smart drug delivery systems to overcome drug resistance in cancer immunotherapy. Cancer Biol. Med. 2023 May 2;20(4):248–267. doi: 10.20892/j.issn.2095-3941.2023.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Messaoudi K., Clavreul A., Lagarce F. Toward an effective strategy in glioblastoma treatment.Part Ⅰ:Resistance mechanisms and strategies to overcome resistance of glioblastoma to temozolomide. Drug Discov. Today. 2015 Jul;20(7):899–905. doi: 10.1016/j.drudis.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 91.Li H., Yuan X., Yan D., et al. Long non-coding RNA MAL AT1 decreases the sensitivity of resistant glioblastoma cell lines to Temozolomide. Cell. Physiol. Biochem. 2017;42(3):1192–1201. doi: 10.1159/000478917. [DOI] [PubMed] [Google Scholar]
- 92.Aryankalayil M.J., Chopra S., Levin J., et al. Radiation-induced long noncoding RNAs in a mouse model after whole-body irradiation. J. Radiat. Res. 2018 Mar;189(3):251–263. doi: 10.1667/RR14891.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li J., Ji X., Wang H. Targeting long noncoding RNA HMMRAS1 suppresses and radiosensitizes glioblastoma. Neoplasia. 2018 May;20(5):456–466. doi: 10.1016/j.neo.2018.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.He C., Jiang B., Ma J., et al. Aberrant NEAT1 expression is associated with clinical outcome in high grade glioma patients. Apmis. 2016 Mar;124(3):169–174. doi: 10.1111/apm.12480. [DOI] [PubMed] [Google Scholar]
- 95.Hu L., Lv Q., Chen S., et al. Up-regulation of long noncoding RNA AB073614 predicts a poor prognosis in patients with glioma. Int. J. Environ. Res. Publ. Health. 2016 Apr 19;13(4):433. doi: 10.3390/ijerph13040433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhou K., Zhang C., Yao H., et al. Knockdown of long noncoding RNA NEAT1 inhibits glioma cell migration and invasion via modulation of SOX2 targeted by miR-132. Mol. Cancer. 2018 Jul 27;17(1):105. doi: 10.1186/s12943-018-0849-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhi F., Wang Q., Xue L., et al. The use of three long noncoding RNAs as potential prognostic indicators of astrocytoma. PLoS One. 2015 Aug 7;10(8) doi: 10.1371/journal.pone.0135242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011 Mar 4;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 99.Jain R.K., Di Tomaso E., Duda D.G., et al. Angiogenesis in brain tumors. Nat. Rev. Neurosci. 2007 Aug;8(8):610–622. doi: 10.1038/nrn2175. [DOI] [PubMed] [Google Scholar]
- 100.Knizhnik A.V., Roos W.P., Nikolova T., et al. Survival and death strategies in glioma cells: autophagy, senescence and apoptosis triggered by a single type of temozolomide-induced DNA damage. PLoS One. 2013;8(1) doi: 10.1371/journal.pone.0055665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shimizu A., Nakayama H., Wang P., et al. Netrin-1 promotes glioblastoma cell invasiveness and angiogenesis by multiple pathways including activation of RhoA, cathepsin B, and cAMP-response element-binding protein. J. Biol. Chem. 2013 Jan 25;288(4):2210–2222. doi: 10.1074/jbc.M112.397398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Iwadate Y. Epithelial-mesenchymal transition in glioblastoma progression. Oncol. Lett. 2016 Mar;11(3):1615–1620. doi: 10.3892/ol.2016.4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chang Y., Wu Q., Tian T., et al. The influence of SRPK1 on glioma apoptosis, metastasis, and angiogenesis through the PI3K/Akt signaling pathway under normoxia. Tumour Biol. 2015 Aug;36(8):6083–6093. doi: 10.1007/s13277-015-3289-2. [DOI] [PubMed] [Google Scholar]
- 104.Bauer H., Traweger A. Tight junctions of the blood brain barrier-a molecular gatekeeper. CNS Neurol. Disord. - Drug Targets. 2016;15(9):1016–1029. doi: 10.2174/1871527315666160915142244. [DOI] [PubMed] [Google Scholar]
- 105.Luissint A.C., Artus C., Glacial F., et al. Tight junctions at the blood brain barrier physiological architecture and disease associated dysregulation. Fluids Barriers CNS. 2012 Nov 9;9(1):23. doi: 10.1186/2045-8118-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ma J, Wang P, Yao Y, et al. Knockdown of long non-coding RNA MALAT1 increases the blood-tumor barrier permeability by up-regulating miR-140. Biochim. Biophys. Acta. 016 Feb;1859(2):324-338. doi: 10.1016/j.bbagrm.2015.11.008. [DOI] [PubMed]
- 107.Guo J., Cai H., Zheng J., et al. Long non-coding RNA NEAT1 regulates permeability of the blood-tumor barrier via miR-181d-5p-mediated expression changes in ZO-1, occludin, and claudin-5. Biochim. Biophys. Acta. 2017 Sep;1863(9):2240–2254. doi: 10.1016/j.bbadis.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 108.Cai H., Xue Y., Wang P., et al. The long noncoding RNA TUG1 regulates blood-tumor barrier permeability by targeting miR-144. Oncotarget. 2015 Aug 14;6(23):19759–19779. doi: 10.18632/oncotarget.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.A L., Li Y., Zhao L., et al. The role of HOTAIR/miR-148b-3p/USF1 on regulating the permeability of BTB. Front. Mol. Neurosci. 2017 Jun 28;10:194. doi: 10.3389/fnmol.2017.00194. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 110.Shen S., Yu H., Liu X., et al. PIWIL1/piRNA-DQ593109 regulates the permeability of the blood-tumor barrier via the MEG3/miR-330-5p/RUNX3 axis. Mol. Ther. Nucleic Acids. 2018 Mar 2;10:412–425. doi: 10.1016/j.omtn.2017.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]