Abstract
Oxidative stress is a common phenomenon in poultry production. Several molecules, including antioxidant genes, miRNAs, and gut microbiota metabolites, have been reported to participate in redox regulation. Lactiplantibacillus plantarum P8 (P8) was shown to improve the antioxidant capacity of chickens, but the specific molecular mechanisms remain unclear. In this study, 400 broilers were allocated to 4 treatment groups: control diet (Con group), control diet + dexamethasone injection (DEX group), control diet containing 1 × 108 CFU/g P8 (P8 group), and control diet containing 1 × 108 CFU/g P8 + DEX injection (DEX_P8 group). Integrated analysis of the microbiome, metabolomics, and miRNAomics was conducted to investigate the roles of P8 in oxidative stress in broilers. Results demonstrated that P8 supplementation significantly improved growth performance, jejunal morphology, and antioxidant function in DEX-treated broilers. Analysis of the gut microbiota revealed a higher abundance of Barnesiella (P = 0.01) and Erysipelatoclostridium (P = 0.05) in the DEX_P8 group than in the DEX group. Functional prediction indicated that certain pathways, including the phenylacetate degradation pathway, were enriched in the DEX_P8 group compared to the DEX group. Metabolites in the cecal contents were distinct between the groups. P8 supplementation increased the content of metabolites with antioxidant capacity, e.g., urobilinogen (P < 0.01), and decreased that of metabolites related to oxidative stress, e.g., genistein (P < 0.01). Functional prediction indicated that metabolites that differed between the DEX_P8 and DEX groups were enriched in pathways including “tryptophan metabolism” and “primary bile acid biosynthesis”. The miRNAomics analysis further showed that, compared to the DEX group, several miRNAs in the jejunum, such as gga-miR-21-3p (P = 0.03), were increased, whereas gga-miR-455-3p (P = 0.02) was decreased in the DEX_P8 group. The PI3K-Akt, Ras, and Rap1 signaling pathways were enriched in the DEX_P8 group compared to the DEX group through KEGG analysis. Correlation analysis revealed potential interactions between growth performance, oxidation/antioxidation, jejunal morphology, gut microbiota, cecal content metabolites, and jejunal miRNAs. Overall, our results indicate that P8 supplementation may improve the growth performance, jejunal morphology and antioxidant capacity of DEX-treated broilers by regulating gut microbiota, its metabolites, and intestinal miRNAs.
Keywords: Broiler, Oxidative stress, Lactiplantibacillus plantarum, Microbiome, Metabolomics, miRNAomics
1. Introduction
In commercial settings, poultry are exposed to a range of stressors, such as infection, high-density housing, and feed contamination. Protection against stress has gained much attention, not only because stress is associated with depressed production but also because consumer acceptance of poultry is influenced by animal welfare (Scanes, 2016). It has been reported that glucocorticoids are secreted by the adrenal gland when animals are under stress (Siegel, 1980). Excessive glucocorticoid levels are hallmarks of stress (Chrousos and Gold, 1992). Dexamethasone (DEX) is a synthetic glucocorticoid widely used to establish stress models (Gao et al., 2010; Osho and Adeola, 2020). Previous studies have shown that DEX induces redox imbalance (Gao et al., 2010), impairs immune function (Liu et al., 2021), and damages intestinal mucosal integrity (Barekatain et al., 2019) as well as the gut microbiota (Zhao et al., 2020).
The gut microbiota plays a crucial role in host health, including the regulation of antioxidant function (Marciano and Vajro, 2017; Dam et al., 2019). Thus, increasing interest has emerged in the prevention or treatment of oxidative stress through the regulation of gut microbiota. Probiotics are known to have a potent ability to improve gut microbiota (Gareau et al., 2010). The antioxidant properties of probiotics may be related to the regulatory effects of the gut microbiota. Feng et al. (2020) reported that Pediococcus acidilactici BT36 protects mice against Cr(VI)-induced oxidative stress by regulating the gut microbiota. Zhao et al. (2018) found that in D-galactose-induced aging mice, Lactiplantibacillus plantarum CCFM10 alleviates oxidative stress and restores gut microbiota. Wang et al. (2018) also suggested that L. plantarum ZDY2013 and Bifidobacterium bifidum WBIN03 ameliorate ulcerative colitis by modifying the gut microbiota to regulate oxidative stress. L. plantarum P8 (P8) is considered as a probiotic strain (Wang et al., 2021b). Studies have demonstrated that P8 plays beneficial roles in improving lipid metabolism in rats (Bao et al., 2012) and regulating intestinal morphology in juvenile turbots (Wang et al., 2016). Moreover, our group recently found that, in broilers, P8 ameliorates Eimeria-induced intestinal oxidative stress by regulating the gut microbiota (Wang et al., 2021b) and attenuates DEX-induced immunological stress (Liu et al., 2021).
Through metabolite production, the gut microbiota modulates the signaling pathways involved in the homeostasis of the intestinal mucosa. Thus, metabolomics has been applied to obtain detailed information on gut metabolic pathways (Pamela et al., 2016). miRNAs are a class of short, endogenously initiated non-coding RNAs (Cai et al., 2009). miRNAomics is the dominant cellular tool that adjusts the levels of all RNA classes in post-transcriptional processes through RNA silencing and target genes (Shin et al., 2021). Compared to conventional methods, single omics methods are more helpful for the in-depth analysis of biological phenotypes, however, the integration of multi-omics methods allows a more comprehensive analysis (Shin et al., 2018; Sun et al., 2019).
In this study, we evaluated the effects of P8 on oxidative stress and intestinal health in a broiler model treated with DEX. Furthermore, microbiome, metabolomics, and miRNAomics analyses were used to investigate the underlying mechanisms at multiple levels of molecules.
2. Materials and methods
2.1. Animal ethics statement
The experimental protocols of this study were approved by the Institutional Animal Care and Use Committee of Qingdao Agricultural University, Qingdao, China (approval number: 20200813065). Animal experiments were approved and performed in accordance with the guidelines of the Ethics and Animal Welfare Committee of Qingdao Agricultural University.
2.2. Chemicals and reagents
P8 powder (1 × 1011 CFU/g) was purchased from Beijing Scitop Bio-tech Co., Ltd. (Beijing, China). DEX injection (5 mg/mL, Veterinary Drugs Production License No. 080061147) was purchased from the Beian Feilong Animal Pharmaceutical Factory (Heilongjiang, China).
2.3. Experimental design and sample collection
A total of 400 male Cobb 500 broilers at 1 d old were randomly allotted to 4 treatment groups (Con, DEX, P8, and DEX_P8 groups) with 10 replicates per group (10 broilers per replicate). Broilers in the control and DEX groups were fed a basal diet. Broilers in the P8 and DEX_P8 groups were fed a basal diet containing 1 × 108 CFU/g P8. Broilers were given free access to water and feed. At 16 d of age, broilers in the DEX and DEX_P8 groups were injected with 3 mg/kg body weight DEX (200 μL) (Wang, 2012), whereas broilers in the Con group were injected with an equal volume of saline. The experiment lasted for 21 d and the basal diet was formulated to meet the requirements of the National Research Council (NRC, 1994; Table 1). Fresh water and feed were provided ad libitum. The temperature of the room was set to 33 to 35 °C in the first week and then decreased to 2 °C every week up to 24 °C.
Table 1.
Composition and nutrient levels of basal diets (%, air-dry basis).
| Item | Content |
|---|---|
| Ingredients | |
| Corn | 61.38 |
| Soybean meal | 31.00 |
| Fish meal | 2.00 |
| Soybean oil | 2.00 |
| Limestone | 1.50 |
| CaHPO4 | 1.30 |
| DL-Met | 0.22 |
| NaCl | 0.30 |
| Premix1 | 0.20 |
| Choline chloride | 0.10 |
| Total | 100.00 |
| Nutrient levels2 | |
| ME, MJ/kg | 12.47 |
| CP | 20.55 |
| CF | 4.59 |
| Ca | 0.95 |
| AP | 0.46 |
| Lys | 1.10 |
| Met | 0.55 |
| Total sulfur amino acids | 0.84 |
| Try | 0.26 |
| Thr | 0.79 |
The premix provided the following per kilogram of diets: vitamin A 8,000 IU, vitamin B1 4.2 mg, vitamin B2 4 mg, vitamin B6 4.5 mg, vitamin B12 0.02 mg, vitamin D3 3,000 IU, vitamin E 20 IU, vitamin K3 2 mg, biotin 0.15 mg, folic acid 1.0 mg, D-pantothenic acid 11 mg, nicotinic acid 10 mg, Cu (as copper sulfate) 10 mg, Fe (as ferrous sulfate) 80 mg, Mn (as manganese sulfate) 80 mg, Zn (as zinc sulfate) 75 mg, I (as potassium iodide) 0.40 mg, Se (as sodium selenite) 0.30 mg.
The nutrient levels were calculated values.
2.4. Growth performance
Broilers were weighed at 1, 16 and 21 d of age, and feed intake was recorded. Average daily feed intake (ADFI), average daily gain (ADG) and feed:gain ratio (F:G) were calculated.
2.5. Sample collection
At the end of the experiment and after fasting for 12 h, one chicken per replicate was randomly selected. Blood samples were collected from the wing vein into vacuum tubes containing an anticoagulant and then centrifuged at 3,000 × g for 10 min at 4 °C for plasma separation, and stored at −20 °C. After blood collection, chickens were sacrificed by cervical dislocation. The segments of jejunum were collected and fixed in 10% buffered formaldehyde for 24 h. The mucosa was scraped from 10 cm of the jejunum using a sterile glass slide (5 cm proximal to the Meckel's diverticulum). The cecal lobes were excised and the cecal contents were aseptically squeezed and pooled into sterile recipients for Illumina sequencing. Samples, except the intestine segments in buffered formaldehyde, were immediately refrigerated on ice and stored at −80 °C until further analysis.
2.6. Intestinal morphology
The intestinal segments were embedded in paraffin, and sections of each sample were stained with hematoxylin and eosin (H&E). Villi were observed under a microscope (OLYMPUS, Japan) using the HMIAS-2000 image analysis system. Villus height was measured from the top of the villus to the villus crypt junction, and crypt depth was measured as the depth of invagination between adjacent villi.
2.7. Analysis of biochemical indices
The contents of malondialdehyde (MDA) and the activities of catalase (CAT), superoxide dismutase (SOD), and glutathione peroxidase (GPX) in the plasma and jejunal mucosa were detected using ELISA kits (Shanghai Enzyme-linked Biotechnology Co., Ltd.) according to the manufacturer's protocol (Sun et al., 2020). The absorbance was read using a microplate reader (SpectraMax iD3, Molecular Devices).
2.8. Reactive oxygen species (ROS) detection
Frozen jejunal sections were incubated with 5 mol/L dihydroethidium (Sigma–Aldrich, United States) for 30 min at 37 °C. After washing with PBS, ROS levels in the tissue were measured using confocal microscopy (Leica, Germany). The fluorescence intensity was quantified using ImageJ software (National Institutes of Health, USA) (Zhang et al., 2022).
2.9. Determination of protein expression levels by western blotting
A nuclear and cytoplasmic protein extraction kit (Beyotime Biotechnology, China) was used to extract proteins from the mucosa of the jejunum according to the manufacturer's protocol. Equal amounts of protein from each sample were subjected to SDS-PAGE, and the proteins on the gel were transferred to a polyvinylidene difluoride (PVDF) membrane. Membranes were blocked with 5% skimmed milk and then incubated with the primary antibodies (anti-β-actin, anti-Nrf2, anti-Keap1, and anti-HO-1) overnight at 4 °C. After washing with Tris-buffered saline Tween, the membranes were incubated with a secondary antibody adjusted with horseradish peroxidase (Beyotime Biotechnology, China). The blots were then developed using an electrochemiluminescence detection system, according to the manufacturer's instructions. Quantification of band intensities was performed using the ImageJ software (National Institutes of Health, USA) (Wang et al., 2021a).
2.10. DNA extraction and microbiomic analysis
The TIANamp stool DNA kit (Tiangen Biotech Co., Ltd., Beijing) was used for bacterial DNA extraction. DNA samples were quantified using a Qubit 2.0 Fluorometer (Invitrogen, USA) and DNA quality was confirmed using 0.8% agarose gel electrophoresis. The purified DNA was sent to Lianchuan Biotechnology Co., Ltd. (Hangzhou, China) for 16S rRNA analysis using the Illumia MiSeq platform. DNA samples were amplified by PCR using bar-coded primers flanking the V3–V4 region of the 16S rRNA genes. The V3–V4 hypervariable region of the 16S rRNA gene was amplified using the primer pair 341F (5′-CCTACGGGNGGCWGCAG-3′) and 805R (5′-GACTACHVGGGTATCTAATCC-3′). The thermocycle protocol includes 1 pre-denaturation cycle of 4 min at 94 °C, 25 cycles of 30 s at 94 °C, annealing for 45 s at 50 °C, elongation for 30 s at 72 °C, and 1 post-elongation cycle of 5 min at 72 °C. High-quality clean tags were generated by quality filtering of the raw tags based on QIIME2 (https://qiime2.org). Operational taxonomic units (OTUs) were clustered at 97% sequence similarity using uCLUST (version 1.2.22), and representative sequences of each cluster were used to assign taxonomy through annotation against the SILVA database (Release 132, https://www.arb-silva.de/documentation/release-132/). Indices of alpha diversity (Chao1, goods_coverage, observed_otus, Shannon, Simpson) of the samples were evaluated. Principal coordinate analysis (PCoA) was undertaken for all samples following analysis through application of Bray–Curtis dissimilarity and unweighted UniFrac using R version 3.5.1.
Taxonomic classification was achieved based on homology (>97% identity) between queried and reference sequences from the Greengenes database v13.8 (https://mothur.org/wiki/greengenes-formatted_databases). Subsequently, the OTU table was normalized using Langille Lab Online Galaxy Instance (http://galaxy.morganlangille.com), followed by metagenome functional prediction based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Significant differences in gene function among the groups were revealed by the Kruskal–Wallis test. All DNA datasets were submitted to the NCBI Sequence Read Archive database under BioProject ID PRJNA827668.
2.11. Metabolomic analysis
Metabolomic analysis of the cecal content was performed as described by Ma et al. (2018). Briefly, the cecal content was lyophilized, ground, and crushed. A total of 300 μL of pre-cooled methanol was added to the 50 mg cecal content sample, which was pre-cooled at −20 °C for 1 h and then homogenized at 60 Hz for 2 min. The sample was then extracted with ultrasonication for 30 min and centrifuged for 10 min at 4 °C and 12,000 × g. After centrifugation, the supernatant was filtered using a 0.22-μm filter membrane, and 200 μL of supernatant was transferred to a storage vial and stored at −80 °C until use for LC-MS analysis (Thermo, Ultimate 3000LC, Q Exactive HF).
Chromatographic separation was carried out using a Zorbax Eclipse C18 column (i.d. 1.8 μm, 100 mm × 2.1 mm, Agilent Technology). The mobile phase consisted of 0.1% formic acid in water (solvent A) and acetonitrile (solvent B). Metabolites were eluted stepwise, at a flow rate of 0.3 mL/min: 0 to 2 min, 5% B; 2 to 7 min, 30% B; 7 to 14 min, 78% B; 14 to 20 min, 95% B; and 20 to 25 min, 5% B. The column temperature was 30 °C, and the injection volume was 2 μL. High resolution Q Exactive HF mass spectrometry was carried out in positive and negative ion modes of electrospray ionization (ESI) with the following settings: spray voltage, 3.50 kV; capillary temperature (ion transfer tube), 330 °C; heater temperature, 325 °C. Compound Discoverer 3.1 was used for retention time correction, peak recognition, peak extraction, peak integral, peak alignment, etc. Simultaneously, the Thermo mzCloud online database, the Thermo mzValut local database, and the ChemSpider database were used for substance identification. The quality control (QC) sample was prepared by mixing the sample extract and was used to analyze the repeatability of the sample using the same treatment method. During instrumental analysis, two QC samples were inserted into each test group to investigate the repeatability of the analysis process. Orthogonal partial least squares discriminant analysis (OPLS-DA) was performed using SIMCA software (V14.1; MKS Data Analytics Solutions, Umea, Sweden). The differential metabolites were screened by combining the P-value of Student's t-test with multiple differences (fold-change). Screening criteria: P < 0.01, metabolites with fold-change ≥2 and ≤0.5. The KEGG database was used to annotate the differential metabolites. MetaboAnalystR was used for the enrichment analysis based on the annotation results.
2.12. miRNA sequencing
Total RNA from the jejunal mucosa was isolated using TRIzol reagent (Tiangen Biochemical Technology, Beijing), and RNA purity and integrity were evaluated using a spectrophotometer (NanoDrop 2000c, Thermo Scientific, USA) and agarose gel electrophoresis. Extracted RNA was sent to Sci-Tech Testing (Qingdao, China) for miRNA sequencing. Briefly, for all generated raw reads, adaptor sequences, low-quality reads (Sanger base quality < 20), and contaminant reads were removed using Fastx-Toolkit software (http://hannonlab.cshl.edu/fastx_toolkit/) to obtain clean reads. Clean reads from 18 to 32 nt were counted and identical sequences were eliminated. Subsequently, the assembled unique sequences were identified for the type and number of small RNA (rRNAs, tRNA, sn/snoRNAs, miRNAs, and other noncoding RNAs) using the Rfam databases 14.7 (http://rfam.xfam.org/). The sequences were mapped to the chicken genome to annotate the location in the chromosomes using Bowtie (http://bowtie-bio.sourceforge.net/index.shtml). Finally, the remaining sequences were analyzed by BLASTing miRbase 22.1 (http://www.mirbase.org/) to identify known miRNAs in chickens. Novel miRNAs were predicted using miRDeep2 (https://www.mdc-berlin.de/8551903/en/) from the unannotated sRNAs. Both known and novel miRNAs were used to predict potential miRNA targets using RNAhybrid (http://bibiserv.techfak.uni-biele.org.de/rnahybrid/) and miRDB (http://mirdb.org), respectively. False discovery rate (FDR) was used for multiple test correction in miRNA sequencing analyses and the adjusted P-value < 0.05 was considered statistically significant. All target genes of differentially expressed miRNAs were subjected to KEGG pathway enrichment analysis using DAVID 6.7 Functional Annotation Tool (http://david.abcc.ncifcrf.gov/) (Ouyang et al., 2015). All DNA datasets were submitted to the NCBI Sequence Read Archive database under BioProject ID PRJNA824247.
2.13. Statistics
Oxidative stress-related parameters, jejunal morphology, and protein expression were assessed by one-way ANOVA using SPSS 20.0. When treatment effects were significant (P < 0.05), Tukey's multiple range test was used to compare pairs of means, and Duncan's multiple comparison test was used to compare differences between the 4 groups. The tabulated results are shown as means with SEM derived from the ANOVA error mean square. Spearman's correlation between each pair was tested, and the FDR correction was adopted to adjust the P-values. Clustering Spearman's correlation heatmap with signs was performed using the OmicStudio tools at https://www.omicstudio.cn (FDR < 0.05).
3. Results
3.1. P8 improved the growth performance of broilers
The effects of P8 on the growth performance of broilers with DEX challenge are shown in Table 2. There were no significant differences among groups in terms of ADFI, ADG and F:G ratio during d 1 to 16 (P > 0.05). During d 17 to 21, no significant differences were observed in the ADFI among groups. Moreover, compared to the Con group, DEX treatment significantly decreased the ADG (P < 0.01) and increased the F:G ratio (P < 0.01). Compared to the DEX group, DEX_P8 treatment significantly increased ADG (P < 0.01) and decreased the F:G ratio (P < 0.01). Furthermore, during d 1 to 21, there were no significant differences in ADFI among groups (P > 0.05). Compared to the Con group, DEX treatment significantly decreased ADG (P < 0.01) and increased the F:G ratio (P < 0.01). DEX_P8 treatment significantly increased the ADG (P < 0.01) and decreased the F:G ratio (P < 0.01) compared to the DEX group.
Table 2.
Effects of Lactiplantibacillus plantarum P8 on the growth performance of broilers.
| Item | Con | DEX | P8 | DEX_P8 | SEM | P-value |
|---|---|---|---|---|---|---|
| Days 1 to 16 | ||||||
| ADFI, g | 35.54 | 36.45 | 35.90 | 36.65 | 0.52 | 0.14 |
| ADG, g | 26.09 | 26.08 | 26.42 | 26.42 | 0.19 | 0.12 |
| F:G ratio, g/g | 1.36 | 1.40 | 1.36 | 1.39 | 0.02 | 0.17 |
| Days 17 to 21 | ||||||
| ADFI, g | 73.45 | 71.92 | 73.52 | 72.59 | 0.31 | 0.15 |
| ADG, g | 45.17a | 32.61c | 45.56a | 40.66b | 0.29 | <0.01 |
| F:G ratio, g/g | 1.63c | 2.24a | 1.61c | 1.81b | 0.02 | <0.01 |
| Days 1 to 21 | ||||||
| ADFI, g | 45.79 | 45.33 | 45.65 | 44.68 | 0.633 | 0.28 |
| ADG, g | 33.80a | 29.05c | 33.97a | 31.82b | 0.629 | <0.01 |
| F:G ratio, g/g | 1.36b | 1.56a | 1.35b | 1.41b | 0.030 | <0.01 |
Con = control diet; DEX = control diet plus dexamethasone injection; P8 = control diet containing 1 × 108 CFU/g Lactiplantibacillus plantarum P8; DEX_P8 = control diet containing 1 × 108 CFU/g L.plantarum P8 plus dexamethasone injection.
a,b,cMean values within a row with no common superscripts differ significantly (P < 0.05).
3.2. P8 improved the jejunal morphology and attenuated the oxidative stress of broilers
Compared to the Con group, DEX treatment significantly decreased the villus height and the ratio of villus height to crypt depth (V:C) (P < 0.01), and P8 treatment had no significant influence on villus height, crypt depth, and V:C ratio (P > 0.05). Moreover, DEX_P8 treatment led to increased villus height and V:C ratio (P < 0.01) compared to the DEX group (Fig. 1A).
Fig. 1.
Effects of Lactiplantibacillus plantarum P8 (P8) on the jejunal morphology and antioxidant capacity of oxidatively stressed broilers. (A) Effects of P8 on the jejunal morphology of oxidatively stressed broilers. (B) Effects of P8 on biochemical indices in the plasma and jejunal mucosa of oxidatively stressed broilers. (C) Effects of P8 on ROS in the jejunal tissue of oxidatively stressed broilers. Con = control diet; DEX = control diet plus dexamethasone injection; P8 = control diet containing 1 × 108 CFU/g Lactiplantibacillus plantarum P8; DEX_P8 = control diet containing 1 × 108 CFU/g L.plantarum P8 plus dexamethasone injection; V:C ratio = villus height to crypt depth ratio; SOD = superoxidase; CAT = catalase; MDA = malondialdehyde; GPX = glutathione peroxidase. a–cMeans with no common superscripts differ significantly (P < 0.05).
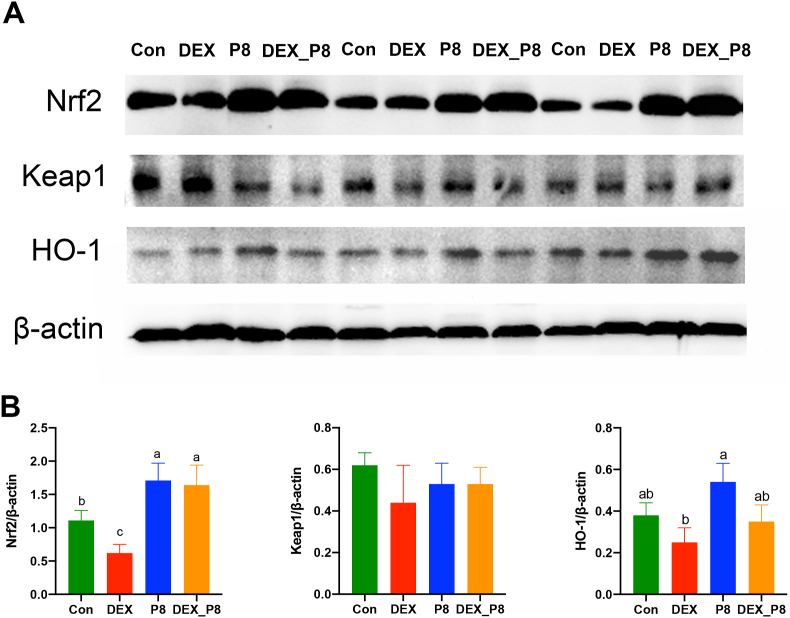
In plasma, DEX injection significantly decreased the level of CAT (P = 0.02), which was significantly increased by DEX_P8 treatment (P = 0.02). However, CAT levels were not significantly altered by P8 treatment compared to those in the Con group (P > 0.05). Moreover, there were no significant differences in the levels of SOD, GPX, or MDA among the groups (P > 0.05). In the jejunal mucosa, compared to the Con group, DEX treatment significantly decreased the levels of SOD (P = 0.01) and GPX (P = 0.01) and increased the levels of MDA (P < 0.01), whereas P8 treatment did not significantly influence the levels of SOD, CAT, GPX, or MDA (P > 0.05). Compared to the DEX group, the levels of SOD (P = 0.01) and GPX (P = 0.01) in jejunum were significantly elevated, and MDA levels in jejunum were significantly decreased in boilers receiving DEX_P8 (P < 0.01) (Fig. 1B). Additionally, ROS fluorescence showed that DEX treatment led to increased ROS production (P < 0.01), but P8 treatment did not significantly alter ROS levels (P > 0.05) in comparison to the Con group. In addition, compared with the DEX group, DEX_P8 treatment significantly lowered ROS production (P < 0.01) (Fig. 1C). As for the Keap1-Nrf2-HO-1 antioxidant signaling pathway, we found that Nrf2 expression was significantly downregulated in the DEX group compared to that in the Con group (P < 0.01). Moreover, P8 treatment significantly upregulated the expression of Nrf2 compared to that in the Con group (P < 0.01). Furthermore, compared to the DEX group, DEX_P8 treatment significantly upregulated the expression of Nrf2 (P < 0.01) (Fig. 2).
Fig. 2.
Effects of Lactiplantibacillus plantarum P8 on the Keap1-Nrf2-HO-1 antioxidant signaling pathway in the jejunal mucosa of oxidatively stressed broilers. Con = control diet; DEX = control diet plus dexamethasone injection; P8 = control diet containing 1 × 108 CFU/g L. plantarum P8; DEX_P8 = control diet containing 1 × 108 CFU/g L.plantarum P8 plus dexamethasone injection; Nrf2 = nuclear factor erythroid 2-related factor 2; Keap1 = kelch-like ECH-associated protein 1; HO-1 = heme oxygenase 1. a–cMeans with no common superscripts differ significantly (P < 0.05).
3.3. P8 regulated the gut microbiota of broilers
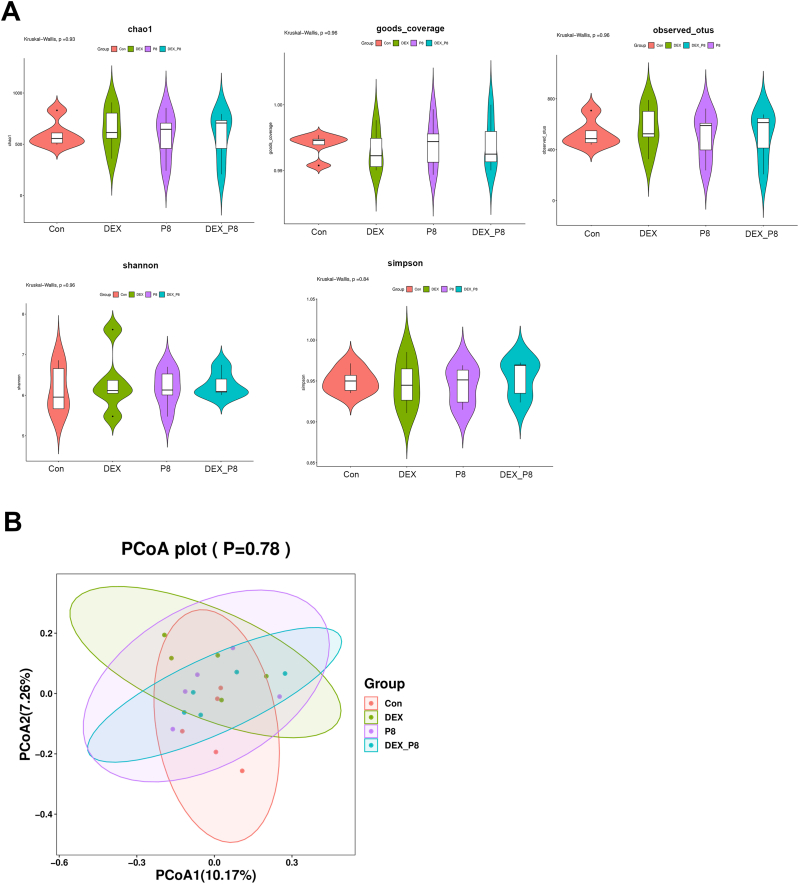
Alpha diversity of the gut microbiota was determined using the Chao1, goods_coverage, observed_otus, Shannon, and Simpson indices. As shown in Fig. 3, there were no significant differences in the α-diversity indices between the groups (P > 0.05). Moreover, β-diversity analysis demonstrates that the gut microbiota was not distinct among the different groups (P > 0.05). Nevertheless, specific differences in the bacterial phyla, families and genera were observed (Table 3, Table 4, Table 5, Fig. 4A). At the phylum level, the relative abundance of Bacteroidetes was significantly decreased in the DEX group compared with that in the Con group (P = 0.03). At the family level, the relative abundance of Barnesiellaceae (family) in broilers of the DEX (P = 0.01) and P8 groups (P = 0.05) was lower than that in the Con group. However, Barnesiellaceae (family) was more abundant in the DEX_P8 group than in the DEX group (P = 0.01). As for the top 30 genera, DEX treatment led to decreased relative abundance of Barnesiella (P = 0.01). P8 treatment reduced the relative abundance of Barnesiella (P = 0.05) and Clostridiales_unclassified (P = 0.02) and increased the abundance of Enterococcus (P = 0.05). Moreover, compared to broilers in the DEX group, those in the DEX_P8 group had a higher abundance of Barnesiella (P = 0.01) and Erysipelatoclostridium (P = 0.05). Linear discriminant analysis (LDA) effect size (LEfSe) analysis was performed to identify taxonomic biomarkers in the gut microbiota. According to the results, the relative abundance of Barnesiellaceae (family), Barnesiella (genus) and Ruminococcus_torques_group (genus) was increased in the Con group. The relative abundance of Holdemania (genus) was increased in the DEX group. The relative abundance of Enterobacter (genus), Enterobacterales (order), Enterobacteriaceae (family), and Marvinbryantia (genus) increased in the P8 group. Moreover, the relative abundance of Anaerostipes (genus) and Flavonifractor (genus) was increased in the DEX_P8 group (Fig. 4B).
Fig. 3.
Effects of Lactiplantibacillus plantarum P8 on the α-diversity (A) and β-diversity (B) of microflora of oxidatively stressed broilers. Con = control diet; DEX = control diet plus dexamethasone injection; P8 = control diet containing 1 × 108 CFU/g L. plantarum P8; DEX_P8 = control diet containing 1 × 108 CFU/g L.plantarum P8 plus dexamethasone injection.
Table 3.
Significantly altered bacterial abundance between the DEX and Con groups.
| Bacteria | Up/down | log2FC | P-value |
|---|---|---|---|
| p__Bacteroidetes | down | −1.49 | 0.03 |
| f__Barnesiellaceae | down | −1.49 | 0.01 |
| g__Holdemania | up | Inf | 0.01 |
| g__Barnesiella | down | −3.98 | 0.01 |
| g__Ruminococcaceae_UCG-013 | up | 1.24 | 0.01 |
| g _Coprobacter | down | −4.94 | 0.02 |
| g__Anaerostipes | up | 2.59 | 0.02 |
| g__Eubacterium]_brachy_group | up | Inf | 0.02 |
| g__Lachnospiraceae_FCS020_group | up | 2.53 | 0.03 |
| g__Family_XIII_AD3011_group | up | 1.96 | 0.05 |
| g__Merdibacter | up | 2.99 | 0.05 |
| g__Marvinbryantia | down | −1.77 | 0.05 |
Con = control diet; DEX = control diet plus dexamethasone injection; FC = fold change.
Table 4.
Significantly altered bacterial abundance between the P8 and Con groups.
| Bacteria | Up/down | log2FC | P-value |
|---|---|---|---|
| f__Rhodospirillaceae | down | −2.43 | 0.02 |
| f__Clostridiales_unclassified | down | −1.51 | 0.02 |
| f__Sporichthyaceae | up | Inf | 0.02 |
| f__Enterococcaceae | up | 1.34 | 0.05 |
| f__Barnesiellaceae | down | −1.30 | 0.05 |
| g__Clostridiales_unclassified | down | −1.51 | 0.02 |
| g__Enterobacter | up | Inf | 0.02 |
| g__Holdemania | up | Inf | 0.02 |
| g__hgcI_clade | up | Inf | 0.02 |
| g__Thalassospira | down | −1.94 | 0.03 |
| g__Coprobacter | down | −2.50 | 0.04 |
| g__Enterococcus | up | 1.34 | 0.05 |
| g__Barnesiella | down | −1.26 | 0.05 |
| g__Merdibacter | up | 2.12 | 0.05 |
Con = control diet; P8 = control diet containing 1 × 108 CFU/g Lactiplantibacillus plantarum P8; FC = fold change.
Table 5.
Significantly altered bacterial abundance between the DEX_P8 and DEX groups.
| Bacteria | Up/down | log2FC | P-value |
|---|---|---|---|
| f__Barnesiellaceae | up | 3.50 | 0.01 |
| g__Barnesiella | up | 3.49 | 0.01 |
| g__Enterobacter | up | Inf | 0.02 |
| g__Family_XIII_AD3011_group | down | −2.54 | 0.03 |
| g__Family_XIII_UCG-001 | down | −1.27 | 0.03 |
| g__Erysipelatoclostridium | up | 1.06 | 0.05 |
| g__Defluviitaleaceae_UCG-011 | up | 1.88 | 0.05 |
DEX = control diet plus dexamethasone injection; DEX_P8 = control diet containing 1 × 108 CFU/g Lactiplantibacillus plantarum P8 plus dexamethasone injection; FC = fold change.
Fig. 4.
Effects of Lactiplantibacillus plantarum P8 (P8) on the gut microbiota of oxidatively stressed broilers. (A) Effects of P8 on the abundance of bacterial phylum, family and genus of oxidatively stressed broilers. (B) Effects of P8 on linear discriminant analysis of microflora of oxidatively stressed broilers. Con = control diet; DEX = control diet plus dexamethasone injection; P8 = control diet containing 1 × 108 CFU/g L. plantarum P8; DEX_P8 = control diet containing 1 × 108 CFU/g L.plantarum P8 plus dexamethasone injection.
PICRUSt analysis was conducted to investigate the potential function of the gut microbiota and predict their classification based on the KEGG pathways. Based on Fig. 5, compared to the Con group, the “gluconeogenesis I” pathway was less enriched in the DEX group. Moreover, 6 pathways, including “methanogenesis from acetate”, “superpathway of L-arginine, putrescine, and 4-aminobutanoate degradation”, “superpathway of L-arginine and L-ornithine degradation”, “norspermidine biosynthesis”, “mycothiol biosynthesis” and “mono-trans, poly-cis decaprenyl phosphate biosynthesis” increased, whereas 3 pathways, including “glutaryl-CoA degradation”, “chlorophyllide a biosynthesis III (aerobic, light independent)” and “chlorophyllide a biosynthesis II (anaerobic)” decreased in the P8 group compared to the Con group. In addition, in comparison to the DEX group, 5 pathways, including the “phenylacetate degradation I (aerobic)”, “superpathway of L-arginine, putrescine, and 4-aminobutanoate degradation”, “superpathway of L-arginine and L-ornithine degradation”, “norspermidine biosynthesis” and “4-hydeoxyphenylacetate degradation” were elevated in the DEX_P8 group (Fig. 5).
Fig. 5.
Effects of Lactiplantibacillus plantarum P8 on the functional prediction of microflora of oxidatively stressed broilers. Con = control diet; DEX = control diet plus dexamethasone injection; P8 = control diet containing 1 × 108 CFU/g L. plantarum P8; DEX_P8 = control diet containing 1 × 108 CFU/g L.plantarum P8 plus dexamethasone injection.
3.4. P8 regulated the metabolites of the gut microbiota of broilers
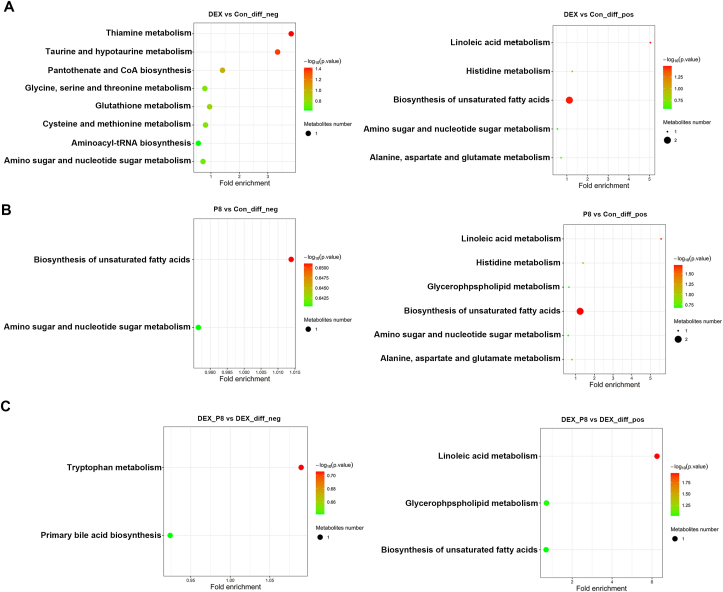
A total of 1,258 metabolites in positive mode and 1,262 metabolites in negative mode were identified in the cecal contents from the Con, DEX, P8, and DEX_P8 groups. To investigate the differences between groups, the OPLS-DA model was used to evaluate the metabolites. We observed a clear separation from the OPLS-DA score plots between the Con and DEX groups, Con and P8 groups, and DEX and DEX_P8 groups (Fig. 6A). Heatmap analysis was used to further investigate metabolite differences. We observed significant different metabolites both in positive and negative modes between Con and DEX groups, Con and P8 groups, DEX and DEX_P8 groups (Fig. 6B). The top 20 metabolites (10 in positive mode and 10 in negative mode) with multiple differences between groups are displayed in Table 6, Table 7, Table 8. As for the metabolites in the negative mode, compared to the Con group, DEX treatment led to increased levels of 9,10-dihydroxystearic acid, chrysin, and 7-deoxy-trans-dihydronarciclasine triacetate (P < 0.01), and decreased levels of pimelic acid and urobilinogen (P < 0.01) (Table 6); P8 treatment led to increased levels of apratoxin E, N-acetyl-D-galactosamine 4-sulfate, and deoxycholic acid (P < 0.01), and decreased levels of pimelic acid and (−)-stercobilin (P < 0.01) (Table 7). Compared to the DEX group, DEX_P8 treatment resulted in higher levels of urobilinogen, 4-chlorobenzamidoxime and N-acetyl-D-galactosamine 4-sulfate (P < 0.01), and lower levels of melilotocarpan C, pantoyltaurine, and mephenesin (P < 0.01) (Table 8). As for the metabolites in positive mode, compared to the Con group, DEX treatment led to increased levels of genistein, glycitein, and daidzein (P < 0.01) (Table 6); P8 treatment led to increased levels of linoleic acid, D-glucosamine 6-phosphate, and 3-ketocholanic acid (P < 0.01) (Table 7). Compared to the DEX group, DEX_P8 treatment resulted in higher levels of LPC 18:1, 3-ketocholanic acid, and docosahexaenoic acid ethyl ester (P < 0.01), and lower levels of genistein, 4-hexyn-1-ylbenzene and 4-(1,2-dihydroxyethyl)-2,6-piperidinedione (P < 0.01) (Table 8). KEGG analysis revealed that, in negative mode, the different metabolites between DEX and Con groups were enriched in the pathways including “thiamine metabolism”, “taurine and hypotaurine metabolism”, “panthothenate and CoA biosynthesis”, “glycine, serine and threonine metabolism”, “glutathione metabolism”, “cysteine and methionine metabolism”, “aminoacyl-tRNA biosynthesis” and “amino sugar and nucleotide sugar metabolism”. In the positive mode, the different metabolites between DEX and Con groups were enriched in the pathways including “linoleic acid metabolism”, “histidine metabolism”, “biosynthesis of unsaturated fatty acids”, “amino sugar and nucleotide sugar metabolism”, and “alanine, aspartate and glutamate metabolism”. Moreover, in negative mode, the different metabolites between P8 and Con groups were enriched in pathways including “biosynthesis of unsaturated fatty acids” and “amino sugar and nucleotide sugar metabolism”. In the positive mode, the different metabolites between P8 and Con groups were enriched in the pathways including “linoleic acid metabolism”, “histidine metabolism”, “glycerphospholipid metabolism”, “biosynthesis of unsaturated fatty acids”, “amino sugar and nucleotide sugar metabolism” and “alanine, aspartate and glutamate metabolism”. Furthermore, in negative mode, the different metabolites between the DEX_P8 and DEX groups were enriched in pathways including “tryptophan metabolism” and “primary bile acid biosynthesis”. In the positive mode, the different metabolites between DEX_P8 and DEX groups were enriched in the pathways including “linoleic acid metabolism”, “glycerphospholipid metabolism” and “biosynthesis of unsaturated fatty acids” (Fig. 7).
Fig. 6.
Effects of Lactiplantibacillus plantarum P8 on the metabolites of the gut microbiota of oxidatively stressed broilers. (A) OPLS-DA model was used to evaluate metabolites and explore differences between groups. (B) Significantly different metabolites both in positive and negative modes between groups. Con = control diet; DEX = control diet plus dexamethasone injection; P8 = control diet containing 1 × 108 CFU/g Lactiplantibacillus plantarum P8; DEX_P8 = control diet containing 1 × 108 CFU/g L.plantarum P8 plus dexamethasone injection.
Table 6.
Top 10 differential metabolites in negative and positive modes between the DEX and Con groups.
| Metabolites in negative mode | Up/down | log2FC | P-value |
|---|---|---|---|
| 9,10-Dihydroxystearic acid | up | 6.55 | <0.01 |
| Chrysin | up | 5.46 | <0.01 |
| 7-Deoxy-trans-dihydronarciclasine triacetate | up | 3.52 | <0.01 |
| 1-{3-Azido-2,3-dideoxy-5-O-[(4-hydroxybutoxy)carbonyl]pentofuranosyl}-5-methyl-2,4(1H,3H)-pyrimidinedione | up | 3.51 | <0.01 |
| (2S,3S,4S,5R,6R)-6-[[(3S,4S,6aR,6bS,8aR,9R,12aS,14bR)-9-hydroxy-4-(hydroxymethyl)-4,6a,6b,8a,11,11,14b-heptamethyl-1,2,3,4a,5,6,7,8,9,10,12,12a,14,14a-tetradecahydropicen-3-yl]oxy]-5-[(2S,3R,4S,5R,6R)-4,5-dihydroxy-6-(hydroxymethyl)-3-[(2S,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methyloxan-2-yl]oxyoxan-2-yl]oxy-3,4-dihydroxyoxane-2-carboxylic acid | up | 3.34 | <0.01 |
| Ricinelaidic acid | up | 3.23 | <0.01 |
| 3-Hydroxy-2-methyl-2-[(sulfooxy)methyl]propanoic acid | up | 2.71 | <0.01 |
| Deoxycholic acid | up | 2.56 | <0.01 |
| Pimelic acid | down | −4.79 | <0.01 |
| Urobilinogen |
down |
−2.95 |
<0.01 |
| Metabolites in positive mode | |||
| Genistein | up | 7.19 | <0.01 |
| Glycitein | up | 5.62 | <0.01 |
| Daidzein | up | 5.16 | <0.01 |
| Linoleic acid | up | 3.5 | <0.01 |
| D-Glucosamine 6-phosphate | up | 3.2 | <0.01 |
| MFCD00020106 | up | 3.09 | <0.01 |
| N-Acetyl-L-2-aminoadipic acid | up | 2.62 | <0.01 |
| Docosahexaenoic acid ethyl ester | up | 2.49 | <0.01 |
| (S)-1-(4-Methoxyphenyl)ethanol | up | 2.46 | <0.01 |
| N-Acetyl-DL-glutamic acid | up | 2.26 | <0.01 |
Con = control diet; DEX = control diet plus dexamethasone injection; FC = fold change.
Table 7.
Top 10 differential metabolites in negative and positive modes between the P8 and Con groups.
| Metabolites in negative mode | Up/down | log2FC | P-value |
|---|---|---|---|
| Apratoxin E | up | 4.71 | <0.01 |
| N-Acetyl-D-galactosamine 4-sulfate | up | 4.23 | <0.01 |
| 1-{3-Azido-2,3-dideoxy-5-O-[(4-hydroxybutoxy)carbonyl]pentofuranosyl}-5-methyl-2,4(1H,3H)-pyrimidinedione | up | 3.14 | <0.01 |
| 7-Deoxy-trans-dihydronarciclasine triacetate | up | 3.12 | <0.01 |
| 3-GluA Soyasapogenol B | up | 3.12 | <0.01 |
| N-Gluconyl ethanolamine phosphate | up | 2.97 | <0.01 |
| Deoxycholic acid | up | 2.72 | <0.01 |
| N-(2-morpholinoethyl)-4-(1H-pyrazol-1-yl)benzamide | up | 2.48 | <0.01 |
| Pimelic acid | down | −5.85 | <0.01 |
| (−)-Stercobilin |
down |
−4.31 |
<0.01 |
| Metabolites in positive mode | |||
| MFCD00020106 | up | 2.85 | <0.01 |
| D-Glucosamine 6-phosphate | up | 2.74 | <0.01 |
| Linoleic acid | up | 2.58 | <0.01 |
| 3-Ketocholanic acid | up | 2.48 | <0.01 |
| Docosahexaenoic acid ethyl ester | up | 2.48 | <0.01 |
| N-Acetyl-L-2-aminoadipic acid | up | 2.32 | <0.01 |
| (S)-1-(4-Methoxyphenyl)ethanol | up | 2.27 | <0.01 |
| N-Acetyl-DL-glutamic acid | up | 2.02 | <0.01 |
| Trigonelline | up | 2.02 | <0.01 |
| 7-Methyl-3-methylene-6-(3-oxobutyl)-3,3a,4,7,8,8a-hexahydro-2H-cyclohepta[b]furan-2-one | up | 1.95 | <0.01 |
Con = control diet; P8 = control diet containing 1 × 108 CFU/g Lactiplantibacillus plantarum P8; FC = fold change.
Table 8.
Top 10 differential metabolites in negative and positive modes between the DEX_P8 and DEX groups.
| Metabolites in negative mode | Up/down | log2FC | P-value |
|---|---|---|---|
| Methyl (9alpha,11alpha,13E,15S)-11,15-diacetoxy-9-hydroxyprost-13-en-1-oate | up | 2.38 | <0.01 |
| Urobilinogen | up | 2.33 | <0.01 |
| 4-Chlorobenzamidoxime | up | 1.95 | <0.01 |
| N-Acetyl-D-galactosamine 4-sulfate | up | 1.79 | <0.01 |
| (2S,5R)-N′-Acetyl-7-oxo-6-(sulfooxy)-1,6-diazabicyclo[3.2.1]octane-2-carbohydrazide | up | 1.74 | <0.01 |
| Melilotocarpan C | down | −2.82 | <0.01 |
| 8-[(2-Aminoethyl)sulfanyl]-1,3-dimethyl-2-thioxo-1,2,3,7-tetrahydro-6H-purin-6-one | down | −2.42 | <0.01 |
| Pantoyltaurine | down | −2.27 | <0.01 |
| 1-[(5-Ethyl-1,3,4-oxadiazol-2-yl)methyl]-6-indolinesulfonamide | down | −1.98 | <0.01 |
| Mephenesin |
down |
−1.79 |
<0.01 |
| Metabolites in positive mode | |||
| LPC 18:1 | up | 1.48 | <0.01 |
| 3-Ketocholanic acid | up | 1.33 | <0.01 |
| Docosahexaenoic acid ethyl ester | up | 1.32 | <0.01 |
| 4-(1,2-Dihydroxyethyl)-2,6-piperidinedione | down | −1.65 | <0.01 |
| (1R,3S,5S)-3-Methyl-8-oxa-6-azabicyclo[3.2.1]octane | down | −1.59 | <0.01 |
| 4-Hexyn-1-ylbenzene | down | −1.45 | <0.01 |
| (1S,2S,5S,6S,7R,8S,9R,12R)-2,6,10,10-Tetramethyl-11-oxatricyclo[7.2.1.0∼1,6∼]dodecane-2,5,7,8,12-pentol | down | −1.42 | <0.01 |
| 2,6-Di-tert-butyl-1,4-benzoquinone | down | −1.4 | <0.01 |
| 2-(6-Hepten-1-yl)-3-(2,4-pentadiyn-1-yl)oxirane | down | −1.39 | <0.01 |
| Genistein | down | −1.38 | <0.01 |
DEX = control diet plus dexamethasone injection; DEX_P8 = control diet containing 1 × 108 CFU/g Lactiplantibacillus plantarum P8 plus dexamethasone injection; FC = fold change.
Fig. 7.
KEGG enrichment analysis of the different metabolites between Con and DEX groups (A), Con and P8 groups (B), and DEX and DEX_P8 groups (C). Con = control diet; DEX = control diet plus dexamethasone injection; P8 = control diet containing 1 × 108 CFU/g Lactiplantibacillus plantarum P8; DEX_P8 = control diet containing 1 × 108 CFU/g L.plantarum P8 plus dexamethasone injection.
3.5. P8 regulated the intestinal miRNA profiles of broilers
miRNA sequencing was performed to identify the intestinal miRNA profiles of broilers. In the present study, different miRNA profiles were observed following different treatments. The Venn diagram show that 194 miRNAs were detected in all 4 groups, 202 in both the Con and P8 groups, 217 in both the Con and DEX groups, and 244 in both the DEX and DEX_P8 groups (Fig. 8A). Among the differentially expressed miRNAs, 23 miRNAs (e.g., gga-let-7k-3p, gga-miR-451, and gga-miR-3538) were upregulated and 10 miRNAs (e.g., gga-miR-6668-p3 and gga-miR-1457-p5) were downregulated in the DEX group compared to the Con group (P < 0.05) (Table 9). Seven miRNAs were upregulated (e.g., gga-miR-3528-p3, gga-miR-148a-3p, and gga-miR-143-3p) and 11 miRNAs (e.g., gga-miR-15a, gga-miR-15b-5p, and gga-miR-147) were downregulated in the P8 group compared to the Con group (P < 0.05) (Table 10). Moreover, 9 miRNAs were upregulated (e.g., gga-miR-21-3p and hsa-miR-27a-3p) and 24 miRNAs (e.g., gga-miR-455-3p, gga-miR-193b-3p, and gga-miR-425-5p) were downregulated in the DEX_P8 group compared with those in the DEX group (P < 0.05) (Fig. 8B, Table 11).
Fig. 8.
Effects of Lactiplantibacillus plantarum P8 (P8) on the intestinal miRNA profiles of oxidatively stressed broilers. (A) Venn diagrams between groups. (B) Differentially expressed miRNAs between Con and DEX groups, Con and P8 groups, and DEX and DEX_P8 groups. Con = control diet; DEX = control diet plus dexamethasone injection; P8 = control diet containing 1 × 108 CFU/g L. plantarum P8; DEX_P8 = control diet containing 1 × 108 CFU/g L.plantarum P8 plus dexamethasone injection.
Table 9.
Top 10 differentially expressed miRNAs between the DEX and Con groups.
| miRNA name | Up/down | log2FC | P-value |
|---|---|---|---|
| gga-let-7k-3p | up | 2.38 | 0.04 |
| gga-miR-451 | up | 2.07 | 0.04 |
| gga-miR-3538 | up | 1.35 | 0.02 |
| gga-miR-148a-3p | up | 1.25 | 0.04 |
| PC-5p-58322_31 | down | −3.30 | 0.04 |
| mmu-miR-1983-p5 | down | −2.36 | 0.04 |
| gga-miR-6668-p3 | down | −1.79 | 0.05 |
| PC-3p-296_8651 | down | −1.62 | 0.02 |
| gga-miR-1457-p5 | down | −1.55 | 0.03 |
| PC-3p-9481_310 | down | −1.40 | 0.02 |
Con = control diet; DEX = control diet plus dexamethasone injection; FC = fold change.
Table 10.
Top 10 differentially expressed miRNAs between the P8 and Con groups.
| miRNA name | Up/down | log2FC | P-value |
|---|---|---|---|
| gga-miR-3528-p3 | up | 1.89 | 0.03 |
| has-miR-200b-3p | up | 1.71 | 0.04 |
| gga-miR-148a-3p | up | 1.11 | 0.05 |
| gga-mir-143-3p | up | 1.10 | 0.04 |
| mmu-miR-6240-p5 | up | 0.86 | 0.04 |
| gga-miR-15a | down | −1.79 | 0.02 |
| gga-miR-15b-5p | down | −0.91 | 0.04 |
| bta-miR-194 | down | −0.87 | 0.02 |
| gga-miR-147 | down | −0.76 | 0.01 |
| gga-miR-375 | down | −0.75 | 0.03 |
Con = control diet; P8 = control diet containing 1 × 108 CFU/g Lactiplantibacillus plantarum P8; FC = fold change.
Table 11.
Top 10 differentially expressed miRNAs between the DEX_P8 and DEX groups.
| miRNA name | Up/down | log2FC | P-value |
|---|---|---|---|
| gga-miR-21-3p | up | 1.57 | 0.03 |
| has-miR-27a-3p | up | 1.37 | 0.02 |
| mmu-miR-3968 | down | −2.06 | 0.01 |
| sha-miR-125a | down | −1.92 | 0.03 |
| cgr-miR-1260 | down | −1.86 | 0.03 |
| pal-miR-9995-3p | down | −1.69 | 0.00 |
| gga-miR-455-3p | down | −1.48 | 0.02 |
| gga-miR-193b-3p | down | −1.32 | 0.04 |
| gga-miR-425-5p | down | −1.28 | 0.04 |
| has-miR-7977 | down | −1.22 | 0.01 |
DEX = control diet plus dexamethasone injection; DEX_P8 = control diet containing 1 × 108 CFU/g Lactiplantibacillus plantarum P8 plus dexamethasone injection; FC = fold change.
To understand the functions and mechanisms of action of the differentially expressed miRNAs, we performed target gene prediction and KEGG pathway analyses. KEGG pathway analysis provided information on gene functions. The most enriched pathways between the DEX and Con groups included “endocytosis”, “ubiquitin-mediated proteolysis”, and “apelin signaling pathway”. The most enriched pathways between P8 and Con groups included “FoxO signaling pathway”, “fatty acid degradation”, and “dopaminergic synapse”. In addition, the most enriched pathways between DEX_P8 and DEX groups included “pathways in cancer”, “PI3K-Akt signaling pathway” and “Ras signaling pathway” and “Rap1 signaling pathway” (Fig. 9).
Fig. 9.
KEGG enrichment analysis of the differentially expressed miRNAs between Con and DEX groups, Con and P8 groups, and DEX and DEX_P8 groups. Con = control diet; DEX = control diet plus dexamethasone injection; P8 = control diet containing 1 × 108 CFU/g Lactiplantibacillus plantarum P8; DEX_P8 = control diet containing 1 × 108 CFU/g L.plantarum P8 plus dexamethasone injection.
3.6. Correlation analysis between biochemical indices, growth parameters and microbiota
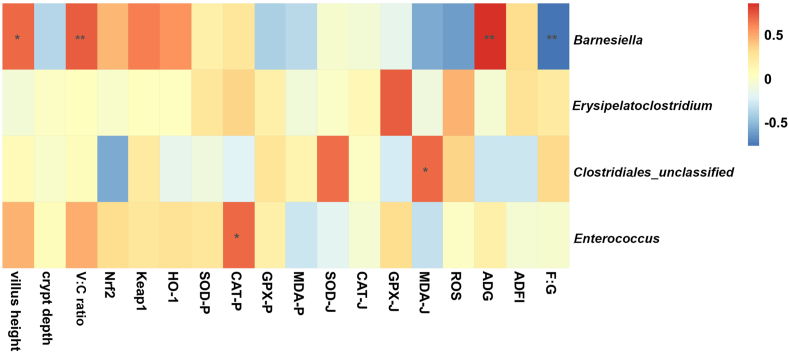
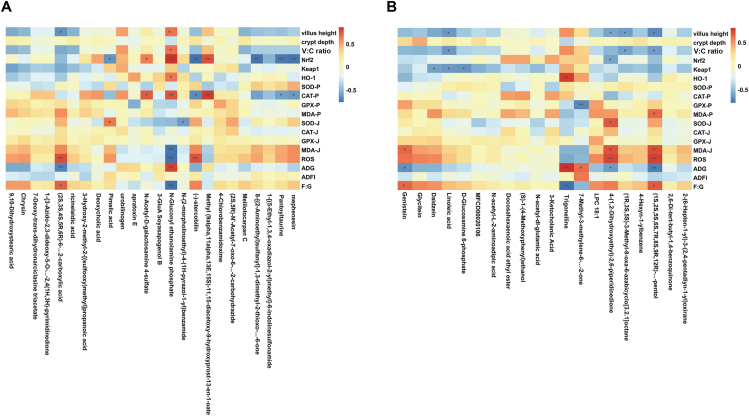
A correlation analysis between the biochemical indices, growth parameters (d 1 to 21), and top 30 genera with significant differences in abundance is shown in Fig. 10. Results with a correlation coefficient (r) larger than 0.6 or less than −0.6 were selected. We found that Barnesiella abundance was positively correlated with V:C ratio (r = 0.69, P = 0.01), villus height (r = 0.65, P = 0.01) and ADG (r = 0.83, P < 0.01) and negatively correlated with F:G ratio (r = −0.81, P < 0.01). Enterococcus abundance was positively correlated with plasma CAT levels (r = 0.66, P = 0.04). Clostridiales_unclassified was positively correlated with jejunal MDA (r = 0.65, P = 0.03) (Table 12, Tables S1–S2).
Fig. 10.
Correlation analysis between biochemical parameters, growth indices and gut microbiota. Asterisk (∗) means FDR-adjusted P-values smaller than 0.05. Double asterisks (∗∗) mean FDR-adjusted P-values smaller than 0.01. MDA-J, SOD-J, GPX-J, and CAT-J indicates jejunal MDA, SOD, GPX, and CAT, respectively. MDA-P, SOD-P, GPX-P, and CAT-P indicates plasma MDA, SOD, GPX, and CAT, respectively. V:C ratio = villus height to crypt depth ratio; MDA = malondialdehyde; SOD = superoxide dismutase; GPX = glutathione peroxidase; CAT = catalase.
Table 12.
Correlation analysis between biochemical indices, growth parameters and microbiota.
| Item | Villus height | V:C ratio | CAT-P | MDA-J | ADG | F:G ratio |
|---|---|---|---|---|---|---|
| Barnesiella | r = 0.65, P = 0.01 | r = 0.69, P = 0.01 | r = 0.83, P < 0.01 | r = −0.81, P < 0.01 | ||
| Clostridiales_unclassified | r = 0.65, P = 0.03 | |||||
| Enterococcus | r = 0.66, P = 0.04 |
V:C ratio = villus height to crypt depth ratio; CAT-P = plasma catalase; MDA-J = jejunal malondialdehyde.
3.7. Correlation analysis between biochemical indices, growth parameters and metabolites
A correlation analysis between biochemical indices, growth parameters (d 1 to 20), and top 20 significantly different metabolites (10 in positive mode and 10 in negative mode) in pairwise comparisons is shown in Fig. 11. Results with a correlation coefficient (r) larger than 0.6 or less than −0.6 were selected. The correlation between biochemical parameters and metabolites in negative mode demonstrated that N-gluconyl ethanolamine phosphate was positively correlated with villus height (r = 0.64, P = 0.02), V:C ratio (r = 0.65, P = 0.02), Nrf2 (r = 0.85, P < 0.01), HO-1 (r = 0.64, P = 0.03), plasma CAT (r = 0.72, P = 0.01), and ADG (r = 0.79, P < 0.01), and negatively correlated with jejunal MDA (r = −0.76, P < 0.01), ROS (r = −0.79, P < 0.01) and F:G ratio (r = −0.78, P < 0.01). Methyl (9alpha,11alpha,13E,15S)-11,15-diacetoxy-9-hydroxyprost-13-en-1-oate was positively correlated with plasma CAT (r = 0.80, P < 0.01) and Nrf2 (r = 0.75, P = 0.01) levels. We found that (−)-stercobilin was negatively correlated with Nrf2 (r = −0.78, P < 0.01), and plasma CAT (r = −0.68, P = 0.01) and positively correlated with ROS (r = 0.75, P = 0.01) levels. Mephenesin was negatively correlated with Nrf2 (r = −0.77, P < 0.01) and plasma CAT (r = −0.63, P = 0.03) levels. Additionally, (2S,3S,4S,5R,6R)-6-[[(3S,4S,6aR,6bS,8aR,9R,12aS,14bR)-9-Hydroxy-4-(hydroxymethyl)-4,6a,6b,8a,11,11,14b-heptamethyl-1,2,3,4a,5,6,7,8,9,10,12,12a,14,14a-tetradecahydropicen-3-yl]oxy]-5-[(2S,3R,4S,5R,6R)-4,5-dihydroxy-6-(hydroxymethyl)-3-[(2S,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methyloxan-2-yl]oxyoxan-2-yl]oxy-3,4-dihydroxyoxane-2-carboxylic acid was positively correlated with ROS levels (r = 0.76, P < 0.01) and F:G ratio (r = 0.69, P = 0.01) and negatively correlated with ADG (r = −0.72, P = 0.01) and villus height (r = −0.66, P = 0.02). Further, 8-[(2-aminoethyl)sulfanyl]-1,3-dimethyl-2-thioxo-1,2,3,7-tetrahydro-6H-purin-6-one was negatively correlated with Nrf2 (r = −0.75, P < 0.01) level. Pantoyltaurine was negatively correlated with Nrf2 (r = −0.74, P = 0.01) and plasma CAT levels (r = −0.60, P = 0.04). N-acetyl-D-galactosamine 4-sulfate was positively correlated with plasma CAT (r = 0.71, P = 0.01) and Nrf2 (r = 0.62, P = 0.03) levels. Pimelic acid was negatively correlated with Nrf2 (r = −0.65, P = 0.02) and positively correlated with jejunal SOD (r = 0.62, P = 0.03) (Fig. 11A, Table 13, Tables S3–S4).
Fig. 11.
Correlation analysis between biochemical parameters, growth indices and metabolites. (A) Correlation between biochemical parameters, growth indices and metabolites in negative mode. (B) Correlation between biochemical parameters, growth indices and metabolites in positive mode. Asterisk (∗) means FDR-adjusted P-values smaller than 0.05. Double asterisks (∗∗) means FDR-adjusted P-values smaller than 0.01. MDA-J, SOD-J, GPX-J, and CAT-J indicates jejunal MDA, SOD, GPX, and CAT, respectively. MDA-P, SOD-P, GPX-P, and CAT-P indicates plasma MDA, SOD, GPX, and CAT, respectively. V:C ratio = villus height to crypt depth ratio; MDA = malondialdehyde; SOD = superoxide dismutase; GPX = glutathione peroxidase; CAT = catalase.
Table 13.
Correlation between biochemical parameters, growth indices and metabolites in negative mode.
| Item | Villus height | V:C ratio | Nrf2 | HO-1 | CAT-P | SOD-J | MDA-J | ROS | ADG | F:G ratio |
|---|---|---|---|---|---|---|---|---|---|---|
| (2S,3S,4S,5R,6R)-6-[[(3S,4S,6aR,6bS,8aR,9R,12aS,14bR)-9-hydroxy-4-(hydroxymethyl)-4,6a,6b,8a,11,11,14b-heptamethyl-1,2,3,4a,5,6,7,8,9,10,12,12a,14,14a-tetradecahydropicen-3-yl]oxy]-5-[(2S,3R,4S,5R,6R)-4,5-dihydroxy-6-(hydroxymethyl)-3-[(2S,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methyloxan-2-yl]oxyoxan-2-yl]oxy-3,4-dihydroxyoxane-2-carboxylic acid | r = −0.66, P = 0.02 | r = 0.76, P < 0.01 | r = −0.72, P = 0.01 |
r = 0.69, P = 0.01 |
||||||
| Pimelic acid | r = −0.65, P = 0.02 | r = 0.62, P = 0.03 | ||||||||
| N-Acetyl-D-galactosamine 4-sulfate | r = 0.62, P = 0.03 | r = 0.71, P = 0.01 | ||||||||
| N-Gluconyl ethanolamine phosphate | r = 0.64, P = 0.02 | r = 0.65, P = 0.02 | r = 0.85, P < 0.01 | r = 0.64, P = 0.03 | r = 0.72, P = 0.01 | r = −0.76, P < 0.01 | r = −0.79, P < 0.01 | r = 0.79, P < 0.01 | r = −0.78, P < 0.01 | |
| N-(2-morpholinoethyl)-4-(1H-pyrazol-1-yl)benzamide | r = −0.62, P = 0.03 | |||||||||
| (−)-Stercobilin | r = −0.78, P < 0.01 | r = −0.68, P = 0.01 | r = 0.75, P = 0.01 | |||||||
| Methyl (9alpha,11alpha,13E,15S)-11,15-diacetoxy-9-hydroxyprost-13-en-1-oate | r = 0.75, P = 0.01 | r = 0.80, P < 0.01 | ||||||||
| 8-[(2-Aminoethyl)sulfanyl]-1,3-dimethyl-2-thioxo-1,2,3,7-tetrahydro-6H-purin-6-one | r = −0.75, P < 0.01 | |||||||||
| Pantoyltaurine | r = −0.74, P = 0.01 | r = −0.60, P = 0.04 | ||||||||
| Mephenesin | r = −0.77, P < 0.01 | r = −0.63, P = 0.03 |
V:C ratio = villus height to crypt depth ratio; Nrf2 = nuclear factor erythroid 2-related factor 2; HO-1 = heme oxygenase 1; CAT-P = plasma catalase; SOD-J = jejunal superoxidase; MDA-J = jejunal malondialdehyde; ROS = reactive oxygen species.
The correlation between metabolites in the positive mode and biochemical parameters demonstrated that trigonelline was positively correlated with HO-1 levels (r = 0.76, P < 0.01) and ADG (r = 0.79, P < 0.01), and negatively correlated with F:G ratio (r = −0.80, P < 0.01). (1S,2S,5S,6S,7R,8S,9R,12R)-2,6,10,10-Tetramethyl-11-oxatricyclo[7.2.1.0∼1,6∼]dodecane-2,5,7,8,12-pentol was positively correlated with ROS (r = 0.73, P = 0.01), jejunal MDA (r = 0.70, P = 0.01), plasma MDA (r = 0.63, P = 0.03), F:G ratio (r = 0.65, P = 0.02), and negatively correlated with villus height (r = −0.70, P = 0.01), V:C ratio (r = −0.63, P = 0.03) and ADG (r = −0.75, P = 0.01). 4-(1,2-Dihydroxyethyl)-2,6-piperidinedione was positively correlated with ROS (r = 0.73, P = 0.01), jejunal MDA (r = 0.68, P = 0.01), and jejunal SOD (r = 0.67, P = 0.02) levels and negatively correlated with ADG (r = −0.63, P = 0.03), villus height (r = −0.60, P = 0.04) and Nrf2 (r = −0.60, P = 0.04). 7-Methyl-3-methylene-6-(3-oxobutyl)-3,3a,4,7,8,8a-hexahydro-2H-cyclohepta[b]furan-2-one was negatively correlated with plasma GPX levels (r = −0.72, P = 0.01) and positively correlated with ADG (r = 0.60, P = 0.04). D-glucosamine 6-phosphate was negatively correlated with Keap1 levels (r = −0.67, P = 0.02). Linoleic acid was negatively correlated with villus height (r = −0.66, P = 0.02), Keap1 level (r = −0.66, P = 0.02), and V:C ratio (r = −0.65, P = 0.02). (1R,3S,5S)-3-Methyl-8-oxa-6-azabicyclo[3.2.1]octane was negatively correlated with villus height (r = −0.63, P = 0.03) and V:C ratio (r = −0.62, P = 0.03). Genistein was positively correlated with jejunal MDA levels (r = 0.62, P = 0.03) and F:G ratio (r = 0.62, P = 0.03), and negatively correlated with ADG (r = −0.68, P = 0.02). Daidzein was negatively correlated with Keap1 levels (r = −0.61, P = 0.03) (Fig. 11B, Table 14, Tables S5–S6).
Table 14.
Correlation between biochemical parameters, growth indices and metabolites in positive mode.
| Item | Villus height | V:C ratio | Nrf2 | Keap1 | HO-1 | GPX-P | MDA-P | SOD-J | MDA-J | ROS | ADG | F:G ratio |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genistein | r = 0.62, P = 0.03 | r = −0.68, P = 0.02 | r = 0.62, P = 0.03 | |||||||||
| Daidzein | r = −0.61, P = 0.03 | |||||||||||
| Linoleic acid | r = −0.66, P = 0.02 | r = −0.65, P = 0.02 | r = −0.66, P = 0.02 | |||||||||
| D-Glucosamine 6-phosphate | r = −0.67, P = 0.02 | |||||||||||
| Trigonelline | r = 0.76, P < 0.01 | r = 0.79, P < 0.01 | r = −0.80, P < 0.01 | |||||||||
| 7-Methyl-3-methylene-6-(3-oxobutyl)-3,3a,4,7,8,8a-hexahydro-2H-cyclohepta[b]furan-2-one | r = −0.72, P = 0.01 | r = 0.60, P = 0.04 | ||||||||||
| 4-(1,2-Dihydroxyethyl)-2,6-piperidinedione | r = −0.60, P = 0.04 | r = −0.60, P = 0.04 | r = 0.67, P = 0.02 | r = 0.68, P = 0.01 | r = 0.73, P = 0.01 | r = −0.63, P = 0.03 | ||||||
| (1R,3S,5S)-3-Methyl-8-oxa-6-azabicyclo[3.2.1]octane | r = −0.63, P = 0.03 | r = −0.62, P = 0.03 | ||||||||||
| (1S,2S,5S,6S,7R,8S,9R,12R)-2,6,10,10-Tetramethyl-11-oxatricyclo[7.2.1.0∼1,6∼]dodecane-2,5,7,8,12-pentol | r = −0.70, P = 0.01 | r = −0.63, P = 0.03 | r = 0.63, P = 0.03 | r = 0.70, P = 0.01 | r = 0.73, P = 0.01 | r = −0.75, P = 0.01 | r = 0.65, P = 0.02 |
V:C ratio = villus height to crypt depth ratio; Nrf2 = nuclear factor erythroid 2-related factor 2; Keap1 = kelch-like ECH-associated protein 1; HO-1 = heme oxygenase 1; GPX-P = plasma glutathione peroxidase; MDA-P = plasma malondialdehyde; SOD-J = jejunal superoxidase; MDA-J = jejunal malondialdehyde; ROS = reactive oxygen species.
3.8. Correlation analysis between biochemical indices, growth parameters and miRNAs
A correlation analysis between biochemical indices, growth parameters (d 1 to 21) and top 10 differentially expressed miRNAs in pairwise comparisons is shown in Fig. 12 and Tables S7–S8. Results with a correlation coefficient larger than 0.6 or less than −0.6 were selected. The results show that PC-3p-9481_310 was positively correlated with ADG (r = 0.73, P = 0.01) and negatively correlated with ROS levels (r = −0.80, P < 0.01) and F:G ratio (r = −0.76, P < 0.01). gga-miR-15a was positively correlated with jejunal MDA (r = 0.78, P < 0.01), ROS (r = 0.74, P = 0.01) levels and F:G ratio (r = 0.61, P = 0.04), and negatively correlated with V:C ratio (r = −0.68, P = 0.02), villus height (r = −0.65, P = 0.02) and ADG (r = −0.68, P = 0.02). PC-3p-296_8651 was positively correlated with ADG (r = 0.63, P = 0.03) and negatively correlated with ROS levels (r = −0.72, P = 0.01) and F:G ratio (r = −0.64, P = 0.02). hsa-miR-7977 was positively correlated with ADG (r = 0.63, P = 0.03) and negatively correlated with F:G ratio (r = −0.64, P = 0.03). The expression of pal-miR-9995-3p was negatively correlated with F:G ratio (r = −0.61, P = 0.04). The expression of gga-miR-15b-5p was negatively correlated with plasma CAT levels (r = −0.72, P = 0.01). The expression of gga-let-7k-3p was negatively correlated with Keap1 levels (r = −0.70, P = 0.01) and villus height (r = −0.62, P = 0.03). The expression of gga-miR-455-3p was negatively correlated with villus height (r = −0.68, P = 0.02) and V:C ratio (r = −0.68, P = 0.02). The expression of gga-miR-193b-3p was positively correlated with crypt depth (r = 0.66, P = 0.02) and negatively correlated with V:C ratio (r = −0.64, P = 0.02) and HO-1 levels (r = −0.63, P = 0.03). The expression of has-miR-200b-3p was positively correlated with HO-1 levels (r = 0.64, P = 0.02). The expression of gga-miR-375 was positively correlated with plasma SOD levels (r = 0.64, P = 0.03). The expression of gga-miR-425-5p was negatively correlated with plasma CAT levels (r = −0.64, P = 0.03). The expression of gga-miR-147 was positively correlated with plasma GPX levels (r = 0.62, P = 0.03). The expression of gga-miR-3528-p3 was negatively correlated with plasma SOD levels (r = −0.64, P = 0.03; Table 15).
Fig. 12.
Correlation analysis between biochemical parameters, growth indices and miRNAs. Asterisk (∗) means FDR-adjusted P-values smaller than 0.05. Double asterisks (∗∗) means FDR-adjusted P-values smaller than 0.01. MDA-J, SOD-J, GPX-J, and CAT-J indicates jejunal MDA, SOD, GPX, and CAT, respectively. MDA-P, SOD-P, GPX-P, and CAT-P indicates plasma MDA, SOD, GPX, and CAT, respectively. V:C ratio = villus height to crypt depth ratio; MDA = malondialdehyde; SOD = superoxide dismutase; GPX = glutathione peroxidase; CAT = catalase.
Table 15.
Correlation analysis between biochemical parameters, growth indices and miRNAs.
| Item | Villus height | Crypt depth | V:C ratio | Keap1 | HO-1 | SOD-P | CAT-P | GPX-P | MDA-J | ROS | ADG | F:G ratio |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gga-let-7k-3p | r = −0.62, P = 0.03 | r = −0.70, P = 0.01 | ||||||||||
| gga-miR-15a | r = −0.65, P = 0.02 | r = −0.68, P = 0.02 | r = 0.78, P < 0.01 | r = 0.74, P = 0.01 | r = −0.68, P = 0.02 | r = 0.61, P = 0.04 | ||||||
| gga-miR-15b-5p | r = −0.72, P = 0.01 | |||||||||||
| gga-miR-147 | r = 0.62, P = 0.03 | |||||||||||
| gga-miR-193b-3p | r = 0.66, P = 0.02 | r = −0.64, P = 0.02 | r = −0.63, P = 0.03 | |||||||||
| gga-miR-375 | r = 0.64, P = 0.03 | |||||||||||
| gga-miR-425-5p | r = −0.64, P = 0.03 | |||||||||||
| gga-miR-455-3p | r = −0.68, P = 0.02 | r = −0.68, P = 0.02 | ||||||||||
| gga-mir-3528-p3 | r = −0.64, P = 0.03 | |||||||||||
| hsa-miR-200b-3p | r = 0.64, P = 0.02 | |||||||||||
| hsa-miR-7977 | r = 0.63, P = 0.03 | r = −0.64, P = 0.03 | ||||||||||
| pal-miR-9995-3p | r = −0.61, P = 0.04 | |||||||||||
| PC-3p-296_8651 | r = −0.72, P = 0.01 | r = 0.63, P = 0.03 | r = −0.64, P = 0.02 | |||||||||
| PC-3p-9481_310 | r = −0.80, P < 0.01 | r = 0.73, P = 0.01 | r = −0.76, P < 0.01 |
V:C ratio = villus height to crypt depth ratio; Keap1 = kelch-like ECH-associated protein 1; HO-1 = heme oxygenase 1; SOD-P = plasma superoxidase; CAT-P = plasma catalase; GPX-P = plasma glutathione peroxidase; MDA-J = jejunal malondialdehyde; ROS = reactive oxygen species.
4. Discussion
In the present study, we examined the interrelations among gut microbial composition, cecal content metabolite levels, intestinal miRNA profiles, dietary P8 supplementation, growth performance, jejunal morphology, and oxidative status in broilers challenged with DEX. Our findings indicate that the impaired growth performance, jejunal morphology and decreased levels of antioxidases in the DEX-challenged broilers were improved by DEX_P8 treatment. The elevated jejunal MDA and ROS levels induced by DEX were reduced by DEX_P8 treatment. Moreover, the inactivated Keap1-Nrf2-HO-1 antioxidant signaling pathway in the DEX group was activated by DEX_P8 treatment. Similar results were observed in previous studies, in which dietary supplementation with probiotics also alleviated oxidative stress and improved the growth performance as well as intestinal morphology of chickens (de Souza et al., 2020; Sampath et al., 2021; Wu et al., 2019).
Significant taxonomic changes were observed in the current study. Compared to the Con group, DEX treatment led to decreased abundance of Barnesiellaceae (family) and Barnesiella (genus), whereas P8 treatment also contributed to decreased abundance of Barnesiellaceae (family), Barnesiella (genus) and Clostridiales_unclassified (genus), and increased abundance of Enterococcus (genus). Compared to the DEX group, DEX_P8 treatment led to increased abundance of Barnesiellaceae (family), Barnesiella (genus) and Erysipelatoclostridium (genus). The genus Barnesiella identified in the present study is a member of the phylum Bacteroidetes. Intestinal Barnesiella spp. are dietary polyphenol-targeting bacteria that can eliminate harmful bacteria from the gut and produce short-chain fatty acids (Wei et al., 2018). Barnesiella can use fucosyllactose as energy source to colonize the intestine and improve its anti-inflammatory ability (Weiss et al., 2014). In a study by Tan et al. (2016), the inclusion of konjac flour in the diet alleviated oxidative stress and increased the abundance of Barnesiella in sows during gestational oxidative stress. Thus, elevated levels of Barnesiella in the DEX_P8 group may imply an improvement in broiler health. However, we also noticed that the abundance of Barnesiellaceae (family) and Barnesiella (genus) was significantly decreased in the P8 group. Similar to our results, by using Clostridium butyricum- and Bacillus spp.-based probiotics, Zeng et al. (2021) also found that supplementation with probiotics dramatically decreases the relative abundance of Barnesiella and improves the growth performance of chickens. Moreover, it has been demonstrated that the abundances of Barnesiella and Enterococcus have opposite trends (Narushima et al., 2006). In the present study, contrary to the Barnesiella alteration, Enterococcus abundance was significantly increased in P8 group. Beneficial roles of Enterococcus strains were reported in regulating health (Carasi et al., 2014; Švec and Franz, 2014). With the change of the gut microbiota structure, the metabolites will also alter. Here, the metabolomics showed that the deoxycholic acid was increased in broilers of P8 group. Deoxycholic acid was found to have beneficial roles in anti-inflammation (Chen et al., 2002; Wang et al., 2019). One study also demonstrated that deoxycholic acid was positively correlated with Enterococcus abundance and negatively correlated with Barnesiella (Narushima et al., 2006). Thus, the decreased Barnesiella and increased Enterococcus abundances in the P8 group may be associated with the altered gut microbiota metabolites. However, the more obviously decreased Barnesiella abundance in the DEX group may be due to the dysbiosis caused by oxidative stress. Unfortunately, no other studies have reported data on Barnesiella under oxidative stress in broilers to serve for comparison with our results; thus, the specific effects of Barnesiella need to be studied further. Erysipelatoclostridium abundance was elevated in the DEX_P8 group compared to that in the DEX group. Erysipelatoclostridium is a part of the normal gut microbiota but can become an opportunistic pathogen (Khan and Chousalkar, 2020). Nevertheless, recent studies have suggested potential health benefits of Erysipelatoclostridium. Chai et al. (2021) revealed that the relative abundance of Erysipelatoclostridium decreases in constipated mice. Nair et al. (2021) showed that the relative abundance of Erysipelatoclostridium increases when the dairy-originated probiotic Propionibacterium freudenreichii subsp. freudenreichii B3523 is supplemented in turkeys. In addition, our KEGG results of gut microbiota indicate that some pathways, including the phenylacetate degradation pathway, were upregulated in the DEX_P8 group compared to the DEX group. Reports have indicated that phenylacetate degradation is required to withstand oxidative stress (Green et al., 2020). Moreover, in the present study, we performed a correlation analysis between the gut microbiota and biochemical parameters. The results show that Barnesiella abundance was positively correlated with ADG, V:C ratio, villus height, and negatively correlated with F:G ratio. Enterococcus abundance was positively correlated with plasma CAT levels. Clostridiales_unclassified abundance was positively correlated with jejunal MDA levels. The above findings imply that gut microbiota had strong correlation with growth performance, oxidative stress and jejunal morphology in P8-treated oxidatively stressed broilers.
Probiotics can exert their antioxidant effects by regulating gut microbiota metabolites (Ho et al., 2019; Saeedi et al., 2020). In this study, metabolites in the cecal contents were also altered by DEX and/or P8 treatments. OPLS-DA analysis clearly discriminated metabolites between the treatments. As for the metabolites in both negative mode and positive mode, the concentrations of many metabolites were significantly altered with P8 or DEX treatments, including 9,10-dihydroxystearic acid, urobilinogen, genistein, glycitein, docosahexaenoic acid ethyl ester, etc. Based on these data, we noticed an increase of glycitein and daidzein content in the DEX group; both metabolites have been reported to protect cells against oxidative stress (Dong and Yang, 2022; Meng et al., 2017), implying that higher levels of glycitein and daidzein play important roles in protecting broilers against DEX-induced oxidative stress. Besides, as mentioned above, the P8 treatment led to increased deoxycholic acid content, which was reported to be positively correlated with Enterococcus abundance and negatively correlated with Barnesiella (Narushima et al., 2006). In addition, pantoyltaurine was found to be decreased in the DEX_P8 group. Pantoyltaurine is a taurine derivative and has been reported to inhibit the growth of several pathogens, including streptococci and Plasmodium relictum (Evans et al., 2002). Thus, the decreased pantoyltaurine levels may indicate less pathogenic bacteria in the cecal content. Furthermore, the decrease in urobilinogen content in the DEX group was increased by DEX_P8 treatment, whereas the increase in genistein content in the DEX group was reduced by DEX_P8 treatment. Urobilinogen is a reductive metabolite of bilirubin, a breakdown product of heme produced by the microbiota (Fahmy et al., 1972). Nakamura et al. (2006) showed that the DPPH radical scavenging activity of urobilinogen is higher than that of other antioxidants (α-tocopherol, bilirubin, and β-carotene). Moreover, the formation of hydroperoxides of linoleic acid with 2,4-dimethylvaleronitrile in methanol is suppressed by the addition of the synthesized urobilinogen, indicating the antioxidant function of urobilinogen. Genistein, a naturally available soy isoflavone, has various medical advantages, owing to its numerous natural properties. Some studies have demonstrated that genistein protects against oxidative stress (Gupta et al., 2015; Weng et al., 2019). However, varying results have been obtained. Simperova et al. (2016) suggested that genistein supplementation prevents weight gain in obese mice but promotes oxidative stress. In addition, genistein was also reported to promote a decrease of MnSOD, CuZnSOD and TrxR mRNA expressions (Prietsch et al., 2014). Thus, the elevated levels of urobilinogen and decreased genistein in the DEX_P8 group implied alleviation of oxidative stress. KEGG analysis of metabolite indicates that the different metabolites between DEX_P8 and DEX groups were enriched in the pathways including “tryptophan metabolism”, “primary bile acid biosynthesis”, “linoleic acid metabolism”, “glycerphospholipid metabolism” and “biosynthesis of unsaturated fatty acids”. Some of these pathways are involved in the regulation of oxidative stress. For example, some tryptophan metabolites have antioxidant properties (Christen et al., 1990; Xu et al., 2018). Heat stress causes a drop in the primary bile acid content in broilers and influences their heat stress resistance capability (Yin et al., 2021). Linoleic acid potentiates oxidative stress (Niki, 2008; Toborek et al., 1997). Thus, KEGG analysis also implied the elevated antioxidant capacity of broilers in the DEX_P8 group. In addition, we found obvious correlations between the altered metabolites and biochemical parameters. For instance, mephenesin and pantoyltaurine were negatively correlated with Nrf2 and plasma CAT levels. N-Acetyl-D-galactosamine 4-sulfate was positively correlated with the plasma levels of CAT and Nrf2. Trigonelline was negatively correlated with F:G ratio, and positively correlated with HO-1 and ADG. Genistein was positively correlated with jejunal MDA, ADG, and F:G ratio. The above findings imply that the metabolites of cecal contents had strong correlations with growth performance, oxidative stress and jejunal morphology in P8-treated oxidatively stressed broilers.
Despite their marked effect on the gut microbiota and its metabolites, probiotics have also been reported to play a role in intestinal miRNA profiles (Zhu et al., 2017; Wang et al., 2021a). In the current study, jejunal miRNA profiles were also influenced by DEX and/or P8 treatment. Here, we found that 23 miRNAs (e.g., gga-let-7k-3p, gga-miR-451, and gga-miR-3538) were upregulated and 10 miRNAs (e.g., gga-miR-6668-p3 and gga-miR-1457-p5) were downregulated in the DEX group compared to those in the Con group. Seven miRNAs were upregulated (e.g., gga-miR-3528-p3, gga-miR-148a-3p, and gga-miR-143-3p) and 11 miRNAs (e.g., gga-miR-15a, gga-miR-15b-5p, and gga-miR-147) were downregulated in the P8 group compared to those in the Con group. Moreover, 9 miRNAs were upregulated (e.g., gga-miR-21-3p and has-miR-27a-3p) and 24 miRNAs (e.g., gga-miR-455-3p, gga-miR-193b-3p, and gga-miR-425-5p) were downregulated in the DEX_P8 group compared with those in the DEX group. Reports suggest that the overexpression of miR-21-3p inhibits the apoptosis of neural cells (Tian et al., 2021), and targets Nox mRNA to repress its expression, thereby improving the redox state (Yan et al., 2015). Moreover, the expression of gga-miR-455-3p is upregulated in broilers with wooden breasts (Shu et al., 2021), which is related to increased oxidative stress (Abasht et al., 2016). Thus, the alteration of miRNA expression in the DEX_P8 group may also contribute to the relief of oxidative stress. Proteins were analyzed using the KEGG database. The most enriched pathways between the DEX and Con groups included “endocytosis”, “ubiquitin-mediated proteolysis” and the “apelin signaling pathway”. The most enriched pathways between P8 and Con groups included “FoxO signaling pathway”, “fatty acid degradation” and “dopaminergic synapse”. The most enriched pathways between the DEX_P8 and DEX groups included the “pathways in cancer”, “PI3K-Akt signaling pathway”, “Ras signaling pathway” and “Rap1 signaling pathway”. It is demonstrated that the inhibition of Nrf2 may also depend upon ubiquitin-mediated proteolysis (Cullinan et al., 2004), thus the enriched ubiquitin-mediated proteolysis pathway between DEX and Con groups was in accordance with the down-regulated Nrf2 expression in DEX group. Moreover, the FoxO signaling pathway, which supports oxidative stress resistance (Akasaki et al., 2014), was enriched between the P8 and Con groups. Additionally, the PI3K-Akt, Ras, and Rap1 signaling pathways have been reported to participate in the inhibition of oxidative stress (Remans et al., 2004; Yang et al., 2017; Yuan et al., 2020). Therefore, the above analysis implies the regulatory effects of P8 on oxidative stress in broilers. Correlation analysis demonstrated that many downregulated miRNAs, such as gga-miR-455-3p, gga-miR-425-5p, and gga-miR-193b-3p, were negatively correlated with antioxidant parameters, jejunal morphology or growth performance indices.
In summary, our data show a significant improvement in growth performance, jejunal morphology and antioxidant capacity of dietary P8 in DEX-challenged broilers. By taking advantage of microbiome analysis, metabolomics, and miRNAomics, we found significant reshaping of the gut microbiota, especially Barnesiella, in the DEX_P8 group, along with changes in the metabolites of cecal contents (including urobilinogen and genistein) and jejunal miRNAs (including miR-455-3p and miR-425-5p). Moreover, significant correlations were observed between the biochemical parameters, growth indices and omics results, implying that the gut microbiota, cecal content metabolites, and jejunal miRNAs play vital roles in regulating antioxidation, jejunal morphology and growth performance in P8-treated oxidatively stressed broilers (Fig. 13). Taken together, the multi-omics analysis in the present study identified key bacteria and molecules for oxidative stress regulation in oxidatively stressed broilers treated with P8 and provides a valuable resource for comparative studies with other experimental datasets.
Fig. 13.
Integrative diagram of the effects of Lactiplantibacillus plantarum P8 (P8) on the growth performance, jejunal morphology and antioxidant capacity of broilers. DEX_P8 = control diet containing 1 × 108 CFU/g L. plantarum P8 plus dexamethasone injection.
Author contributions
Jinshan Zhao: Conceptualization, Writing-Original Draft. Fan Zhao: Investigation. Xuemin Li: Investigation. Junmeng Yuan: Visualization, Investigation. Kai Zhang: Methodology. Huawei Liu: Investigation. Yang Wang: Conceptualization, Writing-Review & Editing, Funding acquisition.
Declaration of competing interest
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, and there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the content of this paper.
Acknowledgments
The authors acknowledge the National Natural Science Foundation of China (Grant No. 32102586) and the Qingdao Science and Technology Program (Grant No. 22-3-7-xdny-11-nsh).
Footnotes
Peer review under responsibility of Chinese Association of Animal Science and Veterinary Medicine.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aninu.2023.06.002.
Appendix supplementary data
The following is the Supplementary data to this article:
References
- Abasht B., Mutryn M.F., Michalek R.D., Lee W.R. Oxidative stress and metabolic perturbations in wooden breast disorder in chickens. PLoS One. 2016;11(4) doi: 10.1371/journal.pone.0153750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akasaki Y., Alvarez-Garcia O., Saito M., Carames B., Iwamoto Y., Lotz M.K. FoxO transcription factors support oxidative stress resistance in human chondrocytes. Arthritis Rheumatol. 2014;66:3349–3358. doi: 10.1002/art.38868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y., Wang Z., Zhang Y., Zhang J., Wang L., Dong X., et al. Effect of Lactobacillus plantarum P-8 on lipid metabolism in hyperlipidemic rat model. Eur J Lipid Sci Technol. 2012;114:1230–1236. [Google Scholar]
- Barekatain R., Nattrass G., Tilbrook A.J., Chousalkar K., Gilani S. Reduced protein diet and amino acid concentration alter intestinal barrier function and performance of broiler chickens with or without synthetic glucocorticoid. Poult Sci. 2019;98:3662–3675. doi: 10.3382/ps/pey563. [DOI] [PubMed] [Google Scholar]
- Cai Y., Yu X., Hu S., Yu J. A brief review on the mechanisms of miRNA regulation. Genom Proteom Bioinform. 2009;7(4):147–154. doi: 10.1016/S1672-0229(08)60044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carasi P., Jacquot C., Romanin D.E., Elie A., De Antoni G.L., Urdaci M.C., et al. Safety and potential beneficial properties of Enterococcus strains isolated from kefir. Int Dairy J. 2014;39(1):193–200. [Google Scholar]
- Chai M., Wang L., Li X., Zhao J., Zhang H., Wang G., et al. Different Bifidobacterium bifidum strains change the intestinal flora composition of mice via different mechanisms to alleviate loperamide-induced constipation. Food Funct. 2021;12(13):6058–6069. doi: 10.1039/d1fo00559f. [DOI] [PubMed] [Google Scholar]
- Chen X., Mellon R.D., Yang L., Dong H., Oppenheim J., Howard O.M.Z., et al. Regulatory effects of deoxycholic acid, a component of the anti-inflammatory traditional Chinese medicine Niuhuang, on human leukocyte response to chemoattractants. Biochem Pharmacol. 2002;63(3):533–541. doi: 10.1016/s0006-2952(01)00917-0. [DOI] [PubMed] [Google Scholar]
- Christen S., Peterhans E., Stocker R. Antioxidant activities of some tryptophan metabolites: possible implication for inflammatory diseases. Proc Natl Acad Sci U S A. 1990;87(7):2506–2510. doi: 10.1073/pnas.87.7.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrousos G.P., Gold P.W. The concepts of stress and stress system disorders: overview of physical and behavioral homeostasis. JAMA. 1992;267(9):1244–1252. [PubMed] [Google Scholar]
- Cullinan S.B., Gordan J.D., Jin J., Harper J.W., Diehl J.A. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24(19):8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dam B., Misra A., Banerjee S. Springer, Singapore; 2019. Role of gut microbiota in combating oxidative stress; pp. 43–82. (Oxidative stress in microbial diseases). [Google Scholar]
- de Souza M., Baptista A.A.S., Valdiviezo M.J.J., Justino L., Menck-Costa M.F., Ferraz C.R., et al. Lactobacillus spp. reduces morphological changes and oxidative stress induced by deoxynivalenol on the intestine and liver of broilers. Toxicon. 2020;185:203–212. doi: 10.1016/j.toxicon.2020.07.002. [DOI] [PubMed] [Google Scholar]
- Dong N., Yang Z. Glycitein exerts neuroprotective effects in Rotenone-triggered oxidative stress and apoptotic cell death in the cellular model of Parkinson's disease. Acta Biochim Pol. 2022;69(2):447–452. doi: 10.18388/abp.2020_5963. [DOI] [PubMed] [Google Scholar]
- Evans D.A., Wu J., Masse C.E., MacMillan D.W.C. A general method for the enantioselective synthesis of pantolactone derivatives. Org Lett. 2002;4(20):3379–3382. doi: 10.1021/ol026489d. [DOI] [PubMed] [Google Scholar]
- Fahmy K., Gary C.H., Nicholson D.C. The reduction of bile pigments by faecal and intestinal bacteria. Biochim Biophys Acta. 1972;264:85–97. doi: 10.1016/0304-4165(72)90119-5. [DOI] [PubMed] [Google Scholar]
- Feng P., Ye Z., Han H., Ling Z., Zhou T., Zhao S., et al. Tibet plateau probiotic mitigates chromate toxicity in mice by alleviating oxidative stress in gut microbiota. Commun Biol. 2020;3(1):1–12. doi: 10.1038/s42003-020-0968-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Lin H., Wang X., Song Z., Jiao H. Vitamin E supplementation alleviates the oxidative stress induced by dexamethasone treatment and improves meat quality in broiler chickens. Poult Sci. 2010;89:318–327. doi: 10.3382/ps.2009-00216. [DOI] [PubMed] [Google Scholar]
- Gareau M.G., Sherman P.M., Walker W.A. Probiotics and the gut microbiota in intestinal health and disease. Nat Rev Gastroenterol Hepatol. 2010;7(9):503–514. doi: 10.1038/nrgastro.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green E.R., Juttukonda L.J., Skaar E.P. The manganese-responsive transcriptional regulator MumR protects Acinetobacter baumannii from oxidative stress. Infect Immun. 2020;88(3) doi: 10.1128/IAI.00762-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S.K., Dongare S., Mathur R., Mohanty I.R., Srivastava S., Mathur S., et al. Genistein ameliorates cardiac inflammation and oxidative stress in streptozotocin-induced diabetic cardiomyopathy in rats. Mol Cell Biochem. 2015;408(1):63–72. doi: 10.1007/s11010-015-2483-2. [DOI] [PubMed] [Google Scholar]
- Ho S.T., Hsieh Y.T., Wang S.Y., Chen M.J. Improving effect of a probiotic mixture on memory and learning abilities in d-galactose-treated aging mice. J Dairy Sci. 2019;102(3):1901–1909. doi: 10.3168/jds.2018-15811. [DOI] [PubMed] [Google Scholar]
- Khan S., Chousalkar K.K. Salmonella Typhimurium infection disrupts but continuous feeding of Bacillus based probiotic restores gut microbiota in infected hens. J Anim Sci Biotechnol. 2020;11(1):1–16. doi: 10.1186/s40104-020-0433-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Zhao F., Zhang K., Zhao J., Wang Y. Investigating the growth performance, meat quality, immune function and proteomic profiles of plasmal exosomes in Lactobacillus plantarum-treated broilers with immunological stress. Food Funct. 2021;12(23):11790–11807. doi: 10.1039/d1fo01936h. [DOI] [PubMed] [Google Scholar]
- Ma N., Liu X.W., Kong X.J., Li S.H., Jiao Z.H., Qin Z., et al. Aspirin eugenol ester regulates cecal contents metabolomic profile and microbiota in an animal model of hyperlipidemia. BMC Vet Res. 2018;14(1):405. doi: 10.1186/s12917-018-1711-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciano F., Vajro P. Academic Press; 2017. Oxidative stress and gut microbiota; pp. 113–123. (Gastrointestinal tissue). [Google Scholar]
- Meng H., Fu G., Shen J., Shen K., Xu Z., Wang Y., et al. Ameliorative effect of daidzein on cisplatin-induced nephrotoxicity in mice via modulation of inflammation, oxidative stress, and cell death. Oxid Med Cell Longev. 2017;2017 doi: 10.1155/2017/3140680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair D.V.T., Johnson T.J., Noll S.L., Johny A.K. Effect of supplementation of a dairy-originated probiotic bacterium, Propionibacterium freudenreichii subsp. freudenreichii, on the cecal microbiome of turkeys challenged with multidrug-resistant Salmonella Heidelberg. Poult Sci. 2021;100(1):283–295. doi: 10.1016/j.psj.2020.09.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Sato K., Akiba M., Ohnishi M. Urobilinogen, as a bile pigment metabolite, has an antioxidant function. J Oleo Sci. 2006;55(4):191–197. [Google Scholar]
- Narushima S., Itoh K., Miyamoto Y., Park S.H., Nagata K., Kuruma K., et al. Deoxycholic acid formation in gnotobiotic mice associated with human intestinal bacteria. Lipids. 2006;41(9):835–843. doi: 10.1007/s11745-006-5038-1. [DOI] [PubMed] [Google Scholar]
- Niki E. Lipid peroxidation products as oxidative stress biomarkers. Biofactors. 2008;34(2):171–180. doi: 10.1002/biof.5520340208. [DOI] [PubMed] [Google Scholar]
- NRC . Nutrition requirements of poultry. 9th ed. National Academy Press; Washington D.C: 1994. [Google Scholar]
- Osho S.O., Adeola O. Chitosan oligosaccharide supplementation alleviates stress stimulated by in-feed dexamethasone in broiler chickens. Poult Sci. 2020;99:2061–2067. doi: 10.1016/j.psj.2019.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang H., He X., Li G., Xu H., Jia X., Nie Q., et al. Deep sequencing analysis of miRNA expression in breast muscle of fast-growing and slow-growing broilers. Int J Mol Sci. 2015;16(7):16242–16262. doi: 10.3390/ijms160716242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamela V., Federica D.C., Lorenza P. Gut microbiota profiling: metabolomics based approach to unravel compounds affecting human health. Front Microbiol. 2016;7:1144. doi: 10.3389/fmicb.2016.01144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prietsch R.F., Monte L.G., Da Silva F.A., Berra F.T., Del Pino F.A.B., Campos V.F., et al. Genistein induces apoptosis and autophagy in human breast MCF-7 cells by modulating the expression of proapoptotic factors and oxidative stress enzymes. Mol Cell Biochem. 2014;390(1):235–242. doi: 10.1007/s11010-014-1974-x. [DOI] [PubMed] [Google Scholar]
- Remans P.H.J., Gringhuis S.I., van Laar J.M., Sanders M.E., Rosas M., Coffer P.J., et al. Rap1 signaling is required for suppression of Ras-generated reactive oxygen species and protection against oxidative stress in T lymphocytes. J Immunol. 2004;173(2):920–931. doi: 10.4049/jimmunol.173.2.920. [DOI] [PubMed] [Google Scholar]
- Saeedi B.J., Liu K.H., Owens J.A., Hunter-Chang S., Camacho M.C., Eboka R.U., et al. Gut-resident lactobacilli activate hepatic Nrf2 and protect against oxidative liver injury. Cell Metab. 2020;31(5):956–968. doi: 10.1016/j.cmet.2020.03.006. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath V., Koo D.H., Lim C.B., Kim I.H. Evaluation of Lactobacillus plantarum on the growth performance, nutrient digestibility, gas emission, excreta microbiota, and meat quality in broilers. Braz J Poult Sci. 2021;23(4) eRBCA-2021-1514. [Google Scholar]
- Scanes C.G. Biology of stress in poultry with emphasis on glucocorticoids and the heterophil to lymphocyte ratio. Poult Sci. 2016;95(9):2208–2215. doi: 10.3382/ps/pew137. [DOI] [PubMed] [Google Scholar]
- Shin T.H., Lee D., Lee H.Y., Park H.J., Jin M.S., Paik M.A., et al. Integration of metabolomics and transcriptomics in nanotoxicity studies. BMB Rep. 2018;51:14–20. doi: 10.5483/BMBRep.2018.51.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin T.H., Nithiyanandam S., Lee D.Y., Kwon D.H., Hwang J.S., Kim S.G., et al. Analysis of nanotoxicity with integrated omics and mechanobiology. Nanomaterials. 2021;11(9):2385. doi: 10.3390/nano11092385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu J., Liu Y., Shan Y., Ji G., Ju X., Tu Y., et al. Deep sequencing microRNA profiles associated with wooden breast in commercial broilers. Poult Sci. 2021;100(12) doi: 10.1016/j.psj.2021.101496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel H.S. Physiological stress in birds. Bioscience. 1980;30(8):529–534. [Google Scholar]
- Simperova A., Al-Nakkash L., Faust J.J., Sweazea K.L. Genistein supplementation prevents weight gain but promotes oxidative stress and inflammation in the vasculature of female obese ob/ob mice. Nutr Res. 2016;36(8):789–797. doi: 10.1016/j.nutres.2016.03.011. [DOI] [PubMed] [Google Scholar]
- Sun J., Zhou Q., Hu X. Integrating multi-omics and regular analyses identifies the molecular responses of zebrafish brains to graphene oxide: perspectives in environmental criteria. Ecotoxicol Environ Saf. 2019;180:269–279. doi: 10.1016/j.ecoenv.2019.05.011. [DOI] [PubMed] [Google Scholar]
- Sun L., Xu G., Dong Y., Li M., Yang L., Lu W. Quercetin protects against lipopolysaccharide-induced intestinal oxidative stress in broiler chickens through activation of Nrf2 pathway. Molecules. 2020;25(5):1053. doi: 10.3390/molecules25051053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Švec P., Franz C.M.A.P. 2014. The genus Enterococcus; pp. 175–211. (Lactic acid bacteria: biodiversity and taxonomy). [Google Scholar]
- Tan C., Wei H., Ao J., Long A., Peng J. Inclusion of konjac flour in the gestation diet changes the gut microbiota, alleviates oxidative stress, and improves insulin sensitivity in sows. Appl Environ Microbiol. 2016;82(19):5899–5909. doi: 10.1128/AEM.01374-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F., Liu G., Fan L., Chen Z., Liang Y. miR-21-3p alleviates neuronal apoptosis during cerebral ischemia-reperfusion injury by targeting SMAD2. Biocell. 2021;45(1):49. [Google Scholar]
- Toborek M., Blanc E.M., Kaiser S., Mattson M.P., Hennig B. Linoleic acid potentiates TNF-mediated oxidative stress, disruption of calcium homeostasis, and apoptosis of cultured vascular endothelial cells. J Lipid Res. 1997;38(10):2155–2167. [PubMed] [Google Scholar]
- Wang J. Northwest A&F University; [China]: 2012. Dietary selenium and vitamin E alleviate DEX-induced oxidative stress in broilers. [Master's thesis] [Google Scholar]
- Wang L., Zhou H., He R., Xu W., Mai K., He G. Effects of soybean meal fermentation by Lactobacillus plantarum P8 on growth, immune responses, and intestinal morphology in juvenile turbot (Scophthalmus maximus L.) Aquaculture. 2016;464:87–94. [Google Scholar]
- Wang Y., Guo Y., Chen H., Wei H., Wan C. Potential of Lactobacillus plantarum ZDY2013 and Bifidobacterium bifidum WBIN03 in relieving colitis by gut microbiota, immune, and anti-oxidative stress. Can J Microbiol. 2018;64(5):327–337. doi: 10.1139/cjm-2017-0716. [DOI] [PubMed] [Google Scholar]
- Wang H., Latorre J.D., Bansal M., Abraha M., Al-Rubaye B., Tellez-Isaias G., et al. Microbial metabolite deoxycholic acid controls Clostridium perfringens-induced chicken necrotic enteritis through attenuating inflammatory cyclooxygenase signaling. Sci Rep-UK. 2019;9(1):1–12. doi: 10.1038/s41598-019-51104-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Sun Q., Wang J., Qiu X., Qi R., Huang J. Lactobacillus plantarum 299v changes miRNA expression in the intestines of piglets and leads to downregulation of LITAF by regulating ssc-miR-450a. Probiotics Antimicrob Proteins. 2021;13(4):1093–1105. doi: 10.1007/s12602-021-09743-1. [DOI] [PubMed] [Google Scholar]
- Wang Y., Lv X., Li X., Zhao J., Zhang K., Hao X., et al. Protective effect of Lactobacillus plantarum P8 on growth performance, intestinal health, and microbiota in Eimeria-infected broilers. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.705758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Tao J., Xiao S., Jiang S., Shang E., Zhu Z., et al. Xiexin Tang improves the symptom of type 2 diabetic rats by modulation of the gut microbiota. Sci Rep. 2018;8:3685. doi: 10.1038/s41598-018-22094-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss G.A., Chassard C., Hennet T. Selective proliferation of intestinal Barnesiella under fucosyllactose supplementation in mice. Br J Nutr. 2014;111:1602–1610. doi: 10.1017/S0007114513004200. [DOI] [PubMed] [Google Scholar]
- Weng L., Zhang F., Wang R., Ma W., Song Y. A review on protective role of genistein against oxidative stress in diabetes and related complications. Chem Biol Interact. 2019;310 doi: 10.1016/j.cbi.2019.05.031. [DOI] [PubMed] [Google Scholar]
- Wu Y., Wang B., Zeng Z., Liu R., Tang L., Gong L., et al. Effects of probiotics Lactobacillus plantarum 16 and Paenibacillus polymyxa 10 on intestinal barrier function, antioxidative capacity, apoptosis, immune response, and biochemical parameters in broilers. Poult Sci. 2019;98(10):5028–5039. doi: 10.3382/ps/pez226. [DOI] [PubMed] [Google Scholar]
- Xu K., Liu G., Fu C. The tryptophan pathway targeting antioxidant capacity in the placenta. Oxid Med Cell Longev. 2018;2018 doi: 10.1155/2018/1054797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S., Liu G., Pei C., Chen W., Li P., Wang Q., et al. Inhibition of NADPH oxidase protects against metastasis of human lung cancer by decreasing microRNA-21. Anticancer Drugs. 2015;26:388–398. doi: 10.1097/CAD.0000000000000198. [DOI] [PubMed] [Google Scholar]
- Yang X., Huo F., Liu B., Liu J., Chen T., Li J., et al. Crocin inhibits oxidative stress and pro-inflammatory response of microglial cells associated with diabetic retinopathy through the activation of PI3K/Akt signaling pathway. J Mol Neurosci. 2017;61(4):581–589. doi: 10.1007/s12031-017-0899-8. [DOI] [PubMed] [Google Scholar]
- Yin C., Xia B., Tang S., Cao A., Liu L., Zhong R., et al. The effect of exogenous bile acids on antioxidant status and gut microbiota in heat-stressed broiler chickens. Front Nutr. 2021;8 doi: 10.3389/fnut.2021.747136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z., Du W., He X., Zhang D., He W. Tribulus terrestris ameliorates oxidative stress-induced ARPE-19 cell injury through the PI3K/Akt-Nrf2 signaling pathway. Oxid Med Cell Longev. 2020;2020 doi: 10.1155/2020/7962393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Li Q., Yang C., Yu Y., Fu Z., Wang H., et al. Effects of Clostridium butyricum- and Bacillus spp.-based potential probiotics on the growth performance, intestinal morphology, immune responses, and caecal microbiota in broilers. Antibiotics. 2021;10(6):624. doi: 10.3390/antibiotics10060624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Deng Y., Feng L., Su Y., Xu D. Study on alleviate effect of Wuzhi capsule (Schisandra sphenanthera Rehder & EH Wilson extract) against mycophenolate mofetil-induced intestinal injury. J Ethnopharmacol. 2022;288 doi: 10.1016/j.jep.2022.114987. [DOI] [PubMed] [Google Scholar]
- Zhao J., Tian F., Yan S., Zhai Q., Zhang H., Chen W. Lactobacillus plantarum CCFM10 alleviating oxidative stress and restoring the gut microbiota in d-galactose-induced aging mice. Food Funct. 2018;9(2):917–924. doi: 10.1039/c7fo01574g. [DOI] [PubMed] [Google Scholar]
- Zhao H., Jiang X., Chu H. Shifts in the gut microbiota of mice in response to dexamethasone administration. Int Microbiol. 2020;23:1–9. doi: 10.1007/s10123-020-00129-x. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Wang W., Yuan T., Fu L., Zhou L., Lin G., et al. MicroRNA-29a mediates the impairment of intestinal epithelial integrity induced by intrauterine growth restriction in pig. Am J Physiol Gastrointest Liver Physiol. 2017;312(5):G434–G442. doi: 10.1152/ajpgi.00020.2017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.