Summary
Salmonella, a foodborne pathogen, has become a major public health concern because of its widespread drug resistance, including resistance to multiple drugs such as third-generation cephalosporin, ceftriaxone (CRO). However, the metabolic profile changes and associated mechanisms engendered by cephalosporin-resistant mutations remain uncharted. In this study, we have employed the LC-MS/MS metabolomics platform to determine the metabolic profiles of 138 strains of Salmonella. Our results show that metabolic profiles correspond to specific serotypes, sources, processing stages, and antibiotic resistance patterns. Notably, we observed that Salmonella Derby (S. Derby) with drug resistance to CRO has a different metabolic status with changes in glutathione biosynthesis. Specifically, glutathione oxidized (GSSG) and citrulline abundances are greatly suppressed in CRO-resistant S. Derby. Furthermore, exogenous GSSG or citrulline, but not glutathione reduced (GSH), restored the susceptibility of multidrug-resistant S. Derby to CRO. This study establishes a strategy based on functional metabolomics to manage the survival of antibiotic-resistant bacteria.
Subject areas: Immunology, Microbiology, Omics, Metabolomics
Graphical abstract

Highlights
-
•
Salmonella’s metabolic profiles are linked to serotypes
-
•
Glutathione biosynthesis affects Salmonella Derby’s ceftriaxone resistance
-
•
Exogenous glutathione oxidized/citrulline alter susceptibility of S. Derby to CRO
Immunology; Microbiology; Omics; Metabolomics
Introduction
Salmonella has emerged as a foodborne pathogen with far-reaching drug resistance, and the trend toward multiple drug resistance is increasingly alarming.1,2,3 Retail meat product surveys in the US reveal that 82% of Salmonella isolates are resistant to one or more antibiotics.4 Widespread antibiotic use in animal breeding facilitates the emergence and exacerbation of Salmonella drug resistance trends. Cross-contamination of Salmonella throughout breeding, slaughtering, processing stages, storage, transportation, marketing, and food chain transmission provides a vital avenue for drug-resistant strains, creating a possible public health threat.5,6 Escalating bacterial resistance has placed significant pressure on clinical treatment and antibiotic research and development, necessitating the monitoring of foodborne Salmonella resistance.7,8
Cephalosporins, a class of broad-spectrum antibiotics, have not been extensively employed in the aquaculture industry owing to their exorbitant cost, and are largely utilized for the treatment of human diseases. However, cephalosporin resistance remains widespread worldwide. According to reports, during the period between 2014 and 2017, cephalosporin resistance was observed in nearly 26.9% of Enterobacter cases associated with healthcare related infections.9 Monitoring data for two decades indicated that among people infected in sub-Saharan Africa, 85.7% of Klebsiella isolates and 31.7% of Escherichia coli (E. coli) isolates exhibited resistance to third-generation cephalosporins.10 Third-generation cephalosporin ceftriaxone (CRO) is currently considered the first and last line of treatment by institutes recognized by the World Health Organization (WHO) for severe bacterial infections.11,12 Developing new drug preparations to combat antibiotic-resistant pathogens is a time-consuming process and not a viable solution to manage the growing prevalence of multidrug-resistant infectious diseases. Therefore, comprehending the mechanism of antibiotic resistance and controlling these resistant pathogens is the most significant scientific challenge at present.
Currently, the investigation of drug resistance mechanisms primarily centers on the genetic level.13,14 However, recently, the metabolic mechanisms underlying bacterial antibiotic resistance have also come to light. A strain’s antibiotic resistance incites fluctuations in the quantity of both known and unknown metabolites.15 Conversely, bacterial metabolism substantially affects the strain’s susceptibility toward antibiotics. Furthermore, the obtained biomarkers from metabolic analysis display the potential to mitigate antibiotic resistance by reinstating the metabolome of antibiotic-resistant strains to that of antibiotic-sensitive strains.16 Studies have demonstrated a variety of mechanisms by which Salmonella and other bacteria develop resistance to cephalosporins, including modifications in membrane permeability/drug intake, target site inactivation, and β-lactamase hydrolysis of antibiotics.17,18,19,20 However, a comprehensive depiction of the metabolic profile changes and associated mechanisms engendered by cephalosporin-resistant mutations currently remains uncharted.
The susceptibility of bacterial strains to antibiotics is tightly linked to their metabolic status, and particular metabolic profiles correspond to specific antibiotic resistance patterns. Therefore, there exists a potential avenue for reversing antibiotic resistance by targeting the metabolomics profile of an antibiotic-resistant strain to match that of an antibiotic-sensitive strain. By comprehending the mechanisms through which antibiotics interact with bacterial metabolism, we can gain valuable insights into their modes of action and develop more effective therapeutic strategies. Targeting microbial metabolism to augment the bactericidal efficacy of antibiotics may offer a promising approach for mitigating the burden of infections. Considering these observations, it is plausible that administering certain chemicals could help reverse antibiotic resistance through modulation of the metabolomics profile of the resistant strain to closely resemble that of the susceptible strain. It has been demonstrated that the augmentation of reactive oxygen species (ROS) synthesis in E. coli may result in a predictable enhancement of the bacterium’s vulnerability to oxidative onslaught. Correspondingly, escalated production of ROS in E. coli may amplify the efficacy of oxidants and antibiotics in inducing pathogen’s demise.21 Selective metabolic triggers facilitate the eradication of both Gram-negative (E. coli) and Gram-positive (Staphylococcus aureus) persisters through aminoglycoside-mediated action.22
In this investigation, we proposed an approach for identifying metabolic modulators through metabolomic analysis. The identified modulators possess potential therapeutic applications. Specifically, the present research aimed to investigate the spatial distribution of food-borne Salmonella drug resistance employing 138 strains obtained from the entire meat food chain of farming, slaughter, and retail. Importantly, our findings revealed a significant prevalence of cephalosporin resistance. We mitigated potential confounding factors such as strain source and serotype heterogeneity by evaluating the metabolic profile of 90 drug-resistant Salmonella strains precisely selected for the same serotype Derby and the same source (pig origin). The current study provides an innovative framework for exploring drug-resistance mechanism while offering new opportunities for preventing and managing pathogenic infections. Altogether, our results offer promise for identifying metabolic modulators for therapy in the future.
Results
Drug resistance of Salmonella isolates
138 strains of Salmonella were isolated and tested for resistance to 24 commonly used antibiotics (Data S1). Results showed that all strains exhibited certain levels of resistance. Among the isolated Salmonella species, the most serious resistance was found against ciprofloxacin, levofloxacin, and methoxybenzyl pyrimidine/sulfamethoxazole, with a resistance rate of 100% (Figure 1A). Meanwhile, cefazolin and CRO exhibited relatively high resistance rates of 71.82% and 52.49%, respectively. In contrast, cefepime and ceftazidime demonstrated lower resistance rates, ranging from approximately 10–17%. In this article, the degree of drug resistance has been normalized based on the MIC fold value of the drug sensitivity standard set by the American Association of Clinical Laboratory Standards.
Figure 1.
Detection of the drug resistance profile of Salmonella
(A) Analysis of the drug resistance rate of 138 Salmonella strains to 24 antibiotics classified into 12 classes.
(B) Histogram of the number of multi-resistant and susceptible strains of 138 Salmonella strains against 24 antibiotics.
In addition, the situation of multiple drug resistance is also alarming (Figure 1B). Among the 24 commonly used antibiotics, 47.51% of them are resistant to 10–15 types of antibiotics, whereas only 12.02% of them are resistant to 0–5 types of antibiotics. Further data analysis revealed that among the isolates of foodborne Salmonella, 72.68% demonstrated intermediate resistance to 0–5 antibiotics, making them prone to developing into drug-resistant strains and potentially becoming multi-drug resistant in the future.
Multivariate statistical analysis of food-borne Salmonella metabolic profiles
The LC-MS/MS metabolomics platform was utilized to detect metabolites in 138 strains of Salmonella. A total of 301 metabolites were annotated based on the identification guidelines mentioned above (Datas S3 and S4). The 138 Salmonella foodborne strains were classified according to serotype, source, and processing stages (Data S2). Principal component analysis (PCA) was then carried out with the annotated metabolites. By analyzing Salmonella from four different processing stages using PCA, differences in metabolites among Salmonella from different processing stages were found (Figure 2A). The PCA map of Salmonella from pig and chicken also reveals differences in the metabolic characteristics of Salmonella from the two sources (Figure 2B). The environmental water source was removed because it contained only one sample. Furthermore, some serotypes exhibited metabolic differences. For example, the metabolic characteristics of Salmonella Typhimurium and Salmonella Indiana were markedly different, whereas the differences with S. Derby serotypes were relatively small (Figure 2C). These results demonstrate differences in metabolites among different serotypes. We therefore investigated the relationship between drug resistance and Salmonella metabolites by selecting the same serotype and source of Salmonella strains.
Figure 2.
PCA analysis of 301 metabolites from 138 Salmonella strains
(A) Bacterial source: Chicken source (n = 35), Pig source (n = 102).
(B) Processing stages: Farmers market (n = 5), Farms (n = 105), Slaughterhouse (n = 12), Supermarket (n = 16).
(C) Serotype: Derby (n = 90), Kentucky (n = 3), Typhimurium (n = 20), Enteritis (n = 7), Corvallis (n = 4), Schwarzengrund (n = 3), Indiana (n = 4), Mbandaka (n = 3), Weltevreden (n = 4).
Multivariate statistical analysis of the metabolic profile and overall drug resistance of foodborne Salmonella
The drug resistance rates for levofloxacin, ciprofloxacin, and trimethoprim/sulfamethoxazole were found to be 100%, whereas tigecycline, doripenem, and ertapenem showed a 0% resistance rate. Because a comparison of drug resistance in metabolic profiles was not feasible, the remaining 15 antibiotics were chosen to study the relationship between metabolic characteristics and antibiotic resistance, as depicted in Figure 3. Furthermore, the metabolic characteristics varied based on the different antibiotic resistance, thus producing distinctive differences.
Figure 3.
PCA analysis plot of metabolites and different antibiotic resistances of Salmonella
(A) beta-Lactam (doripenem, meropenem, ampicillin, ampicillin sulbactam).
(B) Amino glycosides (amikacin, gentamicin, tobramycin).
(C) Quinolone (levofloxacin, ciprofloxacin).
(D) Four-ring class (tetracycline, minocycline).
(E) Semi-synthetic cephalosporins (cefazolin, ceftazidime).
(F) Cephalosporins (cefepime, ceftriaxone).
Most antibiotics have a singular resistance profile result, with an overall bias toward either resistance or non-resistance, and are not suitable for in-depth histological study (Figures 3A–3D). However, nitrofurantoin, cefepime, and CRO antibiotics displayed significantly different metabolic characteristics between drug-resistant and sensitive strains, whereas the metabolic differences for amikacin and meropenem antibiotics were only slightly different. It can be observed that, among the 138 Salmonella strains analyzed, there was a relatively high number of bacteria exhibiting both resistance and sensitivity to semi-synthetic cephalosporins such as cefazolin and ceftazidime, as well as cephalosporins including cefepime and CRO (Figures 3E and 3F). Particularly for CRO, almost half of the bacteria were found to be resistant whereas the other half were not. Therefore, these 138 Salmonella strains are suitable for investigating the association between resistance and metabolism of cephalosporins.
Metabolic analysis of cephalosporin-resistant S. Derby
To avoid any influence from source factors, serotype factors, and factors in the processing stages on our study of Salmonella resistance and metabolism, we screened single serotype, single source strains for analysis. We found that the highest percentage of the serotype S. Derby consisted of approximately 70 strains. On analyzing the antibiotic resistance of these 70 strains, we discovered that 32 of them had very extreme distribution patterns toward semi-synthetic cephalosporins (cefazolin and ceftazidime) as well as cephalosporins (cefepime and CRO) (Data S6). To further investigate, we screened these 32 S. Derby strains to analyze their metabolic profiles with respect to these same types of antibiotics.
The LC-QE-MS platform was used to carry out untargeted metabolomics studies of these 32 S. Derby strains (Data S5). The resistance profiles of these strains to two types of antibiotics, semi-synthetic cephalosporins (cefazolin, ceftazidime) and cephalosporins (cefepime, CRO), are shown in Figure 4A. For cephalosporins (cefepime, CRO), the resistance profiles among the 32 Salmonella strains were widely distributed, with nearly half being fully resistant and the other half not. PCA was carried out for the annotated 301 metabolites. As shown in Figure 4B, the yellow dot represents the drug-resistant group, the blue dot represents the susceptible group, and each dot represents an S. Derby strain. It can be observed that the Salmonella strains in each group clustered together, indicating that the characteristics of the same resistant phenotype were similar. Some degree of separation was observed between the drug-resistant group and the susceptible group. The observed difference shows that the metabolic profiling between the two groups was inconsistent. The results showed that there were differences in metabolic characteristics between the two groups. To improve the illustration of differential substances that contribute more to the model between the two groups, the S-plot analysis indicates that there is up-regulation of metabolites and some down-regulation of metabolites in S. Derby resistant to cephalosporins compared to S. Derby sensitive. These metabolites include GSH, citrulline, and GSSG as shown in Figure 4C.
Figure 4.
Differential metabolite screening for resistance and susceptibility of S. Derby to cephalosporins-like antibiotics
(A) MIC of resistance of 32 screened S. Derby to 2 classes of 4 antibodies (semi-synthetic cephalosporins (cefazolin, ceftazidime) and cephalosporins (cefepime, ceftriaxone)) analysis, Susceptible (n = 15), Resistant (n = 17).
(B) PCA plot, susceptible (n = 15) and resistant (n = 17) are marked with different colors.
(C) S-plot of Par-scaling OPLS-DA model.
(D) Volcano plot, two-sided Welch’s t-test p < 0.05, Fold change >1 or <0.5.
(E) Volcano plot, Mann-Whitney U test p < 0.05, Fold change >1 or <0.5.
(F) Table of statistically significantly different metabolites, including VIP, two-sided Welch’s t-test p value, Mann-Whitney U test p value, FC.
In univariate statistical analysis, volcano plots are effective in displaying the statistical differences of metabolites between groups. We have analyzed the statistical differences of metabolites between two groups using parametric test (p < 0.05) (Figure 4D) and non-parametric test (p < 0.05) (Figure 4E). We have obtained the difference multiple by calculating the abundance ratio of metabolites between the two groups (FC value >2 or <0.5). Both parametric and non-parametric test-based volcano plots have shown that GSSG, D-fructose, citrulline, GSH, L-pyroglutamic acid, and glutamate were statistically significant metabolites. To demonstrate the statistical properties of these differential substances, a table is provided in Figure 4F including the VIP in OPLS-DA, the p value of the parametric and non-parametric tests in one-way statistical analysis, as well as the fold change value.
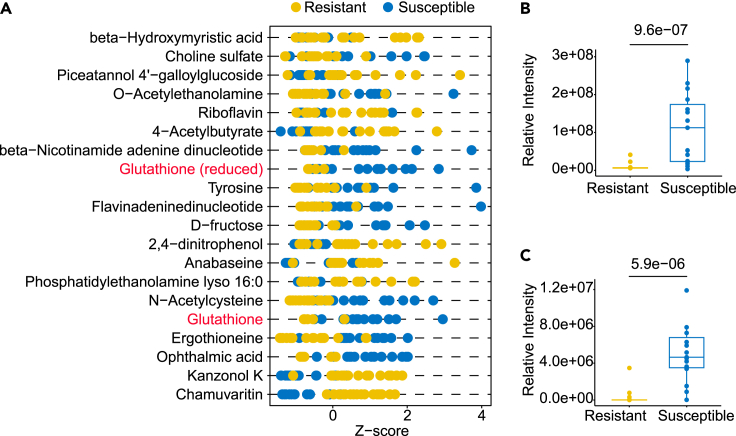
Z score plot is a graphical representation of standardized data used to show the first 20 metabolites from 32 S. Derby strains. As shown in Figure 5A, the yellow dot represents the drug-resistant group (n = 15), whereas the blue dot represents the susceptible group (n = 17). It is an ideal tool for analyzing datasets and identifying potential outliers. Figure 5A illustrates the Z-scores of peak intensities on the x axis and the top 20 metabolites on the y axis. The distribution of quite a few metabolites shows polarization, with GSH and GSSG marked in red. A boxplot is used to display the two metabolites and observe for outliers. Figures 5B and 5C show that GSH and GSSG have significant p values of 9.6e-07 and 5.9e-06, respectively. GSH has no outliers in both groups, whereas GSSG has one outlier in the susceptible group and one in the resistant group.
Figure 5.
Z score plot visualization of statistically significant metabolites
(A) Z score plot of the first 20 metabolites from 32 S. Derby strains, yellow dot represents drug resistant group (n = 15), blue dot represents susceptible group (n = 17). Box and Whisker plots of metabolites.
(B) GSH.
(C) GSSG, with the box ranging from the first quartile to the third whereas the whiskers going from each quartile to the minimum or maximum (resistant, N = 15; susceptible, N = 17), p value was labeled, two-sided Welch’s t-test.
Pathway enrichment of cephalosporin-resistant S. Derby
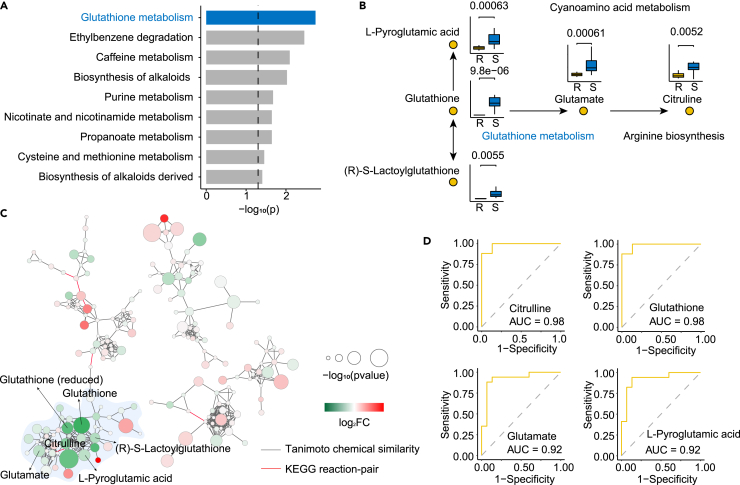
In this study, we performed pathway enrichment on metabolites with significant statistical changes in S. Derby strains. To determine the most relevant pathway affected by cephalosporin resistance, pathway enrichment analysis was performed on the marker differential metabolites. Through analysis and calculation, 26 metabolic pathways were obtained, As shown in Figure 6A, the bar plot visualized the top 9 enriched metabolic pathways. Glutathione metabolism, ranking the first, was the enriched with influence values 0.31, and -log10 p value 2.91, which indicated that the glutathione metabolism is closely related to the drug resistance of foodborne S. Derby to cephalosporins. To better understand the glutathione metabolism pathways, we made a joint pathways map, which concluding citrulline, glutathione, glutamate, L-pyroglutamic acid and (R)-S-lactoylglutathione, displayed in boxplot with p value labeled (Figure 6B). Very interestingly, we found that all these metabolites were down-regulated in drug-resistant bacteria, relative to sensitive strains.
Figure 6.
Pathway analysis of statistically significant changes in metabolites
(A) Metabolic pathway enrichment of statistically significant changes in metabolites of S. Derby strains between resistant and susceptible strains to cephalosporins.
(B) The pathways of glutathione metabolism and their relative pathways and metabolites were displayed as a boxplot with the p value labeled.
(C) MetaMapp visualization of metabolic changes in S. Derby strains relative to metabolite levels different between resistant strains and susceptible strains to cephalosporins. Red nodes represent metabolites with increased signal intensity in resistant strains; green nodes represent metabolites with decreased signal intensity in resistant strains (log2FC). Node sizes scale with -log10p value. Blue edges connect metabolites with Tanimoto chemical similarity 700; red edges connect metabolites from the KEGG RPAIR database.
(D) The ROC curve of citrulline, glutathione, glutamate, L-pyroglutamic acid.
A metabolic network is a visual representation of the interactions between different metabolites involved in various metabolic pathways. It provides a comprehensive overview of the metabolic network and aids in understanding the complex relationships between different biochemical reactions. Metabolic network diagrams are the best way to show significant and non-significant differences are associated through network relationships, including Tanimoto chemical similarity and KEGG RPAIR (Data S7 and S8). Not surprisingly, there are many green circles marked by the light blue section, which means that these metabolites are significantly down-regulated in resistant strains, and these down-regulated metabolites are glutathione metabolism or metabolites in their associated metabolic pathways, including glutathione, glutamate, et al. as mentioned before (Figure 6C). This part of the metabolites is structurally similar or has an enzymatic relationship, which again suggests that the degree of cephalosporin resistance in S. Derby, is related to these pathways or these types of metabolites. To determine whether these metabolites have the predictive potential to distinguish between drug-resistant and sensitive groups, we developed the random forest model based on 10-fold cross validation. Receiver operating characteristic (ROC) curves of the predicting utilities of selected biomarkers are shown in Figure 6D. Four potential biomarkers—citrulline, glutathione, glutamate, and L-pyroglutamic acid—, were found. Their AUC values were 0.98, 0.98, 0.92 and 0.92, respectively.
These results prompted us to explore whether the inhibitory levels of citrulline and glutathione are useful biomarkers of antibiotic resistance and whether the drug resistance phenotype of S. Derby changes in the presence of exogenous citrulline or glutathione (GSSG and GSH).
Exogenous metabolites alter susceptibility of drug-resistant S. Derby to CRO
To visualize more the relationship between different levels of resistance to cephalosporin antibiotics and Salmonella metabolites, linear regression equations were used for visual presentation (Data S9). As shown in Figures 7A–7C, three typical metabolites, including GSH, GSSG, and citrulline, were selected for linear regression analysis with CRO. It is easy to see that in both resistant and sensitive S. Derby, all 3 metabolites show a clear difference in distribution, which was demonstrated in the previous boxplot. For intermediate between sensitive and resistant, i.e., MIC values around 0, in most cases, the abundance of metabolites remains in the middle of resistant and sensitive S. Derby. The overall trend shows a somewhat linear relationship. The exogenous addition experiment was performed to validate whether S. Derby’s resistance to cephalosporins could be modified or reversed by adding supplementation of these three metabolites. As shown in Figure 7D, different concentrations of metabolites and cephalosporins were added to the S. Derby’s resistance to cephalosporins and incubated at 37°C for 6 h. The blank group showed that the addition of GSH, GSSG and citrulline had no effect on the number of bacterial colonies (Figures 7E, 7F and 7H) when no antibiotics were added, excluding the effect of the three metabolites on bacterial survival. However, it is surprising that the addition of 5 mM–80 mM GSH does not weaken bacterial viability. The total number of bacterial colonies is positively correlated with the amount of GSH added. When the amount of GSH added is 80 mM, the total number of bacterial colonies is significantly increased (p < 0.01), reaching 2.94 × 107 CFU/mL, which is 4.62 times that without GSH added (Figure 7E). When grown in the medium supplemented with CRO, the cell survival rate decreased with increasing dose of GSSG or citrulline. The addition of 5 mM GSSG significantly reduced the bacterial count (p < 0.01). When the amount of GSSG reaches 40 mM, the survival rate is only 28.04% (Figure 7G); The bacterial count decreased significantly after the addition of 20 mM citrulline (p < 0.05). In the presence of 80 mM citrulline, the cell survival rate decreased by a factor of 2.02 (Figure 7I).
Figure 7.
Relationship between different levels of cephalosporin antibiotic resistance and Salmonella metabolites
(A–D) Linear regression equation for assessing the relationship between metabolites and the degree of drug resistance, 95% confidence interval, A GSH, B GSSG, C citrulline; D Schematic diagram of CRO-resistant Salmonella metabolism from reprogramming.
(E) Changes in drug resistance of cephalosporin bacteria by exogenous addition of different doses of GSH (n = 3; ∗∗∗, p<0.01).
(F and H) Effects of exogenous addition of different doses of GSSG and citrulline on the viable bacteria without CRO (n = 3; p>0.05).
(G and I) Effects of exogenous addition of different doses of GSSG/citrulline on the viable bacteria with CRO (n = 3; ∗, p<0.05; ∗∗∗, p<0.01).
Discussion
The efficacy of antibiotics against bacteria is strongly influenced by the metabolic state of the latter, and certain metabolic configurations are associated with distinct forms of antibiotic resistance. Identifying and employing compounds capable of modifying the metabolic pattern of a resistant strain to resemble that of a susceptible strain may represent a viable strategy for mitigating antibiotic resistance. Our contention is that specific metabolic profiles are correlated with antibiotic resistance, providing a means to predict a strain’s response to antibiotic treatment. Furthermore, we affirm that compounds capable of modulating the metabolic status of an antibiotic-resistant strain to simulate that of an antibiotic-susceptible one, have the potential to revert the antibiotic resistance phenotype, rendering the bacteria vulnerable to treatment. Our research endeavors are presently directed toward studying the potential efficacy of external metabolites in modifying the metabolic profile of antibiotic-resistant strains to match that of antibiotic-susceptible strains.
In the current investigation, we utilized a metabolomics-based approach to identify potential metabolic modulators as a promising strategy for combating antibiotic resistance. A cohort of 138 Salmonella foodborne strains, randomly selected based on their divergent serotypes, sources, and processing stages, were investigated to evaluate the association between their metabolic profiles and these influential factors. Results indicate that all the aforementioned predictors significantly impact the metabolic profiles of Salmonella. Thereafter, we extended our research by dissecting the resistance profile exhibited by these 138 strains. Intriguingly, we observed that these Salmonella strains displayed substantial resistance to cephalosporin antibiotics, showcasing an unprecedented distribution. After the random selection of these strains one specific serotype, Salmonella Derby with single specie source and food processing stages, was singled out to explore the link between metabolic profile diversity and resistance to antimicrobial drugs.
The metabolic profiling of 32 strains of S. Derby was performed using the LC-MS/MS metabolomics platform, with a specific focus on semi-synthetic cephalosporins (cefazolin and ceftazidime), as well as cephalosporins (cefepime and CRO). The statistical analysis revealed several statistically significant metabolites including GSSG, D-fructose, citrulline, GSH, L-pyroglutamic acid, and glutamate. Furthermore, metabolic pathway enrichment analysis revealed glutathione metabolism to be the most significantly enriched pathway, with an influence value of 0.31, and a -log10 p value of 2.91.
As per the report, the management of the TCA cycle for the enhancement of antibiotic absorption and consequent escalation of destructivity is achieved through the manipulation of glucose and/or alanine, which undermines the resistance and reinforces the penetrative quality of aminoglycoside antibiotics to eradicate strains that are resistant to such medications.23 The susceptibility of bacteria can be affected by the regulation of bacterial-derived exopolysaccharides through fructose and glucose.24 The exogenous glucose stimulates uptake of aminoglycoside antibiotics by E. coli persisters.22 Furthermore, the exogenous glucose and/or alanine are converted to acetyl-CoA and then flux to the TCA cycle, which is regulated by glutamate, aspartate, and glyoxylic acid cycle, indicating that increased activity of a-oxoglutarate dehydrogenase may promote bacterial susceptibility to kanamycin.23
Alanine is catabolized to pyruvate similarly to glucose, and pyruvate is then transformed to acetyl-coenzyme A, which serves as a fundamental precursor molecule for the tricarboxylic acid cycle. The amino group of alanine is transferred to α-ketoglutarate by alanine aminotransferase, generating pyruvate and L-glutamate. The amino acid L-glutamate is unique in that it undergoes rapid oxidative deamination by glutamate dehydrogenase with either NAD or NADP acting as a coenzyme, resulting in the production of α-ketoglutarate. Glutamate, a precursor for the synthesis of glutathione, is involved in the synthesis of glutathione through the γ-glutamyl cycle. Both are essential for the maintenance of cellular redox balance and the detoxification of reactive oxygen and nitrogen species.
We hypothesize that a decrease in citrulline and glutathione levels may underlie the development of drug resistance in CRO. However, previous metabolic studies were unable to differentiate between GSSG and GSH as a biomarker. To address this, we conducted a validation study to investigate the impact of exogenous GSH and GSSG metabolites on the susceptibility of drug-resistant S. Derby to CRO. Our research has uncovered an innovative discovery that the re-programming of glutathione metabolism via GSSG modulation can enhance the resistance of Salmonella against cephalosporins. Notably, we observed that GSH, in contrast, did not demonstrate the same level of modulatory effects. Citrulline has a similar effect to GSSG, modulating bacterial metabolism and altering drug-resistant phenotypes in exogenous addition experiments. In a similar study reported, citrulline and glutamine promoted the TCA cycle, produced more NADH in the bacteria, and increased the proton-motive force, thus promoting apramycin entry into the bacterial cells, and killing the drug-resistant bacteria.25
GSH and GSSG are two forms of the same molecule, glutathione. A reduced ratio of glutathione in the reduced state (GSH) to oxidized state (GSSG) is commonly perceived as an indication of oxidative stress. The alteration in metabolite levels implies enhanced GSH biosynthesis to counterbalance its continuous turnover and consumption because of antioxidant activities. In response to bactericidal antibiotics, an increase in cellular respiration leads to the generation of ROS, whereas the overexpression of Catalase or MutS and the prior antioxidant pretreatment can diminish antibiotic lethality, illustrating the pivotal involvement of ROS in antibiotic-triggered apoptotic processes.26 The glutathione S-transferase plays a vital role in the protection against various types of antibiotics,27 GSH acidity plays a key role in biofilm disruption of many MDR bacterial isolates and enhances antibiotic efficacy.28 A robust correlation exists between the synthesis of GSH and the vulnerability of Enterobacteriaceae toward antibiotics.29
In conclusion, our metabolomics analysis suggests that a reduction in glutathione levels may be primarily attributed to reduced GSSG levels. It is known that GSSG is reduced to GSH under the catalysis of glutathione reductase. Our findings demonstrate that the levels of GSSG are significantly lower in drug-resistant strains, and the supplementation with GSSG enhances bacterial sensitivity to CRO. However, the relationship between GSSG and drug resistance warrants further investigation. Overall, our results suggest that both GSSG and citrulline facilitate effective bacterial killing by CRO, a finding not previously reported in the literature. Our results suggest that GSSG could decipherer the S. Derby metabolic states involved in antibiotic resistance of CRO, targeting glutathione biosynthesis would represent a viable option for tackling antibiotic resistance. This innovative approach has the possibility of expanding therapeutic options and ameliorating the clinical management of bacterial infections. As a result, it could lead to a more favorable future in the field of antibiotic therapy.
Limitations of the study
The article only focused on studying the metabolic differences of CRO-resistant strains of Derby serotype Salmonella, and further validation is required to determine whether the results hold true for other serotypes of Salmonella. The levels of GSSG were found to be significantly lower in drug-resistant S. Derby. In addition, it was observed that supplementing GSSG led to an increase in S. Derby sensitivity to CRO, whereas GSH did not demonstrate such an effect. This phenomenon was not thoroughly investigated in this paper, and therefore, it cannot be deciphered why supplementing GSH fails to increase S. Derby sensitivity to CRO. Future studies could employ transcriptomics and genomics to validate the underlying reasons for GSH’s failure to increase bacterial sensitivity to CRO.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterialand virusstrains | ||
| 90 strains of Salmonella Derby | Pig & Chicken | Farm-pig_1–87; Process _1∼3 |
| 3 strains of Salmonella Kentucky | Chicken | Process _4∼6 |
| 20 strains of Salmonella Typhimurium | Pig & Chicken & Environmental water | Farm-pig_88–102; Process _7∼11 |
| 7 strains of Salmonella Enteritis | Chicken | Process _12∼18 |
| 4 strains of Salmonella Corvallis | Chicken | Process _19∼22 |
| 3 strains of Salmonella Schwarzengrund | Chicken & Anal swab | Process _23∼25 |
| 4 strains of Salmonella Indiana | Chicken & Aquatic product | Process _26∼29 |
| 3 strains of Salmonella Mbandaka | Chicken & Anal swab | Process _30∼32 |
| 4 strains of Salmonella Weltevreden | Chicken & Anal swab | Process _33∼36 |
| Chemicals,peptides,andrecombinant proteins | ||
| Methanol, acetonitrile, isopropanol, chloroform, MTBE | TEDIA Company, Inc. | LC/MS grade |
| GSSG | ShanghaiyuanyeBio-TechnologyCo.,Ltd | LC/MS grade |
| GSH | ShanghaiyuanyeBio-TechnologyCo.,Ltd | LC/MS grade |
| L-Citrulline | ShanghaiyuanyeBio-TechnologyCo.,Ltd | LC/MS grade |
| Critical commercial assays | ||
| Gram Negative MIC plate | Thermo Fisher Scientific | Sensititre™ GN4F |
| Deposited data | ||
| LC-MS/MS raw data | This paper | Metabolight ID MTBLS5183 |
| Software and algorithms | ||
| Image formatting | Adobe Illustrator CS6 | https://www.adobe.com/ |
| LC-MS/MS data processing | MS dial | http://prime.psc.riken.jp/compms/msdial/main.html |
| LC-MS/MS raw data convert | ABF convert | https://www.reifycs.com/AbfConverter/ |
| Data visualization and statistics | R | https://www.r-project.org/ |
| IDE of R | Rstudio | https://www.rstudio.com/categories/rstudio-ide/ |
| Metabolic network calculation | Metamapp | http://metamapp.fiehnlab.ucdavis.edu/ocpu/library/MetaMapp2020/www/ |
| Metabolic network visualization | Cytoscape | https://cytoscape.org/ |
| R code for visulization | Github | Github: JianJi2016/Reprogramming-Salmonella |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jian Ji. E-mail: jijian@jiangnan.edu.cn.
Materials availability
This study did not generate new unique reagents.
Experimental model and study participant details
Strains conditions and bacterial culture
Salmonella strains were provided by Huazhong Agricultural University (Hubei, China). These strains were stored frozen in 20% glycerol at −80°C. Working cultures were prepared by transferring 100 μL samples to 5 mL Luria-Bertani (LB) medium and incubating at 37°C for 24 h. The culture was diluted in Mueller-Hinton broth (MHB) to achieve a final concentration of approximately 6.0 log10 CFU mL−-1 in the reaction mixtures.
Method details
Drug resistance testing
50 μL of broth suspension was transferred to the micropore of the Sensititre Gram-negative plate (GN4F, Thermo Scientific, USA) and incubated at 37°C for 24 h. The positive control pore was Escherichia coli ATCC 25922. Minimal inhibitory concentration (MIC), defined as the lowest concentration of antibiotic required to inhibit visible growth of test microorganisms. The breakpoint MICs of the tested antibiotics against Salmonella were determined using the reference values recommended by the Clinical and Laboratory Standards Institute (CLSI).
Salmonella preparation and metabolite extraction
All samples were harvested after 24 hours of culture and bacterial concentration was measured using a turbidimeter to facilitate normalization in the processing of the metabolic data. The culture tubes were placed on ice for cooling to quench enzyme activity. The supernatant was removed by centrifugation at 12000 rpm at 4°C to collect wet cells. Wash twice with pre-chilled (4°C) PBS, mix by pipetting up and down, then add 2 mL of washed bacterial suspension to centrifuge tubes, centrifuge at 4°C, 12000 r/min, remove supernatant and collect wet bacterial samples.
The collected wet microbial cells were rapidly quenched in liquid nitrogen for 1 min, thawed on ice at 4°C and then extracted. The extraction solvent was prepared from mass spectrometry grade acetonitrile, isopropanol and water in a ratio of 3:3:2 (v : v : v), pre-cooled at −20°C. 0.5 mL of extraction solvent, two 1.5 mm stainless steel balls and one 2 mm stainless steel ball were added to the EP tube for shaking and crushing. Gen-Grinder was used to grind the cells at 1500 rpm for 30 seconds for 8 cycles. The cells were sonicated in an ice bath at 4°C for 5 min. The grinding step was repeated once. After removing the steel balls with a magnet, the homogenate was centrifuged at 13,000 rpm for 2 minutes and 495 μL of supernatant was collected and transferred to a new test tube. Add an aliquot of 0.5 mL extraction solvent to the old tube and grind the cells again. Divide the supernatant into two aliquots of 475 μL, one for analysis and the other for backup.
UHPLC-QE-MS metabolites detection
For UHPLC-MS/MS analysis of metabolites, the chromatographic condition was consistent with the previous report.30 3 μL of the resuspended dried aliquots were injected into a Vanquish UHPLC system (Thermo Scientific, Waltham, MA, USA) equipped with a Waters Acquity UPLC HSS T3 column (1.8 μm, 2.1 × 100 mm) coupled with an additional Waters Acquity VanGuard BEH Amide precolumn (5 mm × 2.1 mm; 1.7 μm). The oven temperature was maintained at 45°C and the flow rate was set at 0.4 mL/min. The chromatographic separations were carried out with the following parameters. In pos mode, solvent A was water with 0.1% formic acid and solvent B was acetonitrile with 0.1% formic acid. In neg mode, solvent A was water with 0.5 mM NH4F and solvent B was acetonitrile. A gradient run was set up as 0–1 min at 99% A, 1–8 min from 99% to 1% A and hold for 2 min, 10–10.1 min from 1% to 90% A and 10.1–12 min re-equilibration at 99% A. Ion source conditions were set as follows: spray voltage, 3.6 kV; sheath gas flow rate, 60 arbitrary units; auxiliary gas flow rate, 25 arbitrary units; sweep gas flow rate, 2 arbitrary units; capillary temperature, 300°C; S-lens RF level, 50; auxiliary gas heater temperature, 370°C. The following acquisition parameters were used for MS1 analysis: resolution, 60000; AGC target, 1e6; maximum IT, 100 ms; scan range, 60–900 m/z; spectrum data type, centroid. Data dependent MS/MS parameters: resolution, 15000; AGC target, 1e5; maximum IT, 50 ms; number of loops, 3; TopN, 3; isolation window, 1.0 m/z; fixed first mass, 70.0 m/z; (N)CE/stepped nce, 20, 30, 40; spectrum data type, centroid; minimum AGC target, 8e3; intensity threshold, 1.6e5; exclude isotopes, on; dynamic exclusion, 3.0 s.
Metabolite profiling analysis
All LC-MS/MS raw data files were converted to ABF format using the ABF converter (https://www.reifycs.com/AbfConverter/). MS-DIAL ver.4.00 software was used for deconvolution, peak picking, alignment, and compound identification.31 The detailed parameter settings were as follows: MS1 tolerance, 0.005 Da; MS2 tolerance, 0.01 Da; minimum peak height, 20000 amplitudes; mass slice width, 0.1 Da; smoothing method, linear weighted moving average; smoothing level, 5 scans; minimum peak width, 10 scans. [M+H]+, [M+NH4]+, [M+Na]+, [2M+H]+, [2M+NH4]+, [2M+Na]+ were included in the adduct ion setting for positive mode metabolomics analysis and [M-H]-, [M+Cl]-, [M+FA-H]-, [2M-H]- for negative mode metabolomics analysis. Compounds were annotated with accurate precursor masses and MS/MS spectra against libraries in MoNA.
Exogenous addition validation experiment
The exogenous addition experiment was performed as published.16 Briefly, a single colony was pre-plated and plated into 130 mL LB broth in 250 mL flasks, placed on a shaker at 200 rpm and incubated overnight at 37°C. After centrifugation at 8000 rpm for 5 min, the samples were washed twice with 30 mL sterile saline and resuspended with M9 minimal medium to bring the MCF to 1.5. Different concentrations of metabolites and antibiotics were added to the reaction samples and incubated at 37°C for 6 h. The use of M9 medium facilitates the testing of the effects of different metabolites on antibiotic resistant strains and avoids the occurrence of confounding factors as in more complex media. After incubation, 1 mL aliquots are periodically removed, serially diluted, and plated (100 μL aliquots) onto Plate Count agar plates. Plates are incubated at 37°C for 18 hours. Only those dilutions yielding 30–300 colonies were counted to calculate colony forming units (CFU). Survival was determined by dividing the CFU obtained from treated samples by that obtained from the control.
Quantification and statistical analysis
Statistical data analysis was conducted utilizing ropls32 R package, which served as the basis for performing a Principal component analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS-DA). Metabolic pathway enrichment was analyzed through the Metaboanalyst 4.0 website.33 Metabolite networks were constructed using a MetaMapp34 approach (web-based portal, version 2023) that calculated biological pathways relevance (KEGG reactant pairs) and chemical structural similarity (Tanimoto coeffificient >0.7) which were further visualized in CytoScape with version 2.8.2 for Window 11. Plots and analyses including Box and Whisker plot, barplot, principal component analysis score and scree plot, variable importance plot, as well as Z score scatterplot were generated in R (version 4.2.2) using packages including textshape (version 1.7.3), ropls (version 1.30.0), ggplot2 (version 3.4.0), ggpubr (version 0.5.0), ggrepel (version 0.9.3), tibble (version 3.1.8), ggsignif (version 0.6.4), plotROC (version 2.3.0), dplyr (version 1.1.0).
Acknowledgments
We thank Prof. Menghong Dai at Huazhong Agricultural University (China) for providing us 3 strains of Salmonella Kentucky, 20 strains of Salmonella Typhimurium, 7 strains of Salmonella Enteritis, 4 strains of Salmonella Corvallis, 3 strains of Salmonella Schwarzengrund, 4 strains of Salmonella Indiana, 3 strains of Salmonella Mbandaka, 4 strains of Salmonella Weltevreden. And Prof. Jianmin Zhang at South China Agricultural University (China) for providing us 90 strains of Salmonella Derby. This work was supported by the National Outstanding Youth Science Foundation Project (32125031), the National Key Research and Development Program of China (2022YFF1101101), 2020-2022 Young Talent Support Project (2020YESS001), the Science and Technology Project of Jiangsu Provincial Bureau of Market Supervision and Administration (M20211001), the Fundamental Research Funds for the Central Universities (JUSRP222001), and the Collaborative Innovation Center of Food Safety and Quality Control.
Author contributions
J.J. and X.S. designed the research; J.J. carried out the analysis, S.W. and Y.Z. performed the experimental studies; X.S. supervised the work. J.J., S.W., and L.S. wrote the article with contributions from all other authors. Y.Y., Y.Z., Y.G., and J.Z. contributed to the statistics and grammar.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: July 3, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107263.
Supplemental information
Data and code availability
-
•
Source data are provided with this paper and deposited on the Metabolight (https://www.ebi.ac.uk/metabolights/) where it has been assigned Project ID MTBLS5183.
-
•
Data used in statistical analyses have been deposited in the paper’s supplemental information and are available as of the date of publication.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
-
•
All original codes are available. R Codes and associated data for reproducing part of the figures in this study have been deposited at GitHub (https://github.com/JianJi2016/Reprogramming-Salmonella).
References
- 1.Algarni S., Ricke S.C., Foley S.L., Han J. The Dynamics of the Antimicrobial Resistance Mobilome of Salmonella enterica and Related Enteric Bacteria. Front. Microbiol. 2022;13:859854. doi: 10.3389/fmicb.2022.859854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dyson Z.A., Klemm E.J., Palmer S., Dougan G. Antibiotic Resistance and Typhoid. Clin. Infect. Dis. 2019;68:S165–S170. doi: 10.1093/cid/ciy1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu S., Yang Y., Wang T., Sun J., Zhang Y., Ji J., Sun X. Effects of acid, alkaline, cold, and heat environmental stresses on the antibiotic resistance of the Salmonella enterica serovar Typhimurium. Food Res. Int. 2021;144 doi: 10.1016/j.foodres.2021.110359. [DOI] [PubMed] [Google Scholar]
- 4.Chen S., Zhao S., White D.G., Schroeder C.M., Lu R., Yang H., McDermott P.F., Ayers S., Meng J. Characterization of Multiple-Antimicrobial-Resistant Salmonella Serovars Isolated from Retail Meats. Appl. Environ. Microbiol. 2004;70:1–7. doi: 10.1128/AEM.70.1.1-7.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nichols M., Gollarza L., Sockett D., Aulik N., Patton E., Watkins L., Gambino-Shirley K.J., Folster J.P., Chen J.C., Tagg K.A. Vol. 19. Foodborne pathogens and disease; 2022. (Outbreak of Multidrug-Resistant Salmonella Heidelberg Infections Linked to Dairy Calf Exposure, United States, 2015–2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu Y., Tao S., Hinkle N., Harrison M., Chen J. Salmonella, including antibiotic-resistant Salmonella, from flies captured from cattle farms in Georgia, U.S.A. Sci. Total Environ. 2018;616–617:90–96. doi: 10.1016/j.scitotenv.2017.10.324. [DOI] [PubMed] [Google Scholar]
- 7.Kuang X., Hao H., Dai M., Wang Y., Ahmad I., Liu Z., Zonghui Y. Serotypes and antimicrobial susceptibility of Salmonella spp. isolated from farm animals in China. Front. Microbiol. 2015;6:602. doi: 10.3389/fmicb.2015.00602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kebede A., Kemal J., Alemayehu H., Habte Mariam S. Isolation, Identification, and Antibiotic Susceptibility Testing of Salmonella from Slaughtered Bovines and Ovines in Addis Ababa Abattoir Enterprise, Ethiopia: A Cross-Sectional Study. Int. J. Bacteriol. 2016;2016 doi: 10.1155/2016/3714785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rizzo K., Horwich-Scholefield S., Epson E. Carbapenem and Cephalosporin Resistance among Enterobacteriaceae in Healthcare-Associated Infections, California, USA. Emerg. Infect. Dis. 2019;25:1389–1393. doi: 10.3201/eid2507.181938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haigh K., Dube Q., Kasambara W., Feasey N.A., Lester R. Cephalosporin resistance in Malawi. Lancet Infect. Dis. 2020;20:285–286. doi: 10.1016/S1473-3099(20)30047-5. [DOI] [PubMed] [Google Scholar]
- 11.Årdal C., Outterson K., Hoffman S.J., Ghafur A., Sharland M., Ranganathan N., Smith R., Zorzet A., Cohn J., Pittet D., et al. International cooperation to improve access to and sustain effectiveness of antimicrobials. Lancet. 2016;387:296–307. doi: 10.1016/S0140-6736(15)00470-5. [DOI] [PubMed] [Google Scholar]
- 12.Musicha P., Cornick J.E., Bar-Zeev N., French N., Masesa C., Denis B., Kennedy N., Mallewa J., Gordon M.A., Msefula C.L., et al. Trends in antimicrobial resistance in bloodstream infection isolates at a large urban hospital in Malawi (1998–2016): a surveillance study. Lancet Infect. Dis. 2017;17:1042–1052. doi: 10.1016/S1473-3099(17)30394-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christou A., Agüera A., Bayona J.M., Cytryn E., Fotopoulos V., Lambropoulou D., Manaia C.M., Michael C., Revitt M., Schröder P., Fatta-Kassinos D. The potential implications of reclaimed wastewater reuse for irrigation on the agricultural environment: The knowns and unknowns of the fate of antibiotics and antibiotic resistant bacteria and resistance genes – A review. Water Res. 2017;123:448–467. doi: 10.1016/j.watres.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 14.Sunuwar J., Azad R.K. A machine learning framework to predict antibiotic resistance traits and yet unknown genes underlying resistance to specific antibiotics in bacterial strains. Briefings Bioinf. 2021;22:bbab179. doi: 10.1093/bib/bbab179. [DOI] [PubMed] [Google Scholar]
- 15.Perry E.K., Meirelles L.A., Newman D.K. From the soil to the clinic: the impact of microbial secondary metabolites on antibiotic tolerance and resistance. Nat. Rev. Microbiol. 2022;20:129–142. doi: 10.1038/s41579-021-00620-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng B., Su Y.B., Li H., Han Y., Guo C., Tian Y.M., Peng X.X. Exogenous alanine and/or glucose plus kanamycin kills antibiotic-resistant bacteria. Cell Metabol. 2015;21:249–262. doi: 10.1016/j.cmet.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 17.Devanga Ragupathi N.K., Muthuirulandi Sethuvel D.P., Shankar B.A., Munusamy E., Anandan S., Veeraraghavan B. Draft genome sequence of blaTEM-1-mediated cephalosporin-resistant Salmonella enterica serovar Typhi from bloodstream infection. J. Glob. Antimicrob. Resist. 2016;7:11–12. doi: 10.1016/j.jgar.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 18.Greninger A.L., Chatterjee S.S., Chan L.C., Hamilton S.M., Chambers H.F., Chiu C.Y. Whole-Genome Sequencing of Methicillin-Resistant Staphylococcus aureus Resistant to Fifth-Generation Cephalosporins Reveals Potential Non-mecA Mechanisms of Resistance. PLoS One. 2016;11 doi: 10.1371/journal.pone.0149541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palzkill T. Structural and Mechanistic Basis for Extended-Spectrum Drug-Resistance Mutations in Altering the Specificity of TEM, CTX-M, and KPC β-lactamases. Front. Mol. Biosci. 2018;5:16. doi: 10.3389/fmolb.2018.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanders C.C. beta-Lactamase stability and in vitro activity of oral cephalosporins against strains possessing well-characterized mechanisms of resistance. Antimicrob. Agents Chemother. 1989;33:1313–1317. doi: 10.1128/AAC.33.8.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brynildsen M.P., Winkler J.A., Spina C.S., MacDonald I.C., Collins J.J. Potentiating antibacterial activity by predictably enhancing endogenous microbial ROS production. Nat. Biotechnol. 2013;31:160–165. doi: 10.1038/nbt.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allison K.R., Brynildsen M.P., Collins J.J. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature. 2011;473:216–220. doi: 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng B., Su Y.-b., Li H., Han Y., Guo C., Tian Y.-m., Peng X.-x. Exogenous Alanine and/or Glucose plus Kanamycin Kills Antibiotic-Resistant Bacteria. Cell Metabol. 2015;21:249–262. doi: 10.1016/j.cmet.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 24.Souza J.G.S., Costa Oliveira B.E., Costa R.C., Bechara K., Cardoso-Filho O., Benso B., Shibli J.A., Bertolini M., Barāo V.A.R. Bacterial-derived extracellular polysaccharides reduce antimicrobial susceptibility on biotic and abiotic surfaces. Arch. Oral Biol. 2022;142 doi: 10.1016/j.archoralbio.2022.105521. [DOI] [PubMed] [Google Scholar]
- 25.Yong Y., Zhou Y., Liu K., Liu G., Wu L., Fang B. Exogenous Citrulline and Glutamine Contribute to Reverse the Resistance of Salmonella to Apramycin. Front. Microbiol. 2021;12:759170. doi: 10.3389/fmicb.2021.759170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dwyer D.J., Belenky P.A., Yang J.H., MacDonald I.C., Martell J.D., Takahashi N., Chan C.T.Y., Lobritz M.A., Braff D., Schwarz E.G., et al. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc. Natl. Acad. Sci. USA. 2014;111:E2100–E2109. doi: 10.1073/pnas.1401876111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pugazhendhi A., Dhanarani S., Shankar C., Prakash P., Ranganathan K., Saratale R.G., Thamaraiselvi K. Electrophoretic pattern of glutathione S-transferase (GST) in antibiotic resistance Gram-positive bacteria from poultry litter. Microb. Pathog. 2017;110:285–290. doi: 10.1016/j.micpath.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 28.Das T., Paino D., Manoharan A., Farrell J., Whiteley G., Kriel F.H., Glasbey T., Manos J. Conditions Under Which Glutathione Disrupts the Biofilms and Improves Antibiotic Efficacy of Both ESKAPE and Non-ESKAPE Species. Front. Microbiol. 2019;10:2000. doi: 10.3389/fmicb.2019.02000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kominkova M., Michalek P., Cihalova K., Guran R., Cernei N., Nejdl L., Smerkova K., Dostalova S., Chudobova D., Heger Z., et al. Study of Linkage between Glutathione Pathway and the Antibiotic Resistance of Escherichia coli from Patients’ Swabs. Int. J. Mol. Sci. 2015;16:7210–7229. doi: 10.3390/ijms16047210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J., Zhang T., Shen X., Liu J., Zhao D., Sun Y., Wang L., Liu Y., Gong X., Liu Y., et al. Serum metabolomics for early diagnosis of esophageal squamous cell carcinoma by UHPLC-QTOF/MS. Metabolomics. 2016;12:116. doi: 10.1007/s11306-016-1050-5. [DOI] [Google Scholar]
- 31.Tsugawa H., Cajka T., Kind T., Ma Y., Higgins B., Ikeda K., Kanazawa M., VanderGheynst J., Fiehn O., Arita M. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods. 2015;12:523–526. doi: 10.1038/nmeth.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thevenot E.A. ropls: PCA, PLS (-DA) and OPLS (-DA) for multivariate analysis and feature selection of omics data. R package version. 2016;1 [Google Scholar]
- 33.Chong J., Soufan O., Li C., Caraus I., Li S., Bourque G., Wishart D.S., Xia J. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46:W486–W494. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barupal D.K., Haldiya P.K., Wohlgemuth G., Kind T., Kothari S.L., Pinkerton K.E., Fiehn O. MetaMapp: mapping and visualizing metabolomic data by integrating information from biochemical pathways and chemical and mass spectral similarity. BMC Bioinf. 2012;13:99. doi: 10.1186/1471-2105-13-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Source data are provided with this paper and deposited on the Metabolight (https://www.ebi.ac.uk/metabolights/) where it has been assigned Project ID MTBLS5183.
-
•
Data used in statistical analyses have been deposited in the paper’s supplemental information and are available as of the date of publication.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
-
•
All original codes are available. R Codes and associated data for reproducing part of the figures in this study have been deposited at GitHub (https://github.com/JianJi2016/Reprogramming-Salmonella).