Abstract
In late 2021, a new variant of SARS-CoV-2 called Omicron emerged, replacing Delta worldwide. Although it has been associated with a lower risk of hospitalization and severe forms of COVID-19, there is little evidence of its relationship with specific symptoms and viral load. The aim of this study was to verify the relationship between Delta and Omicron variants of concern, viral load, and the occurrence of symptoms in individuals with COVID-19. Nasopharyngeal swab samples were collected and sequenced from patients with COVID-19 from the Northeast Region of Brazil between August 2021 and March 2022. The results showed a gradual replacement of the Delta variant by the Omicron variant during the study period. A total of 316 samples (157 Delta and 159 Omicron) were included. There was a higher prevalence of symptoms in Delta-infected individuals, such as coryza, olfactory and taste disturbances, headache, and myalgia. There was no association between viral load and the variants analyzed. The results reported here contribute to the understanding of the symptoms associated with the Delta and Omicron variants in individuals affected by COVID-19.
Keywords: COVID-19, Variants of concern, Delta, Omicron, Symptoms, Viral load
1. Introduction
SARS-CoV-2, the etiologic agent of COVID-19, can acquire mutations that confer immunological, diagnostic, or immune escape advantages and increase the transmissibility and severity of the disease [1]. These mutations occur in key regions of the spike protein and elevate fitness compared to previously circulating strains, which limits natural immunity and may reduce the effectiveness of vaccines [1]. Variants with clinical and epidemiological implications are referred to as variants of concern (VOCs). To date, the main VOCs described are: Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and Omicron (BA.1) [2].
In late 2021, the Omicron variant emerged and quickly replaced Delta worldwide, due to its large number of nonsynonymous mutations and ability to evade the immune system [3]. Despite its high spreading power, previous studies in African, European, and American populations have shown that Omicron is associated with a lower risk of hospitalization [[4], [5], [6], [7]]. However, most large studies lack genomic data at the individual level, as they use information linked to national databases to infer the occurrence of VOCs.
Brazil is one of the countries with the highest numbers of COVID-19 cases worldwide and was strongly affected by the emergence of Omicron in December 2021 [8]. Despite the increasing number of cases of COVID-19, there is still limited data regarding the influence of VOCs at the individual level on the viral load and clinical characteristics of infected individuals, especially in Latin American populations. Thus, the objective of this study was to verify the relationship between Delta and Omicron VOCs, viral load, and the occurrence of symptoms in individuals with COVID-19 from the Northeast Region of Brazil.
2. Methods
2.1. Study population
We included nasopharyngeal swab samples from individuals from 7 municipalities of the VIII Regional Health Management (GERES, acronym in Portuguese) of the state of Pernambuco, located in the Northeast Region of Brazil, collected between August 2021 and March 2022. Samples were received in viral transport medium at the COVID-19 Laboratory of the Dr. Washington Antônio de Barros Teaching Hospital (EBSERH-UNIVASF, acronym in Portuguese) and confirmed by real-time polymerase chain reaction (RT-qPCR). During the study period, suspected cases assisted by the public health system with flu-like symptoms, cases of severe acute respiratory syndrome (SARS), deaths from SARS, contacts of confirmed cases with COVID-19, and patients undergoing elective surgeries were referred for qPCR testing. The sociodemographic and clinical data were obtained through the notification form recorded in the database of the Pernambuco State Department of Health. The data were recorded at the time of sample collection.
Samples identified as Delta (B.1.617.2, AY.*) or Omicron (B.1.1.529, BA.*) by genetic sequencing were included in the analysis. Individuals infected with other strains and those whose clinical data could not be obtained were excluded from the analysis.
This study was approved by the Ethics Committee of the Hospital das Clínicas of the Federal University of Pernambuco (HC/UFPE, acronym in Portuguese) under CAAE: 51751121.0.0000.8807 and was conducted in accordance with the provisions of the Declaration of Helsinki and Good Clinical Practice Guidelines.
2.2. Extraction of genetic material and detection of SARS-CoV-2
Viral RNA was extracted by magnetic beads using the Extracta viral RNA kit in an automatic extractor (Extracta 32, Loccus do Brasil, São Paulo, Brazil), following the manufacturer's recommendations. Immediately afterward, the samples were tested for SARS-CoV-2 by RT-qPCR on the QuantStudio 5 Real-Time PCR System (Thermo Fisher Scientific). During the study period, the following 3 kits were used for detection of SARS-CoV-2 by hydrolysis probes: Allplex SARS-CoV-2 Assay (Seegene), SARS-CoV-2 EDx (Bio-Manguinhos, FIOCRUZ), and the BIOMOL OneStep/COVID-19 kit (Instituto de Biologia Molecular do Paraná - IBMP).
2.3. Sequencing
A random subsample of the positive cases with a threshold cycle value (Ct) < 27 was forwarded for sequencing at the Technology Platform of the Instituto Aggeu Magalhães - Fundação Oswaldo Cruz Pernambuco. Genomic libraries were constructed using the CovidSeq kit (Illumina, San Diego, CA, USA) inserting 3 primer sets described by Naveca et al., 2022 [9], and sequencing was performed using the Miseq Illumina platform with the Miseq V3 150 cycles kit.
2.4. Analysis of the sequencing data
The sequencing data was analyzed with ViralFlow v0.6.0 [10], which comprises the processing of sequencing data, generation of consensus genomes, signature of strains, and obtaining assembly metrics. The versions of each tool used in the steps described below can be checked in the ViralFlow repository (https://github.com/dezordi/ViralFlow).
Briefly, duplicate reads, PCR primers, reads smaller than 75 nucleotides, and regions of reads with an average Phred Score quality lower than 20 were removed with the fastp [11]. The treated reads were mapped against the SARS-CoV-2 reference genome (NC_045512.2) with the BWA tool [12], and consensus was generated using the mapping data in combination with the SAMtools [13] and iVar [14], using a mapping quality threshold of 30, and a minimum depth of 5 reads for identifying single nucleotide variants and indels present as majority alleles. The average sequencing depth of each sample was calculated with the bamdst tool [15], and the exact coverage considering the depth threshold of 5 was calculated with an internal ViralFlow function. The strains were signed with the Pangolin, Nextclade, and Outbreak. info tools [[16], [17], [18]].
2.5. Statistical analysis
Continuous variables were submitted to the Shapiro-Wilk test to verify normal distribution. Comparisons between two groups were made using the Mann-Withney test. Associations between categorical variables were verified using Pearson's chi-square test or Fisher's exact test, when necessary. Statistical analyses were performed using JASP v.0.16.3 software.
3. Results
3.1. Study population
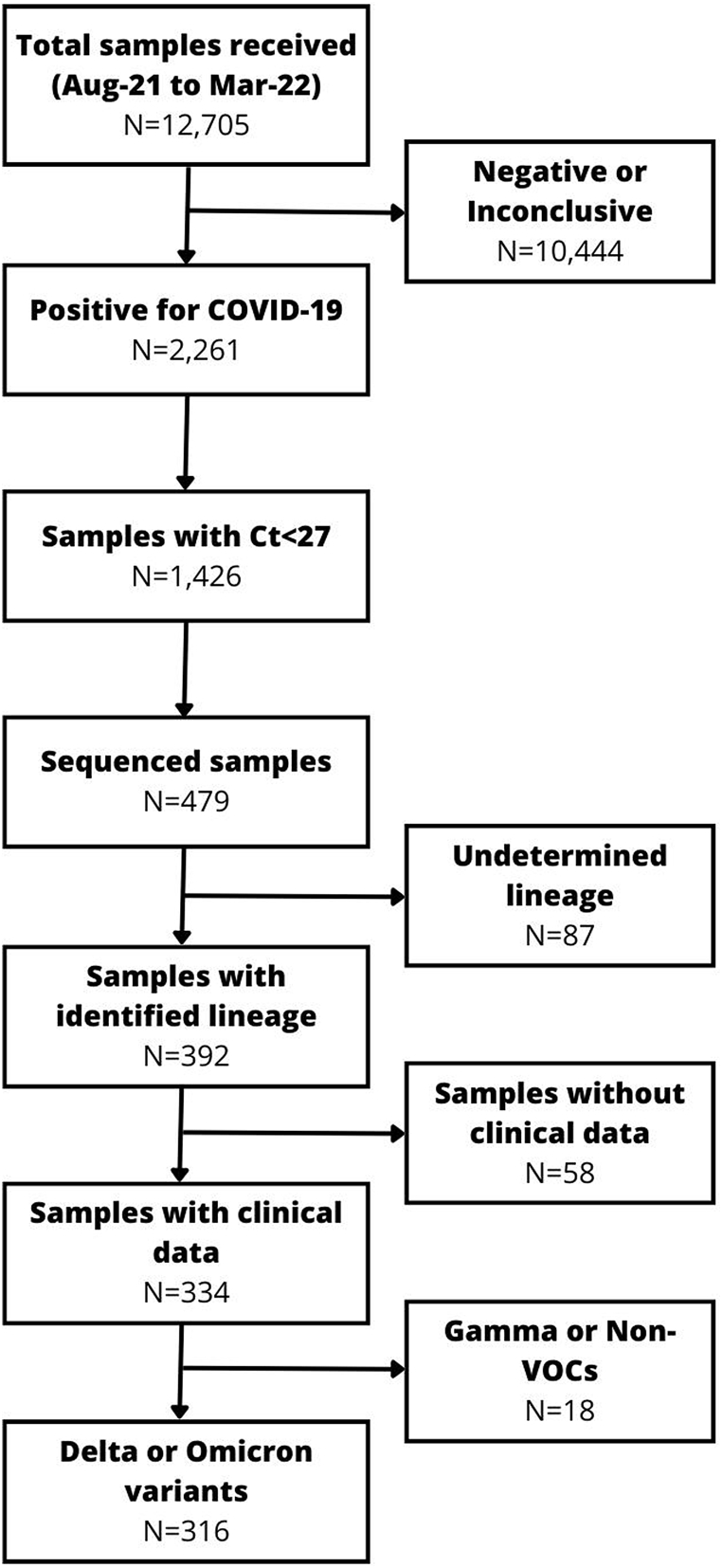
A total of 12,705 samples were received in the COVID-19 Laboratory at UNIVASF between August 2021 and March 2022, among which 2261 (17.8%) were positive for SARS-CoV-2. Among the positive samples, a total of 479 (21.1%) were sequenced. Subsequently, samples in which the viral lineage could not be determined, those with other lineages, and individuals without clinical data were excluded. Consequently, a total of 316 samples were analyzed (157 Delta and 159 Omicron) (Fig. 1).
Fig. 1.
Flow chart of the samples included in the study.
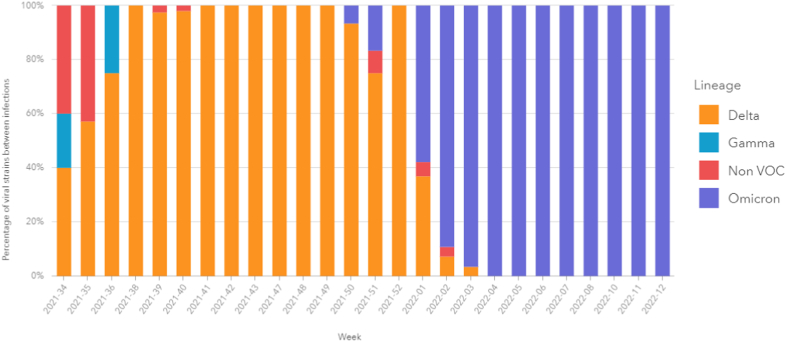
Fig. 2 demonstrates the dynamics of the SARS-CoV-2 strains during the study period. Between epidemiological weeks 34 and 52 of 2021, there was a predominance of the Delta variant in the analyzed samples. At week 50, the first cases of Omicron were detected, and it quickly became the predominant variant from week 1 of 2022 until the end of the analyzed period (Fig. 2). Lineages and sub-lineages are detailed in the supplementary material (Fig. S1).
Fig. 2.
Circulation dynamics of SARS-CoV-2 variants among the samples analyzed from August 2021 to March 2022.
3.2. Association of VOCs with sociodemographic and clinical characteristics
The demographic and clinical data of the patients are summarized in Table 1. There was no significant difference in age and sex distribution between the groups. A total of 10 cases were hospitalized; 5 (3.2%) were Omicron and 5 (3.8%) were Delta. Only 1 death was observed in a patient infected with the Omicron variant. Regarding symptoms, a higher overall frequency of symptoms was observed in individuals infected with the Delta variant. Runny nose, loss of taste and smell, headache, and myalgia were significantly more prevalent symptoms in individuals with the Delta variant (p < 0.05). A higher frequency of asymptomatic individuals was observed in those infected with Omicron (p = 0.003).
Table 1.
Characteristics of Delta and Omicron variant cases included in the study.
| Variables | Delta | Omicron | p-value | OR (CI 95%) |
|---|---|---|---|---|
| N | 157 | 159 | ||
| Age, median (IQR), years | 35 (24–46) | 38 (26–51) | 0.136 | |
| Male sex, n (%) | 81 (51.5) | 67 (42.1) | 0.114 | 0.380 (−0.087–0.849) |
| Symptoms | ||||
| Cough, n (%) | 96 (61.1) | 86 (54.0) | 0.213 | 0.289 (−0.182–0.762) |
| Headache, n (%) | 89 (56.6) | 66 (41.5) | 0.010 | 0.610 (0.142–1.083) |
| Runny nose, n (%) | 79 (50.3) | 58 (36.4) | 0.017 | 0.566 (0.093–1.042) |
| Fever, n (%) | 75 (47.7) | 61 (38.3) | 0.112 | 0.384 (−0.087–0.857) |
| Sore throat, n (%) | 54 (34.3) | 62 (38.9) | 0.416 | −0.198 (−0.683–0.286) |
| Myalgia, n (%) | 34 (21.6) | 10 (6.2) | <0.001 | 1.411 (0.635–2.272) |
| Loss of taste, n (%) | 39 (24.8) | 14 (8.8) | <0.001 | 1.227 (0.539–1.966) |
| Loss of smell, n (%) | 29 (18.4) | 12 (7.5) | 0.004 | 1.018 (0.266–1.827) |
| Dyspnea, n (%) | 12 (7.6) | 15 (9.4) | 0.688 | −0.229 (−1.117–0.637) |
| Weakness, n (%) | 12 (7.6) | 9 (5.6) | 0.507 | 0.321 (−0.663–1.341) |
| Stuffy nose, n (%) | 6 (3.8) | 2 (1.2) | 0.172 | 1.134 (−0.607–3.464) |
| O2 saturation <95%, n (%) | 5 (3.1) | 4 (2.5) | 0.749 | 0.242 (−1.315–1.879) |
| Diarrhea, n (%) | 4 (2.5) | 8 (5.0) | 0.378 | −0.704 (−2.238–0.638) |
| Respiratory distress, n (%) | 4 (2.5) | 6 (3.7) | 0.750 | −0.404 (−1.997–1.057) |
| Vomiting, n (%) | 2 (1.2) | – | 0.246 | – |
| Arthralgia, n (%) | 1 (0.6) | 1 (0.6) | 1.000 | 0.013 (−4.356–4.381) |
| Asymptomatic, n (%) | 12 (7.6) | 30 (18.8) | 0.004 | −1.030 (−1.836–−0.283) |
| Hospitalization, n (%) | 5 (3.8) | 5 (3.2) | 1.000 | 0.154 (−1.340–1.647) |
| Death, n (%) | – | 1 (0.6) | 1.000 | – |
Significant associations are in bold.
3.3. Viral load analysis
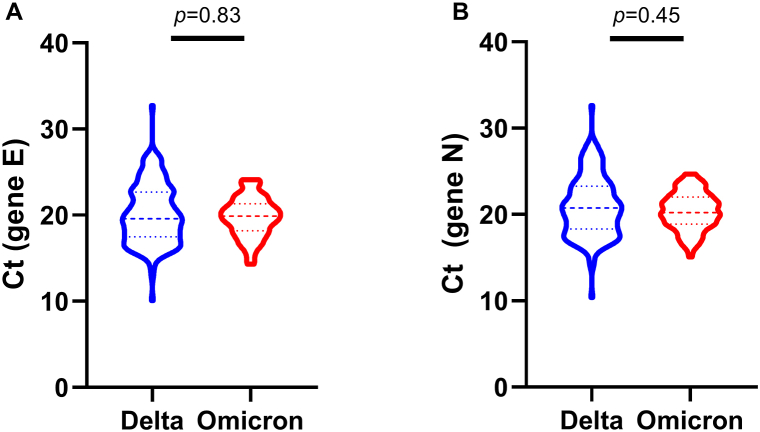
To avoid a possible bias due to the use of different SARS-CoV-2 detection kits during the study period, we decided to analyze the relationship of VOCs with viral load measured through Ct, only of those samples evaluated through the Allplex kit (Seegene), which corresponded to 83.5% (n = 264) of the total samples analyzed. There was no significant difference between the viral load of individuals with Delta or Omicron for the E gene (p = 0.83) (Fig. 3A) or the N gene (p = 0.45) (Fig. 3B).
Fig. 3.
Threshold cycle (Ct) values between Delta and Omicron variants of SARS-CoV-2 in nasopharyngeal swab specimens. A) gene E, B) gene N. Comparison performed by Mann-Whitney test.
4. Discussion
The present study evaluated a cohort of individuals infected with the Delta and Omicron VOCs between the months of August 2021 and March 2022, at which time the Delta variant was replaced by the Omicron variant. Our findings showed that the Omicron variant was associated with a lower occurrence of symptoms compared to Delta.
Characteristics of a VOC include increased transmissibility, a change in the clinical presentation of the disease, or a decrease in the effectiveness of disease control measures, such as increased escape of vaccine-generated antibodies [2]. Delta and Omicron variants are considered VOCs because they have mutations that increase the ability to transmit from person to person, and they more easily escape antibodies generated from previous infections by other variants or antibodies generated from vaccines produced with older strains [19,20].
Previous studies have demonstrated that individuals infected with the Omicron variant have a lower risk of hospitalization, a lower risk of intensive care unit admission, and a shorter length of stay than individuals infected with the Delta variant [4,6,7,[21], [22], [23]]. Most of these studies were conducted with large cohorts of hospitalized individuals in European and North American populations, and their results cannot be directly compared with ours. Our cohort is mainly composed of individuals with mild COVID-19, where only 10 patients were hospitalized and 1 died, making it impossible for us to draw conclusions about the role of Delta and Omicron variants in the risk of hospitalization. This low prevalence of severe cases in our study, especially among the Delta-infected patients, is related to the lower impact that this variant has had on health services in Brazil compared with other countries. In the first half of 2021, Brazil faced one of the largest waves of new infections and the highest number of deaths recorded since the pandemic began, during circulation of the Gamma variant [24]. This wave of cases caused by the Gamma variant may have generated a cross-immunity that made it possible for Brazil to be less impacted by Delta, as observed in other countries. This would explain the low number of hospitalizations and deaths observed during our study.
We observed a higher frequency of symptoms in individuals infected with the Delta variant compared to those with Omicron. Among the symptoms, headache, coryza, myalgia, and taste and smell disturbances were significantly more prevalent in those with Delta. Previous studies have also reported higher symptom frequency in individuals with the Delta variant, including among those who have been vaccinated [[25], [26], [27]]. Menni et al., 2022, evaluating self-reported data from 4990 English subjects with COVID-19, reported that the following 12 symptoms were significantly more prevalent among infected subjects during Delta circulation compared to Omicron: loss of smell, altered sense of smell, sneezing, runny nose, brain fog, eye soreness, headache, fever, hair loss, blistering on feet, ear ringing, and dizzy or light headed, with loss of sense of smell being the most striking difference [27]. The lower prevalence of olfactory and taste disorders among Omicron-infected individuals when compared to other previous variants has been reported previously [[28], [29], [30], [31], [32], [33]]. A recent meta-analysis including 62 studies and 626,035 patients reported that olfactory disturbances caused by Omicron are about 2–10 times less common than observed with the Alpha or Delta variants [34]. Features associated with the mechanism of Omicron entry into the host cell may explain, at least in part, a lower efficiency in infecting olfactory epithelial cells [28]. Additionally, our findings indicated that individuals with Delta were 1.4 times more likely to develop myalgia when compared to Omicron. These findings corroborate data from a recent study from the United Kingdom that showed a reduction in reported symptoms in individuals infected with Omicron compared to Delta, including loss of taste, loss of smell, shortness of breath, myalgia, fatigue/weakness, and headache [33].
In the present study, we found no significant difference in viral load between Delta and Omicron variants. The results in the literature are conflicting. Some studies have observed no difference in viral load between Omicron and Delta [[35], [36], [37]]; others have reported higher viral load in individuals infected by Delta [22,38,39], while others have reported higher viral load in individuals with Omicron [40,41]. It is challenging to make direct comparisons between the studies, since viral load can be influenced by several factors, including equipment model, target gene, probe type, sample type, days of symptoms, extraction method, vaccine status, and others. It is also important to highlight that, due to the cutoff point defined for sending samples for sequencing, samples with high Ct (low viral load) were not included in the study, which may have contributed to the absence of association in this study and in others that use next-generation sequencing to determine SARS-CoV-2 variants.
Our study has strengths and limitations. The first strength is the use of VOC data at individual level through the next-generation sequencing technique; several previously published studies make inferences about the variants based on the period of circulation of VOCs, which may cause misclassification due to the co-circulation of other variants. Second, the use of high-depth sequencing data decreases misclassification errors. Third, the use of a period with co-circulation of Delta and Omicron minimizes temporal biases. Fourth, the study groups have similar sociodemographic characteristics, such as sex, age, and geographical origin. Among the limitations, we highlight, first, the use of secondary data from notification forms, which may generate biases such as filling errors or incompleteness of the data, and, second, the absence of data on vaccination status and other clinical characteristics.
In conclusion, our results have shown a decrease in the prevalence of symptoms during circulation of the Omicron variant compared to Delta. Headache, runny nose, myalgia, and taste and smell disorders were significantly less frequent in those infected with the Omicron variant, with emphasis on taste and smell disorders that decreased considerably during Omicron circulation. No significant difference was observed between viral load measured by Ct between the two VOCS. Our findings contribute to the understanding of the symptoms associated with the Delta and Omicron VOCs among individuals with COVID-19. The decreased frequency of olfactory and taste disturbances, a feature with high predictive potential for COVID-19, among Omicron cases may make clinical suspicion of COVID-19 more difficult and may be more easily confused with other respiratory viruses.
Ethics statement
This study was approved by the Ethics Committee of the Hospital das Clínicas of the Federal University of Pernambuco (HC/UFPE, acronym in Portuguese) under CAAE: 51751121.0.0000.8807 and was conducted in accordance with the provisions of the Declaration of Helsinki and Good Clinical Practice Guidelines.
Author contribution statement
Sávio Luiz Pereira Nunes: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Chirles Araújo de França, Gabriela Dias Rocha, Samily Aquino de Sá Oliveira, Mariana Ramos Freitas, Gustavo Barbosa de Lima, Raul Emídio de Lima, Matheus Filgueira Bezerra: Performed the experiments.
Eliane Oliveira da Silva, Katia Sampaio Coutinho, Aline Silva Jerônimo, Marcelo Henrique Santos Paiva: Contributed reagents, materials, analysis tools or data.
Filipe Zimmer Dezordi: Analyzed and interpreted the data.
Gabriel da Luz Wallau: Contributed reagents, materials, analysis tools or data; Wrote the paper.
Carlos Dornels Freire de Souza: Analyzed and interpreted the data; Wrote the paper.
Anderson da Costa Armstrong, Rodrigo Feliciano do Carmo: Conceived and designed the experiments; Wrote the paper.
Data availability statement
Data will be made available on request.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:Rodrigo Feliciano do Carmo reports financial support was provided by Fundação de Amparo à Ciência e Tecnologia do Estado de Pernambuco (FACEPE). Associate Editor of Heliyon - Rodrigo Feliciano do Carmo.
Acknowledgments
The authors would like to thank the Fundação de Amparo à Ciência e Tecnologia do Estado de Pernambuco (FACEPE) for the financial support (APQ-0723-4.06/21), as well as the Instituto Aggeu Magalhães - FIOCRUZ Pernambuco, the Hospital de Ensino Dr. Washington Antônio de Barros (EBSERH-UNIVASF) and the Secretaria de Saúde do Estado de Pernambuco for the support to the execution of this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e18994.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Dubey A., Choudhary S., Kumar P., Tomar S. Emerging SARS-CoV-2 variants: genetic variability and clinical implications. Curr. Microbiol. 2022;79(1) doi: 10.1007/s00284-021-02724-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Tracking SARS-CoV-2 Variants. Accessed January 31, 2023.https://www.who.int/activities/tracking-SARS-CoV-2-variants.
- 3.Mannar D., Saville J.W., Zhu X., et al. SARS-CoV-2 omicron variant: antibody evasion and cryo-EM structure of spike protein-ACE2 complex. Science. 2022;375(6582):760–764. doi: 10.1126/science.abn7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greene S.K., Levin-Rector A., Kyaw N.T.T., et al. Influenza Other Respir Viruses. Published online 2022; New York City: August 2021–January 2022. Comparative Hospitalization Risk for SARS-CoV-2 Omicron and Delta Variant Infections, by Variant Predominance Periods and Patient-Level Sequencing Results. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolter N., Jassat W., Walaza S., et al. Early assessment of the clinical severity of the SARS-CoV-2 omicron variant in South Africa: a data linkage study. Lancet. 2022;399(10323):437–446. doi: 10.1016/S0140-6736(22)00017-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hyams C., Challen R., Marlow R., et al. Severity of Omicron (B.1.1.529) and Delta (B.1.617.2) SARS-CoV-2 infection among hospitalised adults: a prospective cohort study in Bristol, United Kingdom. The Lancet regional health Europe. 2023;25 doi: 10.1016/J.LANEPE.2022.100556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrigan S.P., Wilton J., Chong M., et al. Clinical severity of Omicron SARS-CoV-2 variant relative to Delta in British Columbia, Canada: a retrospective analysis of whole genome sequenced cases. Clin. Infect. Dis. 2022;9:13. doi: 10.1093/CID/CIAC705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arantes I., Bello G., Nascimento V., et al. Comparative epidemic expansion of SARS-CoV-2 variants Delta and Omicron in Amazonas, a Brazilian setting with high levels of hybrid immunity. medRxiv. 2022;21 doi: 10.1101/2022.09.21.22280193. Published online September. 2022.09.21.22280193. [DOI] [Google Scholar]
- 9.Naveca F.G., Nascimento V., Souza V., et al. Spread of Gamma (P.1) sub-lineages carrying spike mutations close to the furin cleavage site and deletions in the N-terminal domain drives ongoing transmission of SARS-CoV-2 in amazonas, Brazil. Microbiol. Spectr. 2022;10(1) doi: 10.1128/SPECTRUM.02366-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dezordi F.Z., da Silva Neto A.M., de Lima Campos T., et al. ViralFlow: a versatile automated workflow for SARS-CoV-2 genome assembly, lineage assignment, mutations and intrahost variant detection. Viruses. 2022;14(2) doi: 10.3390/v14020217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884. doi: 10.1093/BIOINFORMATICS/BTY560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H., Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25(14):1754. doi: 10.1093/BIOINFORMATICS/BTP324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H., Handsaker B., Wysoker A., et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078. doi: 10.1093/BIOINFORMATICS/BTP352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grubaugh N.D., Gangavarapu K., Quick J., et al. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 2019;20(1) doi: 10.1186/S13059-018-1618-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shiquan. GitHub - bamdst_ a lightweight bam file depth statistical tool.Accessed January 31, 2023. https://github.com/shiquan/bamdst.
- 16.O'Toole Á., Scher E., Underwood A., et al. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 2021;7(2) doi: 10.1093/VE/VEAB064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aksamentov I., Roemer C., Hodcroft E.B., et al. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J. Open Source Softw. 2021;6(67):3773. doi: 10.21105/joss.03773. [DOI] [Google Scholar]
- 18.Gangavarapu K., Latiff A.A., Mullen J.L., et al. Outbreak.info genomic reports: scalable and dynamic surveillance of SARS-CoV-2 variants and mutations. Nat. Methods. 2023;20:512–522. doi: 10.1038/s41592-023-01769-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang P., Nair M.S., Liu L., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593(7857):130–135. doi: 10.1038/s41586-021-03398-2. 2021 593:7857. [DOI] [PubMed] [Google Scholar]
- 20.Nemet I., Kliker L., Lustig Y., et al. Third BNT162b2 vaccination neutralization of SARS-CoV-2 omicron infection. N. Engl. J. Med. 2022;386(5):492–494. doi: 10.1056/NEJMC2119358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leiner J., Pellissier V., Hohenstein S., et al. Characteristics and outcomes of COVID-19 patients during B.1.1.529 (Omicron) dominance compared to B.1.617.2 (Delta) in 89 German hospitals. BMC Infect. Dis. 2022;22(1):1–8. doi: 10.1186/S12879-022-07781-W/TABLES/3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewnard J.A., Hong V.X., Patel M.M., Kahn R., Lipsitch M., Tartof S.Y. Clinical outcomes associated with SARS-CoV-2 Omicron (B.1.1.529) variant and BA.1/BA.1.1 or BA.2 subvariant infection in Southern California. Nat. Med. 2022;28(9):1933–1943. doi: 10.1038/s41591-022-01887-z. 2022 28:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson M.L., Morris C.P., Betz J.F., et al. Impact of SARS-CoV-2 variants on inpatient clinical outcome. Clin. Infect. Dis. 2022 doi: 10.1093/CID/CIAC957. Published online December 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faria N.R., Mellan T.A., Whittaker C., et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 1979;2021(6544):372. doi: 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghosh A.K., Landt O., Yeasmin M., et al. Clinical presentation of COVID-19 and antibody responses in Bangladeshi patients infected with the delta or omicron variants of SARS-CoV-2. Vaccines. 2022;10:1959. doi: 10.3390/VACCINES10111959. 2022;10(11):1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai A., Bergna A., della Ventura C., et al. Epidemiological and clinical features of SARS-CoV-2 variants circulating between april–december 2021 in Italy. Viruses. 2022;14(11):2508. doi: 10.3390/V14112508/S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Menni C., Valdes A.M., Polidori L., et al. Symptom prevalence, duration, and risk of hospital admission in individuals infected with SARS-CoV-2 during periods of omicron and delta variant dominance: a prospective observational study from the ZOE COVID Study. Lancet. 2022;399(10335):1618–1624. doi: 10.1016/S0140-6736(22)00327-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Butowt R., Bilinska K., von Bartheld C.S. Olfactory dysfunction in COVID-19: new insights into the underlying mechanisms. Trends Neurosci. Published online January. 2022;1 doi: 10.1016/j.tins.2022.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boscolo-Rizzo P., Tirelli G., Meloni P., et al. Coronavirus disease 2019 (COVID-19)–related smell and taste impairment with widespread diffusion of severe acute respiratory syndrome–coronavirus-2 (SARS-CoV-2) Omicron variant. Int Forum Allergy Rhinol. 2022 doi: 10.1002/alr.22995. Published online October 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cardoso C.C., Rossi Á.D., Galliez R.M., Faffe D.S., Tanuri A., Castiñeiras T.M.P.P. Olfactory dysfunction in patients with mild COVID-19 during Gamma, delta, and omicron waves in Rio de Janeiro, Brazil. JAMA. 2022;328(6):582. doi: 10.1001/JAMA.2022.11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akaishi T., Kushimoto S., Katori Y., et al. COVID-19-Related symptoms during the SARS-CoV-2 omicron (B.1.1.529) variant surge in Japan. Tohoku J. Exp. Med. 2022;258(2):103–110. doi: 10.1620/tjem.2022.J067. [DOI] [PubMed] [Google Scholar]
- 32.Morioka S., Tsuzuki S., Suzuki M., et al. Post COVID-19 condition of the Omicron variant of SARS-CoV-2. J. Infect. Chemother. 2022;28(11):1546–1551. doi: 10.1016/j.jiac.2022.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vihta K.D., Powels K.B., Peto T.E., et al. Omicron-associated changes in SARS-CoV-2 symptoms in the United Kingdom. Clin. Infect. Dis. 2022;76(3):e133–e141. doi: 10.1093/cid/ciac613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.von Bartheld C.S., Wang L., von Bartheld C. Prevalence of olfactory dysfunction with the omicron variant of SARS-CoV-2: a systematic review and meta-analysis. medRxiv. 2022;19(2023) doi: 10.1101/2022.12.16.22283582. Published online January. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rattan A., Joerger J., Williams D., Pollock N.R. Similar SARS-CoV-2 Ct value distributions in anterior nares versus nasopharyngeal samples from symptomatic children during delta and omicron surges. J Pediatric Infect Dis Soc. Published online December. 2022;15 doi: 10.1093/JPIDS/PIAC130. [DOI] [PubMed] [Google Scholar]
- 36.Yuasa S., Nakajima J., Takatsuki Y., et al. Viral load of SARS‐CoV‐2 Omicron is not high despite its high infectivity. J. Med. Virol. 2022;94(11):5543. doi: 10.1002/JMV.27974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laitman A.M., Lieberman J.A., Hoffman N.G., Roychoudhury P., Mathias P.C., Greninger A.L. The SARS-CoV-2 omicron variant does not have higher nasal viral loads compared to the delta variant in symptomatic and asymptomatic individuals. J. Clin. Microbiol. 2022;60(4) doi: 10.1128/JCM.00139-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ergoren M.C., Komurcu K., Tuncel G., et al. Impact of SARS-CoV-2 Delta and Omicron variants on viral burden and cycle threshold in BNT162b2-vaccinated 12–18 years group. Braz. J. Microbiol. 2022;53(4):1937–1940. doi: 10.1007/S42770-022-00820-3/TABLES/1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sentis C., Billaud G., Bal A., et al. SARS-CoV-2 omicron variant, lineage BA.1, is associated with lower viral load in nasopharyngeal samples compared to delta variant. Viruses. 2022;14(5):919. doi: 10.3390/V14050919/S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salmona M., Chaix M.L., Feghoul L., et al. Detection of SARS-CoV-2 in saliva and nasopharyngeal swabs according to viral variants. Microbiol. Spectr. 2022;10(6) doi: 10.1128/SPECTRUM.02133-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rinaldo H., Tryland M., Granerud B.K., et al. Omicron variant generates a higher and more sustained viral load in nasopharynx and saliva than the delta variant of SARS-CoV-2. Viruses. 2022;14:2420. doi: 10.3390/V14112420. 2022;14(11):2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.