Abstract
Background
Laryngeal squamous cell carcinoma (LSCC) is a common cancer of the head and neck in humans. The 5-years survival rate of patients with LSCC have declined in the past four decades. microRNAs (miRNAs) has been reported to be capable of predicting the prognosis outcomes of patients with different cancers. However, there are no reports on the usage of multi-miRNAs model as signature for the diagnosis or prognosis of LSCC.
Methods
To establish the miRNAs expression-associated model for diagnosis, prognosis prediction and aided therapy of patients with LSCC, the present study enrolled 107 patients with LSCC in clinic and obtained 117 LSCC samples data from TCGA database for evaluation, respectively. Next generation sequencing (NGS), raw data processing, the least absolute shrinkage and selection operator algorithm, Cox regression analysis, construction of nomogram and cell function assays (including proliferation, migration and invasion assays) were sequentially performed.
Results
There were massively dysregulated miRNAs in the LSCC compared to normal tissues. A six-miRNAs signature consists of miR-137–3p, miR-3934–5p, miR-1276, miR-129–5p, miR-7-5p and miR-105–5p was built for prognosis prediction of LSCC patients. The six-miRNAs signature is strongly associated with the poor overall survival (OS, p = 2.5e-05, HR: 4.30 [2.20–8.50]), progression free interval (PFI, p = 0.025, HR: 1.94 [1.08–3.46]) and disease specific survival (DSS, p = 1.1e-05, HR: 5.00 [2.50–10.00]). A nomogram for prediction of 2-, 3- and 5-years OS was also developed based on the six-miRNAs signature and clinical features. Furthermore, blocking the function of each of the six miRNAs inhibited proliferation, invasion and migration of LSCC cells.
Conclusions
The performance of six-miRNAs signature described in the current study demonstrated remarkable potential for progression assessment of LSCC. Moreover, the six-miRNAs signature may serve as predictive tool for prognosis and therapeutic targets of LSCC in clinic.
Keywords: MicroRNA, Molecular biomarkers, Otorhinolaryngology Head and Neck Surgery, Laryngeal squamous cell carcinoma, Prognosis prediction, Overall survival, Oncogene
Abbreviations
- LSCC
Laryngeal squamous cell carcinoma
- miRNAs
microRNAs
- OS
Overall survival
- LASSO
The least absolute shrinkage and selection operator
- PFI
Progression free interval
- DSS
Disease specific survival
- TNM
The tumor-node-metastasis staging
- RPM
The former steps. Reads per million miRNA mapped
- TCGA
The Cancer Genome Atlas
- PCA
Principal component analysis
- GSEA
Gene set enrichment analysis
- MSigDB
Molecular signatures database
- AUC
The area under the curve
- DMEM
Dulbecco's modified Eagle's medium
- BEGM
Bronchial epithelial cell growth medium
- CCK8
Cell counting Kit-8
- PCR
Polymerase chain reaction
- qPCR
Quantitative PCR
- SDMis
Significantly dysregulated miRNAs set.
1. Background
Larynx cancers remain one of the most common malignancies of the head and neck. Globally, it had an estimated 0.211 million (95% UI: 0.206–0.216 million) incident cases in 2017 [1,2]. In China, incidence and death of larynx cancers accounted for 0.67 and 0.57% of the total cancer cases in 2018, respectively [3]. It has been found that 60% of patients with larynx cancers have the advance (stage III or IV) disease at the time of diagnosis [2]. Furthermore, although there are continuous advances in the methods of diagnosis and treatment, the 5-years survival rate of larynx cancers has decreased over the last 40 years [2]. Approximately 90% of larynx cancers are diagnosed as laryngeal squamous cell carcinoma (LSCC) [4,5]. The proneness of local invasion and cervical lymph node metastasis is the major risk factor of poor prognosis of patients with LSCC [6]. To improve the survival time and life quality of patients with LSCC, there is are urgent needs for early and accurate diagnosis as well as prediction of prognosis.
microRNAs (miRNAs) are small noncoding RNAs of 19–25 nucleotides, which have been found to regulate tumor-associated biological process, such as proliferation, apoptosis, migration, invasion, differentiation of stem cells, formation of cancer stem cell and acquisition of the epithelial-mesenchymal transition phenotype [7]. Recently, various studies have focused on the effects of aberrant expression of miRNAs on initiation and progression of cancers [8,9]. Many miRNAs have been reported as potential biomarkers or signatures for diagnosis, prognosis and personalized therapy of cancers, such as renal cancer, esophageal squamous cell carcinoma, colorectal cancer and breast cancer [[9], [10], [11], [12]]. However, there are no reports of multi-miRNAs panel as a signature for the diagnosis and prognosis of LSCC.
To develop a multi-miRNAs signature for prognosis prediction and aided diagnosis in patients with LSCC, we analyzed the miRNA expression data of two independent cohorts of patients with LSCC: TCGA cohort (miRNA expression and clinical data of 117 LSCC and 12 normal tissues from The Cancer Genome Atlas Program) and Shanxi cohort (miRNA sequencing data of 107 pairs of LSCC and matched adjacent normal mucosa tissues collected from The First Hospital of Shanxi Medical University, Shanxi province, China). A six-miRNAs signature was developed using the least absolute shrinkage and selection operator (LASSO) and Cox regression analysis for prediction of overall survival (OS) in patients with LSCC. In addition, a nomogram for prediction of 2-, 3- and 5-years OS was also built based on the six-miRNAs signature and clinical features. Furthermore, the biological roles of six miRNAs in LSCC cells were explored by loss-of-function experiments.
2. Methods
2.1. Tissue samples
A total of 107 pairs of LSCC and matched adjacent normal mucosa (ANM) tissues were collected from surgery of patients with LSCC at Department of Otolaryngology Head and Neck Surgery, The First Hospital of Shanxi Medical University. None of patients underwent radiotherapy or chemotherapy before surgery. The clinical features in this study were obtained from inpatient medical records. The 7th edition of the American Joint Committee on Cancer (AJCC) Staging Manual was used to guide the tumor-node-metastasis (TNM) staging.
2.2. RNA isolation, library preparation and small RNA-sequencing
Next generation sequencing (NGS) technology was used for small RNA-sequencing. The total RNAs were extracted from fresh tissues specimen using TRIzol reagent (Ambion, TX, USA) according to the manufacturer's instructions. For each specimen, 3 μg RNA was used for the library preparation using NEBNext® Multiplex Small RNA Library Prep Set for Illumina® kit (NEB, Ipswich, MA, USA) according to the manufacturer's instructions. Briefly, NEB 3′SR adaptor was first specifically ligated to 3′end of miRNA, siRNA and piRNA. The NEB SR RT primer was then hybridized to the free 3′ SR adaptor. The 5′SR adaptor was ligated to 5′ ends of RNAs, then M-MuLV reverse transcriptase (RNase H) was used for the synthesis of the first strand cDNA. Polymerase chain reaction (PCR) amplification was performed using LongAmp Taq 2 × Master Mix, SR Primer for Illumina and index (X) primer. The clustering of index-coded samples was performed using TruSeq SR Cluster Kit v3-cBot-HS (Illumina, San Diego, CA, USA) as per the manufacturer's recommendations. After generation of cluster, the libraries were sequenced on an Illumina Hiseq 2000 sequencing platform (Novogene, Beijing, China) and 50 bp single-end reads were generated. All the obtained data of sequencing were deposited in GEO database (accession number: GSE132222, GSE133632).
2.3. Data analysis
Raw reads of fastq format were first processed through custom perl (v5.26.2) and python (v3.7.6) scripts. The small RNA tags were mapped to GRCh37 version of reference sequence obtained from Ensembl (http://www.ensembl.org/) using the bowtie (v1.2.2) software. miRBase (v20.0, http://www.mirbase.org/) database were used as the reference data to look for the known miRNA. RepeatMasker (v4.0.6) and Rfam database (v13.0, https://rfam.xfam.org/) were used to remove tags originating from protein-coding genes, repeat sequences, rRNA, tRNA, snRNA, and snoRNA. miREvo (v1.2) and mirdeep2 (v0.0.5) were also integrated to predict novel miRNAs through exploring the secondary structure, the Dicer cleavage site and the minimum free energy of the small RNA tags unannotated in the former steps. Reads per million miRNAs mapped (RPM) as the miRNA's expression measurement unit for deeper bioinformatics analysis. Differential expression analysis between LSCC and normal tissue was also performed using the DESeq2 R package (v1.26.0) with the cutoff of p-value <0.05 and |log2(Fold change) | ≥ 1. miRNAs expression matrix of LSCC in TCGA were downloaded from the TCGA portal website (https://portal.gdc.cancer.gov/) and corresponding clinical features were downloaded from UCSC Xena (https://xenabrowser.net/). Small RNA-sequencing data of Shanxi cohort were downloaded from GEO database (https://www.ncbi.nlm.nih.gov/geo/). In this study, Shanxi cohort were used as a validation cohort to verify analysis results from TCGA cohort.
2.4. Unsupervised analysis
The complete-linkage clustering is one of agglomerative hierarchical clustering. In current study, the complete-linkage clustering was used to analyze the expression distribution of miRNAs among clinical features in heat map. The heat map of miRNA expression was implemented by pheatmap (v1.0.12) R package. Principal component analysis (PCA) is a dimensionality-reduction method of the true eigenvector-based multivariate analyses. PCA was performed to analyze the difference between LSCC and normal tissues based on expression of miRNA in this study. The 3D visualization of PCA was realized using pca3d (v0.10.1) R package.
2.5. Function pathway analysis
Function pathway analysis of the target protein-coding RNAs of significantly dysregulated miRNA in LSCC was implemented through Gene set enrichment analysis (GSEA) software (v3.0) with Molecular Signatures Database (MSigDBv7.0). Briefly, dysregulated miRNA intersection between TCGA and Shanxi cohort was obtained using Venn analysis. Target protein-coding RNAs of each dysregulated miRNA were predicted using miRanda algorithm (v3.3a), miRWalk database (http://mirwalk.umm.uni-heidelberg.de/), TargetScan database (http://www.targetscan.org/vert_72/). The ten target protein-coding RNAs for each dysregulated miRNA were obtained through intersection of the above prediction tools. A total of 1138 target protein-coding RNAs used as input data were analyzed using GSEA software in TCGA and Shanxi cohort. The shared function pathways in both TCGA and Shanxi cohorts were obtained using a cutoff of p-value <0.001. Furthermore, 1750 target protein-coding RNAs of six-miRNAs signature were obtained from the union of miRanda algorithm, TargetScan and miRDB database (http://mirdb.org/). Next, Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis for target protein-coding RNAs of six-miRNAs signature were performed used clusterProfiler (v4.0.0).
2.6. Survival analysis
Cox regression analysis is a statistical method for assessing the association between one or more variables and survival outcomes. Firstly, dysregulated miRNAs were divided into upregulation (log2 (Fold change) > 0 and p-value <0.05) and downregulation sets (log2 (Fold change) < 0 and p-value <0.05) based the fold change of each miRNA in TCGA cohort. The univariate Cox regression analysis was then performed to identify the miRNAs associated with survival time. The value of p-value <0.05 was as a cutoff threshold to defined significant high-risk miRNAs for LSCC patient. To look for more representative high-risk signatures of miRNAs, LASSO algorithm with a penalty parameter of 1000 and 10-fold cross-validation was performed to analyze outputs of the univariate Cox regression analysis. The LASSO algorithm was implemented using glmnet (v2.0-16) R package. To quantify the risk levels of prognostic prediction signatures model of miRNAs for OS, a formula of risk score model for patient with LSCC was calculated by following formula: Risk score = . indicates the correlation coefficient of miRNAi in risk score model, and is the related expression level of risk factor miRNAi, n is the numbers of miRNAs in the risk score model. The patients with LSCC were then classified into the high or low risk groups based on the median value of risk score. Kaplan-Meier method with the log-rank test was used to validate the difference of OS, progression free interval (PFI) and disease specific survival (DSS) in LSCC patients between high and low risk group. Survival (v3.2.3) and survminer (v0.4.4) packages were used to realize the Kaplan-Meier method and draw the survival curve on R programming language. The sensitivity and specificity of the six-miRNAs signature on survival prediction were analyzed using time-dependent receiver operating characteristic (ROC) curve based on time ROC (v0.3) R package. The area under the curve (AUC) of ROC was compared among the 2-, 3- and 5-year survival length prediction. Lastly, a prognostic nomogram was built to predict probabilities of patients with LSCC on 2-, 3- and 5-year OS. The nomogram was realized using rms (v5.1-3) R package and evaluated by calibration curves in both TCGA and Shanxi cohorts.
2.7. Cell culture and transfection
Human LSCC cell line FD-LSC-1 was maintained in Bronchial Epithelial Cell Growth Medium (BEGM) (Lonza, Walkersville, MD, USA) supplemented with 10% fetal bovine serum (FBS) (Biological Industries, CT, USA) [13]. Human HNSCC cell line AMC–HN–8 (Bioleaf Biotech, Shanghai, China) was cultured in Dulbecco's Modified Eagle's Medium (DMEM) (Gibco, Grand Island, NY, USA) supplemented with 10% FBS. Human head and neck cancer cell lines CAL-27, Detroit 562, FaDu and human embryonic kidney 293 T (HEK239T) cell line were obtained from CCTCC (Wuhan, China), and these cell lines were cultured according to manufacturer's instructions. All the cells were incubated in an incubator with a humid atmosphere of 5% CO2 at 37 °C. miRNA inhibitors prevent the miRNA from binding to its intended target gene via forming a duplex with the miRNA guide strand [14]. 2′-O-methyl RNA (2′-OMe) is a nontoxic nucleic acid with a high binding affinity for RNA that provides the resistance to endonucleases [15]. In this study, 2′-OMe-modified miRNA inhibitors and NC inhibitors were synthesized by GenePharma Co., Ltd. (Shanghai, China). The cells (2.0 × 105 cells per well) were seeded in six-well plates for transfection using Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Expression levels of miRNAs were determined by qPCR analysis. The expression level of RNA U6 was used as internal control. The primer sequences for qPCR analysis were listed in Supplementary Table S1.
2.8. Cell proliferation, migration and invasion assays
Cell proliferation ability was determined using the CCK8 kit. Briefly, 1 × 104 cells per well were seeded into 96‐well plates and incubated using CCK8 kit (YEASEN, Shanghai, China) at 37 °C for 4 h. The absorbance values at OD 450 nm were measured every 24 h using a Spectra Max i3x Multifunctional microplate detection system (Molecular Devices, San Jose, CA, USA). The 24-well Transwell plates (Falcon, Franklin Lakes, NJ, USA) were used for migration and invasion assay. For migration assay, 4 × 104 cells per well with 200 μL Serum-free DMEM were added to the upper chamber of Transwell plates, and 500 μL DMEM medium supplemented with 20% FBS was then added to the lower chamber. After 24 h of incubation at 37 °C with 5% CO2, cotton swabs were used to gently remove cells in the upper chamber, then fixation using 4% paraformaldehyde for 20 min and staining with 0.1% crystal violet for 10 min. Finally, images of cells were captured using microscope. For invasion assay, serum-free DMEM was first added to Matrigel (BD Biosciences, San Jose, CA, USA) in a ratio of 1:3, and then the mixture was added to upper chamber of Transwell plates and coagulated at 37 °C for 2 h. The rest of the operations were as same as the migration assay.
3. Results
3.1. Patient features
A total of 107 pairs of LSCC and matched ANM tissues from patients in Shanxi cohort, and 117 LSCC and 12 normal tissues from patients in TCGA cohort were included. Clinical features, including gender, age, vital status, pathologic T stage, clinical N stage, clinical stage, alcohol and cigarettes history were extracted to analyze the potential connection between expression levels of miRNA and progression of LSCC (Table 1). It was observed that there was no significant difference in age structure between TCGA and Shanxi cohort (p = 0.4622). Further, it was found that the occurrence of LSCC is more common in male than in female [2]. Number of male patients with LSCC in both TCGA (82.91%) and Shanxi cohorts (93.46%) were more than female patients. It is evident that the increased risk of laryngeal cancer in smokers [16]. 68.38% of LSCC patients in TCGA cohort and 85.98% of LSCC patients in Shanxi cohort have cigarettes history.
Table 1.
Clinical features of LSCC patients in TCGA and Shanxi cohorts.
| Parameters | Number of Cases |
|
|---|---|---|
| TCGA cohort | Shanxi cohort | |
| Gender | ||
| Male | 97 (82.91%) | 100 (93.46%) |
| Female | 20 (17.09%) | 7 (6.54%) |
| Age(Years) | ||
| ≤60 | 49 (41.88%) | 56 (52.34%) |
| >60 | 68 (58.12%) | 51 (47.66%) |
| Vital Status | ||
| Alive | 67 (57.26%) | 62 (57.94%) |
| Dead | 50 (42.74%) | 17 (15.89%) |
| Unknown | 0 (0.00%) | 28 (26.17%) |
| Pathologic T Stage | ||
| T1 | 7 (5.98%) | 30 (28.04%) |
| T2 | 14 (11.97%) | 28 (26.17%) |
| T3 | 26 (22.22%) | 28 (26.17%) |
| T4 | 55 (47.01%) | 21 (19.62%) |
| TX | 13 (11.11%) | 0 (0.00%) |
| Unknown | 2 (1.71%) | 0 (0.00%) |
| Clinical N Stage | ||
| N0 | 56 (47.86%) | 80 (74.78%) |
| N+ | 55 (47.01%) | 27 (25.22%) |
| NX | 5 (4.27%) | 0 (0.00%) |
| Unknown | 1 (0.86%) | 0 (0.00%) |
| Clinical Stage | ||
| Stage I | 3 (2.56%) | 29 (27.10%) |
| Stage II | 12 (10.26%) | 24 (22.43%) |
| Stage III | 27 (23.08%) | 24 (22.43%) |
| Stage IV | 71 (60.68%) | 30 (28.04%) |
| Unknown | 4 (3.42%) | 0 (0.00%) |
| Alcohol History | ||
| Yes | 76 (64.96%) | 32 (29.91%) |
| No | 39 (33.33%) | 75 (70.09%) |
| Unknown | 2 (1.71%) | 0 (0.00%) |
| Cigarettes History | ||
| Yes | 80 (68.38%) | 92 (85.98%) |
| No | 37 (31.62%) | 15 (14.02%) |
3.2. miRNA expression profile
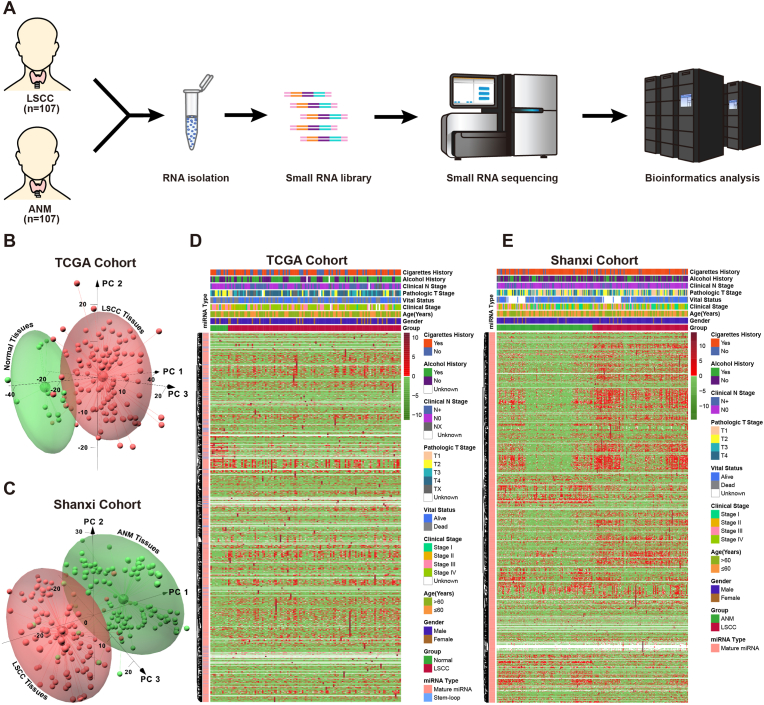
To identify the differentially expressed miRNAs between LSCC and normal tissues, total RNAs of 107 pairs LSCC and matched ANM were sequenced by NGS technology (Fig. 1A). Furthermore, LSCC miRNAs expression data were obtained from TCGA database (TCGA cohort). To evaluate the overall difference of miRNAs expression between LSCC and normal tissues, PCA method was used to reduce the data dimensionality while retaining most of the variation in the miRNAs expression matrix (Fig. 1B and C). Results of this study showed that samples in both TCGA and Shanxi cohorts were significantly divided into two group based on LSCC and normal tissues. The clinical features-associated heat map was built to visualize the expression profile of miRNAs in TCGA and Shanxi cohorts (Fig. 1D and E). Results of heat map based on hierarchical clustering algorithm revealed that there were a significant number of differentially expressed miRNAs in LSCC in comparison with the normal tissue, however, expression pattern of miRNAs had no clear difference amongst different clinical features in the view of overall expression profile in both TCGA and Shanxi cohorts.
Fig. 1.
Construction of miRNAs expression profile in LSCC. (A) Schematic representation of small RNA-Seq workflow. (B) PCA analysis of miRNAs between LSCC and normal tissues in TCGA cohort, red: LSCC tissue; green: normal tissue. (C) PCA analysis of miRNAs between LSCC and ANM in Shanxi cohort, red: LSCC tissue; green: ANM tissue. (D-E) Heat map of the correlation between miRNAs expression levels and clinical features in TCGA (D) and Shanxi (E) cohort.
3.3. Target protein-coding RNAs of dysregulated miRNAs were enriched into cell cycle pathway
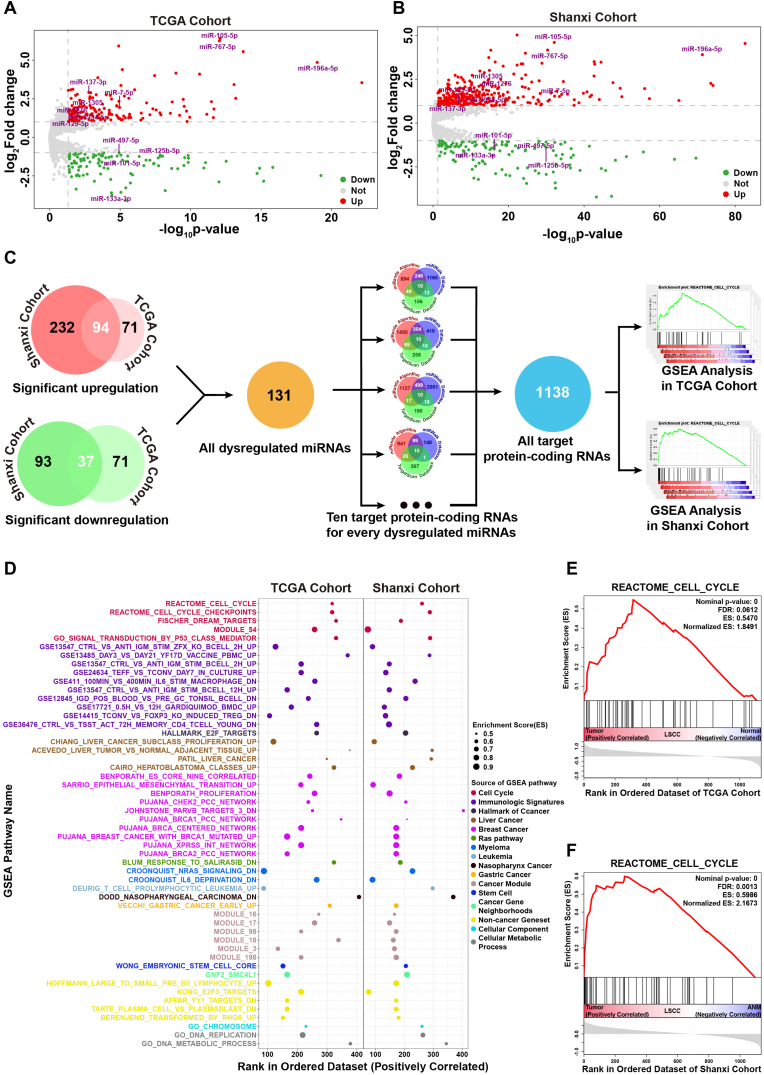
To mine significantly dysregulated miRNAs in LSCC, differential analysis of miRNAs expression was performed in both TCGA and Shanxi cohorts. Under a cutoff of p-value <0.05 and Fold change ≥2 or ≤0.25, it was found that 165 and 108 miRNAs were upregulated and downregulated in TCGA cohort, respectively, whereas 326 and 130 miRNAs were upregulated and downregulated in Shanxi cohort, respectively (Fig. 2A and B). Venn analysis revealed that 94 miRNAs were upregulated and 37 miRNAs were downregulated in both TCGA and Shanxi cohorts. These dysregulated miRNAs constituted a significantly dysregulated miRNAs set (SDMis) for prediction of target protein-coding RNAs. Based on the intersection of prediction results of miRanda, miRWalk and TargetScan, a total of 1138 target protein-coding RNAs of SDMis were obtained to analyze the biological function pathway (Fig. 2C). There were 52 shared functional pathways (p-value ≤0.001) in both two cohorts after GSEA analysis. Most of the shared functional pathways were related with initiation and progression of cancer (Fig. 2D). Cell cycle disturbance cause unscheduled proliferation of tumor cell, thereby propelling the tumor cell and its progeny into uncontrolled expansion and invasion [17]. The cell cycle pathways clearly appeared in shared functional pathways of TCGA and Shanxi cohorts (Fig. 2E and F). The results of GSEA analysis revealed that dysregulated miRNAs may play a critical role in regulating the initiation and progression of LSCC through regulation of target protein-coding genes in cell cycle pathways.
Fig. 2.
Function pathway analysis of significantly dysregulated miRNAs in LSCC. (A-B) Volcano plots of differentially expressed miRNAs in TCGA (A) and Shanxi (B) cohort. (C) Schematic representation for target protein-coding RNA prediction of dysregulated miRNAs in TCGA and Shanxi cohort. (D) Tumor-associated functional pathways were significantly enriched by GSEA analysis in both TCGA and Shanxi cohort. (E-F) Enrichment plot of cell cycle pathway in TCGA (E) and Shanxi (F) cohort.
3.4. Screening of miRNAs significantly associated with OS in patients with LSCC
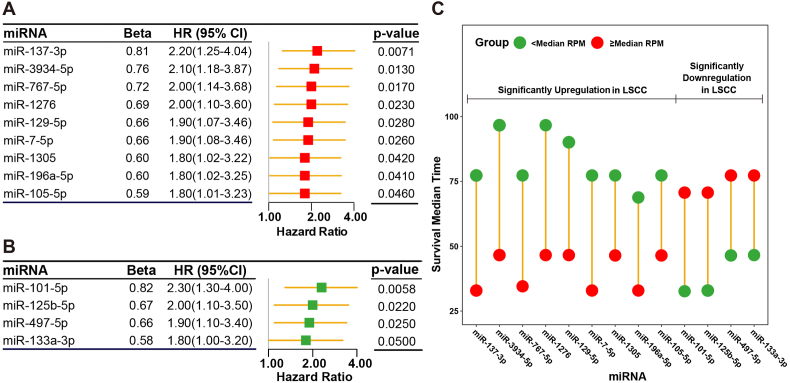
All the miRNAs in SDMis were screened to determine those that associated with poor OS of patients with LSCC. After the univariate Cox regression analysis, a total of nine upregulated miRNAs (miR-137–3p, miR-3934–5p, miR-767–5p, miR-1276, miR-129–5p, miR-7-5p, miR-1305, miR-196a-5p and miR-105–5p) and four downregulated miRNAs (miR-101–5p, miR-125b-5p, miR-479–5p and miR-133a-5p) were identified to be significantly related with OS of patients with LSCC (Fig. 3A and B). Furthermore, screening results of the univariate Cox regression analysis were validated using Kaplan-Meier method (Supplementary Fig. S1). In the set with the nine upregulated miRNAs, the survival time of the patients who had a relatively high miRNA expression level (≥median RPM) was shorter than those who had a relatively low miRNA expression level (<median RPM). On the contrary, the survival time of patients who had a relatively low miRNA expression level was shorter than those who had a relatively high miRNA expression level in four downregulated miRNAs set (Fig. 3C). The results indicated that increased expression levels of the nine upregulated miRNAs or decreased expression levels of the four downregulated miRNAs were significantly associated with shorten OS time of patients with LSCC.
Fig. 3.
Cox regression analysis of significantly dysregulated miRNAs. (A) Forest plot of upregulated miRNAs that were significantly associated with the OS in TCGA cohort. (B) Forest plot of downregulated miRNAs that were significantly associated with the OS in TCGA cohort. (C) Difference of median survival time between high and low miRNAs expression levels for patients in TCGA cohort.
3.5. Identification of a six-miRNAs risk stratification signature for LSCC patients with poor prognosis
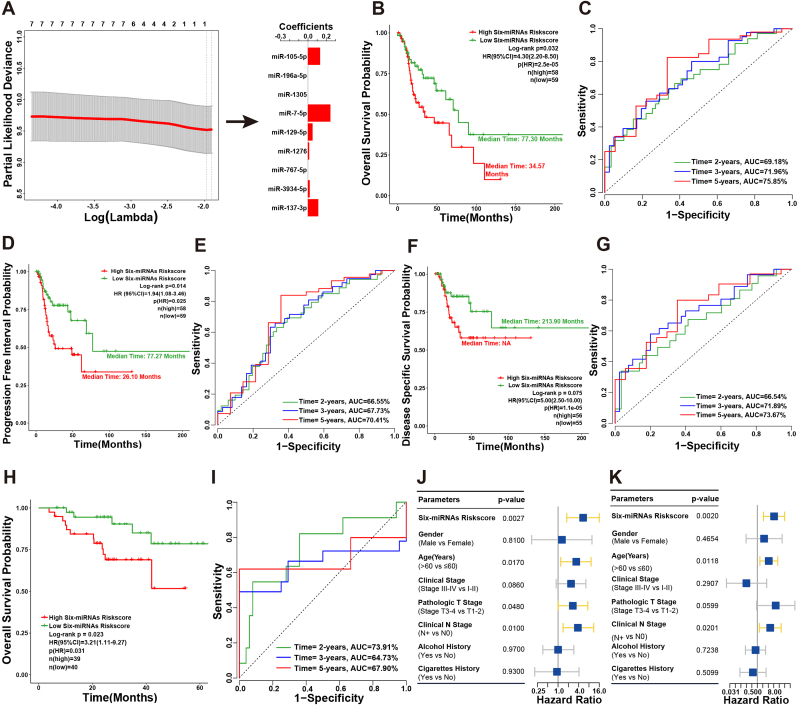
The nine upregulated miRNAs set was analyzed using the LASSO penalized algorithm in the TCGA cohort. A six-miRNAs signature that significantly associated with poor OS in patients with LSCC was identified as follows: Risk score = 0.1125 × + 0.0187 × + 0.0127 × + 0.0500 × + 0.2448 × + 0.1324 × (Fig. 4A). Each miRNA in the six-miRNAs signature had a relatively high expression level in both TCGA and Shanxi cohorts (Supplementary Fig. S2). The median survival time of patients in high six-miRNAs risk score group (34.57 months) was significantly lower than that of low six-miRNAs risk score group (77.30 months, Fig. 4B). The diagnostic accuracy of the six-miRNAs signature in prediction of 2-, 3- and 5-years OS in TCGA cohort yielded the AUC value of 69.18%, 71.96% and 75.85%, respectively (Fig. 4C). In addition, PFI (p = 0.025, HR: 1.94 [1.08–3.46]) and DSS (p = 1.1e-05, HR: 5.00 [2.50–10.00]) for six-miRNAs signature high and low-risk score patients had significant discrimination (Fig. 4D and F). The AUC of 2-, 3- and 5-years for PFI and DSS reached 66.55%, 67.73% and 70.41%, 66.54%, 71.89% and 73.67%, respectively (Fig. 4E and G).
Fig. 4.
Construction of six-miRNAs signature for prediction of LSCC patient's prognosis. (A) Schematic representation of LASSO algorithm workflow for construction of six-miRNAs signature. (B) Kaplan-Meier survival curve of six-miRNAs signature risk score for the OS of TCGA cohort patients. (C) The ROC curve of six-miRNAs signature risk score for predicting the OS of TCGA cohort patients in 2-, 3- and 5-years. (D) Kaplan-Meier survival curve of six-miRNAs signature risk score for the PFI of TCGA cohort patients. (E) The ROC curve of six-miRNAs signature risk score for predicting the PFI of TCGA cohort patients in 2-, 3- and 5-years. (F) Kaplan-Meier survival curve of six-miRNAs signature risk score for the DSS of TCGA cohort patients. (G) The ROC curve of six-miRNAs signature risk score for predicting the DSS of TCGA cohort patients in 2-, 3- and 5-years. (H) The Kaplan-Meier estimates of the patients' OS time using six-miRNAs signature risk score cutoff that divided Shanxi cohort patients into low-risk and high-risk groups. (I) The ROC curve of six-miRNAs signature risk score for predicting the OS of Shanxi cohort patients in 2-, 3- and 5-years. (J) Forest plot of univariate Cox regression analysis based on six-miRNAs signature risk score and clinical features in Shanxi cohort. (K) Forest plot of multivariate Cox regression analysis based on six-miRNAs signature risk score and clinical features in Shanxi cohort.
These findings were subsequently validated using Shanxi cohort. It was evident that the 5-years OS rate of patients who had a six-miRNAs signature high-risk score exhibited substantially low survival outcomes compared to the low-risk score patients (p = 0.031, HR: 3.21 [1.11–9.27]; Fig. 4H). The AUC of ROC for 2-, 3- and 5-years OS in Shanxi cohort were 73.91%, 64.73% and 67.90%, respectively (Fig. 4I). Furthermore, the univariate and multivariate Cox regression analysis were also performed to evaluate the association among six-miRNAs signature, clinical features (gender, age, pathologic T stage, clinical N stage, clinical stage, alcohol and cigarettes history) and OS in Shanxi cohort. In the univariate Cox regression analysis, six-miRNAs signature risk core (p = 0.0027, HR: 5.40 [1.80–16.00]), age (p = 0.0170, HR: 5.40 [1.80–16.00]), pathologic T stage (p = 0.0480, HR: 2.70 [1.00–7.40]) and clinical N stage (p = 0.0100, HR: 3.80 [1.40–11.00]) emerged as the significant predictors of poor OS in patients with LSCC (Fig. 4J). Multivariate Cox regression analysis was performed using some factors that were statistically significant in the univariate Cox regression analysis. Results of this study revealed that the six-miRNAs signature risk core (p = 0.0020, HR: 8.31 [2.17–31.81]), age (p = 0.0118, HR: 4.26 [1.38–13.18]) and clinical N stage (p = 0.0201, HR: 5.12 [1.29–20.32]) were independent risk factors for predicting poor OS in patients with LSCC (Fig. 4K). In addition, the patients in high six-miRNAs signature risk core group (>median) had poorer survival outcomes as compared with those who in low six-miRNAs signature risk core group (≤median) in both TCGA and Shanxi cohorts (Supplementary Fig. S3). The results evidently showed that the six-miRNAs signature could be as a quite robust and superior independent predictor of survival in patients with LSCC.
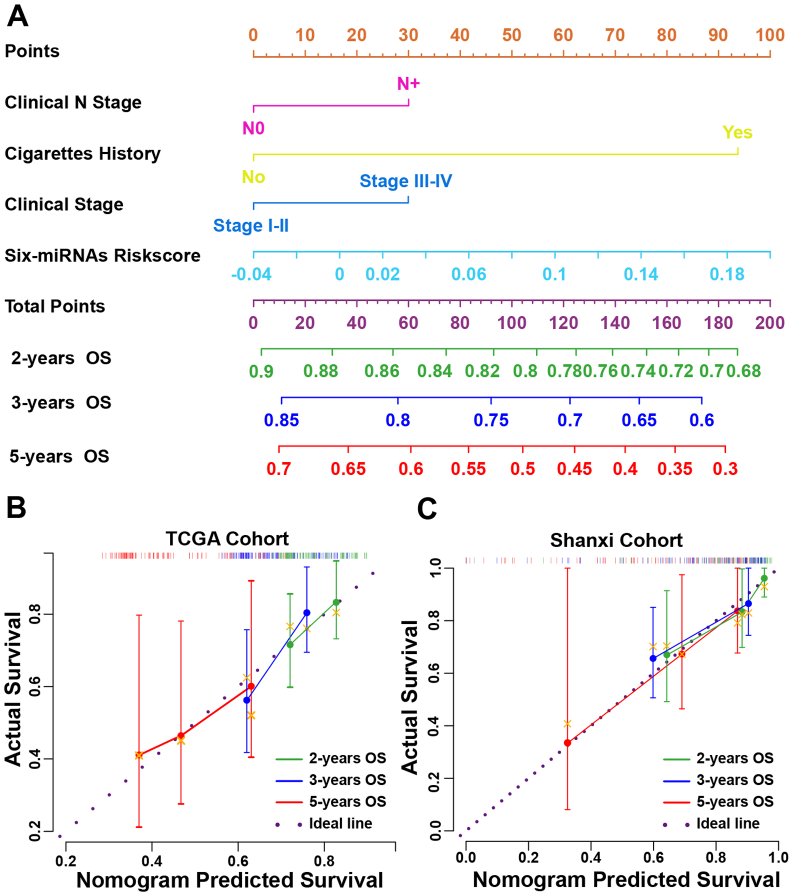
3.6. Nomogram construction
To enhance the predictive ability of the six-miRNAs signature in clinic, a nomogram for prediction of 2-, 3-and 5-years OS was built in TCGA cohort by integrating the six-miRNAs signature risk core and clinical features, including clinical N stage, clinical stage and cigarettes history (Fig. 5A). The calibration curve was used to evaluate the prediction value of the nomogram. The 45-degreedotted lines indicated the ideal predictive ability of the nomogram. Bootstrapped calibration curves showed that the nomogram performed well-compared with the performance of an ideal model at 2-, 3- and 5-years (Fig. 5B). Furthermore, calibration curves in Shanxi cohort also validated the excellent predictive ability of the nomogram for 2-, 3- and 5-years OS (Fig. 5C).
Fig. 5.
Construction of the prognostic nomogram for LSCC patient's OS. (A) Prognostic nomogram consisting of six-miRNAs signature risk score and clinical features, including clinical N stage, clinical stage and cigarettes history, for predicting LSCC patient's OS in 2-, 3- and 5-years. (B) Calibration curves of the prognostic nomogram estimating probability of OS in 2-, 3- and 5-years in TCGA cohort. (C) Calibration curves of the prognostic nomogram estimating probability of OS in2-, 3- and 4.25-years in Shanxi cohort.
3.7. Silencing each miRNA of the six-miRNAs signature suppresses proliferation, invasion and migration of LSCC cells
To investigate the functional effects of six miRNAs on cancer cells, 1750 protein-coding genes of six miRNAs obtained from miRanda program, TargetScan and miRAB database were used for KEGG enrichment analysis (Supplementary Fig. S4A). The interactions among six miRNAs and the 1750 protein-coding genes were analyzed by constructing the miRNA-target network (Supplementary Fig. S4B). There were some intersections between RNAs sets from target protein-coding RNAs of six-miRNAs, however, there were no shared target protein-coding RNAs amongst all six-miRNAs (Supplementary Fig. S5A). Enrichment analysis results revealed that target genes of six-miRNAs were involved in cancer related signaling pathways, such as mTOR, FoxO, EGFR, PI3K-Akt, immune responses, HIF-1, Wnt, and VEGF pathway (Supplementary Fig. S5B).
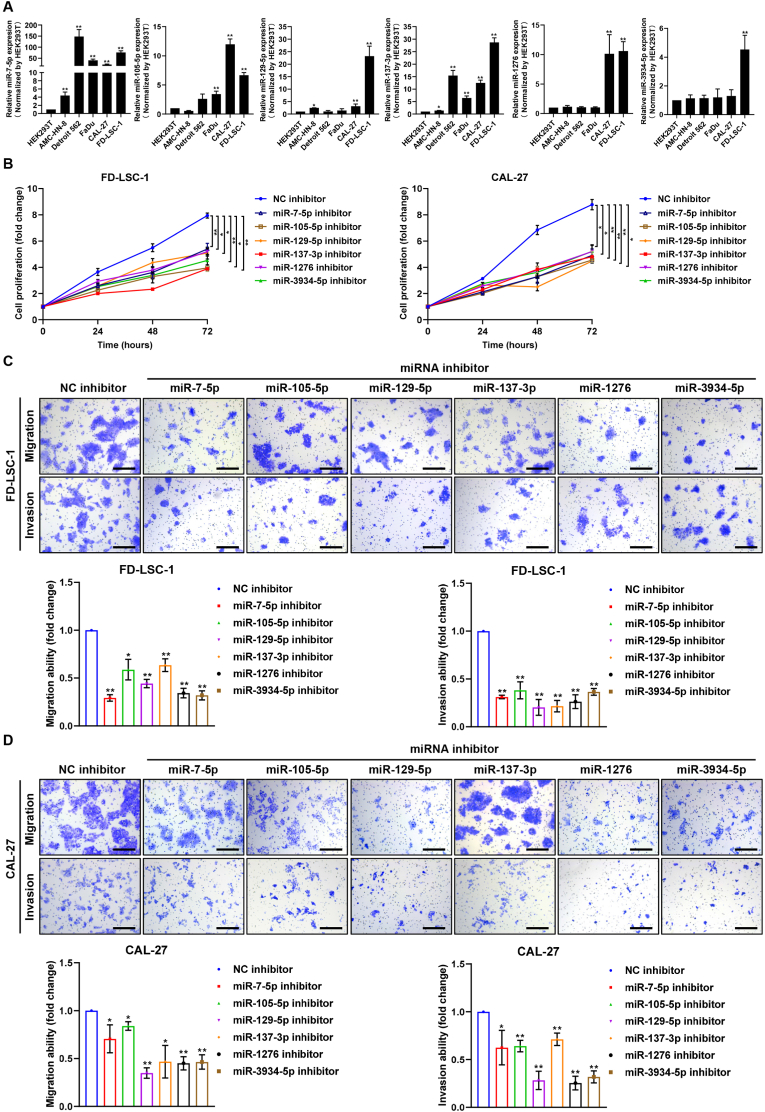
Next, we tested the expression levels of the six miRNAs in LSCC cells FD-LSC-1, head and neck cancer cells (CAL-27, AMC–HN–8, FaDu, Detroit 562) and HEK293T cells by qPCR analysis. Results showed that expression of these six miRNAs were significantly upregulated in FD-LSC-1 and oral squamous cell carcinoma CAL-27 cells compared with HEK239T cells (Fig. 6A). CCK8 assays demonstrated that blocking each of the six miRNAs using miRNA inhibitor suppressed the proliferation of FD-LSC-1 and CAL-27 cells (Fig. 6B). Transwell assays revealed that transfection with miRNA inhibitor of each of the six miRNAs inhibited migration and invasion abilities of FD-LSC-1 and CAL-27 cells (Fig. 6C and D). Taken together, these results indicated that these six miRNAs function as oncogene in LSCC.
Fig. 6.
miRNAs of the six-miRNAs signature promote proliferation, migration and invasion of LSCC cells. (A) Expression levels of six miRNAs in HEK293T, Detroil562, FaDu, CAL-27F, AMC–HN–8 and FD-LSC-1 cells were determined by qPCR analysis. (B) Proliferation assay of cells transfected with indicated miRNA inhibitor or NC inhibitor was performed using CCK8 kit. (C-D) Transwell migration and invasion assays of FD-LSC-1 (C) and CAL27 (D) cells transfected with indicated miRNA inhibitor or NC inhibitor. Scale bar, 200 μm *p < 0.05, **p < 0.01.
4. Discussion
Laryngeal squamous cell carcinoma (LSCC) is the second highest incidence disease in head and neck cancer. Patients with LSCC often have symptoms of dysphonia, dyspnea, and swallowing dysfunction whereby the life quality is severely affected [2,18]. Further, several studies have reported a decline in survival of patients in the past decades because of the easy metastasis and relapse of LSCC [[4], [5], [6]]. Although various therapeutic strategies have been applied in LSCC, the survival outcome is still unsatisfactory, especially for patients in advanced stage of the cancer [5]. Therefore, it is crucial to verify the accurate prognosis predictors and develop new diagnosis and treatment strategies for LSCC. Recently, increasing studies have reported that aberrant expression of miRNAs, such as miR-145–5p, miR-1252–5p, miR-3148, and miR-1207–5p, was closely associated with proliferation, migration, and invasion of LSCC [[18], [19], [20]]. In addition, several studies have also shown that miRNAs could reverse radio-resistance or drug resistance of human LSCC cell lines [[21], [22], [23]]. These studies have revealed that miRNAs could be as the important biomarkers or regulator of drug resistance and radio-resistance for diagnosis and treatment in patients with LSCC. Furthermore, due to the strong association between abnormal expression of miRNAs and prognosis outcome of the patients with cancer, some miRNAs have been thought to have potential for prediction of survival outcomes in different cancers [10,11]. Some miRNAs have also been utilized to predict the prognosis of patients with head and neck cancers [24,25]. However, currently, there are no reports about multiple miRNAs-based signature for prediction of prognosis in patients with LSCC.
In previous studies for miRNAs biomarker in patients with LSCC, only one data set was analyzed and no validation set [26,27]. In the present study, LSCC and matched ANM tissues of 107 patients from Shanxi cohort were sequenced using NGS technology and as an independently validated cohort. The TCGA cohort from TCGA database was the independently training cohort. miRNAs with the same dysregulated expression trend in both cohorts were also mined to analyze the biological function. Disturbance of cell cycle caused unscheduled proliferation and cancerization of cells [17]. GSEA analysis showed that the common differentially expressed miRNAs in TCGA cohort and Shanxi cohort were mainly enriched in cell cycle related pathways, indicating that dysregulated miRNAs may play a critical role in regulating the initiation and progression of LSCC through targeting protein-coding genes in these pathways.
Recent studies investigated the effect of abnormal expression of miRNAs on survival outcomes in patients with LSCC. Dysregulated expression of miR-21, miR-375, miR-23a, miR-101 and miR-9 are strongly associated with poor prognosis of patients with LSCC [26]. In addition, upregulation of miR-23a or downregulation of miR-449a was associated with lymph node metastases of LSCC [26,27]. These studies built the prognosis prediction model of patients with LSCC using single dysregulated miRNA only. However, the available published literatures shows that multi-genes signature had a significant advantage in the prognosis prediction [28].
In this study, a six-miRNAs signature for prediction of OS was developed using LASSO-Cox regression analysis based on aberrantly expressed miRNAs that significant associated with poor prognosis of patients with LSCC. Furthermore, most of previous studies only made attempts to build prediction models for OS based on aberrant expression of miRNAs [9,24,25]. In the present study, six-miRNAs signature included miR-137–3p, miR-3934–5p, miR-1276, miR-129–5p, miR-7-5p and miR-105–5p, was built to predict the outcomes of OS, PFI and DSS in TCGA cohort to verify the prediction ability of miRNAs for prognosis of patients with LSCC. A risk score calculated by six-miRNAs signature classified the patients with LSCC into high and low risk group. The survival time of OS, PFI and DSS of the patients in high risk group was obviously shorter than those in low risk group. In addition, the prediction ability of the six-miRNAs signature for OS was also validated in Shanxi cohort. Furthermore, a nomogram for prediction of 2-, 3- and 5-years OS was also developed based on six-miRNAs signature and clinical features. Calibration curves in both TCGA and Shanxi cohorts showed that the nomogram performed better compared with the performance of an ideal model.
The miR-137–3p was widely studied in cancers, such as colon cancer, colorectal cancer, pancreatic cancer, breast cancer, and lung adenocarcinoma [29]. miR-137–3p directly targeted JNK3 (MAPK10) and thus suppressed the cell proliferation, migration and invasion of prostate cancer [30]. The interaction between miR-137–3p and JNK3 was also predicted in our study. Downregulation of miR-3934–5p enhanced the cisplatin sensitivity of lung carcinomas cells and inhibited the proliferation of neuroblastoma cells [31,32]. It was evident that lncRNA HCG11 could target miR-1276 to promote proliferation and migration of gastric cancer cells [33]. Furthermore, downregulation of miR-1276 contributed to TNFα-Promoted apoptosis of HeLa and HepG2 cells [34]. miR-129–5p targeted Wnt5a to block the proliferation, invasion, migration, angiogenesis, neuro-sphere formation and the resistance to temozolomide of glioblastoma multiforme cells [[35], [36], [37]]. miR-129–5p could target ADAM, HGMB1, PBX3, IPO7, COL1A1, SOX6, FMR1, COL1A1, APC, PAX6, SOX4, ETV1, CAMK4, CBX4, DLK1, CAMK2N1, SOX2, HOXC10, and G3BP1, and thus regulated the development of various diseases [38,39]. Some target genes of miR-7-5p that predicted in this study were reported in published literatures, such as EGFR, NOVA2, REGγ (PSME3) [[40], [41], [42]]. Downregulation of miR-105–5p could contribute to the proliferation of glioma and liver cancer cells and decrease the immunogenicity of gastric cancer cells [43,44]. These studies have demonstrated that miRNAs of the six-miRNAs signature have shown promising potential in regulation of cancer activity and as the signature for prediction of cancers progression. However, there was obvious differences between current and previous study. On one hand, although each miRNA of six-miRNAs signature was investigated in various cancers, studies in LSCC have not yet been reported. On the other hand, miRNAs in six-miRNAs signature was downregulated (e.g., miR-137–3p, miR-1276, miR-129–5p, and miR-105–5p) in most of cancers and played a role of tumor suppressor [29,33,35,36,43]. However, these six miRNAs were upregulation and contributed to proliferation, migration and invasion of cells in LSCC.
Furthermore, based on KEGG enrichment analysis, six-miRNAs signature might affect the malignant progression of LSCC through well-known signaling pathways, such as, EGFR, PI3K-Akt, immune responses, HIF-1, Wnt and VEGF pathway. Most of previous studies did not conduct a functional verification for miRNAs signatures [10,11,24,25]. In this study, the function of each miRNA of the six-miRNAs signature was explored in LSCC cells.
RNA therapeutics are becoming an important alternative plan in the treatment of ‘undruggable’ medical conditions, allowing them to circumvent the obstacles exist in other treatment manners [45,46]. In theory, miRNAs can bind to around 60% of all protein-coding genes with an average of 200 targets per miRNA, hence indicating the therapeutic potential of miRNAs [47]. Our data demonstrated that blocking the function of each of the six miRNAs by miRNA inhibitors significantly reduced proliferation, invasion, and migration of LSCC cells, this indicating these miRNAs are highly attractive therapeutic targets, and highlighting the potential value of miRNA-targeting strategy in the treatment of LSCC.
5. Conclusion
A novel six-miRNAs signature was identified and validated using a systematic and comprehensive signature discovery strategy in the present study. High level of the six-miRNAs signature was associated with shorter survival time of OS, PFI and DSS of patients with LSCC. Inhibition of the function of the six miRNAs suppressed the proliferation, invasion and migration of LSCC cells. However, the detailed mechanisms of the six miRNAs in LSCC have not been analyzed in the present study. Therefore, further studies are required to reveal the mechanisms of these six miRNAs in the future. Collectively, the present study provided potential biomarker for identifying high-risk patients with LSCC and predicting their prognosis. Moreover, our findings highlighted the value of miRNA-targeted strategy for LSCC therapy.
Ethical approval
This study was approved by the ethics committee of the First Hospital of Shanxi Medical University. Informed consent Informed consent was obtained from all individual participants included in the study.
Funding
This work was supported by the National Natural Science Foundation of China (No. 82073101), Basic and Applied Basic Research Foundation of Guangdong Province (No. 2023A1515010342, 2021A1515012161), Shenzhen Science and Technology Innovation Program (No. RCJC20210706091950028, JCYJ20220531102815036, JCYJ20220530153604010), Shenzhen Key Medical Discipline Construction Fund (No. SZXK039), Scientific Research Project of Education Department of Liaoning Province (No. JYTQN2020032), Natural Science Foundation of Zhejiang Province (No. LY22H160017), Research Project Supported by Shanxi Scholarship Council of China (No. 2020165), Fund for the Scientific Activities of Selected Return Overseas Professionals in Shanxi Province (No. 20200034), Natural Science Foundation of Shanxi Province (No. 20210302124088, 201805D211007), Research Project of Shanxi Province Health and Family Planning Commission (No. 2019033), Stable Support Plan Program of Shenzhen Natural Science Fund (No.20220810151804002), Shenzhen International Cooperative Research Project (No. GJHZ20200731095210030).
Data availability
MicroRNA sequencing raw data and normalized results were deposited at GEO database (GSE132222, and GSE133632, https://www.ncbi.nlm.nih.gov/geo/) and The Cancer Genome Atlas (https://cancergenome.nih.gov/) databases. R code and local data in this article are available from the corresponding author upon reasonable request.
CRediT authorship contribution statement
Linshi Zhang: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – review & editing, All authors reviewed and approved the final manuscript. Zhe Zhang: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – review & editing, All authors reviewed and approved the final manuscript. Xiwang Zheng: Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Validation, Visualization, Writing – review & editing, All authors reviewed and approved the final manuscript. Yan Lu: Formal analysis, Investigation, Methodology, Validation, Visualization, All authors reviewed and approved the final manuscript. Li Dai: Investigation, Validation, All authors reviewed and approved the final manuscript. Wenqi Li: Investigation, Validation, All authors reviewed and approved the final manuscript. Hui Liu: Formal analysis, Methodology, All authors reviewed and approved the final manuscript. Shuxin Wen: Formal analysis, Methodology, Supervision, Writing – review & editing, All authors reviewed and approved the final manuscript. Qiuping Xie: Methodology, Writing – review & editing, All authors reviewed and approved the final manuscript. Xiangmin Zhang: Methodology, Supervision, All authors reviewed and approved the final manuscript. Ping Wang: Conceptualization, Project administration, Supervision, Writing – review & editing, All authors reviewed and approved the final manuscript. Yongyan Wu: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Visualization, Writing – review & editing, All authors reviewed and approved the final manuscript. Wei Gao: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Visualization, All authors reviewed and approved the final manuscript.
Declaration of competing interest
The authors declare that they have no competing interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ncrna.2023.08.001.
Contributor Information
Ping Wang, Email: p.wang@zju.edu.cn.
Yongyan Wu, Email: wuyongyan@sxent.org.
Wei Gao, Email: gaoweisxent@sxent.org.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Global Burden of Disease Cancer Collaboration. Fitzmaurice C., Abate D., Abbasi N., Abbastabar H., Abd-Allah F., Abdel-Rahman O., Abdelalim A., Abdoli A., Abdollahpour I., Abdulle A.S.M., et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2017: a systematic analysis for the global burden of disease study. JAMA Oncol. 2019;5:1749–1768. doi: 10.1001/jamaoncol.2019.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steuer C.E., El‐Deiry M., Parks J.R., Higgins K.A., Saba N.F. An update on larynx cancer. CA Cancer J. Clin. 2017;67:31–50. doi: 10.3322/caac.21386. [DOI] [PubMed] [Google Scholar]
- 3.Qi H., Chen W., Zhang C., Zheng X., Peng C., Zhao Q., Guo Y., Wu Y., Gao W., Wang B. Epidemiological Analysis of 1234 Cases of Laryngeal Cancer in Shanxi Province, China. Cancer Control. 2021;28 doi: 10.1177/10732748211041236. 10732748211041236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Z., Huang C., Zhang A., Lu C., Liu L. Overexpression of circRNA_100290 promotes the progression of laryngeal squamous cell carcinoma through the miR-136-5p/RAP2C axis. Biomed. Pharmacother. 2020;125 doi: 10.1016/j.biopha.2020.109874. [DOI] [PubMed] [Google Scholar]
- 5.Megwalu U.C., Sikora A.G. Survival outcomes in advanced laryngeal cancer. JAMA Otolaryngol. Head Neck Surg. 2014;140:855–860. doi: 10.1001/jamaoto.2014.1671. [DOI] [PubMed] [Google Scholar]
- 6.Wu Y., Zhang Y., Zheng X., Dai F., Lu Y., Dai L., Niu M., Guo H., Li W., Xue X., et al. Circular RNA circCORO1C promotes laryngeal squamous cell carcinoma progression by modulating the let-7c-5p/PBX3 axis. Mol. Cancer. 2020;19:99. doi: 10.1186/s12943-020-01215-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rohan T.E., Wang T., Weinmann S., Wang Y., Lin J., Ginsberg M., Loudig O. A miRNA expression signature in breast tumor tissue is associated with risk of distant metastasis. Cancer Res. 2019;79:1705–1713. doi: 10.1158/0008-5472.CAN-18-2779. [DOI] [PubMed] [Google Scholar]
- 8.Mitra R., Adams C.M., Jiang W., Greenawalt E., Eischen C.M. Pan-cancer analysis reveals cooperativity of both strands of microRNA that regulate tumorigenesis and patient survival. Nat. Commun. 2020;11:968. doi: 10.1038/s41467-020-14713-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo Y., Chen L., Wang G., Xiao Y., Ju L., Wang X. Identification of a three-miRNA signature as a novel potential prognostic biomarker in patients with clear cell renal cell carcinoma. J. Cell. Biochem. 2019;120:13751–13764. doi: 10.1002/jcb.28648. [DOI] [PubMed] [Google Scholar]
- 10.Wen J., Wang G., Xie X., Lin G., Yang H., Luo K., Liu Q., Ling Y., Xie X., Lin P., et al. Prognostic value of a four-miRNA signature in patients with lymph node positive locoregional esophageal squamous cell carcinoma undergoing complete surgical resection. Ann. Surg. 2021;273:523–531. doi: 10.1097/SLA.0000000000003369. [DOI] [PubMed] [Google Scholar]
- 11.Kandimalla R., Gao F., Matsuyama T., Ishikawa T., Uetake H., Takahashi N., Yamada Y., Becerra C., Kopetz S., Wang X., et al. Genome-wide discovery and identification of a novel miRNA signature for recurrence prediction in stage II and III colorectal cancer. Clin. Cancer Res. 2018;24:3867–3877. doi: 10.1158/1078-0432.CCR-17-3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H., Liu J., Chen J., Wang H., Yang L., Chen F., Fan S., Wang J., Shao B., Yin D., et al. A serum microRNA signature predicts trastuzumab benefit in HER2-positive metastatic breast cancer patients. Nat. Commun. 2018;9:1614. doi: 10.1038/s41467-018-03537-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu C., Zhou L., Gong H., Du H., Tian J., Sun S., Li J. Establishment and characterization of a novel HPV-negative laryngeal squamous cell carcinoma cell line, FD-LSC-1, with missense and nonsense mutations of TP53 in the DNA-binding domain. Cancer Lett. 2014;342:92–103. doi: 10.1016/j.canlet.2013.08.041. [DOI] [PubMed] [Google Scholar]
- 14.Bhatnagar D., Ladhe S., Kumar D. Discerning the prospects of miRNAs as a multi-target therapeutic and diagnostic for alzheimer’s disease. Mol. Neurobiol. 2023:1–21. doi: 10.1007/s12035-023-03446-0. [DOI] [PubMed] [Google Scholar]
- 15.Lennox K.A., Vakulskas C.A., Behlke M.A. Non-nucleotide modification of anti-miRNA oligonucleotides. Methods Mol. Biol. 2017;1517:51–69. doi: 10.1007/978-1-4939-6563-2_3. [DOI] [PubMed] [Google Scholar]
- 16.Kuper H., Boffetta P., Adami H.O. Tobacco use and cancer causation: association by tumour type. J. Intern. Med. 2002;252:206–224. doi: 10.1046/j.1365-2796.2002.01022.x. [DOI] [PubMed] [Google Scholar]
- 17.Evan G.I., Vousden K.H. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411:342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 18.Gao W., Zhang C., Li W., Li H., Sang J., Zhao Q., Bo Y., Luo H., Zheng X., Lu Y., et al. Promoter methylation-regulated miR-145-5p inhibits laryngeal squamous cell carcinoma progression by targeting FSCN1. Mol. Ther. 2019;27:365–379. doi: 10.1016/j.ymthe.2018.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen S., Yang M., Wang C., Ouyang Y., Chen X., Bai J., Hu Y., Song M., Zhang S., Zhang Q. Forkhead box D1 promotes EMT and chemoresistance by upregulating lncRNA CYTOR in oral squamous cell carcinoma. Cancer Lett. 2021;503:43–53. doi: 10.1016/j.canlet.2020.11.046. [DOI] [PubMed] [Google Scholar]
- 20.Wu Y., Dai F., Zhang Y., Zheng X., Li L., Zhang Y., Cao J., Gao W. miR-1207-5p suppresses laryngeal squamous cell carcinoma progression by downregulating SKA3 and inhibiting epithelial-mesenchymal transition. Mol Ther Oncolytics. 2021;22:152–165. doi: 10.1016/j.omto.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu L., Chen Z., Xue F., Chen W., Ma R., Cheng S., Cui P. MicroRNA-24 inhibits growth, induces apoptosis, and reverses radioresistance in laryngeal squamous cell carcinoma by targeting X-linked inhibitor of apoptosis protein. Cancer Cell Int. 2015;15:61. doi: 10.1186/s12935-015-0217-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin X., Liu H., Li P., Wang H., Yang A., Di J., Jiang Q., Yang Y., Huang J., Yuan M., et al. miR-936 suppresses cell proliferation, invasion, and drug resistance of laryngeal squamous cell carcinoma and targets GPR78. Front. Oncol. 2020;10:60. doi: 10.3389/fonc.2020.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lapa R.M.L., Barros-Filho M.C., Marchi F.A., Domingues M.A.C., de Carvalho G.B., Drigo S.A., Kowalski L.P., Rogatto S.R. Integrated miRNA and mRNA expression analysis uncovers drug targets in laryngeal squamous cell carcinoma patients. Oral Oncol. 2019;93:76–84. doi: 10.1016/j.oraloncology.2019.04.018. [DOI] [PubMed] [Google Scholar]
- 24.Hess J., Unger K., Maihoefer C., Schüttrumpf L., Wintergerst L., Heider T., Weber P., Marschner S., Braselmann H., Samaga D., et al. A five-MicroRNA signature predicts survival and disease control of patients with head and neck cancer negative for HPV infection. Clin. Cancer Res. 2019;25:1505–1516. doi: 10.1158/1078-0432.CCR-18-0776. [DOI] [PubMed] [Google Scholar]
- 25.Jamali Z., Aminabadi N.A., Attaran R., Pournagiazar F., Oskouei S.G., Ahmadpour F. MicroRNAs as prognostic molecular signatures in human head and neck squamous cell carcinoma: a systematic review and meta-analysis. Oral Oncol. 2015;51:321–331. doi: 10.1016/j.oraloncology.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 26.Takeuchi T., Kawasaki H., Luce A., Cossu A.M., Misso G., Scrima M., Bocchetti M., Ricciardiello F., Caraglia M., Zappavigna S. Insight toward the MicroRNA profiling of laryngeal cancers: biological role and clinical impact. Int. J. Mol. Sci. 2020;21:3693. doi: 10.3390/ijms21103693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawasaki H., Takeuchi T., Ricciardiello F., Lombardi A., Biganzoli E., Fornili M., De Bortoli D., Mesolella M., Cossu A.M., Scrima M., et al. Definition of miRNA signatures of nodal metastasis in LCa: miR-449a targets notch genes and suppresses cell migration and invasion. Mol. Ther. Nucleic Acids. 2020;20:711–724. doi: 10.1016/j.omtn.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi Y., Li Y., Lin J., Zheng L., Zhu Y.M., Huang J. The discovery of a novel eight-mRNA-lncRNA signature predicting survival of hepatocellular carcinoma patients. J. Cell. Biochem. 2019;120:7539–7550. doi: 10.1002/jcb.28028. [DOI] [PubMed] [Google Scholar]
- 29.Mahmoudi E., Cairns M.J. MiR-137: an important player in neural development and neoplastic transformation. Mol. Psychiatr. 2017;22:44–55. doi: 10.1038/mp.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zang Y., Zhu J., Li Q., Tu J., Li X., Hu R., Yang D. miR-137-3p modulates the progression of prostate cancer by regulating the JNK3/EZH2 Axis. OncoTargets Ther. 2020;13:7921–7932. doi: 10.2147/OTT.S256161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ren A., Wen Z., Zheng L. Downregulation of miR-3934-5p enhances A549 cell sensitivity to cisplatin by targeting TP53INP1. Exp. Ther. Med. 2019;18:1653–1660. doi: 10.3892/etm.2019.7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye W., Liang F., Ying C., Zhang M., Feng D., Jiang X. Downregulation of microRNA-3934-5p induces apoptosis and inhibits the proliferation of neuroblastoma cells by targeting TP53INP1. Exp. Ther. Med. 2019;18:3729–3736. doi: 10.3892/etm.2019.8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang H., Huang H., Xu X., Wang H., Wang J., Yao Z., Xu X., Wu Q., Xu F. LncRNA HCG11 promotes proliferation and migration in gastric cancer via targeting miR-1276/CTNNB1 and activating Wnt signaling pathway. Cancer Cell Int. 2019;19:350. doi: 10.1186/s12935-019-1046-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou F., Li Y., Huang Y., Wu J., Wu Q., Zhu H., Wang J. Upregulation of CASP9 through NF-κB and its target MiR-1276 contributed to TNFα-promoted apoptosis of cancer cells induced by doxorubicin. Int. J. Mol. Sci. 2020;21:2290. doi: 10.3390/ijms21072290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao N., Fu Y., Chen L., Liu Z., He J., Zhu Y., Xia T., Wang S. Long non-coding RNA NONHSAT101069 promotes epirubicin resistance, migration, and invasion of breast cancer cells through NONHSAT101069/miR-129-5p/Twist1 axis. Oncogene. 2019;38:7216–7233. doi: 10.1038/s41388-019-0904-5. [DOI] [PubMed] [Google Scholar]
- 36.Yu D., Han G., Zhao X., Liu X., Xue K., Wang D., Xu C. MicroRNA-129-5p suppresses nasopharyngeal carcinoma lymphangiogenesis and lymph node metastasis by targeting ZIC2. Cell. Oncol. 2020;43:249–261. doi: 10.1007/s13402-019-00485-5. [DOI] [PubMed] [Google Scholar]
- 37.Gebhardt K., Edemir B., Groß E., Nemetschke L., Kewitz-Hempel S., Moritz R.K.C., Sunderkötter C., Gerloff D. BRAF/EZH2 signaling represses miR-129-5p inhibition of SOX4 thereby modulating BRAFi resistance in melanoma. Cancers. 2021;13:2393. doi: 10.3390/cancers13102393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu S., Li W., Wu J., Lu Y., Xie M., Li Y., Zou J., Zeng T., Ling H. The role of miR-129-5p in cancer: a novel therapeutic target. Curr. Mol. Pharmacol. 2022;15:647–657. doi: 10.2174/1874467214666210914122010. [DOI] [PubMed] [Google Scholar]
- 39.Boicean A., Birsan S., Ichim C., Boeras I., Roman-Filip I., Blanca G., Bacila C., Fleaca R.S., Dura H., Roman-Filip C. Has-miR-129-5p’s involvement in different disorders, from digestive cancer to neurodegenerative diseases. Biomedicines. 2023;11:2058. doi: 10.3390/biomedicines11072058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X., Niu W., Mu M., Hu S., Niu C. Long non-coding RNA LPP-AS2 promotes glioma tumorigenesis via miR-7-5p/EGFR/PI3K/AKT/c-MYC feedback loop. J. Exp. Clin. Cancer Res. 2020;39:196. doi: 10.1186/s13046-020-01695-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao H. MiR-7-5p suppresses tumor metastasis of non-small cell lung cancer by targeting NOVA2. Cell. Mol. Biol. Lett. 2019;24:60. doi: 10.1186/s11658-019-0188-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi Y., Luo X., Li P., Tan J., Wang X., Xiang T., Ren G. miR-7-5p suppresses cell proliferation and induces apoptosis of breast cancer cells mainly by targeting REGγ. Cancer Lett. 2015;358:27–36. doi: 10.1016/j.canlet.2014.12.014. [DOI] [PubMed] [Google Scholar]
- 43.Suo S.T., Gong P., Peng X.J., Niu D., Guo Y.T. Knockdown of long non-coding RNA VIM-AS1 inhibits glioma cell proliferation and migration, and increases the cell apoptosis via modulation of WEE1 targeted by miR-105-5p. Eur. Rev. Med. Pharmacol. Sci. 2020;24:6834–6847. doi: 10.26355/eurrev_202006_21673. [DOI] [PubMed] [Google Scholar]
- 44.Miliotis C., Slack F.J. miR-105-5p regulates PD-L1 expression and tumor immunogenicity in gastric cancer. Cancer Lett. 2021;518:115–126. doi: 10.1016/j.canlet.2021.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang C., Zhang B. RNA therapeutics: updates and future potential. Sci. China Life Sci. 2023;66:12–30. doi: 10.1007/s11427-022-2171-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crooke S.T., Witztum J.L., Bennett C.F., Baker B.F. RNA-targeted therapeutics. Cell Metabol. 2018;27:714–739. doi: 10.1016/j.cmet.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 47.de Rooij L.A., Mastebroek D.J., Voorde N.T., van der Wall E., van Diest P.J., Moelans C.B. The microRNA lifecycle in Health and cancer. Cancers. 2022;14:5748. doi: 10.3390/cancers14235748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MicroRNA sequencing raw data and normalized results were deposited at GEO database (GSE132222, and GSE133632, https://www.ncbi.nlm.nih.gov/geo/) and The Cancer Genome Atlas (https://cancergenome.nih.gov/) databases. R code and local data in this article are available from the corresponding author upon reasonable request.