Abstract
Microtubules are dynamic, non-covalent polymers consisting of α- and β-tubulin subunits that are involved in a wide range of intracellular processes. The polymerization and dynamics of microtubules are regulated by many factors, including small molecules that interact with different sites on the tubulin dimer. Colchicine binding site inhibitors (CBSIs) destabilize microtubules and inhibit tubulin polymerization, leading to cell cycle arrest. Because of their therapeutic potential, the molecular mechanism of CBSI function is an area of active research. Nevertheless, important details of this mechanism have yet to be resolved. In this study, we use atomistic molecular dynamics simulations to show that the binding of CBSIs to the tubulin heterodimer leads to the weakening of tubulin intersubunit interaction. Using atomistic molecular dynamics simulations and binding free energy calculations, we show that CBSIs act as protein–protein interaction inhibitors and destabilize interlinkage between α and β subunits, which is crucial for longitudinal contacts in the microtubule lattice. Our results offer new insight into the mechanisms of microtubule polymerization inhibition by colchicine and its analogs.
Significance Statement
Colchicine binding site inhibitors (CBSIs) lead to cell cycle arrest inhibiting polymerization of tubulin heterodimers and have important therapeutic potential. We demonstrate that the molecular mechanism of CBSIs is based on the destabilization of intersubunit interaction between α and β tubulins.
1. Introduction
Microtubules are a principal part of the eukaryotic cytoskeleton and are involved in a wide range of essential cellular processes, including chromosome segregation, cell shape morphogenesis, and intracellular transport.1,2 Tubulin heterodimers consist of α and β subunits that, in the presence of GTP/Mg2+, polymerize in a head-to-tail manner and form protofilaments that are later assembled into microtubules.3,4 In this way, the microtubule forms a stiff, hollow structure with the lattice stabilized by lateral and longitudinal contacts.5 Environmental conditions like temperature, GTP/Mg2+ concentration, microtubule-associated proteins (MAPs), and small molecules interacting with specific sites on the tubulin dimer can all regulate tubulin polymerization.6−8 The ability of these ligands to stabilize and destabilize microtubules has been used in medicine for cancer therapy.9,10
Colchicine is one of the first compounds identified as a tubulin targeting agent.11 Colchicine treatment destroys microtubules and inhibits the tubulin polymerization process. The first crystallographic structure of the tubulin–colchicine complex revealed conformational changes in the tubulin after colchicine binding.12 Colchicine and its functional analogs bind in the interface of the tubulin subunits, though the binding pocket is generally buried in the β-subunit. The colchicine binding site (CBS) is bounded by β-strains S8 and S9, α-helices H7 H8, loop T5 of the beta subunit, and loop T7 of the α-subunit. Several studies have explored the use of colchicine as an anticancer therapeutic, but the high toxicity of the drug has limited its use.13 Currently, colchicine is an FDA-approved drug and is used to mitigate familial Mediterranean fever (FMF) symptoms and in the treatment of gout.14,15 Due to the potential therapeutic role of colchicine, many other colchicine functional analogs have been developed.16−18 More than 50 colchicine analogs have solved X-ray structures in complex with tubulin. We will refer to these compounds as colchicine binding site inhibitors (CBSIs).
In recent years, the mechanisms of CBSI interaction with tubulin have been intensively studied using structural biology, molecular modeling, and computational chemistry methods.12,18,19 Nevertheless, some principles behind tubulin polymerization inhibition by CBSIs remain elusive. Importantly, the effect of CBSIs on tubulin intersubunit interaction has yet to be documented. It is well known that small molecules that interact with the CBS inhibit the polymerization process and destabilize microtubules. However, this effect is not necessarily due to destabilizing interactions between α and β subunits and the binding free energy between the subunits may be either increased or decreased upon CBSI binding. In the first crystallography structure, it was shown that the binding of colchicine changes the tubulin conformation from straight to curved.12 These conformational changes in the tubulin heterodimer lead to the loss of contacts in the microtubule lattice and destabilize them.6,20 Nonetheless, it remains unclear how the binding free energy between tubulin subunits is changed upon CBSI binding.
Understanding how the CBSIs change the binding free energy between tubulin subunits sheds light on the underlying molecular mechanisms of microtubule dynamics. In this work, we used all-atom molecular dynamics (MD) simulations combined with free energy calculation methods to study tubulin intersubunit interaction. The results of our study provide new insights into tubulin polymerization inhibition by small molecules interacting with CBS.
2. Methods
2.1. MM/PBSA
We used five crystallographic tubulin structures for MD simulations (Table 1). In all cases, molecules of GTP and GDP with their Mg2+ ions were in complex with the tubulin heterodimer. We used the CHARMM36m force field for protein and CGenFF to parametrize small molecules.21−23 The net charge for GTP was set to −4, for GDP -3, and a neutral charge for colchicine and its analogs was used. Amber and AmberTools (v2020) were used for MD simulation and MM/PBSA calculation.24,25 Complexes were solvated with TIP3P water molecules and Na/Cl ions at 150 mM concentration in a triclinic box with sides 12 × 8.5 × 8.5 nm containing ∼25,000 water molecules 85,000 atoms (Jorgensen et al., 1983). The systems were minimized and equilibrated, gradually releasing position restraints for 12 ns. We used the Monte Carlo barostat with reference pressure at 1 bar and the Langevin thermostat with collision frequency (gamma_ln) 2 ps–1 to keep the temperature at 310 K. Particle mesh Ewald (PME) with electrostatic interactions cut off at 1.0 nm was used for the long-range electrostatic interactions.26 Bonds involving hydrogens were constrained using the SHAKE algorithm, and the 2 fs integration step was used.27
Table 1. Physical–Chemical Properties of Colchicine Binding Site Inhibitors.
| name | COL | C-A4 | POD’ | NOC |
|---|---|---|---|---|
| PDB ID | 5xiw | 5lyj | 5jcb | 5ca1 |
| resolution (Å) | 2.90 | 2.40 | 2.30 | 2.40 |
| mol weight (g/mol) | 399.4 | 316.3 | 497.5 | 301.32 |
| N atoms | 54 | 43 | 58 | 32 |
| N heavy atoms | 29 | 23 | 35 | 21 |
| N rotatable bond | 5 | 6 | 6 | 3 |
| log P | 1 | 3.7 | 3.3 | 2.8 |
| N H-bond D | 1 | 1 | 1 | 2 |
| N H-bond A | 6 | 5 | 10 | 5 |
| polar surface area (Å2) | 83.1 | 57.2 | 139 | 112 |
We ran three 100 ns MD simulations with different seeds using the abovementioned X-ray structures. Binding free energy calculations were performed with the MMPBSA.py program using 500 snapshots collected from each run.
2.2. Umbrella Sampling
Non-equilibrium pulling and umbrella sampling were performed using GROMACS-2021.4 with the same CHARMM36m and CgenFF force fields.28 Tubulin heterodimers were solvated in a triclinic box with sides 18x8x8 nm containing ∼38,000 water molecules and ∼ 130,000 atoms. We minimized and equilibrated the systems for 5 ns and performed MD simulations at a constant temperature of 310 K and pressure at 1 bar, using the V-rescale algorithm for temperature coupling and the Parrinello–Rahman barostat for pressure coupling. Van der Waals interactions were cutoff from 0.8 to 1.2 nm with a force switch, and long-range electrostatic interactions were calculated using particle mesh Ewald (PME) summation with a 1.2 nm real space cutoff. All hydrogen bonds were constrained using the LINCS algorithm, and a 2 fs integration time step was used.29
Harmonic potential with a pull rate of 0.002 nm/ps and 1000 kJ mol–1 nm–2 was applied to the center of mass of the α-subunit, and at the same time, position restraints of 1000 kJ/mol/nm were applied to the GDP and neighbor residues within 0.5 nm. To perform umbrella sampling with a series of configurations along reaction coordinate ζ that corresponded to the center-of-mass distance between α/β subunits, we selected 30 snapshots from the pulling trajectory with a spacing of 0.2 nm and added missing windows if the sampling was insufficient. Next, we performed independent simulations within each sampling window and used the weighted histogram analysis method (WHAM) to extract the potential of mean force (PMF).30 Statistical uncertainty was estimated with 250 bootstrap samples. The ΔG was calculated as the difference between the highest and lowest values of the PMF curve.
ICM-pro and VMD were used for electrostatic surface calculations and visualizations.
3. Results
We selected a range of colchicine binding site inhibitors (CBSIs) to understand how CBSI binding affects the interaction between tubulin subunits. As subjects of our study, we selected colchicine (COL) and combretastatin-A4 (C-A4) as natural tubulin polymerization inhibitors,15,31 4β-(1,2,4-triazole-3-ylthio)-4-deoxypodophyllotoxin (POD’) as a derivative of the natural microtubule destabilizing agent podophyllotoxin,32 and nocodazole (NOC) as a synthetic chemical with no natural analogs.33 These compounds were selected as representatives of CBSIs with diverse scaffolds and origins. There are X-ray structures of all of these compounds in complex with tubulin, which we used for MD simulations. We also performed control simulations with the tubulin dimer in a complex with GTP, GDP, and Mg2+ but without any small molecules in the CBS (PDB ID: 4I4T), which we will refer to as free tubulin. Physical–chemical properties of the studied compounds, their chemical structures, and information about the X-ray structures are presented in Table 1 and Figure 1C.
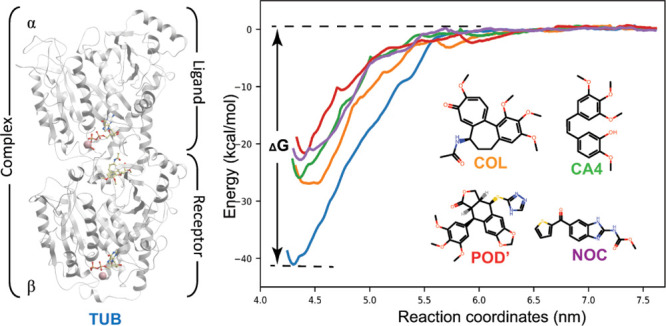
Figure 1.
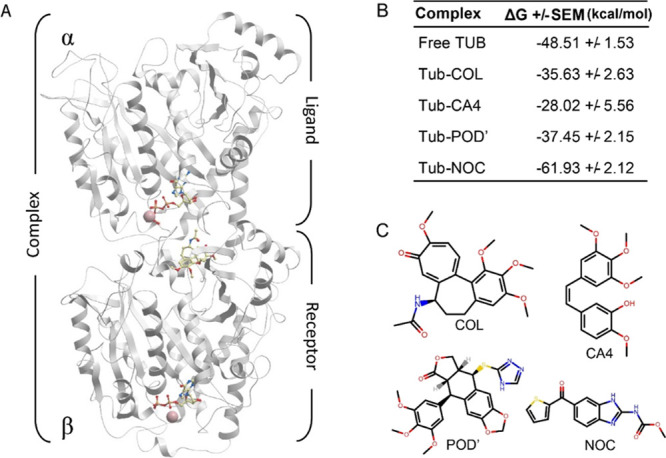
Structure of the tubulin heterodimer and the effect of CBSIs on intersubunit interaction. (A) Structure of tubulin subunits represented as receptor (β) and ligand (α) as it was used for MM/PBSA calculations. (B) Intersubunit binding free energy calculated with MM/PBSA, showing the mean values with standard error (SE). (C) Chemical structures of CBSIs used in this study.
3.1. Intersubunit Binding Free Energy Calculated with MM/PBSA
We ran three independent molecular dynamics (MD) simulations for each complex in an explicit water environment and collected snapshots from each run for binding free energy calculation calculations, as described in the Methods section. Tubulin heterodimers during the MD simulations were stable, and no essential conformational changes were observed in the ligand binding mod or protein structure (Figure S1). In the molecular mechanics Poisson–Boltzmann surface area (MM/PBSA) calculations, the β-subunit with CBSI and GDP/Mg2+ was considered as a receptor, and the α-subunit with GTP/Mg2+ was considered as a ligand (Figure 1A).
According to our MM/PBSA calculations, binding free energy between subunits of the tubulin heterodimer in the free form, without any CBSI in the complex, is −48.51 kcal/mol. In the presence of colchicine, the binding free energy is −35.63 kcal/mol (Figure 1B), which means that the interaction between α/β subunits in the presence of colchicine is less favorable than in the free form. Similar changes were observed for C-A4 and the POD’; both compounds reduce the binding free energy between subunits with a calculated ΔG of −28.02 and −37.45 kcal/mol, respectively. However, according to MM/PBSA calculations, nocodazole is an exception among the studied compounds. In contrast to the previous ligands, it has the opposite effect and strengthens the intersubunit interaction. Binding free energy between subunits in the presence of nocodazole is −61.93 kcal/mol, which is stronger than the intersubunit binding free energy of the free tubulin (Figure 1B).
3.2. Intersubunit Binding Free Energy Calculated with Umbrella Sampling
We carried out additional free energy calculations employing steered molecular dynamics (SMD) simulations and umbrella sampling (US) to reduce the possible bias of our calculations and confirm the results of the MM/PBSA calculations. We applied a pull force to the center of mass of the α-subunit and pulled it out from the positionally restrained β-subunit (Figure 2A, Movies S1–S5). To adequately represent conformational changes on the interface of α/β subunits upon pulling, we restrained only amino acids around the GDP/Mg2+ in the β-subunit. We used the trajectories obtained after pulling to perform umbrella sampling (US) and extract the potential of mean force (PMF).
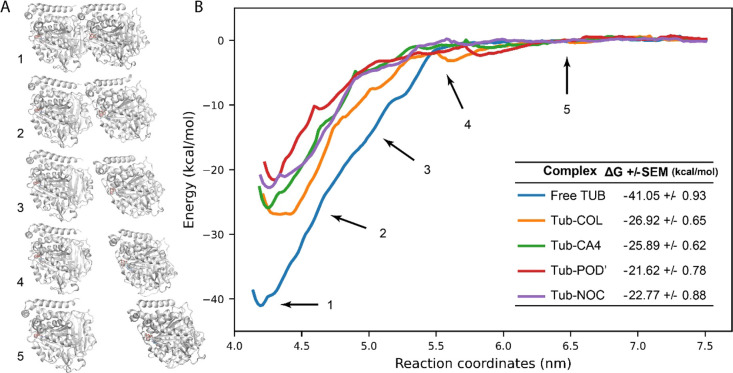
Figure 2.
Tubulin intersubunit interaction during the center of mass pulling. (A) Tubulin subunits with different distances between COMs. Numbers show the distances between centers of mass indicated on panel (B). (B) Potential of mean force (PMF) across reaction coordinates (ζ) derived from umbrella sampling.
Results obtained with the US calculation are generally comparable with the MM/PBSA calculation. For free tubulin, α/β subunits binding ΔG is −41.05 kcal/mol. As expected, in the presence of CBSIs in complex with tubulin heterodimers, binding free energy between tubulin subunits was reduced. Colchicine weakened the interaction between the subunits, reducing binding ΔG to −26.92 kcal/mol. Combretastatin and the podophyllotoxin derivative similarly reduced intersubunit interaction as was observed in the MM/PBSA calculations; ΔG values for tubulin dimers in complex with these compounds are −25.89 and −21.62 kcal/mol, respectively. Interestingly, according to the free energy calculation with umbrella sampling, nocodazole also weakens the interaction between α/β subunits, resulting in a ΔG of −27.53 kcal/mol, which contrasts with the effect observed in the MM/PBSA analysis (Figure 2B). Thus, in the presence of CBSI, we observe a reduction of intersubunit binding free energy. Free energy calculations with umbrella sampling showed a reduction of the intersubunit binding energy of 34–47% and 23–42% in the case of MM/PBSA, excluding nocodazole results.
Although umbrella sampling is accepted to be a more accurate approach for binding free energy calculations than MM/PBSA, the contradiction between these methods in the case of nocodazole merits some discussion.34 Among the studied compounds, only nocodazole has a synthetic origin and a different binding mode compared to the other compounds. In crystallographic structures, it can be clearly distinguished that nocodazole is buried deeper into the tubulin β-subunit, and several amino acids interact with nocodazole but do not interact with other CBSI (Figure S2).35 Furthermore, contrary to the other CBSIs, nocodazole does not interact with the tubulin T5-loop of the α-subunit (αT5). Nocodazole may be indeed an exception; however, it seems more plausible that MM/PBSA calculation is biased, especially in the case when the ligand does not directly interact with the α-subunit and US calculation provides more accurate information.
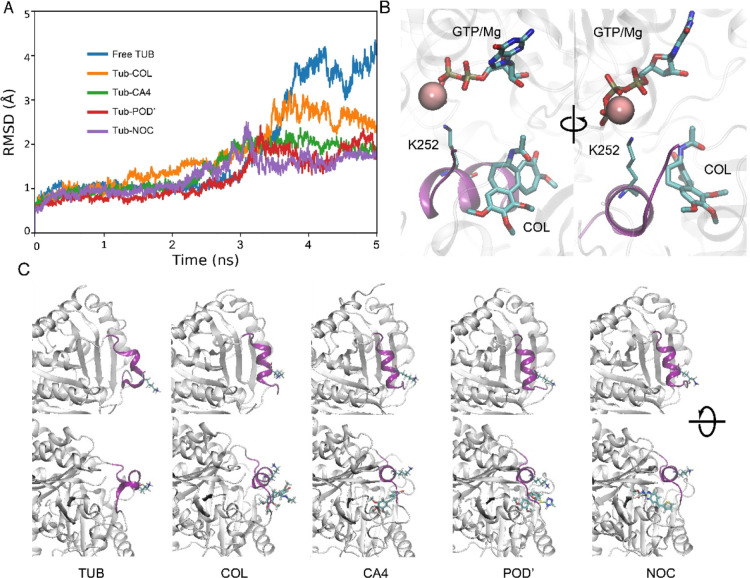
Snapshots collected without large-scale conformational changes in the equilibrium stage for MM/PBSA analyses can be insufficient to accurately estimate binding free energy in complex systems like the tubulin heterodimer when two interacting subunits contain charged nucleotides in complex with Mg2+. In the SMD, we can see that subunits do not simply disassociate from each other, but conformational changes also accompany that process. This is especially clear in helix H8 of the β-subunit, which forms a part of the CBS (Figure 3). This helix is tightly bound to the α-subunit, and during the SMD, this helix is pulled away from the β-subunit (Figure 3, Movies S1–S5). Interaction between helix H8 and the α-subunit occurs through the interlinkage of Lys252 and the α-subunit’s natural cofactor GTP/Mg2+ (Figure 3B). Per-residue binding free energy decomposition shows that Lys252 has the highest contribution in the intersubunit interaction. The positively charged NH3 group of the lysine side chain interacts with the negatively charged GTP. The center of mass pulling leads to deformation of the CBS and helix H8, where Lys252 is located (Figure 3A). In the absence of colchicine and other ligands, helix H8 is pulled away and unfolded, whereas in the presence of CBSIs, there are fewer conformational changes (Figure 3A,C). We can speculate that CBSIs interacting with the binding site stabilize it and make it more rigid. Tubulin flexibility is crucial for microtubule assembly, and a lack of flexibility in the interface where subunits interact can lead to faster destabilization of microtubules.20
Figure 3.
Conformational changes in the tubulin heterodimer interface during subunit disassociation. (A) Root mean squared deviation of helix H8. (B) Interaction of α and β subunits in the CBS region and interlinkage between Lys252 and GTP/Mg2+. (C) Helix H8 conformation (colored purple) after COM pulling, where CBSIs and Lys252 are displayed as stick models.
4. Discussion
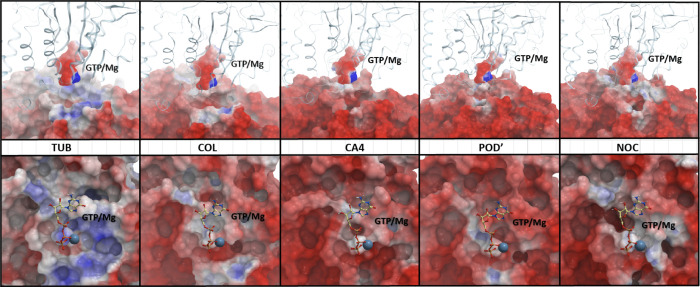
It seems likely that the observed intersubunit interaction weakening mechanism is caused by changes on the electrostatic surface of the β-subunit. Analyzing the electrostatic surface of the tubulin in the free form and in complex with the studied small molecules, we noticed that the region of the CBS on the β-subunit is neutral or positively charged and becomes charged negatively after binding by a CBSI (Figure 4). MMPBSA energy component analyses also supported this observation and showed higher electrostatic repulsion after ligand binding, which is positive even in the ligand-free tubulin dimer (Table S1). These changes are probably caused by the rearrangement of polar residues βGLN-247 and βASN-249 located in the T5 loop. In the free tubulin, these residues are “buried” in the CBS but are relocated to the surface after binding of a CBSI. The GTP/Mg2+ complex and its nearest-neighbor amino acids in the α-subunit are also negatively charged, and two surfaces with the same charge should repulse. This repulsion is not enough to entirely destabilize intersubunit interaction, but it can reduce binding free energy between subunits.
Figure 4.
Changes on the electrostatic surface of tubulin upon CBSI binding. Top panel: side view of the tubulin heterodimer α-subunit is shown as ribbons, for the β-subunit and GTP/Mg2+ the electrostatic surface is visualized. Bottom panel: top view of the electrostatic surface of the β-subunit. GTP/Mg2+ is visualized as balls and sticks.
The tubulin lattice of microtubules is stabilized with lateral and longitudinal contacts.36 Microtubule assembly is accompanied by GTP hydrolysis, resulting in conformational changes in the tubulin dimer from a kinked to a straight conformation. These conformational changes are crucial for microtubule dynamics and stability. In the presence of colchicine and its analogs, tubulin does not switch to the straight conformation because of steric clashes caused by the ligand binding.12 Thus, tubulin dimers cannot establish the lateral contacts necessary for microtubule stabilization in the curved conformation. However, we showed that CBSIs also act as protein–protein interaction inhibitors and interrupt intersubunit interactions, thereby destabilizing longitudinal contacts. It is noteworthy that microtubule assembly starts with the formation of protofilaments, which are chains of αβ tubulins connected with longitudinal contacts. Our observations suggest that CBSIs may weaken links in those chains. Thus, in the presence of CBSIs, a part of the protofilaments may become ″defective″ and cannot support microtubule assembly, leading to polymerization inhibition or complete microtubule destruction depending on the concentration of inhibitors.
5. Conclusions
In this study, we provide new insights into the molecular mechanisms underlying the inhibition of tubulin polymerization by CBSIs using MD simulations and free energy calculations. Colchicine and its functional analogs with different scaffolds interacting with CBS weaken intersubunit interaction between α and β tubulins. This effect along with other conformational changes that take place in the tubulin heterodimer upon CBSIs explains why these compounds inhibit microtubule polymerization. Though we cannot conclude that the described mechanism is common to all CBSIs based on these 4 cases, it is worth noting that these compounds have diverse structures, physical–chemical properties, and different origins (Table 1, Figure 1C). In this light, it seems plausible that binding by the majority of CBSIs leads to less favorable intersubunit interactions in tubulin dimers.
Acknowledgments
We are grateful to the NIH Fellows Editorial Board for help and assistance.
Data Availability Statement
Steered Molecular Dynamics simulation trajectories are available at https://doi.org/10.5281/zenodo.7297043
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c02979.
Author Present Address
‡ Current address: National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, Maryland 20894, United States
Author Contributions
# A.S. and H.S. contributed equally.
Author Contributions
Participated in research design: A.S., H.S., and K.N.; conducted experiments: A.S. and H.S.; performed data analysis: H.S. and K.N.; wrote or contributed to the writing of the manuscript: A.S., H.S., and K.N.
The work was supported by the Science Committee of RA, in the frames of the research project no 21AG-1F057.
The authors declare no competing financial interest.
Supplementary Material
References
- Brouhard G. J.; Rice L. M. Microtubule dynamics: an interplay of biochemistry and mechanics. Nat Rev Mol Cell Biol. 2018, 19, 451–463. 10.1038/s41580-018-0009-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E. Structural Insights into Microtubule Function. Annu. Rev. Biochem. 2000, 69, 277–302. 10.1146/annurev.biochem.69.1.277. [DOI] [PubMed] [Google Scholar]
- Howard J.; Hyman A. A. Growth, fluctuation and switching at microtubule plus ends. Nat Rev Mol Cell Biol. 2009, 10, 569–574. 10.1038/nrm2713. [DOI] [PubMed] [Google Scholar]
- Michaels T. C.; Feng S.; Liang H.; Mahadevan L. Mechanics and kinetics of dynamic instability. eLife. 2020, 9, e54077 10.7554/eLife.54077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross R. A. Microtubule lattice plasticity. Curr. Opin. Cell Biol. 2019, 56, 88–93. 10.1016/j.ceb.2018.10.004. [DOI] [PubMed] [Google Scholar]
- Akhmanova A.; Steinmetz M. O. Control of microtubule organization and dynamics: two ends in the limelight. Nat Rev Mol Cell Biol. 2015, 16, 711–726. 10.1038/nrm4084. [DOI] [PubMed] [Google Scholar]
- Alushin G. M.; Lander G. C.; Kellogg E. H.; Zhang R.; Baker D.; Nogales E. High-resolution microtubule structures reveal the structural transitions in αβ-tubulin upon GTP hydrolysis. Cell 2014, 157, 1117–1129. 10.1016/j.cell.2014.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A.; Mitchison T. J. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 1997, 13, 83–117. 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- Dumontet C.; Jordan M. A. Microtubule-binding agents: a dynamic field of cancer therapeutics. Nat Rev Drug Discov. 2010, 9, 790–803. 10.1038/nrd3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prota A. E.; Bargsten K.; Zurwerra D.; Field J. J.; Díaz J. F.; Altmann K.-H.; et al. Molecular Mechanism of Action of Microtubule-Stabilizing Anticancer Agents. Science 2013, 339, 587–590. 10.1126/science.1230582. [DOI] [PubMed] [Google Scholar]
- Weisenberg R. C.; Borisy G. G.; Taylor E. W. Colchicine-binding protein of mammalian brain and its relation to microtubules. Biochemistry 1968, 7, 4466–4479. 10.1021/bi00852a043. [DOI] [PubMed] [Google Scholar]
- Ravelli R. B. G.; Gigant B.; Curmi P. A.; Jourdain I.; Lachkar S.; Sobel A.; et al. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature 2004, 428, 198–202. 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
- Lu Y.; Chen J.; Xiao M.; Li W.; Miller D. D. An Overview of Tubulin Inhibitors That Interact with the Colchicine Binding Site. Pharm. Res. 2012, 29, 2943–2971. 10.1007/s11095-012-0828-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgeb B.; Kornreich D.; McGuinn K.; Okon L.; Brownell I.; Sackett D. L. Colchicine: an ancient drug with novel applications. Br J Dermatol. 2018, 178, 350–356. 10.1111/bjd.15896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung Y. Y.; Yao Hui L. L.; Kraus V. B. Colchicine—Update on mechanisms of action and therapeutic uses. Seminars in Arthritis and Rheumatism. 2015, 45, 341–350. 10.1016/j.semarthrit.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Roche N. M.; Mühlethaler T.; Di Martino R. M. C.; Ortega J. A.; Gioia D.; Roy B.; et al. Novel fragment-derived colchicine-site binders as microtubule-destabilizing agents. Eur. J. Med. Chem. 2022, 241, 114614 10.1016/j.ejmech.2022.114614. [DOI] [PubMed] [Google Scholar]
- McLoughlin E. C.; O’Boyle N. M. Colchicine-Binding Site Inhibitors from Chemistry to Clinic: A Review. Pharmaceuticals. 2020, 13, 8. 10.3390/ph13010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prota A. E.; Danel F.; Bachmann F.; Bargsten K.; Buey R. M.; Pohlmann J.; et al. The Novel Microtubule-Destabilizing Drug BAL27862 Binds to the Colchicine Site of Tubulin with Distinct Effects on Microtubule Organization. J. Mol. Biol. 2014, 426, 1848–1860. 10.1016/j.jmb.2014.02.005. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Zhang H.; Gigant B.; Yu Y.; Wu Y.; Chen X.; et al. Structures of a diverse set of colchicine binding site inhibitors in complex with tubulin provide a rationale for drug discovery. FEBS J. 2016, 283, 102–111. 10.1111/febs.13555. [DOI] [PubMed] [Google Scholar]
- Igaev M.; Grubmüller H. Microtubule assembly governed by tubulin allosteric gain in flexibility and lattice induced fit. eLife. 2018, 7, e34353 10.7554/eLife.34353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.; Rauscher S.; Nawrocki G.; Ran T.; Feig M.; de Groot B. L.; et al. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanommeslaeghe K.; MacKerell A. D. Automation of the CHARMM General Force Field (CGenFF) I: Bond Perception and Atom Typing. J. Chem. Inf. Model. 2012, 52, 3144–3154. 10.1021/ci300363c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanommeslaeghe K.; Raman E. P.; MacKerell A. D. Automation of the CHARMM General Force Field (CGenFF) II: Assignment of Bonded Parameters and Partial Atomic Charges. J. Chem. Inf. Model. 2012, 52, 3155–3168. 10.1021/ci3003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearlman D. A.; Case D. A.; Caldwell J. W.; Ross W. S.; Cheatham T. E.; DeBolt S.; Ferguson D.; Seibel G.; Kollman P. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput. Phys. Commun. 1995, 91, 1–41. 10.1016/0010-4655(95)00041-D. [DOI] [Google Scholar]
- Miller B. R.; McGee T. D.; Swails J. M.; Homeyer N.; Gohlke H.; Roitberg A. E. MMPBSA.py : An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- Darden T.; York D.; Pedersen L. Particle mesh Ewald: An N ·log( N ) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. 10.1063/1.464397. [DOI] [Google Scholar]
- Ryckaert J.-P.; Ciccotti G.; Berendsen H. J. C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. 10.1016/0021-9991(77)90098-5. [DOI] [Google Scholar]
- Abraham M. J.; Murtola T.; Schulz R.; Páll S.; Smith J. C.; Hess B.; et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015, 1-2, 19–25. 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- Hess B.; Bekker H.; Berendsen H. J. C.; Fraaije J. G. E. M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. . [DOI] [Google Scholar]
- Hub J. S.; de Groot B. L.; van der Spoel D. g_wham—A Free Weighted Histogram Analysis Implementation Including Robust Error and Autocorrelation Estimates. J. Chem. Theory Comput. 2010, 6, 3713–3720. 10.1021/ct100494z. [DOI] [Google Scholar]
- Tron G. C.; Pirali T.; Sorba G.; Pagliai F.; Busacca S.; Genazzani A. A. Medicinal Chemistry of Combretastatin A4: Present and Future Directions. J. Med. Chem. 2006, 49, 3033–3044. 10.1021/jm0512903. [DOI] [PubMed] [Google Scholar]
- Zhao W.; Zhou C.; Guan Z.-Y.; Yin P.; Chen F.; Tang Y.-J. Structural Insights into the Inhibition of Tubulin by the Antitumor Agent 4β-(1,2,4-triazol-3-ylthio)-4-deoxypodophyllotoxin. ACS Chem. Biol. 2017, 12, 746–752. 10.1021/acschembio.6b00842. [DOI] [PubMed] [Google Scholar]
- De Clerck F.; De Brabander M. Nocodazole, a new synthetic antimitotic agent, enhances the production of plasminogen activator by cells in culture. Thromb. Res. 1977, 11, 913–914. 10.1016/0049-3848(77)90120-7. [DOI] [PubMed] [Google Scholar]
- Ngo S. T.; Vu K. B.; Bui L. M.; Vu V. V. Effective Estimation of Ligand-Binding Affinity Using Biased Sampling Method. ACS Omega 2019, 4, 3887–3893. 10.1021/acsomega.8b03258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufareva I.; Ilatovskiy A. V.; Abagyan R. Pocketome: an encyclopedia of small-molecule binding sites in 4D. Nucleic Acids Res. 2012, 40, D535–D540. 10.1093/nar/gkr825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaev M.; Grubmüller H. Microtubule instability driven by longitudinal and lateral strain propagation. PLoS Comput. Biol. 2020, 16, e1008132 10.1371/journal.pcbi.1008132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Steered Molecular Dynamics simulation trajectories are available at https://doi.org/10.5281/zenodo.7297043